Abstract
The dendritic cell (DC)-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin binding receptor (DC-SIGN) was shown to bind human immunodeficiency virus type 1 (HIV-1) viral envelope protein gp120 and proposed to function as a Trojan horse to enhance trans-virus infection to host T cells. To better understand the mechanism by which DC-SIGN and DC-SIGNR selectively bind HIV-1 gp120, we constructed a series of deletion mutations in the repeat regions of both receptors. Different truncated receptors exist in different oligomeric forms. The carbohydrate binding domain without any repeats was monomeric, whereas the full extracellular receptors existed as tetramers. All reconstituted receptors retained their ability to bind gp120. The dissociation constant, however, differed drastically from micromolar values for the monomeric receptors to nanomolar values for the tetrameric receptors, suggesting that the repeat region of these receptors contributes to the avidity of gp120 binding. Such oligomerization may provide a mechanism for the receptor to selectively recognize pathogens containing multiple high-mannose-concentration carbohydrates. In contrast, the receptors bound to ICAMs with submicromolar affinities that are similar to those of two nonspecific cell surface glycoproteins, FcγRIIb and FcγRIII, and the oligomerization of DC-SIGNR resulted in no increase in binding affinity to ICAM-3. These findings suggest that DC-SIGN may not discriminate other cell surface glycoproteins from ICAM-3 binding. The pH dependence in DC-SIGN binding to gp120 showed that the receptor retained high-affinity gp120 binding at neutral pH but lost gp120 binding at pH 5, suggesting a release mechanism of HIV in the acidic endosomal compartment by DC-SIGN. Our work contradicts the function of DC-SIGN as a Trojan horse to facilitate HIV-1 infection; rather, it supports the function of DC-SIGN/R (a designation referring to both DC-SIGN and DC-SIGNR) as an antigen-capturing receptor.
Dendritic cells (DC) are professional antigen presenting cells that capture and present pathogens to cells of the immune system. It has been proposed that viruses, such as human immunodeficiency virus type 1 (HIV-1), have adapted their infection strategy to evade the recognition, trafficking, and antigen presentation machinery of DC to facilitate viral access to target cells (20). For example, immature DC pulsed with HIV particles have been found to bind virus with high affinity and are capable of infecting T cells with high efficiency at suboptimal levels of virus titer (10, 21). While several receptors on the DC surface may be responsible for the binding of HIV viral particles, the DC-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN) has been studied intensively as a prototypic receptor capable of transmitting HIV-1 in trans (10). It is a type II transmembrane protein and a member of the calcium-dependent lectin-like receptor family, which recognizes carbohydrate moieties found on ICAMs, HIV-1 envelope protein gp120, and Ebola virus envelope glycoprotein 1 (gp1) (1, 27). In addition to DC-SIGN, the mannose receptor, Langerhan, and other cell surface C-type lectin receptors have also been found to bind HIV particles (31, 32). It remains controversial whether DC-SIGN-mediated viral recognition leads to amplified viral transmission and hence acts as a Trojan horse receptor for viral infection or antigen presentation.
DC-SIGNR, also a type II C-type lectin receptor, shares 77% sequence identity with DC-SIGN (22, 28). The sequence of both molecules can be divided into four domains, an intracellular domain that is responsible for signaling and internalization, a transmembrane domain, a repeat domain that contains seven 23-amino-acid repeats and one 15-amino-acid repeat, and a carbohydrate recognition domain (CRD) specific for high-mannose carbohydrate. The main difference between DC-SIGNR and DC-SIGN is their distribution on different cell types. DC-SIGN is expressed on DC, whereas DC-SIGNR is expressed primarily on sinusoidal and endothelial cells (2, 22, 28). The physiological role of DC-SIGN was proposed to be an adhesion receptor that recognizes ICAM molecules and thus promotes initial contacts between DC and T cells (11). However, it is unclear what determines DC-SIGN specificity to ICAM molecules from other cell surface glycoproteins and whether the polypeptide of ICAM molecules contributes to the receptor binding.
Our aim is to understand the mechanism by which DC-SIGN/R (a designation referring to both DC-SIGN and DC-SIGNR) selectively recognizes both the physiological ligands, ICAMs, and the HIV-1 envelope glycoprotein gp120. Using recombinant DC-SIGN/R constructs that vary in the number of repeats, we show that the repeat region of DC-SIGN/R determines the receptor oligomerization state, as well as gp120 binding affinity. In contrast, DC-SIGN/R recognition of ICAMs appears to be of a lower-affinity type and independent of the receptor oligomerization state. Furthermore, the measured binding affinity between DC-SIGN/R and ICAMs is similar to that between DC-SIGN/R and other nonspecific glycoproteins, such as Fcγ receptors. Thus, the ICAM recognition of DC-SIGN/R appears to be less specific than previously proposed. Finally, to address the fate of HIV-1 virus upon internalization by DC-SIGN, we measured the pH dependence of DC-SIGN/R and gp120 interaction and showed that the binding of HIV-1 gp120 by DC-SIGN/R decreased drastically at acidic pH.
MATERIALS AND METHODS
Reagents.
ICAM-FC-3, -2, and -1 fusion proteins along with respective antibodies were purchased from R&D Systems. Protein A was purchased from Amersham Bisociences. Cyanovirin-N (CV-N), a carbohydrate specific gp120 binding protein, was provided by James McMahon; the monoclonal antibody 2G12, a conformationally sensitive gp120-specific antibody that recognizes high-mannose carbohydrates on gp120, was obtained from Herman Kattinger; the IIIB strain of HIV-1 gp120 was obtained from Bernard Moss. All other gp120s were CHO cell-expressed recombinant proteins derived from NL4-3, a T-cell-line-adapted clade B strain; 92Ug21-9 and 92Ug037, clade A Uganda strains; and Th14-12, a clade B Thailand strain.
Monoclonal antibodies II.1 and I.13 are raised by immunizing mice with a bacterially expressed recombinant DC-SIGN CRD. Both I.13 and II.1 stained DC-SIGN-transfected Jurkat cells but not untransfected controls by fluorescence-activated cell sorter analysis (data not shown). They also stained immature monocyte-derived DC better than mature monocyte-derived DC (data not shown), consistent with the fact that DC-SIGN expression is downregulated upon DC maturation.
Recombinant DNA expression constructs and purification.
Soluble DC-SIGN and DC-SIGNR with various numbers of repeats were produced by PCR and cloned into the pET22b expression vector (Novagen). They were designated DC-SIGNR CRD (residues 265 to 399) for the CRD-only construct and R8 (residues 249 to 399), R7 (residues 226 to 399), R5 (residues 180 to 399), and R1 (residues 88 to 399) for the DC-SIGNR constructs starting at the eighth, seventh, fifth, and first repeat, respectively. Similarly, DC-SIGN CRD (residues 253 to 404), R8 (residues 237 to 404), and R1 (residues 76 to 404) were also made. All constructs were expressed as inclusion bodies in Escherichia coli. Recombinant proteins were refolded by rapid dilution into a buffer consisting of 0.5 M arginine, 5 mM cysteamine (Sigma), 1 mM cystamine (Sigma), and 0.1 M Tris-HCl (pH 8.0). The refolded receptors were purified with an ion-exchange column source (15Q) and a Superdex 200 HR 16/60 size exclusion column (Amershan Bioscience). Constructs DC-SIGNR R1, DC-SIGN R1, DC-SIGN R8, and DC-SIGN CRD also contained a six-His tag; only DC-SIGN CRD incorporated the use of the six-His tag for purification.
Size exclusion and native gels.
Oligomer determination was performed by the injection of purified refolded protein onto a size exclusion Superdex S200 HR 16/30 column with 0.2 M NaCl and 50 mM Tris-HCl (pH 8.0) as the running buffer. Size exclusion standards (Amersham Bioscience) were injected under identical conditions. Both the native and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses were run on a 10 to 15% polyacrylamide criterion gel (Bio-Rad Laboratories). Samples for SDS-PAGE were reduced with 5 mM dithiothreitol (DTT) and boiled for 10 min. In addition to SDS-PAGE analysis, mass spectrometry and N-terminal amino acid sequencing were used to confirm each construct.
Analytical ultracentrifugation (AUC).
Sedimentation equilibrium experiments were conducted with a Beckman Optima XL-I/A analytical ultracentrifuge (Beckman Coulter, Fullerton, Calif.). The protein was dissolved in 20 mM HEPES (pH 7.5) and 50 mM NaCl, and 180 μl of samples at concentrations between 0.05 and 0.9 mg/ml was loaded in double-sector centerpieces. Sedimentation equilibrium profiles were acquired at 4°C in three rotor speeds of 10,000 to 25,000 rpm. Protein extinction coefficients at 280 nm and partial specific volumes were estimated from amino acid composition with the software SEDNTERP, kindly provided by J. Philo (15). Extinction coefficients at 230 and 250 nm were determined with a spectrophotometer. The receptor oligomerization was modeled as a reversible monomer-dimer or monomer-dimer-tetramer equilibrium. Sedimentation velocity experiments were performed at a rotor speed of 50,000 rpm and a temperature of 23°C, with 400 μl of proteins at the same concentrations and buffer conditions as used for the equilibrium sedimentation. Interference optical fringe displacement data were acquired in intervals of 30 s. Data were analyzed by direct boundary modeling with continuous distributions of Lamm equation solutions with SEDFIT software. Diffusion coefficients were measured with a dynamic light-scattering instrument (Brookhaven Instruments, Holtsville, N.Y.) at 20°C. Scattered light at 514 nm was collected at a 90° angle and analyzed as a single-species model with SEDFIT software.
Surface plasmon resonance.
Binding studies were performed with a BIAcore 3000 instrument (BIAcore AB). Proteins were immobilized at pH 5.5 for albumin (Sigma) and DC-SIGN/R; at pH 4.5 for ICAM-2-Fc and ICAM-3-Fc (ICAM-3-Fc); at pH 5.5 for FcγRIIb (CD32), FcγRIII (CD16), and protein A; and at pH 3.5 for all gp120 strains onto a carboxymethylated dextran (CM5) surface with standard amine coupling by a targeted immobilization procedure (14). In addition to direct coupling, ICAM binding experiments were also performed by capture immobilization in which ICAM-Fc fusion proteins were captured by amine-coupled protein A (Amersham Pharmacia Biosciences). For these experiments, immobilized Fc fragment of a human immunoglobulin G (IgG) was used as a subtracted reference. Injections were performed at a flow rate of 20 μl/min with commercially available HBS-P buffers (BIAcore) supplemented with 2 mM CaCl2 (Quality Biological). Control experiments were performed with HBS-P and HBS-EP buffers. Surfaces were regenerated using 10 mM glycine (BIAcore), at pH 2.0 or by removal of Ca2+ ions from the HBS-P buffer. Dissociation constants (KD) were determined from double reciprocal (1/Req versus 1/[Analyte]) or Scatchard (bound/free versus bound) plots using the equilibrium binding response at the end of injection as Req and the concentration of injected analyte or by fitting binding chromatogram data with BIAcore evaluation software. Due to the intrinsic high-affinity nature of CV-N-gp120 (CV-N-gp120) interaction, the CV-N-gp120 surface was regenerated with two injections of 50 mM sodium hydroxide in the pH-dependent binding experiment. To overcome the incomplete regeneration of the CV-N-gp120 surface at neutral pH, the CV-N injections were performed starting with acidic pH 4.5, proceeding to more basic pH 6.5.
Deglycosylation of gp120.
Deglycosylation of gp120 strains IIIB, MW959, and TH14-12 was performed using the glycosidase enzyme PNGase-F (New England Biolabs) at 37°C in 50 mM Na phosphate at pH 7.5. The reaction consisted of 3.4 μg of native gp120 and 25 U of PNGase-F. The extent of deglycosylation was analyzed by SDS-PAGE. To denature gp120, the soluble sample was boiled for 5 min in a buffer consisting of 50 mM 2-mercaptoethanol (Sigma), 0.75% Triton X-100 (Sigma), and 0.1% SDS (Sigma). For denaturation, the immobilized gp120 was treated with 6 N guanidinium hydrochloride (Sigma) and 5 mM 1,4-dithio-dl-threitol (Sigma) for 10 min.
RESULTS
Expression and refolding of recombinant DC-SIGN and DC-SIGNR.
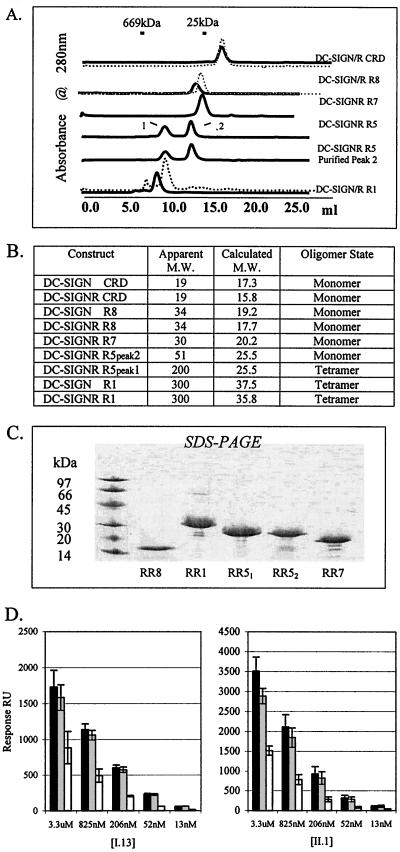
To investigate the functional role of the DC-SIGN and DC-SIGNR repeat domain, we constructed a series of recombinant soluble receptors varying in the number of repeats (Fig. 1). All constructs were expressed in bacteria, refolded by a limited dilution method, and purified by ion-exchange and size exclusion chromatography. Both SDS-PAGE and mass spectrometry confirmed the molecular weights of the constructs (Fig. 2). The function integrity of the recombinant receptors was evaluated with three DC-SIGN and DC-SIGNR dual-specific antibodies, I.13, II.1, and mAB1621 (clone 120612; R&D Systems), as well as a DC-SIGN-specific antibody mAB161 (clone 120507 from R&D Systems). Both the recombinant R1 and CRD constructs of DC-SIGN were recognized by mAB1621 with affinities of 18 and 11 nM, respectively, and by mAB161 with affinities of 0.1 and 1 nM, respectively. The recombinant DC-SIGNR R1 bound to mAB1621 with an affinity of 1.2 nM, but not to mAB161. In addition, both I.13 and II.1 bound to the recombinant R1, R8 and CRD constructs of DC-SIGNR (Fig. 2). The specific recognition between the antibodies and recombinant receptors suggests that these bacterially expressed DC-SIGN/R constructs possess a functional epitope.
FIG. 1.
Depiction of DC-SIGN and DC-SIGNR functional domains, including N-terminal cytoplasmic tail, transmembrane, eight copies of a 23-amino-acid repeat (R), and the CRD. Deletion constructs include the entire CRD plus a number of repeat sequences. DC-SIGN R1 and DC-SIGNR R1 contain the CRD and repeat sequences 1 to 8 (repeat 8 comprises a half repeat), DC-SIGNR R5 contains CRD and repeats 5 to 8, DC-SIGNR R7 contains the CRD and repeat sequences 7 and 8, DC-SIGN R8 and DC-SIGNR R8 contain the CRD and repeat 8, and DC-SIGN CRD and DC-SIGNR CRD contain only the CRD.
FIG. 2.
(A) Size exclusion chromatograms of DC-SIGN (dotted gray) CRD and DC-SIGNR (solid black) CRD, DC-SIGN and DC-SIGNR R8, DC-SIGNR R7, DC-SIGNR R5, and DC-SIGN R1 and DC-SIGNR R1. DC-SIGN and DC-SIGNR CRD, R8, and R7 exhibit single monomeric species. DC-SIGN, DC-SIGNR CRD, and DC-SIGN R8 chromatogram profiles show a total elution volume of 30 ml; the remaining profiles show a total volume of 25 ml. DC-SIGNR R5 before fractionation has oligomer peak 1 and monomer peak 2 labeled. The second DC-SIGNR R5 chromatogram is the fraction peak 2 from a previous size exclusion profile fractioned, concentrated, and separated by size exclusion. DC-SIGNR R1 and DC-SIGN R1 are shown only as oligomer species. Standards shown are thyroglobulin (669 kDa) and chymotrypsinogen (25 kDa). Chromatograms are representative of at least three independent experiments. (B) Table estimating receptor oligomer state. The apparent molecular weight (M.W.) was obtained from the size exclusion elution volume and compared to the elutionvolume of the standards. The calculated molecular weights were determined by predicted molecular weight and verified by mass spectrometry and SDS-PAGE. Oligomer state estimates are based on size exclusion chromatography results, analytical centrifugation data for DC-SIGNR R5 and DC-SIGNR R1, and observed binding affinity. (C) SDS-PAGE analysis (10 to 15% gradient PAGE gel) of purified DC-SIGNR constructs. Lane 1, molecular mass marker; lane 2, DC-SIGNR R8 (∼17 kDa); lane 3, DC-SIGNR R1 (∼34 kDa); lane 4DC-SIGNR R5 peak 1, fractionated (∼26 kDa); lane 5, DC-SIGNR R5 peak 2, fractionated (∼26 kDa); lane 6, DC-SIGNR R7 (∼20 kDa). All samples were treated with 5 mM DTT and incubated at 100°C for 5 min. (D) Surface plasmon resonance binding of DC-SIGN antibodies II.1 and I.13 to immobilized DC-SIGNR R8 (black), DC-SIGNR R5 (gray), and DC-SIGNR R1 (white). Each binding experiment was performed with serial 4× dilutions of the analyte and is representative of averaged triplicate injections.
The repeat domain of DC-SIGN/R contributes to receptor oligomerization.
Depending on the number of repeats, the recombinant receptors displayed different oligomerization states, as shown by their gel filtration profiles (Fig. 2). Both DC-SIGN and DC-SIGNR CRD constructs eluted with an apparent molecular mass of ∼19 kDa by gel filtration and are most likely monomers. The R7 and R8 constructs, which start at the seventh and eighth repeats, respectively, displayed apparent molecular masses of 30 and 34 kDa. The most interesting construct is DC-SIGNR R5, which possesses the last four repeats (repeats five through eight). It existed in two forms with apparent molecular masses of 51 and 200 kDa by gel filtration. Moreover, when the fraction-purified 51- and 200-kDa forms were injected onto the gel filtration column, each form redistributed with an equal partition into the same 51- and 200-kDa peaks (Fig. 2). This suggests that the 51- and 200-kDa forms of DC-SIGNR R5 exist in an interchangeable equilibrium. No intermediate oligomeric species were observed. The longest constructs, DCSIGN R1 and DC-SIGNR R1 (which contains all eight repeats), elute with an apparent molecular mass of 300 kDa.
To further define the oligomeric state of DC-SIGN, an AUC method was also used to measure the oligomeric association of soluble DC-SIGNR R1 and R5 receptors (Fig. 3). Under the conditions of the sedimentation equilibrium experiment, the full extracellular DC-SIGNR R1 construct was found to exist predominantly as a tetramer. The tetrameric state of the receptor was also confirmed by its diffusion coefficient, obtained from the dynamic light-scattering measurement (D20 = 2.94 × 10−7 cm2/s) and the sedimentation velocity (Fig. 3) measurements, showing an estimated receptor molar mass of 4.2 times that of the monomer. The DC-SIGNR R5 construct was observed to be mostly monomeric under the AUC experimental conditions. However, the measured absorbance distributions could be modeled as a reversible weak monomer-tetramer self-association. The weaker self-assembly of the molecule in sedimentation compared to the results of gel filtration may be due to differences in the experimental conditions, such as the pH of the buffer. These results demonstrate that the function of the DC-SIGNR repeats is to form a stable receptor tetramer. They also suggest that different repeats contribute differently in the receptor oligomerization. In particular, the fifth repeat appears to be critical in formation of the tetramer.
FIG. 3.
Sedimentation equilibrium analysis. The radial absorbance distribution of R1 DC-SIGNR at 230 nm is shown at 20,000 rpm (squares) and 25,000 rpm (diamonds). The radial absorbance distribution of DC-SIGNR R5 at 280 nm is shown at the same rotor speeds (20,000 rpm [circles] and 25,000 rpm [triangles]). The solid lines are part of a global fit joint with similar data at loading concentrations from 0.08 to 0.8 mg/ml, scanned at wavelengths of 230, 250, and 280 nm at rotor speeds of 10,000, 15,000, and 20,000 rpm (for DC-SIGNR R1) and 15,000, 20,000, and 25,000 rpm (for DC-SIGNR R5). For DC-SIGNR R1, the data were modeled with a reversible monomer-dimer-tetramer self-association equilibrium (resulting in a monomer-dimer association constant of 1.7 × 106 M−1 and a monomer-tetramer association constant of 2.8 × 1019 M−3 or half-KD values for the monomer-dimer and dimer-tetramer transition of 0.6 and 0.1 μM, respectively), whereas for DC-SIGNR R5, a model for reversible monomer-dimer equilibrium was used (resulting in a monomer-dimer association constant of 5.0 × 102 M−1). The inset shows the sedimentation coefficient distributions of DC-SIGNR R1 at 0.8 mg/ml (solid line) and R5 at 0.9 mg/ml (dotted line).
DC-SIGN/R recognize diverse strains of gp120.
We measured the binding of DC-SIGN/R receptors to gp120 envelope proteins from several strains of HIV-1 with either X4 or R5 tropism (Table 1). To compare the binding among various gp120 proteins, the recombinant extracellular full-length DC-SIGN, DC-SIGNR, and an Fc fusion construct of DC-SIGNR were immobilized on a CM5 sensor chip with primary amine attachment. The analyte consisted of individual gp120 protein at concentrations from 0.04 to 0.8 μM in HBS buffer. All recombinant CHO cell-expressed gp120 tested here bound to immobilized DC-SIGN, DC-SIGNR, and an Fc fusion DC-SIGNR with nanomolar affinity. The close agreement in gp120 binding affinities between the bacterially expressed DC-SIGNR and DC-SIGNR-Fc suggests that the refolded DC-SIGNR is fully functional. Although different strains of gp120 bound to the receptors with different affinities, similar affinities were observed between DC-SIGN and DC-SIGNR for all gp120s tested. The affinity did not correlate with the viral tropism. For example, while both DC-SIGN and DC-SIGNR recognized the R5 tropic Uganda strain of gp120, 92Ug037, with an affinity of about 5 nM, they recognized an R5 Thailand strain of gp120, Th14-12, with 10 times weaker affinity. In contrast, both receptors bound to an X4 tropic of gp120, NL4-3, with affinity similar to that of 92Ug037.
TABLE 1.
DC-SIGN/R affinities to various gp120 strains
| gp120 straina | Tropism | Affinity KD (μM)
|
||
|---|---|---|---|---|
| DC-SIGN | DC-SIGNR | DC-SIGNR-Fc | ||
| NL4-3 | X4 | 0.003 | 0.007 | 0.021 |
| 92Ug21-9 | X4 | 0.023 | 0.027 | 0.026 |
| Th14-12 | R5 | 0.061 | 0.075 | 0.077 |
| 92Ug037 | R5 | 0.003 | 0.005 | 0.002 |
NL4-3 is a T-cell line adapted clade B strain, 92Ug21-9 and 92Ug037 are clade A Uganda strains, and Th14-12 is a clade B Thailand strain. DC-SIGN R1 and DC-SIGNR R1 were immobilized onto the CM5 surface, and DC-SIGNR R1-Fc was captured with immobilized protein A. gp120 strains were injected over immobilized DC-SIGN/Rs. Dissociation constants are represented as 1 × 10 M−6 (μM) unless otherwise indicated. Each binding experiment is representative of either multiple single-injection experiments or averaged triplicate injections.
DC-SIGN/R oligomerization contributes to gp120 recognition.
It is known that some C lectins, such as the macrophage mannose receptor, utilize their receptor oligomerization as a mechanism to increase ligand binding affinity (4, 6, 29). To examine the effect of DC-SIGN/R oligomerization on the binding of the HIV viral envelope protein gp120, we measured the KD values for DC-SIGN/R CRD, R8, R5, and R1 receptors binding to gp120 with a BIAcore 3000 instrument. The results showed that different-length soluble DC-SIGN and DC-SIGNR bound to the same strain of gp120 with different affinities (Table 2). For example, the CRD, R8, and R5 constructs of DC-SIGNR displayed KD values of 3 to 10 μM to the IIIB and Th14-12 strains of gp120 binding. In contrast, the full-length extracellular receptor DC-SIGNR R1, which contains exclusively the tetramer form of the receptor, bound to the same strains of gp120 with an affinity of about 5 nM. Interestingly, DC-SIGNR R5, which existed as a mixture of monomer and tetramer in gel filtration but mostly as a monomer in the AUC experiment, exhibited an affinity similar to those of DC-SIGNR CRD and R8, with an overall KD value of 10 μM. The correlation between the gp120 binding affinity and the oligomerization state of DC-SIGNR suggests that the observed nanomolar binding of DC-SIGNR requires the formation of a stable tetramer. The higher-affinity gp120 binding of DC-SIGNR R1 is also evident from its slower dissociation rate than other receptor constructs (Fig. 4). Similarly, the full-length R1 construct of DC-SIGN bound to immobilized NL4-3 20 to 40 times better than the CRD and R8 form of DC-SIGN, although the affinity difference was less drastic than DC-SIGNR. The observed lower affinity between DC-SIGN R1 and NL4-3 when the gp120 was immobilized than when the receptor was immobilized may be due to a partial blockage of DC-SIGN but not DC-SIGNR binding sites on gp120 as the result of gp120 immobilization. This partial inactivation of immobilized NL4-3 resulted in lower binding affinity to all three forms of DC-SIGN compared to DC-SIGNR. Since all forms of DC-SIGN and DC-SIGNR can be recognized by their specific antibodies, the decrease in gp120 binding affinity as a result of the removal of the receptor repeat domain cannot be explained by the potential disruption in the structure of CRD due to the deletion of the repeat region. Rather, the results suggest that the repeat region of DC-SIGN/R contribute to the avidity of gp120 binding.
TABLE 2.
Summary of DC-SIGN/R binding experimentsa
| Analyte | Oligomer state | Ligand | Analyte (μM) | Measured KD | Monomer adjusted KD |
|---|---|---|---|---|---|
| DC-SIGN R CRD | Monomer | gp120 (Th14-12) | 50.0-0.097 | 2.9 | 2.9 |
| DC-SIGN R R8 | Monomer | gp120 (IIIB) | 3500-0.488 | 3.9 | 3.9 |
| DC-SIGN R R5 | Mono/Tetra | gp120 (IIIB) | 1000-0.96 | 10.4 | 10.4 |
| DC-SIGN R R1 | Tetramer | gp120 (IIIB) | 50-0.0097 | 0.008 | 0.002 |
| DC-SIGN R R1 | Tetramer | gp120 (Th14-12) | 0.8-0.025 | 0.022 | 0.005 |
| DC-SIGN R R1 | Tetramer | gp120 (NL4-3) | 0.8-0.025 | 0.029 | 0.007 |
| DC-SIGN R R1 | Tetramer | gp120(92Ug21-9) | 0.8-0.025 | 0.031 | 0.008 |
| DC-SIGN CRD | Monomer | gp120 (NL4-3) | 500-1.95 | 50.8 | 50.8 |
| DC-SIGN R8 | Monomer | gp120 (NL4-3) | 500-0.488 | 86.5 | 86.5 |
| DC-SIGN R1 | Tetramer | gp120 (NL4-3) | 50-0.97 | 9 | 2.3 |
| DC-SIGN R R8 | Monomer | ICAM-3-Fc | 500-0.488 | 54 | 54 |
| DC-SIGN R R5 | Mono/Tetra | ICAM-3-Fc | 1000-0.96 | 100-200 | 100-200 |
| DC-SIGN R R1 | Tetramer | ICAM-3-Fc | 50.0-0.0097 | 62 | 15 |
| DC-SIGN R R1 | Tetramer | ICAM-2-Fc | 250-0.097 | ND | ND |
| DC-SIGN R R1 | Tetramer | ICAM-1-Fc | 250-0.097 | ND | ND |
| DC-SIGN CRD | Monomer | ICAM-3-Fc | 500-1.95 | 331 | 29.1 |
| DC-SIGN R8 | Monomer | ICAM-3-Fc | 500-0.488 | >500 | >500 |
| DC-SIGN R1 | Tetramer | ICAM-2-Fc | 50-6.25 | ND | ND |
| DC-SIGN R1 | Tetramer | ICAM-3-Fc | 63-3.91 | 4.6 | 1.1 |
| DC-SIGN CRD | Monomer | FcγRIII (CD16) | 500-0.488 | 10.5 | 10.5 |
| DC-SIGN R1 | Tetramer | FcγRIII (CD16) | 250-0.488 | 421.5 | 16.0 |
| DC-SIGN R R1 | Tetramer | FcγRIII (CD16) | 250-3.9 | 15.8 | 3.95 |
| DC-SIGN R R1 | Tetramer | FcγRIIb (CD32) | 250-0.487 | 33.5 | 8.37 |
Ligand immobilization levels and concentration range of injected analyte are indicated. For comparison, monomer KD values were divided by respective receptor oligomerization states. All concentrations and dissociation constants represented as 1 × 10 M−6 (μM) unless otherwise indicated. Each binding experiment was performed with serial 2× dilutions of analyte and is representative of either multiple single-injection experiments or averaged triplicate injections. ND, nondetectable; Mono/Tetra, monomer-tetramer.
FIG. 4.
Surface plasmon binding response curves between immobilized ligand gp120 IIIB and injected DC-SIGNR analyte constructs. Serial dilution injection concentrations and gp120 immobilization densities, as well as additional data for DC-SIGN constructs binding to gp120, are summarized in Table 2. The data are representative of at least three independent experiments. For some injections of DC-SIGNR R5 and DC-SIGNR R8, a portion of the dissociation phase was omitted for clarity; this portion of the data was not used for calculating KD values.
DC-SIGNR recognizes denatured but not deglycosylated gp120.
The binding of DC-SIGN to ICAMs has been shown to be mediated solely by carbohydrates located on ICAM molecules. In contrast, DC-SIGN binding to gp120 has been proposed to depend on both the polypeptide component of gp120 and the high-mannose carbohydrates (12). To investigate the role of carbohydrate in gp120 recognition by DC-SIGNR and whether the polypeptide conformation of gp120 is important for the receptor binding, we examined the binding between the R1 construct of DC-SIGNR and different forms of gp120, including the native, denatured, and denatured and deglycosylated forms of gp120 (Fig. 5A). To deglycosylate gp120, the protein was treated with PNGase-F and the extent of the deglycosylation reaction was evaluated by SDS-PAGE, followed by silver staining or Western blotting and by the binding of deglycosylated gp120 to the mannose-specific gp120-neutralizing antibody 2G12, whose binding to gp120 was shown to be sensitive to both PNGase-F treatment and denaturation of gp120 (Fig. 5B) (24, 25, 30). For denaturation, the immobilized gp120 was treated with 6 N guanidinium hydrochloride and 5 mM DTT for 10 min. As expected, the deglycosylated gp120 did not bind to 2G12. Interestingly, the denatured gp120 retained nearly 40% of 2G12 binding (Fig. 5A). DC-SIGNR R1 bound to both the native and denatured gp120 with similar affinity but did not bind to the denatured and deglycosylated gp120. This suggests that the DC-SIGNR binding to gp120 does not depend on gp120 polypeptide conformation. To further explore the conformational state of the denatured gp120, the binding between gp120 and a conformation-sensitive gp120-specific monoclonal antibody, F105, and a soluble CD4 were also measured (Fig. 5A). The antibodies F105 and CD4 bound to the native gp120 but not to the denatured gp120 (Fig. 5A), indicating a loss of the conformation-sensitive epitopes upon denaturation. The binding of a gp120-specific but conformation-independent antibody SF2 was then tested to verify the activity of all immobilized gp120s. The SF2 antibody bound to all immobilized gp120 regardless of the denaturation state. The results indicate that the denatured but fully glycosylated gp120 bound DC-SIGNR R1 with an affinity similar to that of the native gp120. However, the denatured and deglycosylated gp120 lost nearly all of its ability to bind the receptor (Fig. 5A), indicating that the structural conformation of gp120 does not contribute significantly to DC-SIGNR binding and that the presence of multiple carbohydrates is sufficient to mediate its high-affinity interaction with DC-SIGNR.
FIG. 5.
(A) Surface plasmon resonance binding of DC-SIGNR R1 oligomer, antibody 2G12, soluble CD4, antibody F105, and antibody SF2 to native gp120 (black bars), denatured gp120 (gray bars), denatured and degylcosylated gp120 IIIB (white bars), mock-treated ICAM-3-Fc (bar with black vertical stripes), or native deglycosylated (PNGase F [PNGF]) ICAM-3-Fc (bar with black diagonal stripes). Binding is shown as a percentage compared with binding of native gp120 or ICAM-3 with each respective analyte. The data shown are representative of three independent experiments. (B) Deglycosylation of gp120 IIIB and control protein Fc-Ig; SDS-PAGE with silver staining. Unnumbered lanes, reading from left to right, are as follows: lanes 1 and 2, markers; lane 3, mock-treated gp120; lane 4, gp120 with PNGF and no denaturation; lane 5, gp120 with PNGF and denaturation; lane 6, control Fc-Ig with no PNGF; lane 7, Fc-Ig with PNGF and denaturation.
Binding of DC-SIGN and DC-SIGNR to ICAM-1, -2, and -3.
Intercellular adhesion molecules (ICAMs) are the presumed physiological ligands of DC-SIGN/R receptors. Similar to gp120 recognition, DC-SIGN/R constructs recognize the glycosylation moieties of ICAMs via the function of its CRD. To evaluate the affinity of the ICAM binding and the contribution of DC-SIGN/R repeats to this interaction, we measured the KD values for DC-SIGN/R CRD, R8, R5, and R1 binding to immobilized ICAM-1, -2, and -3 with a BIAcore 3000 instrument (Table 2). To our surprise, only ICAM-3, among the three ICAMs, showed detectable binding in solution to DC-SIGNR, although all ICAMs bound to their conformation-sensitive antibodies with high affinities (data not shown). The KD value between ICAM-3 and DC-SIGN/R was 1 to 10 μM. The binding was of lower affinity than that of gp120 binding and was mediated by the receptor CRD as mannose, EDTA, or deglycosylation of ICAM-3 abolished the receptor interaction. Similar to gp120 binding, denaturating ICAM-3 by urea and reducing agents did not affect DC-SIGNR binding (data not shown), consistent with the carbohydrate on ICAM-3 as the receptor binding epitope. This suggests that DC-SIGN binding to ICAM-3 primarily depends on the distribution of glycosylation sites on ICAM-3. To further evaluate the specificity of DC-SIGN/R and ICAM-3 binding, we investigated the receptor binding to other cell surface-glycosylated proteins, such as FcγRIIb and FcγRIII. To our surprise, the binding between DC-SIGN/R and the soluble FcγRIIb and FcγRIII were both blocked by EDTA and resulted in KD values of 5 to 15 μM (Fig. 6 and Table 2).
FIG. 6.
Metal-dependent binding of DC-SIGNR R1 (black bars) or CD11a (gray bars) to FcγRIIb, ICAM-3-Fc, ICAM-2-Fc, or ICAM-1-Fc in the presence or absence of metal chelator EDTA or EGTA. For DC-SIGNR R1 binding to FcγRIIb, EDTA was incubated with DC-SIGNR R1 (black diagonal bars). For CD11a binding to ICAMs, the metal chelator EGTA was incubated with CD11a (gray diagonal bars).
pH dependence of DC-SIGN/R and gp120 interaction.
Since DC-SIGN-bound antigen is known to be internalized to endosomal compartments, the question of whether DC-SIGN-mediated HIV internalization leads to later cell surface expression of the virus or degradation remains controversial. To address this issue, we explored the pH dependence of DC-SIGN/R and gp120 interactions. Binding experiments under various pH values were performed with running buffers consisting of 200 mM sodium chloride, 2 mM calcium choride, and 50 mM sodium acetate (pH adjusted to 6.5, 5.5, and 4.5). Experiments at pH 7.4 were performed with HBS-P supplemented with 2 mM calcium chloride. A fixed concentration of 0.8 μM NL4-3 gp120 in the same running buffers was injected over immobilized DC-SIGN/R. The binding was also measured with immobilized NL4-3 and 1 μM DC-SIGNR in the same running buffers as analytes. For comparison, the binding of CV-N to the same immobilized gp120 was measured under the same pH conditions. The results showed that DC-SIGNR binding to gp120 was pH dependent. While high-affinity gp120 binding was observed at pH 7.4 and 6.5, binding was reduced drastically at pH 5.5 and 4.5 (Fig. 7A and B). The reduced receptor binding at lower pHs cannot be attributed to the low-pH effect on gp120 carbohydrate ligands, since CV-N binding to gp120 was retained. Instead, the reduced gp120 binding at low pH is most likely due to the pH-induced loss of function in DC-SIGN/R receptors.
FIG. 7.
(A) Surface plasmon resonance binding of gp120 NL4-3 to immobilized DC-SIGN CRD (white bars), DC-SIGN R1 (gray diagonal bars), and DC-SIGNR R1 (black bars) at pH 7.5, 6.5, 5.5, and pH 4.5. The surface was regenerated with two injections of 20 mM EDTA (pH 8.0). (B) Surface plasmon resonance binding of DC-SIGNR R1 (black bars) and CV-N (bars with gray vertical stripes) to immobilized gp120 under various pH conditions. The experiments were performed starting with acidic pH 4.5, then pH 5.5, and pH 6.5. The increased binding of CV-N to gp120 at lower pHs is probably due to an incomplete regeneration of the gp120-CV-N interaction. DC-SIGNR R1 data are the average of three injections. CV-N data are representative of multiple experiments. (C) Surface plasmon resonance showing percent retention of immobilized protein surface and the pH stability of DC-SIGN R1 and DC-SIGNR R1 oligomers. Buffer solutions at pH 7.5, 6.5, 5.5, 4.5, 3.5 (protein A-captured DC-SIGNR-Fc only), and 2.0 (protein A-captured DC-SIGNR-Fc only) containing no protein were injected onto a CM5 surface immobilized with DC-SIGN CRD (white bars), DC-SIGN R1 (black bars with white dots), DC-SIGNR R1 (black bars), and DC-SIGNR R1-Fc bound to immobilized protein A (bars with black horizontal lines). Protein A-immobilized DC-SIGNR-Fc was used as a control for the loss of surface density upon low-pH treatment.
To address whether the loss of binding to gp120 at pH 5.5 and lower was due to the destabilization of the receptor oligomer or to the loss of function of the CRD, we again immobilized DC-SIGN CRD and DC-SIGNR R1 onto a CM5 sensor chip. The immobilized surfaces were then pulsed with buffers at pH 7.5, 6.5, 5.5, and 4.5, and the surface densities were monitored after each pH treatment (Fig. 7C). Since DC-SIGN/R receptor tetramerization is through noncovalent interactions, a low-pH-induced dissociation of the tetramer would predict a loss in surface density of the immobilized receptor as a result of the low pH treatment. In contrast, the surface density of the monomeric CRD construct of DC-SIGN is predicted to be resistant to the low-pH treatment. Thus, a differential reduction in surface density of immobilized R1 versus CRD of DC-SIGN/R following a low-pH treatment would indicate the dissociation of the tetramer under low-pH conditions. The results show that the surface densities of both the CRD and R1 of the receptors appear to resist low-pH treatment, suggesting that the receptor oligomer was stable at pH 5.5 or 4.5. Therefore, the loss of gp120 binding at low pH is likely due to the loss of the bound functional Ca2+ in the CRD that resulted in the loss of gp120 binding. The strong pH dependence of the receptor-gp120 interaction suggests a release mechanism of HIV virus upon DC-SIGN-mediated internalization to the more-acidic endosomal compartments.
DISCUSSION
DC-SIGN, found on professional antigen-presenting DC, was originally identified as an HIV gp120 binding protein (5). More recently, DC-SIGN has been proposed to function as an adhesion receptor for ICAM-3 and ICAM-2 and may be involved in T-cell priming and antigen presentation (10, 12). Here, we showed that the repeat domains of DC-SIGN/R are responsible for oligomerizing the receptor. Previously it was shown by chemical cross-linking that DC-SIGN forms tetramers (18). We observed two distinct states among the truncated receptors, the monomeric CRD, R8, R7, and R5 constructs of DC-SIGN and DC-SIGNR, as well as the tetrameric full-length extracellular construct, by using both size exclusion chromatography and the analytic ultracentrifugation technique. DC-SIGN and DC-SIGNR have been shown to selectively bind branched high-mannose carbohydrate (8, 9). The increased gp120 binding affinity of the tetrameric DC-SIGN/R compared to the shorter and monomeric receptors is most likely an avidity effect in which the tetrameric receptor is capable of recognizing multiple high-mannose carbohydrates found on the heavily glycosylated target molecule HIV-1 gp120.
Recently, a number of DC-SIGN and DC-SIGNR genes in both mice and primates have been identified (3, 19). Overall, they have been shown to be highly homologous in sequence and similar in function to the human receptors. However, one particular clone, CD209L2, found in Old World monkeys and nonhuman primates is missing nearly the entire 23-amino-acid repeat region (3). HIV capture and transmission studies with this gene showed that it is less efficient at binding and transmitting virus. In addition, a mouse DC-SIGN homologue has been identified with only 2.5 repeats; and it is also less efficient at transmitting HIV (19). It is conceivable that receptors with fewer repeats form smaller oligomers and are less effective in binding to viruses. Receptor oligomerization may also help to cluster the cytoplasmic internalization motifs leading to a more efficient signaling platform for internalization.
The role of gp120 polypeptide conformation in DC-SIGN binding remains controversial (12). To further investigate the role of glycosylation in the recognition of gp120 by DC-SIGNR, we examined the binding properties of deglycosylated gp120 to DC-SIGNR. Our results showed that DC-SIGNR binds with similar affinity to both the native and denatured gp120 but not to the deglycosylated gp120. The gp120 epitope recognized by 2G12 has been extensively mapped and shown to bind specific mannose carbohydrates found on gp120 (24, 25). The partial binding of 2G12 to denatured gp120 showed that both structural and carbohydrate-dependent gp120 epitopes are recognized by 2G12, suggesting that the binding epitopes of 2G12 may be more promiscuous than previously thought.
DC-SIGN/R displayed no measurable affinity to ICAM-1 or -2 and micromolar affinity to ICAM-3. In addition, DC-SIGN/R bound ICAM-3 with an affinity similar to that of Fcγ receptors, which contain two potential N-linked glycosylation sites with one or both high-mannose-type carbohydrates (23). The number of high-mannose carbohydrates found on these molecules is lower than that found on gp120 (16, 24). The similar DC-SIGN/R binding affinity to ICAM-3 and FcγR suggests that ICAM-3 may be less unique as a ligand to DC-SIGN than previously proposed and that any cell surface glycoprotein with the proper distribution of glycosylation sites may be recognized by DC-SIGN receptors. This is inconsistent with the proposed function of DC-SIGN as an adhesion receptor to promote cellular contact between T cells and DC. We have observed binding for DC-SIGN/R that is sensitive to both the oligomerization state of the receptor and the type and density of carbohydrates located on target molecules like HIV-1 gp120. These results argue that the function of DC-SIGN is to specifically capture low levels of glycan-containing antigens. Results from recent experiments examining N-linked glycosylation of HIV and Ebola virus as well as DC-mediated HIV transmission and T cells also support the hypothesis that DC-SIGN functions primarily as an antigen-capturing receptor (17).
It has been shown that immature DCs bearing DC-SIGN can internalize HIV-1 virus to endosomal compartments to evade the host immune surveillance and present the virus to T cells at a later time point. However, DC-SIGN was also shown to be capable of processing bound antibody as an antigen to major histocompatibility complex class II loading compartments (7, 13, 26). Furthermore, upon internalization of antigens, DC down regulate the expression of DC-SIGN and other C-type lectins while migrating to secondary lymphoid tissues (31, 32). To further address whether DC-SIGN-mediated gp120 internalization leads to viral representation or antigen processing, we measured the pH dependence of the DC-SIGN/R R1 to gp120. The results show that DC-SIGN/R R1 displayed a marked reduction in affinity to gp120 at an acidic pH close to that of an endosome. The reduced affinity is likely due to a loss of the bound Ca2+ in the CRD of the receptor. These results are more consistent with a receptor that binds antigens at the cell surface, internalizes them to a low-pH endosomal compartment, and then releases them for degradation and loading onto major histocompatibility complex molecules. It suggests that the representation of HIV-1 is most likely independent of DC-SIGN-mediated binding and internalization.
Acknowledgments
We thank C. Hammer for MS measurements, M. Garfield for N-terminal amino acid sequencing, and Biniam Hagos for protein expression and refolding. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: antiserum to HIV-1SF2 gp120 from Kathelyn Steimer, Chiron Corporation, and HIV-1 gp120 monoclonal antibody (F105) from Marshall Posner and Lisa Cavacini.
REFERENCES
- 1.Alvarez, C. P., F. Lasala, J. Carrillo, O. Muniz, A. L. Corbi, and R. Delgado. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76:6841-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashirova, A. A., T. B. Geijtenbeek, G. C. Van Duijnhoven, S. J. Van Vliet, J. B. Eilering, M. P. Martin, L. Wu, T. D. Martin, N. Viebig, P. A. Knolle, V. N. KewalRamani, Y. Van Kooyk, and M. Carrington. 2001. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 193:671-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashirova, A. A., L. Wu, J. Cheng, T. D. Martin, M. P. Martin, R. E. Benveniste, J. D. Lifson, V. N. KewalRamani, A. Hughes, and M. Carrington. 2003. Novel member of the CD209 (DC-SIGN) gene family in primates. J. Virol. 77:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beavil, R. L., P. Graber, N. Aubonney, J. Y. Bonnefoy, and H. J. Gould. 1995. CD23/Fc epsilon RII and its soluble fragments can form oligomers on the cell surface and in solution. Immunology 84:202-206. [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis, B. M., S. Scharnowske, and A. J. Watson. 1992. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 89:8356-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dierks, S. E., W. C. Bartlett, R. L. Edmeades, H. J. Gould, M. Rao, and D. H. Conrad. 1993. The oligomeric nature of the murine Fc epsilon RII/CD23. Implications for function. J. Immunol. 150:2372-2382. [PubMed] [Google Scholar]
- 7.Engering, A., T. B. Geijtenbeek, S. J. Van Vliet, M. Wijers, E. van Liempt, N. Demaurex, A. Lanzavecchia, J. Fransen, C. G. Figdor, V. Piguet, and Y. Van Kooyk. 2002. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 168:2118-2126. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg, H., D. A. Mitchell, K. Drickamer, and W. I. Weis. 2001. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294:2163-2166. [DOI] [PubMed] [Google Scholar]
- 9.Frison, N., M. E. Taylor, E. Soilleux, M. T. Bousser, R. Mayer, M. Monsigny, K. Drickamer, and A. C. Roche. 2003. Oligolysine-based oligosaccharide clusters: selective recognition and endocytosis by the mannose receptor and dendritic cell-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin. J. Biol. Chem. 278:23922-23929. [DOI] [PubMed] [Google Scholar]
- 10.Geijtenbeek, T. B., D. S. Kwon, R. Torensma, S. J. Van Vliet, G. C. Van Duijnhoven, J. Middel, I. L. Cornelissen, H. S. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor, and Y. Van Kooyk. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587-597. [DOI] [PubMed] [Google Scholar]
- 11.Geijtenbeek, T. B., R. Torensma, S. J. Van Vliet, G. C. Van Duijnhoven, G. J. Adema, Y. Van Kooyk, and C. G. Figdor. 2000. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100:575-585. [DOI] [PubMed] [Google Scholar]
- 12.Geijtenbeek, T. B., G. C. Van Duijnhoven, S. J. Van Vliet, E. Krieger, G. Vriend, C. G. Figdor, and Y. Van Kooyk. 2002. Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for intercellular adhesion molecule 3 and HIV-1. J. Biol. Chem. 277:11314-11320. [DOI] [PubMed] [Google Scholar]
- 13.Geijtenbeek, T. B., S. J. Van Vliet, E. A. Koppel, M. Sanchez-Hernandez, C. M. Vandenbroucke-Grauls, B. Appelmelk, and Y. Van Kooyk. 2003. Mycobacteria target DC-SIGN to suppress dendritic cell function. J. Exp. Med. 197:7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnsson, B., S. Lofas, and G. Lindquist. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 198:268-277. [DOI] [PubMed] [Google Scholar]
- 15. Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 16.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 17.Lin, G., G. Simmons, S. Pohlmann, F. Baribaud, H. Ni, G. J. Leslie, B. S. Haggarty, P. Bates, D. Weissman, J. A. Hoxie, and R. W. Doms. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J. Virol. 77:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276:28939-28945. [DOI] [PubMed] [Google Scholar]
- 19.Park, C. G., K. Takahara, E. Umemoto, Y. Yashima, K. Matsubara, Y. Matsuda, B. E. Clausen, K. Inaba, and R. M. Steinman. 2001. Five mouse homologues of the human dendritic cell C-type lectin, DC-SIGN. Int. Immunol. 13:1283-1290. [DOI] [PubMed] [Google Scholar]
- 20.Piguet, V., and A. Blauvelt. 2002. Essential roles for dendritic cells in the pathogenesis and potential treatment of HIV disease. J. Investig. Dermatol. 119:365-369. [DOI] [PubMed] [Google Scholar]
- 21.Pohlmann, S., G. J. Leslie, T. G. Edwards, T. Macfarlan, J. D. Reeves, K. Hiebenthal-Millow, F. Kirchhoff, F. Baribaud, and R. W. Doms. 2001. DC-SIGN interactions with human immunodeficiency virus: virus binding and transfer are dissociable functions. J. Virol. 75:10523-10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohlmann, S., E. J. Soilleux, F. Baribaud, G. J. Leslie, L. S. Morris, J. Trowsdale, B. Lee, N. Coleman, and R. W. Doms. 2001. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 98:2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radaev, S., and P. Sun. 2002. Recognition of immunoglobulins by Fcγ receptors. Mol. Immunol. 38:1073-1083. [DOI] [PubMed] [Google Scholar]
- 24.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlan, C. N., R. Pantophlet, M. R. Wormald, S. E. Ollmann, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schjetne, K. W., K. M. Thompson, T. Aarvak, B. Fleckenstein, L. M. Sollid, and B. Bogen. 2002. A mouse Cκ-specific T cell clone indicates that DC-SIGN is an efficient target for antibody-mediated delivery of T cell epitopes for MHC class II presentation. Int. Immunol. 14:1423-1430. [DOI] [PubMed] [Google Scholar]
- 27.Simmons, G., J. D. Reeves, C. C. Grogan, L. H. Vandenberghe, F. Baribaud, J. C. Whitbeck, E. Burke, M. J. Buchmeier, E. J. Soilleux, J. L. Riley, R. W. Doms, P. Bates, and S. Pohlmann. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115-123. [DOI] [PubMed] [Google Scholar]
- 28.Soilleux, E. J., R. Barten, and J. Trowsdale. 2000. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 165:2937-2942. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, M. E., K. Bezouska, and K. Drickamer. 1992. Contribution to ligand binding by multiple carbohydrate-recognition domains in the macrophage mannose receptor. J. Biol. Chem. 267:1719-1726. [PubMed] [Google Scholar]
- 30.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turville, S. G., J. Arthos, K. M. Donald, G. Lynch, H. Naif, G. Clark, D. Hart, and A. L. Cunningham. 2001. HIV gp120 receptors on human dendritic cells. Blood 98:2482-2488. [DOI] [PubMed] [Google Scholar]
- 32.Turville, S. G., P. U. Cameron, A. Handley, G. Lin, S. Pohlmann, R. W. Doms, and A. L. Cunningham. 2002. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 3:975-983. [DOI] [PubMed] [Google Scholar]