Abstract
Single nucleotide variants in the general population are common genomic alterations, where the majority are presumed to be silent polymorphisms without known clinical significance. Using human induced pluripotent stem cell (hiPSC) cerebral organoid modeling of the 1.4 megabase Neurofibromatosis type 1 (NF1) deletion syndrome, we previously discovered that the cytokine receptor-like factor-3 (CRLF3) gene, which is co-deleted with the NF1 gene, functions as a major regulator of neuronal maturation. Moreover, children with NF1 and the CRLF3L389P variant have greater autism burden, suggesting that this gene might be important for neurologic function. To explore the functional consequences of this variant, we generated CRLF3L389P-mutant hiPSC lines and Crlf3L389P-mutant genetically engineered mice. While this variant does not impair protein expression, brain structure, or mouse behavior, CRLF3L389P-mutant human cerebral organoids and mouse brains exhibit impaired neuronal maturation and dendrite formation. In addition, Crlf3L389P-mutant mouse neurons have reduced dendrite lengths and branching, without any axonal deficits. Moreover, Crlf3L389P-mutant mouse hippocampal neurons have decreased firing rates and synaptic current amplitudes relative to wild type controls. Taken together, these findings establish the CRLF3L389P variant as functionally deleterious and suggest that it may be a neurodevelopmental disease modifier.
Keywords: CRLF3, single nucleotide variant, neuron deficits, neurodevelopment, dendrites, cerebral organoids
Introduction
Single nucleotide variants are common in the general population, where their functional and clinical significance are unknown. These variants are often classified using predictive algorithms without experimental validation. One such variant was identified in a subset of children with the neurofibromatosis type 1 (NF1) neurogenetic syndrome. As such, individuals with NF1 harboring a 1.4 megabase (Mb) total gene deletion (TGD) of the NF1 locus have intellectual disability, autism symptomatology, and severe neurodevelopmental delays [1–3]. Human induced pluripotent stem cell (hiPSC)-derived cerebral organoids and primary neuronal cultures from children with this deletion exhibit deficits in neuronal maturation and dendritic extension [4]. The responsible gene contained within the deleted NF1 locus was found to be the cytokine receptor-like factor-3 (CRLF3) gene, since genetic CRLF3 silencing alone recapitulated the neuronal abnormalities observed in cerebral organoids and neurons derived from hiPSCs with the 1.4 Mb NF1 TGD.
CRLF3 was originally discovered as an orphan class I cytokine receptor in insects. In these eumetazoan animals, CRLF3 is related to the erythropoietin receptor [5] and can function to transduce erythropoietin neuroprotection signaling [6, 7]. Additionally, CRLF3 has been implicated in neuronal escape from cell death [8], as well as a major contributor to platelet genesis [9, 10] in zebrafish and mice. Unfortunately, little is known about CRLF3 function in mammalian brain cells, other than that its haploinsufficiency in hiPSC-derived cerebral organoids from children with the NF1 TGD syndrome is sufficient to elicit neurodevelopmental abnormalities in vitro [4].
Analysis of a small group of children with NF1 who harbor intragenic germline NF1 gene mutations (e.g. missense, nonsense mutations) demonstrated that those with a missense mutation in the CRLF3 gene (CRLF3L389P) had higher autism burden than those without this variant. While these findings were provocative, this particular missense CRLF3 mutation is common in the general population (15%) [gnomAD; https://gnomad.broadinstitute.org/]. Using hiPSCs and mice with the CRLF3L389P mutation, we now demonstrate that this missense alteration reduces neuronal maturation, dendritic arborization, and activity levels.
Results
The CRLF3L389P variant does not alter gene expression, brain cell populations, or behavior in mice
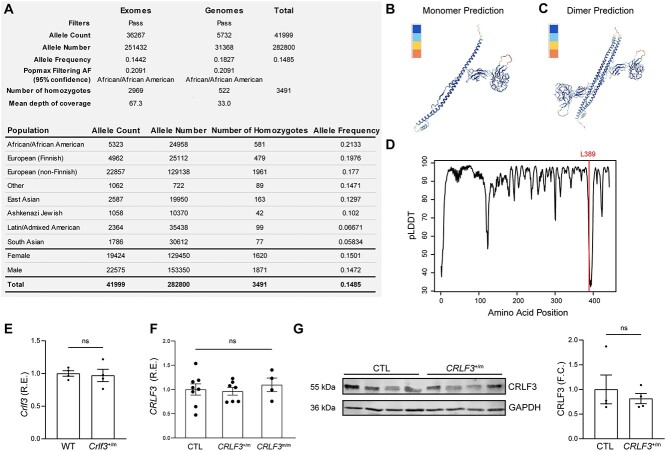
The CRLF3L389P variant is detected in 14.4% and 18.3% of all exomes and genomes, respectively, in the general population (Fig. 1A). Although there is some variation in variant frequencies amongst different ethnic/racial populations, with the highest frequencies noted in African/American and European and lower frequencies in South Asian and Latin/Admixed American cohorts, there were no observed differences between males and females.
Figure 1.
The CRLF3 L389P mutation is a common SNV that does not significantly reduce CRLF3 gene expression. (A) Population frequencies of SNV 17-29111368-A-G from gnomAD reports an overall allele frequency of 14.85%. (B) Predicted CRLF3 3D monomer structure (pLDDT = 90.1). (C) Predicted CRLF3 3D dimer structure (iptm+ptm = 0.685). (D) Prediction confidence of different amino acid positions, indicating low confidence (pLDDT < 40) in the protein region where residue 389 resides. (E) RT-qPCR of 4-week-old mouse hippocampi showing similar Crlf3 RNA expression in Crlf3+/m and WT littermate mice. Each data point represents bilateral hippocampi from one mouse. (F) RT-qPCR of hiPSC-derived NPCs showing similar CRLF3 RNA expression in CRLF3+/m, CRLF3m/m and control cells. Each data point represents an independent biological replicate. (G) Western blot showing similar CRLF3 protein levels in CRLF3+/m and control NPCs. Each data point represents an independent biological replicate. Values for RT-qPCR and WB are reported as fold changes from the control average. All data are shown as the mean ± SEM. Statistical analysis by unpaired, two-tailed t-test with Welch’s correction (E, G), or one-way ANOVA with Dunnett’s multiple comparisons test (F). R.E., relative expression; ns, not significant.
Initially, to estimate the effect of the CRLF3L389P variant on CRLF3 protein structure, we used the AlphaFold algorithm and obtained a highly confident 3D structure prediction of CRLF3 (pLDDT = 90.1) in both its monomer and dimeric forms (Fig. 1B–C). However, as the mutation locus resides within a specific region of the protein with low structure prediction confidence, we could not establish the exact conformational consequence of the leucine 389 to proline substitution (Fig. 1D).
We next sought to ascertain whether this CRLF3L389P variant resulted in reduced CRLF3 protein expression. To this end, we leveraged both genetically engineered mice harboring the Crlf3L389P germline mutation, CRISPR/Cas9-engineered human iPSCs with the CRLF3L389P mutation. Both Crlf3L389P mouse hippocampi, as well as heterozygous and homozygous CRLF3L389P-mutant hiPSC-derived neural progenitor cells (NPCs), exhibited no differences in Crlf3 (Fig. 1E, Fig. S1A) or CRLF3 expression (Fig. 1F), respectively, at the RNA level. While commercially available anti-Crlf3 antibodies do not reliably detect mouse Crlf3 protein, human CRLF3L389P NPCs exhibited similar levels of CRLF3 protein expression relative to their isogenic controls (Fig. 1G).
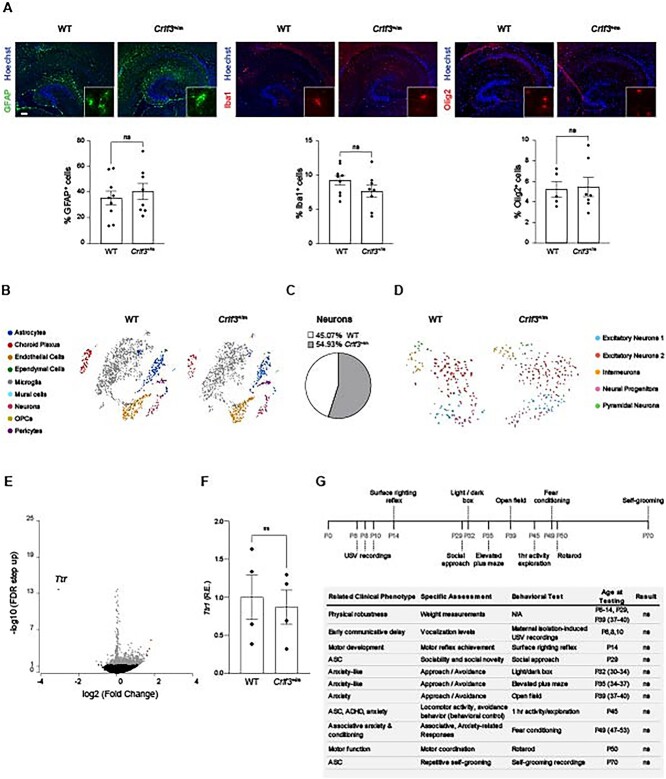
Crlf3 L389P mutant mice lack brain cell composition, transcriptomal or behavioral abnormalities
To determine whether mice harboring the Crlf3L389P variant exhibit changes in their overall brain cell composition, defects at the tissue level, or behavioral deficits, we performed three sets of analyses. First, we analyzed the cellular composition of hippocampi from wild type (WT) and heterozygous Crlf3L389P-mutant (Crlf3+/m) mice using cell type-specific antibodies. No differences in astrocyte (GFAP+ cells), microglia (Iba1+ cells) or oligodendrocyte (Olig2+ cells) content were noted (Fig. 2A, Fig. S1B). We conclude that these populations were not primarily affected by Crlf3 mutation, focusing our attention on neurons (see below). Second, we employed single cell RNA sequencing of Crlf3+/m and WT hippocampi as an additional method to query brain cell populations, and found no reproducible differences in brain cell populations (Fig. 2B), total number of neurons, or neuronal cell subpopulations (Fig. 2C–D). Moreover, analysis of differentially expressed genes within the hippocampus (Fig. 2E; Tables S1–S2) revealed only a single gene had greater than 3-fold differential expression: while transthyretin (Ttr) was 8-fold reduced in Crlf3+/m mice using scRNAseq, Ttr1 RNA expression was similar in independently acquired WT control and Crlf3+/m mouse hippocampi. Third, we performed an age-dependent (postnatal day (PND) 6-PND70) battery of behavioral tests on WT and Crlf3+/m mice aimed at analyzing physical and social developmental milestones, but identified no differences between genotypes (Fig. 2G, Fig. S2). Of note, no sex differences were observed.
Figure 2.
The Crlf3 L389P variant does not alter mouse brain cellular composition or mouse behavior. (A) WT and Crlf3+/m mouse hippocampi have similar numbers of astrocytes (GFAP+ cells), microglia (Iba1+ cells) and oligodendrocytes (Olig2+ cells). (B) t-SNE plot analyses of single-cell sequencing of 4-week-old WT and Crlf3+/m mouse hippocampi reveal similar brain cell populations across genotypes. (C-D) Focused analysis of the WT and Crlf3+/m single-cell sequenced neurons alone shows similar percentages of neurons represented by the two genotypes (C), as well as similar neuronal subpopulations between genotypes as shown in the t-SNE plot (D). (E) Volcano plot comparing gene expression in WT and Crlf3+/m mice hippocampi. Blue indicates downregulated genes (n = 1) in Crlf3+/m samples, grey indicates no difference, and red indicates upregulated genes (n = 4). Ttr is the only gene downregulated greater than 3-fold relative to WT controls. (F) Ttr1 RNA expression is similar in WT and Crlf3+/m mice hippocampi. (G) Schematic and table summarizing the behavioral analyses performed on WT and Crlf3+/m mice, indicating no significant differences in weight, motor development, or behavior tested. Values for RT-qPCR are reported as fold changes from the control average. All data are shown as the mean ± SEM. Statistical analysis by unpaired, two-tailed t-test. ns, not significant.
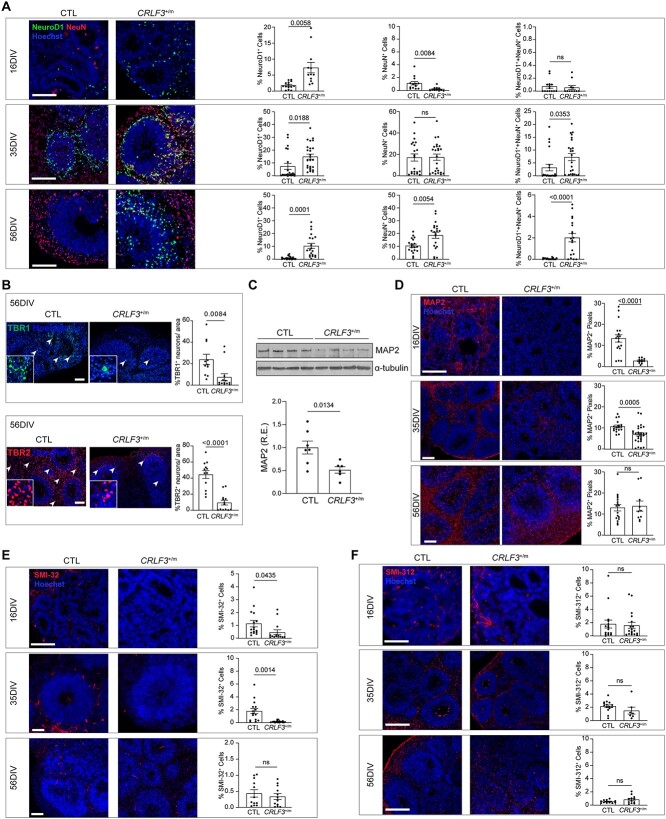
CRLF3 L389P mutation impairs neuronal and dendritic development
To determine the potential impact of the CRLF3L389P mutation on human neuronal development, we generated human cerebral organoids (hCOs) from two separate isogenic hiPSC lines, derived from two independent clones, engineered to harbor a heterozygous CRLF3L389P missense mutation (CRLF3+/m) and compared them to isogenic control BJFF.6 hiPSC-derived hCOs. Similar to shCRLF3 knockdown hCOs [4], CRLF3+/m hCOs had unaltered neural stem cell (NSC) proliferation relative to controls (Fig. S3). We then examined neuronal maturation within the hCOs at 16, 35, and 56 days in vitro (DIV), and found that CRLF3+/m hCOs had an increase in total number of immature neurons (%NeuroD1+ cells) relative to controls at all time points examined (Fig. 3A), mirroring our prior observations using shCRLF3 hCOs [4]. In addition, CRLF3+/m hCOs had reduced numbers of mature neurons (%NeuN+ cells) at 16DIV and increased numbers of intermediate maturity neurons (%NeuN and NeuroD1+ cells) at 35 and 56 DIV, Fig. 3A), as well as reduced number of mature TBR1+ and TBR2+ neurons at 56DIV (Fig. 3B) relative to control hCOs. In addition to these neuronal maturation defects, CRLF3+/m hCOs exhibited reduced dendrite development as indicated by reduced MAP2 (20–80% reduction, Fig. 3C–D) and SMI-32 (50–80% reduction, Fig. 3E) immunostaining at 16 and 35 DIV compared to controls. This deficit was limited to dendrites, as there were no differences in axon staining (SMI-312, Fig. 3F) across all time points analyzed.
Figure 3.
CRLF3 +/m hCOs demonstrate dendritic and maturation defects. (A) Representative images and quantification of immature (NeuroD1+ cells, green), mature (NeuN+ cells, red), and intermediate (NeuroD1+ cells and NeuN+ cells) neurons in the proliferative zones of CTL and CRLF3+/m hCOs at 16, 35, and 56 DIV. In CRLF3+/m hCOs, there are more immature neurons at all times examined, fewer mature neurons at 16 DIV, and more intermediate neurons at 35 and 56 DIV. (B) Representative images and quantification of mature TBR1+ (green) and TBR2+ (red) neurons in 56 DIV hCOs. CRLF3+/m hCOs have reduced TBR1 and TBR2 expression relative to controls. (C) Reduced MAP2 (C/D isoform) protein levels in 16DIV CRLF3+/m hCOs relative to 16 DIV CTL hCOs by Western blot. Each data point represents 5–6 pooled 16 DIV hCOs, 2 experimental replicates. Values reported as fold change from control average. (D) Reduced numbers of MAP2+ dendrites were identified in CRLF3+/m, relative to control, hCOs at 16 and 35 DIV. (E) Fewer SMI-32+ dendrites were found in CRLF3+/m, relative to control, hCOs at 16 and 35 DIV. (F) No changes in SMI-312+ axons were identified in CRLF3+/m hCOs at 16, 35, and 56 DIV. (F–G) Each data point represents the mean of 2 technical replicates of 1 hCO, N = 7–29 hCOs per genotype. Scale bars: 100 μm. All data are shown as the mean ± SEM. Statistical analysis used unpaired, two-tailed t-tests with Welch’s correction. ns, not significant.
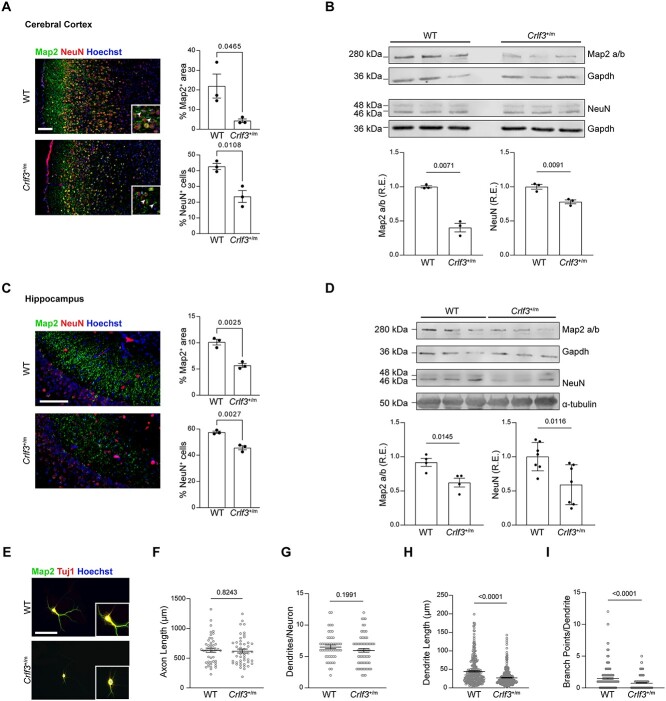
Crlf3+/m mice exhibit dendritic defects
Since hCOs are primarily composed of forebrain cortical cells [4], we examined neurons in the anterior cerebral cortex of Crlf3+/m and WT littermate mice. Similarly to CRLF3+/m hCOs, the outer cortical layers of Crlf3+/m mice had decreased dendrite (Map2) and mature neuron (NeuN) staining relative to WT littermate controls at PND7, as measured by both immunohistochemistry (Fig. 4A; Map2, 80.5% reduction; NeuN, 44.7% reduction), and western blotting (Fig. 4B; Map2 a/b isoform, 60% reduction; NeuN, 22% reduction).
Figure 4.
Crlf3 +/m mice have dendritic defects. (A) Representative images and quantification of mature neurons (NeuN; red) and dendrites (Map2; green) in the outer layers of the cerebral cortex of PND7 mouse Crlf3+/m mice relative to WT littermate controls. (B) Western blot demonstrates reduced Map2 (a/b isoform) and NeuN protein expression in the cerebral cortex of PND7 Crlf3+/m mice relative to WT littermate controls. (C) Representative images and quantification of mature neurons (NeuN; red) and dendrites (Map2; green) in the hippocampus of PND7 Crlf3+/m mice relative to WT littermate controls. (D) Western blotting demonstrates reduced Map2 a/b and NeuN, protein expression in the hippocampus of PND7 Crlf3+/m mice relative to WT littermate controls. (E) Representative images of 14 DIV mouse hippocampal neurons immunolabeled for MAP2 (green) and TUJ1 (red). (F) Axon length is unchanged in Crlf3+/m neurons relative to WT neurons. (G) The number of dendrites per neuron is unchanged in Crlf3+/m neurons relative to WT neurons. (H) Crlf3+/m hippocampal neurons show a reduction in average dendrite length per neuron relative to WT neurons. (I) Crlf3+/m neurons show a reduction in average numbers of branch points per dendrite relative to WT neurons. WB values are reported as fold changes from the control average. All data are shown as the mean ± SEM. Statistical analysis by unpaired, two-tailed t-test with Welch’s correction. ns, not significant. Scale bars: 100 μm.
Similarly, we observed a comparable reduction in dendrite (Map2; 44.6% reduction) staining in the CA1 region (Fig. 4C) and a 32% reduction in Map2 (a/b) protein expression by Western blotting in Crlf3+/m mice relative to WT littermate controls at PND7 (Fig. 4D) Although there was a modest decrease in NeuN+ cell staining (20.5% reduction), we noted a 41% reduction in NeuN protein expression in the hippocampus between Crlf3+/m mice and controls (Fig. 4D).
Given the impairment in Crlf3+/m mouse neurogenesis and the dendritic defect in CRLF3+/m hiPSC-neurons, we leveraged primary Crlf3+/m and WT littermate control hippocampal neurons maintained for 14 days in vitro to allow for the establishment of neuronal polarity and dendrite development (Fig. 4E, Fig. S4A) [11, 12]. Although Crlf3+/m neurons had similar total axon lengths (Fig. 4F, Fig. S4B) and numbers of primary dendrites per neuron (Fig. 4G) compared to WT littermate controls, Crlf3+/m neurons demonstrated a 39% reduction in dendrite length (Fig. 4H, Fig. S4C) and a 48% reduction in the number of branch points per dendrite (Fig. 4I). Together with the hCO data, these findings firmly establish the CRLF3L389P as a regulator of both human and mouse neuronal maturation and dendritic outgrowth.
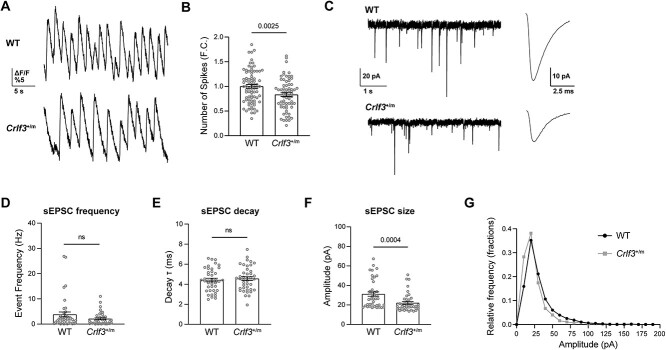
Crlf3+/m neurons exhibit electrophysiologic defects
Since dendrites are key regulators of neuronal electrochemical signal transduction, we next sought to determine whether Crlf3+/m neurons have electrophysiologic defects. Crlf3+/m hippocampal neurons demonstrated a 17% (calcium imaging; Fig. 5A–B) and 49% (multi-electrode array [MEA] recording; Fig. S5) reduction in their firing/spiking frequency relative to WT littermate controls. Firing was insensitive to picrotoxin in both genotypes (Fig. S5), suggesting that GABAA receptor-mediated inhibition is not an important driver of the phenotypic difference in firing. We thus next used whole-cell voltage-clamp recording (Fig. 5C) to measure the excitatory postsynaptic currents (EPSPCs) in individual hippocampal neurons. While there were no differences in sEPSC frequency (Fig. 5D) or decay (Fig. 5E), there was a reduction in the amplitude of sEPSCs in Crlf3+/m neurons (Fig. 5F; 29% reduction), as well as a left-shift in the relative frequency of small amplitude sEPSCs in Crlf3+/m neurons relative to WT controls (Fig. 5G). Collectively, these findings demonstrate that the Crlf3L389P mutation impairs hippocampal neuron function, possibly arising from dendritic and related synaptic deficits in glutamate signaling.
Figure 5.
Crlf3 +/m neurons demonstrate electrophysiological defects. (A) Representative Ca2+ imaging traces of Crlf3+/m and WT hippocampal neurons over 30s of recording. (B) The firing rates were reduced in Crlf3+/m hippocampal neurons relative to WT controls. Data was normalized, with the WT average set as 1. Statistical analysis was performed using unpaired, two-tailed t-tests with Welch’s correction. (C) Representative 5 s whole-cell patch clamp recordings, demonstrating spontaneous EPSCs, and averaged spontaneous EPSC waveforms from the entire recording from WT and Crlf3+/m hippocampal neurons. (D) sEPSC frequency (Hz) is similar in WT and Crlf3+/m neurons. Statistical analysis by Mann–Whitney test. (E) sEPSC decay (т; ms) is similar in WT and Crlf3+/m neurons. (F) sEPSC size (pA) is decreased in Crlf3+/m neurons relative to WT neurons. Statistical analysis by Mann–Whitney test. (G) Relative frequency histogram of sEPSC amplitude (pA) is left shifted in Crlf3+/m neurons (grey) relative to WT neurons (black). Bars and errors denote mean ± SEM. Each point represents a recording of a single neuron. ns, not significant.
Discussion
CRLF3 is an orphan cytokine receptor, best studied in invertebrates as a mediator of neuronal survival. In insects (beetles, locusts), it can bind erythropoietin and exert a neuroprotective effect [6–8], whereas in fish, it acts to promote early hematopoiesis [9] and negatively regulate type 1 interferon production following an experimental anti-viral immunity challenge [13]. In mice, CRLF3 is necessary for platelet generation through hippo pathway-mediated microtubule stabilization [10]. However, little is known about CRLF3 function in the mammalian brain. Herein, we demonstrate that a common CRLF3 missense variant leads to delayed neuronal maturation, reduced dendrite extension, and impaired neuronal function. These findings raise several key points.
First, human iPSC-derived cerebral organoids and neurons, as well as murine neurons containing this mutation closely resemble those with shCRLF3-induced near complete loss of CRLF3 expression [4], suggesting that this variant represents a loss of function mutation. This likely occurs because CRLF3, similar to other cytokine receptors, forms hetero- and homo-dimers [5, 14]. While the mutation occurs in a region in which the structure cannot be accurately predicted and does not result in reduced protein expression, it is possible that this amino acid substitution impairs dimerization, rather than stability, leading to impaired function of all cytokine receptor dimers containing CRLF3-containing monomers.
Second, whereas the ligand for CRLF3 in mammalian cells remains unknown, our prior studies did not support erythropoietin as the responsible driver for CRLF3-dependent neuronal defects [4]. Instead of regulating JAK/STAT signaling [7], we previously showed that loss of CRLF3 expression in human iPSC-derived organoids resulted in reduced Rac1/RhoA signaling, which was similarly observed in Crlf3L389P-mutant brains (Wilson A, unpublished observations). This impairment in Rac1/RhoA function is consistent with the known role for these small GTPases in neuronal maturation [15–17], dendritic maturation [18–22], and synaptic function [23–25]. Further work beyond the scope of this study will be required to identify the responsible CRLF3 ligand and define the mechanism by which CRLF3 regulates dendrite maturation and neuronal physiology in neurons.
Third, the finding of a missense variant common in individuals without known neurological deficits suggests that this mutation, while pathogenic at the level of the early developing brain or primary neurons in isolation, is not sufficient by itself to recapitulate clinical symptomatology. In this regard, Crlf3L389P-mutant mice lack behavioral or motor deficits, suggesting that this variant may function as a modifier in combination with other genetic mutations, such as a germline NF1 mutation, in the setting of the Neurofibromatosis Type 1 neurodevelopmental syndrome. Additionally, it is possible that these subclinical abnormalities will only manifest as behavioral deficits in the setting of other insults, such as brain injury, infection, or stress. Future studies will focus on intercrossing these Crlf3L389P-mutant mice with Nf1- and other mutant mouse strains, as well as exploring the impact of other factors on mouse brain function and behavior.
Taken together, this study expands our understanding of the consequence of subtle single nucleotide variants in the genome and their contribution to human neurological disease. As more of these variants are identified through genome sequencing, it will become increasingly important to determine which alterations are drivers of clinical phenotypes and which act as modifiers that operate to change the risk of specific neuropsychiatric disorders.
Materials and Methods
Mice
All experiments were performed under active Animal Studies Committee protocols at Washington University School of Medicine (Washington University Institutional Animal Care and Use Committee). Mice were maintained on a 12 h light/dark cycle in a barrier facility, at 21°C and 55% humidity, with ad libitum access to food and water.
Heterozygous Crlf3 c.1166 T > C; p.Leu389Pro-mutant (L389P) mice were generated by CRISPR/Cas9 engineering. A mixture of validated gRNAs (GGAACTTCTAACAGCCATGANGG and GATATCGAAGCTGTGACTCTNGG) with > 99% non-homologous end-joining frequencies, a single-stranded oligodeoxynucleotides (ssODNs) donor template, and Cas9 protein was electroporated into C57Bl/6 J mouse zygotes, which were later transferred to pseudopregnant females. Successful transfers resulted in mice with one WT Crlf3 allele and one missense Crlf3L389P allele. The mutation was confirmed by next-generation sequencing. Crlf3L389P-mutant mice (Crlf3+/m) were backcrossed to C57Bl/6 J mice, while WT littermates were used as controls. Mice of both sexes were randomly assigned to all experimental groups without bias, and the investigators were blinded until final data analysis.
Mouse behavior
All behavioral data were collected in the Animal Behavior Subunit of the Intellectual and Developmental Disabilities Research Center at Washington University. All experimental protocols were approved by and performed in accordance with the relevant guidelines and regulations of the Institutional Animal Care and Use Committee of Washington University. Three independent cohorts were used for behavioral analysis. Cohort 1 comprised 23 WT (11 female and 12 male) and 23 Crlf3+/m (11 female and 12 male) mice, and were assessed for developmental trajectories during the first two weeks of life. Cohorts 2 (WT, n = 10 females and 15 males; Crlf3+/m, n = 13 females and 12 males) and 3 (WT, n = 10 females and 10 males; Crlf3+/m, n = 16 females and 18 males) were assessed for clinically-relevant behavioral phenotypes beginning as juveniles through early adulthood. For all tasks, mice were acclimated to the testing room for at least 30 min prior to the start of testing. All assays were conducted by the same female experimenters blinded to experimental group designations during testing, which occurred during the light phase. Mice in cohorts 2 and 3 were weighed and assessed for physical health prior to behavioral assessments following previously published methods [26]. Order of tests for each cohort were chosen to minimize effects of stress. Unless otherwise indicated, all equipment was cleaned between animals with a 0.02% chlorhexidine diacetate solution. All behavioral methods are described in detail in the Supplemental Methods section.
gnomAD analysis
Data on the population frequencies for the CRLF3Leu389Pro variant was obtained from the gnomAD online database (https://gnomad.broadinstitute.org/).
3D structure prediction
We predicted the 3D structure of CRLF3 using AlphaFold v2 with the RefSeq (NP_057070.3) fasta sequence as input. The max_template_date used was 2022-03-31 and the database preset used was reduced_dbs. The best ranking 3D structure (ranked 0) was used in this study. In addition, we predicted the 3D structure of a dimer, trimer, and tetramer of CRLF3 using AlphaFold v2 with the RefSeq (NP_057070.3) fasta sequence, respectively. The max_template_date used was 2022-03-31, the database preset used was reduced_dbs, and the model preset was multimer. The best ranking 3D structure (ranked 0) was used in this study.
Human iPSC culture and differentiation
A single CRLF3 gene mutation (Transcript ID CRLF3-201; ENST00000324238.7; c.1166 T > C, p.Leu389Pro) was engineered using CRISPR/Cas9 technology into a single commercially available male control human iPSC line (BJFF.6) by the Washington University Genome Engineering and iPSC Core (GEiC) facility. The heterozygous CRLF3L389P mutation was confirmed by NGS sequencing, and two different clones were expanded to generate the two CRLF3+/mut iPSC lines. hCOs were generated as previously described [4, 27]. Once generated, hCOs were maintained for up to 84DIV in hCO differentiation medium (125 ml DMEM-F12, 125 ml Neurobasal medium, 1.25 ml N2 supplement, 62.5 μl insulin, 2.5 ml GlutaMAX supplement, 1.25 ml MEM-NEAA, 2.5 ml B27 supplement, 2.5 ml penicillin–streptomycin, 87.5 μl of a 1:100 dilution of 2-mercaptoethanol in DMEM-F12) on an orbital shaker rotating at 80 rpm. Media was changed every 3 days. Neural progenitor cells (NSCs) and non-specific induced neurons (iN) were generated according to previously described methods [4, 28].
Primary hippocampal neuron cell culture
Primary neuron cultures were generated from postnatal day 5–7 WT and Crlf3+/m mice. Bilateral hippocampi were quickly (less than one minute exposed) dissected out and placed in ice-cold Hibernate-A (Gibco). Primary hippocampal neurons were dissociated using the Papain Dissociation System Kit (Worthington Biochemicals) largely according to manufacturer’s instructions. Briefly, tissue samples were gently minced by pipetting once with a P200 pipette, and incubated in papain with DNase (in EBSS) for 90 min at 37°C with constant agitation (175 rpm). Following incubation, samples were mechanically dissociated by pipetting up and down 10 times with a P200 pipette, transferred to a new tube, and centrifuged at 300 g for 5 min at room temperature. The cell pellet was resuspended in an albumin-ovomucoid inhibitor and DNase solution in EBSS, and the resulting cell suspension was layered over a BSA-ovomucoid inhibitor (Worthington Biochemicals) before being centrifuged at 100 g for 6 min (deceleration 0) to isolate dissociated cells in the pellet via a discontinuous density gradient. Pelleted cells were resuspended in neurobasal growth medium (Gibco Neurobasal with B27, L-glutamine, and penicillin/streptomycin) and plated at assay-dependent densities on poly-D-lysine (Sigma)/laminin (STEMCELL) plates. Neurons were grown for 7 to 14 days depending on the experiment, and media was refreshed every other day.
Calcium imaging of hippocampal neurons
Primary hippocampal neurons were plated at 100 000 cells/well onto poly-D-lysine and laminin-coated 96 well plates and grown for 7 days. At 7 days, the cells were treated with Fluo-8/AM (1345980-40-6, AAT Bioquest), PowerLoad (P10020, ThermoFisher), and Probenecid (P36400; ThermoFisher) for 30 min at 37°C and for another 30 min at room temperature. Neurons were subsequently washed with HBSS and incubated for a minimum of 10 min in fresh culture medium supplemented with 5% neuro-background suppressor (F10489; ThermoFisher). Neurons were imaged on a Nikon spinning disk upright epi-fluorescence confocal microscope equipped with a × 10 dry objective, and a 488 nm wavelength laser was used for wide-field imaging. Neurons were stimulated by a Ti LAPP DMD (Deformable Mirror Device) LED source for ultrafast photo-stimulation, with 0.1 mW applied during each recording for Fluo-8 excitation. Fluo-8 images were collected at 15 Hz (2048 × 2048 pixels, 1 × 1 mm) and the duration of each region of interest (ROI) was limited to 10 min. The fluorescence intensity and optical response to depolarizing membrane potential transients (ΔF/F) were calculated in a Matlab programming environment to generate single-neuron activity traces. The ΔF/F threshold was set at 4 standard deviation beyond baseline fluorescence. Neurons from each animal were seeded in six wells and a minimum of three neurons were recorded per well. Data recorded from a minimum of 18 neurons per animal were averaged into a single point. Each data point graphed represents a single animal.
Multi-electrode Array recordings and analyses of hippocampal neurons
Primary hippocampal (300 000 cells/well) from WT and Crlf3+/m mice were plated on AXION Biosystems 48-well MEA plates and grown for 14 days. Neurons from a minimum of seven separate WT or Crlf3+/m mice were plated, from a minimum of two independent litters. Neurons isolated from each animal were plated in a minimum of three technical replicate wells within the MEA plate. Activity of the same neurons was recorded both prior to 40 μM picrotoxin (PTX) administration and immediately following PTX treatment. All neurons were recorded for 3 min at a five standard deviation threshold level and 5000 Hz as a digital filter using AXION Biosystems integrated studio (AxIS) version 2.5.1 software. Spike rates were calculated from the total number of spikes/3 min and are represented as spikes/sec, only accounting for active electrodes within each well. Each data point graphed represents the average of all technical replicates for each given animal. Representative traces of spikes were extracted using the AXION Biosystems neural metric tool.
Whole-cell patch-clamp recordings of hippocampal neurons
Experiments on primary hippocampal neurons were conducted using standard whole-cell patch techniques from 11 to13 days after plating. The extracellular bath solution contained (in mM): 138 NaCl, 4 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, 0.02 gabazine, and 10 HEPES; pH 7.25. Patch pipettes were filled with a solution containing in mM: 130 cesium methanesulfonate, 4 NaCl, 0.5 CaCl2, 5 EGTA, 10 HEPES, pH 7.25. When filled with this solution, pipette tip resistance was 3-6 MΩ. Cells were clamped at −70 mV with an Axoclamp 200B amplifier (Molecular Devices, San Jose, CA). Recordings were performed in in gap-free mode at 5 kHz sampling rate and filtered at 2 kHz to capture spontaneous excitatory synaptic currents (EPSCs). All recordings were performed at room temperature. The analysis of spontaneous EPSCs was conducted using pClamp 9.0 software (Molecular Devices). Individual EPSCs were identified and counted using a template matching algorithm from a 2-min continuous record. Decay (tau) values were estimated using a standard exponential fitting algorithm in PClamp. Approximately 5 cells were recorded per animal culture, from a total of 8 Crlf3+/m and 8 WT mice, respectively.
Immunohistochemistry and immunocytochemistry
For immunohistochemical staining of human cerebral organoids, tissues were fixed, embedded, cryosectioned at 10 μm, and immunostained as previously described at 16, 36 and 56 DIV [27]. For immunohistochemical analyses of mouse brains, whole brains were fixed in 4% paraformaldehyde in water for 24 h at 4°C, rinsed with PBS, and transferred to 30% sucrose in PBS. Tissues remained in this solution until equalized, at which point they would sink to the bottom of the tube (2 to 4 days). Brains were then embedded and immunostained in the same manner as the hCOs [27]. Briefly, all tissues on slides were permeabilized in 0.5% Triton-X in PBS for 5 min, washed in PBS, and blocked in 10% goat serum for one hour at room temperature. If a mouse primary antibody was to be used on mouse brain cryosections, an additional anti-mouse blocking solution (1:200 in PBS, cat#115-007-003, Jackson) was applied for one hour at room temperature prior to primary antibody application. Primary antibodies were diluted in 2% goat serum and were applied overnight at 4°C (Table S3). After primary antibody incubation, slides were washed with PBS, and labeled with appropriate Alexa-fluor-conjugated secondary antibodies (1:200, Invitrogen) diluted in 2% goat serum for one hour at room temperature. Hoechst-33258 (Invitrogen) was used as a nuclear counterstain.
For immunocytochemical analyses of primary mouse hippocampal neurons, neurons were fixed in 4% paraformaldehyde for 15 min before permeabilization in 0.5% Triton-X for 5 min. Neurons were washed in PBS and blocked in a 10% goat serum in PBS solution for one hour at room temperature. Primary antibodies in a 2% GS serum were applied overnight at 4°C (Table S1). After primary antibody incubation, neurons were washed three times with PBS and labeled with appropriate Alexa-Fluor-conjugated secondary antibodies (AlexaFluor488/568, 1:200, Invitrogen) diluted in 2% goat serum for one hour at room temperature. After three washes with PBS, nuclei were counterstained with Hoechst-33258 (Invitrogen). Dendrites were determined as the neurites with stronger Map2 staining relative to Tuj1 staining, after excluding the longest neurite as the axon. Only intact neurons that had not formed networks with other cells were analyzed. Dendrite lengths were measured manually using ImageJ, with tracing starting at the outer edge of the cell body to not skew measurements. Primary dendrites were defined as neurites initiating from the cell body, and their length was determined by tracing from the cell body edge to the furthest terminal, not including branch points. The dendrite number was determined by the number of measurements taken per neuron, and branch points per dendrite were counted manually. Axons were determined as the longest Tuj1+ neurite, and their length was measured using the NeuronJ plugin for ImageJ.
Western blot analysis
Human NSCs, hCOs, and mouse brain tissue samples were collected and sonicated in RIPA buffer (89 900, Thermo Scientific) supplemented with 2 μg/ml aprotinin (ab146286, Abcam), 1 μg/ml leupeptin (L2884, Sigma-Aldrich), and 1 mM PMSF (10 837 091 001, Sigma-Aldrich). Total protein concentrations were determined using a Pierce BCA protein assay kit (23 225, Thermo Scientific). Samples were diluted so that equal amounts of protein (30 to 45 μg) would be loaded per well, and were reduced and denatured in Laemmli buffer (1 610 747, Bio-Rad) supplemented with β-mercaptoethanol at 99°C for 5 min. Equal amounts of sample were loaded into each well of 8% or 10% SDS-PAGE gels and run for 1 to 2 h at 120 V, depending on the protein of interest. Proteins were then transferred to Immobilon-FL polyvinylidene fluoride membranes (IPFL0010, Millipore) at 4°C overnight at 30 V in transfer buffer (10% methanol, 0.6% TRIS. 2.85% glycine in water) using a Criterion blotter (1 704 071, Bio-Rad). The membranes were blocked for 1 h in 5% milk in Tris-buffered saline (TBS), followed by overnight incubation at 4°C with primary antibodies diluted in TBS (Table S3). After washing with TBS, the membranes were incubated with a 1:5000 dilution of goat anti-rabbit IRDye 680RD (926-68 071, LI-COR Biosciences) and goat anti-mouse IRDye 800CW (925-32 210, LI-COR Biosciences) secondary antibodies for one hour at room temperature. Development of the immunoblots was performed using a LI-COR Odyssey Fc imaging system (LI-COR Biosciences), and protein bands were quantified using Fiji for ImageJ Software 1.53 t. Experimental protein values were first normalized to GAPDH or α-Tubulin internal loading controls.
Real-time quantitative PCR
Total RNA was extracted from mouse brain tissue, human NPCs or hCOs following the manufacturer’s instructions (NucleoSpin RNA Plus, 74098_4.50, Takara Bio) and reverse-transcribed using a high-capacity cDNA reverse transcription kit (4 374 967, Applied Biosystems). Quantitative PCR was performed using TaqMan gene expression assays against Crlf3 and CRLF3 (Table S4) and TaqMan Fast Advanced Master Mix (Applied Biosystems) according to the manufacturer’s instructions. All reactions were performed using the Applied Biosystems QuantStudio 3 system and the QuantStudio Design & Analysis Software v1.5.2. Gene expression levels of biological and technical replicates were estimated by ΔΔCt method Gapdh and GAPDH, respectively, as reference genes (Table S4). A minimum of three independently generated samples was used for each condition.
RNA sequencing and analysis
Crlf3L389P-mutant and WT hippocampi were collected from four 4-week-old mice (4 hippocampi/2 mice per genotype) and pooled to prepare single-cell isolations for each region, as previously reported [29]. After collection, the brain tissues were dissociated using the neural dissociation kit (Miltenyi Biotec, 130-095-942) as per manufacturer’s instructions followed by debris (Miltenyi Biotech, 130-107-677) and myelin (Miltenyi Biotec, 130-096-733) removal. The cells were then stained with 7-AAD and sorted using the MO-FLO. Live cells were collected and sent for sequencing. For all samples, raw sequencing data were processed using the 10X Genomics Cell Ranger pipeline (version 6.1.1) to generate gene count matrices, aligned to the mouse reference genome (mm10-2020-A), and analyzed in Partek® Flow® software (Partek Inc. (2020), version 10.0.23, https://www.partek.com/partek-flow). Cell quality was evaluated by the number of reads, detected gene counts, and percentage of mitochondrial reads, followed by CPM (counts per million) normalization and PCA analysis. To account for library chemistry and other possible unknown differences between experiments, cells were integrated using the Seurat3 integration method to produce batch-corrected data. The subsequent graph-based clustering using Louvain algorithm, and t-SNE plot generation employing Euclidean distance, were conducted on the first 20 principal components of the integrated expression data. Differential gene expression analyses (Fig. 2E, Table S1) were performed using the Hurdle model (MAST) to analyze for differentially expressed genes between Crlf3mut and WT samples, and the results were filtered for only those genes with FDR < 0.05, and fold changes either less than or equal to −3 or greater than or equal to 3. For differential pathway analyses (Table S2), we used the DAVID pathway analysis platform [30, 31], analyzing differentially expressed genes between Crlf3mut and WT samples with FDR < 0.05, and fold change is either less than or equal to −2 or greater than or equal to 2, and subsequently filtered the resulting pathways at a significance level of FDR < 0.1.
Statistical analyses
Statistical analyses and data visualization for behavioral tests were conducted using IBM SPSS Statistics (v.28). Prior to analyses, data were screened for missing values and fit of distributions with assumptions underlying univariate analysis. This included the Shapiro–Wilk test on z-score-transformed data and qqplot investigations for normality, Levene’s test for homogeneity of variance, and boxplot and z-score (± 3.29) investigation for identification of influential outliers. Means and standard errors were computed for each measure. Linear modeling including mixed designs and analysis of variance (ANOVA), including repeated measures and covariate designs, were used to analyze data where appropriate, and simple main effects were used to dissect significant interactions. Sex was included as a biological variable in all analyses across all experiments. Where appropriate, the Greenhouse–Geisser or Huynh-Feldt adjustment was used to protect against violations of sphericity for repeated measures designs. Multiple pairwise comparisons were subjected to Bonferroni correction or the Benjamini-Hochberg method for false discovery rate, where appropriate. For data that did not fit univariate assumptions, non-parametric tests were used to confirm effects. Sex*genotype effects are reported where significant, otherwise data are discussed and visualized collapsed for sex, with details of all statistical tests and results reported in Fig. 2G. The critical alpha value for all analyses was p < 0.05 unless otherwise stated. Pearson’s correlation was used to assess relationships between continuous outcomes and one-sample t-tests were used to assess differences from chance levels. The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Statistical tests and visualization for all other analyses were conducted using GraphPad Prism 9.4.1. Statistical significance was set to p < 0.05. All pairwise comparisons underwent 2-tailed student’s t-tests, while all group comparisons underwent one-way ANOVA with appropriate (Dunnet’s, Tukey’s) post-test corrections. The exact tests utilized are indicated within each figure.
Supplementary Material
Acknowledgements
We thank Chloe M. Kernan, Sotaro Atsumi and Taylor John-Lewis for their help with immunohistochemistry, imaging and sectioning. We thank Sara Conyers and Katherine B. McCullough for the behavioral data collection. We thank the Genome Engineering and iPSC Center at Washington University for generating the human iPSCs and the Pediatric Disease Mouse Models Core (CDICORE2019820) supported by the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital for the production of the Crlf3L389P-mutant mice. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis, MO., for the use of the Siteman Flow Cytometry, which provided cell sorting service. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant #P30 CA091842. This work was partially supported by a Research Program Award from the National Institute of Neurological Disorders and Stroke to D.H.G. (R35-NS097211), Just-in-Time grants (JIT820; JIT962H) from the Washington University Institute of Clinical and Translational Sciences (UL-TR002345), a National Cancer Institute R50 grant to C.A. (R50-CA233164), the Taylor Institute for Innovative Psychiatric Research (S.J.M), the National Institute of Mental Health to S.J.M (R01-MH123748 and P50-MH122379), and funding for the Intellectual and Developmental Disabilities Research Center as well as the Animal Behavior Subunit by the Eunice Kennedy Shriver National Institute of Child Health & Human Development (P50 HD103525).
Conflict of interest statement: None of the authors have relevant conflicts of interest to disclose.
Contributor Information
Anna F Wilson, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Rasha Barakat, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Rui Mu, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Leah L Karush, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Yunqing Gao, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Kelly A Hartigan, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Ji-Kang Chen, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Hongjin Shu, Department of Psychiatry, Washington University School of Medicine, Box 8134, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Tychele N Turner, Department of Genetics, Washington University School of Medicine, Box 8232, 660 South Euclid Avenue, St. Louis, MO 63110, United States; Intellectual and Developmental Disabilities Research Center, Washington University School of Medicine, Box 8504, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Susan E Maloney, Department of Psychiatry, Washington University School of Medicine, Box 8134, 660 South Euclid Avenue, St. Louis, MO 63110, United States; Intellectual and Developmental Disabilities Research Center, Washington University School of Medicine, Box 8504, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Steven J Mennerick, Department of Psychiatry, Washington University School of Medicine, Box 8134, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
David H Gutmann, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Corina Anastasaki, Department of Neurology, Washington University School of Medicine, Box 8111, 660 South Euclid Avenue, St. Louis, MO 63110, United States.
Funding
This work was partially supported by a Research Program Award from the National Institute of Neurological Disorders and Stroke to D.H.G. (R35-NS097211), Just-in-Time grants (JIT820; JIT962H) from the Washingt on University Institute of Clinical and Translational Sciences (UL-TR002345), a National Cancer Institute R50 grant to C.A. (R50-CA233164), the Taylor Institute for Innovative Psychiatric Research (S.J.M), the National Institute of Mental Health to S.J.M (R01-MH123748 and P50-MH122379), and funding for the Intellectual and Developmental Disabilities Research Center as well as the Animal Behavior Subunit by the Eunice Kennedy Shriver National Institute of Child Health & Human Development (P50 HD103525).
Data availability
The single cell RNA sequencing datasets were deposited in GEO (accession number GSE235594).
References
- 1. Brunetti-Pierri N, Grange DK, Ou Z. et al. Characterization of de novo microdeletions involving 17q11.2q12 identified through chromosomal comparative genomic hybridization. Clin Genet 2007;72:411–9. [DOI] [PubMed] [Google Scholar]
- 2. Kehrer-Sawatzki H, Kluwe L, Salamon J. et al. Clinical characterization of children and adolescents with NF1 microdeletions. Childs Nerv Syst 2020;36:2297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Venturin M, Guarnieri P, Natacci F. et al. Mental retardation and cardiovascular malformations in NF1 microdeleted patients point to candidate genes in 17q11.2. J Med Genet 2004;41:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wegscheid ML, Anastasaki C, Hartigan KA. et al. Patient-derived iPSC-cerebral organoid modeling of the 17q11.2 microdeletion syndrome establishes CRLF3 as a critical regulator of neurogenesis. Cell Rep 2021;36:109315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boulay JL, O'Shea JJ, Paul WE. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity 2003;19:159–63. [DOI] [PubMed] [Google Scholar]
- 6. Hahn N, Knorr DY, Liebig J. et al. The insect Ortholog of the human orphan cytokine receptor CRLF3 is a neuroprotective erythropoietin receptor. Front Mol Neurosci 2017;10:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hahn N, Buschgens L, Schwedhelm-Domeyer N. et al. The orphan cytokine receptor CRLF3 emerged with the origin of the nervous system and is a neuroprotective erythropoietin receptor in locusts. Front Mol Neurosci 2019;12:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knorr DY, Schneider K, Buschgens L. et al. Protection of insect neurons by erythropoietin/CRLF3-mediated regulation of pro-apoptotic acetylcholinesterase. Sci Rep 2022;12:18565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taznin T, Perera K, Gibert Y. et al. Cytokine receptor-like factor 3 (CRLF3) contributes to early zebrafish hematopoiesis. Front Immunol 2022;13:910428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bennett C, Lawrence M, Guerrero JA. et al. CRLF3 plays a key role in the final stage of platelet genesis and is a potential therapeutic target for thrombocythemia. Blood 2022;139:2227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci 1988;8:1454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci 1994;17:267–310. [DOI] [PubMed] [Google Scholar]
- 13. Yan X, Zheng W, Geng S. et al. Cytokine receptor-like factor 3 negatively regulates antiviral immunity by promoting the degradation of TBK1 in teleost fish. J Virol 2023;97:e0179222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liongue C, Ward AC. Evolution of class I cytokine receptors. BMC Evol Biol 2007;7:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duncan BW, Mohan V, Wade SD. et al. Semaphorin3F drives dendritic spine pruning through rho-GTPase Signaling. Mol Neurobiol 2021;58:3817–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stankiewicz TR, Linseman DA. Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front Cell Neurosci 2014;8:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vadodaria KC, Brakebusch C, Suter U. et al. Stage-specific functions of the small Rho GTPases Cdc42 and Rac1 for adult hippocampal neurogenesis. J Neurosci 2013;33:1179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schulz J, Schumacher S. Methods to study the roles of rho GTPases in dendritic tree complexity. Methods Mol Biol 2018;1821:297–317. [DOI] [PubMed] [Google Scholar]
- 19. Schulz J, Franke K, Frick M. et al. Different roles of the small GTPases Rac1, Cdc42, and RhoG in CALEB/NGC-induced dendritic tree complexity. J Neurochem 2016;139:26–39. [DOI] [PubMed] [Google Scholar]
- 20. Park AR, Oh D, Lim SH. et al. Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a substrate of PTPRT. J Cell Sci 2012;125:4518–31. [DOI] [PubMed] [Google Scholar]
- 21. Gualdoni S, Albertinazzi C, Corbetta S. et al. Normal levels of Rac1 are important for dendritic but not axonal development in hippocampal neurons. Biol Cell 2007;99:455–64. [DOI] [PubMed] [Google Scholar]
- 22. Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci 2000;20:5329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pyronneau A, He Q, Hwang JY. et al. Aberrant Rac1-cofilin signaling mediates defects in dendritic spines, synaptic function, and sensory perception in fragile X syndrome. Sci Signal 2017;10:eaan0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hedrick NG, Harward SC, Hall CE. et al. Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature 2016;538:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang MG, Guo Y, Huganir RL. AMPA receptor and GEF-H1/Lfc complex regulates dendritic spine development through RhoA signaling cascade. Proc Natl Acad Sci U S A 2009;106:3549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J, Lambo ME, Ge X. et al. A MYT1L syndrome mouse model recapitulates patient phenotypes and reveals altered brain development due to disrupted neuronal maturation. Neuron 2021;109:3775–3792.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anastasaki C, Wilson AF, Chen AS. et al. Generation of human induced pluripotent stem cell-derived cerebral organoids for cellular and molecular characterization. STAR Protoc 2022;3:101173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anastasaki C, Wegscheid ML, Hartigan K. et al. Human iPSC-derived neurons and cerebral organoids establish differential effects of germline NF1 gene mutations. Stem Cell Reports 2020;14:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Andrade Costa A, Chatterjee J, Cobb O. et al. RNA sequence analysis reveals ITGAL/CD11A as a stromal regulator of murine low-grade glioma growth. Neuro-Oncology 2022;24:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 31. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell RNA sequencing datasets were deposited in GEO (accession number GSE235594).