Abstract
Bladder cancer and osteosarcoma are 2 types of cancers that originate from epithelial tissues inside the bladder and bone or muscle tissues. Ultrasound-guided biopsies provide crucial support for the diagnosis and treatment of bladder cancer and osteosarcoma. However, the relationship between myosin light chain kinase (MYLK) and caldesmon (CALD1) and bladder cancer and osteosarcoma remains unclear. The bladder cancer datasets GSE65635 and GSE100926, the osteosarcoma dataset GSE39058, were obtained from gene expression omnibus. Differentially expressed genes (DEGs) were screened and weighted gene co-expression network analysis was performed. The construction and analysis of protein-protein interaction network, functional enrichment analysis, gene set enrichment analysis. Gene expression heat map was drawn and immune infiltration analysis was performed. The comparative toxicogenomics database analysis were performed to find disease most related to core gene. Western blotting experiments were performed. TargetScan screened miRNAs that regulated central DEGs. We obtained 54 DEGs. Functional enrichment analysis revealed significant enrichment in terms of cellular differentiation, cartilage development, skeletal development, muscle actin cytoskeleton, actin filament, Rho GTPase binding, DNA binding, fibroblast binding, MAPK signaling pathway, apoptosis, and cancer pathways. Gene set enrichment analysis indicated that DEGs were primarily enriched in terms of skeletal development, cartilage development, muscle actin cytoskeleton, MAPK signaling pathway, and apoptosis. The immune infiltration analysis showed that when T cells regulatory were highly expressed, Eosinophils exhibited a similar high expression, suggesting a strong positive correlation between T cells regulatory and Eosinophils, which might influence the disease progression in osteosarcoma. We identified 6 core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1). MYLK and CALD1 were significantly correlated with survival rate and exhibited lower expression in bladder cancer and osteosarcoma samples compared to normal samples. Comparative toxicogenomics database analysis results indicated associations of core genes with osteosarcoma, bladder tumors, bladder diseases, tumors, inflammation, and necrosis. The results of Western blotting showed that the expression levels of MYLK and CALD1 in bladder cancer and osteosarcoma were lower than those in normal tissues. MYLK and CALD1 likely play a role in regulating muscle contraction and smooth muscle function in bladder cancer and osteosarcoma. The lower expression of MYLK and CALD1 is associated with poorer prognosis.
Keywords: biomarker, CALD1, carcinoma of urinary bladder, MYLK, osteosarcoma, ultrasound-guided
1. Introduction
Bladder cancer is a relatively common cancer with varying incidence rates in different countries. It typically originates from the mucous membrane inside the bladder. The most common symptoms include hematuria (blood in the urine), urinary frequency, urgency, and lower abdominal pain. Bladder cancer can be categorized into different subtypes, including non-muscle-invasive and muscle-invasive types. In the early stages, bladder cancer may not present obvious symptoms, but over time, it can progressively invade deeper layers of the bladder wall and even spread to surrounding tissues and lymph nodes.[1] In advanced stages, bladder cancer can also metastasize to distant organs such as the lungs, liver, and bones, increasing the complexity of treatment and the risk of survival. Patients with bladder cancer often experience discomfort in urination, frequent urination, urgency, lower abdominal pain, and hematuria, which can impact their quality of life and cause pain.[2] The treatment of bladder cancer typically involves surgical procedures, which may require partial or complete removal of the bladder, significantly affecting patients lifestyles. Even after initial control of bladder cancer, it has a high recurrence rate, necessitating regular follow-up and monitoring to ensure disease stability.[3,4]
Osteosarcoma is a rare bone tumor that primarily affects children and young adults. Its incidence is relatively low. Osteosarcoma is a highly aggressive bone tumor commonly found in the proximal ends of long bones.[5] It is typically characterized by bone fractures, bone pain, and the formation of masses. Patients with osteosarcoma usually experience symptoms such as bone pain, swelling, joint inflammation, and limited mobility. These symptoms may worsen at night. Osteosarcoma originates from primitive osteoblasts or stem cells and is pathologically characterized by highly heterogeneous cellular tissue.[6] Histological analysis is often used to determine the tumor type. Osteosarcoma typically grows rapidly, leading to the rapid destruction of affected bones, which can result in fractures, mass formation, and pain.[7] Pain is one of the most common symptoms of osteosarcoma, and it often worsens at night or during activity, causing severe discomfort and significantly impacting the quality of life.[8]
The exact etiology of bladder cancer and osteosarcoma remains incompletely understood, with both diseases potentially being associated with genetic factors, chromosomal abnormalities, gene fusions, and other factors. Therefore, research into early diagnostic targets is crucial, as early diagnosis can enhance treatment success rates and patient survival chances. In-depth molecular biology and genetics studies can help unveil the pathogenesis of these 2 cancers and may aid in the development of more precise treatment approaches.
Ultrasound-guided biopsies play a vital role in helping physicians obtain tissue samples from suspicious tumors, which can be subjected to pathological analysis, leading to a definitive diagnosis and aiding in the early detection of bladder cancer and osteosarcoma.[9] Biopsies provide insights into the nature and characteristics of tumors, facilitating the selection of the most suitable treatment methods and improving treatment efficacy. For known patients with bladder cancer or osteosarcoma, biopsies can also be employed to monitor disease progression and treatment responses, as well as to assess the risk of recurrence.[10]
Various genomic techniques such as whole genome sequencing, exon sequencing, and single-cell sequencing have been widely applied in genomics research, enabling the exploration of gene-disease associations. Hence, utilizing bioinformatics can be instrumental in analyzing gene expression patterns in tissue samples of bladder cancer and osteosarcoma, allowing for the identification of potential biomarkers, aiding in subtype differentiation, and predicting disease progression and treatment response.[11]
Myosin light chain kinase (MYLK) is an enzyme that controls muscle contraction and relaxation by regulating the phosphorylation of the light chain of myosin in muscle cells. The protein encoded by caldesmon (CALD1) is caldesmon, which is also related to muscle contraction. However, the current understanding of the relationship between MYLK, CALD1, and bladder cancer and osteosarcoma remains unclear. Therefore, this study aims to utilize bioinformatics techniques to unearth the core genes differentiating bladder cancer and osteosarcoma from normal tissue. Enrichment analysis and pathway analysis will be conducted, and the significant roles of MYLK and CALD1 in bladder cancer and osteosarcoma will be validated using public datasets. Additionally, basic cell experiments will be applied to validate these findings.
2. Methods
2.1. Bladder cancer and osteosarcoma datasets
In this study, bladder cancer datasets GSE65635 and GSE100926, as well as the osteosarcoma dataset GSE39058 profiles, were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) generated from GPL14550 and GPL14951, respectively. GSE65635 includes 8 bladder cancer and 4 normal tissue samples, GSE100926 includes 3 bladder cancer and 3 normal tissue samples, and GSE39058 includes 37 osteosarcoma and 5 normal tissue samples. These datasets were used to identify differentially expressed genes (DEGs) in bladder cancer and osteosarcoma.
2.2. DEG selection
First, we performed log2 transformation on bladder cancer dataset GSE65635, GSE100926, and osteosarcoma dataset GSE39058. Multivariate linear regression was conducted using the lmFit function, and empirical Bayesian adjustments were made by shrinking the standard errors towards a common value to calculate the moderated t statistics, moderated f statistics, and the log-odds of differential expression using the R package “limma”. Probe summarization and background correction were performed for GSE65635 and GSE100926. The Benjamini–Hochberg method was used to adjust the raw P values. Fold change was calculated using the false discovery rate (FDR). The cutoff criteria for DEGs were P < .05 and fold change > 1.5. A volcano plot was generated to visualize the differential significance of each gene. Subsequently, the DEGs from GSE65635, GSE100926, and GSE39058 were intersected to obtain a common set of DEGs.
2.3. Weighted gene co-expression network analysis (WGCNA)
To create a co-expression network, we first merged the gene expression matrices of bladder cancer datasets GSE65635 and GSE100926, calculated the median absolute deviation for each gene, and removed the bottom 50% of genes with the lowest median absolute deviation. Outliers were eliminated using the good Samples Genes function from the R package WGCNA. A scale-free co-expression network was constructed by computing Pearson correlation matrices for all gene pairs and raising them to a power using the soft-thresholding parameter (β). Afterward, the adjacency matrix was transformed into a topological overlap matrix, which measures the network connectivity of a gene. Hierarchical clustering based on topological overlap matrix-based dissimilarity measures was performed to classify genes into modules, with a minimum module size of 30 genes. A sensitivity value of 3 was set. To further analyze the modules, the dissimilarity of module feature genes was computed, a cut line was chosen for the module dendrogram, and some modules were merged. Additionally, modules with a distance < 0.25 were merged, with the gray module being considered a gene set that could not be assigned to any specific module.
2.4. Functional enrichment analysis
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analyses were used to evaluate gene functions and biological pathways. The differential gene list was input into the KEGG API (https://www.kegg.jp/kegg/rest/keggapi.html) to obtain the latest KEGG Pathway gene annotations, which served as the background. Gene enrichment analysis was performed using the R package cluster Profiler (version 3.14.3), with a minimum gene set size of 5 and a maximum gene set size of 5000. A P value of < .05 and a FDR of < 0.25 were considered statistically significant.
Furthermore, the Metascape database was used to provide comprehensive gene list annotations and analysis resources, which were visualized and exported for functional enrichment analysis of the differential gene list.
2.5. Gene set enrichment analysis (GSEA)
For GSEA, the 2 groups of samples for bladder cancer and normal tissues were created from the bladder cancer dataset. The Molecular Signatures Database (c2.cp.kegg.v7.4.symbols.gmt subset) was downloaded from GSEA (DOI: 10.1073/pnas.0506580102, http://software.broadinstitute.org/gsea/index.jsp). GSEA software (version 3.0, https://www.gsea-msigdb.org/gsea/index.jsp) was obtained from the GSEA website. Based on gene expression profiles and phenotype grouping, GSEA was performed to evaluate relevant pathways and molecular mechanisms. The minimum gene set size was set to 5, the maximum gene set size to 5000, with 1000 permutations. A P value of < .05 and an FDR of < 0.25 were considered statistically significant. Additionally, GO and KEGG analyses for the whole genome were conducted using GSEA.
2.6. Immune infiltration analysis
CIBERSORT (http://CIBERSORT.stanford.edu/) is a widely used method for calculating immune cell infiltration. The LM22 gene signature file was used to define 22 immune cell subtypes. Integrated bioinformatics methods were applied to analyze the gene expression matrix of the osteosarcoma dataset GSE39058 using the CIBERSORT package. Linear support vector regression was used to deconvolute the expression matrix of immune cell subtypes and estimate immune cell abundances. Samples with a confidence score of P < .05 were retained.
2.7. Protein-protein interaction (PPI) network construction and analysis
The STRING database (http://string-db.org/) was employed to collect, score, and integrate protein-protein interaction information from various sources and predict additional interactions. The differential gene list was input into the STRING database to construct a predicted PPI network of core genes (confidence > 0.4). Cytoscape software (http://www.cytoscape.org/) was used for biological network analysis and 2D visualization. The PPI network from the STRING database was imported into Cytoscape for visualization and the identification of core genes. MCODE was used to find the best modules with the highest relevance. Five algorithms (MCC, MNC, DMNC, Closeness, Betweenness) were utilized to calculate the top 10 genes with the highest relevance for visualization. The resulting core gene list was exported.
2.8. Survival analysis
Clinical survival data for bladder cancer were obtained from TCGA. The R package maxstat (version: 0.7–25) was used to calculate the optimal cutoff value for the Risk Score of core genes. Groups were established based on this optimal cutoff value, with a minimum sample size in each group > 25% and a maximum sample size < 75%. Survival analysis was conducted using the survfit function from the R package survival. The log-rank test was employed to assess the significance of survival differences between different groups. Additionally, a forest plot was created using the R package forest to observe the impact of each individual core gene on bladder cancer prognosis.
2.9. Gene expression heatmaps
Heatmaps of core gene expression in bladder cancer datasets GSE65635 and GSE100926, as well as the osteosarcoma dataset GSE39058, were generated using the R package heatmap. These heatmaps visually represented the expression differences of core genes between osteosarcoma, bladder cancer, and normal tissue samples.
2.10. Comparative toxicogenomics database (CTD) analysis
The CTD integrates a wealth of data on interactions between chemicals, genes, functions, phenotypes, and diseases. The core genes were input into the CTD website to identify the most relevant diseases associated with the core genes. Radar plots were created in Excel to illustrate the expression differences for each gene associated with specific diseases.
2.11. Western blotting
Western blotting, also known as immunoblotting, is a method to detect the expression of a certain protein in complex samples according to the specific binding of antigens and antibodies, and can qualitatively and semi quantitatively analyze proteins. Total protein was extracted and the protein content was determined. After SDS-PAGE electrophoresis and membrane transfer, the protein samples were blocked with 5% skim milk for 1 hour at room temperature, shaken with Tris Buffered Saline Tween at high speed on a shaker, washed for 5 minutes, and repeated 3 times. The primary antibody was added and incubated overnight at 4° C, followed by TBST shaking 3 times (5 minutes each time) and TBST shaking 3 times (5 minutes each time). The results were analyzed after chemiluminescence development.
2.12. miRNA
Target Scan (www.targetscan.org) is an online database used for the analysis and prediction of miRNA and target genes. In our study, Target Scan was used to screen for miRNAs that regulate the identified DEGs.
3. Results
3.1. Differential gene analysis
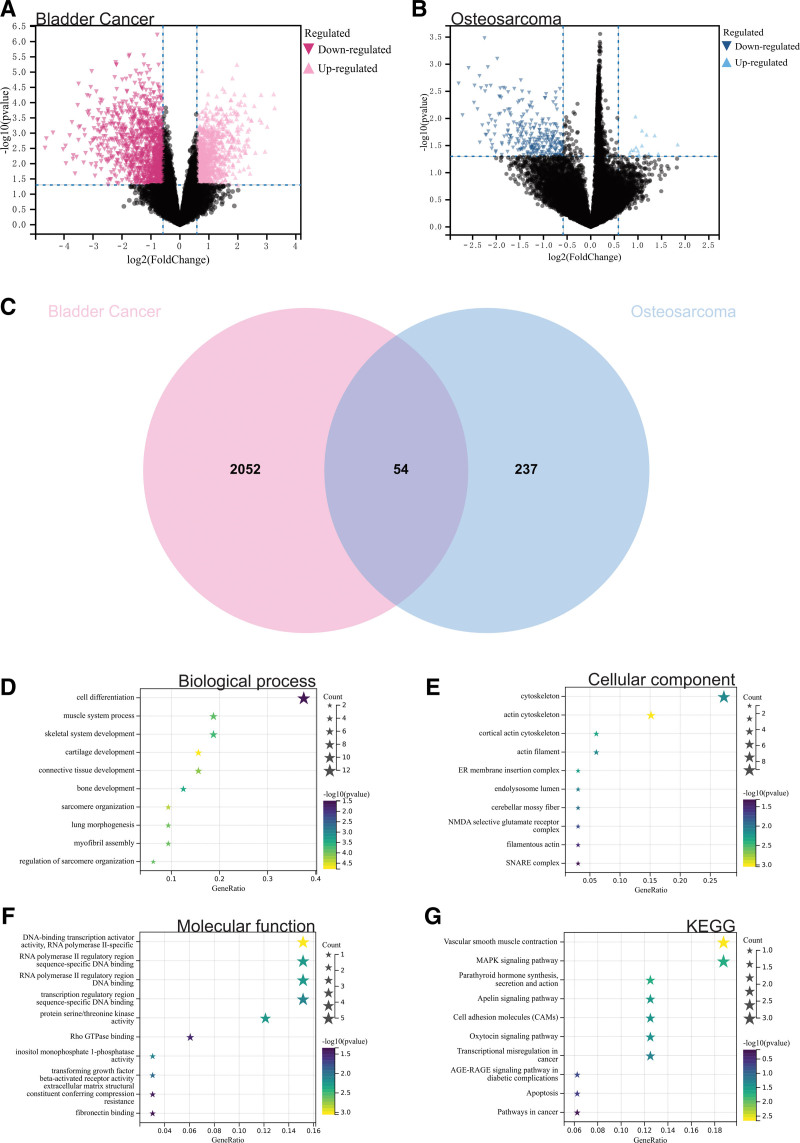
In this study, differential gene expression was identified in the gene expression matrices of bladder cancer datasets GSE65635 and GSE100926 (Fig. 1A) and the osteosarcoma dataset GSE39058 (Fig. 1B) using predefined cutoff values. Venn diagrams were used to determine the intersection of differentially expressed genes between the 2 diseases, resulting in a total of 54 DEGs (Fig. 1C).
Figure 1.
Analysis of differentially expressed genes. (A) Gene expression matrices for bladder cancer datasets GSE65635 and GSE100926 identified differentially expressed genes. (B) Gene expression matrix for the osteosarcoma dataset GSE39058 identified differentially expressed genes. (C) A total of 54 DEGs were identified. (D) Functional enrichment analysis, GOBP analysis. (E) GOCC analysis. (F) GOMF analysis. (G) KEGG analysis. DEGs = differentially expressed genes, KEGG = Kyoto encyclopedia of genes and genomes.
3.2. Functional enrichment analysis
3.2.1. DEGs.
We conducted GO and KEGG analyses for these differentially expressed genes. According to the results of GO analysis, in the biological process category, DEGs were significantly enriched in cell differentiation, cartilage development, and skeletal development (Fig. 1D). In the cellular component category, they were mainly enriched in actin cytoskeleton, and actin filaments (Fig. 1E). In the molecular function category, they were concentrated in Rho GTPase binding, DNA binding, and fibronectin binding (Fig. 1F). In the KEGG analysis, they were mainly enriched in the MAPK signaling pathway, apoptosis, and cancer pathways (Fig. 1G).
3.2.2. GSEA.
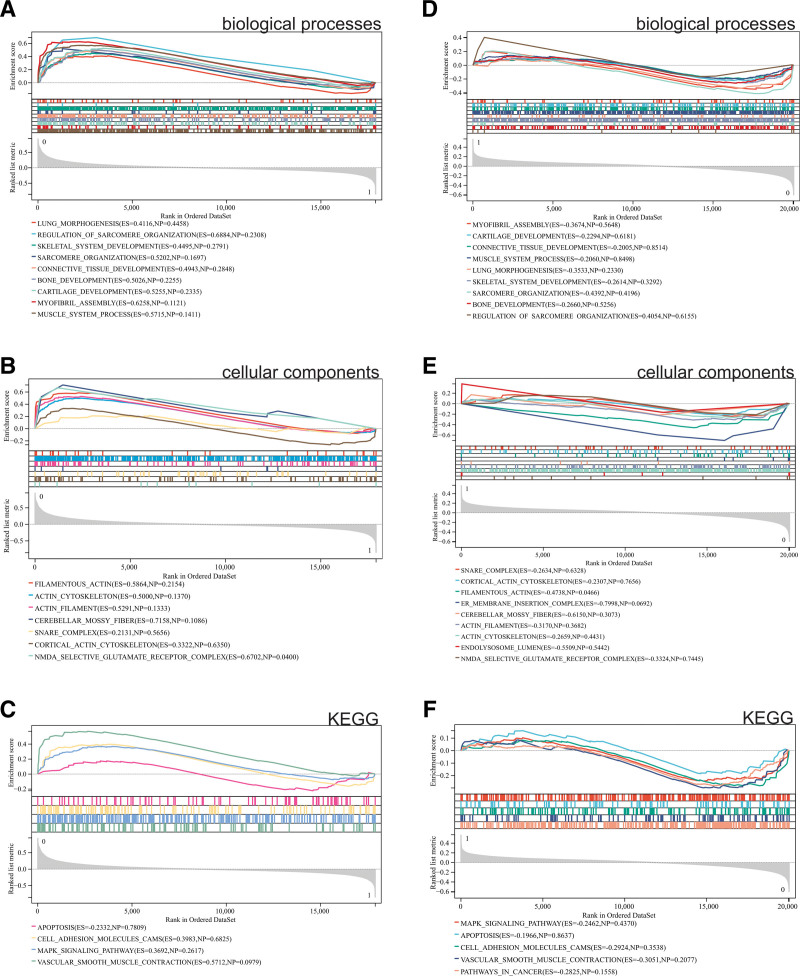
Additionally, we performed GSEA enrichment analysis for the whole genome to identify potential enrichment items in non-differentially expressed genes and validate the results of differentially expressed genes. The intersection of enrichment terms between GO and KEGG terms for differentially expressed genes is shown in the results. Enrichment results for both bladder cancer datasets GSE65635 and GSE100926 (Fig. 2A–C) and the osteosarcoma dataset GSE39058 (Fig. 2D–F) indicated that differentially expressed genes were primarily enriched in skeletal development, cartilage development, actin cytoskeleton, MAPK signaling pathway, and apoptosis.
Figure 2.
GSEA. (A, B, C) Differentially expressed genes in bladder cancer datasets GSE65635 and GSE100926 primarily enriched in skeletal development, cartilage development, cytoskeleton of muscle cells, MAPK signaling pathway, and apoptosis, (D, E, F) differentially expressed genes in the osteosarcoma dataset GSE39058 mainly enriched in skeletal development, cartilage development, cytoskeleton of muscle cells, MAPK signaling pathway, and apoptosis. GSEA = gene set enrichment analysis.
3.2.3. Metascape enrichment analysis.
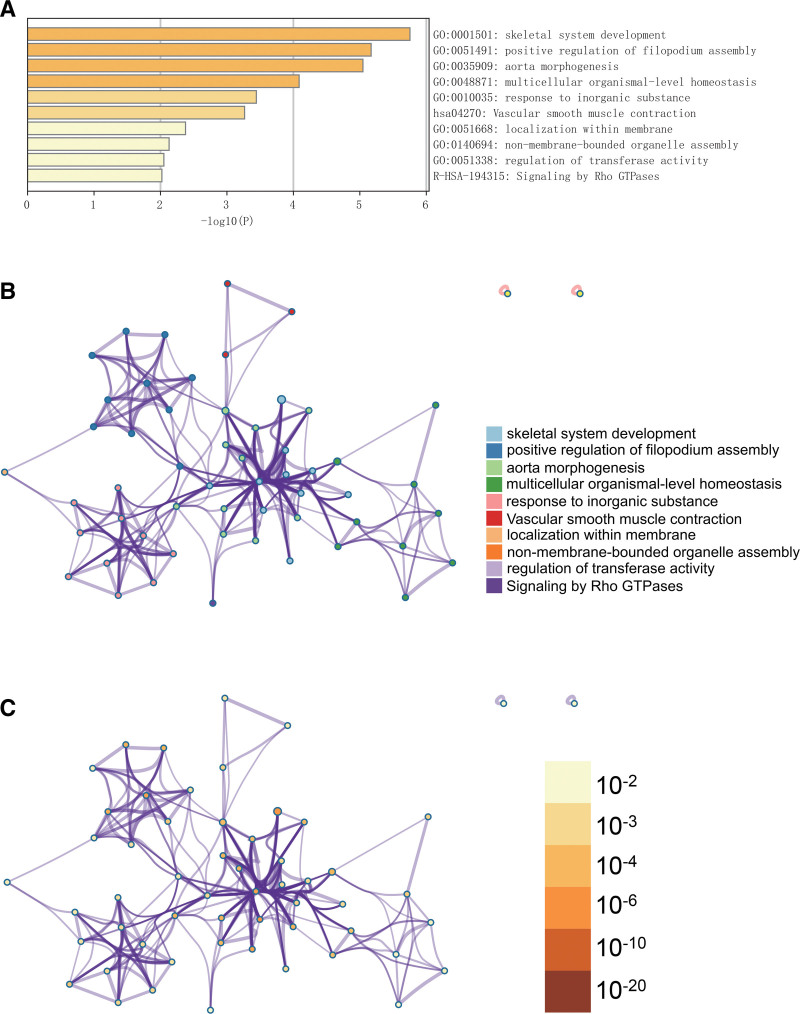
In the Metascape enrichment analysis, the GO enrichment item “bone system development” was observed (Fig. 3A). We also produced enrichment networks color-coded with enrichment terms and P values (Fig. 3B and C), providing visual representation of the associations and confidence of various enrichment items.
Figure 3.
Metascape enrichment analysis. (A) GO enrichment items show enrichment in skeletal system development. (B, C) Enrichment networks colored by enrichment terms and P values. GO = gene ontology.
3.3. Immune infiltration analysis
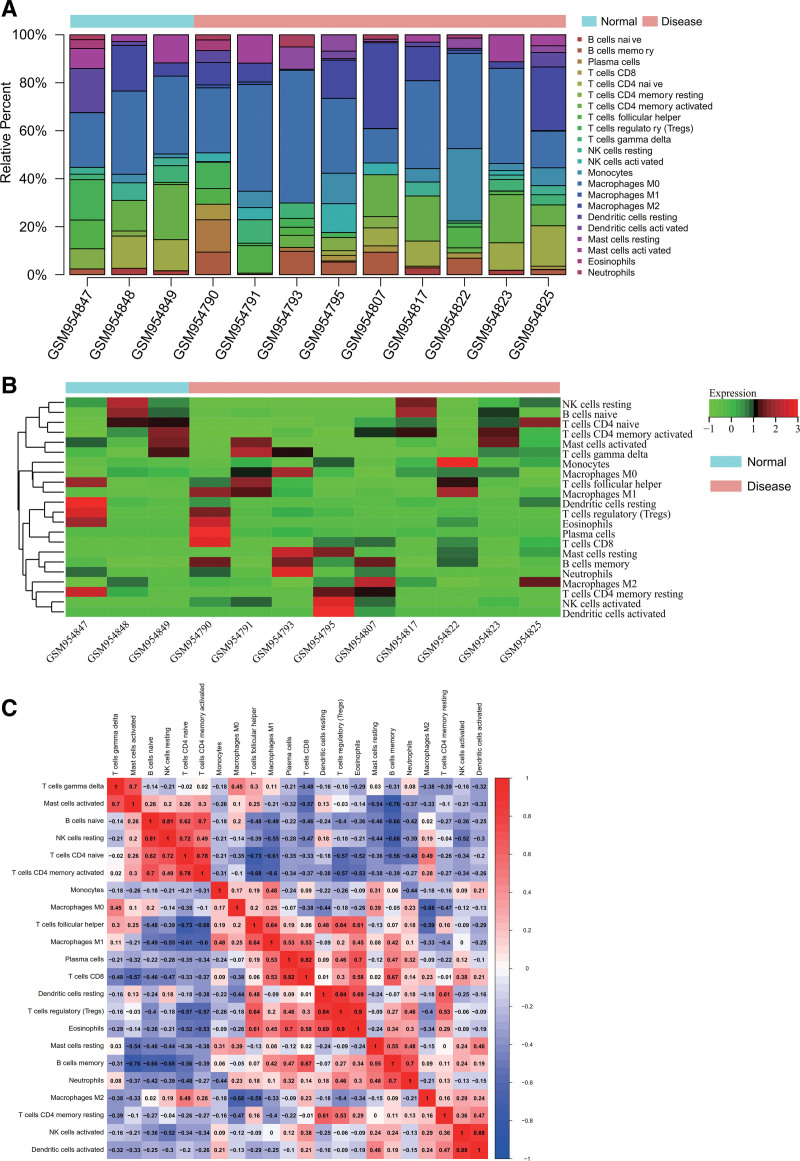
We employed the CIBERSORT package to analyze the gene expression matrix of the osteosarcoma dataset GSE39058. At a 95% confidence level, we obtained the proportions of immune cell types within the entire gene expression matrix. The results indicated a relatively high proportion of Macrophages M0 in the samples (Fig. 4A). An immune cell expression heatmap for the dataset was also generated (Fig. 4B). Furthermore, we performed correlation analysis of infiltrating immune cells to identify co-expression patterns among immune cell components. The results suggested that when T cells regulatory were highly expressed, Eosinophils also exhibited high expression, indicating a strong positive correlation between T cells regulatory and Eosinophils, which might influence the disease progression of osteosarcoma (Fig. 4C).
Figure 4.
Immune infiltration analysis. (A) Samples with a high proportion of macrophages M0. (B) Heatmap of immune cell expression levels in the dataset. (C) Co-expression pattern of immune cell components.
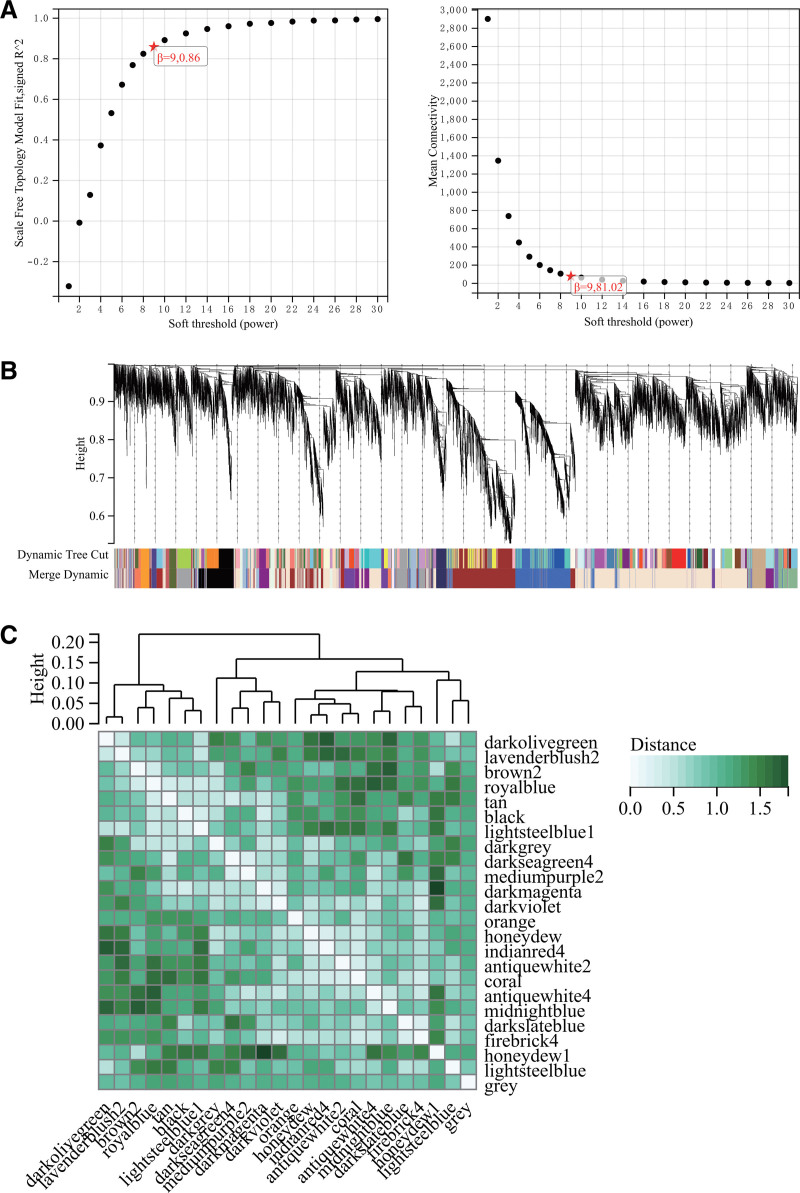
3.4. WGCNA
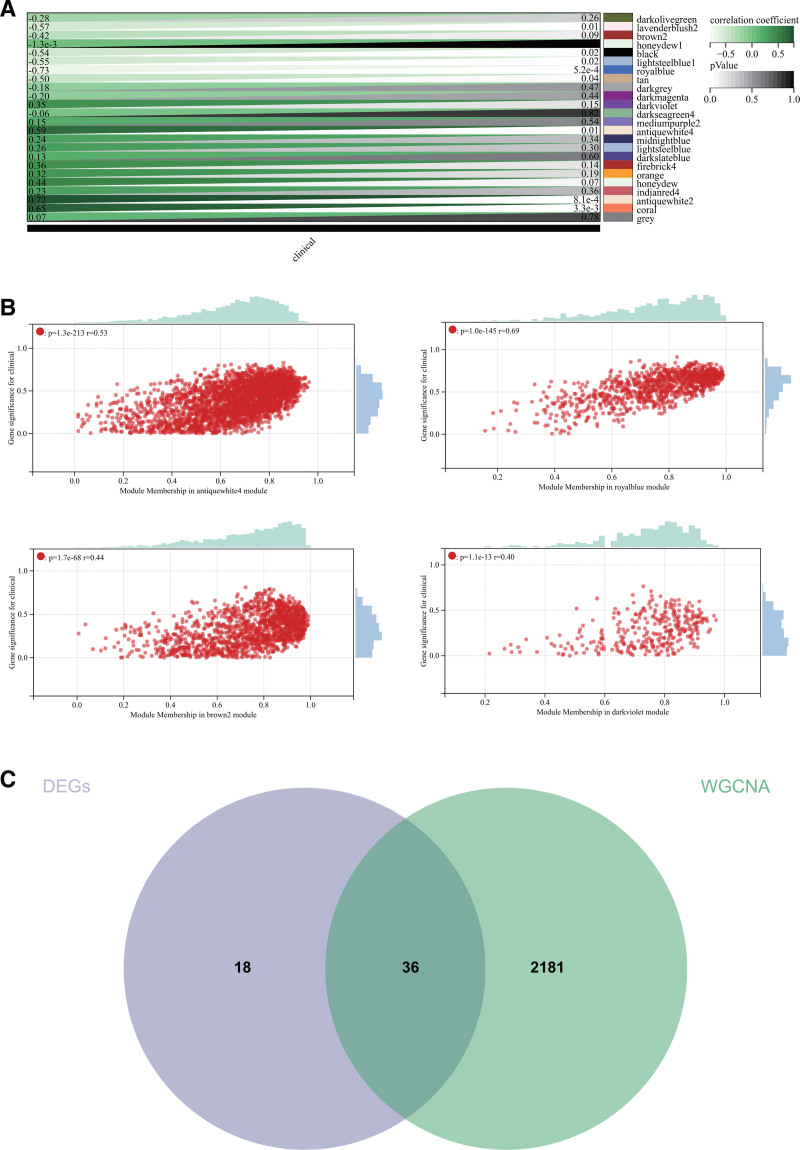
The selection of a soft threshold power is a critical step in WGCNA analysis. A network topology analysis was performed to determine the soft threshold power, which was set to 9 (Fig. 5A). A total of 24 modules were constructed based on all genes (Fig. 5B), and the interactions between important modules were analyzed (Fig. 5C). Module-phenotype correlation heatmaps were generated (Fig. 6A), and a correlation scatterplot of gene significance and module membership (MM) for relevant hub genes was produced (Fig. 6B). We computed the module eigengenes correlation with gene expression to obtain the MM, and using a cutoff criterion (|MM| > 0.8), 4 highly connected genes were identified as hub genes in clinically significant modules.
Figure 5.
WGCNA. (A) β = 9, 0.86, β = 9, 81.02. (B) Gene hierarchical clustering tree generated 24 modules. (C) Interaction between important modules. WGCNA = weighted gene co-expression network analysis.
Figure 6.
WGCNA. (A) Heatmap of module correlation with phenotypes. (B) Scatter plot showing the correlation between GS and MM for related hub genes. (C) Venn diagram showing intersections. GS = gene significance, MM = module membership, WGCNA = weighted gene co-expression network analysis.
We further created a Venn diagram by intersecting the DEGs obtained from WGCNA and the DEGs, resulting in the identification of common genes (Fig. 6C).
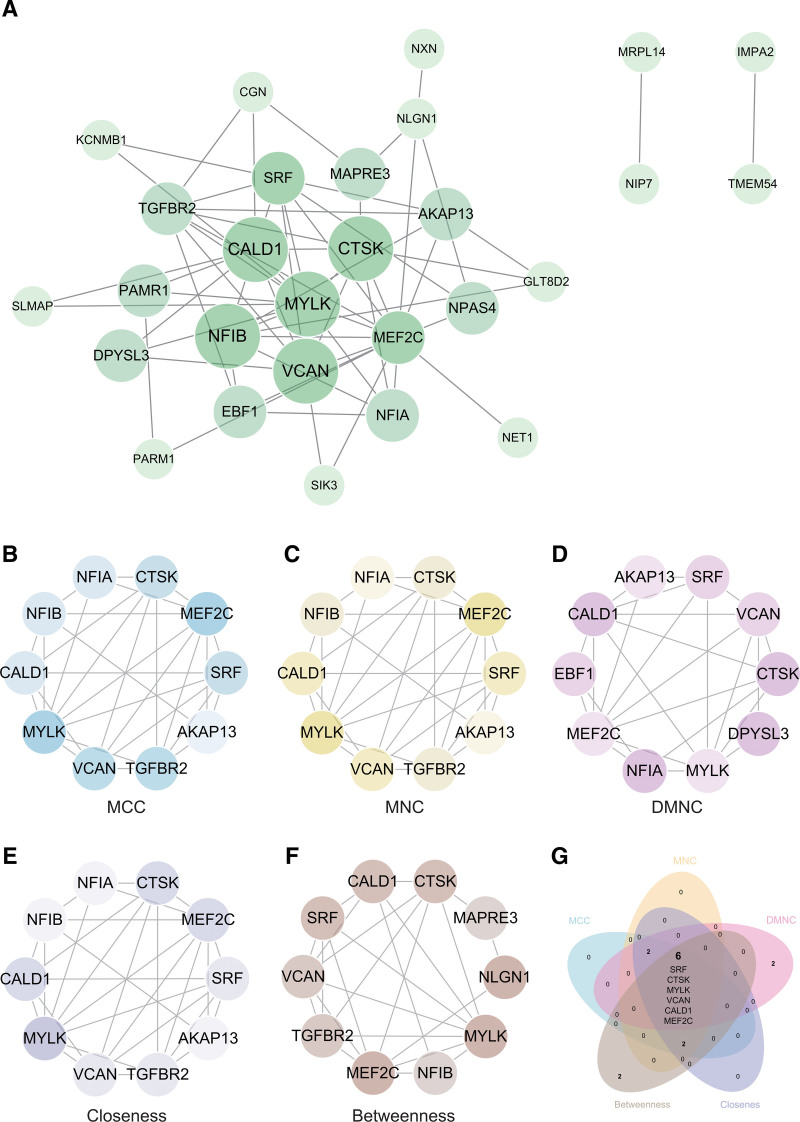
3.5. PPI network construction and analysis
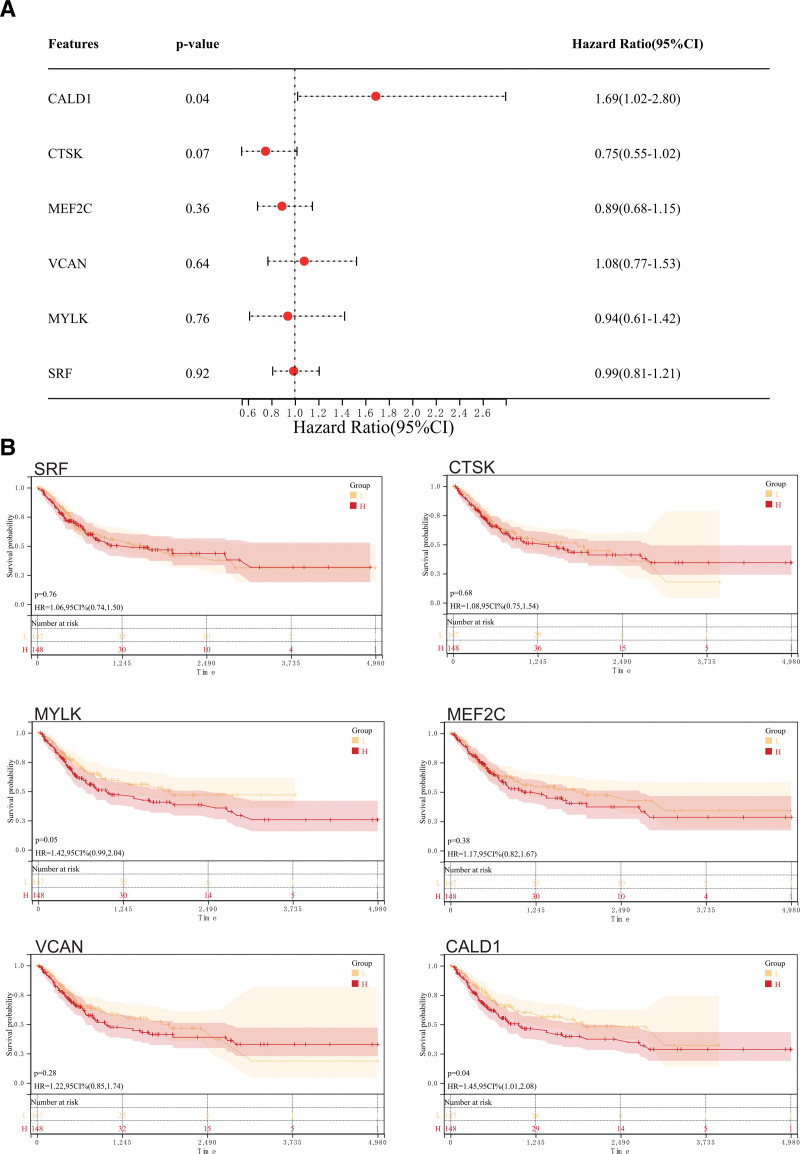
The PPI network of DEGs was constructed using the STRING online database and analyzed using Cytoscape software (Fig. 7A). Central genes were identified using 5 algorithms: MCC, MNC, DMNC, Closeness, and Betweenness (Fig. 7B–F). The intersection of the results from these algorithms was visualized using a Venn diagram, yielding the core genes (Fig. 7G). Ultimately, 6 core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1) were obtained.
Figure 7.
Construction and analysis of protein-protein interaction (PPI) network. (A) PPI network. (B, C, D, E, F) Identification of hub genes using five algorithms: MCC, MNC, DMNC, closeness, and betweenness. (G) Venn diagram to obtain the union as core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1). CALD1 = caldesmon, MYLK = myosin light chain kinase.
3.6. Survival analysis
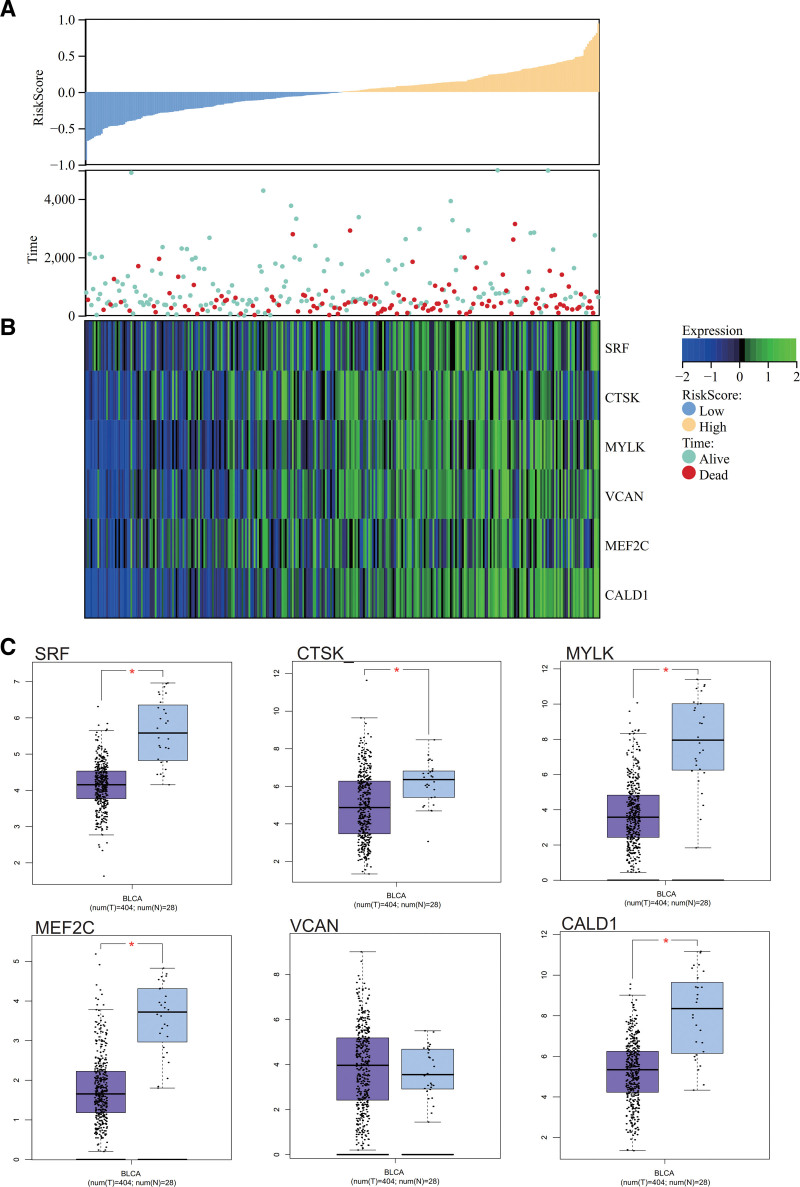
We obtained bladder cancer survival data from TCGA and created a Risk Score relationship plot. It was observed that as the risk score increased, the survival rate of patients significantly decreased, with the low-risk group showing markedly higher survival time and rate than the high-risk group (Fig. 8A). A heatmap of core gene expression in bladder cancer survival data revealed that the core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1) acted as risk factors, exhibiting an upregulated trend with increasing risk scores (Fig. 8B). Box plots were generated to illustrate the significant differences in core gene expression (SRF, CTSK, MYLK, MEF2C, CALD1) between bladder cancer and normal samples (Fig. 8C).
Figure 8.
Survival analysis. (A) Relationship between prognosis score. (B) Visualization of core gene expression heatmap in bladder cancer survival data. (C) Box plot of core gene expression in bladder cancer.
We also obtained forest plots (Fig. 9A) and Kaplan–Meier survival curves (Fig. 9B) associated with the core genes, demonstrating a significant association between core genes (MYLK, CALD1) and survival rates in bladder cancer patients.
Figure 9.
Survival analysis. (A) Forest plot of core genes associated with bladder cancer. (B) KM survival curves of core genes.
3.7. Core gene expression heatmaps
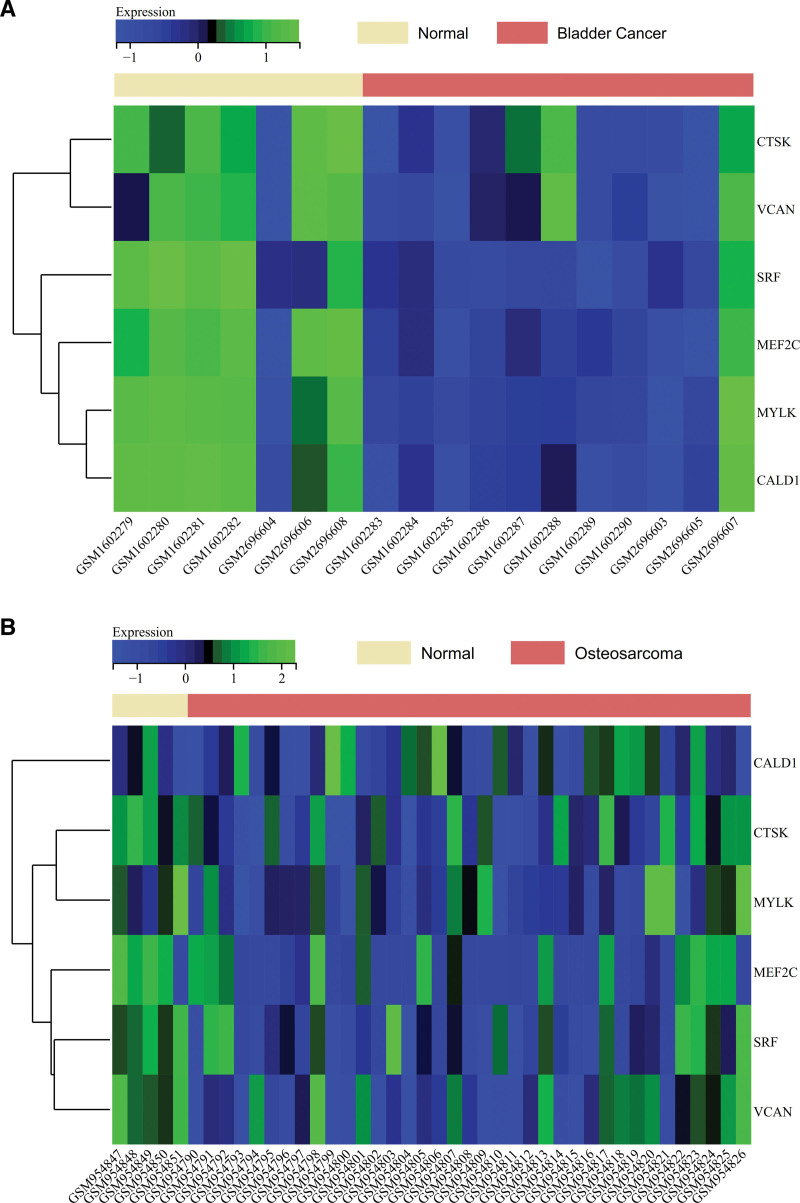
We visualized the expression of core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1) in the merged matrices of bladder cancer datasets GSE65635 and GSE100926 (Fig. 10A) and the osteosarcoma dataset GSE39058 (Fig. 10B). The heatmaps indicated that core genes exhibited lower expression levels in bladder cancer and osteosarcoma samples compared to normal samples, suggesting that core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1) may play a regulatory role in bladder cancer.
Figure 10.
Core gene expression heatmap. (A) Visualization of core gene expression heatmap in the combined matrix for bladder cancer datasets GSE65635 and GSE100926. (B) Visualization of core gene expression heatmap in the osteosarcoma dataset GSE39058.
3.8. CTD analysis
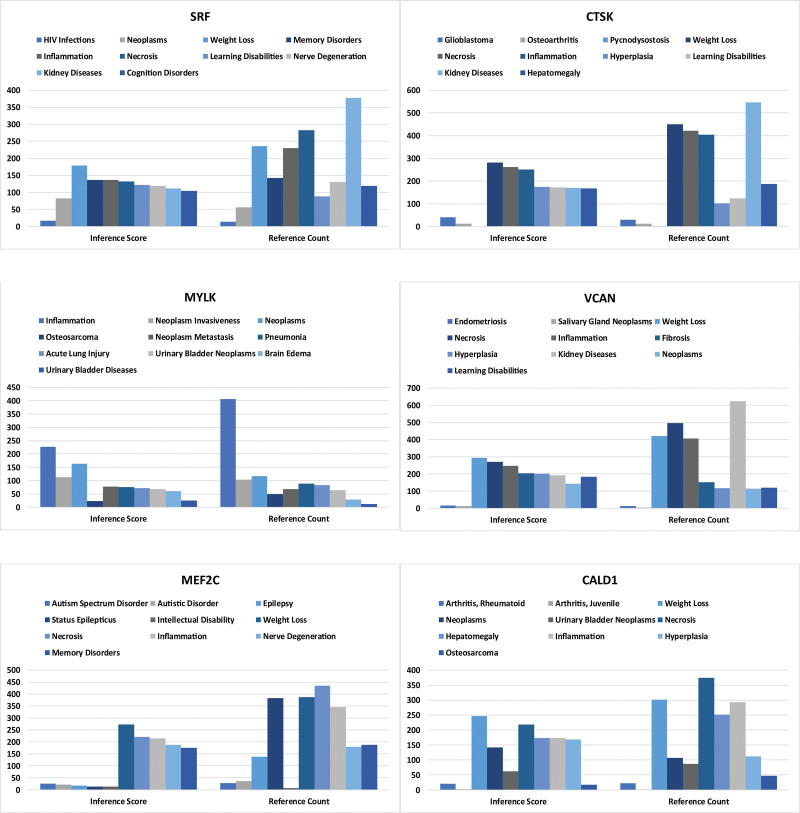
In this study, we input the hub gene list into the CTD website to identify diseases associated with the core genes. This enhanced our understanding of the relationships between genes and diseases. The core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1) were found to be associated with osteosarcoma, bladder neoplasms, bladder diseases, tumors, inflammation, and necrosis (Fig. 11).
Figure 11.
CTD analysis. Core genes (SRF, CTSK, MYLK, VCAN, MEF2C, CALD1) are associated with osteosarcoma, bladder tumors, bladder diseases, tumors, inflammation, and necrosis. CALD1 = caldesmon, CTD = comparative toxicogenomics database, MYLK = myosin light chain kinase.
3.9. Western blotting
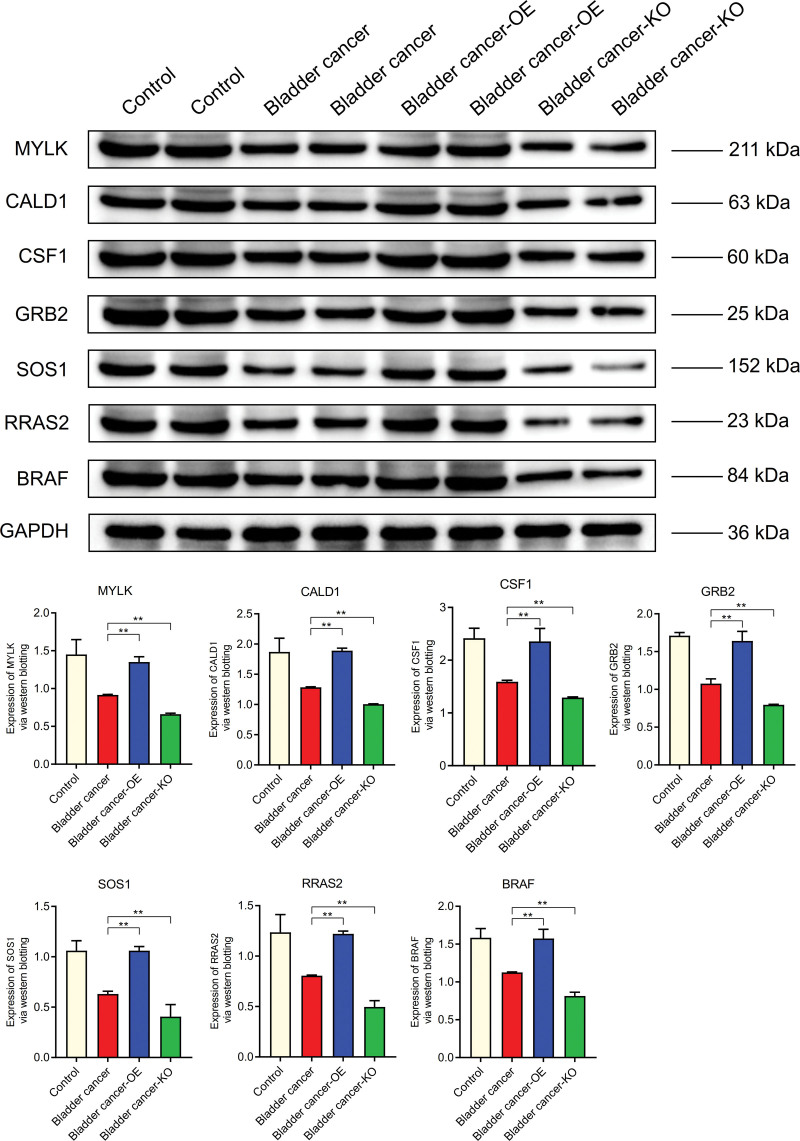
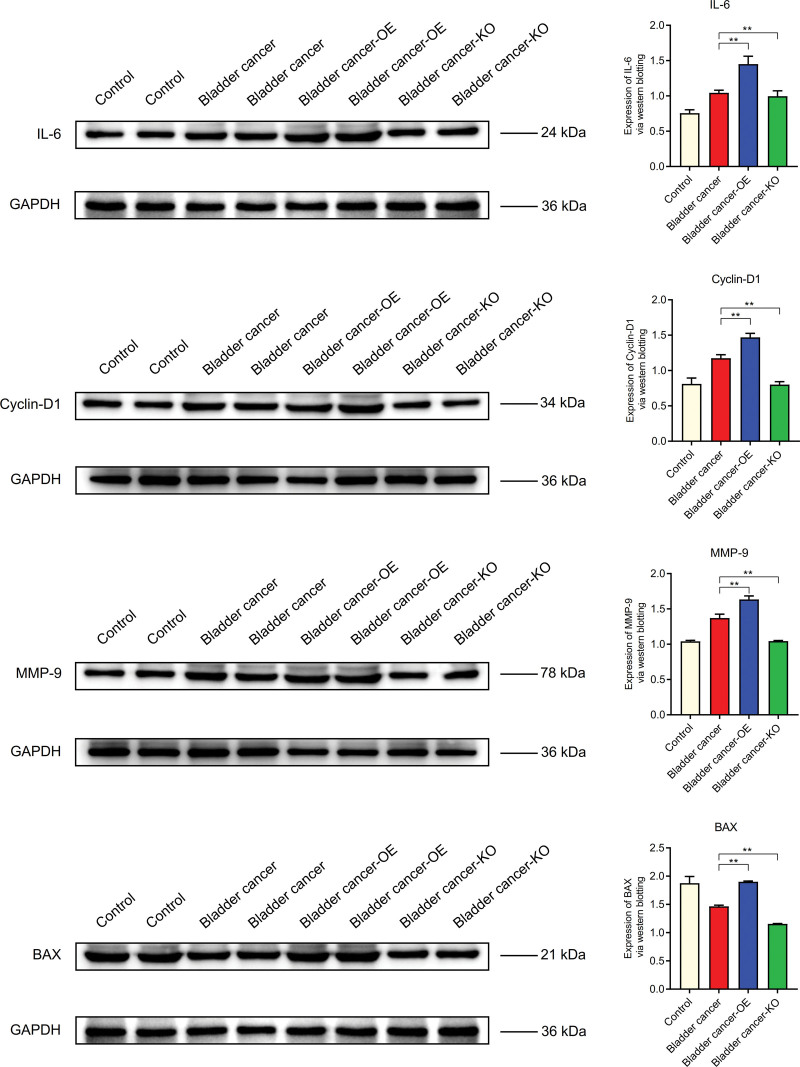
The expression levels of MYLK, CALD1, CSF1, GRB2, SOS1, RRAS2, and BRAF in bladder cancer were lower than those in normal control group, but higher than those in Bladder cancer-KO group. It is higher in Bladder cancer - OE group than in bladder cancer group (Fig. 12). The expression levels of IL-6, Cyclin-D1, and MMP-9 in Bladder cancer were higher than those in normal group, but lower in Bladder cancer-KO group than those in bladder cancer group. It was higher in Bladder cancer - OE group than in bladder cancer. However, the expression level of BAX in bladder cancer was lower than that in normal control group (Fig. 13).
Figure 12.
WB. The expression levels of MYLK, CALD1, CSF1, GRB2, SOS1, RRAS2, and BRAF in bladder cancer were lower than those in normal control group. CALD1 = caldesmon, MYLK = myosin light chain kinase, WB = Western blotting.
Figure 13.
WB. The expression levels of IL-6, cyclin-D1 and MMP-9 in Bladder cancer were higher than those in normal group. However, the expression level of BAX in bladder cancer was lower than that in normal control group. WB = Western blotting.
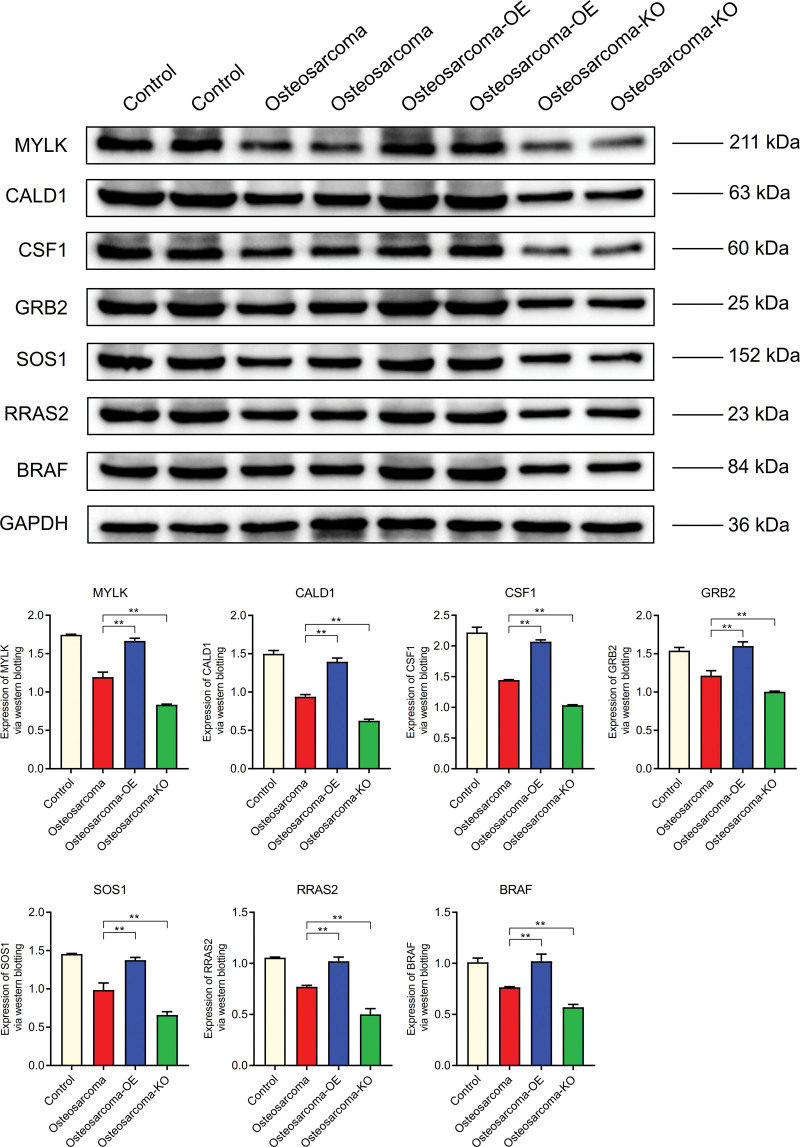
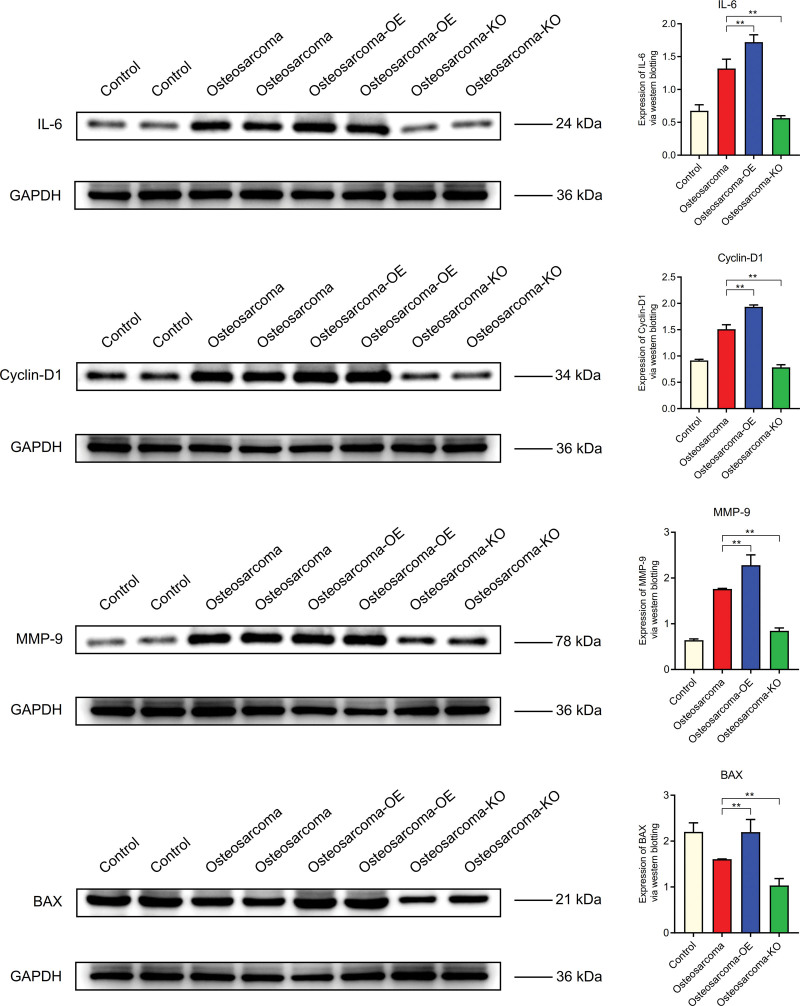
The expression levels of MYLK, CALD1, CSF1, GRB2, SOS1, RRAS2, and BRAF in osteosarcoma were lower than those in normal control group, but higher than those in osteosarcoma-KO group. It is higher in osteosarcoma - OE group than in osteosarcoma group (Fig. 14). The expression levels of IL-6, Cyclin-D1 and MMP-9 in B osteosarcoma were higher than those in normal group, but lower in osteosarcoma -KO group than those in osteosarcoma group. It was higher in osteosarcoma - OE group than in bladder cancer. However, the expression level of BAX in osteosarcoma was lower than that in normal control group (Fig. 15).
Figure 14.
WB. The expression levels of MYLK, CALD1, CSF1, GRB2, SOS1, RRAS2, and BRAF in osteosarcoma were lower than those in normal control group. CALD1 = caldesmon, MYLK = myosin light chain kinase, WB = Western blotting.
Figure 15.
WB. The expression levels of IL-6, cyclin-D1 and MMP-9 in B osteosarcoma were higher than those in normal group. However, the expression level of BAX in osteosarcoma was lower than that in normal control group. WB = Western blotting.
3.10. Prediction and functional annotation of miRNAs related to hub genes
In this study, we input the hub gene list into Target Scan to search for related miRNAs, improving our understanding of gene expression regulation (Table 1). We identified that SRF gene-related miRNA is hsa-miR-9-5p; CTSK gene-related miRNAs include hsa-miR-185-5p, hsa-miR-4306, and hsa-miR-4644; MYLK gene-related miRNA is hsa-miR-129-5p; VCAN gene-related miRNA is hsa-miR-203a-3p.1; MEF2C gene-related miRNAs include hsa-miR-551b-3p and hsa-miR-551a; CALD1 gene-related miRNAs are hsa-miR-19a-3p and hsa-miR-19b-3p.
Table 1.
A summary of miRNAs that regulate hub genes.
| Gene | Predicted MiR | |||
|---|---|---|---|---|
| 1 | SRF | hsa-miR-9-5p | ||
| 2 | CTSK | hsa-miR-185-5p | hsa-miR-4306 | hsa-miR-4644 |
| 3 | MYLK | hsa-miR-129-5p | ||
| 4 | VCAN | hsa-miR-203a-3p.1 | ||
| 5 | MEF2C | hsa-miR-551b-3p | hsa-miR-551a | |
| 6 | CALD1 | hsa-miR-19a-3p | hsa-miR-19b-3p | |
CALD1 = caldesmon, MYLK = myosin light chain kinase.
4. Discussion
Bladder cancer and osteosarcoma are 2 highly debilitating types of cancer that pose significant challenges in their development and treatment. Bladder cancer is an invasive cancer with a high rate of recurrence even after initial control, necessitating regular follow-up and monitoring to ensure disease stability.[12] Treatment methods for bladder cancer include radiation therapy, chemotherapy, and immunotherapy, which may lead to side effects such as nausea, vomiting, fatigue, and immune system suppression.[13,14] Individuals with bladder cancer may face emotional stress, anxiety, and depression, in addition to challenges related to work, family, and social life.[15,16] Treatment of osteosarcoma typically requires surgical intervention, which may involve tumor resection and the removal of affected bone tissue.[17] In some cases, amputation may be necessary, posing significant physical and psychological challenges to patients. Osteosarcoma carries a high-risk of lung metastasis, which greatly increases the complexity of treatment and impacts patient prognosis. Osteosarcoma treatment often includes chemotherapy and radiation therapy, which can result in side effects such as nausea, vomiting, fatigue, and immune system suppression.[18] The diagnosis and treatment of osteosarcoma may have negative effects on patients emotional well-being and social life, leading to anxiety, depression, and emotional stress.[19–21]
This study found an association between the low expression of MYLK and CALD1 in bladder cancer and osteosarcoma with worsened prognosis. This suggests that patients with low expression of MYLK and CALD1 may have a poorer survival outlook. The decreased expression of these proteins may be linked to tumor growth, invasiveness, and treatment resistance, thus holding significant clinical relevance for disease progression and treatment responses. This discovery provides new leads for targeted therapeutic research, with the potential to improve patient prognosis. Therefore, in-depth investigation of the roles and regulatory mechanisms of MYLK and CALD1 is crucial for the management and treatment of bladder cancer and osteosarcoma.
In addition, ultrasound-guided biopsies play a crucial role in the diagnosis and treatment of bladder cancer and osteosarcoma. Ultrasound-guided biopsies assist physicians in early diagnosis by obtaining tissue samples for pathological analysis to determine the nature of lesions. Early cancer detection often leads to better treatment outcomes.[22] Analyzing tissue samples allows physicians to gain a better understanding of the molecular characteristics of tumors, aiding in the development of personalized treatment plans and the selection of the most suitable treatment strategies for patients.[23] Ultrasound-guided biopsies are typically a relatively noninvasive method that, when compared to traditional surgery, can reduce patient discomfort and recovery time.[24]
MYLK is an enzyme protein. MYLK functions by adding phosphate groups to myosin light chains, initiating muscle fiber shortening and, subsequently, muscle contraction. This process is crucial for the contraction of smooth muscle, cardiac muscle, and skeletal muscle.[25] MYLK’s activity is calcium-regulated and depends on cytoplasmic calcium concentration within muscle cells. Increased calcium levels lead to elevated MYLK activity, resulting in myosin light chain phosphorylation and muscle contraction.[26] This process is extensively involved in the regulation of smooth muscle, such as in the contraction and relaxation of vascular smooth muscle. MYLK’s function is not only vital in normal physiological processes but also plays a role in disease development. For instance, abnormal MYLK activity or expression is associated with cardiovascular diseases, smooth muscle disorders, and specific muscle diseases. Shi et al[27] found that adipocyte-induced SIK2 Ser358 phosphorylation and MYLK Ser343 phosphorylation enhance the motility of ovarian cancer cells, with SIK2 promoting ovarian cancer cell motility and metastasis by phosphorylating MYLK. Teng et al[28] discovered that LncRNA MYLK-AS1 promotes tumor progression and angiogenesis in liver hepatocellular carcinoma by targeting the miR-424-5p/E2F7 axis and activating the VEGFR-2 signaling pathway.
Regarding the relationship between bladder cancer and MYLK, recent research has provided insights. Jiang et al[29] identified hub genes associated with the progression and prognosis of bladder cancer through comprehensive bioinformatics analysis. These genes include MYLK, and the expression of these survival-related genes correlates with grading, staging, and TNM staging. qRT-PCR and the HPA database confirmed that all central genes related to survival were downregulated in bladder cancer tissues. Additionally, research has elucidated that CircMYLK, through competitive binding with miR-34a, upregulates CCND3 levels, promoting the growth, migration, invasion, and survival of bladder cancer cells. The downregulation of CircMYLK inhibits bladder cancer cell migration and invasion while promoting apoptosis and cell cycle arrest. MiR-34a, a target of CircMYLK, is downregulated in bladder cancer tissues and cells. MiR-34a inhibition reverses the effects of CircMYLK downregulation, restoring malignant behavior in bladder cells.[30]
The relationship between osteosarcoma and MYLK is also being unveiled. Yang et al[31] demonstrated through mass spectrometry analysis that MYLK4 interacts with the epidermal growth factor receptor in osteosarcoma cells, promoting growth and metastasis through the epidermal growth factor receptor signaling pathway. Overexpression of MYLK4 accelerates OS growth and metastasis, while silencing MYLK4 expression leads to reduced cell growth and migration. Guo et al[32] used targeted RNA sequencing to diagnose known sarcoma fusions and novel fusion partners. Diagnostic in-frame fusion transcripts were detected in 43% of cases, with 3% (6/184) having new fusion partners, including TRPS1-PLAG1, VCP-TFE3, MYLK-BRAF, FUS-TFCP2, and ACTB-FOSB.
CALD1 is a multifunctional protein present in various types of muscle tissues. Its structure includes an N-terminus and C-terminus, along with an extended central coiled-coil structure. This protein has specific isoforms in smooth muscle, often referred to as smooth muscle CALD1.[33] CALD1 plays a critical role in muscle function, particularly associated with muscle contraction and relaxation. In smooth muscle, CALD1 interacts with myosin and regulates muscle tension. It also interacts with calcium-regulatory proteins, a crucial step in controlling smooth muscle contraction.[34,35] CALD1’s activity is closely tied to smooth muscle function. It plays essential roles in normal physiological processes, such as controlling vasomotor tone for blood flow maintenance and regulating smooth muscle motility in the digestive system to facilitate food passage through the gastrointestinal tract. Additionally, CALD1 is involved in muscle tissue development and repair. Aberrant expression or mutations in CALD1 may be associated with several diseases, including smooth muscle disorders, cardiovascular diseases, and cancers.[36]
In some cancers, both overexpression and under expression of CALD1 are associated with the invasiveness and prognosis of the disease. Studies suggest that CALD1 may impact the progression of glioblastoma by regulating tumor angiogenesis. The tumor microenvironment also indicates that high CALD1 expression samples are infiltrated by more stromal cells, such as endothelial cells and pericytes. Single-cell sequencing analysis reveals a significant upregulation of CALD1 in tumor cells, implicating its involvement in the tumorigenesis of glioblastoma.[37] Du et al[38] found that CALD1 may promote bladder cancer progression by reshaping the tumor microenvironment. CIBERSORT and ESTIMATE algorithms demonstrated a significant correlation between CALD1 and tumor microenvironment components, including CAF, macrophages, T cells, and multiple immune checkpoint-related genes. Cheng et al[39] discovered that CALD1 promotes PD-L1 expression in bladder cancer through the JAK/STAT signaling pathway. In a study by Diao et al[40] aiming to identify biomarkers for metastatic osteosarcoma, MiR-202 and miR-9 were potential key factors influencing OS metastasis. CALD1 and STX1A were suggested as potential therapeutic targets for metastatic OS.
The aforementioned literature reviews align with our research findings. MYLK and CALD1 are 2 proteins associated with muscle contraction and smooth muscle function. Both genes participate in the migration and invasion processes of tumor cells. They may influence the organization of actin filaments and the cytoskeleton within cells, as well as the growth and development of skeletal muscles. Lower expression of these genes is associated with poorer prognosis.
5. Limitations
Despite conducting rigorous bioinformatic analyses in this study, there are still some limitations. The research did not include in vivo experiments involving gene overexpression or knockout to further validate their functions. Therefore, future investigations should delve deeper into this aspect.
6. Conclusion
MYLK and CALD1 may exert their effects in bladder cancer and osteosarcoma by regulating muscle contraction and smooth muscle function. Low expression of MYLK and CALD1 in both diseases is associated with worse prognosis.
Author contributions
Conceptualization: Danyang Ding, Haowen Li, Lei Zhang.
Data curation: Haowen Li, Xiangyi Li, Xingyue Fan, Qian Yang, Chunbo Kang, Lei Zhang, Guihu Lin, Meiyue Cui, Bin Liu, Lingling Wang.
Formal analysis: Haowen Li, Xiangyi Li, Qian Yang, Chunbo Kang, Lei Zhang, Guihu Lin, Meiyue Cui, Lingling Wang, Jianzhi Su.
Investigation: Lingling Wang.
Methodology: Danyang Ding, Haowen Li, Xiangyi Li, Xingyue Fan, Qian Yang, Ye Wang, Chunbo Kang, Lei Zhang, Guihu Lin, Meiyue Cui, Bin Liu, Lingling Wang, Jianzhi Su.
Software: Haowen Li, Xingyue Fan, Ye Wang, Chunbo Kang, Guihu Lin, Bin Liu, Lingling Wang.
Visualization: Guihu Lin.
Writing – original draft: Danyang Ding, Xiangyi Li, Ye Wang, Chunbo Kang, Lei Zhang, Jianzhi Su.
Writing – review & editing: Danyang Ding, Xiangyi Li, Lei Zhang.
Abbreviations:
- CALD1
- caldesmon
- CTD
- comparative toxicogenomics database
- DEGs
- differentially expressed genes
- FDR
- false discovery rate
- GO
- gene ontology
- GSEA
- gene set enrichment analysis
- KEGG
- Kyoto encyclopedia of genes and genomes
- MM
- module membership
- MYLK
- myosin light chain kinase
- PPI
- protein-protein interaction
- WGCNA
- weighted gene co-expression network analysis
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
This study was approved by the Ethics Committee of the Fuxing Hospital Affiliated to Capital Medical University.
The study was funded by the scientific research fund of China Aerospace Science and Industry Corporation 731 Hospital (2021-QSYN-16).
The authors have no conflicts of interest to disclose.
How to cite this article: Li H, Lin G, Cui M, Wang L, Ding D, Li X, Fan X, Yang Q, Wang Y, Kang C, Zhang L, Liu B, Su J. Hub biomarkers in ultrasound-guided bladder cancer and osteosarcoma: Myosin light chain kinase and caldesmon. Medicine 2023;102:48(e36414).
Contributor Information
Haowen Li, Email: lixiangyi113@163.com.
Guihu Lin, Email: linguihu21@sina.com.
Meiyue Cui, Email: lliujjing@sohu.com.
Lingling Wang, Email: 1097615710@qq.com.
Danyang Ding, Email: dingdanyang2023@163.com.
Xingyue Fan, Email: lb20216662021@163.com.
Qian Yang, Email: yangqian03112023@163.com.
Ye Wang, Email: 1097615710@qq.com.
Chunbo Kang, Email: kangchunbo220@163.com.
Lei Zhang, Email: zhanglei46442@163.com.
Bin Liu, Email: liubinsy123@163.com.
Jianzhi Su, Email: jianzhi1288@163.com.
References
- [1].Rhea LP, Mendez-Marti S, Kim D, et al. Role of immunotherapy in bladder cancer. Cancer Treat Res Commun. 2021;26:100296. [DOI] [PubMed] [Google Scholar]
- [2].Tang Q, Zuo W, Wan C, et al. Comprehensive genomic profiling of upper tract urothelial carcinoma and urothelial carcinoma of the bladder identifies distinct molecular characterizations with potential implications for targeted therapy and immunotherapy. Front Immunol. 2022;13:1097730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rouanne M, Bajorin DF, Hannan R, et al. Rationale and outcomes for neoadjuvant immunotherapy in urothelial carcinoma of the bladder. Eur Urol Oncol. 2020;3:728–38. [DOI] [PubMed] [Google Scholar]
- [4].Al-Maghrabi JA, Khabaz MN. Clinical significance of galectin-3 expression in urinary bladder carcinoma. J Int Med Res. 2023;51:3000605231153323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18:609–24. [DOI] [PubMed] [Google Scholar]
- [6].Yang C, Tian Y, Zhao F, et al. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci . 2020;21:6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meltzer PS, Helman LJ. New horizons in the treatment of osteosarcoma. N Engl J Med. 2021;385:2066–76. [DOI] [PubMed] [Google Scholar]
- [8].Eaton BR, Schwarz R, Vatner R, et al. Osteosarcoma. Pediatr Blood Cancer. 2021;68(Suppl 2):e28352. [DOI] [PubMed] [Google Scholar]
- [9].Zhang MW, Zhang Y, Lv SY, et al. Application value of coaxial puncture needle (technique) in ultrasound-guided puncture biopsy of peripheral pulmonary masses. Medicine (Baltim). 2022;101:e31070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lieu D. FNA 2.0: value of cytopathologist-performed ultrasound-guided core-needle biopsy. Semin Diagn Pathol. 2022;39:426–35. [DOI] [PubMed] [Google Scholar]
- [11].Chen C, Hou J, Tanner JJ, et al. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci . 2020;21:2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang Q, Zhang T, Wu J, et al. Prognosis and risk factors of patients with upper urinary tract urothelial carcinoma and postoperative recurrence of bladder cancer in central China. BMC Urol. 2019;19:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nejat RJ, Purohit R, Goluboff ET, et al. Cure of undifferentiated small cell carcinoma of the urinary bladder with M-VAC chemotherapy. Urol Oncol. 2001;6:53–5. [DOI] [PubMed] [Google Scholar]
- [14].Tran L, Xiao JF, Agarwal N, et al. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21:104–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Miyake M, Nishimura N, Fujii T, et al. Recent advancements in the diagnosis and treatment of non-muscle invasive bladder cancer: evidence update of surgical concept, risk stratification, and BCG-treated disease. Int J Urol. 2023;30:944–57. [DOI] [PubMed] [Google Scholar]
- [16].Girardi DM, Ghatalia P, Singh P, et al. Systemic therapy in bladder preservation. Urol Oncol. 2023;41:39–47. [DOI] [PubMed] [Google Scholar]
- [17].Da W, Tao Z, Meng Y, et al. A 10-year bibliometric analysis of osteosarcoma and cure from 2010 to 2019. BMC Cancer. 2021;21:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lilienthal I, Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int J Mol Sci . 2020;21:6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Harris MA, Hawkins CJ. Recent and ongoing research into metastatic osteosarcoma treatments. Int J Mol Sci . 2022;23:3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Benjamin RS. Adjuvant and neoadjuvant chemotherapy for osteosarcoma: a historical perspective. Adv Exp Med Biol. 2020;1257:1–10. [DOI] [PubMed] [Google Scholar]
- [21].Nacev BA, Sanchez-Vega F, Smith SA, et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022;13:3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gangadharan S, Howlett DC. Re: ultrasound-guided lymph node sampling: accuracy of FNAC, end-cutting (Franseen), and side-cutting (Temno) needle biopsy techniques. Clin Radiol. 2022;77:e789–90. [DOI] [PubMed] [Google Scholar]
- [23].Hurry KJ, Karunaratne D, Westley S, et al. Ultrasound-guided core biopsy in the diagnosis of parotid neoplasia: an overview and update with a review of the literature. Br J Radiol. 2022;95:20210972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vlasak P, Bouda J, Kostun J, et al. Diagnostic reliability, accuracy and safety of ultrasound-guided biopsy and ascites puncture in primarily inoperable ovarian tumours. Anticancer Res. 2020;40:3527–34. [DOI] [PubMed] [Google Scholar]
- [25].Qin Y, Tu X, Huang M, et al. Novel long noncoding RNAs, LINC01093 and MYLK-AS1, serve as potential diagnostic and prognostic biomarkers or hepatocellular carcinoma. DNA Cell Biol. 2023;42:488–97. [DOI] [PubMed] [Google Scholar]
- [26].Cao X, Cui H, Ji X, et al. Determining the potential roles of branched-chain amino acids in the regulation of muscle growth in common carp (Cyprinus Carpio) based on transcriptome and MicroRNA sequencing. Aquac Nutr. 2023;2023:7965735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi X, Yu X, Wang J, et al. SIK2 promotes ovarian cancer cell motility and metastasis by phosphorylating MYLK. Mol Oncol. 2022;16:2558–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Teng F, Zhang JX, Chang QM, et al. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang S, Ma J, Wei S, et al. Identification of hub genes associated with progression and prognosis of bladder cancer by integrated bioinformatics analysis. Arch Esp Urol. 2022;75:779–90. [DOI] [PubMed] [Google Scholar]
- [30].Ye W, Chen L, Feng C, et al. CircMYLK promotes the growth, migration, invasion, and survival of bladder cancer cells by upregulating CCND3 level via competitively binding to miR-34a. Drug Dev Res. 2021;82:1206–16. [DOI] [PubMed] [Google Scholar]
- [31].Yang M, Zhang T, Zhang Y, et al. MYLK4 promotes tumor progression through the activation of epidermal growth factor receptor signaling in osteosarcoma. J Exp Clin Cancer Res. 2021;40:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhu G, Benayed R, Ho C, et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2019;32:609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kenney HM, Wu CL, Loiselle AE, et al. Single-cell transcriptomics of popliteal lymphatic vessels and peripheral veins reveals altered lymphatic muscle and immune cell populations in the TNF-Tg arthritis model. Arthritis Res Ther. 2022;24:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu Y, Wu X, Wang G, et al. CALD1, CNN1, and TAGLN identified as potential prognostic molecular markers of bladder cancer by bioinformatics analysis. Medicine (Baltim). 2019;98:e13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yoshida Y, Takahashi M, Yamanishi H, et al. Changes in the expression of smooth muscle cell-related genes in human dermal sheath cup cells associated with the treatment outcome of autologous cell-based therapy for male and female pattern hair loss. Int J Mol Sci . 2022;23:7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Carrasco R, Izquierdo L, van der Heijden AG, et al. Differential gene expression profile between progressive and de novo muscle invasive bladder cancer and its prognostic implication. Sci Rep. 2021;11:6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cheng Q, Tang A, Wang Z, et al. CALD1 modulates gliomas progression via facilitating tumor angiogenesis. Cancers (Basel). 2021;13:2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Du Y, Jiang X, Wang B, et al. The cancer-associated fibroblasts related gene CALD1 is a prognostic biomarker and correlated with immune infiltration in bladder cancer. Cancer Cell Int. 2021;21:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li C, Yang F, Wang R, et al. CALD1 promotes the expression of PD-L1 in bladder cancer via the JAK/STAT signaling pathway. Ann Transl Med. 2021;9:1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Diao CY, Guo HB, Ouyang YR, et al. Screening for metastatic osteosarcoma biomarkers with a DNA microarray. Asian Pac J Cancer Prev. 2014;15:1817–22. [DOI] [PubMed] [Google Scholar]