Abstract
A conserved structural motif in the envelope proteins of several viruses consists of an N-terminal, alpha-helical, trimerization domain and a C-terminal region that refolds during fusion to bind the N-helix trimer. Interaction between the N and C regions is believed to pull viral and target membranes together in a crucial step during membrane fusion. For several viruses with type I fusion proteins, C regions pack as alpha-helices in the grooves between N-helix monomers, and exogenously added N- and C-region peptides block fusion by inhibiting the formation of the six-helix bundle. For other viruses, including influenza virus and murine leukemia virus (MLV), there is no evidence for comparably extended C-region alpha-helices, although a short, non-alpha-helical interaction structure has been reported for influenza virus. We tested candidate N-helix and C-region peptides from MLV for their ability to inhibit cell fusion but found no inhibitory activity. In contrast, intracellular expression of the MLV N-helix inhibited fusion by efficiently blocking proteolytic processing and intracellular transport of the envelope protein. The results highlight another mechanism by which the N-helix peptides can inhibit fusion.
The envelope proteins of retroviruses, orthomyxoviruses, paramyxoviruses, and filoviruses have numerous structural similarities that suggest a common mechanism of membrane fusion (1, 4, 5, 12, 14, 19, 23, 24, 37, 40, 41, 42, 43, 44). Designated type I fusion proteins, they are all synthesized as precursors that translocate to the endoplasmic reticulum (ER), where they trimerize, and then to the Golgi apparatus, where sugars originally attached in the ER are modified and cellular proteases in the furin family cleave the envelope polypeptide into two parts (11, 17, 20). For retroviruses, the amino-terminal segment is designated the surface (SU) subunit, and the carboxy-terminal segment, which anchors the protein in the virus membrane, is designated the transmembrane (TM) subunit. For human immunodeficiency virus (HIV), SU-TM cleavage is required for fusion function (28), presumably because it allows rearrangement of the hydrophobic amino terminus of TM, which is believed to act as a fusion peptide, as well as other conformational changes triggered by envelope binding to the receptor and coreceptor (36).
Downstream of the fusion peptide region of TM is an alpha-helical segment designated the N-helix (NH), N-heptad repeat, or HR1, which trimerizes due to the interaction of hydrophobic residues in the a and d positions in a helical wheel depiction (Fig. 1A and D) (5, 14). The N-helix is followed by a short disulfide loop and then, in the case of HIV type 1 (HIV-1) and closely related viruses, an ∼30-amino-acid alpha-helical segment, the C-helix (CH). Crystal and solution nuclear magnetic resonance structures of TM ectodomains (4, 5, 40) show that the N-helices form an inner trimer, with C-helices packing in an antiparallel orientation in the grooves between N-helix monomers. This structure, which resembles a trimer of hairpins, has been dubbed the six-helix bundle. Formation of the six-helix bundle is believed to pull virus and target membranes together to create a fusion pore. The trimer-of-hairpins structure is stabilized by interactions between hydrophobic amino acids lining the grooves between N-helix monomers (e and g positions in the N-helix [Fig. 1D]) and hydrophobic amino acids in the a and d positions of the C-helix. Peptides from the C-helix of HIV-1 potently inhibit membrane fusion by binding in the grooves of viral N-helix trimers and disrupting the formation of the six-helix bundle. C-terminal region peptides form a new class of drugs approved for treatment of HIV-1 infection (27). N-helix peptides also inhibit HIV-1 fusion, albeit at 10- to 100-fold higher concentrations than those of C-helix peptides. It is thought that N-helix peptides function in two ways: by forming heterotrimers with viral N-helices and by forming peptide homotrimers that bind viral C-helices; both mechanisms block the formation of the normal six-helix bundle (2, 16).
FIG. 1.
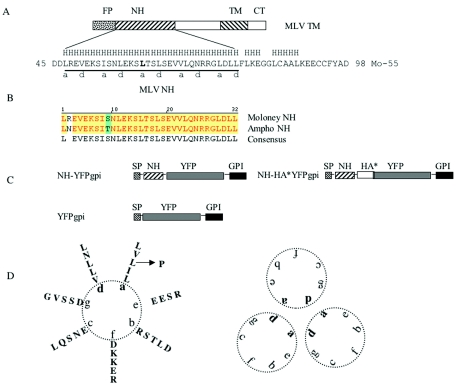
(A) Diagram of Moloney MLV TM. FP, NH, TM, and CT stand for fusion peptide, N-helix, transmembrane domain, and cytoplasmic tail, respectively. The 55-amino-acid sequence of the region that was crystallized is shown (Mo-55). An H above the sequence represents helical prediction (http://www.cmpharm.ucsf.edu/∼nomi/nnpredict.html). The underlined region (MLV NH) formed a trimer in the crystal structure and was tested as peptide or expressed intracellularly in this study. a and d beneath the line indicate helical wheel positions (also shown in panel D). (B) Alignment of Moloney MLV NH and amphotropic MLV NH. (C) Diagram of MLV NH constructs and parental-vector YFPgpi. SP stands for signal peptide; HA* stands for the 9-amino-acid HA epitope followed by two stop codons. (D) Alpha-helical wheel depiction for MLV NH (left) and the trimer structure (right). The leucine (L) shown in bold in panel A and indicated by an arrow in panel D was mutated to proline by site-directed mutagenesis.
N-helix peptides are less potent than C-helix peptides, possibly because (i) they tend to aggregate in aqueous solutions (10), which may restrict their ability to bind C-helices, and (ii) the viral N-helix trimer forms intracellularly, which may limit the ability of exogenously added peptide to form heterotrimers. Both of these limitations might be reversed by expressing N-helix peptides intracellularly.
In the case of murine leukemia virus (MLV), it is not clear if there is a C-helix that functions in a manner structurally analogous to that of the C-helix of HIV. While computer models (e.g., NNPREDICT [http://www.cmpharm.ucsf.edu/∼nomi/nnpredict.html]) predict an ∼25-amino-acid downstream region capable of forming an alpha-helix, this putative C-helix region was not included in the crystallized peptide. The portion of MLV TM that was crystallized formed an N-helix trimer with only a 5-amino-acid segment folded back in a hairpin configuration (14). The e and g positions that line the grooves of the MLV N-helix trimer do not contain hydrophobic amino acids (Fig. 1D), so if the C-helix region of MLV binds in grooves of N-helix trimers, it must do so through nonhydrophobic interactions. The crystal structure of the analogous envelope protein of influenza virus also shows only a short C-terminal segment folded back, which forms an asymmetric hairpin. In the case of influenza virus, a more distal, nonhelical C-terminal segment interacts with a portion of the N-helix trimer (7, 32). Because MLV, like influenza virus, may not use the canonical six-helix bundle, it was of interest to investigate the effect of exogenously added peptides as well as endogenously expressed N-helix.
We started the experiments reported here by testing whether C-helix or N-helix peptides of MLV would inhibit MLV envelope protein-mediated fusion. The peptides that we tested were not inhibitory. Since negative results are difficult to interpret, we then tested whether intracellular expression of the N-helix would inhibit MLV-mediated fusion. This strategy did inhibit fusion. The expressed N-helix formed a mixed oligomer with viral envelope protein, which blocked the proteolytic processing of the envelope protein precursor and its transport to the cell surface. Inhibition of processing as a consequence of hetero-oligomer formation during synthesis demonstrates another mechanism by which N-helix peptides of class I envelope proteins can inhibit fusion.
MATERIALS AND METHODS
Constructs.
The expression vector for MLV N-helix was made as follows. The sequence encoding MLV N-helix was amplified by PCR with the primers 5′ GCGTCGACTGATCTCAGGGAGGTTGAA 3′ and 5′ ACGATGAGTCGACGCGTCACCTCCTAACAAGTCTAGGCCCCTT 3′. The underlined sequences introduced a SalI restriction site for cloning. The PCR product was cut with SalI (New England Biolabs, Beverly, Mass.) and inserted into a unique SalI site between the signal peptide and yellow fluorescent protein (YFP) in plasmid YFP-GL-GPI (22), to generate plasmid NH-YFPgpi (Fig. 1C). NH-HA*YFPgpi was made by replacing the region between KpnI and AgeI of NH-YFPgpi with the following annealed oligonucleotides: 5′ CGTACCCATACGATGTTCCAGATTACGCTTGATAA 3′ and 5′ CCGGTTATCAAGCGTAATCTGGAACATCGTATGGGTACGGTAC 3′. The mutation of leucine 61 to proline in the N-helix was created by site-directed mutagenesis (Stratagene, Cedar Creek, Tex.) with primers 5′ CTCTAACCTCGAGAAGTCTCCCACTTCCCTGTCTG 3′ and 5′ CAGACAGGGAAGTGGGAGACTTCTCGAGGTTAGAG 3′. Two silent mutations (in bold) were introduced to create an XhoI restriction site (underlined) for clone identification. A hemagglutinin (HA) epitope was inserted into the MLV envelope protein by ligating a 5′-end-phosphorylated oligonucleotide adapter (sense strand, 5′ CCGGGATACCCATACGATGTTCCAGATTACGCT 3′, and antisense strand, 5′ CCGGAGCGTAATCTGGAACATCGTATGGGTATC 3′) encoding HA in frame at the unique SgrAI site of pCEETr (33). All the constructs were verified by sequencing. pCEETr and pCEE, which expresses the full-length Moloney MLV envelope protein, and pCAETr, which expresses a fusogenic form of amphotropic MLV envelope, were gifts from J. Ragheb (33). pdl1443 is an expression vector for HIV envelope protein (15). pVSV-G, pTRE-Luc, and pTet-Off encoding the tetracycline transcriptional transactivator (tTA) were purchased from Clontech (Palo Alto, Calif.), and pFB-Luc was from Stratagene.
Reagents.
MG132 was from Calbiochem (San Diego, Calif.). NH4Cl and anti-HA antibody were from Sigma (St. Louis, Mo.). Anti-ecotropic MLV SU goat serum (VR-1521AS-Gt) was from the American Type Culture Collection (Manassas, Va.).
Cell lines.
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (GIBCO-BRL, Grand Island, N.Y.) supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U of penicillin, and 100 U of streptomycin. BHKenv-tTA, U2OSLucCATGFP, and U373-MAGI-CXCR4CEM cells were grown in the same medium plus selection antibiotics (30, 39). GP2-293 (Clontech) cells were grown in the same medium with 0.1 mg of G418 (Invitrogen, Carlsbad, Calif.) per ml. All cell lines were kept at 37°C in a 5% CO2 humidified incubator.
Transfection.
Cells were seeded 1 day before transfection and were about 70% confluent at the time of transfection. For cultures in six-well plates, 3 to 6 μg of plasmid DNA and 6 to 12 μl of Lipofectamine 2000 (Invitrogen) were used per well. Plasmid and Lipofectamine were each diluted in 250 μl of Opti-Mem medium (Invitrogen), mixed well, incubated at room temperature for 20 to 30 min, and then added to cells in 2 ml of antibiotic-free medium.
Cell fusion assay.
Target cells and indicator cells, in some cases transiently transfected 24 to 48 h earlier, were trypsinized and cocultured in triplicate in 24-well plates for about 16 h. Cell fusion was quantified by measuring luciferase activity with a kit from Promega (Madison, Wis.) and a Victor 3 luminometer (Perkin Elmer, Boston, Mass.).
Surface protein labeling.
Thirty-six hours after transfection, the cells were rinsed with phosphate-buffered saline (PBS) buffer, lifted into PBS supplemented with 5 mM EDTA, washed with PBS, resuspended in PBS supplemented with 1 mg of biotinylation reagent (product no. 20338; Pierce, Rockford, Ill.) per ml, and labeled on ice for 1 h. After the labeling, the biotinylation reagent was quenched with 100 mM glycine in PBS buffer. Following the PBS wash, the cells were lysed and biotinylated proteins were purified with a monomeric avidin kit (product no. 20227; Pierce) according to the manufacturer's protocol.
Immunoprecipitation.
Cells were harvested 24 or 48 h after transfection, washed twice with ice-cold PBS, removed from the plates in 5 mM EDTA in PBS, centrifuged to pellet cells, and lysed with 10 volumes of prechilled RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate) containing a protease inhibitor cocktail (Roche, Indianapolis, Ind.) on ice for 30 min, and centrifuged at 10,000 × g in a prechilled centrifuge or rotor (4°C) for 10 to 15 min. The supernatant was collected and precleared with normal serum for 1 h at 4°C with agitation. One-half volume of protein G-Sepharose (Amersham, Piscataway, N.J.) bead slurry (10%, vol/vol, in RIPA buffer containing protease inhibitors) was added and agitated for another hour. The supernatant was collected and immunoprecipitated by adding 5 μl of goat anti-SU serum or 2 μl of monoclonal anti-green fluorescent protein (GFP) antibody per 200 μl of precleared supernatant. After 4 h at 4°C, an equal volume of protein G-Sepharose bead slurry was added and the mixture was rotated at 4°C for an additional 2 h. Beads were washed three to four times in cold lysis buffer. The immunocomplex was eluted by adding 70 μl of 1× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (Invitrogen) to the beads and boiling the mixture for 3 min. Samples were resolved by precast SDS-PAGE gels with 10% or 4 to 12% polyacrylamide (Invitrogen) and analyzed by Western blotting.
Furin cleavage in vitro.
After the last wash, immunoprecipitation beads were resuspended in 50 μl of furin reaction buffer (0.5% Triton X-100, 1 mM CaCl2, 100 mM HEPES, 1 mM β-mercaptoethanol). For each reaction, 2 μl of furin (0.578 mg/ml; R&D Systems) was added to the bead slurry and the mixture was incubated at 37°C for 16 h. The reaction was stopped and protein was eluted by boiling the mixture in 1× SDS gel loading buffer.
Western blot analysis.
Following resolution by SDS-PAGE, proteins were transferred to polyvinylidene difluoride membrane. The membrane was blocked for 1 h at room temperature in 5% nonfat milk in TBST buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Tween 20), and incubated with primary antibodies in 1% nonfat milk in TBST buffer at room temperature for 1 h. After four 10-min washes in TBST buffer, the blots were incubated with secondary antibodies for 1 h at room temperature, followed by another 40-min wash in TBST. Proteins were visualized with the enhanced chemiluminescence detection system (Pierce). The antibodies used were mouse anti-HA and anti-β-actin (Sigma) diluted 1:5,000, mouse anti-GFP (Zymed, South San Francisco, Calif.) diluted 1:4,000, and horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Pierce) diluted 1:5,000.
Confocal fluorescence microscopy.
Cells were plated in eight-well coverglass chambers (Nunc, Rochester, N.Y.) and examined with a Leica TCS-NT/SP confocal microscope (Leica, Wetzlar, Germany) using a 63× objective and Omnichrome argon-krypton lasers emitting at 488 nm.
RESULTS
Lack of inhibition of fusion by C-helix and N-helix peptides of MLV.
We used a sensitive cell fusion assay (30) to test for inhibition by MLV peptides. Briefly, a BHK cell line (BHKenv-tTA) that stably expresses tTA and a fusogenic form of Moloney MLV envelope protein lacking a portion of its cytoplasmic tail (p2−) was cocultivated with U2OSLucCATGFP cells, a human osteosarcoma cell line engineered to stably express the ecotropic MLV receptor mCAT1 and a luciferase reporter gene under the control of the tetracycline response element (pTRE-Luc). Cell fusion leads to tTA transcriptional activation of the luciferase gene and an ∼100- to 1,000-fold increase in luciferase enzyme activity after 16 h of coculture. We compared the luciferase activities in cell lysates after cocultivation in the absence or presence of peptide. Peptides had no effect on luciferase activity at concentrations up to 100 μM for the C-helix and 5 μM for the N-helix. The peptides tested were LVRDSMAKLRERLNQRQKLFESTQ (C-helix) and DLREVEKSISNLEKSLTSLSEVVLQNRRGLDLL (N-helix), modified with acetyl groups at the N terminus and amide groups at the C terminus.
Expression of MLV NH.
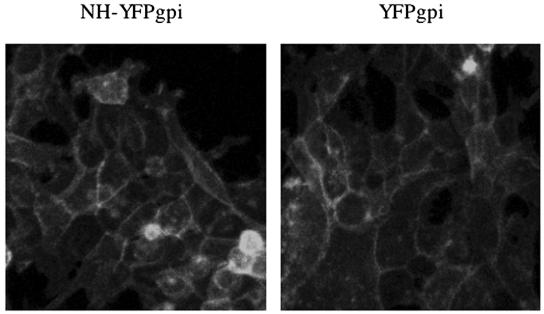
To express MLV N-helix intracellularly, we modified a vector for a cell surface form of YFP (pYFP-GL-GPI [22]). This vector encodes YFP between a signal peptide from lactase-phlorizin hydrolase and a consensus N glycosylation site fused to the glycosylphosphoinositol (gpi) attachment signal from lymphocyte function-associated antigen 3 (LFA-3). For simplicity, we refer to this plasmid as YFPgpi. We inserted the Moloney MLV N-helix coding sequence in frame between the signal peptide and YFP to generate NH-YFPgpi (Fig. 1). Transfection of YFPgpi or NH-YFPgpi led to similar levels of YFP plasma membrane fluorescence (Fig. 2), implying that the N-helix is well expressed by this vector.
FIG. 2.
Expression of MLV NH-YFPgpi. HEK293 cells were transfected with 3 μg of NH-YFPgpi (left) or parental-vector YFPgpi (right) as a control. Twenty-four hours later, the images were obtained using confocal microscopy.
MLV NH inhibits MLV envelope protein-mediated fusion but not that of other viral envelope proteins.
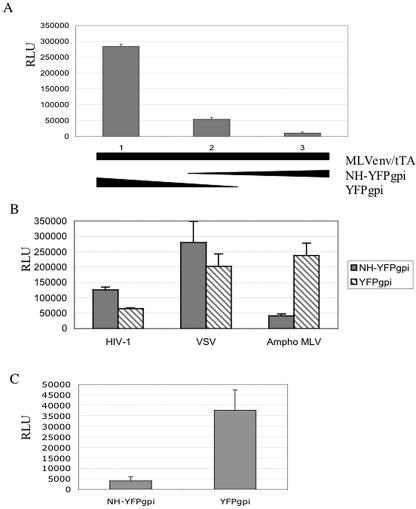
To examine the effect of coexpressed MLV NH on MLV envelope protein-mediated fusion, we cotransfected HEK293 cells with vectors encoding the fusogenic MLV envelope protein (33) and tTA, with various amounts of MLV NH-YFPgpi or the parental vector YFPgpi to normalize the amount of vector-related DNA. Twenty-four hours later, the transfected cells were cocultivated with U2OSLucCATGFP cells. The cocultured cells were harvested after 16 h and assayed for luciferase activity as a marker for cell fusion. NH-YFPgpi inhibited MLV envelope protein-mediated cell fusion in a dose-dependent manner about 8-fold (6- to 10-fold in different experiments) at a 1:1 ratio of NH-YFPgpi to MLV Env DNA (Fig. 3A).
FIG. 3.
MLV NH specifically inhibits MLV envelope protein-mediated fusion. (A) Dose dependence. HEK293 cells were cotransfected with 1.5 μg each of expression vectors for ecotropic MLV envelope protein (pCEETr) and tetracycline transactivator (pTet-Off), along with 0, 1.5, or 3.0 μg of NH-YFPgpi plus 3.0, 1.5, or 0 μg of YFPgpi (lanes 1, 2, and 3, respectively), and cocultured with U2OSLucCATGFP cells. Luciferase activity, measured in relative light units (RLU), was assayed about 16 h after coculture. (B) Specificity. HEK293 cells were cotransfected with 1.5 μg each of expression vectors for HIV-1 envelope protein (pdl1443), amphotropic MLV envelope protein (pCAETr) (Ampho MLV), or VSV-G (pVSV-G), along with a vector for tTA (pTet-Off) and either NH-YFPgpi (filled bars) or YFPgpi (hatched bars). Twenty-four hours later, they were cocultured with U373MAGICXCR4CEM cells transfected with 2 μg of pTRE-Luc (for HIV) or U2OSLucCATGFP cells (for VSV-G and amphotropic MLV). Luciferase activity was measured about 16 h later. (C) Inhibition of pseudotyping. GP2-293 packaging cells were cotransfected with 1.5 μg each of expression vectors for MLV envelope protein (pCEE), a retroviral vector encoding a luciferase reporter gene (pFB-Luc), and either NH-YFPgpi or YFPgpi. Supernatants were collected 48 h later, normalized by reverse transcriptase activity, and used to infect NIH 3T3 cells. The luciferase activity was assayed 48 h later. Error bars indicate the standard deviations of results from triplicate samples in single experiments. The results are representative of at least three independent experiments.
We checked the specificity of the inhibition by comparing the effects of the MLV N-helix on fusion mediated by the HIV-1 envelope protein, vesicular stomatitis virus G protein (VSV-G), or amphotropic MLV envelope protein. Since the MLV N-helix is not expected to interact with HIV envelope protein or VSV-G, it should not inhibit fusion mediated by these envelope proteins. To see if this was the case, we cotransfected vectors encoding tTA, MLV NH-YFPgpi (or YFPgpi as a control), and VSV-G or HIV-1 (strain NL4-3) envelope protein into HEK293 cells and cocultivated them with U2OSLucCATGFP cells (for VSV-G) or U373-MAGI-CXCR4CEM cells (for HIV). The last cells constitutively express the HIV receptor (CD4) and coreceptor (CXCR4) and were transiently transfected with pTRE-Luc 1 day before cocultivation in order to use the same luciferase reporter gene read-out. MLV N-helix did not inhibit fusion mediated by VSV-G or HIV-1 envelope protein (Fig. 3B). The reason for the slight increase in VSV-G and HIV-1 envelope-mediated fusion in the presence of MLV N-helix is not known. In contrast, the N-helix of Moloney MLV was expected to inhibit amphotropic MLV since their TM regions are nearly identical, differing at only two positions in the N-helix, neither of which are on the interacting face (asparagine in place of arginine at b position 2 and threonine in place of serine at b position 9 [Fig. 1B]). We cotransfected HEK293 cells with expression vectors for fusogenic amphotropic MLV envelope (pCAE-Tr [33]), tTA, and MLV NH-YFPgpi or YFPgpi as a control and cocultivated the cells with U2OSLucCATGFP. MLV NH-YFPgpi did inhibit amphotropic MLV envelope-mediated fusion (Fig. 3B), confirming that the inhibitory effect of MLV NH is sequence specific.
Since NH-YFPgpi inhibited MLV envelope protein-mediated cell fusion, we expected that it would also inhibit infection by virus bearing MLV envelope. To test this, we cotransfected a retroviral packaging cell line, GP2-293, with an MLV envelope protein expression vector (pCEE [33]), a retroviral vector encoding a luciferase reporter gene (pFB-Luc; Stratagene), and either NH-YFPgpi or YFPgpi as a control. Forty-eight hours later, the supernatant was collected and assayed for reverse transcriptase and infectivity in a single-round infection assay. NH-YFPgpi did not affect the amount of virus released from the packaging cells as measured by reverse transcriptase activity but did reduce the titer of pseudotyped virus about 90% (Fig. 3C). Thus, the data show that MLV N-helix is a dominant negative inhibitor of virus and cell fusion mediated by ecotropic and amphotropic MLV envelope protein.
MLV NH-YFPgpi forms a hetero-oligomer with MLV envelope protein and blocks proteolytic cleavage to SU/TM.
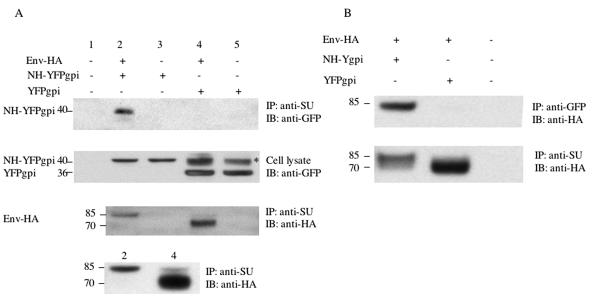
We used coimmunoprecipitation to see if MLV NH-YFPgpi formed a hetero-oligomer with MLV envelope protein. We cotransfected HEK293 cells with expression vectors for MLV envelope protein and NH-YFPgpi (or YFPgpi as a control). Twenty-four hours later, the cells were lysed for immunoprecipitation with anti-MLV SU goat serum. To facilitate detection of envelope protein after immunoprecipitation, we used an HA-tagged version of MLV envelope (Env-HA) in which the HA epitope was inserted at a unique SgrAI site in the proline-rich region of SU, a site previously shown to tolerate small insertions (21). Control experiments showed that Env-HA was functional in the cell fusion assay and that NH-YFPgpi inhibited Env-HA-mediated fusion to the same extent as wild-type envelope (data not shown). To visualize coimmunoprecipitated NH-YFPgpi, we used a monoclonal anti-GFP antibody which detects YFP as well as GFP. NH-YFPgpi was coimmunoprecipitated by the anti-SU antiserum in the presence, but not in the absence, of MLV envelope protein, while the YFPgpi control was not coimmunoprecipitated by the anti-SU antiserum (Fig. 4A, top panel, lanes 2 to 5). Additional controls showed that NH-YFPgpi and YFPgpi were expressed approximately equally well, with or without envelope protein (Fig. 4A, second panel). For unknown reasons, two species of YFPgpi were detected with the anti-GFP antibody, one of which migrated anomalously slowly (Fig. 4A, second panel). To confirm that envelope protein was immunoprecipitated by the anti-SU antiserum in all cases, we blotted with anti-HA after immunoprecipitation with anti-SU. The results show that envelope protein was immunoprecipitated but that it migrated as a 70-kDa species in the presence of YFPgpi, versus an 85-kDa species in the presence of NH-YFPgpi (Fig. 4A, third panel). The MLV envelope protein precursor migrates ∼85 kDa, whereas after SU/TM cleavage, SU migrates ∼70 kDa. To confirm that the envelope protein was predominantly of the uncleaved size when it was expressed in the presence of NH-YFPgpi, we repeated the transfections corresponding to lanes 2 and 4 and show a longer gel run in the fourth panel of Fig. 4A. The third and fourth panels of Fig. 4A also show that there is a moderate decrease in the total amount of envelope protein in cells expressing NH-YFPgpi, compared to that in cells expressing YFPgpi (see below).
FIG. 4.
MLV NH forms a hetero-oligomer with MLV envelope protein and blocks its proteolytic processing. (A) Coimmunoprecipitation of NH-YFPgpi by anti-SU antibody. HEK293 cells were cotransfected with 1.5 μg each of expression vectors for HA-tagged fusogenic MLV envelope protein (Env-HA) and NH-YFPgpi (or YFPgpi as a control) as indicated above each lane. Twenty-four hours later, the cells were lysed for immunoprecipitation with anti-MLV SU goat serum and blotted with anti-GFP (top panel) or anti-HA (third panel). Samples corresponding to those in lanes 2 and 4 in the top three panels but from an independent transfection are shown in the fourth panel with a longer gel run. Cell lysate before immunoprecipitation was blotted with anti-GFP antibody as a control (second panel). Two species of YFPgpi were detected with the anti-GFP antibody, one of which migrated anomalously slowly and is marked with * in the second panel. (B) Coimmunoprecipitation of envelope protein by anti-GFP antibody. The same procedures as described for panel A were carried out except that anti-GFP antibody was used for immunoprecipitation instead of anti-SU goat serum. Molecular mass markers (in kilodaltons) are shown at the left.
To confirm these coimmunoprecipitation results, we reversed the roles of the antibodies, immunoprecipitating with anti-GFP antibody and then blotting with anti-HA antibody. The anti-GFP antibody coimmunoprecipitated envelope protein in the presence of NH-YFPgpi but not YFPgpi (Fig. 4B, lane 1 versus lane 2). A control immunoprecipitation with anti-SU antiserum followed by blotting with anti-HA showed that the envelope protein was present in cell lysates even when it was not coimmunoprecipitated by anti-GFP antibody (Fig. 4B, bottom panel, lane 2). Again, the envelope protein was mainly of the precursor size in the presence of NH-YFPgpi and of the processed size in the presence of YFPgpi, and less total envelope protein was present in cells expressing NH-YFPgpi than in cells expressing YFPgpi (Fig. 4B, bottom, lane 1 versus lane 2).
MLV N-helix inhibits envelope protein transport to the cell surface and distal Golgi apparatus.
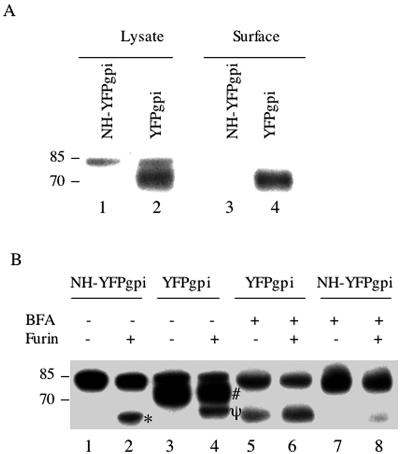
Noncleavage of the MLV envelope precursor protein may be due to conformational changes rendering the hetero-oligomer resistant to proteolytic cleavage or to inhibition of intracellular transport to the distal Golgi apparatus, where cleavage normally occurs (29). We first examined transport to the cell surface by biotinylating cells transfected with the HA-tagged envelope protein expression vector plus pNH-YFPgpi, or pYFPgpi as a control, by using a membrane-impermeable biotinylation reagent. Biotinylated proteins were purified with avidin monomer-agarose and analyzed on a Western blot with antibody to HA. The processed envelope protein was detected on the surfaces of cells cotransfected with pYFPgpi (Fig. 5A, lane 4), whereas neither precursor nor processed envelope protein was detected on the surfaces of cells cotransfected with pNH-YFPgpi (lane 3), despite the presence of the envelope precursor intracellularly in cells expressing N-helix (lane 1) and the presence of approximately equal amounts of total cell surface biotinylated protein from cells expressing, or not expressing, N-helix (data not shown). We conclude that N-helix inhibits envelope protein transport to the cell surface.
FIG. 5.
MLV NH inhibits the transport of MLV envelope protein to the cell surface but not susceptibility to cleavage by furin. (A) HEK293 cells were transfected with expression vectors for MLV envelope protein (Env-HA) and either NH-YFPgpi (lanes 1 and 3) or YFPgpi as a control (lanes 2 and 4). Thirty-six hours later, the cells were surface labeled with biotin and lysed. Biotinylated proteins were purified with avidin monomer-agarose. Total cell lysates (lanes 1 and 2) or biotinylated proteins (lanes 3 and 4) were blotted with anti-HA antibody. (B) HEK293 cells were transfected with expression vectors for MLV envelope protein (Env-HA) and either NH-YFPgpi (lanes 1, 2, 7, and 8) or YFPgpi as a control (lanes 3 to 6). Some cells (lanes 5 to 8) were treated with brefeldin A (BFA; 0.25 μg/ml) added 5 h after transfection and again 21 h after transfection. Twenty-nine hours after transfection, the cells were lysed and envelope protein was purified by cross-immunoprecipitation with anti-GFP antibody (lanes 1, 2, 7, and 8) or anti-SU antiserum (lanes 3 to 6). Aliquots of immunoprecipitated envelope protein were treated with furin (lanes 2, 4, 6, and 8). Samples were analyzed by Western blotting with anti-HA antibody. *, in vitro furin cleavage product of env associated with N-helix-YFPgpi; #, normal SU; ψ (lane 4), in vitro furin cleavage product of env from control cells. See the text for details. Molecular mass markers (in kilodaltons) are shown at the left.
To see if the conformation of envelope precursor in the heterotrimer rendered it resistant to proteolytic cleavage, we purified heterotrimer by immunoprecipitation with anti-GFP antibody and treated the immunoprecipitate with furin enzyme. As a control, we purified wild-type envelope precursor protein from cells treated with brefeldin A, a drug that disrupts the Golgi apparatus (6) and thereby inhibits Golgi apparatus-related proteolytic processing. The envelope precursor protein from cells expressing N-helix was partially cleaved by furin in vitro (Fig. 5B, lane 1 versus lane 2). The envelope precursor from cells not expressing N-helix but treated with brefeldin A was similarly partially cleaved (lane 6). In the absence of furin, brefeldin A-treated cells contain a smaller amount of an envelope fragment (lane 5) the same size as the in vitro cleavage product (lane 2), presumably a result of partial cleavage in vivo despite the presence of brefeldin A. The fact that the cleavage product in the presence of brefeldin A is the same size as the cleavage product in cells expressing N-helix suggests that N-helix arrests the envelope precursor at the same stage of glycosylation as brefeldin A does (see below). The cleavage product in the presence of N-helix or brefeldin A is smaller than the normal SU (lanes 3 and lane 4). This smaller size is likely due to postcleavage modification of sugars in vivo, consistent with the observations that HIV-1 SU and TM are endoglycosidase resistant and that the HIV-1 envelope protein precursor is endoglycosidase sensitive (9, 31). Finally, the furin cleavage product from control cells (lane 4) was intermediate in size, which may be due to subtle glycosylation differences between the envelope precursor in control cells versus cells expressing N-helix or treated with brefeldin A. The simplest interpretation of the combined observations that the heterotrimer (i) does not get to the cell surface, (ii) is susceptible to furin cleavage in vitro, but (iii) remains largely uncleaved in vivo is that N-helix blocks transport of the envelope precursor to its normal cleavage site in the cell.
Accelerated degradation of envelope in the presence of N-helix.
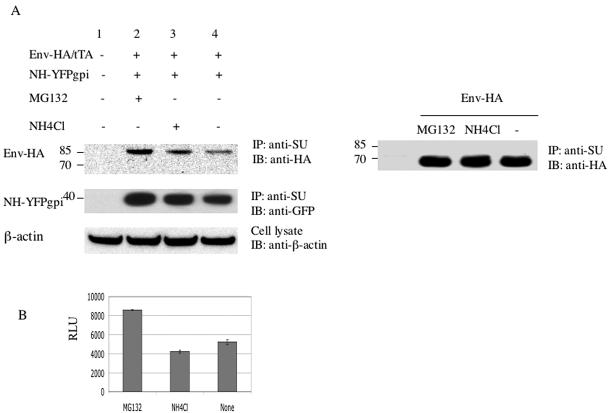
In addition to not being cleaved normally, the total amount of envelope protein was reduced in cells expressing N-helix (Fig. 4). To see if this was due to rapid degradation via the proteasome or lysosome pathways, we treated transfected cells with the proteasome inhibitor MG132, the lysosomal degradation inhibitor NH4Cl, or no inhibitor for 10 h starting 24 h after transfection and then analyzed cell lysates by immunoprecipitation. As expected, anti-SU antiserum coimmunoprecipitated NH-YFPgpi (Fig. 6, middle panel, lanes 2 to 4). When blotted with anti-HA, essentially all of the envelope protein was the size of the uncleaved envelope precursor (Fig. 6A, top panel). When envelope protein was expressed with NH-YFPgpi, slightly more envelope protein was detected in the presence of MG132 and, to a lesser extent, NH4Cl than in the presence of no inhibitor. In contrast, in control cells transfected with the envelope expression vector without NH-YFPgpi (Fig. 6A, right panel), the inhibitors did not result in increased amounts of envelope protein. This result shows that there is more-rapid degradation of envelope protein in cells expressing N-helix and that degradation is primarily through the proteasome pathway. The absence of detectable 70-kDa envelope protein species in the presence of the inhibitors argues against the possibility that the absence of the 70-kDa envelope species is due to rapid degradation of the cleavage product. Portions of the cells transfected with NH-YFPgpi in these experiments were cocultured with U2OSLucCATGFP cells for functional cell fusion assays (Fig. 6B). The proteasome inhibitor MG132 enhanced fusion slightly (a little less than twofold), consistent with the slight increase in total envelope protein.
FIG. 6.
MLV NH slightly increases the level of degradation of MLV envelope protein, mainly through the proteasome pathway. (A) Two micrograms each of expression vectors for Env-HA and tTA were cotransfected into HEK293 cells with (left) or without (right) NH-YFPgpi (2 μg). Twenty-four hours later, these cells were treated with the proteasome inhibitor MG132 (20 μM), treated with the lysosomal degradation inhibitor NH4Cl (20 mM), or not treated. After 10 h, cells were lysed, and the lysate was immunoprecipitated with anti-SU goat serum and blotted with anti-HA (top left and right) or anti-GFP (middle left) antibody. An aliquot of cell lysate that was not immunoprecipitated was blotted with anti-β-actin antibody (bottom left) as a control. Molecular mass markers (in kilodaltons) are shown at the left. IP, immunoprecipitation; IB, immunoblotting. (B) Transfected HEK293 cells corresponding to lanes 2 to 4 in panel A were cocultivated with U2OSLucCATGFP cells and assayed for luciferase activity. The x-axis label indicates inhibitor treatment. Error bars indicate the standard deviations of results with triplicate samples in a representative experiment of at least three independent transfections. RLU, relative light units.
A helix-destabilizing mutation eliminates the inhibitory effect of NH-YFPgpi.
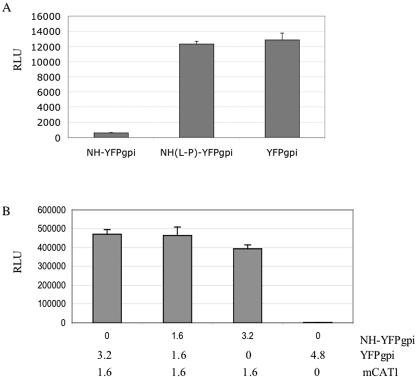
To see if hetero-oligomerization of NH-YFPgpi was responsible for its inhibitory effect on fusion, we mutated NH-YFPgpi by replacing leucine in position 61, an a position residue in the middle of the helix, with proline, a typical helix terminator (Fig. 1A and D). The mutant NH-YFPgpi was compared with wild-type NH-YFPgpi or parental-vector YFPgpi for inhibition of fusion. As shown in Fig. 7A, the L-to-P mutation eliminated the inhibitory effect on fusion. This result shows that when the ability to form an extended helix is compromised, the inhibitory effect on fusion is lost.
FIG. 7.
Evidence that MLV NH-mediated inhibition of fusion involves hetero-oligomerization with envelope protein. (A) Mutation that disrupts the alpha-helical structure of MLV NH causes the loss of its inhibitory activity. HEK293 cells were cotransfected with 1.5 μg each of expression vectors for fusogenic MLV envelope protein (pCEETr), pTet-Off, and either wild-type NH-YFPgpi, the leucine-to-proline mutant NH(L-P)-YFPgpi, or the parental-vector YFPgpi. Twenty four hours later, the transfected cells were cocultured with U2OSLucCATGFP for 16 h and assayed for luciferase activity. (B) The expression of MLV NH in receptor-bearing cells does not inhibit MLV envelope protein-mediated fusion. U2OSLuc cells were cotransfected with 1.6 μg of mCAT1 and 0.0, 1.6, or 3.2 μg of NH-YFPgpi plus 3.2, 1.6, or 0.0 μg of YFPgpi, respectively, and cocultivated with HEK293 cells cotransfected with 2 μg each of pCEETR and pTet-Off. Error bars indicate the standard deviations of results with triplicate samples in a representative experiment of two performed. RLU, relative light units.
Expression of NH-YFPgpi in receptor-bearing cells does not inhibit fusion.
The expression of HIV-1 C-helix on the surfaces of receptor-bearing cells inhibits cell fusion, presumably by disrupting six-helix bundle formation in opposed envelope-expressing cells (13, 18). To see if NH-YFPgpi could block fusion in a similar fashion, we expressed NH-YFPgpi in receptor-bearing cells rather than envelope protein-expressing cells. There was almost no inhibition of fusion when MLV NH-YFPgpi was expressed in receptor-bearing cells (Fig. 7B). This result is consistent with our failure to detect inhibition with soluble N-helix peptide and provides additional evidence that the mechanism of inhibition by MLV N-helix is primarily through heterotrimerization during envelope protein synthesis rather than interaction with a C-terminal portion of envelope during fusion.
The inhibitory effect of N-helix is unrelated to YFP or the gpi membrane linkage in the chimeric protein.
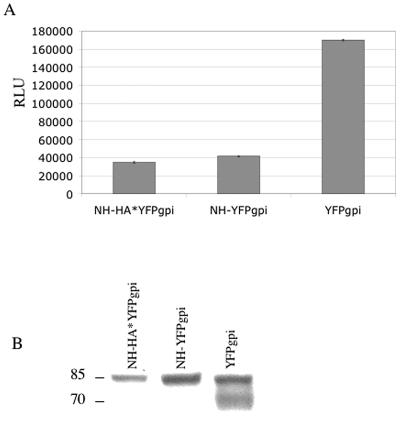
GFP and related proteins have a weak tendency to dimerize, which in the context of heterotrimerization might contribute to the misfolding, inhibit the trafficking, or accelerate the degradation of envelope protein. Also, the gpi membrane attachment signal may contribute to poor proteolytic processing or inhibition of fusion (34). To see if these structural features were important, we inserted the coding sequence for the 9-amino-acid HA epitope followed by two stop codons upstream of YFP in the NH-YFPgpi construct (Fig. 1C). When cotransfected with the envelope protein expression vector, pNH-HA*YFPgpi inhibited cell fusion (Fig. 8A) and proteolytic processing (Fig. 8B) at least as effectively as pNH-YFPgpi, indicating that the inhibitory effect is unrelated to YFP or the gpi membrane linkage.
FIG. 8.
MLV N-helix without YFPgpi inhibits MLV envelope protein-mediated fusion and its proteolytic processing. HEK293 cells were transfected with expression vectors for MLV envelope protein (Env-HA) plus NH-HA*YFPgpi, NH-YFPgpi, or YFPgpi, as indicated. Twenty-four hours later, aliquots of these cells were cocultured with U2OSLucCATGFP cells or lysed. (A) Luciferase activity in the cocultures was measured about 16 h later. RLU, relative light units. (B) Envelope protein was immunoprecipitated from the lysates with anti-SU antiserum and blotted with anti-HA antibody. Molecular mass markers (in kilodaltons) are shown at the left.
DISCUSSION
We found that expression of an MLV N-helix chimera in cells expressing MLV envelope inhibited envelope-mediated membrane fusion but that the same N-helix peptide added exogenously was not inhibitory. The intracellularly expressed N-helix chimera formed a hetero-oligomer with newly synthesized envelope protein, and hetero-oligomerization was apparently required for inhibition since a mutant version of the N-helix with a proline substitution on the trimer interface eliminated its inhibitory effect. Exogenously added N-helix presumably does not cross the cell membrane and therefore does not have a chance to form a hetero-oligomer during synthesis. Failure of the N-helix peptide to inhibit fusion suggests that the peptide does not exchange efficiently with envelope monomers in mature MLV envelope protein.
Hetero-oligomerization between wild-type and mutant envelope proteins is well documented for retroviruses. Direct evidence comes from the ability to cross-immunoprecipitate physically distinguishable envelope proteins; indirect evidence is provided by observations that some mutants dominantly interfere with wild-type envelope protein, while others can complement different envelope mutations or coreceptor restrictions (3, 8, 25, 26, 35, 38, 45). Previous experiments used whole virus envelope genes, which leaves open the question of whether regions other than the N-helix contribute to hetero-oligomerization. As the only portion of envelope protein contained in our NH-YFPgpi construct is the N-helix, our experiments show that this motif alone is sufficient to form mixed oligomers with full-length envelope protein in vivo.
In the system described here, hetero-oligomerization inhibited fusion mainly by trapping envelope protein in an early stage of the secretory pathway, presumably the ER. For envelope protein to be fusogenic on cells or virus, it must be transported to the cell surface and it must be proteolytically processed, both of which were inhibited by N-helix chimeras. In addition, coexpression of N-helix enhanced degradation of envelope protein mainly via the proteosome pathway, although quantitatively the reduction in envelope protein was small compared to the inhibition of processing and transport. Hetero-oligomerization with full-length mutant envelope proteins can inhibit fusion by inhibiting processing (26). Our study shows that a large heterotrimer partner is not required for the dominant negative effect; the N-helix itself is sufficient. It is possible that the outer surface of the N-helix is important for its inhibitory effect; this possibility should be amenable to study since the e and g positions of the helix can be modified without affecting trimerization potential (2). It will also be of interest to determine if the N-helix regions of other viruses are, by themselves, inhibitory. Preliminary experiments with HIV-1 show that its N-helix is inhibitory in the context of the YFPgpi chimera.
A distinct mechanism of inhibition by N-helix peptides for viruses with class I fusion proteins involves homotrimerization and binding to viral C-helices (16). There appear to be two classes of viruses with class I fusion proteins, those like HIV-1 and paramyxoviruses, for which the TM region folds into a six-helix bundle with an extended C-helical region and for which N- and C-helix peptides are inhibitory, and viruses like influenza virus, which have less-ordered distal TM regions with only short segments of alpha-helix. MLV may resemble influenza virus in this regard, based on its published crystal structure. Since N- or C-region peptide inhibitors have not been reported for influenza virus, it may be that an extended interface between N- and C-helices is required for their activities. This possibility would provide an explanation for the lack of effect of the MLV peptide inhibitors that we tested. For influenza virus, the interaction of a few amino acids at the beginning of the N-helix and the end of the C-terminal region, close to the viral membrane, has recently been shown to be crucial for pulling the otherwise disordered C-terminal segment into an extended hairpin (7, 32). If the same is true for MLV, the peptides that we tested might have been inactive because they lacked crucial terminal amino acids.
Our findings that NH-YFPgpi and NH-HA coexpressed with envelope protein inhibited fusion by blocking proteolytic processing and that exogenously added peptides were not inhibitory show that intracellular expression of viral peptides can have different effects from exogenous peptides and provide a rationale for attempting to develop drugs that target the trimerization domain of envelope proteins intracellularly.
Acknowledgments
We thank Jack Ragheb for providing plasmids pCEE, pCEETR, and pCAETR and Patrick Keller for the YFP-GL-GPI plasmid. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: U373-MAGI-CXCR4CEM cells from Michael Emerman.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Bewley, C. A., J. M. Louis, R. Ghirlando, and G. M. Clore. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238-14245. [DOI] [PubMed] [Google Scholar]
- 3.Buchschacher, G. L., Jr., E. O. Freed, and A. T. Panganiban. 1995. Effects of second-site mutations on dominant interference by a human immunodeficiency virus type 1 envelope glycoprotein mutant. J. Virol. 69:1344-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, A. M. Gronenbom, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 6.Chardin, P., and F. McCormick. 1999. Brefeldin A: the advantage of being uncompetitive. Cell 97:153-155. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., J. J. Skehel, and D. C. Wiley. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 96:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S. S. L., S. F. Lee, C. K. Chuang, and V. S. Raj. 1999. trans-Dominant interference with human immunodeficiency virus type 1 replication and transmission in CD4+ cells by an envelope double mutant. J. Virol. 73:8290-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewar, R. L., M. B. Vasudevachari, V. Natarajan, and N. P. Salzman. 1989. Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: effects of monensin on glycosylation and transport. J. Virol. 63:2452-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwyer, J. J., A. Hasan, K. L. Wilson, J. M. White, T. M. Matthews, and M. K. Delmedico. 2003. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry 42:4945-4953. [DOI] [PubMed] [Google Scholar]
- 11.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 13.Egelhofer, M., G. Brandenburg, H. Martinius, P. Schult-Dietrich, G. Melikyan, R. Kunert, C. Baum, I. Choi, A. Alexandrov, and D. van Laer. 2004. Inhibition of human immunodeficiency virus type 1 entry in cells expressing gp41-derived peptides. J. Virol. 78:568-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 Å resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 15.Felser, J., T. Klimkait, and J. Silver. 1987. A syncytia assay for human immunodeficiency virus type I (HIV-I) envelope protein and its use in studying HIV-I mutations. Virology 170:566-570. [DOI] [PubMed] [Google Scholar]
- 16.He, Y., R. Vassell, M. Zaitseva, N. Nguyen, Y. Zhongning, Y. Weng, and C. D. Weiss. 2003. Peptides that trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, L. E., R. Sowder, T. D. Copeland, G. Smythers, and S. Oroszlan. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 52:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildinger, M., M. T. Dittmar, P. Schult-Dietrich, B. Fehse, B. S. Schnierle, S. Thaler, G. Stiegler, R. Welker, and D. von Laer. 2001. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J. Virol. 75:3038-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallel to influenza virus hemagglutinin and HIV gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 20.Kamps, C. A., Y. C. Lin, and P. K. Wong. 1991. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology 184:687-694. [DOI] [PubMed] [Google Scholar]
- 21.Kayman, S. C., H. Park, M. Saxon, and A. Pinter. 1999. The hypervariable domain of the murine leukemia virus surface protein tolerates large insertions and deletions, enabling development of a retroviral particle display system. J. Virol. 73:1802-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller, P., D. Toomre, E. Diaz, J. White, and K. Simons. 2001. Multicolor imaging of post-Golgi sorting and trafficking in live cells. Nat. Cell Biol. 3:140-1149. [DOI] [PubMed] [Google Scholar]
- 23.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malashkevich, V. N., M. Singh, and P. S. Kim. 2001. The trimer-of-hairpins motif in membrane fusion: Visna virus. Proc. Natl. Acad. Sci. USA 98:8502-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matano, T., T. Odawara, M. Ohshima, H. Yoshikura, and A. Iwamoto. 1993. trans-Dominant interference with virus infection at two different stages by a mutant envelope protein of Friend murine leukemia virus. J. Virol. 67:2026-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matano, T., T. Odawara, M. Ohshima, A. Iwamoto, and H. Yoshikura. 1994. Interaction between the dominant negative mutant and the wild-type envelope proteins of Friend murine leukemia virus. J. Virol. 68:6079-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and D. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3:215-225. [DOI] [PubMed] [Google Scholar]
- 28.McCune, J. M., L. B. Rabin, M. B. Feinberg, M. Lieberman, J. C. Kosek, G. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gf160 is required for the activation of human immunodeficiency virus. Cell 53:55-67. [DOI] [PubMed] [Google Scholar]
- 29.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 30.Ou, W., Y. Xiong, and J. Silver. 2004. Quantification of virus envelope-mediated cell fusion using a tetracycline transcriptional transactivator: fusion does not correlate with syncytium formation. Virology 324:263-272. [DOI] [PubMed] [Google Scholar]
- 31.Pal, R., G. M. Hoke, and M. G. Sarngadharan.1989. Role of oligosaccharides in the processing and maturation of envelope glycoproteins of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:3384-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, H., J. A. Gruenke, and J. M. White. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 10:1048-1053. [DOI] [PubMed] [Google Scholar]
- 33.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragheb, J. A., and W. F. Anderson. 1994. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J. Virol. 68:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salzwedel, K., and E. A. Berger. 2000. Cooperative subunit interactions within the oligomeric envelope glycoprotein of HIV-1: functional complementation of specific defects in gp120 and gp41. Proc. Natl. Acad. Sci. USA 97:12794-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Si, Z., N. Madani, J. M. Cox, J. J. Chruma, J. C. Klein, A. Schorn, N. Phan, L. Wang, A. C. Biorn, S. Cocklin, I. Chaiken, E. Freire, A. B. I. Smith, and J. Sodroski. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. USA 101:5036-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, G. W., Y. Gao, and A. D. Sanders. 2001. Fv-4: identification of the defect in env and the mechanism of resistance to ecotropic murine leukemia virus. J. Virol. 75:11244-11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233:193-198. [DOI] [PubMed] [Google Scholar]
- 40.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 41.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 42.Weissenhorn, W., A. Dessen, I. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 43.Yu, M., E. Wang, Y. Liu, D. Cao, N. Jin, C. W. H. Zhang, M. Bartlam, Z. Rao, P. Tien, and G. F. Gao. 2002. Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus. J. Gen. Virol. 83:623-629. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, Y., S. Lee, and W. F. Anderson. 1997. Functional interactions between monomers of the retroviral envelope protein complex. J. Virol. 71:6967-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]