Abstract
In the effort to identify deubiquitinating enzymes required for the growth of colorectal cancer (CRC) cells, we found that OTUB2 knockdown markedly inhibited the viability of these cancer cells in culture and in xenografted mice. It was also found that the level of OTUB2 was elevated in primary CRCs, and its high expression was a poor prognostic indicator for the patients. Interestingly, immunoprecipitation and LC-MS/MS analyses suggested that β-Catenin was an OTUB2-interacting protein, and there was a positive correlation between OTUB2 and β-Catenin expression in both CRC tissues and cell lines. We then performed reciprocal co-immunoprecipitations and demonstrated that OTUB2 and β-Catenin bound to each other. Enforced expression of OTUB2 decreased ubiquitination of β-Catenin and increased the half-life and intracellular level of β-Catenin, whereas the catalytic inactive OTUB2 did not. OTUB2 also enhanced β-Catenin-mediated transactivation as measured by TCF-luciferase and expression of endogenous CCND1 and MYC in CRC cells. These results indicated that OTUB2 was a potential target for therapeutic intervention for CRC.
Keywords: OTUB2, β-Catenin, colorectal cancer, deubiquitinating enzyme, target
Introduction
Most biological processes of human body are controlled by the ubiquitination of proteins, and protein ubiquitination occurs through an elaborate three-step enzymatic cascade mediated by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin ligase (E3) [1]. E1 activates the ubiquitin molecule through an ATP-dependent manner, then activated ubiquitin is transferred to an E2, and finally ubiquitin is ligated onto a specific substrate protein with the effect of an E3 [2]. However, protein ubiquitination is reversible, and ubiquitin can be removed from the substrate proteins by the effects of deubiquitinating enzymes (DUBs) [3]. DUBs, a key component of the ubiquitin-proteasome system, are also very important for most cellular processes, such as cell cycle, proliferation, growth, migration and invasion, and targeting DUBs has been also shown potential in preclinical tumor treatment [4]. Therefore, discovering functional DUBs in tumors is particularly important for the development of new anti-cancer drugs.
DUBs can be classified into major six distinct families, including ubiquitin-specific proteases (USPs), ovarian tumor-like proteases (OTUs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin disease proteases (MJDs), motif interacting with Ub-containing novel DUB family (MINDYs) and JAB1/MPN/MOV34 metalloenzymes (JAMMs) [5]. We recently performed a high-content screening in colorectal cancer (CRC), and identified several functional DUBs, including OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2) [6]. OTUB2 belongs to the OTU family proteins, and has a profound impact on regulating many biological events, including cellular antiviral responses, beta cell function, DNA repair, and signal transduction [7,8]. Recently, OTUB2 has been also reported to be involved in cancer development. OTUB2 was proven to be a key mediator of Hippo signaling by activating YAP/TAZ through its own SUMOylation, and promoted tumor metastasis [9]. OTUB2 was also reported to be upregulated in esophageal squamous cell carcinoma (ESCC), and promoted ESCC tumor development by regulating the protein expression of YAP/TAZ [10]. In non-small cell lung cancer (NSCLC), OTUB2 was also reported to stabilize U2AF2 protein, activate AKT/mTOR pathway and enhance the Warburg effect, which indicated that OTUB2 could be as a novel potential therapeutic target for the treatment of NSCLC [7].
β-Catenin is an adhesive junction protein that can regulate cell growth and intercellular adhesion, and it is a key component of the Wnt/β-Catenin signaling pathway [11]. The transcription factors TCF/LEF can be activated by binding to β-Catenin and then promote target genes’ transcription, such as CCND1 and MYC [12]. In CRC, mutations of APC or β-Catenin can stabilize β-Catenin, and the abnormal accumulation of β-Catenin can ultimately lead to the development of tumors [13]. Therefore, to further investigate the ubiquitination mechanism of β-Catenin is also important for the treatment of CRC.
In this study, we confirmed that OTUB2 was elevated in primary CRCs, and promoted the growth of CRC cells. We also found that OTUB2 and β-Catenin bound to each other in CRC cells, and OTUB2 stabilized β-Catenin by reducing its poly-ubiquitination. Furthermore, the chemical compound I-BET726 was also found exert its anti-tumor activity by inhibiting OTUB2/β-Catenin axis in CRC cells.
Methods
Cells, tissues and reagents
The CRC cell lines (HCT116, HT29, SW480, SW620 and SW948) were purchased from ATCC, Manassas, VA. HEK293T cell line was maintained in our laboratory. Cells were cultured in DMEM medium (Hyclone) with 10% fetal bovine serum (Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (Beyotime, Beijing, China).
Twenty pairs of primary CRC para-cancerous and cancerous tissues and the tissue arrays containing 169 CRC tumor tissues and paired para-cancerous tissues were collected from the Department of Colorectal Surgery, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China. The collection and use of the human tissues were approved by the Ethics Committee of Xinhua Hospital and informed consent was obtained for all the collections. Wnt3a was purchased from Sigma-Aldrich, St. Louis, MO. I-BET726 and puromycin were obtained from Selleck Chemicals, Houston, Texas, USA.
High-content screening
High-content screening was performed as described previously [6]. In brief, HCT116 cells with GFP were constructed by lentivirus, and then HCT116-GFP cells were infected with lentivirus-delivered shRNAs against 20 DUBs (including OTUB2, USP1, USP5, USP7, USP15, USP3, OTUD4, Mpnd, USP24, USP21, USP33, PAN2, USP9X, USP34, USP6, USP29, CYLD, UCHL5, OTUD7B and USP32) [6]. To ensure the efficiency of gene interference, three RNA interference targets were designed for each gene, and three plasmids carrying different targets were mixed in equal proportions for lentivirus packaging. Then, cell photography and counting were conducted by Celigo® Image Cytometer at indicated times, and growth curves were drawn in cells.
Preparation of lentivirus-delivered shRNAs, lentiviral infection
The negative (shNC) and lentivirus-delivered shRNAs against OTUB2 (shOTUB2) were synthesized by GeneChem, Shanghai, China. The target sequences for shOTUB2 were as follows: 5’-CATCCCACTACAACATCCTTT-3’ (shOTUB2#1), 5’-CGAGATGGATACCGCCCTGAA-3’ (shOTUB2#2) and 5’-TGTGGTGGAACTGGTAGAGAA-3’ (shOTUB2#3). The shRNAs and packaging plasmids were co-transfected into HEK293T cells to produce the lentivirus, and the viral particles were prepared following the manufacturer’s protocol (GeneChem, Shanghai, China).
The lentiviral particles containing shNC, shOTUB2#1 and shOTUB2#2 were then used to infect HCT116 or SW480 cells. Following two rounds of puromycin selection, the survival cells were considered stable-expressing cells, and the efficacy of OTUB2 knockdown was verified by western blot analysis.
Western blot analysis
Western blot was carried out as previously reported [14]. In brief, cells were lysed by using RIPA lysis buffer (Beyotime, Beijing, China), and total protein was extracted for following SDS-PAGE analysis. The primary antibodies against Myc-tag and Flag-tag were received from Medical & Biological Laboratories, Tokyo, Japan. Anti-β-Catenin and anti-phospho-β-Catenin (Ser675) antibodies were purchased from Cell Signaling Technology, Danvers, MA. The primary antibody against ubiquitin (Ub) was purchased from Santa Cruz Biotechnology, Santa Cruz, CA. The anti-OTUB2 antibody was bought from Absin, Shanghai, China, and the anti-GAPDH antibody was bought from Proteintech Group, Wuhan, China. HRP-labeled Goat Anti-Mouse IgG (H+L) and HRP-labeled Goat Anti-Rabbit IgG (H+L) were purchased from Beyotime, Beijing, China. The images of western blot were visualized with an ECL-chemiluminescence detection system.
Cell growth, viability, migration and colony formation
CRC cells were cultured for indicated times, and viable cells were evaluated by trypan blue exclusion staining (Beyotime, Beijing, China) or Cell Counting Kit-8 (CCK-8) assay (Selleck Chemicals, Houston, Texas, USA) following the manufacturers’ protocols. Cell migration and colony formation assays were stated in the Supplementary Methods.
Xenograft models
Eighteen female BALB/C nude mice (6~8 weeks old; 20~25 g) were used to establish the xenograft models. Mice were randomly divided into three groups (shNC group, shOTUB2#1 group and shOTUB2#2 group; n = 6 mice per group), and CRC cells infected with indicated lentivirus were subcutaneously injected into indicated mice (3 × 106 cells per mouse). Five days later, the tumor volumes were measured with a vernier caliper every three days for continuously two weeks. The tumor volumes were calculated as follows: V = 1/2 × (L × W × W). On Day 20, mice were sacrificed and tumors were excised for weighing.
Database analysis
GEPIA matched with TCGA normal and GTEx data (http://gepia2.cancer-pku.cn) was used to analyze the expression of OTUB2 in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ). GEO profiles (GDS5364/ILMN_1799198, GDS5365/ILMN_1799198) were used to evaluate the regulatory effects of I-BET726 on OTUB2 expression.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed as reported previously [14]. In brief, total RNA was extracted by using RNAiso Plus, Takara Bio Group, Japan. Then, RNA was reversely transcribed into cDNA, and qRT-PCR was conducted by using SYBR Green qPCR Master Mix (Takara Bio Group, Japan). The primers for qRT-PCR (synthesized by GENEWIZ, Suzhou, China) were as follows: OTUB2, forward, 5’-ATCCGCAAGACCAAAGGG-3’, reverse, 5’-CAAAGCCAGCAGCCAGAA-3’; CCND1, forward, 5’-CTCTAAGATGAAGGAGACCAT-3’, reverse, 5’-TTGGAGAGGAAGTGTTCAA-3’; MYC, forward, 5’-CGTCTCCACACATCAGCACAA-3’, reverse, 5’-TCTTGGCAGCAGGATAGTCCTT-3’; GAPDH, forward, 5’-GCACCGTCAAGGCTGAGAAC-3’, reverse, 5’-TGGTGAAGACGCCAGTGGA-3’.
Immunohistochemistry
Immunohistochemistry (IHC) was conducted as reported previously [6]. The tissue arrays containing 169 CRC tumor tissues and paired para-cancerous tissues were used for IHC analysis, and the case information was shown in our previous work [6]. IHC was performed with the anti-OTUB2 antibody (Absin, Shanghai, China). The staining of OTUB2 in the tissues was scored on a semi-quantitative score: 0, negative; 1, weak; 2, moderate; 3, strong.
Co-immunoprecipitation
Co-immunoprecipitation (Co-IP) was carried out as reported previously [15]. Cells were lysed with cell lysis buffer (Beyotime, Beijing, China), and whole cell lysates were prepared for incubating with indicative primary antibodies overnight at 4°C (anti-Myc-tag and anti-Flag-tag antibodies were from Medical & Biological Laboratories; anti-β-Catenin antibody was from Cell Signaling Technology; anti-OTUB2 antibody was from Novus Biologicals). Then, above mixture was incubated with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, USA) for 2 h, followed by washing, denaturation and western blot analysis.
Plasmids construction and transfection
The human full-length cDNA sequences of OTUB2 and β-Catenin were amplified by PCR and cloned into pcDNA3.1 vector with a Flag or Myc tag, and the catalytically inactive mutant of OTUB2 (C51S) was constructed with a PCR-based site-directed mutagenesis kit [7]. Plasmids were transfected into cells by using Lipofectamine®3000 following the manufacturer’s protocol (Invitrogen).
Cycloheximide chase assay
Empty vector or Flag-OTUB2 plasmids were transfected into HCT116 cells for 24 h. Then, cells were incubated with 50 μg/ml cycloheximide (CHX, Sigma-Aldrich) for indicated times (0, 6, 12 h), followed by western blot analysis.
Luciferase assay
The luciferase report genes Top-Flash and Fop-Flash were received from Addgene, and luciferase assay was performed by using the Dual-Luciferase® Reporter Assay System (Promega, USA) as described previously [16].
Statistical analysis
GraphPad Prism 5 was used to draw pictures and analyze data. The Kaplan-Meier method and Log-rank test were used for survival analysis. When comparing two groups, the student’s t test was used. An ANOVA was used to compare three or more groups. A p value less than 0.05 was considered to be statistically significant.
Results
OTUB2 is required for the growth of colorectal cancer
Through high-content screening based on cell growth, we found several functional DUBs in CRC, including OTUB2 (Figure 1A and 1B). Then, OTUB2 was knocked down by shRNAs in HCT116 and SW480 cells (Figure 1C), and we confirmed that OTUB2 knockdown significantly inhibited the growth of CRC cells in both of HCT116 and SW480 cells (Figure 1D and 1E). In contrast, overexpression of OTUB2 promoted CRC cell growth (Figure S1). Knockdown of OTUB2 also inhibited the migration and colony formation of CRC cells (Figure S2). Furthermore, cell-derived xenografts were established and knockdown of OTUB2 also significantly suppressed the tumor growth of CRC in vivo (Figure 1F-H). Taken together, these results showed that OTUB2 exerted tumor-promoting effects in CRC cells.
Figure 1.
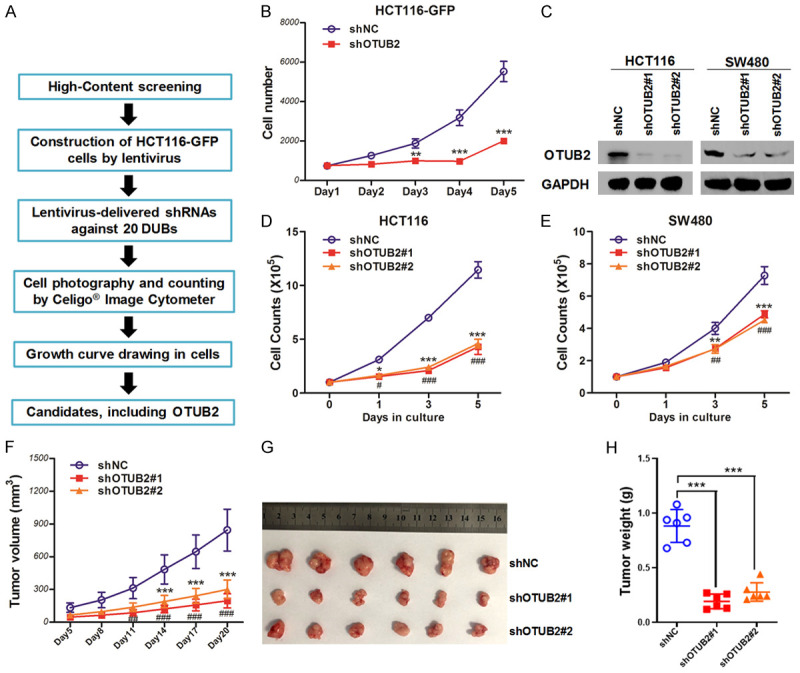
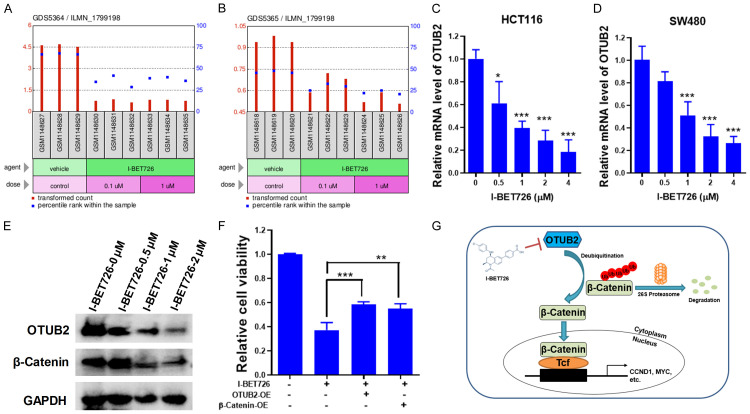
OTUB2 is required for the growth of colorectal cancer. (A and B) Flow chart of the high-content screening (A), and the cell growth curves of the candidate OTUB2 after its silencing by using shOTUB2-derived lentivirus (lentivirus packaged in a mixed ratio of shOTUB2#1, shOTUB2#2 and shOTUB2#3) (B). (C) Silencing efficiency of indicated shRNAs by western blot. (D and E) HCT116 (D) and SW480 (E) cells were infected with indicated shRNAs-derived lentivirus for indicated times, followed by trypan blue exclusion staining, and cells were counted. (F) Tumor growth curves. (G and H) At the end of the animal study, tumors were excised, taken photos (G) and weighed (H). *P<0.05, **P<0.01, ***P<0.001; ##P<0.01, ###P<0.001.
Elevated OTUB2 predicts a poor prognosis for colorectal cancer patients
Subsequently, the expression of OTUB2 was analyzed by the public cancer database, and the results showed that OTUB2 was significantly upregulated in both of colon adenocarcinoma and rectum adenocarcinoma (Figure 2A). Then, twenty pairs of human primary CRC para-cancerous and cancerous tissues were collected and prepared for qRT-PCR analysis, and the results showed that the mRNA levels of OTUB2 were significantly upregulated in CRC tumor tissues (Figure 2B). To further confirm the expression of OTUB2 in CRC tissues, the tissue arrays containing 169 pairs of CRC para-cancerous and cancerous tissues were used for IHC analysis, and OTUB2 was found elevated in CRC cancerous tissues (Figure 2C and 2D; Table S1). Based on the expression levels of OTUB2 in CRC tumor tissues, the Kaplan-Meier survival analysis showed that patients with high expression of OTUB2 had a shorter overall survival than the patients with low expression (Figure 2E). Taken together, these results indicated that OTUB2 was elevated in CRC and its high expression was involved in the poor prognosis of CRC patients.
Figure 2.
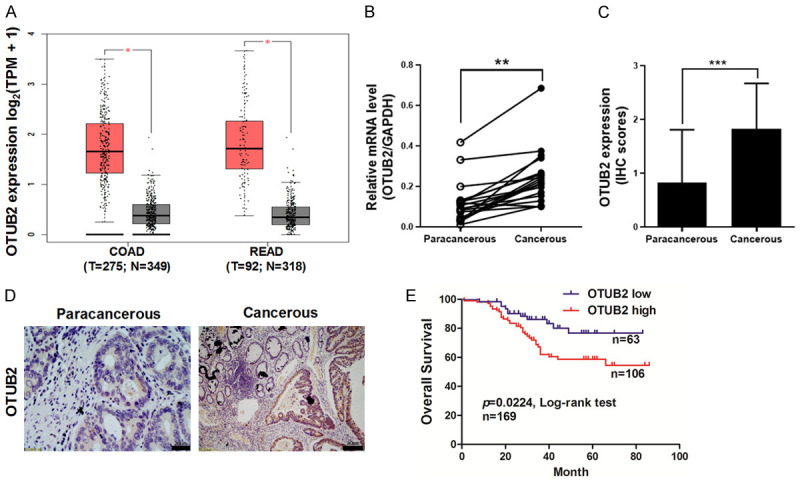
Elevated OTUB2 predicts a poor prognosis for colorectal cancer patients. (A) The expression of OTUB2 in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) analyzed by GEPIA matched with TCGA normal and GTEx data (http://gepia2.cancer-pku.cn). The red boxes represent the tumor tissues; the gray boxes represent the normal tissues. *P<0.01. (B) Twenty pairs of para-cancerous and cancerous tissues of colorectal cancer (CRC) were prepared for qRT-PCR to detect the mRNA levels of OTUB2. **P<0.01. (C and D) The tissue chips embedding 169 CRC para-cancerous and cancerous tissues were prepared for immunohistochemical analysis of OTUB2, and the immunohistochemical scores were calculated (C). The representative images of immunohistochemistry (D). ***P<0.001. (E) Survival curve analysis based on immunohistochemistry results.
OTUB2 interacts with β-Catenin in colorectal cancer
Our results of western blot revealed a positive correlation between OTUB2 protein level and β-Catenin protein level in CRC tissues and cell lines (Figure 3A-C). The relevant evidence that OTUB2 may interact with β-Catenin was also observed from the LC-MS/MS results (Figure S3; Table S2). To further confirm the protein interaction between OTUB2 and β-Catenin, Co-IPs were conducted. As shown in Figure 3D, the reciprocal Co-IPs showed that exogenous OTUB2 interacted with exogenous β-Catenin. Then, CRC cells were also lysed for Co-IPs. As shown in Figure 3E and 3F, the reciprocal Co-IPs showed that endogenous OTUB2 interacted with endogenous β-Catenin in CRC cells. Above results revealed that OTUB2 and β-Catenin bound to each other in CRC cells.
Figure 3.
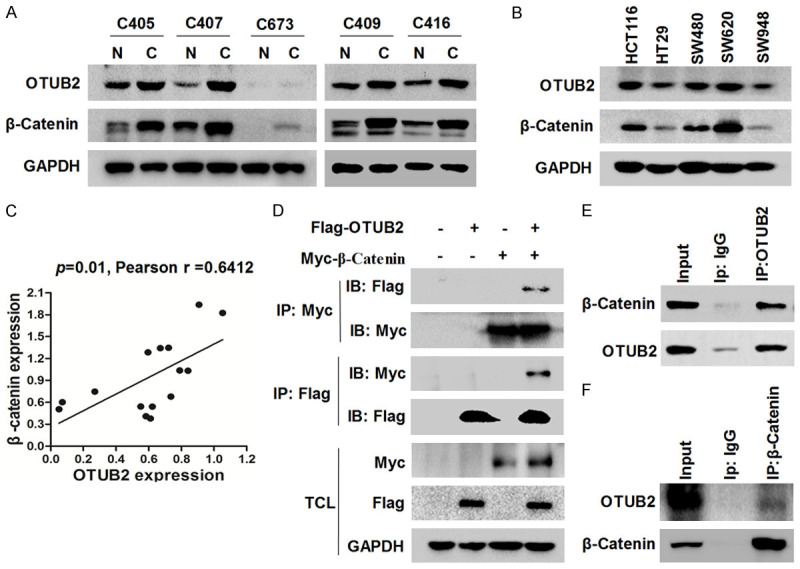
OTUB2 interacts with β-Catenin in colorectal cancer. (A-C) Five pairs of CRC para-cancerous and cancerous tissues (A) and five CRC cell lines (B) were prepared for western blot against OTUB2, β-Catenin and GAPDH. Correlation analysis between OTUB2 and β-Catenin in the tissues and cells were calculated (C). (D) The reciprocal co-immunoprecipitation of exogenous OTUB2 and β-Catenin was performed and analyzed by western blot. (E and F) HCT116 cells were prepared for reciprocal co-immunoprecipitation of endogenous OTUB2 and β-Catenin.
OTUB2 stabilizes β-Catenin by reducing its poly-ubiquitination
OTUB2 is a deubiquitinating enzyme, so we next evaluated whether it could regulate the protein expression of β-Catenin. As shown in Figure 4A, the western blot analyses showed that overexpression of the wild-type OTUB2 markedly increased the protein expression of β-Catenin in a dose-dependent manner, but the catalytically inactive mutant of OTUB2 (C51S) lost its up-regulatory effect. It was worth noting that OTUB2 C51S down-regulated β-Catenin, likely through competing the substrate with endogenous wild-type OTUB2 in CRC cells (Figure 4A). To further confirm whether OTUB2 regulated the protein stability of β-Catenin, CHX chase assay was conducted. As shown in Figure 4B and 4C, the CHX chase assay showed that OTUB2 overexpression significantly prolonged the half-life of β-Catenin protein. Due to the ability of β-Catenin to degrade through the ubiquitin-proteasome system, we next examined the effect of OTUB2 on its ubiquitination level. As shown in Figure 4D, the Co-IP showed that overexpression of OTUB2 markedly reduced the poly-ubiquitination of β-Catenin. Taken together, these results indicated that OTUB2 stabilized β-Catenin protein through inhibiting its poly-ubiquitination.
Figure 4.
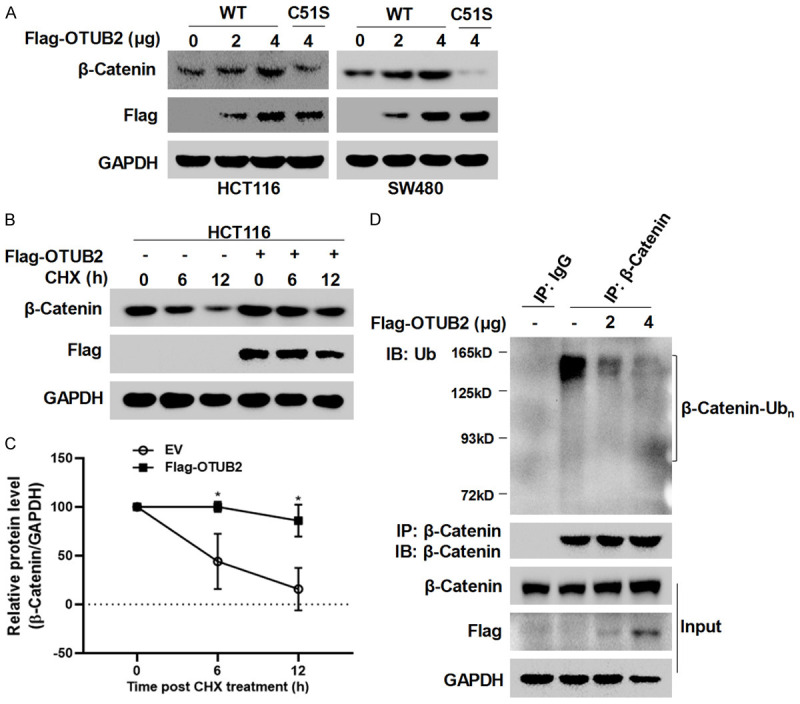
OTUB2 stabilizes β-Catenin by reducing its poly-ubiquitination. (A) Wild-type and catalytic-site mutated OTUB2 plasmids with a Flag tag were transfected into HCT116 and SW480 cells for 48 hours, followed by western blot analysis. (B and C) Cycloheximide (CHX) chase assay was performed and detected by western blot (B). Optical density was also measured, and statistical analysis was conducted (C). (D) Co-immunoprecipitation (Co-IP) was performed to evaluate the ubiquitination of β-Catenin.
OTUB2 regulates β-Catenin signaling in colorectal cancer cells
Next, the effect of OTUB2 on β-Catenin signaling was further investigated. Firstly, the TCF/LEF-driven luciferase reporter gene Top-Flash (indicating the transcriptional activity of β-Catenin) and the negative control Fop-Flash were prepared for luciferase assay. As shown in Figure 5A, overexpression of OTUB2 significantly enhanced the luciferase activity of Top-Flash in CRC cells, but had no effects on Fop-Flash. Conversely, knockdown of OTUB2 downregulated the luciferase activity of Top-Flash with or without Wnt3a stimulation (Figure 5B), which further indicated that OTUB2 mediated Wnt/β-Catenin signaling in CRC cells. Meanwhile, western blot was also carried out to detect the activation of β-Catenin in CRC cells. As shown in Figure 5C, overexpression of OTUB2 markedly upregulated the expression of phospho-β-Catenin in CRC cells. In contrast, knockdown of OTUB2 downregulated the expression of phospho-β-Catenin (Figure 5D). To further verify the effects of OTUB2 on Wnt/β-Catenin signaling, qRT-PCR analysis was carried out to detect the expression of Wnt-related downstream genes, including classical CCND1 and MYC. Our results of qRT-PCR confirmed that knockdown of OTUB2 inhibited the expression of CCND1 and MYC in CRC cells (Figure 5E and 5F), and overexpression of OTUB2 increased the mRNA levels of CCND1 and MYC (Figure 5G and 5H).
Figure 5.
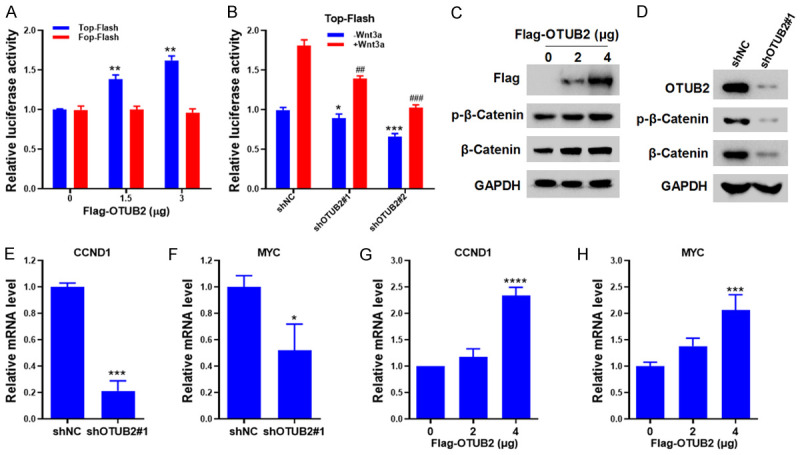
OTUB2 regulates β-Catenin signaling in colorectal cancer cells. (A) Top-Flash and Fop-Flash plasmids together with indicated concentrations of OTUB2 plasmids were transfected into HCT116 cells for 48 hours, followed by luciferase assay. (B) HCT116 cells transfected with Top-Flash plasmids were infected with indicative shRNAs-derived lentivirus or incubated with Wnt3a, followed by luciferase assay. (C and D) HCT116 cells overexpressed (C) or silenced with OTUB2 (D) were lysed for western blot. (E and F) HCT116 cells infected with indicated shRNAs-derived lentivirus were prepared for qRT-PCR analysis to evaluate the mRNA levels of CCND1 (E) and MYC (F). (G and H) HCT116 cells were transfected with increasing doses of Flag-OTUB2 plasmids (0, 2, 4 μg) for 48 hours, followed by qRT-PCR analysis to evaluate the mRNA levels of CCND1 (G) and MYC (H). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; ##P<0.01, ###P<0.001.
I-BET726 inhibits OTUB2 expression in colorectal cancer cells
From the GEO profiles, OTUB2 was downregulated by the chemical compound I-BET726 (Figure 6A and 6B). Then, to verify it, CRC cells were incubated with I-BET726, and qRT-PCR analysis was conducted. As shown in Figure 6C and 6D, the expression of OTUB2 was dramatically inhibited by I-BET726 in CRC cells. Meanwhile, the viability of CRC cells was also inhibited by the treatment of I-BET726 (Figure S4). In addition, I-BET726 inhibited the expression of β-Catenin in CRC cells (Figure 6E), and the expression levels of downstream genes CCND1 and MYC were also significantly downregulated by I-BET726 in a dose-dependent manner (Figure S5). Interestingly, overexpression of OTUB2 or β-Catenin could significantly attenuate the antitumor activity of I-BET726 in CRC cells (Figure 6F), which further indicated that I-BET726 exerted its anti-CRC activity partly by inhibiting OTUB2 (Figure 6G).
Figure 6.
I-BET726 inhibits OTUB2 expression in colorectal cancer cells. (A) The effect of I-BET726 on MYCN-amplified neuroblastoma cell line CHP-212 (GEO profile: GDS5364/ILMN_1799198). (B) The effect of I-BET726 on non-MYCN-amplified neuroblastoma cell line SK-N-SH (GEO profile: GDS5365/ILMN_1799198). (C and D) HCT116 (C) and SW480 (D) cells were incubated with indicated concentrations of I-BET726 for 24 hours, followed by qRT-PCR to detect the mRNA levels of OTUB2. (E) HCT116 cells were incubated with indicated concentrations of I-BET726 for 24 hours, followed by western blot analysis. (F) HCT116 cells overexpressed OTUB2 or β-catenin were incubated with 2 μM I-BET726 or vehicle for 24 hours, followed by CCK-8 assay to detect the cell viability. (G) Schematic model showing that OTUB2 deubiquitinates β-Catenin and mediates β-Catenin signaling in CRC cells, which is abolished by I-BET726 treatment through inhibiting the transcription of OTUB2. *P<0.05, **P<0.01, ***P<0.001.
Discussion
As the key component of the ubiquitin-proteasome system, DUBs have been proved to be functionally involved in lots of cellular processes, and more and more evidences indicate that DUBs devote to the development of tumors, which suggests that developing inhibitors of DUBs could be a novel strategy for the therapy of cancers [17,18]. Although scientists have made tremendous efforts in the development of DUB inhibitors, the clinical development of DUB inhibitors continues to be hindered [19]. One reason is the low selectivity of the compound itself; another reason is that our understanding of DUBs’ biology is still poor, especially regarding the specificity of DUBs’ substrate proteins [20]. Therefore, identifying functional DUBs in specific tumor types and discovering their specific substrate proteins are crucial for the successful development of DUB inhibitors.
Our previous studies have identified some functional DUBs in CRC through a high-content screening, and their respective functions and possible mechanisms have been reported by us. For example, we reported that the deubiquitinase USP5 promoted CRC cell growth and resistance to chemotherapeutics by stabilizing TUFM [6]. USP1 was also reported by us to be elevated in CRC and inhibiting USP1 sensitized CRC cells to DNA-targeting chemotherapeutics, which suggested that USP1 was a promising target for anti-CRC chemotherapy [14,21]. We also discovered the role of USP47 in CRC chemotherapy that USP47 attenuated chemotherapeutic doxorubicin-induced cell pyroptosis and apoptosis by deubiquitinating and stabilizing TCEA3 [15]. Our research object OTUB2 in this study was also a potent candidate screened out from our previous high-content screening [6], and our investigations confirmed that elevated OTUB2 promoted CRC cell growth and predicted as a negative index for CRC patients. Strikingly, OTUB2 has been recently reported to be upregulated in CRC by other researchers, which was consistent with our results [8]. OTUB2 was also found upregulated in other tumors, including NSCLC [7], hepatocellular carcinoma [22], gastric cancer [23] and esophageal squamous cell carcinoma [10]. These results further proved that OTUB2 exerted tumor-promoting effects in these cancers, and targeting OTUB2 could be a promising strategy for the therapy of CRC.
Then, our further investigations showed that OTUB2 stabilized β-Catenin and participated in regulating β-Catenin signaling in CRC cells. The Wnt/β-Catenin signaling has been regarded as a key driver for the carcinogenesis of CRC, and β-Catenin is a functional effector molecule of the signaling [24,25]. The post-modification and degradation of β-Catenin have been proved as the key events of the Wnt signaling and involved in the pathogenesis of CRC, suggesting that further understanding the ubiquitination mechanism of β-Catenin is crucial for the development of anti-CRC drugs [26]. Interestingly, our study showed that OTUB2 interacted with β-Catenin, and stabilized β-Catenin by reducing its polyubiquitination, which further indicated that suppressing Wnt/β-Catenin signaling by inhibiting OTUB2 may be a useful strategy for the treatment of CRC. However, recent studies also showed that β-Catenin was reported as a substrate for some other DUBs, such as USP37 in CRC [27], USP8 in hepatocellular carcinoma [28], and USP4 in CRC [29,30]. Therefore, the physiological and/or pathological importance of OTUB2 for β-Catenin is still obscure, which will be studied in our future work.
Through the GEO database, the chemical compound I-BET726 was found reduce the expression of OTUB2, and then we confirmed that I-BET726 decreased OTUB2 expression in CRC cells and exerted its anti-CRC activity by suppressing OTUB2/β-Catenin axis. I-BET726 has been reported to be a novel BRD4 inhibitor, and potently inhibited cell survival, proliferation and migration in skin squamous cell carcinoma [31]. I-BET726 was also reported to have anti-tumor activity in neuroblastoma by inhibiting MYCN and BCL-2 expression [32]. However, the toxic effect of I-BET726 on CRC cells has not been reported yet, and based on our current study, we speculate that I-BET726 may also exert anti-CRC effects by inhibiting OTUB2, whose further investigations will be tested in our future work.
Conclusion
Our current study presents the potential evidence for an OTUB2-mediated mechanism to regulate β-Catenin levels in CRC cells, but the precise mechanism that OTUB2 stabilizes β-Catenin still needs to be elucidated in our future work. Our current limited experimental results suggest that inhibiting OTUB2-β-Catenin axis may be a new strategy for the treatment of CRC.
Acknowledgements
We are grateful to Shanghai GeneChem for providing us with the high-content screening service. This study was supported by the National Natural Sciences Foundation of China (NSF 81973358).
All patients signed written informed consent.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Bonacci T, Emanuele MJ. Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin Cancer Biol. 2020;67:145–158. doi: 10.1016/j.semcancer.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng L, Meng T, Chen L, Wei W, Wang P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther. 2020;5:11. doi: 10.1038/s41392-020-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov. 2018;17:57–78. doi: 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farshi P, Deshmukh RR, Nwankwo JO, Arkwright RT, Cvek B, Liu J, Dou QP. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opin Ther Pat. 2015;25:1191–1208. doi: 10.1517/13543776.2015.1056737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–88. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, Huang A, Cui X, Han K, Hou X, Wang Q, Cui L, Yang Y. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9:4208–4220. doi: 10.7150/thno.33803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L, Geng Q, Pan H, Yan M, Yao M. OTUB2 stabilizes U2AF2 to promote the Warburg effect and tumorigenesis via the AKT/mTOR signaling pathway in non-small cell lung cancer. Theranostics. 2019;9:179–195. doi: 10.7150/thno.29545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S, Zang W, Qiu Y, Liao L, Zheng X. Deubiquitinase OTUB2 exacerbates the progression of colorectal cancer by promoting PKM2 activity and glycolysis. Oncogene. 2022;41:46–56. doi: 10.1038/s41388-021-02071-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Du J, Wang S, Shao L, Jin K, Li F, Wei B, Ding W, Fu P, van Dam H, Wang A, Jin J, Ding C, Yang B, Zheng M, Feng XH, Guan KL, Zhang L. OTUB2 promotes cancer metastasis via Hippo-independent activation of YAP and TAZ. Mol Cell. 2019;73:7–21. e7. doi: 10.1016/j.molcel.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Cheng H, Ji M, Su L, Lu Z, Hu X, Guan Y, Xiao J, Ma L, Zhang W, Pu H. OTUB2 regulates YAP1/TAZ to promotes the progression of esophageal squamous cell carcinoma. Biol Proced Online. 2022;24:10. doi: 10.1186/s12575-022-00169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novellasdemunt L, Foglizzo V, Cuadrado L, Antas P, Kucharska A, Encheva V, Snijders AP, Li VSW. USP7 is a tumor-specific WNT activator for APC-mutated colorectal cancer by mediating beta-Catenin deubiquitination. Cell Rep. 2017;21:612–627. doi: 10.1016/j.celrep.2017.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Zhang Y, Wong CC, Zhang J, Dong Y, Li X, Kang W, Chan FKL, Sung JJY, Yu J. RNF6 promotes colorectal cancer by activating the Wnt/beta-Catenin pathway via ubiquitination of TLE3. Cancer Res. 2018;78:1958–1971. doi: 10.1158/0008-5472.CAN-17-2683. [DOI] [PubMed] [Google Scholar]
- 13.Shi J, Liu Y, Xu X, Zhang W, Yu T, Jia J, Liu C. Deubiquitinase USP47/UBP64E regulates beta-Catenin ubiquitination and degradation and plays a positive role in Wnt signaling. Mol Cell Biol. 2015;35:3301–11. doi: 10.1128/MCB.00373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Mei X, Han K, Wu G, Li R, Yang Y. The deubiquitinating enzyme USP1 is auto-ubiquitinated and destabilized by ML323 in colorectal cancer cells. Eurasian J Med Oncol. 2023;7:174–179. [Google Scholar]
- 15.Hou X, Xia J, Feng Y, Cui L, Yang Y, Yang P, Xu X. USP47-mediated deubiquitination and stabilization of TCEA3 attenuates pyroptosis and apoptosis of colorectal cancer cells induced by chemotherapeutic doxorubicin. Front Pharmacol. 2021;12:713322. doi: 10.3389/fphar.2021.713322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, Han K, Tang X, Zeng Y, Lin X, Zhao Y, Zhang Z, Cao B, Wu D, Mao X. The ring finger protein RNF6 induces leukemia cell proliferation as a direct target of pre-B-cell leukemia homeobox 1. J Biol Chem. 2016;291:9617–28. doi: 10.1074/jbc.M115.701979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 18.Schauer NJ, Magin RS, Liu X, Doherty LM, Buhrlage SJ. Advances in discovering deubiquitinating enzyme (DUB) inhibitors. J Med Chem. 2020;63:2731–2750. doi: 10.1021/acs.jmedchem.9b01138. [DOI] [PubMed] [Google Scholar]
- 19.Caba C, Mohammadzadeh A, Tong Y. On the study of deubiquitinases: using the right tools for the job. Biomolecules. 2022;12:703. doi: 10.3390/biom12050703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan WC, Liu X, Magin RS, Girardi NM, Ficarro SB, Hu W, Tarazona Guzman MI, Starnbach CA, Felix A, Adelmant G, Varca AC, Hu B, Bratt AS, DaSilva E, Schauer NJ, Jaen Maisonet I, Dolen EK, Ayala AX, Marto JA, Buhrlage SJ. Accelerating inhibitor discovery for deubiquitinating enzymes. Nat Commun. 2023;14:686. doi: 10.1038/s41467-023-36246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Li S, Cui X, Han K, Wang J, Hou X, Cui L, He S, Xiao J, Yang Y. Inhibition of ubiquitin specific protease 1 sensitizes colorectal cancer cells to DNA-damaging chemotherapeutics. Front Oncol. 2019;9:1406. doi: 10.3389/fonc.2019.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu G, Yang J, Zhang H, Huang Z, Yang H. OTUB2 promotes proliferation and migration of hepatocellular carcinoma cells by PJA1 deubiquitylation. Cell Mol Bioeng. 2022;15:281–292. doi: 10.1007/s12195-022-00720-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang S, Zeng Z, Liu Z, Zhang Z, Sun J, Wang X, Ma M, Ye X, Yu J, Kang W. OTUB2 regulates KRT80 stability via deubiquitination and promotes tumour proliferation in gastric cancer. Cell Death Discov. 2022;8:45. doi: 10.1038/s41420-022-00839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Ming T, Tang S, Ren S, Yang H, Liu M, Tao Q, Xu H. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer. 2022;21:144. doi: 10.1186/s12943-022-01616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the Wnt/beta-catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–481. doi: 10.1016/j.biopha.2018.11.082. [DOI] [PubMed] [Google Scholar]
- 26.Ye D, Wang S, Wang X, Lin Y, Huang Y, Chi P. Overexpression of OTU domain-containing ubiquitin aldehyde-binding protein 1 exacerbates colorectal cancer malignancy by inhibiting protein degradation of beta-Catenin via Ubiquitin-proteasome pathway. Bioengineered. 2022;13:9106–9116. doi: 10.1080/21655979.2022.2057897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong D, Yuan J, Wang Z, Yu E, E J. USP37 promotes angiogenesis and metastasis in colorectal cancer by facilitating beta-catenin stability. Am J Cancer Res. 2023;13:2323–2341. [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J, Long G, Xiao L, Zhou L. USP8 positively regulates hepatocellular carcinoma tumorigenesis and confers ferroptosis resistance through beta-catenin stabilization. Cell Death Dis. 2023;14:360. doi: 10.1038/s41419-023-05747-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun SI, Kim HH, Yoon JH, Park WS, Hahn MJ, Kim HC, Chung CH, Kim KK. Ubiquitin specific protease 4 positively regulates the WNT/beta-catenin signaling in colorectal cancer. Mol Oncol. 2015;9:1834–51. doi: 10.1016/j.molonc.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen HH, Kim T, Nguyen T, Hahn MJ, Yun SI, Kim KK. A selective inhibitor of ubiquitin-specific protease 4 suppresses colorectal cancer progression by regulating beta-catenin signaling. Cell Physiol Biochem. 2019;53:157–171. doi: 10.33594/000000127. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Li P, Yang YQ, Cai S, Lin X, Chen MB, Guo H. I-BET726 suppresses human skin squamous cell carcinoma cell growth in vitro and in vivo. Cell Death Dis. 2020;11:318. doi: 10.1038/s41419-020-2515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyce A, Ganji G, Smitheman KN, Chung CW, Korenchuk S, Bai Y, Barbash O, Le B, Craggs PD, McCabe MT, Kennedy-Wilson KM, Sanchez LV, Gosmini RL, Parr N, McHugh CF, Dhanak D, Prinjha RK, Auger KR, Tummino PJ. BET inhibition silences expression of MYCN and BCL2 and induces cytotoxicity in neuroblastoma tumor models. PLoS One. 2013;8:e72967. doi: 10.1371/journal.pone.0072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.