Abstract
This study aimed to investigate the impact of IGF2BP3, a well-known m6A modification-related protein, on the metabolic and immune microenvironment of human cervical cancer. Bioinformatics analysis was performed to analyze the expression of IGF2BP3 in various databases, and its findings were validated using human cervical cancer tissue microarrays. We conducted a study to investigate the impact of IGF2BP3 on glutamine metabolism in cervical cancer cells through the application of metabolomics and metabolic flow analysis. Additionally, we explored how cervical cancer cells promote immune escape by secreting glutamine-derived lactate in a 3D culture setting. To identify the specific targets of IGF2BP3 that influence glutamine metabolism in cervical cancer, we employed RIP-seq analysis. IGF2BP3 exhibited high expression levels in multiple cervical cancer datasets, and its expression was significantly associated with the prognosis of cervical cancer patients. In mixed 3D cell cultures of cervical cancer and T cells, IGF2BP3 was found to enhance glutamate and glutamine metabolism in cervical cancer cells by up regulating the expression of GLS and GLUD1 genes. Moreover, it influenced the differentiation of Treg cells by promoting lactate production and secretion in cervical cancer, leading to immune escape. Mechanistic analysis revealed that IGF2BP3 stabilized the mRNA of GLS and GLUD1 genes through m6A modification, thereby facilitating glutamate and glutamine metabolism in cervical cancer cells and regulating lactate production. Additionally, we investigated the correlation between GLS, GLUD1 protein expression, and IGF2BP3 expression in human cervical cancer through multicolor immunofluorescence staining. The relevance of IGF2BP3 in the context of Treg cell-associated immune escape in cervical cancer was also confirmed. IGF2BP3 exhibits high expression in human cervical cancer and plays a crucial role in stabilizing the mRNA of GLS and GLUD1 genes, key metabolic enzymes in glutamate and glutamine metabolism, through m6A modification. This process leads to immune escape in cervical cancer by promoting lactate production and secretion.
Keywords: IGF2BP3, cervical cancer, glutamine metabolism, M6A modification
Introduction
m6A modification, also known as N6-methyladenosine, is a prevalent RNA modification found in eukaryotic cells. It involves the addition of a methyl group to the nitrogen atom at the sixth position of adenosine within the RNA molecule. This modification is catalyzed by the “m6A writer complex”, which includes methyltransferases such as METTL3, METTL14, and WTAP. m6A modification occurs in various types of RNA, including messenger RNA (mRNA), long noncoding RNA (lncRNA), and microRNA (miRNA) [1,2]. It plays a critical role in RNA metabolism and function by influencing mRNA stability, splicing, export, translation efficiency, and RNA-protein interactions. The dynamic regulation of m6A modification involves “m6A erasers”, such as FTO and ALKBH5, which remove the methyl group, and “m6A readers”, such as YTH domain-containing proteins, that recognize and bind to m6A-modified RNAs. The interaction between m6A readers and the modified RNA affects RNA processing, localization, and translation. m6A modification is involved in essential biological processes, including embryonic development, cell differentiation, tissue homeostasis, and stress responses [3]. Dysregulation of m6A modification has been implicated in various diseases, including cancer, neurological disorders, and metabolic diseases [4].
Insulin-like Growth Factor 2 mRNA-Binding Protein 3 (IGF2BP3) is an RNA-binding protein that plays a crucial role in post-transcriptional gene regulation. It is involved in various cellular processes, including stabilizing RNA molecules, regulating their translation, and facilitating their localization within the cell [5]. In cancer research, IGF2BP3 has garnered significant attention due to its dysregulation in several cancer types. One important aspect of IGF2BP3’s function is its interaction with m6A-modified RNA molecules. m6A modification refers to the addition of a methyl group to adenosine residues within RNA molecules. IGF2BP3 is implicated in the regulation of m6A-modified transcripts, influencing their stability and efficiency of translation. By binding to specific m6A-modified regions within target mRNAs, IGF2BP3 can affect the fate and function of these transcripts. This suggests a potential interplay between IGF2BP3 and the m6A modification machinery in the post-transcriptional regulation of gene expression [6]. Further research is necessary to fully elucidate the functional significance and regulatory mechanisms underlying the interplay between IGF2BP3 and m6A modifications [7-9].
The m6A modification can affect the stability, splicing, and translation efficiency of target mRNAs. Specific m6A “reader” proteins, such as YTHDF2 (YTH domain-containing family 2), recognize and bind to m6A-modified transcripts, leading to various downstream effects. In the context of glutamine metabolism, m6A modifications within target mRNAs can influence their stability and translation, thereby impacting the levels and activity of enzymes involved in glutamine utilization. Moreover, m6A modification has been linked to the regulation of genes associated with glutamine transporters, such as SLC1A5 and ASCT2, which are involved in the uptake of extracellular glutamine into cells [10,11]. By modulating the m6A status of these genes, m6A writers, erasers, and readers can affect their expression and ultimately influence cellular glutamine levels. The interplay between m6A modification and glutamine metabolism in cancer cells highlights the complexity of post-transcriptional gene regulation in cellular metabolism. Dysregulation of m6A-modified transcripts involved in glutamine metabolism can contribute to the rewiring of metabolic pathways, promoting cancer cell growth, survival, and progression.
In this study, our findings demonstrated the significant role of IGF2BP3 in promoting glutamine metabolism in cervical cancer cells. We observed that IGF2BP3 plays a crucial role in facilitating the utilization of glutamine by cancer cells, thereby supporting their metabolic needs. Furthermore, we discovered that IGF2BP3 enhances the differentiation of Treg cells by promoting the production and secretion of lactate within cervical cancer cells. This lactate production, induced by IGF2BP3, contributes to the immune escape in human cervical cancer cells.
Materials and methods
Clinical specimens and cell lines
Paraffin-embedded human cervical cancer specimens were obtained from 160 patients diagnosed between 2016 and 2018, with written informed consent obtained from all participants. This study was approved by the Ethics in Research Committee of People’s Hospital of Lianyungang, China. The clinical information of cervical cancer patients were listed in Table 1. The Hela and CaSki cell lines were procured from Procell Life Science & Technology Co., Ltd. (Wuhan, China). Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 100 U/mL penicillin G at 37°C in a 5% CO2 atmosphere.
Table 1.
The relationship between IGF2BP3 expression and clinical features of human cervical cancer
| IGF2BP3High (n=85) | IGF2BP3Low (n=75) | P Value* | |
|---|---|---|---|
| Age | 0.19 | ||
| ≥ 50 | 45 | 32 | |
| < 50 | 40 | 43 | |
| HPV status | 0.24 | ||
| HPV- | 16 | 20 | |
| HPV+ | 69 | 55 | |
| Differentiation grade | 0.0003 | ||
| G1 | 16 | 30 | |
| G2 | 28 | 25 | |
| G3 | 21 | 18 | |
| Gx | 20 | 2 | |
| Lymph invasion | 0.3469 | ||
| Yes | 46 | 35 | |
| No | 39 | 40 | |
| Vaginal invasion | 0.55 | ||
| Yes | 46 | 37 | |
| No | 39 | 38 | |
| Tumor size, cm | < 0.0001 | ||
| < 4 | 20 | 48 | |
| ≥ 4 | 65 | 27 | |
| FIGO stage | 0.001 | ||
| < IIA | 48 | 23 | |
| ≥ IIB | 37 | 52 |
By chi-square test.
Reagents
Lactic Acid (LA) Content Assay Kit (AKAC001M, Boxbio, Beijing, China), Glutamine Colorimetric Assay Kit (K556-100, Biovision), Glutamate Assay Kit (ab83389, Abcam).
Cell transduction and 3D culture
Lentiviral vectors for IGF2BP3 knockdown were obtained from GeneChem (Shanghai, China), and an empty vector was used as the negative control (NC), as specified in Table S1. Stable transduced cells were selected using puromycin. To reduce batch-to-batch variation, each comparison involved pairs of samples from the same batch. All transfections were performed according to the manufacturers’ instructions.
We utilized a micro-tissue provided in vitro 3D culture system to combine human cervical cancer cells with CD4+ T cells at a ratio of 10:1 for a duration of 7 days. During this period, TGF-b was introduced at a final concentration of 1 ng/mL to induce Treg cell. After 7 days, the 3D spheres were dissociated for single cell suspension or fixed using paraformaldehyde. Flow cytometry and multispectral staining techniques were employed to determine the proportion of Treg cells.
Quantitative RT-PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Subsequently, complementary DNA (cDNA) was synthesized using the PrimeScript TM RT Master Mix Kit (TaKaRa, Dalian, China). Real-time PCR analysis was conducted using SYBR Premix Ex Taq TM (Tli RNaseH Plus) (TaKaRa, Dalian, China). The expression levels were normalized to the housekeeping control GAPDH. The amplification primers are provided in Table S1.
Immunoblot and multiple immunohistochemistry (mIHC) assay
Total protein was extracted from cultured cells using a Protein Extraction Kit (Kaiji, Nanjing, China). The proteins were separated on SDS-PAGE gels and then transferred onto PVDF membranes (0.45-μm pore, Millipore). mIHC staining of tissues section was performed using the PANO 5-plex immunohistochemistry kit (Cat#10080100100, Panovue) according to manufacturer’s instructions. In brief, paraffin slices were incubated with one primary antibody after microwave heat treatment, then incubated them with horseradish peroxidase (HRP) conjugated secondary antibodies and amimide signal amplification. The antibodies used were as follows: anti-IGF2BP3 (14642-1-AP, Proteintech), anti-CD4 (67786-1-Ig, Proteintech), anti-Foxp3 (22228-1-AP, Proteintech), anti-m6A (68055-1-Ig, Proteintech), anti-GLS (81486-1-RR, Proteintech), anti-GLUD1 (14299-1-AP, Proteintech) and anti-α-Tubulin (11224-1-AP, Proteintech). Prior to ECL detection, anti-rabbit or anti-mouse HRP-conjugated secondary antibodies were applied. Multi-spectral images were obtained by scanning stained slides using Vectra 3 System (PerkinElmer, Waltham, MA, USA) with the same exposure time. On the basis of full film scanning, three high magnification fields were randomly selected from each tissue point for image capture and captured with the same exposure time. The cell counts and fluorescence intensity were determined by InForm Tissue Finder (PerkinElmer, Waltham, MA, USA). Automated tissue segmentation identified tissue and blank regions.
RNA sequencing analysis
Total RNA was isolated from IGF2BP3 knockdown and control CaSki cells using TRIzol Reagent. RNA sequencing (RNA-seq) analysis was performed at LC Biotech Inc. (Guangzhou, China).
Metabolite-level measurements
Metabolite-level measurements were conducted using GC-MS analysis. To analyze intracellular metabolites, Hela and Hela-IGF2BP3 shRNA cells (1 × 107/sample) were quenched, and the metabolites were derivatized using methoxyamine (15 mg/mL in pyridine) for 90 minutes at 37°C. Subsequently, the derivatives were treated with Bis(trimethylsilyl)trifluoroacetamide (1% chlorotrimethylsilane) and 20 μl of n-hexane for 60 minutes at 70°C.
For metabolic flux analysis, Hela and Hela-IGF2BP3 shRNA cells (5 × 106/sample) were cultured to 80% confluence in complete media. The media was then replaced with glutamine-free RPMI 1640 supplemented with 2 mM [U-13 C5] glutamine and 10% dialyzed FBS for 24 hours. After quenching the cells, the metabolites were derivatized with methoxyamine (15 mg/mL in pyridine) for 90 minutes at 37°C, followed by treatment with N-(ter-Butyldimethylsilyl)-N-methyltrifluoroacetamide at 55°C for 60 minutes.
Metabolomics instrumental analysis was performed using an Agilent 7890A gas chromatography system coupled to an Agilent 5975 C inert MSD system. A HP-5ms fused-silica capillary column (30 m × 0.25 mm × 0.25 μm) from Agilent J&W Scientific was used for separation of the derivatives. Mass spectra were collected in the m/z range of 50-600 under the selected reaction monitoring mode. To visualize the metabolic alterations among the experimental groups, Partial Least Squares-Discriminant Analysis (PLS-DA) was conducted on the intracellular metabolite data. The Variable Importance in the Projection (VIP) method was used to rank the overall contribution of each variable to the PLS-DA model. Significantly different metabolites were identified based on a combination of two criteria: a p-value < 0.05 from t-tests and a VIP score > 1 from PLS-DA.
For stable isotope-tracing analysis, the measured distribution of mass isotopomers was corrected for the natural abundance of isotopes using the IsoCor software. Labeled metabolite data were expressed as a percentage of the total pool or relative ion abundances. Metabolite levels were normalized to the internal standard and cell number from parallel plates.
RNA immunoprecipitation (RIP) assay
The RIP assay was performed according to the instructions of the Geneseed RIP Kit (Guangzhou, China). Briefly, magnetic beads were mixed with anti-m6A/IGF2BP3/IgG antibodies before the addition of cell lysates. Then, the bound complexes were thoroughly washed, eluted, purfied, and analyzed by RT-qPCR. Enrichment of precipitated RNAs was normalized relative to input controls.
mRNA stability assay
Cells were seeded in 6-well plates and were then treated with 5 μg/mL actinomycin D for 0 h, 3 h, or 6 h prior to RNA extraction. The transcript level of GLS and GLUD1 mRNA were estimated as the half-life of the mRNA and normalized to ACTIN as the standard.
Statistical analysis
Statistical analyses were conducted as described using R language (version 3.5.2) or GraphPad Prism (version 9.0.0). Two-tailed Student’s t-tests were employed for comparing two groups, while the chi-square test was used for categorical data assessment. Survival curves were generated using the Kaplan-Meier method, and survival data were evaluated through univariate and multivariate Cox regression analyses. Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and gene set enrichment analysis (GSEA) were performed using Rlanguage (version 3.5.2). Statistical significance was considered at P < 0.05.
Results
IGF2BP3, a gene involved in M6A RNA modification, exhibits high expression in human cervical cancer tissues
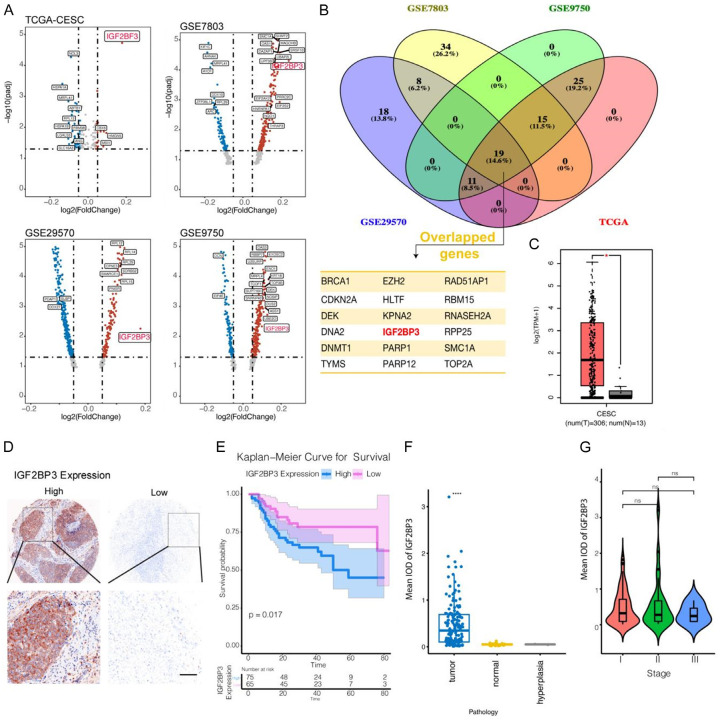
To investigate the expression of m6A-related genes in human cervical cancer, we conducted a comprehensive analysis using public databases, including TCGA and three human cervical cancer datasets from the GEO database (GES7803, GSE29570, and GSE9750). Initially, we examined the expression of m6A-related genes in these datasets. Additionally, we explored the GOMF_RNA_BINDING gene set to identify RNA-binding genes. Through a differential gene analysis of cancer and paracancerous tissues, we identified significantly differentially expressed genes (adjusted P value < 0.05) represented in Figure 1A using a volcano plot. Furthermore, employing Venny analysis (Figure 1B), we discovered that 19 RNA-binding genes exhibited significant differential expression across all four databases. Notably, among these genes, IGF2BP3 was the sole gene associated with RNA M6 modification and showed significantly high expression in tumor tissues within all four public datasets (Figure 1C).
Figure 1.
IGF2BP3, a gene involved in M6A RNA modification, exhibits high expression in human cervical cancer tissues. A: Volcano plot depicting the expression levels of differentially expressed RNA-binding proteins in tumor and normal tissues, based on RNA-seq data from TCGA and GEO datasets associated with human cervical cancer. B: Venn diagram illustrating the overlapping differential RNA-binding protein genes identified in the TCGA and GEO datasets. C: Comparative analysis of IGF2BP3 gene expression between cancerous and paracancerous tissues using data from the TCGA database. D: Immunohistochemical staining (X100 for upper panel, and X400 for lower panel) showing the protein expression of IGF2BP3 in human cervical cancer tissues. E: Survival analysis conducted for patients with cervical cancer based on the expression levels of IGF2BP3. F, G: IGF2BP3 protein expression in normal tissues, hyperplasia, and different stages of cervical cancer tissues. All experiments were performed in triplicate, and statistical significance was considered at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
To validate the findings from public databases, we conducted an IHCexamination of IGF2BP3 expression using tissue microarrays comprising 160 tumor tissues and 30 normal tissues. Our analysis revealed heterogeneous expression of IGF2BP3 in human cervical cancer. Specifically, positive tissues exhibited predominant cytoplasmic distribution of the IGF2BP3 protein within tumor cells (Figure 1D), whereas the expression of IGF2BP3 in normal cervical glandular tissues was significantly lower than that in tumor tissues (P < 0.05) (Figure 1E). The 140 tumor tissues were subsequently categorized into three levels based on TMN staging, and the expression of IGF2BP3 did not differ significantly across these three levels (Figure 1F). Moreover, through survival analysis, we observed that patients with cervical cancer and low IGF2BP3 expression exhibited a longer survival rate compared to those with high IGF2BP3 expression in tumor tissues (Figure 1G). Further analysis of variance (ANOVA) was employed to examine the correlation between IGF2BP3 expression and different characteristics of cervical cancer. The findings revealed a significant association between IGF2BP3 expression and cervical cancer differentiation, tumor size, and FIGO grade, while no significant correlations were observed with patient age, HPV infection, lymphatic infiltration, or vaginal invasion (Table 1).
IGF2BP3 promotes Treg cell-mediated immune escape through the regulation of glutamine metabolism in cervical cancer cells
Given the established association between m6A and cervical cancer malignancy, this study aimed to explore the relationship between IGF2BP3, cervical cancer cell metabolism, and tumor immunity. Initially, we divided all cervical cancer tissues into IGF2BP3 high and low expression groups based on the expression levels in tumor tissues, using TCGA data. Utilizing GSVA analysis, we assessed the expression of 87 metabolism-related gene sets in cervical cancer tumor tissues. Figure S1A displays the differentially expressed gene sets, with significant differences observed in unsaturated fatty acid synthesis, D-glutamate and D-glutamine metabolism, oxidative phosphorylation, and neomycin kanamycin synthesis between the two IGF2BP3 expression groups.
Furthermore, leveraging a dataset related to the tumor immune microenvironment, we employed GSVA to calculate the infiltration differences of various immune cells in cervical cancer tissues with different IGF2BP3 expression. The results demonstrated significant disparities in effector memory CD4T cells and Treg cells within cervical cancer tissues based on IGF2BP3 expression (Figure S1B). Additionally, by employing gene sets associated with Treg and CD4+ memory T cells from The Molecular Signatures Database (MSigDB), validated through the GSVA approach, we found a close association between cervical cancer and the infiltration of these two cell groups mediated by IGF2BP3 (Figure S1C).
Moreover, we conducted multiple correlation analyses on immune and metabolic-related gene sets. Notably, D-glutamate and D-glutamine metabolism displayed a close correlation with the infiltration of Treg cells within tumor tissues (Figure S1D, S1E). Furthermore, we conducted a comparison of the expression levels of the constituent genes within both gene sets. This analysis revealed a significantly higher surface expression of GLS, GLS2, GLUD1, and other genes in tissues with high IGF2BP3 expression compared to tissues with low IGF2BP3 expression. In contrast, among the genes associated with Treg cells, CCR1, CCR8, IL-10, IL-6R, and other genes exhibited significantly higher expression in tissues with high IGF2BP3 expression (Figure S1F, S1G).
IGF2BP3 plays a role in promoting glutamate and glutamine metabolism in cervical cancer cells by enhancing the transcription of GLS and GLUD1 gene
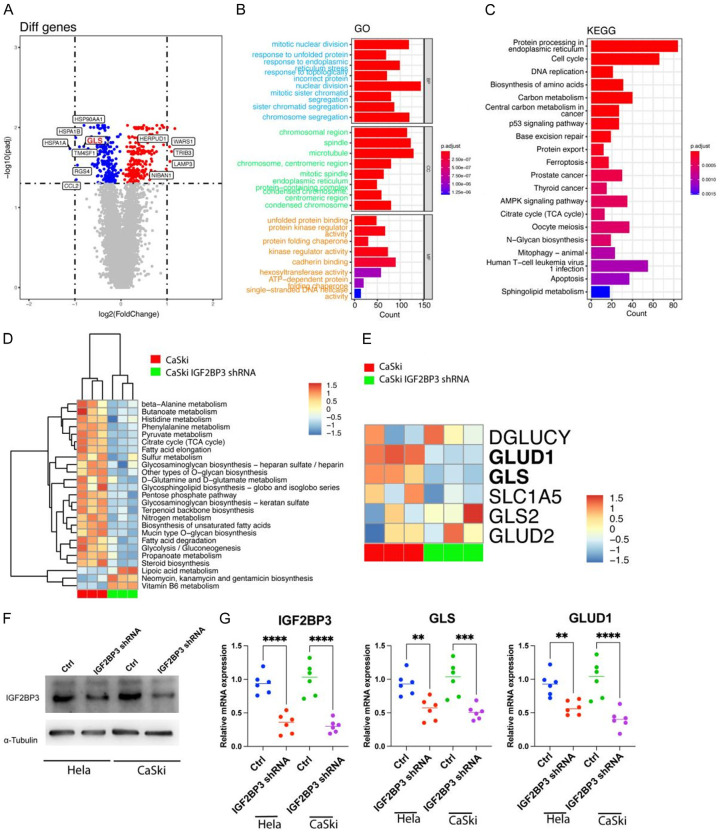
To investigate the impact of IGF2BP3 on gene transcription in cervical cancer cells, we employed specific shRNAs to knock down IGF2BP3 expressioninCaSkicell, followed by RNA-seq analysis to identify differentially expressed genes. Our findings revealed that IGF2BP3 knockdown led to the up-regulation of 1586 genes and down-regulation of 1523 genes (adj. P Value < 0.05) (Figure 2A). To further analyze these differential genes, we performed GO and KEGG enrichment analyses. The results demonstrated that IGF2BP3 knockdown primarily affected tumor endoplasmic reticulum-related organelles and biological processes, such as endoplasmic reticulum-related proteins and protein folding within the endoplasmic reticulum (Figure 2B, 2C). Additionally, by employing GSVA to assess 87 metabolism-related gene sets, we observed that IGF2BP3 knockdown had a significant impact on various metabolic processes, including D-glutamate and D-glutamine metabolism (Figure 2D). Further examination of the component genes within this gene set revealed that IGF2BP3 primarily influenced the transcripts of GLUD1 and GLS genes through differential gene analysis (Figure 2E).
Figure 2.
IGF2BP3 plays a role in promoting glutamate and glutamine metabolism in cervical cancer cells by enhancing the transcription of GLS and GLUD1 gene. A: Volcano plot displaying differential gene expression in RNA-seq analysis of wild-type and IGF2BP3 knockdown cervical cancer cells. B, C: Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses conducted using the differentially expressed genes. D: Heat map illustrating the results of gene set variation analysis (GSVA) based on RNA-seq data from wild-type and IGF2BP3 knockdown cervical cancer cells. E: Heat map presenting the differential expression of genes associated with the glutamine gene set. F: Western blot assay depicting the protein expression of IGF2BP3 in wild-type and IGF2BP3 knockdown cervical cancer cells, with α-Tubulin serving as an internal reference gene. G: Real-time PCR analysis showing the transcript levels of IGF2BP3, GLS, and GLUD1 genes in wild-type and IGF2BP3 knockdown cervical cancer cells. All experiments were performed in triplicate, and statistical significance was considered at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
To validate our findings, we conducted western blot analysis, confirming the knockdown of the IGF2BP3 gene in CaSki and Hela cells (Figure 2F). Furthermore, using Real-time PCR, we detected the transcription levels of GLS and GLUD1 genes. The results indicated that IGF2BP3 downregulation correlated with a significant decrease in GLS and GLUD1 gene expression (Figure 2G). Therefore, based on our analysis utilizing TCGA data and our own RNA-seq analysis following IGF2BP3 knockdown, we propose that IGF2BP3 may regulate glutamate and glutamine metabolism in cervical cancer by modulating the expression of GLS and GLUD1 genes.
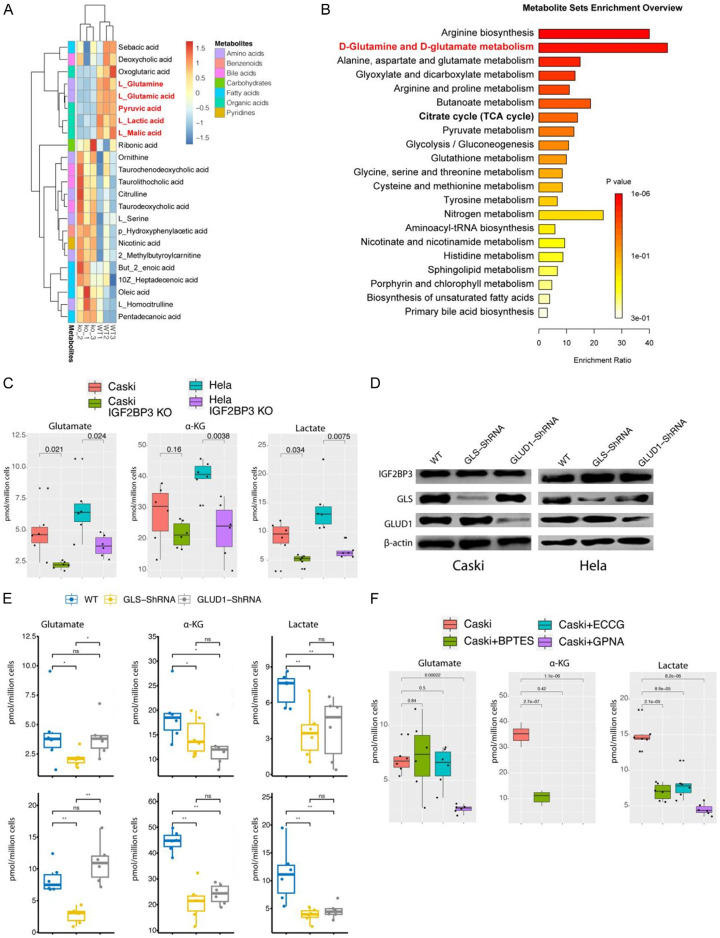
Following the indications from RNA-seq results that IGF2BP3 knockdown disrupted the metabolism of cervical cancer cells, we conducted metabolomics analysis to further investigate the metabolic changes in IGF2BP3 knockdown CaSki. The findings revealed significant alterations in 23 metabolites, including L-glutamate, L-glutamine, L-lactate, L-malate, and pyruvate, all of which exhibited a notable decrease (Figure 3A). MSEA analysis of the metabolites that displayed alterations indicated enrichment in arginine anabolism, D-glutamate, and D-glutamine metabolism (Figure 3B). Furthermore, we performed assays using glutamate, α-ketoglutarate, and lactate kits, which confirmed a significant decrease in these metabolites as a consequence of IGF2BP3 knockdown in two cervical cancer cell lines (Figure 3C). Meanwhile, to provide additional confirmation that IGF2BP3 exerts specific regulatory control over glutamine metabolism via its modulation of GLS and GLUD1 enzymes, we separately knockdown GLS and GLUD1 in two distinct types of cervical cancer cells overexpression of IGF2BP3 (Figure 3D). Subsequent analysis of the mentioned metabolites revealed that IGF2BP3 indeed exerts precise control over glutamine metabolism within cervical cancer cells through the regulation of GLS and GLUD1 (Figure 3E). To investigate the mechanism by which IGF2BP3 regulates lactate production, we conducted experiments using various inhibitors targeting different aspects of glutamine metabolism. These inhibitors included BPTEs, which are GLS inhibitors, ECCG, which inhibits the conversion of glutamate to alpha-ketoglutarate, and GPNA, an inhibitor of the glutamine transporter ADCT2. We examined intracellular levels of glutamine, glutamate, and lactate and observed that GPNA reduced the cellular uptake of glutamine. Additionally, BPTEs inhibited the conversion of glutamine to glutamate, while ECCG did not affect the intracellular levels of glutamine and glutamate. All three inhibitors effectively reduced intracellular lactate production (Figure 3F). These findings suggest that lactate in cervical cancer cells can be derived from glutamine metabolism.
Figure 3.
IGF2BP3 plays a role in promoting glutamate and glutamine metabolism in cervical cancer cells. A: Heat map illustrating the levels of differentially expressed metabolites in wild-type and IGF2BP3 knockdown cervical cancer cell lines. B: Enrichment analysis using differential metabolites through Metabolite Set Enrichment Analysis (MSEA). C: Measurement the levels of glutamate, α-ketoglutarate, and lactate in wild-type and IGF2BP3 knockdown cervical cancer cell lines. D: IGF2BP3, GLS and GLUD1 protein expression detected by using western-blot in cervial cancer cell lines treated indicated in the figure. E: Measurement the levels of glutamate, α-ketoglutarate, and lactate in cervial cancer cell lines treated indicated in the figure. F: Measurement the levels of glutamate, α-ketoglutarate, and lactate in Caski cells treated indicated in the figure. All experiments were performed in triplicate, and statistical significance was considered at P < 0.05.
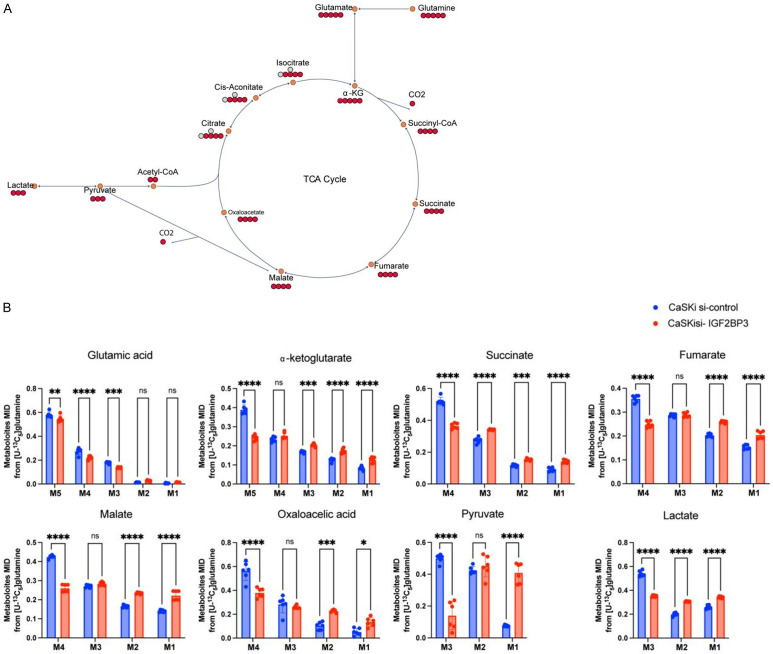
To further demonstrate the impact of IGF2BP3 on lactate production through glutamine metabolism, we treated CaSki with varying levels of IGF2BP3 expression using C13-labeled glutamine. The distribution of C13 in each metabolite was analyzed using metabolic flow analysis (Figure 4A). The results showed that when IGF2BP3 was knocked down, the distribution of C13 in each metabolite, including pyruvate and lactate in the TCA cycle, was significantly lower compared to wild-type cells. This reduction was observed in M5 glutamate, alpha-ketoglutarate, M4 fumarate, malate, oxaloacetic acid, and M3 pyruvate and lactate (Figure 4B). These results provide further evidence that IGF2BP3 can regulate lactate production mediated by glutamine metabolism.
Figure 4.
IGF2BP3 plays a role in promoting glutamate and glutamine metabolism in cervical cancer cells. A: Schematic model of glutamine metabolism in cancer cells. Red circles represent carbons derived from [U-13 C5] glutamine, and hollow circles are unlabeled. The black arrows indicate oxidative carboxylation flux from glutamine. B: Mass isotopologue distributions of glutamate in TCA cycle metabolites including α-KG, succinate, fumarate, malate, pyruvate and lactate in Caski cells treated with the control siRNA or IGF2BP3 siRNAs. Cells were cultured in [U-13 C5] glutamine for 24 h before metabolites extraction and GC-MS analysis; n=6, data in d-h are shown as the mean ± SD. statistical significance was considered at P < 0.05.
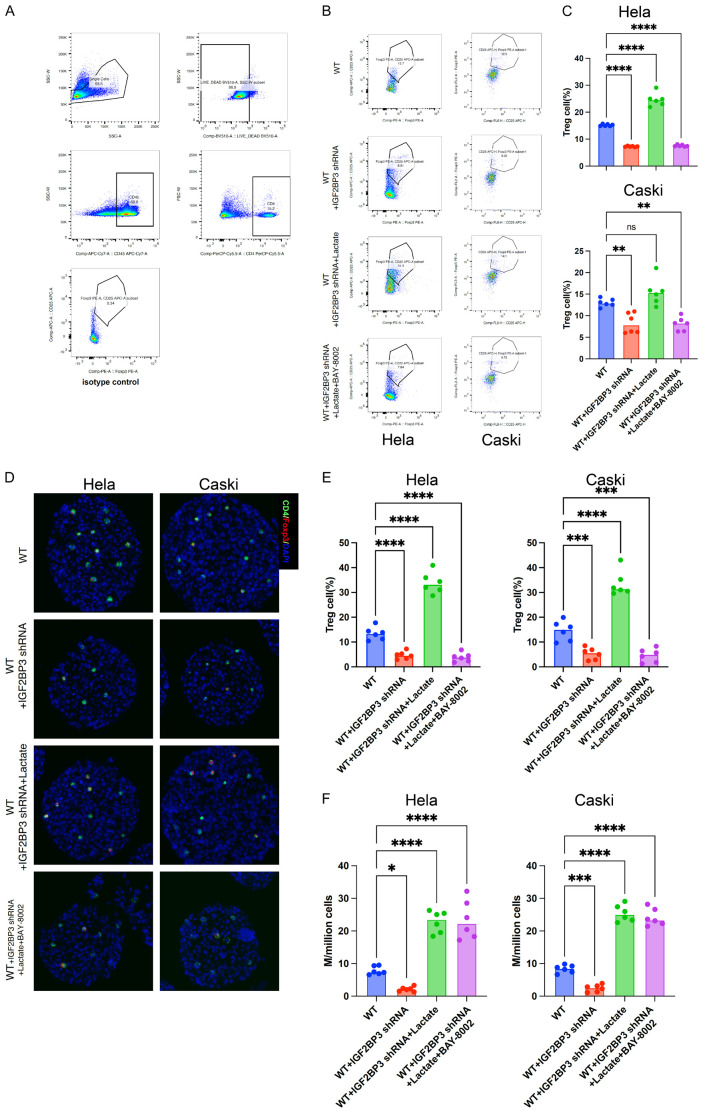
IGF2BP3 facilitates the differentiation of Treg cells by enhancing lactate production through the promotion of glutamine metabolism
To investigate the potential of IGF2BP3 in promoting Treg cell differentiation in cervical cancer cells, we conducted experiments using the 3D culture method. Firstly, we co-cultured cervical cancer cells with Naive T cells in the presence of TGF-β. After 7 days of mixed culture, we treated the 3D cultured cells with a single cell suspension and utilized flow cytometry to detect the Treg cells within the mixed culture, as illustrated in Figure 5A (gating strategy using flow cytometry for Treg cell detection). Our findings demonstrated that knockdown of IGF2BP3 significantly reduced the proportion of Treg cells in the 3D histospheres compared to wild-type cells. Conversely, the addition of Lactate to IGF2BP3 knockdown cells restored the proportion of Treg cells. Notably, this restorative effect was inhibited by the addition of BAY-8002, an inhibitor of the lactate receptor MCT1 (Figure 5B, 5C). Immunofluorescence staining of 3D spheres provided similar results, directly confirming the reduction of Treg cells (CD4+Foxp3+ cells) in IGF2BP3 knockdown cells (Figure 5D, 5E). Furthermore, we examined lactate levels in the culture supernatants and observed significantly reduced lactate levels in the supernatants of IGF2BP3 knockdown cervical cancer cells compared to wild-type cells (Figure 5F). Collectively, these in vitro studies suggest that IGF2BP3 may enhance lactate production in tumor cells by promoting glutamine metabolism. Consequently, this increased lactate production may contribute to the promotion of Treg cell differentiation through the lactate receptors present on the surface of T cells.
Figure 5.
IGF2BP3 facilitates the differentiation of Treg cells by enhancing lactate production through the promotion of glutamine metabolism. A: Gating strategy employed for the detection of Treg cells in 3D cell spheres. B, C: Flow cytometry analysis depicting the impact of various treatments on the proportion of Treg cells in 3D cell spheres, as illustrated in the Figure. D, E: Immunofluorescence staining (×200) showcasing the influence of different treatments on the proportion of Treg cells in 3D cell spheres. F: Lactate determination of supernatant of cell culture treated differently indicated in the figure. All experiments were performed in triplicate, and statistical significance was considered at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
IGF2BP3 regulates the expression of GLS and GLUD1 in an m6A-dependent mechanism
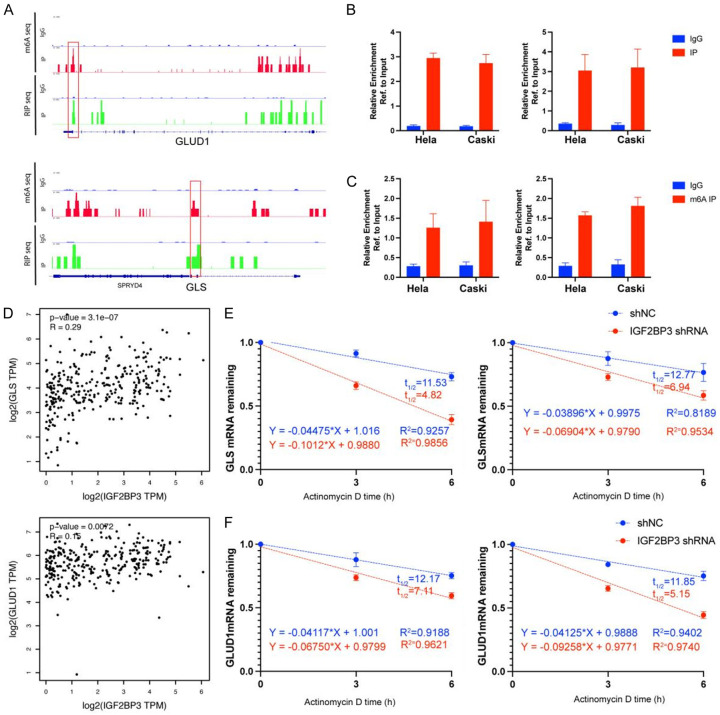
To elucidate the regulatory mechanism by which IGF2BP3 controls GLS and GLUD1, we conducted IGF2BP3 RIP-seq and m6A RIP-seq experiments separately. Notably, as depicted in Figure 6A, only GLS and GLUD1 exhibited a direct association with metabolism, particularly glutamine metabolism. Further analysis of the IGF2BP3 RIP-seq and m6A RIP-seq findings uncovered that IGF2BP3 could regulate the expression of GLS and GLUD1 genes through an m6A-dependent mechanism, thereby enhancing their stability. This was validated through IP/Real-time PCR assays, which confirmed the m6A-mediated binding of IGF2BP3 to GLS and GLUD1 mRNAs (Figure 6B, 6C). Moreover, analysis of the TCGA database demonstrated a significant and positive correlation between IGF2BP3 and the expression of GLS and GLUD1 genes (Figure 6D). Lastly, we investigated the mRNA stability of these two genes in both wild-type and IGF2BP3 knockdown cells. The results revealed a significant decrease in mRNA stability of GLS and GLUD1 genes upon IGF2BP3 knockdown (Figure 6E, 6F). These collective findings strongly suggest that IGF2BP3 regulates the expression of GLS and GLUD1 genes through an m6A-dependent mechanism.
Figure 6.
IGF2BP3 regulates the expression of GLS and GLUD1 in an m6A-dependent mechanism. A: Integrative Genomics Viewer (IGV) tracks of m6A and IGF2BP3 RIP-seq of GLUD1 and GLS. B, C: Real-time PCR analysis depicting the expression levels of IGF2BP3 and the enrichment of GLS and GLUD1 gene mRNA through m6A RIP-seq. D: TCGA-CESC dataset demonstrating a linear correlation between IGF2BP3 and the genes GLUD1 and GLS. E, F: Evaluation of the impact of IGF2BP3 loss on the stability of GLUD1 and GLS in HL-60 and KG-1 cells. Transfected cells were treated with 5 µg/ml actinomycin D for 0 h, 3 h, or 6 h prior to RNA extraction. All experiments were performed in triplicate, and statistical significance was considered at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
IGF2BP3 regulation of GLS and GLUD1 expression and promotes Treg-induced immune escape in human cervical cancer
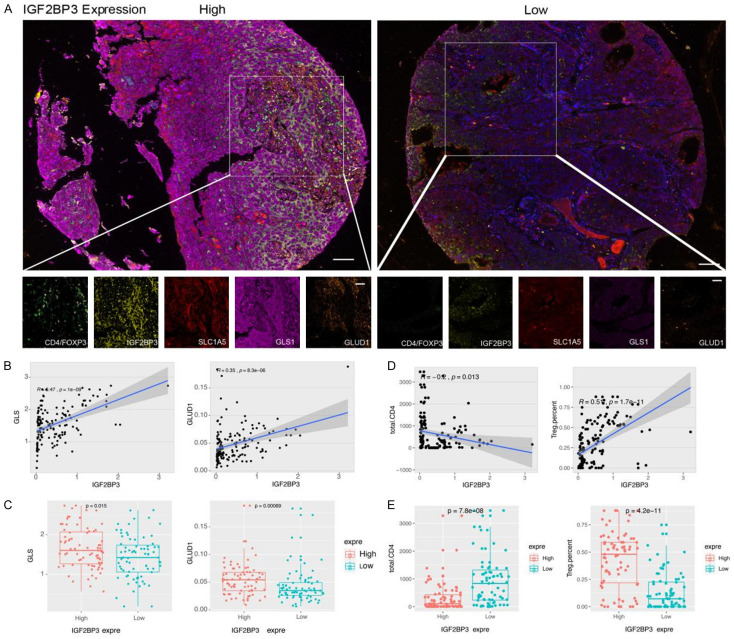
To validate the aforementioned findings in human cervical cancer tissues, we conducted an analysis to examine the expression of IGF2BP3, GLS, GLUD1, CD4, and Foxp3 in cervical cancer tissue microarrays using multispectral staining (Figure 7A). We categorized 140 cervical cancer tissues into high and low expression groups based on the levels of IGF2BP3 expression. Our results revealed a positive correlation between IGF2BP3 expression and the expression of GLS and GLUD1 in cervical cancer tissues (IGF2BP3 vs. GLS, R=0.45, P < 0.001, IGF2BP3 vs. GLUD1, R=0.35, P < 0.01) (Figure 7B). Moreover, the expression levels of GLS and GLUD1 were significantly higher in cervical cancer tissues with high IGF2BP3 expression compared to those with low IGF2BP3 expression (Figure 7C).
Figure 7.
IGF2BP3 regulation of GLS and GLUD1 expression and promotes Treg-induced immune escape in human cervical cancer. A: Multicolor immunostaining illustrating the protein expression of IGF2BP3, GLS, GLUD1, CD4, and Foxp3 in human cervical cancer tissues (× 100 for main figure, × 200 for enlarged figure). B: Linear correlation analysis demonstrating the relationship between GLUD1 and GLS protein expression levels and IGF2BP3 in human cervical cancer tissues. C: Comparison of GLUD1 and GLS protein expression in cervical cancer tissues with high and low levels of IGF2BP3. D: Linear correlation analysis between the number of CD4+ T cells, Treg cells, and the protein expression levels of IGF2BP3 in human cervical cancer tissues. E: Comparison of the number of CD4+ T cells and Treg cells in cervical cancer tissues with high and low expression levels of IGF2BP3. All experiments were performed in triplicate, and statistical significance was considered at P < 0.05. *P < 0.05; **P < 0.01; ***P < 0.001.
Furthermore, we observed a negative correlation between the expression level of IGF2BP3 and the number of CD4 cells in cervical cancer tissues (R=-0.2, P=0.013). Conversely, a positive correlation was found between the expression level of IGF2BP3 and the number of Treg cells (R=0.51, P < 0.001) (Figure 7D). Notably, the tissues exhibiting high IGF2BP3 expression displayed a significantly lower number of CD4 cells compared to the low expression group, while the number of Treg cells was significantly higher in the high expression group compared to the low expression group (Figure 7E).
Discussion
While previous studies have highlighted the role of IGF2BP3 in facilitating tumor progression in malignancy [12], our study presents a novel contribution by demonstrating the association of this m6A reader gene with the metabolic and immune microenvironment of cervical cancer. This is the first report to establish such a connection. Notably, m6A modification represents a prevalent molecular alteration in human cervical cancer [13,14]. Numerous studies have documented significant deviations in m6A modification patterns in cervical cancer cells when compared to normal cervical tissues [13]. These alterations in m6A modification have the capacity to impact the stability, translation, and splicing of RNA molecules, consequently leading to abnormal gene expression, exerting different biological process including pivotal cellular processes, including cell proliferation, apoptosis, and metastasis. Next, m6A impact on Human Papillomavirus (HPV) Infection, research suggests that m6A modification may play a role in HPV-related cervical carcinogenesis [15]. The interplay between m6A regulators and HPV viral RNA can potentially influence viral replication, persistence, and the host immune response. However, our study showed that IGF2BP3 expression was not associated with HPV infection. In recent years, an increasing number of studies have shown that m6A modification is closely related to tumor metabolism.
The relationship between m6A modifications and cancer metabolism is an emerging area of research with compelling evidence indicating their close association [16-18]. Several aspects demonstrate this connection. Firstly, cancer metabolic reprogramming, characterized by distinct metabolic patterns in tumors, is accompanied by concurrent alterations in m6A modifications. Additionally, m6A modifications play a regulatory role in metabolic genes [19]. Specifically, they exert control over the expression of metabolic genes in cancer cells by affecting the stability, translation, and splicing of RNA molecules. This regulatory influence extends to key metabolic pathways, including glycolysis, the tricarboxylic acid (TCA) cycle, fatty acid synthesis, and amino acid metabolism. Notably, enzymes responsible for m6A modifications, such as writers, erasers, and readers, interact with crucial metabolic pathways in cancer. For example, the m6A writer METTL3 is implicated in glycolysis regulation, while the m6A reader YTHDF2 participates in lipid metabolism. Dysregulation of these m6A regulators disrupts the metabolic homeostasis of cancer cells, further emphasizing the interplay between m6A modifications and cancer metabolism.
The relationship between m6A modification and glutamine metabolism involves a reciprocal regulatory mechanism. Firstly, m6A modification plays a role in the regulation of genes associated with glutamine metabolism. By modifying RNA molecules, m6A can modulate the stability, translation, and splicing of transcripts involved in the glutamine metabolic pathway [10,11,20]. Consequently, this regulation can impact the expression levels of critical enzymes and transporters involved in glutamine utilization and conversion [11]. For instance, our study demonstrates that IGF2BP3 can regulate the key metabolic enzymes GLS and GLUD1 in the context of glutamine metabolism through m6A modification. Secondly, the cellular concentration of glutamine also influences the level of m6A modifications. Glutamine availability has been shown to affect the extent of m6A modification. Experimental evidence indicates that reduced glutamine levels lead to alterations in the pattern of m6A modification. This observation suggests a potential link between cellular nutrient status, particularly the availability of glutamine, and m6A modification. Thirdly, m6A modification exerts regulatory effects on the epigenetic and post-transcriptional aspects of genes involved in glutamine metabolism. Through epigenetic mechanisms, m6A modifications can modulate chromatin structure and gene accessibility, thereby influencing the expression of genes related to glutamine metabolism. Additionally, m6A modifications on RNA molecules can govern their stability, translation efficiency, and alternative splicing, further impacting the expression of genes associated with glutamine metabolism.
Glutamine plays a pivotal role as a nutrient for tumor cells, and its metabolism within the tumor microenvironment is influenced by nutrient availability and competition. Tumor cells exhibit heightened uptake and utilization of glutamine to meet their energy demands, facilitate biosynthesis, and maintain redox homeostasis. Moreover, glutamine metabolism holds significant importance in immune cell function within the tumor microenvironment. Immune cells, including T cells and natural killer cells, heavily rely on glutamine as a vital energy source and for the synthesis of crucial metabolites like nucleotides and antioxidants. Consequently, glutamine depletion in the tumor microenvironment hampers immune cell function and undermines effective antitumor immune responses. Furthermore, glutamine serves as a nitrogen donor for nucleotide synthesis and contributes to the tricarboxylic acid (TCA) cycle by replenishing intermediates through anaplerotic reactions. Additionally, glutamine-derived metabolites promote fatty acid synthesis, maintain redox homeostasis, and contribute to the production of integrative metabolites that impact cellular metabolism and tumor development. Hypoxia, a hallmark of the tumor microenvironment, exerts an influence on glutamine metabolism. Under hypoxic conditions, there is an upregulation of glutamine transporters and glutaminase, facilitating increased glutamine uptake and utilization by tumor cells. This adaptive response enables tumor cells to sustain energy production and biosynthesis even in the presence of limited oxygen availability.
Our study highlights the significant expression of IGF2BP3, an m6A reader, in human cervical cancer cells, and its close association with glutamine metabolism and local immune evasion in tumors. This finding adds valuable insights to the understanding that m6A modification plays a regulatory role in tumor metabolism and the metabolism-related immune microenvironment in human cervical cancer. Specifically, the presence of IGF2BP3 in our study stabilizes the mRNAs of key metabolic enzymes involved in glutamine metabolism, namely GLS and GLUD1, subsequently enhancing glutamine metabolism and promoting increased lactate production and release from tumor cells. These alterations contribute to Treg cell-mediated immune evasion, identifying potential crucial targets for future therapeutic interventions in human cervical cancer, particularly immunotherapy.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83:428–441. doi: 10.1016/j.molcel.2023.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Tang J, Wang X, Xiao D, Liu S, Tao Y. The chromatin-associated RNAs in gene regulation and cancer. Mol Cancer. 2023;22:27. doi: 10.1186/s12943-023-01724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulias K, Greer EL. Biological roles of adenine methylation in RNA. Nat Rev Genet. 2023;24:143–160. doi: 10.1038/s41576-022-00534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Yang D, Liu T, Chen J, Yu J, Yi P. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol Med. 2023;29:454–467. doi: 10.1016/j.molmed.2023.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Zhu X, Fu H, Sun J, Xu Q. Interaction between N6-methyladenosine (m6A) modification and environmental chemical-induced diseases in various organ systems. Chem Biol Interact. 2023;373:110376. doi: 10.1016/j.cbi.2023.110376. [DOI] [PubMed] [Google Scholar]
- 6.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Huttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du M, Peng Y, Li Y, Sun W, Zhu H, Wu J, Zong D, Wu L, He X. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022;8:53. doi: 10.1038/s41420-022-00844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang H, Sun Q, Zhou J, Zhang H, Song Q, Zhang H, Yu G, Guo Y, Huang C, Mou Y, Jia C, Song Y, Liu A, Song K, Lu C, Tian R, Wei S, Yang D, Chen Y, Li T, Wang K, Yu Y, Lv Y, Mo K, Sun P, Yu X, Song X. m(6)A methylation reader IGF2BP2 activates endothelial cells to promote angiogenesis and metastasis of lung adenocarcinoma. Mol Cancer. 2023;22:99. doi: 10.1186/s12943-023-01791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Q, Li L, Wang Y, Wu P, Hou X, Ouyang J, Fan C, Li Z, Wang F, Guo C, Zhou M, Liao Q, Wang H, Xiang B, Jiang W, Li G, Zeng Z, Xiong W. RNA modifications in cancer. Br J Cancer. 2023;129:204–221. doi: 10.1038/s41416-023-02275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Liang C, Zhang M, Xu Y, Weng Y, Li X, Yu W. N6-methyladenosine modification and metabolic reprogramming of digestive system malignancies. Cancer Lett. 2022;544:215815. doi: 10.1016/j.canlet.2022.215815. [DOI] [PubMed] [Google Scholar]
- 11.Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y, Zhou K, Li W, Hu J, Fu C, Aziz T, Li H, Li J, Yang Y, Han L, Zhang S, Ma Y, Sun M, Wu H, Zhang Z, Wunderlich M, Robinson S, Braas D, Hoeve JT, Zhang B, Marcucci G, Mulloy JC, Zhou K, Tao HF, Deng X, Horne D, Wei M, Huang H, Chen J. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40:1566–1582. e10. doi: 10.1016/j.ccell.2022.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Chen J, Chen W, Xu Y, Shen Y, Xu X. Targeting IGF2BP3 in cancer. Int J Mol Sci. 2023;24:9423. doi: 10.3390/ijms24119423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu K, Gao T, Wang Z, Zhang L, Tan K, Lv Z. RNA N6-methyladenosine reader IGF2BP3 interacts with MYCN and facilitates neuroblastoma cell proliferation. Cell Death Discov. 2023;9:151. doi: 10.1038/s41420-023-01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu TY, Hong LL, Ling ZQ. Oncofetal protein IGF2BPs in human cancer: functions, mechanisms and therapeutic potential. Biomark Res. 2023;11:62. doi: 10.1186/s40364-023-00499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Zhang T, You Z, Zhang Y. Positive surgical margin, HPV persistence, and expression of both TPX2 and PD-L1 are associated with persistence/recurrence of cervical intraepithelial neoplasia after cervical conization. PLoS One. 2015;10:e0142868. doi: 10.1371/journal.pone.0142868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jo H, Shim K, Jeoung D. Roles of RNA methylations in cancer progression, autophagy, and anticancer drug resistance. Int J Mol Sci. 2023;24:4225. doi: 10.3390/ijms24044225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petri BJ, Klinge CM. m6A readers, writers, erasers, and the m6A epitranscriptome in breast cancer. J Mol Endocrinol. 2023;70:e220110. doi: 10.1530/JME-22-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Zhou J, Zhang H, Ge L, Li J, Wang H. RNA m(6) A methylation in cancer. Mol Oncol. 2023;17:195–229. doi: 10.1002/1878-0261.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng K, Liu S, Li C, Zhao Y, Wang Q. IGF2BP3/HIF1A/YAP signaling plays a role in driving acute-on-chronic liver failure through activating hepatocyte reprogramming. Cell Signal. 2023;108:110727. doi: 10.1016/j.cellsig.2023.110727. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Wu Z, Zhang Y, Lu X, Miao L. Glutamine metabolism genes prognostic signature for stomach adenocarcinoma and immune infiltration: potential biomarkers for predicting overall survival. Front Oncol. 2023;13:1201297. doi: 10.3389/fonc.2023.1201297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.