Abstract
The first common enzyme of isoleucine and valine biosynthesis, acetolactate synthase (ALS), is specifically inhibited by the herbicide sulfometuron methyl (SM). To further understand the physiological consequences of flux alterations at this point in metabolism, Escherichia coli genes whose expression was induced by partial inhibition of ALS were sought. Plasmid-based fusions of random E. coli DNA fragments to Photorhabdus luminescens luxCDABE were screened for bioluminescent increases in actively growing liquid cultures slowed 25% by the addition of SM. From more than 8,000 transformants, 12 unique SM-inducible promoter-lux fusions were identified. The lux reporter genes were joined to seven uncharacterized open reading frames, f253a, f415, frvX, o513, o521, yciG, and yohF, and five known genes, inaA, ldcC, osmY, poxB, and sohA. Inactivation of the rpoS-encoded sigma factor, ςS, reduced basal expression levels of six of these fusions 10- to 200-fold. These six genes defined four new members of the ςS regulon, f253a, ldcC, yciG, and yohF, and included two known members, osmY and poxB. Furthermore, the weak acid salicylate, which causes cytoplasmic acidification, also induced increased bioluminescence from seven SM-inducible promoter-lux fusion-containing strains, namely, those with fusions of the ςS-controlled genes and inaA. The pattern of gene expression changes suggested that restricted ALS activity may result in intracellular acidification and induction of the ςS-dependent stress response.
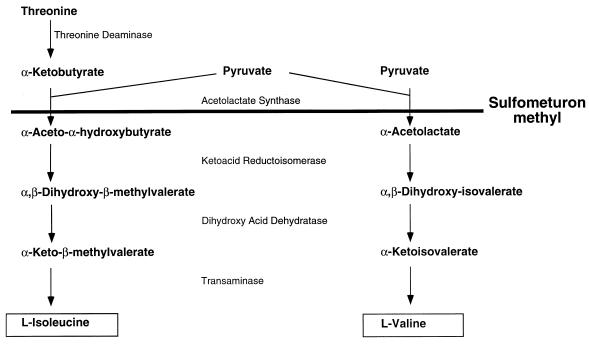
Sulfometuron methyl (SM) is a potent and specific inhibitor of the first common enzyme of isoleucine and valine biosynthesis (Fig. 1) in bacteria, fungi, and plants (23, 24, 41). Thus, SM is a useful tool for localized constriction of metabolic flux (26). Such inhibition of acetolactate synthase (ALS; EC 4.1.3.18) by SM results in starvation for isoleucine and valine as well as accumulation of its substrates, the α-ketoacids pyruvate and α-ketobutyrate (13, 27). These and other α-ketoacids, and their acyl coenzyme A derivatives, are important central metabolites, as they account for about 70% of carbon flux in Escherichia coli (17). Hence, changes in intracellular levels of α-ketoacids resulting from metabolic perturbations such as ALS inhibition may have multiple physiological consequences.
FIG. 1.
The biosynthetic pathway for synthesis of l-isoleucine and l-valine. ALS catalyzes the decarboxylation of pyruvate and condensation of the resultant hydroxyethyl-thiamine pyrophosphate moiety with α-ketobutyrate in isoleucine biosynthesis and condensation with another molecule of pyruvate in valine biosynthesis. In E. coli K-12, two ALS isozymes are normally expressed. In the strain used in this study, ALS I, which is insensitive to SM, was not expressed. Catalysis of this step was by ALS III, which is inhibited by SM.
Accumulation of α-ketobutyrate plays an important role in the deleterious biological effects of ALS inhibition in bacteria. This has been demonstrated in studies using an isoleucine feedback-resistant mutant threonine deaminase, which catalyzes conversion of threonine to α-ketobutyrate. Although SM-mediated growth inhibition of wild-type Salmonella typhimurium is fully reversed by isoleucine and valine addition, growth inhibition of the mutant is not fully alleviated by this addition (27). Thus, continued synthesis of α-ketobutyrate accounts for the isoleucine- and valine-independent SM-mediated inhibition. Such growth inhibition of the mutant strain is alleviated by the immediate biosynthetic precursor of valine, α-ketoisovalerate, suggesting the deleterious effects of competition between these two α-ketoacids (55). Furthermore, a number of SM-hypersensitive S. typhimurium mutants are defective in α-ketobutyrate degradation (27, 56). Lack of these α-ketobutyrate catabolic pathways, such as that mediated by acetate kinase and phosphotransacetylase (54), is suggested to result in higher levels of either this toxic intermediate or a by-product. However, there is not a close correlation at sublethal doses of SM between the degree of growth inhibition of S. typhimurium caused by SM and the accumulation of α-ketobutyrate (13). Likewise, the role of α-ketobutyrate accumulation in the phytotoxicity of ALS inhibition in plants is not certain (47). Thus, other approaches that may yield further insights into the physiological ramifications of flux alterations at this key point in intermediary metabolism are needed.
In this study, we analyzed alterations in gene expression induced by sublethal doses of SM that partially constrict flux through ALS. Typically, bacteria regulate transcription in response to conditions that alter cellular physiology. Often the set of proteins induced by a particular stress, a stimulon (37), includes some that eliminate the stress and others that are important for maintenance of cellular homeostasis. Thus, the profile of gene expression changes induced by any agent will reveal the nature of and responses to the stress condition. Such an approach may be particularly useful in understanding the biological consequences of metabolic flux alterations, such as that mediated by SM.
Reporter genes are commonly used to discover and characterize bacterial promoters activated by environmental stresses. Of the various reporter systems available, bacterial bioluminescence has the unique advantage that gene expression can be monitored in real time without cell lysis. Moreover, if a five-gene luxCDABE reporter is used, all of the agents required for bioluminescence, the five Lux polypeptides, O2, ATP, reduced flavin mononucleotide, and NADPH, are present in aerobically grown cells (32). In this work, we used a moderate-copy-number promoter probe vector, pDEW201, that contains a multiple cloning site between transcriptional terminators and a luxCDABE reporter gene complex from Photorhabdus luminescens. The expressed Lux proteins are stable at temperatures up to 45°C (52). Such a plasmid-based system can identify expression changes in essential genes because the chromosome remained unaltered. In addition, amplification of weak transcriptional signals may be important for detection of transcriptional activity from promoter fusions that would be undetected in single copy. Our experience has been that plasmid-based lux genetic fusions respond to the same regulatory controls as do chromosomal genes for one negatively (60) and several positively (8, 12, 50) controlled regulatory circuits.
We describe the use of such plasmid-based lux fusions to E. coli promoters to characterize gene expression changes following imposition of a metabolic perturbation. Partial inhibition of ALS by SM led to moderate increases in bioluminescence from E. coli strains containing SM-induced (smi) promoters controlling expression of luxCDABE. The majority of the 12 identified smi-luxCDABE fusions were induced by weak acid treatment and regulated by ςS. These results suggested that the physiological consequences of partial inhibition of ALS activity may be intracellular acidification and induction of the ςS-dependent stress response. This work thus provides an example of the interplay between metabolic flux alterations and global control of gene expression.
MATERIALS AND METHODS
E. coli strains and plasmids.
E. coli W3110 (14) was used for isolation of chromosomal DNA. E. coli DPD1675 [ilvB2101 ara thi Δ(proAB-lac) tolC::miniTn10] (58) was used as the host strain for screening for induction of bioluminescence from the chromosomal-luxCDABE genetic fusions. Strains isolated from this screening are listed in Table 1. Additional E. coli strains used were the otherwise isogenic pair MP180 (HfrH thi-1) and UM122 (HfrH thi-1 rpoS13::Tn10) (30) and an otherwise isogenic set: GC4468 (F− Δlac-4169 rpsL) (3), N7840 [F− Δlac4169 rpsL Δ(mar sad)1738] (43), N8452 [F− Δlac-4169 rpsL Δ(mar sad)1738 rob::kan] (from J. L. Rosner), and DPD2209 [F− Δlac-4169 rpsL Δ(mar sad)1738 rob::kan rpoS13::Tn10]. The latter strain was constructed by generalized transduction using phage P1clr100 (34) grown on strain UM122 as the donor and strain N8452 as the recipient, selecting for tetracycline resistance.
TABLE 1.
Characteristics of E. coli strains containing smi promoter-luxCDABE fusions
| Strain | Plasmid | Gene fused to luxCDABE | Basal RLUa | SM response ratiob | No. foundc |
|---|---|---|---|---|---|
| DPD2081 | pDEW213 | f415 | 0.2 | 1.83 ± 0.14 | 3 |
| DPD2084 | pDEW215 | yciG | 0.27 | 1.58 ± 0.22 | 1 |
| DPD2087 | pDEW218 | inaA | 9.8 | 1.57 ± 0.40 | 1 |
| DPD2088 | pDEW219 | yohF | 0.14 | 1.71 ± 0.12 | 2 |
| DPD2089 | pDEW220 | o82/o521 | 1.4 | 1.54 ± 0.11 | 1 |
| DPD2090 | pDEW221 | osmY | 0.08 | 1.78 ± 0.10 | 1 |
| DPD2092 | pDEW223 | o513 | 5.1 | 1.35 ± 0.19 | 1 |
| DPD3501 | pDEW301 | frvX | 13.4 | 3.09 ± 0.96 | 1 |
| DPD3505 | pDEW305 | f253a | 0.9 | 1.52 ± 0.29 | 2 |
| DPD3507 | pDEW307 | sohA | 62.4 | 1.20 ± 0.06 | 1 |
| DPD3509 | pDEW309 | poxB | 1.5 | 1.72 ± 0.09 | 1 |
| DPD3512 | pDEW312 | ldcC | 0.5 | 2.10 ± 0.22 | 1 |
| DPD2083 | pDEW201 | None | 0.002 |
The RLU reading from the culture grown in the wells of a microplate, untreated with SM, at the initial time point of the primary screen was recorded as the basal light output. This value was expected to roughly correlate to the amount of transcription initiation at promoters upstream of luxCDABE.
Calculated by dividing the RLU of the SM-treated culture by the RLU of the untreated culture at about 180 min (except the ratio for strain DPD3507, which was calculated at 120 min) after addition of cells to SM in the second screening test. Values are means and standard deviations of the triplicate cultures.
Number of independent plasmids found with the same fusion of E. coli chromosomal DNA to luxCDABE as in the representative strain listed. In each case when multiple hits were obtained, the joint of lux to the chromosomal segment was identical.
Plasmid pDEW201 (58) has the origin of replication and bla gene conferring ampicillin resistance from pBR322 and four transcription terminators upstream of promoterless P. luminescens luxCDABE genes. The multiple cloning site that lies between the terminators and lux contains unique EcoRI, BamHI, KpnI, and SacI sites. A plasmid, pDEW207, that contained the grpE heat shock promoter driving expression of luxCDABE was constructed by ligating the 0.6-kb BamHI fragment from plasmid pGrpELux5 (57) into BamHI-digested and calf intestinal alkaline phosphatase-treated pDEW201. The correct orientation of the grpE promoter was confirmed by HindIII digestion of the resultant plasmid. E. coli DPD2077 is a transformant of DPD1675 that contains pDEW207.
Growth media and chemicals.
The defined growth medium was Vogel-Bonner medium (11), with glucose as a carbon source, supplemented with thiamine, uracil, and proline. Ampicillin was added at either 25 or 10 μg/ml to this medium. The rich medium was LB (34) to which ampicillin was added at 150 or 50 μg/ml. SM was obtained from the Agricultural Products Department of the DuPont Company. A 2-mg/ml solution of SM in 0.01 N NaOH was prepared and stored at −20°C. Dilution of this SM stock to 32 μg/ml or less into the Vogel-Bonner medium did not affect the resultant pH. A 1 M stock solution of sodium salicylate, purchased from EM Science, in water was stored at −20°C.
Turbidity measurements and growth rate determinations in microplates.
Culture turbidity was routinely measured with a Klett-Summerson colorimeter by using the red filter. For measurement of growth rate inhibition by SM and ethanol, E. coli DPD1675 was grown at 37°C in the defined medium in a flask to early exponential phase (8 to 20 Klett units). Then 50 μl of this culture was placed in the wells of a sterile, clear microplate (Falcon Microtest III 96-well, flat-bottom tissue culture plate with low-evaporation lid) containing 50 μl of medium with various concentrations of the chemicals. The covered plate was incubated at 37°C. At various times after inoculation, the plate was shaken and the optical densities at 650 nm of the cultures in the wells of uncovered microplates were measured with a Molecular Devices 96 well plate reader. The background optical density at 650 nm from wells containing 100 μl of medium only was subtracted from all readings prior to plotting and calculation of growth rates.
Gene fusion library generation.
Chromosomal DNA isolated from E. coli W3110 was partially digested with Sau3A1 and size fractionated by agarose gel electrophoresis. A fraction with an average size of approximately 1.8 kb was ligated to pDEW201 that had previously been digested with BamHI and treated with calf intestinal alkaline phosphatase. The ligation products were used to transform ultracompetent E. coli XL2Blue cells (Stratagene) to ampicillin resistance, using the protocol provided by Stratagene. Preliminary characterization of individual random XL2Blue transformants indicated that all (16 of 16) contained insert DNA with sizes ranging from 0.9 to 3.0 kb. Approximately 24,000 of these transformants were pooled and used as a source of heterogeneous plasmid DNA isolated by using Qiagen tip20 columns. This plasmid DNA pool was used to transform (38) E. coli DPD1675, selecting for ampicillin resistance and using a 30-min phenotypic expression time to minimize the presence of siblings. Individual transformants were used to inoculate the 96-well sterile Falcon Microtest III tissue culture plates containing 190 μl of the defined medium with 25 μg of ampicillin per ml. These plates were covered and incubated overnight at 37°C.
Bioluminescence analysis.
The overnight cultures in 96-well plates were used for both permanent cryogenic storage (33) and dilution and regrowth to exponential phase in the defined medium containing 10 μg of ampicillin per ml. A 15-μl aliquot of the overnight culture was added to 150 μl of prewarmed medium in microplates and incubated at 37°C without shaking for 3 h. In the primary screen, these actively growing cultures were divided into SM-treated and untreated wells of sterile white microplates (Microlite; Dynex). Addition of 50 μl of the culture to 50 μl of fresh prewarmed medium lacking ampicillin but containing 4 μg of SM per ml yielded a final SM concentration of 2 μg/ml. For each culture, the untreated control was in the same microplate. Light production was measured in a Dynatech (now Dynex) ML3000 luminometer at 0, 90, and 180 min of incubation at 37°C after addition of cells to chemical. The dimensionless units of light production, relative light units (RLU), are obtained by comparison with the light reading from an internal light-emitting diode. The levels of light production of the SM-treated and untreated wells were compared for each culture. A ranged set of criteria was used to identify putative SM-inducible fusions. These criteria, which considered the increase in expression as calculated by both a difference in light production (ΔRLU = RLU [SM treated] − RLU [control]) and the ratio of light production (ratio = RLU [SM treated]/RLU [control]), were as follows: for ΔRLU between 0.02 and 0.1, the ratio was required to be ≥1.5; for ΔRLU between 0.1 and 1.0, the ratio was required to be ≥1.35; for ΔRLU between 1.0 and 10.0, the ratio was required to be ≥1.25; and for ΔRLU of >10.0, the ratio was required to be ≥1.20. Due to variabilities inherent in growing cells in microplates and the narrow range of SM resulting in induction of bioluminescence, it was likely that the number of SM-inducible genetic fusions identified represents an underestimate of the actual proportion of smi promoters in the E. coli chromosome.
Putative SM-inducible transformants were reisolated from the appropriate wells of the duplicate cultures stored at −80°C and retested in triplicate under the same conditions as above except that data were semicontinuously collected by using the cycle mode of the ML3000 luminometer, similar to previous descriptions (57). Those that showed SM-induced bioluminescence increases in the secondary screen were grown to exponential phase in a flask and then tested a third time at a variety of SM concentrations, using the cycle mode of the ML3000 luminometer. For all experiments, the actively growing culture was divided at the time of SM addition to ensure identical populations when the stress was imposed. Response ratios were calculated by dividing the RLU of the SM-treated culture by the RLU of the untreated control culture at each time point. Because SM reduced the growth rate, the increase of bioluminescence was an underestimate of the fold increase in light production per cell, as there were fewer cells in the SM-treated cultures than the untreated cultures at the end of the experiments.
The effects of other chemicals on bioluminescence were tested by using cultures that were grown to exponential phase at 37°C and divided at the time of chemical addition. Bioluminescence was quantitated in the cycle mode by using the ML3000 luminometer, and response ratios were calculated.
Plasmid isolation and insert analysis.
Plasmid DNA was isolated by using Qiagen tip100 columns and the protocol provided by the manufacturer. The size of the insert DNA of individual clones was estimated by digesting with EcoRI and SacI, followed by agarose gel electrophoresis and comparison with markers. DNA sequence data were obtained by using ABI Prism dye terminator cycle sequencing kits with AmpliTaq DNA polymerase and oligonucleotide primers specific for the regions of pDEW201 flanking the multiple cloning site in the transcription terminator region (5′-GGATCGGAATTCCCGGGGAT-3′) and in the luxC region (5′-CTGGCCGTTAATAATGAATG-3′). The sequence reactions were run on ABI 373A and 377 sequencers. DNA homologies with the entire E. coli genome sequence (9) were determined with the program BLAST (1) on the NCBI database. The ECDC database (20, 61) was used to determine the genetic map positions of the genes fused to lux.
RESULTS
Identification of smi promoters.
The ideal E. coli strain for these studies contains both ilvB and tolC mutations. An ilvB mutation eliminates the SM-resistant ALS isozyme I (25), and a tolC mutation results in lack of an outer membrane channel for efflux pumps (15, 19), making the cells sensitive to growth inhibition at reduced chemical concentrations (46). The growth of strain DPD1675, which contains both of these key mutations, was inhibited by addition of 1, 2, 4, or 8 μg of SM per ml. The treated cultures maintained exponential growth but at decreasing rates with increasing doses of SM (data not shown). For subsequent screening, 2 μg/ml (5.5 μM), which resulted in 25% growth rate inhibition, was used. This SM concentration also resulted in a partial decrease in the bioluminescence from strain DPD2077, which carries a plasmid with the E. coli heat shock promoter, grpE, driving P. luminescens luxCDABE (data not shown).
A screening protocol was used to identify isolates of strain DPD1675 containing rare E. coli promoter-luxCDABE fusions that, in contrast to the grpE-luxCDABE fusion and about 99% of the fusions in the library, yielded an increase in bioluminescence upon treatment with the sublethal dose of SM. Individual transformants of E. coli DPD1675 containing fusions of random E. coli chromosomal DNA to the P. luminescens luxCDABE were challenged with SM at 2 μg/ml while actively growing in microplates. Of 8,066 individual cultures screened, the bioluminescence from 19 strains was reproducibly SM inducible. Thus, the chromosomal DNA upstream of the luxCDABE reporter in these 19 fusion plasmids was presumed to contain an smi promoter.
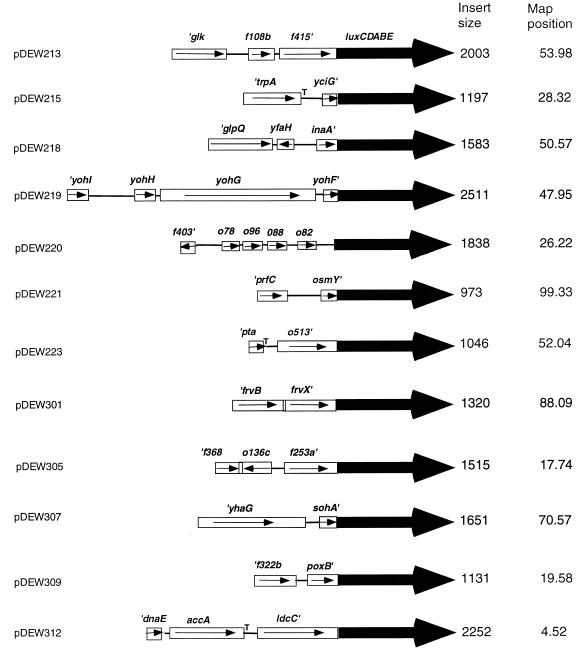
The identity of the E. coli chromosomal DNA in each of the 19 plasmids containing smi promoters was determined by DNA sequencing of each end of the inserted DNA followed by comparison to the complete E. coli genome sequence (9). Of these 19 plasmids, 3 contained regions of DNA from differing distal portions of the E. coli chromosome, most likely due to insertion of two or more independent Sau3A1 fragments into one plasmid. These were not further considered. Of the remaining 16, there were 12 unique chromosomal regions represented. Figure 2 shows the structures of these regions and their fusion point to the lux operon. In 11 of the cases, the lux operon was inserted within the coding sequences of genes or open reading frames (ORFs). In each of these cases, the direction of transcription of that gene or ORF was the same as that of the lux operon. Our assumption was that the promoter that drives the expression of the gene into which lux is inserted also controls lux operon expression. One exception was found in plasmid pDEW220, where the lux operon was inserted in an intergenic space. The direction of transcription of the nearest upstream ORF, o82, and the nearest downstream ORF, o521, was the same as for the lux operon. Since 201 of the 290 bp of the intergenic space separating o82 and o521 were present in this plasmid, it was not clear whether the promoter that drives o82 expression or that driving o521 expression was responsible for the lux operon expression.
FIG. 2.
Structure of smi-luxCDABE gene fusions (drawn approximately to scale). Known or proposed terminators (T) are shown. The designations for ORFs are from ECDC, release 28 (20, 61), as are the map positions in minutes for the gene proximal to luxC.
Characterization of 12 smi promoter-lux fusions.
Table 1 summarizes the basal, uninduced bioluminescence and the SM-induced response ratio of strain DPD1675 containing each of the 12 unique smi fusions. Also shown in Table 1 is the number of times each chromosomal segment was found in these screens. Saturation of the genome was clearly not reached because most smi fusions were found only once. Although this survey was not exhaustive, the genes found should be representative of the types of genes that are induced by SM-mediated inhibition of ALS. The basal level of light production from strain DPD1675 containing each fusion was substantially greater than that of strain DPD1675 containing plasmid pDEW201. This was consistent with the presence of promoter sequences in each of the DNA inserts. Furthermore, the range of promoter strengths among these 12 smi promoters was large; the uninduced bioluminescent activities differed by a factor of more than 500. Upon treatment with the sublethal SM dose of 2 μg/ml, the induction responses observed were modest, ranging from 20% increases to threefold increases.
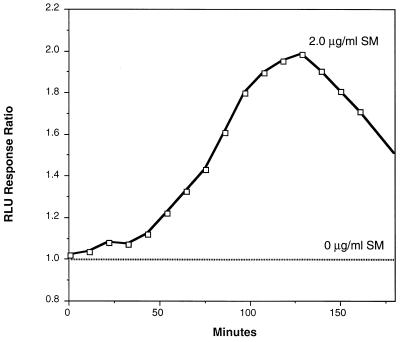
The time course of bioluminescence induction of one such smi fusion is shown in Fig. 3. These kinetics represent a typical response in that there was lag time with little change in bioluminescence, followed by an increase in bioluminescence relative to the control untreated sample. The lag time presumably represents the time required for the stress response to be initiated and for transcription and translation of the luxCDABE reporter complex.
FIG. 3.
Kinetics of bioluminescent induction by SM treatment of E. coli DPD2088 containing plasmid pDEW219. The actively growing culture in defined medium was treated with 2 μg of SM per ml at time zero. The response ratio is the RLU of the SM-treated culture divided by the RLU of the untreated culture at each time point. The average of duplicates for both treated and untreated cultures was plotted.
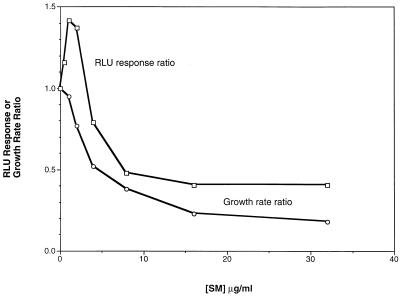
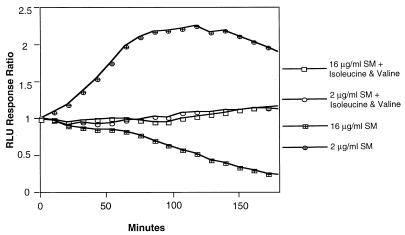
Dose-response curves for SM effects on bioluminescence revealed that a narrow range of SM concentrations yielded an induction response. Figure 4 shows a representative example of the strain containing pDEW213. At SM concentrations that resulted in growth rate inhibition of 50% or more, the light production was less than that of the parallel, untreated culture. This “lights-off” response at higher SM concentrations was likely due to an insufficiency of isoleucine and valine that limited formation of the Lux proteins. As expected, the SM effects of increased bioluminescence at lower concentrations and the lights-off response at higher concentrations upon each of the 12 smi fusions were overcome by isoleucine and valine supplementation. An example is shown in Fig. 5. Light production in the presence of 16 μg of SM per ml from the strain containing pDEW305 was less than that from the untreated control but was restored by isoleucine and valine addition. Likewise, the induction of bioluminescence by SM at 2 μg/ml observed in the absence of isoleucine and valine was prevented by their addition. Thus, adequate SM needed to be added for a response to be induced, but higher concentrations precluded a bioluminescent report of the response.
FIG. 4.
Dose response to SM of strain DPD2081 containing plasmid pDEW213. The bioluminescence response ratio was calculated by dividing the RLU of each SM-treated culture by the RLU of the untreated culture at 165 min after SM addition, using the average of duplicate cultures. The growth rate of DPD2081 in defined medium growing at 37°C in microplates was determined from the slope of the exponential curve fit of optical density versus time. The growth rate reduction ratio was calculated by dividing the growth rate of each SM-treated culture by the growth rate of the control untreated culture.
FIG. 5.
Prevention of toxic and inducing effects of SM by addition of isoleucine and valine in strain DPD3505 containing plasmid pDEW305. A culture was grown to exponential phase in defined medium without isoleucine or valine. Addition of these two amino acids (each at 42 μg/ml) was made at the same time as addition of SM to the divided culture. The response ratios were calculated at each time point by dividing the RLU from the SM-treated culture without isoleucine and valine by the RLU from the otherwise identical culture lacking SM. Likewise, the response ratio for the SM-treated culture with isoleucine and valine was calculated from the control with isoleucine and valine addition but lacking SM. The average of duplicates was used for each data point.
Specificity of smi promoter induction.
To test if the induction of any of these promoters was due simply to partial growth rate inhibition, the responses to an unrelated growth inhibitory chemical were tested. Cultures growing in defined medium in microplates were stressed by ethanol additions at the sublethal concentrations of 2 and 4% (44). These concentrations of ethanol reduced the growth rate of strain DPD1675 in microplates by 8 and 12%, respectively. The bioluminescence from strain DPD2077 with the E. coli heat shock grpE promoter driving P. luminescens luxCDABE was increased 1.3-fold at 50 min after addition of either 2 or 4% ethanol. These concentrations of ethanol, however, did not induce increased bioluminescence in 11 of the 12 distinct smi fusions. The one exception was strain DPD2081 containing plasmid pDEW213. While the addition of 4% ethanol did not result in bioluminescence of DPD2081 greater than that of the untreated control, the bioluminescence was increased 1.4-fold at 60 min after addition of 2% ethanol. The bioluminescence of DPD2081 was also noted to be induced by a wide variety of growth-inhibiting chemicals (data not shown). Thus, activation of the promoter driving expression of the f415′-luxCDABE fusion in DPD2081 may be tied to growth rate reduction. SM induction of the other 11 smi promoters, however, was not simply due to reduction in growth rate.
Promoters regulated by ςS.
What are the physiological consequences of partial ALS inhibition? Two possibilities were suggested by the identified smi promoter-luxCDABE fusions. One was that treatment with SM may result in induction of the ςS-dependent stress response. This regulatory circuit is induced by numerous stresses, including entry into stationary phase. At least two genes, poxB (10) in pDEW309 and osmY (21, 63) in pDEW221, regulated by ςS were among the 12 smi fusions. Furthermore, the bioluminescence of several of the smi fusion strains, including the poxB and osmY fusions, appeared to increase dramatically as the culture in the microplates approached stationary phase (data not shown), suggesting that others among the smi fusions may be controlled by ςS. This was tested by placing each of the 12 plasmids in a pair of E. coli strains, containing either an rpoS+ or nonfunctional rpoS allele but otherwise isogenic. As shown in Table 2, five plasmids (pDEW213, pDEW218, pDEW223, pDEW301, and pDEW307) expressed bioluminescence which was not substantially altered by the two rpoS alleles. In contrast, the basal bioluminescence expressed from another group of six plasmids was dramatically (11- to 188-fold) depressed in the rpoS mutant. Such a result was expected for loss of an element required for transcription. Thus, these results suggested a strong ςS dependence of the promoters driving transcription of the luxCDABE fusions to poxB (pDEW309), osmY (pDEW221), yciG (pDEW215), yohF (pDEW219), f253a (pDEW305), and ldcC (pDEW312). For one plasmid, pDEW220, there was an intermediate (sixfold) reduction attributed to the rpoS mutation. This plasmid may have more than one promoter in the cloned region. Thus, of 12 smi fusions, the expression of 6 was clearly controlled by ςS.
TABLE 2.
Effect of rpoS mutation on smi promoter-luxCDABE fusionsa
| Plasmid | Gene fused to luxCDABE | RLU inb:
|
rpoS de- pendence ratioc | |
|---|---|---|---|---|
| E. coli MP180 (rpoS+) | E. coli UM122 (rpoS) | |||
| pDEW213 | f415 | 0.0033 ± 0.0014 | 0.0079 ± 0.0070 | 0.42 |
| pDEW215 | yciG | 1.11 ± 0.09 | 0.0059 ± 0.0020 | 188 |
| pDEW218 | inaA | 2.44 ± 0.43 | 2.60 ± 0.55 | 0.94 |
| pDEW219 | yohF | 0.455 ± 0.040 | 0.00888 ± 0.00114 | 51 |
| pDEW220 | o82/0521 | 3.82 ± 0.86 | 0.642 ± 0.040 | 6.0 |
| pDEW221 | osmY | 0.362 ± 0.073 | 0.0081 ± 0.0018 | 45 |
| pDEW223 | o513 | 7.09 ± 1.77 | 9.61 ± 0.74 | 0.74 |
| pDEW301 | frvX | 0.161 ± 0.034 | 0.146 ± 0.013 | 1.1 |
| pDEW305 | f253a | 0.947 ± 0.077 | 0.0187 ± 0.005 | 51 |
| pDEW307 | sohA | 30.0 ± 3.0 | 24.1 ± 2.5 | 1.2 |
| pDEW309 | poxB | 2.38 ± 0.30 | 0.0241 ± 0.0121 | 99 |
| pDEW312 | ldcC | 1.69 ± 0.20 | 0.148 ± 0.014 | 11 |
Visual inspection in a dark room of the bioluminescence of hundreds of transformants on petri plates confirmed the quantitative results presented. Either there was a dramatic loss of bioluminescence in the rpoS host or there was no visually discernible difference between two host strains.
Cultures of two transformants were grown in LB medium containing 150 μg of ampicillin per ml overnight at 37°C, diluted into LB medium containing 50 μg of ampicillin per ml, and grown to log phase (20 to 24 Klett units) at 37°C. The bioluminescence from two 100-μl aliquots of each of the duplicate cultures was measured. Values are means and standard deviations of the four measurements.
Calculated as follows: RLU [transformants of MP180 (rpoS+)]/RLU [transformants of UM122 (rpoS)].
Weak acid-inducible promoters.
Another possible stress sustained by the cell when ALS is partially inhibited is cytoplasmic acidification. Included in the set of smi fusions was inaA, a known acid-inducible gene (49, 62), and ldcC, encoding a lysine decarboxylase (18). The function of inaA is not known (43). In contrast, the activity of lysine decarboxylase in converting lysine to cadaverine, an alkaline molecule, could neutralize acids. A prediction of the SM-mediated acidification hypothesis was that other promoters responsive to cytoplasmic acidification may be among the set of smi fusions. This was explored by testing for induction by salicylate, a membrane-permeant weak acid that results in cytoplasmic acidification and potent induction of inaA expression (49). As expected, the bioluminescence from the strain containing the inaAluxCDABE fusion was induced 17-fold after treatment with 5 mM sodium salicylate and 21-fold after treatment with 10 mM sodium salicylate (Table 3). There was also a strong 13-fold induction of bioluminescence upon treatment with 10 mM sodium salicylate of the strain containing the poxB-luxCDABE fusion (Table 3). In addition, moderate (two- to fivefold) increases in bioluminescence were induced by 10 mM salicylate for the strains containing the yciG, yohF, osmY, f253a, and ldcC fusions (Table 3). Yet addition of salicylate can be ruled out as having a general enhancing effect on bioluminescence because there were several fusions that were not induced by salicylate addition (Table 3). The genes represented by these fusions, o513, frvX, and sohA, are probably not involved in a response to internal acidification.
TABLE 3.
Effects of 10 mM salicylate on bioluminescence induction responses in mar+ rob+ and Δmar rob hosts
| Plasmid | Gene fused to luxCDABE | Salicylate response ratioa in:
|
|
|---|---|---|---|
| GC4468 (mar+ rob+) | N8452 (Δmar rob) | ||
| pDEW213 | f415 | NDb | ND |
| pDEW215 | yciG | 3.4 | 5.3 |
| pDEW218 | inaA | 20.6 | 0.92 |
| pDEW219 | yohF | 5.4 | 5.2 |
| pDEW220 | o82/o521 | 1.8 | 4.0 |
| pDEW221 | osmY | 2.7 | 3.9 |
| pDEW223 | o513 | 0.05 | 0.21 |
| pDEW301 | frvX | 0.62 | 0.45 |
| pDEW305 | f253a | 2.6 | 1.8 |
| pDEW307 | sohA | 1.1 | 1.0 |
| pDEW309 | poxB | 12.8 | 4.3 |
| pDEW312 | ldcC | 3.8 | 3.2 |
The cultures were grown to log phase (30 to 42 Klett units) in LB medium containing 50 μg of ampicillin per ml. The culture was then split, and 50 μl was added to 50 μl of LB medium containing or lacking 20 mM sodium salicylate, yielding a final concentration of 10 or 0 mM. The response ratio, which was the RLU of the salicylate-treated culture divided by the RLU of the untreated culture, was calculated from the mean of duplicate cultures at 30 min after combining the cells and salicylate.
ND, not determined. The bioluminescence from E. coli GC4468 or N8452 containing pDEW213 was very low and was reduced below the detection limit by addition of 10 mM salicylate.
The salicylate-mediated induction of inaA is known to be partially regulated by the multiple antibiotic resistance (mar) stress response system (43) through binding of salicylate to the repressor of the mar operon, MarR (31). As in published results (43), placement of pDEW218 containing the inaA-luxCDABE fusion into a Δmar strain greatly decreased (to 2.4-fold) but did not eliminate induction of bioluminescence upon addition of 5 mM salicylate. However, placement of this plasmid in a strain lacking both mar and rob, which encodes a DNA binding protein (48) that can also activate transcription of inaA (3), had a more substantial effect on bioluminescence induction. There was no increase upon addition of 10 mM salicylate (Table 3), and the induction by 5 mM salicylate was reduced to 1.2-fold. Similar results on the effects of a strain carrying both mar and rob mutations have also been obtained for a chromosomal inaA-lacZ transcriptional fusion (42). Interestingly, the effect of the double-mutant host strain differed for the other smi fusions. The salicylate-mediated induction of the group of moderately induced smi fusions remained in the two- to fivefold range in the mar rob double mutant (Table 3). In contrast to both inaA-luxCDABE and the moderately induced fusions, the salicylate-mediated induction of the poxB-luxCDABE fusion was substantially decreased but not eliminated in the double mutant (Table 3). The residual degree of salicylate induction of this fusion was similar to that of the moderately induced fusions, all of which were controlled by ςS. In a triple mutant host strain lacking function of mar, rob, and rpoS, the salicylate induction of poxB-luxCDABE was eliminated; the response ratio to 10 mM salicylate was 0.45, and that to 5 mM salicylate was 0.65.
DISCUSSION
Specific chemical inhibitors of metabolic enzymes provide a useful mechanism for flux alteration, allowing analysis of actively growing cultures abruptly stressed by constricted flux at a precise point. We used sublethal doses of SM and found unexpected changes in gene expression induced by partial inhibition of ALS, the first common step of branched-chain amino acid biosynthesis. Promoters associated with the amino acid starvation response were not found in this survey, which likely indicates that the level of starvation for isoleucine and valine was not severe when SM inhibited the growth rate by 25%. Likewise, this level of ALS inhibition did not induce the heat shock-controlled grpE promoter, indicating that the level of amino acid limitation was not severe enough to cause substantial amounts of nonnative proteins to accumulate. Thus, the promoters activated by this flux constriction represent responses to other, perhaps more subtle, physiological perturbations. That these perturbations were not severe may be reflected in the relatively modest degree of inductions observed. Yet these were clearly due to SM-mediated inhibition of branched-chain amino acid biosynthesis rather than an unknown effect of SM because the presence of isoleucine and valine prevented SM-mediated induction of all the identified smi-lux fusions.
An interesting and coherent picture emerged from the pattern of promoters found to be activated by partial ALS inhibition. The majority of the smi promoters were controlled by ςS and also induced by weak acid treatment. The latter observation suggested the possibility that inhibition of ALS in E. coli results in cytoplasmic acidification. Although such acidification may not have been previously considered as an immediate consequence of partial ALS inhibition, it is plausible because of the considerable flux through the branched-chain amino acid biosynthetic pathway. For example, a maximal synthesis rate of α-ketobutyrate in S. typhimurium is estimated at 6 nmol/min/109 cells from the rates of its accumulation and degradation (27). Furthermore, in accordance with inhibition of ALS leading to acidification, overexpression of ALS in E. coli has been used to direct metabolism away from production of acidic by-products (2). Induction of acid-responsive gene products may allow the cell to combat this acidification stress by neutralization or other strategies.
The majority of the acid-responsive smi promoters found were members of the ςS regulon. This included the known ςS regulon genes, osmY and poxB, and several newly identified members of the ςS regulon, f253a, ldcC, yciG, and yohF. The sublethal SM treatment may have resulted in increased cellular levels of ppGpp, a positive effector of ςS levels (16, 22). Alternatively, acidification stress may be the trigger that initiates the ςS-dependent stress response. A connection between ςS and acid stress responses has been characterized in E. coli and S. typhimurium (6). Weak acid treatment, which results in cytoplasmic acidification, induces expression of rpoS in E. coli (45). In S. typhimurium, rpoS mutants do not have the acid-inducible resistance to weak acids characteristic of the wild type (5). It has also been shown that ςS is acid inducible and controls expression of at least eight other acid-inducible proteins (29). These ςS-dependent acid-inducible proteins include the S. typhimurium homolog of OsmY (6), which was found in this study to have an smi promoter. Furthermore, another S. typhimurium ςS-dependent acid-inducible protein is encoded by gene orf3 of the tonB-trpA region or yciE (6) that is nearby and possibly cotranscribed with yciG (51), another of the E. coli smi promoters. The acid induction of five proteins is negatively regulated by mviA in S. typhimurium (7). The product of rssB (or sprE), the E. coli mviA homolog, plays a similar role in regulating RpoS stability (36, 40). Whether cytoplasmic acidification is one signal that the cell uses to induce the ςS-dependent stress response generally or whether there are specific ςS-regulated genes that also respond to acidification remains to be clarified.
The global transcriptional regulator MarA controls responses to some weak acids such as salicylate (35). Here we showed Mar regulation of salicylate induction of two smi promoters: inaA, a known Mar regulon gene, and poxB, not previously known to be a member of the Mar regulon. These two promoters differed in that inaA was unaffected by an rpoS mutation, while poxB was strongly affected by loss of rpoS function. Our results are consistent with the salicylate induction of poxB expression being under dual regulation by mar and rpoS. We have also found that expression of the poxB- luxCDABE fusion is induced by methyl viologen treatment under the control of soxRS (4). Furthermore, expression of poxB is affected by mutations in the regulatory genes, lrp and hns (28).
The multiple regulation of poxB suggests a key cellular role for its product, pyruvate oxidase. However, the phenotypic consequences of poxB mutations are difficult to ascertain (10). Likewise, pyruvate oxidase induction by SM treatment and the reaction that it catalyzes, the conversion of pyruvate to acetate and CO2, suggest a role for it in response to ALS inhibition. However, a poxB mutant of E. coli is not altered in sensitivity to SM (59). Either the induction of pyruvate oxidase plays a minor adaptive role in SM stress or this induction may not be a specific defense response. Possibly the cell responds to certain stresses by inducing a gene expression pattern similar to that found in the stationary phase. Notably, the degree of growth rate reduction by SM was much less severe than that found to be necessary in chemostats for induction of ςS-controlled genes (39). The induction of the ςS-mediated stress response may reflect an alternative survival strategy to that afforded from induction of proteins specific for combating the adverse effects of a stress.
The use of an easily assayed transcriptional reporter was critical for our random screening using cells growing in liquid medium. Thus, the five-gene luxCDABE operon that does not require breaking cells or substrate addition was chosen. The sensitivity of the lux reporter was important in the identification the modestly activated promoters described here. Such threefold or less SM-induced increases in reporter gene activity may not have been readily uncovered in assays using standard reporters with petri plate-based methods or by newer approaches based on cell sorting (53). Furthermore, the advantage of the large dynamic range of the luxCDABE reporter was demonstrated by the identification of promoters of widely disparate strengths under identical screening conditions.
ACKNOWLEDGMENTS
We thank S. Stack for sequencing, M. Bailey for oligonucleotide synthesis, P. Loewen for E. coli strains, and J. L. Rosner for E. coli strains and discussions.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aristidou A A, San K Y, Bennett G N. Metabolic engineering of Escherichia coli to enhance recombinant protein production through acetate reduction. Biotechnol Prog. 1995;11:475–478. doi: 10.1021/bp00034a019. [DOI] [PubMed] [Google Scholar]
- 3.Ariza R R, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayers B L. B.S. thesis. Swarthmore, Pa: Swarthmore College; 1997. [Google Scholar]
- 5.Baik H S, Bearson S, Dunbar S, Foster J W. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology. 1996;142:3195–3200. doi: 10.1099/13500872-142-11-3195. [DOI] [PubMed] [Google Scholar]
- 6.Bearson S, Bearson B, Foster J W. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 7.Bearson S, Benjamin W H, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkin S, Smulski D R, Vollmer A C, Van Dyk T K, LaRossa R A. Oxidative stress detection with Escherichia coli harboring a katG′::lux fusion. Appl Environ Microbiol. 1996;62:2252–2256. doi: 10.1128/aem.62.7.2252-2256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 10.Chang Y-Y, Wang A Y, Cronan J E., Jr Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol Microbiol. 1994;11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 11.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 12.Dukan S, Dadon S, Smulski D R, Belkin S. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl Environ Microbiol. 1996;62:4003–4008. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epelbaum S, Chipman D M, Barak Z. Metabolic effect of inhibitors of two enzymes of the branched-chain amino acid pathway in Salmonella typhimurium. J Bacteriol. 1996;178:1187–1196. doi: 10.1128/jb.178.4.1187-1196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernsting B R, Atkinson M R, Ninfa A J, Matthews R G. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J Bacteriol. 1992;174:1109–1118. doi: 10.1128/jb.174.4.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentry D R, Hernandez V J, Nguyen L H, Jensen D B, Cashel M. Synthesis of the stationary-phase sigma factor ςS is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingraham J L, Maaloe O, Neidhardt F C. Growth of the bacterial cell. Sunderland, Mass: Sinauer Associates, Inc.; 1983. pp. 124–132. [Google Scholar]
- 18.Kikuchi Y, Kojima H, Tanaka T, Takatsuka Y, Kamio Y. Characterization of a second lysine decarboxylase isolated from Escherichia coli. J Bacteriol. 1997;179:4486–4492. doi: 10.1128/jb.179.14.4486-4492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 20.Kröger M, Wahl R. Compilation of DNA sequences of Escherichia coli K12; description of the interactive databases ECD and ECDC (update 1996) Nucleic Acids Res. 1997;25:39–42. doi: 10.1093/nar/25.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange R, Barth M, Hengge-Aronis R. Complex transcriptional control of the ςS-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggest novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J Bacteriol. 1993;175:7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the ςS subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaRossa R A, Falco S C. Amino acid biosynthetic enzymes as targets of herbicide action. Trends Biotechnol. 1984;2:158–161. [Google Scholar]
- 24.LaRossa R A, Schloss J V. The sulfonylurea herbicide sulfometuron methyl is an extremely potent and selective inhibitor of acetolactate synthase in Salmonella typhimurium. J Biol Chem. 1984;259:8753–8757. [PubMed] [Google Scholar]
- 25.LaRossa R A, Smulski D R. ilvB-encoded acetolactate synthase is resistant to the herbicide sulfometuron methyl. J Bacteriol. 1984;160:391–394. doi: 10.1128/jb.160.1.391-394.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaRossa R A, Van Dyk T K. Utilization of sulfometuron methyl, an acetolactate synthase inhibitor, in molecular biological and metabolic studies of plants and microbes. Methods Enzymol. 1988;166:97–107. doi: 10.1016/s0076-6879(88)66015-0. [DOI] [PubMed] [Google Scholar]
- 27.LaRossa R A, Van Dyk T K, Smulski D R. Toxic accumulation of α-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J Bacteriol. 1987;169:1372–1378. doi: 10.1128/jb.169.4.1372-1378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurent-Winter C, Ngo S, Danchin A, Bertin P. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response. Identification of targets by two-dimensional electrophoresis. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 29.Lee I S, Lin J, Hall H K, Bearson B, Foster J W. The stationary-phase sigma factor ςS(RpoS) is required for a sustained acid tolerance response in virulent Samonella typhiumurium. Mol Microbiol. 1995;17:155–167. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- 30.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meighen E A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menzel R. A microtiter plate-based system for the semiautomated growth and assay of bacterial cells for β-galactosidases activity. Anal Biochem. 1989;181:40–50. doi: 10.1016/0003-2697(89)90391-6. [DOI] [PubMed] [Google Scholar]
- 34.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 35.Miller P F, Sulavik M C. Overlaps and parallels in the regulation of intrinsic multiple-antibiotic resistance in Escherichia coli. Mol Microbiol. 1996;21:441–448. doi: 10.1111/j.1365-2958.1996.tb02553.x. [DOI] [PubMed] [Google Scholar]
- 36.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςS subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 37.Neidhardt F C, Savageau M A. Regulation beyond the operon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1310–1324. [Google Scholar]
- 38.Nishimura A, Morita M, Nishimura Y, Sugino Y. A rapid and highly efficient method for preparation of competent Escherichia coli cells. Nucleic Acids Res. 1990;18:6169. doi: 10.1093/nar/18.20.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray T. Site of action of chlorsulfuron. Plant Physiol. 1984;75:827–831. doi: 10.1104/pp.75.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosner, J. L. Personal communication.
- 43.Rosner J L, Slonczewski J L. Dual regulation of inaA by the multiple antibiotic resistance (Mar) and superoxide (SoxRS) stress response systems of Escherichia coli. J Bacteriol. 1994;176:6262–6269. doi: 10.1128/jb.176.20.6262-6269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rupani S P, Gu M B, Konstantinov K B, Dhurjati P S, Van Dyk T K, LaRossa R A. Characterization of the stress response of a bioluminescent biological sensor in batch and continuous cultures. Biotechnol Prog. 1996;12:387–392. doi: 10.1021/bp960015u. [DOI] [PubMed] [Google Scholar]
- 45.Schellhorn H E, Stones V L. Regulation of katF and katE in Escherichia coli by weak acids. J Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnaitman C. Improved strains for target-based chemical screening. ASM News. 1991;57:612. [Google Scholar]
- 47.Shaner D L, Singh B K. Phytotoxicity of acetohydroxyacid synthase inhibitors is not due to accumulation of 2-ketobutyrate and/or 2-aminobutyrate. Plant Physiol. 1993;103:1221–1226. doi: 10.1104/pp.103.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skarstad K, Thony B, Hwang D S, Kornberg A. A novel binding protein of the origin of the Escherichia coli chromosome. J Biol Chem. 1993;268:5365–5370. [PubMed] [Google Scholar]
- 49.Slonczewski J L, Gonzalez T N, Bartholomew F M, Holt N J. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987;169:3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smulski, D. R., and R. A. LaRossa. Unpublished data.
- 51.Stoltzfus A, Leslie J F, Milkman R. Molecular evolution of the Escherichia coli chromosome. I. Analysis of structure and natural variation in a previously uncharacterized region between trp and tonB. Genetics. 1988;120:345–358. doi: 10.1093/genetics/120.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szittner R, Meighen E. Nucleotide sequence, expression, and properties of luciferase coded by lux genes from a terrestrial bacterium. J Biol Chem. 1990;265:16581–16587. [PubMed] [Google Scholar]
- 53.Valdivia R H, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 54.Van Dyk T K, LaRossa R A. Involvement of ack-pta operon products in α-ketobutyrate metabolism by Salmonella typhimurium. Mol Gen Genet. 1987;207:435–440. doi: 10.1007/BF00331612. [DOI] [PubMed] [Google Scholar]
- 55.Van Dyk T K, LaRossa R A. Prevention of endogenous 2-ketobutyrate toxicity in Salmonella typhimurium. In: Barak Z, Chipman D M, Schloss J V, editors. Biosynthesis of branched chain amino acids. New York, N.Y: VCH and Balaban Publishers; 1990. pp. 123–130. [Google Scholar]
- 56.Van Dyk T K, LaRossa R A. Sensitivity of a Salmonella typhimurium aspC mutant to sulfometuron methyl, a potent inhibitor of acetolactate synthase II. J Bacteriol. 1986;165:386–392. doi: 10.1128/jb.165.2.386-392.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati P S, LaRossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Dyk T K, Rosson R A. Photorhabdus luminescens luxCDABE promoter probe vectors. Methods Mol Biol. 1998;102:85–95. doi: 10.1385/0-89603-520-4:85. [DOI] [PubMed] [Google Scholar]
- 59.Van Dyk T K, Smulski D R, Chang Y-Y. Pleiotropic effects of poxA regulatory mutations of Escherichia coli and Salmonella typhimurium, mutations conferring sulfometuron methyl and α-ketobutyrate hypersensivtiy. J Bacteriol. 1987;169:4540–4546. doi: 10.1128/jb.169.10.4540-4546.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vollmer A C, Belkin S, Smulski D R, Van Dyk T K, LaRossa R A. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux or alkA′::lux reporter plasmids. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wahl R, Kröger M. ECDC—a totally integrated and interactively usable genetic map of Escherichia coli K12. Microbiol Res. 1995;150:7–61. doi: 10.1016/S0944-5013(11)80034-0. [DOI] [PubMed] [Google Scholar]
- 62.White S, Tuttle F E, Blankenhorn D, Dosch D C, Slonczewski J. pH dependence and gene structure of inaA in Escherichia coli. J Bacteriol. 1992;174:1537–1543. doi: 10.1128/jb.174.5.1537-1543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yim H H, Brems R L, Villarego M. Molecular characterization of the promoter of osmY, an rpoS-dependent gene. J Bacteriol. 1994;176:100–107. doi: 10.1128/jb.176.1.100-107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]