SUMMARY
Mechanosensitive hair cells in the cochlea are responsible for hearing, but are vulnerable to damage by genetic mutations and environmental insults. The paucity of human cochlear tissues makes it difficult to study cochlear hair cells. Organoids offer a compelling platform to study scarce tissues in vitro; however, derivation of cochlear cell types has proven non-trivial. Here, using 3D cultures of human pluripotent stem cells, we sought to replicate key differentiation cues of cochlear specification. We found that timed modulations of Sonic Hedgehog and WNT signalling promote ventral gene expression in otic progenitors. Ventralized otic progenitors subsequently give rise to elaborately patterned epithelia containing hair cells with morphology, marker expression, and functional properties consistent with both outer and inner hair cells in the cochlea. These results suggest that early morphogenic cues are sufficient to drive cochlear induction and establish an unprecedented system to model the human auditory organ.
eTOC Blurb
Moore and colleagues present a protocol for generating cochlear organoids from human PSCs by treating otic progenitors with ventralization signals. Derived hair cells in cochlear organoids exhibit molecular, structural and functional features of cochlear hair cells, and serve as a model to study how sound-sensing hair cells in humans develop.
Graphical Abstract
INTRODUCTION
The human inner ear is among the most elaborate organs in nature; featuring the spiral-shaped cochlea with its orderly rows of sound-detecting sensory cells and the intricate labyrinth of the vestibular organs that confer the sense of balance. The astonishing complexity of inner ear morphogenesis is orchestrated by a symphony of interwoven signalling events during fetal development that proceeds with remarkable fidelity 1–3. Indeed, while almost 10% of adults have moderate-to-profound hearing loss, the incidence at birth is less than 0.2%, meaning that the vast majority of sensorineural hearing loss results from the post-natal death or dysfunction of cells within a developmentally pristine inner ear 4.
To recapitulate the intricate process of human inner ear development in vitro, we and others previously established defined 3D culture systems to generate inner ear sensory epithelia from aggregates of mouse or human pluripotent stem cells 5–10. These so-called “inner ear organoids” harbour a layer of supporting cells and functional hair cells that are innervated by sensory-like neurons. Although these organoids consistently generate hair cells with structural, biochemical and functional properties comparable to native vestibular hair cells, they lack any cochlear cell types. To address the need for better tools to study human inner ear development and disease, we sought to establish a new organoid system that gives rise to cochlear cell types, including inner and outer hair cells, which are the functional unit of auditory mechanosensation.
RESULTS
A multiplex reporter hESC line provides enhanced culture optimization
To monitor the derivation of otic progenitors and hair cells in culture and improve the efficiency of our previously published protocols 5,8 we generated a PAX2-2A-nGFP/POU4F3-2A-ntdTomato reporter human embryonic stem cell (hESC) line using the CRISPR/Cas9 genome engineering technology. PAX2 is an early marker of otic progenitors in vivo, while POU4F3 expression is highly specific to hair cells and provides a late-stage readout of culture efficiency 11–13. We first confirmed that PAX2b is the most abundantly expressed PAX2 isoform in human inner ear organoid tissues (Figures S1A–B). Using a high-fidelity Cas9 14 and a sgRNA targeting the stop codon of this isoform, we knocked in a 2A-nGFP cassette immediately downstream of the endogenous PAX2 coding sequence (Figures 1A, S1C–D; Table S1). A 2A-ntdTomato cassette was similarly knocked in to the POU4F3 locus downstream of the POU4F3 stop codon using the established PAX2-2A-nGFP hESCs as a parental cell line (Figures 1A, S1G–H; Table S1). The resulting multiplex reporter cell line expressing multiple pluripotency markers (Figure S1E) labels the PAX2+ otic progenitors with nuclear GFP starting from around organoid culture day 12 (D12), and the POU4F3+ hair cells with nuclear tdTomato starting from around D35 (Figures 1B–L, S1F, I–L”; Video S1). Using the hESC reporter line we had established, we systematically changed basal media, timings and durations of small molecule treatment to optimize otic induction (Figure 2A). Following optimization, the number of POU4F3-positive aggregates more than doubled vs. our previous protocols 8,15, and the average POU4F3-positive area per aggregate increased ~20-fold (Figures 2B and 2C).
Figure 1. PAX2-2A-nGFP/POU4F3-2A-ntTomato (PAX2nG/POU4F3nT) multiplex reporter hESCs faithfully recapitulate otic progenitor and hair cell differentiations in inner ear organoids.
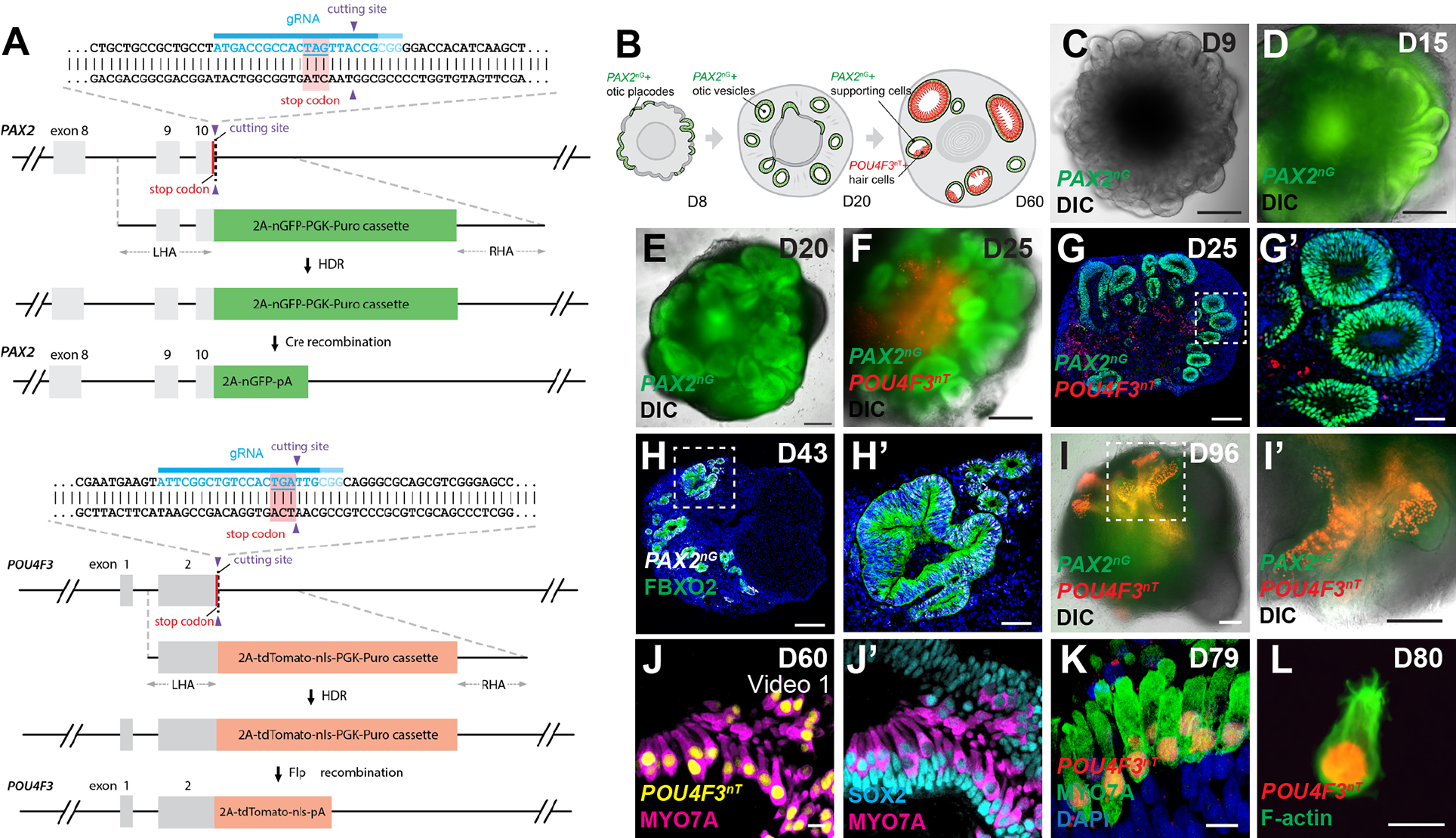
(A) Schematic illustrations of the PAX2-2A-nGFP and POU4F3-2A-ntdTomato CRISPR design.
(B) Schematic of PAX2nG and POU4F3nT reporter expression during inner ear organoid development.
(C-F) Live images of whole aggregates containing multiple developing inner ear organoids show the spatio-temporal progression of PAX2nG reporter expression and early morphogenesis of PAX2+ epithelium.
(G-H) Representative images of hESC-derived aggregates showing PAX2nG+ epithelium organized into vesicles that co-express the otic-specific marker FBXO2, but devoid of POU4F3nT expression.
(I-I’) Live images of late-stage (D96) aggregates showing intense POU4F3nT+ puncta localized to epithelial vesicles.
(J-K) POU4F3nT+ cells in inner ear organoids also express the hair cell markers MYO7A and SOX2, and are located on the luminal surface of SOX2+ supporting epithelia.
(L) Fixed cell suspension of dissociated POU4F3nT+ cells isolated from D80 inner ear organoids reveals tdTomato+ nuclei and F-actin+ membranes and stereocilia.
Scale bars, 200 μm (C-G, H, I, I’), 50 μm (G’, H’), 10 μm (J-L).
Figure 2. Optimization of inner ear organoid derivation protocol.
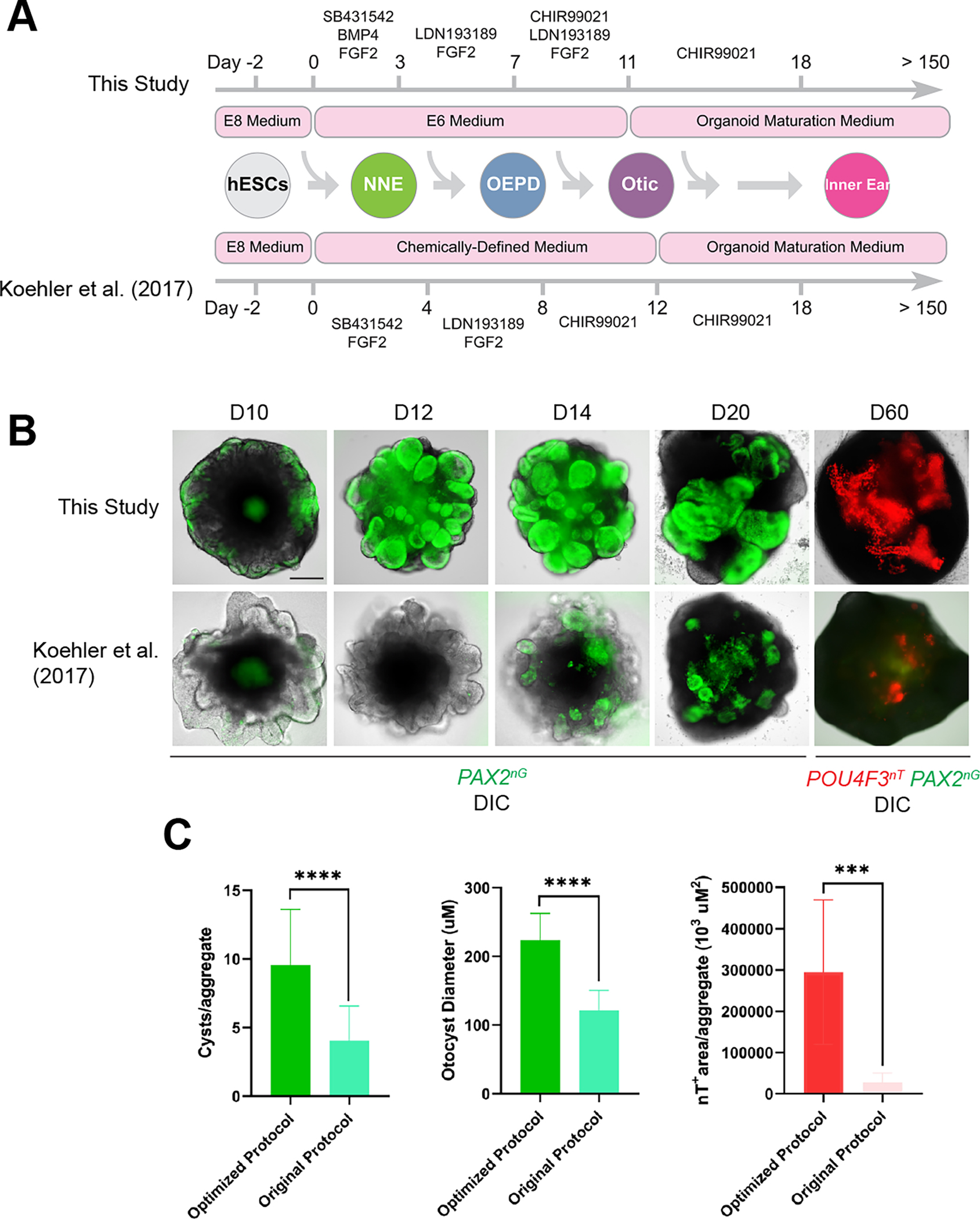
(A) Schematic comparison of our original vs. optimized protocol.
(B) Live images of whole cell aggregates containing inner ear organoids derived from our PAX2nG/POU4F3nT multiplex reporter hESC line cultured under the original vs. optimized protocol.
(C) Quantitative comparison of culture outcomes in optimized vs. original culture protocol on D60. n = 20 (green histograms), 13 (red histogram) biological samples from separate experiments per group. Welch’s two-sided t-test ***P=0.000132, ****P=0.000013 (#cysts) P<0.000001 (diameter).
Scale bars, 200 μm.
Sequential modulations of Sonic Hedgehog and WNT signaling promote ventralization of inner ear organoids
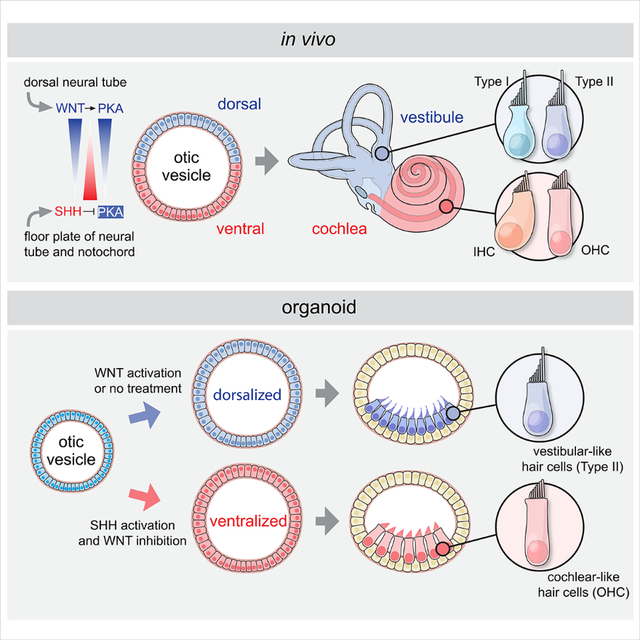
During inner ear development, the cochlear structure is derived from the most ventral region of the otic vesicle, whereas the vestibular structures originate from a more dorsal otic region 1 (Figure 3A). Our culture platform relies in part on self-guided differentiation and patterning, and similar to previously reported inner ear organoid protocols 8,9,15,16, our optimized basal control (CTRL) culture system consistently generates hair cells with a vestibular phenotype. We hypothesized that, as in embryonic development, signals exogenous to the otocyst may be necessary for cochlear induction. Sonic Hedgehog (SHH), a signaling molecule secreted from the floor plate of the neural tube and underlying notochord, is essential for patterning of the ventral otic vesicle. SHH is necessary to ventralize the otocyst and derive cochlear structures in mice and chicks 17,18. In contrast, the WNT signaling pathway has been shown to play a role in induction of dorsal gene expression during inner ear development 19,20.
Figure 3. PUR+IWP2 treatment promotes ventralization of otic progenitors in human inner ear organoids.
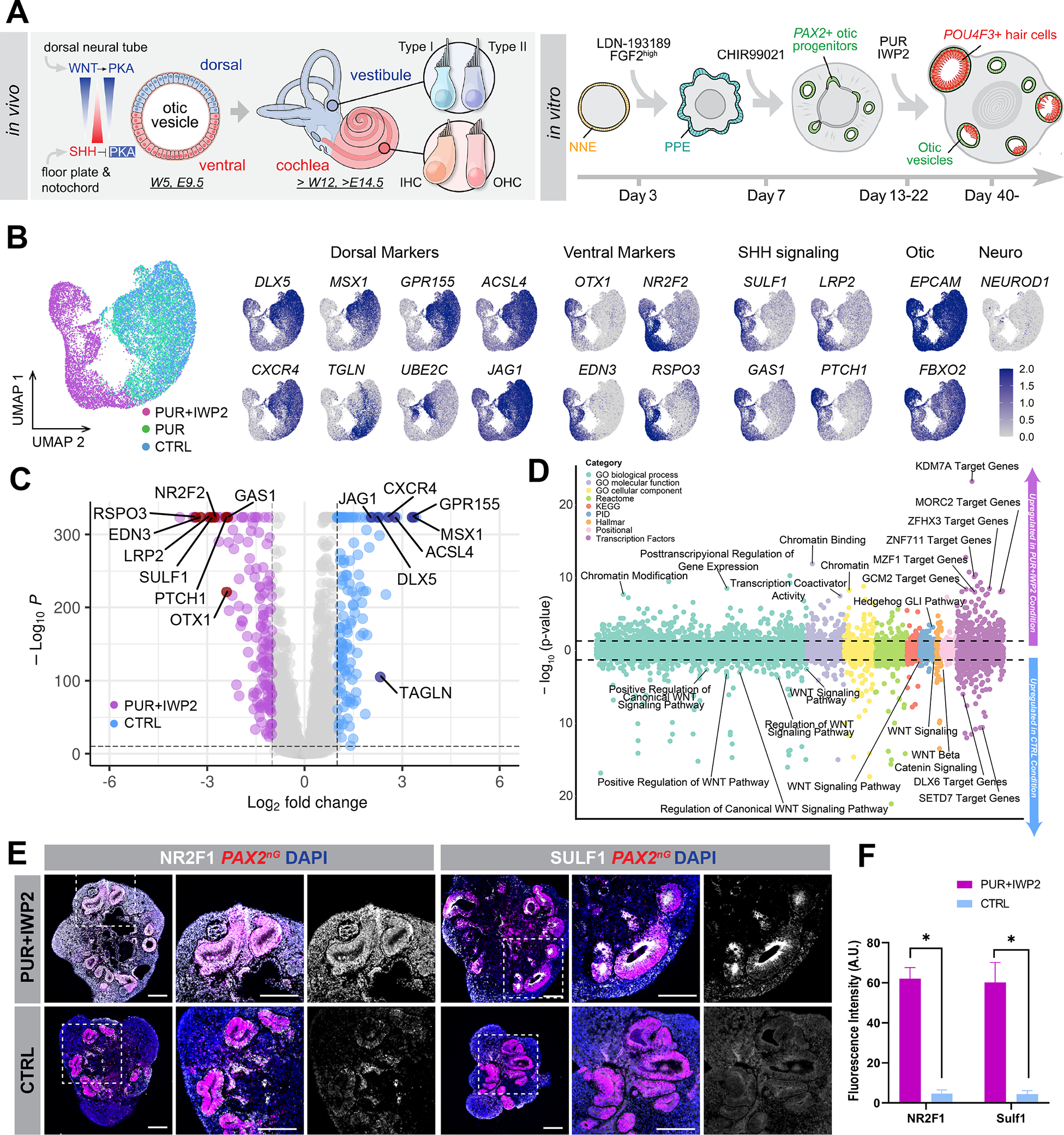
(A) Schematic illustration of known ventralization and dorsalization signals during mouse inner ear development and application of this principle to the human inner ear organoid system. PKA: protein kinase A, W: human gestational week, E: mouse embryonic day.
(B-D) D20 scRNA-seq analysis of FACS-sorted PAX2nG+ otic progenitors in human inner ear organoids. UMAP projections of otic progenitors from PUR+IWP2, PUR and CTRL samples (B). Feature plots demonstrate that dorsal otic markers are predominantly expressed in PUR and CTRL otic progenitors, while ventral otic markers and SHH signaling components are confined largely to PUR+IWP2 cells. Consistent with this, the volcano plot (C) shows differentially expressed dorsal and ventral otic marker genes between PUR+IWP2 and CTRL otic progenitors. Gene-set enrichment analysis of genes upregulated in the PUR+IWP2 and CTRL otic progenitors (above and below 0 in the bubble plot, respectively) (D) shows genes associated with posttranscriptional regulation of gene expression, chromatin modifications and Hedge Hog signaling are enriched in ventralized otic progenitors in inner ear organoids.
(E) Representative images of D25 samples show notably higher expression of NR2F1 and SUlF1 in PUR+IWP2 organoids vs. CTRL organoids.
(F) Quantitative analysis of vesicles co-expressing PAX2 and NR2F1 (or SULF1) in PUR+IWP2 vs. CTRL organoids; n = 8 biological samples from separate experiments per group; Welch’s two-sided t-test *P=0.0013 (NR2F1), *P=0.0089 (SULF1); values are mean ± SEM.
Scale bars, 200 μm.
Based on these previous mouse genetics studies, we hypothesized that timed activation of the SHH pathway, with concomitant suppression of WNT signaling may promote upregulation of ventral otic genes, leading to generation of cochlear hair cells in long-term culture (Figure 3A). To examine this hypothesis, we cultured hESC-derived organoids containing otic progenitors in the presence of the small molecule SHH agonist purmorphamine (PUR) alone, in combinations with the WNT inhibitor IWP2 (PUR+IWP2), or neither (CTRL) (Figure 3A). All conditions followed our optimized differentiation protocol to obtain otic progenitors, and differed only in the presence or absence of the noted small molecules.
We performed single-cell RNA-sequencing (scRNA-seq) analysis on D20 PAX2-nGFP+ sorted cells from aggregates generated under CTRL, PUR or PUR+IWP2 conditions (Figure S2A–E). Each condition was analysed separately, one batch per condition. A total of 37,073 cells were collected, and the cells from each batch were merged and clustered together. Unbiased clustering, as implemented in Seurat v3.2, revealed that EPCAM+/FBXO2+ otic progenitors from the PUR+IWP2 condition formed a distinct cluster, whereas PUR and CTRL otic progenitors clustered together (Figure 3B). Otic progenitors from all three conditions were isolated, and subsequent differential expression analysis among conditions revealed that dorsal otic markers such as DLX5, MSX1, GPR166, and ACSL4, were largely confined to PUR and CTRL treated cells, whereas ventral otic markers, including OTX1/2, NR2F1/2, EDN3, and RSPO3 were concentrated in the PUR+IWP2 condition. Moreover, genes involved in the SHH-pathway, such as SULF1, LRP2, GAS1, and PTCH1 were highly expressed in the PUR+IWP2-treated cells, but their expression was attenuated in CTRL and PUR-treated progenitors (Figure 3C). Consistent with this, Gene Set Enrichment Analysis (GSEA) 21 revealed enrichment of gene sets associated with the Hedgehog pathway and downregulation of gene sets associated with the canonical WNT pathway in PUR+IWP2 treated otic progenitors when compared to untreated CTRL otic progenitors. Additionally, analysis of PUR+IWP2 treated otic progenitors revealed multiple enriched gene sets for posttranscriptional regulation of gene expression, chromatin modifications, and gene sets composed of the targets of known hereditary deafness genes coding for transcription factors such as ZNF711, MORC2, GCM2 and BARHL1 (Figure 3D). These finding are suggestive of a developmental trajectory that is divergent from CTRL and PUR conditions, with enrichment for genetic programs linked to establishment and maintenance of hearing. We substantiated the scRNA-seq data with immunofluorescence for NR2F1, SULF1, OTX2 and DLX5 using D25 organoids (Figures 3E and S2F–G). We concluded that a more efficient ventralization of hESC-derived otic progenitors after sequential treatment with PUR and IWP2 is due to, at least partially, modulation of SHH down-stream gene targets. Since protein kinase A (PKA) has been shown to act as a negative SHH mediator 18, we examined effects of the small molecule H89, a cell-permeable PKA inhibitor, alone or in combination with PUR and IWP2, but none of these treatments were efficacious in deriving hair cells (Figure S3).
Otic progenitors in ventralized inner ear organoids give rise to cochlear hair cells
PUR+IWP2-treated and CTRL aggregates were grown in defined culture medium devoid of exogenous signalling molecules or growth factors from D22 onward. To compare the transcriptional profile between PUR+IWP2-treated and CTRL hair cells, D80 and −109 POU4F3+ sorted cells were analysed via scRNA-seq (Figures 4, S4). We employed a non-stringent FACS gating strategy to collect high and low POU4F3-expressing cells, which resulted in recovering sequence data from 3,332 hair cells (18.9% of total 17,668 cells) and 4,582 hair cells (28.6% of total 16,044 cells) for D80 and −109 samples, respectively. Similar to D20 scRNA-seq data, PUR+IWP2 hair cells separated out from CTRL hair cells when the cells were subjected to unbiased clustering (Figures 4B–E, S4A, C, E). The volcano plot showing differentially expressed genes between the two conditions identified several known cochlear hair cell markers 22–25, including GATA3, INSM1, HES6, TMPRSS3 and GNG8, whose expression was significantly higher in PUR+IWP2-treated vs. CTRL hair cells at D109 (Figures 5A–B). Consistent with our scRNA-seq data, immunofluorescence of PUR+IWP2-treated organoids showed that POU4F3+ cells express significantly higher levels of cochlear hair cell marker proteins, such as NR2F1 and GATA3 (Figures 5C–D; Video S2), which was confirmed in hair cells of fetal human cochleae at gestation week (GW) 18 (Figure 5E).
Figure 4. Differential gene expression in POU4F3nT+ cells of D109 ventralized vs. control inner ear organoids.
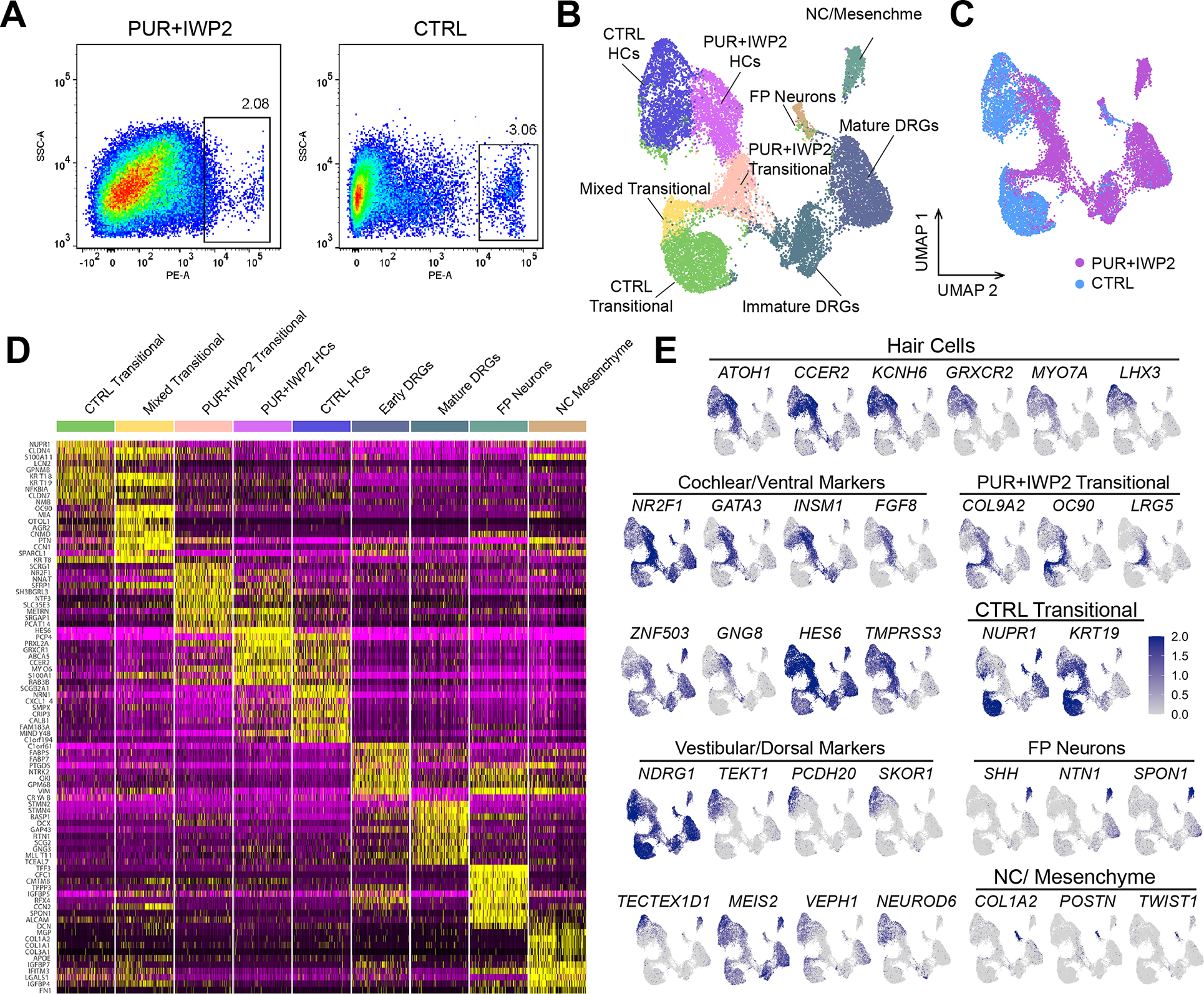
(A) FACS gating strategy used to isolate POU4F3nT+ cells from dissociated D109 inner ear organoids in PUR+IWP2 and CTRL conditions.
(B-C) UMAP plot of POU4F3nT+ cells isolated from D109 PUR+IWP2 and CTRL inner ear organoids (B) and UMAP plot grouped by experimental conditions (C). The number of sequenced cells: 7,968 (CTRL), 11,107 (PUR+IWP2). The number of cells after QC filtering: 5,971 (CTRL), 10,073 (PUR-IWP2).
(D) Heat map showing gene expression across clusters.
(E) Feature plots showing the distributions of marker genes across the cluster map and those showing differential expression of known cochlear and vestibular marker genes in annotated hair cell populations.
DRG; dorsal root ganglion, FP; floor plate, NC; neural crest.
Figure 5. POU4F3nT+ cells in ventralized inner ear organoids express cochlear hair cell markers.
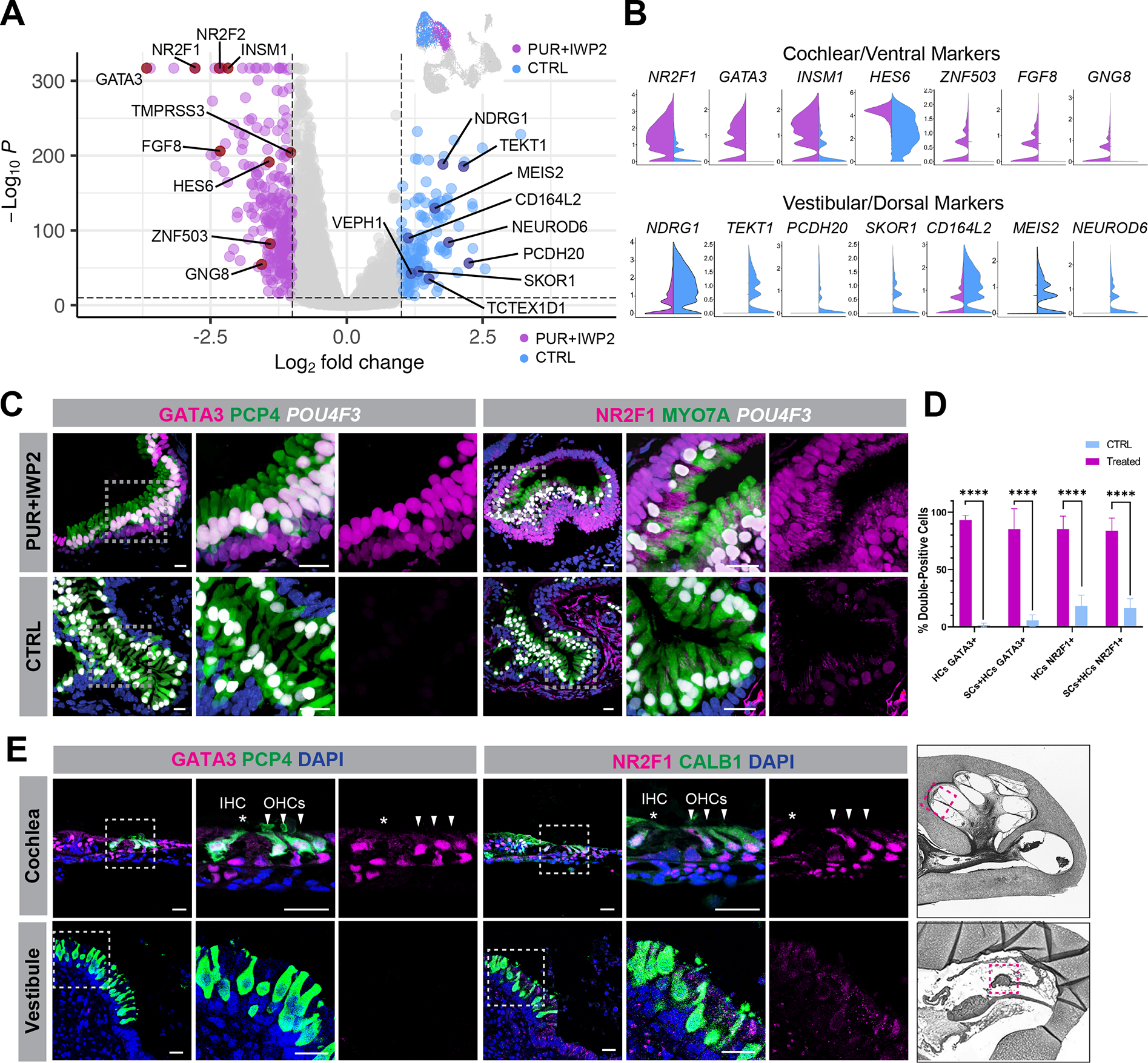
(A-B) Volcano plot (A) together with violin plots (B) confirm differentially expressed cochlear and vestibular hair cell marker genes between PUR+IWP2 and CTRL hair cells. Additionally, previously unrecognized genes, such as NR2F1, TMPRSS3, CD164L2, ZBBX and SKOR1, are differentially expressed between PUR+IWP2 and CTRL hair cells.
(C) Representative immunohistochemistry validates differential expression of NR2F1 and GATA3 between PUR+IWP2 and CTRL organoids.
(D) Comparison of the percentage of NR2F1- or GATA3-positive hair cells (HCs: POU4F3nT-labeled) and supporting cells (SCs: SOX2-labeled) in PUR+IWP2 vs. CTRL immunofluorescent images of inner ear organoids; n = 9 biological samples from separate experiments; Welch’s two-sided t-test ****P<0.000001; values are mean ± SEM.
(E) NR2F1 and GATA3 are expressed in cochlear hair cells, but absent in vestibular hair cells of the crista ampulla in the GW18 human inner ear.
Scale bars, 20 μm (C, E).
We used scanning electron microscopy to compare the structural properties of hair bundles on the apical surface of derived hair cells between PUR+IWP2 and CTRL organoids (Figures 6A–H’). Structural development of the hair bundle - from the emergence of a kinocilium in the middle of the cuticular plate to development of a stair-cased pattern of stereocilia 26, typically seen in the mouse inner ear, was observed in organoid hair cells (Figures S5A–C). We also detected tip-link-like structures between individual stereocilia (Figure S5C’–C”). These transitional properties recapitulate development of the hair bundle in mouse cochleae. In CTRL organoids at D110–200, we consistently observed long pointed arrangement of hair bundles, characteristic of vestibular hair cells. In striking contrast, the length of hair bundles was significantly shorter in PUR+IWP2-treated samples and stereocilia were often arranged in a linear or concave geometry, reminiscent of stereocilia on the apical surface of inner hair cells in the mouse cochlea (Figures 6A–H and S5H–I). High-resolution confocal microscopy substantiated these results by showing differences in the arrangement and overall length of stereocilia between PUR+IWP2-treated and untreated CTRL hair cells (Figures 6I–L and S5E–F, L–M). Native cochlear hair bundles feature narrow stereocilia in the front rows with larger diameters in the middle and back rows, while vestibular stereocilia are more uniform. PUR+IWP2-treated hair bundles also featured a gradient of stereocilia diameters, while CTRL stereocilia were uniform along their varying lengths s (Figures 6M, S5D–D’, J–K). These structural differences in the hair bundle between PUR+IWP2 and control hair cells are consistent with structural differences observed between cochlear and vestibular hair cells in the mammalian inner ear. TUJ1+ neurite processes infiltrating sensory epithelia and contacting the basolateral region of hair cells were observed in both PUR+IWP2 and control organoids (Figures S5G, N), suggesting development of neural innervation in these organoids.
Figure 6. Hair cells derived from ventralized inner ear organoids exhibit structural properties of cochlear hair cells.
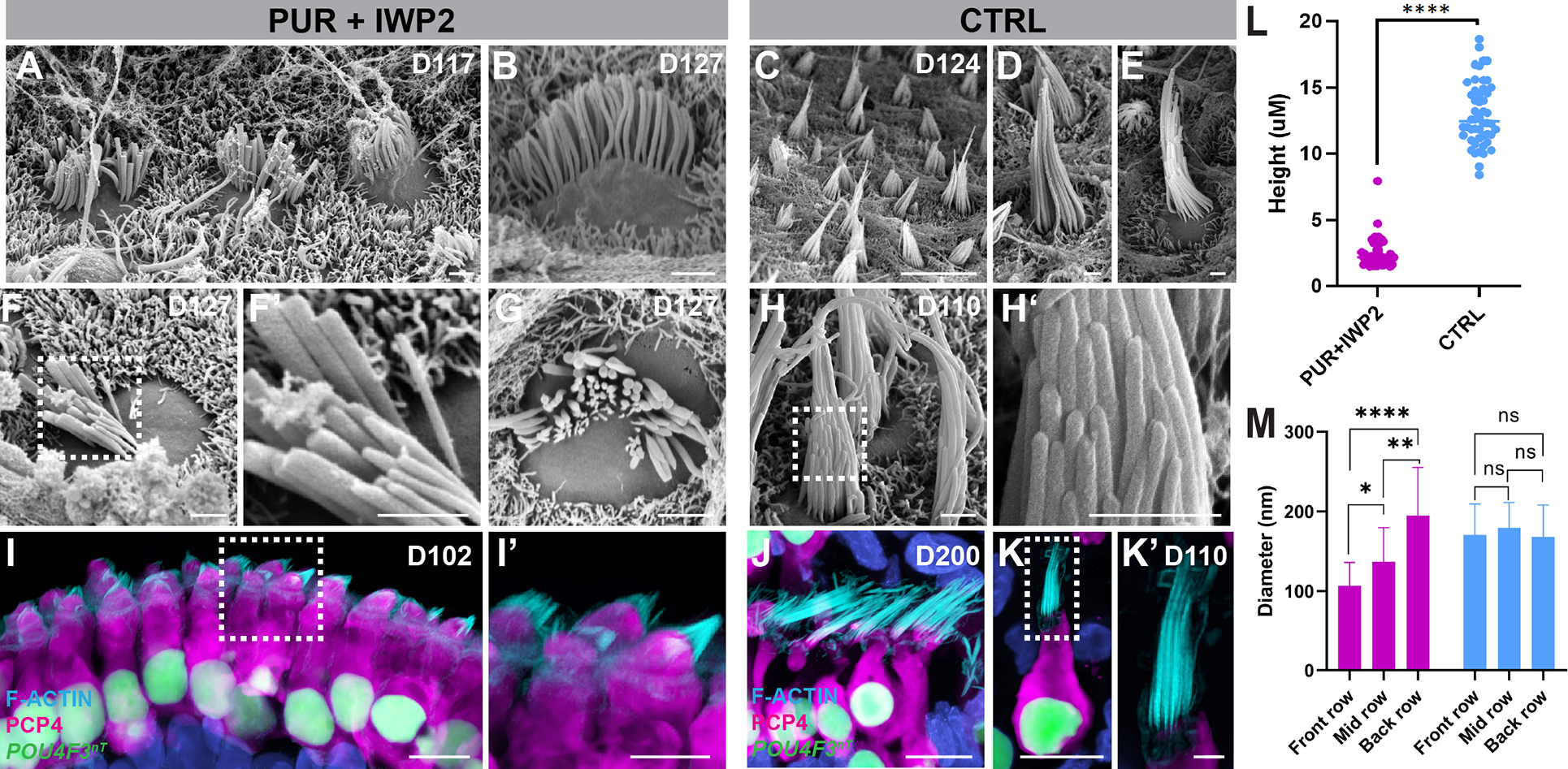
(A-H’) Scanning electron micrographs of PUR+IWP2 hair bundles (A-B, F-G) reveal relatively short stereocilia organized into concave rows of increasing height and diameter characteristic of a cochlear hair cell phenotype. In contrast, scanning electron micrographs of CTRL hair bundles (C-E, H-H’) reveal elongated stereocilia organized into convex rows that are of equivalent diameter characteristic of native vestibular hair cells. i-K’, Confocal microscopic images of PUR+IWP2-treated hair cells (I-I’) reveal short F-actin+ hair bundles and rectangular soma with basally-positioned nuclei, whereas those of CTRL hair cells.
(J-K’) reveal elongated F-actin+ hair bundles and an often-bulbous or flask-shaped soma. CTRL hair cells retain vestibular morphology even at D200 (J).
(L-M) Quantitative analysis of the stereocilia height and the diameter of individual stereocilia in PUR+IWP2 vs. CTRL hair cells; n = 50 biological samples from separate experiments: L, Welch’s two-sided t-test; M, One-way ANOVA with Tukey’ ****P<0.00001; values are mean ± SEM.
Scale bars, 10 μm (C, I, J, K), 1 μm (A, B, D-H, K’).
We tested the reproducibility of our new protocol using a human induced pluripotent stem cell (iPSC) line (mND2–0). As previously described8, this line required a higher BMP4 concentration (2 ng/mL vs 1 ng/mL for the WA25 hESC line) to consistently produce epithelial vesicles (Figure S6A) due probably to the variability in endogenous BMP concentrations inherent to different pluripotent stem cell lines. Treating iPSC-derived aggregates with PUR and IWP2 resulted in abolished DLX5 expression and increased NR2F1 expression in PAX2+/PAX8+ vesicular cells in organoids at D25 (Figure S6B). At D82, all PCP4+ cells (presumably hair cells) and the surrounding cells (presumably supporting cells) expressed GATA3 in PUR+IWP2 samples, which was in striking contrast with a total lack of GATA3+ cells in PCP4+ vesicles of CTRL samples (Figure S6C). Similarly, NR2F1 was detected in all PCP4+ and surrounding cells in PUR+IWP2 organoids, but was undetectable in those in CTRL organoids. Confocal microscopy of D113 PUR+IWP2 hair cells uniformly exhibited short hair bundles (Figure S6D), consistent with our results obtained from hESC-derived organoids.
To assess the identity of PUR+IWP2-treated hair cells, we examined expression of PRESTIN, a hallmark of cochlear outer hair cells 27,28. Membrane-localized PRESTIN was detectible in some samples as early as D102 (0.33% of hair cells), and expression becomes more widespread as cultures age. On D150 and 200, 12.6% and 16.8% of all PUR+IWP2-treated hair cells, respectively, express membrane-localized PRESTIN (Figures 7A–B). In contrast, PRESTIN was undetectable in any CTRL hair cells, up to D200 (Figure 7A). To assess functional development of PUR+IWP2-derived hair cells, we performed conventional whole cell patch clamp recordings of the voltage-activated ion conductances in the cells exhibiting strong tdTomato reporter signal (Figure 7C). Two functionally distinct types of cells were identified with K+-based intracellular solution. The cells of the first group (type A) were observed more often (78%) and were characterized by fast outward currents and virtually no inward currents, similar to the mature outer cochlear hair cells 29. Less frequent (22%) cells (type B) exhibited slow outward currents and prominent fast inward currents, reminiscent of K+ and Na+ currents in immature inner hair cells 29. Both types of cells had similar reversal potential (mean = −34.7 mV, range from −17.5 to −55.5 mV) and showed no evidence of KCNQ-type fast K+ current activated at resting potential 30.
Figure 7. A subpopulation of hair cells derived from ventralized organoids express PRESTIN and exhibit voltage-gated currents characteristic of cochlear outer hair cells.
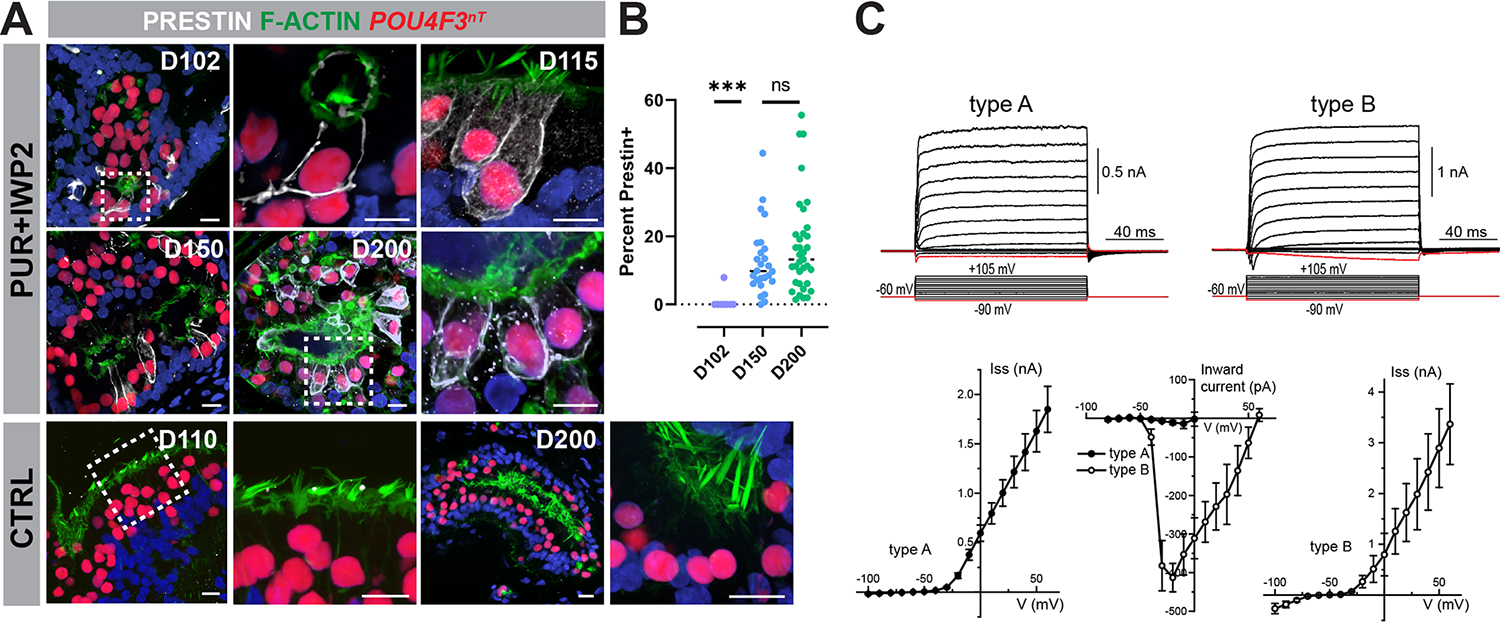
(A-B) PRESTIN+ hair cells increase over time in PUR+IWP2 inner ear organoids. Representative immunohistochemistry for PRESTIN in PUR+IWP2 samples at D102, −150, and −200, along with the comparison of the percentage of hair cells expressing PRESTIN among the different age groups, reveals an increasing number of hair cells expressing membranous PRESTIN over time in culture. In contrast, PRESTIN is undetectable in CTRL hair cells at D110 or −200.
(C) Voltage-gated currents in hESC-derived hair cells. Typical whole cell current responses (top traces) to the voltage step protocol (bottom traces) in type A and type B cells. Average steady-state current amplitude at the end of voltage step in type A and type B cells. The peak amplitude of the negative inward current. All data are shown as Mean ± Standard Error. Age of the cells: d138-d164.
Scale bars,10 μm (A).
To determine the identity of other cell populations arising in cochlear organoids, we performed scRNA-seq analysis of EPCAM+ and EPCAM− cell populations isolated from D81 PUR+IWP2 organoids. This approach increased the percentage of EPCAM+ cells, including hair cells and supporting cells, of all sampled cells for sequencing, which allowed for high-resolution analysis and clustering of otic epithelial cells. The merged dataset identified several clusters, including hair cell, supporting cell, otic epithelium, neuron and Schwann cell clusters (Figure S7A–D). The majority of cells in the hair cell, supporting cell and otic epithelial cell clusters were EPCAM-positive, while the other clusters contained both EPCAM-positive and -negative cells. Subclustering of the supporting cell cluster identified 2 cell populations with one representing BRICD5+/SPARCL1+ supporting cells and another cell population expressing the stria vascularis markers CLDN11, LRP2, DACH1 and PTGDS 31,32 (Figure S7E). Immunofluorescence for CLDN11 revealed its expression in epithelial regions of vesicles containing POU4F3+ hair cells (Figure S7F). The endolymphatic duct marker LMX1A 33,34 was absent in PUR+IWP2 organoids, but present in POU4F3− ductal structures of CTRL organoids (Figure S7G–G’).
DISCUSSION
Existing protocols for inner ear organoids yield exclusively hair cells with structural, molecular, and physiological properties resembling those of vestibular hair cells. Thus, the goal of the present study was to develop a means to derive cochlear hair cell types in organoids. Since a gradient SHH concentration across the dorso-ventral axis of otic vesicles is considered a primary ventralizing cue during inner ear development, we first examined the effects of the potent smoothened agonist PUR on changes in gene expression in D20 organoids, however augmentation of the SHH pathway alone was not sufficient to promote ventral otic differentiation. In parallel, we co-treated hESC-derived aggregates with the WNT inhibitor IWP2 beginning five days into treatment with PUR. This treatment resulted in a significant upregulation of ventral otic markers, while suppressing dorsal otic markers. We also observed significant upregulation of SHH down-stream effectors, such as SULF1, LRP2, GAS1 and PTCH1, in PUR+IWP2-treated samples, but not in CTRL or PUR-treated samples, suggesting that ventralization of otic progenitors requires a threshold of SHH pathway activation that includes upregulation of ligand receptors and down-stream effectors that are antagonized by WNT signaling.
Our scRNA-seq analyses of D80 and −109 samples revealed that PUR+IWP2 treated and untreated control hair cells exhibit distinctive transcriptional profiles. Of the differentially expressed genes between these two cell populations, NR2F1 and GATA3 were among the most highly expressed in PUR+IWP2 treated hair cells at both D80 and −109, suggesting these two genes as potential candidates for core elements in the transcriptional pathway leading to cochlear differentiation. GATA3 has been shown to be predominantly expressed in cochlear vs. vestibular tissues 35–37, but less is known about the role for the orphan nuclear receptors NR2F1/2 in cochlear specification. Targeted inactivation of NR2F1 results in supernumerary hair cells and supporting cells in the developing mouse cochlea, which is associated with dysregulation of Notch signaling components 38. NR2F1/2 are thought to function as ligand-dependent transcription factors, and as such might act as master drivers to confer multipotent otic progenitors with competence to differentiate into cochlear cell types. It is worth noting, however, that NR2F1/2 have been shown to play an essential role in dorso-ventral patterning of the optic vesicle through direct regulation of OTX2 expression 39. We consistently observed that the first sign of POU4F3+ cells in PUR+IWP2 treated organoids approximately 10 days later than that in control organoids. The delayed emergence of cochlear hair cells compared to the timing of vestibular hair cell emergence in organoids is consistent with the temporal order of hair cell differentiation in the mouse inner ear 40,41. This suggests that vestibular and cochlear tissue differentiation is temporally controlled by yet-to-be defined mechanisms and that GATA3 and/or NR2F1/2 could be involved in temporal control of cochlear differentiation.
Our present results showing the differential expression of the stria vascularis marker CLDN11 in PUR+IWP2 organoids and the endolymphatic duct marker LMX1A in CTRL organoids suggest that nonsensory cell types in the cochlear or vestibular epithelia can be selectively derived by our PUR+IWP2 or CTRL organoid protocol. The stria vascularis, a vascularized tissue located in the lateral wall of the cochlea, acts to maintain cochlear homeostasis through CLDN11-dependent basal cell tight junctions and is essential for hearing 31,42,43. Thus, establishing human cochlear epithelia containing not only hair cells and supporting cells, but also stria vascularis cells, would have a major impact on hearing research in the future, as they could serve as a more faithful model system to recapitulate development of sensory transduction in the cochlea. However, conformation of the presence of the stria vascularis in POU4F3+ vesicles of PUR+IWP2 organoids awaits further investigation on cellular composition and functional assessment. The presence of LMX1A in ductal structures of CTRL organoids and its absence in PUR+IWP2 organoids is consistent with the notion that the endolymphatic duct arises from the dorsal part of the otic vesicle during development 44,45.
We sought to further classify derived hair cells in ventralized inner ear organoids as either outer or inner cochlear hair cells. Structurally, these hair cells exhibit U-shaped hair bundles with short stereocilia bearing varying diameters, as typically seen in inner hair cells of the mammalian cochlea 46,47. However, INSM1, a Zinc finger transcription factor essential for outer hair cell differentiation in the mouse 48, is predominantly expressed in hair cells of PUR+IWP2 treated organoids, while expression of TBX2, a pioneering factor for inner hair cell differentiation 49 is not significantly different between PUR+IWP2 and control hair cells. Additionally, our electrophysiological recordings identified two distinctive hair cell populations in D138–164 PUR+IWP2 organoids. Approximately 78% of these hair cells were characterized by fast outward currents and virtually no inward currents, which is similar to native outer but not inner hair cells in the mouse cochlea. Although this proportion is much higher than the 1.6% of hair cells that we detect expressing the outer hair cell marker SLC26A5 (PRESTIN) in our scRNA-seq dataset at D109, immunofluorescence data suggests that the number of PRESTIN+ hair cells increase dramatically over time in PUR+IWP2 organoids from D102 to 200 (from 0.33% to 16.8%), which may explain the higher percentage of outer hair cell-like cells in our patch clamp recordings at D138-D164. This also indicates that derived hair cells may not have reached phenotypic maturation when the scRNA-seq experiments were performed. While we found multiple hallmarks of the outer hair cell identity, our data pointing to the inner hair cell identity are limited. Investigation is underway to determine if bona fide inner hair cells arise concomitantly with outer hair cells in these organoids.
Based on our transcriptional and structural analysis, the time course of hESC-derived cochlear organoid development closely parallels that of human cochlear development, with D110 inner ear organoids corresponding approximately to human fetal inner ears at GW18. Moreover, the order of marker expression is also faithfully recapitulated in inner ear organoids with ATOH1 expression preceding MYO7A expression. PRESTIN is detected in outer hair cells of the human fetal cochlea at GW18, but absent at GW 13. Likewise, PRESTIN is detected in approximately 16% of hair cells in PUR+IWP2 treated organoids at D200, but is barely detectable at D102. These results suggest that the cochlear organoids we have established in the present study can reach a developmental stage comparable to the human cochlea well into the third trimester.
In conclusion, we have demonstrated that precisely timed modulations of the SHH and WNT pathways confer multipotent otic progenitors with a ventral otic phenotype, some of which subsequently give rise to hair cells with structural, transcriptional and functional properties of the two types of cochlear hair cells, inner and outer hair cells and mature in vitro well into a stage corresponding the third trimester of human fetal development. Additionally, scRNA-seq analysis identifies NR2F1 as a previously unrecognized candidate for the key transcriptional pathway essential for cochlear and vestibular diversification. Further investigation is required to elucidate the mechanisms underlying the cross talk between the transcriptional pathways and structural development, and establish a means to control inner vs. outer hair cell generation. The cochlear organoids we have established in this study are expected to serve as a powerful human model to investigate the biology of human cochlear development, elucidate pathogenesis of hereditary hearing loss, and identify therapeutic targets to treat profound hearing loss.
Limitations of the Study
While the cochlear organoid system we have established in the present study is a valuable tool for studying human cochlear development, it lacks patterning pertinent to the snail-shaped cochlear organ. Moreover, the current system contains sensory epithelia and connecting neurons, but lacks entirely central nervous system components. Future studies overcoming these limitations are needed to develop a next-generation model system that recapitulates aspects of patterning and regional specificity as well as auditory neural circuitry.
STAR methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eri Hashino (ehashino@iu.edu)
Material Availability
Research reagents generated in this study will be distributed upon request to other investigators under MTA.
Data and code availability
Single-cell RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CALB1 | SWANT | Cat#300 |
| CLDN11 | Invitrogen | Cat#36-4500 |
| DLX3 | Santa Cruz | Cat#sc-514094 |
| DLX5 | Novus Biologicals | Cat#NBP1-85793 |
| FBXO2 | Santa Cruz | Cat#sc-398111 |
| GATA3 | R&D Systems | Cat#AF2605 |
| GFP | Thermo Fisher | Cat#A-11120 |
| INSM1 | Santa Cruz | Cat#sc-271408 |
| LMOD3 | Proteintech | Cat#14948-1-AP |
| LMX1A | Sigma-Aldrich | Cat#AB10533 |
| MYO7A | Proteus | Cat#256790 |
| MYO7A | Santa Cruz | Cat#sc-74516 |
| NDGR1 | Abcam | Cat#ab-124689 |
| NR2F1 | Invitrogen | Cat#ab-124689 |
| NR2F1 | R&D Systems | Cat#PP-H8132-00 |
| OCT4 (conjugated to AF488) | eBioscience | Cat#53584182 |
| OTX2 | R&D Systems | Cat#AF1979 |
| PAX2 | Abnova | Cat#H00005076-M01 |
| PAX8 | Abcam | Cat#AB97477 |
| PCP4 | Santa Cruz | Cat#sc-74816 |
| PCP4 | Invitrogen | Cat#PA5-52209 |
| phalloidin (conjugated to AF488) | Thermo Fisher | Cat#A12379 |
| POU4F3 | Santa Cruz | Cat#sc-81980 |
| Prestin | Generous gift of Drs. Jing Zheng and Kazuaki Homma et al.28 | NW-802 |
| Sox10 | eBioscience | Cat#14-5923-82 |
| Sox2 | BD Pharmingen | Cat#561469 |
| SSEA4 (conjugated to AF594) | BioLegend | Cat#330414 |
| SULF1 | Invitrogen | Cat#PA5-113112 |
| TUBA4A | Sigma-Aldrich | Cat#T6793 |
| TUJ1 (TUBB3) | Biolegend | Cat#801201 |
| Bacterial and virus strains | ||
| DH5a | New England Biolabs | Cat#C2987H |
| Biological samples | ||
| GW 13 Human fetal cochlea | This paper | N/A |
| GW 18 Human fetal cochlea | This paper | N/A |
| GW 18 Human fetal vestibule | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Accutase | Thermo Fisher Scientific | Cat# A1110501 |
| Advanced DMEM/F12 | Thermo Fisher Scientific | Cat#12634028 |
| Alt-R™ S.p. Cas9 Nuclease V3 | Integrated DNA technologies (IDT) | Cat#1081058 |
| B27 (minus vitamin A) | Thermo Fisher Scientific | Cat#12587010 |
| β-Mercaptoethanol | Thermo Fisher Scientific | Cat#21985023 |
| BMP4 | Reprocell | Cat#03-0007 |
| CHIR99021 | Reprocell | Cat#04-0004-10 |
| DMEM/F12 +HEPES | Thermo Fisher Scientific | Cat#11320033 |
| DPBS | Thermo Fisher Scientific | Cat#14040133 |
| Essential 6 (E6) medium | Thermo Fisher Scientific | Cat#A1516401 |
| Essential 8 (E8) Flex medium | Thermo Fisher Scientific | Cat#A2858501 |
| 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat#15575020 |
| FGF2 | Stem Cell Technologies | Cat#780003 |
| GFR Matrigel | Corning | Cat#354230 |
| GlutaMAX | Thermo Fisher Scientific | Cat#35050061 |
| Glutaldehyde | Electron Microscopy Sciences (EMS) | Cat#16537-15 |
| IWP-2 | Tocris | Cat#3533 |
| LDN193189 | Stemgent | Cat#04-0074-02 |
| L-15 cell culture medium | Thermo Fisher Scientific | Cat#21083027 |
| Neurobasal medium | Thermo Fisher Scientific | Cat#21103049 |
| Normocin | Invivogen | Cat# ant-nr-1 |
| N2 | Thermo Fisher Scientific | Cat#17502048 |
| Osmium tetroxide | Electron Microscopy Sciences (EMS) | Cat#19150 |
| Propidium Iodide | Invitrogen | Cat#P1304MP |
| Purmorphamine | Stemgent | Cat#04-0009) |
| RevitaCell | Thermo Fisher Scientific | Cat#A2644501 |
| SB431542 | Stemgent | Cat#04-0010-05 |
| Scr7 | Xcessbio | Cat#M60082-2S |
| TRIzol | Thermo Fisher Scientific | Cat#15596026 |
| TrypLE | Thermo Fisher Scientific | Cat#A1217701 |
| Vitronectin-N | Thermo Fisher Scientific | Cat#A14700 |
| Y-27632 | Stemgent | Cat#04-0012-02 |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell Kit v3 | 10x Genomics | Cat#1000075 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE200629 |
| Experimental models: Cell lines | ||
| WA25 | WiCell | (NIHhESC-12-0196; RRID:CVCL_E080) |
| mND2-0 | WiCell | mND2-0 (MIRJT7i-mND2-0,RRID:CVCL_U173) |
| Oligonucleotides | ||
| PAX2 RT assay Fwd: CCTGGCAGGAATGGTGCCT | This paper | N/A |
| PAX2 RT assay Rev: GTCAGACGGGGACGATGTGG | This paper | N/A |
| Primers for PAX2 donor vector: see supplementary table 2 | This paper | N/A |
| Primers for POU4F3 donor vector: see supplementary table 2 | This paper | N/A |
| PAX2 gRNA: ATGACCGCCACTAGTTACCG | This paper | N/A |
| POU4F3-targeting sgRNA for CRISPR/Cas9 genome editing: ATTCGGCTGTCCACTGATTG | This paper | N/A |
| Recombinant DNA | ||
| SpCas9-HF1 vector | Kleinstiver et al.14 | Addgene#72247 |
| U6 expression vector | Slaymaker et al.55 | Addgene #71814 |
| Cre expression vector | Matsuda et al.57 | Addgene #13775 |
| FLPo expression vector | Raymond et al.58 | Addgene #13793 |
| 2A-eGFP-PGK-Puro vector | Hockemeyer et al.54 | Addgene #31938 |
| Software and algorithms | ||
| ImageJ | Schneider et al.59 | https://imagej.nih.gov/ij/ |
| Flow Jo | FlowJo LLC | https://www.flowjo.com/ |
| Prism 9 | GraphPad Software | https://graphpad.com |
| Imaris 8 | Bitplane | https://Imaris.oxinst.com/ |
| R | Team R C. R: A language and environment for statistical computing[J]. 2013. | https://cran.r-project.org/bin/macosx/ (macOS)https://cran.r-project.org/bin/windows/base/ (Windows)https://cran.r-project.org/bin/linux/ (Linux) |
| R studio | Team R S. Rstudio: integrated development for R[J]. Rstudio, Inc., Boston, MA URL http://www.Rstudio.com, 2015, 42: 14. | https://www.rstudio.com/products/rstudio/download/ |
| R package Seurat | https://github.com/satijalab/seurat | |
| DESeq2 | Love et al.62 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| zingeR | Love et al.62 | https://github.com/statOmics/zingeR/blob/master/R/methods.R |
| iDEA | Ma et al.21 | https://xzhoulab.github.io/iDEA/ |
| Custom code | This paper | https://github.com/HashinoLab/Moore_et¬_al_Cochlear_Organoids |
| Nikon Elements microscope imaging software | Nikon Corporation | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer |
All original code has been deposited at the GitHub repository and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODELS AND STUDY PARTICIPANT DETAILS
Human cell lines
We used the female human embryonic stem cell line WA25 (NIHhESC-12-0196, RRID:CVCL_E080) obtained from WiCell Research Institute as a parental cell line to generate the PAX2-2A-nGFP/POU4F3-2A-ntdTomato reporter stem cell line used throughout this study. We also used the male human induced pluripotent stem cell line mND2–0 (MIRJT7i-mND2-0,RRID:CVCL_U173) to repeat critical experiments. Cells were maintained and passaged in Essential 8 Flex Medium (Thermo Fisher, A2858501) supplemented with 100 μg/ml Normocin (E8fn) on recombinant human Vitronectin-N (Thermo Fisher, A14700)-coated 6-well plates at 37 C with 5% CO2. Media was replaced every other day and cells were passaged every 5–6 days using 0.5mM EDTA (Thermo Fisher). Cell lines were quality controlled with karyotyping, immunofluorescent labeling of pluripotency markers, and sequencing of predicted off-target regions after CRISPR/Cas9 gene editing.
Human tissues
Human fetal cochleae were collected and processed at the Leiden University Medical Center (Netherland) from tissues obtained from elective abortion using vacuum aspiration. Prior to the procedure, obstetric ultrasonography was performed to determine the gestational weeks (GW) and days. One GW 13 cochlear, one GW18 cochlear and one GW 18 vestibular samples were used in this study. The use of human fetal tissues was approved by the Medical Ethical Committee of the Leiden University Medical Center (Protocol Number 08.087). Informed written consents were obtained in accordance with the WMA Declaration of Helsinki guidelines.
METHOD DETAILS
Establishment of PAX2/POU4F3 reporter hESC lines with CRISPR/Cas9
To monitor the derivation of otic progenitors in 3D culture, we generated PAX2 reporter hESC lines by integrating a 2A-eGFP-nls (2A-nGFP) fluorescence reporter at the endogenous PAX2 locus using the CRISPR/Cas9 genome engineering technology. Based on the locations of the stop codons, PAX2 splicing variants can be grouped as Types PAX2b, PAX2c, and PAX2d 50–52. Total RNA was isolated from D40 inner ear organoids by homogenizing whole aggregates in ice-cold TRIzol (Fisher, cat. no. 15596026) followed by chloroform extraction and isopropanol precipitation. A cDNA library was constructed using iScript cDNA synthesis kit following the manufacturer’s suggested protocol (Biorad, Cat. No. 1708891). Standard PCR was performed on the cDNA library as well as on PAX2b, PAX2c, and PAX2d synthetic cDNA gBlocks (IDT) for size reference using primers 5’CCTGGCAGGAATGGTGCCT3’ and 5’GTCAGACGGGGACGATGTGG3’ targeting a common exon and the 3’UTR of PAX2. Agarose gel electrophoresis of the PCR amplicons followed by Sanger sequencing verification of the extracted brightest band showed that PAX2b was the most abundant type of PAX2 isoforms in human inner ear organoids. Therefore, gRNA and homology arms were designed to target the PAX2b stop codon locus. To construct the PAX2-2A-eGFP-nls-pA-loxP-PGK-Puro-pA-loxP donor vector, two 1 kb homology arms flanking the PAX2b stop codon were PCR amplified from WA25 hESC (WiCell, derived from a female donor) genomic DNA. Gibson assembly 53 was used to connect the two homology arms with 2A-eGFP 8,54, nls-stop-bGH polyA (gBlock, IDT), and loxP-PGK-Puro-pA-loxP DNA fragments 8,54, as well as a linearized pUC19 plasmid backbone into the final donor vector. gRNA (5’- ATGACCGCCACTAGTTACCG −3’) targeting the PAX2b stop codon was cloned into an expression plasmid under the control of a U6 promoter (Addgene #71814) 55. The PAX2-2A-nGFP donor vector, the PAX2b gRNA plasmid, as well as a high fidelity Cas9 expression plasmid (SpCas9-HF1, Addgene #72247) 14 were transfected into WA25 hESCs with 4D Nucleofector (Lonza) using the P3 Primary Cell 4D-Nucleofector X kit and Program CB-150. After nucleofection, cells were plated in E8fn medium containing 1× RevitaCell (Thermo Fisher) for improved cell survival rate, and 1 μM of Scr7 (Xcessbio) for higher HDR efficiency 56. 0.5 μg/mL puromycin selection was performed for 14 days starting from 48 h post-nucleofection. The PGK-Puro sub-cassette flanked by two loxP sites was removed from the genome after puromycin selection by nucleofection of a Cre recombinase expressing vector (Addgene #13775)57. Clonal cell lines were established by low-density seeding (1–3 cells/cm2) of dissociated single hESCs followed by isolation of hESC colonies after 5–7 d of expansion. Genotypes of the clonal cell lines were analysed by PCR amplification of genomic DNA isolated using the QuickExtract DNA Extraction Kit (Fischer Scientific, NC9904870), followed by gel electrophoresis, and by Sanger sequencing of total PCR amplicons or individual PCR amplicons cloned into TOPO vectors (Thermo Fisher). Cell lines with bi-allelic 2A-nGFP integration were used for subsequent experiments. Top 10 predicted off-target sites of the gRNA were PCR amplified (~1 kb) from the genomic DNA of the established cell lines and were Sanger sequenced to test for off-target mutations.
To build a PAX2-2A-nGFP/POU4F3-2A-ntdTomato multiplex reporter cell line, the POU4F3-2A-ntdTomato knockin CRISPR was performed on the PAX2-2A-nGFP genetic background. The POU4F3-2A-tdTomato-nls-bGH polyA-frt-PGK-Puro-pA-frt donor plasmid was constructed by connecting the following DNA fragments via Gibson assembly: Two 1 kb homology arms flanking the POU4F3 stop codon amplified from WA25 genomic DNA, 2A-tdTomato gBlock DNA, nls-stop-bGH pA gBlock DNA, frt-PGK-Puro-frt gBlock DNA (IDT), and a linearized pUC19 backbone. The completed donor plasmid was transfected into the PAX2-2A-nGFP parental hES cell line along with a ribonucleoprotein complex composed of a synthetic sgRNA (5’- ATTCGGCTGTCCACTGATTG - 3’) (Synthego) targeting the POU4F3 stop codon locus, and a high fidelity Cas9 protein (HiFi Cas9 v3, IDT). The transfection was also performed with a 4D Nucleofector (Lonza) using the P3 Primary Cell 4D-Nucleofector X kit and Program CB-150. After nucleofection, cells were plated in E8fn medium containing 1× RevitaCell (Thermo Fisher) and 1 μM of Scr7 (Xcessbio). 0.5 μg/mL puromycin selection was performed for 9 days starting from 48 h post-nucleofection. The frt-flanked PGK-Puro cassette was removed via FLPo transfection (Addgene #13793)58. Clonal cell line isolation and genotyping procedures were the same as PAX2-2A-nGFP CRISPR. The final multiplex cell line chosen for downstream experiments had bi-allelic 2A-nGFP knockin at the PAX2 locus and bi-allelic 2A-ntdTomato knockin at the POU4F3 locus, and was karyotyped by the KaryoLogic Inc. (Research Triangle Park, North Carolina).
Organoid culture
PAX2-2A-nGFP/POU4F3-2A-ntdTomat hESCs (passages 22–50) or mND2–0 human iPSCs (WiCell, derived from a male donor, passages 33–50) were maintained and passaged in Essential 8 Flex Medium (Thermo Fisher, A2858501) supplemented with 100 μg/ml Normocin (E8fn) on recombinant human Vitronectin-N (Thermo Fisher, A14700)-coated 6-well plates. Human inner ear organoids were derived from hESCs based on our previous protocol 8,15 with major modifications. Briefly, hESCs were dissociated with StemPro Accutase (Thermo Fisher, A1110501) and distributed at 3500 cells per well onto low-adhesion 96-well U-bottom plates in 100 μL E8fn containing 20 μM Y-27632 (Stemgent, 04–0012-02) and centrifuged at 120 g for 5 minutes. 100 μL of E8fn was added to each well following 4h of incubation. After 48h of incubation, induction of non-neural epithelium (differentiation day zero), proceeded as follows: aggregates were transferred to a 10 cm dish using a multichannel pipette, the plate was gently swirled to collect aggregates in the center, then samples were transferred to a 2 mL tube using a wide-orifice P1000 pipette. Aggregates were then washed 3 times in DMEM:F12 with HEPES buffer (Thermo Fisher, 11320033) to remove all E8fn, and a further 3 times with new differentiation media made from E6 medium (Thermo Fisher, A1516401) with Normocin (E6n) containing 4 ng mL−1 FGF-2 (StemCell Technologies, 780003), 10 μM SB-431542 (Stemgent, 04–0010-05), and 2% growth factor reduced (GFR) Matrigel (Corning, 354230), and 1–2 ng mL−1 BMP4 (Reprocell, 03–0007), then transferred to new 96-well U-bottom plates in 100 μL of the same new differentiation media. On day 3 of differentiation, an additional 25 μL of E6n containing 250 ng/mL FGF-2, 200 nM LDN-193189 (Stemgent, 04–0074-02) was added to each well (125 μL total media/well). Culture medium was changed completely on days 7 and 9 by washing as before, 3 times with DMEM:F12+HEPES and 3 times with E6n containing 3 μM CHIR 99021 (Reprocell, 04–0004-10), 200 nM LDN, and 50 ng/mL FGF-2, then transferred to new 96-well U-bottom plates in 250 uL of new media. On day 11, aggregates were washed as before, then 10 aggregates per well were transferred to a Nunc Delta surface 6-well culture dish in Organoid Maturation Medium (OMM) containing 1% GFR Matrigel and 3 μM CHIR99021. OMM consists of a 50:50 mixture of Advanced DMEM:F12 (Thermo Fisher, 12634028) and Neurobasal Medium (Thermo Fisher, 21103049) supplemented with 0.5× N2 Supplement (Thermo Fisher, 17502048), 0.5× B27 minus Vitamin A (Thermo Fisher, 12587010), 1× GlutaMAX (Thermo Fisher, 35050061), 0.1 mM β-Mercaptoethanol (Thermo Fisher, 21985023), and Normocin. Until day 13, CTRL and cochlear cultures received identical treatment. Day 13 cultures were split into CTRL or cochlear as follows: CTRL cultures were maintained in OMM + 3 μM CHIR99021 until day 18 with media changes on days 13 and 15 (to refresh media, washes are not necessary as the composition of the media does not change). On day 18, CTRL cultures were washed and transferred to a 10 cm culture dish in OMM (NO CHIR99021) and received twice-weekly media changes using fresh OMM for the remainder of the culture. On day 13 and 15, PUR+IWP2 culture media was changed to OMM + 3 μM CHIR99021 + 1 μM Purmorphamine (Stemgent, 04–0009). On day 18, PUR+IWP2 aggregates were washed to remove CHIR99021, and the media was changed to OMM + 3 μM IWP-2 (Tocris, 3533) +1 μM Purmorphamine. The media was changed on day 20 with fresh identical media. On day 22, PUR+IWP2 cultures were washed and transferred to a 10 cm culture dish in OMM-only, and received twice-weekly media changes using fresh OMM for the remainder of the culture.
Immunohistochemistry
Aggregates were fixed with 4% paraformaldehyde for 30 min at room temperature or at 4 °C overnight. The fixed specimens were cryoprotected with a graded series of sucrose and then embedded in tissue-freezing medium. Frozen tissue blocks were sectioned into 12-μm cryosections on a Leica CM-1860 cryostat. For immunostaining, a 10% goat or horse serum in 0.1% Triton X-100 1× PBS solution was used for blocking, and a 3% goat or horse serum in 0.1–1% Triton X-100 1× PBS solution was used for primary/secondary antibody incubations. Primary antibodies used in this study are listed in Supplementary Table 1. Alexa Fluor conjugated antimouse, rabbit, or goat IgG (Thermo Fisher) were used as secondary antibodies. ProLong Gold Antifade Reagent with DAPI (Thermo Fisher) was used to mount the samples and visualize cellular nuclei.
For whole-mount immunofluorescence, the AbScale tissue-clearing protocol (Hama et al., 2015) was applied to aggregate samples. Incubation and washing steps were performed on a rotor, and incubation steps were performed at 37°C on a rotor unless otherwise noted. After fixation of samples with 4% paraformaldehyde overnight, the samples were incubated for 6 h in Scale S0 solution, followed by incubation in Scale A2 for 16 h, ScaleB4 solution for 24 h, and ScaleA2 solution for 8 h. Thereafter the samples were incubated in 0.1M PBS for 4 h at room temperature, and blocking was performed with 10% normal horse serum (Vector Laboratories) in AbScale solution for 16 h. Incubation with primary antibodies in AbScale solution containing 3% normal horse serum was performed for 48 h, followed by sequential washing with AbScale solution for 15, 30, 60, and 120 min and incubation with fluorophore-conjugated secondary antibodies in AbScale solution containing 3% normal horse serum for 24 h. After rinsing three times with AbScale at RT for 30 min and twice with AbScale Rinse solution at RT for 30 min, the samples were re-fixed with 4% paraformaldehyde at RT for 1 h, washed twice with 0.1M PBS, and incubated in ScaleS4 solution for 16 h. The stained samples were mounted with a small amount of ScaleS4 solution on poly-L-ornithine coated coverslips with silicone gaskets (Fisher Scientific). Imaging of the samples was carried out on a Leica Dive Confocal/Multiphoton Microscope or a Nikon A1R HD25 confocal microscope. Three-dimensional reconstructions were performed using the Imaris 8 software package (Bitplane) and the NIS Elements Advanced Research application (Nikon).
Quantification of immunohistochemistry data
Treated and control samples were processed for immunostaining simultaneously using identical reagents and protocols for each comparison. Images were acquired using either a Nikon A1R-HD25 confocal microscope, or a Leica DMI8 widefield fluorescent microscope using identical image acquisition parameters between conditions. Raw images were analysed using Nikon GI3 suite or exported in TIFF format and analysed using ImageJ59. Co-expression analysis was performed by analysing regions for coincident signal corresponding to a labelled tissue-specific marker and a gene of interest. Mean grey value and total colocalized area were collected. Statistical analysis was performed in GraphPad Prism 9.
Scanning electron microscopy
Aggregates with abundant POU4F3-ntdTomato+ puncta between D81 and 141 of differentiation were fixed with 2.5% glutaraldehyde in sodium cacodylate buffer (Electron Microscopy Sciences) at 4°C overnight. The fixed samples were dissected under a Nikon SMZ18 stereo fluorescence microscope to expose the luminal surface of vesicles containing ntdTomato+ cells and post-fixed with 1% osmium tetroxide (Electron Microscopy Sciences) at RT for 1 h. The samples were then dehydrated through a graded series of ethanol and transferred into a Leica EM CPD300 critical-point dryer. After critical point drying, the samples were mounted on aluminium stubs and sputter coated with a Denton Vacuum Desk V sample preparation system. The samples were viewed on a JEOL JSM-7800F field emission scanning electron microscope at an accelerating voltage of 5 kV.
Representative data and reproducibility
Unless stated otherwise, images are representative of specimens obtained from at least three separate experiments. For immunohistochemical analysis of aggregates, we typically sectioned 6–15 aggregates from each condition in each experiment.
scRNA-seq and raw data processing
PAX2nG+ (D20 samples) or POU4F3nT+ (D80 and −109 samples) cells were isolated and used for scRNA-seq analyses. Thirty to 45 aggregates per condition were washed with 0.5 mM EDTA (Thermo Fisher) in DPBS, then incubated with 1.1 mM EDTA in 1X TrypLE (Life Technologies) in DPBS on an orbital shaker at 37°C for 30 min followed by 40–50 min without shaking. During incubation, the samples were mechanically dissociated on Nunclon Sphera 24-well plates (Thermo Fisher) with shaking and occasional gentle pipetting with P1000 tips. After confirming that most cells, especially those with reporter expression, were completely dissociated, they were filtered sequentially through a 100-μm and a 40-μm cell strainer (Falcon) and then transferred into 2-ml tubes (Eppendorf). After spinning down at 100 × g for 5 min, dissociated cells were resuspended in FACS buffer consisting of 2% fetal bovine serum (Thermo Fisher) in 1X DPBS. Cells were again filtered through a 40-μm cell strainer and collected into a 5-ml round-bottom polypropylene tube (Falcon). Dead cells were stained with propidium iodide (PI, Invitrogen) diluted at 1:500. Simultaneously, size control cells without PI staining were prepared using aggregates made from wild-type hESCs using the above protocol. Cells were stored on ice and protected from light prior to sorting. PI-negative and GFP (or tdTomato)-positive populations were collected and cleaned using a SORP Aria (BD Biosciences).
For collecting EPCAM-positive and -negative cells, dissociated D81 organoid cells were stained with 1:100 PE-conjugated EPCAM antibody (BioLegend) for 40 min prior to FACS sorting. PE-EPCAM+ and PE-EPCAM– populations were collected in separate tubes for separate downstream scNA-seq reactions. Since POU4F3nT+ hair cells also express EPCAM on their cell surfaces, these hair cells exhibit both tdTomato and PE fluorescence signals, both of which are red. As a result, hair cells were FACS-isolated as a sub-population of PE-EPCAM+ cells.
scRNA-seq for all samples was performed using the 10X Genomics Chromium 3’ v3 platform for cDNA library construction and the NovaSeq 6000 system (Illumina) for sequencing. For each sample, between 12,000 and 18,000 cells were added to the single cell master mix, following the Chromium NextGEM Single Cell 3’ Reagent Kits User Guide. The single cell master mix, along with the single-cell gel beads and partitioning oil, was dispensed onto a Single Cell Chip G and the chip was loaded into the Chromium Controller for barcoding and cDNA synthesis. The resulting cDNA library was sequenced with the NovaSeq 6000 system running a custom program for 28-bp plus 91-bp paired-end sequencing, resulting in a read depth of more than 40,000 reads per cell.
Illumina’s CellRanger v4.0.0 program was used to generate BCL files, which were de-mutiplexed and converted to FASTQ files via bcl2fastq Conversion Software (Illumina). The FASTQ files were then aligned to the GRCh38–3.0.0 reference genome using the STAR (Spliced Transcripts Alignment to a Reference) aligner. Mapped reads were grouped by cell barcode, single-cell gene expression was quantified using unique molecular identifiers (UMIs), and the resulting filtered gene-barcode (count) matrices were used as input for downstream analysis.
scRNA-seq data analysis
The filtered count matrices were preprocessed individually for each dataset, removing cells where the number of mitochondrial gene counts exceeded 12.5 percent of the total number of molecules detected. A second preprocessing step removed cells with anomalous housekeeping gene expression: Cells with log-transformed RPL27 expression ± 2 standard deviations away from the mean were removed. After preprocessing, datasets were merged across conditions, resulting in one combined dataset for each timepoint. After merging, the gene expression levels were normalized using the SCTransform function in Seurat 60, which transforms raw count data into Pearson residuals, effectively controlling for technical variation resulting from heterogenous sequencing depth. Over fifty different cluster partitions were generated using Seurat’s unsupervised clustering workflow by varying the values of the resolution and k.param parameters required for clustering and shared nearest neighbor graph construction, respectively. Then, the cluster partition with the highest silhouette index was selected for further analysis 61. Cluster identity was manually determined based on the expression of canonical markers.
Relevant populations (e.g. otic progenitors, hair cells) were isolated on the basis of cluster identity and comparisons between treatment groups were performed using DESeq2 and zingeR 62. Cell-level weights – which are required to adjust for drop-out events endemic to scRNA-sequencing – were calculated with zingeR. Differential expression between treatment groups was determined using DESeq2, and genes were considered statistically significant with a Benjamini-Hochberg P < 0.05. The results of the DESeq2 analysis were passed to iDEA 21, a platform for gene set enrichment analysis, and differentially expressed genes were compared to gene sets taken from MSigDB databases 63. The Gene Transcription Regulation Database (GTRD) was used to define the 573 transcription factor target gene sets our data were compared against 64. While these gene sets are well defined, they do not constitute an exhaustive list of all possible transcription factors.
Electrophysiological analyses
D138–164 hESC-derived organoids were first imaged in bright field and epifluorescent illumination (TE2000-U, Nikon) to determine localization of otic vesicles with tdTomato positive cells. Then, the organoids were sectioned with a diamond knife to expose otic vesicles. The section was placed into the custom-made recording chamber, where it was held by two strands of dental floss. The recordings were performed at room temperature (20–25°C) in Leibovitz’s L-15 cell culture medium (Cat #21083027, Gibco/ThermoFisher, USA) containing the following inorganic salts (in mM): NaCl (137), KCl (5.4), CaCl2 (1.26), MgCl2 (1.0), Na2HPO4 (1.0), KH2PO4 (0.44), MgSO4 (0.81). The cells were viewed with an upright microscope (E600FN, Nikon), equipped with a high numerical aperture (NA) objective (60x, 1.0 NA) and epifluoresent attachment. Only the cells with bright tdTomato signal were chosen for recordings. During recordings, the organoid sections were continuously perfused with L-15 medium. Pipettes for whole-cell patch-clamp recordings were filled with intracellular solution containing (in mM): KCl(12.6), KGlu(131.4), MgCl2(2), EGTA(0.5), K2HPO4 (8), KH2PO4 (2), Mg2-ATP (2), and Na4-GTP(0.2). The solution was adjusted to pH 7.3–7.4 with KOH and to 320 mOsm with D-glucose. The uncompensated pipette resistance was typically 5–8 MOhm when measured in the bath. Whole cell current responses were recorded with a MultiClamp 700B patch clamp amplifier controlled by pClamp software (Molecular Devices, USA).
Human tissue ethics, collection and immunofluorescence
Human fetal cochleae were collected and processed at the Leiden University Medical Center (Netherland) from tissues obtained from elective abortion using vacuum aspiration. Prior to the procedure, obstetric ultrasonography was performed to determine the gestational weeks (GW) and days. One GW 13 cochlear, one GW18 cochlear and one GW 18 vestibular samples were used in this study. The fresh samples were collected in PBS, fixed in 4% paraformaldehyde in PBS overnight at 4°C, decalcified and embedded in paraffin as previously described 65. The paraffin blocks were cut into 5 μm thick sections through the sagittal plane using a Leica rotary microtome. The serial sections were deparaffinized in xylene, rehydrated in a descending ethanol series (100%, 90%, 70%, 50%), and rinsed in distilled water. Thereafter, the sections were treated in 0.01 M sodium citrate buffer (pH 6.0) for 12 minutes at 98 °C using a boiling pot for antigen-retrieval and then processed for immunofluorescence. Primary antibodies used in this study are listed in Supplementary Table 1. The stained samples were viewed and imaged on a Zeiss LSM900 confocal microscope. The use of human fetal tissues was approved by the Medical Ethical Committee of the Leiden University Medical Center (Protocol Number 08.087). Informed written consents were obtained in accordance with the WMA Declaration of Helsinki guidelines.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± SEM and were derived from at least 3 independent experiments unless otherwise indicated. Statistical analysis was carried out in Graphpad Prism 9. Paired analyses were performed using a two-sided Welch’s t-test. Multiple comparisons were performed using one-way ANOVA with Tukey’s multiple comparisons test. For both tests the cutoff for statistical significance was P<0.01. Information regarding replicates (n), definition of center, statistical tests, and significance notation can be found in the figure legends.
Supplementary Material
Video S1. POU4F3nT+ hair cells in SOX2+ vesicles of a D60 control organoid, Related to Figure 1.
Wholemount aggregate immunostained with antibodies for MYO7A (magenta) and SOX2 (cyan). Reporter tdTomato expression is shown in yellow. The sample was cleared and imaged using confocal microscopy. All tdTomato-positive cells also express MYO7A, a hair cell marker, confirming faithful expression of the POU4F3 reporter.
Video S2. GATA3 expression is confined to a POU4F3nT/PCP4+ hair cell-containing vesicle in a PUR+IWP-2 treated organoid on D105, Related to Figure 5.
Wholemount aggregate immunostained with antibodies for GATA3 (magenta) and PCP4 (green), cleared and imaged with confocal microscopy. Reporter tdTomato expression is shown in white. GATA3 is expressed in both hair cells and non-hair cells in the vesicle.
Highlights.
Timed modulations of SHH and WNT ventralize hPSC-derived inner ear organoids
Cochlear organoids exhibit highly differential expression of GATA3 and NR2F1
Derived hair cells exhibit cochlear-like structural and functional properties
Cochlear organoids give rise to both inner and outer hair cell-like cells
ACKNOWLEDGEMENTS
This work was supported by a Department of Defense U.S. Army Medical Research and Materiel Command Congressionally Directed Medical Research Program grant W81XWH-18-1-0062 (E. H.), National Institute of Health grants R01 DC013294 and R01 DC015788 (E.H.), K08 DC016034 (R.F.N.), JSPS KAKENHI Grant Number JP21K09638 (T.N.), the Indiana University Health-Indiana University School of Medicine Strategic Research Initiative (E.H.), and the Ruth C. Holton Research Funds (E.H.). The authors would like to thank the Center for Contraception, Sexuality and Abortion (CASA) in Leiden and The Hague for the collection of the human fetal material, B. Daye, T. van der Helm, F. Wei, J. Harkin, T. Saeki, M. Jafarkkhani, E. Longworth-Mills, P. McGuire, H. Gao for their expert technical assistance, J. Zheng, K. Homma for antibodies against PRESTIN, and M. W. Kelley for his comments on the manuscript.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
S.T.M. and E.H. are the inventors on a PCT patent application titled “Methods of generating human cochlear hair cells”. The other authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFRENCES
- 1.Barald KF, and Kelley MW (2004). From placode to polarization: new tunes in inner ear development. Development 131, 4119–4130. 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- 2.Groves AK, and Fekete DM (2012). Shaping sound in space: the regulation of inner ear patterning. Development 139, 245–257. 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu DK, and Kelley MW (2012). Molecular mechanisms of inner ear development. Cold Spring Harb Perspect Biol 4, a008409. 10.1101/cshperspect.a008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morton CC, and Nance WE (2006). Newborn hearing screening--a silent revolution. N Engl J Med 354, 2151–2164. 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 5.Koehler KR, Mikosz AM, Molosh AI, Patel D, and Hashino E (2013). Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500, 217–221. 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koehler KR, and Hashino E (2014). 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat Protoc 9, 1229–1244. 10.1038/nprot.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu XP, Koehler KR, Mikosz AM, Hashino E, and Holt JR (2016). Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nat Commun 7, 11508. 10.1038/ncomms11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehler KR, Nie J, Longworth-Mills E, Liu XP, Lee J, Holt JR, and Hashino E (2017). Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat Biotechnol 35, 583–589. 10.1038/nbt.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nie J, Ueda Y, Solivais AJ, and Hashino E (2022). CHD7 regulates otic lineage specification and hair cell differentiation in human inner ear organoids. Nat Commun 13, 7053. 10.1038/s41467-022-34759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong M, O’Reilly M, Kirkwood NK, Al-Aama J, Lako M, Kros CJ, and Armstrong L (2018). Generating inner ear organoids containing putative cochlear hair cells from human pluripotent stem cells. Cell Death Dis 9, 922. 10.1038/s41419-018-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouchard M, de Caprona D, Busslinger M, Xu P, and Fritzsch B (2010). Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol 10, 89. 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang M, Gan L, Li D, Chen ZY, Zhou L, O’Malley BW Jr., Klein W, and Nathans J (1997). Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A 94, 9445–9450. 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yucel R, Frankel WN, Rechavi G, Moroy T, Friedman TB, Kelley MW, and Avraham KB (2004). Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet 13, 2143–2153. 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- 14.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, and Joung JK (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie J, and Hashino E (2020). Generation of inner ear organoids from human pluripotent stem cells. Methods Cell Biol 159, 303–321. 10.1016/bs.mcb.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Ueda Y, Moore ST, and Hashino E (2022). Directed Differentiation of Human Pluripotent Stem Cells into Inner Ear Organoids. Methods Mol Biol 2520, 135–150. 10.1007/7651_2021_448. [DOI] [PubMed] [Google Scholar]
- 17.Riccomagno MM, Martinu L, Mulheisen M, Wu DK, and Epstein DJ (2002). Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev 16, 2365–2378. 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohta S, Wang B, Mansour SL, and Schoenwolf GC (2016). SHH ventralizes the otocyst by maintaining basal PKA activity and regulating GLI3 signaling. Dev Biol 420, 100–109. 10.1016/j.ydbio.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riccomagno MM, Takada S, and Epstein DJ (2005). Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev 19, 1612–1623. 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohta S, Wang B, Mansour SL, and Schoenwolf GC (2016). BMP regulates regional gene expression in the dorsal otocyst through canonical and non-canonical intracellular pathways. Development 143, 2228–2237. 10.1242/dev.137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Sun S, Shang X, Keller ET, Chen M, and Zhou X (2020). Integrative differential expression and gene set enrichment analysis using summary statistics for scRNA-seq studies. Nat Commun 11, 1585. 10.1038/s41467-020-15298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns JC, Kelly MC, Hoa M, Morell RJ, and Kelley MW (2015). Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat Commun 6, 8557. 10.1038/ncomms9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, Palermo AT, So KS, Mays JC, Orvis J, et al. (2020). Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun 11, 2389. 10.1038/s41467-020-16113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheffer DI, Shen J, Corey DP, and Chen ZY (2015). Gene Expression by Mouse Inner Ear Hair Cells during Development. J Neurosci 35, 6366–6380. 10.1523/jneurosci.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elkon R, Milon B, Morrison L, Shah M, Vijayakumar S, Racherla M, Leitch CC, Silipino L, Hadi S, Weiss-Gayet M, et al. (2015). RFX transcription factors are essential for hearing in mice. Nat Commun 6, 8549. 10.1038/ncomms9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barr-Gillespie PG (2015). Assembly of hair bundles, an amazing problem for cell biology. Mol Biol Cell 26, 2727–2732. 10.1091/mbc.E14-04-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Shen W, He DZ, Long KB, Madison LD, and Dallos P (2000). Prestin is the motor protein of cochlear outer hair cells. Nature 405, 149–155. 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 28.Homma K, Miller KK, Anderson CT, Sengupta S, Du GG, Aguinaga S, Cheatham M, Dallos P, and Zheng J (2010). Interaction between CFTR and prestin (SLC26A5). Biochim Biophys Acta 1798, 1029–1040. 10.1016/j.bbamem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kros CJ, Ruppersberg JP, and Rusch A (1998). Expression of a potassium current in inner hair cells during development of hearing in mice. Nature 394, 281–284. 10.1038/28401. [DOI] [PubMed] [Google Scholar]
- 30.Marcotti W, and Kros CJ (1999). Developmental expression of the potassium current IK,n contributes to maturation of mouse outer hair cells. J Physiol 520 Pt 3, 653–660. 10.1111/j.1469-7793.1999.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, Yamauchi D, Marcus DC, and Kachar B (2004). Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neurosci 24, 7051–7062. 10.1523/jneurosci.1640-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korrapati S, Taukulis I, Olszewski R, Pyle M, Gu S, Singh R, Griffiths C, Martin D, Boger E, Morell RJ, and Hoa M (2019). Single Cell and Single Nucleus RNA-Seq Reveal Cellular Heterogeneity and Homeostatic Regulatory Networks in Adult Mouse Stria Vascularis. Front Mol Neurosci 12, 316. 10.3389/fnmol.2019.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, and Fritzsch B (2008). Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell Tissue Res 334, 339–358. 10.1007/s00441-008-0709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koo SK, Hill JK, Hwang CH, Lin ZS, Millen KJ, and Wu DK (2009). Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Dev Biol 333, 14–25. 10.1016/j.ydbio.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo XJ, Deng M, Xie X, Huang L, Wang H, Jiang L, Liang G, Hu F, Tieu R, Chen R, and Gan L (2013). GATA3 controls the specification of prosensory domain and neuronal survival in the mouse cochlea. Hum Mol Genet 22, 3609–3623. 10.1093/hmg/ddt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bardhan T, Jeng JY, Waldmann M, Ceriani F, Johnson SL, Olt J, Rüttiger L, Marcotti W, and Holley MC (2019). Gata3 is required for the functional maturation of inner hair cells and their innervation in the mouse cochlea. J Physiol 597, 3389–3406. 10.1113/jp277997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, de Caprona D, and Fritzsch B (2001). Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J Comp Neurol 429, 615–630. . [DOI] [PubMed] [Google Scholar]
- 38.Tang LS, Alger HM, and Pereira FA (2006). COUP-TFI controls Notch regulation of hair cell and support cell differentiation. Development 133, 3683–3693. 10.1242/dev.02536. [DOI] [PubMed] [Google Scholar]
- 39.Tang K, Xie X, Park JI, Jamrich M, Tsai S, and Tsai MJ (2010). COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development 137, 725–734. 10.1242/dev.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInturff S, Burns JC, and Kelley MW (2018). Characterization of spatial and temporal development of Type I and Type II hair cells in the mouse utricle using new cell-type-specific markers. Biol Open 7. 10.1242/bio.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driver EC, and Kelley MW (2020). Development of the cochlea. Development 147. 10.1242/dev.162263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thulasiram MR, Ogier JM, and Dabdoub A (2022). Hearing Function, Degeneration, and Disease: Spotlight on the Stria Vascularis. Front Cell Dev Biol 10, 841708. 10.3389/fcell.2022.841708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly MC, and Chen P (2009). Development of form and function in the mammalian cochlea. Curr Opin Neurobiol 19, 395–401. 10.1016/j.conb.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brigande JV, Kiernan AE, Gao X, Iten LE, and Fekete DM (2000). Molecular genetics of pattern formation in the inner ear: do compartment boundaries play a role? Proc Natl Acad Sci U S A 97, 11700–11706. 10.1073/pnas.97.22.11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kil SH, and Collazo A (2002). A review of inner ear fate maps and cell lineage studies. J Neurobiol 53, 129–142. 10.1002/neu.10127. [DOI] [PubMed] [Google Scholar]
- 46.Vélez-Ortega AC, Freeman MJ, Indzhykulian AA, Grossheim JM, and Frolenkov GI (2017). Mechanotransduction current is essential for stability of the transducing stereocilia in mammalian auditory hair cells. Elife 6. 10.7554/eLife.24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hadi S, Alexander AJ, Vélez-Ortega AC, and Frolenkov GI (2020). Myosin-XVa Controls Both Staircase Architecture and Diameter Gradation of Stereocilia Rows in the Auditory Hair Cell Bundles. J Assoc Res Otolaryngol 21, 121–135. 10.1007/s10162-020-00745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiwatpanit T, Lorenzen SM, Cantú JA, Foo CZ, Hogan AK, Márquez F, Clancy JC, Schipma MJ, Cheatham MA, Duggan A, and García-Añoveros J (2018). Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1. Nature 563, 691–695. 10.1038/s41586-018-0570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.García-Añoveros J, Clancy JC, Foo CZ, García-Gómez I, Zhou Y, Homma K, Cheatham MA, and Duggan A (2022). Tbx2 is a master regulator of inner versus outer hair cell differentiation. Nature 605, 298–303. 10.1038/s41586-022-04668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dressler GR, and Douglass EC (1992). Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci U S A 89, 1179–1183. 10.1073/pnas.89.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tavassoli K, Rüger W, and Horst J (1997). Alternative splicing in PAX2 generates a new reading frame and an extended conserved coding region at the carboxy terminus. Hum Genet 101, 371–375. 10.1007/s004390050644. [DOI] [PubMed] [Google Scholar]
- 52.Ward TA, Nebel A, Reeve AE, and Eccles MR (1994). Alternative messenger RNA forms and open reading frames within an additional conserved region of the human PAX-2 gene. Cell Growth Differ 5, 1015–1021. [PubMed] [Google Scholar]
- 53.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 54.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29, 731–734. 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, and Zhang F (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, and Ploegh HL (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33, 538–542. 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuda T, and Cepko CL (2007). Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A 104, 1027–1032. 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raymond CS, and Soriano P (2007). High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One 2, e162. 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rousseeuw PJ (1987). Silhouettes - a Graphical Aid to the Interpretation and Validation of Cluster-Analysis. J Comput Appl Math 20, 53–65. doi:Doi 10.1016/0377-0427(87)90125-7 (1987). [DOI] [Google Scholar]
- 62.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolmykov S, Yevshin I, Kulyashov M, Sharipov R, Kondrakhin Y, Makeev VJ, Kulakovskiy IV, Kel A, and Kolpakov F (2021). GTRD: an integrated view of transcription regulation. Nucleic Acids Res 49, D104–d111. 10.1093/nar/gkaa1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Locher H, Frijns JH, van Iperen L, de Groot JC, Huisman MA, and Chuva de Sousa Lopes SM (2013). Neurosensory development and cell fate determination in the human cochlea. Neural Dev 8, 20. 10.1186/1749-8104-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. POU4F3nT+ hair cells in SOX2+ vesicles of a D60 control organoid, Related to Figure 1.
Wholemount aggregate immunostained with antibodies for MYO7A (magenta) and SOX2 (cyan). Reporter tdTomato expression is shown in yellow. The sample was cleared and imaged using confocal microscopy. All tdTomato-positive cells also express MYO7A, a hair cell marker, confirming faithful expression of the POU4F3 reporter.
Video S2. GATA3 expression is confined to a POU4F3nT/PCP4+ hair cell-containing vesicle in a PUR+IWP-2 treated organoid on D105, Related to Figure 5.
Wholemount aggregate immunostained with antibodies for GATA3 (magenta) and PCP4 (green), cleared and imaged with confocal microscopy. Reporter tdTomato expression is shown in white. GATA3 is expressed in both hair cells and non-hair cells in the vesicle.
Data Availability Statement
Single-cell RNA-seq data have been deposited at the Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CALB1 | SWANT | Cat#300 |
| CLDN11 | Invitrogen | Cat#36-4500 |
| DLX3 | Santa Cruz | Cat#sc-514094 |
| DLX5 | Novus Biologicals | Cat#NBP1-85793 |
| FBXO2 | Santa Cruz | Cat#sc-398111 |
| GATA3 | R&D Systems | Cat#AF2605 |
| GFP | Thermo Fisher | Cat#A-11120 |
| INSM1 | Santa Cruz | Cat#sc-271408 |
| LMOD3 | Proteintech | Cat#14948-1-AP |
| LMX1A | Sigma-Aldrich | Cat#AB10533 |
| MYO7A | Proteus | Cat#256790 |
| MYO7A | Santa Cruz | Cat#sc-74516 |
| NDGR1 | Abcam | Cat#ab-124689 |
| NR2F1 | Invitrogen | Cat#ab-124689 |
| NR2F1 | R&D Systems | Cat#PP-H8132-00 |
| OCT4 (conjugated to AF488) | eBioscience | Cat#53584182 |
| OTX2 | R&D Systems | Cat#AF1979 |
| PAX2 | Abnova | Cat#H00005076-M01 |
| PAX8 | Abcam | Cat#AB97477 |
| PCP4 | Santa Cruz | Cat#sc-74816 |
| PCP4 | Invitrogen | Cat#PA5-52209 |
| phalloidin (conjugated to AF488) | Thermo Fisher | Cat#A12379 |
| POU4F3 | Santa Cruz | Cat#sc-81980 |
| Prestin | Generous gift of Drs. Jing Zheng and Kazuaki Homma et al.28 | NW-802 |
| Sox10 | eBioscience | Cat#14-5923-82 |
| Sox2 | BD Pharmingen | Cat#561469 |
| SSEA4 (conjugated to AF594) | BioLegend | Cat#330414 |
| SULF1 | Invitrogen | Cat#PA5-113112 |
| TUBA4A | Sigma-Aldrich | Cat#T6793 |
| TUJ1 (TUBB3) | Biolegend | Cat#801201 |
| Bacterial and virus strains | ||
| DH5a | New England Biolabs | Cat#C2987H |
| Biological samples | ||
| GW 13 Human fetal cochlea | This paper | N/A |
| GW 18 Human fetal cochlea | This paper | N/A |
| GW 18 Human fetal vestibule | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Accutase | Thermo Fisher Scientific | Cat# A1110501 |
| Advanced DMEM/F12 | Thermo Fisher Scientific | Cat#12634028 |
| Alt-R™ S.p. Cas9 Nuclease V3 | Integrated DNA technologies (IDT) | Cat#1081058 |
| B27 (minus vitamin A) | Thermo Fisher Scientific | Cat#12587010 |
| β-Mercaptoethanol | Thermo Fisher Scientific | Cat#21985023 |
| BMP4 | Reprocell | Cat#03-0007 |
| CHIR99021 | Reprocell | Cat#04-0004-10 |
| DMEM/F12 +HEPES | Thermo Fisher Scientific | Cat#11320033 |
| DPBS | Thermo Fisher Scientific | Cat#14040133 |
| Essential 6 (E6) medium | Thermo Fisher Scientific | Cat#A1516401 |
| Essential 8 (E8) Flex medium | Thermo Fisher Scientific | Cat#A2858501 |
| 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | Cat#15575020 |
| FGF2 | Stem Cell Technologies | Cat#780003 |
| GFR Matrigel | Corning | Cat#354230 |
| GlutaMAX | Thermo Fisher Scientific | Cat#35050061 |
| Glutaldehyde | Electron Microscopy Sciences (EMS) | Cat#16537-15 |
| IWP-2 | Tocris | Cat#3533 |
| LDN193189 | Stemgent | Cat#04-0074-02 |
| L-15 cell culture medium | Thermo Fisher Scientific | Cat#21083027 |
| Neurobasal medium | Thermo Fisher Scientific | Cat#21103049 |
| Normocin | Invivogen | Cat# ant-nr-1 |
| N2 | Thermo Fisher Scientific | Cat#17502048 |
| Osmium tetroxide | Electron Microscopy Sciences (EMS) | Cat#19150 |
| Propidium Iodide | Invitrogen | Cat#P1304MP |
| Purmorphamine | Stemgent | Cat#04-0009) |
| RevitaCell | Thermo Fisher Scientific | Cat#A2644501 |
| SB431542 | Stemgent | Cat#04-0010-05 |
| Scr7 | Xcessbio | Cat#M60082-2S |
| TRIzol | Thermo Fisher Scientific | Cat#15596026 |
| TrypLE | Thermo Fisher Scientific | Cat#A1217701 |
| Vitronectin-N | Thermo Fisher Scientific | Cat#A14700 |
| Y-27632 | Stemgent | Cat#04-0012-02 |
| Critical commercial assays | ||
| Chromium Next GEM Single Cell Kit v3 | 10x Genomics | Cat#1000075 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE200629 |
| Experimental models: Cell lines | ||
| WA25 | WiCell | (NIHhESC-12-0196; RRID:CVCL_E080) |
| mND2-0 | WiCell | mND2-0 (MIRJT7i-mND2-0,RRID:CVCL_U173) |
| Oligonucleotides | ||
| PAX2 RT assay Fwd: CCTGGCAGGAATGGTGCCT | This paper | N/A |
| PAX2 RT assay Rev: GTCAGACGGGGACGATGTGG | This paper | N/A |
| Primers for PAX2 donor vector: see supplementary table 2 | This paper | N/A |
| Primers for POU4F3 donor vector: see supplementary table 2 | This paper | N/A |
| PAX2 gRNA: ATGACCGCCACTAGTTACCG | This paper | N/A |
| POU4F3-targeting sgRNA for CRISPR/Cas9 genome editing: ATTCGGCTGTCCACTGATTG | This paper | N/A |
| Recombinant DNA | ||
| SpCas9-HF1 vector | Kleinstiver et al.14 | Addgene#72247 |
| U6 expression vector | Slaymaker et al.55 | Addgene #71814 |
| Cre expression vector | Matsuda et al.57 | Addgene #13775 |
| FLPo expression vector | Raymond et al.58 | Addgene #13793 |
| 2A-eGFP-PGK-Puro vector | Hockemeyer et al.54 | Addgene #31938 |
| Software and algorithms | ||
| ImageJ | Schneider et al.59 | https://imagej.nih.gov/ij/ |
| Flow Jo | FlowJo LLC | https://www.flowjo.com/ |
| Prism 9 | GraphPad Software | https://graphpad.com |
| Imaris 8 | Bitplane | https://Imaris.oxinst.com/ |
| R | Team R C. R: A language and environment for statistical computing[J]. 2013. | https://cran.r-project.org/bin/macosx/ (macOS)https://cran.r-project.org/bin/windows/base/ (Windows)https://cran.r-project.org/bin/linux/ (Linux) |
| R studio | Team R S. Rstudio: integrated development for R[J]. Rstudio, Inc., Boston, MA URL http://www.Rstudio.com, 2015, 42: 14. | https://www.rstudio.com/products/rstudio/download/ |
| R package Seurat | https://github.com/satijalab/seurat | |
| DESeq2 | Love et al.62 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| zingeR | Love et al.62 | https://github.com/statOmics/zingeR/blob/master/R/methods.R |
| iDEA | Ma et al.21 | https://xzhoulab.github.io/iDEA/ |
| Custom code | This paper | https://github.com/HashinoLab/Moore_et¬_al_Cochlear_Organoids |
| Nikon Elements microscope imaging software | Nikon Corporation | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer |
All original code has been deposited at the GitHub repository and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


