Abstract
Vibrio cholerae is known to persist in aquatic environments under nutrient-limiting conditions. To analyze the possible involvement of the alternative sigma factor encoded by rpoS, which is shown to be important for survival during nutrient deprivation in several other bacterial species, a V. cholerae rpoS homolog was cloned by functional complementation of an Escherichia coli mutant by using a wild-type genomic library. Sequence analysis of the complementing clone revealed an 1.008-bp open reading frame which is predicted to encode a 336-amino-acid protein with 71 to 63% overall identity to other reported rpoS gene products. To determine the functional role of rpoS in V. cholerae, we inactivated rpoS by homologous recombination. V. cholerae strains lacking rpoS are impaired in the ability to survive diverse environmental stresses, including exposure to hydrogen peroxide, hyperosmolarity, and carbon starvation. These results suggest that rpoS may be required for the persistence of V. cholerae in aquatic habitats. In addition, the rpoS mutation led to reduced production or secretion of hemagglutinin/protease. However, rpoS is not critical for in vivo survival, as determined by an infant mouse intestinal competition assay.
Vibrio cholerae O1, the causative agent of Asiatic cholera, has been isolated from coastal and estuarine environmental samples, both as free-living bacteria and in association with phytoplankton, zooplankton, crustaceans, and mollusks (26). These observations and the capacity of the organism to adaptively respond to changes in salinity, temperature, and the availability of nutrients (48, 49) have led to the idea that this species can successfully occupy one or more ecological niches in a variety of aquatic habitats. If this is so, then it seems likely that the physiology and structure of V. cholerae O1 in environmental reservoirs might differ in fundamental ways from these features of the organism in the intestine.
Recent studies support this view. Upon nutrient deprivation, comma-shaped V. cholerae cells become coccoid, significantly decrease their volume (4), and reduce their lipid, carbohydrate, RNA, and protein contents (22). Changes of this kind to nutrient deprivation have been observed in other bacterial species as well, most notably Escherichia coli, where, in addition to metabolic and structural changes, nutrient starvation also induces resistance to heat shock, oxidative stress, osmotic shock, and acid (27, 28, 31, 33, 35, 36).
The global response by E. coli to starvation and stress requires rpoS, which is induced to express an alternative sigma factor, ςS, during nutrient deprivation or the stationary phase of growth (31, 36). In turn, ςS governs the expression of multiple genes that mediate the observed changes in physiology and structure. These properties of the E. coli rpoS system have led us, in the work reported here, to examine the possibility that a V. cholerae homolog of rpoS exists and is required for survival of the organism under conditions of growth that it might encounter in environmental aquatic habitats. Here we report the cloning and characterization of V. cholerae rpoS (rpoSvc), the phenotype of a rpoSvc knockout mutant, and the unexpected finding that this mutant is markedly deficient in its capacity to produce and/or secrete an extracellular protease.
MATERIALS AND METHODS
Media and growth conditions.
All strains were maintained at −80°C in Luria-Bertani (LB) broth supplemented with glycerol (15%, vol/vol). For the experiments described below, the cells were grown aerobically at 37°C in LB broth, on MacConkey plates (Difco, Detroit, Mich.), in M9 minimal medium (37) with either no glucose or 0.02% (wt/vol) glucose, in artificial sea water (ASW; Sigma Chemical Co., St. Louis, Mo.), and on thiosulfate-citrate-bile salts-sucrose (TCBS; Difco) agar. The following supplements were added as appropriate: ampicillin (100 μg/ml), kanamycin (35 μg/ml), chloramphenicol (5 μg/ml), rifampin (100 μg/ml), and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml).
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are described in Table 1. Plasmids were introduced into E. coli strains by transformation and into V. cholerae strains through electroporation or through mating with E. coli S17-1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| V. cholerae | ||
| 92A1552 | El Tor, Inaba, wild type, Rifs | Laboratory collection |
| 92A1552-Rifr | El Tor, Inaba, wild type, Rifr | This study |
| FY1 | 92A1552-Rifr, rpoS::cm | This study |
| 3083 | El Tor, Ogawa, wild type | 18 |
| HAP-1 | El Tor, Ogawa, HA/protease-negative derivative of 3083 | 18 |
| E. coli | ||
| ZK918 | W3110 ΔlacU169 tna-2 rpoS::kan bolA::lacZ | 6 |
| DH5α | F′ endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 (φ80dlacZΔM15) | Laboratory collection |
| S17-1 (λ pir) | Tpr SmrrecA thi pro rK− mK+RP4:2-Tc:MuKm Tn7 λ pir | Laboratory collection |
| Plasmids | ||
| pBluescriptKS+ | Apr, ColE1, high-copy-number cloning vector | Stratagene |
| pGEM-7 ZF+ | Apr | Promega |
| pUC19/18 (HEH) | Apr tandem pUC19/18 multiple cloning site | Laboratory collection |
| pHP45Ω-Cm | Apr, Cmr, source of the cat gene | 12 |
| pKNG101 | Smr, oriR6K, mobRK2, sacB, suicide vector | 44 |
| pMMB67EH | Apr, IncQ broad-host-range cloning vector | 38 |
| pWKS130 | Kanr, low-copy-number cloning vector | Laboratory collection |
| pFY1 | pBluescriptKS+ with 4-kb V. cholerae Sau3A fragment containing the complete coding sequence of rpoS, 464 bp of nlpD, and 2,364 bp of mutS | This study |
| pFY2 | pBluescriptKS+ with 1.698-kb XbaI fragment containing complete coding sequence of rpoS, 464 bp of nlpD, and 44 bp of mutS | This study |
| pFY3 | pGEM-7 ZF+ with 1.698-kb XbaI fragment of pFY1 | This study |
| pFY4 | pUC19/18 with 1-kb PCR-amplified rpoS coding region | This study |
| pFY5 | pFY4 with rpoS coding region interrupted with cm cassette (rpoS::cm) | This study |
| pFY6 | pKNG101 containing rpoS::cm at SalI site | This study |
| pFY7 | pMMB67EH containing 1.698-kb XbaI fragment of pFY1 in opposite orientation to tac promoter | This study |
| pFY8 | pBluescriptKS+ containing 4-kb EcoRI fragment spanning whole coding region of nlpD and rpoS flanked with surE and mutS | This study |
| pFY9 | pWKS130 containing 4-kb EcoRI fragment spanning whole coding region of nlpD and rpoS flanked with surE and mutS | This study |
| pFY10 | pBluescriptKS+ containing 3-kb BamHI fragment spanning whole coding region of nlpD, its upstream region, and first 500 bp of rpoS | This study |
| pDEB2 | pUC19 containing E. coli rpoS | 6 |
DNA manipulations and analysis.
Plasmid DNA and chromosomal DNA preparation, DNA ligation, bacterial transformation, agarose gel electrophoresis, and Southern blotting were carried out by standard techniques described by Sambrook et al. (46). Restriction enzymes and DNA modification enzymes were purchased from New England Biolabs, Inc. (Beverly, Mass.). [α-32P]dCTP was obtained from Amersham Life Science Inc. (Arlington Heights, Ill.). Oligonucleotides were purchased from Operon Technologies, Inc.
Genomic library construction.
Wild-type V. cholerae 92A1552 DNA was partially digested with Sau3A and size selected by agarose gel electrophoresis using SeaKem (FMC Bioproducts, Rockland, Maine). DNA fragments of 3 to 6 kb were pooled and ligated into a multicopy vector (pBluescriptKS+) that had been digested with BamHI and treated with bacterial alkaline phosphatase. The ligation mixture was transformed into E. coli DH5α. The colonies were pooled, and the plasmid DNA from these pools was isolated. A V. cholerae genomic library was also constructed in λ DashII (Stratagene, La Jolla, Calif.), using wild-type 92A1552 genomic DNA which was partially digested with Sau3A and treated with bacterial alkaline phosphatase before being ligated into BamHI-digested λ DashII. The ligation mixture was packaged by using Gigapack II (Stratagene). The library was then screened with the rpoS gene to obtain clones that carry rpoS and nlpD and the upstream promoter region of nlpD. The positive clones were mapped by standard molecular techniques, and a 4-kb EcoRI fragment that contains whole rpoS and nlpD genes as well as its upstream region was identified. This fragment was cloned into pBluescriptKS+ and pWKS130 which had been digested with EcoRI, yielding pFY8 and pFY9, respectively.
Isolation of V. cholerae rpoS by functional complementation.
The wild-type V. cholerae Sau3A recombinant library in pBluescriptKS+ was introduced into E. coli ZK918 by transformation. Transformants were selected on MacConkey-lactose plates containing ampicillin (100 μg/ml) and kanamycin (35 μg/ml), on which colonies of the parent strain are white. Red colony transformants were tested for catalase activity, using H2O2 on LB plates. Plasmid DNA was isolated from red, catalase-positive transformants and analyzed by restriction enzyme digestions. Plasmids with overlapping fragments that were able to complement E. coli ZK918 were identified. A 1.698-kb XbaI fragment from pFY1 that complemented both lactose fermentation and catalase production in E. coli ZK918 was selected for sequence analysis.
DNA sequence analysis.
The 1.698-kb XbaI fragment was cloned into the XbaI site of pGEM-7 ZF+ (Promega, Madison, Wis.), generating plasmid pFY3. A series of plasmids containing nested deletions of the insert DNA, generated by using an Erase-a-Base kit (Promega), were used as a template for sequence analysis. Double-stranded DNA sequencing was performed with an Applied Biosystems 373A automated DNA sequencer. The remaining gaps in the sequence were filled, using specifically designed oligonucleotide primers. Sequence analysis and database searches were performed with IntelliGenetics (Menlo Park, Calif.) and Genetics Computer Group (Madison, Wis.) software packages and the NCBI BLAST server.
Construction of insertion mutants in rpoS.
A mutant strain, derived from V. cholerae 92A1552, which carries a chloramphenicol resistance (cm) cassette in the rpoS coding sequence was constructed as follows. A 1.022-bp PCR fragment containing the rpoS coding sequence was generated by using pFY2 as the template and the primers 5′-CGCGTCGACATGAGTGTCAGCAATACC-3′ and 5′-CGCGTCGACGTTGTCGTATTCGACGTT-3′. This fragment contained a 1-kb region from the rpoS start codon to the rpoS stop codon, which is flanked by SalI restriction sites which were used to clone into pUC19/18 that had been digested with SalI. The resulting plasmid, pFY4, was digested with BamHI and ligated to a cm cassette contained within a BamHI restriction fragment of pHP45WΩ-Cm (3.6 kb). The resulting plasmid, pFY5, was digested with SalI, and the resulting 4.6-kb fragment was cloned into pKNG101, yielding pFY6. FY6 was mobilized into the wild-type V. cholerae strain from E. coli S17-1 λ pir by conjugation.
Conjugal transfer was performed by mixing 1-ml aliquots of both donor and recipient cell cultures and then immediately concentrating the cells by filtration through a 0.45-μm-pore-size filter (Nalgene, Rochester, N.Y.). The filters were then placed onto LB plates and, after incubation at 37°C for 5 h, resuspended in LB and plated onto selective plates (LB agar plates supplemented with rifampin [100 μg/ml], chloramphenicol [5 μg/ml], and streptomycin [100 μg/ml]). Cointegrate conjugants containing no plasmid and representing a single homologous recombination were isolated, and the genotype was confirmed by Southern analysis. The strains were subjected to sucrose selection, and clones with characteristics indicative of a double homologous recombination event (streptomycin sensitive and sucrose resistant) were identified and further screened by Southern analysis for replacement of the wild-type rpoS sequence with the rpoS::cm construction.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis.
Total cell lysates were prepared from overnight grown cultures by sonication. Protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.). Lysates containing 25 μg of protein were separated on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels and directly electroblotted onto Immobilon-P membranes (Millipore, Bedford, Mass.). Blots were blocked and then incubated with an antiserum to E. coli RpoS followed by incubation with an anti-rabbit horseradish peroxide-conjugated secondary antibody. The blots were developed with the Amersham enhanced chemiluminescence system.
Survival assays.
Long-term starvation survival was analyzed in M9 medium supplemented with 0.02% glucose, in M9 without any carbon source, and in nutrient-free ASW basal salts medium. For these experiments, cultures were grown overnight in LB, centrifuged, washed, resuspended in 25 ml of the appropriate medium at a density of approximately 106 to 107 CFU ml−1, and incubated at 37°C aerobically (200 rpm). Immediately after inoculation and for up to 5 and 30 days, viable cell numbers were determined on LB plates after appropriate dilution. For hydrogen peroxide (15 mM) and osmolarity (NaCl, 2.4 M) survival studies, the overnight grown cultures in LB were collected by centrifugation, resuspended, and incubated under the indicated conditions. At the indicated time points, samples were taken and plated onto LB agar plates for determination of viable cell numbers.
2D gel electrophoresis.
Two-dimensional (2D) gel electrophoresis was performed by the method of O’Farrell (42). Cells were grown in M9 minimal medium supplemented with 0.2% glucose to mid-logarithmic phase and to stationary phase. For logarithmically grown cultures, 50 μCi of l-[35S]methionine (Du Pont NEN, Wilmington, Del.) was used to label a 1-ml sample for 2 min followed by a 1-min chase with 0.2 mM unlabeled methionine. Labeling time was increased to 30 min in the case of stationary-phase cultures, followed by a 15-min chase with unlabeled methionine. The samples were washed twice with phosphate-buffered saline. The amount of l-[35S]methionine incorporated was determined by trichloroacetic acid (10%) precipitation and liquid scintillation counting of a small portion of the cell lysate. Equal amounts of counts (106 cpm) were loaded onto the tube gels. 2D gel electrophoresis was carried out with a Protean II (Bio-Rad, Hercules, Calif.) device as directed by the manufacturer. Labeled proteins were visualized by autoradiography.
Protease assay and hemagglutinin/protease (HA/protease) Western analysis.
The initial screen for protease production was done on LB agar plates supplemented with 1% skim milk, where protease clearance zones of strains were analyzed. Following the initial screen, a fluorescence-based EnzCheck assay (Molecular Probes, Eugene, Oreg.) was used to determine protease activity in the samples. The BODIPY-casein substrate was incubated in a 96-well microplate with culture supernatants or with cell sonicates for 1 h at 37°C; fluorescence was measured (485 and 530 nm for excitation and emission, respectively) with a fluorescence microplate reader.
For analysis of the HA/protease by Western blotting, 5 ml of culture supernatant from overnight grown cultures was concentrated 25-fold by trichloroacetic acid precipitation, and 100 μl of the concentrated sample was separated on SDS–12% polyacrylamide gels and directly electroblotted onto Immobilon-P membranes (Millipore). Blots were blocked and then incubated with HA/protease-specific antiserum followed by incubation with an anti-rabbit horseradish peroxide-conjugated secondary antibody. The blots were developed with the Amersham enhanced chemiluminescence system.
In vivo assay for V. cholerae intraintestinal growth.
A competition assay for intraintestinal growth in 4- to 5-day-old CFW suckling mice was performed by administering mixtures containing equal numbers of the rpoS::cm mutant and wild-type V. cholerae parent, by gavage, into 4- to 5-day-old CFW mice. After 24 h, the small intestine was removed and intestinal homogenates were serially diluted and plated onto TCBS agar. Enumeration of mutant strain viable counts was performed after replica plating of recovered cells onto TCBS-chloramphenicol agar plates. Then the ratio of values for the mutant and wild-type strains was determined and used to derive the competitive index (15).
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this study is AF000945.
RESULTS
Cloning and characterization of V. cholerae rpoS.
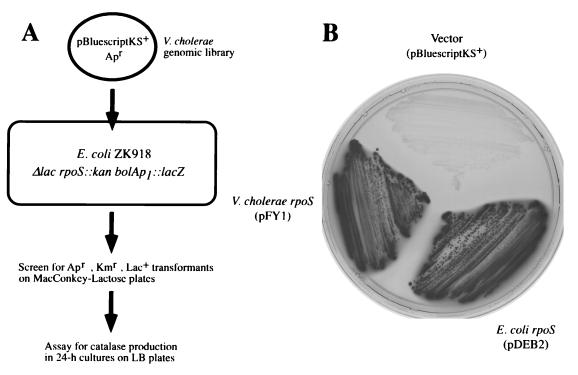
The V. cholerae O1 functional and structural homolog of E. coli rpoS was cloned by complementation using E. coli ZK918, a strain with an insertionally inactivated rpoS gene, a deletion in the lac operon, and a chromosomal bolAp1-lacZ fusion (6). bolAp1 requires rpoS for expression; it encodes a morphogene which is necessary for changes in bacterial shape that occur during starvation (6, 30). Lactose fermentation by this strain, as determined by the production of red colonies on MacConkey-lactose agar, does not normally occur but is regained when a functional copy of rpoS is provided. To clone rpoSvc, a genomic library was prepared in pBluescriptKS+ with DNA from V. cholerae 92A1552, a wild-type strain (El Tor biotype, Inaba serotype) isolated in 1992 from a traveler from Latin America (Table 1), and this library was introduced into E. coli ZK918. Red colonies were picked and tested for catalase production, since expression of katE, the HPII catalase structural gene that confers resistance to H2O2, is also controlled by rpoS. This complementation strategy is shown in Fig. 1A. When a plasmid containing either the E. coli rpoS gene (pDEB2) or the V. cholerae rpoS gene (pFY1) is present in E. coli ZK918, the recombinant ferments lactose, imparting a red color to the corresponding colony on MacConkey indicator plates (Fig. 1B). In contrast, the same host carrying the cloning vector (pBluescriptKS+) does not ferment lactose. The same recombinants shown in Fig. 1B were also tested for catalase production on LB agar; all but the strain containing the cloning vector tested positive.
FIG. 1.
(A) Schematic diagram of the complementation strategy. E. coli ZK918 carries an rpoS::cm insertion mutation and a bolAp1::lacZ fusion and has a deletion of the lac operon. This strain was transformed with a V. cholerae genomic library, and transformants that were lac+ were selected on MacConkey agar and then assayed for catalase activity. (B) Complementation by V. cholerae rpoS. E. coli ZK918 was transformed with a clone harboring V. cholerae rpoS (pFY1), vector alone (pBluescriptKS+), or E. coli rpoS clone (pDEB2). Ampicillin- and kanamycin-resistant transformants were streaked onto MacConkey-lactose plates, yielding lactose-positive (red) colonies (which appear darker).
This method led to the identification of four complementing plasmids; each was found to contain overlapping fragments, as determined by Southern analysis using a 4-kb insert of pFY1 as a probe (data not shown). The smallest of the subcloned complementing fragments, a 1.678-bp XbaI insert in pFY1, was cloned into pGEM-7 ZF+ for sequence analysis. Three open reading frames (ORFs) were identified and are shown schematically in Fig. 2A. One of these is predicted to contain a complete 1.008-bp coding sequence that would specify a 336-amino-acid protein with a calculated molecular mass of 37 kDa. Its predicted translation product was found to have homology with ςS sequences from the following species, here indicated by the percent similarity and percent identity, respectively: E. coli (77.5 and 70.9) (39), Salmonella typhimurium (77.8 and 71.1) (43), Shigella flexneri (75.2 and 68.0) (50), Yersinia enterocolitica (78.6 and 71.3) (25), Erwinia carotovora (77.1 and 70.1), and Pseudomonas aeruginosa (70.0 and 63.3) (51).
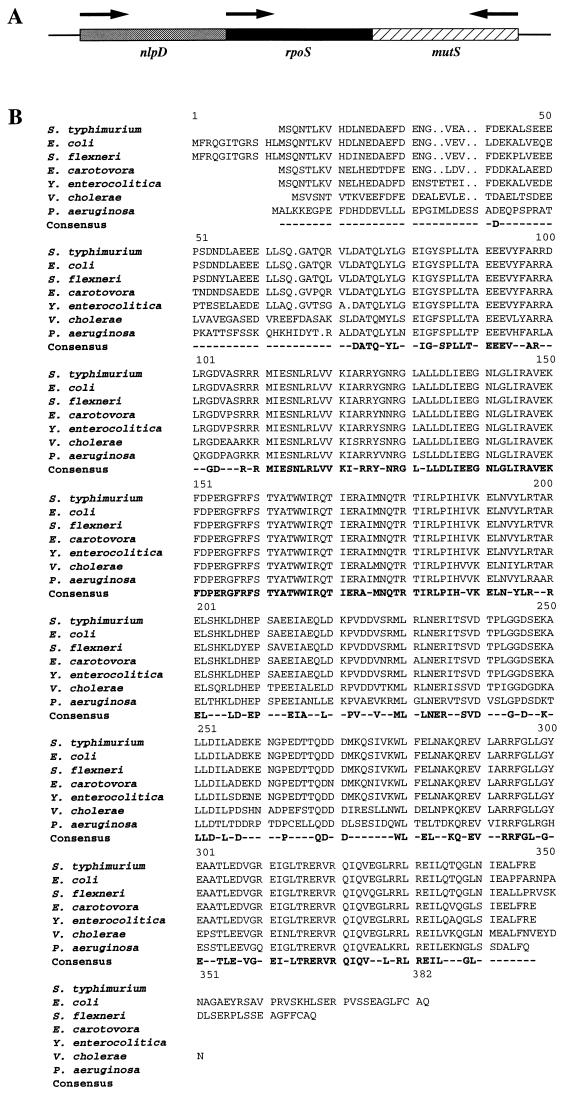
FIG. 2.
(A) Genomic organization of rpoS and its flanking genes on the V. cholerae O1 El Tor chromosome. rpoS is surrounded by nlpD and mutS, based on homologous sequences that flank E. coli rpoS. The transcriptional orientation of each gene is indicated by an arrow. (B) Alignment of amino acid sequences of RpoS from S. typhimurium (S47534), E. coli (Z14969), S. flexneri (U00119), E. carotovora (U66542), Y. enterocolitica (P47765), V. cholerae (this study), and P. aeruginosa (P45684). The amino acid sequences were retrieved from the databases via the accession numbers indicated in parentheses, and alignment was determined by the PileUp program of the Genetics Computer Group. Amino acids which are identical in all of these sequences are shown in the consensus. The V. cholerae RpoS translated sequence overall identity ranges from 60 to 71% for the other RpoS sequences shown.
The E. coli rpoS gene is reported to be flanked by nlpD and mutS, which encode a novel lipoprotein and a methyl-directed mismatch repair protein, respectively (24, 32, 34). Inspection of the cloned V. cholerae sequence shows an identical arrangement (Fig. 2A). Immediately upstream of rpoSvc and in the same orientation, we identified an incomplete ORF that is predicted to encode the carboxy-terminal 153 amino acids of a protein which exhibits 60.0% identity and 65.8% similarity to the NlpD protein of E. coli. The stop codon of nlpDvc is located 73 bp upstream of the predicted rpoS start codon. Directly downstream of rpoSvc and in the opposite orientation, a second incomplete ORF was identified; it is predicted to encode the carboxy-terminal 14 amino acids of a protein. To provide additional sequence, we identified an extra 71 amino acids in the carboxy-terminal region of the protein by using a pFY1 template and a specifically designed oligonucleotide primer. This led to the identification of this ORF by demonstrating that the carboxy-terminal 85 amino acids of the protein are 52% identical and 56% similar to the carboxy-terminal end of the MutS E. coli homolog. The stop codon of mutSvc is separated by 92 bp from the predicted stop codon of rpoSvc.
Insertional inactivation of rpoS.
To identify rpoS-associated phenotypes, we inactivated the rpoS gene of the wild-type V. cholerae strain 92A1552 by inserting the cm cassette at the BamHI site of a PCR-generated 1.002-kb fragment corresponding to the rpoS coding region. The resulting construct, rpoS::cm, interrupts translation at amino acid 149 of ςS and prevents transcription downstream of the insertion site. rpoS::cm was then cloned into suicide vector pKNG101, yielding plasmid pFY6. This plasmid was introduced into V. cholerae 92A1552 by conjugation, and Rifr, Smr, and Cmr transconjugates were selected. Two cointegrate transconjugants containing no plasmid and representing a single homologous recombination event were identified and used for sucrose selection to induce second-site recombination.
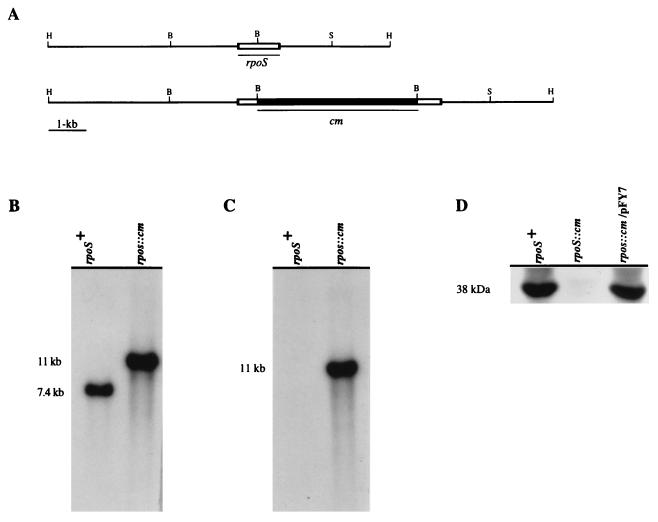
Southern analysis of one such mutant and of the wild-type parent strain is depicted in Fig. 3. Hybridization of a HindIII digest of chromosomal DNA using rpoSvc as a probe shows a single 7.4-kb fragment (Fig. 3B). This fragment was replaced with an 11-kb hybridizing fragment in the rpoS::cm mutant (Fig. 3B), consistent with the size expected from insertion of the 3.6-kb cm resistance cassette into the 7.4-kb fragment containing the native rpoS gene. When the cm cassette was used to probe the same digest, an 11-kb fragment was also detected in the rpoS::cm mutant (Fig. 3C), providing additional evidence that this fragment contains rpoS::cm.
FIG. 3.
Confirmation of the structure of the rpoS insertion mutation. (A) Physical maps of the rpoS regions of the chromosomes from the rpoS+ wild-type parent strain and the rpoS::cm mutant derivative. The two probes that were used for the hybridization studies shown in panels B and C are indicated under the restriction maps as rpoS and cm, respectively. Restriction enzyme sites: H, HindIII; S, SalI; B, BamHI. (B and C) Southern analysis of the parent and rpoS::cm insertion mutant. Chromosomal DNA was digested with HindIII, separated on a 0.8% agarose gel, blotted, and hybridized with the rpoS (B) or cm (C) probe. (D) Immunoblot analysis to detect of RpoS production by the parent strain (rpoS+), mutant (rpoS::cm), and complemented mutant, using pFY7 (rpoS::cm/pFY7). Total-cell extracts from the indicated strains were separated by SDS-PAGE (12% gel), blotted, and then probed with an antiserum to the RpoS protein of E. coli.
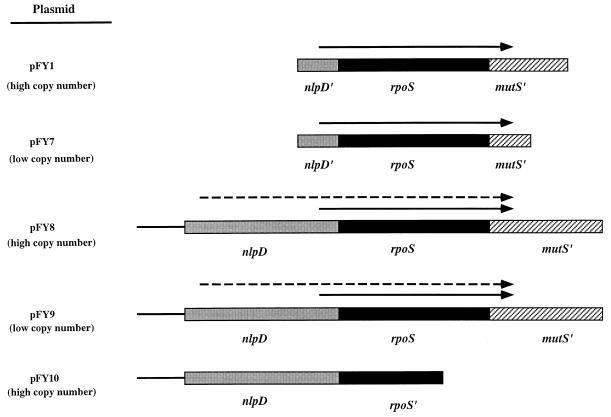
To determine if the rpoS-dependent phenotypes described below were attributable to disruption of the rpoS coding sequence or were instead due to another unrecognized mutation, we prepared four rpoS-containing plasmids to test if a normal rpoS gene could complement the rpoS::cm mutation. Two of these plasmids, designated pFY1 and pFY7, contain the entire rpoS structural gene and the stationary-phase inducible promoter region, by analogy to E. coli rpoS promoter structure analysis (Fig. 4). The other plasmids, pFY8 and pFY9, harbor a fragment that contains complete copies of rpoS and nlpD and the sequence upstream of nlpD (Fig. 4). By analogy with the E. coli rpoS promoter structure, pFY8 and pFY9 should contain both the inducible stationary-phase and exponential-phase promoters of rpoSvc. Both pFY1 and pFY8 are high-copy-number plasmids, whereas pFY7 and pFY9 are low-copy-number plasmids. To determine if nlpD, in the absence of an intact rpoS gene, could complement the rpoS mutant, a plasmid containing the entire coding sequence of nlpD, the sequence upstream of nlpD, and the first 500 bp of the rpoS gene was constructed. This plasmid, designated pFY10, was used to distinguish the rpoS-mediated complementation phenotype from a complemented phenotype that might come from provision of nlpD.
FIG. 4.
Diagram of the inserts used to construct the plasmids used for complementation analysis. pFY8 and pFY9 carry complete nlpD and rpoS genes and thus, by analogy to E. coli promoter analysis (29, 32), are predicted to carry both the logarithmic- and stationary-phase-driven promoters of rpoS, indicated by dashed and straight arrows, respectively. Both pFY1 and pFY7 carry only 464 bp of the nlpD gene and thus are predicted to encompass only the stationary-phase-driven promoter, which is indicated by the arrow. pFY7 and pFY9 are low-copy-number plasmids, whereas both pFY1 and pFY8 are high-copy-number plasmids. pFY10 harbors the entire coding sequence of nlpD, its upstream region, and the first 500 bp of rpoS. Construction of these plasmids is described in Materials and Methods.
The wild-type parent strain, the rpoS::cm mutant, and the mutant carrying complementing plasmid pFY7 were further characterized by Western blotting using an antiserum to the E. coli ςS protein. This antiserum cross-reacted with a ca. 38-kDa antigen in the lysate of V. cholerae 92A1552 (Fig. 3D, lane 1). The size of this protein is in good agreement with the molecular weight predicted from the translated sequence of rpoSvc and suggests that the ςS homologs of E. coli and V. cholerae O1 El Tor share antigenic determinants. In contrast, this antigen is not detected in a lysate of the rpoS::cm mutant (Fig. 3D, lane 2), providing further evidence that the rpoS coding sequence has been disrupted. Complementation was demonstrated with plasmid pFY7, which restored production of the 38-kDa antigen by the mutant (Fig. 3D, lane 3). These strains were then used to examine the function of rpoSvc in the studies reported below.
Survival of the rpoS::cm mutant during environmental stress.
rpoS mutants of E. coli, S. typhimurium, Y. enterocolitica, and S. flexneri have been studied by other investigators and found to be more susceptible than the corresponding wild-type strain to a variety of physicochemical conditions that are thought to simulate different host or natural environmental conditions of growth (3, 10, 25, 31, 35, 47, 50, 53). Among the tested conditions were carbon starvation, hyperosmolarity, heat shock, low pH, and oxidative stress. From these results, we reasoned that the V. cholerae rpoS::cm mutant might survive less well than the parent strain under conditions of nutrient deprivation (in particular, carbon starvation), hyperosmolarity, and oxidative stress. To exclude the possibility that the lack of ςS-mediated functions might affect the general fitness of the mutant during nonstressed cultivation, we compared its growth to that of the parent strain in LB broth at 37 and 30°C. No differences in viable plate counts were observed during the exponential or stationary phase of growth (data not shown).
Survival in nutrient-free ASW, adjusted to contain 2.5% (0.43 M) NaCl, was tested because of evidence indicating that V. cholerae can persist in estuarine and marine environments (23, 48, 49). When cells were cultivated in ASW at 37°C, the effect of the rpoS mutation was observable within 24 h, at which point survival of the rpoS::cm mutant was determined to be 6,000-fold less (based on viable plate counts) than that of the wild-type and complemented strains (Fig. 5A), indicating that ςS significantly enhances short-term survival of V. cholerae in seawater. The effect of the mutation on survival in ASW continued to be evident at 120 h, the last tested time point, at which time the percent survival of the wild-type and complemented strains remained approximately 200-fold higher than that of the mutant strain. Because the osmolarity of ASW is greater than optimal (2.5 to 5.0 mM in nutrient broth) for many strains of V. cholerae O1 (45), the result depicted in Fig. 5A may have been due to hyperosmolarity, carbon starvation, or a combined effect of the two. To address this question, the experiment was repeated with M9 medium without a carbon source. When incubated in M9 medium without a carbon source, both the wild-type parent and the complemented rpoS::cm mutant strain carrying pFY7 exhibited a reproducible initial increase in viable plate count during the first 24 h of incubation (Fig. 5B). In contrast, the viable plate count of the uncomplemented rpoS::cm mutant strain steadily decreased during the first 24 h, at which time its survival was approximately fivefold lower than that of the wild-type strain. These survival profiles suggest that an important contribution to the difference between the wild type and mutant during the first 24 h of carbon starvation in M9 medium is the failure of the mutant to transiently increase in apparent cell number, a phenomenon previously recognized in starvation studies of a marine vibrio strain designated Vibrio sp. strain S14 that has been attributed to starvation-induced reductive cell division (40, 41).
FIG. 5.
Effect of rpoS::cm on resistance to carbon starvation, hyperosmolarity, and H2O2. Stress survival of stationary-phase V. cholerae wild-type (open squares), rpoS::cm (filled squares), and rpoS::cm/pFY7 (open circles) bacteria was tested by incubating the cultures in carbon-free ASW (A), in M9 medium without added glucose (B), at high osmolarity (2.4 M NaCl, in LB) (C), and in the presence of H2O2 (15 mM) (D). At indicated times, the viable cell count was determined by plating dilutions onto LB; 100% survival corresponds to the viable cell count determined just prior to exposure to the indicated stress.
Comparison of the ASW and M9-without-glucose survival profiles depicted in Fig. 5A and B shows that the mutant survives less well than the wild-type strain in both media and that the effect of the rpoS::cm mutation is most evident during the first 24 h and is more pronounced in ASW than in M9 medium without glucose. These data suggest that both carbon starvation and hyperosmolarity affect the survival of the mutant. The significance of the former is suggested by a longer-term survival experiment in which the wild type, mutant, and complemented mutant were incubated for 30 days in M9 medium containing 0.02% glucose. Each of these strains survived for the 30-day period of observation, but the viable plate count of the rpoS mutant on day 30 was only 6.6% of that of its wild-type parent (data not shown). These findings suggest that the effect of carbon source exhaustion during the course of this 30-day experiment may have had a greater impact on the viable plate count of the mutant strain than of the counts of the wild-type and complemented strains.
To further examine the effect of the rpoS mutation on nutrition-dependent survival versus osmolarity-dependent survival, viable plate counts were determined for each of the three strains incubated in iso-osmolar LB broth or LB broth containing 14.1% (2.4 M) NaCl. The viable plate counts of all three strains declined rapidly in this high-salt solution, but at the end of the 60-min test period, survival of the rpoS::cm mutant was 2.5-fold less than that of the wild-type strain or the complemented mutant (Fig. 5C).
Finally, resistance to H2O2 was investigated to examine the role of V. cholerae rpoS in the oxidative stress response. Resistance to H2O2-mediated killing may be particularly important for aquatic organisms because H2O2 is generated by the action of UV radiation on water and therefore is expected to be produced within sun-illuminated aquatic habitats (2, 5). Incubation in 15 mM H2O2 for 30 min caused the percent survival of the rpoS mutant to decline by 99%. In contrast, percent survival of the wild-type strain and the complemented mutant declined by only 25% during the same time period (Fig. 5D).
rpoS-dependent protein expression.
To identify changes in the protein synthesis pattern caused by the rpoS::cm mutation, cultures were intrinsically labeled with [35S]methionine during both the exponential and stationary phases of growth, and expression of labeled proteins was determined by 2D gel electrophoresis.
Figure 6A and B shows the protein expression patterns from exponentially grown bacteria and reveal the absence of 11 proteins in the rpoS::cm mutant. In contrast, three proteins are expressed only by the mutant, and the expression of two more proteins is increased in the mutant compared to the wild-type strain. Differences in the patterns of expressed proteins were greater in stationary-phase cultures (Fig. 6C and D). Twenty-five proteins that are expressed by the wild-type strain were not detected in lysates of the stationary-phase rpoS::cm mutant; an additional two proteins are more strongly expressed by the wild-type than by the mutant. However, 14 proteins that are expressed by the rpoS::cm mutant during stationary phase were not identified among the proteins expressed by the wild-type parent. Thus, rpoS appears to positively and negatively affect the expression of multiple proteins, and while this effect is most evident during the stationary phase, it operates in the exponential phase as well.
FIG. 6.
2D gel electrophoresis pattern of [35S]methionine-labeled proteins in lysates of wild-type V. cholerae and rpoS::cm strains. Shown are protein expression patterns of the wild type during logarithmic growth (A), the rpoS::cm mutant during logarithmic growth (B), the wild type during stationary phase (C), and the rpoS::cm mutant during stationary phase (D). Bacteria were labeled with [35S]methionine either during logarithmic growth phase or during late stationary phase. Processing of cell extracts and 2D gel electrophoresis were performed as described in Materials and Methods. Proteins expressed only in rpoS+ strains during logarithmic growth or in stationary phase are shown by arrows in panels A and C, respectively. Proteins expressed only by the rpoS::cm mutant during logarithmic (B) or during stationary phase (D) are also indicated by arrows. Proteins whose expression is increased compared to that of the other strain during the specific growth condition are circled. IEF, isoelectric focusing. Sizes are given in kilodaltons on the left of each panel.
Because the lipopolysaccharide (LPS) molecule that carries the O1 antigen is a surface-accessible feature of the V. cholerae outer membrane and thus might interact with components of the environment that affect survival, we also qualitatively analyzed the LPS contents of the parent and rpoS::cm mutant strains. No differences were detected, each strain producing identical LPS ladders that are typical of wild-type V. cholerae O1 LPS when analyzed on silver-stained SDS-polyacrylamide gels (data not shown).
Effect of rpoS on the production of the HA/protease.
During the course of these experiments, we observed that SDS-PAGE analysis of V. cholerae outer membrane proteins revealed marked degradation of many proteins in lysates of the wild-type strain but not in lysates prepared with the rpoS::cm mutant. We reasoned that the rpoS mutation may have affected either the secretion or production of one or more of the previously described V. cholerae O1 proteases (55). This possibility was examined by measuring the protease activity secreted into culture supernatants by the wild-type strain, the rpoS::cm mutant, and the mutant with complementing plasmids. The results, shown in Table 2, demonstrate that the rpoS::cm mutant produces and/or secretes ∼108-fold less protease activity than the wild-type parent strain. The absence of protease activity in the mutant was partially, but not completely restored, by providing either pFY1, pFY8, or pFY9; the resulting complemented strains then exhibited 37.8-, 5.9-, and 9.8-fold, respectively, less protease activity than the parent strain. In contrast, pFY7, which had complemented the stress-related phenotypes shown in Fig. 5, did not complement the protease-deficient phenotype of the mutant. This result points to a possible difference between the transcriptional control of rpoS for genes conferring resistance to stress versus genes required for protease production or secretion. Alternatively, the difference between the protease complementation capacities of pFY1 and pFY7 may have been due to differences in copy number between the two plasmids: pFY1 is present in high copy number, whereas pFY7 is present in low copy number. As expected, when only the cloning vectors pBluescript KS+ and pWKS130 were provided to the mutant strain, no increase in protease activity was detected.
TABLE 2.
Effect of rpoS::cm on protease production
| Strain | Plasmid | Protease productiona
|
|
|---|---|---|---|
| Medium | Cells | ||
| 92A1552 (wild type) | None | 1,028.56 ± 58.60 | 9.76 ± 0.29 |
| 92A1552 (rpoS::cm) | None | 9.5 ± 2.1 | 10.29 ± 0.25 |
| pBKS | 7.29 ± 3.78 | 10.60 ± 0.14 | |
| pFY1 | 27.17 ± 7.90 | 11.60 ± 0.25 | |
| pFY7 | 5.37 ± 3.59 | 12.56 ± 0.33 | |
| pFY8 | 173.48 ± 18.45 | 10.63 ± 0.36 | |
| pFY9 | 104.83 ± 13.77 | 11.52 ± 0.28 | |
| pFY10 | 7.95 ± 4.23 | 10.91 ± 0.31 | |
| pWKS130 | 5.17 ± 3.52 | 8.55 ± 0.32 | |
Means ± standard deviations from the means (n = 4); determined by measurement of fluorescence (per A600 unit) of BODIPY FL peptides which are generated by hydrolysis of BODIPY FL casein. Supernatants of cultures grown in LB for 24 h and the cell sonicates of them were used for analysis.
To determine if the failure to provide complete complementation of protease activity by pFY8 or pFY9 might have been due to an unrecognized mutation of nlpD, arising as an unintentioned complication of the mutational methods used, complementation was also attempted with pFY10, which carries a complete copy of nlpD but only the 5′ end of rpoS. The result of this study, shown in Table 2, indicates that nlpD by itself is unable to complement the protease-deficient phenotype of the mutant. Thus, only plasmids containing a complete rpoS gene provided complementation.
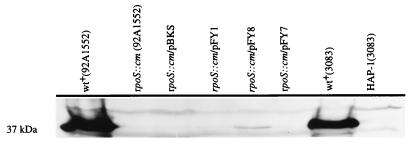
The protease assay that we used in these experiments did not specifically identify the affected protease as the zinc- and calcium-dependent HA/protease described by Hase and Finkelstein (18, 19). We addressed this issue by Western blot analysis of the culture supernatants, using an antiserum to the purified HA/protease (Fig. 7). The supernatant fraction of the wild-type strain contained a prominent 37-kDa antigen that was not detected in the supernatant of the mutant strain. In addition, the amount of the 37-kDa antigen was greatly diminished in the mutant strain carrying pFY8 and was below detection in the mutant strain carrying pFY1 and pFY7, results that are in agreement with the protease activity measurements reported in Table 2. The estimated size of this protein (37 kDa) is close to, but not identical with, the reported size of the fully processed and secreted HA/protease (32 kDa) (18, 19). However, we were able to attribute this discrepancy in size to probable technical differences in the SDS-PAGE methods used because an HA/protease-negative mutant (HAP-1) demonstrated the selective absence of a protein antigen of the same estimated molecular mass (Fig. 7). Taken together, these results show that the culture supernatant of the rpoS::cm mutant contains substantially less of the HA/protease than the wild-type strain.
FIG. 7.
HA/protease production and/or secretion. Immunoblot analysis using an antiserum to the HA/protease was performed on concentrated culture supernatants of the wild-type (wt+) strain, the rpoS::cm mutant, and the mutant strain carrying rpoS-complementing plasmids. HA/protease-negative mutant (HAP-1) was included as a negative control together with its wild-type parent (3083), which served as a positive control.
We have not definitively determined if the decreased protease activity of the mutant is due to decreased production or decreased secretion. However, we have measured the ratio of protease activity in the cell pellet and in the supernatant of the wild-type parent strain, of the rpoS mutant, and of each of the complemented mutants. The results of this experiment are depicted in Table 2. For each of the tested strains, only basal levels of protease activity were detected in the cell pellet. In particular, no accumulation of protease activity was detected in the cell pellet of the rpoS mutant. This result suggests that the rpoS mutation primarily effects production of the HA/protease. However, it is also possible that autodigestion of nonsecreted protease occurred, leading to the appearance of basal levels of enzyme activity in the cell pellet of the mutant strain.
Intraintestinal survival of the V. cholerae rpoS mutant.
To analyze the importance of RpoS for intraintestinal survival, we tested the intestinal colonizing capacity of the mutant and the wild-type parent strain. Equal numbers (3.7 × 105 CFU) of the wild-type parent and the rpoS::cm mutant were combined and administered by gavage to 4- to 5-day-old CFW mice. Twenty hours later, the animals were sacrificed, the small intestine was removed and homogenized, and dilutions of the homogenate were plated onto TCBS agar plates. Individual colonies were then replicate plated onto LB agar containing chloramphenicol (to select for the rpoS::cm mutant) and LB agar without antibiotic (for growth of both the antibiotic-sensitive parent strain and the rpoS mutant). The output ratio of the wild type to the rpoS mutant strain surviving in the murine intestine was then calculated from these plate counts. Using an initial inoculum containing a ratio of wild type to rpoS mutant of 1.0, the output ratio was found to be 0.97 ± 0.49 (average ± standard deviation, n = 12). This result indicates that rpoS is not required for intraintestinal survival under the conditions tested.
DISCUSSION
We have shown that an rpoS mutant of a recently isolated V. cholerae O1 El Tor strain is less resistant than its wild-type parent to carbon starvation, hyperosmolarity, and oxidative stress. Further, based on the 2D gel electrophoresis results shown in Fig. 6, we have demonstrated that rpoSvc seemingly regulates the expression of 25 or more genes during stationary-phase growth. In these respects, the function of rpoSvc appears to be similar to that of the well-studied rpoS homolog of E. coli, which also regulates the expression of multiple genes, some of which participate in the general stress response (katE, xth, appA, and mcc) (33), are required for starvation-induced changes in cell shape (bolA) (30), or confer an adaptive response to hyperosmolarity (osmB, osmE, otsAB, and treA) (20, 21).
In other respects, the role of rpoS in V. cholerae seems to differ significantly from its apparent role in two enteric pathogenic genera, Salmonella and Shigella. This is exemplified by our finding that the V. cholerae rpoS mutant and the parent strain survive equally well in the murine small intestine. In contrast, an S. typhimurium rpoS mutant was found to be attenuated for virulence in mice (11, 54), probably because it controls the expression of spv plasmid virulence genes that are induced during stationary-phase growth and in the intracellular environment (11). However, further analysis of these contrasting in vivo results reveals that they may not be due to essential differences in the physiological role of rpoS in the two species but rather may have resulted from differences in how each species interacts with the murine intestinal tract. V. cholerae does not normally enter host cells during an infectious episode. Consequently, our use of the murine model was intended to measure rpoS-dependent adaptation to an extracellular, intraintestinal environment that is rich in nutrients, is iso-osmolar, and probably lacks oxidative stress. In contrast, S. typhimurium causes murine enteric fever, which requires survival within host macrophages, an environment that entails oxidative stress through the production of oxygen free radicals. Thus, these seemingly different results may in fact point to a common function for rpoS because resistance to oxidative stress, whether generated by UV radiation in an aquatic environment (2), as may be the case for V. cholerae, or by the oxidative burst of a phagocyte, in both species may require rpoS.
The Shigella rpoS homolog has also been assigned a role in pathogenesis that does not seem to be shared by the V. cholerae homolog. rpoS is required for the resistance of Shigella to acid and thus is plausibly responsible for the small inoculum required to produce shigellosis in humans by stimulating a protective response by organisms transiting the acidic compartment of the stomach (50, 53). We could identify no such function for rpoSvc (data not shown), perhaps in part because V. cholerae is intrinsically only very weakly resistant to acid.
The most striking functional difference between the rpoS homologs of V. cholerae and most other enteric pathogens is the requirement of rpoSvc for the production and/or exportation of the HA/protease. We could find only one other example of this function for rpoS in another species, namely, the requirement of rpoS for the expression by Y. enterocolitica of yst, which encodes a secreted heat-stable enterotoxin (25). In the case of V. cholerae, we have not elucidated the mechanism by which rpoSvc subserves this function. Possibilities that are now under study include a direct effect by rpoS on the transcription of the HA/protease coding sequence or through an indirect effect on the transcription of the hap gene by affecting the levels of its transcriptional activator(s).
We have no explanation for the capacity of pFY1, pFY7, pFY8, and pFY9 to complement the rpoS::cm mutant for resistance to H2O2, carbon starvation, and hyperosmolarity but to compensate rather poorly for the effect of the rpoS mutation on the production and/or secretion of the HA/protease (Table 2). In trying to understand this difference, we reasoned that the promoter of rpoS that activates its transcription during the exponential phase (by analogy to the E. coli rpoS promoter) also might be required to complement the HA/protease phenotype. In contrast, the clone carrying less of the 5′ upstream promoter region, which by analogy to E. coli would encompass the stationary-phase promoter (carried by pFY7), might suffice to complement those functions most directly related to survival during stress and carbon starvation. Although the longer clone (carried by pFY8 and pFY9), which we predicted would contain the exponential-phase promoter of rpoSvc, provided better complementation of the HA/protease phenotype, the levels of complementation attained with these plasmids were still 6- to 10-fold less than that for the parent strain. We further ruled out plasmid copy number as a probable cause for partial complementation, since pFY9, which is a low-copy-number plasmid, also provided only incomplete complementation of the protease activity. Explanations for these discrepant results that are now under investigation include the possible differences in the expression of chromosomal versus episomal copies of the rpoS gene and a possible growth phase control over either the secretion or production of HA/protease.
The effect of rpoS on the secretion and/or production of the HA/protease suggests that this sigma factor could be important for the pathogenesis of cholera. Cholera toxin is the quintessential virulence determinant of V. cholerae O1, and its A subunit may need to be activated by the HA/protease before it can function as a secretogogue (14). Therefore, in this manner, its biological activity, if not its production and secretion, may require HA/protease. HA/protease was once considered to be a virulence determinant in its own right because of its capacity to degrade mucin, fibronectin, and lactoferrin (14, 17). However, because an HA/protease-negative mutant was found to be as virulent as the parent strain when tested in an infant rabbit model, it is now believed to function in vivo to detach V. cholerae from epithelial cell surfaces by degrading the receptors for bacterial adhesins (13). This “detachase” role would allow the bacteria to exit an infected host and reenter environmental reservoirs. Alternatively, and in accord with our starvation survival results which suggest a role for rpoS in the survival of V. cholerae in aquatic habitats, another potential function of the HA/protease is the digestion of proteinaceous dissolved organic matter in water, leading to the liberation of amino acid nutritional substrates. Indeed, studies of a marine vibrio have shown that it secretes proteases under conditions of carbon starvation (1), but the possible rpoS dependency of this phenomenon has not been reported. It will be interesting to explore this possibility by comparing the survival of V. cholerae strains mutated in rpoS or the HA/protease gene with the wild-type parent strains in laboratory microcosms that contain complex macromolecules as dissolved organic matter, in concentrations found in natural aquatic habitats, as the only extrinsic source of carbon and nitrogen.
We undertook this study with the preconception that the life style of V. cholerae takes place in one of two loci: the nutrient-rich intestinal lumen or in nutrient-poor aquatic habitats. We further hypothesized that many of the organism’s most highly regulated genes would belong either to the pathogenic set of genes (expressed mainly in the intestine) or the environmental set of genes (expressed mainly in contaminated water). Those genes regulated by ToxR, including the tcp operon and ctxAB, would surely belong to the former set (7–9, 16, 52). On balance, rpoS belongs more in the latter set, an assignment supported by its effect on carbon starvation survival, resistance to hyperosmolarity, and H2O2, and its apparent lack of effect on intraintestinal survival, and its far more potent effect on gene expression in stationary-phase growth than in exponential-phase growth.
ACKNOWLEDGMENTS
We thank Chon Martinez and Roberto Kolter for providing E. coli ZK918 and pDEB2; A. C. Matin for providing RpoS antibody; Richard Finkelstein for providing V. cholerae 3083 and HAP-1 and HA/protease antibody; and Chengyen Wu for help with HA/protease Western analysis.
REFERENCES
- 1.Albertson N H, Nystrom T, Kjelleberg S. Exoprotease activity of two marine bacteria during starvation. Appl Environ Microbiol. 1990;56:218–223. doi: 10.1128/aem.56.1.218-223.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arana I, Muela A, Iriberri J, Egea L, Barcina I. Role of hydrogen peroxide in loss of culturability mediated by visible light in Escherichia coli in a freshwater ecosystem. Appl Environ Microbiol. 1992;58:3903–3907. doi: 10.1128/aem.58.12.3903-3907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badger J L, Miller V L. Role of rpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J Bacteriol. 1995;177:5370–5373. doi: 10.1128/jb.177.18.5370-5373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker R M, Singleton F L, Hood M A. Effects of nutrient deprivation on Vibrio cholerae. Appl Environ Microbiol. 1983;46:930–940. doi: 10.1128/aem.46.4.930-940.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barcina I, Gonzalez J M, Iriberri J, Egea L. Effect of visible light on progressive dormancy of Escherichia coli cells during the survival process in natural fresh water. Appl Environ Microbiol. 1989;55:246–251. doi: 10.1128/aem.55.1.246-251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohannon D E, Connell N, Keener J, Tormo A, Espinosa-Urgel M, Zambrano M M, Kolter R. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of sigma 70. J Bacteriol. 1991;173:4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 8.DiRita V J. Multiple regulatory systems in Vibrio cholerae pathogenesis. Trends Microbiol. 1994;2:37–38. doi: 10.1016/0966-842x(94)90122-8. [DOI] [PubMed] [Google Scholar]
- 9.DiRita V J, Parsot C, Jander G, Mekalanos J J. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenstark A, Calcutt M J, Becker-Hapak M, Ivanova A. Role of Escherichia coli rpos and associated genes in defense against oxidative damage. Free Radical Biol Med. 1996;21:975–993. doi: 10.1016/s0891-5849(96)00154-2. [DOI] [PubMed] [Google Scholar]
- 11.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein R A, Boesman-Finkelstein M, Chang Y, Hase C C. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect Immun. 1992;60:472–478. doi: 10.1128/iai.60.2.472-478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkelstein R A, Boesman-Finkelstein M, Holt P. Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F. M. Burnet revisited. Proc Natl Acad Sci USA. 1983;80:1092–1095. doi: 10.1073/pnas.80.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freter R, O’Brien P C. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: fitness and virulence of nonchemotactic Vibrio cholerae mutants in infant mice. Infect Immun. 1981;34:222–233. doi: 10.1128/iai.34.1.222-233.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardel C L, Mekalanos J J. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 17.Hase C C, Finkelstein R A. Bacterial extracellular zinc-containing metalloproteases. Microbiol Rev. 1993;57:823–837. doi: 10.1128/mr.57.4.823-837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hase C C, Finkelstein R A. Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J Bacteriol. 1991;173:3311–3317. doi: 10.1128/jb.173.11.3311-3317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hase C C, Finkelstein R A. Comparison of the Vibrio cholerae hemagglutinin-protease and the Pseudomonas aeruginosa elastase. Infect Immun. 1990;58:4011–4015. doi: 10.1128/iai.58.12.4011-4015.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 21.Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J Bacteriol. 1993;175:259–265. doi: 10.1128/jb.175.1.259-265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood M A, Guckert J B, White D C, Deck F. Effect of nutrient deprivation on lipid, carbohydrate, DNA, RNA, and protein levels in Vibrio cholerae. Appl Environ Microbiol. 1986;52:788–793. doi: 10.1128/aem.52.4.788-793.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huq A, West P A, Small E B, Huq M I, Colwell R R. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar O1 associated with live copepods in laboratory microcosms. Appl Environ Microbiol. 1984;48:420–424. doi: 10.1128/aem.48.2.420-424.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichikawa J K, Li C, Fu J, Clarke S. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam M S, Drasar B S, Sack R B. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 27.Jenkins D E, Schultz J E, Matin A. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J Bacteriol. 1988;170:3910–3914. doi: 10.1128/jb.170.9.3910-3914.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 29.Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 32.Lange R, Hengge-Aronis R. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol Microbiol. 1994;13:733–743. doi: 10.1111/j.1365-2958.1994.tb00466.x. [DOI] [PubMed] [Google Scholar]
- 33.Loewen P C, Hengge-Aronis R. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 34.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 36.McCann M P, Kidwell J P, Matin A. The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 38.Morales V M, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene (Amsterdam) 1991;97:39–48. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 39.Mulvey M R, Loewen P C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 1989;17:9979–9992. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nystrom T, Flardh K, Kjelleberg S. Responses to multiple-nutrient starvation in marine Vibrio sp. strain CCUG 15956. J Bacteriol. 1990;172:7085–7097. doi: 10.1128/jb.172.12.7085-7097.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nystrom T, Kjelleberg S. Role of protein synthesis in the cell division and starvation induced resistance to autolysis of a marine Vibrio during the initial phase of starvation. J Gen Microbiol. 1989;135:1599–1606. [Google Scholar]
- 42.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 43.Prince R W, Xu Y, Libby S J, Fang F C. Cloning and sequencing of the gene encoding the rpoS (katF) sigma factor from Salmonella typhimurium 14028. Biochim Biophys Acta. 1994;1219:198–200. doi: 10.1016/0167-4781(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 44.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 45.Reichelt J L, Baumann P. Effect of sodium chloride on growth of heterotrophic marine bacteria. Arch Microbiol. 1974;97:329–534. doi: 10.1007/BF00403071. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Seymour R L, Mishra P V, Khan M A, Spector M P. Essential roles of core starvation-stress response loci in carbon-starvation-inducible cross-resistance and hydrogen peroxide-inducible adaptive resistance to oxidative challenge in Salmonella typhimurium. Mol Microbiol. 1996;20:497–505. doi: 10.1046/j.1365-2958.1996.5451068.x. [DOI] [PubMed] [Google Scholar]
- 48.Singleton F L, Attwell R, Jangi S, Colwell R R. Effects of temperature and salinity on Vibrio cholerae growth. Appl Environ Microbiol. 1982;44:1047–1058. doi: 10.1128/aem.44.5.1047-1058.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singleton F L, Attwell R W, Jangi M S, Colwell R R. Influence of salinity and organic nutrient concentration on survival and growth of Vibrio cholerae in aquatic microcosms. Appl Environ Microbiol. 1982;43:1080–1085. doi: 10.1128/aem.43.5.1080-1085.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Small P, Blankenhorn D, Welty D, Zinser E, Slonczkewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka K, Takahashi H. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 52.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2839. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterman S R, Small P L C. Identification of sigma-S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 54.Wilmes-Riesenberg M R, Foster J W, Curtiss R., III An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young D B, Broadbent D A. Biochemical characterization of extracellular proteases from Vibrio cholerae. Infect Immun. 1982;37:875–883. doi: 10.1128/iai.37.3.875-883.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]