Abstract
This research is dedicated to investigating the mechanism of programmed cell death ligand 1 (PD-L1) and tumor protein 53 target gene 1 (TP53TG1) in immune regulation of colon cancer (CC). Expressions of TP53TG1, PD-L1 and signal transducers and activators of transcription (STATs) in CC and their correlation were detected through bioinformatics analysis. Effects of PD-L1 and TP53TG1 on the CC were assessed by in vivo and in vitro experiments. Herein, PD-L1 level was negatively correlated with TP53TG1 expression, but was positively correlated with the levels of STATs. Both overexpressed TP53TG1 and PD-L1 antibody reversed the effects of CT26 cells on inhibiting cell proliferation, cytokine secretion and PD-L1 level, and enhancing the cytotoxicity of NK cells and CD8+ T cells. TP53TG1 reduced PD-L1 level by inactivating STATs pathway. Downregulation of PD-L1 increased cytokine secretion and T lymphocyte killing ability, promoted tumor cell apoptosis, and inhibited the tumor growth. Altogether, TP53TG1/STAT axis regulates the immunomodulatory mechanism of CC by reducing PD-L1 expression.
Keywords: PD-1/PD-L1, TP53TG1, immune escape, STAT pathway, colon cancer
Introduction
Colon cancer is one of the most frequent malignant tumors in digestive tract, which is attributed to the interaction of multiple factors such as heredity, environment, diet, and intestinal microecology [1,2]. In recent years, the incidence of colon cancer has been increasing [3]. This disease is often difficult to be diagnosed at the early stage due to its insidious onset [4,5].
Immunotherapy, including non-specific immunotherapy, tumor vaccine and monoclonal antibody therapy, is a recently emerging treatment option [6,7]. T cells (cytotoxic T cells (Tc), T helper, regulatory T cells (Treg) and natural killing (NK) T cells) are important immune cells and play different roles in the process of mediated immunity [8,9]. The key to inducing effective anti-tumor immunity is the activation of effector T cells [10]. Programmed cell death ligand 1 (PD-L1), a molecule from B7 family, is also known as B7-H1 or CD274 [11,12]. Immunotherapy targeting PD-1/PD-L1 is a new progress in the field of tumor therapy in recent years [13,14]. Studies have found that the expression of PD-L1 is up-regulated in lung cancer, breast cancer, pancreatic cancer, hepatocellular carcinoma, melanoma and other tumor tissues [15,16]. PD-L1 can inhibit the proliferation and activation of T cells by binding to its receptor programmed cell death 1 (PD-1) and negatively regulate the immune response process of the body, thus mediating the immune escape of tumor and promoting tumor growth [17,18]. There is a breakthrough in the study of PD-1/PD-L1 pathway, unveiling that antagonists targeting PD-1/PD-L1 pathway can relieve their inhibitory effects on T cell function and restore the anti-tumor immunity of the body [19,20].
Long noncoding RNAs (lncRNAs) are RNA transcripts of non-coding proteins with a length of more than 200 nucleotides [21]. In some cancers, a series of lncRNAs have been found to have carcinogenic or anticancer functions. For example, colon cancer-associated transcript-1 (CCAT1) has been evidenced to be a carcinogenic lncRNA in gastric cancer, colorectal cancer and hepatocellular carcinoma [22]. In colon cancer, lncRNA BC200 is found to be a new oncogene and a new therapeutic target [23]. Tumor protein 53 target gene 1 (TP53TG1), which is a p53-induced lncRNA, also plays an important role in the progression of cancer [24]. Besides, the deletion of TP53TG1 affects the expressions of metabolism-related genes in glioma and can promote cell proliferation and migration [25]. Angel Diaz-Lagaresa et al. have confirmed the anticancer activity of TP53TG1 and demonstrated that TP53TG1 is involved in the regulatory network of cancer cells [26]. However, the role of TP53TG1 in colon cancer has not been reported.
In this study, the correlation of PD-L1 and TP53TG1 in colon cancer was detected through in vivo and in vitro experiments, and the potential regulatory mechanism was further investigated.
Materials and methods
Bioinformatics analysis
The expression correlations between TP53TG1/STAT1 and PD-L1 in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), bladder urothelial carcinoma (BLCA), colon adenocarcinoma (COAD), liver hepatocellular carcinoma (LIHC), mesothelioma (MESO), kidney renal clear cell carcinoma (KIRC), prostate adenocarcinoma (PRAD), pancreatic adenocarcinoma (PAAD), thyroid carcinoma (THCA), stomach adenocarcinoma (STAD), esophageal carcinoma (ESCA), lung adenocarcinoma (LUAD), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), head and neck squamous cell carcinoma (HNSC), or breast invasive carcinoma (BRCA) were analyzed using StarBase (https://starbase.sysu.edu.cn/index.php).
Ethical approval and consent to participate
Animal experiments were approved by the Animal Ethics Committee of the First Hospital of Shanxi Medical University (Z20190523X).
Cell collection, culture and treatment
The mouse colon cancer cell line CT26 (CL-0071) was purchased from Procell (Wuhan, China) and cultured in RPMI-1640 medium (PM150110, Procell) containing 10% fetal bovine serum (FBS; 164210, Procell) at 37°C with 5% CO2. Then, CT26 cells were collected for subsequent in vitro and in vivo cellular and animal experiments with or without transfection.
For murine splenic T cells, a total of 10 BALB/c mice (SPF, 4-6 weeks old, 20±2 g, male) were obtained from CAVENS (Changzhou, China). The mice were intraperitoneally injected with 50 mg/kg sodium pentobarbital (2%, B005, Jiancheng Bioengineering, Nanjing, China) and then euthanized by cervical dislocation. Following disinfection with 75% alcohol, the mouse spleens were collected, ground and then filtered by sieve mesh. The mononuclear cells were obtained by Ficoll density gradient centrifugation, and the cell concentration was adjusted to 2×106 cell/ml. Next, cells were isolated by Easy Sep Mouse T cell Isolation Kit (#19851A, STEMCELL, Vancouver, Canada) according to the manufacturer’s instructions, subsequent to which the mouse T cells with CD3+ T >90% were obtained. T cells were re-cultured in RPMI1640 lymphocyte medium at 37°C with 5% CO2. In the following in vitro experiments, the proliferation of T cells was detected by cell counting kit-8 (CCK-8) assay and flow cytometry, and the levels of cytokines secreted by T cells were assessed by enzyme-linked immunosorbent assay (ELISA). T cells were randomly assigned into the following groups: mIgG+T group (T cells+mIgG); α-CD3+T group (T cells+α-CD3 mAb); α-CD3+T+CT26 group (T cells+α-CD3 mAb+CT26 cells); α-CD3+mIgG+T+CT26 group (T cells+α-CD3 mAb+mIgG+CT26 cells); α-CD3+α-PD-L1+T+CT26 group (T cells+α-CD3 mAb+α-PD-L1 mAb+CT26 cells); α-CD3+T+mock group (T cells+α-CD3 mAb+CT26 cells transfected with mock); α-CD3+T+TP53TG1 group (T cells+α-CD3 mAb+CT26 cells transfected with TP53TG1).
For CD8+ T/natural killer (NK) cells, the spleens of the mice were ground to obtain single-cell suspension. The erythrocyte lysate (C3702, Beyotime, Shanghai, China) was added to the single-cell suspension and lysed at room temperature for 5 minutes (min). Next, the cells were mixed with the diluted fluorescent dye-labeled antibodies and incubated at 4°C for 30 min. Anti-mouse CD8-FITC antibody (ab237367, Abcam, Cambridge, UK) was applied to label CD8+ T cells, while anti-mouse NK1.1-APC antibody (50 μl, 1805-11, SouthernBiotech, Birmingham, Alabama, USA) was used to mark NK cells. The positive cells were collected and sorted into the collection tube by a flow cytometer (Epics-XLII, Beckman, Bria, California, USA). The cytotoxicity of CD8+ T cells and NK cells targeting CT26 cells was detected by LDH nonradioactive assay. CT26 cells were randomly assigned into control group (CT26 cells+CD8+ T cells), mIgG group (mIgG+CT26 cells+CD8+ T cells), α-PD-L1 group (α-PD-L1+CT26 cells+CD8+ T cells), mock group (CT26 cells transfected with mock+CD8+ T cells), TP53TG1 group (CT26 cells transfected with TP53TG1 overexpression plasmid+CD8+ T cells), siTP53TG1 group (CT26 cells transfected with small interfering RNA (siRNA) against TP53TG1 (siTP53TG1)+CD8+ T cells), and siTP53TG1+α-PD-L1 group (α-PD-L1+CT26 cells transfected with siTP53TG1+CD8+ T cells). NK cells were allocated into groups below: control group (CT26 cells+NK cells); mIgG group (mIgG+CT26 cells+NK cells); α-PD-L1 group (α-PD-L1+CT26 cells+NK cells); mock group (CT26 cells transfected with mock+NK cells); TP53TG1 group (CT26 cells transfected with TP53TG1 overexpression plasmid+NK cells); siTP53TG1 group (CT26 cells transfected with siTP53TG1+NK cells); siTP53TG1+α-PD-L1 group (α-PD-L1+CT26 cells transfected with siTP53TG1+NK cells).
Transfection
TP53TG1 overexpression plasmid and pc-DNA3.1 vector were obtained from Youbio Biotech (Changsha, China). siTP53TG1, siSTAT1, siSTAT2, siSTAT3 and corresponding siRNA negative control (siNC) were synthesized by GenePharma (Shanghai, China) and used for the transfection of CT26 cells. According to the instructions of Lipofectamine 2000 reagent (Thermo, Waltham, Massachusetts, USA), Lipofectamine 2000 reagent and the plasmids were separately diluted by Gibco™Opti-MEM™IReducedSerumMedium, and then the two solutions were mixed for further 5 min of incubation. The diluted Lipofectamine 2000 reagent was mixed with the diluted plasmids and incubated for 20 min. Next, the mixture was added to the cells and cultivated in an incubator at 37°C with 5% CO2 for 48 hours (h). Untreated cells were assigned into a control group. After 48 h, cells were collected for later experiments. The sequences used in the transfection were listed in Table 1.
Table 1.
The sequences for transfection
| Name | Sequence: 5’--3’ |
|---|---|
| siTP53TG1 | CUACUUCCUGAAAACAACG |
| siSTAT1 | CCCUAGAAGACUUACAAGAUGAAUA |
| siSTAT2 | GAAGUGAAUGCAGAGCUCUUGUUAG |
| siSTAT3 | CCAACGACCUGCAGCAAUA |
T cell proliferation detection
The 96-well plates were encapsulated by anti-CD3 mouse monoclonal antibody (α-CD3 mAb, 0.5 μg/ml, UCHT1, 217570, Merck, St. Louis, Missouri, USA) at 4°C overnight. Next, the concentration of transfected or un-transfected CT26 cells was adjusted to 1×105 cell/ml using RPMI-1640 medium and then cells were inoculated into 96-well plates. Thereafter, CT26 cells were mixed with the purified mouse T cells (5×104 cell/ml) at the ratio of 1:5. Subsequently, PD-L1 antibody (α-PD-L1 mAb, 0.5 μg/ml, 12-5983-42, Invitrogen, Waltham, Massachusetts, USA) was added to block the PD-1/PD-L1 signaling pathway. Herein, 0.5 µg/ml mouse IgG (mIgG) antibody (08-6599, Invitrogen) was adopted as the corresponding isotype control. Afterwards, the cells were placed in incubators at 37°C with 5% CO2 for 3-5 days, and then collected for follow-up experiments.
To detect T cell proliferation, CCK-8 assay was implemented. Specifically, CCK-8 solution (C0037, Beyotime) was added into each well to further incubate with cells for 4-6 h at 37°C with 5% CO2. Afterwards, the optical density (OD) in each well was measured by a microplate reader (Model 680, Bio-Rad, Hercules, California, USA) at the wavelength of 450 nm.
To determine the cell cycle, flow cytometry was exploited. Simply put, cells were fixed with 70% ethanol at 4°C overnight, re-suspended with FBS and then cultured with RNaseA at 37°C for 10 min. Next, the propidium iodine (PI, ST511, Beyotime) solution was added to stain the cells for 20 min in the dark. Finally, the cell cycle distribution was analyzed under the help of the flow cytometer (Epics-XLII, Beckman) and Mod Fit LT software V2.0 (Becton Dickinson).
Cytotoxicity assay
According to the manufacturer’s specification of LDH kit (CK12, Dojindo, Tokyo, Japan), the LDH non-radioactive cytotoxicity was adopted to detect the cytotoxicity of CD8+ T cells and NK cells to target cells. Herein, CT26 cells were used as the target cells, and CD8+ T cells and NK cells were employed as the effector cells. CT26 cells (both transfected and un-transfected) in α-PD-L1 group and siTP53TG1+α-PD-L1 group were incubated with α-PD-L1 for 15 min, whereas cells in mIgG group were treated with the control mIgG. CD8+ T cells and NK cells were mixed with CT26 cells (5,000 cells/ml) at the effector-target ratio of 5:1, and subjected to 4-h incubation at 37°C with 5% CO2. Thereafter, the working solution was added to further cultivate the cells for 30 min. Later, the stop solution was applied to terminate the reaction. Finally, OD value was recorded at a wavelength of 490 nm by a microplate reader, followed by the calculation of cytotoxicity index (%). The specific formula of cytotoxicity index was listed below: Cytotoxicity (%) = [(Experimental value - LDH Effector cells spontaneous control - Target cells spontaneous control)/(Target cells maximum control - Target cells spontaneous control)] × 100%.
Animal model establishment and treatment
In model mice, a total of 36 BALB/c mice (SPF, 4-6 weeks old, 20±2 g, and male) were obtained from CAVENS. The mice were raised in an environment at 20-26°C in 40-70% humidity under a 12 h light/12 h dark cycle and fed by a normal diet. The mice were divided into 6 groups (n = 6), namely normal group, control group, α-PD-L1 group, mock group, siTP53TG1+α-PD-L1 group and siTP53TG1 group.
Colon cancer mouse models were established in all groups, except normal group. Concretely, 2 ml (0.2×107 cells) of CT26 cell suspension was subcutaneously injected into the armpit of the right forelimb of each mouse. 3 to 4 days after the subcutaneous inoculation, the tumor at a diameter of 4-5 mm could be touched under the armpit of mice, indicating the successful inoculation. After 2 weeks, the colon cancer mouse model was established. In control group, mice were intraperitoneally injected with 300 μg of physiological saline on the 8th, 10th and 12th days. Moreover, in α-PD-L1 group, each mouse was intraperitoneally injected with 300 μg of α-PD-L1 on the 8th, 10th and 12th days. Mice in the mock group were subcutaneously injected with CT26 cells (0.2×107 cells) transfected with empty vectors to construct colon cancer models, and then intraperitoneally injected with 300 μg of physiological saline on the 8th, 10th and 12th days. Mice in siTP53TG1 group and siTP53TG1+α-PD-L1 group were subcutaneously injected with siTP53TG1-transfected CT26 cells (0.2×107 cells) to construct colon cancer models. In siTP53TG1 group, each mouse was intraperitoneally injected with 300 μg of physiological saline on the 8th, 10th and 12th days. Similarly, mice in siTP53TG1+α-PD-L1 group were subjected to intraperitoneal injection of α-PD-L1 (300 μg) on the 8th, 10th and 12th days. Mice in normal group were normally fed and intraperitoneally injected with 300 μg of physiological saline on the 8th, 10th and 12th days.
14 days later, mice were injected with 50 mg/kg sodium pentobarbital and euthanized. Then one of their eyes was removed with tweezers, and their blood was dropped into a prepared EP tube containing EDTA anticoagulant for follow-up experiments. Subsequently, mice were euthanized by cervical dislocation, and their tumors and spleens were dissected and isolated. After these, the weight, length and width of mouse tumors in each group were measured, and the tumor volumes were calculated with the following formula: tumor volume = ((width)2 × length)/2. Peripheral blood, spleens and tumors of mice were applied for the subsequent flow cytometry to determine the proportion of immune cell subsets in each group. Tumor tissues were extracted for quantitative real-time polymerase chain reaction (qRT-PCR), Western blot, terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) and immunohistochemistry, and tumor supernatant was used for ELISA.
TUNEL assay
The apoptosis was detected using the TUNEL kit (C1091, Beyotime). The tumor tissue samples of the mice were paraffin-embedded and sectioned into 4-μm-thick slices, followed by digestion with protease K for 30 min. The slices were placed in a wet box and incubated with TUNEL reaction fluid at 37°C for 1 h. After the addition of streptavidin-horseradish peroxidase (HRP) working solution, the slices were incubated at room temperature for another 30 min. Next, the slices were stained with 3,3’-diaminobenzidine (DAB) chromogen and counterstained with hematoxylin, followed by being dehydrated, cleared and sealed. Finally, the slices were observed under an optical microscope (BX40, Olympus, Japan). The nuclei of apoptotic cells were brown-yellow or brownish, while the nuclei of normal cells were blue.
ELISA
The levels of interleukin (IL)-2, IL-4, IL-10, and interferon-γ (IFN-γ) secreted by murine splenic T cells and mouse tumor tissues were detected using ELISA. Moreover, the levels of IFN-γ, IL-10 and tumor necrosis factor-β (TNF-β) in tumor tissue supernatant were determined by ELISA as well. The specific steps were performed according to the instructions of ELISA kit. Briefly, the standard substances and samples were placed into 96-well plates and cultured at room temperature for 30 minutes (min), after which detection agent was added to each well for 1-h cultivation in a humid incubator at 37°C. Next, the enzymatic solution was added and placed at room temperature for 30 min. Eventually, OD value was measured at a wavelength of 450 nm under the help of a microplate reader (Model 680, Bio-Rad, USA). Herein, IFN-γ ELISA Kit (PI508), IL-2 ELISA Kit (PI575), IL-4 ELISA Kit (PI612), and IL-10 ELISA Kit (PI522) were purchased from Beyotime. TNF-β ELISA Kit (MM-0131M2) was bought from MEIMIAN (Jiangsu, China).
qRT-PCR
Total RNA was extracted by Trizol reagent (R1100, Solarbio), and the concentration and purity of total RNA were determined by Nano Drop 2000 spectrophotometer (Thermo). cDNA synthesis kit (AE301-02, TransGen Biotech, Shanghai, China) was used to reversely transcribe total RNA (1 µg). QPCR was conducted with QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Waltham, Massachusetts, USA) and SYBR Premix Ex Taq (Takara), and PCR amplification was performed as follows: pretreatment at 94°C for 5 min, and 35 cycles of 94°C for 45 seconds (s), 58°C for 1 min and 72°C for 1 min, followed by extension at 72°C for 5 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The expression of relative gene mRNA was measured by 2-ΔΔCT method [27]. The primers were listed in Table 2.
Table 2.
The primer sequences for qRT-PCR
| Forward Prime: 5’-3’ | Reverse Prime: 5’-3’ | |
|---|---|---|
| TP53TG1 | CAGCTTCCTGCATGATGCTG | GACCTGCCAGCTCTCAGAG |
| PD-L1 | GCTATGGTGGTGCCGACTAC | TTGGTGGTGGTGGTCTTACC |
| CCL17 | GGGAGTGCTGCCTGGAGTAC | TACAAAAACGATGGCATCCCT |
| CCL22 | TGCCGTGATTACGTCCGTTA | AAGGTTAGCAACACCACGCC |
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
Western blot
Total protein was extracted by Radio-Immunoprecipitation Assay (RIPA) Lysis Buffer (P0013C, Beyotime), and the protein concentration was measured by Bicinchoninic Acid (BCA) Protein Assay Kit (P0009, Beyotime). Specifically, the total proteins (20 μg) were electrophoresed through SDS-PAGE (P0052B, Beyotime) and transferred to PVDF membrane (ISEQ00010, Solarbio). After being blocked in 5% non-fat milk for 1 h, the membrane was probed first with primary antibodies at 4°C overnight and then with secondary antibodies for 2 h. Enhanced chemiluminescence substrate kit (PE0010, Solarbio) was used to stain the protein membrane, and the protein bands on the membrane were analyzed by Image Lab Software (Bio-Rad). The primary antibodies used for Western blot included those against PD-L1 (ab233482, 1:100, Abcam), GAPDH (ab181602, 1:2000, Abcam), STAT1 (ab3987, 1:2000, Abcam), phosphorylated (p)-STAT1 (phospho Y701; ab30645, 1:500, Abcam), STAT2 (ab32367, 1:2000, Abcam), p-STAT2 (phospho Y690; ab53132, 1:300, Abcam), p-STAT3 (phospho Y705; ab76315, 1:2000, Abcam) and STAT3 (ab119352, 1:5000, Abcam). The secondary antibodies adopted here comprised goat anti-rabbit IgG H&L (HRP, ab205718, 1:2000, Abcam) and goat anti-mouse IgG H&L (HRP, ab205719, 1:2000, Abcam) antibodies. GAPDH was used as an internal standard.
Statistical analysis
All experiments were repeated at least three times. SPSS19.0 (SPSS Inc., USA) was employed for statistical analysis. All data were expressed as the mean ± standard deviation. Analysis of variance (ANOVA) was applied for evaluating significant differences in all groups. P<0.05 was considered to be statistically significant.
Results
TP53TG1 was negatively correlated with PD-L1
To explore the relationship between PD-L1 and TP53TG1, bioinformatics analysis was conducted. The results showed that there was a negative correlation between TP53TG1 and PD-L1 in DLBC, BLCA, COAD, LIHC, MESO, KIRC, PRAD, PAAD and THCA (Figure 1A-I, P<0.05). Thus, the up-regulation of PD-L1 in colon cancer tissues was associated with the down-regulation of TP53TG1.
Figure 1.
Correlation analysis between TP53TG1 and PD-L1 (CD274) in cancer. TP53TG1 was negatively correlated with PD-L1 in lymphoid neoplasm diffuse large B-cell lymphoma (A), bladder urothelial carcinoma (B), colon adenocarcinoma (C), liver hepatocellular carcinoma (D), mesothelioma (E), kidney renal clear cell carcinoma (F), prostate adenocarcinoma (G), pancreatic adenocarcinoma (H) and thyroid carcinoma (I).
TP53TG1 up-regulation inhibited STAT pathway and PD-L1 level in tumor cells
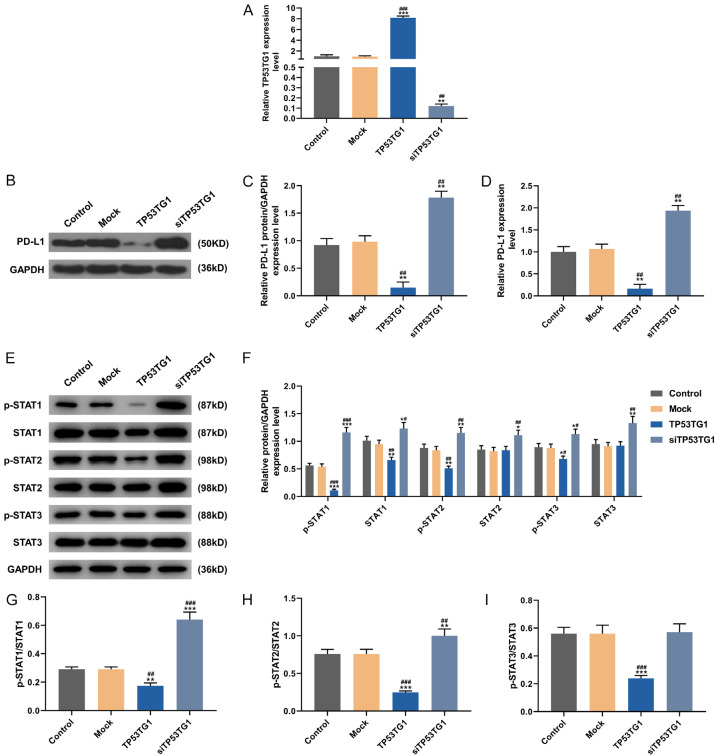
Subsequently, in vitro cell experiments were carried out to further determine the regulatory relationship between TP53TG1 and PD-L1 and delve into the corresponding mechanism. CT26 cells were transfected with TP53TG1 overexpression plasmid and siTP53TG1. Strikingly, TP53TG1 expression was higher in TP53TG1 group, while being slightly lower in siTP53TG1 group, as compared with that in control group and mock group (Figure 2A, P<0.001). After the transfection, the expression of PD-L1 in each group was also detected by qRT-PCR and Western blot. Compared with those in control group and mock group, the protein (Figure 2B, 2C) and mRNA (Figure 2D) levels of PD-L1 were noticeably decreased in TP53TG1 group, but were explicitly increased in siTP53TG1 group (P<0.001).
Figure 2.
Silencing of TP53TG1 inhibited PD-L1 expression and the activation of STAT pathways in CT26 cells. A: The transfection efficiency of TP53TG1 overexpression plasmid was evaluated by qRT-PCR in CT26 cells. B-D: The protein and mRNA expressions of PD-L1 in different groups were detected by Western blot and qRT-PCR. E-I: The protein levels of p-STAT1, STAT1, p-STAT2, STAT2, p-STAT3 and STAT3 in different groups were determined by Western blot. *P<0.05, **P<0.01 vs. Control; #P<0.05, ##P<0.01 vs. Mock.
Likewise, Western blot was performed to detect the activation of STAT pathway. As compared with those in control group and mock group, the expression levels of p-STAT1, STAT1, p-STAT2, and p-STAT3 were down-regulated in TP53TG1 group, while being up-regulated in siTP53TG1 group (Figure 2E, 2F, P<0.05). Besides, STAT2 and STAT3 levels in siTP53TG1 group were also higher than those in control group and mock group (Figure 2F, P<0.05). Relative to those in control group and mock group, the levels of p-STAT1/STAT1 (Figure 2G) and p-STAT2/STAT2 (Figure 2H) were remarkable lower in TP53TG1 group, but were higher in siTP53TG1 group (P<0.05); while the level of p-STAT3/STAT3 (Figure 2I) was also markedly lower in TP53TG1 group, but didn’t differ in siTP53TG1 group (P<0.05). Collectively, overexpression of TP53TG1 inhibited while knockdown of TP53TG1 promoted p-STAT expression. These results suggested that the inhibition of STAT pathway was possibly related to the overexpression of TP53TG1.
Likewise, the expressions of PD-L1, p-STAT and STAT were measured by Western blot after the transfection of siTP53TG1, siSTAT1, siSTAT2, siSTAT3 and corresponding siNCs into cells. Compared with those in mock+siNC group, the levels of PD-L1, p-STAT1, p-STAT2, p-STAT3, STAT1, STAT2, STAT3, p-STAT1/STAT1 and p-STAT2/STAT2 were evidently up-regulated in siTP53TG1+siNC group, but the levels of PD-L1, p-STAT1 and STAT1 in mock+STAT1 group, the levels of p-STAT2 and STAT2 in mock+STAT2 group, as well as the levels of PD-L1, p-STAT3, STAT3 and p-STAT3/STAT3 in mock+STAT3 group were all down-regulated (Figure 3A-I, P<0.05). Meanwhile, p-STAT2/STAT2 level in mock+siSTAT2 group was higher than that in mock+siNC group (P<0.05). In siTP53TG1+siSTAT1 group, we observed that the levels of PD-L1, p-STAT1, STAT1 and p-STAT1/STAT1 were lower than those in siTP53TG1+siNC group, yet they were higher than that in mock+siSTAT1 group (Figure 3A-C, P<0.05). In siTP53TG1+siSTAT2 group, the levels of p-STAT2 and STAT2 were lower than those in siTP53TG1+siNC group, whereas the level of PD-L1 was higher than that in mock+siSTAT2 group (Figure 3D-F, P<0.05). In addition, p-STAT2/STAT2 level in siTP53TG1+siSTAT2 group was lower than that in mock+siSTAT2 group, but was higher than that in siTP53TG1+siNC group (Figure 3D-F, P<0.05). In siTP53-TG1+siSTAT3 group, the levels of PD-L1, p-STAT3 and STAT3 were lower than those in siTP53TG1+siNC group, while the levels of PD-L1, p-STAT3 and p-STAT3/STAT3 were higher than those in mock+siSTAT3 group (Figure 3G-I, P<0.05). Meanwhile, siTP53TG1-induced activation of STAT pathway and the up-regulation of PD-L1 could be partly reversed by silencing STAT1 and STAT3. Interestingly, siSTAT1 had a stronger effect on the expression of PD-L1 than siSTAT3, yet siSTAT2 exerted no significant effect on the expression of PD-L1.
Figure 3.
STAT silencing partially reversed the effect of siTP53TG1 on STAT pathways. A-C: The effects of siTP53TG1 and siSTAT on the expressions of PD-L1, p-STAT1 and STAT1 were detected by Western blot and qRT-PCR. D-F: The expressions of PD-L1, p-STAT2 and STAT2 were determined by Western blot and qRT-PCR. G-I: The expressions of PD-L1, p-STAT3 and STAT3 were detected by Western blot and qRT-PCR. *P<0.05, **P<0.01 vs. Mock+siNC; #P<0.05, ##P<0.01 vs. siTP53TG1+siNC; ^P<0.05, ^^P<0.01 vs. Mock+siSTAT1; ∆P<0.05, ∆∆P<0.01 vs. Mock+siSTAT2; ‡P<0.05, ‡‡P<0.01 vs. Mock+siSTAT3.
In view of the above experimental results, the relationship between PD-L1 and STAT1 in different cancers was explored by bioinformatics analysis. The data exhibited that there was a remarkable positive correlation between PD-L1 with STAT1 in COAD, BLCA, BRCA, CESC, STAD, ESCA, HNSC, LIHC and LUAD (Figure 4A-I). Taken together, increasing the TP53TG1 expression could suppress STAT pathway to reduce the expression of PD-L1 in tumor cells.
Figure 4.
Correlation analysis between STAT1 and CD274 in cancer. PD-L1 was positively correlated with STAT1 in colon adenocarcinoma (A), bladder urothelial carcinoma (B), breast invasive carcinoma (C), cervical and endocervical cancers (D), stomach adenocarcinoma (E), esophageal carcinoma (F), head and neck squamous cell carcinoma (G), liver hepatocellular carcinoma (H) and lung adenocarcinoma (I).
The anti-tumor effect of TP53TG1 may be enhanced by promoting the proliferation of T cells and the secretion of related cytokines
To investigate the effect of PD-L1 expression on T cells, CD3+T cells from the spleens of mice were isolated and then incubated with CT26 cells in the presence or absence of α-PD-L1 and TP53TG1. CCK-8 assay and flow cytometry were used to detect the T cell viability and cell cycle. As shown in Figure 5A, compared with that in mIgG+T group, the cell viability in α-CD3+T group was increased (P<0.05). However, the viability of α-CD3-induced T cells could be reduced by CT26 cells (Figure 5A, P<0.05). In the meantime, the cell viability in α-CD3+α-PD-L1+T+CT26 group was stronger than that in α-CD3+T+CT26 group and α-CD3+mIgG+T+CT26 group (Figure 5A, P<0.05). Similarly, the cell viability in α-CD3+T+TP53TG1 group was elevated as compared with that in α-CD3+T+mock group (Figure 5A, P<0.05). Besides, α-CD3+T group presented a lower proportion of cells at G1 phase and a higher proportion of cells at S and G2 phases relative to mIgG+T group (Figure 5B, 5C, P<0.05). Compared with α-CD3+T group, increased proportion of cells at G1 phase but decreased proportion of cells at G2 phase were observed in α-CD3+T+CT26 group (Figure 5B, 5C, P<0.05). In α-CD3+α-PD-L1+T+CT26 group, the proportion of cells at G1 phase was less but the proportion of cells at G2 phase was more than those in α-CD3+T+CT26 group and α-CD3+mIgG+T+CT26 group (Figure 5B, 5C, P<0.05). In α-CD3+T+TP53TG1 group, the proportion of cells at G1 phase was less yet the proportion of cells at G2 phase was more than those in α-CD3+T+mock group (Figure 5B, 5C, P<0.05). Notably, α-PD-L1 could promoted T cell viability and partially reversed the inhibiting effect induced by co-treatment of α-CD3 and CT26 cells; overexpressed TP53TG1 also led to promoted T cell viability and partially reversed the inhibiting effect induced by co-treatment of α-CD3 and mock. These data signified that overexpressed TP53TG1 and PD-L1 antibody regulated cell cycle, thereby rescuing the viability of T cells suppressed by tumor cells.
Figure 5.
Effects of cancer cells, PD-L1 monoclonal antibody (α-PD-L1 mAb) and TP53TG1 on T cells. A: The viability of T cells was detected by CCK-8 assay. B, C: The flow cytometer was used to determine cell cycle, and the proportion of cells at G1, S and G2 phases in different groups was quantified. D-G: The levels of IL-2, IL-4, IL-10 and IFN-γ in each group were detected by ELISA. T cells were obtained from spleens of mice. *P<0.05, **P<0.01 vs. mIgG+T; #P<0.05, ##P<0.01 vs. α-CD3+T; ^P<0.05, ^^P<0.01 vs. α-CD3+T+CT26; ΔP<0.05, ΔΔP<0.01 vs. α-CD3+mIgG+T+CT26; ‡P<0.05, ‡‡P<0.01 vs. α-CD3+T+mock.
To measure the levels of IL-2, IL-4, IL-10 and IFN-γ, ELISA was performed. Compared with those in mIgG+T group, the levels of IL-2 (Figure 5D), IL-4 (Figure 5E), IL-10 (Figure 5F) and IFN-γ (Figure 5G) in α-CD3+T group were up-regulated (P<0.05), and the increased levels induced by α-CD3 could be partly down-regulated by CT26 cells (P<0.05). In α-CD3+α-PD-L1+T+CT26 group, the levels of IL-2, IL-4, IL-10 and IFN-γ were higher than those in α-CD3+T+CT26 and α-CD3+mIgG+T+CT26 groups (P<0.05). Similarly, α-CD3+T+TP53TG1 group had higher levels of IL-2, IL-4, IL-10 and IFN-γ relative to α-CD3+T+mock group (P<0.05). In light of the above information, it was plausible to conclude that PD-L1 antibody and overexpressed TP53TG1 partially reversed the effects of CT26, and up-regulated the expression levels of IL-2, IL-4, IL-10 and IFN-γ.
The cytotoxic effects of CD8+ T cells and NK cells on CT26 cells could be enhanced by inhibiting PD-L1 expression with overexpressed TP53TG1 and PD-L1 antibody
To investigate whether PD-L1 affected the cytotoxicity of CD8+ T and NK cells to CT26 cells, flow cytometry was used to separate CD8+ T and NK cells from the spleens of the mice. Subsequently, the cytotoxicity of CD8+ T cells (Figure 6A) and NK cells (Figure 6B) targeting CT26 cells was detected by LDH non-radioactive assay. Compared with that in control group and mIgG group, the cytotoxicity was increased in α-PD-L1 group and TP53TG1 group, but was decreased in siTP53TG1 group (Figure 6A, 6B, P<0.05). In addition, the cytotoxicity in mock group was lower than that in TP53TG1 group, but was higher than that in siTP53TG1 group (P<0.05). In siTP53TG1+α-PD-L1 group, the cytotoxicity was lower than that in α-PD-L1 group, while being higher than that in siTP53TG1 group (P<0.05). These results signified that PD-L1 antibody and overexpressed TP53TG1 improved while siTP53TG1 repressed the cytotoxic effects of CD8+ T cells and NK cells on CT26 cells.
Figure 6.

Cytotoxic effects of CD8+ T and NK cells on CT26 cells. A: The effects of α-PD-L1 and TP53TG1 on the cytotoxicity of CD8+ T cells to CT26 were evaluated by LDH nonradioactive assay. CD8+ T cells from spleens of mice were selected by flow cytometry, CT26 cells served as the target cells, and CD8+ T cells acted as effector cells (effector-target ratio = 5:1). B: The effects of α-PD-L1 and TP53TG1 on the cytotoxicity of NK cells to CT26 were detected by LDH nonradioactive assay. NK cells from spleens of mice were selected by flow cytometry, CT26 cells acted as the target cells, and NK cells functioned as effector cells (effector-target ratio = 5:1). *P<0.05, **P<0.01 vs. Control; #P<0.05, ##P<0.01 vs. mIgG; ^P<0.05, ^^P<0.01 vs. α-PD-L1; ΔP<0.05, ΔΔP<0.01 vs. TP53TG1; ‡P<0.05, ‡‡P<0.01 vs. siTP53TG1.
The tumor growth induced by CT26 cells and siTP53TG1 was dampened by inhibiting PD-L1 expression via PD-L1 antibody
The specific mechanism of PD-L1 affecting the occurrence and development of colon cancer was analyzed via animal experiments. A total of 24 BALB/c mice were divided into 6 groups (n = 6): normal group, control group, α-PD-L1 group, mock group, siTP53TG1+α-PD-L1 group and siTP53TG1 group. After 14 days, the average tumor volume of the mice in each group was 100 mm3, indicating the successful establishment of the model (Figure 7A).
Figure 7.
Roles of α-PD-L1 and siTP53TG1 in tumor growth. A: The mouse model was successfully constructed, and the tumor was excised and assigned into different groups. B: The tumor volume of mice in each group. C: The tumor weight of mice in each group. *P<0.05, **P<0.01 vs. Control; #P<0.05, ##P<0.01 vs. α-PD-L1; ^P<0.05, ^^P<0.01 vs. mock; ∆P<0.05, ∆∆P<0.01 vs. siTP53TG1.
After the mice were euthanized, the tumor weight and volume of mice in each group were measured and compared (Figure 7B, 7C). We found that the tumor weight and volume were decreased in α-PD-L1 group, but were increased in siTP53TG1 group, as compared with those in control group (Figure 7B, 7C, P<0.05). Moreover, the tumor weight and volume in siTP53TG1 group were higher than those in mock group (P<0.05). In siTP53TG1+α-PD-L1 group, the tumor weight and volume were higher than those in α-PD-L1 group, but were lower than those in siTP53TG1 group (P<0.05). These findings suggested that PD-L1 antibody inhibited the tumor growth, and partially reversed the promoting effect of siTP53TG1 on tumor growth.
The proportion of CD8+ T cells and NK cells reduced by siTP53TG1 was reversed by inhibiting PD-L1 expression via PD-L1 antibody, and the trafficking of Tregs regulated by siTP53TG1 was counteracted via PD-L1 antibody
The peripheral blood and tumors of mice in each group were isolated, and the proportion of immune cell subsets in peripheral blood and tumors was detected by flow cytometry. As denoted in Tables 3, 4, in comparison with normal group, the proportion of CD8 +CD3 +/CD3 + was higher in control group, while the proportion of NK cells (Table 4) was lower in control group (P<0.05). However, compared with that in control group, the proportions of CD8 +CD3 +/CD3 + and NK cells were increased in α-PD-L1 group (P<0.05), but were decreased in siTP53TG1 group (P>0.05). Furthermore, the proportions of CD8 +CD3 +/CD3 + and NK cells in siTP53TG1+α-PD-L1 group were more than that in siTP53TG1 group (P<0.05).
Table 3.
Percentage of CD8+CD3+/CD3+ in different position
| Group | Peripheral Blood | Tumor |
|---|---|---|
| Normal | 25.6±1.2 | - |
| Control | 31.5±1.6‡‡ | 11.6±0.8 |
| α-PD-L1 | 38.2±1.4** | 20.2±1.4** |
| Mock | 30.9±1.4 | 11.4±0.9 |
| siTP53TG1 | 28.4±1.3 | 9.7±0.7 |
| siTP53TG1+α-PD-L1 | 37.0±1.5ΔΔ | 18.8±1.1ΔΔ |
P<0.01 vs. normal;
P<0.01 vs. control;
P<0.01 vs. siTP53TG1.
Table 4.
Percentage of NK cells in different position
| Group | Peripheral Blood | Tumor |
|---|---|---|
| Normal | 4.11±0.46 | - |
| Control | 3.17±0.41‡ | 4.21±0.36 |
| α-PD-L1 | 4.02±0.40* | 5.16±0.52* |
| Mock | 3.22±0.45 | 4.35±0.38 |
| siTP53TG1 | 3.00±0.32 | 3.79±0.35 |
| siTP53TG1+α-PD-L1 | 3.80±0.35Δ | 4.84±0.46Δ |
P<0.05 vs. normal;
P<0.05 vs. control;
P<0.05 vs. siTP53TG1.
In peripheral blood, the proportion of Tregs in control group was less than that in normal group. When compared with that in control group, the proportion of Tregs in α-PD-L1 group was increased significantly, but was slightly decreased in siTP53TG1 group. Besides, the proportion of Tregs in siTP53TG1+α-PD-L1 group was lower than that in α-PD-L1 group (Table 5, P<0.05). In tumor, the proportion of Tregs was the lowest in α-PD-L1 group while being the highest in siTP53TG1 group as comparison with control, mock and siTP53TG1+α-PD-L1 groups (P<0.05). Apart from these, we found that PD-L1 antibody increased the proportion of CD8+ T cells, NK cells and peripheral blood Tregs, yet decreased the proportion of Tregs in tumor. Notably, siTP53TG1 had no significant inhibitory effect on the proportion of CD8+ T cells and NK cells, but reduced the proportion of Tregs in the peripheral blood while promoting the proportion of Tregs in tumor. Moreover, such effects of siTP53TG1 could be reversed by PD-L1 antibody.
Table 5.
Percentage of Tregs in different position
| Group | Peripheral Blood | Tumor |
|---|---|---|
| Normal | 2.35±0.24 | - |
| Control | 1.42±0.25‡‡ | 3.94±0.27 |
| α-PD-L1 | 1.93±0.21* | 2.17±0.29** |
| Mock | 1.38±0.21 | 4.15±0.28 |
| siTP53TG1 | 1.23±0.26 | 6.03±0.45**,^^ |
| siTP53TG1+α-PD-L1 | 1.28±0.25# | 5.14±0.37##,Δ |
P<0.01 vs. normal;
P<0.05 vs. control;
P<0.01 vs. control;
P<0.05 vs. α-PD-L1
P<0.01 vs. α-PD-L1;
P<0.01 vs. mock;
P<0.05 vs. siTP53TG1.
Decreased cytokine expressions and increased chemokine expressions in tumor tissues caused by siTP53TG1 were reversed by PD-L1 antibody
Expressions of IFN-γ, TNF-β and IL-10 in tumor tissues of mice in each group were detected by ELISA. As shown in Figure 8A-C, the levels of IFN-γ, TNF-β and IL-10 were up-regulated in α-PD-L1 group, but were down-regulated in siTP53TG1 group compared with those in control group (P<0.05). Meanwhile, the levels of IFN-γ, TNF-β and IL-10 in siTP53TG1 group were lower than those in mock group (Figure 8A-C, P<0.05). In comparison with those in siTP53TG1+α-PD-L1 group, the levels of IFN-γ, TNF-β and IL-10 were higher in α-PD-L1 group, but were lower in siTP53TG1 group (Figure 8A-C, P<0.05). From the data above, it could be found that siTP53TG1 inhibited while PD-L1 antibody promoted the expressions of IFN-γ, TNF-β and IL-10, and PD-L1 antibody reversed the effects produced by siTP53TG1.
Figure 8.
Effects of α-PD-L1 and siTP53TG1 on cytokine expressions and apoptosis. A-C: The levels of IFN-γ, TNF-β and IL-10 in each group were detected by ELISA. D: The levels of CCL17 and CCL22 in each group were quantified by qRT-PCR. E: The level of TP53TG1 in each group was detected by qRT-PCR. F, G: The apoptosis was detected by TUNEL. *P<0.05, **P<0.01 vs. Control; #P<0.05, ##P<0.01 vs. α-PD-L1; ^P<0.05, ^^P<0.01 vs. mock; ΔP<0.05, ΔΔP<0.01 vs. siTP53TG1.
The expressions of CCL17 and CCL22 in tumor tissues of mice in each group were detected by qRT-PCR. Compared with those in control group, the levels of CCL17 and CCL22 were diminished in α-PD-L1 group, but were boosted in siTP53TG1 group (Figure 8D, P<0.05). Besides, siTP53TG1 group exhibited higher levels of CCL17 and CCL22 than mock group (Figure 8D, P<0.001). In siTP53TG1+α-PD-L1 group, the levels of CCL17 and CCL22 were higher than those in α-PD-L1 group, but were lower than those in siTP53TG1 group (Figure 8D, P<0.001). From these results, it could be seen that TP53TG1 silencing promoted while PD-L1 antibody inhibited the expressions of CCL17 and CCL22 in tumor tissues, and PD-L1 antibody reversed the effect produced by siTP53TG1.
The expression of TP53TG1 in tumor tissues of each group was quantified by qRT-PCR. We found that TP53TG1 level in siTP53TG1 group was lower than that in control and mock groups (Figure 8E, P<0.001), and TP53TG1 level in siTP53TG1+α-PD-L1 group was lower than that in α-PD-L1 group (P<0.001). Nevertheless, no significant difference was observed in the level of TP53TG1 between the control and α-PD-L1 groups. The aforementioned findings suggested that PD-L1 antibody had no significant effect on TP53TG1 expression, and TP53TG1 unidirectionally regulated PD-L1 expression.
The inhibitory effect of TP53TG1 knockdown on the apoptosis of mouse tumor cells was possibly achieved via activating STAT pathway to promote PD-L1 expression, while PD-L1 antibody reversed the effect of STAT pathway
The apoptosis was detected by TUNEL assay, where the nucleus of apoptotic cells was brown-yellow or brownish, and the nucleus of normal cells was blue. As delineated in Figure 8F, 8G, fewer apoptotic cells were found in the control group and the siTP53TG1 group. Compared with those in the control group, the apoptotic cells in the α-PD-L1 group were remarkably increased. Furthermore, the apoptotic cells in the siTP53TG1+α-PD-L1 group were more than those in the siTP53TG1 group but were less than those in the α-PD-L1 group. PD-L1 antibody promoted the apoptosis of CT26 cells, but dwindled PD-L1 expression. However, the knockdown of TP53TG1 inhibited the apoptosis, and led to the increased PD-L1 expression and decreased TP53TG1 expression.
The effects of α-PD-L1 and siTP53TG1 on STAT pathway were detected by Western blot. Compared with those in control group, the levels of p-STAT1, p-STAT2 and p-STAT3 were down-regulated in α-PD-L1 group, but were up-regulated in siTP53TG1 group (Supplementary Figure 1A, 1B, P<0.05). Meanwhile, the levels of p-STAT1, p-STAT2 and p-STAT3 in siTP53TG1 group were higher than those in mock group (Supplementary Figure 1A, 1B, P<0.001). In siTP53TG1+α-PD-L1 group, the levels of p-STAT1, p-STAT2 and p-STAT3 were higher than those in α-PD-L1 group, but were lower than those in siTP53TG1 group (Supplementary Figure 1A, 1B, P<0.001). STAT3 level in siTP53TG1+α-PD-L1 group was diminished relative to that in siTP53TG1 group and α-PD-L1 group (Supplementary Figure 1A, 1B, P<0.001). Moreover, the expression levels of p-STAT1/STAT1, p-STAT2/STAT2 and p-STAT3/STAT3 were down-regulated in α-PD-L1 group, but were up-regulated in siTP53TG1 group, as compared with those in control group (Supplementary Figure 1C-E, P<0.001). Additionally, the expression levels of p-STAT1/STAT1, p-STAT2/STAT2 and p-STAT3/STAT3 in siTP53TG1+α-PD-L1 group were lower than those in siTP53TG1 group, but were higher than those in α-PD-L1 group (Supplementary Figure 1C-E, P<0.001). At the same time, the expression levels of p-STAT1/STAT1, p-STAT2/STAT2 and p-STAT3/STAT3 in siTP53TG1 group were elevated as compared with those in mock group (Supplementary Figure 1C-E, P<0.001). These data collectively indicated that PD-L1 antibody inhibited the activation of STAT pathways by decreasing PD-L1 expression, while silencing of TP53TG1 activated STAT pathways. Thus, the inhibitory effect of TP53TG1 knockdown on the apoptosis of mouse tumor cells may be achieved by activating STAT pathway to promote PD-L1 expression.
Discussion
In this study, we found a significant negative correlation between TP53TG1 and PD-L1 in certain cancers, particularly in COAD. This result further proved that TP53TG1 and PD-L1 played important roles in promoting the progression of colon cancer, and there was a correlation between their expression levels in colon cancer. In order to further explore the roles of TP53TG1 and PD-L1 in colon cancer, this study analyzed the regulatory mechanism of TP53TG1 on the expression of PD-L1. Studies have revealed that inhibiting the activation of the JAK/STAT pathway can repress the proliferation and growth of colon cancer cells [28,29]. It has been reported that orexin A suppresses the expression of exosomal PD-L1 in colon cancer by inhibiting JAK2/STAT3 signaling pathway [30], signifying that the expression of PD-L1 is positively correlated with the JAK2/STAT3 pathway. In this study, we also found the expression of PD-L1 was inhibited by STAT3 silencing and the tendency was further strengthened when TP53TG1 was overexpressed in CT26 cells, along with the inactivation of STAT pathway. These findings suggested that TP53TG1 possibly dampened the PD-L1 expression via inhibiting STAT signaling pathway. Apart from these, we found that STAT1 was the main conditional factor.
The occurrence of tumor is closely related to immune imbalance of the body [31,32]. Anti-tumor effector cells play pivotal roles in anti-tumor immune response, among which T cells, NK cells and macrophages are the most important effector cells [33,34]. PD-L1 has been reported to negatively regulate T lymphocytes, as PD-L1 inhibits the proliferation of T cells and the secretion of cytokines, and induces the apoptosis of T cells [35,36]. In this study, we found that CT26 cells inhibited the proliferation of T cells and caused cell cycle arrest at G1 phase, and the changes could be partly rescued by the overexpressed TP53TG1 and PD-L1 antibody. Moreover, under the inhibition of T cell proliferation, the expression levels of IL-2, IL-4, IL-10 and IFN-γ were down-regulated at the same time, but were up-regulated intensively when the cells were transfected with TP53TG1 overexpression plasmid and reacted with PD-L1 antibody. As an important indicator to evaluate the immune function of the cells, IL-2 can potentiate the proliferation of T cells, the activity of NK cells and the killing effect of T cells [37]. IL-4 can promote the proliferation of activated T cells and the immune function of the body [38]. IL-10 exerts inhibiting effects on both immune cells and tumor cells [39-41]. IFN-γ is produced by activated T cells and natural killer cells, and can act as immune regulator owing to its antiviral activity [42]. These above results indicated that the overexpressed TP53TG1 and PD-L1 antibody may promote the proliferation of T cells by regulating the secretion of related cytokines, thus inhibiting the immune escape mediated by tumor cells.
CD8+ T cells and NK cells are the main anti-tumor effector cells and play highly important roles in the anti-tumor immune response [43,44]. CD8+ T cells, which are cytotoxic T lymphocytes, can eliminate cancer cells through antigen-specific killing effect. The activation of CD8+ T cells, together with the production, maturation and differentiation of cytotoxic T cells, depends on the activation of CD4+ T cells and the release of IL-2 and other cytokines [45,46]. NK cells, which can directly kill cancer cells through their cytotoxicity and promote the anti-tumor immune response mediated by effector T cells, are the first-line defense for the anti-tumor immune response of the body [47-49]. NK cells can secrete cytokines (such as TNF-β, IFN-γ, etc.), perforin and serine esterase, thereby playing immunomodulatory roles [50]. Notably, IFN-γ can activate the killing activity of NK cells, whereas IFN-γ and TNF-β can activate caspase-3 cascade reaction and induce apoptosis. It has been evidenced that PD-1/PD-L1 pathway has inhibitory effects on the activation and proliferation of CD8+ T cells and CD4+ T cells, and thus tumor cells can escape from the killing effect of cytotoxic T cells [51,52]. In this study, we found that cytotoxicity of CD8+ T/NK cells to tumor cells was inhibited by siTP53TG, suggesting that the overexpression of PD-L1 could enhance the resistance of cancer cells to CD8+ T/NK cells, thereby promoting tumor progression. However, the cytotoxicity was more intense when PD-L1 expression was inhibited by overexpressed TP53TG and PD-L1 antibody. Moreover, the expressions of IFN-γ, TNF-β and IL-10 in tumor tissues were inhibited by siTP53TG1, which were reversed by PD-L1 antibody. The inhibition of PD-L1 expression could enhance the killing effect of cytotoxic T cells and NK cells to cancer cells, and improve cytokine secretion and the tumor cell apoptosis, thus inhibiting the immune escape of tumor cells.
CD4+ CD25+ Tregs can hinder tumor-specific immunity and cause tumor immune escape, playing vital roles in regulating immune response [53,54]. Trafficking of Tregs provides a solid foundation for realization of immune function [55]. In this study, we found that the trafficking pattern of Tregs in the colon cancer mouse model was aggregated from peripheral blood to tumor tissues and spleen. The main molecules that affect the trafficking of Tregs to tumor sites are chemokines and their receptors as well as some adhesion molecules [56,57]. The increased expressions of CCL17 and CCL22 in tumor tissue can attract Tregs to gather into the tumor tissue, resulting in the immunosuppressive state of the tumor microenvironment [58]. Lynda J. Hatam et al. have shown that CCL17 and CCL22 can recruit Tregs into the tissues in which CCL17 and CCL22 are expressed [59]. Herein, significant up-regulation of CCL17 and CCL22 in tumor tissues was observed when TP53TG1 was lowly expressed, and obvious enrichment of Tregs was also found in tumor and spleen tissues. These results were in accordance with findings as previously reported [58,59]. However, blocking the PD-1/PD-L1 pathway can cause the redistribution of CD4+ CD25+ Tregs in the colon cancer model mice, as manifested by the increased proportion of Tregs in the peripheral blood and the decreased proportion of Tregs in the tumor and spleen tissues. At the same time, we observed that CCL17 and CCL22 expressions were also decreased in tumor tissues, revealing that chemokine expression reduction through inhibition of PD-L1 expression can affect the trafficking of Tregs, contributing to weakened immunosuppressive effects of tumor cells.
In conclusion, we find that PD-L1 is negatively correlated with TP53TG1. TP53TG1 inhibits the expression of PD-L1 by suppressing the activation of STAT pathway. On one hand, the inhibition of PD-L1 expression can promote the proliferation of T lymphocytes and the secretion of cytokine, and enhance the cytotoxicity of CD8+ T cells and NK cells to cancer cells, thus inhibiting the immune escape of tumor cells. On the other hand, the inhibition of PD-L1 expression could enhance the apoptosis of tumor cells while suppressing the migration of Tregs to tumor cells, thereby weakening the immunosuppressive effect of tumor cells, and ultimately restraining the development of colon cancer in mice.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J. Clin. Oncol. 2015;33:1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 2.O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang ZX, Cao JX, Liu ZP, Cui YX, Li CY, Li D, Zhang XY, Liu JL, Li JL. Combination of chemotherapy and immunotherapy for colon cancer in China: a meta-analysis. World J Gastroenterol. 2014;20:1095–1106. doi: 10.3748/wjg.v20.i4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284–8292. doi: 10.3748/wjg.v21.i27.8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aoyagi T, Terracina KP, Raza A, Takabe K. Current treatment options for colon cancer peritoneal carcinomatosis. World J Gastroenterol. 2014;20:12493–12500. doi: 10.3748/wjg.v20.i35.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10:909–915. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 8.Cheng M, Chen Y, Xiao W, Sun R, Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol. 2013;10:230–252. doi: 10.1038/cmi.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishikawa H. Regulatory T cells in cancer immunotherapy. Rinsho Ketsueki. 2014;55:2183–2189. [PubMed] [Google Scholar]
- 10.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronig H, Kremmler L, Haller B, Englert C, Peschel C, Andreesen R, Blank CU. Interferon-induced programmed death-ligand 1 (PD-L1/B7-H1) expression increases on human acute myeloid leukemia blast cells during treatment. Eur J Haematol. 2014;92:195–203. doi: 10.1111/ejh.12228. [DOI] [PubMed] [Google Scholar]
- 12.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee LH, Cavalcanti MS, Segal NH, Hechtman JF, Weiser MR, Smith JJ, Garcia-Aguilar J, Sadot E, Ntiamoah P, Markowitz AJ, Shike M, Stadler ZK, Vakiani E, Klimstra DS, Shia J. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol. 2016;29:1433–1442. doi: 10.1038/modpathol.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morse MA. The outlook for immune checkpoint targeting strategies in colorectal cancer. Curr Colorectal Cancer Rep. 2016 [Google Scholar]
- 15.Chen XY, Zhang J, Hou LD, Zhang R, Chen W, Fan HN, Huang YX, Liu H, Zhu JS. Upregulation of PD-L1 predicts poor prognosis and is associated with miR-191-5p dysregulation in colon adenocarcinoma. Int J Immunopathol Pharmacol. 2018;32:2058738418790318. doi: 10.1177/2058738418790318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamanishi J, Mandai M, Matsumura N, Abiko K, Baba T, Konishi I. PD-1/PD-L1 blockade in cancer treatment: perspectives and issues. Int J Clin Oncol. 2016;21:462–473. doi: 10.1007/s10147-016-0959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balar AV, Weber JS. PD-1 and PD-L1 antibodies in cancer: current status and future directions. Cancer Immunol Immunother. 2017;66:551–564. doi: 10.1007/s00262-017-1954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Incecco A, Andreozzi M, Ludovini V, Rossi E, Capodanno A, Landi L, Tibaldi C, Minuti G, Salvini J, Coppi E, Chella A, Fontanini G, Filice ME, Tornillo L, Incensati RM, Sani S, Crino L, Terracciano L, Cappuzzo F. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer. 2015;112:95–102. doi: 10.1038/bjc.2014.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125:3384–3391. doi: 10.1172/JCI80011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Yuan R, Song W, Sun J, Liu D, Li Z. PD-1, PD-L1 (B7-H1) and tumor-site immune modulation therapy: the historical perspective. J Hematol Oncol. 2017;10:34. doi: 10.1186/s13045-017-0403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Jing H, Li H. A novel cuproptosis-related lncRNA signature to predict prognosis and immune landscape of lung adenocarcinoma. Transl Lung Cancer Res. 2023;12:230–246. doi: 10.21037/tlcr-22-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49:255–260. doi: 10.1111/cpr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu K, Xu K, Liu K, Huang J, Chen J, Zhang J, Zhang N. Long noncoding RNA BC200 regulates cell growth and invasion in colon cancer. Int J Biochem Cell Biol. 2018;99:219–225. doi: 10.1016/j.biocel.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Xiao H, Liu Y, Liang P, Wang B, Tan H, Zhang Y, Gao X, Gao J. TP53TG1 enhances cisplatin sensitivity of non-small cell lung cancer cells through regulating miR-18a/PTEN axis. Cell Biosci. 2018;8:23. doi: 10.1186/s13578-018-0221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Gao Y, Li D, Cao Y, Hao B. LncRNA-TP53TG1 participated in the stress response under glucose deprivation in glioma. J Cell Biochem. 2017;118:4897–4904. doi: 10.1002/jcb.26175. [DOI] [PubMed] [Google Scholar]
- 26.Diaz-Lagares A, Crujeiras AB, Lopez-Serra P, Soler M, Setien F, Goyal A, Sandoval J, Hashimoto Y, Martinez-Cardus A, Gomez A, Heyn H, Moutinho C, Espada J, Vidal A, Paules M, Galan M, Sala N, Akiyama Y, Martinez-Iniesta M, Farre L, Villanueva A, Gross M, Diederichs S, Guil S, Esteller M. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc Natl Acad Sci U S A. 2016;113:E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3:71–85. [PMC free article] [PubMed] [Google Scholar]
- 28.Sun KX, Xia HW, Xia RL. Anticancer effect of salidroside on colon cancer through inhibiting JAK2/STAT3 signaling pathway. Int J Clin Exp Pathol. 2015;8:615–621. [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, Guo Q, Xie J, Jin D, Zhu Y. Combination of MEK inhibitor and the JAK2-STAT3 pathway inhibition for the therapy of colon cancer. Pathol Oncol Res. 2019;25:769–775. doi: 10.1007/s12253-019-00592-6. [DOI] [PubMed] [Google Scholar]
- 30.Wen J, Chang X, Bai B, Gao Q, Zhao Y. Orexin A suppresses the expression of exosomal PD-L1 in colon cancer and promotes T cell activity by inhibiting JAK2/STAT3 signaling pathway. Dig Dis Sci. 2022;67:2173–2181. doi: 10.1007/s10620-021-07077-0. [DOI] [PubMed] [Google Scholar]
- 31.Lizee G, Overwijk WW, Radvanyi L, Gao J, Sharma P, Hwu P. Harnessing the power of the immune system to target cancer. Annu Rev Med. 2013;64:71–90. doi: 10.1146/annurev-med-112311-083918. [DOI] [PubMed] [Google Scholar]
- 32.Moritz J, Stoffels I, Helfrich I, Bekeschus S. Plasma-derived oxidants and the modulation of immunsuppression in melanoma. Clinical Plasma Medicine. 2018;9:11–12. [Google Scholar]
- 33.Liu YC, Xiao YF. Macroscopic mechanical systems are entering the quantum world. Nat Sci Rev. 2015;2:9–10. [Google Scholar]
- 34.Zhang X, Liu Q, Liao Q, Zhao Y. Potential roles of peripheral dopamine in tumor immunity. J Cancer. 2017;8:2966–2973. doi: 10.7150/jca.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viry E, Noman MZ, Arakelian T, Lequeux A, Chouaib S, Berchem G, Moussay E, Paggetti J, Janji B. Hijacker of the antitumor immune response: autophagy is showing its worst facet. Front Oncol. 2016;6:246. doi: 10.3389/fonc.2016.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naidoo J, Page DB, Wolchok JD. Immune modulation for cancer therapy. Br J Cancer. 2014;111:2214–2219. doi: 10.1038/bjc.2014.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skrombolas D, Frelinger JG. Challenges and developing solutions for increasing the benefits of IL-2 treatment in tumor therapy. Expert Rev Clin Immunol. 2014;10:207–217. doi: 10.1586/1744666X.2014.875856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Boer BA, Fillie YE, Kruize YC, Yazdanbakhsh M. Antigen-stimulated IL-4, IL-13 and IFN-gamma production by human T cells at a single-cell level. Eur J Immunol. 1998;28:3154–3160. doi: 10.1002/(SICI)1521-4141(199810)28:10<3154::AID-IMMU3154>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 39.Joss A, Akdis M, Faith A, Blaser K, Akdis CA. IL-10 directly acts on T cells by specifically altering the CD28 co-stimulation pathway. Eur J Immunol. 2000;30:1683–1690. doi: 10.1002/1521-4141(200006)30:6<1683::AID-IMMU1683>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 40.Gu T, De Jesus M, Gallagher HC, Burris TP, Egilmez NK. Oral IL-10 suppresses colon carcinogenesis via elimination of pathogenicCD4(+) T-cells and induction of antitumor CD8(+) T-cell activity. Oncoimmunology. 2017;6:e1319027. doi: 10.1080/2162402X.2017.1319027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis KL, Blatner NR, Gounari F, Khazaie K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr Opin Oncol. 2013;25:637–645. doi: 10.1097/CCO.0000000000000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, Chen PL, Hwu P, Allison JP, Futreal A, Wargo JA, Sharma P. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404. e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara HMCS, Signori E, Honoki K, Georgakilas AG, Amin A, Helferich WG, Boosani CS, Guha G, Ciriolo MR, Chen S, Mohammed SI, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Fujii H, Aquilano K, Ashraf SS, Nowsheen S, Yang X, Choi BK, Kwon BS. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35(Suppl):S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Gao X, Wang X, Yang Q, Zhao X, Wen W, Li G, Lu J, Qin W, Qi Y, Xie F, Jiang J, Wu C, Zhang X, Chen X, Turnquist H, Zhu Y, Lu B. Tumoral expression of IL-33 inhibits tumor growth and modifies the tumor microenvironment through CD8+ T and NK cells. J Immunol. 2015;194:438–445. doi: 10.4049/jimmunol.1401344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li K, Baird M, Yang J, Jackson C, Ronchese F, Young S. Conditions for the generation of cytotoxic CD4(+) Th cells that enhance CD8(+) CTL-mediated tumor regression. Clin Transl Immunology. 2016;5:e95. doi: 10.1038/cti.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umeshappa CS, Nanjundappa RH, Xie Y, Freywald A, Xu Q, Xiang J. Differential requirements of CD4+ T-cell signals for effector cytotoxic T-lymphocyte (CTL) priming and functional memory CTL development at higher CD8+ T-cell precursor frequency. Immunology. 2013;138:298–306. doi: 10.1111/imm.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberg J, Huang J. CD8(+) T cells and NK cells: parallel and complementary soldiers of immunotherapy. Curr Opin Chem Eng. 2018;19:9–20. doi: 10.1016/j.coche.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sconocchia G, Eppenberger S, Spagnoli GC, Tornillo L, Droeser R, Caratelli S, Ferrelli F, Coppola A, Arriga R, Lauro D, Iezzi G, Terracciano L, Ferrone S. NK cells and T cells cooperate during the clinical course of colorectal cancer. Oncoimmunology. 2014;3:e952197. doi: 10.4161/21624011.2014.952197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurioka A, Klenerman P, Willberg CB. Innate-like CD8+ T-cells and NK cells: converging functions and phenotypes. Immunology. 2018;154:547–556. doi: 10.1111/imm.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 51.Pauken KE, Nelson CE, Martinov T, Spanier JA, Heffernan JR, Sahli NL, Quarnstrom CF, Osum KC, Schenkel JM, Jenkins MK, Blazar BR, Vezys V, Fife BT. Cutting edge: identification of autoreactive CD4+ and CD8+ T cell subsets resistant to PD-1 pathway blockade. J Immunol. 2015;194:3551–3555. doi: 10.4049/jimmunol.1402262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen L, Gao Y, Liu Y, Zhang B, Liu Q, Wu J, Fan L, Ou Q, Zhang W, Shao L. PD-1/PD-L pathway inhibits M. tb-specific CD4(+) T-cell functions and phagocytosis of macrophages in active tuberculosis. Sci Rep. 2016;6:38362. doi: 10.1038/srep38362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shu Y, Hu Q, Long H, Chang C, Lu Q, Xiao R. Epigenetic variability of CD4+CD25+ tregs contributes to the pathogenesis of autoimmune diseases. Clin Rev Allergy Immunol. 2017;52:260–272. doi: 10.1007/s12016-016-8590-3. [DOI] [PubMed] [Google Scholar]
- 54.Whiteside TL. Induced regulatory T cells in inhibitory microenvironments created by cancer. Expert Opin Biol Ther. 2014;14:1411–1425. doi: 10.1517/14712598.2014.927432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mailloux AW, Young MR. Regulatory T-cell trafficking: from thymic development to tumor-induced immune suppression. Crit Rev Immunol. 2010;30:435–447. doi: 10.1615/critrevimmunol.v30.i5.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adeegbe DO, Nishikawa H. Regulatory T cells in cancer; can they be controlled? Immunotherapy. 2015;7:843–846. doi: 10.2217/imt.15.52. [DOI] [PubMed] [Google Scholar]
- 57.Ondondo B, Jones E, Godkin A, Gallimore A. Home sweet home: the tumor microenvironment as a haven for regulatory T cells. Front Immunol. 2013;4:197. doi: 10.3389/fimmu.2013.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bayry J. Regulatory T cells as adjuvant target for enhancing the viral disease vaccine efficacy. Virusdisease. 2014;25:18–25. doi: 10.1007/s13337-013-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatam LJ, Devoti JA, Rosenthal DW, Lam F, Abramson AL, Steinberg BM, Bonagura VR. Immune suppression in premalignant respiratory papillomas: enriched functional CD4+ Foxp3+ regulatory T cells and PD-1/PD-L1/L2 expression. Clin Cancer Res. 2012;18:1925–1935. doi: 10.1158/1078-0432.CCR-11-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.