Abstract
Objective
To determine if patients with systemic lupus erythematosus (SLE), a disease characterised by elevated type I interferons reminiscent of anti-viral immunity, have expression of human endogenous retrovirus K (HERV-K) proviruses capable of producing envelope (Env) protein, as well as associated autoantibodies against the Env protein.
Methods
ELISAs were conducted with recombinant Env protein and sera from SLE patients with active (n=60) or inactive (n=49) disease, healthy controls (n=47), other rheumatic disorders (n=59), as well as plasma from paediatric lupus patients with active (n=30) or inactive (n=30) disease, and 17 healthy children. Antibody reactivity was evaluated for correlations with clinical and laboratory parameters of the patients. Expression of HERV-K transcripts were profiled in SLE leukocytes by RNA-Seq.
Results
Both adult and paediatric SLE patients had autoantibodies against HERV-K Env with higher titers than healthy controls or patients with Sjögren’s syndrome, small- or large-vessel vasculitis, or psoriatic arthritis. Transcripts from only two HERV-K loci capable of producing Env, HERV-K102 and -K108, were detected among the 10 expressed loci in SLE patients.
Conclusion
Our data reveal that HERV-K proviruses are expressed in SLE and that the HERV-K-encoded Env protein elicits an immune response in patients, particularly during active disease.
Keywords: systemic lupus erythematosus, autoantibodies, endogenous retrovirus K
Introduction
More than 30 years ago, several groups reported that sera from patients with SLE (1 2), Sjögren’s syndrome (3), or other autoimmune disorders (4, 5), contained antibodies reactive with human immunodeficiency virus (HIV) proteins, such as p24gag, even if the patients did not have a history of HIV. These findings ignited a search for a causative infectious retrovirus, which was not successful (6). It was later discovered that the anti-HIV antibodies likely represented antibodies against proteins encoded by human endogenous retroviruses (HERVs), sequences of which constitute as much as 8% of our genome. Although the overall homology between HIV and HERVs is limited, there are conserved motifs and structural features that they share.
Early studies on HERV expression patterns that relied on polymerase chain reaction (PCR) amplification and biochemical assays of retroviral reverse transcriptase activity were positive in samples from the lung (7), immune cells (8), and skin (9) of lupus patients, but the exact nature of the expressed HERVs remained unclear. The advent of high-throughput RNA sequencing (RNA-Seq) in combination with bioinformatics tools like ERVmap (10) and Telescope (11), enabled a much deeper analysis of the repertoire of expressed HERVs in SLE and other conditions. These studies have revealed that numerous HERV sequences are indeed expressed at altered levels in SLE (10, 11). However, it is important to recognise that none of these expressed loci can produce fully infectious retroviruses and that over 99% of them are unable to produce retroviral proteins. In contrast, several different fully infectious retroviruses are active in mouse models of lupus-like disease (12-14), which also have autoantibodies reactive with retroviral envelope proteins (15, 16). Lupus-prone mouse strains generally express many more endogenous retroviruses than other strains. This is at least in part due to impaired epigenetic control of retroviral loci in these mice (17). A recent paper (18) reported that the lupus-related gene sgp3 encodes a Kruppel-associated box zinc-finger protein that binds to the retroviral long terminal repeat (LTR) to silence their expression. Homozygous deletion of Sgp3 resulted in elevated production of retrovirus and manifest lupus nephritis in susceptible mice (18). They also found that increased HERV expression in human SLE correlated inversely with three Sgp3-related proteins, suggesting that dysregulated HERV expression may contribute to the pathogenesis of SLE.
Here, we expand on this previous work by showing that both adult and paediatric SLE patients have IgG autoantibodies that recognise the envelope (Env) protein of HERV-K, the youngest subgroup of HERVs, while several other rheumatological conditions do not. By RNA sequencing we find that SLE patient leukocytes express ten HERV-K proviruses of which only two still have an intact open reading frame for the retroviral Env protein.
Materials and methods
Adult SLE patients.
A cohort of SLE patients with active disease (n=60) and with disease in remission (n=49), and age- and gender-matched healthy individuals (n=28), were from the Department of Rheumatology, Skåne University Hospital, Lund, Sweden. The study was approved by Lund University local ethics board (LU06014520, and LU 378-02). Informed written consent was obtained from all participants according to the Declaration of Helsinki. This patient cohort has been described in detail previously (19-21).
Additional adult patients with SLE (n=3) for fresh blood samples were recruited through the University of Washington, Division of Rheumatology Biorepository to participate in research studies at the University of Washington Medical Center, Seattle, WA. Serum samples from patients with primary Sjögren’s syndrome (SjS), anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV), large-vessel vasculitis (LVV), psoriatic arthritis (PsA), and additional healthy controls (n=19) were also obtained from this Biorepository. Our study was approved by the University of Washington Medical Center ethics board (STUDY00006196), and informed written consent was obtained from all participants.
pSLE patients and healthy children
A cohort of paediatric lupus (pSLE) patients (n=30) were recruited at Seattle Children’s Hospital, from each of whom plasma samples were collected once during active disease and once when the disease was inactive (SLEDAI <4). In 13 cases the first sample was taken at the time of diagnosis. The clinical characteristics of this cohort have been reported before (Ukadike et al., submitted). Exclusion criteria included severe anaemia and inability to consent. An additional exclusion criterion for healthy children (n=17) was any history of immune-mediated disorder. The study was approved by the Seattle Children’s Research Hospital Human Subjects Committee (PIROSTUDY14045). Informed written consent was obtained from the parents or guardians of all participants according to the Declaration of Helsinki.
Recombinant Env
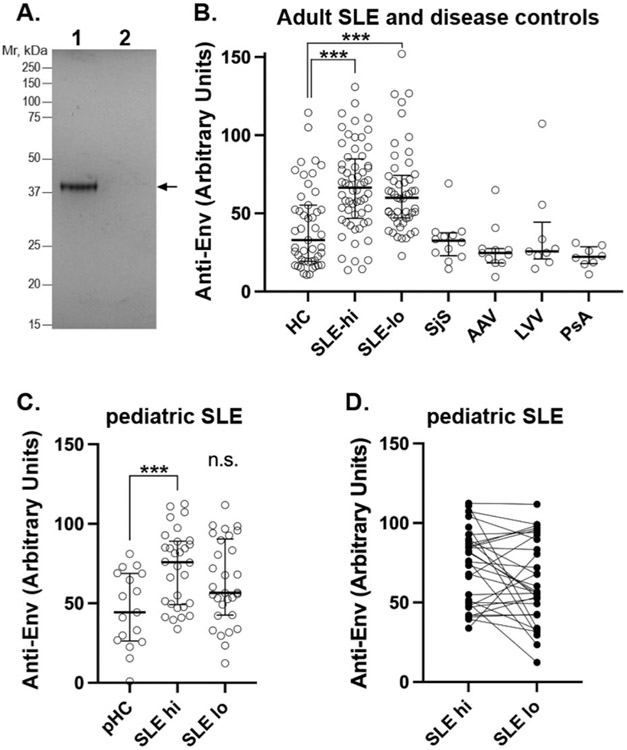
The amino acid sequence of Env from the seemingly intact HERV-K provirus at Xq21.33 was used to design a prokaryotic expression plasmid (Twist Biosciences) for amino acid residues 114-465, corresponding to the sequence shared between type 1 and type 2 HERV-K proviruses from the end of the leader sequence to the protease cleavage site (22). The 42-kDa recombinant protein was expressed in E. coli and purified by Ni2+-agarose affinity chromatography by Olympic Protein Technologies (Seattle, WA). This protein is 99% identical with K102 and K108, which also are 99% identical to each other. Its purity is illustrated in Figure 1A.
Fig. 1. Autoantibodies against HERV-K Env in patients with SLE, but not other rheumatological diseases.
A: Coomassie staining of the recombinant Env protein used in the ELISAs.
B: ELISA for IgG anti-Env in healthy controls (HC) and SLE patients with active (SLE-hi) or inactive (SLE-lo) disease, and in patients with Sjögren’s syndrome (SjS), ANCA-associated vasculitis (AAV), large-vessel vasculitis (LVV), or psoriatic arthritis (PsA).
C: ELISA for IgG anti-Env in healthy children (pHC) and paediatric SLE patients with active (SLE-hi) or inactive (SLE-lo) disease.
D: The data points in C for each individual connected with lines.
Statistical significance in indicated by *** for p<0.0001 and all experiments were repeated at least 3 times.
ELISAs
50 ng/well of purified Env protein was adsorbed onto 96-well polystyrene plates in 0.1 M bicarbonate (pH 9.6) buffer overnight, washed in phosphate-buffered saline with 0.05% Tween, and blocked in 2% bovine serum albumin (BSA) in phosphate-buffered saline for 2 h. Control wells lacked Env protein, but were blocked the same way. Serum or plasma was added at 0.5% in blocking buffer (with BSA) for 2 h at 4°C, washed extensively, and then incubated with 1:2,000 dilution of horse radish peroxidase-conjugated anti-human IgG. The reaction was then washed, and developed with 3,3’,5,5’-tetra-methylbenzidine (TMB, BioLegend), with the color reaction terminated with 2N sulfuric acid, and the absorbance measured at 450 nm using a plate reader. A dilution series of serum from a selected rheumatoid arthritis patient with high anti-Env reactivity was included on every plate to ensure consistency between plates. The reactivity of this standard (at the same dilution as test samples) was designated as 100 U/ml and all other samples normalised accordingly.
Immune cell isolation
Polymorphonuclear (PMN) and peripheral blood mononuclear cells (PBMC) were isolated from freshly drawn venous blood by gradient centrifugation on PolymorphPrep according to the manufacturer’s instructions. Cells were washed and suspended in Hanks’ buffered salt solution at 107/ml.
RNA isolation and RNA-Seq
Extraction of RNA from patient (n=3) or healthy control (n=3) leukocytes (lymphocytes and neutrophils) was accomplished by using a hybrid protocol of Trizol Reagent (Invitrogen cat# 15596026) and RNeasy Micro Kit (Qiagen cat. no. 74004). Briefly, the cells were pelleted, then lysed and homogenised with Trizol Reagent. Chloroform was added to layer out the DNA and proteins from the RNA to which β-mercaptoethanol was added for additional RNase inhibition. The RNA was precipitated with 70% ethanol, passed through a RNeasy MinElute spin column, and treated with DNase I on the column. The spin column was subsequently washed extensively with the kit-supplied proprietary wash buffers, followed by additional conditioning with 70% ethanol before eluting the RNA with RNase-free water.
For RNA-Seq, sample RNA purity was verified by measuring their concentrations and A260/A280 using BioTek Take3 microplate reader. RNA integrity was verified by performing non-denaturing gel electrophoresis on 1% agarose gel, confirming the presence of the 28S and 18S rRNAs, before sending to BGI Genomics for RNA sequencing. They subjected the samples to further quality control testing using Agilent 2100 Bioanalyzer instrument to verify RNA integrity, RNA quality and other measures before performing the RNA sequencing using their DNBseq NGS platform. Over 40 million clean reads were obtained from each sample.
Analysis of HERV-K transcripts
All transcripts annotated as derived from HERV-K (or any of its synonyms) were segregated from the total pool of transcripts, and their respective matching genomic loci examined by extracting their nucleotide sequence, including 5’ and 3’ flanking sequences, and translating them into amino acid sequence in all three reading frames. All resulting polypeptides over 50-amino acid residues in length were subjected to BLAST searches for homology with HERV-K proteins.
Statistical analyses
For non-paired sample sets with non-Gaussian distribution, Mann-Whitney U-test and Spearman’s correlation test were used, as applicable. For paired sample sets, Wilcoxon matched-pairs signed rank test was used. GraphPad Prism (version 9) and IBM SPSS were used for the analyses. All analyses were considered statistically significant at p<0.05.
Results
IgG autoantibodies against Env in SLE patients, but not in other diseases
ELISAs performed with adsorbed recombinant Env protein revealed a broad range reactivity in the serum of SLE patients with active disease (n=60), a somewhat lower reactivity in patients in remission (n=49) and only a low to medium reactivity in the serum from healthy donors (Fig. 1B). The difference between controls and active patients was statistically significant (p<0.0001), as was the difference between patients in remission and healthy controls (p<0.0001), while the difference between active disease and remission was not. Serum from patients with other rheumatological conditions also did not contain anti-Env reactivity different from healthy controls (Fig. 1B). These included Sjögren’s syndrome, ANCA-associated vasculitis, large-vessel vasculitis, and psoriatic arthritis.
Anti-Env in paediatric SLE patients
Since children with newly diagnosed paediatric lupus are, on average, closer to the beginning of their disease, we assessed a cohort of 30 paediatric lupus patients for IgG autoantibodies against HERV-K Env. This cohort includes two samples for each patient, one taken during active disease with an average SLEDAI of 10.6±4.8 (standard deviation), and another sample during inactive disease with an average SLEDAI of 2.4±1.8. Compared to healthy children, the active disease samples had a statistically significantly (p=0.0009) higher reactivity against Env (Fig. 1C), while the inactive disease samples did not (p=0.067) using the Mann-Whitney test. Assessing whether titres of anti-Env fluctuated within patients, comparing the two data points for each individual patient reveals that anti-Env titers declined from active to inactive disease in nearly half the patients, but increased in 7 or remained stable in 7 (Fig. 1D).
Expression of HERV-K proviruses in SLE leukocytes
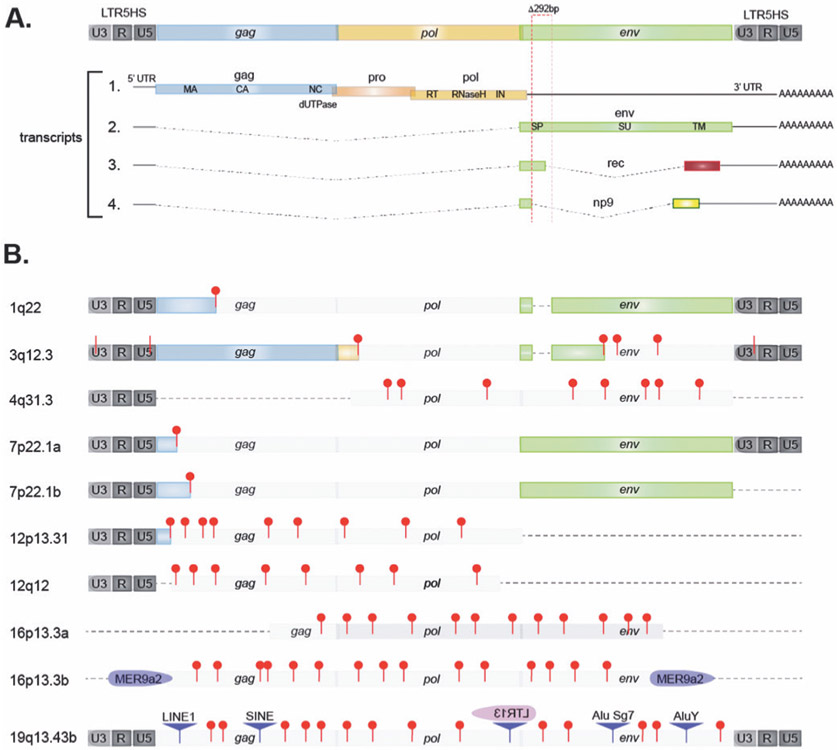
The alternative splicing and translation of transcripts from an intact HERV-K provirus is illustrated in Fig. 2A. The 10 genomic HERV-K proviruses from which transcripts were detected in the leukocytes from SLE patients (Table I) are represented schematically in Fig. 2B. Notably, only transcripts from HERV-K102 (1q22) and the tandem duplicated HERV-K108 (7p22.1) can be translated into Env protein from mRNA spliced as transcript 2 in Fig. 2A, while all the other HERV-K proviruses expressed in SLE leukocytes have numerous stop codons, insertions, or lack the env region altogether. Hence, only HERV-K102 and K108 can produce the immunogenic Env that elicits an antibody response.
Fig. 2. Structure of HERV-K loci transcribed in RA patients.
A: Schematic representation of an intact HERV-K/HML-2 provirus and the 4 transcripts it can generate through alternative splicing. Note that only transcript 2 can be translated into Env. The 292-bp sequence missing in type 1 and present in type 2 proviruses is indicated with dotted red box.
B: the HERV-K proviruses from which transcripts were detected at higher levels in SLE patients than in healthy donors. Red pins indicate stop codons, including those following frame-shifts (approximate locations and numbers). Pale grey boxes represent elements that cannot be translated into full-length protein. A dotted line represents missing sequence. Disrupting insertions of other retroelements are indicated as purple triangles, LTRs in purple of pink.
Note that the inserted LTR13 in 19q13.43b is in the opposite direction (other DNA strand).
Table I.
HERV-K proviruses expressed at elevated levels in SLE.
| Chromosomal location | Strand | Transcript designation | Synonyms | Comments |
|---|---|---|---|---|
| 1q22a (155,599,135–155,604,668) | − | HERVK-int_dup15 | K102, ERVK7 | full-length type 1 provirus |
| 3q12.3 (101,411,706–101,417,184) | + | HERVK-int_dup37 | ERVK5 | Gag, but not Pol or Env |
| 4q31.3 (154,610,027–154,615,070) | − | HERVK9-int_dup190 | NA | incomplete, short ORFs |
| 7p22.1a (4,623,025–4,630,560) | − | HERVK-int_dup102 | K108L, ERVK6 | full-length type 2 provirus |
| 7p22.1b (4,631,529–4,639,063) | − | HERVK-int_dup103 | K108R, ERVK6 | full-length type 2 provirus |
| 12p13.31 (8,711,047–8,715,066) | − | HERVK22-int_dup314 | NA | 3’ truncated, stop codons |
| 12q12 (42,837,368–42,840,271) | + | HERVK22-int_dup317 | periphilin1 | 3’ truncated, stop codons |
| 16p13.3a (2,711,289–2,718,374) | − | HERVK13-int_dup43 | NA | 3’,5’truncated, stop codons |
| 16p13.3b (3,127,354–3,131,057) | − | HERVK9-int_dup555-558 | NA | insertions, stop codons |
| 19q13.43c (58,824,957–58,825,710) | + | HERVK3-int_dup298 | NA | insertions, stop codons |
Discussion
The presence of autoantibodies that recognise HERV-K Env suggests that this protein is, or was, expressed in patients and that it is sufficiently immunogenic to elicit an antibody response. Among the HERV-K transcripts we detect in patient leukocytes only two proviruses, K102 and K108, contain an intact open reading frame for Env. Hence, these two proviruses must be the source of the immunogenic Env. They belong to the HML-2 subgroup of HERV-K, are human-specific, and considerably less than a million years old as indicated by their near identical 5’ and 3’ LTRs. In contrast, the vast majority of other HERVs in our genome are much older and have sustained extensive deletions, insertions, frame-shift mutations and amino acid substitutions and, as a result, any transcripts from them can only be translated into short peptides devoid of original functions. The few non-HERV-K env-derived polypeptides that have been reported (e.g. syncytin) have minimal homology with HERV-K Env and it is very unlikely that the anti-Env autoantibodies we detect would cross-react with any of them.
Two previous papers (10, 11) reported that several HERVs were overexpressed in SLE leukocytes, including the HERV-K proviruses 4q31.3 and 19q13.42, which we also see. However, these papers did not include any HERV-K proviruses able to produce Env, perhaps due to stricter criteria for statistical significance of differential expression. It is also notable that none of the approximately 10 other HERV-K/HML-2 proviruses in our genome with an intact open reading frame for Env were detected in either these previous papers (10, 11). However, after the completion of the work described herein, Tokuyama and coworkers (35) published their findings that HERV-K102 is expressed in SLE patients, resulting in a robust anti-Env autoantibody response, in good agreement with our findings. They reported that the increased expression of K102 correlated closely with reduced expression of repressive epigenetic factors like TRIM28 and that the anti-Env autoantibodies correlated positively with the type I interferon gene signature in the blood of the patients. We did not observe this correlation. Another difference between our study and theirs is that they did not see a statistically significant difference in K108 expression between patients and healthy controls. They also detected transcripts from HERV-K106 and K115, which we did not see. These differences between our studies may relate to the relatively small numbers of patients in both studies, the insertional polymorphisms (e.g. K115 exists in <10% of individuals) of these highly homologous human-specific proviruses, which are the result of repeated re-infections of the germline by the same exogenous retrovirus. Hence, it is not clear if these subtle differences between our studies are of any particular relevance.
While epigenetically silenced in healthy individuals, HERV-K proviruses are known to be transcribed during early embryonic development (23), in malignant cells (24), in HIV-infected individuals (25-33), and in blood and synovial fluid from RA patients (34). SLE is now added to this list. Interestingly, the set of transcriptionally active HERV-K proviruses is different in different cancer lines and distinct from any of them in SLE leukocytes. It also appears that healthy individuals can develop low to moderate levels of anti-Env antibodies, suggesting that some expression can occur during normal life without causing autoimmune disease. The absence of statistically significant correlations with specific symptoms, organ manifestations, or laboratory measures in SLE (data not shown), also suggests that HERV-K expression and anti-Env autoantibodies are more likely a consequence of epigenetic dysregulation in SLE, rather than causative in its pathogenesis.
Even if HERV-K reactivation may not have a causative role in SLE pathogenesis, it is still possible that increased transcription of HERV-K loci that have retained the ability to encode functional proteins, such as a transmembrane, proteolytically processed, and glycosylated Env protein that can trimerise, traffic to the plasma membrane, assemble with intracellular N-terminally myristoylated Gag proteins (from the same or a different locus), and perhaps even bud off as a virus-like particle, gives HERV-K Env a modulating role in the pathogenesis of SLE (36). We would also argue that antibodies against Env encoded by HERV-K already represent a form of ‘auto’ immunity since even the youngest HERV-K loci have resided in our genome subject to Mendelian inheritance for tens of thousands of years. They have effectively become ‘self’. One would also predict that such anti-Env autoantibodies can bind surface Env expressed by immune cells and that such binding events may have functional consequences. Tokuyama et al. propose that these antibodies can activate neutrophils (35), a notion that we agree with because Env is expressed in neutrophils. It is also conceivable that anti-Env antibodies may mediate complement activation, or engage antibody-dependent cellular cytotoxicity, resulting in the killing of cells exposing Env on their surface. Finally, since fully functional Env has affinity for heparan sulfate-containing surface proteins, which gave the original exogenous HERV-K retrovirus its cellular tropism (37), the mere expression of Env could increase leukocyte adhesion and lead to inflammation. These, and other possible contributions to SLE pathogenesis (38, 39), will be challenging to prove or disprove until clinical trials can be conducted with agents that halt critical steps of HERV-K expression or function.
Acknowledgements
This work was supported by National Institutes of Health (T32 AR007108 to KCU, R21 AR077266, R21 AR075134, and R01 AR074939 to T. Mustelin), Lupus Research Alliance grant 519414 (to C. Lood).
Footnotes
Competing interests: T. Mustelin reports personal consulting fees from Kiniksa, from Cugene, from QiLu Biopharma, and from Miro Bio, all outside the scope of this paper. A. Stevens is an employee of Janssen Research & Development. The other co-authors have declared no competing interests.
References
- 1.TALAL N, GARRY RF, SCHUR PH et al. : A conserved idiotype and antibodies to retroviral proteins in systemic lupus erythematosus. J Clin Invest 1990; 85: 1866–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.BLOMBERG J, NIVED O, PIPKORN R, BENGTSSON A, ERLINGE D, STURFELT G: Increased antiretroviral antibody reactivity in sera from a defined population of patients with systemic lupus erythematosus. Correlation with autoantibodies and clinical manifestations. Arthritis Rheum 1994; 37: 57–66. [DOI] [PubMed] [Google Scholar]
- 3.TALAL N, DAUPHINEE MJ, DANG H, ALEXANDER SS, HART DJ, GARRY RF: Detection of serum antibodies to retroviral proteins in patients with primary Sjögren’s syndrome (autoimmune exocrinopathy). Arthritis Rheum 1990; 33: 774–81. [DOI] [PubMed] [Google Scholar]
- 4.DANG H, DAUPHINEE MJ, TALAL N et al. : Serum antibody to retroviral gag proteins in systemic sclerosis. Arthritis Rheum 1991; 34: 1336–7. [DOI] [PubMed] [Google Scholar]
- 5.FRAZIANO M, MONTESANO C, LOMBARDI VR et al. : Epitope specificity of anti-HIV antibodies in human and murine autoimmune diseases. AIDS Res Hum Retroviruses 1996; 12: 491–6. [DOI] [PubMed] [Google Scholar]
- 6.PELTON BK, NORTH M, PALMER RG et al. : A search for retrovirus infection in systemic lupus erythematosus and rheumatoid arthritis. Ann Rheum Dis 1988; 47: 206–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.TAMURA N, SEKIGAWA I, HASHIMOTO H, YAMAMOTO N, KIRA S: Syncytial cell formation in vivo by type C retroviral particles in the systemic lupus erythematosus (SLE) lung. Clin Exp Immunol 1997; 107: 474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MCINTOSH RS, HUGGINS ML, KONTOGIANNIS V, TIGHE PJ, POWELL RJ: Retroviral activity in Behçet’s syndrome and systemic lupus erythematosus detected by a PCR-based reverse transcriptase assay. Rheumatol Int 2001; 21: 53–7. [DOI] [PubMed] [Google Scholar]
- 9.PROKOP J, JAGODZINSKI PP: Identification of retroviral DNA sequences in serum of cutaneous forms of lupus erythematosus patients. Eur J Dermatol 2003; 13: 354–8. [PubMed] [Google Scholar]
- 10.TOKUYAMA M, KONG Y, SONG E, JAYEWICKREME T, KANG I, IWASAKI A: ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proc Natl Acad Sci USA 2018; 115: 12565–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.INIGUEZ LP, DE MULDER ROUGVIE M, STEARRETT N et al. : Transcriptomic analysis of human endogenous retroviruses in systemic lupus erythematosus. Proc Natl Acad Sci USA 2019; 116: 21350–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GOURLEY MF, KISCH WJ, MOJCIK CF, KING LB, KRIEG AM, STEINBERG AD: Molecular aspects of systemic lupus erythematosus: murine endogenous retroviral expression. DNA Cell Biol 1992; 11: 253–7. [DOI] [PubMed] [Google Scholar]
- 13.KRIEG AM, GOURLEY MF, STEINBERG AD: Association of murine lupus and thymic full-length endogenous retroviral expression maps to a bone marrow stem cell. J Immunol 1991; 146: 3002–5. [PubMed] [Google Scholar]
- 14.KRIEG AM, STEINBERG AD: Analysis of thymic endogenous retroviral expression in murine lupus. Genetic and immune studies. J Clin Invest 1990; 8): 809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.TABATA N, MIYAZAWA M, FUJISAWA R, TAKEI YA, ABE H, HASHIMOTO K: Establishment of monoclonal anti-retroviral gp70 autoantibodies from MRL/lpr lupus mice and induction of glomerular gp70 deposition and pathology by transfer into non-autoimmune mice. J Virol 2000; 74: 4116–26- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.BAUDINO L, YOSHINOBU K, MORITO N et al. : Dissection of genetic mechanisms governing the expression of serum retroviral gp70 implicated in murine lupus nephritis. J Immunol 2008; 181: 2846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.BAUDINO L, YOSHINOBU K, DUNAND-SAUTHIER I, EVANS LH, IZUI S: TLR-mediated up-regulation of serum retroviral gp70 is controlled by the Sgp loci of lupus-prone mice. J Autoimmun 2010; 35: 153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.TREGER RS, POPE SD, KONG Y, TOKUYAMA M, TAURA M, IWASAKI A: The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity 2019; 50: 334–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LOOD C, ERIKSSON S, GULLSTRAND B et al. : Increased C1q, C4 and C3 deposition on platelets in patients with systemic lupus erythematosus--a possible link to venous thrombosis? Lupus 2012; 21: 1423–32. [DOI] [PubMed] [Google Scholar]
- 20.LOOD C, TYDEN H, GULLSTRAND B et al. : Platelet-derived S100A8/A9 and cardiovascular disease in systemic lupus erythematosus. Arthritis Rheumatol 2016; 68: 1970–80. [DOI] [PubMed] [Google Scholar]
- 21.LOOD C, TYDEN H, GULLSTRAND B et al. : Decreased platelet size is associated with platelet activation and anti-phospholipid syndrome in systemic lupus erythematosus. Rheumatology (Oxford) 2017; 56: 408–16. [DOI] [PubMed] [Google Scholar]
- 22.BEIMFORDE N, HANKE K, AMMAR I, KURTH R, BANNERT N: Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 2008; 371: 216–25. [DOI] [PubMed] [Google Scholar]
- 23.GROW EJ, FLYNN RA, CHAVEZ SL et al. : Intrinsic retroviral reactivation in human pre-implantation embryos and pluripotent cells. Nature 2015; 522: 221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.GOERING W, RIBARSKA T, SCHULZ WA: Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 2011; 32: 1484–92.. [DOI] [PubMed] [Google Scholar]
- 25.HARDWAJ N, MALDARELLI F, MELLORS J, COFFIN JM: HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J Virol 2014; 88: 11108–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.BOWEN LN, TYAGI R, LI W et al. : HIV-associated motor neuron disease: HERV-K activation and response to antiretroviral therapy. Neurology 2016; 87: 1756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CONTRERAS-GALINDO R, ALMODOVAR-CAMACHO S, GONZALEZ-RAMIREZ S, LORENZO E, YAMAMURA Y: Comparative longitudinal studies of HERV-K and HIV-1 RNA titers in HIV-1-infected patients receiving successful versus unsuccessful highly active antiretroviral therapy. AIDS Res Hum Retroviruses 2007; 23: 1083–6. [DOI] [PubMed] [Google Scholar]
- 28.CONTRERAS-GALINDO R, GONZALEZ M, ALMODOVAR-CAMACHO S, GONZALEZ-RAMIREZ S, LORENZO E, YAMAMURA Y: A new Real-Time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J Virol Methods 2006; 136: 51–7. [DOI] [PubMed] [Google Scholar]
- 29.CONTRERAS-GALINDO R, KAPLAN MH, MARKOVITZ DM, LORENZO E, YAMAMURA Y: Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res Hum Retroviruses 2006; 22: 979–84. [DOI] [PubMed] [Google Scholar]
- 30.CONTRERAS-GALINDO R, LOPEZ P, VELEZ R, YAMAMURA Y: HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res Hum Retroviruses 2007; 23: 116–22. [DOI] [PubMed] [Google Scholar]
- 31.DE MULDER M, SENGUPTA D, DEEKS SG et al. : Anti-HERV-K (HML-2) capsid antibody responses in HIV elite controllers. Retrovirology 2017; 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.EHLHARDT S, SEIFERT M, SCHNEIDER J, OJAK A, ZANG KD, MEHRAEIN Y: Human endogenous retrovirus HERV-K(HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J Rheumatol 2006; 33: 16–23. [PubMed] [Google Scholar]
- 33.GONZALEZ-HERNANDEZ MJ, SWANSON MD, CONTRERAS-GALINDO R et al. : Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol 2012; 86: 7790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.REYNIER F, VERJAT T, TURREL F et al. : Increase in human endogenous retrovirus HERV-K (HML-2) viral load in active rheumatoid arthritis. Scand J Immunol 2009; 70: 295–9. [DOI] [PubMed] [Google Scholar]
- 35.TOKUYAMA M, GUNN BM, VENKATARAMAN A et al. : Antibodies against human endogenous retrovirus K102 envelope activate neutrophils in systemic lupus erythematosus. J Exp Med 2021; 218: e20191766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.ZUCCHI D, ELEFANTE E, CALABRESI E, SIGNORINI V, BORTOLUZZI A, TANI C: One year in review 2019: systemic lupus erythematosus. Clin Exp Rheumatol 2019; 37: 715–22 [PubMed] [Google Scholar]
- 37.ROBINSON-McCARTHY LR, McCARTHY KR, RAABEN M et al. : Reconstruction of the cell entry pathway of an extinct virus. PLoS Pathog 2018; 14: e1007123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UKADIKE KC, MUSTELIN T: Implications of endogenous retroelements in the etiopathogenesis of systemic lupus erythematosus. J Clin Med 2021; 10: 856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MUSTELIN T, UKADIKE KC: How retroviruses and retrotransposons in our genome may contribute to autoimmunity in rheumatological conditions. Front Immunol 2020; 11: 593891. [DOI] [PMC free article] [PubMed] [Google Scholar]