Through single-cell transcriptomic analysis, Feng et al. identified a novel T follicular helper cell subset with stem cell–like transcriptional features and long-term survival ability, which gave rise to effector Tfh cells in recall immune responses.
Abstract
T follicular helper (Tfh) cells, essential for germinal center reactions, are not identical, with different phenotypes reported. Whether, when, and how they generate memory cells is still poorly understood. Here, through single-cell RNA-sequencing analysis of CXCR5+Bcl6+ Tfh cells generated under different conditions, we discovered, in addition to PD-1hi effector Tfh cells, a CD62L+PD1low subpopulation. CD62L-expressing Tfh cells developed independently from PD-1+ cells and not in direct contact with B cells. More importantly, CD62L+ Tfh cells expressed memory- and stemness-associated genes, and with better superior long-term survival, they readily generated PD-1hi cells in the recall response. Finally, KLF2 and IL7R, also highly expressed by CD62L+ Tfh cells, were required to regulate their development. Our work thus demonstrates a novel Tfh memory-like cell subpopulation, which may benefit our understanding of immune responses and diseases.
Introduction
Follicular helper T (Tfh) cells are essential in the regulation of germinal center (GC) reactions. They were originally defined by the expression of CXCR5, which facilitates CD4+ T cell migration into B cell follicles (Ansel et al., 1999; Breitfeld et al., 2000; Schaerli et al., 2000; Kim et al., 2001). Subsequently, Tfh cells were found to highly express PD-1, ICOS, or BTLA, but not CD62L, CCR7, and PSGL1, markers for naïve T cells, and were proposed to be distinct from other helper T cell subsets (Yu and Vinuesa, 2010; Suan et al., 2015; Crotty, 2014; Nurieva et al., 2008). In 2009, Bcl6 was defined as the lineage-specific transcription factor for Tfh cells; lack of Bcl6 in CD4+ T cells impaired Tfh cell development and the GC reaction (Nurieva et al., 2009; Yu et al., 2009; Johnston et al., 2009). Using a Bcl6-RFP reporter mouse, we previously showed that CXCR5+Bcl6+ T cells were functionally polarized Tfh cells and were sometimes called GC-Tfh cells (Liu et al., 2012; Crotty, 2011). However, Tfh cells are not phenotypically and functionally identical. Heterogeneity of CXCR5+Bcl6+ GC-Tfh cells has been reported, characterized by expression of IFN-γ (Velu et al., 2016; Liang et al., 2019), IL-4 (Weinstein et al., 2016), or IL-13 (Gowthaman et al., 2019). On the other hand, Tfh cells circulating in human peripheral blood were classified into Tfh1, Tfh2, and Tfh17 subpopulations based on their CXCR3 and CCR6 expression (Morita et al., 2011; Schmitt et al., 2009). Thus, single-cell analysis of Tfh cells is needed to better understand their functional heterogeneity in immune responses.
Memory T cells are generated as a result of immune responses. After the peak of the immune response, most effector cells undergo apoptosis during the contraction phase, leaving a small population of long-lived memory cells, which can maintain quiescent states but are also capable of self-renewal and survival without further antigenic stimulation (Seder and Ahmed, 2003). Memory T cells have been defined as central memory (Tcm), effector memory (Tem), and tissue-resident memory (Trm cells) T cells based on the expression of surface molecules and functions (Sallusto et al., 2004). Tem cells express homing receptors that facilitate their migration to peripheral tissues and produce a variety of cytokines to perform rapid effector functions. Tcm cells rarely produce any effector cytokine, but express CD62L and CCR7, which regulate cell circulation through lymph nodes (Sallusto et al., 2004). Trm cells express CD103 and CD69 and are specifically localized at the inflamed tissues (Mueller et al., 2013). Recent studies on chronic infection and cancer models have identified a subset of CD8+ lymphocytes with progenitor-like features, including self-renewal and the capacity to generate long-lived Tcm cells (Leong et al., 2016; He et al., 2016; Im et al., 2016; Gattinoni et al., 2017). This population of cells is sensitive to the immune checkpoint blockade therapy in cancer and thus became a promising immunotherapeutic target (Im et al., 2016; Utzschneider et al., 2016; Brummelman et al., 2018). Though CD8+ memory T cells have been characterized, their CD4+ T cell counterparts have not been well studied, especially with regard to their generation and function. In particular, the ontogeny of memory Tfh cells remains poorly understood, although we and others showed that CXCR5+ or Bcl6+ effector Tfh cells could generate memory Tfh cells (Lüthje et al., 2012; Liu et al., 2012).
In this study, we conducted single-cell RNA-sequencing (RNA-seq) analysis on CXCR5+Bcl6+ Tfh cells generated under several conditions. In addition to the previously characterized PD-1+ effector Tfh cells, we discovered a novel Tfh subset with memory cell features. This population of cells exhibits increased survival and gives rise to effector cells in the recall response. Our results thus reveal Tfh cell heterogeneity and offer new insights into memory Tfh cell development and regulation.
Results
Single-cell RNA-seq analysis of Tfh cell heterogeneity
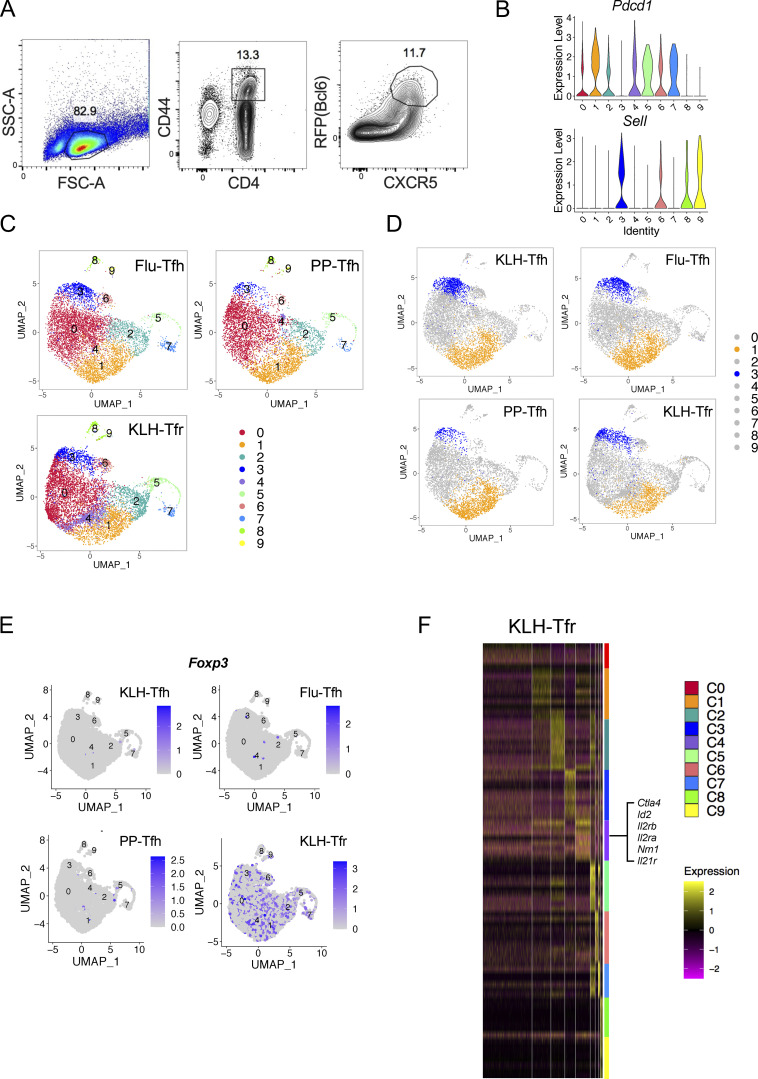
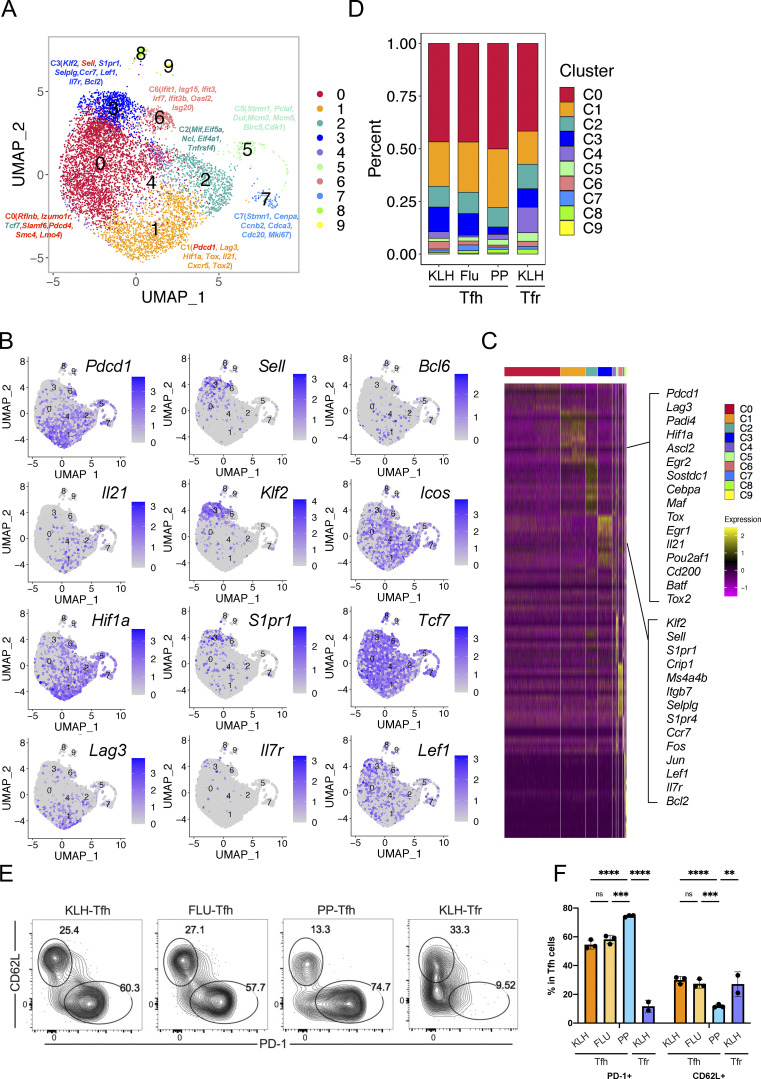
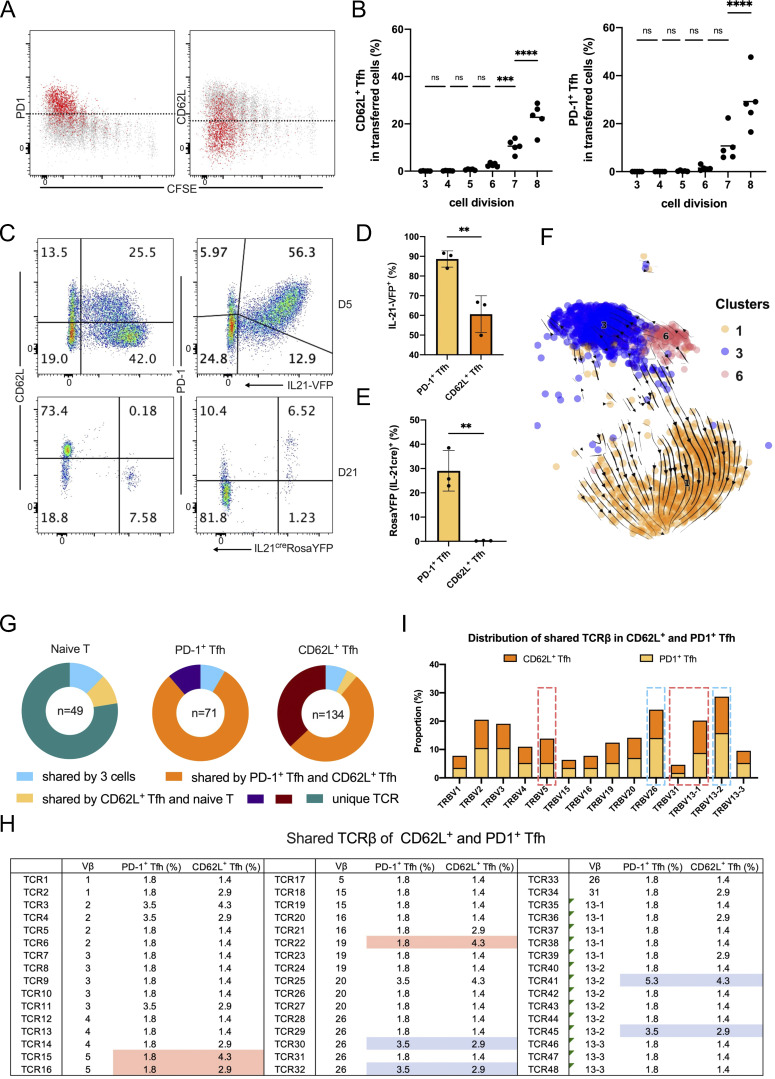
To characterize the heterogeneity of Tfh cells, we performed single-cell RNA-seq analysis, using the 10x Genomics platform, on live Tfh cells isolated from keyhole limpet hemocyanin (KLH)–immunized Bcl6RFP × Foxp3GFP reporter mice based on the CD4+CD44hiCXCR5+Bcl6-RFP+Foxp3-GFP−B220− gating (Fig. S1 A). After concatenation of data and quality control, a total of 5,410 cells were captured with 1,251 median genes per cell. Bioinformatic analysis of gene expression via unsupervised clustering identified 10 major cell clusters (Fig. 1 A). To define the identity of each cell cluster, we defined the most significantly differentially expressed genes (DEGs) in each cluster. DEGs in cluster 1 (C1) exhibited the characteristics of conventional effector Tfh cells, with high levels of expression in classical Tfh cell signature genes including Pdcd1, Lag3, Hif1a, Tox, Il21, Cxcr5, and Tox2. We also found several previously unidentified Tfh cell subtypes: Sell+ central memory-like (Pepper and Jenkins, 2011) (C3), Ifit1+ interferon-stimulated (C6), and Stmn1+ highly proliferating (C5 and C7). The lack of signature gene transcripts precluded the definition of C0 and C2 clusters (Fig. 1 A). Due to the existence of duplicates and multiplets, cells of C8 and C9 were likely macrophages and B cells, respectively, based on DEGs, and were excluded from further analysis. Interestingly, the expression of Pdcd1 and Sell in Tfh cells was mutually exclusive (Fig. S1 B). Pdcd1 expression was mostly detected in C1, C5, and C7, but barely in C3, whereas Sell was highly expressed in C3 and C6 (Fig. S1 B). Il21, Hif1a, and Lag3 expression correlated well with that of Pdcd1, while Klf2, S1pr1, and Il7r shared similar expression patterns with Sell. On the other hand, the expression of Icos and Tcf7 was widely distributed in each cluster (Fig. 1 B).
Figure S1.
Heterogeneity of Tfh and Tfr cells revealed by single-cell RNA-seq. (A) The gating strategy of GC-Tfh cells for single-cell RNA-seq. Live CD4+CD44hiCXCR5+RFP+GFP−B220− cells were sorted from draining lymph nodes of Bcl6RFP × Foxp3GFP reporter mice on day 8 after KLH immunization, and single-cell RNA-seq (10x platform) was performed. (B) Violin plots of Pdcd1 and Sell expressions in each cluster. (C) Live CD4+CD44hiCXCR5+RFP+GFP−B220− (GC-Tfh) cells were sorted from PP of Bcl6RFP × Foxp3GFP reporter mice under steady state and draining lymph nodes on day 8 after KLH immunization (KLH), day 9 after influenza infection (Flu) and were used for single-cell RNA-seq (10x platform). Unsupervised clustering of single-cell RNA-seq data of each sample was performed on a UMAP. (D) Cell distribution patterns in each model were shown by UMAP visualization. C1 and C3 were visualized in orange and blue, respectively. (E) Foxp3 expressions in indicated samples. (F) Heatmaps of DEGs in Tfr cells of each cluster. Selected marker genes of C4 were highlighted.
Figure 1.
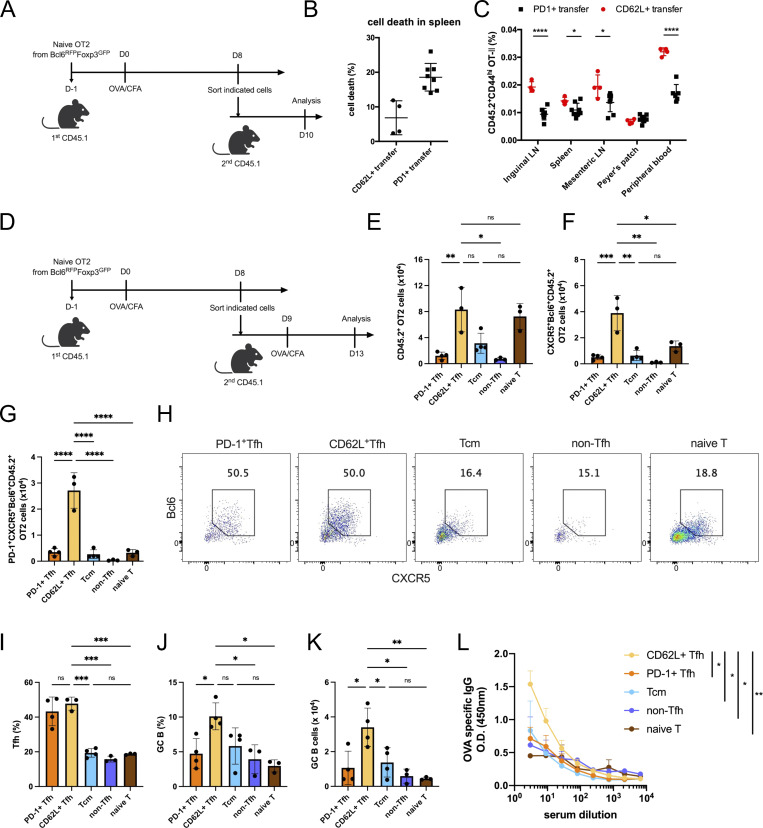
Identification of CD62L-expressing Tfh cells through single-cell transcriptomic analysis. (A) Unsupervised clustering of KLH immunization–derived Tfh single-cell RNA-seq data was performed on a UMAP. Selected DEGs of the indicated cluster were highlighted in the UMAP. (B) UMAP visualization showing gene expressions of Pdcd1, Il21, Hif1a, Lag3, Sell, Klf2, S1pr1, Il7r, Bcl6, Icos, Tcf7, Lef1. (C) DEGs of each cluster were shown by heatmaps. Selected genes of marker genes in C1 and C3 were highlighted. (D) Cluster distributions of each single-cell sample were shown by stacked bars. (E) Flow cytometry analysis of PD-1 and CD62L expression in Tfr cells derived from draining lymph nodes on day 8 after KLH immunization and GC-Tfh cells derived from draining lymph nodes at day 8 after KLH immunization (KLH), day 9 after influenza infection (Flu), as well as PP at steady state. (F) Statistical analysis of PD1+ and CD62L+ percentages in indicated cell types. Data of E and F represent two independent experiments. Data are shown as mean ± SD; two-tailed t test; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
We then further focused on DEGs in C1 (Pdcd1hi) and C3 (Sellhi). Many genes previously characterized as important in Tfh cell generation and function, including Ascl2, Egr2, Maf, Egr1, Pou2af1, Cd200, Batf, and Tox2 (Liu et al., 2012, 2014; Xu et al., 2019), were highly expressed in C1 (Fig. 1 C). In addition, several unidentified genes including Srgn, Padi4, Tbc1d4, and Angptl2, involved in cell metabolic pathways and inflammatory responses (Suzuki et al., 2003; Moltke et al., 2014; Tabata et al., 2009; Chen et al., 2020), were also found in DEGs of C1. C1 cells thus likely represented effector Tfh cells. In the meantime, cells in C3 specifically upregulated expression of genes related to memory T cell formation, such as Sell, Lef1, and Ccr7 (Seder and Ahmed, 2003; Sallusto et al., 2004; Mueller et al., 2013; Zhao et al., 2010), as well as genes involved in cellular localization and migration, including S1pr1, Itgb7, Vim, Selplg, S1pr4, Il7r, and Cytip (Rivera et al., 2008; Neri et al., 2011; Mueller et al., 2013); anti-apoptosis and cell stemness–related genes, such as Klf2, Klf6, and Bcl2 (Bai et al., 2007; Wang et al., 2016; Carlson et al., 2006; García-Sáez, 2012); and cell cycle inhibitors, including Ms4a4b, Cdkn2d, and Lsp1 (Baumgartner and Harper, 2003; Xu et al., 2010) (Fig. 1 C).
The above data suggest that Bcl6+ Tfh cells may be heterogenous. To examine if they were not limited to KLH immunization, we also conducted single-cell RNA-seq of Tfh cells in draining lymph nodes of influenza-infected mice and Peyer’s patches (PP) of naïve mice using the same gating strategy. Tfr cells in draining lymph nodes of KLH-immunized Bcl6RFP × Foxp3GFP reporter mice were also isolated based on CD4+CXCR5+Bcl6-RFP+Foxp3-GFP+ gating and analyzed similarly. The Uniform Manifold Approximation and Projection (UMAP) analysis showed generally consistent cell cluster subtypes in these Tfh and Tfr cells as Tfh cells in the KLH model (Fig. S1 C), although the ratios between the cell clusters were different depending on the immune contexts and cell subtypes (Fig. 1 D). Tfh cells in PP showed relatively higher frequencies of Pdcd1hi cluster (C1) and lower frequencies of Sellhi cluster (C3) compared to those from KLH immunization and influenza infection (Fig. 1 D and Fig. S1 D). Furthermore, C4 in Tfr cells derived from KLH immunization was unique (Fig. S1 C), and almost not detected in Tfh cells under all conditions (Fig. 1 D). Although the majority of the Tfr cells express Foxp3 (Fig. S1 E), genes with the highest levels of expression in C4 of Tfr cells were Ctla4, Id2, Foxo1, Il2ra, Il2rb, and Nrn1, all related to regulatory T cell development and function (Fig. S1 F).
To validate the above single-cell RNA-seq results, we analyzed PD-1 and CD62L protein expression by flow cytometry in CD4+CD44hiCXCR5+Bcl6+ Tfh cells in draining lymph nodes of KLH-immunized mice. We found ∼60% GC-Tfh cells expressed PD-1 and ∼25% CD62L+ (Fig. 1, E and F). In addition, we also examined Tfh cells in draining lymph nodes of influenza virus-infected mice and in PP in unchallenged mice at steady states. Similar cell distribution patterns were observed (Fig. 1, E and F), though the ratio of PD1+ versus CD62L+ Tfh cells in PP was significantly higher compared with that in KLH immunization and influenza infection (Fig. 1, E and F). Consistent with the single-cell RNA-seq results, CD4+CXCR5+Bcl6+ follicular regulatory (Tfr) cells from KLH immunization also contained PD1+ and CD62L+ subpopulations but the ratio of PD1+ versus CD62L+ in Tfr cells were relatively lower than those in Tfh cells (Fig. 1 F and Fig. S1 D).
Taken from the above data, it appears that during the immune response, Tfh cells, and perhaps Tfr cells, contain several distinct subtypes, especially with a CD62L-expressing population, in addition to the conventional PD-1hi effector Tfh cells.
Increased percentages of CD62L+ Tfh cells along with immune response progression
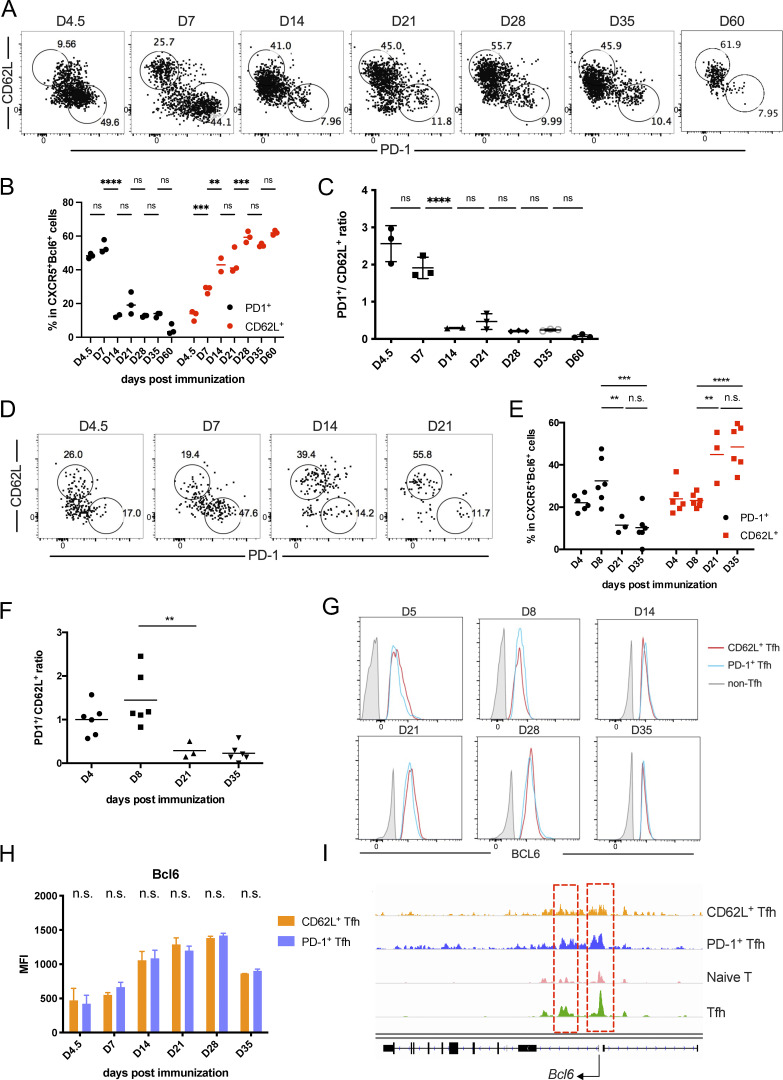
The above single-cell transcriptomic analysis has revealed a CD62L-expressing Tfh cell population. To characterize these cells further, we first examined Tfh cells in B6 mice analyzed at different time points after KLH immunization. PD-1+ Tfh cells were the predominant cell population at the early stages of the immune response but sharply decreased 14 days after immunization (Fig. 2, A–C). In contrast, CD62L+ Tfh cells increased in percentages over time and became the major composition of Tfh cells 28 days after immunization (Fig. 2, A–C), suggesting that they may indeed represent the memory cells.
Figure 2.
Increased percentages of CD62L+ Tfh cells during the progression of immune responses. (A and B) Kinetic expression of PD-1 and CD62L in CD4+CD44hiCXCR5+Bcl6+ GC-Tfh cells after KLH immunization at indicated time points. (C) The ratios of PD1+ versus CD62L+ Tfh cell percentages at indicated time points. (D and E) Kinetic expression of PD-1 and CD62L in adoptive-transferred CD45.1+CD44hiCXCR5+Bcl6+ OT-II T cells after OVA immunization at indicated time points. (F) The ratios of PD1+ versus CD62L+ cell percentages in transferred CD45.1+CD44hiCXCR5+Bcl6+ OT-II T cells at indicated time points. (G) Representative Bcl6 intracellular staining levels in PD-1+, CD62L+ Tfh cells, and non-Tfh cells at indicated time points. (H) Mean fluorescence intensities (MFI) of Bcl6 expression in CD62L and PD-1+ Tfh cells at each time point. (I) Chromatin accessibility of the Bcl6 gene locus of CD62L+ Tfh, PD-1+ Tfh, and naïve CD4+ T cells were analyzed in parallel. Red lines highlight the differentially accessible regions of Bcl6 gene within different cells. Data of A–G represent two independent experiments. Data are shown as mean or mean ± SD; two-tailed t test; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
We also analyzed antigen-specific Tfh cells using OT-II TcR transgenic T cells. Similar to KLH immunization, naïve OT-II T cells transferred into CD45.1 recipient mice developed into PD-1+ and CD62L+ Tfh cell populations after OVA immunization (Fig. 2, D–F). The ratios of PD-1+ versus CD62L+ Tfh cells were firstly increased upon antigen immunization and then decreased after the peak of the immune response (Fig. 2 F).
Although in single-cell analysis, Bcl6 transcripts were not been well detected in Tfh cells (Fig. 1 B), possibly due to limitation of sensitivity, through Bcl6 intracellular staining, we found that Bcl6 protein expression in CD62L+ Tfh cells and PD-1+ Tfh cells were comparable (Fig. 2, G and H). Meanwhile, the overall chromatin accessibilities at the Bcl6 gene locus had no significant difference between CD62L+ Tfh cells and PD-1+ Tfh cells (Fig. 2 I). Taken together, we believe that CD62L+ Tfh cells discovered in single-cell RNA-seq were indeed bona fide Tfh cells.
Ontogeny of CD62L-expressing Tfh cells
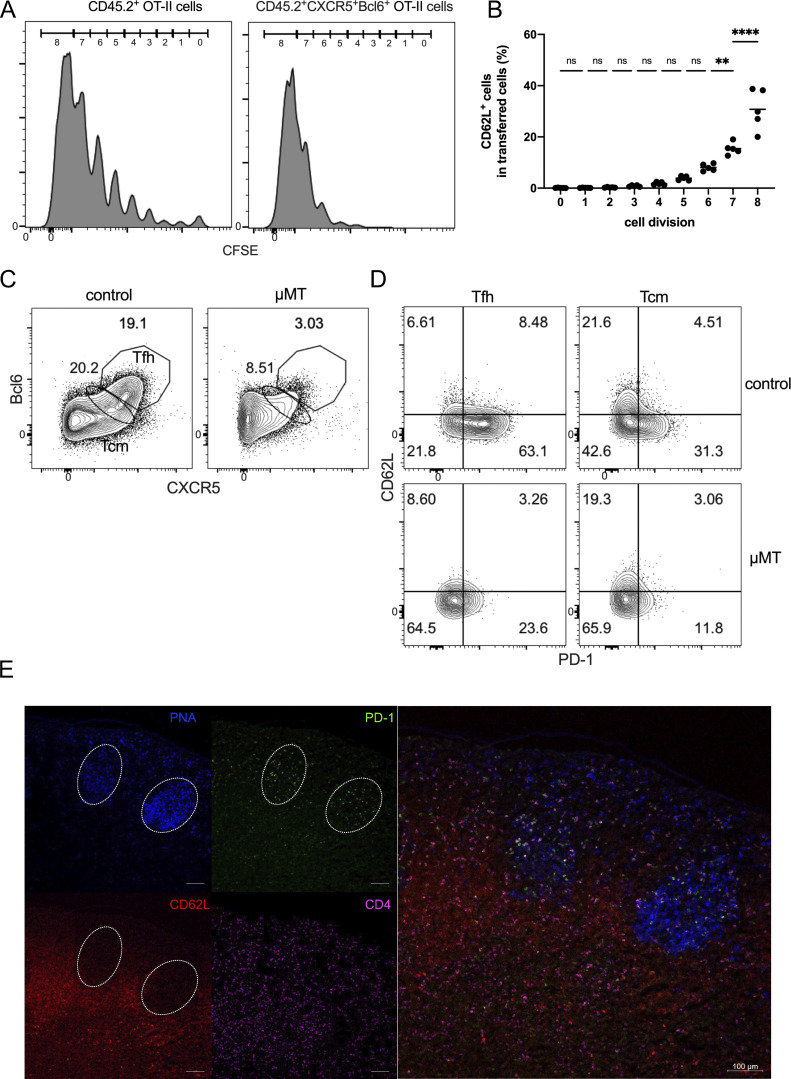
To analyze if CD62L-expressing Tfh cells develop as a function of cell divisions, we transferred CFSE-labeled-naïve OT-II T cells from Bcl6RFP × Foxp3GFP × OT-II mice into CD45.1 transgenic mice and immunized them with OVA protein. Since CD45.2+CD44hiCXCR5+Bcl6+ Tfh cells were barely detected in the first three cell divisions at day 4 after OVA immunization (Fig. S2 A), we mainly analyzed the expressions of PD-1 and CD62L in Tfh cells having divided above three times. After OVA immunization, T cells, even without divisions, rapidly downregulated CD62L expressions and some gradually regained the expressions of CD62L after divisions (Fig. S2 B). More importantly, Tfh cells simultaneously acquired PD-1 and CD62L expression along with the cell divisions (Fig. 3, A and B), indicating that CD62L+ and PD-1+ Tfh cells were generated simultaneously at early phases of immune responses.
Figure S2.
Developmental features of PD-1+ and CD62L+ Tfh cells. (A and B) Naïve OT-II labeled with CFSE were adoptively transferred into CD45.1 recipient mice, which were subsequently immunized subcutaneously with OVA. Draining lymph nodes were isolated for analysis on day 4 after immunization. (A) Immunofluorescence intensity of CFSE in each cell division of indicated cell populations. (B) CD62L+ cell percentages in transferred CD45.2+ OT-II cells. (C and D) μMT and control mice were subcutaneously immunized with KLH. 7 days later, draining lymph nodes were isolated for analysis. (C) Representative flow cytometry analysis of CXCR5 and Bcl6 expression in CD4+CD44hi cells of μMT and control mice. (D) Representative flow cytometry analysis of PD-1 and CD62L expression in Tfh (CXCR5hiBcl6hi) and Tcm (CXCR5midBcl6mid) cells of μMT and control mice. (E) C57BL/6 mice were subcutaneously immunized with KLH, and the draining lymph nodes were isolated, embedded in OCT, and subjected to cryosectioning for subsequent immunofluorescence staining. Representative immunofluorescence staining of PNA (blue), PD-1 (green), CD62L (red), and CD4 (violet) and four-color overlay image of the frozen sections. The length of scale bars represents 100 µm, unless labeled in the image. Data of A–E represent two independent experiments. Data are shown as mean; two-tailed t test; **, P < 0.01; ****, P < 0.0001 ns, no significance.
Figure 3.
Ontogeny of CD62L-expressing Tfh cells in immune responses. (A and B) Naïve OT-II labeled with CFSE were adoptively transferred into CD45.1 recipient mice, which were subsequently immunized subcutaneously with OVA. Mouse draining lymph nodes were isolated for analysis at day 4 after immunization. (A) Representative flow cytometry staining of PD-1 or CD62L in each cell division of CXCR5+Bcl6+CD45.2+ OT-II cells. (B) Statistical analysis of (left) CD62L+CXCR5+Bcl6+ and (right) PD1+ CXCR5+Bcl6+ cell percentages in transferred CD45.2+ OT-II cells. (C–E) IL21-VFP and Il21creR26YFP mice were subcutaneously immunized with KLH. 5 or 21 days later, mice were sacrificed for analysis. (C) Representative flow cytometry analysis of PD-1 and CD62L versus IL-21-VFP or YFP expression in CD4+CD44hiCXCR5+ cells at day 5 after KLH immunization of IL21-VFP mice or day 21 after KLH immunization of Il21creR26YFP mice, respectively. (D and E) Statistical analysis of IL-21-VFP+ or YFP+ percentages in PD1+ or CD62L+ Tfh cells, respectively. (F) RNA velocity analysis of cells in C1, C3, and C6 projected onto the UMAP plot. Arrows indicated the direction and speed of each cell. (G–I) Bcl6RFPFoxp3GFP mice were immunized with KLH, 8 days later, PD-1+ Tfh, CD62L+ Tfh, and naïve T cells were isolated into 96-well plates, and the TCRβ chain mRNA sequences were amplified and sequenced. (G) TCRβ clonotypes in PD-1+ Tfh, CD62L+ Tfh, and naïve T cells. (H) The distribution of shared TCRβ sequences in CD62L+ and PD1+ Tfh cells. Blue and red boxes indicate the uneven presence of indicated TCRβ clonotypes in PD1+ and CD62L+ Tfh cells, respectively. (I) The proportions of shared TCR in each TCR Vb subtype between CD62L+ and PD1+ Tfh cells. Blue and red lines indicate higher percentages of indicated TCR Vb in PD1+ and CD62L+ Tfh cells, respectively. Data above are representative of two independent experiments. Data of A–E represent two independent experiments. Data are shown as mean or mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
To explore the ontogeny of CD62L+ Tfh cells, especially to understand if they are derived from PD-1+ Tfh cells, we further employed a new Il21 fate-mapping mice in our analysis. In the single-cell analysis, Il21 expression was restricted in PD1+, but not CD62L+ Tfh cells (Fig. 1 B). Using IL21VFP reporter mice, we confirmed that PD-1 was coexpressed with IL-21 in CXCR5+ cells after KLH immunization (Fig. 3, C and D). By utilizing a new Il21creRosaYFP lineage-tracing mouse, we found that at the immune contraction phase (day 21) after KLH immunization, memory CD62L+ Tfh cells had not expressed IL-21 (Fig. 3, C and E), indicating that the majority of memory CD62L+ Tfh cells were not derived from IL-21–expressing PD-1+ Tfh cells. In the meantime, we also observed a fraction of CD62L+ Tfh cells expressing intermediate levels of IL-21 at the early stage, but these cells were not detected at the immune contraction phase (Fig. 3 C), which indicates that they were not long-lived and may not serve as the Tfh memory precursor cells. Meanwhile, through the prediction of cell developmental trajectory calculated by the RNA velocity analysis, a strong developmental path from Sellhi toward Pdcd1hi Tfh cells, but not the reverse, was observed (Fig. 3 F).
To better characterize CD62L+ Tfh cells, we performed TCR clonotype analysis on PD-1+ and CD62L+ Tfh cells generated after KLH immunization with naïve T cells as controls. Based on the TCRβ chain sequences, as expected, TCR clonotypes were distinct between naïve T cells and two subsets of Tfh cells (Fig. 3 G). Importantly, CD62L+ and PD1+ Tfh cells shared many TCR sequences, which suggests that PD1+ and the majority of CD62L+ cells were derived from the same progenitors (Fig. 3 G). This result is consistent with that obtained with OT-II T cells, and they together indicate a stochastic process in the generation of antigen-specific CD62L+ and PD-1+ Tfh cells. However, we still observed a fraction of CD62L+ Tfh cells carrying unique TCRs, the basis of which is still unknown (Fig. 3 G). These CD62L+ Tfh cells, as one possibility, may derive from pre-existing or bystander memory T cells. When we further analyzed the shared TCRβ sequences between PD-1+ and CD62L+ Tfh cells, we found that certain TCRs showed differential distributions between two groups of cells (Fig. 3, H and I), suggesting that TCR signaling may impact the development of PD-1+ and CD62L+ Tfh cells.
Taken together, CD62L+ Tfh cells were generated at an early stage during development, simultaneously with PD-1+ Tfh cells derived from the same progenitors with biased TCR preferences. Moreover, long-lived CD62+ Tfh cells were not derived from IL-21–expressing PD-1+ Tfh cells.
CD62L-expressing Tfh cells are not preferentially in contact with B cells
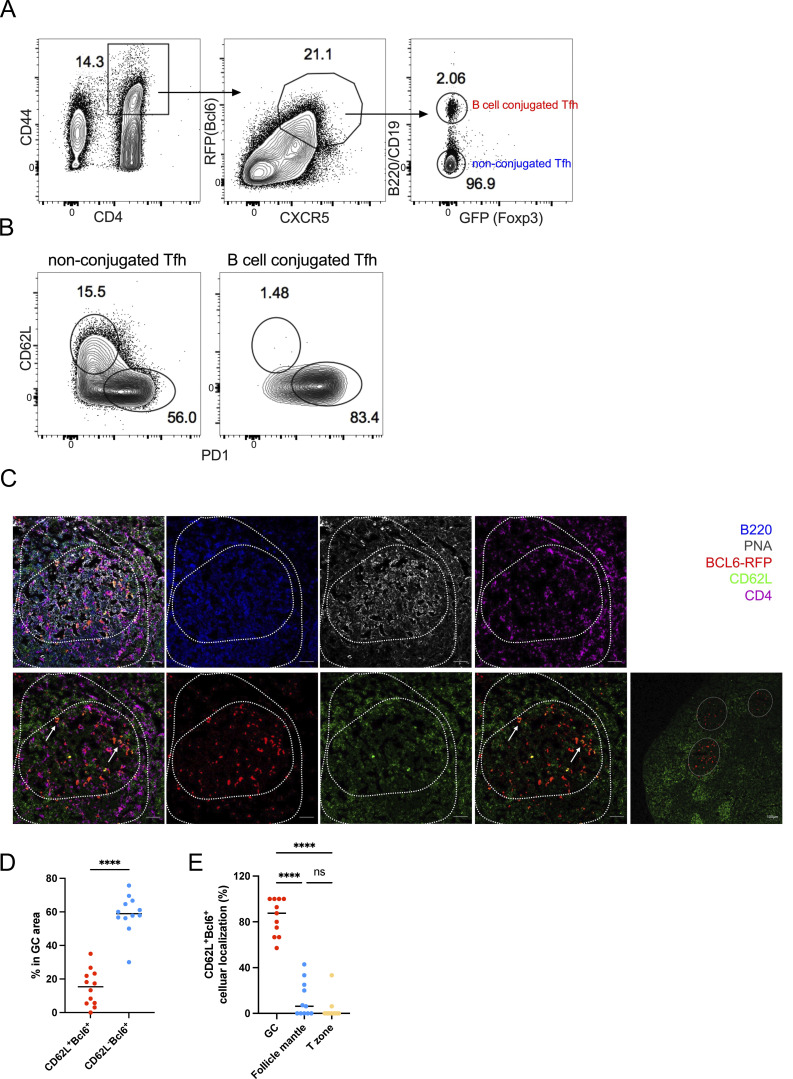
Tfh cells are in close contact with cognate B cells to provide “help” in B cell proliferation and differentiation. Thus, we compared PD-1– and CD62L-expressing Tfh cells in forming conjugates with B cells. B cell–conjugated Tfh cells were sorted from KLH-immunized Bcl6RFP × Foxp3GFP reporter mice (Fig. 4 A) and disassociated in vitro. The majority of B cell–conjugated Tfh cells were PD-1+, but barely expressed CD62L (Fig. 4 B), suggesting that CD62L-expressing Tfh cells were not preferentially in contact with B cells. However, we observed that both PD-1– and CD62L-expressing Tfh cells required B cells for their generation, as KLH-immunized B cell–deficient μMT mice showed the absence of both PD-1+ and CD62L+ Tfh cells (Fig. S2, C and D). These results suggest that CD62L+ Tfh cells, though not preferentially in contact with B cells, depended on B cells for their development. To further characterize the localization of CD62L+ Tfh cells, we then performed immunofluorescence. By immunizing Bcl6-RFP reporter mice, we observed notably reduced proportions of CD62L-expressing Bcl6+ T cells within the GC area when compared with CD62L−Bcl6+ T cells (Fig. 4, C and D). This observation suggests that the majority of Bcl6-expressing CD4 T cells within the GC, very likely expressing PD-1 (Fig. S2 E), lacked CD62L expression. Furthermore, our comprehensive analysis of the overall cellular distribution of CD62L+BCL6−RFP+ cells elucidated their predominant localization within PNA+B220− GC areas, with only a sparse presence detected in the PNA−B220+ follicle mantle and T cell zone (Fig. 4 E). These discerning findings effectively distinguish CD62L−Bcl6+ Tfh cells discovered in our study from the previously characterized follicle mantle Tfh cells. Based on the above observation that CD62L+ Tfh cells highly expressed T cell migration markers, such as S1pr1, S1pr4, etc. (Fig. 1, B and C), CD62L+ Tfh cells are likely to circulate into the periphery.
Figure 4.
CD62L-expressing Tfh cells are not in contact with B cells, and prone to circulate. (A) The gating strategy used for isolating B cell–conjugated GC-Tfh cells. Cells were obtained from the draining lymph nodes of Bcl6RFPxFoxp3GFP double reporter mouse on day 8 after KLH immunization. (B) Representative flow cytometry staining of PD-1 and CD62L expression on B cell–conjugated and unconjugated Tfh cells after EDTA treatment. (C–E) BCL6-RFP reporter mice were subcutaneously immunized with KLH, and the draining lymph nodes were isolated, embedded in OCT, and subjected to cryosectioning for subsequent immunofluorescence staining. (C) Representative immunofluorescence staining of B220 (blue), PNA (gray), BCL6-RFP (red), CD62L (green), and CD4 (violet), and multicolor overlapping images of the frozen sections. The upper panels represent five-color overlapping, B220, PNA, and CD4 staining (left to right); the bottom panels represent three-color overlapping (CD62L+Bcl6−RFP+CD4), Bcl6−RFP, CD62L, two-color overlapping (CD62L+Bcl6−RFP) of single GC, and entire follicles (left to right). White arrows indicate CD62L+BCL6-RFP+ CD4 T cells in the GC area. The length of scale bars represents 50 µm, unless labeled in the image. (D) Statistical analysis of CD62L+BCL6-RFP+ and CD62L−BCL6-RFP+ CD4 T cells in GC areas. (E) Statistical analysis of CD62L+BCL6-RFP+ CD4 T cells localized in GC area (PNA+B220low), follicle mantle (PNA−B220+), and T cell zone (PNA−B220−CD4+). Data of A–E represent two independent experiments. Data are shown as mean. Paired t test; ****, P < 0.0001, ns, no significance.
CD62L-expressing Tfh cells exhibit memory cell features
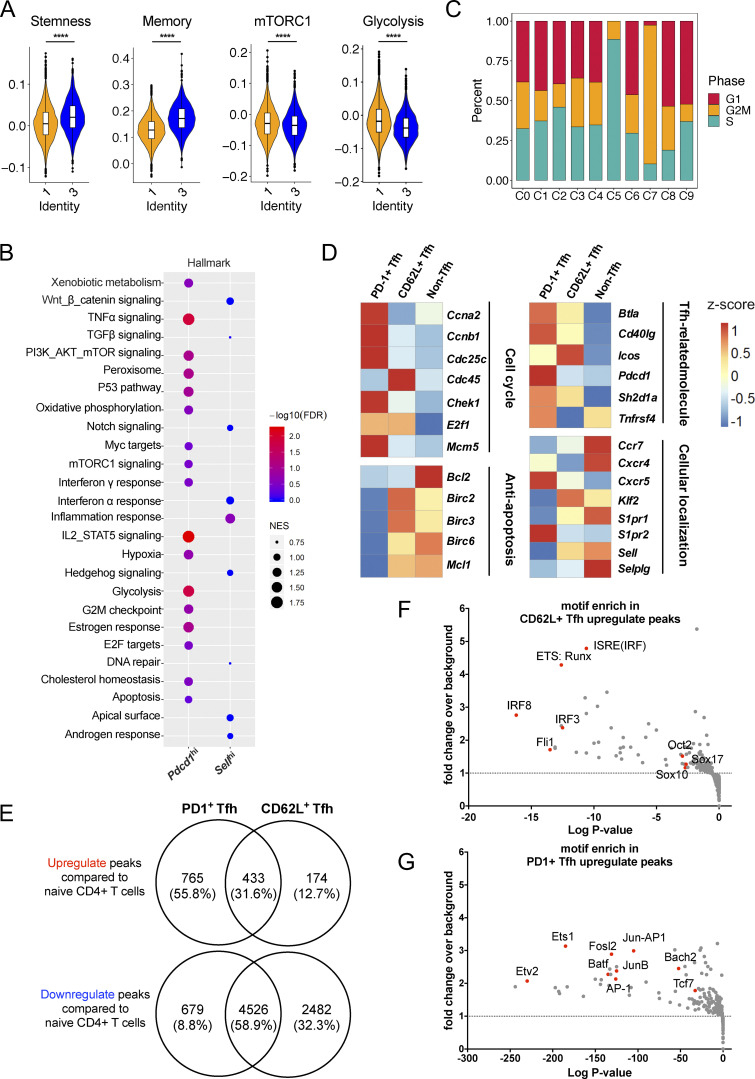
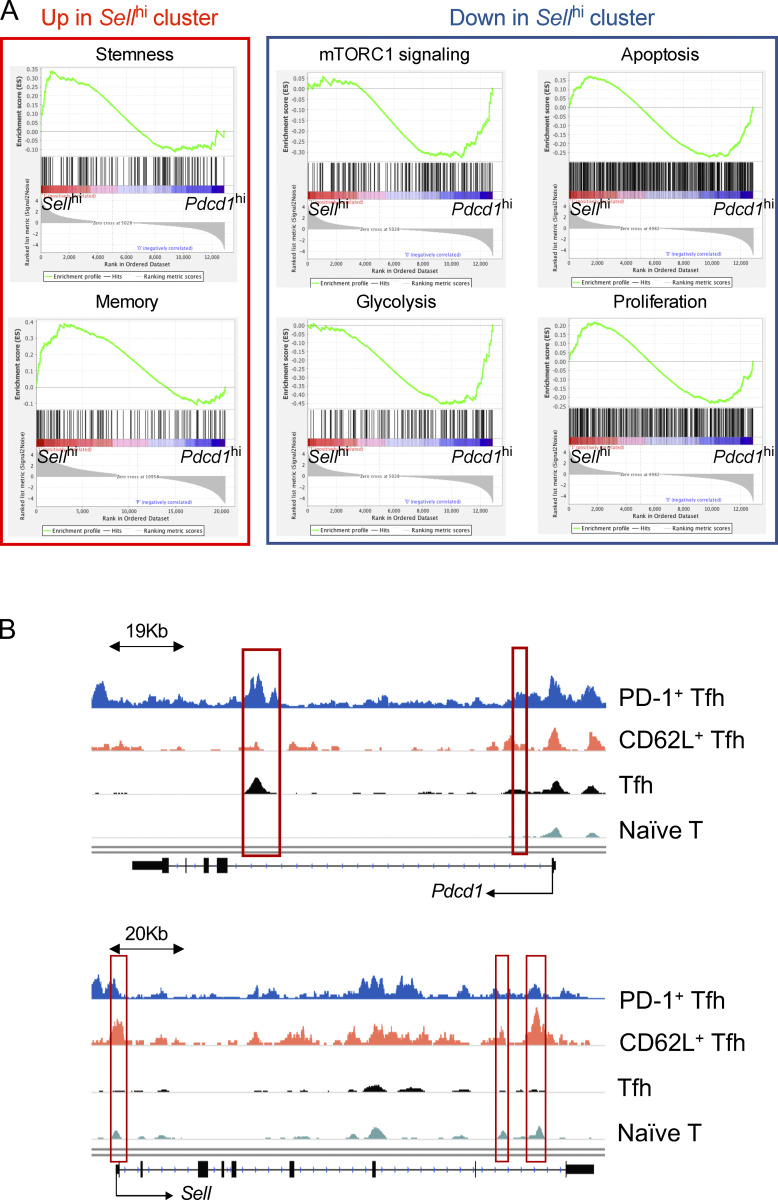
To better analyze the characteristics of CD62L+ Tfh cells, we compared Pdcd1hi cell cluster (C1) and Sellhi cluster (C3) in their DEGs based on our single-cell RNA-seq results. The genes specifically expressed in Sellhi cluster were related to pathways associated with cell stemness (GO: 0019827; GO: 0035019) and T cell memory (Hale et al., 2013) from published gene sets, but less correlated with genes involved in mTCRC1 signaling and glycolysis processes (Fig. 5 A and Fig. S3 A). Gene set enrichment analysis (GSEA) also revealed that CD62L+ Tfh cells were characterized by transcripts associated with Wnt, Notch, and Hedgehog signaling pathways, whereas PD-1+ Tfh cells were characterized by transcripts associated with cell cycle, apoptosis, and oxidative phosphorylation (Fig. 5 B). Moreover, Pdcd1 expression was mostly enriched in C1, C2, C5, and C7 (Fig. 1 C). Although the cell cycle did not have any significant difference between C1 and C3, C5 and C7 represented distinct cell status and proliferation phases (Fig. 5 C), indicating that Pdcd1-expressing cells were more proliferative.
Figure 5.
CD62L-expressing Tfh cells exhibit stem-like memory precursor transcriptional features. (A) Enrichment scores of gene sets associated with stemness, memory, mTORC1 signaling, and glycolysis of C1 (orange color) and C3 (blue color). (B) Pathways enriched in Pdcd1hi and Sellhi clusters as analyzed by GSEA. (C) Stacked bars showing cell cycle phases in each cluster. (D) Heatmaps showing genes associated with cell cycle, anti-apoptosis, Tfh cell signature, and cellular localization expression in CD62L+ Tfh, PD-1+ Tfh, and non-Tfh cells derived from draining lymph nodes of Bcl6RFP × Foxp3GFP double reporter mice at day 8 after KLH immunization. (E) Chromatin accessible regions of CD62L+ Tfh, PD-1+ Tfh, and naïve CD4+ T cells were analyzed in parallel. Venn map shows the overlap of upregulated peaks (top) of CD62L+ Tfh and PD-1+ Tfh cells compared with naïve CD4+ T cells or downregulated peaks (bottom) of CD62L+ Tfh and PD-1+ Tfh cells compared with naïve CD4+ T cells. (F and G) CD62L+ and PD-1+ Tfh cells derived from draining lymph nodes of Bcl6RFP × Foxp3GFP double reporter mice on day 8 after KLH immunization were isolated and ATAC-seq was performed. Binding motif of gene or gene complex enriched in CD62L+ Tfh cells (F) and (G) PD-1+ Tfh cells upregulated peaks. Specific genes were highlighted in red dots. ****, P < 0.0001.
Figure S3.
Transcriptional and epigenetic features of CD62L+ Tfh cells. (A) GSEA of gene sets associated with stemness, memory, mTORC1 signaling, glycolysis, apoptosis, and proliferation in Sellhi and Pdcd1hi clusters. (B) Chromatin regions of Pdcd1 and Sell in CD62L+ Tfh cells, PD-1+ Tfh cells, naïve CD4+ T cells and in vitro differentiated Tfh cells differentially accessible regions were highlighted in red.
To further validate the observations in single-cell RNA-seq analysis, we compared bulk RNA-seq data of CD62L+ and PD-1+ Tfh cells, as well as non-Tfh cells isolated based on CD4+CD44hiCXCR5−Bcl6-RFP−Foxp3-GFP−. Consistent with single-cell RNA-seq data, CD62L+ Tfh cells exhibited increased expression of genes associated with anti-apoptosis and lymphocytes egress and decreased expression in those associated with cell cycles and Tfh effector function compared with PD-1+ Tfh cells (Fig. 5 D).
To understand if PD-1+ and CD62L+ Tfh are distinct at epigenetic levels, we performed an assay for transposase-accessible chromatin with high throughput sequencing (ATAC-seq) to compare the open chromatin landscapes of CD62L+ and PD-1+ Tfh cells and analyzed differentially accessible regions. Naïve CD4+ T cells and in vitro differentiated Tfh-like cells were analyzed in parallel as controls. Overall, 174 genes were more accessible and 2,482 genes were less accessible in chromatins of CD62L+ Tfh cells, while 765 genes were more accessible and 679 genes less accessible in chromatins of PD-1+ Tfh cells, both compared to naïve CD4+ T cells (Fig. 5 E). The total open chromatin regions in CD62L+ Tfh cells were significantly less than in PD-1+ Tfh cells. As expected, the Pdcd1 and Sell loci showed increased accessibility in PD-1+ and CD62L+ Tfh cells, respectively (Fig. S3 B). Further analysis on the open chromatin regions in PD-1+ and CD62L+ Tfh cells revealed that binding motifs of genes associated with cell stemness, like Fli1, Sox10, Sox17, Oct2, were strongly associated with the accessible chromatin regions of CD62L+ Tfh cells (Fig. 5 F). In contrast, motifs related to T cell effector function, especially those of Jun-AP1 complex and AP-1 family members, were enriched in PD-1+ Tfh cells (Fig. 5 G). The chromatin accessibility data support that CD62L+ Tfh cells exhibit quiescent, stem-like, and reduced chromatin accessibility.
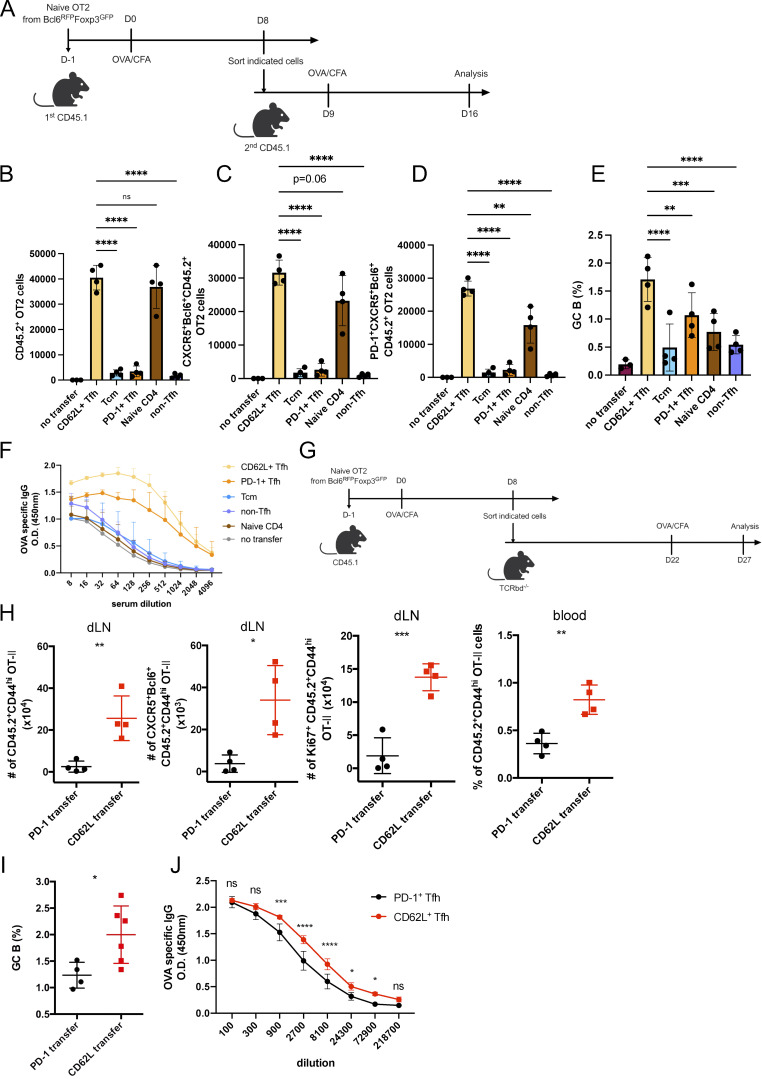
To verify the memory and stem-like features of CD62L+ Tfh cells, we compared the survival of CD62L+ and PD-1+ Tfh cells. We adoptively transferred CD62L+ and PD-1+ Tfh cells derived from OVA-immunized CD45.1 mice receiving naïve CD4+ T cells from Bcl6RFP × Foxp3GFP OT-II transgenic mice (Fig. 6, A and B). Without antigen stimulation, PD-1+ Tfh cells went apoptotic quickly; meanwhile, CD62L+ Tfh cells showed lower cell death rates (Fig. 6 B) and migrated into peripheral lymphoid organs through blood (Fig. 6 C). More importantly, upon antigen rechallenge (Fig. 6 D), CD62L+ Tfh cells proliferated as well as naïve T cells (Fig. 6 E), but much more rapidly than PD-1+ Tfh cells, CXCR5−Bcl6−CD62L− non-Tfh cells, or CXCR5intBcl6−CD62L+ Tcm cells (Fig. 6 E). Notably, CD62L+ Tfh cells gave rise to secondary CXCR5+Bcl6+ Tfh, especially PD-1+ effector Tfh cells, more efficiently compared with all other cell types (Fig. 6, E and F). Similarly, CD62L+ and PD-1+ Tfh cells largely maintained CXCR5+Bcl6+ Tfh phenotypes in the recall response (Fig. 6, H and I), in contrast to naïve T cell and Tcm cell controls (Fig. 6, H and I). These results indicate that CD62L+ Tfh represents effector memory Tfh cells, but not multipotent memory CD4+ T cells, like Tcm cells. Of note, GC B cell responses and the production of antigen-specific antibodies were higher in mice receiving CD62L+ Tfh cells (Fig. 6, J–L). Similar observations were also made at 1 wk after secondary immunization (Fig. S4, A–F). GC B development and antigen-specific antibody production were highest in the recipients of CD62L+ Tfh cells (Fig. S4, E and F). Intriguingly, at this time point, the total cell proliferation and Tfh development were relatively similar in the recipients of CD62L+ Tfh cells and of naïve OT-II cells (Fig. S4, B and C), which contrasts with day 4 (Fig. 6, E and F). These findings suggest that CD62L+ Tfh cells act as quicker responders upon recall response and provide efficient help to B cells.
Figure 6.
CD62L-expressing Tfh cells give rise to effector Tfh cells in the secondary immune response. (A) Schematic diagram of PD-1+ or CD62L+ Tfh cell adoptive transfer. (B) Cell death rates in CD45.2+ OT-II+ CD4+ T cells in the spleen from CD45.1 recipient mice adoptively transferred with PD-1+ or CD62L+ Tfh cells. (C) CD45.2+ OT-II+ CD4+ T cell percentages in indicated tissues from CD45.1 recipient mice adoptively transferred with PD-1+ or CD62L+ Tfh cells. (D) Schematic diagram of PD-1+ or CD62L+ Tfh cell adoptive transfer and OVA immunization. (E–G) Absolute numbers of (E) CD45.2+OT-II+CD4+ T cells, (F) CD45.2+OT-II+ CD4+CXCR5+Bcl6+ Tfh cells, or (G) PD-1+CD45.2+OT-II+ CD4+CXCR5+Bcl6+ Tfh cells in mice adoptively transferred with indicated cells on day 5 after OVA immunization. (H) Representative flow cytometry staining of CXCR5 and Bcl6 in CD45.2+OT-II+ T cells of mice receiving indicated cell types. (I) Statistical analysis of CXCR5+Bcl6+ cell percentages in CD45.2+ OT-II+ T cell derived from indicated transferred mice. (J and K) Statistical analysis of GC B cell percentages (J) and absolute number of mice received indicated cells (K). (L) OVA-specific IgG levels in serum of mice receiving indicated cells were measured by ELISA. Data above are representative of two independent experiments. Data of A–C represent two independent experiments. Data of D–L represent at least three independent experiments. Data are shown as mean ± SD; two-tailed t test (C, E–G, and I–K) or two-way ANOVA (L); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
Figure S4.
CD62L-expressing Tfh cells promote recall responses. (A) Schematic diagram of adoptive transfer with indicated T cell populations and OVA immunization. (B–D) Absolute numbers of (B) CD45.2+OT-II+CD4+ T cells, (C) CD45.2+OT-II+ CD4+CXCR5+Bcl6+ Tfh cells, (D) PD-1+CD45.2+OT-II+ CD4+CXCR5+Bcl6+ Tfh cells, in mice adoptively transferred with indicated cells on day 7 after OVA immunization. (E) Statistical analysis of GC B cell percentages of mice receiving indicated cells. (F) OVA-specific IgG levels in sera of mice receiving indicated cells were measured by ELISA. (G) Schematic diagram of PD-1+ or CD62L+ Tfh cell adoptive transfer experiment with OVA immunization. (H) Absolute numbers of CD45.2+ OT-II T cells, CD45.2+ CD4+CXCR5+Bcl6+ OT-II Tfh cells, or Ki67+ CD45.2+ OT-II T cells in draining lymph nodes or circulating blood from recipient mice adoptively transferred with PD-1+ or CD62L+ Tfh cells on day 5 after OVA reimmunization. (I) GC B cell percentages in draining lymph nodes from recipient mice were adoptively transferred with PD-1+ or CD62L+ Tfh cells on day 5 after OVA reimmunization. (J) Antigen-specific IgG level in serum from recipient mice receiving PD-1+ or CD62L+ Tfh cells. Data above are representative of two independent experiments. Data of A–J represent two independent experiments. Data are shown as mean ± SD; two-tailed t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
In addition, when CD62L+ and PD-1+ Tfh cells were transferred into Tcrbd−/− mice and reimmunized with OVA antigen 14 days later (Fig. S4 G), CD62L+ Tfh cells exhibited increased survival rates and proliferation in response to antigen exposure, while PD-1+ Tfh cells were almost incapable of producing more effector Tfh cells (Fig. S4 H). Moreover, both GC B cell responses and antigen-specific IgG production in mice receiving CD62L+ Tfh cells were significantly higher compared with those transferred with PD-1+ Tfh cells (Fig. S4, I and J).
Collectively, these data together indicate that CD62L+ Tfh cells are relatively resistant to cell apoptosis, and with stem-like phenotypes, can give rise to effector cells efficiently in response to secondary immune response.
Transcriptional factor KLF2 regulates the development of CD62L-expressing Tfh cells
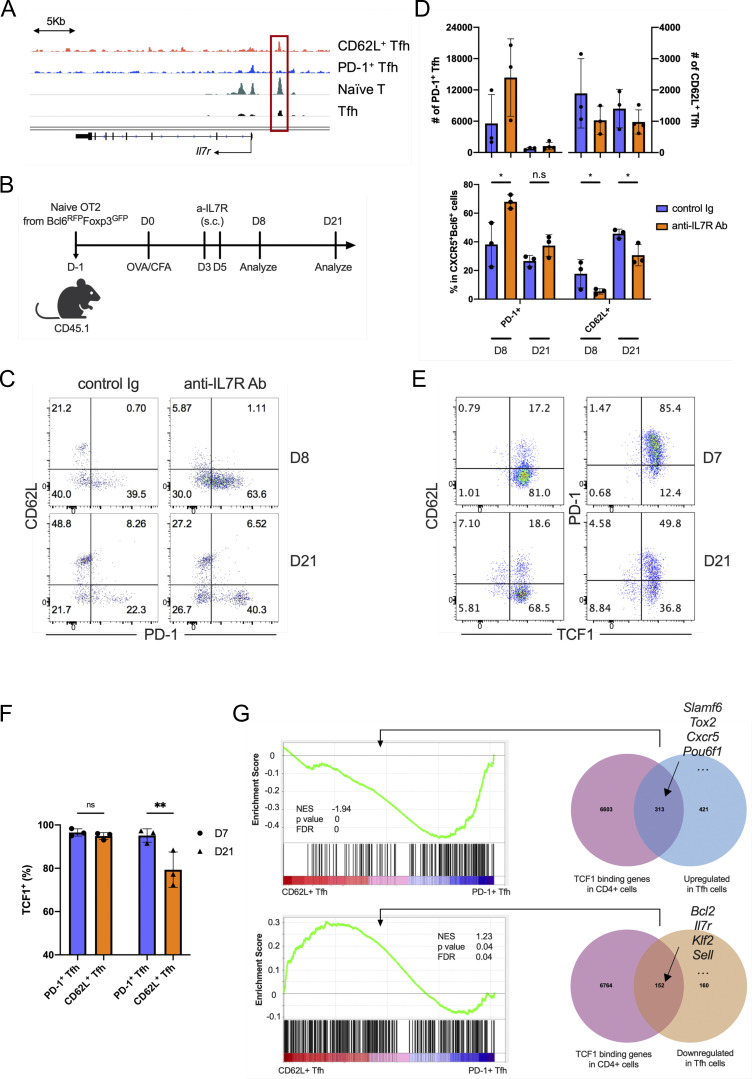
In the single-cell RNA-seq data, Il7r expression correlated with Sell, but not Pdcd1 expression in Tfh cells (Fig. 1 B). The ATAC-seq analysis also revealed that the Il7r gene locus was more accessible in CD62L+ than PD-1+ Tfh cells (Fig. S5 A), suggesting that the IL7R signaling, an important regulator in memory T cell generation and maintenance (Seddon et al., 2003; Kondrack et al., 2003; Li et al., 2003; Kitano et al., 2011; McDonald et al., 2016), may contribute to the regulation of CD62L+ Tfh cells. We therefore administrated anti-IL7R blocking antibody to CD45.1 recipient mice transferred with naïve CD4+ T cells from Bcl6RFP × Foxp3GFP OT-II transgenic mice and immunized with OVA (Fig. S5 B). IL7R blockade increased PD-1 expression and decreased CD62L expression in Tfh cells at both peak and late stage of immune response (Fig. S5, C and D). These data indicate that IL7R signaling regulates the development of CD62L+/PD-1+ Tfh cells.
Figure S5.
IL7R signaling regulates CD62L+ Tfh cell development. (A) Mean ATAC-seq coverage at the Il7r gene locus in CD62L+ Tfh cell, PD1+ Tfh, naïve T cell, and in vitro induced Tfh-like cells. (B) Schematic diagram of IL-7R blocking antibody treatment in mice adoptively transferred with naïve OT-II+ T cells. (C) Representative flow cytometry plots of CD62L and PD-1 expression in the transferred CD45.2+ OT-II+ CD4+CXCR5+Bcl6+ Tfh cells from control and IL-7R blocking antibody-treated recipient mice. (D) Statistical analysis of PD-1+ and CD62L+ cell numbers (upper) and percentages (bottom) in Tfh cells of control and IL-7R blocking antibody-treated recipient mice at indicated time points. (E and F) B6 mice were subcutaneously immunized with KLH and analyzed on day 8 and 21 after immunization. (E) Representative flow cytometry staining of TCF1 versus CD62L and PD-1 in CD4+CD44hiCXCR5+Bcl6+ cells. (F) TCF1+ percentages in PD-1+ and CD62L+ Tfh cells. (G) GSEA indicated the genes upregulated and downregulated by TCF7 in CD62L+ Tfh cells and PD1+ Tfh cells. Data of B–F represent two independent experiments. Data are shown as mean ± SD; two-tailed t test; *, P < 0.05; **, P < 0.01; ns, no significance.
Given the established roles of TCF1 in stem-like and central memory CD8+ T cells (Jeannet et al., 2010; Pais Ferreira et al., 2020; Zhao et al., 2010; Choi et al., 2015; Xu et al., 2015), we further analyzed its expression in different Tfh cell subsets. The vast majority of the CXCR5+Bcl6+ Tfh cells at day 7 after KLH immunization expressed TCF1, consistent with the previous reports about the essential roles of TCF1 in the development of Tfh cells. Unlike PD-1+ Tfh cells, which ubiquitously expressed TCF1, there was a fraction of CD62L+ Tfh cells that did not express TCF1 at day 21 after immunization, which indicated that TCF1 may not be necessary for the maintenance of CD62L+ Tfh cells (Fig. S5, E and F). Moreover, by integrative analysis of TCF1-bound genes in CD4+ T cells in public, TCF1 chromatin immunoprecipitation sequencing dataset (GSE103385), and genes upregulated or downregulated during Tfh cell development (GSE40068), we have defined TCF1-regulated genes in Tfh cells (Fig. S5 G). TCF1 directly bound to plenty of genes highly expressed in PD-1+ Tfh cells, mainly associated with effector functions of Tfh cells, which suggests the key roles of TCF1 in regulating PD-1+ Tfh cells (Fig. S5 G). TCF1 also could bind to several feature genes of CD62L+ Tfh cells, including Sell, Klf2, Il7r, Bcl2, etc. (Fig. S5 G). Thus, TCF1 may not specifically regulate PD-1+ or CD62L+ Tfh cells alone.
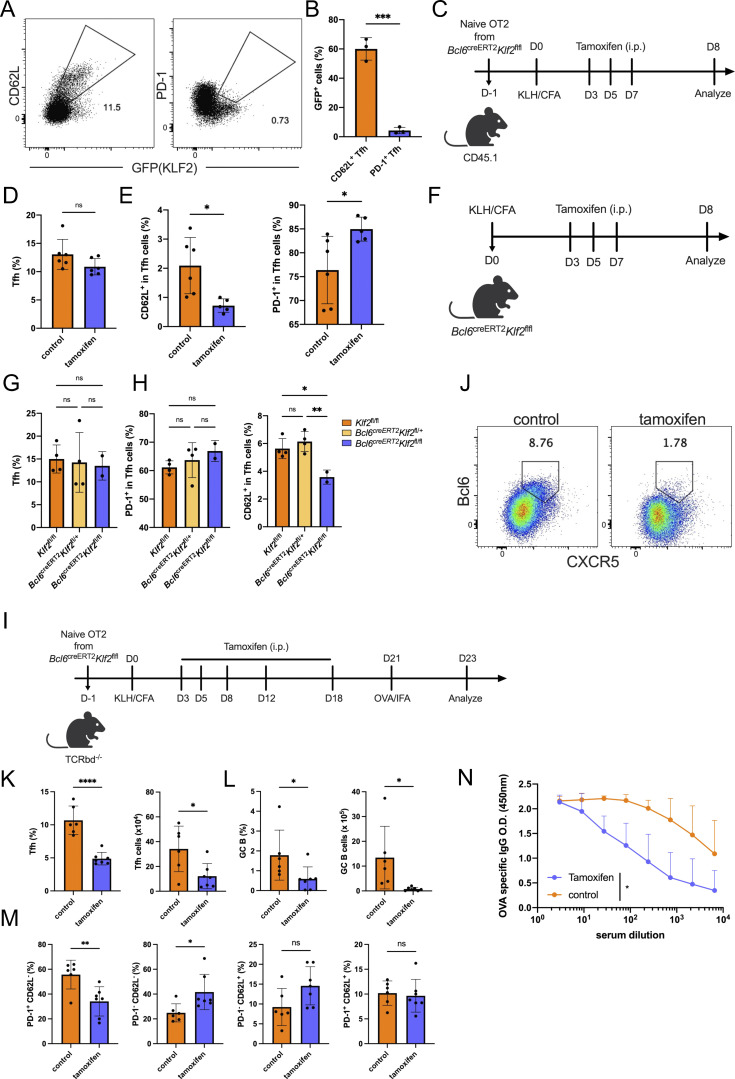
To understand the regulation of CD62L+ Tfh cells, we thus turned to another transcription factor, KLF2. Klf2 expression was restricted within Sell-expressing, but not Pdcd1-expressing Tfh cells in single-cell RNA-seq data (Fig. 1 B), raising a possibility that KLF2 may regulate CD62L+ Tfh cells. Utilizing KLF2GFP reporter mice, we found obvious coexpression of GFP with CD62L, but not PD-1 in Tfh cells after KLH immunization (Fig. 7, A and B). Then we generated Bcl6creERT2Klf2flfl × OT-II mice, which allowed specific knockout of Klf2 in Bcl6-expressing Tfh cells after the tamoxifen treatment without interfering CD4+ T cell homeostasis. Naïve OT-II cells from Bcl6creERT2Klf2flfl × OT-II mice were transferred into CD45.1 recipient mice followed by OVA immunization, and tamoxifen or control corn oil were intraperitoneally injected in recipient mice on days 3, 5, and 7 after immunization (Fig. 7 C). Total CXCR5+Bcl6+ Tfh cell percentages were not affected by Klf2 deficiency (Fig. 7 D); however, both PD-1+ and CD62L+ Tfh cells were significantly altered (Fig. 7 E). Although a previous study suggested that KLF2 inhibited Tfh cell differentiation (Lee et al., 2015), our data demonstrated that deficiency in Klf2 in committed Bcl6+ Tfh cells did not impair Tfh cells in general, but mainly increased the PD1+ versus CD62L+ Tfh cell ratios. Similar results were also observed in the KLH-immunized Bcl6creERT2Klf2flfl mice (Fig. 7, F–H). Deficiency in Klf2 in Tfh cells suppressed CD62L+, but not total Tfh cell populations (Fig. 7, G and H). Furthermore, we observed that Icos expression was relatively higher in KLF2hiCD62L+ Tfh cells compared with PD-1+ Tfh cells (Fig. 5 D). This finding contradicts a previous work that the blockade of ICOS signaling favors Klf2 expression in CD4+ T cells (Weber et al., 2015). These results support the differential roles of Klf2 at different stages of Tfh cell development. Moreover, the function of Klf2 in regulating CD62L+ Tfh cells was dose-dependent since minimal effects were detected in Bcl6creERT2Klf2fl/+ mice (Fig. 7 H).
Figure 7.
KLF2 regulate the development of CD62L-expressing Tfh cells. (A) Representative expression of KLF-GFP with CD62L and PD-1 in Tfh cells on day 14 after KLH immunization of Klf2GFP Bcl6RFP mice. (B) KLF2-GFP+ cell percentages in CD62L+ and PD-1+ Tfh cells on day 14 after KLH immunization of Klf2GFP Bcl6RFP mice. (C) Experimental workflow of Bcl6creERT2Klf2fl/fl OT-II cell adoptive transfer and immunization experiment. (D and E) Tfh cell percentages in CD4+CD44hi cells (D), and (E) percentages of PD-1+ (left) and CD62L+ (right) in Tfh cells of vehicle- and tamoxifen-treated mice. (F) Experimental workflow of KLH immunization of Bcl6creERT2Klf2fl/fl and control Klf2fl/fl WT mice. (G and H) Tfh cell percentages in CD4+CD44hi cells (G), and (H) the percentages of PD-1+ (left) and CD62L+ (right) cells in Tfh cells of KLH-immunized WT, Bcl6creERT2Klf2fl/+, and Bcl6creERT2Klf2fl/fl mice. Data above are representative of two independent experiments. (I) Experimental workflow of the recall immunization. (J) Representative flow cytometry staining of CXCR5+Bcl6+ cells in CD45.2+CD44hi OT-II cells derived from draining lymph nodes of control and tamoxifen-treated recipient mice. (K and L) Percentages and absolute numbers of (K) CXCR5+Bcl6+ Tfh cells and (L) GL7+Fas+ GC B cells. (M) Proportions of indicated cell populations among secondary CXCR5+Bcl6+ Tfh cells. (N) OVA-specific IgG production in serum from control and tamoxifen-treated recipient mice. Data of A, B, and F–N represent two independent experiments. Data of C–E represent three independent experiments. Data are shown as mean ± SD; two-tailed t test (B, D, E, G, H, and K–M) or two-way ANOVA (N); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
In addition, we further investigated the effect of Klf2 deficiency in Bcl6-fated cells in the recall immune response. Naïve OT-II cells from Bcl6creERT2Klf2flfl OT2 transgenic mice were transferred into Tcrbd−/− recipient mice followed by OVA immunization and tamoxifen-induced Klf2 deletion (Fig. 7 I). OVA/IFA reimmunization was conducted on day 21 after primary immunization. CXCR5+Bcl6+ Tfh cells were greatly decreased in tamoxifen-treated than control mice (Fig. 7, J and K), especially PD-1hi effector Tfh cells (Fig. 7 M). Additionally, loss of Klf2 in Tfh cells resulted in the accumulation of non-functional PD-1− CD62L− Tfh cells during secondary immune response (Fig. 7 M). Moreover, GC B cell responses and antigen-specific IgG production were also substantially reduced due to Klf2 deficiency (Fig. 7, L and N). These results indicate that the deletion of Klf2 in Bcl6-expressing Tfh cells impaired memory Tfh cell formation, but not simply affecting L-selection expression in this population.
These data overall support that Klf2, specifically expressed in CD62L+ Tfh cells, is required for the generation of these cells, and more importantly, these cells are required in Tfh cell responses in the memory phase.
Discussion
In this work, through single-cell RNA-seq analysis of CXCR5+Bcl6+ Tfh cells, we discovered a new subset, which highly expressed CD62L but not PD-1. These cells were induced starting at early stages and gradually became the predominant Tfh population at the memory phase of the immune response. The CD62L+ Tfh cells, with memory cell–like transcriptional features, exhibited increased survival and efficiently gave rise to effector Tfh cells in secondary immune response.
How memory T cells are generated remains unclear and may vary in different effector cell types. A small fraction of CD8+ T cells in lymphocytic choriomeningitis virus (LCMV) or Listeria monocytogenes infection acquired memory precursor characteristics with increased expression of IL-7 receptor subunit-α (IL-7Rα), CD27, and B cell lymphoma 2 (BCL-2), and decreased expression of killer cell lectin-like receptor G1 (KLRG1), distinct from the KLRG1hiIL7RαlowCD27hiBCL-2hi terminal effector cells (Kaech et al., 2003; Joshi et al., 2007; Joshi and Kaech, 2008; Sarkar et al., 2008; Schluns et al., 2000). Memory CD8+ T cells in circulation have been classified as Tcm or Tem cells. Tcm cells typically express lymph node homing markers, L-selectin (CD62L), and CCR7, and exhibit self-renewal and proliferation capacity. Subsequently, a small fraction of Tcm cells was identified and termed as stem cell memory T cell (Tscm), which coexpress stem cell markers such as Sca1 in addition to CD62L and CCR7 and represent a multipotent memory T cell precursor to give rise to other memory subsets (Gattinoni et al., 2011; Lugli et al., 2013). The identification of Tscm strongly supports a progressive model for CD8+ memory T cell differentiation, suggesting that naïve T cells, upon activation, further differentiate into Tscm and progressively reconstitute the entire memory cell pool. Moreover, a CD62L-expressing Tpex population was recently identified with long-term proliferative potential, multipotency, and repopulation of exhausted T cells in chronic infection. While Tpex shares transcriptional similarity with Tfh cells, they are fundamentally distinct populations with differing roles and regulation mechanisms (Tsui et al., 2022). Therefore, the pathways of memory CD4+ T cell generation have not been well elucidated. There is evidence which shows that lineage-committed effector CD4+ T cells, such as IFN-γ–producing Th1 effector cells with transgenic TCR specific for LCMV peptide, can return to quiescent state with IL-7 receptor but not CD62L expression and produce IFN-γ during secondary responses, therefore acting like Tem cells (Harrington et al., 2008). In the literature, a population of T-betlowCXCR5+CCR7+PD-1− T cells were shown to be generated during infection and subsequently gave rise to Tfh and Th1 cells, which fit the definition of Tcm cells with the potential of differentiating into different types of T cells in the recall response (Pepper et al., 2011). Subsequently, more studies showed that long-lived CXCR5+ T cells with memory and stem cell features had the ability to generate Tfh cells upon rechallenge (Hale et al., 2013; Robinson et al., 2022; Xia et al., 2022). However, these populations of T cells, notably with a gradient of Bcl6 expression levels from none to high, were likely a mixture. Indeed, we reported previously that using our Bcl6 reporter mice, CXCR5+Bcl6− cells exhibited non-polarized gene expression patterns (Liu et al., 2012). In contrast, our current study specifically focused on Bcl6+CXCR5+ Tfh cells and identified a distinct CD62L+ subpopulation, which served as effector memory Tfh precursors. Importantly, we conducted a side-by-side comparison of different T cell populations during secondary immune response. Our findings revealed that CD62L+Bcl6+CXCR5+ T cells predominantly maintained their Tfh identity during the transfer experiment. In contrast, only a small fraction of the progenies of CXCR5+Bcl6− Tcm cells became Tfh cells, with the majority of this cell population adopting non-Tfh cells containing multiple cell phenotypes (Fig. 6, H and I). These results showed the distinct nature of CD62L+Bcl6+CXCR5+ Tfh cells studied in our research compared with the cell population discussed in previous literature.
There are rising discussions about the contribution of peripheral CXCR5+ Tfh-like cells in Tfh cell memory. The characteristics of circulating CXCR5+ Tfh-like cells and distinct CCR7lowPD1hi cells and CCR7lowPD1hi cell subpopulations were defined with resting and effector phenotypes, respectively (He et al., 2013). However, due to the lack of Bcl6 expression in blood CXCR5+ Tfh-like cells, whether circulating CXCR5+ Tfh-like cells are derived from Bcl6+ Tfh cells was unclear. We have proved that blood CXCR5+ Tfh-like cells contain both Bcl6-expressed and non-expressed cell populations, which indicated mixed origins (data not shown). Moreover, although CXCR5+CCR7hiPD1low Tfh-like cells expressed several molecules including CD62L, CCR7, and IL7Rα, similar to our CD62L+ Tfh cells, they were poorer in the induction of GC responses compared with CXCR5+CCR7lowPD1hi cells, which is opposite to CD62L+ Tfh cells in our research. Therefore, circulating CXCR5+CCR7hiPD1low T cells are different from the CD62L+ Tfh cells we analyzed in our study.
In this study, we showed that CD4+ T cells, after activation, differentiate into CXCR5+Bcl6+PD-1+ effector and CD62L+CXCR5+Bcl6+ memory-like Tfh cells simultaneously. CD62L+CXCR5+Bcl6+ memory-like cells exhibited an increased ability to circulate and efficiently generate effector cells in the secondary response, somewhat similar, but superior to Tcm cells. Our finding thus revealed the development of memory Tfh cells, filling the gaps between naïve, effector, and memory T cells.
There are currently debates regarding the relationships between GC-Tfh and memory Tfh cells. Some regard memory Tfh cells as the descendants of GC-Tfh cells. In this opinion, the majority of effector Tfh cells undergo apoptosis as the GC reaction terminates, leaving behind a fraction of cells that survive and transition into memory Tfh cells. Long-lived CXCR5+PD-1+ Tfh cells have been reported recently in the context of NAD-induced cell death blockade (Künzli et al., 2020). However, these cells differ from CD62L+ effector memory Tfh precursor cells discussed in our manuscript in their multipotent capacity and elevated glycolysis activity in the secondary response, suggesting a mixed cell population with both effector and memory Tfh cells, including Tcm cells. The second possibility is that a proportion of memory Tfh cells may be generated very early during an immune response, and the development of memory Tfh cells is in parallel with that of effector Tfh cells (Crotty, 2018). Our data showed simultaneous induction of CD62L+ memory Tfh precursor cells at an early stage of the immune response with PD-1+ Tfh cells, supporting the bifurcation of Tfh cells into memory and effector Tfh cells with similar TCRs during differentiation.
The cellular metabolism is associated with the quiescent and activation states of T cells. T cell activation requires catabolic and anabolic metabolisms that facilitate energy consumption and biosynthesis (Chapman et al., 2020). The stem cell–like memory T cells maintain a quiescent state with a paused cell cycle; in the meantime, the requirements for nutrient uptake and mitochondrial metabolism are quite low in these cells (Gattinoni et al., 2017; Galletti et al., 2020). Through transcriptional profiling of CD62L+ Tfh cells, we found that the glycolysis process and mitochondrial metabolism, mainly oxidative phosphorylation, are largely decreased in CD62L+ compared with PD-1+ Tfh cells. In the meantime, the cell cycle– and proliferation-related genes are also downregulated, whereas the anti-apoptosis genes are highly expressed in CD62L+ Tfh cells, indicating that CD62L+ Tfh cells maintain a quiescent, low metabolic activity, but have strong survival, in line with Tscm CD8+ T cells. However, when encountering an antigen again, CD62L+ Tfh cells can efficiently exit the quiescent state to enter the activation state and differentiate into PD-1+ Tfh cells, as well as maintain a CD62L+ Tfh cell pool.
In conclusion, through single-cell RNA-seq analysis, we discovered a novel Tfh subpopulation with CD62L expression. This population has stem cell–like and memory precursor features, and with long-term survival ability, can give rise to effector Tfh cells in the recall immune response. The exploration of CD62L+ Tfh cells gives further insights into memory Tfh cell development and regulation.
Materials and methods
Mice
C57BL/6, OT-II, μMT, TCRbd−/−, and CD45.1 congenic mice were maintained in-house. The Bcl6RFP mice were generated previously (Liu et al., 2012) and were backcrossed with C57BL/6 mice at least eight times. The Bcl6RFPmice were crossed with Foxp3GFP mice (Lin et al., 2007) to generate Bcl6RFP × Foxp3GFP double reporter mice. Bcl6creERT2 and Il21cre mice were generated as described (Feng et al., 2022). KLF2-GFP mice and Klf2flox mice are kind gifts of Dr. Stephen C. Jameson of the University of Minnesota Medical School (Minneapolis, MN). IL21-VFP mice (JAX 030295) were purchased from the Jackson Laboratory. All the mice were maintained under specific pathogen–free conditions in the Animal Facility of Tsinghua University. All the animal protocols used in this study were accredited by the American Association for Accreditation of Laboratory Animal Care and Institutional Animal Care and Use Committee of Tsinghua University.
Animal models
Immunization
For KLH immunization, the mice were subcutaneously immunized with KLH (100 µg/mouse) mixed with complete Freund’s adjuvant (CFA, 1 mg/ml; Sigma-Aldrich) near the base of the tail and sacrificed and analyzed on day 7 after immunization.
For chicken OVA immunization, the mice were subcutaneously immunized with OVA (150 µg/mouse) mixed with CFA (1.5 mg/ml; Sigma-Aldrich) near the base of the tail and sacrificed and analyzed on day 7 or indicated time point after immunization.
Infection
Influenza virus A/Puerto Rico/8 (PR8, H1N1) was generated by Dr. Hong Tang and his colleague at CAS Key Laboratory of Infection and Immunity. The mice were anesthetized and intranasally infected with 0.1 half lethal dose (LD50) of H1N1 PR8 influenza virus and were sacrificed and analyzed on day 9 after infection.
Flow cytometry and cell sorting
For surface staining, cells were first incubated with anti-CD4 (17-0042-83; eBioscience), anti-CD45.2 (56-0454-82; eBioscience), anti-TCRvβ5.1/5.2 (46-5796-82; eBioscience), anti-CD44 (103030; BioLegend; 563058, BD), biotinylated anti-CXCR5 (551960; BD), followed by streptavidin-BV421 (405225; BioLegend), anti-B220 (11-0452-82; eBioscience), anti-CD279 (17-9982-82; eBioscience), anti-CD62L (47-0621-82; eBioscience), anti-CD127 (560733; BD), anti-CD19 (550992, 553088; BD), anti-IgD (405708; BioLegend), anti-GL7 (48-5902-80; eBioscience), and anti-CD95 (53-0951; eBioscience) staining. Dead cells were excluded by cell viability dye staining (Fixable viability dye eF505; eBioscience). To trace the cellular division, cells were prelabeled with (CFSE (565082; BD).
For intracellular staining, cells were fixed and permeabilized with the Foxp3 transcription factor staining buffer kit (eBioscience), followed by incubation with anti-BCL6 (562401; BD), anti-Foxp3 (11-5773-82; eBioscience), and anti-TCF1 (6444S; CST). Cells were analyzed on LSRFortessa (BD) flow cytometer and analyzed using FlowJo.
For cell sorting, cells were sorted on FACSAria (BD) cell sorter with indicated surface marker staining.
Adoptive cell transfer
Flow cytometric sorting was used for the isolation of various T cell populations before adoptive transfer. For analysis of cell survival ability, TCRvβ5.1/5.2+CD44hiCXCR5+Bcl6-RFP+Foxp3−PD1+CD62L− PD-1+ Tfh cells and TCRvβ5.1/5.2+CD44hiCXCR5+Bcl6-RFP+Foxp3−PD1−CD62L+ CD62L+ Tfh cells from OVA-immunized CD45.1+ recipient mice receiving naïve Bcl6RFP × Foxp3GFP OT-II cells were sorted with FACSAria flow cytometer at day 8 after immunization. 105 sorted cells were then intravenously injected into TCRbd−/− recipients respectively. 3 days later, the mice were sacrificed and analyzed as described above.
For analysis of memory potential in recall response, TCRvβ5.1/5.2+CD44−Bcl6-RFP−Foxp3-GFP−CD62Lhi naïve OT-II T cells from age- and sex-matched naïve Bcl6RFP × Foxp3GFP OT-II transgenic mice, as well as TCRvβ5.1/5.2+CD44hiCXCR5−Bcl6-RFP−Foxp3−CD62L− non-Tfh cells, TCRvβ5.1/5.2+CD44hiCXCR5midBcl6-RFP−Foxp3−CD62L+ Tcm cells, TCRvβ5.1/5.2+CD44hiCXCR5+Bcl6-RFP+Foxp3−PD1+CD62L− PD-1+ Tfh cells, and TCRvβ5.1/5.2+CD44hiCXCR5+Bcl6-RFP+Foxp3−PD1−CD62L+ CD62L+ Tfh cells from OVA-immunized CD45.1+ recipients previously transferred with naïve OT-II T cells were sorted with FACSAria flow cytometer at day 8 after immunization. 104 sorted cells were then intravenously injected into new CD45.1+ recipients respectively. On day 4 or day 7 after OVA immunization, the mice were sacrificed and analyzed as described above.
Single-cell RNA-seq
Tfh cells were isolated from PP at steady state, as well as draining lymph nodes of KLH immunized (8 days after immunization) or H1N1 PR8 infected (9 days after immunization). Bcl6RFP × Foxp3GFP double reporter mice based on the gating of CD4+CD44hiCXCR5+Bcl6-RFP+Foxp3-GFP−B220−. Tfr cells were isolated from KLH immunized (8 days after immunization). Bcl6RFP × Foxp3GFP double reporter mice based on the gating of CD4+CXCR5+ Bcl6-RFP+ Foxp3-GFP+B220−. All mice used for single-cell sorting were aged between 6 and 8 wk. Sorted cells were resuspended in PBS containing 0.5% FBS at 106/ml concentration. Approximately 10,000 cells from each sample were loaded into one channel of Single Cell Chip A using a Chromium Single Cell 3′ v2 Reagent Kit (10x Genomics). After cell capture and lysis, complementary DNA was synthesized and amplified for 12 cycles according to the manufacturer’s protocol (10x Genomics). Libraries were synthesized and amplified for 10 cycles according to the manufacturer’s protocol (10x Genomics). The library quantity and quality were examined by Agilent 2100 Bioanalyzer and Qubit (Thermo Fisher Scientific). Sequencing was performed using Hiseq-PE150 (Illumina) by Novogene.
Single-cell RNA-seq data analysis
Sample demultiplexing, barcode processing, and unique molecular identifier (UMI) counting were performed using Cell Ranger version 3.0 (10x Genomics). Pooled data were imported into R version 3.6 using Seurat version 3.2.2. Cells with unique feature counts <200 or >3,000 were excluded for further analysis, along with cells containing mitochondrial counts above 10%. Samples with the same identities, i.e., two batches of Tfh cells sorted from the KLH immunization model, were combined into one dataset with “Merge” function in Seurat version 3.2.2. Datasets with different identities were then integrated for analysis using “FindIntegrationAnchors” and “IntegrateData” functions in Seurat version 3.2.2. For gene expression profiles, UMIs were normalized across cells per 10,000 and log-transformed using “NormalizeData” function in Seurat version 3.2.2 before integration. Then the values were converted to z-scores using “ScaleData” function in Seurat version 3.2.2 on the integrated dataset. For dimensional reduction, a nonlinear dimensional reduction technique, i.e., UMAP, was chosen. Data were then subjected to clustering using “FindNeighbors” and “FindClusters” functions in Seurat version 3.2.2 (resolution 0.3). DEGs for each cluster were identified using “FindMarkers” function in Seurat version 3.2.2 (Stuart et al., 2019). “AddModuleScore” function was used to calculate module scores for selected features. For calculating cell cycle phase scores and assigning cell phase, “CellCycleScoring” was used.
Due to the inevitable existence of doublets and multiplets, cells used for clustering were not pure Tfh cells. According to the DEGs in C8 and C9, we thought those cells were likely macrophages and B cells respectively based on the differential expressions of Lyz2, Apoe or Cd79a, Cd79b, Iglc2. However, because the proportion of those cells was very limited and barely influenced the further analysis, we thus did not exclude those cells from the total cell pool. But we won’t pay attention to these two clusters in the following part.
For RNA velocity analysis, python package “Velocyto” (https://github.com/velocyto-team/velocyto.py) was used to calculate spliced reads and unspliced reads from previously aligned bam files (from Cell Ranger). Loom files were generated from this step for further analysis. Then metadata from the Seurat object including cell clustering and cell embedding information was integrated into the loom file. The RNA velocity was estimated by the python package “scVelo” based on the loom object.
Bulk RNA-seq
PD-1+ Tfh (CD4+CD44hiCXCR5+RFP+PD-1+GFP−B220−) cells, CD62L+ Tfh (CD4+CD44hiCXCR5+RFP+CD62L+GFP−B220−) cells, and non-Tfh (CD4+CD44hiCXCR5−RFP−GFP−B220−) cells were isolated from draining lymph nodes of KLH immunized Bcl6RFP × Foxp3GFP double reporter mice, and CXCR5+ CD4+ T cells from peripheral blood were isolated from the same batch of mice. The sorted cells were then lysed by RLT buffer (Qiagen) and RNA was extracted by using RNeasy Plus Mini Kit (Qiagen). RNA quality was examined by Agilent 2100 Bioanalyzer. Libraries were prepared using SMART-Seq Ultra Low Input RNA Kit (Clontech). Quality for the cDNA library was examined by Agilent 2100 Bioanalyzer. Sequencing was performed using Hiseq-SE50 (Illumina) by BGI.
Bulk RNA-seq data analysis
The clean reads were aligned to mm10 by Bowtie2 version 2.2.9 with default parameters and then uniquely mapping reads were summarized by “featureCounts” from Subread package. DEGs were identified by at least twofold change and adjusted P value (false discovery rate [FDR]) <0.01. “ClusterProfiler” function in R version 3.6 was used for pathway analysis.
ATAC-seq
PD-1+ Tfh cells and CD62L+ Tfh cells were isolated from draining lymph nodes of KLH immunized Bcl6RFP × Foxp3GFP double reporter mice and used for ATAC-seq experiments. ATAC-seq library was constructed using TruePrep DNA library Prep Kit V2 and TruePrep Index Kit V2 for Illumina. Basically, 50,000 cells were resuspended in 35 μl of lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and incubated on ice for 10 min with three times vortex. DNA fragmentation was performed by adding 10 μl TruePrep Tagment Buffer L and 5 μl TruePrep Tagment Enzyme to the system and the mixture was incubated and shaken at 37°C for 30 min. The fragmented DNA was then purified by using 1.8× AMPure Beads, barcoded with dual indexes and PCR amplified. Size selection and library purification were conducted by using 0.4× and 1.6× AMPure beads, respectively. The library quantity and quality were examined by Agilent 2100 Bioanalyzer and Qubit (Thermo Fisher Scientific). Sequencing was performed using Hiseq-PE150 (Illumina) by Novogene.
ATAC-seq data analysis
Low-quality reads and adaptor sequences were removed by Trim Galore version 0.4.4 with parameters “-q 10 -length 30 -stringency 50.” Paired-end reads were aligned to the mm10 using Bowtie2 version 2.2.9 with -X 2,000 (Langmead and Salzberg, 2012). Reads that mapped to the mitochondrial genome were filtered and PCR duplicates were removed using “picard MarkDuplicates.” To identify peaks, the bam files containing unique, non-chrM reads were used to call peaks with MACS2 using parameters “-nomodel-keep-dup all -q 0.01 -g mm” (Zhang et al., 2008). For differential coverage, corresponding bam files were merged to call peaks to get a union peak set. For each peak in the peak set, raw ATAC-seq reads were counted using “featureCounts.” Differentially expressed peaks were identified by at least 1.5-fold change and adjusted P value (FDR) <0.1. For downstream analysis, HOMER was used to find transcriptional factor motifs (http://homer.ucsd.edu/homer).
Immunofluorescence
Tissues were isolated and fixed in 4% paraformaldehyde, then sliced to 8 µm. The frozen sections were stained overnight at 4°C with primary antibodies: biotinylated anti-PNA (B-1075; Vector lab), anti-PD-1 (11-9981-85; eBioscience), anti-CD62L (104407; BioLegend), and anti-CD4 (553051; BD). The sections were then incubated with streptavidin-BV421 (405225; BioLegend) at 37°C for 2 h. The images were acquired by confocal microscope LSM 780 (Zeiss) and processed using ZEN imaging software (Zeiss).
Statistical analysis
The statistics were performed with two-tailed unpaired Student’s t test or two-way ANOVA (Graphpad Prism 6), when indicated. Differences with P value <0.05 were considered significant: *, P < 0.05, **, P < 0.01, ***, P < 0.001, ns, no significant difference.
Online supplemental material
Fig. S1 shows the heterogeneity of Tfh and Tfr cells revealed by single-cell RNA-seq (related to Fig. 1). Fig. S2 shows developmental features of PD-1+ and CD62L+ Tfh cells (related to Figs. 3 and 4). Fig. S3 shows transcriptional and epigenetic features of CD62L+ Tfh cells (related to Fig. 5). Fig. S4 shows CD62L-expressing Tfh cells promote recall responses (related to Fig. 6). Fig. S5 shows IL7R signaling regulates CD62L+ Tfh cell development (related to Fig. 7).
Acknowledgments
We thank the Tsinghua University Institute for Immunology Core facility for FACS sorting.
The work was supported in part by the National Natural Science Foundation of China (31630022, 31821003, and 31991170 to C. Dong) and Beijing Municipal Science and Technology (Z181100001318007 and Z171100000417005 to C. Dong).
Author contributions: H. Feng designed and performed the experiments and wrote the manuscript. X. Zhao and L. Zheng performed the bioinformatic analysis. W. Fu, Z. Zhao, X. Zhao, and X. Bai performed the experiments. X. Wang supervised the experiments. Z. Zhang supervised the experiments and guided the bioinformatic analysis. C. Dong designed and supervised the project and edited the manuscript.
Data availability
Expression data have been deposited in the Gene Expression Omnibus database under the Super Series accession number GSE230438. This Super Series includes data for single-cell RNA-seq, bulk RNA-seq, and ATAC-seq in this paper.
References
- Ansel, K.M., McHeyzer-Williams L.J., Ngo V.N., McHeyzer-Williams M.G., and Cyster J.G.. 1999. In vivo-activated CD4 T cells upregulate CXC chemokine receptor 5 and reprogram their response to lymphoid chemokines. J. Exp. Med. 190:1123–1134. 10.1084/jem.190.8.1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, A., Hu H., Yeung M., and Chen J.. 2007. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J. Immunol. 178:7632–7639. 10.4049/jimmunol.178.12.7632 [DOI] [PubMed] [Google Scholar]
- Baumgartner, B., and Harper J.W.. 2003. Deafening cycle. Nat. Cell Biol. 5:385–387. 10.1038/ncb0503-385 [DOI] [PubMed] [Google Scholar]
- Breitfeld, D., Ohl L., Kremmer E., Ellwart J., Sallusto F., Lipp M., and Förster R.. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192:1545–1552. 10.1084/jem.192.11.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelman, J., Mazza E.M.C., Alvisi G., Colombo F.S., Grilli A., Mikulak J., Mavilio D., Alloisio M., Ferrari F., Lopci E., et al. 2018. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J. Exp. Med. 215:2520–2535. 10.1084/jem.20180684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C.M., Endrizzi B.T., Wu J., Ding X., Weinreich M.A., Walsh E.R., Wani M.A., Lingrel J.B., Hogquist K.A., and Jameson S.C.. 2006. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 442:299–302. 10.1038/nature04882 [DOI] [PubMed] [Google Scholar]
- Chapman, N.M., Boothby M.R., and Chi H.. 2020. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20:55–70. 10.1038/s41577-019-0203-y [DOI] [PubMed] [Google Scholar]
- Chen, Y., Li H., Lai L., Feng Q., and Shen J.. 2020. Identification of common differentially expressed genes and potential therapeutic targets in ulcerative colitis and rheumatoid arthritis. Front. Genet. 11:572194. 10.3389/fgene.2020.572194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.S., Gullicksrud J.A., Xing S., Zeng Z., Shan Q., Li F., Love P.E., Peng W., Xue H.-H.H., and Crotty S.. 2015. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16:980–990. 10.1038/ni.3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty, S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity. 41:529–542. 10.1016/j.immuni.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S. 2018. Do memory CD4 T cells keep their cell-type programming: Plasticity versus fate commitment? Complexities of interpretation due to the heterogeneity of memory CD4 T cells, including T follicular helper cells. Cold Spring Harb. Perspect. Biol. 10:a032102. 10.1101/cshperspect.a032102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H., Zhao X., Xie J., Bai X., Fu W., Chen H., Tang H., Wang X., and Dong C.. 2022. Pathogen-associated T follicular helper cell plasticity is critical in anti-viral immunity. Sci. China Life Sci. 65:1075–1090. 10.1007/s11427-021-2055-x [DOI] [PubMed] [Google Scholar]
- Galletti, G., De Simone G., Mazza E.M.C., Puccio S., Mezzanotte C., Bi T.M., Davydov A.N., Metsger M., Scamardella E., Alvisi G., et al. 2020. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 21:1552–1562. 10.1038/s41590-020-0791-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sáez, A.J. 2012. The secrets of the Bcl-2 family. Cell Death Differ. 19:1733–1740. 10.1038/cdd.2012.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni, L., Lugli E., Ji Y., Pos Z., Paulos C.M., Quigley M.F., Almeida J.R., Gostick E., Yu Z., Carpenito C., et al. 2011. A human memory T cell subset with stem cell-like properties. Nat. Med. 17:1290–1297. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni, L., Speiser D.E., Lichterfeld M., and Bonini C.. 2017. T memory stem cells in health and disease. Nat. Med. 23:18–27. 10.1038/nm.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowthaman, U., Chen J.S., Zhang B., Flynn W.F., Lu Y., Song W., Joseph J., Gertie J.A., Xu L., Collet M.A., et al. 2019. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science. 365:eaaw6433. 10.1126/science.aaw6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, J.S., Youngblood B., Latner D.R., Mohammed A.U.R., Ye L., Akondy R.S., Wu T., Iyer S.S., and Ahmed R.. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 38:805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.E., Janowski K.M., Oliver J.R., Zajac A.J., and Weaver C.T.. 2008. Memory CD4 T cells emerge from effector T-cell progenitors. Nature. 452:356–360. 10.1038/nature06672 [DOI] [PubMed] [Google Scholar]
- He, J., Tsai L.M., Leong Y.A., Hu X., Ma C.S., Chevalier N., Sun X., Vandenberg K., Rockman S., Ding Y., et al. 2013. Circulating precursor CCR7(lo)PD-1(hi) CXCR5⁺ CD4⁺ T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 39:770–781. 10.1016/j.immuni.2013.09.007 [DOI] [PubMed] [Google Scholar]
- He, R., Hou S., Liu C., Zhang A., Bai Q., Han M., Yang Y., Wei G., Shen T., Yang X., et al. 2016. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. 537:412–428. 10.1038/nature19317 [DOI] [PubMed] [Google Scholar]
- Im, S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannet, G., Boudousquié C., Gardiol N., Kang J., Huelsken J., and Held W.. 2010. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl. Acad. Sci. USA. 107:9777–9782. 10.1073/pnas.0914127107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.S., and Kaech S.M.. 2008. Effector CD8 T cell development: A balancing act between memory cell potential and terminal differentiation. J. Immunol. 180:1309–1315. 10.4049/jimmunol.180.3.1309 [DOI] [PubMed] [Google Scholar]
- Kaech, S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., and Ahmed R.. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kim, C.H., Rott L.S., Clark-Lewis I., Campbell D.J., Wu L., and Butcher E.C.. 2001. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J. Exp. Med. 193:1373–1381. 10.1084/jem.193.12.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., and Okada T.. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 34:961–972. 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Kondrack, R.M., Harbertson J., Tan J.T., McBreen M.E., Surh C.D., and Bradley L.M.. 2003. Interleukin 7 regulates the survival and generation of memory CD4 cells. J. Exp. Med. 198:1797–1806. 10.1084/jem.20030735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzli, M., Schreiner D., Pereboom T.C., Swarnalekha N., Litzler L.C., Lötscher J., Ertuna Y.I., Roux J., Geier F., Jakob R.P., et al. 2020. Long-lived T follicular helper cells retain plasticity and help sustain humoral immunity. Sci. Immunol. 5:eaay5552. 10.1126/sciimmunol.aay5552 [DOI] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Skon C.N., Lee Y.J., Oh S., Taylor J.J., Malhotra D., Jenkins M.K., Rosenfeld M.G., Hogquist K.A., and Jameson S.C.. 2015. The transcription factor KLF2 restrains CD4⁺ T follicular helper cell differentiation. Immunity. 42:252–264. 10.1016/j.immuni.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, Y.A., Chen Y., Ong H.S., Wu D., Man K., Deleage C., Minnich M., Meckiff B.J., Wei Y., Hou Z., et al. 2016. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17:1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Li, J., Huston G., and Swain S.L.. 2003. IL-7 promotes the transition of CD4 effectors to persistent memory cells. J. Exp. Med. 198:1807–1815. 10.1084/jem.20030725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H., Tang J., Liu Z., Liu Y., Huang Y., Xu Y., Hao P., Yin Z., Zhong J., Ye L., et al. 2019. ZIKV infection induces robust Th1-like Tfh cell and long-term protective antibody responses in immunocompetent mice. Nat. Commun. 10:3859. 10.1038/s41467-019-11754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W., Haribhai D., Relland L.M., Truong N., Carlson M.R., Williams C.B., and Chatila T.A.. 2007. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 8:359–368. 10.1038/ni1445 [DOI] [PubMed] [Google Scholar]
- Liu, X., Chen X., Zhong B., Wang A., Wang X., Chu F., Nurieva R.I., Yan X., Chen P., van der Flier L.G., et al. 2014. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 507:513–518. 10.1038/nature12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Yan X., Zhong B., Nurieva R.I., Wang A., Wang X., Martin-Orozco N., Wang Y., Chang S.H., Esplugues E., et al. 2012. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 209:1841–52, S1-24. 10.1084/jem.20120219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli, E., Dominguez M.H., Gattinoni L., Chattopadhyay P.K., Bolton D.L., Song K., Klatt N.R., Brenchley J.M., Vaccari M., Gostick E., et al. 2013. Superior T memory stem cell persistence supports long-lived T cell memory. J. Clin. Invest. 123:594–599. 10.1172/JCI66327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje, K., Kallies A., Shimohakamada Y., Belz G.T., Light A., Tarlinton D.M., and Nutt S.L.. 2012. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 13:491–498. 10.1038/ni.2261 [DOI] [PubMed] [Google Scholar]
- Moltke, I., Grarup N., Jørgensen M.E., Bjerregaard P., Treebak J.T., Fumagalli M., Korneliussen T.S., Andersen M.A., Nielsen T.S., Krarup N.T., et al. 2014. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 512:190–193. 10.1038/nature13425 [DOI] [PubMed] [Google Scholar]
- McDonald, P.W., Read K.A., Baker C.E., Anderson A.E., Powell M.D., Ballesteros-Tato A., and Oestreich K.J.. 2016. IL-7 signalling represses Bcl-6 and the TFH gene program. Nat. Commun. 7:10285. 10.1038/ncomms10285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, R., Schmitt N., Bentebibel S.E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N., et al. 2011. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 34:108–121. 10.1016/j.immuni.2010.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S.N., Gebhardt T., Carbone F.R., and Heath W.R.. 2013. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31:137–161. 10.1146/annurev-immunol-032712-095954 [DOI] [PubMed] [Google Scholar]
- Neri, P., Ren L., Azab A.K., Brentnall M., Gratton K., Klimowicz A.C., Lin C., Duggan P., Tassone P., Mansoor A., et al. 2011. Integrin β7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood. 117:6202–6213. 10.1182/blood-2010-06-292243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva, R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.H., Watowich S.S., Jetten A.M., Tian Q., and Dong C.. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149. 10.1016/j.immuni.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva, R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.-H.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais Ferreira, D., Silva J.G., Wyss T., Fuertes Marraco S.A., Scarpellino L., Charmoy M., Maas R., Siddiqui I., Tang L., Joyce J.A., et al. 2020. Central memory CD8+ T cells derive from stem-like Tcf7hi effector cells in the absence of cytotoxic differentiation. Immunity. 53:985–1000.e11. 10.1016/j.immuni.2020.09.005 [DOI] [PubMed] [Google Scholar]
- Pepper, M., and Jenkins M.K.. 2011. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 12:467–471. 10.1038/ni.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera, J., Proia R.L., and Olivera A.. 2008. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat. Rev. Immunol. 8:753–763. 10.1038/nri2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, A.M., Higgins B.W., Shuparski A.G., Miller K.B., Mcheyzer-Williams L.J., and Mcheyzer-Williams M.G.. 2022. Evolution of antigen-specific follicular helper T cell transcription from effector function to memory. Sci. Immunol. 7:eabm2084. 10.1126/sciimmunol.abm2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto, F., Geginat J., and Lanzavecchia A.. 2004. Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu. Rev. Immunol. 22:745–763. 10.1146/annurev.immunol.22.012703.104702 [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., and Ahmed R.. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli, P., Willimann K., Lang A.B., Lipp M., Loetscher P., and Moser B.. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 192:1553–1562. 10.1084/jem.192.11.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns, K.S., Kieper W.C., Jameson S.C., and Lefrançois L.. 2000. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 1:426–432. 10.1038/80868 [DOI] [PubMed] [Google Scholar]
- Schmitt, N., Morita R., Bourdery L., Bentebibel S.E., Zurawski S.M., Banchereau J., and Ueno H.. 2009. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 31:158–169. 10.1016/j.immuni.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon, B., Tomlinson P., and Zamoyska R.. 2003. Interleukin 7 and T cell receptor signals regulate homeostasis of CD4 memory cells. Nat. Immunol. 4:680–686. 10.1038/ni946 [DOI] [PubMed] [Google Scholar]
- Seder, R.A., and Ahmed R.. 2003. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 4:835–842. 10.1038/ni969 [DOI] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suan, D., Nguyen A., Moran I., Bourne K., Hermes J.R., Arshi M., Hampton H.R., Tomura M., Miwa Y., Kelleher A.D., et al. 2015. T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity. 42:704–718. 10.1016/j.immuni.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Yamada R., Chang X., Tokuhiro S., Sawada T., Suzuki M., Nagasaki M., Nakayama-Hamada M., Kawaida R., Ono M., et al. 2003. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 34:395–402. 10.1038/ng1206 [DOI] [PubMed] [Google Scholar]
- Tabata, M., Kadomatsu T., Fukuhara S., Miyata K., Ito Y., Endo M., Urano T., Zhu H.J., Tsukano H., Tazume H., et al. 2009. Angiopoietin-like protein 2 promotes chronic adipose tissue inflammation and obesity-related systemic insulin resistance. Cell Metab. 10:178–188. 10.1016/j.cmet.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Tsui, C., Kretschmer L., Rapelius S., Gabriel S.S., Chisanga D., Knöpper K., Utzschneider D.T., Nüssing S., Liao Y., Mason T., et al. 2022. MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature. 609:354–360. 10.1038/s41586-022-05105-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider, D.T., Charmoy M., Chennupati V., Pousse L., Ferreira D.P., Calderon-Copete S., Danilo M., Alfei F., Hofmann M., Wieland D., et al. 2016. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 45:415–427. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Velu, V., Mylvaganam G.H., Gangadhara S., Hong J.J., Iyer S.S., Gumber S., Ibegbu C.C., Villinger F., and Amara R.R.. 2016. Induction of Th1-biased T follicular helper (Tfh) cells in lymphoid tissues during chronic simian immunodeficiency virus infection defines functionally distinct germinal center Tfh cells. J. Immunol. 197:1832–1842. 10.4049/jimmunol.1600143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Zhou Y., Yu D., and Zhu H.. 2016. Klf2 contributes to the stemness and self-renewal of human bone marrow stromal cells. Cytotechnology. 68:839–848. 10.1007/s10616-014-9837-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, J.P., Fuhrmann F., Feist R.K., Lahmann A., Al Baz M.S., Gentz L.J., Vu Van D., Mages H.W., Haftmann C., Riedel R., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 212:217–233. 10.1084/jem.20141432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, J.S., Herman E.I., Lainez B., Licona-Limón P., Esplugues E., Flavell R., and Craft J.. 2016. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 17:1197–1205. 10.1038/ni.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., Sandor K., Pai J.A., Daniel B., Raju S., Wu R., Hsiung S., Qi Y., Yangdon T., Okamoto M., et al. 2022. BCL6-dependent TCF-1+ progenitor cells maintain effector and helper CD4+ T cell responses to persistent antigen. Immunity. 55:1200–1215.e6. 10.1016/j.immuni.2022.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Yan Y., Williams M.S., Carey G.B., Yang J., Li H., Zhang G.X., and Rostami A.. 2010. MS4a4B, a CD20 homologue in T cells, inhibits T cell propagation by modulation of cell cycle. PLoS One. 5:e13780. 10.1371/journal.pone.0013780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Cao Y., Xie Z., Huang Q., Bai Q., Yang X., He R., Hao Y., Wang H., Zhao T., et al. 2015. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat. Immunol. 16:991–999. 10.1038/ni.3229 [DOI] [PubMed] [Google Scholar]
- Xu, W., Zhao X., Wang X., Feng H., Gou M., Jin W., Wang X., Liu X., and Dong C.. 2019. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 51:826–839.e5. 10.1016/j.immuni.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Yu, D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468. 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Yu, D., and Vinuesa C.G.. 2010. The elusive identity of T follicular helper cells. Trends Immunol. 31:377–383. 10.1016/j.it.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D.-M., Yu S., Zhou X., Haring J.S., Held W., Badovinac V.P., Harty J.T., and Xue H.-H.. 2010. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J. Immunol. 184:1191–1199. 10.4049/jimmunol.0901199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Expression data have been deposited in the Gene Expression Omnibus database under the Super Series accession number GSE230438. This Super Series includes data for single-cell RNA-seq, bulk RNA-seq, and ATAC-seq in this paper.