Abstract
Gene expression signatures provide valuable information to guide postoperative treatment in breast cancer (BC) patients. However, genetic tests are prohibitively expensive for the majority of BC patients. Immunohistochemical staining (IHC) subtype classification system has been widely used for treatment guideline and is affordable to most BC patients. We aimed to revise immunohistochemical staining (IHC) subtyping to better match gene expression-based Prediction Analysis of Microarray 50 (PAM50) subtyping. Real world data of 372 BC patients were recruited in the Tri-Service General Hospital between Jan 2019 and Dec 2021. Clinical pathological information, blood, twelve pathological tissue slide samples, and fresh surgical tumor specimens were collected to examine IHC and PAM50. Current IHC subtyping (cIHC) tends to misclassify PAM50-based luminal A (lum A) to luminal B (lum B) by 35.81%, PAM50-lum B to PAM50-lum A by 9.09%, PAM50-Her2-enriched to lum B by 61.11%, PAM50-based Her2-enriched to lum B by 61.11%, and PAM50-based basal-like to lum B by 33.33%. We used random forest to identify estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (Her2), and Ki-67 status as the best indicators for revised IHC subtyping (rIHC4) and revised the classification rules by stratified analysis and prediction efficacy. rIHC4 increased the concordance rate for PAM50 subtypes from 68.3% to 74.7%. Both sensitivity and precision increased in most rIHC4 subtypes. Sensitivity increased from 33.3% to 87.4% in the Her2-enriched subtype; precision increased more evidently in the basal-like and lum B subtypes, from 71.4% to 83.3% and 57% to 65.1%, respectively. Our rIHC4 subtyping improved consistency with the PAM50 subtype, which could improve clinical management of BC patients without increasing medical expense.
Keywords: Breast cancer, IHC4, PAM50, molecular subtypes
Introduction
Breast cancer (BC) is the most common malignant tumor in women worldwide in terms of morbidity and mortality [1,2]. The precise classification of BCs into clinically relevant subtypes is crucial for making optimal therapeutic decisions, as it is believed that BCs with different histological and biological characteristics exhibit varying treatment responses. Therefore, there is an urgent need for such classification [3,4].
According to the National Comprehensive Cancer Network® guidelines, surrogate (molecular-like) BC subtyping is assessed using immunohistochemical (IHC) staining. Lum A-like BCs show expression of hormone receptor (HR), no overexpression of Her2, low Ki-67 expression (and a low/intermediate tumor grade). Lum B-like Her2-negative tumors display HR positivity and Her2 negativity but high Ki-67 (and/or high-grade morphology), whereas lum B-like Her2-positive BCs express both HRs and Her2 independently of the proliferation rate (tumor grade: low, intermediate or high). Her2-positive (non-luminal) BCs show Her2 positivity but HR negativity. As mentioned above, triple-negative breast cancers (TNBCs) display both negative HR and Her2 statuses [5-9]. Therapy options include surgical and radiation therapies as well as systemic therapy, such as endocrine therapy and chemotherapy. The treatment strategy is determined individually for each breast cancer patient based on the biology of both the tumor and patient, as well as according to international and national guidelines [7-9]. BC patients with lum A have a very good prognosis and benefit in a clinically relevant dimension from endocrine therapy alone but not from chemotherapy [10]. However, due to endocrine resistance, patients with lum B tumors might have a poorer prognosis if treated with endocrine therapy alone [11] but might profit from chemotherapy [12,13]. Her2-enriched BCs are highly sensitive to anti-Her2 agents [14]. Finally, patients with basal-like BC benefit from chemotherapy [15]. In patients with HR+/Her2- BC who have an intermediate risk of recurrence, as estimated using conventional clinical and pathological risk factors, the decision of whether to use adjuvant chemotherapy is very challenging for clinicians.
However, accumulating evidence suggests that standard IHC-based classifications are less effective than newly developed gene expression-based molecular subtyping. Intrinsic subtyping, based on high-throughput technologies, suggests similar tumors follow the same path and require treatment [3]. RNA profiling of tumor tissues can reflect more complex characteristics of breast cancer and provide better prediction of breast cancer outcomes [16-18]. Multiple molecular-based prognostic tools have been developed for breast cancer, including Mamma Print®, Oncotype DX®, EndoPredict® and Prosigna® [19-22]. In particular, the PAM50 (Prediction Analysis of Microarray 50)-based Prosigna® (NanoString nCounter platform) classification has been shown to be a powerful tool for predicting the risk of recurrence and provides guidance for adjuvant treatment decisions [23-25].
An increasing number of studies have been performed to explore the relationship of IHC markers to molecular subtypes [26-29]. Approximately 20%-38.4% of cases were discordant between the IHC-based subtype and the PAM50 intrinsic subtype, and the most disagreement was in luminal (HR+, Her2-) subtype breast cancer. A significant proportion of patients had a mismatch between their IHC subtype and their PAM50 intrinsic subtype. The survival analysis revealed that the current IHC-based classification may mislead treatment and result in a poor outcome. Current IHC guidelines may be revised accordingly. However, the clinical use of the PAM50 assay remains limited due to its high cost and technical requirements. Discordant therapy choices resulting from PAM50 intrinsic subtyping and risk of recurrence (PAM50-ROR) use result in quality losses. This case study illustrates that value-based payment models need safeguards to avoid the potentially suboptimal substitution of preferred interventions [30]. PAM50 costs approximately 5,300 US dollars per session in Taiwan. It leads to a substantial medical cost burden at the national level and is not affordable for most patients who are from low-income countries. In addition, it remains controversial whether genomic assays should be applied routinely. If we can lower the discrepancy between IHC subtype and PAM50, that would be an instant solution to this issue without adding any medical expense; moreover, this solution would be affordable by most BC patients. We aim to revise the current IHC subtype classification to increase the concordance with PAM50 and compare the classification performance with the current IHC subtype classification.
Methods and materials
Study samples
Between Jan 2019 and Dec 2021, we recruited breast cancer patients from the Tri-Service General Hospital. The inclusion criteria were as follows: 1) age greater than or equal to 20 years old when signing the agreement; 2) diagnosed with invasive breast cancer (unlimited period); 3) the largest diameter of the tumor must be greater than or equal to 1.0 cm before surgery; and 4) understand and be willing to sign the informed consent form. The exclusion criteria were as follows: 1) diagnosis of carcinoma in situ breast cancer; 2) patients who received anticancer treatment such as preoperative chemotherapy, hormone therapy, or targeted therapy; and 3) previous history of invasive breast cancer.
A total of 372 patients were enrolled in the study. We collected clinical pathological information, blood, twelve pathological tissue slide samples, and fresh surgical tumor specimens. The clinical and pathological data included diagnosed age, operation types, grading, IHC expression of ER, PR, Her2, and Ki-67 status, tumor size, lymph node status, PAM50 intrinsic subtypes, and IHC subtypes. All study participants signed informed consent forms, and the Tri-Service General Hospital’s institutional ethical committees approved the study’s protocols (TSGH IRB no. 2-107-05-141).
Immunohistochemical subtyping
IHC subtypes were categorized into four classes, including lum A, lum B, Her2-enriched, and TNBC (corresponding to PAM50-based basal-like). All slides were prepared at our institution and read by three pathologists. For the purposes of this study, ER/PR positivity was determined by IHC analysis of the number of positively stained nuclei (>1%), and Her2 positivity was defined as IHC 3+ or fluorescence in situ hybridization (FISH) positive.
RNA preparation, qRT-PCR, and assignment of biological subtype
For all subjects, one piece of 4-5 μM and 2 to 6 pieces (depending on the size of the sampled tumor tissue) of 10 μM tumor pathology slides were analyzed by Nanostring technology for messenger RNA (mRNA) extraction and PAM50 classification detection.
PAM50 intrinsic subtype prediction
The NanoString nCounter Breast Cancer 360™ Panel, which could analyze a total of 770 genes, was used to analyze the gene expression of breast tumors in the study. Then, we used PAM50 to classify the intrinsic subtypes of breast tumors. PAM50, a 50-gene subtype predictor, is a standardized classification method. According to the RT-qPCR quantitative results of 50 classified genes and 5 control genes, breast tumors can be categorized into four intrinsic subtypes: lum A, lum B, Her2-enriched, and basal-like.
Random forests
Random forests or random decision forests are ensemble learning methods for classification, regression and other tasks that operates by constructing a multitude of decision trees at training time. Random forests deals with the problem of overfitting by creating multiple trees, with each tree trained slightly differently so that it overfits differently. It can create a less biased model and is useful in feature selection. We used random forests to select the important clinical features for PAM50 classification.
Receiver operating characteristic (ROC) curve
The ROC curve is a dichotomous classifier, defining sensitivity (true positive rate, TPR) on the y-axis and (1 - specificity) (false positive rate, FPR) on the x-axis. The curve could be useful in determining the optimal cutoff point and evaluating the discriminatory ability of measuring tools. Here, we used the R package ‘cutpoint’ to plot ROC curves and to estimate the optimal cutoff point of Ki67 using Youden’s J statistic (also called Youden’s index). The formula is J = sensitivity + specificity - 1. The index is defined for all points of an ROC curve, and the maximum value of the index was defined as a criterion for selecting the optimum cutoff point.
Results
As shown in Table 1, 372 breast cancer patients were included in the analysis. Their ages ranged from 33 to 93 years old, and the average age was 58 years old. We stratified the patients by PAM 50 subtyping, e.g., lum A (n=148), lum B (n=143), Her2-enriched (n=54) and basal-like (n=27). The majority of patients were lum A or lum B, which accounted for 78.2% of all patients. The distribution of the operation type and lymph node status was not significantly different among PAM50-based subtypes. The statuses of ER, PR, Her2, grade, Ki67, and tumor size were related to PAM50. The current IHC subtyping was associated with the PAM50 subtyping.
Table 1.
The characteristics of breast cancer patients grouped by PAM50
| PAM50 | |||||
|---|---|---|---|---|---|
|
| |||||
| Luminal A (n=148) | Luminal B (n=143) | Her2-enriched (n=54) | Basal-like (n=27) | P-value | |
| Diagnosed age | 0.27 | ||||
| Mean (SD) | 55.8 (11.5) | 58.2 (12.1) | 58.4 (13.3) | 58.3 (10.3) | |
| Median [Min, Max] | 53.5 [33.0, 90.0] | 57.0 [33.0, 93.0] | 58.5 [24.0, 82.0] | 58.0 [40.0, 78.0] | |
| Operation type | 0.46 | ||||
| Breast-conserving surgery | 72 (48.6%) | 70 (49.0%) | 20 (37.0%) | 12 (44.4%) | |
| Modified radical mastectomy | 76 (51.4%) | 73 (51.0%) | 34 (63.0%) | 15 (55.6%) | |
| ER | 5.00E-04 | ||||
| - | 4 (2.7%) | 0 (0%) | 24 (44.4%) | 21 (77.8%) | |
| + | 144 (97.3%) | 143 (100%) | 30 (55.6%) | 6 (22.2%) | |
| PR | 2.50E-23 | ||||
| - | 15 (10.1%) | 15 (10.5%) | 30 (55.6%) | 21 (77.8%) | |
| + | 133 (89.9%) | 128 (89.5%) | 24 (44.4%) | 6 (22.2%) | |
| Her2 | 8.10E-41 | ||||
| - | 144 (97.3%) | 127 (88.8%) | 8 (14.8%) | 24 (88.9%) | |
| + | 4 (2.7%) | 16 (11.2%) | 46 (85.2%) | 3 (11.1%) | |
| Grade | 5.00E-04 | ||||
| 1 | 38 (25.7%) | 5 (3.5%) | 1 (1.9%) | 0 (0%) | |
| 2 | 99 (66.9%) | 81 (56.6%) | 12 (22.2%) | 3 (11.1%) | |
| 3 | 11 (7.4%) | 57 (39.9%) | 41 (75.9%) | 24 (88.9%) | |
| Ki67 | 1.80E-52 | ||||
| Mean (SD) | 12.2 (8.92) | 28.4 (14.7) | 43.1 (18.1) | 54.3 (20.5) | |
| Median [Min, Max] | 10.0 [1.00, 60.0] | 25.0 [5.00, 75.0] | 40.0 [15.0, 80.0] | 60.0 [20.0, 90.0] | |
| Ki67 (cutoff point =14) | 9.20E-33 | ||||
| - | 96 (64.9%) | 13 (9.1%) | 0 (0%) | 0 (0%) | |
| + | 52 (35.1%) | 130 (90.9%) | 54 (100%) | 27 (100%) | |
| Ki67 (cutoff point =20) | 5.20E-30 | ||||
| - | 114 (77.0%) | 35 (24.5%) | 4 (7.4%) | 0 (0%) | |
| + | 34 (23.0%) | 108 (75.5%) | 50 (92.6%) | 27 (100%) | |
| Tumor size | 1.80E-03 | ||||
| Mean (SD) | 1.95 (0.972) | 2.29 (1.09) | 2.49 (1.04) | 2.44 (1.08) | |
| Median [Min, Max] | 1.80 [0.600, 5.80] | 2.10 [0.400, 7.00] | 2.20 [0.700, 5.10] | 2.20 [1.10, 6.00] | |
| Tumor size (cut 2 cm) | 0.012 | ||||
| Small | 84 (56.8%) | 66 (46.2%) | 20 (37.0%) | 8 (29.6%) | |
| Large | 64 (43.2%) | 77 (53.8%) | 34 (63.0%) | 19 (70.4%) | |
| Lymph node number | 0.77 | ||||
| Mean (SD) | 1.72 (5.14) | 1.86 (4.26) | 2.09 (3.48) | 2.70 (5.30) | |
| Median [Min, Max] | 0 [0, 37.0] | 0 [0, 33.0] | 1.00 [0, 15.0] | 0 [0, 24.0] | |
| Lymph node | 0.074 | ||||
| - | 101 (68.2%) | 89 (62.2%) | 26 (48.1%) | 16 (59.3%) | |
| + | 47 (31.8%) | 54 (37.8%) | 28 (51.9%) | 11 (40.7%) | |
| IHC subtypes (Ki67 cutoff point =14) | 5.00E-04 | ||||
| Luminal A-like | 91 (61.5%) | 13 (9.1%) | 0 (0%) | 0 (0%) | |
| Luminal B-like | 53 (35.8%) | 130 (90.9%) | 33 (61.1%) | 9 (33.3%) | |
| Her2-enriched | 1 (0.7%) | 0 (0%) | 18 (33.3%) | 3 (11.1%) | |
| TNBC | 3 (2.0%) | 0 (0%) | 3 (5.6%) | 15 (55.6%) | |
Molecular subtyping has been used as a tool for precise subtyping in clinical practice. Additionally, PAM50 subtypes correspond with IHC-based subtypes. Therefore, we compared the concordance between molecular PAM50 subtyping and nonmolecular IHC subtyping. As shown in Figure 1, although IHC subtyping was associated with PAM50 subtyping, it presented a substantial discordance with PAM50 subtyping that hindered the application of IHC-based subtypes in precise treatment strategies for breast cancer patients. Current IHC subtyping (cIHC) tended to misclassify PAM50-based lum A to lum B by 35.81%, PAM50-lum B to lum A by 9.09%, PAM50-Her2-enriched to lum B by 61.11%, PAM50-based Her2-enriched to lum B by 61.11%, and PAM50-based basal-like to lum B by 33.33%.
Figure 1.
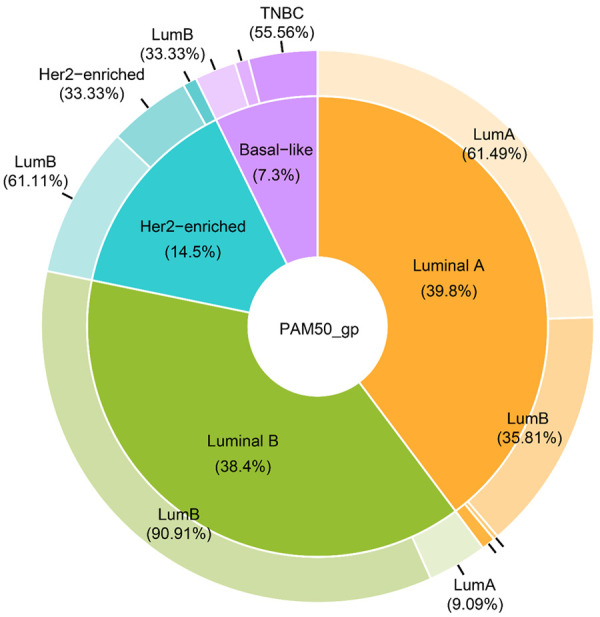
PieDonut chart of current IHC-based subtypes versus PAM50-based subtypes of breast cancer patients.
We explored the new classification rules for BC patients with affordable IHC4 indicators and current clinical indicators. We used random forests, an ensemble learning method for classification, to select the important features for predicting PAM50 subtypes. Figure 2 shows the importance of relative clinical indicators. The top five were Ki67, Her2, ER, and grade PR. Except for grade, the other four are the same indicators of IHC subtyping. We used these indicators to re-evaluate the classification rules of IHC subtyping. Either ER or PR positivity was used to define cIHC4-based lum A and cIHC4-based lum B. In the case of losing possible valuable information for developing new classification rules, we treated them as an independent indicator.
Figure 2.
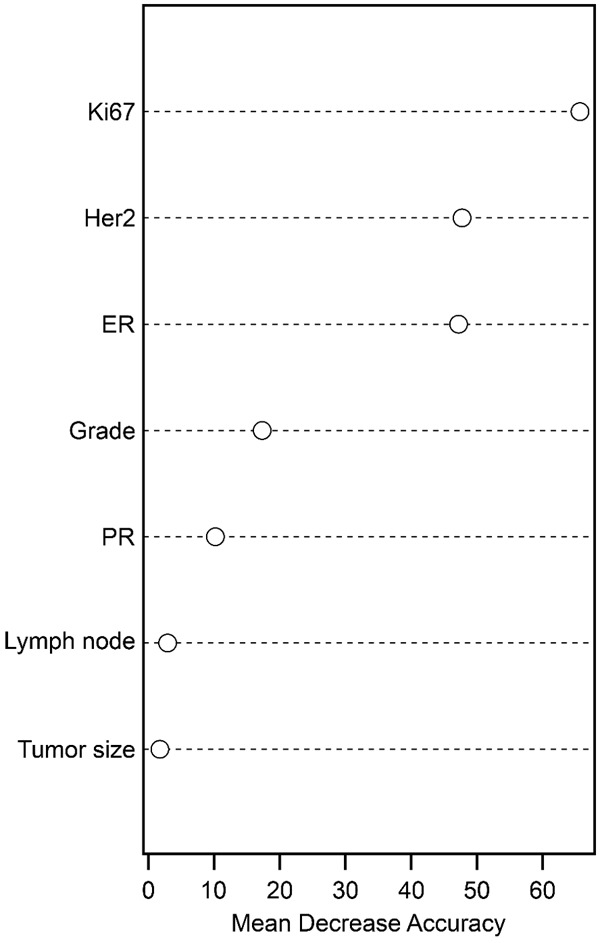
Feature importance for PAM50 subtypes using the random forest model. The higher the mean decrease accuracy is, the more important the feature. The top five features were Ki67, Her2, ER, grade and PR.
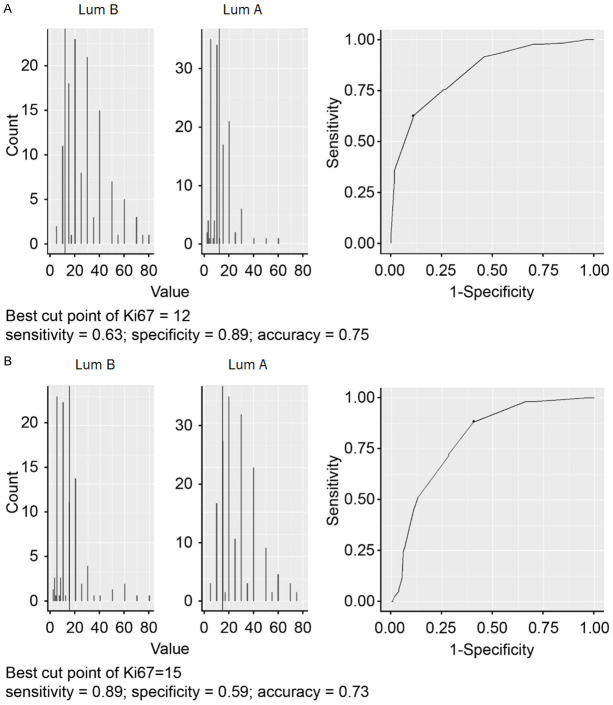
The distribution of PAM50-based subtypes of BC patients stratified by ER, PR, Her2, Ki67 and grade is shown in Figure 3. PAM50-based lum A and lum B were hard to discriminate perfectly in patients with ER+/PR+Her2- at a Ki67 value of approximately 14 (Figure 4). The grades did not seem to contribute to classification. We used the ROC curve to choose the optimal cutoff point of Ki67 for discriminating between PAM50-based lum A and PAM50-based lum B using the Youden index. The result is shown in Figure 4. For identifying PAM50-based lum A, the optimal cutoff point of Ki67 was 12. The sensitivity, specificity, and accuracy were 0.63, 0.89, and 0.78, respectively. For identifying PAM50-based lum B, the optimal cutoff point of Ki67 was 15. The sensitivity, specificity and accuracy were 0.89, 0.59 and 0.73, respectively. We leveraged the best cut among 12-15 for the whole classification accuracy and found that there were no differences among Ki67 cutoff points 12, 13 and 14, but the accuracy was worse for Ki67 cutoff point 15 (data not shown). We maintained the Ki67 cutoff point at 14 (current clinical standard) to discriminate lum A (Ki67≤14) and lum B (Ki67>14) in ER+/PR+Her2- patients.
Figure 3.
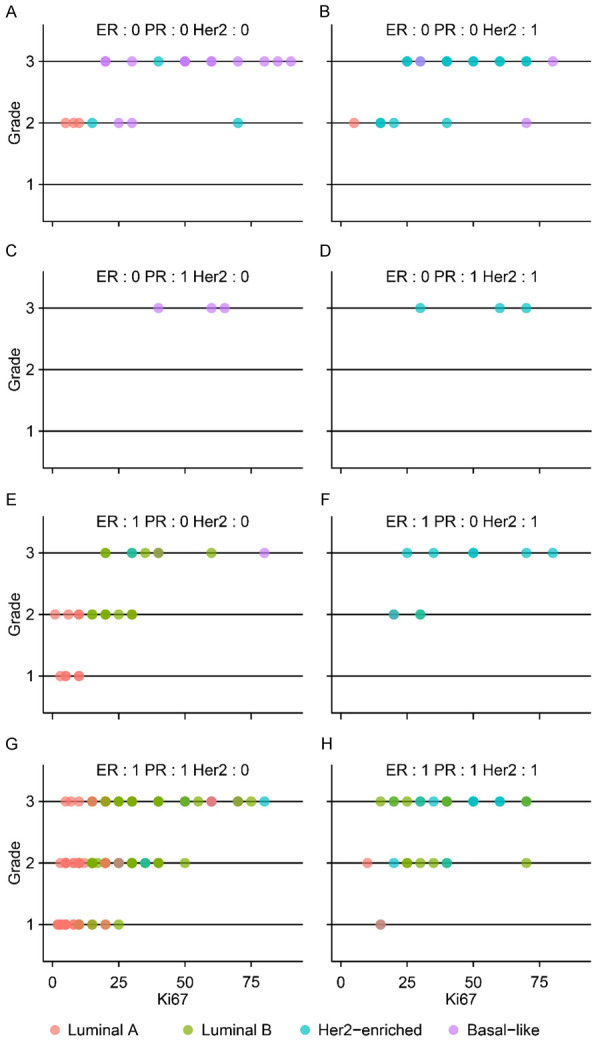
Scatter plots of breast cancer patients colored by PAM50 subtype and stratified by ER(+/-), PR(+/-) and Her2(+/-). X denotes Ki67, and Y denotes the grade.
Figure 4.
Exploring the best cutoff point of Ki67 for discriminating between luminal A (Lum A) and luminal B (Lum B) in ER+/PR+/Her2- breast cancer patients using an ROC curve. The best cutoff point was defined by the Youden index.
Lum B- and Her2-enriched patients were hard to discriminate from ER+/PR+/Her2+ BC patients (Figure 3H). Among these patients, we found that major PAM50-based Her2-enriched patients were Her2+ (3+) and major PAM50-based lum B patients were Her2+ (2+). The detailed Her2 test results (2+ and 3+) could decrease the misclassification of PAM50-based Her2-enriched patients via IHC (Figure 5). This indicates that ER+/PR+/Her2+ (2+) should be reclassified as lum B.
Figure 5.
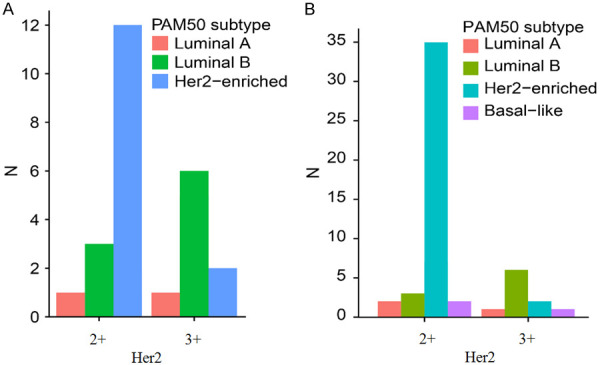
The distribution of PAM 50 subtypes in (A) ER+/PR+/Her2+ and (B) Her2+ breast cancer patients. (A) Her2-enriched patients were mostly Her2+ (3+), and lum B patients were mostly Her2+ (2+). (B) Her2-enriched patients were mostly Her2+ (3+), and lum B patients were mostly Her2+ (2+).
In Figure 3, we learned that Her2-enriched patients appeared in ER+ or PR+ subgroups, contradictory to the current IHC4 classification of Her2-enriched patients whose ER and PR have to be negative. This classification should be redefined. The PAM50 Her2-enriched patients accounted for 82% of all Her2+ (3+) patients (n=52) (Figure 5B). This result indicated that Her2+ (3+) status was important for defining Her2-enriched patients.
In the current IHC4 classification, ER-/PR-/Her2- patients were defined as TNBC (equal to basal-like). As shown in Figure 1, we observed that basal-like patients existed not only in ER-/PR-/Her2- patients but also in ER-/PR+/Her2-, ER-/PR-/Her2+ and ER+/PR-/Her2- patients. The Ki67 cutoff point of 14 decreased the misclassification of basal-like patients. In addition, all ER-/PR+/Her2- patients were basal-like. In summary, the revised classification of basal-like patients with ER-/PR-/Her2-/Ki67≥14 and ER-/PR+/Her2- could increase the classification accuracy.
Using the original indicators ER, PR, Her2, and Ki67, we revised the IHC4 classification to be more consistent with PAM50 subtypes. Table 2 lists the revised rules. The confusion matrix of rIHC4 and PAM50 is shown in Table 3. When we compared the classification accuracy of our revised IHC4 (rIHC4) and current IHC4 (cIHC4), we discovered that rIHC4 outperformed cIHC4 and was more similar to the PAM50 classification. Using rIHC4, the total accuracy increased from 68.3% to 74.7%, with a total increase of 6.4%. The sensitivity of Her2-enriched subtyping increased most from 33.3% to 81.5%, with a total increase of 48.2%. Except for lum B, the sensitivities of the other three subtypes were better than or no less than those of cIHC4. Regarding precisions, rIHC4 outperformed cIHC. Lum B and basal-like patients’ recall rates rose dramatically, from 57% to 65.1% and 71.4% to 83%, respectively. Overall, the rIHC4 and current IHC4 recall rates were 75% and 68%, respectively. In conclusion, rIHC4 outperformed cIHC4 and was more in line with PAM50 (Figure 6).
Table 2.
The table of revised IHC and PAM50 subtypes
| Revised IHC subtypes | Definition |
|---|---|
| IHC-based Lum A | * ER-/PR-/Her2-/ki67<14 |
| (ER+/PR+)/Her2-/ki67<14 | |
| IHC-based Lum B | (ER+/PR+)/Her2-/ki67≥14 |
| * ER+/PR+/Her2+ (2+ and FISH+) | |
| * ER-/PR>20/Her2- | |
| IHC-based Her2-enriched | ER-/PR-/Her2+ |
| * ER+/PR+/Her2+ (3+) | |
| * ER+/PR-/Her2+ | |
| * ER-/PR+/Her2+ | |
| IHC-based Basal-like | * ER-/PR≤20/Her2- |
| ER-/PR-/Her2-/* Ki67≥14 |
denotes the difference from the current IHC4 classification and is marked in bold font.
Table 3.
The confusion matrix of revised IHC4-based and PAM50-based subtypes
| PAM50-based subtypes | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Lum A (n=148) | Lum B (n=143) | Her2-enriched (n=54) | Basal-like (n=27) | Overall (n=372) | ||
| Revised IHC4-based subtypes | Lum A | 94 (63.5%) | 13 (9.1%) | 0 (0%) | 0 (0%) | 107 (28.8%) |
| Lum B | 51 (34.5%) | 125 (87.4%) | 7 (13.0%) | 9 (33.3%) | 192 (51.6%) | |
| Her2-enriched | 3 (2.0%) | 5 (3.5%) | 44 (81.5%) | 3 (11.1%) | 55 (14.8%) | |
| TNBC | 0 (0%) | 0 (0%) | 3 (5.6%) | 15 (55.6%) | 18 (4.8%) | |
Figure 6.
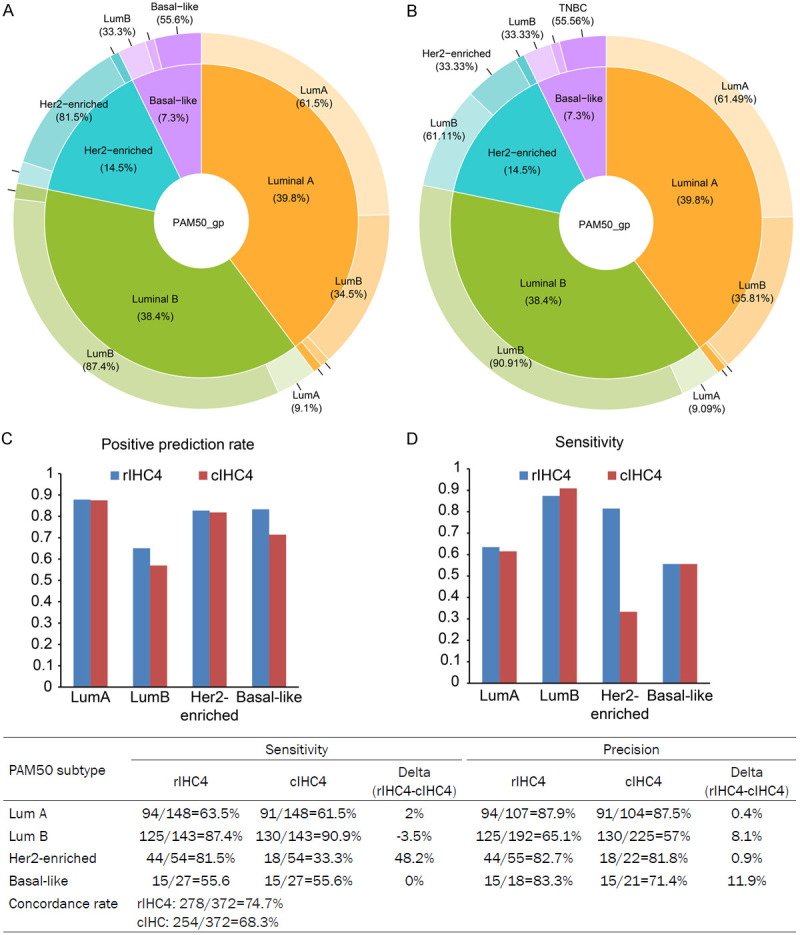
Comparison of (A) revised IHC4 (rIHC4), (B) current IHC4 (cIHC4) and PAM 50 classifications. The rIHC4 outperforms the cIHC4 and is more concordant with PAM50. The precisions and sensitivities of predicting PAM50 subtypes using rIHC4 and IHC4 were shown in (C) and (D) plots. Concordance rate = Total correct prediction number/Total number.
Discussion
In this study, we developed revised IHC4 classification guidelines using the same clinical indicators (ER, PR, Her2 and Ki67) but amended decision rules to make the classification of lum A, lum B, Her2-enriched and Basal-like subtypes more concordant with PAM50. This is the first study to discuss the concordance of IHC subtypes and PAM50 subtypes by modifying the IHC classification rules. In addition, we included a large number of Taiwanese breast cancer patients for the comparison of IHC and PAM50 subtypes. The total concordance rate between our rIHC4 and PAM50 increased from 68.3% (cIHC4) to 74.7%, which represented a 6.4% increase. The sensitivity of rIHC-based Her2-enriched subtypes increased from 33.3% to 81.5%, which represented a 48.2% increase. Compared with cIHC, our rIHC4 provided more precise subtyping for treatment strategy without increasing extra medical expenses. It has a significant clinical impact, particularly for BC patients who are unable to afford the PAM50 test.
A previous study demonstrated that both IHC-based and PAM50-based subtypes are predictive of breast cancer mortality but exhibit discordance that should be modified [31]. PAM50 subtyping can improve risk prediction for ER+/PR+ tumors, especially for ER+/PR+/Her2- tumors; it can be implemented as an additional tool for this subtype to improve disease management. Because the technological complexity and high operating costs have limited the use of molecular detection, the ability of PAM50 to accurately predict prognosis in patients with breast cancer has not been compared with that of fresh tissue. To date, there is limited real-world data evaluating PAM50. The availability of PAM50 results increased the confidence of treating physicians in their adjuvant treatment decisions and led to a 24% [(53+1+3+13)/(148+143)%] change in the chemotherapy treatment plan (from adjuvant chemotherapy to no adjuvant chemotherapy or vice versa) in our study.
The discordance between PAM50 subtyping and IHC4 (ER, PR, Her2, Ki67) subtyping indicates that the two methods may not agree on the classification of the cancer or the recommended treatment approach. This discordance can occur for several reasons. They are 1) different cutoff values for determining positive or negative expression of ER, PR, Her2, and Ki-67, which can lead to different classifications of the cancer; 2) PAM50 takes into account a large number of genes and the interactions among them, while IHC4 focuses on a limited number of proteins; 3) sampling errors, where the tissue analyzed by IHC4 may not represent the entire tumor; and 4) technical errors, as a result of variations in staining or interpretation of the results. It is important to consider that discordance between PAM50 and IHC4 can occur and may require further investigation or additional testing to confirm the classification and treatment approach. Both methods can provide valuable information, and it is important to evaluate them together to determine the most accurate picture of the patient’s cancer.
The 2011 St. Gallen guidelines recommend that breast cancer be treated according to the pathologic determination of ER, PR, Her2, and Ki-67 [32,33]. Ki-67 and the IHC-based subtypes are helpful for accurate classification and contribute to the low concordance between the two classifications. We observed the importance of Ki67 in the classification of IHC-based lum A, lum B, and basal-like patients. Discrepancies in surrogate subtyping were due to significant differences in both Ki-67 expression values and tumor grade assessments. These findings confirm that, to date, the distinction between lum A-like and lum B-like tumors by IHC is still problematic and controversial. Some people suggest emphasizing grade and PR expression with regard to luminal subtype distinction [22,34].
In our study, we separated the HR+, Ki67, and grade to find out how they related to the luminal subtype. We then linked them to PAM50 to get the most accurate results. However, we found that the grade did not obviously improve the IHC subtyping. In addition, we also used two Ki67 cutoff points (14% and 20%), and the 14% cutoff point yielded better classification accuracy than the 20% cutoff point. Una et al. [34] concluded that if the tumor grade was 2 or the Ki67 score was intermediate, the PAM50 test should be used to check if the patient has the lum A or B subtype. We also discovered that in ER+/PR+/Her2-/Grade 2 breast cancer patients, if Ki67 was less than 14, the possibility of lum A was high, similar to PAM50 lum A, and if Ki67 was greater than 30, the possibility of lum B was good, similar to PAM50 lum B (Figure S1). If Ki67 was between 14 and 30 in ER+/PR+/Her2-/Grade 2 breast cancer patients, PAM50 was suggested to determine the molecular subtype for further management.
Several pieces of evidence show that endocrine therapy works better on PR-positive tumors that have been treated with neoadjuvant therapy. Prat et al. [17] reported that the disease-free survival of patients with high PR expression (>20%) was significantly longer than that of patients with low PR expression (≤20%). Li et al. [35] showed that in luminal-type patients, there was no statistically significant difference in the clinicopathological features between patients with low PR expression (1% to 19%) and those without PR expression. These results suggested that 20% should be used as the PR cutoff between lum A and lum B. Similar to our results, ER-, low PR expression (<10%), and Her2- were included in a PAM50-based basal-like group.
Intriguingly, if Ki67 is less than 14 in IHC-based basal-like patients, the PAM50 result is lum A, whereas if Ki67 is greater than 14, the PAM50 result is genuinely basal-like. This may explain why some patients with TNBC have a very excellent prognosis after adequate treatment, but some patients still experience recurrence or metastasis after chemotherapy. In addition, androgen receptor (AR) expression is negatively correlated with Ki-67 expression in TNBC [36]. A higher Ki-67 expression or a lower AR expression was associated with an increased risk of metastasis. According to the findings of Hon et al. [37], TNBC expressing AR may benefit from therapies targeting AR or other luminal pathways.
Many studies have shown that cases with low ER expression (1% to 9%) are similar to ER-negative cases; it is difficult for these patients to benefit from endocrine therapy, and their recurrence-free survival was significantly shorter than that of ER>10% patients [38,39]. The 2015 St. Gallen consensus suggested that ER expression ranging from 1% to 9% of breast cancer is a hormone receptor uncertainty status, and the decision of whether to use endocrine therapy cannot depend on the results of IHC analysis and requires comprehensive consideration [32]. Our results showed that in IHC-based lum A patients, if Ki67 was less than 14, the PAM50 result was basal-like. However, PCR was not sensitive enough to detect cases with low HR expression, which may be related to the insufficient number of cases with low HR expression in our research.
Furthermore, agreement between PAM50 Her2-enriched tumors and the Her2-positive subtype defined by standard IHC/FISH is not always given [40,41], which might lead to confusion and changes in definition. Approximately 15 to 20% of breast cancers are Her2+, and it is independently associated with a higher grade, a more aggressive phenotype, and a poorer prognosis [42]. In our revised IHC subtyping, ER+/PR+/Her2 2+/FISH+ was classified as lum B, and the PAM50 Her2-enriched group included ER+/PR+/Her2 3+, ER-/PR-/Her2+, ER+/PR-/Her2+, and ER-/PR+/Her2+. Although the new definition did not alter the current treatment plan (chemotherapy plus targeted therapy), it demonstrated that the tumor exited heterogeneity and that possible targeted therapy did not have a good response in the lum B subtype compared to the Her2-enriched subtype. New therapies continue to emerge, such as anti-Her2 agents and immunotherapy medications combined with anti-Her2 agents. A more precise subclassification of the Her2-positive subtype was required for precise medicine [42].
Limitations
Limitations in this study that should be acknowledged include the lack of detailed cost-benefit estimations, which are ongoing. Furthermore, no follow-up is available yet, although it is likely that the results of similar demographics would be comparable [34]. Although the study population was limited, this study specifically studied the clinical results of the PAM50 and compared them with currently used treatment guidelines, with the aim of allowing clinicians to make more informed treatment decisions.
Conclusions
Our rIHC4 subtyping improved consistency with the PAM50 subtype, which could improve clinical management of BC patients without increasing medical expense. The proposed classification rules may help improve future versions of the treatment guidelines.
Acknowledgements
This work was supported by the Tri-Service General Hospital, National Defense Medical Center (grant number 801GB112190).
All study participants signed informed consent forms.
Disclosure of conflict of interest
None.
Abbreviations
- BC
breast cancer
- IHC
immunohistochemical staining
- HR
hormone receptor
- PAM50
intrinsic subtyping and risk of recurrence
- FISH
fluorescence in situ hybridization
- PAM50
Prediction Analysis of Microarray 50
- lum A
luminal A
- lum B
luminal B
- ER
estrogen receptor
- PR
progesterone receptor
- Her2
human epidermal growth factor receptor 2
- TNBC
triple-negative breast cancers
Supporting Information
References
- 1.Łukasiewicz S, Czeczelewski M, Forma A, Baj J, Sitarz R, Stanisławek A. Breast cancer-epidemiology, risk factors, classification, prognostic markers, and current treatment strategies-an updated review. Cancers (Basel) 2021;13:4287. doi: 10.3390/cancers13174287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du XL, Li Z. Incidence trends in triple-negative breast cancer among women in the United States from 2010 to 2019 by race/ethnicity, age and tumor stage. Am J Cancer Res. 2023;13:678–691. [PMC free article] [PubMed] [Google Scholar]
- 3.Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, Shi B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 4.Li C, Tao Y, Chen Y, Wu Y, He Y, Yin S, Xu S, Yu Y. Development of a metabolism-related signature for predicting prognosis, immune infiltration and immunotherapy response in breast cancer. Am J Cancer Res. 2022;12:5440–5461. [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF American Society of Clinical Oncology; College of American Pathologists. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 7.Gradishar WJ, Moran MS, Abraham J, Aft R, Agnese D, Allison KH, Anderson B, Burstein HJ, Chew H, Dang C, Elias AD, Giordano SH, Goetz MP, Goldstein LJ, Hurvitz SA, Isakoff SJ, Jankowitz RC, Javid SH, Krishnamurthy J, Leitch M, Lyons J, Mortimer J, Patel SA, Pierce LJ, Rosenberger LH, Rugo HS, Sitapati A, Smith KL, Smith ML, Soliman H, Stringer-Reasor EM, Telli ML, Ward JH, Wisinski KB, Young JS, Burns J, Kumar R. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20:691–722. doi: 10.6004/jnccn.2022.0030. [DOI] [PubMed] [Google Scholar]
- 8.Giordano SH, Franzoi MAB, Temin S, Anders CK, Chandarlapaty S, Crews JR, Kirshner JJ, Krop IE, Lin NU, Morikawa A, Patt DA, Perlmutter J, Ramakrishna N, Davidson NE. Systemic therapy for advanced human epidermal growth factor receptor 2-positive breast cancer: ASCO guideline update. J. Clin. Oncol. 2022;40:2612–2635. doi: 10.1200/JCO.22.00519. [DOI] [PubMed] [Google Scholar]
- 9.National Health Commission Of The People’s Republic Of China. National guidelines for diagnosis and treatment of breast cancer 2022 in China (English version) Chin J Cancer Res. 2022;34:151–175. doi: 10.21147/j.issn.1000-9604.2022.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao JJ, Swain SM. Luminal A breast cancer and molecular assays: a review. Oncologist. 2018;23:556–565. doi: 10.1634/theoncologist.2017-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 12.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 13.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schettini F, Prat A. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast. 2021;59:339–350. doi: 10.1016/j.breast.2021.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yerushalmi R, Gelmon K. Chemotherapy for oestrogen-receptor-negative breast cancer. Lancet. 2008;371:4–5. doi: 10.1016/S0140-6736(08)60043-4. [DOI] [PubMed] [Google Scholar]
- 16.Cejalvo JM, Pascual T, Fernández-Martínez A, Brasó-Maristany F, Gomis RR, Perou CM, Muñoz M, Prat A. Clinical implications of the non-luminal intrinsic subtypes in hormone receptor-positive breast cancer. Cancer Treat Rev. 2018;67:63–70. doi: 10.1016/j.ctrv.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 17.Prat A, Cheang MC, Martín M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO, Perou CM. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J. Clin. Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: past, present and future. Semin Cancer Biol. 2018;52:56–73. doi: 10.1016/j.semcancer.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Piccart M, van’t Veer LJ, Poncet C, Lopes Cardozo JMN, Delaloge S, Pierga JY, Vuylsteke P, Brain E, Vrijaldenhoven S, Neijenhuis PA, Causeret S, Smilde TJ, Viale G, Glas AM, Delorenzi M, Sotiriou C, Rubio IT, Kümmel S, Zoppoli G, Thompson AM, Matos E, Zaman K, Hilbers F, Fumagalli D, Ravdin P, Knox S, Tryfonidis K, Peric A, Meulemans B, Bogaerts J, Cardoso F, Rutgers EJT. 70-gene signature as an aid for treatment decisions in early breast cancer: updated results of the phase 3 randomised MINDACT trial with an exploratory analysis by age. Lancet Oncol. 2021;22:476–488. doi: 10.1016/S1470-2045(21)00007-3. [DOI] [PubMed] [Google Scholar]
- 20.Crager M, Wijayawardana SR, Gruver AM, Blacklock A, Russell C, Baehner FL, Sapunar F. Population-based estimate for the correlation of the oncotype Dx breast recurrence score® result and Ki-67 IHC MIB-1 pharmDx in HR+, HER2-, node-positive early breast cancer. Breast Cancer Res. 2022;24:74. doi: 10.1186/s13058-022-01571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinh P, Graham JD, Elder EN, Kabir M, Doan TB, French J, Meybodi F, Hui R, Wilcken NR, Harnett PR, Hsu J, Stuart KE, Wang T, Ahern V, Brennan M, Fox SB, Dear RF, Lim E, White M, Mann GB, Pathmanathan N. Impact of the EndoPredict genomic assay on treatment decisions for oestrogen receptor-positive early breast cancer patients: benefits of physician selective testing. Breast Cancer Res Treat. 2022;191:501–511. doi: 10.1007/s10549-021-06456-5. [DOI] [PubMed] [Google Scholar]
- 22.Erber R, Angeloni M, Stöhr R, Lux MP, Ulbrich-Gebauer D, Pelz E, Bankfalvi A, Schmid KW, Walter RFH, Vetter M, Thomssen C, Mayr D, Klauschen F, Sinn P, Sotlar K, Stering K, Stenzinger A, Wunderle M, Fasching PA, Beckmann MW, Hoffmann O, Kimmig R, Harbeck N, Wuerstlein R, Ferrazzi F, Hartmann A. Molecular subtyping of invasive breast cancer using a PAM50-based multigene expression test-comparison with molecular-like subtyping by tumor grade/immunohistochemistry and influence on oncologist’s decision on systemic therapy in a real-world setting. Int J Mol Sci. 2022;23:8716. doi: 10.3390/ijms23158716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohnstad HO, Borgen E, Falk RS, Lien TG, Aaserud M, Sveli MAT, Kyte JA, Kristensen VN, Geitvik GA, Schlichting E, Wist EA, Sørlie T, Russnes HG, Naume B. Prognostic value of PAM50 and risk of recurrence score in patients with early-stage breast cancer with long-term follow-up. Breast Cancer Res. 2017;19:120. doi: 10.1186/s13058-017-0911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pu M, Messer K, Davies SR, Vickery TL, Pittman E, Parker BA, Ellis MJ, Flatt SW, Marinac CR, Nelson SH, Mardis ER, Pierce JP, Natarajan L. Research-based PAM50 signature and long-term breast cancer survival. Breast Cancer Res Treat. 2020;179:197–206. doi: 10.1007/s10549-019-05446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agostinetto E, Rediti M, Fimereli D, Debien V, Piccart M, Aftimos P, Sotiriou C, de Azambuja E. HER2-low breast cancer: molecular characteristics and prognosis. Cancers (Basel) 2021;13:2824. doi: 10.3390/cancers13112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundgren C, Bendahl PO, Church SE, Ekholm M, Fernö M, Forsare C, Krüger U, Nordenskjöld B, Stål O, Rydén L. PAM50 subtyping and ROR score add long-term prognostic information in premenopausal breast cancer patients. NPJ Breast Cancer. 2022;8:61. doi: 10.1038/s41523-022-00423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ming W, Zhu Y, Bai Y, Gu W, Li F, Hu Z, Xia T, Dai Z, Yu X, Li H, Gu Y, Yuan S, Zhang R, Li H, Zhu W, Ding J, Sun X, Liu Y, Liu H, Liu X. Radiogenomics analysis reveals the associations of dynamic contrast-enhanced-MRI features with gene expression characteristics, PAM50 subtypes, and prognosis of breast cancer. Front Oncol. 2022;12:943326. doi: 10.3389/fonc.2022.943326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren X, Song Y, Zhang Y, Wu H, Chen L, Pang J, Zhou L, Shen S, Liang Z. Prognostic significance of different molecular typing methods and immune status based on RNA sequencing in HR-positive and HER2-negative early-stage breast cancer. BMC Cancer. 2022;22:548. doi: 10.1186/s12885-022-09656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Chen Y, Xie Z, Luo J, Wang Y, Zhou J, Huang L, Li H, Wang L, Liu P, Shu M, Zhang W, Ke Z. Comparison of immunohistochemistry and RT-qPCR for assessing ER, PR, HER2, and Ki67 and evaluating subtypes in patients with breast cancer. Breast Cancer Res Treat. 2022;194:517–529. doi: 10.1007/s10549-022-06649-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim HK, Park KH, Kim Y, Park SE, Lee HS, Lim SW, Cho JH, Kim JY, Lee JE, Ahn JS, Im YH, Yu JH, Park YH. Discordance of the PAM50 intrinsic subtypes compared with immunohistochemistry-based surrogate in breast cancer patients: potential implication of genomic alterations of discordance. Cancer Res Treat. 2019;51:737–747. doi: 10.4143/crt.2018.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Li Q, Aushev VN, Neugut AI, Santella RM, Teitelbaum S, Chen J. PAM50- and immunohistochemistry-based subtypes of breast cancer and their relationship with breast cancer mortality in a population-based study. Breast Cancer. 2021;28:1235–1242. doi: 10.1007/s12282-021-01261-w. [DOI] [PubMed] [Google Scholar]
- 32.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn HJ Panel Members. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 34.Kjällquist U, Acs B, Margolin S, Karlsson E, Kessler LE, Garcia Hernandez S, Ekholm M, Lundgren C, Olsson E, Lindman H, Foukakis T, Matikas A, Hartman J. Real world evaluation of the prosigna/PAM50 test in a node-negative postmenopausal swedish population: a multicenter study. Cancers (Basel) 2022;14:2615. doi: 10.3390/cancers14112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li AQ, Zhou SL, Li M, Xu Y, Shui RH, Yu BH, Yang WT. Clinicopathologic characteristics of oestrogen receptor-positive/progesterone receptor-negative/Her2-negative breast cancer according to a novel definition of negative progesterone receptor status: a large population-based study from China. PLoS One. 2015;10:e0125067. doi: 10.1371/journal.pone.0125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutton LM, Cao D, Sarode V, Molberg KH, Torgbe K, Haley B, Peng Y. Decreased androgen receptor expression is associated with distant metastases in patients with androgen receptor-expressing triple-negative breast carcinoma. Am J Clin Pathol. 2012;138:511–516. doi: 10.1309/AJCP8AVF8FDPTZLH. [DOI] [PubMed] [Google Scholar]
- 37.Hon JD, Singh B, Sahin A, Du G, Wang J, Wang VY, Deng FM, Zhang DY, Monaco ME, Lee P. Breast cancer molecular subtypes: from TNBC to QNBC. Am J Cancer Res. 2016;6:1864–1872. [PMC free article] [PubMed] [Google Scholar]
- 38.Balduzzi A, Bagnardi V, Rotmensz N, Dellapasqua S, Montagna E, Cardillo A, Viale G, Veronesi P, Intra M, Luini A, Pruneri G, Mastropasqua G, Goldhirsch A, Colleoni M. Survival outcomes in breast cancer patients with low estrogen/progesterone receptor expression. Clin Breast Cancer. 2014;14:258–264. doi: 10.1016/j.clbc.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 39.Yoder R, Kimler BF, Staley JM, Schwensen K, Wang YY, Finke K, O’Dea A, Nye L, Elia M, Crane G, McKittrick R, Pluenneke R, Madhusudhana S, Beck L, Shrestha A, Corum L, Marsico M, Stecklein SR, Godwin AK, Khan QJ, Sharma P. Impact of low versus negative estrogen/progesterone receptor status on clinico-pathologic characteristics and survival outcomes in HER2-negative breast cancer. NPJ Breast Cancer. 2022;8:80. doi: 10.1038/s41523-022-00448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweeney C, Bernard PS, Factor RE, Kwan ML, Habel LA, Quesenberry CP Jr, Shakespear K, Weltzien EK, Stijleman IJ, Davis CA, Ebbert MT, Castillo A, Kushi LH, Caan BJ. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2014;23:714–724. doi: 10.1158/1055-9965.EPI-13-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreutzfeldt J, Rozeboom B, Dey N, De P. The trastuzumab era: current and upcoming targeted HER2+ breast cancer therapies. Am J Cancer Res. 2020;10:1045–1067. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.