Abstract
The clinical significance and prognostic role of whole-blood EBV-DNA in EBV-associated nasopharyngeal carcinoma (NPC) have not been thoroughly investigated. This study aims to explore the diagnostic and prognostic value of EBV-DNA for NPC in a non-endemic region of China. We enrolled patients with chronic active EBV infection (CAEBV), nasopharyngitis (NA), extranodal NK/T-cell lymphoma, nasal type (ENKTCL-NT), and NPC. Demographic and clinical data were collected and the diagnostic and prognostic values of EBV-DNA were analyzed. Immunohistochemistry was performed to detect EBV-encoded small ribonucleic acids (EBER), as well as the expression of p53, Ki-67, and epidermal growth factor receptor (EGFR). The levels of pretreatment Epstein-Barr virus DNA (preEBV-DNA) in new NPC cases were found to differ from those in other diseases and exhibited varying age distributions. The threshold value of preEBV-DNA for distinguishing NPC from CAEBV and NA was determined. We confirmed that epistaxis, diabetes mellitus, T3N2 or T4N0-2 stage, and IgM positivity were associated with higher levels of preEBV-DNA, and identified risk factors associated with the prognosis of locoregionally advanced NPC (La-NPC). Patients with intermittently or persistently positive EBV-DNA (IPCP), higher preEBV-DNA levels, and positive Epstein-Barr virus-encoded small RNA (EBER) status (EBERpos) had worse survival. New cases of NPC with elevated levels of EBV in the whole-blood and positive EBER status were shown to have a poor prognosis upon progression to La-NPC. EBV-DNA was found to be an indicator for predicting prognosis in La-NPC and could also be used to distinguish new NPC cases.
Keywords: Epstein-Barr virus, nasopharyngeal carcinoma, NPC, whole-blood EBV-DNA
Introduction
Nasopharyngeal carcinoma (NPC) is not only a common malignant disease of the head and neck, but also presented as locoregionally advanced NPC at diagnosis with poor prognosis. The NPC accounts for approximately 60% of the 200,000 new tumor cases [1]. Unlike other cancers, NPC has a distinct regional distribution and it is also highly prevalent in East and Southeast Asia, particularly in southern China [2]. China has an age-standardized mortality rate of 1.6/100,000, which is approximately twice the global NPC average for NPC [3].
Approximately 70% of patients were diagnosed with locoregionally advanced NPC (La-NPC) (stage III-IVa) and have an unsatisfactory prognosis [4]. The NPC is ubiquitously associated with the Epstein-Barr virus (EBV), and the presence of EBV-encoded RNA (EBER) has been consistently demonstrated in this cancer [5]. These EBV-encoded latent genes can interact with host oncogenes, leading to a disturbed host cell cycle and promoting the development of EBV-associated epithelial neoplasms [6]. Even though NPC is a highly radio-sensitive and curable disease, most people diagnosed with NPC tend to be in the middle or late stages of the disease, when symptoms are much more pronounced. Furthermore, loco-regional relapses following definitive therapy contribute significantly to cancer-specific mortality among NPC patients [7].
To date, there is no single laboratory blood test that provides a highly sensitive and specific results for the screening and diagnosis of NPC. However, strong relationships between higher EBV serological markers and EBV-DNA load levels and NPC risk have been documented in populations from various endemic locations [8,9]. Studies have further claimed that the progressive accumulation of genomic instability and persistent EBV infection have encouraged the development of NPC [10]. Continued EBV infection enhances the resilience of NPC cells in the tumor microenvironment. NPC cells acquired the capacity for malignant biological behaviors and eventually formed remote metastasis [11]. Previous work reported that higher levels of EBV-DNA have been considered a useful indicator in assessing NPC risk [12]. Moreover, different EBV-DNA results might represent two distinct subtypes of the disease [13]. However, the intrinsic features of NPC associated with the status of whole-blood EBV-DNA have not been elucidated in detail.
In this study, we investigated the variations in preEBV-DNA levels at diagnosis and performed a comprehensive comparison of these levels across several diseases presenting similar symptoms. Our aim is to evaluate the potential of preEBV-DNA as a screening tool for NPC. In addition, we analyzed the patterns of variation of posEBV-DNA changes over time and hypothesized that these distinct patterns of dynamic variability could be used to predict the prognosis of La-NPC. The findings of our study have important implications for clinical decision-making and personalized therapy in La-NPC patients.
Materials and methods
Patient selection
The study included patients who met the following eligibility requirements: age over 18 years, confirmed diagnosis of nasopharyngeal carcinoma (NPC) through pathological examination or clinical diagnosis of CAEBV, NA, or ENKTCL-NT. Patients with secondary cancer, pregnancy or nursing care were excluded.
For the survival analysis of locally advanced nasopharyngeal carcinoma (La-NPC), patients were required to meet additional criteria: confirmation of stages III-IVA NPC based on the 8th TNM staging, treatment with three cycles of induction chemotherapy, treatment with intensity-modulated radiotherapy (IMRT) and two cycles of platinum-based concurrent chemoradiotherapy, availability of EBV-DNA data at first diagnosis and after induction chemotherapy (monthly for at least six months), and adequate hematologic, liver, and renal function. Patients who received adjuvant chemotherapy or did not complete radiotherapy were excluded. The study protocol was approved by the institutional review board and the research ethics committee at the participating sites, and all patients provided written informed consent for exploratory biomarker analysis.
The whole-blood EBV-DNA detection and immunofluorescence assays
Viral DNA was extracted from EDTA whole blood and quantified using quantitative real-time polymerase chain reaction (PCR) (reported as copies/mL). DNA extraction was performed with the EBV PCR Fluorescence Quantitative Diagnostic Kit (Daan Gene Co., Guangzhou, China). Real-time quantitative PCR was carried out using an ABI PRISM 7500 instrument (Applied Biosystems, Foster City, CA, USA). The cutoff for viral load was set at 500 copies/mL [14]. Indirect immunofluorescence was used to measure the presence of IgM and IgG antibodies against EBV-VCA. Samples were processed on a fully automated random-access analyzer, and chemiluminescent signals were measured by a photomultiplier tube and expressed as relative light units (RLU). Finally, the system calculates each result by using the ratio of the sample RLU to the cutoff RLU (s/co) [15].
Immunohistochemistry and in situ hybridization
Immunohistochemistry was performed on formalin-fixed paraffin-embedded (FFPE) tissue sections. The immunoreactivity of Ki-67 (UMAB-107, 1:1200; Leica) was judged positive when more than 10% of the cell nuclei showed strong intensity staining. The immunoreactivity of p53 (DO-7, 1:200; Roche) and EGFR (EP-22, 1:300; Leica) was judged as positive when more than 1% of the cell nuclei showed strong intensity staining. In general, p53 and EGFR are considered positive if the tumor cell shows nuclear staining, and the Ki-67 proliferation index is calculated as the percentage of positive cells. To detect EBV, in situ hybridization for EBV-encoded small RNA (EBER-ISH) (EBER PNA Probe, ISH-7001UM; ZSGB-BIO, Beijing, China) was performed according to the manufacturer’s instructions using a fluorescein-conjugated EBER oligonucleotide probe and the purified IgG fraction of a mouse monoclonal anti-fluorescein antibody. EBER-positive neoplastic cells exceeded 50 per high-power field (HPF) (> 50/HPF) and were defined as EBER-positive, and for other positive results, please refer to our previous studies [16]. At least 1000 dysplastic cells were counted, and ten high-powered fields (magnification ×400) were assessed. The results are presented in units of cells per square millimeter.
Definitions of EBV-DNA dynamic change and follow-up
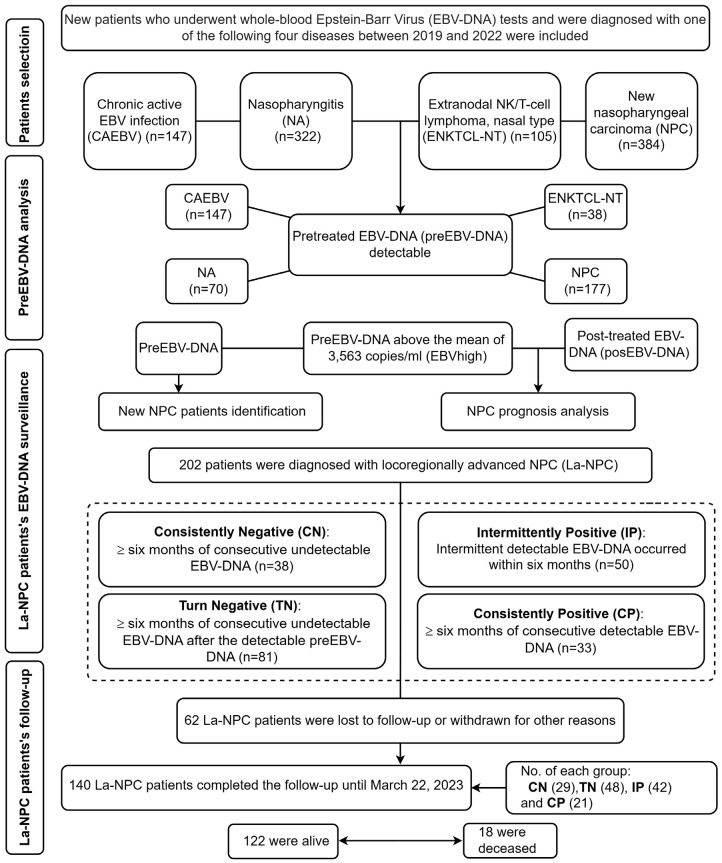
The first EBV-DNA load after induction chemotherapy (posEBV-DNA) was recorded. Follow-up assessments also include head and neck examinations, an MRI of the nasopharynx, a chest CT scans, and an ultrasound or CT scan of the neck and abdomen every six months. Patients with La-NPC were followed from diagnosis until March 22, 2023. EBVneg was defined as having a negative preEBV-DNA results, while EBVlow was defined as having preEBV-DNA levels below the mean of 3,563 copies/ml. EBVhigh, on the other hand, is defined as having a preEBV-DNA level above the mean. The dynamic changes in EBV-DNA were categorized as follows: (1) Consistently Negative (CN): more than six months of consecutive undetectable EBV-DNA; (2) Turn Negative (TN): more than six months of consecutive undetectable EBV-DNA after detectable preEBV-DNA; (3) Intermittent Positivity (IP): intermittent detectable EBV-DNA occurred within six months; and (4) Consistently Positive (CP): more than six months of consecutive detectable EBV-DNA (Figure 1).
Figure 1.
The flowchart illustrates the experimental design of this study.
Clinical relevance analysis
We first performed a regression analysis to identify the relevant risk factors for EBVhigh-NPC. Points were then assigned to each item based on patient information, EBV-DNA testing, and histological examination. Factors such as comorbidities (epistaxis) or underlying diseases (diabetes mellitus and hypertension), stage, preEBV-DNA, and EBV-DNA change patterns of IPCP were considered to have a direct impact on prognosis analysis and were assigned higher scores. The scoring rules for each item were as follows: Cus (exist = 1; other = 0), EBVhigh (EBVhigh = 1; EBVneg = 0), IPCP (Yes = 1; No = 0), Stage (T3N2 or T4Nx = 1; other = 0) (Nx, N0-2), Age (≥ 65 = 0.5; other = 0), Ki-67 (≥ 60% = 0.5; other = 0), and the status of EBER/EGFR/p53 (positive = 0.5; negative = 0). Patients with a score above the mean were classified into the high-risk group.
Statistical analysis
We utilized the “ggpubr” package in R software to compare the differences in EBV-DNA load levels. To distinguish NPC from CAEBV, NA, and ENKTCL-NT, we performed receiver operating characteristic (ROC) curve analysis using the “pROC” package in R software. In addition, ROC curve analysis was performed to determine the optimal threshold to distinguish NPC from CAEBV and NA. Univariate and multivariate logistic regression analyses were used to identify the variables associated with EBVhigh-NPC. In the multivariable analysis, variables with P < 0.05 in the univariable analysis were retained using forward selection. Sankey plots were generated using the R “ggalluvial” package to visualize the relationship between patients in the high and low-risk groups. Furthermore, a nomogram was constructed based on the Sankey plot results using the “rms” package in R software to provide a visual toolkit for assessing the risk of La-NPC. Lastly, overall survival (OS) analysis was conducted using the “survival” and “survminer” packages in R software. All statistical tests were two-sided, and statistical significance was determined at P < 0.05.
Results
Patient information
Out of the 1,160 eligible patients, the median age was 57 years (IQR: 48, 64). Of these, 147 had CAEBV, 322 had NA, 105 had ENKTCL-NT, 384 had newly diagnosed NPC, and 202 had La-NPC. The male-to-female ratio among new NPC patients was 2.24:1. At diagnosis, 177 new NPC patients tested positive for preEBV-DNA (Figure 1).
Differences in preEBV-DNA levels at diagnosis among patients with diseases of similar symptoms
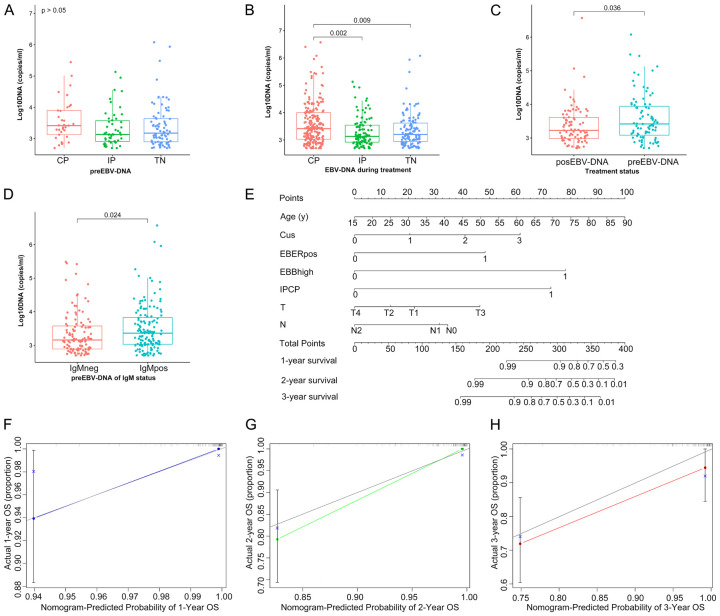
Figure 2A showed that the preEBV-DNA levels for CAEBV (n = 147), NA (n = 70), ENKTCL-NT (n = 38) and new NPC (n = 177) were 4,310, 6,858, 4,634, and 2,050 copies/ml, respectively. Among these groups, new NPC had the lowest preEBV-DNA levels (P < 0.001) (Figure 2A). Figure 2B indicates that there were no differences in preEBV-DNA levels between males and females among new NPC patients (P = 0.298) (Figure 2B). In comparison, preEBV-DNA levels were 843, 3,480, 2,940, and 2,590 copies/ml for patients ages 18 to 35, 36 to 45, 56 to 65, and 65 and older, respectively. Overall, the median preEBV-DNA levels of new NPC patients at diagnosis were significantly various in different age groups (P = 0.003) (Figure 2C).
Figure 2.
Difference and diagnostic performance at diagnosis in whole blood EBV-DNA. A. Box plot showing preEBV-DNA levels at diagnosis among patients with similar symptoms. B. Box plot showing preEBV-DNA levels at diagnosis between two groups of new NPC patients. C. Box plot showing preEBV-DNA levels at diagnosis among different age groups of new NPC patients. D. Receiver operating characteristic (ROC) curve analysis comparing NPC with CAEBV, ENKTCL-NT, and NA (n = 432). E. ROC curve analysis comparing NPC with CAEBV and NA using optimal cutoff values (n = 394). NPC: nasopharyngeal carcinoma; CAEBV: chronic active EBV infection; NA: nasopharyngitis; ENKTCL-NT: extranodal NK/T-cell lymphoma, nasal type.
Diagnostic ability of preEBV-DNA levels using the corresponding thresholds in diseases with similar symptoms
Receiver operating characteristic (ROC) curve analyses were performed (Figure 2D and 2E). To account for the influence of gender and age on the ROC analysis, we first conducted Chi-square tests for the differences in the composition ratio of gender and age in each group. The results showed that there were no statistically significant differences in the composition ratio of gender and age (Supplementary Table 1). The ROC results demonstrated that the differences in preEBV-DNA levels between NPC and CAEBV, NA and ENKTCL-NT were statistically significant (all P < 0.01), with an area under the curve (AUC) ranging from 0.626 to 0.661 (Figure 2D). Notably, the AUC of CAEBV was 0.718 (95% CI: 0.65 to 0.786, P < 0.001) using a threshold of 2,748 copies/ml. When the threshold was set at 4,920 copies/ml, the AUC of NA was 0.857 (95% CI: 0.809 to 0.905, P < 0.001) (Figure 2E).
Regression analysis of risk stratification in new EBVhigh-NPC patients and La-NPC patients
In the univariable analysis, it was found that new NPC patients with epistaxis and diabetes mellitus, T3N2 or T4Nx stage, and positive IgM were significantly associated with EBVhigh (all P < 0.05). The final multivariate logistic regression analysis revealed that patients with all these risk factors were more likely to be diagnosed with EBVhigh-NPC (Figure 3A and 3B).
Figure 3.
Regression analysis for new EBV high-NPC patients and risk stratification for La-NPC patients. (A) Forest plot of univariate analysis and (B) multivariate regression analysis in the new NPC cohort (n = 256, odds ratio with 95% confidence interval). (C) Sankey diagram illustrating the risk stratification of La-NPC patients. Cus: comorbidities (epistaxis) or underlying diseases (diabetes mellitus and/or hypertension); EBERpos/EGFRpos/p53pos: positive EBER/EGFR/p53 status; EBVhigh: preEBV-DNA levels above the mean of 3,563 copies/ml; Ki-67high: Ki-67 index above 60%; T: tumor size; N: nearby lymph nodes with cancer (Nx, N0-2); IP: Intermittently Positive; CP: Consistently Positive.
Based on the multivariate analysis, La-NPC patients were stratified into high-risk and low-risk groups using the mean risk score. The risk score was calculated as follows: Risk score = N × 100/449, where N represents the total score of each patient. The Sankey diagram showed that patients in the high-risk group, as well as those with age above 65, Cus, EBERpos status, EBVhigh, IPCP, and T3N2 or T4Nx stage subtype, had worse prognosis (Figure 3C).
EBV-DNA levels for different patterns and nomogram constructions based on risk analysis
Among the 202 patients diagnosed with La-NPC, ongoing EBV-DNA monitoring was performed monthly. The number of La-NPC cases in the CN, TN, IP, and CP groups were 38, 81, 50, and 33, respectively (Figure 1). No significant differences in preEBV-DNA were found among the TN, IP, and CP groups (P = 0.119) (Figure 4A). However, when considering all EBV-DNA results during treatment, the median EBV-DNA of the CP group (2,590 copies/ml) was significantly higher than that of the TN (1,580 copies/ml) and IP groups (1,350 copies/ml) (P = 0.003) (Figure 4B). Supplementary Figure 1 demonstrated the dynamic change model of EBV-DNA for the corresponding group (Supplementary Figure 1). The preEBV-DNA level (2,610 copies/ml) was higher than the posEBV-DNA level (1,680 copies/ml) (P = 0.036) (Figure 4C). Furthermore, the preEBV-DNA level of the positive IgM group (2,305 copies/ml) was higher than that of the negative group (1,441 copies/ml) (P = 0.024) (Figure 4D). We constructed a nomogram, based on the multivariate Cox regression results for age, Cus, EBERpos, EBVhigh, IPCP, and pathological stage in the La-NPC cohort (Figure 4E). This nomogram demonstrated a relatively high level of accuracy predicting the overall survival (OS) of La-NPC patients, as evidenced by the calibration curves (Figure 4F-H).
Figure 4.
EBV-DNA levels, nomogram construction and risk analysis. A. Box plot showing pre-treatment EBV-DNA levels at diagnosis for each group. B. Box plot showing pre-treatment EBV-DNA levels during treatment for each group. C. Box plot showing EBV-DNA levels for different treatment statuses. D. Box plot showing EBV-DNA levels for different IgM statuses. E. Nomogram for predicting 1-year, 2-year, and 3-year survival probabilities. F-H. Calibration curves of the nomogram in the La-NPC cohort. The x-axis represents the predicted probability of the nomogram and the y-axis represents the actual survival probability. TN: Turn Negative; preEBV-DNA: pretreatment EBV-DNA; posEBV-DNA: post-treatment EBV-DNA; IgMneg: negative IgM results; IgMpos: positive IgM results.
Overall survival in La-NPC patients
Out of the 140 La-NPC patients who completed the final follow-up, 18 (12.86%) died, while 122 (87.14%) remained alive at the end of the follow-up period (Figure 1). Using the Kaplan-Meier method with a log-rank test, we found that OS was significantly lower in patients exhibiting EBV-DNA patterns of IPCP compared to those with CNTN (P = 0.005) (Figure 5A). Figure 5B showed worse OS in patients with EBVhigh-NPC than those with EBVneg-NPC (P = 0.02). Similarly, Figure 5E indicated worse OS in patients with EBERpos compared to the negative group (P = 0.023). However, when we stratified by p53, Ki-67, and EGFR, log-rank analyses did not reveal significant survival differences among patients with positive p53 and EGFR statuses and Ki-67 expressions above 60%, as compared to their respective control groups (P > 0.05) (Figure 5C, 5D and 5F).
Figure 5.
Overall survival analysis based on different factors for La-NPC patients. A. Overall survival comparison between CNTN and IPCP. B. Overall survival comparison between high and negative EBV-DNA levels. C. Overall survival comparison between positive and negative p53 status. D. Overall survival comparison between high and low Ki-67 index. E. Overall survival comparison between positive and negative EBER status. F. Overall survival comparison between positive and negative EGFR status. CN: consistently negative; EBVneg: negative preEBV-DNA; EBERneg/EGFRneg/p53neg: negative EBER/EGFR/p53 status; Ki-67low: Ki-67 index is below 60%.
Discussion
Humans are the host of EBV, and most cases are asymptomatic infection and self-limiting [17]. Chan et al. conducted a large-scale prospective trial to evaluate the use of EBV-based biomarkers for NPC screening among asymptomatic individuals in Southern China. They defined participants as screen-positive if both their baseline and retest EBV results were positive [18]. Despite the prevalence of EBV infection in humans, a detailed description of any differences in EBV-DNA load between NPC and other diseases with similar symptoms has yet to be documented. Most studies to date have primarily focused on optimizing the detection limit [19] and selecting optimal time points for EBV-DNA tests [20]. Consequently, there are limited studies that have executed cross-sectional comparisons of whole-blood EBV-DNA, a method that holds potential to effectively eliminate malignancy risks in negative tests as recommended by the WHO [21]. This type of comparison is significant given the profound anatomical structure of NPC patients, their atypical early symptoms, and the reality that most new NPC cases are already in the terminal stage at diagnosis [22]. Furthermore, the ease of collecting peripheral blood samples enhances the appeal of this approach [23].
Despite the positive diagnosis of preEBV-DNA, we observed significant differences between the new NPC and other similar diseases. The preEBV-DNA level of new NPC was found to be 2,050 copies/ml, which was comparatively lower than that of other diseases (Figure 2A). This finding sets the stage for the potential use of preEBV-DNA as a diagnostic marker for new NPC. Our results indicate that the preEBV-DNA levels of CAEBV and NA were double and triple the level of new NPC respectively (Figure 2A). It is important to note that NA patients not only exhibit positive EBV-DNA results, but also present similar nasal symptoms prior to diagnosis. Following, we perform ROC analysis among NPC, CAEBV, and NA. The results revealed that the AUC of CAEBV was 0.718 (95% CI: 0.65 to 0.786) when the preEBV-DNA fell below 2,748 copies/ml, and the AUC of NA was 0.857 (95% CI: 0.809 to 0.905) when the preEBV-DNA exceeded 4,920 copies/ml (Figure 2E). Although clinical pathologists may easily distinguish NPC from NA histologically, the early diagnosis and differentiation from NA remain problematic because of the lack of sensitivity in routine screening methods in clinical practice [24]. Early symptoms of the condition often lead to confusion among patients about the source of their nasal discomfort. In this study, we have confirmed the diagnostic value of preEBV-DNA at diagnosis through ROC analysis. Our findings allow for the exclusion of a subset of participants not at high risk by identifing a preEBV-DNA range in the new NPC screening. Additionally, studies have concluded that EBV-DNA detection is more sensitive in whole-blood samples compared to plasma samples [25]. Our study findings suggest that a sensitive range of preEBV-DNA loadings could serve as a promising biomarker for early detection of new NPC. Further studies are needed to validate these findings and establish the clinical utility of this biomarker in new NPC screening.
Multiple reports have suggested that dynamic changes in EBV-DNA can predict long-term survival outcomes and accurately stratify the risk in NPC [9,20,26], and our study confirms these findings. However, previous studies have primarily focused on specific time points [20,27]. In our study, we investigated the prognostic value of EBV-DNA at monthly intervals and examined the patterns of dynamical variability. We found that continuous monitoring of EBV-DNA predicted long-term survival outcomes and accurately stratified risk in La-NPC. By categorizing EBV-DNA changes into four patterns (CN, TN, IP, and CP), we observed that patients with patterns of IPCP had worse OS compared to those with CNTN patterns (Figure 5A). This conclusion aligns with other studies [20,28].
Furthermore, we demonstrated that patients with high EBV levels in La-NPC had a worse OS compared to those with negative EBV (Figure 5B). This suggests that preEBV-DNA levels can accurately predict the outcome of La-NPC, which is consistent with previous studies [29]. Additionally, we found that pre EBV-DNA levels can be used to evaluate the efficacy of NPC treatment, as patients who responded well to treatment showed a decrease in posEBV-DNA levels (Figure 4C). There were no significant differences in preEBV-DNA levels among the TN, IP, and CP groups (Figure 4A). However, we observed differences among these groups during treatment, with the CP group showing higher levels compared to the IP and TN groups (Figure 3B). These findings further support the notion that multiple EBV-DNA tests can provide valuable prognostic information for La-NPC [30]. Previous studies have shown that NPC patients with persistently detectable posEBV-DNA have a higher rate of treatment failure and poorer prognosis [31]. It has been suggested that early reduction of posEBV-DNA levels indicates treatment effectiveness, which highlights the importance of long-term monitoring of EBV-DNA during and after treatment [20,31]. Among various non-invasive specimens, whole-blood samples have been found to be the most convenient and have higher diagnostic accuracy for NPC compared to other specimens such as plasma [25,32]. However, more efficient nucleic acid extraction and detection methods are needed to optimize the detection limit and increase the clinical application of EBV-DNA for NPC [9].
Studies have consistently shown that overexpression of Ki-67 is associated with poorer survival outcomes [33]. Additionally, inhibiting p53 overexpression has been found to reduce the risk of NPC [34]. However, when we stratified the data based on p53 and Ki-67 expression levels, we did not observe a significant correlation between Ki-67 expression and p53 status with survival (Figure 5C and 5D). This finding differs from previous studies that have reported a significant association between positive Ki-67 [33,35], p53 [36] and EGFR [37] expression and poor survival. The discrepancy may be attributed to the use of different cut-off values to define high and low expressions for Ki-67 and positive p53 status [38]. Also, the differences could be due to variations in patient characteristics or the relatively small sample size of our study. We speculate that these differences may also be influenced by genetic mutations in signaling pathways among NPC patients from different regions, leading to more complex clinical outcomes [39]. Therefore, further studies with larger sample sizes and standardized cutoff values are necessary to investigate the prognostic value of Ki-67 expression and positive p53 and EGFR status. Furthermore, consistent with previous reports, we found that EBER expression is a potential prognostic factor for NPC and is highly negatively correlated with disease progression [40].
There are several limitations to the present study. Firstly, the retrospective nature of this study may introduce selection bias. We retrieved and re-stained specimens from the tissue bank, which may have led to some bias in the staining results compared to fresh specimens at the time of diagnosis. In addition, we only performed qualitative evaluation of p53, EGFR and EBER-ISH, rather than relative quantification. These factors may contribute to the observed differences compared to findings from other studies. Secondly, not all patients completed the EBV-DNA testing according to the frequency of our study, resulting in a small sample size as we only selected patients with complete data. Moreover, the follow-up period was relatively short, and long-term survival data were lacking. Lastly, although patients were treated under a fully uniform regimen as recommended by institutional guidelines, it was not possible to objectively assess the effects of other factors other than medical treatment that were associated with clinical outcomes. In general, these limitations should be taken into account when interpreting the results of this study.
Conclusions
New cases of NPC exhibiting higher levels of EBV in the whole blood and positive status for EBER have been shown to have a poor prognosis upon progression to La-NPC. Monitoring of EBV-DNA levels on a monthly basis has been found to be a reliable indicator of prognosis in La-NPC. In addition, the EBV-DNA levels at the time of diagnosis have the potential to be used as a complementary method to distinguish NPC from other diseases.
Acknowledgements
The authors acknowledge the Hangzhou Center for Diseases Control and Prevention for its participation in this study.
Disclosure of conflict of interest
None.
Supplementary Table 1
Supplementary Figure 1
References
- 1.Li W, Duan X, Chen X, Zhan M, Peng H, Meng Y, Li X, Li XY, Pang G, Dou X. Immunotherapeutic approaches in EBV-associated nasopharyngeal carcinoma. Front Immunol. 2023;13:1079515. doi: 10.3389/fimmu.2022.1079515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Pan JJ, Ng WT, Zong JF, Chan LL, O’Sullivan B, Lin SJ, Sze HC, Chen YB, Choi HC, Guo QJ, Kan WK, Xiao YP, Wei X, Le QT, Glastonbury CM, Colevas AD, Weber RS, Shah JP, Lee AW. Proposal for the 8th edition of the AJCC/UICC staging system for nasopharyngeal cancer in the era of intensity-modulated radiotherapy. Cancer. 2016;122:546–558. doi: 10.1002/cncr.29795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui KF, Chan TF, Yang W, Shen JJ, Lam KP, Kwok H, Sham PC, Tsao SW, Kwong DL, Lung ML, Chiang AKS. High risk Epstein-Barr virus variants characterized by distinct polymorphisms in the EBER locus are strongly associated with nasopharyngeal carcinoma. Int J Cancer. 2019;144:3031–3042. doi: 10.1002/ijc.32049. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Lui KS, Ye Z, Fung TY, Chen L, Sit PY, Leung CY, Mak NK, Wong KL, Lung HL, Tanaka Y, Cheung AKL. EBV latent membrane protein 1 augments gammadelta T cell cytotoxicity against nasopharyngeal carcinoma by induction of butyrophilin molecules. Theranostics. 2023;13:458–471. doi: 10.7150/thno.78395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang YM, Gong GZ, Qiu QT, Han YW, Lu HM, Yin Y. Radiomics for diagnosis and radiotherapy of nasopharyngeal carcinoma. Front Oncol. 2022;11:767134. doi: 10.3389/fonc.2021.767134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha S, Dickey BL, Coghill AE. Utility of Epstein-Barr virus (EBV) antibodies as screening markers for nasopharyngeal carcinoma: a narrative review. Ann Nasopharynx Cancer. 2022;6:6. doi: 10.21037/anpc-21-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Q, Zhou Y, Zhou J, Zhao D, Li L, Li X, Huang Y, Wang Q, Zou H, Zhang K, Li Y, Wang Z, Deng Y, Meng F, Ying B, Yang M, Wang D. Clinical relevance of plasma EBV DNA as a biomarker for nasopharyngeal carcinoma in non-endemic areas: a multicenter study in southwestern China. Clin Chim Acta. 2023;541:117244. doi: 10.1016/j.cca.2023.117244. [DOI] [PubMed] [Google Scholar]
- 10.Hau PM, Lung HL, Wu M, Tsang CM, Wong KL, Mak NK, Lo KW. Targeting Epstein-Barr virus in nasopharyngeal carcinoma. Front Oncol. 2020;10:600. doi: 10.3389/fonc.2020.00600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Deng Y, Huang Y, Ye J, Xie S, He Q, Chen Y, Lin Y, Liang R, Wei J, Li Y, Zhang J. Nasopharyngeal carcinoma progression: accumulating genomic instability and persistent Epstein-Barr virus infection. Curr Oncol. 2022;29:6035–6052. doi: 10.3390/curroncol29090475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel PD, Alghareeb R, Hussain A, Maheshwari MV, Khalid N. The association of Epstein-Barr virus with cancer. Cureus. 2022;14:e26314. doi: 10.7759/cureus.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li WZ, Wu HJ, Lv SH, Hu XF, Liang H, Liu GY, Lu N, Bei WX, Lv X, Guo X, Xia WX, Xiang YQ. Assessment of survival model performance following inclusion of Epstein-Barr virus DNA status in conventional TNM staging groups in Epstein-Barr virus-related nasopharyngeal carcinoma. JAMA Netw Open. 2021;4:e2124721. doi: 10.1001/jamanetworkopen.2021.24721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Qin S, Jin Z, Chen Q, Xing L, Qiu T, Xia Y, Liang J, Zhu H, Wang L, Fan L, Xu W, Li J, Miao Y. The clinical significance and prognostic role of whole-blood Epstein-Barr virus DNA in lymphoma-associated hemophagocytic lymphohistiocytosis. J Clin Immunol. 2023;43:1302–1310. doi: 10.1007/s10875-023-01493-9. [DOI] [PubMed] [Google Scholar]
- 15.Corrales I, Gimenez E, Navarro D. Evaluation of the architect Epstein-Barr virus (EBV) viral capsid antigen (VCA) IgG, VCA IgM, and EBV nuclear antigen 1 IgG chemiluminescent immunoassays for detection of EBV antibodies and categorization of EBV infection status using immunofluorescence assays as the reference method. Clin Vaccine Immunol. 2014;21:684–688. doi: 10.1128/CVI.00104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng M, Jia Q, Chen J, Xu L, Xie L, Cheng Q, Li Q, Xiao M, Fang Z. High plasma EBV-DNA load and positive EBER status associated with viral recurrence and persistent infection in early treatment of lymphoma. Clin Exp Med. 2023;23:1307–1316. doi: 10.1007/s10238-022-00900-6. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Guo X, Guan H. Serious consequences of Epstein-Barr virus infection: hemophagocytic lymphohistocytosis. Int J Lab Hematol. 2022;44:74–81. doi: 10.1111/ijlh.13736. [DOI] [PubMed] [Google Scholar]
- 18.Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, Chu SWI, Mak C, Tse IOL, Leung SYM, Chan G, Hui EP, Ma BBY, Chiu RWK, Leung SF, van Hasselt AC, Chan ATC, Lo YMD. Analysis of plasma Epstein-Barr Virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377:513–522. doi: 10.1056/NEJMoa1701717. [DOI] [PubMed] [Google Scholar]
- 19.Lertbutsayanukul C, Kannarunimit D, Netsawang B, Kitpanit S, Chakkabat C, Hansasuta P, Prayongrat A. Optimal plasma pretreatment EBV DNA cut-off point for nasopharyngeal cancer patients treated with intensity modulated radiation therapy. Jpn J Clin Oncol. 2018;48:467–475. doi: 10.1093/jjco/hyy027. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Chen J, Liang B, Li Z, Li J, Yuan X, Wu S, Zeng F, Peng X, Li Y, Lu J, Zhao F, Liu X. Long-term monitoring of dynamic changes in plasma EBV DNA for improved prognosis prediction of nasopharyngeal carcinoma. Cancer Med. 2021;10:883–894. doi: 10.1002/cam4.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludvigsen LUP, Andersen AS, Hamilton-Dutoit S, Jensen-Fangel S, Bottger P, Handberg KJ, Ivarsen P, d’Amore F, Bibby BM, Albertsen BK, Jespersen B, Thomsen MK. A prospective evaluation of the diagnostic potential of EBV-DNA in plasma and whole blood. J Clin Virol. 2023;167:105579. doi: 10.1016/j.jcv.2023.105579. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Zhang B, Zhang X, Wang C, Xu Y. Identification of a 3-miRNA signature associated with the prediction of prognosis in nasopharyngeal carcinoma. Front Oncol. 2022;11:823603. doi: 10.3389/fonc.2021.823603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song J, Zhu K, Wang X, Yang Q, Yu S, Zhang Y, Fu Z, Wang H, Zhao Y, Lin K, Yuan G, Guo J, Shi Y, Liu C, Ai J, Zhang H, Zhang W. Utility of clinical metagenomics in diagnosing malignancies in a cohort of patients with Epstein-Barr virus positivity. Front Cell Infect Microbiol. 2023;13:1211732. doi: 10.3389/fcimb.2023.1211732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Tian Y, Di H, Xue C, Zheng Y, Hu B, Lin Q, Yan X. Noninvasive diagnosis of nasopharyngeal carcinoma based on phenotypic profiling of viral and tumor markers on plasma extracellular vesicles. Anal Chem. 2022;94:9740–9749. doi: 10.1021/acs.analchem.2c01311. [DOI] [PubMed] [Google Scholar]
- 25.Rzepka M, Depka D, Gospodarek-Komkowska E, Bogiel T. Diagnostic value of whole-blood and plasma samples in Epstein-Barr virus infections. Diagnostics (Basel) 2023;13:476. doi: 10.3390/diagnostics13030476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan SK, Chan SY, Choi HC, Tong CC, Lam KO, Kwong DL, Vardhanabhuti V, Leung TW, Luk MY, Lee AW, Lee VH. Prognostication of half-life clearance of plasma EBV DNA in previously untreated non-metastatic nasopharyngeal carcinoma treated with radical intensity-modulated radiation therapy. Front Oncol. 2020;10:1417. doi: 10.3389/fonc.2020.01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee VH, Kwong DL, Leung TW, Choi CW, Lai V, Ng L, Lam KO, Ng SC, Sze CK, Tong CC, Ho PP, Chan WL, Wong LS, Leung DK, Chan SY, Khong PL. Prognostication of serial post-intensity-modulated radiation therapy undetectable plasma EBV DNA for nasopharyngeal carcinoma. Oncotarget. 2017;8:5292–5308. doi: 10.18632/oncotarget.14137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen FP, Huang XD, Lv JW, Wen DW, Zhou GQ, Lin L, Kou J, Wu CF, Chen Y, Zheng ZQ, Li ZX, He XJ, Sun Y. Prognostic potential of liquid biopsy tracking in the posttreatment surveillance of patients with nonmetastatic nasopharyngeal carcinoma. Cancer. 2020;126:2163–2173. doi: 10.1002/cncr.32770. [DOI] [PubMed] [Google Scholar]
- 29.Pramanik R, Arora S, Sharma P, Biswas A, Nayak B, Thakar A, Sharma A, Ghose S. Cell-free EBV DNA as a biomarker during clinical management of nasopharyngeal carcinoma in a nonendemic region. J Med Virol. 2022;94:720–728. doi: 10.1002/jmv.27440. [DOI] [PubMed] [Google Scholar]
- 30.Lu H, Guo S, Liu L, Chen Q, Liang Y, Liu S, Sun X, Tang Q, Li X, Guo L, Mo H, Tang L, Mai H. Prognostic significance of a combined and controlled nutritional status score and EBV-DNA in patients with advanced nasopharyngeal carcinoma: a long-term follow-up study. Cancer Biol Med. 2021;19:551–564. doi: 10.20892/j.issn.2095-3941.2020.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang WY, Lin TY, Twu CW, Tsou HH, Lin PJ, Liu YC, Huang JW, Hsieh HY, Lin JC. Long-term clinical outcome in nasopharyngeal carcinoma patients with post-radiation persistently detectable plasma EBV DNA. Oncotarget. 2016;7:42608–42616. doi: 10.18632/oncotarget.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W, Chen G, Gong X, Wang Y, Zheng Y, Liao X, Liao W, Song L, Xu J, Zhang X. The diagnostic value of EBV-DNA and EBV-related antibodies detection for nasopharyngeal carcinoma: a meta-analysis. Cancer Cell Int. 2021;21:164. doi: 10.1186/s12935-021-01862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Yue L, Li Y, Zhang Q, Liang X. Prognostic value of Ki-67 in nasopharyngeal carcinoma: a meta-analysis. Biosci Rep. 2021;41:BSR20203334. doi: 10.1042/BSR20203334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou R, Liu X, Yang H, Deng S, Cheng C, Liu J, Li Y, Zhang Y, Jiang J, Zhu Z, Su Y, Wu L, Xie Y, Li X, Li W, Liu Z, Fang W. Chemically synthesized cinobufagin suppresses nasopharyngeal carcinoma metastasis by inducing ENKUR to stabilize p53 expression. Cancer Lett. 2022;531:57–70. doi: 10.1016/j.canlet.2022.01.025. [DOI] [PubMed] [Google Scholar]
- 35.Shi Z, Jiang W, Chen X, Xu M, Wang X, Zha D. Prognostic and clinicopathological value of Ki-67 expression in patients with nasopharyngeal carcinoma: a meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920951346. doi: 10.1177/1758835920951346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Liu Y, Zhang Z, Sun H, Ji X, Li B, Zhou X, Gai P. Prognostic value of the Epstein-Barr virus and tumor suppressor gene p53 gene in nasopharyngeal squamous cell carcinoma. J Cancer Res Ther. 2019;15:426–436. doi: 10.4103/jcrt.JCRT_750_18. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Youhong T, Tan Y, He Y, Ban Y, Cai J, Li X, Xiong W, Zeng Z, Li G, Yi M, Liu W, Xiang B. EGFR-PKM2 signaling promotes the metastatic potential of nasopharyngeal carcinoma through induction of FOSL1 and ANTXR2. Carcinogenesis. 2020;41:723–733. doi: 10.1093/carcin/bgz180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang GD, Wang ZC, Chen QY, Zhang HL, Lin XG, Huang TJ, Qian CN, Huang BJ. p53, latent membrane protein 1, bcl-2, and prognosis in nasopharyngeal carcinoma: a meta-analysis. Histol Histopathol. 2019;34:103–110. doi: 10.14670/HH-18-032. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, He Q, Ma H, Zhang L, Liu F, Han Y. Relationship of PI3K-Akt/mTOR/AMPK signaling pathway genetic mutation with efficacy and prognosis in nasopharyngeal carcinoma. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2022;47:165–173. doi: 10.11817/j.issn.1672-7347.2022.200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng Z, Fan S, Zhang X, Li S, Zhou M, Xiong W, Tan M, Zhang W, Li G. Epstein-Barr virus-encoded small RNA 1 (EBER-1) could predict good prognosis in nasopharyngeal carcinoma. Clin Transl Oncol. 2016;18:206–211. doi: 10.1007/s12094-015-1354-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.