Abstract
Wild-type equine herpesvirus 1 (EHV-1) strains express a large (250-kDa) glycoprotein, gp2, that is encoded by EUs4 (gene 71) located within the unique short region of the genome. DNA sequence analysis revealed that EUs4 of the pathogenic EHV-1 strain RacL11 is an open reading frame of 2,376 bp that encodes a protein of 791 amino acids. The attenuated EHV-1 vaccine strain KyA harbors an in-frame deletion of 1,242 bp from bp 222 to 1461 and expresses a truncated gp2 of 383 amino acids. To determine the relative contribution of gp2 to EHV-1 pathogenesis, we compared the course of respiratory infection of CBA mice infected with either wild-type RacL11, attenuated KyA, or a recombinant KyA that expresses the full-length gp2 protein (KyARgp2F). Mice infected with KyA lost a negligible amount of body weight (0.18% total weight loss) on day 1 postinfection and regained weight thereafter, whereas mice infected with KyARgp2F or RacL11 steadily lost weight beginning on day 1 and experienced a 20 and 18% loss in body weight, respectively, by day 3. Immunohistochemical and flow cytometric analyses revealed higher numbers of T and B lymphocytes and an extensive consolidation consisting of large numbers of Mac-1-positive cells in the lungs of animals infected with KyARgp2F compared to animals infected with KyA. RNase protection analyses revealed increased expression of numerous cytokines and chemokines, including interleukin-1β (IL-1β), IL-6, tumor necrosis factor alpha, macrophage inflammatory protein 1α (MIP-1α), MIP-1β, MIP-2, interferon γ-inducible protein, monocyte chemotactic protein 1, and T-cell activation gene 3 at 12 h postinfection with KyARgp2F. Three independent DNA array experiments confirmed these results and showed a 2- to 13-fold increase in the expression of 31 inflammatory genes at 8 and 12 h postinfection with KyARgp2F compared to infection with KyA. Taken together, the results indicate that expression of full-length gp2 is sufficient to restore full respiratory virulence to the attenuated KyA strain and raise caution concerning the inclusion of full-length gp2 in the development of EHV-1 vaccines.
Equine herpesvirus 1 (EHV-1), a member of the subfamily Alphaherpesvirinae, is one of the most prevalent respiratory pathogens of equine populations worldwide, and outcomes of infection include abortion in pregnant mares and respiratory rhinopneumonitis with occasional neurological sequelae (2, 4, 7, 11, 18, 19, 25-27). EHV-1 typically enters the body via respiratory mucosal surfaces and establishes latency in peripheral blood mononuclear cells. Currently available EHV-1 vaccines provide little, if any, long-term immunity and require repeated administration to elicit protective responses (5, 6, 16). The fully sequenced EHV-1 genome is comprised of at least 76 genes, all of which are shared with its close relative, EHV-4 (36, 37). EHV-1 encodes 13 glycoproteins, 11 of which are conserved in other Alphaherpesvirinae viruses, including herpes simplex virus 1 and varicella-zoster virus (37, 28). EHV-1 encodes an additional glycoprotein, gp2 or gp300 (3, 40), expressed from gene 71 (EUs4) (8, 36, 35, 39), which maps within the unique short (Us) genomic segment. Besides EHV-1, homologues of gp2 are present only in two other equid alphaherpesviruses, EHV-4 and asinine herpesvirus 3 (10). EHV-1 gp2 is expressed as a ∼797-amino-acid precursor (36) that is endoproteolytically cleaved by staggered processing immediately following adjacent arginine residues 506 and 507 (21, 39), separating the protein into a cysteine-rich 42-kDa C-terminal subunit containing the transmembrane anchor sequence and a serine/threonine-rich N-terminal region that is highly glycosylated with O-linked carbohydrates (40). Although gp2 is not essential for virus growth in cell culture (35) or in the mouse (24), deletion of gp2 impairs virus cell-to-cell spread, readsorption (31, 34), and secondary envelopment (unpublished data). The EUs4 gene of the attenuated vaccine candidate strain Kentucky A (KyA) harbors a deletion of 1,242 bp and encodes a truncated gp2 of only 383 amino acids (8, 9) in contrast to gp2 of 797 and 791 amino acids of the two pathogenic strains, Ab4 (36) and RacL11 (this study), respectively. This truncated gp2 expressed by KyA is incorporated into extracellular virions (38). In addition to the internal deletion within EUs4, KyA has undergone the complete deletion of nonessential Us genes EUs6 and EUs7 that encode glycoprotein I (gI) and gE, respectively (12). Previously, we demonstrated that the restoration of gI and gE to attenuated KyA partially restored virulence (15) in the CBA mouse model of respiratory infection (33, 32, 42). However, the deletion of EUs6 and EUs7 from pathogenic RacL11 did not significantly reduce clinical signs of respiratory disease in CBA mice (15), indicating that a virulence factor(s), still present in RacL11 but absent in KyA contributed to viral pathogenesis in the murine lung. Given that the only other major deletion within the KyA genome compared to characterized wild-type EHV-1 strains is the large in-frame deletion within EUs4, we hypothesized that expression of full-length gp2 contributes to the virulence phenotype of RacL11 observed in the absence of gI and gE.
To assess the contribution of gp2 to the virulence phenotype in CBA mice, we utilized a recombinant KyA (KyARgp2F) that expresses the full-length EUs4 gene product of the pathogenic RacL11 strain (38). These studies demonstrated that the expression of full-length gp2 in an otherwise-attenuated KyA genomic background is sufficient to restore full respiratory virulence to the attenuated strain and raise concerns regarding the inclusion of full-length gp2 in the development of EHV-1 vaccines.
MATERIALS AND METHODS
Virus and cell culture.
EHV-1 strain KyA and recombinant KyARgp2F (38) used for all intranasal (i.n.) infections were propagated in NBL-6 equine dermal cells (American Type Culture Collection, Manassas, Va.). Methods for propagating and titer determinations of viral stocks have been described previously (1, 13, 20, 33). NBL-6 cells were maintained at 37°C, 5% CO2 in Eagle's minimal essential medium supplemented with penicillin (100 U per ml), streptomycin (100 μg per ml), nonessential amino acids, and 20% fetal calf serum (FCS). RK13 cells were maintained at 37°C, 5% CO2 in Eagle's minimal essential medium supplemented with penicillin (100 U per ml), streptomycin (100 μg per ml), nonessential amino acids, and 10% FCS.
DNA sequencing.
The EUs4 gene was amplified from RacL11 DNA by PCR using forward and reverse primers based on the fully sequenced Ab4 strain (36). The forward primer was engineered to contain a BamHI restriction enzyme site (5′-GGATCC-3′) immediately before 5′-GAAACATACGACACCATCCGC-3′ (corresponding to bp 128,547 to 128,568 of Ab4, beginning 549 bp upstream of the EUs4 start codon). The reverse primer was engineered to contain an EcoR1 site (5′-GAATTC-3′) preceding 5′-TTAAACCAGGTGAGTCTGGCG-3′ (complementary to nucleotides 131,943 to 131,922 of Ab4, beginning 433 bp downstream of the EUs4 stop codon). PCR products were digested for 2 h each with BamH1 and EcoR1 (New England Biolabs, Beverly, Mass.) according to the manufacturer's protocol and electrophoresed on a 0.8% agarose gel. The appropriate fragment was excised and purified using the QIAquick gel extraction kit (QIAGEN, Valencia, Calif.) and ligated using T4 ligase (Invitrogen, Carlsbad, Calif.) into donor plasmid pEGFP-N1 (BD/Pharmingen, San Diego, Calif.). The presence of the EUs4 gene was verified by restriction enzyme digestion, PCR, and Southern blot analyses. Plasmid DNA of the final construct (Racgp2pEGFP) was purified using the QIAprep spin miniprep kit (QIAGEN) and sequenced (SeqWright, Houston, Tex.). The resulting RacL11 EUs4 sequence was deposited with GenBank (accession number AY702017).
Mice.
Female CBA mice, 3 to 4 weeks of age, obtained from Harlan Sprague Dawley (Indianapolis, Ind.), were maintained in filter-topped cages within the Animal Resource Facility of the Louisiana State University Health Sciences Center, Shreveport. The Animal Resource Facility is certified by the Association for Assessment and Accreditation of Laboratory Animal Care International. All procedures were approved precedently by the University Animal Care Committee. All mice were rested for a period of 5 to 7 days prior to use. All experimental groups consisted of five mice each, and all experiments were repeated a minimum of three times.
Infection and assessment of respiratory disease.
CBA mice were anesthetized with halothane (Sigma Chemical Co, St. Louis, Mo.) inhalation and inoculated i.n. with 4 × 105 PFU of KyA, recombinant KyARgp2F, or wild-type RacL11 in a total inoculum of 50 μl. Mice were observed daily following infection for clinical signs of respiratory disease, including labored breathing, ruffled fur, and huddling. For weight loss experiments, mice were weighed individually immediately prior to infection and daily thereafter at the same time each day. Animals exhibiting a profound wasting or inability to move or function were humanely sacrificed to prevent undue suffering.
Isolation of mRNA and RPA.
At the indicated times postinfection, mice were sacrificed by prolonged halothane inhalation and the lungs were removed. Total lung RNA was obtained by grinding the lungs in glass tissue grinders in the presence of TRIzol (Life Technologies, Grand Island, N.Y.) according to the manufacturer's protocol. Poly(A)+ RNA was isolated from total RNA using the FastTrack 2.0 kit (Invitrogen) according to the manufacturer's instructions. The resulting poly(A)+ RNA was quantitated and analyzed by RNase protection analyses (RPA), which were performed using the 32P-based Ribo-Quant multiprobe RPA system (BD/Pharmingen) as previously described (15, 32, 41). Hybridized samples were electrophoresed in 5% acrylamide gels, transferred to filter paper, and exposed to a phosphor screen. The screen was scanned with a Molecular Imager FX (Bio-Rad, Hercules, Calif.) and analyzed using Quantity One 1-D analysis software (Bio-Rad).
Flow cytometric analyses.
CBA mice were infected with 4 × 105 PFU of KyA or KyARgp2F, and on days 3 and 4 lungs were removed and single-cell suspensions were obtained as follows. Lungs from each group of five mice were diced into small pieces with scissors, pressed through a 60-gauge screen, and digested for 90 min with collagenase (250 U per ml) and DNase I (50 U per ml.) A single-cell suspension, free of tissue debris, was obtained by low-speed centrifugation at 228 × g for 10 min. Resulting cells were briefly exposed to the ammonium chloride lysing reagent BD PharM Lyse (BD/Pharmingen) to lyse erythrocytes and were resuspended in the same volume (3 ml) of culture medium to allow accurate counting of total cells per lung. One hundred microliters of cells from each group was placed in wells of a 96-well round-bottom plate (Corning, Corning, N.Y.) and stained for T lymphocytes with biotin-conjugated monoclonal antibodies (MAb) specific for murine CD4 or CD8 (BD/Pharmingen), followed by fluorescein isothiocyanate (FITC)-conjugated streptavidin (SA) and a phycoerythrin (PE)-conjugated MAb specific for murine CD3ɛ (BD/Pharmingen). Macrophages were stained with a FITC-conjugated MAb specific for murine Mac-1 (CD11b; BD/Pharmingen) and a PE-conjugated MAb specific for murine major histocompatibility complex class II IEk (BD/Pharmingen). B lymphocytes were stained with a biotin-conjugated MAb specific for murine B220 (BD/Pharmingen) followed by FITC-SA. All samples were stained with allophycocyanin-conjugated MAb specific for mouse leukocyte common antigen CD45 (BD/Pharmingen). All antibodies were diluted in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline [PBS] containing 1% FCS and 0.1% sodium azide), and the final volume was adjusted to 400 μl with FACS buffer. Immediately prior to analysis, 40 μl of a known concentration of fluorescent counting beads (Caltag, Burlingame, Calif.) was added. Following analysis, the absolute cell count for each stained population was determined according to the manufacturer's protocol. Experimental means were tested for significance by the two-tailed Student's t test.
Histopathology.
Lungs from mice infected with 4 × 105 PFU of KyA, KyARgp2F, or RacL11 were harvested and processed for histopathological analysis as follows. Mice were sacrificed by prolonged halothane inhalation, and the pleural cavity was exposed. A smooth-tipped 20-gauge needle was inserted into the trachea just below the larynx, and the lungs were infused with approximately 1 ml of OCT (Tissue-Tek/Sakura, Tokyo, Japan). The trachea was then tied with waxed dental floss, and the lungs were removed and placed in a 25- by 20-mm cryomold (Tissue-Tek/Sakura). For hematoxylin and eosin (H & E) staining, the cryomold containing the lung was filled completely with OCT and frozen by “canoeing,” placing the cryomold into a precooled plastic petri dish floating on liquid nitrogen until completely frozen. For immunohistochemical staining, the cryomold containing the lung was dipped into a beaker containing methyl-2-butane precooled to temperature by partial submersion in liquid nitrogen. After snap-freezing, the cryomold was removed, residual methyl-2-butane was evaporated, and the cryomold was filled with OCT and frozen by the canoeing method described above.
Sections 6 μm thick were cut using a Leica CM1850 cryostat (Leica, Bannockburn, Ill.), placed onto precleaned superfrost-plus slides (VWR, West Chester, N.Y.), air dried overnight, and stored at −20°C until use. For H & E staining, slides were fixed for 10 min in 10% buffered formalin and stained using a standard H & E protocol. For immunohistochemistry, slides were fixed for 20 min in an ice-cold mixture of 75% acetone-25% ethanol, rinsed gently with distilled H2O, blocked for 15 min with FCS, rinsed gently with distilled H2O, stained for 30 min with a biotin-conjugated MAb specific for murine CD4, CD8, CD11b, or B220, and incubated for 30 min at room temperature in a humidified chamber. Following incubation, slides were rinsed gently with distilled H2O and incubated for 30 min at room temperature with alkaline phosphatase-conjugated SA in a humidified chamber. Stained antigens were visualized using the 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium alkaline phosphatase chromogen kit (Biomeda, Foster City, Calif.) with the addition of (−)-tetramisole hydrochloride (Sigma) to a final concentration of 2 mM to block endogenous alkaline phosphatase activity. Slides were then counterstained for 3 min at room temperature with a solution consisting of 1% methyl green (Sigma) and 1% acetic acid in double-distilled H2O. Coverslips were mounted using a single drop of Entellan mounting medium (EM Science, Gibbstown, N.J.). Slides were viewed and photographed using a Nikon DXM 1200F digital camera-equipped Nikon Eclipse TE300 inverted microscope (Nikon, Tokyo, Japan).
Microarray analysis.
Purified mRNA for microarray analysis was isolated at the indicated times from the lungs of mice infected with 4 × 105 PFU of KyA or KyARgp2F as described above. The resulting mRNA was quantified photospectrometrically and independently assessed for degradation with an Agilent 2100 Bioanalzyer (Agilent Technologies, Palo Alto, Calif.). cDNA for each sample was synthesized from approximately 1.5 μg of mRNA using a Superscript cDNA synthesis kit (Invitrogen) in combination with a T7-(dT)24 primer. Biotinylated cRNA was transcribed in vitro using the BioArray HighYield RNA transcript labeling kit (ENZO Biochem, New York, N.Y.) and purified using the GeneChip sample cleanup module (Affymetrix, Santa Clara, Calif.). Twenty micrograms of purified cRNA was fragmented by incubation in fragmentation buffer (200 mM Tris-acetate [pH 8.1], 500 mM potassium acetate, 150 mM magnesium acetate) at 94°C for 35 min and chilled on ice. Fifteen micrograms of fragmented, biotin-labeled cRNA was hybridized to the MOE 430A array (Affymetrix), interrogating a total of 22,626 murine gene cDNA probes, for 16 h at 45°C with constant rotation (60 rpm). The arrays were washed and then stained for 10 min at 25°C with 10 μg of streptavidin-R phycoerythrin (Vector Laboratories)/ml followed by 3 μg of biotinylated goat antistreptavidin antibody (Vector Laboratories)/ml for 10 min at 25°C. Arrays were then stained once again with streptavidin-R-phycoerythrin for 10 min at 25°C. After washing and staining, the arrays were scanned using a GeneArray scanner (Agilent Technologies) at 570 nm. Pixel intensities were measured, expression signals were analyzed, and features were extracted using a commercial software package (Microarray Suite version 5.0; Affymetrix). Data mining and statistical analyses were performed with Data Mining Tool version 3.0 (Affymetrix) algorithms. Arrays were globally scaled to a target intensity value of 2,500 in order to compare individual experiments. The absolute determination (present, marginal, or absent) of each gene expression in each sample, as well as the direction of change and fold change of gene expression between samples, was identified using the above-mentioned software. Using GeneSifter.Net (VizX Labs, Seattle, Wash.), statistics for the three replicates of each gene were calculated independently. Genes were selected that were differentially expressed twofold in pairwise comparisons between sample x (KyA) and sample y (KyARgp2F) and showed corresponding two-sided P values of <0.05 (derived from a t test analysis assuming equal variances and six degrees of freedom). For each probe, the gene name and functional summary were obtained from public databases using GeneSifter.Net. Functional classification of the genes was performed using Spotfire DecisionSite for Functional Genomics (Spotfire, Somerville, Mass.) which uses the Gene Ontology database.
Nucleotide sequence accession numbers.
The EUs4 sequences of RacL11 and KyA have been deposited with GenBank and assigned accession numbers AY702017 and AY762536, respectively.
RESULTS
Sequences of EUs4 cloned from RacL1l, KyARgp2F, and KyA.
The generation of recombinant KyARgp2F that expresses the EUs4 gene of pathogenic RacL11 has been described previously (38). In that study, the correct insertion of the full-length EUs4 gene was confirmed by restriction enzyme digestion, PCR, and Southern blot analyses, and gp2 expression was confirmed by Western blot analysis. Although the sequences of EUs4 of the EHV-1 Ab4 strain (36) and KyA (8) have been reported, the EUs4 sequence from RacL11 had never been determined. A recent report demonstrated that EUs4 was highly polymorphic due to variances in the numbers of reiterated sequences located toward the 5′ end of the EUs4 open reading frame (17), corresponding to amino acids 185 to 260 (Fig. 1). To compare EUs4 of RacL11 with that of the fully sequenced Ab4 strain and to confirm the correct insertion of RacL11 EUs4 into recombinant KyARgp2F, the EUs4 genes from both the parental RacL11 and KyARgp2F were cloned and sequenced. The EUs4 gene of KyA was previously reported (8), but it was sequenced again for this study to ensure that significant changes had not occurred. A comparison of the EUs4 sequences of Ab4, RacL11, KyARgp2F, and KyA is presented in Fig. 1. The EUs4 sequence of KyARgp2F is identical to that of parental RacL11 and consists of 2,373 bp and encodes a predicted gp2 of 791 amino acids, in contrast to the 797-amino-acid gp2 of Ab4. The differences between the EUs4 sequences of Ab4 and RacL11 mapped almost entirely within the polymorphic reiterated repeat region (17, 36) (Fig. 1). In addition, there were two differences between gp2 of Ab4 and RacL11 at amino acid residues 430 and 441. These results confirmed that KyARgp2F expresses a full-length gp2 identical to that of the pathogenic RacL11 and that this protein differs from the fully sequenced Ab4 strain (with the exception of changes at amino acids 430 and 441) within the highly variable reiterated repeat region in the N-terminal half of the protein. The EUs4 gene of the attenuated KyA (1,149 bp) was confirmed to be less than half of the size of its counterparts in the pathogenic Ab4 and RacL11 strains and to encode a predicted gp2 of only 383 amino acids (Fig. 1). It is noteworthy that KyA gp2 lacks the repeat sequences TAATT and SSATTAATT present in full-length gp2; the significance of these repeats and their potential role in determining a pathogenic phenotype, however, remain to be determined.
FIG. 1.
Comparison of the amino acid sequences of gp2 isolated from parental EHV-1 Ab4, RacL11, KyA, and recombinant KyARgp2F. (a) Amino acid numbers relative to the EHV-1 Ab4 sequence, with the underlined residue corresponding to the number above. (b) Deletion of amino acids 75 to 489 from gp2 of KyA. (c) Beginning of the reiterated repeat region (highlighted in bold italics) consisting of TAATT (amino acid residues 185 to 224), SSATTAATT (residues 225 to 234), and TAATT (residues 234 to 260). (d) Site for staggered endoproteolytic cleavage (↓) following arginine residues 506 and 507. *, single amino acid change between the compared sequences. (e) Putative transmembrane anchor sequence.
Assessment of clinical disease following i.n. infection of CBA mice.
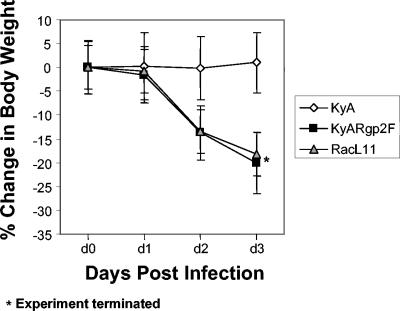
CBA mice were infected i.n. with 4 × 105 PFU of EHV-1 KyA, KyARgp2F, or wild-type RacL11 and observed daily for clinical signs of virus-induced respiratory disease, including crouching, huddling behavior, ruffled fur, and loss of body weight. While mice infected with KyA showed none of the clinical signs of disease, mice infected with KyARgp2F or RacL11 began exhibiting a ruffled appearance and huddling behavior as early as day 2 postinfection. All mice were weighed immediately prior to infection and daily thereafter to monitor body weight loss. The results demonstrated that KyA-infected mice exhibited a negligible weight loss by day 2 postinfection (0.18%) and began gaining weight thereafter (Fig. 2). In contrast, mice infected with KyARgp2F or RacL11 began losing weight on day 1 postinfection and continued losing body weight to day 3 (20 and 18.18% mean body weight, respectively), at which time the experiment was terminated to prevent undue suffering due to the severe clinical signs of both KyARgp2F- and RacL11-infected mice (Fig. 2).
FIG. 2.
Assessment of mean body weight. CBA mice in groups of five were weighed immediately prior to i.n. infection with 4 × 105 PFU of EHV-1 KyA, EHV-1 recombinant KyARgp2F, or wild-type pathogenic RacL11 and weighed each subsequent day through day 3. Each data point shown represents the mean body weight for the indicated group at that particular time point, with error bars indicating the standard deviations from the means. *, the experiment was terminated at day 3 due to the physical condition of KyARgp2F- and RacL11-infected mice to prevent undue suffering.
Examination of inflammatory infiltration in infected lungs.
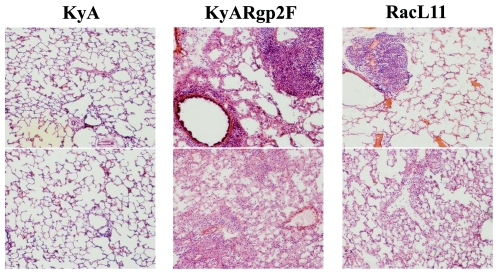
Lungs were removed from mice infected with KyA, KyARgp2F, and RacL11 at various times postinfection, including days 4 and 5, when clinical signs were most severe, and examined macroscopically for signs of inflammatory infiltration and consolidation (data not shown). While the lungs of mice infected with KyA had the typical healthy pink appearance throughout infection, KyARgp2F- and RacL11-infected lungs exhibited a severe consolidation involving greater than 90% of the lung tissue by days 4 and 5, consistent with previous observations of mice infected with RacL11 (32). Histological studies showed that while lungs of animals infected with KyA exhibited an almost-undetectable inflammatory infiltration at day 4 postinfection, those of mice infected with KyARgp2F and RacL11 exhibited extensive perivascular and peribronchial cuffing as well as interstitial inflammatory infiltration (Fig. 3). Taken together, the almost-identical pattern of severe weight loss and clinical signs observed following infection with recombinant KyARgp2F and wild-type RacL11 (Fig. 2) and the similar patterns and degree of inflammatory infiltration into the infected lungs (Fig. 3) suggest that the expression of the full-length EUs4 gene product in the KyA genomic background restored full respiratory virulence to the otherwise-fully attenuated KyA.
FIG. 3.
Histological sections of infected lungs. CBA mice were infected intranasally with 4 × 105 PFU of KyA, KyAgp2F, or RacL11. On day 4 postinfection, the mice were sacrificed and the lungs were infused with OCT and removed. The lung tissue was mounted in tissue molds, covered in OCT, cryostat sectioned, fixed, and stained with H & E as described in Materials and Methods. Resulting slides were visualized and photographed at 10× magnification.
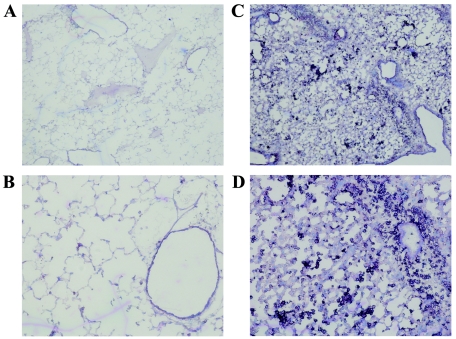
To begin to characterize the inflammatory response elicited by gp2 in the CBA lung, frozen cryostat sections from KyA- and KyARgp2F-infected lungs were stained for a variety of different cell surface molecules, including Mac-1, CD4, CD8, and B220. Immunohistochemical staining revealed large numbers of Mac-1-positive cells present in the lungs of KyARgp2F-infected mice on days 3, 4, and 5 postinfection (Fig. 4), but Mac-1-positive cells were rare in lungs of mice infected with KyA and numbers were comparable to those in lungs of mock-infected mice (data not shown). Staining for CD4- or CD8-positive T lymphocytes and B220-positive B lymphocytes revealed higher numbers of both cell types in lungs of mice infected with KyARgp2F compared to KyA (data not shown). To examine more closely and to quantify the phenotypes of cells present in infected lungs, single-cell suspensions were assessed by flow cytometric analyses. Significantly higher numbers of CD3/CD4, CD3/CD8, and B220 lymphocytes were observed in the lungs of KyARgp2F-infected mice on days 3 and 4 postinfection compared to cell numbers in lungs of mice infected with KyA (Table 1). These results confirm those obtained after immunohistochemical staining of lung tissues. In addition, a striking difference between the two groups was the total number of Mac-1/IEk-positive cells: 1,432,760 versus 653,862 cells on day 3 and 1,689,738 versus 259,598 cells on day 4 postinfection in KyARgp2F- and KyA-infected lungs, respectively (Table 1). Since Mac-1 is also present on some granulocytes, double staining for Mac-1 and major histocompatibility complex class II (IEk) was used to more accurately determine the number of macrophages present in the infected lung and exclude cross-reacting granulocytes. These results document that large numbers of macrophages infiltrate the lungs of KyARgp2F-infected mice (Fig. 4), consistent with the previous observation of macrophages in the RacL11-infected lung (32).
FIG. 4.
Immunohistochemical staining of lung sections at 4 days postinfection. CBA mice were infected intranasally with 4 × 105 PFU of KyA (A and B) or KyAgp2F (C and D). On day 4 postinfection, the mice were sacrificed and the lungs were infused with OCT, removed, and sectioned. The sections were fixed for 20 min in an ice-cold mixture of 75% acetone-25% ethanol, stained with antibody specific for Mac-1, and counterstained with a solution containing 1% methyl green as described in Materials and Methods. Resulting slides were visualized and photographed at 4× (A and C) or 10× (B and D) magnification.
TABLE 1.
Flow cytometric analysis of cells isolated from KyARgp2F- and KyA-infected lungsa
| Group | Absolute cell no. per lungb
|
|||
|---|---|---|---|---|
| CD3/CD4 | CD3/CD8 | B220 | Mac-1/IEk | |
| Day 3 | ||||
| KyARgp2F | 490,357c | 272,956 | 1,836,238 | 1,446,924 |
| KyA | 276,802 (1.77)d | 156,842 (1.74) | 641,583 (2.86) | 470,858 (3.06) |
| Day 4 | ||||
| KyARgp2F | 616,178 | 331,567 | 1,540,434 | 1,260,110 |
| KyA | 308,962 (1.99) | 160,768 (2.06) | 602,095 (2.55) | 295,794 (4.26) |
Cells were isolated from total lung tissue by enzymatic digestion on days 3 and 4 postinfection as described in Materials and Methods.
Absolute cell numbers per lung were determined using a known concentration of fluorescent counting beads as described in Materials and Methods.
Mean value from three replicate experiments. Experimental means were tested for significance by the two-tailed Student t test. All P values were ≤0.05 except for day 4 CD3/CD8 and B220, which were 0.08 and 0.06, respectively.
Ratio of means comparing KyARgp2F to KyA.
Analysis of cytokine-chemokine profiles in lungs of KyA- and KyARgp2F-infected mice.
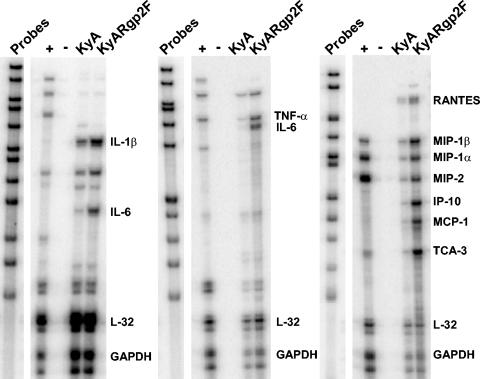
Previously, we reported a close correlation between the severe respiratory inflammatory response following infection of CBA mice with pathogenic RacL11 and the increased expression of mRNA specific for proinflammatory factors tumor necrosis factor alpha (TNF-α), macrophage inflammatory protein 1α (MIP-1α), MIP-1β, and MIP-2 in the bronchoalveolar lavage fluid (BAL) (32). To determine if similar cytokine profiles correlated with the severe respiratory inflammatory response following KyARgp2F infection and to extend this analysis beyond only those cells obtainable in the BAL, RPA were performed on poly(A)+ RNA isolated from the entire lung at various times postinfection. As early as 8 h (data not shown) and 12 h (Fig. 5) postinfection, an upregulation of poly(A) + RNA specific for interleukin-1β (IL-1β), IL-6, MIP-1β, interferon γ-inducible protein (IP-10), monocyte chemotactic protein 1 (MCP-1), and T-cell activation gene 3 (TCA-3) was observed in the lungs of KyARgp2F-infected mice compared to mice infected with attenuated KyA. The levels of transcripts encoding these proinflammatory factors reached a peak at 12 h postinfection and returned to levels equivalent to those seen in KyA-infected mice by 24 h postinfection (data not shown). The pattern of a transient increase in expression of these proinflammatory factors was similar to that observed in the BAL for TNF-α, MIP-1α, MIP-1β, and MIP-2, which reached a peak on day 3 postinfection in RacL11-infected mice and decreased to levels equivalent to those in KyA-infected mice by day 4 (32).
FIG. 5.
Detection of chemokine and cytokine mRNA by RPA. CBA mice were infected i.n. with 4 × 105 PFU of KyA or KyAgp2F. At 12 h postinfection, mice were sacrificed and total RNA was isolated followed by poly(A)+ RNA isolation as described in Materials and Methods. 32P-labeled probes specific for various chemokine and cytokine mRNAs were generated by in vitro transcription using the probe sets mCK-2b (left), mCK-3b (middle), and mCK-5c (right). RPA were performed using the 32P-based Ribo-Quant multiprobe RPA system (BD/Pharmingen) as per the manufacturer's protocol. Control lanes included the labeled probes alone, pooled mouse RNA (+), and yeast tRNA (−). Visible bands not differentially expressed twofold or greater are not labeled.
DNA microarray analysis of host gene expression in the lungs of KyA- and KyARgp2F-infected mice.
Time course analysis of chemokine-cytokine profiles by RPA at 8, 12, 24, and 48 h postinfection revealed an early increase in proinflammatory factors in KyARgp2F-infected mice as early as 8 h postinfection, which increased and reached a peak at 12 h postinfection (Fig. 5). Therefore, the 8- and 12-h time points were chosen to analyze global gene expression in the infected lungs by DNA array analyses using the Affymetrix mouse expression set 430A, which assesses the expression of 22,626 genes and expressed sequence tags (EST). Genes of interest were those important in innate immunity and inflammation as classified by Spotfire DecisionSite for Functional Genomics, as described in Materials and Methods. A total of 121 genes were assessed based on this functional classification.
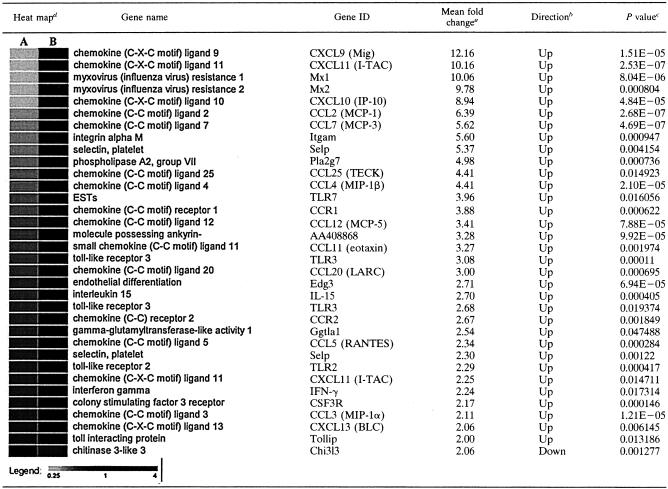
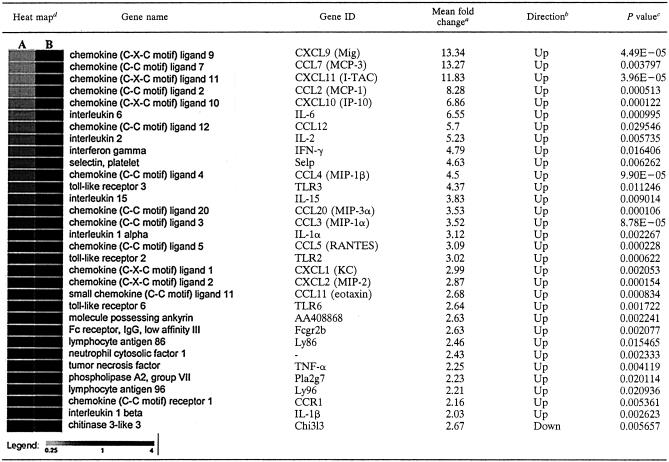
The results of analyses of poly(A)+ RNA isolated at 8 and 12 h postinfection from lungs infected with either KyA or KyARgp2F are presented in Tables 2 and 3. Each table shows the data from three independent experiments, each comparing those transcripts differing by twofold or greater in lungs of KyARgp2F-infected mice relative to those of KyA-infected animals. As early as 8 h postinfection, transcripts from 31 of the 121 genes analyzed for functionality in innate immunity and inflammation were increased twofold or greater in KyARgp2-infected lungs relative to those with KyA. These genes included CCL3 (MIP-1α), CCL4 (MIP-1β), CXCL10 (IP-10), CCL5 (RANTES), CXCL9 (Mig), CXC11 (I-TAC), CCL2 (MCP-1), CCL7 (MCP-3), CCL12 (MCP-5), CCL25 (TECK), CCL11 (eotaxin), CCL20 (LARC), and CXCL13 (BLC). Cytokine transcripts encoding IL-15 and gamma interferon (IFN-γ) were also increased greater than twofold in the lungs of KyARgp2F-infected animals compared with KyA-infected mice (Table 2). Transcripts for only one, chitinase 3-like protein (chi3l3), were decreased twofold in KyARgp2F-infected lungs at both 8 and 12 h postinfection (Tables 2 and 3). DNA array analyses at 12 h postinfection (Table 3) revealed the increased expression of transcripts from 31 genes of interest, including CCL5 (RANTES), TNF-α, CCL3 (MIP-1α), CCL4 (MIP-1β), CXCL2 (MIP-2), CXCL10 (IP-10), and CCL2 (MCP-1). These findings confirmed the results of the RPA (Fig. 5). Chemokines CXCL9 (Mig), CCL7 (MCP-3), CCL12 (MCP-5), CXCL11 (I-TAC), CCL20 (MIP-3α), CXCL1 (KC), and CCL11 (eotaxin) and proinflammatory cytokines IFN-γ, IL-6, IL-15, IL-1α, and IL-1β were expressed greater than twofold in lungs of mice infected with KyARgp2F-infected mice relative to responses in KyA-infected mice (Table 3). In addition to the increase in mRNAs specific for these proinflammatory chemokines and cytokines, we also observed higher levels of transcripts specific for pattern recognition receptors TLR2, TLR3, and TLR6, colony-stimulating factor 3 receptor (CSF3R), and chemokine receptors CCR1 and CCR2 (Tables 2 and 3). Taken together, these results demonstrated that, in contrast to the parental KyA strain expressing truncated gp2, the expression of full-length gp2 by recombinant KyARgp2F is sufficient to elicit a complex and extensive chemotactic and inflammatory milieu in the lung at early times postinfection, consistent with the subsequent inflammatory infiltration into the lung by days 3 and 4 resulting in a fatal outcome.
TABLE 2.
DNA array analayses of KyARgp2F- and KyA-infected lungs 8 h postinfection
Presented as the mean fold change in values from three replicate experiments comparing KyARgp2F and KyA.
Direction of fold change of KyARgp2F relative to KyA.
Statistics for the three replicates of each gene were calculated independently comparing the mean signals of KyARgp2F and KyA using a t test analysis assuming equal variances and six degrees of freedom.
Heat map plots were generated by comparing the signal values of KyA (A) and KyARgp2F (B) using GeneSifter, as described in Materials and Methods.
TABLE 3.
DNA array analayses of KyARgp2F- and KyA-infected lungs 12 h postinfection
Presented as the mean fold change from three replicate experiments comparing KyARgp2F and KyA.
Direction of fold change of KyARgp2F relative to KyA.
Statistics for the three replicates of each gene were calculated independently comparing the mean signals of KyARgp2F and KyA using a t test analysis assuming equal variances and six degrees of freedom.
Heat map plots were generated by comparing the signal values of KyA (A) and KyARgp2F (B) using GeneSifter, as described in Materials and Methods.
DISCUSSION
In the present study, experiments were conducted to assess the potential role of the EUs4 gene product gp2 in the severe respiratory inflammatory response observed following intranasal infection of CBA mice with pathogenic EHV-1. We report here that the expression of the full-length RacL11 gp2 (791 amino acids) by a recombinant KyA virus completely restores the ability of this otherwise-fully attenuated KyA strain to elicit fatal immunopathological responses in the lower respiratory tract, indicating a vital role of gp2 in EHV-1 pathogenesis.
Our hypothesis that gp2 may play an important role in EHV-1 pathogenesis came from studies using a recombinant RacL11 (RacL11ΔgIgE) with glycoproteins gI and gE deleted and a recombinant KyA (KgIgE), in which gI and gE were restored to the attenuated KyA background (15). In that study, the expression of gI and gE by KyA did not restore full virulence as KgIgE-infected mice exhibited only a moderate weight loss and, in contrast to RacL11-infected mice, did not succumb to infection by day 6 postinfection. The lack of severe respiratory illness and survival of these mice, however, finally resulted in spread of KgIgE to the brain, an outcome directly attributed to the expression of gI and gE (14). Surprisingly, the deletion of gI and gE from RacL11 (RacL11ΔgIΔgE) resulted in a course of respiratory illness virtually indistinguishable from that following infection with the pathogenic RacL11 strain (15). Taken together, these results suggested that RacL11 expresses a virulence factor(s) that is not present in KyA and is capable of eliciting severe respiratory disease in the absence of gI and gE expression.
Over years in cell culture, KyA has undergone several genomic deletions, the most prominent in the unique-short genomic region being deletions of EUs6 (gI), EUs7 (gE), and more than half of the EUs4 open reading frame (8, 12). The failure of KgIgE (lacking full-length gp2) to elicit severe respiratory illness and the ability of RacL11ΔgIΔgE that expresses gp2 of 791 amino acids to elicit severe respiratory illness led to the hypothesis that the expression of full-length gp2 was a critical factor in the induction of severe respiratory immunopathology. The results presented here strongly support this hypothesis.
The loss of 20% total body weight and the severe clinical signs observed in KyARgp2F-infected mice are similar to those following infection with the highly pathogenic RacL11 (this study and references 9 and 15). The massive influx of inflammatory cells into KyARgp2F-infected lungs, which was preceded by increased levels of mRNA specific for MIP-1α, MIP-1β, MIP-2, and TNF-α, mimics the steps of pathogenesis observed in RacL11 infection (32). The observation that greater than 30 transcripts encoding factors that function in innate immunity and inflammation were increased in lungs of mice infected with KyARgp2F versus KyA (Fig. 5; Tables 2 and 3) precedent to the influx of large numbers of macrophages strongly suggests a role for gp2 in this immunopathology. That lower levels of transcripts encoding chi313, an inhibitor of IL-1 and TNF-α (23) and a potent recruiter of eosinophils (29, 30), were present in KyARgp2F-infected lungs is consistent with the absence of eosinophilia following infection with either RacL11 (32) or KyARgp2F (data not shown). In the context of the release of potent proinflammatory and inflammatory factors, it is noteworthy that gp2 has been shown to be cleaved into two subunits, the 42-kDa C-terminal membrane-bound moiety and a larger secreted form. It is conceivable that soluble gp2 may function as a signaling molecule, and this function is currently being addressed by generating mutant viruses that are unable to express the secreted form of the protein.
Previous reports assessing the role of gp2 in EHV-1 virulence yielded conflicting results. In one study the deletion of EUs4 from the pathogenic Ab4 strain reduced virulence, as infected mice exhibited no clinical signs and lost only moderate amounts of body weight (24), suggesting a role for gp2 in respiratory virulence. The finding of a complete reduction of virulence was corroborated in another study using gp2-negative RacL11, which was completely apathogenic for BALB/c mice. However, in the same study, infection of mice with KyARgp2F expressing the full-length RacL11 gp2 failed to result in a significant weight loss (38), raising questions regarding the role of gp2 in EHV-1 pathogenesis. There are three significant differences in the previous studies and the work presented here. First, the dosage applied to mice here was fourfold higher. Second, in contrast to the use of BALB/c mice in the two studies mentioned above, this investigation employed CBA mice which were found to be more susceptible to EHV-1-mediated disease than either the C57BL/6 or BALB/c mouse strains (references 14, 15, 32, and 33 and personal observation). To directly address this question, 3- to 4-week-old BALB/c mice were weighed immediately prior to infection with the higher dose of 4 × 105 PFU of KyA, KyARgp2F, or RacL11, weighed daily thereafter, and monitored for clinical signs of respiratory disease. In a pattern of weight loss almost identical to that observed in CBA mice, KyARgp2F- and RacL11-infected BALB/c mice exhibited a mean total body weight loss of 21 and 22%, respectively, on day 4 postinfection, when the experiment was terminated (data not shown). BALB/c mice survived 1 day longer than CBA mice infected with the same dose, indicating a possible influence in the strain of mouse used. These results strongly suggested that the higher dose of 4 × 105 PFU/mouse in this study versus 105 PFU/mouse in the previous study (38) may be the critical factor responsible in the discrepancies in outcomes described in these two studies.
Important questions remain to be answered regarding the role of gp2 in EHV-1 virulence. First, it is not known whether cleavage of full-size gp2 to a highly O-glycosylated N-terminal region and a 42-kDa C-terminal subunit (39, 40) is required for virulence. Second, the specific region(s) of full-size gp2 essential for virulence has yet to be identified. A recent study comparing gp2 of various strains and isolates of EHV-1 and EHV-4 noted extensive polymorphism (17) in the reiterated repeat region toward the N terminus (Fig. 1). Our findings that gp2 of two pathogenic strains, RacL11 and its counterpart in Ab4 (36), both harbor this polymorphic region and that gp2 of attenuated KyA lacks the entire variable region are consistent with the hypothesis that this polymorphic region may play a role in determining EHV-1 biological properties.
A recent study reported that inoculation of BALB/c mice with DNA encoding gp2 resulted in a slight reduction in viral lung titers upon subsequent virus challenge and suggested the inclusion of gp2 in a subunit vaccine and its importance as a component of whole-virus vaccines (22). The results in the present study indicate caution regarding the inclusion of full-length gp2 in EHV-1 vaccines until a better understanding is obtained concerning the role of this large glycoprotein in infection and the mechanism(s) by which it elicits inflammation in the respiratory tract.
Acknowledgments
We thank Suzanne Zavecz for excellent technical assistance and Deborah Chervenak and Paula Polk of the LSUHSC Research Core Facility for their invaluable assistance with flow cytometric and DNA array analyses, respectively.
This study was supported by research grant AI-22001 from the National Institute of Allergy and Infectious Diseases and by center grant 1P20RR18724 through the COBRE Program of The National Center for Research Resources, National Institutes of Health (D.J.O.) and by grants from the Morris Animal Foundation (D03-EQ74) and the Harry M. Zweig Fund for Equine Research to N.O.
REFERENCES
- 1.Albrecht, R. A., S. K. Kim, Y. Zhang, Y. Zhao, and D. J. O'Callaghan. 2004. The equine herpesvirus 1 EICP27 protein enhances gene expression via an interaction with TATA box-binding protein. Virology 324:311-326. [DOI] [PubMed] [Google Scholar]
- 2.Allen, G. P., and J. T. Bryans. 1986. Molecular epizootiology, pathogenesis, and prophylaxis of equine herpesvirus-1 infections. Prog. Vet. Microbiol. Immunol. 2:78-144. [PubMed] [Google Scholar]
- 3.Allen, G. P., and M. R. Yeargan. 1987. Use of lambda gt11 and monoclonal antibodies to map the genes for the six major glycoproteins of equine herpesvirus 1. J. Virol. 61:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, G. P., M. R. Yeargan, and J. T. Bryans. 1983. Alterations in the equine herpesvirus 1 genome after in vitro and in vivo virus passage. Infect. Immun. 40:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burki, F., W. Rossmanith, N. Nowotny, C. Pallan, K. Mostl, and H. Lussy. 1990. Viremia and abortions are not prevented by two commercial equine herpesvirus-1 vaccines after experimental challenge of horses. Vet. Q. 12:80-86. [DOI] [PubMed] [Google Scholar]
- 6.Burrows, R., D. Goodridge, and M. S. Denyer. 1984. Trials of an inactivated equid herpesvirus-1 vaccine: challenge with a subtype-1 virus. Vet. Res. 114:369-374. [DOI] [PubMed] [Google Scholar]
- 7.Carrol, C. L., and H. A. Westbury. 1985. Isolation of equine herpesvirus type 1 from the brain of a horse affected with paresis. Aust. Vet. J. 62:345-346. [DOI] [PubMed] [Google Scholar]
- 8.Colle, C. F., III, C. C. Flowers, and D. J. O'Callaghan. 1992. Open reading frames encoding a protein kinase, homolog of glycoprotein gX of pseudorabies virus, and a novel glycoprotein map within the unique short segment of equine herpesvirus type 1. Virology 188:545-557. [DOI] [PubMed] [Google Scholar]
- 9.Colle, C. F., III, E. B. Tarbet, W. D. Grafton, S. R. Jennings, and D. J. O'Callaghan. 1996. Equine herpesvirus-1 strain KyA, a candidate vaccine strain, reduces viral titers in mice challenged with a pathogenic strain, RacL. Virus Res. 43:111-124. [DOI] [PubMed] [Google Scholar]
- 10.Crabb, B. S., G. P. Allen, and M. J. Studdert. 1991. Characterization of the major glycoproteins of equine herpesviruses 4 and 1 and asinine herpesvirus 3 using monoclonal antibodies. J. Gen. Virol. 72:2075-2082. [DOI] [PubMed] [Google Scholar]
- 11.Crabb, B. S., and M. J. Studdert. 1995. Equine herpesviruses 4 (equine rhinopneumonitis virus) and 1 (equine abortion virus). Adv. Virus Res. 45:153-190. [DOI] [PubMed] [Google Scholar]
- 12.Flowers, C. C., and D. J. O'Callaghan. 1992. The equine herpesvirus type 1 (EHV-1) homolog of herpes simplex virus type 1 US9 and the nature of a major deletion within the unique short segment of the EHV-1 KyA strain genome. Virology 190:307-315. [DOI] [PubMed] [Google Scholar]
- 13.Frampton, A. R., W. F. Goins, J. B. Cohen, N. Osterrieder, D. J. O'Callaghan, and J. C. Glorioso. 2005. Equine herpesvirus type 1 utilizes a novel herpesvirus entry receptor. J. Virol. 79:3169-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frampton, A. R., Jr., P. M. Smith, Y. Zhang, W. D. Grafton, T. Matsumura, N. Osterrieder, and D. J. O'Callaghan. 2004. Meningoencephalitis in mice infected with an equine herpesvirus 1 strain KyA recombinant expressing glycoprotein I and glycoprotein E. Virus Genes 29:9-17. [DOI] [PubMed] [Google Scholar]
- 15.Frampton, A. R., Jr., P. M. Smith, Y. Zhang, T. Matsumura, N. Osterrieder, and D. J. O'Callaghan. 2002. Contribution of gene products within the unique short segment of equine herpesvirus 1 to virulence in a murine model. Virus Res. 90:287-301. [DOI] [PubMed] [Google Scholar]
- 16.Hannant, D., D. M. Jessett, T. O'Neill, C. A. Dolby, R. F. Cook, and J. A. Mumford. 1993. Responses of ponies to equid herpesvirus-1 ISCOM vaccination and challenge with virus of the homologous strain. Res. Vet. Sci. 54:299-305. [DOI] [PubMed] [Google Scholar]
- 17.Huang, J. A., N. Ficorilli, C. A. Hartley, G. P. Allen, and M. J. Studdert. 2002. Polymorphism of open reading frame 71 of equine herpesvirus-4 (EHV-4) and EHV-1. J. Gen. Virol. 83:525-531. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, T. A., and J. W. Kendrick. 1971. Paralysis of horses associated with equine herpesvirus 1 infection. J. Am. Vet. Med. Assoc. 158:1351-1357. [PubMed] [Google Scholar]
- 19.Jackson, T. A., B. I. Osburn, D. R. Cordy, and J. W. Kendrick. 1977. Equine herpesvirus 1 infection of horses: studies on the experimentally induced neurologic disease. Am. J. Vet. Res. 38:709-719. [PubMed] [Google Scholar]
- 20.Kim, S. K., R. A. Albrecht, and D. J. O'Callaghan. 2004. A negative regulatory element (base pairs −204 to −177) of the EICP0 promoter of equine herpesvirus 1 abolishes the EICP0 protein's trans-activation of its own promoter. J. Virol. 78:11696-11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Learmonth, G. S., D. N. Love, J. R. Gilkerson, J. E. Wellington, and J. M. Whalley. 2002. The C-terminal regions of the envelope glycoprotein gp2 of equine herpesvirus 1 and 4 are antigenically distinct. Arch. Virol. 147:607-615. [DOI] [PubMed] [Google Scholar]
- 22.Learmonth, G. S., D. N. Love, J. R. Gilkerson, J. E. Wellington, and J. M. Whalley. 2003. Inoculation with DNA encoding the glycoprotein gp2 reduces the severity of equine herpesvirus 1 infection in a mouse respiratory model. Arch. Virol. 148:1805-1813. [DOI] [PubMed] [Google Scholar]
- 23.Ling, H., and A. D. Recklies. 2004. The chitinase 3-like protein human cartilage glycoprotein 39 inhibits cellular responses to the inflammatory cytokines interleukin-1 and tumour necrosis factor-α. J. Biochem. 380:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall, K. R., Y. Sun, S. M. Brown, and H. J. Field. 1997. An equine herpesvirus-1 gene 71 deletant is attenuated and elicits a protective immune response in mice. Virology 231:20-27. [DOI] [PubMed] [Google Scholar]
- 25.Meyer, H., P. Thein, and P. Hubert. 1987. Characterization of two equine herpesvirus (EHV) isolates associated with neurological disorders in horses. Zentralbl Veterinarmed. B 34:545-548. [DOI] [PubMed] [Google Scholar]
- 26.Mumford, J. A., D. A. Hannant, D. M. Jessett, T. O'Neill, K. C. Smith, and E. N. Ostlund. 1995. Abortagenic and neurological disease caused by infection with equid herpesvirus-1, p. 261-275. In H. Nakajima and W. Plowright (ed.), Proceedings of the 7th International Conference of Equine Infectious Diseases. R & W Publishers, Newmarket, United Kingdom.
- 27.O'Callaghan, D. J., and N. Osterrieder. 1999. Equine herpesviruses, p. 508-515. In R. Webster, and A. Granoff (ed.), Encyclopedia of virology. Academic Press Ltd., London, United Kingdom.
- 28.Osterrieder, N., O. R. Kaaden, and A. Neubauer. 1999. Structure and function of equine herpesvirus glycoproteins-a review, p. 111-118. In H. Nakajima and W. Plowright (ed.), Proceedings of the 7th International Conference of Equine Infectious Diseases. R & W Publications, Newmarket, United Kingdom.
- 29.Owhashi, M., H. Arita, and N. Hayai. 2000. Identification of a novel eosinophil chemotactic cytokine (ECF-L) as a chitinase family protein. J. Biol. Chem. 14:1279-1286. [DOI] [PubMed] [Google Scholar]
- 30.Owhashi, M., H. Arita, and A. Niwa. 1998. Production of eosinophil chemotactic factor by CD8+ T-cells in Toxocara canis-infected mice. Parasitol. Res. 84:136-138. [DOI] [PubMed] [Google Scholar]
- 31.Rudolph, J., D. J. O'Callaghan, and N. Osterrieder. 2002. Cloning of the genomes of equine herpesvirus type 1 (EHV-1) strains KyA and RacL11 as bacterial artificial chromosomes (BAC). J. Vet. Med. B 49:31-36. [DOI] [PubMed] [Google Scholar]
- 32.Smith, P. M., Y. Zhang, W. D. Grafton, S. R. Jennings, and D. J. O'Callaghan. 2000. Severe murine lung immunopathology elicited by the pathogenic equine herpesvirus 1 strain RacL11 correlates with the early production of macrophage inflammatory proteins 1α, 1β, and 2 and tumor necrosis factor alpha. J. Virol. 74:10034-10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, P. M., Y. Zhang, S. R. Jennings, and D. J. O'Callaghan. 1998. Characterization of the cytolytic T-lymphocyte response to a candidate vaccine strain of equine herpesvirus 1 in CBA mice. J. Virol. 72:5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, Y., A. R. MacLean, J. D. Aitken, and S. M. Brown. 1996. The role of the gene 71 product in the life cycle of equine herpesvirus 1. J. Gen. Virol. 77:493-500. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Y., and S. M. Brown. 1994. The open reading frames 1, 2, 71, and 75 are nonessential for replication of equine herpesvirus type 1 in vitro. Virology 199:448-452. [DOI] [PubMed] [Google Scholar]
- 36.Telford, E. A., M. S. Watson. K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 37.Telford, E. A., M. S. Watson, J. Perry, A. A. Cullinane, and A. J. Davison. 1998. The DNA sequence of equine herpesvirus-4. J. Gen. Virol. 79:1197-1203. [DOI] [PubMed] [Google Scholar]
- 38.von Einem, J., J. Wellington, J. M. Whalley, K. Osterrieder, D. J. O'Callaghan, and N. Osterrieder. 2004. The truncated form of glycoprotein gp2 of equine herpesvirus type 1 (EHV-1) vaccine strain KyA is not functionally equivalent to full-length gp2 encoded by EHV-1 wild-type strain RacL11. J. Virol. 78:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wellington, J. E., G. P. Allen, A. A. Gooley, D. N. Love, N. H. Packer, J. X. Yan, and J. M. Whalley. 1996. The highly O-glycosylated glycoprotein gp2 of equine herpesvirus 1 is encoded by gene 71. J. Virol. 70:8195-8198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittaker, G. R., L. A. Wheldon, L. E. Giles, J. M. Stocks, I. W. Halliburton, R. A. Killington, and D. M. Meredith. 1990. Characterization of the high Mr glycoprotein (gp300) of equine herpesvirus type 1 as a novel glycoprotein with extensive O-linked carbohydrate. J. Gen. Virol. 71:2407-2416. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y., P. M. Smith, A. R. Frampton, N. Osterrieder, S. R. Jennings, and D. J. O'Callaghan. 2003. Cytokine profiles and long-term virus-specific antibodies following immunization of CBA mice with equine herpesvirus 1 and viral glycoprotein D. Viral Immunol. 16:307-320. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y., P. M. Smith, S. R. Jennings, and D. J. O'Callaghan. 2000. Quantitation of virus-specific classes of antibodies following immunization of mice with attenuated equine herpesvirus 1 and viral glycoprotein D. Virology 268:482-492. [DOI] [PubMed] [Google Scholar]