Abstract
The Yersinia enterocolitica inv gene encodes the primary invasion factor invasin, which has been previously shown to be critical in the initial stages of infection. The expression of inv is influenced by growth phase and temperature and is maximal during late exponential-early stationary phase at 23°C. In addition, motility of Y. enterocolitica is regulated by temperature. Y. enterocolitica cells are motile when grown at lower temperatures (30°C or below), while bacteria grown at 37°C are nonmotile. This study was initiated to determine the molecular basis for the temperature regulation of inv expression. Two mutants were isolated that both showed a significant decrease in invasin expression but are hypermotile when grown at 23°C. The first mutant (JB1A8v) was a result of a random mTn5Km insertion into the uvrC gene. The uvrC mutant JB1A8v demonstrated a significant decrease in inv and an increase in fleB (encodes flagellin) expression. These results suggest that expression of inv and flagellin genes is coordinated at the level of transcription. The second regulatory mutant, JB16v, was a result of a targeted insertion into a locus similar to sspA which in E. coli encodes a stationary-phase regulator. The E. coli sspA gene was cloned and assayed for complementation in both of the regulatory mutants. It was determined that E. coli sspA restored invasin expression in both the uvrC mutant and the sspA mutant. In addition, the complementing clone decreased flagellin levels in these mutants.
Yersinia enterocolitica is a common human enteropathogen that causes gastrointestinal syndromes ranging from mild diarrhea to mesenteric adenitis resembling appendicitis (7). The principal mode of transmission for Y. enterocolitica is consumption of contaminated food or water, although person-to-person transmission can occur through the fecal-oral route (13, 19, 59). After ingestion, the bacteria migrate through the gastrointestinal tract to the terminal ileum. It is at the terminal ileum where the bacteria attach and subsequently invade the mucosal epithelial cells or M cells overlying the lymphoid follicles (Peyer’s patches) (18, 22, 60). Three genes whose products contribute to attachment and invasion of cultured cells by enteropathogenic Yersinia species are inv, ail, and yadA (6, 25, 40, 44, 65). The Y. enterocolitica chromosomal gene inv encodes the primary invasion factor invasin (44). Invasin is a 92-kDa outer membrane protein that has been demonstrated to bind several members of the integrin family and to be important for penetration of the intestinal epithelium by Y. enterocolitica (24, 44, 67). Once Y. enterocolitica organisms enter host tissue, the capacity of the bacteria to survive and replicate is associated with the presence of a 70-kb virulence plasmid (pYV) (10, 21, 47). Encoded on the virulence plasmid are YadA and a set of secreted proteins termed Yops (Yersinia outer proteins) which mediate functions such as adherence, internalization, serum resistance, autoagglutinability, resistance to phagocytosis, and cytotoxicity (6, 9, 30, 33, 34, 37, 42, 48, 51, 54, 65). Ail functions both as an adhesin and as an invasion factor for Y. enterocolitica (40). In addition, Ail has been shown to protect against the bactericidal activity of human serum (5, 46, 61).
Many identified Y. enterocolitica virulence factors are expressed in a temperature-dependent manner in vitro. For example, ail, myfA (encodes fibrillar surface antigen), yadA, and the yop genes are most prominently expressed at 37°C (11, 12, 23, 46, 57). In contrast, inv and yst (encodes the enterotoxin Yst) are maximally expressed at 23°C (8, 43, 45). In addition, motility is maximally expressed at 23°C (28, 50). We recently reported that regulation of inv expression is also affected by growth phase and a variety of environmental conditions, including low pH and Na+ concentration. Expression is maximal in late exponential phase to early stationary phase at 23°C and pH 7.5; however, significant expression also occurs at 37°C in late exponential phase to early stationary phase if the pH is lowered to pH 5.5 (43). Although the effect of temperature on the expression of inv has been well characterized, the molecular basis for this regulation has not been elucidated. We report here the identification and characterization of two Y. enterocolitica mutants in which the expression of both inv and flagellin is affected at 23°C.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Y. enterocolitica 8081v and JB580v (nalidixic acid [NAL] resistant) are referred to as the wild type. The latter is an R− M+ derivative of 8081v (31). JB580v retains full virulence in BALB/c mice (3). The v designation refers to Y. enterocolitica strains harboring virulence plasmid pYV8081. Expression of the inv gene was examined in Y. enterocolitica strains grown aerobically with shaking or on a roller for 16 to 18 h at 23 or 37°C in L broth (LB). Escherichia coli SM10λpir (56) was used to deliver mobilizable plasmids into Y. enterocolitica, and when necessary, pRK2013 was used as a conjugation helper plasmid (16). Antibiotics were used as required at the following concentrations (in micrograms per milliliter): NAL, 25; chloramphenicol (CM), 25; tetracycline, 7.5; kanamycin, 50; ampicillin, 100.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Y. enterocolitica 8081v | Serogroup O:8; pYV8081 Nalr | R. Martinez |
| Y. enterocolitica JB580v | Derivative of 8081v; R− M+ Nalr | 31 |
| Y. enterocolitica JB41v | JB580v wild-type inv; inv::phoA Nalr Cmr | This study |
| Y. enterocolitica JB1A8v | mTn5Km derivative of JB41v; invasin expression regulatory mutant; Nalr Cmr Kmr | This study |
| Y. enterocolitica JB16v | JB580v with integrated pJB207; same phenotype as JB1A8v; Nalr Cmr | This study |
| E. coli SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-TC::Mu; Kmr | 32, 56 |
| E. coli DH5α | supE44 ΔlacU169φ80dlacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 20 |
| Plasmids | ||
| pINP41 | pEP184 containing inv Δ412::phoA Cmr | 43 |
| pmTn5Km | mTn5Km on delivery plasmid pUT; Kmr | 14 |
| pTM100 | Mobilizable derivative of pACYC184; Tetr | 38 |
| pCR2.1 | PCR product cloning vector; Ampr Kmr | Invitrogen |
| pEP185.2 | Mobilizable suicide vector; Cmr | 31 |
| pRK2013 | ColE1 Tra (RK2)+; Kmr | 16 |
| pUC19 | Cloning vector; Apr | NEB |
| pJB15 | pUC19 containing 6.0-kb mTn5Km transposon-chromosome junction in SacI site; Apr | This study |
| pJB16 | pUC19 containing 28-kb mTn5Km transposon-chromosome junction in XbaI site; Apr Kmr | This study |
| pKVS2 | pBluescript containing 2.2-kb SacII fragment of 3′ end of fleA, all of fleB, and 5′ of fleC; Apr | 28 |
| pJB207 | pEP185.2 containing DNA derived from pJB16 used to disrupt sspA locus in JB16v; Cmr | This study |
| pJB309 | pTM100 containing E. coli sspA; Tetr | This study |
Construction of reporter strain JB41v.
Y. enterocolitica JB41v was constructed as previously described (43). Briefly, mobilizable suicide vector pIN41, which contains the promoter region and the region encoding the first 40 amino acids of inv fused to phoA (Cmr) (44), was mated with Y. enterocolitica JB580v. Y. enterocolitica transconjugants were selected on M63 agar plates supplemented with NAL and CM. Transconjugants were screened for integration at the inv locus by Southern analysis by using an inv sequence-specific probe (data not shown). The resulting strain, JB41v, is merodiploid at the inv locus; JB41v contains a wild-type copy of inv and the inv::phoA fusion separated by the suicide vector (Cmr).
Isolation of mutant JB1A8v.
Transposon mTn5Km (14) was used to mutagenize Y. enterocolitica JB41v essentially as previously described (14). E. coli SM10λpir (pmTn5Km) was filter mated with JB41v on LB plates for 6 to 8 h. The mating mixture was then plated onto M63 agar containing NAL (to select against the donor E. coli), CM (to maintain the inv::phoA fusion on the JB41v chromosome), kanamycin (to select for the transposon), and 4-chloro-3-indolyl-β-d-phosphate (XP; to screen for insertions that result in altered alkaline phosphatase [AP] activity). The plates were incubated at either 23 or 37°C. Colonies showing decreased inv::phoA expression (white or light blue on LB-XP plates) were purified and individually tested in AP assays.
DNA manipulations.
Plasmid DNA was isolated by the alkaline lysis method (35) or with Wizard Minipreps (Promega, Madison, Wis.). DNA fragments used in plasmid construction and probe preparation were prepared by digestion with the appropriate restriction endonuclease. After digestion, the resulting fragments were gel purified by using Gene Clean (Bio 101, Inc., La Jolla, Calif.). The purified fragments used as probes were labeled with [32P]dATP by the random primer method as previously described (15). DNA restriction enzymes, T4 DNA ligase, T4 kinase, T4 DNA polymerase, and Klenow fragment were purchased from NEB (Beverly, Mass.) and used in accordance with the manufacturer’s instructions.
The transposon (mTn5Km)-chromosome junction was cloned from JB1A8v as follows. JB1A8v chromosomal DNA was digested with XbaI (which does not cut within the transposon), and the resulting DNA fragments were ligated into pUC19 digested with XbaI. The ligation pool was transformed into DH5α, and Apr Kmr transformants were selected. One clone, pJB16, was isolated and determined to contain a 28-kb DNA insert. It was subsequently determined that pJB16 contained two noncontiguous XbaI DNA fragments; a 10-kb XbaI fragment 5′ of the transposon and a 3′ 18-kb XbaI fragment that contains DNA from the transposon (mTn5Km)-chromosome junction to beyond the sspA gene (see Fig. 3). The transposon (mTn5Km)-chromosome clone pJB15, used for sequence analysis of the transposon-chromosome junction, was constructed as follows. Chromosomal DNA of JB1A8v was digested with SacI (which cuts within the transposon) and ligated into pUC19 digested with SacI. The resulting ligation pool was transformed into DH5α. Clones carrying the transposon-chromosome junction were identified by colony hybridization using kan sequences homologous to mTn5Km as a probe. One positive clone, pJB15, that contained 6.0 kb of partial mTn5Km sequences and chromosomal DNA adjacent to the insertion site, was isolated.
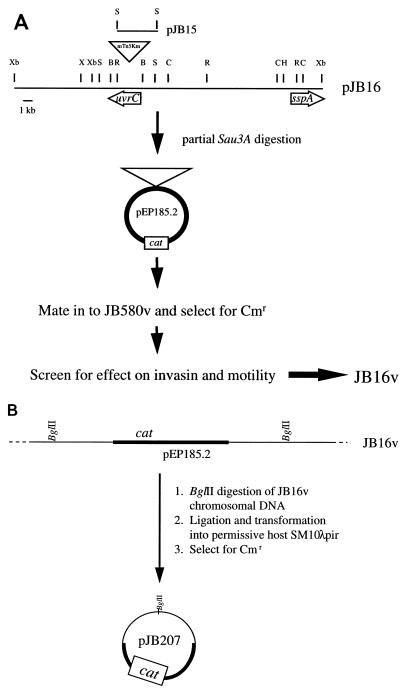
FIG. 3.
pJB16 physical map and schematic diagrams for construction of JB16v and cloning of pJB207. (A) Physical map of pJB16 and schematic diagram for construction of regulatory mutant JB16v. (B) Schematic diagram for cloning of pJB207. Restriction endonuclease sites indicated are not inclusive or unique. B, BamHI; C, ClaI; H, HindIII; R, EcoRI; S, SacI; Xb, XbaI; X, XhoI.
The transposon (mTn5Km)-chromosome junction sequence from pJB15 was determined by using oligonucleotide mTn5 (5′-AAAACGGGAAAGGTTC-3′), which is homologous to sequences within mTn5Km. pJB207 was sequenced by using T7 and T3 primers (NEB) that hybridize within the multiple cloning site of pEP185.2. The nucleotide sequence was obtained by the dideoxynucleotide chain termination method (53) with the Sequenase sequencing kit (U.S. Biochemical Corp.). Some of the sequencing was performed by the Murdock Molecular Biology automated sequencing facility (University of Montana). Sequences were analyzed with the BLAST program and University of Wisconsin sequence analysis software (Genetics Computer Group).
To clone E. coli sspA, oligonucleotide primers ssp1 (5′ CCGGCATCGACTCACCAC 3′) and ssp3 (5′ CTGATAGAATGCACGCAGC 3′), based on sequences 5′ and 3′ of sspA, respectively, were synthesized and used to amplify sspA from the E. coli DH5α chromosome by PCR. The PCR product, 900 bp, was cloned into pCR2.1 (Invitrogen Co.), resulting in plasmid pJB308. pJB308 was digested with EcoRI, and insert DNA isolated and ligated into pTM100 digested with EcoRI. The resulting plasmid, designated pJB309, was introduced into the various strains and assayed for complementation. The nucleotide sequence of the 900-bp insert in pJB309 was determined and found to be 100% identical to the published sequence of E. coli sspA (data not shown).
Isolation of JB16v and cloning of pJB207.
Mutant JB16v was constructed as follows. Suicide vector pEP185.2 carrying Sau3AI fragments derived from pJB16 were transformed into SM10λpir. The resulting transformants were pooled and mated into JB580v. Y. enterocolitica transconjugants were selected on M63 agar plates supplemented with NAL and CM. Transconjugants were screened by Western analysis for decreased invasin production (see below). One mutant (designated JB16v) was further characterized. Integration of the plasmid into the chromosome was confirmed by Southern analysis (data not shown); JB16v had an insertion at only one site. The plasmid, along with flanking DNA, was recovered from JB16v chromosomal DNA by plasmid rescue using BglII, essentially as previously described (66); this plasmid was designated pJB207.
Western analysis.
All of the strains analyzed in this study had the same growth rates, and comparable culture densities were reached after overnight growth. Whole-cell lysates were prepared from bacteria grown to stationary phase in LB at either 23 or 37°C as previously described (43). Equal amounts of whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Blots were then examined by Western immunoblot analysis with polyclonal anti-invasin antibody as the primary antibody and visualized as described previously (2, 43). Coomassie-stained gels were run in parallel to confirm that the same amount of protein was loaded in each lane. Relative invasin levels, as detected by immunoblot analysis, were estimated by densitometry with UVP gel documentation system GDS2000 (UVP International).
Cell surface-associated flagellin was detected by immunoblotting with monoclonal antibody 15DB (anti-E. coli flagellin) (Igen, Inc.) used at a dilution of 1:2,000. Briefly, Y. enterocolitica cells grown at 23°C overnight on LB plates supplemented with appropriate antibiotics were suspended in phosphate-buffered saline, and flagella were sheared from the cells by vortexing them twice for 30 s. Cells were removed by centrifugation at 10,000 × g for 10 min. Proteins in the cell-free supernatants were concentrated by acetone precipitation, and protein from equivalent whole-cell OD600 (optical density at 600 nm) amounts were analyzed by SDS–12.5% PAGE, and examined by Western analysis as described above by using antibody 15DB as the primary antibody. Proteins were visualized by the Amersham ECL Western Blotting Detection System (Amersham Corp., Arlington Heights, Ill.).
Motility assays.
Motility assays were conducted in LB with 0.3% agar and appropriate antibiotics. Motility agar plates were point inoculated from a colony by stabbing the agar with a toothpick and incubated at 23°C for approximately 16 h.
Invasion assays.
Invasion assays were performed with bacterial cultures grown aerobically for 16 to 18 h in LB at 23 or 37°C, as indicated in figure legends. Bacteria were added to subconfluent human laryngeal epithelial (HEp-2) cells at a multiplicity of infection of 100, and the invasion assay was performed as previously described (40). Results are expressed as percent invasion = 100 × (number of bacteria recovered/number of bacteria added).
AP assays.
AP activity was measured in permeabilized cells, and results are expressed in enzyme units per OD600 unit as previously described (36). Assays were performed on duplicate cultures grown for 16 to 18 h aerobically in LB. Y. enterocolitica 8081v and JB580v were always assayed and had low but detectable background levels of AP activity (∼10 U) that were subtracted from the values presented (except as shown in Fig. 1).
FIG. 1.
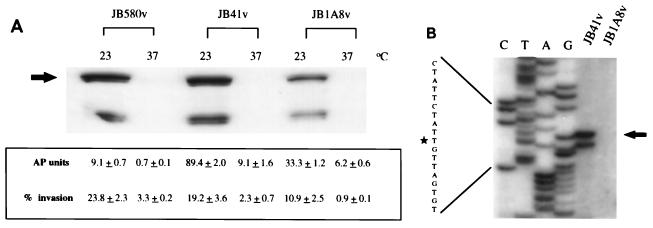
Analysis of invasin expression in regulatory mutant JB1A8v. (A) Analysis of invasin protein levels and activity. Whole-cell extracts of the indicated strains grown at the indicated temperatures were separated by SDS-PAGE, and invasin was detected by Western analysis. The arrow points to the band representing full-length invasin. AP activities were assayed in duplicate and are represented as means and ranges. Invasion assays with HEp-2 cells were performed in duplicate, and the data are presented as means and ranges. (B) Comparison of inv mRNA levels in Y. enterocolitica JB41v and JB1A8v. Reverse transcriptase and labeled primer INV1 were used to primer extend total cellular RNAs from JB41v and JB1A8v grown at 23°C to late-log phase. The arrow points to the product of the primer extension reactions and corresponds to nucleotide −103 (star) with respect to the inv open reading frame. The data shown are from a single experiment and are representative of numerous experiments performed with similar results.
Northern analysis and primer extension.
Total cellular RNA was analyzed by primer extension using a primer to inv as previously described (43). Electrophoresis of RNA and Northern hybridizations were conducted as previously described (52). A pKVS2 SacII DNA fragment, which contains the 5′ end of fleA, all of fleB, and the 3′ end of fleC (28), was used as a probe to detect fleB mRNA.
RESULTS
Construction of an inv::phoA fusion and isolation of an inv regulatory mutant in Y. enterocolitica.
To understand the molecular basis of the observed environmental regulation of Y. enterocolitica inv expression, a genetic approach was taken. Earlier studies suggested that temperature regulation of inv expression occurs at the transcriptional level (43, 45). Therefore, a strain carrying a reporter fusion was first constructed to monitor the loss or alteration in temperature regulation of inv expression. An inv::phoA translational fusion, consisting of the promoter region and coding sequences for the first 40 amino acids of inv fused to phoA, was introduced into the chromosome of Y. enterocolitica JB580v. The resulting strain, JB41v, is genotypically and phenotypically Inv+, which allowed levels of inv expression to be monitored via AP activity, Western analysis, primer extension, and tissue culture invasion assays. Analysis of inv expression in JB41v by any of these assays showed the same pattern; expression of the inv gene was elevated at 23°C, while expression at 37°C was reduced (Fig. 1).
The AP activity of JB41v was used to identify mutations affecting temperature regulation of inv expression. JB41v colonies are blue on plates containing the AP chromogenic indicator XP when grown at 23°C but white when grown at 37°C. Transposon mTn5Km insertion mutants of JB41v were generated and screened on LB-XP plates at both 23 and 37°C for altered inv expression. Of approximately 2,000 transposon insertion mutants, one was identified that showed a significant decrease in inv::phoA expression at 23°C; this mutant was designated JB1A8v. No mTn5Km insertion mutants exhibiting altered invasin expression at 37°C or increased expression at 23°C were found. Southern hybridization analysis indicated that JB1A8v had a single transposon insertion that was not within the inv structural gene or upstream sequences (data not shown). When the bacteria were grown at 23°C, JB41v had 89.4 U of AP activity while JB1A8v showed 33.3 U of AP activity (Fig. 1A). Similarly, the ability of JB1A8v to invade tissue culture cells (TCI phenotype) was reduced compared to that of parental strains JB580v and JB41v (Fig. 1A). Loss of proper temperature regulation of JB1A8v inv expression was verified at the protein level. Immunoblot analysis with a polyclonal anti-invasin antibody showed that JB1A8v produced three- to fourfold less invasin than did strains JB580v and JB41v when grown at 23°C, as quantitated by densitometry of Western blots (Fig. 1A). These results suggest that the factor affected by the mTn5Km insertion in JB1A8v acts in trans because it affects the expression of both inv and inv::phoA.
To determine if the reduction of inv expression was at the transcriptional level, total mRNA from early stationary-phase cells grown at 23°C was analyzed by primer extension. JB1A8v produced significantly less inv-specific product than did wild-type JB41v (Fig. 1B). As previously shown (43), the transcriptional start site corresponded to nucleotide −103. As a control for RNA preparation, primer extension reactions were performed with a primer hybridizing to the 5′ end of ail. ail transcript products were detected at the same levels for the mutant JB1A8v and wild-type JB41v (data not shown). This is consistent with the observation that JB41v and JB1A8v produce similar amounts of Ail (data not shown). These results are consistent with the hypothesis that the mutation in JB1A8v affects inv expression at the transcriptional level, by affecting either transcriptional initiation or message stability.
The motility phenotype of JB1A8v.
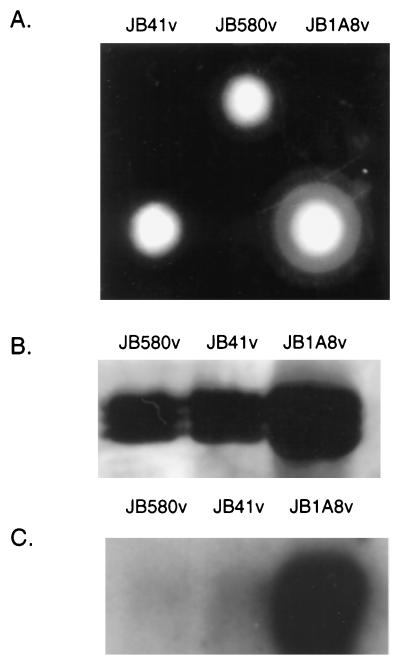
In addition to inv, there are several other identified factors that are most prominently expressed at 23°C. In vitro, Y. enterocolitica bacteria are motile when grown at 23°C yet nonmotile at 37°C. Thus, if inv and motility are coordinately regulated, one may expect JB1A8v to be nonmotile at 23°C. Interestingly, the transposon mutant JB1A8v appeared hypermotile in soft agar plates at 23°C compared to strains JB580v and JB41v (Fig. 2A); both strains were nonmotile at 37°C (data not shown). In addition, Western analysis with monoclonal antiflagellin antibody showed that flagellin levels were significantly increased in JB1A8v compared to those in wild types JB580v and JB41v (Fig. 2B). Similarly, Northern analysis demonstrated that fleB (a gene encoding flagellin) mRNA was increased in JB1A8v compared to that in the wild type (Fig. 2C). In wild-type Y. enterocolitica, expression of flagellin and that of invasin are similarly affected by temperature, yet the mutation in JB1A8v has the opposite effect on the expression of inv and flagellar genes. Nevertheless, in both cases, the effect appears to be at the transcriptional level.
FIG. 2.
Motility phenotypes of JB1A8v. (A) Motility assays were performed with JB580v, JB41v, and JB1A8v in 0.3% soft agar plates incubated at 23°C for 16 h. (B) Western analysis of flagellin preparations probed with monoclonal antibody 15D8 as described in Materials and Methods. (C) Northern analysis of total cellular RNA isolated from mid-log-phase cultures probed with fleB-specific DNA. The data shown are from a single experiment and are representative of numerous experiments performed with similar results.
Localization of the transposon insertion within JB1A8v and regeneration of the JB1A8v phenotype.
To determine what gene was disrupted by the mTn5Km insertion in JB1A8v, the transposon-chromosome junction from JB1A8v was cloned (pJB15) (Fig. 3A). The nucleotide sequence of the junction region was determined, and sequence analysis revealed that mTn5Km had inserted into a gene similar to E. coli uvrC. In E. coli, uvrC has been shown to encode a subunit of UV DNA damage repair enzyme exinuclease ABC. It seems unlikely that UvrC plays a direct role in Y. enterocolitica inv expression. mTn5Km insertions have been reported to have strong polar effects (14), and it is possible that this is the case with the mTn5Km insertion in JB1A8v. To further characterize the nature of the mutation in JB1A8v responsible for the observed phenotypes, we attempted to obtain a complementing clone. For this purpose, sequences immediately adjacent to the insertion site (pJB15 insert DNA) were used as a probe against a Y. enterocolitica cosmid library. Two separate clones (cloned in low-copy cosmid vector pLAFR2) were obtained but proved to be highly unstable in E. coli LE392 and DH5α. Several different manipulations and attempts were made to propagate these probe-positive clones, without success. In addition, attempts to isolate smaller clones from subgenomic libraries constructed in high- and low-copy-number vectors were also unsuccessful. We also tried using PCR to clone the wild-type copy of uvrC; primers were derived from the E. coli uvrC sequence and used in PCR of Y. enterocolitica chromosomal DNA. An appropriate-size fragment was amplified, but attempts to clone this fragment were unfruitful.
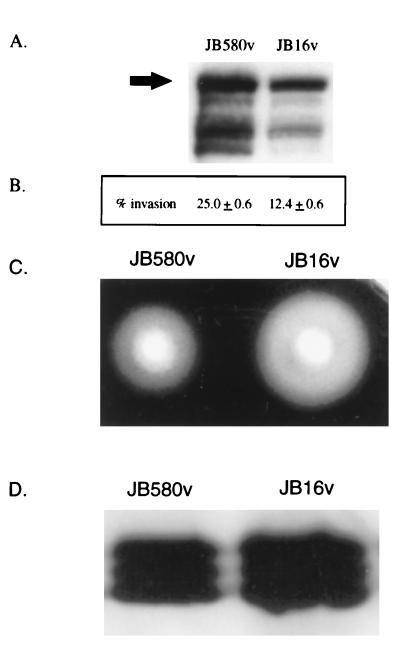
Without a complementing clone, it was difficult to ascertain if the phenotype of JB1A8v was due to the loss of uvrC, to polar effects of mTn5Km, or to an unidentified, unlinked mutation. To rule out the latter possibility, we tried to recreate the phenotype of JB1A8v. If a mutation in a gene immediately downstream of uvrC has the same phenotype as JB1A8v, this would support the idea that the mTn5Km insertion of JB1A8v had a polar effect and that the phenotype of JB1A8v is linked to the transposon insertion. To recreate the JB1A8v phenotype, a clone derived from JB1A8v, pJB16, which contains the mTn5Km insertion and flanking chromosomal DNA of JB1A8v (Fig. 3A), was partially digested with Sau3A1 and the resulting fragments were ligated into the suicide vector pEP185.2. Conjugation of these plasmids into wild-type strain JB580v and selection for the presence of the plasmid selects for strains in which the plasmid has integrated into the chromosome by homologous recombination. This can cause disruption of the locus at the site of recombination. Several mutants were analyzed, and one, JB16v, was found to have a phenotype similar to that of JB1A8v. JB16v had reduced amounts of invasin and showed reduced invasion of tissue culture cells when bacteria were grown at 23°C (Fig. 4A and B). In addition, JB16v was hypermotile in soft agar and had increased amounts of flagellin (Fig. 4C and D).
FIG. 4.
Phenotype of reconstructed mutant JB16v. (A) Invasin expression in regenerated regulatory mutant JB16v. Whole-cell extracts of JB580v and JB16v grown at 23°C were separated by SDS-PAGE, and invasin levels were determined by Western analysis. The arrow points to the band representing full-length invasin. (B) Invasion phenotype exhibited by JB580v and JB16v. Invasion assays with HEp-2 cells were performed in duplicate, and the data presented are means and ranges. (C) Motility phenotype of regenerated regulatory mutant JB16v. Motility assays were performed with JB580v and JB16v in 0.3% soft agar plates incubated at 23°C for 16 h. (D) Western analysis of flagellin preparations probed with monoclonal antibody 15D8 as described in Materials and Methods. The data shown are from a single experiment and are representative of numerous experiments performed with similar results.
Complementation and identification of the locus disrupted in Y. enterocolitica JB16v.
To identify the disrupted gene in JB16v responsible for the mutant phenotypes, the integrated plasmid pEP185.2, along with flanking chromosomal DNA, was cloned by plasmid rescue (66) (Fig. 3B). DNA flanking the pEP185.2 insertion site in JB16v maps 16 kb from the site of the mTn5Km insertion within JB1A8v (Fig. 3A). To identify the locus where pEP185.2 had integrated in JB16v, the nucleotide sequence of the DNA junction was determined. Sequence analysis revealed that the locus disrupted in JB16v is similar to an E. coli gene encoding the stationary-phase regulator SspA (50% predicted amino acid identity over 40 amino acids; data not shown). While it is possible that the phenotype of JB1A8v is due to polar effects of the mTn5Km insertion on sspA expression, this seems unlikely given that 16 kb separate sspA and mTn5Km. An alternative explanation is that this region of the Y. enterocolitica chromosome has more than one locus that affects expression of inv and motility.
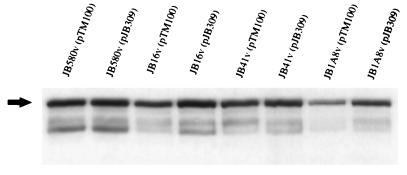
To ascertain if the phenotypes of JB16v were due to the loss of sspA or due to polar effects of the integrated plasmid, we sought to determine if the E. coli sspA locus could complement the mutation in JB16v. For this purpose, oligonucleotide primers homologous to DNA flanking the E. coli sspA gene were synthesized and used to amplify the E. coli sspA locus by PCR. The resulting PCR product was subcloned into pTM100 and designated pJB309. pJB309 was introduced into mutant strains JB1A8v and JB16v to determine if it could complement the decrease in invasin and hypermotile phenotypes. When bacteria were grown at 23°C, pJB309 restored invasin expression to wild-type levels in mutants JB16v and JB1A8v, as assayed by Western blot analysis (Fig. 5). In addition, the complementing clone pJB309 restored the normal motility phenotype and flagellin production in mutants JB1A8v and JB16v (Fig. 6 and data not shown). The vector pTM100 had no effect on the invasin or motility phenotypes of the strains assayed. Interestingly, pJB309 lowered flagellin levels and motility in wild-type strains JB580v and JB41v but invasin levels were not affected (Fig. 5).
FIG. 5.
Complementation of JB1A8v and JB16v invasin levels with pJB309. Western analysis of invasin expression by JB580v, JB16v, JB41v, and JB1A8v harboring either pTM100 (cloning vector) or pJB309 (E. coli sspA). Whole-cell extracts of the indicated strains grown at 23°C were separated by SDS-PAGE, and invasin levels were determined by Western analysis. The arrow points to the band representing full-length invasin. The data shown are from a single experiment and are representative of numerous experiments performed with similar results.
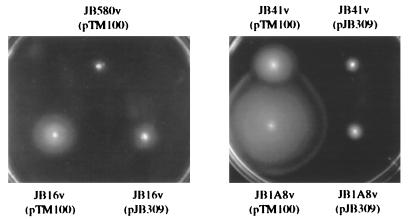
FIG. 6.
Complementation of JB1A8v and JB16v motility phenotype with pJB309. Motility assays were performed with the strains indicated and 0.3% soft agar plates incubated at 23°C for 16 h. The data shown are from a single experiment and are representative of numerous experiments performed with similar results.
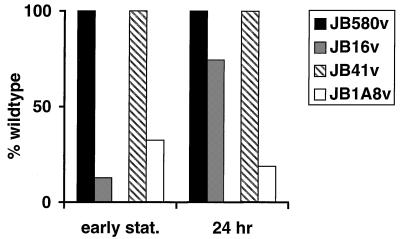
In E. coli, SspA positively and negatively regulates several genes in stationary phase. In addition, it has been shown that sspA expression is induced upon entry into stationary phase (64). Because inv is analogously expressed in stationary phase, we sought to determine if sspA mutant JB16v and uvrC mutant JB1A8v were altered in invasin expression in a growth phase-dependent manner. For this purpose, bacterial cultures were assayed for invasin expression by Western analysis at designated time points in the growth phase. Invasin levels were too low in mid-log- and late-log-phase cultures to determine significant and reproducible differences between the mutant and wild-type strains. However, levels of invasin in early stationary phase showed a significant difference; the sspA mutant JB16v demonstrated 13% of wild-type JB580v invasin levels, while JB1A8v showed 32% of wild-type JB41v invasin levels. In contrast, 24-h cultures demonstrated no significant difference in invasin levels between the wild-type JB580v strain and the sspA mutant JB16v; whereas the uvrC mutant JB1A8v maintained its phenotype of decreased invasin levels (19% of wild-type JB41v invasin levels) (Fig. 7). These results suggest that the sspA mutation, but not the uvrC mutation, affects invasin expression in a growth phase-dependent way.
FIG. 7.
Effect of growth phase on mutants JB16v and JB1A8v. Cultures grown for 16 h at 23°C were diluted to an OD600 of 0.1 and grown at 23°C in LB, and aliquots were taken at the indicated growth phase points. Invasin levels detected by immunoblot analysis were estimated by densitometry. The data shown are from a single experiment and are representative of numerous experiments performed with similar results. stat., stationary phase.
DISCUSSION
In this study, we identified two Y. enterocolitica mutants affected in both inv and fleABC (flagellin) expression. Mutants JB1A8v and JB16v show a significant decrease in invasin expression but are hypermotile compared to the wild type. Mutant JB1A8v was created by mTn5Km transposon insertion mutagenesis, while JB16v was created by targeted insertion mutagenesis. uvrC and sspA are disrupted in JB1A8v and JB16v, respectively; these genes are separated by approximately 16 kb. It is not known how the mTn5Km insertion mutation results in the observed phenotypes of JB1A8v due to an inability to clone the wild-type locus. The mTn5Km insertion within JB1A8v could exert a polar effect on a downstream gene(s); there may be one or more additional, uncharacterized mutations separate from the insertion site that is responsible for the mutant phenotype; or uvrC may have heretofore unrecognized properties. There are two lines of evidence that suggest that the mutation in JB1A8v does not exert its effect, either directly or indirectly, through sspA. The phenotype of the JB1A8v mutant, unlike that of the JB16v mutant, is not growth phase dependent. In addition, the JB1A8v mutation appears to exert its effect on inv and fle expression at the transcript level, whereas the JB16v mutation exerts its effect posttranscriptionally for inv (1).
In E. coli, the uvrC gene is upstream of pgsA (encodes phosphatidylglycerolphosphate synthase) and downstream of uvrY. While the function of uvrY has not been elucidated in E. coli, an allele of uvrY, called sirA, has been identified in Salmonella typhimurium. sirA has been shown to be necessary for efficient hilA expression (27). hilA encodes a transcriptional regulator which activates the expression of several operons specific for S. typhimurium invasion (4). The predicted amino acid sequences of S. typhimurium SirA and E. coli UvrY suggest that these proteins are response regulators of two-component regulatory systems (4). If the genetic organization of the Y. enterocolitica uvrC region is conserved, it is possible that the mTn5Km insertion in uvrC exerts upstream polar effects on a gene similar to S. typhimurium sirA. However, the uvrC mutant JB1A8v and JB41v harboring a S. typhimurium sirA clone showed no effect on invasin levels (39). Nonetheless, there are several caveats for these results. It is possible that the S. typhimurium sirA clone is not properly expressed in Y. enterocolitica due to cloning vector effects or other factors needed for S. typhimurium sirA expression. Alternatively, S. typhimurium SirA may differ functionally from the putative Y. enterocolitica SirA/UvrY homolog. Lastly, the mTn5Km insertion in the uvrC mutant JB1A8v may have a polar effect on multiple genes necessary for invasin expression, such that supplying only sirA in trans does not complement the mutation.
The gene disrupted in JB16v that causes a reduction in invasin levels and increased amounts of flagellin encodes a homolog of SspA (stringent starvation protein). E. coli SspA was originally identified as a protein that copurifies with RNA polymerase and subsequently was shown to directly bind RNA polymerase (26, 63). In addition, SspA is the predominant protein synthesized in E. coli undergoing the stringent response to amino acid starvation (49). In E. coli, sspA is the first gene of the sspAB operon; the function of the sspB gene product is not known (62, 64). It has been shown that the sspAB operon is involved in both positive and negative regulation of 11 genes during the exponential and postexponential growth phases (64). Two of the proteins whose expression was shown to be under the influence of SspA include Hns (a histone-like protein) and an alternative nucleoside diphosphate kinase. An E. coli sspA mutant shows a 5- to 10-fold increase in Hns expression, in addition to exhibiting a decrease in levels of an undefined alternative nucleoside diphosphate kinase (55, 64). In addition, the expression of sspA itself is induced by entry into stationary phase, as well as starvation for phosphate, glucose, nitrogen, and amino acids (64). The exact mechanism for SspA regulation is unknown but has been hypothesized to be related to the ability of SspA to bind RNA polymerase (i.e., a sigma factor-like component) (63).
It is intriguing that E. coli sspA and Y. enterocolitica inv are analogously growth phase regulated in vitro, such that both transcripts are maximally expressed in early stationary phase. Furthermore, it is interesting that the sspA mutation affects invasin expression in a growth phase-dependent manner. The observation that invasin levels in the sspA mutant do not rise until later than normal suggests that the sspA mutation causes a delay in sensing the transition to stationary phase. Given previous evidence that sspA encodes an RNA polymerase binding protein involved in the positive and negative regulation of many genes in E. coli, one could speculate that the Y. enterocolitica SspA homolog is a positive and negative regulator of the expression of invasin and flagellin, respectively. The fact that sspA complements both the JB1A8v and JB16v mutant phenotypes suggests that the role of SspA in the regulation of invasin and flagellin expression is farther downstream than the effects of the uvrC mutation in JB1A8v. Alternatively, there may be parallel pathways for the coordinate regulation of invasin and motility, such that providing sspA in trans in the uvrC mutant JB1A8v upregulates the SspA-dependent pathway.
It is interesting that pJB309 (low-medium-copy-number plasmid carrying E. coli sspA) complements the decrease in invasin levels in sspA mutant JB16v and uvrC mutant JB1A8v yet lowers flagellin or motility levels in all of the strains tested, including the wild-type strains. There are several possible explanations for this observed phenomenon. The simplest explanation may be that the SspA-dependent repression of flagellin or motility may be more sensitive to the amount of SspA in the cell, while the SspA-dependent expression of invasin may not be as sensitive to SspA or there may be a threshold for maximal SspA-dependent expression of invasin. Alternatively, E. coli SspA may differ functionally from Y. enterocolitica SspA, such that E. coli SspA functions properly to complement invasin expression yet does not function properly for expression of flagellin or motility. It is also possible that expression of E. coli sspA in Y. enterocolitica is not controlled normally (i.e., E. coli promoter sequences or uncontrolled plasmid-driven expression of sspA) such that aberrant expression of sspA affects flagellin synthesis or motility more than invasin levels. Lastly, expression of invasin may require only SspA whereas proper expression of flagellin or motility may require both SspA and SspB. Given that in E. coli, sspA is within the operon sspAB, one could envision that providing SspA in trans may alter the stoichiometry of SspA and SspB molecules, thus resulting in an altered expression pattern of flagellin or motility.
The coordinate regulation of invasin and motility in Y. enterocolitica is reminiscent of what has been observed for Vibrio cholerae, the causative agent of the diarrheal disease cholera. Many factors have been identified that play a role in V. cholerae virulence, including cholera toxin, toxin-coregulated pili (TCP), accessory colonization factors, outer membrane proteins, proteases, and hemolysins (29). A subset of these virulence properties is coordinately regulated by ToxR. ToxR is a 32-kDa transmembrane transcriptional regulatory protein responsible for environmental sensing and signal transduction that leads to virulence gene expression (41). Recently, it has been observed that alterations in V. cholerae motility phenotypes (i.e., bacteria are either hypermotile or nonmotile) correlates with the expression of many ToxR-regulated and non-ToxR-regulated virulence determinants. Hypermotile mutants of V. cholerae were shown to be defective in the production of cholera toxin, TCP, and hemolysin, yet these mutants showed increased production of protease and hemagglutinin (17). Conversely, isolated nonmotile V. cholerae had increased production of cholera toxin, TCP, and hemolysin while showing decreased levels of hemagglutinin (17). In addition, it has been noted that V. cholerae toxR null mutants are hypermotile (58). Taken together, these results suggest that the motility phenotype of V. cholerae is tightly coupled to the expression of multiple ToxR-regulated and other non-ToxR-regulated virulence properties.
Before Y. enterocolitica bacteria infect a host, they must survive nutrient-limiting environments that are generally moist and at ambient temperatures. After ingestion, the bacteria adapt to the mammalian host temperature of 37°C, a myriad of environmental assaults, and the immune system. Therefore, Y. enterocolitica must optimize the regulation of virulence factor expression in all phases of its life cycle to survive in nature, as well as within its mammalian host. In addition, the synthesis and assembly of flagellin require a large investment of energy and may consequently provide antigenic targets to the host immune system; thus, expression of the motility regulon is also subject to stringent control. One can readily imagine overlapping sets of conditions that would influence the synthesis of both invasin and flagella. However, there are also probably conditions in the natural environment or in a mammalian host under which expression of one phenotype but not the other is needed. Thus, rigorous control of the expression of these phenotypes may be important for the long-term survival and persistence of Y. enterocolitica.
ACKNOWLEDGMENTS
We especially thank Jeff F. Miller for his support and advice, as well as for the use of his lab during part of this project. We thank S. Minnich for providing pKVS2. We thank G. Young and P. Cotter for critically reading the manuscript.
This work was supported in part by National Institutes of Health grants AI-27342 and AI-01230 to V.L.M. J.L.B. is a recipient of the UCPF award.
REFERENCES
- 1.Badger, J. L., and V. L. Miller. The co-ordinate regulation of invasin and motility in Yersinia enterocolitica is dependent on ClpB and the alternative sigma factor FliA. Submitted for publication.
- 2.Badger J L, Miller V L. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J Bacteriol. 1995;177:5370–5373. doi: 10.1128/jb.177.18.5370-5373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badger, J. L., and V. L. Miller. Unpublished results.
- 4.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 5.Bliska J, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci USA. 1992;89:3561–3565. doi: 10.1073/pnas.89.8.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bliska J B, Compass M C, Falkow S. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect Immun. 1993;61:3914–3921. doi: 10.1128/iai.61.9.3914-3921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottone E J. Yersinia enterocolitica. Boca Raton, Fla: CRC Press, Inc.; 1981. [Google Scholar]
- 8.Boyce J M, Evans E J, Jr, Evans D G, DuPont H L. Production of heat-stable, methanol-soluble enterotoxin by Yersinia enterocolitica. Infect Immun. 1979;25:532–537. doi: 10.1128/iai.25.2.532-537.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.China B, N’guyen B T, DeBruyere M, Cornelis G R. Role of YadA in resistance of Yersinia enterocolitica to phagocytosis by human polymorphonuclear leukocytes. Infect Immun. 1994;62:1275–1281. doi: 10.1128/iai.62.4.1275-1281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G, Laroche Y, Balligand G, Sory M-P, Wauters G. Y. enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis G, Sory M-P, Laroche Y, Derclaye I. Genetic analysis of the plasmid region controlling virulence in Y. enterocolitica O:9 by mini-Mu insertions and lac gene fusions. Microb Pathog. 1986;1:349–359. doi: 10.1016/0882-4010(86)90067-7. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis G, Vanooteghem J-C, Sluiters C. Transcription of the yop regulon from Y. enterocolitica requires trans-acting pYV and chromosomal genes. Microb Pathog. 1987;2:367–379. doi: 10.1016/0882-4010(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 13.Cover T L, Aber R C. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grutzkau A, Hanski C, Hahn H, Riecken E O. Involvement of M cells in the bacterial invasion of Peyer’s patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut. 1990;31:1011–1015. doi: 10.1136/gut.31.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutman L T, Ottesen E A, Quan T J, Noce P S, Katz S L. An inter-familial outbreak of Yersinia enterocolitica enteritis. N Engl J Med. 1973;288:1372–1377. doi: 10.1056/NEJM197306282882604. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Hanski C, Kutschka U, Schmoranzer H P, Naumann M, Stallmach A, Hahn H, Menge H, Riecken E O. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O:8 with intestinal mucosa during experimental enteritis. Infect Immun. 1989;57:673–678. doi: 10.1128/iai.57.3.673-678.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanski C, Naumann M, Hahn H, Riecken E O. Determinants of invasion and survival of Yersinia enterocolitica in intestinal tissue: an in vivo study. Med Microbiol Immunol. 1989;178:289–296. doi: 10.1007/BF00191063. [DOI] [PubMed] [Google Scholar]
- 23.Iriarte M, Vanooteghem J C, Delor I, Diaz R, Knutton S, Cornelis G R. The Myf fibrillae of Yersinia enterocolitica. Mol Microbiol. 1993;9:507–520. doi: 10.1111/j.1365-2958.1993.tb01712.x. [DOI] [PubMed] [Google Scholar]
- 24.Isberg R R. Mammalian cell adhesion functions and cellular penetration of enteropathogenic Yersinia species. Mol Microbiol. 1989;3:1449–1453. doi: 10.1111/j.1365-2958.1989.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 25.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 26.Ishihama A, Saitoh T. Subunits of RNA polymerase in function and structure. IX. Regulation of RNA polymerase activity by stringent starvation protein (SSP) J Mol Biol. 1979;129:517–530. doi: 10.1016/0022-2836(79)90466-2. [DOI] [PubMed] [Google Scholar]
- 27.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 28.Kapatral V, Minnich S A. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol Microbiol. 1995;17:49–56. doi: 10.1111/j.1365-2958.1995.mmi_17010049.x. [DOI] [PubMed] [Google Scholar]
- 29.Kaper J B, Morris J G, Jr, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:46–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapperud G, Namork E, Skurnik M, Nesbakken T. Plasmid-mediated surface fibrillae of Yersinia pseudotuberculosis and Yersinia enterocolitica: relationship to the outer membrane protein YOP1 and possible importance for pathogenesis. Infect Immun. 1987;55:2247–2254. doi: 10.1128/iai.55.9.2247-2254.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 32.Kolter R, Inuzuka M, Helinski D R. Transcomplementation-dependent replication of a low molecular weight origin fragment from plasmid RK6. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 33.Laird W J, Cavanaugh D C. Correlation of autoaggultination and virulence of yersiniae. J Clin Microbiol. 1980;11:430–432. doi: 10.1128/jcm.11.4.430-432.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung K Y, Reisner B S, Straley S C. YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect Immun. 1990;58:3262–3271. doi: 10.1128/iai.58.10.3262-3271.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 36.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez R J. Thermoregulation-dependent expression of Yersinia enterocolitica protein 1 imparts serum resistance to Escherichia coli K-12. J Bacteriol. 1989;171:3732–3739. doi: 10.1128/jb.171.7.3732-3739.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michiels T, Cornelis G R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, V. L. Unpublished results.
- 40.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 42.Pai C H, DeStephano L. Serum resistance associated with virulence in Yersinia enterocolitica. Infect Immun. 1982;35:605–611. doi: 10.1128/iai.35.2.605-611.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pepe J C, Badger J L, Miller V L. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 44.Pepe J C, Miller V L. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc Natl Acad Sci USA. 1993;90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierson D, Falkow S. Nonpathogenic isolates of Yersinia enterocolitica do not contain functional inv-homologous sequences. Infect Immun. 1990;58:1059–1064. doi: 10.1128/iai.58.4.1059-1064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierson D E, Falkow S. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect Immun. 1993;61:1846–1852. doi: 10.1128/iai.61.5.1846-1852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Portnoy D A, Martinez R J. Role of a plasmid in the pathogenicity of Yersinia species. Curr Top Microbiol Immunol. 1985;118:29–51. doi: 10.1007/978-3-642-70586-1_3. [DOI] [PubMed] [Google Scholar]
- 48.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–792. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeh S, Pedersen S, Friesen J D. Biosynthetic regulation of individual proteins in relA+ and relA strains of Escherichia coli during amino acid starvation. Mol Gen Genet. 1976;149:279–289. doi: 10.1007/BF00268529. [DOI] [PubMed] [Google Scholar]
- 50.Rohde J R, Fox J M, Minnich S A. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 51.Rosqvist R, Forsberg A, Rimpilainen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E J, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulze-Koops H, Burkhardt H, Heesemann J, Kirsch T, Swoboda B, Bull C, Goodman S, Emmrich F. Outer membrane protein YadA of enteropathogenic yersiniae mediates specific binding to cellular but not plasma fibronectin. Infect Immun. 1993;61:2513–2519. doi: 10.1128/iai.61.6.2513-2519.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shankar S, Schlictman D, Chakrabarty A M. Regulation of nucleoside diphosphate kinase and an alternative kinase in Escherichia coli: role of the sspA and mk genes in nucleoside triphosphate formation. Mol Microbiol. 1995;17:935–943. doi: 10.1111/j.1365-2958.1995.mmi_17050935.x. [DOI] [PubMed] [Google Scholar]
- 56.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 57.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 58.Strauss E J. Bacterial pathogenesis. When a turn off is a turn on. Curr Biol. 1995;5:706–709. doi: 10.1016/s0960-9822(95)00139-4. [DOI] [PubMed] [Google Scholar]
- 59.Toivanen P, Toivanen A, Olkkonen L, Aantaa S. Hospital outbreak of Yersinia enterocolitica infection. Lancet. 1973;i:801–803. doi: 10.1016/s0140-6736(73)90601-6. [DOI] [PubMed] [Google Scholar]
- 60.Une T. Studies on the pathogenicity of Yersinia enterocolitica. I. Experimental infection in rabbits. Microbiol Immunol. 1977;21:341–363. [PubMed] [Google Scholar]
- 61.Wachtel M R, Miller V L. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect Immun. 1995;63:2541–2548. doi: 10.1128/iai.63.7.2541-2548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams M D, Fuchs J A, Flickinger M C. Null mutation in the stringent starvation protein of Escherichia coli disrupts lytic development of bacteriophage P1. Gene. 1991;109:21–30. doi: 10.1016/0378-1119(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 63.Williams M D, Ouyang T X, Flickinger M C. Glutathione S-transferase–sspA fusion binds to E. coli RNA polymerase and complements delta sspA mutation allowing phage P1 replication. Biochem Biophys Res Commun. 1994;201:123–127. doi: 10.1006/bbrc.1994.1677. [DOI] [PubMed] [Google Scholar]
- 64.Williams M D, Ouyang T X, Flickinger M C. Starvation-induced expression of SspA and SspB: the effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol Microbiol. 1994;11:1029–1043. doi: 10.1111/j.1365-2958.1994.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 65.Yang Y, Isberg R R. Cellular internalization in the absence of invasin expression is promoted by the Yersinia pseudotuberculosis yadA product. Infect Immun. 1993;61:3907–3913. doi: 10.1128/iai.61.9.3907-3913.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 67.Young V B, Miller V L, Falkow S, Schoolnik G K. Sequence, localization and function of the invasin protein of Yersinia enterocolitica. Mol Microbiol. 1990;4:1119–1128. doi: 10.1111/j.1365-2958.1990.tb00686.x. [DOI] [PubMed] [Google Scholar]