Abstract
KIAA1429, an important component of the N6-methyladenine methyltransferase complex, is involved in the pathology of many types of cancer. In this study, the mechanisms through which KIAA1429 promotes non-small cell lung cancer (NSCLC) progression were explored using in vitro and in vivo experiments. Additionally, bioinformatics analysis of publicly available data was used to determine the relationship between KIAA1429 expression and NSCLC patient survival. The results showed that KIAA1429 was upregulated in NSCLC tissues and cells, and its high expression level was associated with low overall survival. Transcriptome analysis of KIAA1429-silenced NSCLC cells identified 346 differentially expressed genes, which were enriched in ferroptosis and the p53 signaling pathway. KIAA1429 silencing using small interfering (si) RNA promoted erastin-induced ferroptosis in NSCLC cells and activated the p53 signaling pathway. Moreover, si-KIAA1429 inhibited the proliferative, migratory, and invasive abilities of NSCLC cells in vitro and tumor growth in vivo. These in vitro effects were weakened by pifithrin-μ, a p53 inhibitor. Therefore, given its effects on ferroptosis and the p53 signaling pathway, targeting KIAA1429 could be an effective strategy for treating NSCLC.
Keywords: Non-small cell lung cancer, KIAA1429, ferroptosis, N6-methyladenine, p53 signaling pathway
Introduction
Non-small cell lung cancer (NSCLC) is a common form of lung cancer, the three main subtypes of which are adenocarcinoma, squamous cell carcinoma, and large-cell carcinoma [1]. As a malignant tumor with a high incidence and mortality rate, NSCLC is a serious threat to human health [2]. Clinically, the treatment of NSCLC depends mainly on a combination of surgery and adjuvant chemotherapy [3]. Although tyrosine kinase inhibitors and immunotherapy have improved the survival of NSCLC patients [3,4], the overall cure and survival rates remain unsatisfactory, particularly in patients with metastases [5]. Nonetheless, an increasing number of molecular targets with therapeutic potential against NSCLC have been discovered, providing additional possibilities for improving patient outcomes [6,7].
The transcriptional modification of eukaryotic mRNAs is important for ensuring the proper splicing, editing, stability, and degradation of gene transcripts. The most commonly occurring post-transcriptional modification is the methylation of the sixth nitrogen atom of adenine, otherwise known as N6-methyladenine (m6A), which is carried out by a methyltransferase complex [8]. Functionally, mRNAs with m6A participate in various pathophysiological processes, including adipogenesis, cell differentiation, proliferation, and apoptosis, and immune and inflammatory responses [9]. Notably, m6A-modified mRNAs are involved in the development of lung cancer [10]. Evidence has demonstrated that diverse m6A-related factors can regulate the malignant behavior of NSCLC cells by acting as oncogenes (e.g., ALKBH5) or anti-oncogenes (e.g., FTO, METTL3, HNRNPA2B1, and YTHDF1) [11]. Among the main components of the methyltransferase complex, KIAA1429 (also known as protein virilizer homolog (VIRMA)) is the largest methyltransferase involved in m6A methylation [12]. KIAA1429 plays a key regulatory role in the tumorigenesis and progression of liver [12], breast [13], gastric [14], and colorectal cancers [15]. Additionally, KIAA1429-mediated m6A modification promotes the progression of NSCLC and accelerates the resistance of the cancer cells to gefitinib [16,17]. However, the mechanism of action of KIAA1429 in NSCLC remains unclear.
Ferroptosis, which is a specific type of iron-related programmed cell death, is involved in various human diseases, including ischemia-reperfusion injury, degenerative diseases, cardiovascular diseases, and several types of cancer [18,19]. Many potential molecular targets are involved in the progression of NSCLC through their regulation of ferroptosis, including SLC7A11 [20], MT1DP [21], AKR1C1 [22], miR-302a-3p [23], and circDTL [24]. Ferroptosis can also be mediated by m6A modification [25,26]. Song et al. showed that miR-4443 enhanced cisplatin resistance in NSCLC by regulating FSP1 in an m6A-like manner [27]. Additionally, the p53 signaling pathway, which is a crucial regulator of NSCLC, participates in the regulation of ferroptosis [28,29].
In this study, we explored the mechanisms downstream of KIAA1429 in NSCLC using transcriptome sequencing. We found that differentially expressed genes (DEGs) in KIAA1429-silenced NSCLC cells were highly enriched in the p53 signaling pathway and ferroptosis process. Therefore, we used in vivo and in vitro experiments to further explore the potential mechanism by which KIAA1429 regulates ferroptosis and the p53 signaling pathway in NSCLC.
Materials and methods
Bioinformatics data analysis
KIAA1429 expression in 59 normal and 515 lung adenocarcinoma (LUAD) tissues was analyzed using The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN, https://ualcan.path.uab.edu/index.html) based on The Cancer Genome Atlas database. The correlation between the KIAA1429 expression level and the survival prognosis of 239 patients with NSCLC was analyzed using the Gene Expression Profiling Interactive Analysis portal (http://gepia.cancer-pku.cn/). Additionally, datasets related to “lung cancer” were searched on the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) to verify the expression of KIAA1429 in human lung cancer and normal tissues. The microarray dataset GSE18842 was selected, and the expression of KIAA1429 was analyzed using Oncomine (https://www.oncomine.org/).
Cell culture and transfection
BEAS-2B (a normal human lung epithelial cell line) and A549 and H1299 (two human NSCLC cell lines) were purchased from the American Type Culture Collection (Manassas, VA, USA). The NSCLC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin, at 37°C under 5% CO2. The BEAS-2B cells were cultured in RPMI-1640 medium and supplemented with the same components at 37°C under 5% CO2.
Small-interfering RNAs (siRNAs) targeting KIAA1429 (si-KIAA1429-1, -2, and -3) and the corresponding negative control (si-NC) were purchased from RiboBio (Guangzhou, China) and transfected into the NSCLC cells using Lipofectamine 3000 for 48 h. Additionally, H1299 and A549 cells were treated with the p53 inhibitor pifithrin-μ (PFT, 10 μM), the ferroptosis activator erastin (10 μM), or the ferroptosis inhibitor ferrostatin-1 (Fer-1, 2 μM) for 48 h. The siRNA sequences are listed in Table S1.
Quantitative reverse transcription-polymerase chain reaction
Total RNA was extracted from the NSCLC cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) and reverse transcribed into cDNA. The quantitative reverse transcription-polymerase chain reaction (RT-qPCR) was performed on a 7300PLUS instrument (Applied Biosystems, Foster City, CA, USA) using the SYBR Green qPCR Kit (Thermo Fisher Scientific). The reaction procedure was as follows: pre-denaturation at 95°C for 10 min, followed by 36 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 30 s. The relative mRNA expression level of KIAA1429 was calculated using the 2-∆∆Ct method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal control. The primer sequences are listed in Table S2.
Cell counting kit-8 assay
Cell viability was measured using the Cell Counting Kit-8 (CCK8) assay (Solarbio, Beijing, China). In brief, transfected NSCLC cells (2 × 104 cells/well) were seeded into a 96-well plate and cultured at 37°C for 24, 48, and 72 h. Subsequently, the cells were incubated with 10 μL of CCK8 reagent for 2 h. Finally, to determine the optimal value, the optical density (OD) at 450 nm was recorded every 1 h using a microplate reader (Multiskan SkyHigh, Thermo Fisher Scientific).
EdU staining
The Click-iT™ Plus EdU Cell Proliferation Kit (Thermo Fisher Scientific) was used to detect the proliferation of the NSCLC cells. In brief, the transfected cells (1 × 105 cells/well) were seeded into 24-well plates. After 2 h of incubation with 50 μM EdU reagent, the cells were first fixed with paraformaldehyde (4%) and then incubated with Triton X-100 (0.5%). Subsequently, the cells were incubated with the Click-iT Cocktail (Alexa Fluor 555; Thermo Fisher Scientific) for 30 min, followed by 4’,6-diamidino-2-phenylindole solution for 10 min in the dark. The stained cells were photographed under a fluorescence microscope (BX53; Olympus, Tokyo, Japan).
Wound-healing assay
A wound-healing assay was performed to measure cell migration. In brief, transfected cells were first cultured to 90% confluency, and a wound gap was then created on the cell layer using a pipette tip. After wash with phosphate-buffered saline to remove cell debris, the cells were cultured for 24 h. The wound gap was photographed and measured under a microscope (Olympus) at 0 and 24 h post-wounding.
Transwell assay
Transwell chambers (Corning, Kennebunk, ME, USA) were used to conduct the transwell assay for detecting cell invasion. In brief, the transwell chamber was coated with Matrigel (BD, Franklin Lakes, NJ, USA), after which 2 × 104 transfected cells were seeded into the upper chamber and DMEM containing 20% FBS was added to the lower chamber. After 24 h, the cells in the lower chamber were fixed with paraformaldehyde (4%) and stained with crystal violet (0.5%). The stained cells were photographed under a microscope (Olympus).
Enzyme-linked immunosorbent assay
The cell culture supernatants were collected for measurement of their malondialdehyde (MDA), iron ion (Fe2+), glutathione peroxidase (GSH), and reactive oxygen species (ROS) levels using enzyme-linked immunosorbent assay kits (Esebio, Shanghai, China). In brief, 50 μL of diluted sample was added to each well of a 96-well plate, followed by 100 μL of horseradish peroxidase (HRP)-labeled antibody, and the samples were incubated for 60 min at 37°C. Within 15 min after incubation, the OD value at 450 nm was measured using a microplate reader.
Transcriptome sequencing
H1299 cells transfected with either si-KIAA1429 or si-NC were used for transcriptome sequencing. In brief, mRNA was purified from the total RNA (poly(A) selection) isolated from the cells and randomly fragmented into 200-300 bp pieces using metal ion-catalyzed hydrolysis. Subsequently, a cDNA library was constructed and sequenced on an Illumina NovaSeq 6000 platform. After sequencing, the DEGs were analyzed using DESeq2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (http://david.abcc.ncifcrf.gov/).
Tumor formation in a mouse model
Female BALB/c nude mice (age: 4-5 weeks; Antaik, Beijing, China) were used to establish a xenograft tumor model (n = 6). H1299 cells (1 × 106) that had been stably transfected with KIAA1429-shRNA (LV-shKIAA1429) or NC-shRNA (LV-shNC) were subcutaneously injected into the right dorsal flank of the mice. The tumor volume was measured every 7 days. After 28 days, the mice were anesthetized with an intraperitoneal injection of pentobarbital sodium (50 mg/kg) and euthanized by cervical dislocation. The excised tumor xenografts were weighed and then used to detect KIAA1429 protein expression. The animal experiments were approved by the local ethics committee and conducted in accordance with the Guide for the Care and Use of Laboratory Animals.
Hematoxylin-eosin staining
Fresh tissues were fixed with paraformaldehyde (4%) and embedded in paraffin, after which the paraffin block was sectioned into thin slices. The dried tissue sections were immersed sequentially in xylene and gradient ethanol solutions. Subsequently, the slices were stained with hematoxylin and eosin and then dehydrated using an ethanol gradient. Finally, the slices were sealed with neutral balsam and examined and photographed under an inverted biological microscope.
Immunohistochemistry
The paraffin-embedded tissue sections were dewaxed and rehydrated, and ethylenediaminetetraacetic acid was then added for antigen retrieval. Subsequently, hydrogen peroxide (3%) was added to inactivate endogenous peroxidase at ambient temperature. The slices were incubated with anti-Ki67 antibody (ab15580; Abcam, Cambridge, MA, USA) at 4°C overnight, followed by HRP-labeled secondary antibody at 37°C for 1 h. Finally, the slices were stained with 3,3’-diaminobenzidine and hematoxylin and photographed under a microscope.
Western blot assay
Cells or tissues were lysed in radioimmunoprecipitation assay buffer to extract their total proteins, which were then separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane (Millipore, Danvers, MA, USA). The membrane was blocked with nonfat milk and incubated with primary antibodies raised against KIAA1429, prostaglandin-endoperoxide synthase 2 (PTGS2), phospholipid hydroperoxide glutathione peroxidase (GPX4), ferritin heavy chain (FTH1), protein p53, phosphorylated p53 (p-p53), and β-actin (all 1:1000 dilutions; Abcam) for 12 h. Subsequently, the membrane was incubated with HRP-conjugated IgG secondary antibody (Abcam) for 2 h. The protein bands were visualized using enhanced chemiluminescence, after which they were quantified using a Gel-Pro analyzer (Media Cybernetics, Rockville, MD, USA).
Statistical analysis
All data are presented as the mean ± standard deviation and were processed using Prism 7.0 (GraphPad, San Diego, CA, USA). The t-test was used for comparisons between two groups, whereas one-way analysis of variance and Tukey’s test were used for comparisons among multiple groups. A P-value of < 0.05 was regarded as statistically significant.
Results
KIAA1429 is upregulated in NSCLC
The expression levels of KIAA1429 in the 59 normal and 515 LUAD tissue samples were analyzed using UALCAN and were significantly higher in the cancerous tissues than in the normal controls (P < 0.01; Figure 1A). Survival analysis using GEPIA revealed that the overall survival period of patients with low KIAA1429 expression levels was significantly longer than that of patients with high KIAA1429 expression levels (P = 0.022; Figure 1B), demonstrating that high KIAA1429 expression is related to a poor prognosis. Analysis of the GSE18842 dataset, which contains 46 lung cancer and 45 normal tissue samples, revealed that KIAA1429 was substantially higher in the cancerous samples (Figure 1C). Additionally, KIAA1429 was significantly more highly expressed in the H1299 and A549 cells than in the BEAS-2B cells (P < 0.01; Figure 1D).
Figure 1.
The up-regulation of KIAA1429 in NSCLC. A. The expression of KIAA1429 in LUAD tissues was analyzed using the UALCAN database. B. The correlation of KIAA1429 expression with overall survival in patients with LUAD was analyzed using GEPIA database. C. Oncomine was applied to evaluate KIAA1429 expression between cancer and normal tissues in microarray dataset (GSE18842). D. The expression of KIAA1429 in NSCLC cell lines (H1299 and A549 cells) and a normal lung epithelial cell line (BEAS-2B cells) was measured by qRT-PCR. **P < 0.01 vs. BEAS-2B.
Knockdown of KIAA1429 inhibits the proliferative, migratory, and invasive abilities of NSCLC cells
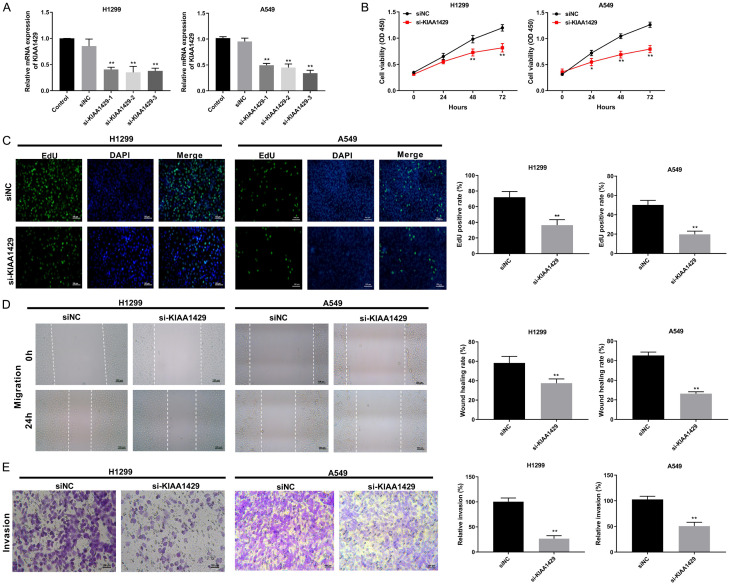
To explore the role of KIAA1429 in NSCLC, KIAA1429 expression was silenced in the two NSCLC cell lines. The transfection of si-KIAA1429-1, -2, and -3 into H1299 and A549 cells was verified using RT-qPCR, and si-KIAA1429-3 was used for the subsequent experiments because it had the highest silencing efficiency of the three siRNAs (P < 0.01; Figure 2A). The viability of the si-KIAA1429-transfected H1299 and A549 cells was significantly lower than that of the si-NC-transfected cells (P < 0.01; Figure 2B). Under the microscope, a lower EdU positivity rate was observed in the si-KIAA1429 groups than in the si-NC groups (P < 0.01; Figure 2C). Additionally, we found that si-KIAA1429 also inhibited the migratory and invasive abilities of the H1299 and A549 cells (P < 0.01; Figure 2D and 2E).
Figure 2.
Knockdown of KIAA1429 inhibited the proliferation, migration, and invasion of NSCLC cells. A. The expression level of KIAA1429 in NSCLC cells transfected with si-KIAA1429-1/-2-/-3/si-NC was detected by qRT-PCR. B. The viability of transfected NSCLC cells was measured by CCK8 assay. C. EdU positive cells were detected by EdU assay (200×). D. The migration of transfected NSCLC cells was measured by wound healing assay. E. The invasion of transfected NSCLC cells was measured by Transwell assay (200×). *P < 0.05, **P < 0.01 vs. siNC.
Knockdown of KIAA1429 inhibits tumor growth in mice
To determine the role of KIAA1429 in NSCLC in vivo, H1299 cells transfected with either NC-shRNA or KIAA1429-shRNA were injected into mice. The tumor volume was lower in the LV-shKIAA1429 group than in the LV-shNC group on the 14th day post-inoculation (P < 0.05; Figure 3A and 3B). The LV-shKIAA1429 group also had significantly low tumor weights on the 28th day post-inoculation (P < 0.01; Figure 3C). The western blotting results verified the downregulation of KIAA1429 expression in the tumor tissues of mice in the LV-shKIAA1429 group (P < 0.01; Figure 3D). Moreover, the knockdown of KIAA1429 significantly reduced the degree of tumor malignancy (Figure 3E) and decreased the proliferation of the cancer cells (Figure 3F).
Figure 3.
Knockdown of KIAA1429 inhibits tumor growth in mice. A. The morphology of xenograft tumors in mice. B. The volume of xenograft tumors. C. The weight of xenograft tumors. D. The expression of KIAA1429 in xenograft tumors. E. Hematoxylin-eosin staining was performed to observe the histological changes of tumors. F. Immunohistochemistry was executed to observe the proliferation of cancer cells. *P < 0.05, **P < 0.01 vs. LV-shNC.
Knockdown of KIAA1429 promotes ferroptosis in NSCLC
si-KIAA1429 significantly reduced the viability of the erastin-treated NSCLC cells (H1299 and A549), and the inhibitory effect was greater with increasing erastin concentrations (Figure 4A). This inhibitory effect was reversed by adding the ferroptosis inhibitor Fer-1 (Figure 4B). Moreover, the knockdown of KIAA1429 markedly increased the levels of ROS and Fe2+ and reduced those of GSH and MDA in the erastin-treated cells (Figure 4C-F). Additionally, erastin treatment markedly increased the protein expression of PTGS2 in the si-NC group and decreased that of GPX4 and FTH1 (Figure 4G, 4H), similar to the si-KIAA1429 group. In summary, KIAA1429 knockdown promoted erastin-mediated ferroptosis.
Figure 4.
Knockdown of KIAA1429 promoted the ferroptosis in NSCLC cells. A. CCK-8 was executed to evaluate cell viability in A549 cells after erastin treatment. B. CCK-8 was executed to evaluate cell viability in H1299 and A549 cells after erastin (10 μM) ± ferrostatin-1 (2 μM). C-F. The levels of Fe2+ and lipid peroxidation indexes ROS, GSH and MDA in H1299 and A549 cells were detected by ELISA. G, H. Western-blot was used to detect the expression levels of ferroptosis-related proteins (PTGS2, GPX4, and FTH1) in H1299 and A549 cells. **P < 0.01 vs. si-NC.
Transcriptome sequencing analysis
The molecular mechanisms downstream of KIAA1429 silencing were explored in NSCLC cells. In total, 346 DEGs were identified, 196 of which were upregulated and 150 were downregulated (Figure 5A and 5B). GO enrichment analysis revealed that the DEGs were enriched in various biological functions, including autolysosome, negative regulation of endoplasmic reticulum unfolded protein response, and secondary lysosome (rich > 0.2) (Figure 6A). Moreover, KEGG analysis showed that the DEGs were enriched in many pathways, including those related to galactose metabolism, mineral absorption, nicotine addiction, p53 signaling, and ferroptosis (Figure 6B).
Figure 5.

Differentially expressed genes (DEGs) in KIAA1429-silenced H1299 cells. A. A heatmap of DEGs in different groups. B. A Volcano map of DEGs.
Figure 6.
Functional enrichment of differentially expressed genes isolated from KIAA1429-silenced H1299 cells. A. Bar plot of GO enrichment analysis. B. Bubble plots of KEGG enrichment analysis.
Knockdown of KIAA1429 activates the p53 signaling pathway to inhibit NSCLC progression
The mechanism through which KIAA1429 regulates the p53 signaling pathway was further analyzed using H1299 cells. The western blot results revealed that the p-p53/p53 level was higher in the si-KIAA1429-transfected cells than in their si-NC-transfected counterparts (P < 0.01; Figure 7A). The p53 inhibitor PFT blocked the p53 signaling pathway by decreasing p-p53/p53 expression, whereas si-KIAA1429 reversed this effect (Figure 7B). Furthermore, functional assays showed that PFT significantly promoted the proliferative, migratory, and invasive abilities of the NSCLC cells relative to its effects on the si-NC group (P < 0.01, Figure 7C-F). Additionally, PFT weakened the inhibitory effects of si-KIAA1429 on NSCLC cell proliferation, migration, and invasion (P < 0.01, Figure 7C-F).
Figure 7.
Knockdown of KIAA1429 activated the p53 signaling pathway to inhibit NSCLC progression. A. The expression of the p53 pathway-related proteins in H1299 cells was detected by Western blot. B. After PFT treatment, the expression of the p53 pathway-related proteins was detected by Western blot. C. The viability of transfected/treated H1299 cells was measured by CCK8 assay. D. EdU positive cells was detected by EdU assay (200×). E. The migration of transfected/treated H1299 cells was measured by wound healing assay. F. The invasion of transfected/treated H1299 cells was measured by Transwell assay (200×). **P < 0.01 vs. siNC; ##P < 0.01 vs. si-KIAA1429.
Discussion
NSCLC remains a leading cause of death worldwide, and effective treatments for improving the poor outcomes of affected patients remain a challenge [30]. Several lines of evidence have shown that the m6A modification can influence diverse tumor characteristics, including the maintenance of proliferation signals, evasion of growth inhibition, resistance to apoptosis, induction of angiogenesis, activation of metastasis, changes in energy metabolism, and immune escape [31,32]. KIAA1429 is the largest of the methyltransferases involved in m6A modification [15]. KIAA1429 has multiple roles in various tumors. For example, KIAA1429 silencing suppresses the proliferation and metastasis of hepatocellular carcinoma cells through the post-transcriptional regulation of GATA3 in an m6A-dependent manner [12]. Furthermore, KIAA1429 enhances breast cancer progression by regulating CDK1 in an m6A-independent manner [13]. Additionally, Xu et al. found that KIAA1429 enhanced the proliferation of NSCLC cells and the growth of NSCLC xenografts via the m6A-dependent degradation of DAPK3 mRNA [17]. In this study, we determined through public database analysis that KIAA1429 expression was upregulated in NSCLC tissues, and its high level correlated positively with poor overall survival. Our in vitro experiments verified that KIAA1429 was upregulated in NSCLC cells and that its knockdown inhibited malignant features at the cellular level. These results were similar to those of previous studies illustrating that KIAA1429 acts as an oncogene in NSCLC. Furthermore, our in vivo experiments verified that KIAA1429 silencing can inhibit the growth of tumor xenografts.
To identify the mechanisms of action downstream of KIAA1429 in NSCLC, transcriptome sequencing was performed on KIAA1429-silenced cells. Ferroptosis was selected as the research target, as it was enriched in the identified DEGs. KIAA1429 silencing enhanced ferroptosis in the NSCLC cells, as evidenced by the increased Fe2+ and ROS and decreased GSH and MDA levels, as well as the upregulation of PTGS2 and downregulation of GPX4 and FTH1 expression. Ferroptosis is an iron-dependent cell death process that acts as a natural barrier to prevent the tumorigenesis, progression, and metastasis of diverse types of cancer [33]. Under oxidative stress conditions, the ferroptosis defense system benefits the survival of tumor cells and their response to various cancer therapies [34]. Huang et al. found that the ferroptosis activator erastin inhibited the proliferation of lung cancer cells by arresting the cell cycle at the G1 phase [35]. Similarly, Chen et al. revealed that erastin suppressed the proliferative and migratory abilities of lung cancer cells by inducing calcium/calmodulin-dependent ferroptosis [36]. Other studies have determined the regulatory role of m6A modification in ferroptosis [25-27]. Combined with the reverse regulatory relationship between KIAA1429 and ferroptosis, we speculate that activating this cell death process by silencing KIAA1429 may inhibit the progression of NSCLC.
In this study, the DEGs in KIAA1429-silenced NSCLC cells were also enriched in the p53 signaling pathway. p53 is an important tumor suppressor that inhibits cancer progression by regulating cell proliferation, apoptosis, and cell-cycle arrest [37]. Molecules such as TNFAIP8 [38], USP22 [39], HAX1 [40], and GINS2 [41] regulate the p53 signaling pathway and, thus, show therapeutic potential against NSCLC. Sang et al. reported that the m6A methyltransferase subunits METTL3 and METTL14 enhanced acute myeloid leukemia by targeting the MDM2/p53 signaling pathway [42]. In our study, KIAA1429 silencing promoted antitumor effects by facilitating the activation of the p53 signaling pathway, whereas PFT (a p53 inhibitor) reversed these effects of si-KIAA1429. These results indicate that the p53 signaling pathway is an active target of KIAA1429 and contributes to the remission of NSCLC. Moreover, p53 also plays a key regulatory role in ferroptosis [28,29]. Huang et al. found that erastin-induced ROS production enhanced ferroptosis and inhibited cell proliferation by activating p53 in lung cancer cells [35]. Meng et al. showed that levobupivacaine inhibited ferroptosis in NSCLC and the progression of the disease by activating p53 [43]. In our study, KIAA1429 silencing promoted erastin-induced ferroptosis and activated the p53 signaling pathway, revealing the potential mechanisms underlying the pathology of NSCLC.
Conclusions
In summary, the expression of the m6A methyltransferase KIAA1429 was elevated in NSCLC tissues and cells. KIAA1429 silencing promoted the p53 signaling pathway and ferroptosis in the NSCLC cells and inhibited their proliferative, migratory, and invasive abilities. KIAA1429 silencing also inhibited the growth of tumor xenografts in mice. However, a limitation of our study is that it lacked human clinical samples for analysis; these should be considered in future studies. Nonetheless, our findings that KIAA1429 regulates the p53 signaling pathway and ferroptosis suggest that this m6A methyltransferase can be a promising molecular target for treatment strategies against NSCLC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25:585–594. doi: 10.1016/j.molmed.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Duma N, Santana-Davila R, Molina JR. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94:1623–1640. doi: 10.1016/j.mayocp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Uprety D, Mandrekar SJ, Wigle D, Roden AC, Adjei AA. Neoadjuvant immunotherapy for NSCLC: current concepts and future approaches. J Thorac Oncol. 2020;15:1281–1297. doi: 10.1016/j.jtho.2020.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Murugesan S, Murugesan J, Palaniappan S, Palaniappan S, Murugan T, Siddiqui SS, Loganathan S. Tyrosine kinase inhibitors (TKIs) in lung cancer treatment: a comprehensive analysis. Curr Cancer Drug Targets. 2021;21:55–69. doi: 10.2174/1568009620666201009130008. [DOI] [PubMed] [Google Scholar]
- 5.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 6.Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194. doi: 10.1016/j.critrevonc.2020.103194. [DOI] [PubMed] [Google Scholar]
- 7.Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med. 2019;27:167–170. [PubMed] [Google Scholar]
- 8.Pian C, Yang Z, Yang Y, Zhang L, Chen Y. Identifying RNA N6-methyladenine sites in three species based on a markov model. Front Genet. 2021;12:650803. doi: 10.3389/fgene.2021.650803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong H, Tang HF, Kai Y. N6-methyladenine RNA modification (m(6)A): an emerging regulator of metabolic diseases. Curr Drug Targets. 2020;21:1056–1067. doi: 10.2174/1389450121666200210125247. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Chen J, Fei X, Wang X, Wang K. N6-methyladenine RNA modification and cancer. Oncol Lett. 2020;20:1504–1512. doi: 10.3892/ol.2020.11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu FS, He JQ, Zhong YS, Guo MY, Yu CH. Implications of m6A methylation and microbiota interaction in non-small cell lung cancer: from basics to therapeutics. Front Cell Infect Microbiol. 2022;12:972655. doi: 10.3389/fcimb.2022.972655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan T, Li H, Zhang D, Xu L, Liu H, Hao X, Yan X, Liao H, Chen X, Xie K, Li J, Liao M, Huang J, Yuan K, Zeng Y, Wu H. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian JY, Gao J, Sun X, Cao MD, Shi L, Xia TS, Zhou WB, Wang S, Ding Q, Wei JF. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38:6123–6141. doi: 10.1038/s41388-019-0861-z. [DOI] [PubMed] [Google Scholar]
- 14.Miao R, Dai CC, Mei L, Xu J, Sun SW, Xing YL, Wu LS, Wang MH, Wei JF. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J Cell Physiol. 2020;235:7420–7432. doi: 10.1002/jcp.29645. [DOI] [PubMed] [Google Scholar]
- 15.Ma L, Lin Y, Sun SW, Xu J, Yu T, Chen WL, Zhang LH, Guo YC, Wang YW, Chen T, Wei JF, Zhu LJ. KIAA1429 is a potential prognostic marker in colorectal cancer by promoting the proliferation via downregulating WEE1 expression in an m6A-independent manner. Oncogene. 2022;41:692–703. doi: 10.1038/s41388-021-02066-z. [DOI] [PubMed] [Google Scholar]
- 16.Tang J, Han T, Tong W, Zhao J, Wang W. N(6)-methyladenosine (m(6)A) methyltransferase KIAA1429 accelerates the gefitinib resistance of non-small-cell lung cancer. Cell Death Discov. 2021;7:108. doi: 10.1038/s41420-021-00488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Chen Y, Yao Y, Xie H, Lu G, Du C, Cheng J, Zhou J. VIRMA contributes to non-small cell lung cancer progression via N(6)-methyladenosine-dependent DAPK3 post-transcriptional modification. Cancer Lett. 2021;522:142–154. doi: 10.1016/j.canlet.2021.08.027. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao N, Sun B, Wang G. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei X, Yi X, Zhu XH, Jiang DS. Posttranslational modifications in ferroptosis. Oxid Med Cell Longev. 2020;2020:8832043. doi: 10.1155/2020/8832043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X, Kang N, Ling X, Pan M, Du W, Gao S. MiR-27a-3p promotes non-small cell lung cancer through SLC7A11-mediated-ferroptosis. Front Oncol. 2021;11:759346. doi: 10.3389/fonc.2021.759346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gai C, Liu C, Wu X, Yu M, Zheng J, Zhang W, Lv S, Li W. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020;11:751. doi: 10.1038/s41419-020-02939-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang F, Zheng Y, Li X, Luo H, Luo L. Ferroptosis-related gene AKR1C1 predicts the prognosis of non-small cell lung cancer. Cancer Cell Int. 2021;21:567. doi: 10.1186/s12935-021-02267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei D, Ke YQ, Duan P, Zhou L, Wang CY, Cao P. MicroRNA-302a-3p induces ferroptosis of non-small cell lung cancer cells via targeting ferroportin. Free Radic Res. 2021;55:821–830. doi: 10.1080/10715762.2021.1947503. [DOI] [PubMed] [Google Scholar]
- 24.Shanshan W, Hongying M, Jingjing F, Yiming Y, Yu R, Rui Y. CircDTL functions as an oncogene and regulates both apoptosis and ferroptosis in non-small cell lung cancer cells. Front Genet. 2021;12:743505. doi: 10.3389/fgene.2021.743505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen M, Li Y, Wang Y, Shao J, Zhang F, Yin G, Chen A, Zhang Z, Zheng S. N(6)-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021;47:102151. doi: 10.1016/j.redox.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, He J, Sun G, Huang N, Bian Z, Xu C, Zhang Y, Cui Z, Xu W, Sun F, Zhuang C, Man Q, Gu S. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med. 2022;12:e778. doi: 10.1002/ctm2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song Z, Jia G, Ma P, Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021;276:119399. doi: 10.1016/j.lfs.2021.119399. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Zhang C, Wang J, Hu W, Feng Z. The regulation of ferroptosis by tumor suppressor p53 and its pathway. Int J Mol Sci. 2020;21:8387. doi: 10.3390/ijms21218387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang CL, Yokomise H, Miyatake A. Clinical significance of the p53 pathway and associated gene therapy in non-small cell lung cancers. Future Oncol. 2007;3:83–93. doi: 10.2217/14796694.3.1.83. [DOI] [PubMed] [Google Scholar]
- 30.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, Li H, Hu D, Wang Y, Shao W, Zhong J, Yang S, Liu J, Zhang J. Insights into N6-methyladenosine and programmed cell death in cancer. Mol Cancer. 2022;21:32. doi: 10.1186/s12943-022-01508-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Gu J, Xu F, Zhu Q, Chen Y, Ge D, Lu C. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief Bioinform. 2021;22:bbaa225. doi: 10.1093/bib/bbaa225. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Zhang Y, Wu X, Xu F, Ma H, Wu M, Xia Y. Targeting ferroptosis pathway to combat therapy resistance and metastasis of cancer. Front Pharmacol. 2022;13:909821. doi: 10.3389/fphar.2022.909821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei G, Zhuang L, Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22:381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C, Yang M, Deng J, Li P, Su W, Jiang R. Upregulation and activation of p53 by erastin-induced reactive oxygen species contribute to cytotoxic and cytostatic effects in A549 lung cancer cells. Oncol Rep. 2018;40:2363–2370. doi: 10.3892/or.2018.6585. [DOI] [PubMed] [Google Scholar]
- 36.Chen P, Wu Q, Feng J, Yan L, Sun Y, Liu S, Xiang Y, Zhang M, Pan T, Chen X, Duan T, Zhai L, Zhai B, Wang W, Zhang R, Chen B, Han X, Li Y, Chen L, Liu Y, Huang X, Jin T, Zhang W, Luo H, Chen X, Li Y, Li Q, Li G, Zhang Q, Zhuo L, Yang Z, Tang H, Xie T, Ouyang X, Sui X. Erianin, a novel dibenzyl compound in dendrobium extract, inhibits lung cancer cell growth and migration via calcium/calmodulin-dependent ferroptosis. Signal Transduct Target Ther. 2020;5:51. doi: 10.1038/s41392-020-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang YT, Chien YC, Lin YH, Wu HH, Lee DF, Yu YL. The function of the mutant p53-R175H in cancer. Cancers (Basel) 2021;13:4088. doi: 10.3390/cancers13164088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing Y, Liu Y, Liu T, Meng Q, Lu H, Liu W, Hu J, Li C, Cao M, Yan S, Huang J, Wang T, Cai L. TNFAIP8 promotes the proliferation and cisplatin chemoresistance of non-small cell lung cancer through MDM2/p53 pathway. Cell Commun Signal. 2018;16:43. doi: 10.1186/s12964-018-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding F, Bao C, Tian Y, Xiao H, Wang M, Xie X, Hu F, Mei J. USP22 promotes NSCLC tumorigenesis via MDMX up-regulation and subsequent p53 inhibition. Int J Mol Sci. 2014;16:307–20. doi: 10.3390/ijms16010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Z, Zhong Y, Meng L, Chen Y, Liu Y, Wu A, Li X, Wang M. HAX1 enhances the survival and metastasis of non-small cell lung cancer through the AKT/mTOR and MDM2/p53 signaling pathway. Thorac Cancer. 2020;11:3155–3167. doi: 10.1111/1759-7714.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi F, Wang Z, Li Y, Chang N. Knockdown of GINS2 inhibits proliferation and promotes apoptosis through the p53/GADD45A pathway in non-small-cell lung cancer. Biosci Rep. 2020;40:BSR20193949. doi: 10.1042/BSR20193949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sang L, Wu X, Yan T, Naren D, Liu X, Zheng X, Zhang N, Wang H, Li Y, Gong Y. The m(6)A RNA methyltransferase METTL3/METTL14 promotes leukemogenesis through the mdm2/p53 pathway in acute myeloid leukemia. J Cancer. 2022;13:1019–1030. doi: 10.7150/jca.60381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng M, Huang M, Liu C, Wang J, Ren W, Cui S, Gu J, Xie J, Ma B, Yang G, He S. Local anesthetic levobupivacaine induces ferroptosis and inhibits progression by up-regulating p53 in non-small cell lung cancer. Aging (Albany NY) 2021 doi: 10.18632/aging.203138. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.