Abstract
Constitutive activation of signal transducer and activator of transcription 1 (STAT1) is a distinctive feature of Epstein-Barr virus (EBV)-immortalized B cells (lymphoblastoid cell lines [LCLs]). The expression of STAT1 in these cells is modulated by the latent membrane protein 1 (LMP1), but the mechanism of STAT1 activation has remained unclear. We demonstrate that the tyrosine phosphorylation of STAT1 in LCLs results from an indirect pathway encompassing an NF-κB-dependent secretion of interferons (IFNs). The cell culture supernatant of LCLs induced tyrosine phosphorylation of STAT1 in cells with no constitutively activated STAT1. Moreover, removal of supernatant from LCLs was sufficient to decrease the phosphorylation of STAT1. Inhibition of NF-κB activity by different pharmacological inhibitors (i.e., parthenolide, MG132 and BAY 11-7082) and by overexpressed mutated IκBα prevented the activation of STAT1. To identify the factors involved, we performed macroarray cDNA profiling with or without inhibition of NF-κB. The expression of several cytokines was NF-κB dependent among those alpha and gamma IFNs (IFN-α and IFN-γ), known activators of STAT1. By real-time PCR and enzyme-linked immunosorbent assay we show that IFN-α and IFN-γ are expressed and released by LCLs in an NF-κB-dependent manner. Finally, the blocking of the IFN-α and IFN-γ by neutralizing antibodies led to the complete inhibition of tyrosine phosphorylation of STAT1. Taken together, our results clearly show that LMP1-induced tyrosine phosphorylation of STAT1 is almost exclusively due to the NF-κB-dependent secretion of IFNs. Whether this response, which is usually considered to be antiviral, is in fact required for the persistence of the virus remains to be elucidated.
The Epstein-Barr virus (EBV) is the causal agent of infectious mononucleosis and is associated with severe infections in immunocompromised patients. In addition, EBV is associated with malignancies such as Burkitt lymphoma, nasopharyngeal carcinoma, and Hodgkin's lymphoma (30).
In primary lymphocyte infection, cell proliferation is stimulated by a set of viral gene products termed the latency III program, which is also characteristic of EBV-transformed lymphoblastoid cell lines (LCLs) (31). Among the latency III viral gene products, the latent membrane protein 1 (LMP1) is essential for the EBV-induced transformation of primary B lymphocytes (22). The oncogenic properties of LMP1 are associated with stimulation of DNA synthesis (28), stimulation of the transcription of antiapoptotic genes (9, 34), and suppression of cellular senescence (37). LMP1 is a transmembrane protein, analogous to a constitutively activated CD40 receptor, although structurally different (18). Specialized regions (CTAR1 and CTAR2) of the cytoplasmic domain of LMP1 recruit components of the TNF-R signaling pathway and activate the transcriptional factor NF-κB. The two regions are not equivalent: CTAR1 operates by recruiting TRAF1, -2, and -3, and CTAR2 operates by recruiting RIP and TRADD (4, 14, 21).
Constitutive STAT1 activation has been observed in EBV-associated tumors, including nasopharyngeal carcinoma (9), and in LCLs (10, 35). The STATs are transcription factors that are activated after triggering of cells with cytokines. Although most STATs, including STAT3, STAT4, and STAT5, are involved in the proliferative response of cells to cytokines, STAT1 and STAT2 are associated with the cellular response to interferons (IFNs), which reduce cell proliferation and increase apoptosis. Thus, in the context of EBV-transformed LCLs, the activation of STAT1 raises the question of its function. We previously observed that inhibition of STAT1 in LCLs by overexpression of an inhibitory form of STAT1 (STAT1β) reduces drug-induced apoptosis and increases the growth rate of cells (3), demonstrating that even in the context of EBV-transformed cells, STAT1 remains an inhibitor of cell proliferation. Nevertheless, expression of LMP1 itself was shown to be sufficient to induce a higher level of STAT1 expression (29), but the mechanism involved remains unclear. A direct activation of STATs was suggested, after interaction of JAK3 with a cytoplasmic region (amino acids 275 to 330) located between CTAR1 and CTAR2 (17). However, neither the association of JAK3 to the putative JAK3-binding domain of LMP1 from positions 232 to 351 nor the activation of JAK3 by this domain could be confirmed in another study (20). Finally, it was observed that the transfection of a mutated LMP1 (LMP1AAAG) containing three point mutations in the CTAR1 and one in the CTAR2 motif into a Burkitt lymphoma cell line abolished the increased expression and activity of STAT1 (4, 29). This suggested a possible role for NF-κB in the activation of STAT1 by LMP1.
EBV-immortalized B lymphocytes express alpha IFN (IFN-α), IFN-β, and IFN-γ, as well as IFN-α/β- and IFN-γ-inducible genes (2, 5, 7, 8, 28), and NF-κB is known to increase the transcription of several genes of cytokines and cytokine receptors (25, 27), including IFN-γ (32), IFN-γRα (7), and IFN-α/β receptor 2 (24). To determine the possible involvement of indirect mechanisms bringing about the activation of STAT1 in cells expressing LMP1, we studied the ability of cell culture supernatants from LCL cells to stimulate STAT1 tyrosine phosphorylation. We found that the LMP1-induced stimulation of the tyrosine phosphorylation of STAT1 is almost exclusively due to an NF-κB-dependent secretion of IFNs.
MATERIALS AND METHODS
Plasmid constructs.
The V1NL IκBαdn vector was derived from the previously used CKR 516 vector (3, 13). The EBNA1 gene was added to ensure episomal replication. The vector contains a bidirectional tetracycline-inducible promoter driving the expression of two independent cDNAs: IκBαdn and a truncated version of the nerve growth factor receptor (NGFR) lacking the cytoplasmic domain and used as a surrogate marker of IκBαdn induction. Induction of the expression of NGF-R is easily verified by flow cytometry and is used to sort cells by magnetic separation. The induction of the expression of IκBαdn was measured by Western blotting.
Cell culture, treatments, transfection, cell sorting, and flow cytometry.
The lymphoblastoid cell lines Co (kindly provided by S. Dupuis and J.-L. Casanova, INSERM U550), PRI and 1602, and the Burkitt lymphoma cell line BL2 were cultured at 37°C in a humidified 5% CO2 atmosphere in RPMI 1640 medium (Eurobio, Les Ulis, France) supplemented with 10% decomplemented fetal calf serum (Dutcher, Brumath, France), 100 U of penicillin/ml, 10 μg of streptomycin (Gibco-BRL-Life Technologies, Cergy-Pontoise, France)/ml, and 2 mM l-glutamine (Eurobio). Where indicated, cells were treated with 25 ng of IFN-γ (Roche Diagnostics, Meylan, France)/ml for 1 h, with brefeldin A (BFA; Sigma, Saint Quentin Fallavier, France), parthenolide (Sigma), MG132 (Sigma), and BAY11-7082 [E-3-(4-methylphenylsulfonyl)-2-propenenitrile; BAY11] (Calbiochem, VWR International SAS, Fontenay-Sous-Bois, France). Parthenolide and BAY11 were reconstituted in dimethyl sulfoxide (DMSO) (Calbiochem). Transfection, selection of hygromycin-resistant cells, and doxycycline treatment were performed as described previously (13). Both NGFR and IκBαdn expression were assessed by flow cytometry and Western blotting, respectively, after 6 weeks of hygromycin selection.
Doxycycline-induced cells were magnetically labeled with anti-LNGFR coupled to microBeads after 24 h of induction and separated by using the MACSelect LNGFR System procedure (Miltenyi Biotech, Paris, France). The induction of the expression of LNGFR was assessed by flow cytometry. Cells were washed in phosphate-buffered saline (bioMérieux, Marcy l'Etoile, France) and incubated with phycoerythrin-labeled NGFR antibody (Becton Dickinson, Morangis, France). Phycoerythrin-positive cells were counted by using the FL2 photomultiplier of a XL Beckman-Coulter counter.
Western blotting.
Viable cells were counted by using the trypan blue method. Total protein extracts were obtained as follows: 1 million cells were resuspended in lysis buffer containing: 0.01% bromophenol blue (Bio-Rad, Marnes la Coquette, France), 50 mM Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate (Bio-Rad), 20% glycerol (Sigma), and 5% 2-mercaptoethanol (Bio-Rad). The lysates were sonicated, boiled, and stored at −80°C. The extracts were separated on a 10% polyacrylamide denaturing gel at 30 mA/gel and transferred (Bio-Rad) to nitrocellulose (Schleicher & Schuell, Dassel, Germany) membranes. After transfer, the membranes were stained with Ponceau red in order to check that equal amounts of protein were present in each lane. After the membranes were blocked for 1 h with 5% dry skimmed milk (Régilait, Saint Martin Belle Roche, France) in Tris-buffered saline (TBS; 20 mM NaCl, 500 mM Tris-HCl [pH 8]; Bio-Rad), the incubation with the first antibody was performed overnight in TBS (Sigma) and 1% dry skimmed milk. The antibodies used were anti-STAT1 (rabbit polyclonal; Cell Signaling, Beverly, Mass.) at 1/1,000 and anti-phosphotyrosine 701 STAT1 (rabbit polyclonal; Cell Signaling) at 1/1,000. After 5 min of washing (three times) in TBS-0.1% Tween, the corresponding horseradish peroxidase-conjugated secondary antibody goat anti-rabbit (Bio-Rad) at 1/5,000 was added for 1 h. After three 5-min washes in TBS-0.1% Tween, visualization was achieved by chemiluminescence (Amersham, Orsay, France) and autoradiography (X-Omat R film; Kodak).
Electrophoretic mobility shift assay.
Nuclear and cytosolic proteins were extracted as described previously (12). A total of 20 μg of nuclear protein extracts in 5 μl of extraction buffer C (20 mM HEPES, 25% glycerol, 0.5 M NaCl, 1.5 mM MgCl2) was mixed with 1 μl of annealed 32P-labeled oligonucleotide containing the κB site (Proligo, Paris, France) (36). Electromobility shift assay was performed as previously described (11). The DNA-protein complexes were separated on a 6% nondenaturating polyacrylamide gel in 0.25× Tris-borate-EDTA buffer (Bio-Rad) by a 4-h migration at 250 V. The gel was dried and exposed to a phosphor imaging screen (Packard Instruments, Meriden, Conn.). The radioactive signal was visualized by using a phosphor system analyzer (Cyclone; Packard Instruments).
Luciferase activity.
To measure the transcriptional activity of STAT1, we cotransfected cells with a TAP1 promoter luciferase-reporter construct in which the κB site had been inactivated (a generous gift of D. Johnson, National Institutes of Health, Bethesda, Md.), together with the bidirectional inducible V1NL IκBαdn plasmid encoding IkBαdn and NGFR. Transfected cells were induced during 24 h with doxycycline, sorted with anti-LNGFR microbeads (Miltenyi Biotech), and lysed for 15 min with the lysis buffer provided in the kit (Tropix Kit) supplemented with 1 mM dithiothreitol. The lysates were clarified by centrifugation, and the luciferase assay was performed by using a luminometer (BCL Book Luminometer).
cDNA macroarrays.
The expression of 112 cytokines was screened by using the cDNA GEArray (SuperArray; Tebu-bio, Le Perray en Yvelines, France). The RNA was prepared from 5 × 106 cells that were treated or not with parthenolide (20 μM) for 4 h. The cDNA was generated by using the Ampolabeling-LPR kit according to the manufacturer's protocol. The membranes were then hybridized as described in the GEArray user manual and, after the washes, incubated in enhanced chemiluminescence reagent and autoradiographed by using Kodak X-Omat AR films (Sigma).
Quantitative reverse transcription-PCR (RT-PCR).
Total RNA was extracted from sorted NGFR-positive and -negative cells by using the Qiagen kit according to the recommendations of the manufacturer. We defined as reference RNA a pool of RNAs extracted from different tonsils, lymph nodes, and spleen with benign reactive follicular hyperplasia.
RNA levels for the IFN-γ and IFN-α gene were quantified in parallel in the different RNA extracts and in the RNA pool on an ABI Prism 7000 Automat by using the TaqMan Assay-on-Demand gene expression reference system (Applied Biosystems; (catalog nos. Hs00174143-m1 and Hs00265051-m1)). The Abl1 gene was used as a reference gene for the control of amplification (catalog no. Hs00245443-m1). RT was performed with 2 μg of total RNA with the Archive Kit RT from Applied Biosystems in a final volume of 50 μl. From this, 1.25 μl of cDNA was used for gene amplification. All of these steps were performed according to the recommendations of the manufacturer.
The relative expression levels of the genes were calculated as previously reported (15), with Abl1 mRNA expression for normalization.
Quantification of cytokines in cell culture supernatants by using ELISA.
Ten to twenty million (0.8 million/ml) cells were grown in culture medium without fetal calf serum for 15 h. Supernatants were concentrated from 5 to 20 ml to obtain 200 μl by using centrifugal filter with a cutoff of 5,000 Da (Millipore, Molsheim, France). Secreted IFN-γ and IFN-α were measured by enzyme-linked immunosorbent assay (ELISA) with a Quantikine ELISA kit (R&D Systems, Lille, France) and the Biotrak ELISA system (Amersham Biosciences, Orsay, France), respectively. The optical density (OD) was determined by using a spectrophotometer (Multiskan EX; Thermo, Cergy-Pontoise, France) set to 450 nm. The ODs were measured by using Ascent Software (Thermo). The average of the duplicate concentrations was calculated.
RESULTS
The cell culture supernatant of LCL induces STAT1 tyrosine phosphorylation.
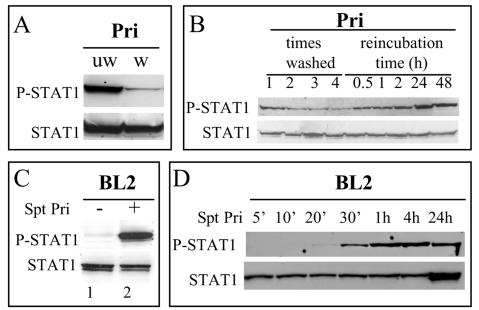
Constitutive phosphorylation of STAT1 on tyrosine has been observed in lymphoblastoid cell lines (LCLs) (10, 17, 35). We hypothesized that cytokines released in the supernatant could be responsible for this phosphorylation. First, we observed that the tyrosine phosphorylation of STAT1 in LCLs was considerably reduced after the cells were washed (Fig. 1A) and that when the LCL cells were reincubated in new medium, a gradual recovery of this phosphorylation was noted after 30 min (Fig. 1B). Second, we found that resuspension of the EBV-negative BL2 cell line, which lacks basal STAT1 activity, in the cell culture supernatant of LCL cells leads to the phosphorylation of STAT1 on tyrosine 701 (Fig. 1C). This phosphorylation became detectable after 20 min of incubation (Fig. 1D). A similar result was obtained by using the supernatant of two other LCL cell lines, 1602 and Co, or the supernatant of the EBV-converted BL2 cells (data not shown). Thus, STAT1-activating factor(s) are released by LCL cells.
FIG. 1.
Detection of a STAT1-activating activity in the cell culture supernatant of the LCL Pri cells. (A) The constitutive activation of STAT1 detected in the LCL Pri cells is almost entirely suppressed by simple washing. Pri cells were either not washed (lane 1) or washed three times in RPMI medium (lane 2), whole-cell extracts were obtained and separated on acrylamide gel and transferred onto nitrocellulose, and the Western blotting performed with anti-phospho Y701 STAT1 or anti-STAT1 (α, β) antibody. (B) After one, two, three, or four washes, the cells were left in their own culture medium for the indicated times; the extracts were then processed as in panel A. (C) STAT1 becomes phosphorylated on tyrosine in BL2 cells after addition of cell culture supernatant from Pri cells. Lane 1, no supernatant added; lane 2, BL2 cells resuspended for 45 min in the supernatant from Pri cells. (D) In a kinetic assay, the phosphorylation of STAT1 in BL2 cells became detectable after 20 min and maximal after 1 h of incubation in the supernatant from Pri cells.
Inhibition of secretion prevents LCL cells supernatants from activating STAT1 tyrosine phosphorylation.
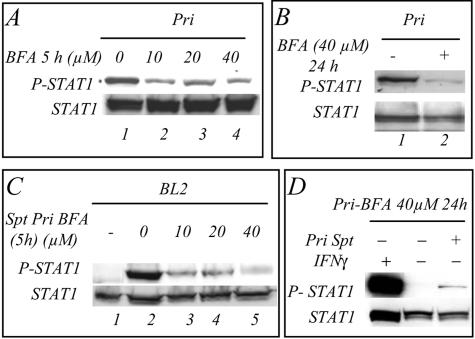
The detection in cell culture supernatants from LCL cells of an activity able to stimulate the tyrosine phosphorylation of STAT1 was indicative of the presence of factor(s) secreted by these cells. We therefore incubated the cells in the presence of different concentrations of BFA, a well-characterized secretion inhibitor which operates by inhibiting protein transport from the endoplasmic reticulum to the Golgi apparatus (23). After 5 h of incubation of the cells in medium containing 10 to 40 μM BFA, a strong inhibition of the tyrosine phosphorylation of STAT1 was noted (Fig. 2A). A nearly complete inhibition was observed after 24 h in 40 μM BFA (Fig. 2B). The cell culture supernatant obtained from BFA-treated LCL cells was used to analyze the stimulation of STAT1 tyrosine phosphorylation in BL2 cells: there was a strong inhibition of the ability of these supernatants to stimulate STAT1 tyrosine phosphorylation, and at 40 μM the inhibition was complete (Fig. 2C, lane 5). Interestingly, the BFA-treated LCL Pri cells were still able to respond to IFN-γ stimulation (Fig. 2D). Thus, treatment of the LCL Pri cells with BFA abolished the release of activator(s) of the tyrosine phosphorylation of STAT1.
FIG. 2.
The cell culture supernatant of BFA-treated LCL Pri cells does not induce tyrosine phosphorylation of STAT1. (A) LCL Pri cells were treated for 5 h with no BFA (lane 1) or with 10 (lane 2), 20 (lane 3), or 40 (lane 4) μM BFA. (B) LCL Pri cells were treated with BFA (40 μM) (lane 2) for 24 h to achieve almost complete inhibition of the tyrosine phosphorylation of STAT1. Lane 1, control nontreated LCL Pri cells. (C) BL2 cells were resuspended and incubated for 45 min in the supernatant of LCL Pri cells that had been treated for 5 h with either 10 (lane 3), 20 (lane 4), or 40 (lane 5) μM. Lane 1, control BL2 cells incubated in their own medium; lane 2, control BL2 cells incubated in the medium from LCL Pri cells that have not been treated with BFA. (D) The BFA-treated Pri cells were either treated for 1 h with IFN-γ (25 ng/ml) (lane 1), not treated (lane 2), or treated with supernatant from control Pri cells.
Inhibition of the NF-κB pathway prevents the phosphorylation of STAT1-induced by cell culture supernatant.
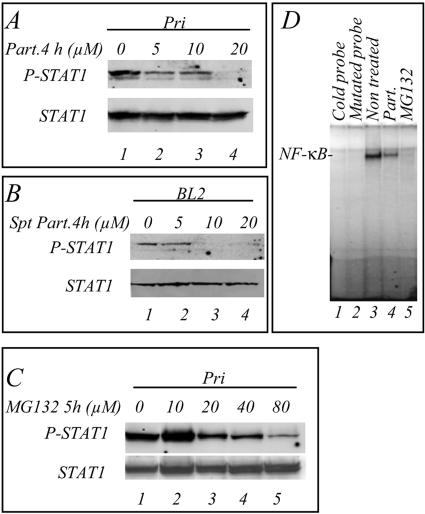
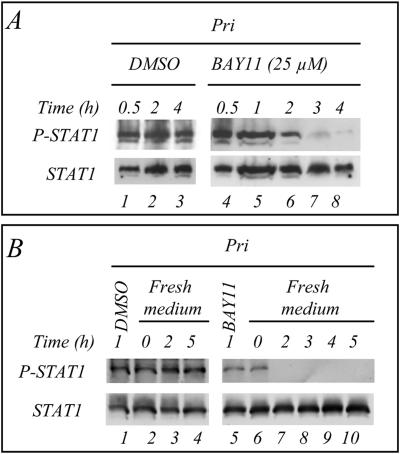
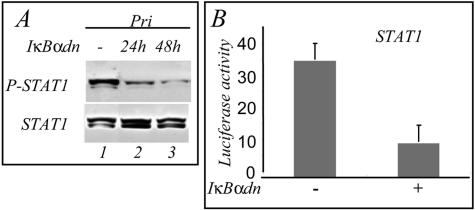
We next addressed the question of the involvement of the NF-κB pathway in the production of the factor(s) bringing about tyrosine phosphorylation of STAT1. To inhibit NF-κB, we incubated the LCL cells with parthenolide, a compound that inhibits the IκB kinase (IKK) (19). The results show that after 4 h of incubation the inhibition of the phosphorylation of STAT1 was detectable with 5 μM parthenolide (Fig. 3A, lane 2) and nearly complete with 20 μM (lane 4). The cell culture supernatants of the LCL cells were then used to resuspend BL2 cells for 45 min and analyze STAT1 tyrosine phosphorylation. Treatment of the Pri LCL cells with parthenolide resulted in a reduced capacity of their supernatant to stimulate STAT1 phosphorylation in BL2 cells (Fig. 3B). We determined that the concentrations of parthenolide used did not interfere with the IFN-γ-induced STAT1 phosphorylation (not shown). MG132, another NF-κB inhibitor, blocks the degradation of IκB by the proteasome (1). We observed that 20 μM MG132 applied for 5 h inhibited the phosphorylation of STAT1 in LCL cells (Fig. 3C, lane 3) and that maximal inhibition was observed with 80 μM (lane 5). We verified by electrophoretic mobility shift assay that these pharmacological compounds were indeed inhibiting NF-κB in our cell system (Fig. 3D). BAY11, a specific NF-κB inhibitor, was previously shown to inhibit rapidly and irreversibly the activity of NF-κB in LCLs (7). In a kinetic study, we incubated the Pri LCL cells with BAY11 (25 μM) and observed that the phosphorylation of STAT1 on tyrosine was inhibited after 2 h and abolished after 3 h (Fig. 4A). In an independent experiment, the cells were incubated with BAY11 (40 μM) for 1 h and then placed in fresh medium for different times: the phosphorylation of STAT1 on tyrosine was completely abolished after 2 h (Fig. 4B, lane 7). To confirm the involvement of NF-κB, we analyzed the phosphorylation of STAT1 on tyrosine in LCL cells transfected with a plasmid expressing dominant-active IκBα (V1NL IκBαdn). After 24 h of induction of IκBαdn, the phosphorylation of STAT1 on tyrosine was inhibited; this inhibition was maximal after 48 h of induction (Fig. 5A). The IκBαdn-transfected cells were also cotransfected with a luciferase reporter plasmid containing the promoter sequence of the STAT1-target gene TAP1 with an inactivated κB site. This resulted in the inhibition of the transcriptional activity of STAT1 (Fig. 5B). Thus, in LCLs, the phosphorylation and the transcriptional activity of STAT1 strongly depends on the constitutive NF-κB activity.
FIG. 3.
The cell culture supernatants of LCL Pri cells treated with parthenolide or MG132 do not induce tyrosine phosphorylation of STAT1. (A) LCL Pri cells were treated with 5 (lane 2), 10 (lane 3), or 20 (lane 4) μM of the IKK inhibitor parthenolide for 4 h, and the phosphorylation of STAT1 was determined by Western blotting. (B) BL2 cells were resuspended and incubated for 45 min in the supernatant of LCL Pri cells that had been treated for 4 h with either 5 (lane 2), 10 (lane 3), or 20 (lane 4) μM parthenolide. Lane 1, control BL2 cells resuspended in supernatant from nontreated LCL Pri cells. (C) LCL Pri cells were treated with a 10 (lane 2), 20 (lane 3), 40 (lane 4), or 80 μM concentration of the proteasome inhibitor MG132 for 5 h, and the phosphorylation of STAT1 determined by Western blotting. Lane 1, control nontreated LCL Pri cells. (D) Analysis of the DNA-binding activity to the κB probe of nuclear extracts from nontreated LCL Pri cells (3), parthenolide-treated cells (2), and MG132-treated cells (1). Nuclear extracts (20 μg) were incubated for 30 min with a 32P-radiolabeled κB probe and separated on gel (controls, cold probe [lane 1] and mutated probe [lane 2]). The gels were exposed to radiosensitive screens, which were then scanned.
FIG. 4.
Kinetics of the inhibition of the tyrosine phosphorylation of STAT1 in LCLs by the NF-κB specific inhibitor BAY11-7082. (A) Pri cells were treated with BAY11 (25 μM) for 30 min or 1, 2, 3, or 4 h (lanes 4 to 8). Control experiments were performed with DMSO (lanes 1 to 3). (B) LCL Pri cells were treated for 1 h with BAY11(40 μM) (lane 5). Cells were then resuspended in fresh medium without inhibitor for 2, 3, 4 or 5 h (lane 7 to 10). Control incubations were performed with DMSO (lanes 1 to 4). In lanes 2 and 6, cells were lysed immediately after resuspension in fresh medium to verify that the change of the medium in itself had no effect on the phosphorylation of STAT1. Similar results were obtained in three independent experiments.
FIG. 5.
The dominant-active form of IκBα blocks the phosphorylation and the transcriptional activity of STAT1 in the LCL Pri cells. LCL Pri cells were transfected with a doxycycline-inducible plasmid expressing a dominant-active form of IκBα (IκBαdn). (A) After induction for 24 (lane 2) or 48 (lane 3) h, the phosphorylation of STAT1 was determined by Western blotting. Lane 1, phosphorylation of STAT1 in control noninduced transfected LCL Pri cells. (B) The LCL Pri cells were cotransfected with the plasmid containing IκBαdn and the TAP1 luciferase reporter plasmid with a mutated κB site, the cells were sorted and, after 24 h of induction, the luciferase activity was measured in a luminometer. The luciferase activity was also measured in control noninduced cells.
Identification of NF-κB-dependently expressed cytokines in LCL cells.
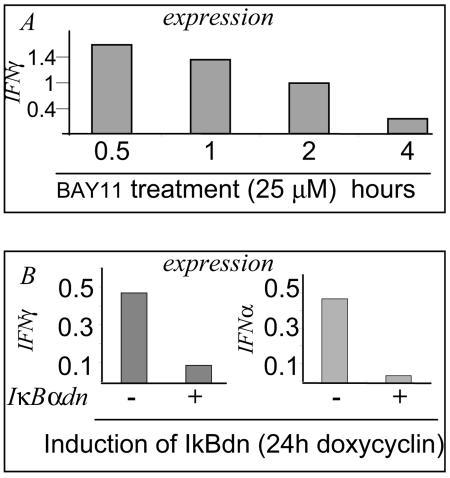
To detect NF-κB-regulated cytokine target genes transcribed in LCL cells, we analyzed the RNAs from control or parthenolide-treated (20 μM, 4 h) cells by using cDNA macroarray membranes spotted with 112 different cytokine probes. The expression of several cytokines was undetectable; other cytokines were detected, but their expression remained unchanged after treatment with parthenolide. The expression of several cytokines was reduced (see Table 1), including interleukin-6 (IL-6), IL-8, IL-12, IL-16, and IFN-γ. There was a slight inhibition of IFN-α isoforms. Among the cytokines the expression of which was reduced by parthenolide, IFN-α and IFN-γ are the ones known to induce STAT1 phosphorylation. To confirm the dependence of IFN-α and IFN-γ expression and secretion on NF-κB activity, we measured their expression by quantitative RT-PCR in BAY11-treated and in sorted induced IκBαdn-transfected LCL cells: there was a significant and gradual reduction of IFN-α and IFN-γ expression after inhibition of NF-κB (Fig. 6).
TABLE 1.
Variation of cytokines expression after treatment of LCL Pri cells with parthenolidea
| Variation in expression (%) | Gene description | GenBank no. |
|---|---|---|
| −97 | IL-16 (lymphocyte chemoattractant factor) | M90391 |
| −91 | IL-12B, p40 | M65272 |
| −86 | Homo sapiens TNF superfamily member LIGHT | AF036581 |
| −81 | Transforming growth factor β3 | NM_003239 |
| −75 | IFN-ω1 | NM_002177 |
| −71 | Insulin-like growth factor 1 (somatomedin C) | M27544 |
| −67 | IL-12A, p35 | M65271 |
| −64 | Vascular endothelial growth factor C | X94216 |
| −64 | Fibroblast growth factor 5 | NM_004464 |
| −63 | IFN-γ | X13274 |
| −63 | Bone morphogenetic protein 2 | NM_001200 |
| −62 | Insulin-like growth factor 2 (somatomedin A) | M29645 |
| −60 | Fibroblast growth factor 3 | X14445 |
| −60 | TNF (TNF superfamily, member 2) | X01394 |
| −59 | Fibroblast growth factor 4h | J02986 |
| −59 | Vascular endothelial growth factor B | U48801 |
| −58 | IL-17 (cytotoxic-T-lymphocyte-associated serine esterase 8) | U32659 |
| −55 | Platelet-derived growth factor alpha polypeptide | X06374 |
| −52 | IL-11 | M57765 |
| −50 | Transforming growth factor β1 | X02812 |
| −49 | Homo sapiens Apo3/DR3 ligand (APO3L) mRNA | AF055872 |
| −45 | IL-19 | NM_013371 |
| −43 | IL-9 | X17543 |
| −43 | Allograft inflammatory factor 1 | NM_001623 |
| −34 | Lymphotoxin alpha (TNF superfamily, member 1) | D12614 |
| −19 | Homo sapiens IFN-α1 | NM_024013 |
| −14 | IFN-α2 | V00544 |
| −14 | Colony-stimulating factor 1 (macrophage) | M37435 |
| −12 | Fibroblast growth factor 6 | NM_020996 |
| −11 | IFN-α4 | M27318 |
| −11 | IFN-α5 | NM_002169 |
| −10 | IL-10 | M57627 |
| −5 | IFN-α7 | M34913 |
| −4 | Colony-stimulating factor 2 (granulocyte-macrophage) | M11734 |
| 2 | IFN-α6 | NM_021002 |
| 93 | Fibroblast growth factor 12 | NM_021032 |
| 97 | Bone morphogenetic protein 8 (osteogenic protein 2) | NM_001720 |
| 108 | Fibroblast growth factor 14 | NM_004115 |
| 146 | IL-17B | AF110385 |
| 165 | Colony-stimulating factor 3 (granulocyte) | X03438 |
| 200 | Fibroblast growth factor 12B | NM_004113 |
The Pri cells were either incubated or not with 20 μM parthenolide for 4 h, the mRNA obtained was transcribed into cDNA in the presence of fluorescent ATP, and the cDNAs were hybridized on commercial prespotted membranes and exposed to radiographic films. The results are expressed as the percent inhibition of gene expression in parthenolide-treated cells relative to control cells.
FIG. 6.
Inhibition of NF-κB activity in the LCL Pri cells results in the repression of mRNA expression of IFN-γ and IFNα. (A) LCL Pri cells were treated with BAY11 (25 μM), mRNAs were extracted at different times of treatment. Quantitative RT-PCR analysis was performed to measure the expression of IFN-γ. (B) Selected V1LNGFR IκBαdn-transfected LCL Pri cells were induced for 24 h. Induced and noninduced cells were sorted by using antibody to LNGFR coupled to magnetic beads. The mRNAs of sorted cells were prepared and the expression of IFN-γ and IFN-α was analyzed by real-time PCR.
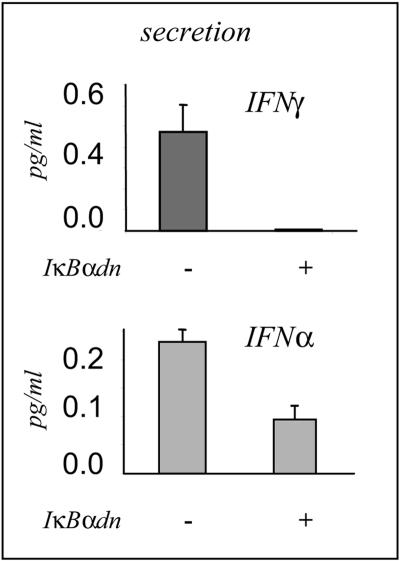
We next measured by ELISA the secretion of IFN-α and IFN-γ in the cell culture supernatants of the sorted induced IκBαdn transfected LCLs. We found that IFN-γ secretion was completely abolished and that IFN-α secretion was partially abolished (Fig. 7). Thus, LCL cells express and secrete several cytokines, including IFN-γ and IFNα, both involved in STAT1 activation.
FIG. 7.
Inhibition of NF-κB activity in the LCL Pri cells inhibits the release of IFN-γ and IFN-α. Selected V1LNGFR IκBαdn-transfected LCL Pri cells were induced for 24 h. The culture media of induced and noninduced cells were analyzed by ELISA for the secretion of IFN-γ (upper panel) and IFN-α (lower panel).
Neutralization of IFN-γ in cell supernatants blocks STAT1 phosphorylation.
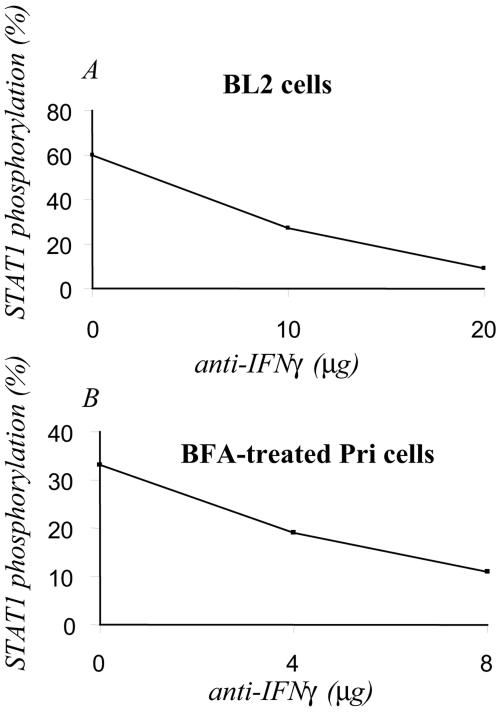
To ascertain that IFN secretion, per se, was responsible for the stimulation of STAT1 phosphorylation, we treated the cell culture supernatants with different concentrations of neutralizing antibodies. The supernatant neutralized with anti-IFN-γ was unable to induce the phosphorylation of STAT1 in BL2 cells (Fig. 8A). Identical results were obtained with anti-IFN-α (not shown). As shown above, in LCLs treated with BFA there was a strong inhibition of the tyrosine phosphorylation of STAT1 (see Fig. 2B). The resuspension of the BFA-treated LCLs with cell culture supernatant from untreated LCLs restored the phosphorylation of STAT1; however, an LCL supernatant neutralized with antibody to IFN-γ was unable to restore this phosphorylation (Fig. 8B). Thus, clearly, the neutralization of IFN-γ in the cell culture supernatant results in the almost complete inhibition of the stimulation of the phosphorylation of STAT1.
FIG. 8.
Treatment of the supernatants of the LCL Pri cells with IFN-γ-neutralizing antibodies blocks their capacity to stimulate STAT1 tyrosine phosphorylation. (A) Samples (1.5 ml) of the supernatants of Pri cells were incubated for 4 h with increasing concentrations (10 to 20 μg) of neutralizing anti-IFN-γ antibody. The neutralized supernatants were assayed on BL2 cells for 1 h. Western blotting was performed to determine the phosphorylation of STAT1. After autoradiography, the bands were quantified by using the Quantity One program. The results are expressed as a percentage of the control. (B) LCL Pri cells that had been preincubated with BFA to block cytokine secretion were resuspended in control supernatant from LCL Pri cells or supernatants that had been neutralized as in panel A.
DISCUSSION
In the present study, we demonstrate that the tyrosine phosphorylation of STAT1 in EBV-immortalized B cells expressing LMP1 is the result of the secretion of IFNs induced by the constitutive activation of NF-κB. The involvement of NF-κB is demonstrated by different pharmacological inhibitors and is confirmed by specific inhibition after the overexpression of a dominant-active form of IκBα. The direct involvement of IFNs is established by using neutralizing antibodies.
The phosphorylation, the activation and the overexpression of STATs have been demonstrated in several studies on EBV-transformed B cells and EBV-infected tissues (10, 16, 35). The overexpression of STAT1 in the EBV context was shown to be strongly associated with the level of LMP1 in the cell and to be dependent on the NF-κB activity (29, 38). Therefore, although these studies clearly indicated that the increased expression of STAT1 in EBV-immortalized cells is dependent on LMP1 and NF-κB, they have not pinpointed the precise mechanism involved and have not explored the mechanism of the activation of STAT1 phosphorylation in cells in which LMP1 is expressed. In the present study, we demonstrate in an unambiguous manner that the secretion of IFNs itself, resulting from the activation of NF-κB by LMP1, is the mechanism that accounts for STAT1 phosphorylation on tyrosine. This mechanism involves the induction of the expression of IFN-γ by NF-κB, the latter being activated by LMP1. Induction of IFN-γ is followed by its secretion, IFN-γ then activates its receptor on the LCL cells, leading to STAT1 tyrosine phosphorylation (Fig. 9). Interestingly, the induction of IRF1, a target of STAT1, is known to result in increased expression of IFN-α (26) the secretion of which stimulates the phosphorylation of STAT1. It is known that IFN-α activates both STAT1 and STAT2 (33) and indeed in our cell system we also find a constitutive activation of STAT2 (data not shown). Thus, the activation of NF-κB by LMP1 stimulates not only IFN-γ secretion but also IFN-α secretion (39). However, although the mechanism described here accounts for the major part of the phosphorylation of STAT1 on tyrosine in the LCL cells, we noted in our experiments that a residual phosphorylation was constantly present. This residual phosphorylation may result from incomplete inhibition of either NF-κB activity or IFN neutralization. Indeed, although the secretion of IFN-γ is completely abolished after the induction of IκΒαdn, the secretion of IFN-α is not. We cannot exclude the possibility that the phosphorylation of STAT1 results in part from an intracellular mechanism involving LMP1, as previously suggested (17). Such mechanism would involve the CTAR1 and CTAR2 motifs of LMP1 rather than the putative JAK3-binding motif from positions 232 to 351 (20).
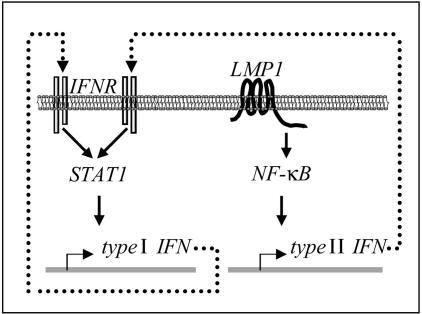
FIG. 9.
Proposed mechanism for the activation of the STAT1 pathway in lymphoblastoid cells. The viral latent membrane protein LMP1 activates the transcription factor NF-κB. IFN-γ, a target gene of NF-κB, is expressed, secreted and stimulates its receptor resulting in the phosphorylation of STAT1. STAT1 activates the transcription of several target genes.
In a previous study we observed that the inhibition of STAT1 by overexpression of the inhibitory isoform STAT1β resulted in an increased growth rate and reduced drug-induced apoptosis of the LCL cells (3), thereby demonstrating that the function of STAT1 in the EBV context was similar to that observed generally in other cell systems. The findings reported in the present study indicate that in LCL cells the constitutive phosphorylation of STAT1 is the result of the pleiotropic action of NF-κB. This transcription factor is known to be involved in cell growth, to reduce apoptosis, and to promote inflammation, and the induction of IFN-γ is in fact a side effect of this proinflammatory action. Although it has been clearly shown that LCL cells require NF-κB for their survival (6, 13), it is not clear whether the STAT1 pathway is required for the EBV to persist in cells. In favor of a requirement is the observations that the promoter region of LMP1 contains a functional STAT-binding site to which STAT1 binds strongly (9). Therefore, the EBV may ultimately require and usurp some of the defenses that were raised by cells against it.
Acknowledgments
We thank D. Johnson (National Institutes of Health, Bethesda, Md.) for providing the luciferase TAP1 reporter plasmid. We thank Stéphanie Le Coquil for skillful technical assistance.
This study was supported by Association de Recherche contre le Cancer grant 5861, the Ligue Nationale contre le Cancer (Labelisation Ligue UMR CNRS 6101, year 2002), the Conseil Régional du Limousin, and grants to G.W.B. from the Deutsche Forschungsgemeinschaft (SFB455) and the Wilhelm-Sander-Foundation. I.N. was supported by the Ministère de l'Éducation Nationale, de l'Enseignement Supérieur, et de la Recherche; I.Y.-M. was supported by the Conseil Régional du Limousin; and C.L.C. was supported by the Ligue contre le Cancer, Comité de la Corrèze.
REFERENCES
- 1.Andela, V. B., and R. N. Rosier. 2004. The proteosome inhibitor MG132 attenuates retinoic acid receptor transactivation and enhances trans-repression of nuclear factor κB: potential relevance to chemo-preventive interventions with retinoids. Mol. Cancer 3:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baran-Marszak, F., R. Fagard, B. Girard, S. Camilleri-Broet, F. Zeng, G. M. Lenoir, M. Raphaël, and J. Feuillard. 2002. Gene array identification of Epstein-Barr virus-regulated cellular genes in EBV-converted Burkitt lymphoma cell lines. Lab. Investig. 82:1463-1479. [DOI] [PubMed] [Google Scholar]
- 3.Baran-Marszak, F., J. Feuillard, I. Najjar, C. Le Clorennec, J. M. Bechet, I. Dusanter-Fourt, G. W. Bornkamm, M. Raphaël, and R. Fagard. 2004. Differential roles of STAT1α and STAT1β in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood 104:2475-2483. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, P., J. E. Floettmann, A. Mehl, M. Jones, and M. Rowe. 2001. Mechanism of action of a novel latent membrane protein-1 dominant negative. J. Biol. Chem. 276:1195-1203. [DOI] [PubMed] [Google Scholar]
- 5.Brewster, F. E., and J. L. Sullivan. 1983. Epstein-Barr virus-infected B lymphoblastoid cell lines: dynamics of interferon and 2′5′-oligoadenylate synthetase activity. Antivir. Res. 3:195-209. [DOI] [PubMed] [Google Scholar]
- 6.Cahir-McFarland, E., and E. Kieff. 2002. NF-κB inhibition in EBV-transformed lymphoblastoid cell lines. Cancer Res. 159:44-48. [DOI] [PubMed] [Google Scholar]
- 7.Cahir-McFarland, E. D., K. Carter, A. Rosenwald, J. M. Giltnane, S. E. Henrickson, L. M. Staudt, and E. Kieff. 2004. Role of NF-κB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J. Virol. 78:4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, K. L., E. Cahir-McFarland, and E. Kieff. 2002. Epstein-Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76:10427-10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., J. M. Lee, Y. Zong, M. Borowitz, M. H. Ng, R. F. Ambinder, and S. D. Hayward. 2001. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 75:2929-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagard, R., H. Mouas, I. Dusanter-Fourt, C. Devillers, P. Bissieres, A. Martin, G. M. Lenoir, H. VanTan, J. Feuillard, and M. Raphaël. 2002. Resistance to fludarabine-induced apoptosis in Epstein-Barr virus-infected B cells. Oncogene 29:4473-4480. [DOI] [PubMed] [Google Scholar]
- 11.Feuillard, J., C. Dargemont, V. Ferreira, N. Tarantino, P. Debré, M. Raphaël, and M. Korner. 1997. Nuclear Rel-A and c-Rel protein complexes are differentially distributed within human thymocytes. J. Immunol. 158:2585-2591. [PubMed] [Google Scholar]
- 12.Feuillard, J., H. Gouy, G. Bismuth, L. M. Lee, P. Debré, and M. Korner. 1991. NF-κB activation by tumor necrosis factor alpha in the Jurkat T-cell line is independent of protein kinase A, protein kinase C, and Ca2+-regulated kinases. Cytokine 3:257-265. [DOI] [PubMed] [Google Scholar]
- 13.Feuillard, J., M. Schuhmacher, S. Kohanna, M. Asso-Bonnet, F. Ledeur, R. Joubert-Caron, P. Bissieres, A. Polack, G. W. Bornkamm, and M. Raphael. 2000. Inducible loss of NF-κB activity is associated with apoptosis and Bcl-2 down-regulation in Epstein-Barr virus-transformed B lymphocytes. Blood 95:2068-2075. [PubMed] [Google Scholar]
- 14.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein-1 (LMP1) C terminus activation region 2 (CTAR2) maps to the far C terminus and requires oligomerisation for NF-B activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 15.Gabert, J., E. Beillard, V. H. J. van der Velden, W. Bi, D. Grimwade, N. Pallisgaard, G. Barbany, G. Cazzaniga, J. M. Cayuela, H. Cavé, F. Pane, J. L. E. Aerts, D. De Micheli, X. Thirion, V. Pradel, M. González, S. Viehmann, M. Malec, G. Saglio, and J. J. M. van Dongen. 2003. Standardization and quality control studies of “real-time” quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia. Leukemia 17:2318-2357. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, J. F., F. I. Camacho, M. Morente, M. Fraga, C. Montalban, T. A. Bellas, Carmen, A. Castano, A. Diez, T. Flores, C. Martin, M. A. Martinez, F. Mazorra, J. Menarguez, M. J. Mestre, M. Mollejo, A. I. Saez, L. Sanchez, and M. A. Piris. 2003. Hodgkin and Reed-Sternberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue microarrays. Blood 101:681-689. [DOI] [PubMed] [Google Scholar]
- 17.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hehner, S. P., T. G. Hofmann, W. Droge, and M. L. Schmitz. 1999. The antiinflammatory sesquiterpene lactone parthenolide inhibits NF-κB by targeting the IκB kinase complex. J. Immunol. 163:5617-5623. [PubMed] [Google Scholar]
- 20.Higuchi, M., E. Kieff, and K. M. Izumi. 2002. The Epstein-Barr virus latent membrane protein 1 putative Janus kinase 3 (JAK3) binding domain does not mediate JAK3 association or activation in B-lymphoma or lymphoblastoid cell lines. J. Virol. 76:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 19:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X., P. E. Massa, A. Hanidu, G. W. Peet, P. Aro, A. Savitt, S. Mische, J. Li, and K. B. Marcu. 2002. IKKα, IKKβ, and NEMO/IKKγ are each required for the NF-κB-mediated inflammatory response program. J. Biol. Chem. 277:45129-45140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, T., S. Sathe, S. M. Swiatkowski, C. V. Hampole, and G. R. Stark. 2004. Secretion of cytokines and growth factors as a general cause of constitutive NF-κB activation in cancer. Oncogene 23:2138-2145. [DOI] [PubMed] [Google Scholar]
- 26.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 27.Pahl, H. L. 1999. Activators and target genes of Rel/NF-B transcription factors. Oncogene 49:6853-6866. [DOI] [PubMed] [Google Scholar]
- 28.Peng, M., and E. Lundgren. 1992. Transient expression of the Epstein-Barr virus LMP1 gene in human primary B cells induces cellular activation and DNA synthesis. Oncogene 7:1775-1782. [PubMed] [Google Scholar]
- 29.Richardson, C., C. Fielding, M. Rowe, and P. Brennan. 2003. Epstein-Barr virus regulates STAT1 through latent membrane protein 1. J. Virol. 77:4439-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, p. 2573-2627. In D. Knipe, P. Howley, D. Griffin, R. Lamb, M. Martin, B. Roizman, and S. Straus (ed.), Virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 31.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In D. M. Knipe, B. N. Fields, and P. M. Howley (ed.), Virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 32.Sica, A., L. Dorman, V. Viggiano, M. Cippitelli, P. Ghosh, N. Rice, and H. A. Young. 1997. Interaction of NF-κB and NFAT with the interferon-gamma promoter. J. Biol. Chem. 272:30412-30420. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi, T., and A. Takaoka. 2001. A weak signal for strong responses: interferon-alpha/beta revisited. Nat. Rev. Mol. Cell Biol. 2:378-386. [DOI] [PubMed] [Google Scholar]
- 34.Wang, S., M. Rowe, and E. Lundgren. 1996. Expression of the Epstein-Barr virus transforming protein LMP1 causes a rapid and transient stimulation of the Bcl-2 homologue Mcl-1 levels in B-cell lines. Cancer Res. 20:4610-4613. [PubMed] [Google Scholar]
- 35.Weber-Nordt, R. M., C. Egen, J. Wehinger, W. Ludwig, V. Gouilleux-Gruart, R. Mertelsmann, and J. Finke. 1996. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood 88:809-816. [PubMed] [Google Scholar]
- 36.Wu, F., J. Garcia, R. Mitsuyasu, and R. Gaynor. 1988. Alterations in binding characteristics of the human immunodeficiency virus enhancer factor. J. Virol. 62:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, X., Z. He, B. Xin, and L. Cao. 2000. LMP1 of Epstein-Barr virus suppresses cellular senescence associated with the inhibition of p16INK4a expression. Oncogene 19:2002-2013. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, L., K. Hong, J. Zhang, and J. S. Pagano. 2004. Multiple signal transducers and activators of transcription are induced by EBV LMP-1. Virology 323:141-152. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, L., and J. S. Pagano. 2002. Structure and function of IRF-7. J. Interferon Cytokine Res. 22:95-101. [DOI] [PubMed] [Google Scholar]