Abstract
The gene rapL lies within the region of the Streptomyces hygroscopicus chromosome which contains the biosynthetic gene cluster for the immunosuppressant rapamycin. Introduction of a frameshift mutation into rapL by ΦC31 phage-mediated gene replacement gave rise to a mutant which did not produce significant amounts of rapamycin. Growth of this rapL mutant on media containing added l-pipecolate restored wild-type levels of rapamycin production, consistent with a proposal that rapL encodes a specific l-lysine cyclodeaminase important for the production of the l-pipecolate precursor. In the presence of added proline derivatives, rapL mutants synthesized novel rapamycin analogs, indicating a relaxed substrate specificity for the enzyme catalyzing pipecolate incorporation into the macrocycle.
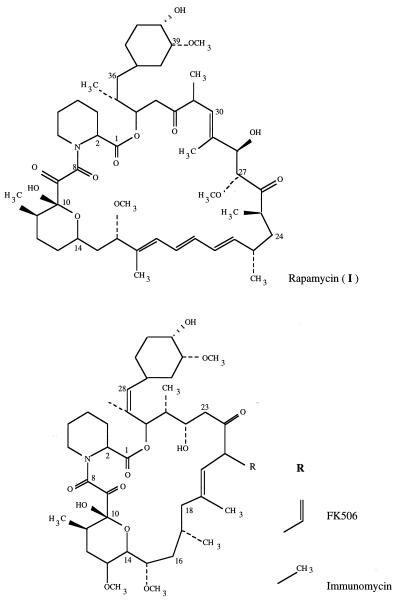
Rapamycin is a 31-member macrocyclic polyketide produced by Streptomyces hygroscopicus NRRL 5491 which, like the structurally related compounds FK506 and immunomycin (Fig. 1), has potent immunosuppressive properties (24). Such compounds are potentially valuable in the treatment of autoimmune diseases and in preventing the rejection of transplanted tissues (16). The biosynthesis of rapamycin requires a modular polyketide synthase, which uses a shikimate-derived starter unit (11, 20) and which carries out a total of fourteen successive cycles of polyketide chain elongation that resemble the steps in fatty acid biosynthesis (2, 27). l-Pipecolic acid is then incorporated (21) into the chain, followed by closure of the macrocyclic ring, and both these steps are believed to be catalyzed by a pipecolate-incorporating enzyme (PIE) (18), the product of the rapP gene (8, 15). Further site-specific oxidations and O-methylation steps (15) are then required to produce rapamycin.
FIG. 1.
Structures of rapamycin, FK506, and immunomycin.
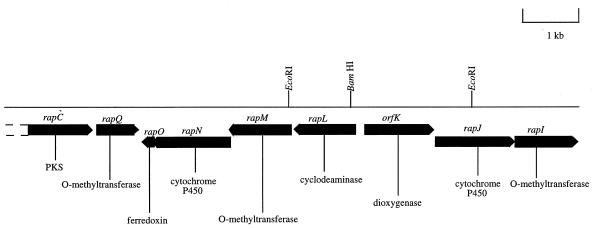
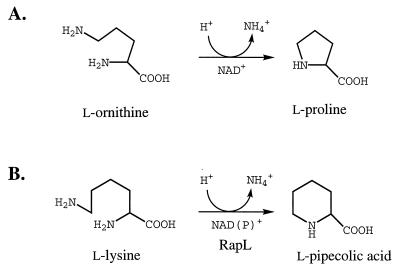
The origin of the pipecolic acid inserted into rapamycin has been previously established (21) to be free l-pipecolic acid derived from l-lysine (although the possible role of d-lysine as a precursor must also be borne in mind) (9). Previous work with other systems has suggested several alternative pathways for pipecolate formation from lysine (22), but the results of the incorporation of labelled lysine into the pipecolate moiety of immunomycin (Fig. 1) clearly indicate loss of the α-nitrogen atom (3). More recently, the sequencing of the rap gene cluster revealed the presence of the rapL gene (Fig. 2), whose deduced gene product bears striking sequence similarity to two isoenzymes of ornithine deaminase from Agrobacterium tumefaciens (25, 26). Ornithine deaminase catalyzes the deaminative cyclization of ornithine to proline, and we have proposed (15) that the rapL gene product catalyzes the analogous conversion of l-lysine to l-pipecolate (Fig. 3).
FIG. 2.
A portion of the rapamycin biosynthetic gene cluster which contains ancillary (non-polyketide synthase) genes (15, 27). PKS, polyketide synthase.
FIG. 3.
(A) The conversion of l-ornithine to l-proline by ornithine cyclodeaminase (17). (B) Proposed conversion of l-lysine to l-pipecolic acid by the rapL gene product.
Here, we report the use of ΦC31 phage-mediated gene replacement (10) to introduce a frameshift mutation into rapL and the ability of the mutant to synthesize rapamycins in the absence or presence of added pipecolate or pipecolate analogs.
MATERIALS AND METHODS
Materials.
All molecular biology enzymes and reagents were from commercial sources. Viomycin was the kind gift of Pfizer, Inc. l-Pipecolic acid, l-proline, 3,4-dehydroproline, picolinic acid, pyrrole-2-carboxylic acid, trans-4-hydroxyproline, cis-4-hydroxyproline, and cis-3-hydroxyproline were obtained from Aldrich Chemical Co.
Bacterial strains, plasmids, and phages.
The bacterial strains, plasmids, and phages used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and phages
| Strain, plasmid, or phage | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli DH10B | F−mcrA Δ(mrr hsdRMS-mcrBC) φ80dlacZΔM15ΔlacX74 deoR recA1 endA1 | Gibco-BRL |
| Streptomyces strains | ||
| S. hygroscopicus NRRL 5491 | Wild type; produces rapamycin | ATCCa |
| S. hygroscopicus LEK111 | rapL gene disrupted | This study |
| S. lividans JI1326 | Host for phage infection | 7 |
| Plasmids | ||
| pUC18 | Cloning vector in E. coli | Sigma Chemical Co. |
| pLK1 | pUC18 with BamHI site removed | This study |
| pLK2 | pLK1 containing a 3-kb EcoRI fragment corresponding to nucleotides 93956 to 96990 of the rap cluster | This study |
| pLK3 | pLK2 with unique BamHI site removed | This study |
| Phages | ||
| KC515 | c+ attP::tsr::vph | 7 |
| ΦRAPL | KC515 containing a 3-kb insert | This study |
ATCC, American Type Culture Collection, Rockville, Md.
Media and growth conditions.
Escherichia coli DH10B (Gibco-BRL) was grown in 2× tryptone-yeast extract medium (23). E. coli transformants were selected with 100 μg of ampicillin per ml. The rapamycin producer S. hygroscopicus NRRL 5491 and its derivatives were maintained on SY agar (8) and cultivated in tryptic soy broth (Difco) with 100 mM MES [2-(N-morpholino)-ethanesulfonic acid]–1.0% glucose, pH 6.0 (TSBGM), supplemented with 10 μg of viomycin per ml as required. The defined medium used was as described by Cheng et al. (6). Streptomyces lividans JI1326 was cultivated in YEME (7) or in tap water medium (0.5% glucose, 1% sucrose, 0.5% tryptone, 0.25% yeast extract, 10 mM EDTA; pH 7.1) (8). Liquid cultures were grown at 30°C in Erlenmeyer flasks shaken at 200 to 250 rpm. Infection with the att mutant actinophage KC515 (4) and its derivative ΦRAPL was carried out on solid DNA medium supplemented with 10 mM MgSO4–8 mM Ca(NO3)2–0.5% (wt/vol) glucose (7).
DNA techniques and plasmid construction.
Procedures for electroporation, PCR, and manipulation of DNA were carried out as described in Sambrook et al. (23). Total DNA from S. hygroscopicus spheroplasts was isolated with DNAzol (Gibco) as described by the manufacturer. Southern hybridization was carried out with probes labelled with digoxigenin by using the DIG DNA labelling kit (Boehringer Mannheim). DNA fragments for labelling and subcloning were isolated with the Qiaex II (Qiagen) gel extraction kit. Protoplasts of S. lividans JI1326 were transfected with KC515 and ΦRAPL as described by Hopwood et al. (7). Recombinant phage was identified by PCR analysis.
Construction of a frameshift mutation in rapL.
The rapL gene was disrupted by introducing a frameshift by KC515 phage-mediated gene replacement as follows. A 3,034-bp EcoRI fragment (nucleotides 93956 to 96990 of the rap cluster) (27) encompassing the entire rapL gene and flanked by rapK and part of the rapM gene (Fig. 2) was cloned into an EcoRI-cut pUC18-derived vector that had been modified by removal of the BamHI site in the polylinker region by BamHI digestion, end filling, and religation to yield pLK2 (Table 1). A unique BamHI site was located in the coding region of the rapL gene 42 bp from the 5′ end. Plasmid pLK2 was digested with BamHI, and the cohesive ends were filled in by treatment with E. coli DNA polymerase I (Klenow fragment) and religated. The ligated plasmid DNA was redigested with BamHI and used to transform E. coli. Ampicillin-resistant transformants were selected, and their plasmid DNA was checked for the removal of the BamHI site by restriction enzyme analysis and then DNA sequencing. The 3-kb insert was excised from the plasmid with EcoRI, the cohesive ends were blunt ended by treatment with E. coli DNA polymerase I (Klenow fragment), and the insert was cloned into PvuII-cut phage KC515 vector. S. lividans JI1326 protoplasts were transfected with the ligation mixture, and the resultant phage plaques were screened by PCR for the presence of rapL-derived sequences. Positive phage ΦRAPL was selected and used to transfect S. hygroscopicus NRRL 5491 as described by Lomovskaya et al. (10) on DNA plates. Lysogens were selected by overlaying the plates with viomycin (50-μg ml−1 final concentration) 24 h after infection. Approximately 2,000 initial transductants were obtained, of which about 200 were stable integrants. Three of these were arbitrarily chosen and used to isolate viomycin-sensitive derivatives that had undergone a second recombination event deleting the integrated phage. These viomycin-sensitive isolates were obtained after three rounds of nonselective growth and sporulation on SY plates. Of the four colonies isolated, two were found to have reverted to the wild type with the BamHI site restored at position 95036, while the other two harbored the desired mutation with the loss of this BamHI site. The insertion and subsequent loss of the phage were confirmed by genomic Southern hybridization.
Precursor feeding and fermentation of the rapL mutant.
S. hygroscopicus LEK111 (Table 1) was cultured in 500-ml Erlenmeyer flasks containing 100 ml of TSBGM. When added, proline or other amino acids were present at a final concentration of 1 mg/ml. In certain experiments, S. hygroscopicus LEK111 was cultivated in 2-liter flasks, each containing 400 ml of a defined medium to which no pipecolic acid was added (6). For larger-scale fermentation, 10 μl of a spore suspension of S. hygroscopicus LEK111 was used to inoculate a 100-ml flask containing 30 ml of TSBGM medium. The culture was incubated at 28°C for 4 days with shaking at 300 rpm. Then, 4 ml of this culture was transferred to a 2-liter flask containing 400 ml of TSBGM medium and the mixture was shaken at 300 rpm at 28°C for 4 days. This culture in turn was transferred to a 20-liter fermenter containing 15 liters of TSBGM medium supplemented with l-trans-4-hydroxyproline. The fermentation was carried out at 28°C for 4 days, with aeration at 15 liters of air/min and an agitation rate of 500 rpm, in the presence of Sigma A antifoam (Aldrich Chemical Co.). The cells were harvested by filtration and extracted with two volumes of methanol overnight at 4°C. Extracts were analyzed by liquid chromatography-mass spectrometry (LC-MS) with a Finnigan MAT (San Jose, Calif.) LCQ linked to a Hewlett-Packard (Böblingen, Germany) 1100 high-pressure liquid chromatograph.
For isolation of metabolites, the extracts were evaporated to dryness and then purified by flash chromatography on silica gel 60 (Merck) eluted with acetone-hexane (1:1 [vol/vol]). The fractions containing rapamycins were further purified by preparative high-pressure liquid chromatography on a reverse-phase C18 column (250 by 20 mm; 5-μm bead diameter) (HPLC Technology, Macclesfield, United Kingdom) (8). Nuclear magnetic resonance (NMR) spectra were acquired on a Bruker DRX 500 spectrometer.
Biological activity of rapamycin analogs.
Human lymphocyte 536 cells (obtained from the human genetic mutant cell repository, Camden, N.J.) were cultured in Iscove’s medium supplemented with 10% (vol/vol) fetal calf serum. For the bioassay (14), cells were seeded into 96-well microtiter plates at 10,000 per well in 100 μl of growth medium. Solutions of each metabolite were prepared in dimethyl sulfoxide (DMSO) (1% [wt/vol]), and further dilutions were made to give a final concentration of 0.1% DMSO in growth medium. Each culture received either a 1 μM, 100 nM, 10 nM, or 1 nM final concentration of each rapamycin or rapamycin analog or a final concentration of 0.1% DMSO in growth medium only. Each culture (in triplicate) was labelled with 1 μCi of tritiated thymidine (Amersham International; specific activity, 70 Ci/mM) per well for 3 h either immediately after addition of the rapamycin analogs (0- to 3-h label) or at 24 h (21- to 24-h label) or at 48 h (45- to 48-h label). After being labelled, the cultures were lysed with water and harvested onto glass fiber paper disks to trap the released DNA. Radioactivity incorporated into the filter disks was counted in a Packard scintillation counter.
RESULTS
Disruption of the rapL gene.
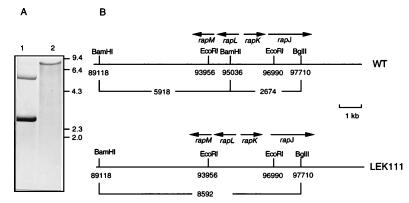
A frameshift mutation was introduced into the rapL gene of S. hygroscopicus NRRL 5491 as described in Materials and Methods. The presence of the frameshift mutation in S. hygroscopicus LEK111 was confirmed by Southern hybridization (Fig. 4). Chromosomal DNA isolated from S. hygroscopicus NRRL 5491 and LEK111 and digested with both BamHI and BglII was probed with a 3-kb EcoRI fragment (between nucleotides 93956 and 96990 of the rap gene cluster). This probe overlaps the BamHI site at position 95036, which was expected to be eliminated in LEK111. Hybridizing bands of 5.9 kb (representing nucleotides 89118 to 95036) and 2.7 kb (representing nucleotides 95036 to 97710) were found for the wild-type strain as expected (Fig. 4). With chromosomal DNA from strain LEK111, only an 8.6-kb BamHI-BglII fragment (representing nucleotides 89118 to 97710) was detected, indicating that the BamHI site at position 95036 had been removed. This was also confirmed by PCR analysis. Chromosomal DNA was subjected to PCR with oligonucleotide primers identical to the sequences from nucleotide 93950 to nucleotide 93968 and from nucleotide 96990 to nucleotide 97010. The expected 3-kb DNA fragment was amplified from wild-type DNA, and following BamHI digestion, two bands, approximately 2 and 1 kb in size, were detected. In samples containing S. hygroscopicus LEK111 chromosomal DNA, the 3-kb PCR product was found to be resistant to BamHI digestion as predicted (data not shown).
FIG. 4.
Southern analysis and restriction map of the rapL region of S. hygroscopicus NRRL 5491 (WT) and S. hygroscopicus LEK111. (A) Genomic DNA from S. hygroscopicus NRRL 5491 (lane 1) and S. hygroscopicus LEK111 (lane 2) was digested with BamHI and probed with the 3-kb EcoRI fragment. The sizes (in kilobases) of the λ-HindIII DNA markers are indicated on the right. (B) Restriction map and gene organization of the rapL region in S. hygroscopicus NRRL 5491 and S. hygroscopicus LEK111.
LEK111 requires added pipecolate for full restoration of rapamycin production.
Wild-type S. hygroscopicus, when grown in a rich medium (TSBGM) for 96 h, produced both rapamycin and prolylrapamycin, a rapamycin analog in which l-proline replaces pipecolate in the macrocyclic ring (Fig. 5), in a ratio of about 20:1 (Table 2). LEK111 under the same conditions produced smaller quantities of the same rapamycins in a ratio of approximately 1:2 to 1:3. In TSBGM medium supplemented with pipecolic acid, LEK111 produced rapamycins at or near wild-type levels and the ratio of rapamycin to prolylrapamycin was also very similar to that for the wild type.
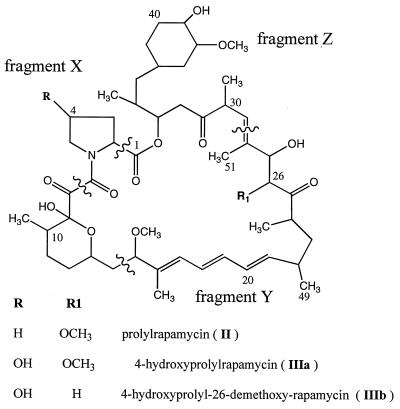
FIG. 5.
Rapamycin analogs produced in this study.
TABLE 2.
Production of metabolites by S. hygroscopicus NRRL 5491 and S. hygroscopicus LEK111
| Strain | Metabolite | Amt of metabolite produced (mg/liter) ina:
|
||||
|---|---|---|---|---|---|---|
| TSBGM | TSBGM + pipecolate | TSBGM + proline | TSBGM + 4-hydroxy-proline | Defined mediumb | ||
| I | 15.2 | 15.9 | 14.4 | 14.0 | 0.12 | |
| Wild type | II | 0.7 | 0.6 | 0.4 | 0.5 | Traces |
| IIIa | N.D.c | N.D. | N.D. | N.D. | N.D. | |
| IIIb | N.D. | N.D. | N.D. | N.D. | N.D. | |
| I | 0.4 | 13.1 | 0.3 | 0.4 | 0.03 | |
| LEK111 | II | 1.1 | 0.6 | 1.3 | 1.4 | 0.05 |
| IIIa | N.D. | N.D. | N.D. | 0.2 | N.D. | |
| IIIb | N.D. | N.D. | N.D. | 1.1 | N.D. | |
Production of novel rapamycin analogs containing 4-hydroxyprolyl moieties.
The production of prolylrapamycin, also previously noted by Nielsen et al. for prolylimmunomycin (18), suggested that LEK111 might incorporate alternative amino acids when added to the medium. When fermentations of S. hygroscopicus LEK111 were supplemented with l-trans-4-hydroxyproline, two new peaks were detected by LC-MS (Table 3). The major peak with the characteristic triene UV absorption at 277 nm contained a compound with an m/z of 908, which would correspond to a 4-hydroxyprolyl–rapamycin analog lacking a methoxy group (Fig. 5, IIIb). Smaller amounts of a compound with an m/z of 938 which would correspond to 4-hydroxyprolyl-rapamycin (Fig. 5, IIIa) were also detected. To obtain enough material for NMR characterization, l-trans-4-hydroxyproline was used to supplement a 15-liter fermentation of S. hygroscopicus LEK111, from which 3 mg of the compound with the m/z of 908 (Fig. 5A, IIIb), together with about 15 mg of pure prolylrapamycin (Fig. 5, II), was extracted as described in Materials and Methods. The NMR data (Table 4) reflected the chemical shifts and couplings expected for the 4-hydroxyproline spin system and also confirmed the absence of the methoxy group at C-26 (corresponding to C-27 in rapamycin), although the l-trans configuration of the 4-hydroxyprolyl moiety could not be directly established. The observed MS fragmentation data (Table 5) confirmed these findings. For the compound labeled I, the loss of 129 atomic mass units (amu) reflects the loss of the pipecolate moiety while in the compounds labeled IIIa and IIIb, the loss of an amino acid of 131 amu confirms the identity of the amino acid moiety (Fig. 5, fragment X) as hydroxyproline. The loss of fragment Y (loss of 294 amu in compound IIIa and loss of 264 amu in compound IIIb) indicated that the methoxy group in position 26 is missing in compound IIIb but not in compound IIIa. Furthermore, the loss of fragment Z (322 amu) can be seen for compounds IIIa and IIIb as well as for compound I, indicating that there is no alteration affecting this region of the molecule. Therefore, compound IIIb (Table 5) with an m/z of 908 is 4-hydroxyprolyl-26-demethoxy-rapamycin and compound IIIa with an m/z of 938 is 4-hydroxyprolyl-rapamycin. In addition to l-trans-4-hydroxyproline, l-cis-4-hydroxyproline and l-cis-3-hydroxyproline were also incorporated, as judged by LC-MS analysis (data not shown). No new products were observed with 3,4-dehydroproline and picolinic acid, while pyrrole-2-carboxylic acid appears to inhibit growth at the concentrations used.
TABLE 3.
Metabolites produced by S. hygroscopicus LEK111 on medium supplemented with pipecolate or pipecolate analogs
| Compound fed | Product
|
||
|---|---|---|---|
| m/z (M + Na+) | LC retention time (min) | Name | |
| l-Pipecolic acid | 936 | 8.84 | Rapamycin |
| l-Proline | 922 | 7.99 | Prolylrapamycin |
| l-trans-4-Hydroxyproline | 938 | 5.35 | Compound IIIaa |
| 908 | 6.29 | Compound IIIba | |
The compound designation is defined in Fig. 5. The structure of the compound was verified by NMR.
TABLE 4.
Selection of 1H and 13C NMR data for 4-hydroxyprolyl-26-demethoxy-rapamycin (compound IIIb [Fig. 5]) and rapamycin (compound I [Fig. 1])a
| Position | 1H δ (ppm) | 13C δ (ppm) |
|---|---|---|
| 1 | 171.30 (169.2) | |
| 2 | 5.24 (5.29) | 58.17 (51.3) |
| 3 | 2.65, 1.69 (2.34, 1.76) | 38.48 (27.0) |
| 4 | 4.38 (1.78, 1.47) | 70.63 (20.6) |
| 5 | 3.37, 2.94 (1.75, 1.48) | 56.53 (25.3) |
| 26 | 3.58 (3.71) | Not determined |
| 27 | 3.89 (4.17) | 70.63 (77.3) |
| 38 | 2.93 (2.93) | 83.90 (84.4) |
| 39 | 3.37 (3.37) | 73.95 (73.9) |
| 40 | 1.99, 1.33 (1.99, 1.38) | 31.22 (31.3) |
| 49 | 3.12 (3.13) | 55.68 (55.8) |
| 51 | 3.39 (3.41) | 56.50 (56.5) |
Data in parentheses are chemical shifts for equivalent positions in rapamycin (13).
TABLE 5.
MS fragmentation analysis of metabolites produced by S. hygroscopicus LEK111 grown in the presence of 4-hydroxyproline
Biological activities of rapamycin analogs.
The immunosuppressant activities of rapamycin, prolylrapamycin, and 4-hydroxy-26-demethoxy-rapamycin were tested with human lymphoblastoid cell line 536 as described in Materials and Methods. Previous experiments have shown that rapamycin is a strong inhibitor of G1 progression in the 536 cell line (14). For all three compounds, the 0- to 3-h drug exposure had no appreciable effect on DNA synthesis at concentrations up to 100 nM (data not shown). This implies that none of the compounds is toxic to the 536 cell line. After 24 and 48 h (Table 6), the 536 cells showed a concentration-dependent inhibition of DNA synthesis, with 50% inhibition at 10 nM rapamycin and at 30 nM prolylrapamycin. 4-Hydroxy-26-demethoxy-rapamycin was also inhibitory, but the inhibition did not reach 50%, even at 1 μM.
TABLE 6.
Inhibition of cell cycle progression by rapamycin and analogsa
| Compound | Incorporation of [3H]dTTP into nucleic acids (%) at indicated time and compound concentration
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 21–24 h
|
45–48 h
|
|||||||
| 1 nM | 10 nM | 100 nM | 1 μM | 1 nM | 10 nM | 100 nM | 1 μM | |
| Rapamycin | 92 | 48 | 27 | 20 | 99 | 46 | 10 | 7 |
| Prolylrapamycin | 98 | 65 | 26 | 21 | 103 | 91 | 22 | 7 |
| 4-Hydroxyprolyl-26-demethoxy-rapamycin | 87 | 86 | 80 | 67 | 111 | 98 | 98 | 71 |
The control experiment was the incorporation of [3H-dTTP] into nucleic acids in the 536 cell line in the presence of 0.1% DMSO (100%).
DISCUSSION
We have previously characterized rapL, a gene within the rapamycin biosynthetic gene cluster which, on the basis of a sequence comparison with authentic cyclodeaminases (25, 26), appears likely to catalyze the cyclodeamination of l-lysine to form l-pipecolate. This, in turn, is required as a specific precursor for the formation of the macrocyclic lactone in rapamycin. The clustering of such a gene for the provision of an essential precursor with the rapamycin biosynthetic genes would be an unusual but not an unprecedented arrangement (1). When in the present work the rapL gene was inactivated by phage-mediated gene replacement (10), the mutant strain, LEK111, was found to produce only minute amounts of rapamycin (Fig. 1, compound I), together with a derivative (Fig. 5, compound II) in which the pipecolate moiety is replaced by proline (Fig. 5; Table 2). Even smaller amounts of these rapamycins were produced when LEK111 was grown in a defined medium in the absence of added pipecolate. Since the frameshift introduced into rapL lies very near the 5′ end of the coding region, the low-level production of rapamycin by LEK111 cannot be due to residual activity of the rapL gene product. Either the media contain unsuspected traces of pipecolate or alternative pathways must exist in S. hygroscopicus for the endogenous production of pipecolic acid. The existence of multiple pathways which generate pipecolate via lysine catabolism has been demonstrated in Pseudomonas spp., Rhizoctania leguminicola, Streptomyces virginiae (3), and β-lactam antibiotic-producing strains of Streptomyces (12). The addition of l-pipecolate to the mutant strain completely restored to wild-type levels its ability to synthesize compound I when grown on either defined medium or TSBGM and restored the wild-type product ratio of compound I to compound II (Table 2). This is fully consistent with our previous proposal that rapL encodes an l-lysine cyclodeaminase, although it does not prove it (15). A complete proof of the putative role of this gene will require the future demonstration that purified RapL catalyzes the deamination together with evidence that the cloned rapL gene can complement the mutation in LEK111. We have no ready explanation for the observation that the mutant strain does not accumulate more of compound II than the wild type in proline-containing media (Table 2). This would have been anticipated from the known substrate specificity of PIE (8, 18). The production of some fully processed rapamycin in LEK111 shows that the effects of the disruption of rapL are not solely mediated by polar effects on the downstream rapM, rapN, and rapO genes.
It has also been demonstrated here that novel rapamycins may be generated by supplementing the fermentation medium of S. hygroscopicus LEK111 with proline or with amino acids that are close structural analogs of proline. In the case of the PIE from immunomycin, 4-methylproline (which is not commercially available) was found to be a better substrate than pipecolate itself for the adenylation reaction in vitro (18). A more extensive survey, therefore, might well uncover other amino acids capable of being incorporated by the PIE of the rapamycin biosynthetic pathway. The rapamycin analog, 4-hydroxyprolyl-26-demethoxy-rapamycin (Fig. 5, compound IIIb) has been characterized extensively by NMR and MS. It appears that the altered macrocyclic compound is not a good substrate for the cytochrome P450 required to initiate modification at the C-26 position. The importance of the missing methoxy group at C-26 for activity has been noted previously (5, 19), and indeed the inhibitory activity of compound IIIb in a proliferation assay was significantly less than that of rapamycin. The successful incorporation of several proline analogs into altered rapamycins indicates that the PIE of the rapamycin producer (8) has a significant tolerance for structural variation in the amino acid substrate.
ACKNOWLEDGMENTS
This work was supported by a project grant from The Wellcome Trust. L. E. Khaw was supported in part by a studentship from the National Kidney Research Fund.
We thank Keith Chater and Celia Bruton for sharing their expert knowledge in ΦC31 and István Molnár, Barrie Wilkinson, and Laurenz Kellenberger for helpful discussions and suggestions.
REFERENCES
- 1.Aharonowitz Y, Cohen G, Martin J F. Penicillin and cephalosporin biosynthetic genes—structure, organization, regulation, and evolution. Annu Rev Microbiol. 1992;46:461–495. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio J F, Molnár I, Schwecke T, König A, Khaw L E, Staunton J, Leadlay P F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene. 1996;169:9–16. doi: 10.1016/0378-1119(95)00800-4. [DOI] [PubMed] [Google Scholar]
- 3.Bryne K M, Shafiee A, Nielsen J B, Arison B H, Monaghan R L, Kaplan L. The biosynthesis and enzymology of an immunosuppressant, immunomycin, produced by Streptomyces hygroscopicus var. ascomyceticus. In: Nash C, Hunter-Cevera J, Cooper R, Eveleih D E, Hamill R, editors. Developments in industrial microbiology series. Vol. 32. Dubuque, Iowa: Wm. C. Brown Publisher; 1993. pp. 29–47. [Google Scholar]
- 4.Chater K F, Lomovskaya N D, Voeykova T A, Sladkova I A, Mkztumian N M, Muravnik G L. Streptomyces ΦC31-like phages: cloning vectors, genome changes and host range. In: Sabo G, Biro S, Goodfellow M, editors. Biological, biochemical and biomedical aspects of actinomycetes. Proceedings of the 6th International Symposium on Actinomycete Biology. Budapest, Hungary: Academic Kvado; 1986. pp. 45–53. [Google Scholar]
- 5.Chen Y, Zhou P, Berova N, Zhang H, Nakanishi K. Conformational changes of rapamycin and analogs upon complexing with FKBP associated with activity: an application of second derivative CD spectroscopy. J Am Chem Soc. 1994;116:2683–2684. [Google Scholar]
- 6.Cheng Y R, Fang A, Demain A L. Effect of amino acids on rapamycin biosynthesis by Streptomyces hygroscopicus. Appl Microbiol Biotechnol. 1995;43:1096–1098. doi: 10.1007/BF00166931. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: The John Innes Foundation; 1985. [Google Scholar]
- 8.König A, Schwecke T, Molnár I, Böhm G A, Lowden P A S, Staunton J, Leadlay P F. The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin. Eur J Biochem. 1997;247:526–534. doi: 10.1111/j.1432-1033.1997.00526.x. [DOI] [PubMed] [Google Scholar]
- 9.Leistner E, Gupta R N, Spenser I D. A general method for the determination of precursor configuration in biosynthetic precursor-product relationships. Derivation of pipecolic acid from d-lysine, and of piperidine alkaloids from l-lysine. J Am Chem Soc. 1973;95:4040–4047. doi: 10.1021/ja00793a035. [DOI] [PubMed] [Google Scholar]
- 10.Lomovskaya N, Fonstein L, Xuan X, Stassi D, Katz L, Hutchinson C R. Gene disruption and replacement in the rapamycin-producing Streptomyces hygroscopicus strain ATCC 29253. Microbiology (Reading) 1997;143:875–883. doi: 10.1099/00221287-143-3-875. [DOI] [PubMed] [Google Scholar]
- 11.Lowden P A S, Böhm G A, Staunton J, Leadlay P F. The nature of the starter unit for the rapamycin polyketide synthase. Angew Chem Int Ed Engl. 1996;35:2249–2251. [PubMed] [Google Scholar]
- 12.Madduri K, Stuttard C, Vining L C. Cloning and location of a gene governing lysine ɛ-aminotransferase, an enzyme initiating β-lactam biosynthesis in Streptomyces spp. J Bacteriol. 1991;173:985–988. doi: 10.1128/jb.173.3.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAlpine J B, Swanson S J, Jackson M, Whittern D N. Revised NMR assignments for rapamycin. J Antibiot. 1991;44:688–690. doi: 10.7164/antibiotics.44.688. [DOI] [PubMed] [Google Scholar]
- 14.Metcalfe S M, Canman C E, Milner J, Morris R E, Goldman S, Kastan M B. Rapamycin and P53 act on different pathways to induce G1 arrest in mammalian cells. Oncogene. 1997;15:1635–1642. doi: 10.1038/sj.onc.1201341. [DOI] [PubMed] [Google Scholar]
- 15.Molnár I, Aparicio J F, Haydock S F, Khaw L E, Schwecke T, König A, Staunton J, Leadlay P F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase genes. Gene. 1996;169:1–7. doi: 10.1016/0378-1119(95)00799-7. [DOI] [PubMed] [Google Scholar]
- 16.Morris R E. Immunopharmacology of the macrolides FK506 and rapamycin: toward the era of rational immunosuppressive drug discovery, development and use. Transplant Proc. 1991;23:2722–2724. [PubMed] [Google Scholar]
- 17.Muth W L, Costilow R N. Ornithine cyclase (deaminating). III. Mechanism of the conversion of ornithine to proline. J Biol Chem. 1974;249:7463–7467. [PubMed] [Google Scholar]
- 18.Nielsen J B, Hsu M-J, Bryne K M, Kaplan L. Biosynthesis of the immunosuppressant immunomycin: the enzymology of pipecolate incorporation. Biochemistry. 1991;30:5789–5796. doi: 10.1021/bi00237a023. [DOI] [PubMed] [Google Scholar]
- 19.Nishida H, Sakakibara T, Aoki F, Saito T, Ichikawa K, Inagaki T, Kojima Y, Yamauchi Y, Huang L H, Guadliana M A, Kaneko T, Kojima N. Generation of novel rapamycin structures by microbial manipulations. J Antibiot. 1995;48:657–666. doi: 10.7164/antibiotics.48.657. [DOI] [PubMed] [Google Scholar]
- 20.Paiva N L, Roberts M F, Demain A L. The cyclohexane moiety of rapamycin is derived from shikimic acid in Streptomyces hygroscopicus. J Ind Microbiol. 1993;12:423–428. [Google Scholar]
- 21.Paiva N L, Demain A L, Roberts M F. The immediate precursor of the nitrogen-containing ring is free pipecolic acid. Enzyme Microb Technol. 1993;15:581–585. [Google Scholar]
- 22.Reynolds K A, Demain A L. Rapamycin, FK506, and ascomycin-related compounds. In: Strohl W R, editor. Drugs and pharmaceutical sciences series. Vol. 82. New York, N.Y: Marcel Dekker Publisher; 1997. pp. 497–520. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Sanglier J J, Baumann G, Dreyfuss M, Fehr T, Traber R, Schreier M H. Immunosuppressants of microbial origin 1. In: Nash C, Hunter-Cevera J, Cooper R, Eveleih D E, Hamill R, editors. Developments in industrial microbiology series. Vol. 32. Dubuque, Iowa: Wm. C. Brown Publisher; 1993. pp. 1–27. [Google Scholar]
- 25.Sans N, Schindler U, Schröder J. Ornithine cyclodeaminase from Ti plasmid C58: DNA sequence, enzyme properties and regulation of activity by arginine. Eur J Biochem. 1988;173:123–130. doi: 10.1111/j.1432-1033.1988.tb13975.x. [DOI] [PubMed] [Google Scholar]
- 26.Schindler U, Sans N, Schröder J. Ornithine cyclodeaminase from octopaline Ti plasmid Ach5: identification, DNA sequence, enzyme properties, and comparison with gene and enzyme from nopaline Ti plasmid C58. J Bacteriology. 1989;171:847–854. doi: 10.1128/jb.171.2.847-854.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwecke T, Aparicio J F, Molnár I, König A, Khaw L E, Haydock S F, Oliynyk M, Caffrey P, Cortés J, Lester J B, Böhm G A, Staunton J, Leadlay P F. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc Natl Acad Sci USA. 1995;92:7839–7843. doi: 10.1073/pnas.92.17.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]