Abstract
To characterize the occurrence, frequency, and kinetics of retroviral recombination in vivo, we intravaginally inoculated rhesus macaques, either simultaneously or sequentially, with attenuated simian immunodeficiency virus (SIV) strains having complementary deletions in their accessory genes and various degrees of replication impairment. In monkeys inoculated simultaneously with SIVmac239Δvpx/Δvpr and SIVmac239Δnef, recombinant wild-type (wt) virus and wild-type levels of plasma viral RNA (vRNA) were detected in blood by 2 weeks postinoculation. In monkeys inoculated first with SIVmac239Δvpx/Δvpr and then with SIVmac239Δnef, recombination occurred but was associated with lower plasma vRNA levels than plasma vRNA levels seen for monkeys inoculated intravaginally with wt SIVmac239. In one monkey, recombination occurred 6 weeks after the challenge with SIVmac239Δnef when plasma SIVmac239Δvpx/Δvpr RNA levels were undetectable. In monkeys inoculated first with the more highly replicating strain, SIVmac239Δnef, and then with SIVmac239Δvpx/Δvpr, wild-type recombinant virus was not detected in blood or tissues. Instead, a virus that had repaired the deletion in the nef gene by a compensatory mutation was found in one animal. Overall, recombinant SIV was eventually found in four of six animals intravaginally inoculated with the two SIVmac239 deletion mutants. These findings show that recombination can occur readily in vivo after mucosal SIV exposure and thus contributes to the generation of viral genetic diversity and enhancement of viral fitness.
The primate immunodeficiency viruses, human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV), employ a multifaceted strategy to achieve the genetic diversity required for survival in the challenging environment of the infected host. High levels of replication achieved by utilizing an error-prone polymerase that lacks a proofreading function to copy the viral genetic material result in production of a breadth of genetic variants (7, 8, 30). Although most mutations are deleterious, occasional genetic changes can confer a selective advantage for a given environment. Genetic recombination occurs frequently during retroviral replication as a consequence of packaging two RNA molecules in one virus particle (14). Recombination provides a complementary mechanism to nucleotide substitution to generate the genetic diversity needed to maintain a persistent infection in the face of host defenses and even antiretroviral drug treatment (16). For retroviruses, the calculated recombination rate is even greater than the point-substitution rate associated with mutation (14). Together, mutation and recombination can produce a large pool of viral variants from which the variants best adapted for a given environment can emerge by selection.
It has been proposed that for RNA viruses, such as HIV and SIV, recombinant viruses with mosaic patterns can increase the diversity of the virus population quickly, leading to enhanced viability (20, 22). This mechanism is potentially significant in light of data that suggest that the HIV type 1 population present shortly after transmission may possess limited diversity (11, 29). It has become clear, as the number of available sequences from cloned full-length proviruses has increased, that chimeric or mosaic viruses, derived by recombination, are common in HIV-infected individuals (22, 25, 28, 31). Both intrasubtype and intersubtype recombination between infecting and superinfecting HIV strains have been documented (3, 12). The frequency of recombinant viruses in some cohorts of HIV-infected individuals where several clades circulate is estimated to be approximately 20% (27). This high frequency suggests that fitness advantages can be conferred by recombination, and it highlights the possible evolutionary significance of the process.
Despite much work investigating the mechanisms of retroviral recombination and reverse transcription in vitro (4, 15), the massive level of recombination that occurs in infected individuals has only recently been appreciated (5, 26). In an earlier study with a nonhuman primate model of AIDS, simultaneous intravenous injections of a rhesus monkey at different sites with two complementary SIV deletion mutants resulted in the production of wild-type (wt) virus derived by recombination, which was isolated from blood 2 weeks after infection (32).
To more closely examine the process of retroviral recombination in vivo and the potential contributions of recombination to AIDS pathogenesis and viral transmission, rhesus macaques were intravaginally inoculated with two replication-attenuated mutants of SIVmac239 having reciprocal deletions in their accessory genes (SIVmac239Δvpx/Δvpr [which carries a wild-type nef gene] and SIVmac239Δnef [which carries a wild-type vpx/vpr gene] sequentially or simultaneously. We then studied the replication patterns and genetic structure of the resulting viral populations in these animals. Recombinant SIV variants, demonstrated by the presence of both wt vpx/vpr and nef genes in a single viral variant, arose quickly in vivo. Thus, recombination can occur readily in vivo following simultaneous or sequential mucosal inoculations, and this process may have implications for the sexual transmission and pathogenesis of AIDS.
MATERIALS AND METHODS
Animals.
Six female, multiparous, regularly cycling rhesus macaques (Macaca mulatta) were used for the vaginal inoculation studies, and two juvenile male rhesus macaques were used for the plasma passage experiment (see below). All animals were housed at the California National Primate Research Center, in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care standards. All animals were negative for antibodies to HIV type 2, SIV, type D retrovirus, and simian T-cell lymphotropic virus type 1 at the time that the study was initiated.
Viruses and inoculation schedules.
SIVmac239Δnef and SIVmac239Δvpx/Δvpr are replication-competent but attenuated variants of SIVmac239 (13). Stocks were produced by inoculating CEMx174 cells with seed stocks of each virus, followed by 7 days of culture. Supernatants were harvested, and 1-ml aliquots were frozen at −135°C until they were used for animal inoculations. By endpoint dilution titration on CEMx174 cells, both stocks had titers of approximately 106 TCID50 per ml. By use of a 1-ml tuberculin syringe without a needle, adult female rhesus macaques were inoculated by atraumatically introducing 1 ml of stock into the vaginal canal.
Figure 1 shows the animal groups and the timing of the intravaginal inoculation schedules with SIVmac239Δvpx/Δvpr and SIVmac239Δnef simultaneously (group I animals 25557 and 27713), SIVmac239Δvpx/Δvpr followed by SIVmac239Δnef (group II animals 25958 and 30854), or SIVmac239Δnef preceding SIVmac239Δvpx/Δvpr (group III animals 30395 and 32816). The SIVmac239Δnef inoculations of group III animals were performed twice in 1 day, as this regimen increases the efficiency of vaginal SIV transmission compared to once-daily inoculations (23); all other inoculations were once a day.
FIG. 1.
SIV inoculation schedule. Group I animals were intravaginally inoculated with a mixture of SIVmac239Δnef and SIVmac239Δvpx/Δvpr. Group II monkeys were intravaginally inoculated with SIVmac239Δvpx/Δvpr first and then with SIVmac239Δnef. Group III monkeys were intravaginally inoculated initially with SIVmac239Δnef and then with SIVmac239Δvpx/Δvpr. The overlapping arrows represent two intravaginal inoculations in 1 day, two single arrows in 1 week represent a single intravaginal inoculation per day on two separate days. Group II monkeys were inoculated with SIVmac239Δnef after systemic SIVmac239Δvpx/Δvpr infection was documented and after the peak of plasma viremia in animal 30854. As the initial SIVmac239Δnef infection did not produce a spike in plasma viremia in macaque 30854, the animal was reinoculated with SIVmac239Δnef. Group III monkeys were inoculated with SIVmac239Δvpx/Δvpr after we documented systemic SIVmac239Δnef infection and after the peak of plasma viremia. To maximize the opportunity for recombination, group III animals were inoculated several times with SIVmac239Δvpx/Δvpr as shown.
Transfer of plasma to determine the replication potential of wt SIV variants in plasma of group II animals in naïve animals.
For the plasma passage experiments, 2 ml of plasma from group II animals taken on the day of necropsy were inoculated intravenously into two naïve juvenile male rhesus macaques. Animal 32908 received 2 ml of plasma from group II animal 30854 (320 viral RNA [vRNA] copies/ml), and animal 33460 received 2 ml of plasma from group II animal 25958 (approximately 2,500 vRNA copies/ml).
Measurement of SIV RNA in plasma.
Virion-associated SIV RNA levels in plasma for animals in this study were measured by use of a real-time reverse transcription (RT)-PCR assay based on a sequence detection system (Prism 7700; Applied Biosystems, Foster City, Calif.) and reported as vRNA copies per milliliter of plasma (19). As used in this study, the lower limit of quantitation for this assay was 100 copy equivalents of vRNA per ml of plasma.
PBMC isolation.
Peripheral blood mononuclear cells (PBMC) were isolated from heparin or EDTA-treated blood samples by using lymphocyte separation media (ICN Biomedicals, Aurora, Ohio). PBMC were frozen in 10% dimethyl sulfoxide (Sigma, St. Louis, Mo.)-90% fetal bovine serum (Gemini BioProducts, Calabasas, Calif.) and stored in liquid nitrogen until future analysis.
Measurement of anti-SIV antibody titers.
Anti-SIV binding antibody titers in plasma were measured by using enzyme-linked immunosorbent assay plates coated with detergent-disrupted SIVmac251 virions as previously described (21). The results of the anti-SIV antibody enzyme-linked immunosorbent assay are reported as the reciprocals of the highest dilutions of samples that produced optical density values above the cutoff value.
Proliferation assay.
SIV-specific T-cell proliferative responses were measured as previously described, by using freshly isolated PBMC (24). AT-2-inactivated SIV, provided by the AIDS Vaccine Program, SAIC Frederick, Inc., National Cancer Institute—Frederick, Frederick, Md., was used for the in vitro stimulation of PBMC cultures. Results reported from this assay represent the highest responses obtained using 125 and 12.5 ng of p27CA equivalents/well. Each PBMC sample was stimulated with concanavalin A as a positive control in every assay. Further, fresh PBMC samples from at least two SIV-naïve monkeys were included as negative controls in every assay. All controls gave appropriate results in all experiments.
IFN-γ ELISPOT assay.
The number of gamma interferon (IFN-γ)-secreting cells in frozen PBMC responding to a SIVmac239 Gag p27 peptide pool was determined by using an IFN-γ monkey cytokine ELISPOT kit (U-CyTech, Utrecht University, Utrecht, The Netherlands) as described previously (1). Negative controls consisted of cells that were cultured in medium only and cells from uninfected monkeys. A sample was considered positive only if the number of IFN-γ-secreting cells per well exceeded 50/106 PBMC and was greater than the mean spot-forming-cell number of the medium-only wells ± 2 standard deviations. Data are reported as the numbers of IFN-γ spot-forming cells per 106 PBMC, and the mean numbers of IFN-γ spots in medium-only wells were subtracted from the mean spot numbers of SIV peptide-stimulated wells. In addition to stimulating each PBMC sample with phorbol myristate acetate-ionomycin, fresh PBMC samples from at least two monkeys infected with SIVmac239Δnef and known to have strong anti-SIV p27-specific IFN-γ responses were included as positive controls in every assay. Further, fresh PBMC samples from at least two SIV-naïve monkeys were included as negative controls in every assay. All the positive and negative controls gave appropriate results in all experiments.
Virus population structure.
To explore the relationship between the viruses used for inoculation and the virus population structures in the animals, we tracked the evolution of viral sequences within the six intravaginally inoculated macaques by examining the vDNA and vRNA sequences spanning the vpx through nef genes amplified by PCR (see below) from PBMC and plasma samples at selected time points (see Fig. 2, 3, and 4). PCR product DNAs from 10 to 28 samples per animal were cloned, and up to 13 clones from each sample were sequenced. Clones were derived from multiple pooled PCR assays. Alignment of the deduced amino acid sequences revealed distinct virus population structures for each group of macaques, indicating the absence of PCR product DNA contamination. In addition, a real-time PCR assay was used to characterize the range of nef and vpx/vpr variants in plasma and PBMC at early time points (see below).
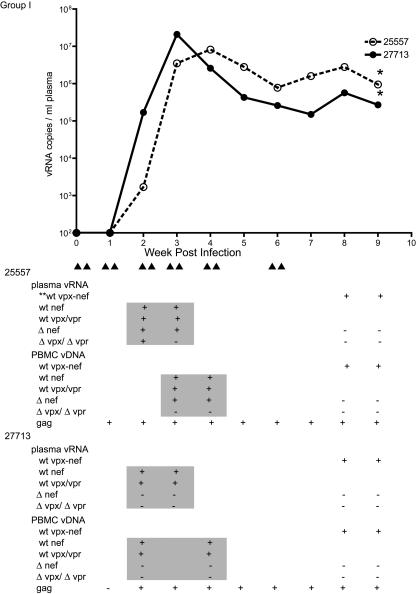
FIG. 2.
Plasma SIV RNA levels and genotypes of viral variants for group I animals. The arrowheads under the x axis of the figure represent viral inoculations as described in the legend to Fig. 1. The table is aligned vertically with the x axis of the graph and indicates the genotype of variants detected in plasma and PBMC of each monkey at the corresponding time point. The shaded areas in the table indicate the results that were generated by the real-time PCR assay that was capable of discriminating vRNA variants in plasma. The nonshaded areas indicate results that were based on cloning, sequencing, and alignment of the 4-kb region that spans the vpx and nef genes. The asterisks in the graph denote the time of necropsy for each monkey. **wt vpx-nef, variants that contain wt vpx/vpr and nef gene sequences, based on sequencing the cloned 4-kb region spanning the vpx/vpr and nef genes. The results of the nested PCR assay for detection of SIVgag sequences in PBMC DNA are shown in the last row of results for each monkey.
FIG. 3.
Plasma SIV RNA levels and genotypes of viral variants for group II animals. The organization of this figure is the same as that of Fig. 2.
FIG. 4.
Plasma SIV RNA levels and genotypes of viral variants for group III animals. The organization of this figure is the same as that of Fig. 2.
PCR assays.
To detect SIV infection, a nested PCR for SIVgag was performed on PBMC DNA samples as previously described (1, 24). To produce the ∼4-kb clones spanning the vpx to nef genes for sequence analysis, viral DNA was isolated from PBMC by using the PureGene DNA isolation kit (Gentra Systems Inc., Minneapolis, Minn.). Viral RNA was isolated from plasma by using the High Pure viral nucleic acid buffer set (Roche Diagnostics, Penzberg, Germany). RNA was reverse transcribed into cDNA by using the Thermoscript RT kit (Invitrogen, San Diego, Calif.) with primer EK7r (5′-CGACCGGCTCCTCCCCAGTA-3′). To determine the genotypes of the infecting viral variants, nested PCR amplification of a 4-kb fragment spanning the vpx to nef genes was performed using the outer primers 5823VPX (5′-CAGGGAGAGAATCCCACCTGG-3′) and 9816FEN3U (5′-GAAGGCCTCTTGCGGTTAGCC-3′) in the first-round reaction and inner primers 5864VPX (5′-TAGGAGAGGCCTTCGAATGGC-3′) and 9788FEN3U (5′-AACCTCTTCCTCTGACAGGCC-3′) in the second-round reaction. The first round of amplification used 2 U of rTth DNA polymerase XL in 1.2 mM Mg(OAc)2, 10 μM concentrations of dNTPs, 0.2 μM concentrations of primers, and either 5 μl of cDNA or 1 μg of genomic DNA template in a total volume of 50 μl. After an initial hot start at 94°C for 1 min, the cycling profile was 94°C for 20 s and 68°C for 5 min 30 s for 35 cycles, with a final extension at 72°C for 20 min. Second-round amplification was performed with high-fidelity platinum Taq polymerase (Invitrogen) by using 5 μl of the first-round PCR with a cycling profile of 94°C for 20 s, 55°C for 30 s, and 68°C for 5 min for 35 cycles and a final extension at 72°C for 30 min. Because of the relatively long length of the viral sequences analyzed in this study, we tested whether the observed recombinants within the macaques were due to in vivo or in vitro recombination. We screened for the production of SIV recombinants by PCR using mixtures of SIVmac239Δvpx/Δvpr and SIVmac239Δnef genomes. Viral sequences spanning the vpx through nef genes were amplified by PCR, and product DNA from 10 clones from each mixture was sequenced. These reconstruction experiments showed that no recombinants were generated in vitro under the PCR conditions used in this study (data not shown).
DNA cloning and sequencing.
PCR product DNA spanning the vpx to nef genes was inserted into pCR-XL-TOPO (Invitrogen), and individual transformed clones were analyzed by dideoxy DNA sequencing using 16 SIV gene-specific primers and T7 forward and M13 reverse primers in the vector. DNA sequencing reactions were analyzed with an ABI Prism 3100 sequencer (Applied Biosystems). Overlapping DNA sequences were aligned and edited by using the SeqMan program (DNASTAR, Inc., Madison, Wis.).
Representation of specific deletion mutants and wild-type SIV in plasma.
To track the SIV variants in plasma, we used a real-time PCR assay to determine the representation of the different forms of the nef and vpx/vpr genes by using an ABI 7900 sequence detection system (Applied Biosystems). The plasma vRNA was reverse transcribed into cDNA by using the Thermoscript RT kit (Invitrogen) with primer EK7r, and then the first PCR amplification of the 4-kb fragment spanning the vpx to nef genes was performed using the primers 5823VPX and 9816FEN3U as described above. To detect the wild-type SIVmac239 nef sequence, we used FC01 forward primer (5′-GGCCAAAAGTTCCCCTAAGAAC-3′) and RB1 reverse primer (5′-ATCCCACTGGGAAGTTTGAGCT-3′) with the probe P0-nef (5′-6-carboxyfluorescein [FAM]-GAAGAAGGCATCATACCAGATTGGCAGGATTAC-xanthylium, 9-[2-[[4-[[(2,5-dioxo-1-pyrrolidinyl)oxy]carbonyl]-1-piperidinyl]sulfonyl]phenyl1]-3,6-bis(methylphenylamino)-, chloride [QSY]-3′). To detect the wild-type SIVmac239vpx/vpr sequence, we used FV01 forward primer (5′-TGGAAGAAAGACCTCCAGAAAATG-3′) and RV01 reverse primer (5′-TTCCAGAACCTCCACTACCCA-FAM-3′) with the probe P1-wt vpx/vpr (5′-QSY-AGGACCACAAAGGGAACCATGGGATG-3′, FAM-QSY). SIVmac239Δnef sequences were measured by using FdelB1 forward primer (5′-TCTTGTGAGGGACAGGTCTCATTT-3′) and RB1 reverse primer (5′-ATCCCACTGGGAAGTTTGAGCT-3′) with the probe P0-nef. Since the viral sequences from one macaque in group III contained both the original SIVmac239Δnef challenge strain and strains with additional deletions that reconstituted a truncated nef open reading frame(data not shown), the additional deletions in nef (ΔΔnef) were measured by the same primers which were used but were used in combination with specific probes capable of detecting the new 148- and 97-bp deletions (P2del-nef-L probe 5′-FAM-AAGGGGGGACTGGAAGGGATTTATCAGATGAG-QSY-3′ and P2del-nef-S probe 5′-FAM-GGAAGGGATTTATTACAGTGCAAGACATTTGGCT-QSY-3′, respectively). To detect the SIVmac239Δvpx/Δvpr sequence, we used F-vpx forward primer (5′-GGGAGCTAATTTTCCAGGTTTG) and R-vpr reverse primer (5′-TCAAGGGTGTCTCCATGTCTATTA-3′) with the probe P-del-vpx/vpr(5-FAM-GGGAATACTGGCATGATGATTGATCCTCGC-QSY-3′). This assay can discriminate among the different SIVmac239 deletion mutants and their wild-type equivalents (data not shown). The cycling profile was 95°C for 10 min and 45 cycles of 95°C for 15 s and 60°C for 1 min. Results were confirmed with a second PCR using the first-round PCR product with primers FA1-nef (5′-GCGCGTGGGGAGACTTATGG-3′) and RA1-nef (5′-CTCTGACAGGCCTGACTTGCTTCC-3′) and primers F-vpx and R-vpr to amplify nef and vpx/vpr, respectively. The cycling profile was 95°C for 12 min, 35 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 45 s and an extension at 72°C for 7 min. PCR product DNA was resolved by electrophoresis on a 1.2% agarose gel.
RESULTS
Virologic findings after simultaneous intravaginal inoculation with SIVmac239Δnef and SIVmac239Δvpx/Δvpr (group I).
In group I macaques, plasma SIV RNA levels increased to a peak at 3 to 4 weeks postinfection (PI) and then declined to a plateau (Fig. 2). SIVgag DNA was initially detected in the PBMC of animal 25557 at 1 week PI and in animal 27713 at 2 weeks PI. The PBMC samples of these animals remained positive for SIV DNA at all subsequent time points (Fig. 2). At weeks 2 and 3 PI, real-time PCR analysis of the plasma vRNA from macaque 25557 detected the original SIVmac239Δnef and SIVmac239Δvpx/Δvpr inoculating strains and wt SIVmac239nef and SIVmac239vpx/vpr (Fig. 2). Results of vDNA analyses of 3- and 4-weeks-PI PBMC samples were similar, although SIVmac239Δvpx/Δvpr sequences were not detected (Fig. 2). At 8 and 9 weeks PI, only wt SIVmac239 sequences were detected in plasma and PBMC analyzed by cloning and sequencing (Fig. 2). By real-time PCR, the other macaque (27713) had wt SIVmac239nef and wt SIVmac239vpx/vpr vRNA in plasma at weeks 2 and 3 PI and vDNA in PBMC at weeks 2 and 4 PI. Wild-type SIVmac239 was the only variant detected in plasma vRNA and PBMC vDNA by cloning and sequencing from blood samples collected at weeks 8 and 9 (Fig. 2). In both animals, wt SIVmac239 was the only variant detected by cloning and sequencing DNA from three lymphoid tissues (for monkey 25557 spleen, 10 of 10 clones; axillary lymph node, 8 of 8 clones; and iliac lymph node, 9 of 9 clones; for monkey 27713 spleen, 8 of 8 clones; axillary lymph node, 7 of 7 clones; and iliac lymph node, 10 of 10 clones) collected at necropsy at week 9 PI.
Virologic findings after sequential intravaginal inoculation with SIVmac239Δnef and SIVmac239Δvpx/Δvpr (groups II and III).
One macaque in group II (30854) had a modest level of SIV RNA in plasma at weeks 4 and 5 after intravaginal SIVmac239Δvpx/Δvpr inoculation, but vRNA levels were near or below the limit of detection between 6 and 17 weeks PI, and then vRNA became intermittently positive at slightly higher levels (Fig. 3). For macaque 30854, SIVgag DNA was initially detected in the 4-weeks-PI PBMC sample, and all subsequent PBMC samples were SIVgag DNA positive (Fig. 3). Plasma SIV RNA in the other group II macaque (25958) was undetectable until a peak (>106 copies/ml) at week 12 PI, 3 weeks after the intravaginal inoculation with SIVmac239Δnef. From weeks 12 to 18 PI, vRNA levels declined, and then, despite no further inoculations, plasma vRNA peaked (>106 copies/ml) again at week 19 PI (Fig. 3). Up to week 11 after the first intravaginal inoculation with SIVmac239Δvpx/Δvpr, SIVgag vDNA was sporadically detectable in PBMC of animal 25958. From week 11 (2 weeks after the intravaginal SIVmac239Δnef inoculation) to the end of the study, all PBMC samples from animal 25958 were SIVgag DNA positive (Fig. 3). The presence of SIV DNA in PBMC after inoculation with SIVmac239Δvpx/Δvpr and before inoculation with SIVmac239Δnef demonstrates that both group II animals became systemically infected after intravaginal inoculation with the highly attenuated SIVmac239Δvpx/Δvpr virus.
Based on the real-time PCR assay, monkey 25958 had SIVmac239Δnef strain sequences in plasma at weeks 12, 18, and 19 PI, but only variants containing wt SIVmac239 vpx/vpr and nef were detected in plasma at week 20 PI (Fig. 3). Using cloning and sequencing, both SIVmac239Δnef and wt SIVmac239 were found in plasma vRNA and PBMC vDNA from monkey 25958 at 23 and 26 weeks PI (Fig. 3). Based on the real-time PCR assays, the other monkey (30854) had SIVmac239Δnef vRNA in plasma at weeks 17, 20, and 21 PI, but only variants containing wt SIVmac239 vpx/vpr and nef were detected in plasma at week 23 PI. Using cloning and DNA sequence analysis of plasma vRNA from macaque 30854, wt SIVmac239 and SIVmac239Δvpx/Δvpr were detected at week 27 PI, but only wt SIVmac239 was detected at week 28 PI (Fig. 3). Wild-type SIVmac239 was the only variant detected in three lymphoid tissues (spleen, 10 of 10 clones; iliac lymph node, 10 of 10 clones; axillary lymph node, 9 of 9 clones) collected at necropsy in week 28 PI from monkey 30854. Wild-type SIVmac239 was the only variant detected in DNA from two of three lymphoid tissues (spleen, 10 of 10 clones; axillary lymph node, 9 of 9 clones), and SIVmac239Δnef was the only variant detected in DNA from one of three tissues (iliac lymph node, 10 of 10 clones) collected at necropsy in week 26 PI from monkey 25958.
As shown in Fig. 3, the viral loads in the group II monkeys were relatively low and undulating throughout the course of the infection, despite the fact that viruses with intact vpx, vpr, env, and nef genes were found in both animals. To determine if these wild-type viruses had the expected replication kinetics in vivo, each of two naïve juvenile rhesus macaques was inoculated with the plasma from one of the group II animals. Both of the naïve juvenile male rhesus macaques inoculated intravenously with plasma collected at necropsy from group II animals developed systemic SIV infections. The plasma vRNA levels of both of the animals had a pattern that was indistinguishable from the plasma vRNA pattern observed for animals inoculated intravenously with wt SIVmac239 (peak, >106 vRNA copies/ml; set point, >105 vRNA copies/ml). Thus, the recombinant SIV variants in the group II animals with wt nef and vpx/vpr nucleotide sequences had wt in vivo replication capacity.
For both group III macaques, plasma SIV vRNA levels increased during the first 3 to 6 weeks PI. For one macaque (30395), vRNA maintained an undulating plateau from week 6 PI to necropsy (Fig. 4). After declining to undetectable levels at week 9 PI, the plasma vRNA levels in macaque 32816 spiked at week 11, 1 week after intravaginal SIVmac239Δvpx/Δvpr inoculation (Fig. 4). PBMC of both macaques in group III were SIVgag vDNA positive at 2 weeks PI, and they remained persistently vDNA positive from that point until necropsy (Fig. 4). From the sequencing analysis, the viral sequences from one macaque (30395) in group III showed the original SIVmac239Δnef challenge strain and strains with an additional deletion (SIVmac239ΔΔnef) that resulted in a truncated nef open reading frame. Using the real-time PCR assays to analyze plasma vRNA sequences from monkey 30395, SIVmac239Δnef and wt SIVmac239vpx/vpr were detected at weeks 2, 3, and 4 PI, while both SIVmac239Δnef and SIVmac239ΔΔnef strain sequences were detected at weeks 4 and 5 PI (Fig. 4). Based on cloning and sequencing of variants, only SIVmac239ΔΔnef was detected at weeks 20 and 22 PI in plasma vRNA, while SIVmac239Δnef was the only variant detected at 20 weeks PI in PBMC vDNA and SIVmac239ΔΔnef was the only variant detected at 22 weeks PI in PBMC vDNA. By use of the real-time PCR assays to analyze plasma vRNA, SIVmac239Δnef and wt SIVmac239vpx/vpr were detected in plasma samples collected at 2 and 10 weeks PI from the other group III monkey (32816), and by cloning and sequencing, only SIVmac239Δnef sequences were detected in PBMC vDNA and plasma vRNA from blood samples collected at 20 and 22 weeks PI from monkey 32816 (Fig. 4). The original SIVmac239Δnef was the only variant detected in DNA from the three lymphoid tissues (spleen, 10 of 10 clones; axillary lymph node, 13 of 13 clones; iliac lymph node, 10 of 10 clones) collected at necropsy in week 28 PI from monkey 32816. The original SIVmac239Δnef was detected in the three lymphoid tissues (spleen, 1 of 8 clones; iliac lymph node, 6 of 8 clones; axillary lymph node, 7 of 10 clones) collected at necropsy in week 26 from monkey 30395. The remaining clones in the spleen and the axillary and iliac lymph nodes of monkey 30395 were SIVmac239ΔΔnef.
Antiviral immune responses after simultaneous or sequential intravaginal inoculation with SIVmac239Δnef and SIVmac239Δvpx/Δvpr (groups I, II, and III).
For both group I animals, low-titer anti-SIV antibodies were detected at 4 weeks PI, after plasma vRNA levels peaked. From 4 weeks PI on, antibody titers gradually increased (Fig. 5). SIV-specific T-cell responses in group I animals were not assessed.
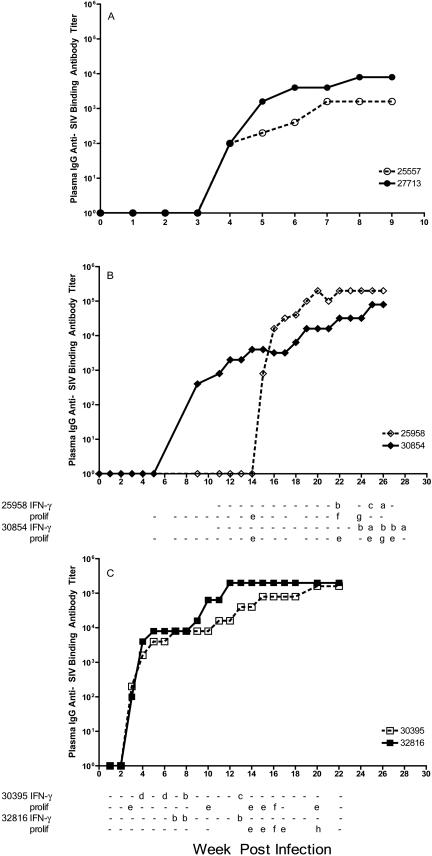
FIG. 5.
Plasma anti-SIV antibody levels and T-cell responses. (A) Group I animals; (B) group II animals; (C) group III animals. Antibody titers are indicated by the line graph. The tables are aligned vertically with the x axis of the graphs and indicate the results of SIV-specific T-cell assays for each monkey at each indicated time point. Blank spaces in the table indicate that samples collected at those time points were not analyzed. Each result of the anti-SIV p27 peptide IFN-γ ELISPOT assay is reported as the number of IFN-γ-secreting T cells/106 PBMC minus the mean number of spots in the medium-only wells. a, 50 to 99 IFN-γ-secreting T cells/106 PBMC; b, 100 to 199 IFN-γ-secreting T cells/106 PBMC; c, 200 to 300 IFN-γ-secreting T cells/106 PBMC; d, >300 IFN-γ-secreting T cells/106 PBMC; prolif, results of the lymphocyte (T-cell) proliferation assay reported as stimulation indices; e, 2 to 4.9; f, 5 to 9.9; g, 10 to 20; h, >20; −, the result was scored negative.
Plasma anti-SIV antibodies were first detected at week 9 PI in group II monkey 30854 (Fig. 5), at a point when the animal had become plasma vRNA negative after having been positive at weeks 4 and 5 PI (Fig. 5). Animal 39854 was inoculated with SIVmac239Δnef at week 10, and anti-SIV antibody titers steadily increased from that point on (Fig. 5), although vRNA levels did not increase dramatically. We measured SIV-specific T-cell responses in group II animals, beginning at week 11 PI (Fig. 5). Anti-SIV T-cell proliferative responses were first detected at 22 weeks PI in monkey 30854, and T-cell proliferative responses were detectable from that point on. Anti-SIV IFN-γ responses were measured beginning at week 5 PI, and monkey 30854 had detectable SIVgag-specific IFN-γ-secreting cells from week 24 to week 28 PI (Fig. 5).
For the other group II monkey (25958), plasma anti-SIV antibodies were first detected at week 15 PI, 6 weeks after this previously vRNA-negative animal was inoculated with SIVmac239Δnef at week 9 PI (Fig. 5). Plasma anti-SIV antibody titers for this animal steadily increased from week 15 on (Fig. 5). For monkey 25958, anti-SIV T-cell proliferative responses were first detected at 13 weeks PI, and SIVgag-specific IFN-γ-secreting T cells were detected at week 17 and weekly from 22 to 26 weeks PI.
Both group III animals were positive for plasma anti-SIV antibodies 3 weeks after intravaginal SIVmac239Δnef inoculation, and anti-SIV antibody titers increased to a plateau by approximately 16 weeks PI. SIV-specific T-cell responses of group III animals were assessed beginning at week 1 PI, and T-cell proliferative responses in monkey 30395 were initially detectable at week 3 PI, and those in monkey 25958 were detectable at week 14 PI (Fig. 5). For both group III monkeys, anti-SIVgag IFN-γ-secreting cells were detectable in PBMC at multiple time points tested, beginning at 4 weeks PI for monkey 30395 and 14 weeks PI for monkey 32816 (Fig. 5).
A consistent finding among the seemingly divergent antiviral immune responses in all six of the animals intravaginally inoculated with the attenuated SIV deletion mutants is that the anti-SIV antibody responses became detectable only after plasma vRNA levels exceeded 103 copies/ml. In contrast, anti-SIV T-cell responses became detectable before vRNA levels reached this level, but they were generally detected after SIVmac239Δnef inoculation. T-cell responses were even found in one animal (30854) after SIVmac239Δnef inoculation, although plasma vRNA levels were <500 copies/ml at the time.
DISCUSSION
To characterize key aspects of the process of retroviral replication in vivo, including its potential contribution to the establishment of systemic infection following sexual transmission, we intravaginally inoculated rhesus macaques simultaneously or sequentially with attenuated SIV strains having complementary deletion mutations in their accessory genes. Under these circumstances, the only plausible mechanism for generating wild-type virus is through recombination. Demonstrating the dynamic and robust nature of the recombination process, chimeric viruses with wt sequences were found in the blood and tissues in four of six rhesus macaques after simultaneous or sequential intravaginal inoculation with SIVmac239Δvpx/Δvpr and SIVmac239Δnef. As both of these strains of virus are impaired in their abilities to replicate in vivo, the recombinant variants with repaired deletions would be expected to have a replicative advantage over both of the deleted parental viruses. In four of six animals, recombination-derived wt SIVmac239 sequences became dominant in blood and lymphoid tissues.
For animals inoculated sequentially, the order of inoculation and the relative extent of virus attenuation may have affected the pattern of recombination observed. Recombination-derived wt virus was detected within 2 weeks of inoculation in both animals simultaneously coinoculated with the two mutant viruses; wt sequences were the only ones detectable in plasma and PBMC by 8 weeks. Recombinant wt sequences were also demonstrated and eventually became dominant in plasma and PBMC in both macaques inoculated initially with SIVmac239Δvpx/Δvpr and then with SIVmac239Δnef. Interestingly, SIV variants with wt nef sequences were seen after SIVmac239Δnef infection of SIVmac239Δvpx/Δvpr-inoculated animal 25958, even though there was minimal evidence of SIVmac239Δvpx/Δvpr replication prior to challenge with the SIVmac239Δnef virus. Despite the eventual emergence of wt viral sequences, plasma viremia was lower than expected for wt virus, perhaps reflecting partial immune control attributable to responses induced prior to the emergence of wt virus, although neither humoral nor cellular immune responses were demonstrable in blood prior to challenge with SIVmac239Δnef (see below). The relatively low plasma vRNA levels in these animals were likely to be due to host factors rather than viral factors, since, for both animals, the inoculation of their plasma into naïve hosts resulted in a wt profile of plasma viremia.
In contrast to the results described above, we failed to detect recombination in monkeys inoculated first with SIVmac239Δnef and then with the more attenuated SIVmac239Δvpx/Δvpr (group III). Neither animal had SIVmac239Δvpx/Δvpr sequences in plasma or PBMC, despite multiple inoculations with this virus given to both SIVmac239Δnef-infected animals. The lack of ready recombination after inoculation with SIVmac239Δvpx/Δvpr in the setting of established SIVmac239Δnef infection may be due to the greater replicative capacity of SIVmac239Δnef relative to that of SIVmac239Δvpx/Δvpr, in combination with the effects of immune responses induced by SIVmac239Δnef infection (6, 9).
In animal 30395, virus replication resulted in the rapid emergence of a variant of SIVmac239Δnef that generated a truncated open reading frame of the nef gene. This variant, SIVmac239ΔΔnef, is similar to previously described and documented variants in monkeys chronically infected with SIVmac239Δnef (6); additional extensive deletions can also occur upon in vivo serial passage of SIVmac239Δnef (2, 17). Thus, rather than repairing the nef deletion in the inoculated SIVmac239Δnef variant through recombination with the wt nef of SIVmac239Δvpx/Δvpr, animals inoculated first with SIVmac239Δnef and then with SIVmac239Δvpx/Δvpr generated SIVmac239Δnef variants with compensatory mutations. These mutant SIVmac239Δnef variants appear to have eventually reached fixation in the in vivo viral population. The SIVmac239ΔΔnef variant (data not shown) that arose in animal 30395 is an extreme example of this type of compensatory mutation, but all the mutant SIVmac239Δnef variants arising in the first few weeks of infection likely contributed to the evolutionary adaptation of the SIV population in the group III animals.
All the monkeys in this study made systemic anti-SIV immune responses that became detectable after plasma vRNA levels exceeded 103 copies/ml. For both group I animals, recombination was detectable in blood by week 2 PI, well before anti-SIV antibodies were detected at 4 weeks PI or before anti-SIV CD8+ T cells could be detected in the tissues of monkeys inoculated intravaginally with wt SIVmac239 at week 3 PI (M. R. Reynolds, E. Rakasz, P. J. Skinner, C. White, K. Abel, M. Ma, L. Compton, G. Napoe, N. Wilson, C. J. Miller, A. Haase, and D. I. Watkins, submitted for publication). Thus, despite little apparent host immune pressure on the inoculated, replication-attenuated SIV deletion mutants, the wt SIVmac239 that arose by recombination quickly overgrew the infecting parental variants. For both group II animals, intravaginal inoculation with SIVmac239Δvpx/Δvpr produced systemic infection, as indicated by the presence of viral DNA in PBMC, but only one of the two animals produced a weak antibody response to this infection. Recombinant wt SIVmac239 in these animals became detectable 10 to 12 weeks after the intravaginal SIVmac239Δnef inoculation. For both group III animals, intravaginal inoculation with SIVmac239Δnef produced systemic infections with strong anti-SIV antibody and T-cell responses. Immune responses induced by the SIVmac239Δnef infection may have increased the resistance of the monkeys to the subsequent intravaginal SIVmac239Δvpx/Δvpr exposures (10 to 16 weeks PI), presumably in a manner similar to live attenuated lentiviral vaccines (1, 9, 10).
Taken together, these data show that recombination can occur readily after the intravaginal SIV inoculation of rhesus monkeys. The ability of recombination to repair damaged genes, thereby conferring a competitive advantage, is demonstrated in this study, although it should be noted that the significant effects on in vivo replicative capacity of the deletion mutants used may have resulted in particularly strong selection. This factor may have exaggerated the frequency or extent of recombination compared to what might be expected for less-impaired viruses. This study clearly demonstrates that the genotype of the virus producing the clinical systemic infection in monkeys intravaginally inoculated with multiple SIVmac variants can be the sum of the genotypes to which these animals were exposed. This finding may be relevant to the sexual transmission of HIV, as recombination may provide a mechanism for rapidly increasing the genetic diversity and potential fitness of virus populations of relatively limited diversity that have been described at early times following sexual transmission (11, 18).
Acknowledgments
The authors thank Lara Compton, Ding Lu, Blia Vang, Kristen Bost, and Rino Dizon of the Immunology Core Laboratory and Primate Services Units at the California National Primate Research Center and Julian Bess, Jr., AIDS Vaccine Program, SAIC Frederick, Inc., National Cancer Institute—Frederick, Frederick, Md., for excellent technical assistance.
This work was supported by Public Health Services grants U51RR00169, from the National Center for Research Resources, and R01 AI51596, from the National Institute of Allergy and Infectious Diseases. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. N01-CO-12400 (article H.36 of the prime contract).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Abel, K., L. Compton, T. Rourke, D. Montefiori, D. Lu, K. Rothaeusler, L. Fritts, K. Bost, and C. J. Miller. 2003. Simian-human immunodeficiency virus SHIV89.6-induced protection against intravaginal challenge with pathogenic SIVmac239 is independent of the route of immunization and is associated with a combination of cytotoxic T-lymphocyte and alpha interferon responses. J. Virol. 77:3099-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, L., P. O. Illyinskii, S. M. Lang, R. E. Means, J. Lifson, K. Mansfield, and R. C. Desrosiers. 2003. Determinants of increased replicative capacity of serially passaged simian immunodeficiency virus with nef deleted in rhesus monkeys. J. Virol. 77:6823-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434-439. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, J. A., R. J. Teufel II, P. D. Yin, and W. S. Hu. 1998. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J. Virol. 72:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bretscher, M. T., C. L. Althaus, V. Muller, and S. Bonhoeffer. 2004. Recombination in HIV and the evolution of drug resistance: for better or for worse? Bioessays 26:180-188. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti, L. A., K. J. Metzner, T. Ivanovic, H. Cheng, J. Louis-Virelizier, R. I. Connor, and C. Cheng-Mayer. 2003. A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Δnef increases viral replication. J. Virol. 77:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J. M. 1992. Genetic diversity and evolution of retroviruses. Curr. Top. Microbiol. Immunol. 176:143-164. [DOI] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1986. Genetic variation in AIDS viruses. Cell 46:1-4. [DOI] [PubMed] [Google Scholar]
- 9.Connor, R. I., D. C. Montefiori, J. M. Binley, J. P. Moore, S. Bonhoeffer, A. Gettie, E. A. Fenamore, K. E. Sheridan, D. D. Ho, P. J. Dailey, and P. A. Marx. 1998. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 72:7501-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 11.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 12.Fang, G., B. Weiser, C. Kuiken, S. M. Philpott, S. Rowland-Jones, F. Plummer, J. Kimani, B. Shi, R. Kaul, J. Bwayo, O. Anzala, and H. Burger. 2004. Recombination following superinfection by HIV-1. AIDS 18:153-159. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 14.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 16.Kellam, P., and B. A. Larder. 1995. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J. Virol. 69:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchhoff, F., H. W. Kestler III, and R. C. Desrosiers. 1994. Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene. J. Virol. 68:2031-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 21.Lu, X., H. Kiyono, D. Lu, S. Kawabata, J. Torten, S. Srinivasan, P. J. Dailey, J. R. McGhee, T. Lehner, and C. J. Miller. 1998. Targeted lymph-node immunization with whole inactivated simian immunodeficiency virus (SIV) or envelope and core subunit antigen vaccines does not reliably protect rhesus macaques from vaginal challenge with SIVmac251. AIDS 12:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malim, M. H., and M. Emerman. 2001. HIV-1 sequence variation: drift, shift, and attenuation. Cell 104:469-472. [DOI] [PubMed] [Google Scholar]
- 23.Marthas, M. L., D. Lu, M. C. Penedo, A. G. Hendrickx, and C. J. Miller. 2001. Titration of an SIVmac251 stock by vaginal inoculation of Indian and Chinese origin rhesus macaques: transmission efficiency, viral loads, and antibody responses. AIDS Res. Hum. Retrovir. 17:1455-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McChesney, M. B., J. R. Collins, D. Lu, X. Lu, J. Torten, R. L. Ashley, M. W. Cloyd, and C. J. Miller. 1998. Occult systemic infection and persistent simian immunodeficiency virus (SIV)-specific CD4+-T-cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 72:10029-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 26.Meyerhans, A., A. Jung, R. Maier, J. P. Vartanian, G. Bocharov, and S. Wain-Hobson. 2003. The non-clonal and transitory nature of HIV in vivo. Swiss Med. Wkly. 133:451-454. [DOI] [PubMed] [Google Scholar]
- 27.Neilson, J. R., G. C. John, J. K. Carr, P. Lewis, J. K. Kreiss, S. Jackson, R. W. Nduati, D. Mbori-Ngacha, D. D. Panteleeff, S. Bodrug, C. Giachetti, M. A. Bott, B. A. Richardson, J. Bwayo, J. Ndinya-Achola, and J. Overbaugh. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14(Suppl. 3):S129-S140. [PubMed] [Google Scholar]
- 29.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 31.Takehisa, J., L. Zekeng, E. Ido, Y. Yamaguchi-Kabata, I. Mboudjeka, Y. Harada, T. Miura, L. Kaptu, and M. Hayami. 1999. Human immunodeficiency virus type 1 intergroup (M/O) recombination in Cameroon. J. Virol. 73:6810-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wooley, D. P., R. A. Smith, S. Czajak, and R. C. Desrosiers. 1997. Direct demonstration of retroviral recombination in a rhesus monkey. J. Virol. 71:9650-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]