Abstract
G protein-coupled receptors (GPCRs) are versatile and vital proteins involved in a wide array of physiological processes and responses, such as sensory perception (e.g., vision, taste, and smell), immune response, hormone regulation, and neurotransmission. Their diverse and essential roles in the body make them a significant focus for pharmaceutical research and drug development. Currently, approximately 35% of marketed drugs directly target GPCRs, underscoring their prominence as therapeutic targets. Recent advances in structural biology have substantially deepened our understanding of GPCR activation mechanisms and interactions with G-protein and arrestin signaling pathways. This review offers an in-depth exploration of both traditional and recent methods in GPCR structure analysis. It presents structure-based insights into ligand recognition and receptor activation mechanisms and delves deeper into the mechanisms of canonical and noncanonical signaling pathways downstream of GPCRs. Furthermore, it highlights recent advancements in GPCR-related drug discovery and development. Particular emphasis is placed on GPCR selective drugs, allosteric and biased signaling, polyphamarcology, and antibody drugs. Our goal is to provide researchers with a thorough and updated understanding of GPCR structure determination, signaling pathway investigation, and drug development. This foundation aims to propel forward-thinking therapeutic approaches that target GPCRs, drawing upon the latest insights into GPCR ligand selectivity, activation, and biased signaling mechanisms.
Introduction
GPCRs, as the largest membrane protein superfamily, are categorized into five distinct subfamilies: the rhodopsin-like family (Class A), the secretin/adhesion family (Class B), the metabotropic family (Class C), the smoothened/frizzled family (Class F), and the taste2 family (Class T). GPCRs play a pivotal role in transducing signals from the extracellular environment to the intracellular environment, regulating a variety of physiological processes. The diversity of signals they relay encompasses odors, light, neurotransmitters, and kinins. All GPCRs adopt the classic seven-transmembrane helix structure, connected by three extracellular loops (ECL1-3) as well as three intracellular loops (ICL1-3). However, each GPCR subfamily exhibits unique structural characteristics and ligand-binding specificities, intricately linked to their physiological roles [1].
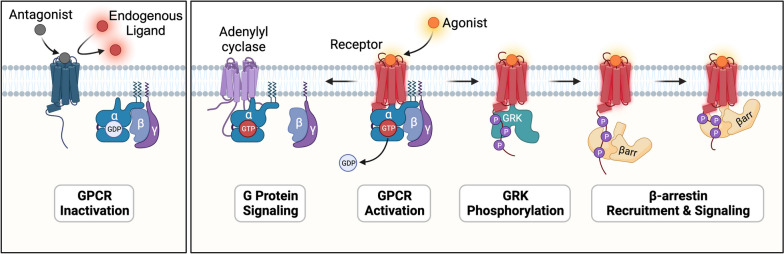
Over recent years, the landscape of GPCR investigation has been revolutionized by breakthroughs in structural biology, particularly with the advent of cryo-electron microscopy (cryo-EM) [2]. These advancements have shed light on the dynamic conformational changes of GPCRs, providing unprecedented insights into their inactivation or activation mechanisms and interactions with intracellular signaling transducers such as G-proteins and arrestins (Fig. 1) [3, 4]. Understanding the GPCR structure alterations, from inactive to active states, has been instrumental in deciphering the nuances of their signal transduction pathways and their implications on cellular responses.
Fig. 1.
Scheme of ligand-mediated GPCR inactivation or activation. Agonists bind to GPCRs and trigger downstream G-protein or β-arrestin signaling. Antagonists occupy the agonism-associated pocket and prevent endogenous ligand binding
Furthermore, a deepened comprehension of GPCR structural biology has significantly accelerated drug discovery endeavors [5]. With a precise grasp of ligand-receptor interactions and activation mechanisms, researchers have been empowered to rationally design and optimize drug candidates targeting GPCRs [6–8]. This has not only facilitated the identification of novel therapeutic agents but also enabled the refinement of existing drugs to enhance their efficacy, selectivity, and safety profiles [9].
Here, we aim to encapsulate the recent strides in GPCR structural elucidation, explore the implications of these findings on our understanding of GPCR-mediated signal transduction, and highlight the emerging opportunities in developing innovative GPCR-targeted drugs. Through a synthesis of current knowledge and prospects, we aspire to underscore the significance and potential of GPCRs in biomedical research and therapeutic development.
Technology progress for GPCR structure determination
GPCRs are dynamic in the cell membrane and respond to different types of ligands, including agonists, antagonists or allosteric modulators. In turn, this characteristic governs various intracellular pathways. Understanding the structure of GPCRs is fundamental for understanding ligand recognition and the mechanism of receptor activation, which could accelerate drug discovery.
Nonetheless, the task of elucidating the GPCR structure has been challenging, primarily owing to their low expression levels and dynamic features. To overcome these challenges, X-ray crystallography and single-particle cryo-EM techniques were subsequently applied for GPCR structure determination. A total of 243 unique structures have been deposited in the Protein Data Bank (PDB), including 188 Class A GPCRs, 28 Class B GPCRs (20 for Class B1 and 8 for Class B2), 19 Class C GPCRs, 5 Class F GPCRs, and 1 Class T GPCR.
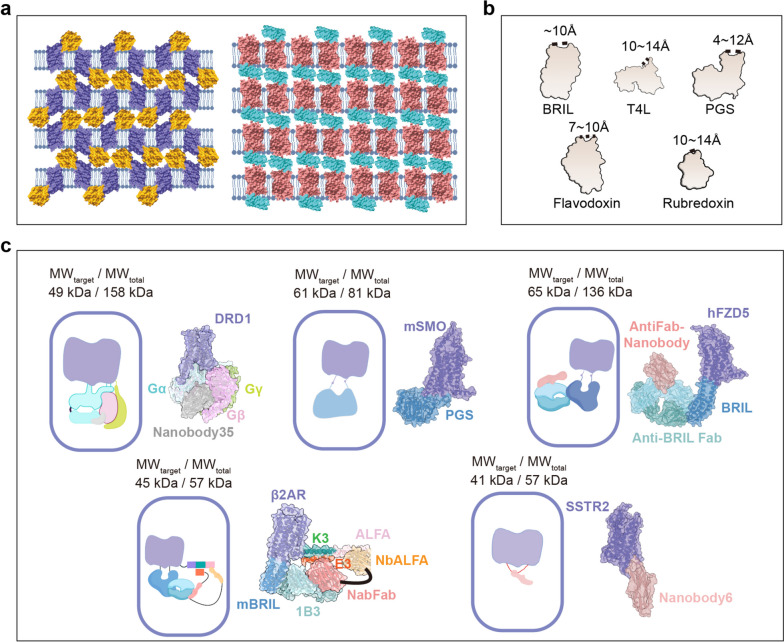
For X-ray crystallography, GPCRs are extracted from the cell membrane using synthetic detergents. This often reduces the hydrophilic surface essential for crystallization packing. To address these challenges, in 2007, Brian K. Kobilka’s team innovated by developing a fusion protein strategy combined with the lipid cubic phase (LCP) method (Fig. 2a, b) [10]. This approach facilitated both the stabilization and crystallization of GPCRs. Moreover, the introduction of specific mutations within the transmembrane domain was reported to enhance the thermostability of the receptor. A pivotal moment in GPCR structural biology arrived in 2011 when Brian K. Kobilka reported the first crystal structure of the agonist-induced β-2 adrenergic receptor (β2AR)-Gs protein signaling complex at near-atom resolution, which unveiled GPCR-mediated transmembrane signal transduction [11]. This breaking work was recognized internationally when Kobilka, along with Robert J. Lefkowitz, was awarded the 2012 Nobel Prize in Chemistry. However, despite these advances, obtaining well-diffracting crystals of GPCRs with ligands is time-consuming and challenging. Additionally, the majority of GPCR crystal structures solved thus far represent either inactive states or conformations that mimic active states rather than truly active ones. On a promising note, advancements in ultrafast time-resolved crystallography have paved the way for deeper insights into GPCR dynamics. A notable example is the utilization of this technology to elucidate the cascade of events through which photoactivated retinal triggers the activation process of rhodopsin, as referenced in [12].
Fig. 2.
Techniques for GPCR structure determination. a Crystal packing for GPCRs in the presence of fusion proteins. GnRH1R-PGS (left, PDB: 7BR3), GnRH1R: purple, PGS: yellow. ghrelin-BRIL (right, PDB: 7F83), ghrelin: red, BRIL: green. b Schematic diagrams of fusion proteins. The dashed line shows the distances between the N-terminus and C-terminus of the fusion proteins. c Strategies for cryo-EM structure determination of GPCRs. The complex of D1R with Gs and Nb35 (PDB: 7CKZ). mSMO with PGS fusion protein (PDB: 8CXO). hFZD5 with BRIL fusion protein binding with anti-BRIL Fab and anti-Fab Nanobody (PDB: 6WW2). β2AR links ICL3 to engaging BRIL mBRIL and the C-terminus to the K3 helix with an ALFA tag. The complex involves an anti-BRIL Fab, along with a bivalent ‘glue’ molecule containing anti-Fab (NbFab) and anti-ALFA (NbALFA) (PDB: 8J7E and 8JJO). SSTR2 is bound to nanobody6 (PDB: 7UL5)
The rise of cryo-EM technique has been a boon for GPCR research, especially for GPCR-signaling complexes, including the complexes of receptors with G-protein, arrestins, and other signaling mediators. It complements traditional techniques and offers new avenues to probe the relationship between GPCR structures and functions. This has been crucial in understanding the full spectrum of GPCR signaling pathways and paving the way for novel therapeutics. Cryo-EM has brought significant advancements to the structural biology of active GPCRs. To date, 151 receptors, which account for 60% of the total GPCR complex structures, have been resolved by the single-particle cryo-EM technique. Nevertheless, solving the cryo-EM structure of inactive GPCRs with antagonists presents inherent challenges. The primary hurdle stems from the relatively low molecular weight of such complexes, which can significantly compromise the signal-to-noise ratio in image processing. As a result, while cryo-EM offers transformative potential, its application for inactive GPCRs, especially those bound solely to antagonists, requires specialized approaches.
Recent strides in protein engineering have made much progress in determining the structures of inactive GPCRs using the cryo-EM method. These advancements can be broadly categorized into two strategic approaches. The first strategy employs the fusion of a rigid protein to the ICL3 region. Notably, Zhang et al. deciphered the structure of Smoothened receptor (SMO) at a remarkable global resolution of 3.7 Å by substituting ICL3 with the fusion protein Pyrococcus abysii glycogen synthase (PGS) (Fig. 2c) [13]. While the extended helix linking SMO and PGS typically lacks structural rigidity, the hydrophobic interactions between the two surprisingly augment the overall structural integrity. In addition, Gabriella Collu’s team introduced a rigid fusion protein, AmpC β-lactamase, to enhance both the molecular weight and stability of the β-1 adrenergic receptor (β1AR) [14].
The alternate strategy is the use of antibodies (nanobody or Fab) that can stabilize receptors. In a pioneering protein engineering study, the cryo-EM structure of the Frizzled-5 receptor (FZD5) was determined using antibodies against apocytochrome b562 RIL (BRIL) and the Fab nanobody (Fig. 2c) [15]. Following this, a research group independently resolved the inactive structure of GPR183 employing a similar approach [16]. Here, the rigidity between the BRIL fusion protein and the receptor, which forms extended helices for TM5 and TM6, proves critical. In a recent study, Guo’s team amalgamated previously mentioned strategies, employing a refined multipoint fusion approach (Fig. 2c) [17]. In the process of protein reconstruction, the BRIL fusion protein was inserted to substitute ICL3, and the ALFA helix tag was fused to the terminus of helix 8 (H8) [17]. A bivalent ‘glue’ molecule containing the anti-BRIL and anti-ALFA nanobodies was added to conjugate BRIL and ALFA and stabilize the cytoplasmic domain of the receptor [17]. To further enhance the rigidity of the intercellular region of the receptor, the E3/K3 coiled coil was introduced into this system [17]. In addition, the Nb6 nanobody was specifically designed to target the ICL3 region of the κ-opioid receptor (κOR) (Fig. 2c) [18]. The ICL3 region replacement at other GPCRs could accelerate GPCR determination.
The structural characteristics of GPCRs for ligand recognition and receptor activation
Canonical and noncanonical activation mechanisms of class A GPCRs
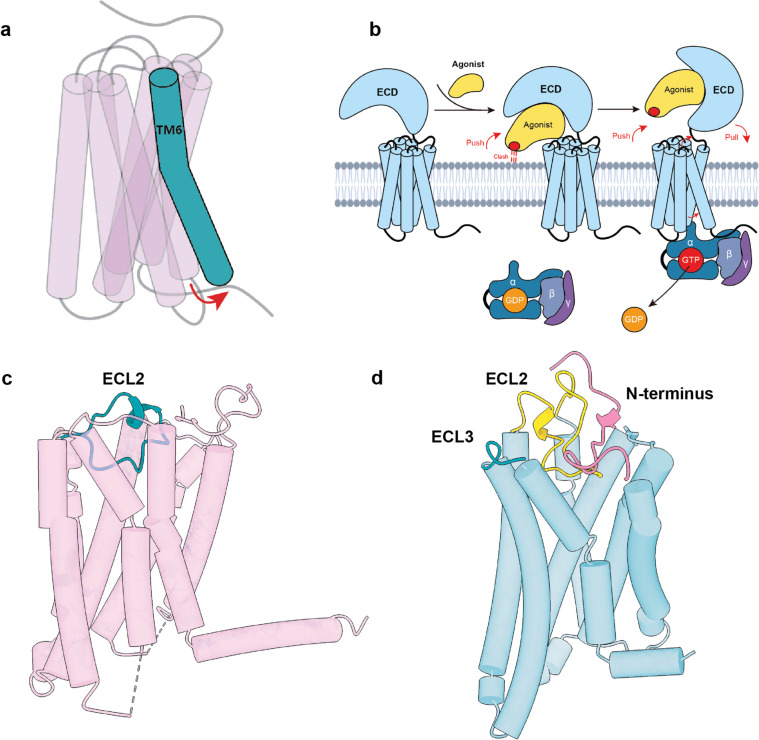
Class A GPCRs, also known as the “rhodopsin-like family”, encompass various subgroups based on their ligand specificity. These subgroups include aminergic, peptide, protein, lipid, melatonin, nucleotide, steroid, alicarboxylic acid, sensory, and olfactory [5]. The pocket formed by the 7TM bundles serves as a binding site for orthosteric ligands, while the intracellular region is responsible for coupling with downstream effector proteins such as G-proteins and arrestins. Additionally, a conserved disulfide bond between ECL2 and TM3 contributes to the structural stability of the GPCR [19]. Upon activation, a serial conserved “micro switch” motif, including CWxP, Na+ pocket, PIF, DRY and NPxxY motif, is observed to exhibit conformation arrangement in GPCR [20–22]. When sensing the agonist, the collapse of the Na+ pocket (D2.50, S3.39, N7.45 and N7.49) in the 7TM core domain (7TMD) occludes the sodium ion, triggering the movement of TM7 toward TM3. In the intracellular region, residue Y7.53 in the NPxxY motif loses contacts with residues in TM1 or H8 and forms new contacts with residues in TM3, strengthening the packing of TM3 and TM7. In addition, the interhelical salt bridge between R3.50 and D3.49 in the DRY motif was disrupted. Collectively, the conformational arrangements of these conserved motifs result in notable outward displacement of TM5 and TM6 with respect to intracellular G-protein coupling (Fig. 3a).
Fig. 3.
The structural features of Class A GPCRs. a Hallmark for Class A GPCR activation. The cytoplasmic region of TM6 moves outward during receptor activation. b The “push-pull” activation model of the glycoprotein hormone receptor subfamily. c The ECL2 region acts as a “built-in” agonist for GPR52 (PDB: 6LI2). d The N-terminus, ECL2 and ECL3 contribute to the activation of OR51E2 (PDB: 8F76)
In addition, certain class A GPCRs exhibit unique characteristics related to ligand binding and activation. A notable example is the glycoprotein hormone receptor subfamily (encompassing FSHR, LHR, and TSHR) of GPCRs [23–26]. These receptors contain a large extracellular domain (ECD) that primarily recognizes endogenous ligands, particularly glycoprotein hormones. When these hormones bind to the receptor, they trigger an upward rotation of the ECD, displaying a “push-and-pull” activation mechanism powered by glycoproteins (Fig. 3b). This dynamic alteration coincides with a significant conformational shift at the interface between the ECD and the 7TM bundles. Within this interface, particular attention is given to the p10 peptide, which acts as an intrinsic agonist. The shifting configuration of the p10 region crafts a space that allows the extracellular region of TM7 to move inward. This causes a distinct kink in the helix at positions M6.48 and D6.44. Notably, these residues are conserved across the glycoprotein hormone receptors and mirror the toggle switch residues W6.48 and F6.44 in classic GPCRs.
Most class A GPCRs are orphan receptors, since their endogenous ligands have not yet been discovered. Orphan GPCRs play important roles in physiological functions and are associated with various human diseases, such as schizophrenia, type 2 diabetes, attention deficit and hyperactivity disorder (ADHD), cognitive impairments, brain malformations, and Alzheimer’s disease. Approximately 75% of these receptors exhibit constitutive activity, and structure determination could help us to understand the mechanism of activation [27]. To date, approximately a dozen structures are available for orphan receptors. For example, the structures of GPR52, GPR21 and GPR17 reveal that the ECL2 region acts as a built-in agonist for receptor activation (Fig. 3c) [28–30]. Compared with G-protein bound A2A or β2 receptors, the cytoplasmic end of TM6 is much shorter in GPR21, which might suggest divergent G-protein coupling for orphan GPCRs [29]. Orphan GPCR research is still in its infancy, but as the number of reported receptor structures and in-depth studies, various mechanisms of constitutive activity and active conformation will continue to be revealed, new opportunities are emerging in this field.
Olfactory receptors sense odorants and can be classified into three subgroups, among which the odorant receptors (ORs) and trace amine-associated receptor (TAAR) families belong to Class A GPCRs. In addition to being discovered in olfactory sensory neurons (OSNs), they are also expressed in extranasal tissues and are involved in diverse biological processes, revealing potential therapeutic and diagnostic targets. Recently, two distinct research groups independently made structural breakthroughs in olfactory receptors that provide insight into odorant recognition and receptor activation. Aashish Manglik’s team reported the structure of Olfactory receptor 51E2 (O51E2R), a member of ORs, as well as mTAAR9 resolved by Sun’s team. The overall structures of these two receptors display similar architecture and activation hallmarks with the canonical Class A GPCRs [31]. However, a unique structural feature in which the N-terminus and ECL2 of the receptor form a conserved disulfide bond has been found in OR51E2 and mTAAR9 (Fig. 3d) [32]. This particular disulfide bond may contribute to odorant binding and play a critical role in receptor activation. In addition to structural features, the activation mechanism of OR51E2 is significantly distinct from that of canonical Class A receptors. Serial conserved “micro switch” motifs, such as CWxP and PIF, are absent in ORs. In the OR51E2 structure, the conserved FYGx6.50 motif in TM6 substituted the canonical CWxP motif, forming an extended hydrogen-bonding network between the residues of Y6.48, S3.40, R4.52 and D5.50 that leads to the outward movement of TM6 in the cytoplasmic end, similar to the arrangement of the canonical PIF motif. Furthermore, the rotation of R6.59, which is absent in Class II ORs adjacent to ECL3, triggers the activation of OR51E2 (Fig. 3d). These two studies elucidate the molecular mechanisms underlying the recognition and activation of two different groups of olfactory receptors, providing a theoretical and structural basis for more in-depth study of olfactory receptors and targeted drug development.
Ligand recognition and receptor activation of class B GPCRs
Class B GPCRs are categorized into two subfamilies: B1 secretin receptors and B2 adhesion receptors. The B1 secretin family comprises 15 members. Their endogenous ligands are peptide hormones. B1 GPCRs play a pivotal role across a spectrum of physiological processes, such as blood sugar regulation (glucagon receptor-like family) [33–37], calcium modulation (parathyroid hormone & calcitonin receptors) [38–41], adrenal hormone control (corticotropin-releasing factor receptors) [42, 43], and gastrointestinal motility and secretion (vasoactive intestinal polypeptide (VIP) & pituitary adenylate cyclase-activating peptide (PACAP) receptors) [44–46].
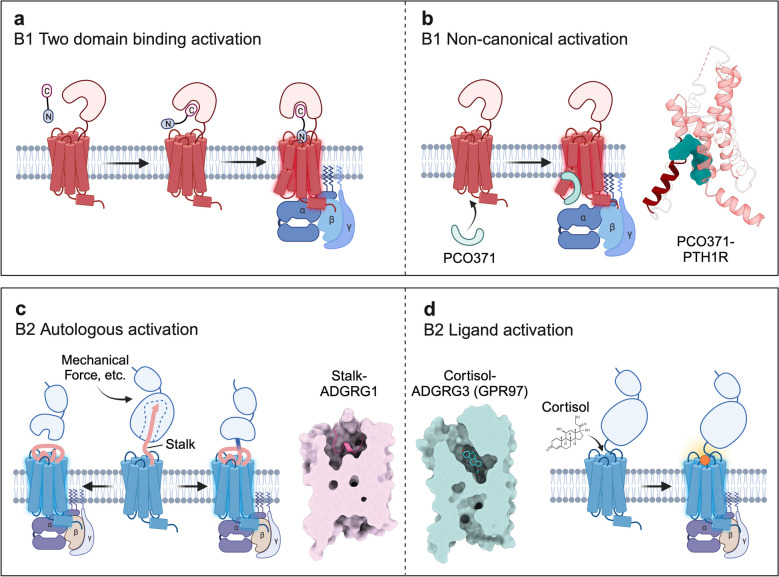
B1 receptors share a common architecture consisting of an extracellular domain (ECD) with 120-160 residues and a transmembrane domain (TMD) formed by seven helices. Both domains collaboratively engage in ligand recognition, which is acknowledged as the ‘two-domain binding model’ of the B1 GPCR activation scheme (Fig. 4a) [33, 47]. The ECD of the receptors rapidly recognizes and binds the C-terminus of the ligand, establishing the initial ligand‒receptor specificity. Subsequently, the N-terminus of the ligand penetrates the orthosteric pocket. This interaction induces receptor conformational changes, facilitating the recruitment of downstream G-proteins, and represents the rate-determining step in receptor activation [48].
Fig. 4.
Activation mechanisms of Class B GPCRs. a Scheme of the “two domain binding model” as the common activation mechanism of Class B1 receptors. b PCO371 binds to the intracellular pocket (e.g., PCO371-PTH1R-Gs complex, PDB: 8GW8) of Class B1 receptors and reveals a noncanonical activation mode. c Stalk undergoes a transition from a β-sheet to a partial α helix when mediating autologous activation of aGPCRs. Binding of the α helical stalk in the ligand pocket of ADGRG1 is from the stalk-ADGRG1-miniG13 complex (PDB: 7SF8). d Cortisol binds to the orthosteric pocket of ADGRG3 (GPR97) and triggers receptor activation as the endogenous ligand. The binding of cortisol in the ligand pocket of ADGRG3 is from the cotisol-ADGRG3-miniGo complex (PDB: 7D77)
Class B1 GPCRs undergo a series of conserved conformational changes within the 7TMD. These changes are characterized by an outward movement of the extracellular side of TM6, TM7, and ECL3, coupled with an inward movement of the extracellular side of TM1, forming a V-shaped pocket conducive for ligand binding [49]. The common motif P6.47b-××-G6.50b (superscripts refer to the Wootten numbering system for class B GPCRs) [50] induces a kink in the middle of TM6 upon activation. This facilitates the outward movement of the intracellular end of TM6, thereby opening the intracellular pocket for G-protein coupling. The B1 receptor family couples to a variety of G-protein subtypes to activate downstream signaling, with Gs being the predominant subtype [51].
A recent study identified a conserved pocket within the intracellular regions of TM2/3/6/7 of Class B1 receptors [52, 53]. The small molecule PCO371 binds to this site, prompting the intracellular side of TM6 to swing outward, thus stabilizing the receptor in an activated state conducive to G-protein coupling even in the absence of orthosteric ligands (Fig. 4b). Notably, 7 out of 15 B1 receptors can be activated by PCO371, indicating the diverse activation mechanisms inherent to the B1 family.
The B2 adhesion G protein-coupled receptor (aGPCR) family comprises 33 members. These receptors participate in fundamental physiological processes such as tissue development, reproduction, cerebrocardiovascular function, and endocrine regulation. Mutations or aberrant expression of these receptors are directly linked to diseases, including reproductive and neurodevelopmental disorders, as well as tumors [54, 55]. For instance, ADGRB3 plays a crucial role in synapses within the hippocampus and cerebellum, and its dysfunction has been proven to be associated with schizophrenia [56]. ADGRD1 (GPR133), ADGRG1 (GPR56), and ADGRG5 (GPR114) sense mechanical forces through their N-terminal extracellular domain, thereby participating in cell-cell contact and maintaining cellular homeostasis [57–59]. ADGRE1 and ADGRE5 are prominently expressed on immune cells and are involved in various immune responses, such as neutrophil migration and phagocytosis [60, 61]. Additionally, associations have been drawn between ADGRF1 (GPR110) and breast cancer progression [62], as well as ADGRL3 and attention-deficit/hyperactivity disorder [63, 64]. Mutations in ADGRV1 have been implicated in Usher syndrome type 2C, leading to deafness and blindness [65]. Thus, understanding the structural characteristics and operational modes of aGPCRs holds promise for revealing the molecular basis of biological processes, disease mechanisms, and the development of novel therapeutic strategies.
aGPCRs exhibit distinctive structural features, encompassing a multidomain N-terminal extracellular region and the GPCR autoproteolysis-inducing (GAIN) domain, which contains the conserved GPCR proteolysis site (GPS) [54, 66].
Predominantly, aGPCRs hydrolyze at the GPS site spontaneously, yielding two distinct fragments: the extracellular N-terminal fragment (NTF) and the C-terminal fragment (CTF), which contains seven transmembrane helices. These fragments, NTF and CTF, are observed to maintain a noncovalent association on the cellular surface post hydrolysis [67–69].
The sequence at the N-terminus of CTF can act as an agonist to activate the receptor and recruit downstream G-proteins. This sequence is referred to as the “Stalk” (also known as “Stachel” or “tethered agonist”) [70–74]. aGPCRs exhibit an inherent capacity for self-activation through Stalk, displaying high constitutive activity. Upon activation, the Stalk sequence undergoes a conformational shift, transitioning from a β-sheet to a partial α-helical loop, which then engages with the orthosteric pocket (Fig. 4c). The Stalk predominantly interacts with the TMD via its hydrophobic residues. Notably, it is not imperative for proteolysis to form a free Stalk sequence. Even while tethered to the GAIN domain, it possesses the capability to activate the receptor in a similar binding fashion [70, 73].
Beyond the intrinsic Stalk-mediated activation triggered by mechanical force, the ADGRG subfamily within aGPCRs demonstrates a capability for steroid recognition, including glucocorticoids, progesterone, and testosterone (Fig. 4c) [75, 76]. This suggests that aGPCRs, in addition to their self-activation mode, are also endowed with endogenous ligands that can engage directly with the 7TM core.
Despite the low sequence similarity among the aGPCR family, there remains a consistent theme in the conformational changes of the transmembrane helices upon activation. The entry of Stalk into the orthosteric pocket induces an outward deflection of the extracellular facets of TM6 and TM7. G6.50b and G7.50b are central amino acids contributing to this conformational change, with W6.53b serving as the activation toggle switch [70–73]. In contrast to autologous activation, the agonism of steroids, such as cortisol, elicits a subtler outward shift of the extracellular aspects of TM6 and TM7, anchoring the receptor in an intermediate active state (Fig. 4d). This is distinct from the Stalk sequence-mediated self-activation, which represents a fully activated state of the receptors, reflecting that the Stalk sequence acts as a full agonist [71, 77].
Architectural features of inactive and active states of class C GPCRs
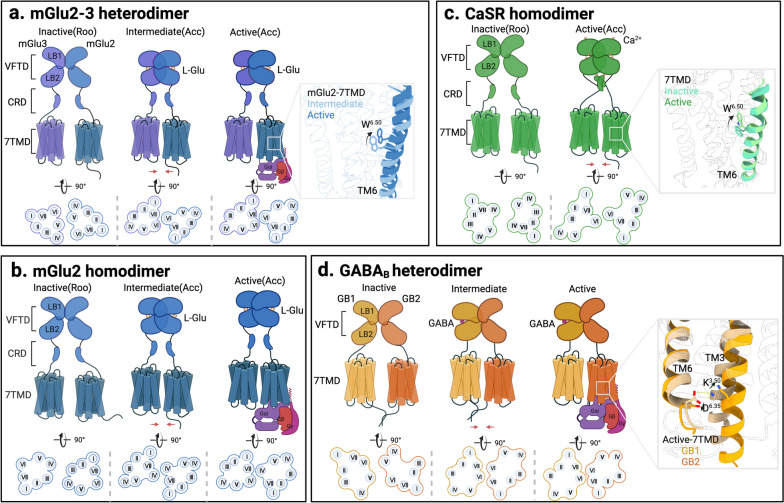
Class C GPCRs are subdivided into four groups based on their specific agonists: calcium-sensing receptor (CaSR), amino acid receptors (including γ-aminobutyric acid receptors GABAB1/GABAB2 and metabotropic glutamate receptors (mGluRs), taste receptors (Taste 1 receptors 1-3), and orphan receptors [78]. The advent of cryo-EM technology has revolutionized our understanding of Class C GPCRs. By offering near-molecular-level resolution, it lays the groundwork for comprehending their mechanisms of activation and functionality. The structural characteristics and signal transduction of CaSR, GABAB, and mGluRs have been more extensively studied, yet there remains much to uncover about Class C GPCR intricate structures and receptor dynamics.
Class C GPCRs are distinguished from other GPCRs by two structural features: large ECDs and constitutive dimerization (Fig. 5a and d) [78]. CaSR, GABAB and mGluRs are characterized by an extracellular domain (ECD) comprised of a large bilobed “clamshell” domain [78]. Their ECD contains a venus fly trap domain (VFTD) for ligand binding, which is further subdivided into upper lobe (LB1) and lower lobe (LB2) domains [78]. For GPR158, the ECD contains a cache domain (a name derived from “calcium channels and chemotaxis receptors”) for ligand binding [79]. For both CaSR and mGluRs, their ECD additionally incorporates a cysteine-rich domain (CRD) that bridges the VFTD and the 7TMD, while GABAB is structured with a VFTD directly connected to the 7TMD by a more rigid linker (Fig. 5a and d) [78].
Fig. 5.
General activation mechanisms of Class C GPCRs. Class C GPCRs are color coded as follows. The mGlu2, mGlu3, CaSR, GB1 and GB2 subunits are blue, light purple, green, yellow and orange, respectively. The endogenous agonists L-Glu, Ca2+, and GABA are indicated by colored balls; the G protein heterotrimers Gαi, Gβ, and Gγ subunits are indicated by purple, red, and fuchsia, respectively. The general conformational alterations of the class C GPCR dimer are shown above, the intracellular view of the TMD is displayed below, and the conformation of key amino acids is presented on the right. a Structures of the mGlu2-3 heterodimer in the inactivated state (PDB: 8JCV), intermediate activated state (PDB: 8JD2), and fully activated state (PDB: 8JD3). b Structures of the mGlu2 homodimer in the inactivated state (PDB: 7EPA), intermediate activated state (PDB: 7EPB), and fully activated state (PDB: 7E9G). c CaSR homodimer in the inactivated state (PDB: 7M3J) and the structure of the activated state (PDB: 7M3G). d Structures of the GABAB heterodimer in the inactivated state (PDB: 6VJM), the intermediate activated state (PDB: 6UO9), and the fully activated state (PDB: 7EB2)
CaSR, GABAB, and mGluR dimers generally share a common activation mechanism [80–84]. In their resting state, the VFTD of these receptors adopts an open conformation. VFTD dimerization is orchestrated by the LB1-LB1 interaction [85–87], while the 7TMD-level dimerization interface is versatile, e.g., the TM5/6-TM5/6 interface for CaSR [80], TM3/5-TM3/5 for GABAB [88], and TM5-TM5 interface for mGluRs [89]. Upon agonist binding to the VFTD cleft, it engages with residues within both LB1 and LB2, prompting closure of the VFTD. This structural shift enhances the LB2-LB2-mediated dimerization interface for CaSR and GABAB [85, 86] or shortens the LB2-LB2 distance for mGluRs [87], inducing rotation and convergence of the two subunits, further leading to the formation of the TM6-TM6 interface (Fig. 5a and d) [80, 82, 89]. This process activates the 7TMD, allowing one of the subunits to couple with the G protein heterotrimer, culminating in the full activation of the receptor.
CaSR stands alone as a unique member of its subfamily. Functionally, it operates as a homodimer and primarily couples with Gq. There are four binding pockets of the agonist Ca2+ on each subunit of the CaSR dimer. Beyond the dual binding sites nested within the VFTD cleft, Ca2+ shows affinity for the apical loop region of LB1 and establishes an association at the LB2-LB2 dimer interface. This binding paradigm bridges the LB2 of one subunit with the CRD of its counterpart, thereby inducing a conformational change in the CRD [90]. The CRD further relays activation signals to the 7TMD via its interplay with ECL2. The key residue of CaSR activation, W8186.50, undergoes a dramatic conformational shift, pivoting its side chain from an external to an internal orientation and gravitating toward the 7TM core (Fig. 5c). Further activation of the 7TMD for G-protein coupling is asymmetric. At the 7TM interface, TM6 of 7TMA sits higher than the opposing TM6 of 7TMB, which is tilted relative to 7TMA (Fig. 5c). It is also reflected by the same 7TM positive allosteric modulators (PAMs), either “evocalcet” or “cinacalcet”, assuming distinct poses in the two protomers. In 7TMA, the PAMs adopt an extended conformation, whereas in 7TMB, they are bent. Experiments also corroborate that the specific conformation of asymmetric 7TMD dimerization favors Gq coupling [80].
GABAB functions as a heterodimer with two subfamily members, GB1 and GB2. While GB1 is responsible for agonist binding, GB2 facilitates coupling with the G protein heterotrimer [81]. Notably, an ionic lock formed between K3.50 and D6.35 in GABAB stabilizes its inactive state [82]. The heterodimer interface between the TM5 and TM3 helices of both subunits embodies the signature of the inactive conformation of the GABAB receptor. This interaction from the TM3 and TM5 helices (H5723.55 and E6735.60 in GABAB1; H5793.55 and E6775.60 in GABAB2), defined as the “intersubunit latch”, preserves the transmembrane orientation of the dual subunits in the inactive state [88]. During activation, the agonist GABA exclusively binds to the VFTD cleft of GB1, triggering the closure of the GB1-VFTD, whereas the GB2-VFTD remains in an open conformation (Fig. 5d) [86]. However, the closure of GB1-VFTD is sufficient to induce a conformational change in GB1-7TMD and GB2, enhancing the LB2-LB2 interaction and establishing the TM6-TM6 dimerization interface (Fig. 5d) [82]. Upon activation of the 7TMD, the ionic lock between K3.50 and D6.35 in GB1 persists, whereas in GB2, this ionic lock is disrupted due to the increased distance between the intracellular ends of TM3 and TM5 upon receptor activation (Fig. 5d) [82]. Nevertheless, the movement of TM3 and TM5 creates ample space to accommodate the G-protein, enabling GB2 to couple with the Gi heterotrimer and activate downstream signal transduction [81, 82].
The mGluR family encompasses eight members, mGluR1-8, and is categorized into three distinct groups. Group I consists of mGluR1 and mGluR5, which mainly couple with Gq protein heterotrimers. Group II comprises mGluR2 and mGluR3, while Group III includes mGluR4, 6, 7, and mGluR8, both of which primarily couple with Gi heterotrimers [91]. Although mGluRs predominantly function as homodimers, functional heterodimers such as mGluR1-5, mGluR2-3, mGluR2-4, and mGluR2-7 have also been identified [92, 93]. When mGluRs bound to an agonist without G-protein coupling, their 7TMD displays a symmetric activated conformation mediated by the TM6-TM6 dimer interface (Fig. 5a and b) [84, 89, 94, 95]. W6.50 acts as the activation switch for mGluRs. Following coupling with G-protein heterotrimers, the 7TMD exhibits a fully activated asymmetric conformation mediated by the TM1/5/6/7-TM6 dimer interface (Fig. 5a and b) [83]. In this scenario, the subunit contributing TM1/5/6/7 to the dimer interface remains uncoupled from the G-protein heterotrimer, whereas the subunit contributing TM6 is coupled to the Gi protein heterotrimer (Fig. 5a and b) [83]. In the context of the heterodimer, exemplified by mGlu2-4 pairing, the agonist-bound state without G-protein coupling reveals an asymmetric dimer interface at the 7TMD, orchestrated through TM1/5/6/7-TM6 interactions [95]. This presents a distinct contrast to the mGluR homodimers devoid of G-protein coupling. However, it bears resemblance to the dimerization interface in Gi-coupled mGlu2 or mGlu4 homodimers, underscoring the absence of a stable symmetric dimerization interface during the activation of heterodimers [95]. The diverse dimeric forms of mGluRs contribute to their intricate functional and pharmacological properties, warranting further exploration in future studies.
Conformation arrangements in Class F GPCR activation
Class F GPCRs comprise one SMO and Frizzled family receptors (FZDs) [96]. SMO is primarily involved in the Hedgehog (Hh) signaling pathway and is essential in homeostasis maintenance and tissue repair [97]. The FZD family consists of 10 receptors, which can be further divided into 5 subfamilies based on sequence homology and their recognition specificity for the endogenous ligand Wnt: FZD1/2/7, FZD3/6, FZD4, FZD5/8, and FZD9/10 [98]. FZDs play a crucial role in embryonic development, stem cell regulation, and tissue homeostasis [99, 100], while their dysfunction has been implicated in various tumors, including colon [101], breast [102, 103], and ovarian cancers [104], highlighting their potential as therapeutic targets.
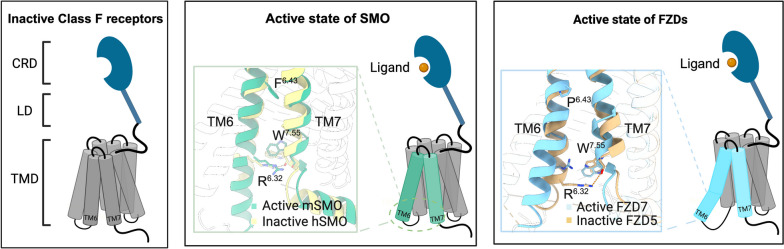
Compared to other GPCRs, Class F GPCRs have a cysteine-rich domain (CRD) at the N-terminus, which is connected to the conserved 7TM domain via a linker (Fig. 6) [96]. Notably, the CRD serves as the recognition region for endogenous ligands (cholesterol for SMO and Wnt for FZDs) and is involved in receptor activation and downstream signaling initiation [96], emerging as a hotspot for drug development [105]. The CRD contains three ligand binding sites. While the lipid-binding groove (site 1) is present in all Class F GPCRs, FZD CRDs also possess two additional ligand-binding sites (sites 2 and 3) that are absent in SMO [96].
Fig. 6.
Conformational alterations during Class F receptor activation. Class F receptors are composed of a cystine-rich domain (CRD), linker domain (LD) and transmembrane domain (TMD), and the inactive state is shown on the left. When activated by the ligand, TM6 in SMO and FZDs undergoes an outward shift, and a hydrogen bond between the conserved residues R6.32 and W7.55 in the inactive receptor is disrupted. The difference between SMO and FZDs is that the TM6 in SMO exhibits a parallel outward shift, while FZDs achieve a similar displacement of its cytoplasmic segment through a helical kink. This difference may be caused by the conserved residue P6.43 in the FZDs (as opposed to F6.43 in SMO). PDB ID of these structures: active mSMO (6O3C), inactive hSMO (5I7D); active FZD7 (7EVW), inactive FZD5 (6WW2)
Several active structures of SMO have been elucidated to date [106–109]. Insights from the complex of Gi heterotrimer-coupled human SMO (hSMO) (PDB: 6OT0) [106] and the agonist SAG21k-bound mouse SMO (mSMO) with a stabilizing nanobody NbSmo8 (PDB: 6O3C) [107] reveal a consistent receptor activation mechanism marked by analogous structural rearrangements and shared molecular switch dynamics.
In comparison to the inactive state, both structures reveal outward movement of the intracellular end of TM6 and inward movement of TM5 in the activated SMO, specifically a 7-8 Å outward shift for TM6 and a 4-5 Å inward shift for TM5 [108]. Additionally, in the active state of mSMO bound to the nanobody NbSmo8, the entire ECL3-TM6 helix shows a 3 Å displacement toward the extracellular side compared to its position in inactive mSMO [107]. The binding of cholesterol in the CRD induces an upward shift in ECL3 toward the CRD and directly relays to TM6 [107]. This provides insight into the role of the CRD in regulating SMO activation. Notably, the activated SMO structure, with its intracellular outward movement of TM6, mirrors similar structural shifts observed in Class A and B GPCRs [11, 40, 110].
The conserved residues R6.32 and W7.55 (superscript numbers refer to the Ballesteros and Weinstein numbering system) in Class F GPCRs were previously considered molecular switches for receptor activation [111], characterized by a hydrogen bond observed between R6.32 and W7.55 in the inactive conformation (Fig. 6). In contrast to the inactive state, the activation of SMO is accompanied by the disruption of polar interactions between R6.32 and W7.55, leading to a conformational rearrangement of TM6 (Fig. 6) [106, 107]. A similar mechanism is also evident in the FZD7-Gs-Nb35 complex [112].
Wnt serves as an endogenous ligand for FZD receptors, orchestrating downstream Wnt/β-catenin signaling pathways through the formation of ternary complexes with coreceptors LRP5/6 [98]. Comparative analyses of the Xenopus Wnt8 (xWnt8) and human Wnt3 (hWnt3)-bound mFZD8 CRD structures [113, 114] revealed that the receptor recognition pattern of hWnt3 is nearly identical to that of xWnt8. Wnt displays a unique dual-domain structure, resembling a “hand” with an outstretched “thumb” and “index finger”, to grasp the two distinct binding sites of FZD8-CRD. One site is dominated by the palmitoylation Ser187, extending from the Wnt “thumb” tip into the deep groove of FZD8-CRD. The second site is situated opposite site 1, where the conserved tip of Wnt’s “index finger” (residues Cys315-Cys325) forms hydrophobic contacts within a recessed region between interhelical loops on the CRD. Within site 2, the finger loop positions hydrophobic residues, including Cys315, Phe317, Trp319, a unique tandem Cys320-Cys321 disulfide bond, and Val323, to establish primary van der Waals interactions with both mainchain and nonpolar residues of FZD8-CRD. The conservation of amino acids across both interfaces appears to facilitate a mechanism that underpins ligand-receptor cross-reactivity. Furthermore, the xWnt8-mFZD8 structure reveals that Wnt and Frizzled CRD can assemble into a 2:2 complex, which potentially has impacts on downstream signaling [114].
In 2021, the constitutively active structure of the FZD7-Gs-Nb35 complex was elucidated [112]. When comparing the FZD7-Gs structure with the inactive FZD4 (PDB: 6BD4) [115] and FZD5 (PDB: 6WW2) [15], there was a notable outward curvature of TM6 and an inward displacement of TM5 on the cytoplasmic side. This conformational change mirrors that seen during SMO activation. The difference is that TM6 in the FZD7-mGs complex achieves a similar displacement of its cytoplasmic segment through a helical kink [112], while TM6 in SMO-Gi exhibits a parallel outward shift compared to its inactive state (Fig. 6) [106, 107]. The MD simulations indicate that this difference may be caused by the conserved residue P6.43 in the 10 FZDs (as opposed to F6.43 in SMO) (Fig. 6) [111, 112]. In conclusion, SMO demonstrates a tendency for a straight TM6 in both ligand binding and functional readouts, whereas FZDs exhibit a kinked TM6 upon activation due to the presence of residue P6.43 [111]. These divergent activation mechanisms may provide insights beneficial for targeting Class F receptors to design drugs with better selectivity and pharmacological attributes.
The activation mechanism of TAS2R46
Gustation is a sensory system used to prevent the intake of harmful substances and consists of five tastes: sour, sweet, bitter, salty, and umami, while the perception of bitter, sweet and umami is mediated by GPCRs [116–118]. Bitter taste recognition primarily involves TAS2Rs, which constitute a distinct class T GPCR subfamily characterized by their low sequence identity compared to other GPCRs [119–122].
TAS2Rs expressed in extraloral tissues represent potential drug targets for addressing conditions such as obesity, asthma, diabetes, and metabolic diseases [123, 124]. Previous research has revealed that TAS2Rs are expressed in enteroendocrine cells and play a pivotal role in appetite reduction. Specifically, TAS2Rs influence the release of orexigenic gut hormones and modulate intestinal movement upon detecting bitter compounds [125, 126]. Moreover, bitter agonists have been found to alleviate certain asthma symptoms by inhibiting the release of inflammatory factors in leukocytes and promoting relaxation of airway smooth muscles [127].
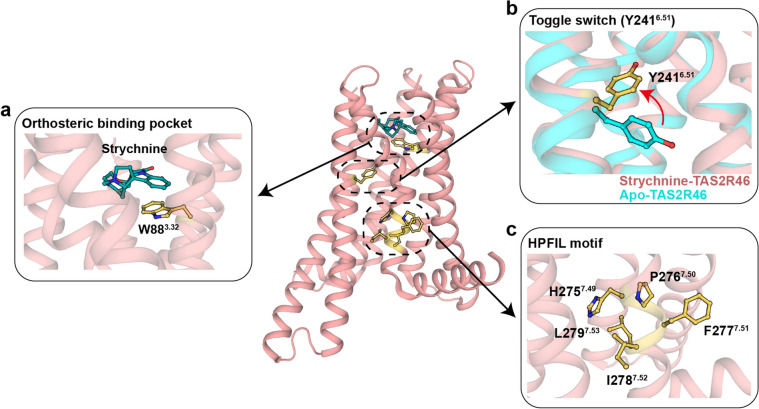
To date, only one TAS2R structure has been reported. Structural analysis of strychnine-bound TAS2R46 and its apo-form indicates that ECL2 adopts a short helical conformation, occupying the orthosteric binding pocket, similar to GPR52 (Fig. 7) [128]. However, there are notable distinctions in the activation mechanism of TAS2R46 compared to Class A GPCRs. Notably, in Class A GPCRs, the W6.48 residue within the CWxP motif serves as a “micro switch” during activation, whereas TAS2R46 features a cysteine at this position, which is not necessary for TAS2R46 activation [128]. Structural superimposition with C-X-C chemokine receptor type 2 (CXCR2), the most structurally similar to TAS2R46 in Class A GPCRs, reveals that the residue corresponding to W6.48 in TAS2R46 is Y2416.51 [128]. During TAS2R46 activation, Y2416.51 acts as a “toggle switch,“ undergoing an approximately 90° rotation, shifting from an outward orientation to pointing toward the core of the transmembrane helix (Fig. 7b) [128]. Although Y2416.51 plays a role akin to W6.48 in TAS2R46 activation, its rotation does not induce the outward movement of TM6, which distinguishes TAS2R46 from Class A GPCRs [128].
Fig. 7.
Structural features of TAS2R46. a Orthosteric binding pocket of TAS2R46 (PDB: 7XP6). The indole ring of W883.32 is horizontally parallel to the benzene ring of strychnine. b The conformational changes of the “toggle switch” Y2416.51 between apo-TAS2R46 (PDB: 7XP4) and strychnine-TAS2R46; its side chain changed from pointing outward to pointing toward the core of the transmembrane helix. c The HPFIL motif in strychnine-TAS2R46
Furthermore, TAS2R46 lacks certain conserved motifs involved in the activation mechanism of Class A GPCRs, such as the NPxxY motif and the DRY motif. Instead, TAS2R46 features the HPFIL motif, and the residues within this motif participate in a hydrophobic interaction network that mediates the packing of TM3, TM6, and TM7 (Fig. 7c). This interaction mode differs from the role of the NPxxY motif in Class A GPCRs [128].
GPCR signal transduction
GPCRs, as their name implies, couple G-proteins in the membrane when bound with agonists. The G-protein is composed of three subunits: Gα and Gβ, Gγ, forming a stable dimer. The activated GPCRs trigger the recruitment of inactive G-protein (the Gα subunit bound with GDP), leading to the exchange of GDP by GTP. Activated Gα dissociates from the Gβγ dimer and from GPCRs. G-proteins can be divided into four subgroups, Gs, Gi, G11/q and G12/13, according to the function of Gα [129, 130]. In contrast to the G-protein signaling pathways, GPCRs also trigger arrestin signaling and other noncanonical pathways. The different signaling pathways meditate distinct physiological or pathological processes.
G-protein-mediated signaling pathway of GPCRs
Typically, Gαs stimulates adenylyl cyclase (AC) and improves the second messenger cyclic adenosine monophosphate (cAMP) level in cells. In contrast, Gαi inhibits adenylyl cyclase (AC) and decreases cAMP levels [131]. Gα11/q activates phospholipase C (PLC), which catalyzes the conversion of phosphatidylinositol bisphosphate (PIP2) into diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) [132]. The released DAG activates protein kinase C (PKC), while IP3 diffuses to the endoplasmic reticulum (ER) and binds to IP3 receptors on ligand-gated calcium channels on the surface of the ER, leading to a massive release of calcium ions into the cytosol [133]. The evaluated levels of Ca2+ activate Ca2+ and calmodulin-dependent protein kinase II (CaMKII) [134]. Gα12/13 stimulates Rho GTPase replacement by second messengers [135]. Meanwhile, the dissociation of the Gβγ dimer can also regulate numerous molecules (e.g., GIRK channels, TRPM3 and CaV) [136].
G-protein subtype selectivity of GPCRs
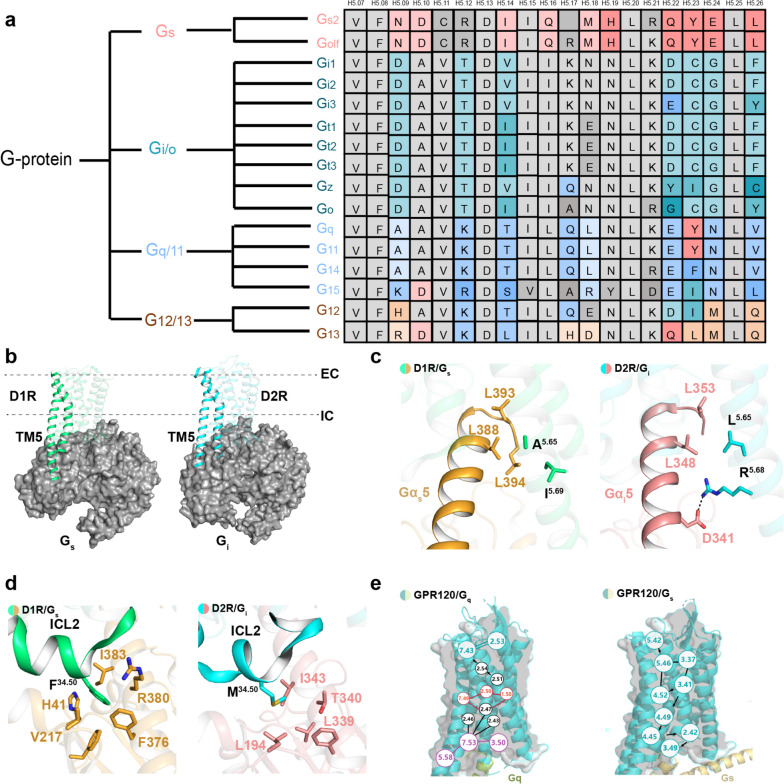
The GPCR complex structures reveal that the regions TM3, TM5-7 and ICL2 are involved in G-protein heterotrimer coupling, especially for the C-terminal α5 helix in Gα. Sequence alignment of the Gα subunit suggests that sequence diversity is observed in the C-terminus. Previous studies reported that the C-terminus, also known as “the wavy hook”, plays a critical role in G-protein selectivity (Fig. 8a) [137–139]. In the α5 helix, the residue is Y in position H5.23 for Gs and Gq and smaller residues for Gi/o and G12/13, suggesting that the α5 helix also contributes to G-protein selectivity. Additionally, previous studies imply that the residues at position 34.51 of ICL2 play a critical role in Gi and Gs selectivity.
Fig. 8.
The elements in receptors determine the G-protein subtype selectivity. a Classification and sequence alignment of the C-terminus of the Gα subunit family. b, c, d Structural comparison of Gs (left)-D1R (PDB: 7CKW) and Gi (right)-D2R (PDB: 7JVR). b TM5 of D1R is longer than D2R in the cytoplasmic region. c. The distinct motifs in TM5 contribute to G-protein subtype selectivity between D1R and D2R. d Detailed interaction between the residue at the 34.51 position of ICL2 and Gα-proteins in Gs (left)- and Gi (right)-bound dopamine receptors. e Schematic diagrams displaying the propagating pathways that contribute to G-protein subtype-biased signal transduction for GPR120
In the dopamine receptor subgroup, D1-like receptors (D1R and D5R) couple to Gs, while D2-like receptors (D2R, D3R and D4R) are recruited to Gi. The antagonistic regulation of intracellular cAMP levels by D1-like and D2-like receptors reflects their distinct roles in modulating physiological functions. Understanding the functional differences and selective recruitment of G-protein subtypes by D1-like and D2-like receptors provides a foundation for designing drugs that can modulate these receptors in a highly specific and controlled manner, potentially leading to more effective treatments for a range of neuropsychiatric and neurological disorders. Structural comparisons of D1R-Gs and D2R-Gi structures may provide insight into Gs and Gi coupling selectivity [140]. In the D1R-Gs complex, TM6 moves outward 8.4 Å more than D2R. The larger outward movement of TM6 allows it to accommodate the bulky amino acid on the α5 helix of Gs, where D2R may form a severe steric clash between TM6 and the α5 helix of Gs [140]. Additionally, compared with D2R, TM5 of D1R extends an additional two and a half helix turns on the cytoplasmic side, directly interacting with the Ras domain of Gs (Fig. 8b) [140]. This characteristic, where TM5 of the Gs-coupled receptor is longer than the Gi-coupled receptor, has been observed in structural comparisons between 5HT4/6/7R-Gs and 5HT1/4R-Gi [141].
Furthermore, sequence alignment of TM5 of the Gs-coupled receptor revealed that the A/V5.65 × 5.69 motif (x is mostly a hydrophobic residue), which is relatively conserved among D1R, D5R, and β2AR, plays an important role in Gs coupling [142, 143]. In the D1R-Gs structure, A5.65 points toward the hydrophobic pocket composed of L388G.H5.20, L394G.H5.25 and L395G.H5.26, while I5.69 forms hydrophobic interactions with the Ras domain of Gs (Fig. 8c) [142]. Notably, mutation of A5.65 into valine has a lesser impact on the activation potency of dopamine, whereas it significantly decreases with leucine substitution due to the side chain of leucine forming steric clashes with the hydrophobic pocket of Gs [142]. In contrast, the corresponding A/V5.65 × 5.69 motif is L5.65xxxR/E5.69 (R5.68 in D2R), which is also found in other Gi-coupled receptors, such as δOR, µOR, and κOR (Fig. 8c) [143]. The residue at position 5.69 is mostly a charged residue that differs from D1R. Previous studies have suggested that the C-terminus of TM5 in Gi-coupled receptors contains charged residues that play a crucial role in Gi coupling selectivity. Substitution of x5.69 in the A/V5.65 × 5.69 motif of D1R to a charged residue significantly affects the potency of dopamine [144].
Additionally, the residue at position 34.51 of ICL2 also contributes to Gs coupling selectivity [143, 145, 146]. The residue corresponding to position 34.51 in D1R is F129ICL2, which interacts with a hydrophobic pocket formed by β1 and β3 strands and the α5 helix of Gs (Fig. 7d). Replacing F129ICL2 with a small side chain, such as leucine and alanine, deeply influences Gs coupling selectivity [143]. A similar pattern can also be observed for β1AR and β2AR [147]. In contrast, the allelic residue in D2R is M140ICL2, which contacts L194G.S3.01 and I343G.H5.15 of Gi through weak hydrophobic interactions (Fig. 8d) [143].
G-protein promiscuity and biased signaling pathway of G-protein subtypes
Most GPCRs recruit a specific subtype G-protein to elicit cytoplasmic signal transduction. However, numerous GPCRs bind to diverse G-protein subfamilies, such as CCK1A [148], NTS1R [149], and GPR120. Each of the G-protein subtype signaling pathways may be correlated with distinct physiological or pathological processes. For instance, EP4, a type of prostaglandin E2 (PGE2) receptor, holds therapeutic potential for various conditions, including kidney injury (KI) and X-linked nephrogenic diabetes insipidus (NDI) [150]. Interestingly, the Gs signaling pathway of the EP4 receptor has shown beneficial effects on KI and NDI, while the Gi signaling pathway can modulate neurotransmitter release and cell migration [151]. Understanding the molecular mechanism for biased G-protein subtype signal transduction can lead to more effective and safer therapeutic interventions for GPCRs.
Recently, the team of Sun Jin-Peng revealed the properties for biased G-protein subtype signal transduction of GPR120. Structural analysis of GPR120 in complex with different G-proteins revealed that the overall architecture of the receptor is equivalent regardless of the G-protein subtype [152]. Among the 24 residues involved in mediating the recruitment of Gi and Gq, 19 residues form conserved interactions with the αN and α5 helices (αH5) of Gi and Gq, whereas 15 residues interact with Gi within the 23 residues of GPR120 in contact with Gs, potentially explaining the G-protein promiscuity of GPR120. The three subtypes of G-proteins (Gi, Gq, and Gs) possess partially distinctive features at the G-protein coupling interface. Through mutation experiments, it has been confirmed that R2405.71, S248ICL3, or D2596.30 in GPR120 is crucial for coupling with Gi but does not contribute to Gq coupling. The diversity of signaling pathways mediates distinct pathophysiological events.
The GPR120 ligands exhibit different signaling pathways and achieve functional selectivity. Structural analysis of GPR120 in complex with diverse ligands, such as the Gq-biased ligand TUG891 and unsaturated FAs, reveals different recognition models for ligands in the pocket, operating distinct propagating paths, which may underlie differential G-protein subtype coupling. Within these diverse signaling pathways, conformational locks located at TM3 and TM4 are responsible for connecting the specific π-π interactions in the GPR120 binding pocket and the structural rearrangements coupled to Gs-protein on the cytoplasmic side. Meanwhile, conformational locks in TM1-TM2 and TM7 are responsible for connecting the ligand pockets with the downstream identification and selective coupling of Gq and Gi. In the Gq signaling propagating path, TUG891 forms π-π interactions with the consecutive residues F882.53 and F3117.43 in the binding pocket, along with the cytoplasmic side hydrophobic packing between Y2275.58 and Y3217.53, enabling an outward tilt of TM7 and an inward tilt of TM1 (Fig. 8e). This allows for a tighter insertion of the α5 helix of Gq, facilitating the crucial cation-π interaction between receptor R1363.50 and Y356G.H5.23 of Gq, which is essential for Gq bias. In contrast, the bias toward Gs is closely related to the propagating path beginning at F2115.42, passing through Y1654.52- L1273.41-I1624.49-L1584.45-L772.42 and leading to a structural rearrangement of E1353.49, ultimately forming a hydrogen bond with Y391G.H5.23 of Gs (Fig. 8e).
The arrestin-mediated signaling pathway of GPCRs
In addition to G-protein coupling, GPCRs can recruit arrestin, representing another vital aspect of GPCR signaling. As one of the core regulators of GPCR signal transduction, arrestins participate in regulating GPCR desensitization, internalization, and intracellular transport. Furthermore, they also function as scaffold proteins to activate downstream effector proteins such as mitogen-activated protein kinases (MAPKs), extracellular signal-regulated kinase 1 and 2 (ERK1/2), and Src family tyrosine kinases, thereby playing a crucial role in cell cycle regulation/proliferation and cell survival/apoptosis signal transduction [153]. In addition, recent studies have also identified that the GPCR-βarr1 complex, in addition to serving as a scaffold protein, can also directly activate the protein kinases Src and C-Raf in an allosteric mode [154–156].
There are four subtypes of arrestin: among them, arrestin1 (arr1) and arrestin4 (arr4) are known as visual arrestins and are primarily distributed in the visual sensory system of animals [157]. Arr1 can combine with the light-activated receptor rhodopsin and inhibit its downstream signal transduction, and arr4 can deactivate color opsins [157]. The other two nonvisual arrestins, arrestin2 and arrestin3, are also known as β-arrestin1 (βarr1) and β-arrestin2 (βarr2) [158]. They are widely expressed in various tissues and can be recruited by phosphorylated receptors and subsequently regulate multiple (patho)physiological processes [157].
Development progression for GPCR-arrestin structure determination
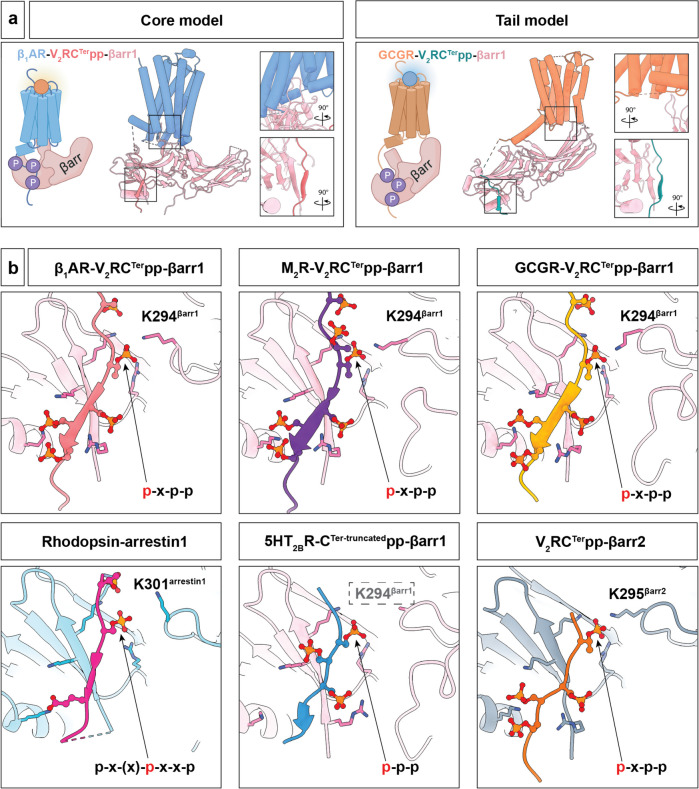
Arrestins are composed of the N-domain and the C-domain, each forming a β-stranded sandwich structure connected by a hinge region [158]. With the development of biological techniques, multiple structures of arrestin-bound receptors have been gradually elucidated. In 2013, the crystal structure of βarr1 and the phosphorylated vasopressin-2 receptor carboxyl tail (V2RC) was elucidated, providing a foundation for understanding the activation of βarr1 by phosphorylated receptor tails and the conformational changes that occur after activation (Fig. 9a) [159]. In 2015, a study utilized X-ray free electron laser (XFEL) technology to report the crystal structure of constitutively active human rhodopsin in complex with active mouse visual arrestin, contributing to the understanding of GPCR-mediated arrestin-biased signal transduction [160]. It is acknowledged that GPCRs can bind to arrestin in two ways: one is called the “core” conformation, where the C-terminus of the receptor and the core region of the receptor’s transmembrane domain bind to arrestin together; the other is the “tail” conformation (also acknowledged as the “hanging” mode), where the C-terminus of the receptor binds to arrestin independently [154]. The different conformations indicated above may lead to different receptor signaling pathways. In 2023, the cryo-EM structure of the glucagon receptor (GCGR)-βarr1 complex was elucidated [161]. Functional experiments revealed that the “tail” conformation of GCGR-βarr1 controls the recruitment of βarr1 to the cell membrane and the internalization of GCGR, which is consistent with the previous conclusion (Fig. 9a).
Fig. 9.
Activation mechanisms of GPCR-arrestin signaling. a Two common GPCR-arrestin binding conformations: “Core” mode (e.g., β1AR-V2RCTerpp-βarr1, PDB: 6TKO) and “Tail” mode (e.g., GCGR-V2RCTerpp-βarr1, PDB: 8JRU). b Arrestin activation mediated by polar interaction between the key phosphorylation of receptor C-terminus and the lysine in the lariat loop. (β1AR-V2RCTerpp-βarr1, PDB:6TKO; M2R-V2RCTerpp-βarr1, PDB:6U1N; GCGR-V2RCTerpp-βarr1, PDB:8JRU; Rhodopsin-arrestin1, PDB: 4ZWJ; 5HT2BR-CTer−truncatedpp-βarr1, PDB: 7SRS; V2RCTerpp-βarr2, PDB: 8I10)
Conformation changes of arrestin activation
The phosphorylation of serine and threonine residues in GPCRs, facilitated by GRKs, is typically a prerequisite for binding to arrestins [162, 163]. Upon forming a complex with the phosphopeptide, arrestins undergo a series of conformational transitions, leading to their activation [159, 164]. A central step in this activation is the disruption of the polar core situated in the N-domain. In the context of βarr1 as an illustrative model, the polar core in its apo state harbors a network of highly conserved ionic interactions [159]. These include D26βarr1 and R169βarr1 in the N-domain, D290βarr1 and D297βarr1 on the lariat loop within the C-domain, and R393βarr1 positioned after the C-terminal strand β20βarr1. In concert with these elements, a salt bridge between R25βarr1 and E389βarr1, coupled with hydrogen bonds uniting strands β1βarr1 and β20βarr1, reinforces the linkage between the two arrestin domains and stabilizes its quiescent conformation. Upon arrestin activation, the emergence of the phosphopeptide-arrestin complex is characterized by the substitution of arrestin’s strand β20 in its intramolecular β-sheet with β1 by the phosphopeptide [165]. This pivotal intermolecular β-strand exchange results in the release of the entire arrestin C-terminus from its core. Subsequently, conformational changes occur within the arrestin crest, particularly in the lariat, finger, and middle loops, followed by a twist between its N- and C-terminal domains [158].
While the exact mechanism by which phosphopeptide attracts arrestin and displaces the C-terminus of arrestin remains to be determined, it is evident that phosphopeptide binding serves as the initiating force for arrestin activation [166–168]. Several studies have identified unique phosphopeptide recognition motifs across different arrestin isoforms. For instance, the p-×-(×)-p-×-×-p motif was proposed from the rhodopsin-arrestin1 structural analysis [164]. Moreover, recent research has highlighted a p-×-p-p phosphorylation motif in GPCRs (Fig. 9b) [167, 168]. This motif interacts with a spatially organized K-K-R-R-K-K sequence present in the N-domains of both βarr1 and βarr2, indicating a shared activation motif of βarrs. Intriguingly, many GPCRs incorporate the p-×-p-p motif, located either in their C-terminus or ICL3, underscoring its widespread role in facilitating activation [167, 169]. In detail, the initial phosphoresidue of the p-×-p-p motif disrupts existing salt bridges between R169βarr1 (R170βarr2) and D290βarr1/D297βarr1 (D291βarr2/D298βarr2) and establishes new intermolecular salt bridges with R25βarr1 (R26βarr2), K11βarr1 (K12βarr2), and K294βarr1 (K295βarr2), effectively engaging the lariat loop. The interaction between the first phosphoresidue and K294βarr1 (K295βarr2) represents a primary driving force, drawing the lariat loop closer to the N-domain and inducing a conformational twist in the C-domain. This observation aligns with findings from the recently characterized GCGR(V2RC)-βarrestin1 complex, suggesting a general mechanism of phosphorylation-driven arrestin activation (Fig. 9b) [161]. Notably, in the ACKR3pp-βarr2 complex, while ACKR3pp extends toward the finger loop with its adoption of a p-×-×-p-×-×-p motif rather than p-×-p-p, its pT342 aligns with the initial phosphoresidue in the p-×-p-p motif, hinting at a consistent mode of arrestin activation [170].
Beyond the central role of the 1st phosphoresidue in the motif, other phosphoresidues also significantly influence arrestin signaling. Specifically, the terminal residue of the p-×-p-p motif appears essential for βarr1 recruitment of CCR5, as evidenced by the notable reduction in efficacy when CCR5T343A is introduced in functional assays [168]. Additional phosphorylation sites are postulated to enhance the affinity of GPCR-arrestin interactions and modulate subsequent signaling activities, in line with previous suggestions. It is noteworthy that CCR5pp with only 3 phosphorylated sites of the p-×-p-p motif prompts a similar βarr1-pp-Fab30 population as observed with V2Rpp and the fully phosphorylated CCR5pp [168]. This suggests that the p-×-p-p motif alone is adequate to form a stable and active βarr1-pp-Fab30 complex. Furthermore, it hints at an evolutionary adaptation wherein an increased number of phosphorylation sites can either enhance the GPCR-arrestin complex affinity or act as redundancy in phosphorylation regulation. It is also worth noting that multiple GPCRs exhibit several instances of the p-×-p-p motif in their C-terminus and ICL3, similar to the p-×-(×)-p-×-×-p patterns as previously described [167]. Some receptors also present with an extended ICL3 but have a short or absent C-terminus [157, 171, 172]. The variance in the lengths of the C-terminal tail and ICL3, combined with the density of phosphorylation sites, highlights the inherent structural diversity within the GPCR-arrestin system, which orchestrates their functional versatility (Fig. 9b).
Another GPCR-arrestin binding interface is between the receptor core and the arrestin central crest loops, which represents the “core” mode [157, 160, 173]. In contrast, this interaction is absent in the “tail” mode of GPCR-arrestin engagement. Among the known structures, only the GCGR(V2RC)-βarr1 complex represents the “tail” conformation [161]. The interactions stem from the C-terminus of GCGR, including both helix VIII and the V2R tail, leaving its intracellular pocket vacant. In contrast, previously elucidated GPCR-arrestin complexes predominantly display a core conformation wherein the finger loop of arrestins penetrates into the intracellular pocket of the receptor’s helical bundle [157, 160, 164, 171–175]. In the GCGR(V2RC)-βarr1 complex, the central crest loops of βarr1, including the finger loop, extensively engage with helix VIII of the GCGR. Notably, while the finger loop does not directly interact with the receptor, removing its entire turn region (residues 64-77) of βarr1 significantly diminishes its recruitment. This may result from an altered conformation within the βarr1 central crest or potentially disrupt an alternative arrestin-binding mode, such as the “core” configuration (Fig. 9b) [161, 176].
Investigations on β2AR, V2R, and GCGR indicate that when βarr1 adopts a tail conformation, it predominantly participates in cellular trafficking [177, 178]. Conversely, the desensitization of G-protein activation is uniquely orchestrated by the arrestins engaged with the receptor core. This engagement is indispensable, as spatial hindrance is needed for effectively inhibiting G-protein coupling. In addition, the “tail” conformation plays a pivotal role in maintaining sustained signaling within endosomes, such as the continuous production of second messenger molecules. This is further supported by the demonstrated existence of the Gs-GCGR-βarr1 megaplex in signaling assays [161]. Such revelations highlight the intricate mechanisms through which arrestin modulates receptor functionality.
The barcode hypothesis for arrestin-mediated signal regulation
Previous studies have already found that phosphorylation of GPCRs in different patterns can lead to different arrestin-mediated signaling effects, referred to as the “barcode hypothesis” [179]. In 2020, molecular dynamics (MD) simulations and site-directed spectroscopy were applied to investigate the impact of GPCR phosphorylation patterns on arrestin binding and conformation [180]. The authors found that phosphopeptides with the same number of phosphates can activate arrestin to diverse degrees. Furthermore, the affinity of phosphopeptides to βarr1 depends on the spatial arrangement of phosphorylated residues rather than their quantity. For example, when phosphorylation occurs solely at S350βarr1 or T360βarr1, the addition of phosphate at S357βarr1 significantly reduces arrestin activity. Similarly, when phosphorylation occurs exclusively at S350βarr1, the addition of phosphate at S362βarr1 significantly decreases binding stability. In addition, the results presented by the site-directed spectroscopy method also indicate that different arrestin structural domains can independently alter conformation. GPCR phosphorylation can affect the conformation of certain arrestin structures without affecting other domains, suggesting that specific conformational changes in arrestin can be induced by specific phosphorylation patterns, thereby exposing certain downstream signal protein binding sites and thus affecting specific downstream effects. Together, these studies reveal the structural basis of the “barcode hypothesis” and highlight its significance in the design of functionally selective GPCR-targeting drugs.
In addition, in the study of the phosphorylation-encoding mechanism of GPCRs, researchers have innovatively proposed the “flute model” theory for receptor phosphorylation [181]. In this article, different phosphorylation barcodes induce distinct structural rearrangements in βarr1, potentially imparting different functions to βarr1 through clathrin, SRC, ERK, or other downstream effector proteins. In 2021, they further analyzed the crystal structure of complexes formed by V2RC with four different phosphorylation patterns and βarr1 [182]. This revealed that a single phosphorylation site defect in GPCR can lead to distinct conformational changes in the distant functional domain of arrestin. Moreover, mutations at different phosphorylation sites in V2RC can result in varying degrees of impact on arrestin recruitment for MEK and c-Raf-1. This study not only reveals the regulatory mechanism of a single phosphorylation site on arrestin function but also discovers the sequential principles in the phosphorylation encoding process, where the binding of phosphorylation sites at certain positions determines whether other positions can bind.
Moreover, studies have also observed a similar degree of desensitization in both wild-type (WT) and GRK (-) (all sites that can be phosphorylated by GRK are mutated) dopamine D2R, suggesting that phosphorylation is not necessary for the arrestin-D2R interaction [183]. However, when detecting the recycling of D2R to the cell surface following agonist-induced endocytosis, the GRK (-) receptor exhibited less recycling than the WT receptor, indicating that phosphorylation can facilitate receptor recycling. While arrestins may not necessarily rely on phosphorylation to exert their functions, phosphorylation remains one of the primary regulatory mechanisms.
Noncanonical GPCR signal transduction
In addition to the G-protein and arrestin pathways, noncanonical GPCR signaling is also involved in various physiological processes, often in a cell type-specific or context-dependent manner. These pathways are particularly essential in cell proliferation/survival, neurotransmission, immune function, and metabolic regulation. Despite the growing appreciation of the importance of noncanonical GPCR signaling, much remains to be learned about the specific roles of these pathways in physiology and disease. The recent determination of the GPR158-RGS7-Gβ5 complex has provided valuable insights into the operation of noncanonical signaling pathways by GPCRs. GPR158 mainly functions as a homodimer and is distinguished by noncanonical signal transduction pathways [79, 184], which employ G-protein-independent modes. In its ligand-free state, the dimer interface is formed by the apical portion of the cache domain, the extracellular ends of TM4-5 and ECL2, along with the intracellular end of TM3 and ICL2 [185]. This consortium asymmetrically couples with the RGS7-Gβ5 heterodimeric complex [185]. Intriguingly, GPR158 does not directly interact with Gβ5 but establishes direct interactions with RGS7 at two distinct sites [185]. The first interface is formed between the C-terminal coiled-coil configuration (CT-CC) of GPR158 and the DEP-DHEX domain of RGS7, facilitated by amphiphilic interactions. The secondary interface is characterized by the engagement of the intracellular facet of one GPR158 subunit, specifically TM3, TM5, and ICL3, with the DHEX domain of RGS7. This interface overlaps with the interaction domains commonly seen in GPCR-G-protein and β-arrestin complexes [185]. Thus, recruitment of RGS7-Gb5 would preclude GPR158 from interacting with the G-protein, supporting a lack of G-protein activation [185].
In 2023, GPR158 was identified as a metabotropic glycine receptor (mGlyR) [79]. Glycine, when bound to the cache domain, acts as an antagonist to the GPR158-RGS7-Gb5 complex. RGS7-Gb5 is a selective guanosine triphosphatase (GTPase)-activating protein (GAP) for Gi/o proteins [79]. Glycine specifically inhibited the GAP activity of RGS7-Gb5 by engaging GPR158 [79]. This, in turn, suppresses the inactivation of Gαo, leading to a subsequent reduction in the secondary messenger cAMP, further eliciting a cellular response and regulating neuronal excitability [79]. These findings highlight that GPR158 is no longer an “orphaned” receptor, with implications for targeted drug research and development from its endogenous ligand “glycine”.
Unveiling the functional and structural characteristics of GRK
G protein-coupled receptor kinases (GRKs) are involved in phosphorylation-dependent or phosphorylation-independent regulation of GPCRs [162, 163, 186], which plays a key role in physiological and pathophysiological processes such as cardiovascular biology, neurodegeneration, and immune response [187, 188]. For instance, overexpression of GRK2 and GRK5 in vivo decreases adrenergic receptor-induced myocardial contractility and cardiac output, whereas inhibition of GRK2, GRK3, and GRK5 counteracts this effect [187, 188]. Phosphorylation of the schizophrenia-associated D3 receptor by GRK2 disrupts the interaction between the receptor and filamin A [188]. High expression of GRK2 and GRK5 in sepsis induces phosphorylation of chemotactic receptors such as CXCR1, thereby inhibiting neutrophil migration [188]. GRK3 inhibits breast cancer metastasis by regulating CXCR4 signaling [187].
Subtype selectivity in GRK
The seven GRKs are grouped into the rhodopsin kinase subfamily (GRK1 and GRK7), the β-adrenergic receptor kinase subfamily (GRK2 and GRK3), and the GRK4 subfamily (GRK4, GRK5 and GRK6) [162, 163]. While all GRKs possess conserved sequence characteristics and structural alignments, they employ distinct mechanisms to regulate GPCRs [162, 163, 189]. GRK1/7 and GRK2/3 are activated only by binding to active GPCRs, whereas GRK5/6 also phosphorylates inactive GPCRs [189, 190]. For instance, GRK2 phosphorylates only the C-terminal Ser residue of Neurotensin Receptor 1 (NTSR1) and is agonist dependent, whereas GRK5 phosphorylates NTSR1 intracellular loop 3 and the C-terminal Ser and Thr residues in an activation-independent manner [190]. The order of GRK-mediated GPCR phosphorylation can be barcoded (receptors responding to a specific agonist are phosphorylated at different sites by different GRKs, creating a “barcode”), sequential (a larger number of serine/threonine residues are phosphorylated first), or hierarchical (specific sequences of serines and threonines are preferentially targeted) [187, 188]. Notably, AT1R recruitment of β-arrestin for Ang II binding relies on both GRK2/3 and GRK5/6. However, binding to the β-arrestin-biased ligand TRV027 solely depends on GRK5/6 [191].
Advancements in the structural understanding of GPCR-GRK complexes
To date, the structures of rhodopsin-GRK1 and NTSR1-GRK2 have been determined, which revealed that the mode of interaction between GRK and GPCR depends on the activation states of GPCR and GRK [162, 163]. Considering GRK2-NTSR1 as an example, compared to the inactive NTSR1 structure, the cytoplasmic ends of TM5, TM6, and TM7 of NTSR1 are shifted by 4.5 Å, 11.3 Å, and 1.5 Å, respectively, and ICL2 adopts an α-helical structure consistent with an active conformation. The GRK2 structure from the NTSR1 complex, compared with the inactive state, contains an N-terminal helix that is packed onto the kinase domain, has a break in the ionic lock between its RHD from the kinase domain, and adopts a closed conformation in its kinase domain that is in the active state. The GRK2-NTSR1 complex has a major interface consisting of the N-terminal helix of GRK2 that inserts into the open TM6 pocket in a manner that overlaps with the finger loop of arrestin and a minor interface consisting of ICL2 of NTSR1 that interacts with the loop between the N-terminal helix and the RHD [163]. However, it is not ICL2 of rhodopsin but ICL1 and ICL3 that interact with GRK1 in the rhodopsin-GRK1 complex [162]. According to the structure of the GPCR-GRK complex, the extended loop of the intracellular third loop (ICL3) or the extended C-terminal tail of the GPCR reaches the active cleft of GRK, allowing GRK to phosphorylate it [163]. The phosphorylation of the GPCR further facilitates the recruitment of arrestin, which prevents G-protein binding and desensitizes G-protein signaling [162, 163].
Although all GRKs have regulator of G-protein signaling homology (RH) domains, it appears that only the RH structural domain of GRK2/3 binds Gαq, whereas the RH structural domains of the other GRKs do not appear to be able to interact with any of the G-proteins because they lack key binding residues [189]. Furthermore, the GRK2/3 subfamily contains a pleckstrin homology (PH) domain that facilitates recruitment to the membrane via interaction with Gβγ subunits [186]. GRK2/3 achieves phosphorylation-independent regulation of GPCRs by binding Gαq and Gβγ, sequestering downstream effectors [163, 189]. For example, GRK2 rapidly and transiently recruits arrestin and induces desensitization without initiating endocytosis by inhibiting Gq coupling when µOR and δOR are unphosphorylated [163]. GRK2, by binding Gβγ, inhibits the Gβγ signaling process of adenosine A1, µ-opioid receptor or κ-opioid receptor [189].
GRKs may also facilitate biased signaling through their key role in arrestin recruitment [163, 186]. The allosteric modulator SBI-553 of NTSR1 promotes the binding of GRK2, reshaping the interface in a manner that is compatible with β-arr2 binding but conflicts with Gαq protein binding [163]. D2R may also directly recruit GRK2 to mediate biased signaling of the arrestin-biased agonist UNC9994 [186]. GRK2 activity was needed for receptor phosphorylation and arrestin recruitment in an arrestin-biased D2R mutant unable to bind G-protein [186]. A G-protein-biased D2R mutant deficient in arrestin recruitment also exhibited reduced GRK2 recruitment [186]. The G-protein-biased β2AR mutant Y129A is unable to recruit arrestin due to a lack of phosphorylation by GRKs [186]. In addition, G-protein bias is induced by mutation of M3AChR phosphorylation sites [186]. These findings suggest that GRK-mediated phosphorylation may serve as an intervening target to regulate biased signaling in GPCRs.
GPCR drug discovery
As an important therapeutic target, the low subtype specificity and significant toxic side effects of GPCR ligands limit their therapeutic potential. Key constraints in GPCR drug development include limited drug selectivity, imprecise modulation of receptor signaling pathways, and issues related to tolerance and desensitization.
Development of GPCR-selective drugs
The pursuit of selective GPCR drugs has garnered significant attention due to its numerous advantages:
Enhanced therapeutic efficacy
Selective drugs can precisely target specific GPCR subtypes or signaling pathways, finely modulate GPCR function and effectively intervene in relevant physiological functions or pathological processes.
Reduced adverse effects
Drugs that act on multiple GPCRs may induce a range of off-target adverse effects. Selective drugs minimize their impact on other subtype GPCRs, thereby preventing such issues.
Enhanced drug safety and predictability
Selective drugs serve as valuable tools in early drug development stages, facilitating the study of GPCR physiological and pathological functions, as well as the understanding of distinct signaling pathways among GPCR subgroups. These insights can inform assessments of pharmacological mechanisms and pharmacokinetic properties, thereby enhancing drug safety and predictability.
The dynamic and plastic properties of the ligand binding pocket of GPCRs present opportunities for identifying new druggable sites, known as extended binding pockets (EBPs). In contrast to the orthosteric binding pocket (OBP), the EBP plays a pivotal role in determining ligand selectivity. In the context of research focused on developing selective drugs targeting the dopamine receptor subfamily, the EBPs for these receptors exhibit distinct characteristics.
The selective agonists SKF83959 and PW0464, which exhibit high affinity for D1R, occupy the EBP composed of extracellular portions of TM2-3 and TM6-7 [143]. In contrast, the D2-like subfamily shares a similar position for the EBP comprised of TM2-3 and ECL1-2 [192–195]. However, despite the common location, the shapes and sizes of these EBPs are distinctive among different receptors within the D2-like subfamily. The unique distinctions in their extracellular binding pocket (EBP) regions enable the selective targeting of each receptor using distinct ligands with specific properties. This selectivity in targeting can have significant implications for drug development and therapeutic interventions related to these receptors.
When designing subtype-selective ligands, it is essential to pay attention to the specific pathway through which the drug enters the receptor, in addition to considering the EBP. For instance, βARs play a crucial role in mediating physiological responses to catecholamines, such as epinephrine and norepinephrine, to regulate cardiovascular, respiratory and metabolic functions. Epinephrine displays equal affinity and occupies an identical orthosteric binding pocket for both β1AR and β2AR. Norepinephrine is slightly smaller than epinephrine, and its binding pockets in β1AR and β2AR are expected to be nearly identical [196]. However, norepinephrine exhibits significantly higher affinity for β1AR than β2AR.
This significant difference in affinity for norepinephrine between β1AR and β2AR can be attributed to their distinct binding kinetics. The binding rate constant (Kon) of norepinephrine to β1AR is approximately 22-fold higher than that to β2AR [196]. On the other hand, the dissociation rate constant (Koff) is approximately 1.5-fold higher for β1AR than β2AR. Consequently, the dissociation constant (Kd), which is a measure of binding affinity (Kd = Koff/Kon), is lower for norepinephrine to β1AR, suggesting stronger binding affinity [196]. Further analysis revealed that norepinephrine accesses the orthosteric binding pockets of β1AR and β2AR through distinct binding pathways [196]. Key amino acids within these pathways play a pivotal role in influencing the binding rate and affinity of norepinephrine, thereby determining its selectivity. In contrast, although epinephrine also follows different binding pathways to interact with β1AR and β2AR, it contains an additional methyl group in its chemical structure relative to norepinephrine [196]. This methyl group introduces electrostatic property differences, rendering epinephrine nonselective for both receptors [196]. These findings may not only contribute to the design of subtype-selective drugs for βARs but also for other GPCRs.
The development of biased drugs for GPCRs
In addition to selective ligands, biased ligands are a promising direction in drug design for GPCRs. Biased signaling, also known as functional selectivity or biased agonism, is a phenomenon in which a ligand selectively activates either the G-protein or the β-arrestin pathway of a GPCR, leading to distinct functional outcomes. This concept offers tremendous potential for drug innovation, as it allows for the design of ligands that specifically target one signaling pathway while avoiding the activation of the other. This precision has the potential to reduce undesirable side effects and enhance the desired therapeutic effects. Many GPCRs have exhibited biased signaling, opening the door to the development of drugs that leverage this characteristic to create more targeted and potent therapeutics.
Several mechanisms have been identified that facilitate biased signaling. Investigation of C5aR1 with the G-protein-biased agonist BM213 has pinpointed the binding pocket and “IWI” motif formed by TM2-ECL1-TM3 as playing an essential role in biased signaling, providing insights into the design of C5aR1-biased molecules [197]. The guanidine of the G-protein-biased bitopic ligand C5 (or C6) guano interacts with the key Asp2.50 in the sodium ion-binding pocket of µOR, which can reduce or even abolish β-arrestin-1 and β-arrestin-2 recruitment compared with the parent ligand fentanyl (Fig. 10) [7].
Fig. 10.
Structural basis of µOR G-protein biased agonism. The interactions between the balanced agonist fentanyl (PDB: 8EF5) or the G-protein-biased ligand C5 guano (PDB: 7U2L) and the involved residues in the µOR orthosteric pocket. The guanidine of C5 guano interacts with the key D2.50 in the Na+ binding pocket. Salt-bridge interactions and cation-π interactions are represented by black and red dashed lines, respectively. Schematic diagrams of cAMP inhibition and β-arrestin-2 recruitment on µOR with balanced or G-protein biased ligands are displayed compared with the reference ligand
In 2020, oliceridine, which is a G-protein-biased agonist for the µ-opioid receptor (µOR), was approved for the management of acute severe pain in adults where alternative treatments fall short. By specifically triggering the Gi signaling pathway, oliceridine aims to achieve effective pain relief while minimizing typical side effects, such as respiratory depression and constipation, which are often linked to the activation of the β-arrestin pathway [198]. Additional G-protein-biased µOR agonists, such as PZM21, have demonstrated better analgesic effects and reduced side effects in preclinical pain studies [199]. In addition, several β-arrestin-biased drugs have been identified and studied. Carvedilol, a β-arrestin-biased agonist of β1AR and β2AR, has been shown to counteract the harmful effects of G-protein-mediated catecholamines while promoting β-arrestin-driven cell survival pathways. It is currently further approved for the management of heart failure [200]. Another example, TRV120027, is a potent β-arrestin-biased ligand for AT1R. It holds the potential to enhance cardiac contractility through β-arrestin pathways and maintain cardiac stroke volume, making it a candidate for acute heart failure treatment [201]. However, while these effects were observed in mouse studies, human clinical trials have yet to meet the set clinical endpoints.
Beyond ligand-induced biased signaling, certain receptors also intrinsically exhibit bias during their evolutionary development. For instance, C5aR2 lacks functional G-protein coupling but demonstrates robust β-arrestin recruitment [202]. Sequence alignment indicates that the residues within the intracellular pocket of C5aR2, which are associated with G-protein coupling, are alanine, differing from typical class A GPCRs with G-protein signaling. This distinction might impede G-protein binding, partially explaining why C5aR2 predominantly mediates β-arrestin signaling.
The current therapeutic landscape boasts a range of biased ligands tailored for specific disease treatments. However, certain receptors, such as GPR120, present an intricate picture. GPR120 can engage multiple G-protein subtypes (Gq/Gi/Gs) and β-arrestin1/2, thereby orchestrating a variety of downstream signaling pathways [152]. Thus, the focus should shift toward creating ligands that show a distinct preference for specific G-protein or β-arrestin subtypes. Such innovations could lead to more nuanced control of signaling pathways, offering potential breakthroughs in precision medicine and novel therapeutic strategies. To achieve this, additional structures of biased ligand-GPCR complexes are needed, which will shed light on the mechanisms of biased signaling and offer a refined molecular foundation for drug design.
Allosteric pharmacology of GPCR and drug discovery
The majority of FDA-approved GPCR drugs on the market target the orthosteric site, which is the same site as the endogenous ligand binding pocket [203]. Although the success of drugs targeting GPCRs has been proven, it is still difficult to design selective ligands or drug candidates for individual GPCRs owing to the high conservation of orthosteric sites among subgroups. Thus, the development of new approaches/strategies for discovering therapeutic agents is imperative. Allosteric modulators, which bind to distinct sites from the orthosteric pocket, have emerged as a promising direction in GPCR drug discovery. They offer unique properties and the potential for increased selectivity. To date, 31 allosteric drugs have entered clinical phases, with six of them receiving FDA approval (Table 1). For instance, Avacopan, a NAM for C5aR, was approved to treat ANCA-associated vasculitis, which is an autoimmune disease, by inhibiting the binding of C5a, thus reducing the inflammatory response and improving autoimmune disease symptoms [204]. Moreover, cinacalcet, which is primarily used to treat hypercalcemia by regulating the secretion of parathyroid hormone to reduce the level of blood calcium, is a PAM for CaSR [205–207].
Table 1.
The allosteric drugs for GPCRs
| Drugs | Target | Action | Phase | Indications | References |
|---|---|---|---|---|---|
| Avacopan | C5aR1 | NAM | Approved | Anti-neutrophil cytoplasmic autoantibodies-associated vasculitis | [208] |
| Cinacalcet | CaSR | PAM | Approved | Hyperparathyroidism and calciphylaxis | [205–207] |
| Ticagrelor | P2Y12 | NAM | Approved | Prevention of thrombosis | [209] |
| Ivermectin | GABAB | PAM | Approved | Parasitic roundworm infections | [210] |
| ATx-201 | NPY4 | PAM | Approved | Viral and bacterial infections;Atopic dermatitis; Cancer;Rheumatoid arthritis; | [211] |
| Maraviroc | CCR5 | NAM | Phase III | HIV infection | [212] |
| Vercirnon | CCR9 | NAM | Phase III | Inflammatory bowel disease | [213] |
| BMS-986,165 | mGluR4 | Unclear | Phase III | Plaque psoriasis; Psoriatic arthritis;Crohn’s disease; Systemic lupus erythematosus | Allosteric database |
| Mavoglurant | mGluR5 | NAM | Phase III | Fragile X syndrome | [214] |
| ADX-48,621 | mGluR5 | NAM | Phase III | Parkinson’s disease levodopainduced dyskinesia | [215] |
| Basimglurant | mGluR5 | NAM | Phase III | Fragile X syndrome | [216] |
| ASP-4345 | D1R | PAM | Phase II | Schizophrenia; Cognitive disorders | [217] |
| LY-315,402 | D1R | PAM | Phase II | Dementia; Parkinson | [218] |
| ADX-10,059 | mGluR5 | NAM | Phase II | Reflux, Gastroesophageal migraines | [219] |
| DT-1687 | mGluR4 | PAM | Phase II | Parkinson’s disease | Allosteric database |
| AZD-8529 | mGluR2 | PAM | Phase II | Smoking cessation therapy; Schizophrenia | [220] |
| ADX-71,149 | mGluR2 | PAM | Phase II | Epilepsy; Anxiety disorder; Schizophrenia | [83, 221] |
| MK-7622 | M1 | PAM | Phase II | Pain; Schizophrenia; Sleep disorder; Dementia, Alzheimer’s type | [222–224] |
| ASP-8302 | M3 | PAM | Phase II | Detrusor underactivity (Underactive bladder) | [225] |
| Emraclidine | M4 | PAM | Phase II | Schizophrenia | [226] |
| T-62 | A1AR | PAM | Phase II | Neuropathic pain; Postherpetic neuralgia (PHN) | [227] |
| HTL-14,242 | mGluR5 | NAM | Phase I | Neurological disorders; Psychiatric disorders | [228] |
| RG-7342 | mGluR5 | PAM | Phase I | Schizophrenia | Allosteric database |
| RGH-618 | mGluR5 | NAM | Phase I | Anxiety disorder | Allosteric database |
| (11 C) JNJ-42,491,293 | mGluR2 | PAM | Phase I | Diagnostics | [229] |
| JNJ-55,375,515 | mGluR2 | NAM | Phase I | Cognitive disorders; Psychosis | Allosteric database |
| TAK-071 | M1 | PAM | Phase I | Lewy body dementia; Neurological Disorders; Dementia;Alzheimer’s type | [230] |
| VU-319 | M1 | PAM | Phase I | Pain; Sleep disorder;Dementia; Alzheimer’s type | Allosteric database |
| (11 C) MK-6884 | M4 | PAM | Phase I | Dementia, Alzheimer’s type | [231] |
| ODM-106 | GABAB | PAM | Phase I | Essential tremor | Allosteric database |
| JNJ-2463 | CB1 | NAM | Phase I | Nonalcoholic steatohepatitis; Nephropathy, diabetic; Nonalcoholic fatty liver disease (NAFLD); Fibrosis; Metabolic Diseases | [232] |
Allosteric modulators offer a valuable alternative in GPCR drug development, allowing for the development of more selective and precise therapeutic agents, which can enhance treatment efficacy while minimizing off-target effects.
Classification of GPCR allosteric modulators
GPCRs have multiple functionally distinct conformation states that display varying affinities for orthosteric ligands and different abilities to engage effectors: G-proteins, arrestins and other signaling molecules. Allosteric modulators stabilize the receptor in a specific conformational state and thus modulate the potency of signal transduction [233, 234]. According to the effect on orthosteric ligand, allosteric ligands can be classified into three groups: positive allosteric modulator (PAM), negative allosteric modulator (NAM) and neutral allosteric ligand (NAL) [235, 236]. PAMs and NAMs can modulate the affinity of orthosteric ligands or affect the intrinsic efficacy of orthosteric agonists (OAs). PAMs enhance signaling by increasing the affinity or efficiency of OAs (Fig. 11a), while NAMs do the opposite (Fig. 11b). NAL can bind to the allosteric site but has no effect on signaling (Fig. 11c).
Fig. 11.
Effects of allosteric modulators on receptor signaling. PAMs (a) and NAMs (b) modulate the effect of orthosteric agonists, while NALs (c) have no influence on receptor signaling mediated by orthosteric agonists. BAMs (d) potentiate signaling pathway a while inhibiting signaling pathway b of the receptor. The dose-dependence curves (below) display the effect of orthosteric agonists in the presence of allosteric modulations. The color saturation indicates the concentrations of allosteric modulators. OA: orthosteric agonist. e The binding sites of BAMs in receptors. SBI-553 (blue) binds to the pocket located at an intracellular region composed of TM6-7 and H8 of NTSR1 (PDB: 8FN0), while compound 9n sits in the upper portion between TM5-6 and ECL2 of HCAR2 (PDB: 8JII)
The diverse effects of allosteric modulators on receptor signal transduction can be described and quantified using the allosteric ternary-complex model (ATCM) [237–240]. This model takes into account the interactions among the receptor, orthosteric agonist (OA), allosteric modulator, and effector protein to determine the direction and magnitude of allosterism. The model employs cooperativity factors to govern these interactions.
In the operational model of ATCM:
The modulation of binding affinity is regulated by the cooperativity factor (α). When α > 1, it indicates positive cooperativity, enhancing the affinity of the allosteric agonist (OA) for the receptor. When 0 < α < 1, it denotes negative cooperativity, reducing the affinity of OA for the receptor. A value of α = 1 represents neutral cooperativity, indicating no effect on the affinity of OA for the receptor.
The modulation of OA signaling efficacy is controlled by the cooperativity factor (β). A value of β > 1 signifies positive cooperativity, enhancing the signaling effect of OA. When 0 < β < 1, it indicates negative cooperativity, diminishing OA’s signaling effect. A value of β = 1 signifies no effect on OA signaling.
These cooperativity factors, α and β, provide a quantitative framework for understanding how allosteric modulators influence receptor binding and signaling in the allosteric ternary complex model.
Advantages of allosteric pharmacology
The complexity and unique characteristics of allosteric modulation provide the predicted advantages over orthosteric ligands for GPCR drug discovery.
Selectivity
One of the primary motivations behind focusing on allosteric compounds in GPCR drug discovery is the hope of developing subtype-selective ligands for specific receptor groups. This is particularly relevant when traditional orthosteric strategies face challenges due to the high similarity in the orthosteric pocket among receptor subtypes. For instance, the high conservation of the orthosteric pocket among cannabinoid receptors implies that it is impossible to design selective orthosteric ligands for CB1 or CB2. Surprisingly, ORG27569, only performed as a NAM for CB1, binds to CB1 in a unique site that is different from CB2 [110, 241]. Meanwhile, ZCZ011, acting as a PAM, displays high subtype selectivity for CB1 [242, 243].
It often appears to be the case that designing a selective allosteric modulator is comparatively easier than developing selective orthosteric ligands. Many allosteric modulators have been identified through screening processes and have shown high subtype selectivity without the need for extensive optimization efforts [244]. This has made allosteric modulation a valuable approach in achieving selectivity for specific GPCR subtypes in drug discovery.
Allosteric agonist and ago-PAMs
The classic allosteric modulators, often referred to as pure allosteric modulators, are compounds that exert their allosteric actions only in the presence of OAs [245, 246]. This characteristic suggests several advantages for allosteric modulators, including spatial and temporal specificity. In tissues where the endogenous ligand is present at higher levels, allosteric drugs can exhibit greater sensitivity and efficacy in modulating the receptor’s activity. This sensitivity is linked to the presence of OA. Additionally, the dependence of pure allosteric modulators on the presence of the endogenous ligand results in a ceiling effect for allosterism. This means that even if a higher dose of the allosteric modulator is administered, the response will eventually reach a maximum level, increasing the safety margin of the candidate agent in overdose situations [244].
In addition to pure allosteric modulators, there are also agonist-PAMs (ago-PAMs). These ligands have the intrinsic ability to activate receptor signaling in the absence of an agonist while simultaneously performing the PAM effect [47, 247]. For mutated GPCRs that have lost the ability to bind endogenous ligands, ago-PAMs can act as supplements to restore or partially restore GPCR function [248]. Furthermore, in cases where endogenous ligand levels are low, ago-PAMs can enhance receptor signaling sufficiently due to their high cooperativity and intrinsic activity. This property makes them valuable in situations where endogenous ligands are limited or compromised.
Biased activation
GPCRs are dynamic and interact with various effectors, including G-proteins (Gs, Gi/o, G11/q, and G12/13) and arrestins (e.g., β-arrestin1 and β-arrestin2), to mediate distinct physiological effects. Biased ligands are compounds that selectively stimulate specific therapeutic pathways while avoiding unwanted on-target side effects [249]. In addition to biased orthosteric agonists, there are also biased allosteric modulators (BAMs) that can trigger biased signal transduction of GPCRs [249, 250]. BAMs offer the advantage of positive PAMs when combined with orthosteric ligands, allowing for fine-tuned and selective modulation of GPCR signaling pathways (Fig. 11d) [249]. This approach holds promise for the development of more precise and effective therapeutics targeting GPCRs.
Biased allosteric pharmacology of GPCR
The binding sites of BAMs (biased allosteric modulators) in GPCRs can vary and are distributed throughout different regions of the receptor (Fig. 11d), including the extracellular region, transmembrane domain (TMD), and cytoplasmic region. These binding sites play a crucial role in allosteric modulation and influence the receptor’s interaction with various signaling pathways. Here are some recent examples of BAM binding sites in GPCRs:
Neurotensin receptor (NTSR1)
A BAM called SBI-553 binds to NTSR1 at an intracellular site that is composed of transmembrane helices TM6 and TM7, as well as helix H8 [149]. The binding of SBI-553 to this unique pocket causes remodeling of the interface between NTSR1 and the Gα protein. This rearrangement affects the conformation of the Gα protein, particularly the wavy hook and α5 helix, which are critical for determining G-protein subgroup selectivity. As a negative allosteric modulator (NAM) for the NTSR G-protein signaling pathway, SBI-553 displays complex allosteric effects with different Gα subtypes (Fig. 11e).
Hydroxycarboxylic acid receptor 2 (HCAR2)
Compound 9n is a BAM that binds to HCAR2 [251, 252]. Its binding site is located in the upper half of transmembrane helices TM5 and TM6, as well as extracellular loop 2 (ECL2). This unique binding site contributes to distinct allosteric effects on HCAR2 signaling through both G-proteins and β-arrestin (Fig. 11e).
These structural determinants of GPCRs with allosteric modulators, especially BAMs, are essential for understanding the molecular mechanisms of biased allosteric modulation and advancing the development of biased allosteric pharmacology and biased allosteric drugs for GPCRs. They provide valuable insights into how BAMs can influence receptor conformation and signaling pathways, ultimately aiding in drug design and therapeutic development.
Poly-pharmacology of GPCR and drug discovery
The drug development strategies mentioned above, including selective ligands, biased ligands, and allosteric ligands, typically focus on targeting specific GPCRs. However, the landscape of drug development is undergoing a paradigm shift, with an increasing emphasis on multitarget drugs. Poly-pharmacology, which refers to drugs that interact with multiple targets, is attributed to complicated biological pathways and consequently to multiple effects [253, 254]. GPCRs frequently share similar structural frameworks, and the key residues within their binding pockets exhibit certain conservation, making them attractive candidates for multitarget drug development [255]. Notably, the interplay of a drug molecule with multiple targets can be a double-edged sword [256]. While it can lead to beneficial outcomes, especially in drug repurposing, it can also result in detrimental off-target effects. Therefore, comprehensive polypharmacological analysis is indispensable in drug development for a holistic understanding of both the desired and potential adverse effects. Here, our focus primarily rests on drugs targeting GPCRs that exhibit therapeutic effects by engaging multiple beneficial targets, sidelining those with predominant side effects.
In the realm of multipharmacological drugs targeting GPCR subfamilies, dual agonists and antagonists often offer enhanced therapeutic outcomes compared with single-target alternatives. A case in point is tirzepatide, a fatty-acid-modified polypeptide. It mirrors the behavior of native GIP at the gastric inhibitory polypeptide receptor (GIPR) and simultaneously showcases a preference for G-protein signaling at GLP-1R [257]. The molecular basis underpinning tirzepatide’s dual agonism has been recently elucidated [258]. Similar to native ligands, tirzepatide adopts an α-helical structure with its N-terminus deeply embedded within the transmembrane core of both receptors. However, a notably compact tirzepatide-GIPR complex has been observed. Specifically, the strong interactions between Tyr1Tzp and other residues within the GIPR core enable tirzepatide to accept fatty acid modifications, thereby achieving an affinity comparable to GIP. In contrast, its high-affinity interaction with the extracellular domain of GLP-1R, combined with the reduced stability from Tyr1Tzp and the lipid moiety, promotes biased signaling and diminishes receptor desensitization. Such bias potentially amplifies the effectiveness of tirzepatide in managing glucose levels and body weight in type 2 diabetes mellitus patients [259, 260]. Furthermore, the therapeutic prospects of tirzepatide extend to conditions such as obesity and nonalcoholic steatohepatitis (NASH), underscoring its versatility in addressing multifaceted diseases [261, 262]. In the context of dual antagonists, bosentan, a compound that concurrently targets both endothelin receptor type A (ETAR) and ETBR, has been approved for addressing pulmonary hypertension [263]. Its interaction with ETAR curtails vasoconstriction, while engagement with ETBR impedes bronchoconstriction [264]. The therapeutic objective is to counteract these constraining effects, facilitating the relaxation of the pulmonary vasculature and thereby attenuating pulmonary pressures and resistance.
While many drugs typically engage in similar binding patterns within the orthosteric pockets of receptors, there are multitarget drugs that operate through diverse pharmacophores. A case in point is sparsentan. This dual antagonist selectively targets both ETAR and AT1R [265]. It is noteworthy for its high affinity for both receptors and was designed by integrating functional structural elements from two separate antagonists, irbesartan (specific to AT1R) and biphenylsulfonamide (specific to ETAR) (Table 2) [266, 267]. This approach epitomizes another dimension in the poly-pharmacological drug design paradigm.
Table 2.
FDA-approved agents targeting GPCRs with polypharmacological characteristics (2010–2023)
| First Approval | Drug | Modes of Action | Indications | References |
|---|---|---|---|---|
| 2023 | Sparsentan | Antagonist of AT1R and ETAR | Immunoglobulin A Nephropathy | [280] |
| 2022 | Tirzepatide | Agonist of GIPR and GLP-1R | Type 2 Diabetes | [281] |
| 2020 | Bencycloquidium Bromide | Antagonist of M1 and M3 | Allergic Rhinitis | [282, 283] |
| 2020 | Ozanimod | Agonist of S1PRs | Crohn’s Disease, Relapsing-Remitting Multiple Sclerosis, Ulcerative Colitis. Multiple Sclerosis | [284–286] |
| 2020 | Fenfluramine | Agonist of 5HT1Rs and 5HT2Rs | Lennox-Gastaut Syndrome, Dravet Syndrome, CDKL5 Deficiency, Seizures | [287–294] |
| 2019 | Siponimod | Agonist of S1PR1 and S1PR3-5 | Secondary Progressive Multiple Sclerosis, Multiple Sclerosis | [286, 295–299] |
| 2019 | Lumateperone | Antagonist of 5HT2Rand D2R | Depression, Schizophrenia | [300, 301] |
| 2018 | Revefenacin | Antagonist of muscarinic receptors | Chronic Obstructive Pulmonary Disease | [302] |
| 2017 | Dinalbuphine Sebacate | Agonist of κOR; Antagonist of µOR | Pain | |
| 2015 | Aripiprazole Lauroxil | Agonist of D2R, 5HT1Rs, 5HT2Rs, and 5HT7R; Antagonist of H1R, D4R, and 5HT6R | Schizophrenia | [303–308] |
| 2015 | Brexpiprazole | Agonist of 5HT1R and D2R | Agitation, Severe Depressive Disorder, Schizophrenia | [309, 310] |
| 2015 | Cariprazine | Agonist of 5HT1R, D2R and D3R; Antagonist of 5HT2R | Severe Depressive Disorder, Bipolar Disorder And Related Disorders, Bipolar 1 Disorder, Schizophrenia | [311–313] |
| 2015 | Eluxadoline | Agonist of µOR and κOR; Antagonist of δOR | Diarrhea-Type Irritable Bowel Syndrome, Irritable Bowel Syndrome (IBS) | [314] |
| 2015 | Flibanserin | Agonist of 5HT1R; Antagonist of 5HT2R and D4R | Psychosexual Dysfunction | [315] |
| 2013 | Vortioxetine | Agonist of 5HT1A - BR; Antagonist of 5HT1DR, 5HT2R, 5HT3R and 5HT7R | Severe Depressive Disorder, Depressive | [315] |
| 2013 | Macitentan | Antagonist of ETAR and ETBR | Connective Tissue Disease, Pulmonary Hypertension | [316] |
| 2012 | Loxapine | Antagonist of H1R, 5HT2Rs, 5HT6R, 5HT7R, and D2R-D4R | Bipolar Disorder, Bipolar 1 Disorder, Schizophrenia | [304, 317–323] |
| 2011 | Motilitone | Agonist of 5HT4R; Antagonist of 5HT3R and D2R | Dyspepsia | |
| 2010 | Lurasidone | Agonist of 5HT1R; Antagonist of 5HT2R, 5HT7R, D2R, Α2AR and Α2CR | Bipolar Disorder, Bipolar 1 Disorder, Schizophrenia | [324] |
| 2010 | Fingolimod | Agonist of S1PR1 and S1PR5; Modulator of S1PR3 and S1PR4 | Relapsing-Remitting Multiple Sclerosis, Multiple Sclerosis, The Relapsing Multiple Sclerosis | [325] |
| 2010 | Buprenorphine | Agonist of µOR; Antagonist of κOR | Opium Dependence, Opioid-Related Disorder, Pain | [326, 327] |
Certain dual-target drugs possess the capability to interact with both GPCR and non-GPCR targets simultaneously. Roluperidone serves as a notable example. As a dual antagonist, it targets the 5HT2AR and σ2 receptors [268]. Clinical trials suggest that roluperidone has therapeutic potential in addressing the negative symptoms associated with schizophrenia [269]. This reflects the extensive nature of polypharmacology and shows the potential for drug possibilities across different types of targets.
When considering three or more targeted drugs of GPCR, it emerges passively during the investigation of the pharmacological properties of the drugs. The aminergic receptors, notably the 5-HT receptors, dopaminergic receptors, and adrenoceptors, stand out due to their ligand promiscuity, shared binding modes, and orthosteric pocket similarities [270]. For instance, ergotamine for migraine treatment has broad interactions with multiple receptors and usually causes serious side effects [271, 272]. As the understanding of the molecular mechanisms of the ligand-receptor recognition and downstream signaling initiation has deepened, a few compounds have been designed to specifically target therapeutic receptors or signaling pathways. Drugs such as brexpiprazole and cariprazine exemplify this approach [273]. They act as 5HT2AR antagonists while concurrently serving as agonists for 5HT1AR and D2R, primarily catering to the treatment of psychiatric conditions.
Another promising frontier in multitarget drug development targets the GLP-1-like receptor subfamily. Prominent examples include the triagonists Retatrutide, HM15211, and MAR423 [274–276]. Taking retatrutide as an illustrative case, it was derived from the GIP architecture, not only mirroring tirzepatide’s dual agonist effects but also introducing enhanced glucagon receptor activation. As of now, Retatrutide is undergoing several phase 3 clinical trials, primarily tending to address cardiovascular and metabolic endocrinological disorders [277, 278].
In summary, the advent of multitarget drugs, derived from a deeper understanding of complicated biological processes, offers a promising avenue for treating a myriad of diseases more holistically and effectively. While the individual activities of multitarget drugs might be subdued compared to their single-target counterparts, their ability to synergistically modulate interconnected disease targets renders them especially advantageous [279]. Such an approach is particularly beneficial for diseases with multiple etiologies, including malignancies, cardiovascular diseases, neurodegenerative disorders, and autoimmune diseases, where poly-pharmacological drugs hold immense prospective value. A list of approved therapeutic polypharmacological agents is shown in Table 2.
The drugs listed above were identified from databases such as guide to pharmacology, Drug Bank and zhihuiya new drug repository.
Antibody drug development for GPCRs
Compared to conventional chemical drugs, antibody-based therapies for GPCRs are preferred by major domestic and international pharmaceutical companies due to their unique properties, which include specificity, high affinity, and longer serum half-life. GPCR antibody drugs can be categorized into three main groups:
Anti-receptor antibody
These antibodies directly target GPCRs themselves, interfering with receptor function or signaling pathways. Currently, there are two FDA-approved antibody drugs targeting GPCRs. Erenumab, a selective monoclonal antibody designed to target the calcitonin gene-related peptide type 1 receptor (CGRPR), received FDA approval in 2018 for the treatment of migraines [328]. Mogamulizumab, a humanized monoclonal antibody for CCR4, is employed in the treatment of two rare cutaneous T-cell lymphomas (Table 3) [329].
Table 3.
The antibody drugs for GPCRs
| Drugs | Target | Phase | Indications | References |
|---|---|---|---|---|
| Erenumab | CGRP | Approved | Migraine,Temporomandibular joint dysfunction syndrome, Headache | [332] |
| Mogamulizumab | CCR4 | Approved | Cancer, Adult T-cell Leukemia/Lymphoma (ATLL), Cutaneous T-cell Lymphoma (CTCL) | [333–336] |
| Leronlimab | CCR5 | Phase III | COVID-19 Pneumonia, Metastatic microsatellite-stabilized colorectal cancer, Nonalcoholic steatohepatitis | [337] |
| Talquetamab | GPRC5D | Phase III | Multiple Myeloma, Plasma cell myeloma refractory, Recurrent Multiple Myeloma | [338, 339] |
| REMD-477 | GCGR | Phase II | Type 1 diabetes, Type 2 diabetes, Glucose intolerance | [340, 341] |
| Plozalizumab | CCR2 | Phase II | Diabetic nephropathy, Melanoma | [342] |
| LY-3,041,658 | CXCR1 and CXCR2 | Phase II | Hidradenitis suppurativa | [343, 344] |
| Avdoralimab | C5aR1 | Phase II | Advanced Solid Tumors, COVID-19 Pneumonia,Herpetic pemphigoid | [345] |
| Volagidemab | GCGR | Phase II | Type 1 diabetes, Type 2 diabetes, Glucose intolerance | [340, 341] |
| AMG-301 | PAC1R | Phase II | Migraine | [346] |
| Getagozumab | ETAR | Phase II | Pulmonary arterial hypertension | [347, 348] |
| Tidutamab | SSTR2 | Phase II | Merkel cell carcinoma, Small Cell Lung Cancer, Gastrointestinal Mesenchymal Tumor, Neuroendocrine Tumor | [349] |
| Nimacimab | CB1 | Phase II | Diabetic gastroparesis, Diabetic nephropathy, Nonalcoholic steatohepatitis, Obesity | [350] |
| Ulocuplumab | CXCR4 | Phase II | Pancreatic Cancer, Multiple Myeloma, Acute Myeloid Leukemia, Macroglobulinemia | [351, 352] |
| LM-305 | GPRC5D | Phase I/II | Multiple Myeloma, Solid Tumors, Hematologic Diseases | [353] |
| TAK-500 | CCR2 | Phase I | Breast Cancer, Esophageal Cancer, Nasopharyngeal Cancer, Solid tumor, Head and Neck Squamous Cell Carcinoma, Gastric cancer | [354] |
| JBH492 | CCR7 | Phase I | B-cell chronic lymphocytic leukemia, Non-Hodgkin’s Lymphoma | [355, 356] |
| HZ-515H7 | CXCR4 | Phase I | Neoplasms | [357, 358] |
| SAR-113,244 | CXCR5 | Phase I | Systemic lupus erythematosus | [359] |
Anti-ligand antibody
These antibodies are designed to block or neutralize ligands (molecules that activate GPCRs) and can indirectly modulate GPCR activity. For instance, anti-calcitonin gene-related peptide (anti-CGRP) monoclonal antibodies such as fremanezumab, ellipsumab-jjmr, and galcanezumab have shown promise in the treatment of migraines [330]. They selectively target both the α and β subunits of human CGRP, leading to the blockade of CGRPP activation. In 2023, a significant milestone was achieved with the FDA approval of Veopoz (pozelimab) [331]. Veopoz is a fully human monoclonal IgG4 antibody with high binding affinity for wild-type and variant human complement C5 proteins and effectively inhibits the activity of complement factor C5 for therapeutic purposes. Veopoz represents a groundbreaking therapy, being the first FDA-approved treatment for CD55-deficient protein-losing enteropathy (CHAPLE) in adults and pediatric patients over the age of 1 year. This achievement highlights the potential of monoclonal antibodies in addressing rare and complex medical conditions.
Antibody-drug conjugates (ADCs) for GPCRs
ADCs are a class of targeted drugs that combine monoclonal antibodies with cytotoxic drugs via linkers. These ADCs leverage the targeting capability of antibodies to deliver toxic drugs specifically to cells expressing the target GPCR, reducing drug toxicity while maintaining therapeutic efficacy. Globally, there are 19 ADC candidate drugs designed to target GPCRs. Among these, 4 are in clinical phase I or II, 6 are in preclinical stages, 2 are in drug discovery phases, and 7 are not in active development. These ADCs are designed to target a range of GPCRs, including CXCR4, CCR5, CCR3, C5AR1, LGR4 and others, presenting a diverse set of potential therapeutic options. Furthermore, significant progress has been made in the development of ADC drugs targeting orphan GPCRs. A noteworthy achievement in this context is the recent FDA approval of the world’s first bispecific antibody targeting GPRC5D/CD3 for the treatment of relapsed/refractory multiple myeloma. This groundbreaking therapy is specifically designed to target the orphan class C GPCR GPRC5D, which exhibits high expression levels in affected patients. This achievement underscores the potential of ADCs in addressing previously untargeted or challenging medical conditions.
The development landscape for GPCR antibody drugs has expanded significantly, encompassing various innovative approaches and combinations to enhance therapeutic efficacy, including bispecific antibodies, nanobodies, and combinations with other therapies such as CAR-T (chimeric antigen receptor T cell), checkpoint inhibitors, chemotherapy drugs, and cell therapies. The growing number of projects in the global research and development pipeline, along with the successful entry of GPCR antibodies into clinical development over the past decade, underscores the importance of targeting GPCRs with monoclonal antibodies in pharmaceutical research. This expanding landscape reflects the pharmaceutical industry’s commitment to exploring innovative approaches to address a wide range of medical conditions and advance the field of GPCR-based therapeutics. A list of the antibody drugs for GPCRs is shown in Table 3.
Future opportunities and summary
The frontier of GPCR investigation is particularly in the realms of biased agonism, allosteric modulation, and compartmentalized signaling [234, 249, 360, 361]. Each of these avenues presents a unique opportunity to deepen our understanding of GPCR functionality. Biased signaling, for instance, allows for the nuanced control of receptor responses, allosteric modulation offers insights into specific receptor activity manipulation, and compartmentalized signaling provides a framework for understanding the spatial and temporal dynamics of GPCR signaling.
As we navigate through this exciting era of GPCR research, artificial intelligence (AI) stands out as an invaluable tool, playing a promising role in elucidating GPCR structures and facilitating the discovery and development of novel drugs [362]. AI computational capabilities allow for the efficient integration and analysis of vast and complex datasets, aiding in the identification of potential therapeutic targets and the optimization of lead compounds for GPCRs [363]. Through AI-driven virtual and experimental screening processes, novel chemotypes and scaffolds are identified, accelerating the pace of drug discovery and bringing us closer to realizing the therapeutic potential of GPCRs.
Collectively, the collaborative efforts of in-depth GPCR research and AI technology are guiding us toward unprecedented breakthroughs in the field. The integration of modern technology marks a transformative period in our pursuit of understanding and effectively targeting GPCRs for therapeutic intervention, heralding a promising future for precision medicine and drug discovery.
Acknowledgements
The structural figures were prepared with PyMOL (https://pymol.org/2/) software and ChimeraX (https://www.cgl.ucsf.edu/chimerax/) software. The schematic diagrams were created with BioRender.com.
Authors' contributions
W.Y. and Z.S. supervised the overall project and designed the manuscript; L.C., F.X., Z.L., C.S., Z.Y., H.H. wrote the manuscript and created the figures and tables with the assistance of S.S., Y.F., X.Y., X.T., and H.Q. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32100988 and 32371288 to W.Y., 31972916 to Z.S., 82271190 and 32100965 to L.C., 82201453 to F.X.), Science and Technology Department of Sichuan Province (2022ZYD0085 to Z.S.), and Ministry of Technology Department of China grant (2019YFA0508800 to Z.S.). 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYYC20023 to Z.S.). Frontiers Medical Center, Tianfu Jincheng Laboratory Foundation (TFJC2023010010 to Z.S.).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare no conflicts of interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lin Cheng, Fan Xia, Ziyan Li, Chenglong Shen, Zhiqian Yang and Hanlin Hou contributed equally to this work.
Contributor Information
Wei Yan, Email: weiyan2018@scu.edu.cn.
Zhenhua Shao, Email: zhenhuashao@scu.edu.cn.
References
- 1.Hauser AS, Kooistra AJ, Munk C, Heydenreich FM, Veprintsev DB, Bouvier M, et al. GPCR activation mechanisms across classes and macro/microscales. Nat Struct Mol Biol. 2021;28(11):879–888. doi: 10.1038/s41594-021-00674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Nafria J, Tate CG. Structure determination of GPCRs: cryo-EM compared with X-ray crystallography. Biochem Soc Trans. 2021;49(5):2345–2355. doi: 10.1042/BST20210431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wingler LM, Lefkowitz RJ. Conformational basis of G protein-coupled receptor signaling versatility. Trends Cell Biol. 2020;30(9):736–747. doi: 10.1016/j.tcb.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, et al. Common activation mechanism of class A GPCRs. Elife. 2019 doi: 10.7554/eLife.50279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang D, Zhou Q, Labroska V, Qin S, Darbalaei S, Wu Y, et al. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther. 2021;6(1):7. doi: 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S, Gumpper RH, Huang XP, Liu Y, Krumm BE, Cao C, et al. Molecular basis for selective activation of DREADD-based chemogenetics. Nature. 2022;612(7939):354–362. doi: 10.1038/s41586-022-05489-0. [DOI] [PubMed] [Google Scholar]
- 7.Faouzi A, Wang H, Zaidi SA, DiBerto JF, Che T, Qu Q, et al. Structure-based design of bitopic ligands for the micro-opioid receptor. Nature. 2023;613(7945):767–774. doi: 10.1038/s41586-022-05588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan AL, Confair DN, Kim K, Barros-Alvarez X, Rodriguiz RM, Yang Y, et al. Bespoke library docking for 5-HT(2A) receptor agonists with antidepressant activity. Nature. 2022;610(7932):582–591. doi: 10.1038/s41586-022-05258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadybekov AA, Sadybekov AV, Liu Y, Iliopoulos-Tsoutsouvas C, Huang XP, Pickett J, et al. Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature. 2022;601(7893):452–459. doi: 10.1038/s41586-021-04220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Sci (New York NY) 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruhl T, Weinert T, Rodrigues MJ, Milne CJ, Ortolani G, Nass K, et al. Ultrafast structural changes direct the first molecular events of vision. Nature. 2023;615(7954):939–944. doi: 10.1038/s41586-023-05863-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang K, Wu H, Hoppe N, Manglik A, Cheng Y. Fusion protein strategies for cryo-EM study of G protein-coupled receptors. Nat Commun. 2022;13(1):4366. doi: 10.1038/s41467-022-32125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collu G, Mohammed I, Lafita A, Bierig T, Poghosyan E, Bliven S, et al. 2021. 10.1101/2021.09.25.461805.
- 15.Tsutsumi N, Mukherjee S, Waghray D, Janda CY, Jude KM, Miao Y, et al. Structure of human Frizzled5 by fiducial-assisted cryo-EM supports a heterodimeric mechanism of canonical wnt signaling. Elife. 2020 doi: 10.7554/eLife.58464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Huang W, Li X. Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response. Structure. 2022;30(7):1016–1024. doi: 10.1016/j.str.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Q, He B, Zhong Y, Jiao H, Ren Y, Wang Q, et al. A method for structure determination of GPCRs in various states. Nat Chem Biol. 2023 doi: 10.1038/s41589-023-01389-0. [DOI] [PubMed] [Google Scholar]
- 18.Robertson MJ, Papasergi-Scott MM, He F, Seven AB, Meyerowitz JG, Panova O, et al. Structure determination of inactive-state GPCRs with a universal nanobody. Nat Struct Mol Biol. 2022;29(12):1188–1195. doi: 10.1038/s41594-022-00859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unal H, Karnik SS. Domain coupling in GPCRs: the engine for induced conformational changes. Trends Pharmacol Sci. 2012;33(2):79–88. doi: 10.1016/j.tips.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30(5):249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Deupi X, Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr Opin Struct Biol. 2011;21(4):541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan J, Xu P, Zhang H, Luan X, Yang J, He X, et al. Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor. Nat Commun. 2023;14(1):519. doi: 10.1038/s41467-023-36170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan J, Xu P, Cheng X, Mao C, Croll T, He X, et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature. 2021;598(7882):688–692. doi: 10.1038/s41586-021-03924-2. [DOI] [PubMed] [Google Scholar]
- 25.Duan J, Xu P, Luan X, Ji Y, He X, Song N, et al. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature. 2022;609(7928):854–859. doi: 10.1038/s41586-022-05173-3. [DOI] [PubMed] [Google Scholar]
- 26.Faust B, Billesbolle CB, Suomivuori CM, Singh I, Zhang K, Hoppe N, et al. Autoantibody mimicry of hormone action at the thyrotropin receptor. Nature. 2022;609(7928):846–853. doi: 10.1038/s41586-022-05159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin AL, Steurer MA, Aronstam RS. Constitutive activity among orphan Class-A G protein coupled receptors. PLoS One. 2015;10(9):e0138463. doi: 10.1371/journal.pone.0138463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X, Li M, Wang N, Wu Y, Luo Z, Guo S, et al. Structural basis of ligand recognition and self-activation of orphan GPR52. Nature. 2020;579(7797):152–157. doi: 10.1038/s41586-020-2019-0. [DOI] [PubMed] [Google Scholar]
- 29.Lin X, Chen B, Wu Y, Han Y, Qi A, Wang J, et al. Cryo-EM structures of orphan GPR21 signaling complexes. Nat Commun. 2023;14(1):216. doi: 10.1038/s41467-023-35882-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye F, Wong TS, Chen G, Zhang Z, Zhang B, Gan S, et al. Cryo-EM structure of G-protein-coupled receptor GPR17 in complex with inhibitory G protein. MedComm. 2022;3(4):e159. doi: 10.1002/mco2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billesbolle CB, de March CA, van der Velden WJC, Ma N, Tewari J, Del Torrent CL, et al. Structural basis of odorant recognition by a human odorant receptor. Nature. 2023;615(7953):742–749. doi: 10.1038/s41586-023-05798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, Cheng J, Lian S, Liu Q, Lu Y, Zheng Y, et al. Structural basis of amine odorant perception by a mammal olfactory receptor. Nature. 2023;618(7963):193–200. doi: 10.1038/s41586-023-06106-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Sun B, Feng D, Hu H, Chu M, Qu Q, et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature. 2017;546(7657):248–253. doi: 10.1038/nature22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun W, Chen LN, Zhou Q, Zhao LH, Yang D, Zhang H, et al. A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor. Cell Res. 2020;30(12):1098–1108. doi: 10.1038/s41422-020-00442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao A, Han S, Li X, Li Z, Zhao P, Dai A, et al. Structural basis of G(s) and G(i) recognition by the human glucagon receptor. Sci (New York NY) 2020;367(6484):1346–1352. doi: 10.1126/scienceaaz5346. [DOI] [PubMed] [Google Scholar]
- 36.Zhao F, Zhang C, Zhou Q, Hang K, Zou X, Chen Y, et al. Structural insights into hormone recognition by the human glucose-dependent insulinotropic polypeptide receptor. Elife. 2021. 10.7554/eLife.68719. [DOI] [PMC free article] [PubMed]
- 37.Dong M, Deganutti G, Piper SJ, Liang YL, Khoshouei M, Belousoff MJ, et al. Structure and dynamics of the active Gs-coupled human secretin receptor. Nat Commun. 2020;11(1):4137. doi: 10.1038/s41467-020-17791-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao LH, Ma S, Sutkeviciute I, Shen DD, Zhou XE, de Waal PW, et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Sci (New York NY) 2019;364(6436):148–153. doi: 10.1126/science.aav7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Cheng X, Zhao L, Wang Y, Ye C, Zou X, et al. Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2. Proc Natl Acad Sci U S A. 2021;118:32. doi: 10.1073/pnas.2101279118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang YL, Khoshouei M, Deganutti G, Glukhova A, Koole C, Peat TS, et al. Cryo-EM structure of the active, G(s)-protein complexed, human CGRP receptor. Nature. 2018;561(7724):492–497. doi: 10.1038/s41586-018-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dal Maso E, Glukhova A, Zhu Y, Garcia-Nafria J, Tate CG, Atanasio S, et al. The molecular control of calcitonin receptor signaling. ACS Pharmacol Transl Sci. 2019;2(1):31–51. doi: 10.1021/acsptsci.8b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma S, Shen Q, Zhao LH, Mao C, Zhou XE, Shen DD, et al. Molecular basis for hormone recognition and activation of corticotropin-releasing factor receptors. Mol Cell. 2020;77(3):669–680. doi: 10.1016/j.molcel.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Liang YL, Belousoff MJ, Zhao P, Koole C, Fletcher MM, Truong TT, et al. Toward a Structural understanding of class B GPCR peptide binding and activation. Mol Cell. 2020;77(3):656–668. doi: 10.1016/j.molcel.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Duan J, Shen DD, Zhou XE, Bi P, Liu QF, Tan YX, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat Commun. 2020;11(1):4121. doi: 10.1038/s41467-020-17933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Feng W, Zhou Q, Liang A, Li J, Dai A, et al. A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2. Nat Commun. 2022;13(1):2272. doi: 10.1038/s41467-022-30041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Song X, Zhang D, Chen X, Li X, Sun Y, et al. Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism. Cell Res. 2020;30(5):436–445. doi: 10.1038/s41422-020-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cong Z, Chen LN, Ma H, Zhou Q, Zou X, Ye C, et al. Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor. Nat Commun. 2021;12(1):3763. doi: 10.1038/s41467-021-24058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Graaf C, Song G, Cao C, Zhao Q, Wang MW, Wu B, et al. Extending the structural view of class B GPCRs. Trends Biochem Sci. 2017;42(12):946–960. doi: 10.1016/j.tibs.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Cary BP, Zhang X, Cao J, Johnson RM, Piper SJ, Gerrard EJ, et al. New insights into the structure and function of class B1 GPCRs. Endocr Rev. 2023;44(3):492–517. doi: 10.1210/endrev/bnac033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wootten D, Simms J, Miller LJ, Christopoulos A, Sexton PM. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc Natl Acad Sci U S A. 2013;110(13):5211–5216. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cong Z, Liang YL, Zhou Q, Darbalaei S, Zhao F, Feng W, et al. Structural perspective of class B1 GPCR signaling. Trends Pharmacol Sci. 2022;43(4):321–334. doi: 10.1016/j.tips.2022.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi K, Kawakami K, Kusakizako T, Tomita A, Nishimura M, Sawada K, et al. Class B1 GPCR activation by an intracellular agonist. Nature. 2023;618(7967):1085–1093. doi: 10.1038/s41586-023-06169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao LH, He Q, Yuan Q, Gu Y, He X, Shan H, et al. Conserved class B GPCR activation by a biased intracellular agonist. Nature. 2023;621(7979):635–641. doi: 10.1038/s41586-023-06467-w. [DOI] [PubMed] [Google Scholar]
- 54.Hamann J, Aust G, Arac D, Engel FB, Formstone C, Fredriksson R, et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev. 2015;67(2):338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langenhan T, Piao X, Monk KR. Adhesion G protein-coupled receptors in nervous system development and Disease. Nat Rev Neurosci. 2016;17(9):550–561. doi: 10.1038/nrn.2016.86. [DOI] [PubMed] [Google Scholar]
- 56.Wong TS, Li G, Li S, Gao W, Chen G, Gan S, et al. G protein-coupled receptors in neurodegenerative Diseases and psychiatric disorders. Signal Transduct Target Ther. 2023;8(1):177. doi: 10.1038/s41392-023-01427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeung J, Adili R, Stringham EN, Luo R, Vizurraga A, Rosselli-Murai LK, et al. GPR56/ADGRG1 is a platelet collagen-responsive GPCR and hemostatic sensor of shear force. Proc Natl Acad Sci U S A. 2020;117(45):28275–28286. doi: 10.1073/pnas2008921117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scholz N, Guan C, Nieberler M, Grotemeyer A, Maiellaro I, Gao S, et al. Mechano-dependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. Elife. 2017 doi: 10.7554/eLife.28360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scholz N, Dahse AK, Kemkemer M, Bormann A, Auger GM, Vieira Contreras F, et al. Molecular sensing of mechano- and ligand-dependent adhesion GPCR dissociation. Nature. 2023;615(7954):945–953. doi: 10.1038/s41586-023-05802-5. [DOI] [PubMed] [Google Scholar]
- 60.Lin HH, Hsiao CC, Pabst C, Hebert J, Schoneberg T, Hamann J. Adhesion GPCRs in regulating immune responses and inflammation. Adv Immunol. 2017;136:163–201. doi: 10.1016/bs.ai.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 61.Leemans JC, te Velde AA, Florquin S, Bennink RJ, de Bruin K, van Lier RA, et al. The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. J Immunol. 2004;172(2):1125–1131. doi: 10.4049/jimmunol17221125. [DOI] [PubMed] [Google Scholar]
- 62.Abdulkareem NM, Bhat R, Qin L, Vasaikar S, Gopinathan A, Mitchell T, et al. A novel role of ADGRF1 (GPR110) in promoting cellular quiescence and chemoresistance in human epidermal growth factor receptor 2-positive Breast cancer. FASEB J. 2021;35(7):e21719. doi: 10.1096/fj.202100070R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arcos-Burgos M, Velez JI, Martinez AF, Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, et al. ADGRL3 (LPHN3) variants predict substance use disorder. Transl Psychiatry. 2019;9(1):42. doi: 10.1038/s41398-019-0396-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Acosta MT, Swanson J, Stehli A, Molina BS, Team MTA, Martinez AF, et al. ADGRL3 (LPHN3) variants are associated with a refined phenotype of ADHD in the MTA study. Mol Genet Genomic Med. 2016;4(5):540–547. doi: 10.1002/mgg3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma J, Ma X, Lin K, Huang R, Bi X, Ming C, et al. Genetic screening of a Chinese cohort of children with hearing loss using a next-generation sequencing panel. Hum Genomics. 2023;17(1):1. doi: 10.1186/s40246-022-00449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yona S, Lin HH, Siu WO, Gordon S, Stacey M. Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem Sci. 2008;33(10):491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 67.Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC, et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31(6):1364–1378. doi: 10.1038/emboj201226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krasnoperov V, Lu Y, Buryanovsky L, Neubert TA, Ichtchenko K, Petrenko AG. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J Biol Chem. 2002;277(48):46518–46526. doi: 10.1074/jbcM206415200. [DOI] [PubMed] [Google Scholar]
- 69.Lin HH, Chang GW, Davies JQ, Stacey M, Harris J, Gordon S. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279(30):31823–31832. doi: 10.1074/jbcM402974200. [DOI] [PubMed] [Google Scholar]
- 70.Qu X, Qiu N, Wang M, Zhang B, Du J, Zhong Z, et al. Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1. Nature. 2022;604(7907):779–785. doi: 10.1038/s41586-022-04580-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barros-Alvarez X, Nwokonko RM, Vizurraga A, Matzov D, He F, Papasergi-Scott MM, et al. The tethered peptide activation mechanism of adhesion GPCRs. Nature. 2022;604(7907):757–762. doi: 10.1038/s41586-022-04575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao P, Guo S, Wen X, He QT, Lin H, Huang SM, et al. Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4. Nature. 2022;604(7907):771–778. doi: 10.1038/s41586-022-04590-8. [DOI] [PubMed] [Google Scholar]
- 73.Ping YQ, Xiao P, Yang F, Zhao RJ, Guo SC, Yan X, et al. Structural basis for the tethered peptide activation of adhesion GPCRs. Nature. 2022;604(7907):763–770. doi: 10.1038/s41586-022-04619-y. [DOI] [PubMed] [Google Scholar]
- 74.Jones DTD, Dates AN, Rawson SD, Burruss MM, Lipper CH, Blacklow SC. Tethered agonist activated ADGRF1 structure and signalling analysis reveal basis for G protein coupling. Nat Commun. 2023;14(1):2490. doi: 10.1038/s41467-023-38083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.An W, Lin H, Ma L, Zhang C, Zheng Y, Cheng Q, et al. Progesterone activates GPR126 to promote Breast cancer development via the Gi pathway. Proc Natl Acad Sci U S A. 2022;119(15):e2117004119. doi: 10.1073/pnas.2117004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin H, Xiao P, Bu RQ, Guo S, Yang Z, Yuan D, et al. Structures of the ADGRG2-G(s) complex in apo and ligand-bound forms. Nat Chem Biol. 2022;18(11):1196–1203. doi: 10.1038/s41589-022-01084-6. [DOI] [PubMed] [Google Scholar]
- 77.Ping YQ, Mao C, Xiao P, Zhao RJ, Jiang Y, Yang Z, et al. Structures of the glucocorticoid-bound adhesion receptor GPR97-G(o) complex. Nature. 2021;589(7843):620–626. doi: 10.1038/s41586-020-03083-w. [DOI] [PubMed] [Google Scholar]
- 78.Ellaithy A, Gonzalez-Maeso J, Logothetis DA, Levitz J. Structural and Biophysical mechanisms of Class C G protein-coupled receptor function. Trends Biochem Sci. 2020;45(12):1049–1064. doi: 10.1016/jtibs202007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Laboute T, Zucca S, Holcomb M, Patil DN, Garza C, Wheatley BA, et al. Orphan receptor GPR158 serves as a metabotropic glycine receptor: mGlyR. Science (New York, NY) 2023;379(6639):1352–1358. doi: 10.1126/scienceadd7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao Y, Robertson MJ, Rahman SN, Seven AB, Zhang C, Meyerowitz JG, et al. Asymmetric activation of the calcium-sensing receptor homodimer. Nature. 2021;595(7867):455–459. doi: 10.1038/s41586-021-03691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen C, Mao C, Xu C, Jin N, Zhang H, Shen DD, et al. Structural basis of GABA(B) receptor-G(i) protein coupling. Nature. 2021;594(7864):594–598. doi: 10.1038/s41586-021-03507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaye H, Ishchenko A, Lam JH, Han GW, Xue L, Rondard P, et al. Structural basis of the activation of a metabotropic GABA receptor. Nature. 2020;584(7820):298–303. doi: 10.1038/s41586-020-2408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin S, Han S, Cai X, Tan Q, Zhou K, Wang D, et al. Structures of G(i)-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature. 2021;594(7864):583–588. doi: 10.1038/s41586-021-03495-2. [DOI] [PubMed] [Google Scholar]
- 84.Seven AB, Barros-Alvarez X, de Lapeyriere M, Papasergi-Scott MM, Robertson MJ, Zhang C, et al. G-protein activation by a metabotropic glutamate receptor. Nature. 2021;595(7867):450–454. doi: 10.1038/s41586-021-03680-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geng Y, Mosyak L, Kurinov I, Zuo H, Sturchler E, Cheng TC, et al. Structural mechanism of ligand activation in human calcium-sensing receptor. Elife. 2016 doi: 10.7554/eLife.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geng Y, Bush M, Mosyak L, Wang F, Fan QR. Structural mechanism of ligand activation in human GABA(B) receptor. Nature. 2013;504(7479):254–259. doi: 10.1038/nature12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407(6807):971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 88.Park J, Fu Z, Frangaj A, Liu J, Mosyak L, Shen T, et al. Structure of human GABA(B) receptor in an inactive state. Nature. 2020;584(7820):304–309. doi: 10.1038/s41586-020-2452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koehl A, Hu H, Feng D, Sun B, Zhang Y, Robertson MJ, et al. Structural insights into the activation of metabotropic glutamate receptors. Nature. 2019;566(7742):79–84. doi: 10.1038/s41586-019-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen X, Wang L, Cui Q, Ding Z, Han L, Kou Y, et al. Structural insights into the activation of human calcium-sensing receptor. Elife. 2021 doi: 10.7554/eLife.68578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ribeiro FM, Vieira LB, Pires RG, Olmo RP, Ferguson SS. Metabotropic glutamate receptors and neurodegenerative Diseases. Pharmacol Res. 2017;115:179–191. doi: 10.1016/j.phrs.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 92.Gregory KJ, Goudet C. International Union of Basic and Clinical Pharmacology. CXI. Pharmacology, Signaling, and physiology of metabotropic glutamate receptors. Pharmacol Rev. 2021;73(1):521–569. doi: 10.1124/pr.119.019133. [DOI] [PubMed] [Google Scholar]
- 93.McCullock TW, Kammermeier PJ. The evidence for and consequences of metabotropic glutamate receptor heterodimerization. Neuropharmacology. 2021 doi: 10.1016/j.neuropharm.2021.108801. [DOI] [PubMed] [Google Scholar]
- 94.Du J, Wang D, Fan H, Xu C, Tai L, Lin S, et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature. 2021;594(7864):589–593. doi: 10.1038/s41586-021-03641-w. [DOI] [PubMed] [Google Scholar]
- 95.Wang X, Wang M, Xu T, Feng Y, Shao Q, Han S, et al. Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers. Cell Res. 2023 doi: 10.1038/s41422-023-00830-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang X, Dong S, Xu F. Structural and Druggability Landscape of Frizzled G protein-coupled receptors. Trends Biochem Sci. 2018;43(12):1033–1046. doi: 10.1016/j.tibs.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 97.Briscoe J, Therond PP. The mechanisms of hedgehog signalling and its roles in development and Disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 98.MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol. 2012;4(12). 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed]
- 99.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 100.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169(6):985–99. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 101.Ueno K, Hazama S, Mitomori S, Nishioka M, Suehiro Y, Hirata H, et al. Down-regulation of frizzled-7 expression decreases survival, invasion and metastatic capabilities of colon Cancer cells. Br J Cancer. 2009;101(8):1374–1381. doi: 10.1038/sjbjc6605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu F, Liu Y, Shen J, Zhang G, Han J. MicroRNA-224 inhibits proliferation and migration of Breast cancer cells by down-regulating fizzled 5 expression. Oncotarget. 2016;7(31):49130–49142. doi: 10.18632/oncotarget9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Corda G, Sala G, Lattanzio R, Iezzi M, Sallese M, Fragassi G, et al. Functional and prognostic significance of the genomic amplification of frizzled 6 (FZD6) in Breast cancer. J Pathol. 2017;241(3):350–361. doi: 10.1002/path.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y, Zhao G, Condello S, Huang H, Cardenas H, Tanner EJ, et al. Frizzled-7 identifies platinum-tolerant Ovarian Cancer cells susceptible to Ferroptosis. Cancer Res. 2021;81(2):384–399. doi: 10.1158/0008-5472.CAN-20-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, et al. Wnt pathway inhibition via the targeting of frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109(29):11717–11722. doi: 10.1073/pnas1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Qi X, Liu H, Thompson B, McDonald J, Zhang C, Li X. Cryo-EM structure of oxysterol-bound human smoothened coupled to a heterotrimeric G(i) Nature. 2019;571(7764):279–283. doi: 10.1038/s41586-019-1286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, et al. Smoothened stimulation by membrane sterols drives hedgehog pathway activity. Nature. 2019;571(7764):284–288. doi: 10.1038/s41586-019-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kozielewicz P, Turku A, Schulte G. Molecular Pharmacology of Class F receptor activation. Mol Pharmacol. 2020;97(2):62–71. doi: 10.1124/mol.119.117986. [DOI] [PubMed] [Google Scholar]
- 109.Qi X, Friedberg L, De Bose-Boyd R, Long T, Li X. Sterols in an intramolecular channel of smoothened mediate hedgehog signaling. Nat Chem Biol. 2020;16(12):1368–1375. doi: 10.1038/s41589-020-0646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shao Z, Yan W, Chapman K, Ramesh K, Ferrell AJ, Yin J, et al. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nat Chem Biol. 2019;15(12):1199–1205. doi: 10.1038/s41589-019-0387-2. [DOI] [PubMed] [Google Scholar]
- 111.Turku A, Schihada H, Kozielewicz P, Bowin CF, Schulte G. Residue 6.43 defines receptor function in class F GPCRs. Nat Commun. 2021;12(1):3919. doi: 10.1038/s41467-021-24004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu L, Chen B, Schihada H, Wright SC, Turku A, Wu Y, et al. Cryo-EM structure of constitutively active human frizzled 7 in complex with heterotrimeric G(s) Cell Res. 2021;31(12):1311–1314. doi: 10.1038/s41422-021-00525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of wnt recognition by Frizzled. Sci (New York NY) 2012;337(6090):59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hirai H, Matoba K, Mihara E, Arimori T, Takagi J. Crystal structure of a mammalian wnt-frizzled complex. Nat Struct Mol Biol. 2019;26(5):372–379. doi: 10.1038/s41594-019-0216-z. [DOI] [PubMed] [Google Scholar]
- 115.Yang S, Wu Y, Xu TH, de Waal PW, He Y, Pu M, et al. Crystal structure of the frizzled 4 receptor in a ligand-free state. Nature. 2018;560(7720):666–70. doi: 10.1038/s41586-018-0447-x. [DOI] [PubMed] [Google Scholar]
- 116.Wong GT, Gannon KS, Margolskee RF. Transduction of bitter and sweet taste by gustducin. Nature. 1996;381(6585):796–800. doi: 10.1038/381796a0. [DOI] [PubMed] [Google Scholar]
- 117.Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444(7117):288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- 118.Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, et al. An amino-acid taste receptor. Nature. 2002;416(6877):199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 119.Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100(6):693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 120.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, et al. T2Rs function as bitter taste receptors. Cell. 2000;100(6):703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 121.Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404(6778):601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 122.Kooistra AJ, Mordalski S, Pandy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, et al. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 2021;49(D1):D335–D343. doi: 10.1093/nar/gkaa1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shaik FA, Singh N, Arakawa M, Duan K, Bhullar RP, Chelikani P. Bitter taste receptors: extraoral roles in pathophysiology. Int J Biochem Cell Biol. 2016;77(Pt B):197–204. doi: 10.1016/j.biocel.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 124.Lee SJ, Depoortere I, Hatt H. Therapeutic potential of ectopic olfactory and taste receptors. Nat Rev Drug Discovery. 2019;18(2):116–138. doi: 10.1038/s41573-018-0002-3. [DOI] [PubMed] [Google Scholar]
- 125.Deloose E, Corsetti M, Van Oudenhove L, Depoortere I, Tack J. Intragastric infusion of the bitter tastant quinine suppresses hormone release and antral motility during the fasting state in healthy female volunteers. Neurogastroenterol Motil. 2018;30(1):e13171. doi: 10.1111/nmo.13171. [DOI] [PubMed] [Google Scholar]
- 126.Deloose E, Janssen P, Corsetti M, Biesiekierski J, Masuy I, Rotondo A, et al. Intragastric infusion of denatonium benzoate attenuates interdigestive gastric motility and hunger scores in healthy female volunteers. Am J Clin Nutr. 2017;105(3):580–588. doi: 10.3945/ajcn116138297. [DOI] [PubMed] [Google Scholar]
- 127.Orsmark-Pietras C, James A, Konradsen JR, Nordlund B, Soderhall C, Pulkkinen V, et al. Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur Respir J. 2013;42(1):65–78. doi: 10.1183/09031936.00077712. [DOI] [PubMed] [Google Scholar]
- 128.Xu W, Wu L, Liu S, Liu X, Cao X, Zhou C, et al. Structural basis for strychnine activation of human bitter taste receptor TAS2R46. Sci (New York NY) 2022;377(6612):1298–1304. doi: 10.1126/science.abo1633. [DOI] [PubMed] [Google Scholar]
- 129.Kamato D, Thach L, Bernard R, Chan V, Zheng W, Kaur H, et al. Structure, function, Pharmacology, and therapeutic potential of the G Protein, Galpha/q,11. Front Cardiovasc Med. 2015;2:14. doi: 10.3389/fcvm.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Avet C, Mancini A, Breton B, Le Gouill C, Hauser AS, Normand C, et al. Effector membrane translocation biosensors reveal G protein and betaarrestin coupling profiles of 100 therapeutically relevant GPCRs. Elife. 2022;11. 10.7554/eLife.74101. [DOI] [PMC free article] [PubMed]
- 131.Serezani CH, Ballinger MN, Aronoff DM, Peters-Golden M, Cyclic AMP. Master regulator of innate immune cell function. Am J Respir Cell Mol Biol. 2008;39(2):127–132. doi: 10.1165/rcmb.2008-0091TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Harden TK, Waldo GL, Hicks SN, Sondek J. Mechanism of activation and inactivation of Gq/phospholipase C-beta signaling nodes. Chem Rev. 2011;111(10):6120–6129. doi: 10.1021/cr200209p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Newton AC, Bootman MD, Scott JD. Second messengers. Cold Spring Harb Perspect Biol. 2016;8. 10.1101/cshperspect.a005926. [DOI] [PMC free article] [PubMed]
- 134.Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB. Ca2+/calmodulin-dependent protein kinase II (CaMKII) is activated by calmodulin with two bound calciums. Proc Natl Acad Sci U S A. 2006;103(38):13968–13973. doi: 10.1073/pnas0606433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fromm C, Coso OA, Montaner S, Xu N, Gutkind JS. The small GTP-binding protein rho links G protein-coupled receptors and Galpha12 to the serum response element and to cellular transformation. Proc Natl Acad Sci U S A. 1997;94(19):10098–10103. doi: 10.1073/pnas941910098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dascal N, Kahanovitch U. The roles of Gbetagamma and Galpha in Gating and Regulation of GIRK channels. Int Rev Neurobiol. 2015;123:27–85. doi: 10.1016/bs.irn.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 137.Liu Q, Yang D, Zhuang Y, Croll TI, Cai X, Dai A, et al. Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor. Nat Chem Biol. 2021;17(12):1238–1244. doi: 10.1038/s41589-021-00841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Suno R, Sugita Y, Morimoto K, Takazaki H, Tsujimoto H, Hirose M, et al. Structural insights into the G protein selectivity revealed by the human EP3-G(i) signaling complex. Cell Rep. 2022;40(11):111323. doi: 10.1016/j.celrep.2022.111323. [DOI] [PubMed] [Google Scholar]
- 139.Andersen HR, Nielsen D, Hansen LG. The normal right chest electrocardiogram. J Electrocardiol. 1987;20(1):27–32. doi: 10.1016/0022-0736(87)90004-5. [DOI] [PubMed] [Google Scholar]
- 140.Zhuang Y, Xu P, Mao C, Wang L, Krumm B, Zhou XE, et al. Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Cell. 2021;184(4):931–942. doi: 10.1016/j.cell.2021.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang S, Xu P, Shen DD, Simon IA, Mao C, Tan Y, et al. GPCRs steer G(i) and G(s) selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors. Mol Cell. 2022;82(14):2681–2695. doi: 10.1016/j.molcel.2022.05.031. [DOI] [PubMed] [Google Scholar]
- 142.Teng X, Chen S, Wang Q, Chen Z, Wang X, Huang N, et al. Structural insights into G protein activation by D1 dopamine receptor. Sci Adv. 2022;8(23):eabo4158. doi: 10.1126/sciadvabo4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xiao P, Yan W, Gou L, Zhong YN, Kong L, Wu C, et al. Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell. 2021;184(4):943–956. doi: 10.1016/j.cell.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu P, Huang S, Mao C, Krumm BE, Zhou XE, Tan Y, et al. Structures of the human dopamine D3 receptor-G(i) complexes. Mol Cell. 2021;81(6):1147–1159. doi: 10.1016/j.molcel.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 145.Moro O, Lameh J, Hogger P, Sadee W. Hydrophobic amino acid in the i2 loop plays a key role in receptor-G protein coupling. J Biol Chem. 1993;268(30):22273–22276. doi: 10.1016/S0021-9258(18)41524-4. [DOI] [PubMed] [Google Scholar]
- 146.Kim HR, Xu J, Maeda S, Duc NM, Ahn D, Du Y, et al. Structural mechanism underlying primary and secondary coupling between GPCRs and the Gi/o family. Nat Commun. 2020;11(1):3160. doi: 10.1038/s41467-020-16975-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhang Y, Yang F, Ling S, Lv P, Zhou Y, Fang W, et al. Single-particle cryo-EM structural studies of the beta(2)AR-Gs complex bound with a full agonist formoterol. Cell Discov. 2020;6:45. doi: 10.1038/s41421-020-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mobbs JI, Belousoff MJ, Harikumar KG, Piper SJ, Xu X, Furness SGB, et al. Structures of the human cholecystokinin 1 (CCK1) receptor bound to Gs and Gq mimetic proteins provide insight into mechanisms of G protein selectivity. PLoS Biol. 2021;19(6):e3001295. doi: 10.1371/journal.pbio.3001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Krumm BE, DiBerto JF, Olsen RHJ, Kang HJ, Slocum ST, Zhang S, et al. Neurotensin receptor Allosterism revealed in complex with a biased allosteric modulator. Biochemistry. 2023;62(7):1233–1248. doi: 10.1021/acsbiochem3c00029. [DOI] [PubMed] [Google Scholar]
- 150.Huang SM, Xiong MY, Liu L, Mu J, Wang MW, Jia YL, et al. Single hormone or synthetic agonist induces G(s)/G(i) coupling selectivity of EP receptors via distinct binding modes and propagating paths. Proc Natl Acad Sci U S A. 2023;120(30):e2216329120. doi: 10.1073/pnas.2216329120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yokoyama U, Iwatsubo K, Umemura M, Fujita T, Ishikawa Y. The prostanoid EP4 receptor and its signaling pathway. Pharmacol Rev. 2013;65(3):1010–1052. doi: 10.1124/pr112007195. [DOI] [PubMed] [Google Scholar]
- 152.Mao C, Xiao P, Tao XN, Qin J, He QT, Zhang C, et al. Unsaturated bond recognition leads to biased signal in a fatty acid receptor. Sci (New York NY) 2023;380(6640):eadd6220. doi: 10.1126/science.add6220. [DOI] [PubMed] [Google Scholar]
- 153.Peterson YK, Luttrell LM. The diverse roles of Arrestin scaffolds in G protein-coupled receptor signaling. Pharmacol Rev. 2017;69(3):256–297. doi: 10.1124/pr.116.013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wess J, Oteng AB, Rivera-Gonzalez O, Gurevich EV, Gurevich VV. Beta-arrestins: structure, function, physiology, and pharmacological perspectives. Pharmacol Rev. 2023;75(5):854–884. doi: 10.1124/pharmrev.121.000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pakharukova N, Masoudi A, Pani B, Staus DP, Lefkowitz RJ. Allosteric activation of proto-oncogene kinase src by GPCR-beta-arrestin complexes. J Biol Chem. 2020;295(49):16773–16784. doi: 10.1074/jbcRA120015400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zang Y, Kahsai AW, Pakharukova N, Huang LY, Lefkowitz RJ. The GPCR-beta-arrestin complex allosterically activates C-Raf by binding its amino terminus. J Biol Chem. 2021;297(6):101369. doi: 10.1016/j.jbc.2021.101369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huang W, Masureel M, Qu Q, Janetzko J, Inoue A, Kato HE, et al. Structure of the neurotensin receptor 1 in complex with beta-arrestin 1. Nature. 2020;579(7798):303–308. doi: 10.1038/s41586-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hilger D, Masureel M, Kobilka BK. Structure and dynamics of GPCR signaling complexes. Nat Struct Mol Biol. 2018;25(1):4–12. doi: 10.1038/s41594-017-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shukla AK, Manglik A, Kruse AC, Xiao K, Reis RI, Tseng WC, et al. Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature. 2013;497(7447):137–141. doi: 10.1038/nature12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523(7562):561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen K, Zhang C, Lin S, Yan X, Cai H, Yi C, et al. Tail engagement of arrestin at the glucagon receptor. Nature. 2023;620(7975):904–910. doi: 10.1038/s41586-023-06420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen Q, Plasencia M, Li Z, Mukherjee S, Patra D, Chen CL, et al. Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1. Nature. 2021;595(7868):600–605. doi: 10.1038/s41586-021-03721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Duan J, Liu H, Zhao F, Yuan Q, Ji Y, Cai X, et al. GPCR activation and GRK2 assembly by a biased intracellular agonist. Nature. 2023;620(7974):676–81. doi: 10.1038/s41586-023-06395-9. [DOI] [PubMed] [Google Scholar]
- 164.Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, et al. Identification of Phosphorylation codes for Arrestin Recruitment by G protein-coupled receptors. Cell. 2017;170(3):457–469. doi: 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Asher WB, Terry DS, Gregorio GGA, Kahsai AW, Borgia A, Xie B, et al. GPCR-mediated beta-arrestin activation deconvoluted with single-molecule precision. Cell. 2022;185(10):1661–1675. doi: 10.1016/j.cell.2022.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gurevich VV, Gurevich EV. The structural basis of the arrestin binding to GPCRs. Mol Cell Endocrinol. 2019;484:34–41. doi: 10.1016/j.mce.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Isaikina P, Petrovic I, Jakob RP, Sarma P, Ranjan A, Baruah M, et al. A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes. Mol Cell. 2023;83(12):2108–2121. doi: 10.1016/j.molcel.2023.05.002. [DOI] [PubMed] [Google Scholar]
- 168.Maharana J, Sarma P, Yadav MK, Saha S, Singh V, Saha S, et al. Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation. Mol Cell. 2023;83(12):2091–2107. doi: 10.1016/j.molcel.2023.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dwivedi-Agnihotri H, Chaturvedi M, Baidya M, Stepniewski TM, Pandey S, Maharana J, et al. Distinct phosphorylation sites in a prototypical GPCR differently orchestrate beta-arrestin interaction, trafficking, and signaling. Sci Adv. 2020;6(37). 10.1126/sciadv.abb8368. [DOI] [PMC free article] [PubMed]
- 170.Min K, Yoon HJ, Park JY, Baidya M, Dwivedi-Agnihotri H, Maharana J, et al. Crystal structure of beta-arrestin 2 in Complex with CXCR7 phosphopeptide. Structure. 2020;28(9):1014–1023. doi: 10.1016/j.str.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cao C, Barros-Alvarez X, Zhang S, Kim K, Damgen MA, Panova O, et al. Signaling snapshots of a serotonin receptor activated by the prototypical psychedelic LSD. Neuron. 2022;110(19):3154–3167. doi: 10.1016/j.neuron.2022.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Staus DP, Hu H, Robertson MJ, Kleinhenz ALW, Wingler LM, Capel WD, et al. Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc. Nature. 2020;579(7798):297–302. doi: 10.1038/s41586-020-1954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yin W, Li Z, Jin M, Yin YL, de Waal PW, Pal K, et al. A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 2019;29(12):971–83. doi: 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee Y, Warne T, Nehme R, Pandey S, Dwivedi-Agnihotri H, Chaturvedi M, et al. Molecular basis of beta-arrestin coupling to formoterol-bound beta(1)-adrenoceptor. Nature. 2020;583(7818):862–866. doi: 10.1038/s41586-020-2419-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bous J, Fouillen A, Orcel H, Trapani S, Cong X, Fontanel S, et al. Structure of the vasopressin hormone-V2 receptor-beta-arrestin1 ternary complex. Sci Adv. 2022;8(35):eabo7761. doi: 10.1126/sciadvabo7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nguyen AH, Thomsen ARB, Cahill TJ, 3rd, Huang R, Huang LY, Marcink T, et al. Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex. Nat Struct Mol Biol. 2019;26(12):1123–1131. doi: 10.1038/s41594-019-0330-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cahill TJ, 3rd, Thomsen AR, Tarrasch JT, Plouffe B, Nguyen AH, Yang F, et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc Natl Acad Sci U S A. 2017;114(10):2562–2567. doi: 10.1073/pnas1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kumari P, Srivastava A, Ghosh E, Ranjan R, Dogra S, Yadav PN, et al. Core engagement with beta-arrestin is dispensable for agonist-induced vasopressin receptor endocytosis and ERK activation. Mol Biol Cell. 2017;28(8):1003. doi: 10.1091/mbc.E16-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tobin AB. G-protein-coupled receptor phosphorylation: where, when and by whom. Br J Pharmacol. 2008;153(Suppl 1):167–176. doi: 10.1038/sj.bjp.0707662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Latorraca NR, Masureel M, Hollingsworth SA, Heydenreich FM, Suomivuori CM, Brinton C, et al. How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell. 2020;183(7):1813–1825. doi: 10.1016/j.cell.2020.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang F, Yu X, Liu C, Qu CX, Gong Z, Liu HD, et al. Phospho-selective mechanisms of arrestin conformations and functions revealed by unnatural amino acid incorporation and (19)F-NMR. Nat Commun. 2015;6:8202. doi: 10.1038/ncomms9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.He QT, Xiao P, Huang SM, Jia YL, Zhu ZL, Lin JY, et al. Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2. Nat Commun. 2021;12(1):2396. doi: 10.1038/s41467-021-22731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Namkung Y, Dipace C, Javitch JA, Sibley DR. G protein-coupled receptor kinase-mediated phosphorylation regulates post-endocytic trafficking of the D2 dopamine receptor. J Biol Chem. 2009;284(22):15038–15051. doi: 10.1074/jbcM900388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jeong E, Kim Y, Jeong J, Cho Y. Structure of the class C orphan GPCR GPR158 in complex with RGS7-Gbeta5. Nat Commun. 2021;12(1):6805. doi: 10.1038/s41467-021-27147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Patil DN, Singh S, Laboute T, Strutzenberg TS, Qiu X, Wu D, et al. Cryo-EM structure of human GPR158 receptor coupled to the RGS7-Gbeta5 signaling complex. Science (New York, NY) 2022;375(6576):86–91. doi: 10.1126/science.abl4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sulon SM, Benovic JL. Targeting G protein-coupled receptor kinases (GRKs) to G protein-coupled receptors. Curr Opin Endocr Metab Res. 2021;16:56–65. doi: 10.1016/j.coemr.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chaudhary PK, Kim S. The GRKs Reactome: Role in Cell Biology and Pathology. Int J Mol Sci. 2021;22(7):3375. doi: 10.3390/ijms22073375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhai R, Snyder J, Montgomery S, Sato PY. Double life: how GRK2 and beta-arrestin signaling participate in Diseases. Cell Signal. 2022;94:110333. doi: 10.1016/j.cellsig.2022.110333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gurevich VV, Gurevich EV. GPCR Signaling Regulation: the role of GRKs and arrestins. Front Pharmacol. 2019 doi: 10.3389/fphar.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Inagaki S, Ghirlando R, Vishnivetskiy SA, Homan KT, White JF, Tesmer JJ, et al. G protein-coupled receptor kinase 2 (GRK2) and 5 (GRK5) exhibit selective phosphorylation of the neurotensin receptor in Vitro. Biochemistry. 2015;54(28):4320–4329. doi: 10.1021/acsbiochem5b00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kawakami K, Yanagawa M, Hiratsuka S, Yoshida M, Ono Y, Hiroshima M, et al. Heterotrimeric Gq proteins act as a switch for GRK5/6 selectivity underlying beta-arrestin transducer bias. Nat Commun. 2022;13(1):487. doi: 10.1038/s41467-022-28056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fan L, Tan L, Chen Z, Qi J, Nie F, Luo Z, et al. Haloperidol bound D(2) dopamine receptor structure inspired the discovery of subtype selective ligands. Nat Commun. 2020;11(1):1074. doi: 10.1038/s41467-020-14884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wang S, Che T, Levit A, Shoichet BK, Wacker D, Roth BL. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature. 2018;555(7695):269–273. doi: 10.1038/nature25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science (New York, NY) 2010;330(6007):1091–1095. doi: 10.1126/science1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang S, Wacker D, Levit A, Che T, Betz RM, McCorvy JD, et al. D(4) dopamine receptor high-resolution structures enable the discovery of selective agonists. Sci (New York NY) 2017;358(6361):381–386. doi: 10.1126/scienceaan5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xu X, Kaindl J, Clark MJ, Hubner H, Hirata K, Sunahara RK, et al. Binding pathway determines norepinephrine selectivity for the human beta(1)AR over beta(2)AR. Cell Res. 2021;31(5):569–579. doi: 10.1038/s41422-020-00424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Feng Y, Zhao C, Deng Y, Wang H, Ma L, Liu S, et al. Mechanism of activation and biased signaling in complement receptor C5aR1. Cell Res. 2023;33(4):312–324. doi: 10.1038/s41422-023-00779-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Soergel DG, Subach RA, Burnham N, Lark MW, James IE, Sadler BM, et al. Biased agonism of the mu-opioid receptor by TRV130 increases analgesia and reduces on-target adverse effects versus morphine: a randomized, double-blind, placebo-controlled, crossover study in healthy volunteers. Pain. 2014;155(9):1829–1835. doi: 10.1016/jpain201406011. [DOI] [PubMed] [Google Scholar]
- 199.Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature. 2016;537(7619):185–190. doi: 10.1038/nature19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Packer M, Bristow MR, Cohn JN, Colucci WS, Fowler MB, Gilbert EM, et al. The effect of carvedilol on morbidity and mortality in patients with chronic heart failure. U.S. carvedilol heart failure study group. N Engl J Med. 1996;334(21):1349–1355. doi: 10.1056/NEJM199605233342101. [DOI] [PubMed] [Google Scholar]
- 201.Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, et al. Selectively engaging beta-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther. 2010;335(3):572–579. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- 202.Pandey S, Maharana J, Li XX, Woodruff TM, Shukla AK. Emerging insights into the structure and function of complement C5a receptors. Trends Biochem Sci. 2020;45(8):693–705. doi: 10.1016/j.tibs.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 203.Casado V, Casado-Anguera V. What are the current trends in G protein-coupled receptor targeted drug discovery? Expert opinion on drug discovery. 2023;18(8):815–20. doi: 10.1080/17460441.2023.2216014. [DOI] [PubMed] [Google Scholar]
- 204.Lee A. Avacopan: First Approval. Drugs. 2022;82(1):79–85. doi: 10.1007/s40265-021-01643-6. [DOI] [PubMed] [Google Scholar]
- 205.Cook AE, Mistry SN, Gregory KJ, Furness SG, Sexton PM, Scammells PJ, et al. Biased allosteric modulation at the CaS receptor engendered by structurally diverse calcimimetics. Br J Pharmacol. 2015;172(1):185–200. doi: 10.1111/bph.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Davey AE, Leach K, Valant C, Conigrave AD, Sexton PM, Christopoulos A. Positive and negative allosteric modulators promote biased signaling at the calcium-sensing receptor. Endocrinology. 2012;153(3):1232–1241. doi: 10.1210/en2011-1426. [DOI] [PubMed] [Google Scholar]
- 207.Leach K, Gregory KJ, Kufareva I, Khajehali E, Cook AE, Abagyan R, et al. Towards a structural understanding of allosteric Drugs at the human calcium-sensing receptor. Cell Res. 2016;26(5):574–592. doi: 10.1038/cr.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li XX, Lee JD, Massey NL, Guan C, Robertson AAB, Clark RJ, et al. Pharmacological characterisation of small molecule C5aR1 inhibitors in human cells reveals biased activities for signalling and function. Biochem Pharmacol. 2020;180:114156. doi: 10.1016/j.bcp.2020.114156. [DOI] [PubMed] [Google Scholar]
- 209.Hoffmann K, Lutz DA, Strassburger J, Baqi Y, Muller CE, von Kugelgen I. Competitive mode and site of interaction of ticagrelor at the human platelet P2Y12 -receptor. J Thromb Haemost. 2014;12(11):1898–1905. doi: 10.1111/jth.12719. [DOI] [PubMed] [Google Scholar]
- 210.Krusek J, Zemkova H. Effect of ivermectin on gamma-aminobutyric acid-induced chloride currents in mouse hippocampal embryonic neurones. Eur J Pharmacol. 1994;259(2):121–128. doi: 10.1016/0014-2999(94)90500-2. [DOI] [PubMed] [Google Scholar]
- 211.Sliwoski G, Schubert M, Stichel J, Weaver D, Beck-Sickinger AG, Meiler J. Discovery of small-molecule modulators of the human Y4 receptor. PLoS ONE. 2016;11(6):e0157146. doi: 10.1371/journal.pone.0157146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wold EA, Chen J, Cunningham KA, Zhou J. Allosteric modulation of class A GPCRs: targets, agents, and emerging concepts. J Med Chem. 2019;62(1):88–127. doi: 10.1021/acs.jmedchem.8b00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Walters MJ, Wang Y, Lai N, Baumgart T, Zhao BN, Dairaghi DJ, et al. Characterization of CCX282-B, an orally bioavailable antagonist of the CCR9 chemokine receptor, for treatment of inflammatory bowel Disease. J Pharmacol Exp Ther. 2010;335(1):61–69. doi: 10.1124/jpet.110.169714. [DOI] [PubMed] [Google Scholar]
- 214.Vranesic I, Ofner S, Flor PJ, Bilbe G, Bouhelal R, Enz A, et al. AFQ056/mavoglurant, a novel clinically effective mGluR5 antagonist: identification, SAR and pharmacological characterization. Bioorg Med Chem. 2014;22(21):5790–5803. doi: 10.1016/j.bmc.2014.09.033. [DOI] [PubMed] [Google Scholar]
- 215.Chae E, Shin YJ, Ryu EJ, Ji MK, Ryune Cho N, Lee KH, et al. Discovery of biological evaluation of pyrazole/imidazole amides as mGlu5 receptor negative allosteric modulators. Bioorg Med Chem Lett. 2013;23(7):2134–2139. doi: 10.1016/jbmcl201301116. [DOI] [PubMed] [Google Scholar]
- 216.Jaeschke G, Kolczewski S, Spooren W, Vieira E, Bitter-Stoll N, Boissin P, et al. Metabotropic glutamate receptor 5 negative allosteric modulators: discovery of 2-chloro-4-[1-(4-fluorophenyl)-2,5-dimethyl-1H-imidazol-4-ylethynyl]pyridine (basimglurant, RO4917523), a promising novel medicine for psychiatric Diseases. J Med Chem. 2015;58(3):1358–1371. doi: 10.1021/jm501642c. [DOI] [PubMed] [Google Scholar]
- 217.Hall A, Provins L, Valade A. Novel strategies to activate the dopamine D(1) receptor: recent advances in Orthosteric Agonism and positive allosteric modulation. J Med Chem. 2019;62(1):128–140. doi: 10.1021/acs.jmedchem.8b01767. [DOI] [PubMed] [Google Scholar]
- 218.Hao J, Beck JP, Schaus JM, Krushinski JH, Chen Q, Beadle CD, et al. Synthesis and pharmacological characterization of 2-(2,6-Dichlorophenyl)-1-((1S,3R)-5-(3-hydroxy-3-methylbutyl)-3-(hydroxymethyl)-1-methyl-3,4-dihydroisoquinolin-2(1H)-yl)ethan-1-one (LY3154207), a potent, Subtype Selective, and orally available positive allosteric modulator of the human dopamine D1 receptor. J Med Chem. 2019;62(19):8711–8732. doi: 10.1021/acs.jmedchem.9b01234. [DOI] [PubMed] [Google Scholar]
- 219.Marin JC, Goadsby PJ. Glutamatergic fine tuning with ADX-10059: a novel therapeutic approach for migraine? Expert Opin Investi Drugs. 2010;19(4):555–61. doi: 10.1517/13543781003691832. [DOI] [PubMed] [Google Scholar]
- 220.Justinova Z, Panlilio LV, Secci ME, Redhi GH, Schindler CW, Cross AJ, et al. The Novel Metabotropic Glutamate receptor 2 positive Allosteric Modulator, AZD8529, decreases Nicotine Self-Administration and Relapse in Squirrel monkeys. Biol Psychiatry. 2015;78(7):452–462. doi: 10.1016/j.biopsych.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lavreysen H, Ahnaou A, Drinkenburg W, Langlois X, Mackie C, Pype S, et al. Pharmacological and pharmacokinetic properties of JNJ-40411813, a positive allosteric modulator of the mGlu2 receptor. Pharmacol Res Perspect. 2015;3(1):e00096. doi: 10.1002/prp2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Beshore DC, Di Marco CN, Chang RK, Greshock TJ, Ma L, Wittmann M, et al. MK-7622: A First-in-Class M(1) Positive Allosteric Modulator Development Candidate. ACS Med Chem Lett. 2018;9(7):652–6. doi: 10.1021/acsmedchemlett.8b00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mandai T, Sako Y, Kurimoto E, Shimizu Y, Nakamura M, Fushimi M, et al. T-495, a novel low cooperative M(1) receptor positive allosteric modulator, improves memory deficits associated with cholinergic dysfunction and is characterized by low gastrointestinal side effect risk. Pharmacol Res Perspect. 2020;8(1):e00560. doi: 10.1002/prp2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Uslaner JM, Kuduk SD, Wittmann M, Lange HS, Fox SV, Min C, et al. Preclinical to human translational pharmacology of the Novel M(1) positive Allosteric Modulator MK-7622. J Pharmacol Exp Ther. 2018;365(3):556–566. doi: 10.1124/jpet.117.245894. [DOI] [PubMed] [Google Scholar]
- 225.Okimoto R, Ino K, Ishizu K, Takamatsu H, Sakamoto K, Yuyama H, et al. Potentiation of Muscarinic M(3) receptor activation through a new allosteric site with a novel positive allosteric modulator ASP8302. J Pharmacol Exp Ther. 2021;379(1):64–73. doi: 10.1124/jpet.121.000709. [DOI] [PubMed] [Google Scholar]
- 226.Krystal JH, Kane JM, Correll CU, Walling DP, Leoni M, Duvvuri S, et al. Emraclidine, a novel positive allosteric modulator of cholinergic M4 receptors, for the treatment of schizophrenia: a two-part, randomised, double-blind, placebo-controlled, phase 1b trial. Lancet. 2022;400(10369):2210–2220. doi: 10.1016/S0140-6736(22)01990-0. [DOI] [PubMed] [Google Scholar]
- 227.Pan HL, Xu Z, Leung E, Eisenach JC. Allosteric adenosine modulation to reduce allodynia. Anesthesiology. 2001;95(2):416–420. doi: 10.1097/00000542-200108000-00025. [DOI] [PubMed] [Google Scholar]
- 228.Christopher JA, Aves SJ, Bennett KA, Dore AS, Errey JC, Jazayeri A, et al. Fragment and structure-based drug Discovery for a class C GPCR: Discovery of the mGlu5 negative allosteric modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile) J Med Chem. 2015;58(16):6653–6664. doi: 10.1021/acsjmedchem5b00892. [DOI] [PubMed] [Google Scholar]
- 229.Andres JI, Alcazar J, Cid JM, De Angelis M, Iturrino L, Langlois X, et al. Synthesis, evaluation, and radiolabeling of new potent positive allosteric modulators of the metabotropic glutamate receptor 2 as potential tracers for positron emission tomography imaging. J Med Chem. 2012;55(20):8685–8699. doi: 10.1021/jm300912k. [DOI] [PubMed] [Google Scholar]
- 230.Sako Y, Kurimoto E, Mandai T, Suzuki A, Tanaka M, Suzuki M, et al. TAK-071, a novel M(1) positive allosteric modulator with low cooperativity, improves cognitive function in rodents with few cholinergic side effects. Neuropsychopharmacology. 2019;44(5):950–960. doi: 10.1038/s41386-018-0168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tong L, Li W, Lo MM, Gao X, Wai JM, Rudd M, et al. Discovery of [(11)C]MK-6884: a Positron Emission Tomography (PET) imaging Agent for the study of M4Muscarinic receptor positive allosteric modulators (PAMs) in neurodegenerative Diseases. J Med Chem. 2020;63(5):2411–2425. doi: 10.1021/acsjmedchem9b01406. [DOI] [PubMed] [Google Scholar]
- 232.Amato G, Khan NS, Maitra R. A patent update on cannabinoid receptor 1 antagonists (2015–2018) Expert Opin Ther Pat. 2019;29(4):261. doi: 10.1080/13543776.2011597851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gao ZG, Jacobson KA. Allosteric modulation and functional selectivity of G protein-coupled receptors. Drug Discov Today Technol. 2013;10(2):e237–243. doi: 10.1016/j.ddtec.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Shen S, Zhao C, Wu C, Sun S, Li Z, Yan W, et al. Allosteric modulation of G protein-coupled receptor signaling. Front Endocrinol (Lausanne) 2023 doi: 10.3389/fendo.2023.1137604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Wootten D, Christopoulos A, Sexton PM. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat Rev Drug Discovery. 2013;12(8):630–644. doi: 10.1038/nrd4052. [DOI] [PubMed] [Google Scholar]
- 236.Wu Y, Tong J, Ding K, Zhou Q, Zhao S. GPCR Allosteric Modulator Discovery. Adv Exp Med Biol. 2019;1163:225–251. doi: 10.1007/978-981-13-8719-7_10. [DOI] [PubMed] [Google Scholar]
- 237.Jakubik J, Randakova A, Chetverikov N, El-Fakahany EE, Dolezal V. The operational model of allosteric modulation of pharmacological agonism. Sci Rep. 2020;10(1):14421. doi: 10.1038/s41598-020-71228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gnacadja G. A method to calculate binding equilibrium concentrations in the allosteric ternary complex model that supports ligand depletion. Math Biosci. 2011;232(2):135–141. doi: 10.1016/j.mbs.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 239.Diaz O, Martin V, Renault P, Romero D, Guillamon A, Giraldo J. Allosteric binding cooperativity in a kinetic context. Drug Discov Today. 2023;28(2):103441. doi: 10.1016/j.drudis.2022.103441. [DOI] [PubMed] [Google Scholar]
- 240.Zhang R, Kavana M. Quantitative analysis of receptor allosterism and its implication for drug discovery. Expert Opin Drug Discov. 2015;10(7):763–780. doi: 10.1517/17460441.2015.1041498. [DOI] [PubMed] [Google Scholar]
- 241.Ahn KH, Mahmoud MM, Kendall DA. Allosteric modulator ORG27569 induces CB1 cannabinoid receptor high affinity agonist binding state, receptor internalization, and Gi protein-independent ERK1/2 kinase activation. J Biol Chem. 2012;287(15):12070–12082. doi: 10.1074/jbc.M111.316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lu D, Immadi SS, Wu Z, Kendall DA. Translational potential of allosteric modulators targeting the cannabinoid CB(1) receptor. Acta Pharmacol Sin. 2019;40(3):324–335. doi: 10.1038/s41401-018-0164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yang X, Wang X, Xu Z, Wu C, Zhou Y, Wang Y, et al. Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1. Nat Chem Biol. 2022;18(8):831–840. doi: 10.1038/s41589-022-01038-y. [DOI] [PubMed] [Google Scholar]
- 244.Conn PJ, Lindsley CW, Meiler J, Niswender CM. Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat Rev Drug Discovery. 2014;13(9):692–708. doi: 10.1038/nrd4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Fasciani I, Petragnano F, Aloisi G, Marampon F, Carli M, Scarselli M, et al. Allosteric modulators of G protein-coupled dopamine and serotonin receptors: a New Class of atypical antipsychotics. Pharmaceuticals (Basel) 2020;13(11):388. doi: 10.3390/ph13110388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Zhou Y, Chun A, Gogliotti RD, Dawson ES, Vinson PN, Niswender CM, et al. Pure Positive Allosteric Modulators (PAMs) of mGlu5 with Competitive MPEP-Site Interaction. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda: National Center for Biotechnology Information (US); 2010. https://www.ncbi.nlm.nih.gov/books/NBK143535. Accessed 31 Oct 2023. [PubMed]
- 247.Foster DJ, Conn PJ. Allosteric modulation of GPCRs: New insights and potential utility for treatment of Schizophrenia and other CNS disorders. Neuron. 2017;94(3):431–446. doi: 10.1016/j.neuron.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Satsu H, Schaeffer MT, Guerrero M, Saldana A, Eberhart C, Hodder P, et al. A sphingosine 1-phosphate receptor 2 selective allosteric agonist. Bioorg Med Chem. 2013;21(17):5373–5382. doi: 10.1016/j.bmc.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Slosky LM, Caron MG, Barak LS. Biased allosteric modulators: New frontiers in GPCR Drug Discovery. Trends Pharmacol Sci. 2021;42(4):283–299. doi: 10.1016/j.tips.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Slosky LM, Bai Y, Toth K, Ray C, Rochelle LK, Badea A, et al. beta-arrestin-biased Allosteric Modulator of NTSR1 selectively attenuates addictive behaviors. Cell. 2020;181(6):1364–1379. doi: 10.1016/j.cell.2020.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zhao C, Wang H, Liu Y, Cheng L, Wang B, Tian X, et al. Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation. Mol Cell. 2023;83(17):3171–3187. doi: 10.1016/j.molcel.2023.07.030. [DOI] [PubMed] [Google Scholar]
- 252.Cheng L, Sun S, Wang H, Zhao C, Tian X, Liu Y, et al. Orthosteric ligand selectivity and allosteric probe dependence at Hydroxycarboxylic acid receptor HCAR2. Signal Transduct Target Ther. 2023;8(1):364. doi: 10.1038/s41392-023-01625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Medina-Franco JL, Giulianotti MA, Welmaker GS, Houghten RA. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov Today. 2013;18(9–10):495–501. doi: 10.1016/j.drudis.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Anighoro A, Bajorath J, Rastelli G. Polypharmacology: challenges and opportunities in drug discovery. J Med Chem. 2014;57(19):7874–7887. doi: 10.1021/jm5006463. [DOI] [PubMed] [Google Scholar]
- 255.Lee Y, Basith S, Choi S. Recent advances in structure-based Drug Design Targeting Class A G protein-coupled receptors utilizing Crystal structures and computational simulations. J Med Chem. 2018;61(1):1–46. doi: 10.1021/acs.jmedchem.6b01453. [DOI] [PubMed] [Google Scholar]
- 256.Reddy AS, Zhang S. Polypharmacology: drug discovery for the future. Expert Rev Clin Pharmacol. 2013;6(1):41–47. doi: 10.1586/ecp.12.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Syed YY. Tirzepatide: first approval. Drugs. 2022;82(11):1213–20. doi: 10.1007/s40265-022-01746-8. [DOI] [PubMed] [Google Scholar]
- 258.Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, et al. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022;119(13):e2116506119. doi: 10.1073/pnas.2116506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rosenstock J, Wysham C, Frias JP, Kaneko S, Lee CJ, Fernandez Lando L, et al. Efficacy and safety of a novel dual GIP and GLP-1 receptor agonist tirzepatide in patients with type 2 Diabetes (SURPASS-1): a double-blind, randomised, phase 3 trial. Lancet. 2021;398(10295):143–155. doi: 10.1016/S0140-6736(21)01324-6. [DOI] [PubMed] [Google Scholar]
- 260.Frias JP, Davies MJ, Rosenstock J, Perez Manghi FC, Fernandez Lando L, Bergman BK, et al. Tirzepatide versus Semaglutide once Weekly in patients with type 2 Diabetes. N Engl J Med. 2021;385(6):503–515. doi: 10.1056/NEJMoa2107519. [DOI] [PubMed] [Google Scholar]
- 261.Jastreboff AM, Aronne LJ, Ahmad NN, Wharton S, Connery L, Alves B, et al. Tirzepatide once Weekly for the treatment of obesity. N Engl J Med. 2022;387(3):205–216. doi: 10.1056/NEJMoa2206038. [DOI] [PubMed] [Google Scholar]
- 262.Hartman ML, Sanyal AJ, Loomba R, Wilson JM, Nikooienejad A, Bray R, et al. Effects of Novel Dual GIP and GLP-1 receptor agonist tirzepatide on biomarkers of nonalcoholic steatohepatitis in patients with type 2 Diabetes. Diabetes Care. 2020;43(6):1352–1355. doi: 10.2337/dc19-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Motte S, McEntee K, Naeije R. Endothelin receptor antagonists. Pharmacol Ther. 2006;110(3):386–414. doi: 10.1016/jpharmthera200508012. [DOI] [PubMed] [Google Scholar]
- 264.Rubin LJ, Badesch DB, Barst RJ, Galie N, Black CM, Keogh A, et al. Bosentan therapy for pulmonary arterial Hypertension. N Engl J Med. 2002;346(12):896–903. doi: 10.1056/NEJMoa012212. [DOI] [PubMed] [Google Scholar]
- 265.Davenport AP, Kuc RE, Southan C, Maguire JJ. New Drugs and emerging therapeutic targets in the endothelin signaling pathway and prospects for personalized precision medicine. Physiol Res. 2018;67(Suppl 1):37–S54. doi: 10.33549/physiolres.933872. [DOI] [PubMed] [Google Scholar]
- 266.Miura S, Karnik SS, Saku K. Review: angiotensin II type 1 receptor blockers: class effects versus molecular effects. J Renin Angiotensin Aldosterone Syst. 2011;12(1):1–7. doi: 10.1177/1470320310370852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Murugesan N, Gu Z, Stein PD, Bisaha S, Spergel S, Girotra R, et al. Biphenylsulfonamide endothelin antagonists: structure-activity relationships of a series of mono- and disubstituted analogues and pharmacology of the orally active endothelin antagonist 2’-amino-N- (3,4-dimethyl-5-isoxazolyl)-4’-(2-methylpropyl)[1, 1’-biphenyl]-2-sulfonamide (BMS-187308) J Med Chem. 1998;41(26):5198–5218. doi: 10.1021/jm970872k. [DOI] [PubMed] [Google Scholar]
- 268.Davidson M, Saoud J, Staner C, Noel N, Werner S, Luthringer E, et al. Efficacy and safety of Roluperidone for the treatment of negative symptoms of Schizophrenia. Schizophr Bull. 2022;48(3):609–619. doi: 10.1093/schbul/sbac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Rabinowitz J, Staner C, Saoud J, Weiser M, Kuchibhatla R, Davidson M, et al. Long-term effects of Roluperidone on negative symptoms of schizophrenia. Schizophr Res. 2023;255:9–13. doi: 10.1016/j.schres.2023.03.028. [DOI] [PubMed] [Google Scholar]
- 270.Peng Y, McCorvy JD, Harpsoe K, Lansu K, Yuan S, Popov P, et al. 5-HT(2 C) receptor structures reveal the structural basis of GPCR Polypharmacology. Cell. 2018;172(4):719–730. doi: 10.1016/j.cell.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Meyler WJ. Side effects of ergotamine. Cephalalgia. 1996;16(1):5–10. doi: 10.1046/j.1468-2982.1996.1601005.x. [DOI] [PubMed] [Google Scholar]
- 272.Tfelt-Hansen P, Saxena PR, Dahlof C, Pascual J, Lainez M, Henry P, et al. Ergotamine in the acute treatment of migraine: a review and European consensus. Brain. 2000;123(Pt 1):9–18. doi: 10.1093/brain/123.1.9. [DOI] [PubMed] [Google Scholar]
- 273.Frankel JS, Schwartz TL. Brexpiprazole and cariprazine: distinguishing two new atypical antipsychotics from the original dopamine stabilizer aripiprazole. Ther Adv Psychopharmacol. 2017;7(1):29–41. doi: 10.1177/2045125316672136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Knerr PJ, Mowery SA, Douros JD, Premdjee B, Hjollund KR, He Y, et al. Next generation GLP-1/GIP/glucagon triple agonists normalize body weight in obese mice. Mol Metabolism. 2022;63:101533. doi: 10.1016/j.molmet.2022.101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Abdelmalek MF, Suzuki A, Sanchez W, Lawitz E, Filozof C, Cho H, et al. A phase 2, adaptive randomized, double-blind, placebo-controlled, multicenter, 52-week study of HM15211 in patients with biopsy-confirmed non-alcoholic steatohepatitis - study design and rationale of HM-TRIA-201 study. Contemp Clin Trials. 2023;130:107176. doi: 10.1016/j.cct.2023.107176. [DOI] [PubMed] [Google Scholar]
- 276.Zhao F, Zhou Q, Cong Z, Hang K, Zou X, Zhang C, et al. Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors. Nat Commun. 2022;13(1):1057. doi: 10.1038/s41467-022-28683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jastreboff AM, Kaplan LM, Frias JP, Wu Q, Du Y, Gurbuz S, et al. Triple-hormone-receptor agonist retatrutide for obesity - A phase 2 trial. N Engl J Med. 2023;389(6):514–526. doi: 10.1056/NEJMoa2301972. [DOI] [PubMed] [Google Scholar]
- 278.Rosenstock J, Frias J, Jastreboff AM, Du Y, Lou J, Gurbuz S, et al. Retatrutide, a GIP, GLP-1 and glucagon receptor agonist, for people with type 2 Diabetes: a randomised, double-blind, placebo and active-controlled, parallel-group, phase 2 trial conducted in the USA. Lancet. 2023;402(10401):529–544. doi: 10.1016/S0140-6736(23)01053-X. [DOI] [PubMed] [Google Scholar]
- 279.Makhoba XH, Viegas C, Jr, Mosa RA, Viegas FPD, Pooe OJ. Potential impact of the Multi-target Drug Approach in the treatment of some Complex Diseases. Drug Des Devel Ther. 2020;14:3235–3249. doi: 10.2147/DDDTS257494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Morphy R, Rankovic Z. Designed multiple ligands. An emerging drug discovery paradigm. J Med Chem. 2005;48(21):6523–6543. doi: 10.1021/jm058225d. [DOI] [PubMed] [Google Scholar]
- 281.Coskun T, Sloop KW, Loghin C, Alsina-Fernandez J, Urva S, Bokvist KB, et al. LY3298176, a novel dual GIP and GLP-1 receptor agonist for the treatment of type 2 Diabetes Mellitus: from discovery to clinical proof of concept. Mol Metabolism. 2018;18:3–14. doi: 10.1016/j.molmet.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Jiang JX, Cao R, Deng WD, Jin F, Dong XW, Zhu Y, et al. Characterization of bencycloquidium bromide, a novel muscarinic M(3) receptor antagonist in guinea pig airways. Eur J Pharmacol. 2011;655(1–3):74–82. doi: 10.1016/j.ejphar.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 283.Sun L, Ding L, Wang Y, Zhou W, Yan Z, Sun W, et al. Pharmacokinetics, safety and tolerability of Bencycloquidium bromide, a novel selective muscarinic M1/M3 receptor antagonist, after single and multiple intranasal doses in healthy Chinese subjects: an open-label, single-center, first-in-human study. Drugs R D. 2012;12(1):17–28. doi: 10.2165/11599330-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Gonzalez-Cabrera PJ, Jo E, Sanna MG, Brown S, Leaf N, Marsolais D, et al. Full pharmacological efficacy of a novel S1P1 agonist that does not require S1P-like headgroup interactions. Mol Pharmacol. 2008;74(5):1308–1318. doi: 10.1124/mol108049783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Scott FL, Clemons B, Brooks J, Brahmachary E, Powell R, Dedman H, et al. Ozanimod (RPC1063) is a potent sphingosine-1-phosphate receptor-1 (S1P1) and receptor-5 (S1P5) agonist with autoimmune disease-modifying activity. Br J Pharmacol. 2016;173(11):1778–1792. doi: 10.1111/bph13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Shimano K, Maeda Y, Kataoka H, Murase M, Mochizuki S, Utsumi H, et al. Amiselimod (MT-1303), a novel sphingosine 1-phosphate receptor-1 functional antagonist, inhibits progress of chronic Colitis induced by transfer of CD4 + CD45RBhigh T cells. PLoS One. 2019;14(12):e0226154. doi: 10.1371/journal.pone.0226154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Fuller RW, Snoddy HD, Robertson DW. Mechanisms of effects of d-fenfluramine on brain serotonin metabolism in rats: uptake inhibition versus release. Pharmacol Biochem Behav. 1988;30(3):715–721. doi: 10.1016/0091-3057(88)90089-5. [DOI] [PubMed] [Google Scholar]
- 288.Griffin A, Hamling KR, Knupp K, Hong S, Lee LP, Baraban SC. Clemizole and modulators of serotonin signalling suppress seizures in Dravet syndrome. Brain. 2017;140(3):669–683. doi: 10.1093/brain/aww342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Martin P, de Witte PAM, Maurice T, Gammaitoni A, Farfel G, Galer B. Fenfluramine acts as a positive modulator of sigma-1 receptors. Epilepsy Behav. 2020;105:106989. doi: 10.1016/j.yebeh.2020.106989. [DOI] [PubMed] [Google Scholar]
- 290.Porter RH, Benwell KR, Lamb H, Malcolm CS, Allen NH, Revell DF, et al. Functional characterization of agonists at recombinant human 5-HT2A, 5-HT2B and 5-HT2C receptors in CHO-K1 cells. Br J Pharmacol. 1999;128(1):13–20. doi: 10.1038/sj.bjp.0702751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Rodriguez-Munoz M, Sanchez-Blazquez P, Garzon J. Fenfluramine diminishes NMDA receptor-mediated seizures via its mixed activity at serotonin 5HT2A and type 1 sigma receptors. Oncotarget. 2018;9(34):23373–23389. doi: 10.18632/oncotarget.25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Schoonjans AS, Ceulemans B. An old drug for a new indication: repurposing fenfluramine from an anorexigen to an antiepileptic drug. Clin Pharmacol Ther. 2019;106(5):929–932. doi: 10.1002/cpt.1469. [DOI] [PubMed] [Google Scholar]
- 293.Sourbron J, Schneider H, Kecskes A, Liu Y, Buening EM, Lagae L, et al. Serotonergic modulation as effective treatment for Dravet Syndrome in a zebrafish mutant model. ACS Chem Neurosci. 2016;7(5):588–598. doi: 10.1021/acschemneuro.5b00342. [DOI] [PubMed] [Google Scholar]
- 294.Sourbron J, Smolders I, de Witte P, Lagae L. Pharmacological analysis of the anti-epileptic mechanisms of fenfluramine in scn1a mutant zebrafish. Front Pharmacol. 2017;8:191. doi: 10.3389/fphar.2017.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Glaenzel U, Jin Y, Nufer R, Li W, Schroer K, Adam-Stitah S, et al. Metabolism and disposition of siponimod, a novel selective S1P(1)/S1P(5) agonist, in healthy volunteers and in vitro identification of human cytochrome P450 enzymes involved in its oxidative metabolism. Drug Metab Dispos. 2018;46(7):1001–1013. doi: 10.1124/dmd.117.079574. [DOI] [PubMed] [Google Scholar]
- 296.Pan S, Gray NS, Gao W, Mi Y, Fan Y, Wang X, et al. Discovery of BAF312 (Siponimod), a potent and selective S1P receptor modulator. ACS Med Chem Lett. 2013;4(3):333–337. doi: 10.1021/ml300396r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Gates E. A complement to care. Nurs Times. 1998;94(9):55–57. [PubMed] [Google Scholar]
- 298.Gergely P, Nuesslein-Hildesheim B, Guerini D, Brinkmann V, Traebert M, Bruns C, et al. The selective sphingosine 1-phosphate receptor modulator BAF312 redirects lymphocyte distribution and has species-specific effects on heart rate. Br J Pharmacol. 2012;167(5):1035–1047. doi: 10.1111/j1476-5381201202061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Urbano M, Guerrero M, Rosen H, Roberts E. Modulators of the sphingosine 1-phosphate receptor 1. Bioorg Med Chem Lett. 2013;23(23):6377–6389. doi: 10.1016/j.bmcl.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Li P, Zhang Q, Robichaud AJ, Lee T, Tomesch J, Yao W, et al. Discovery of a tetracyclic quinoxaline derivative as a potent and orally active multifunctional drug candidate for the treatment of neuropsychiatric and neurological disorders. J Med Chem. 2014;57(6):2670–2682. doi: 10.1021/jm401958n. [DOI] [PubMed] [Google Scholar]
- 301.Snyder GL, Vanover KE, Zhu H, Miller DB, O’Callaghan JP, Tomesch J, et al. Functional profile of a novel modulator of serotonin, dopamine, and glutamate neurotransmission. Psychopharmacology. 2015;232(3):605–621. doi: 10.1007/s00213-014-3704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Hegde SS, Pulido-Rios MT, Luttmann MA, Foley JJ, Hunsberger GE, Steinfeld T, et al. Pharmacological properties of revefenacin (TD-4208), a novel, nebulized long-acting, and lung selective muscarinic antagonist, at human recombinant muscarinic receptors and in rat, guinea pig, and human isolated airway tissues. Pharmacol Res Perspect. 2018;6(3):e00400. doi: 10.1002/prp2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Allen JA, Yost JM, Setola V, Chen X, Sassano MF, Chen M, et al. Discovery of beta-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc Natl Acad Sci U S A. 2011;108(45):18488–18493. doi: 10.1073/pnas.1104807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kroeze WK, Hufeisen SJ, Popadak BA, Renock SM, Steinberg S, Ernsberger P, et al. H1-histamine receptor affinity predicts short-term weight gain for typical and atypical antipsychotic Drugs. Neuropsychopharmacology. 2003;28(3):519–526. doi: 10.1038/sj.npp.1300027. [DOI] [PubMed] [Google Scholar]
- 305.Lawler CP, Prioleau C, Lewis MM, Mak C, Jiang D, Schetz JA, et al. Interactions of the novel antipsychotic aripiprazole (OPC-14597) with dopamine and serotonin receptor subtypes. Neuropsychopharmacology. 1999;20(6):612–627. doi: 10.1016/S0893-133X(98)00099-2. [DOI] [PubMed] [Google Scholar]
- 306.Schetz JA, Benjamin PS, Sibley DR. Nonconserved residues in the second transmembrane-spanning domain of the D(4) dopamine receptor are molecular determinants of D(4)-selective pharmacology. Mol Pharmacol. 2000;57(1):144–152. [PubMed] [Google Scholar]
- 307.Shapiro DA, Renock S, Arrington E, Chiodo LA, Liu LX, Sibley DR, et al. Aripiprazole, a novel atypical antipsychotic drug with a unique and robust pharmacology. Neuropsychopharmacology. 2003;28(8):1400–1411. doi: 10.1038/sjnpp1300203. [DOI] [PubMed] [Google Scholar]
- 308.Zajdel P, Marciniec K, Maslankiewicz A, Grychowska K, Satala G, Duszynska B, et al. Antidepressant and antipsychotic activity of new quinoline- and isoquinoline-sulfonamide analogs of aripiprazole targeting serotonin 5-HT(1)A/5-HT(2)A/5-HT(7) and dopamine D(2)/D(3) receptors. Eur J Med Chem. 2013;60:42–50. doi: 10.1016/jejmech201211042. [DOI] [PubMed] [Google Scholar]
- 309.Maeda K, Sugino H, Akazawa H, Amada N, Shimada J, Futamura T, et al. Brexpiprazole I: in vitro and in vivo characterization of a novel serotonin-dopamine activity modulator. J Pharmacol Exp Ther. 2014;350(3):589–604. doi: 10.1124/jpet.114.213793. [DOI] [PubMed] [Google Scholar]
- 310.Fornaro M, Fusco A, Anastasia A, Cattaneo CI, De Berardis D. Brexpiprazole for treatment-resistant major depressive disorder. Expert Opin Pharmacother. 2019;20(16):1925–1933. doi: 10.1080/14656566.2019.1654457. [DOI] [PubMed] [Google Scholar]
- 311.Kiss B, Horvath A, Nemethy Z, Schmidt E, Laszlovszky I, Bugovics G, et al. Cariprazine (RGH-188), a dopamine D(3) receptor-preferring, D(3)/D(2) dopamine receptor antagonist-partial agonist antipsychotic candidate: in vitro and neurochemical profile. J Pharmacol Exp Ther. 2010;333(1):328–340. doi: 10.1124/jpet.109.160432. [DOI] [PubMed] [Google Scholar]
- 312.Agai-Csongor E, Domany G, Nogradi K, Galambos J, Vago I, Keseru GM, et al. Discovery of cariprazine (RGH-188): a novel antipsychotic acting on dopamine D3/D2 receptors. Bioorg Med Chem Lett. 2012;22(10):3437–3440. doi: 10.1016/jbmcl201203104. [DOI] [PubMed] [Google Scholar]
- 313.Stahl SM. Mechanism of action of cariprazine. CNS Spectr. 2016;21(2):123. doi: 10.1017/S1092852916000043. [DOI] [PubMed] [Google Scholar]
- 314.Breslin HJ, Diamond CJ, Kavash RW, Cai C, Dyatkin AB, Miskowski TA, et al. Identification of a dual delta OR antagonist/mu OR agonist as a potential therapeutic for diarrhea-predominant irritable bowel syndrome (IBS-d) Bioorg Med Chem Lett. 2012;22(14):4869–4872. doi: 10.1016/jbmcl201205042. [DOI] [PubMed] [Google Scholar]
- 315.Borsini F, Evans K, Jason K, Rohde F, Alexander B, Pollentier S. Pharmacology of flibanserin. CNS Drug Rev. 2002;8(2):117–142. doi: 10.1111/j.1527-3458.2002.tb00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Bolli MH, Boss C, Binkert C, Buchmann S, Bur D, Hess P, et al. The discovery of N-[5-(4-bromophenyl)-6-[2-[(5-bromo-2-pyrimidinyl)oxy]ethoxy]-4-pyrimidinyl]-N’-propylsulfamide (Macitentan), an orally active, potent dual endothelin receptor antagonist. J Med Chem. 2012;55(17):7849–7861. doi: 10.1021/jm3009103. [DOI] [PubMed] [Google Scholar]
- 317.Herrick-Davis K, Grinde E, Teitler M. Inverse agonist activity of atypical antipsychotic Drugs at human 5-hydroxytryptamine2C receptors. J Pharmacol Exp Ther. 2000;295(1):226–232. [PubMed] [Google Scholar]
- 318.Kohen R, Metcalf MA, Khan N, Druck T, Huebner K, Lachowicz JE, et al. Cloning, characterization, and chromosomal localization of a human 5-HT6 serotonin receptor. J Neurochem. 1996;66(1):47–56. doi: 10.1046/j.1471-4159.1996.66010047.x. [DOI] [PubMed] [Google Scholar]
- 319.Monsma FJ, Jr, Shen Y, Ward RP, Hamblin MW, Sibley DR. Cloning and expression of a novel serotonin receptor with high affinity for tricyclic psychotropic Drugs. Mol Pharmacol. 1993;43(3):320–327. [PubMed] [Google Scholar]
- 320.Purohit A, Smith C, Herrick-Davis K, Teitler M. Stable expression of constitutively activated mutant h5HT6 and h5HT7 serotonin receptors: inverse agonist activity of antipsychotic Drugs. Psychopharmacology. 2005;179(2):461–469. doi: 10.1007/s00213-004-2057-6. [DOI] [PubMed] [Google Scholar]
- 321.Roth BL, Craigo SC, Choudhary MS, Uluer A, Monsma FJ, Jr, Shen Y, et al. Binding of typical and atypical antipsychotic agents to 5-hydroxytryptamine-6 and 5-hydroxytryptamine-7 receptors. J Pharmacol Exp Ther. 1994;268(3):1403–1410. [PubMed] [Google Scholar]
- 322.Seeman P, Corbett R, Van Tol HH. Atypical neuroleptics have low affinity for dopamine D2 receptors or are selective for D4 receptors. Neuropsychopharmacology. 1997;16(2):93–110; discussion 1–35. doi: 10.1016/S0893-133X(96)00187-X. [DOI] [PubMed] [Google Scholar]
- 323.Shen Y, Monsma FJ, Jr, Metcalf MA, Jose PA, Hamblin MW, Sibley DR. Molecular cloning and expression of a 5-hydroxytryptamine7 serotonin receptor subtype. J Biol Chem. 1993;268(24):18200–18204. doi: 10.1016/S0021-9258(17)46830-X. [DOI] [PubMed] [Google Scholar]
- 324.Ishibashi T, Horisawa T, Tokuda K, Ishiyama T, Ogasa M, Tagashira R, et al. Pharmacological profile of lurasidone, a novel antipsychotic agent with potent 5-hydroxytryptamine 7 (5-HT7) and 5-HT1A receptor activity. J Pharmacol Exp Ther. 2010;334(1):171–181. doi: 10.1124/jpet.110.167346. [DOI] [PubMed] [Google Scholar]
- 325.Hale JJ, Lynch CL, Neway W, Mills SG, Hajdu R, Keohane CA, et al. A rational utilization of high-throughput screening affords selective, orally bioavailable 1-benzyl-3-carboxyazetidine sphingosine-1-phosphate-1 receptor agonists. J Med Chem. 2004;47(27):6662–6665. doi: 10.1021/jm0492507. [DOI] [PubMed] [Google Scholar]
- 326.Toll L, Berzetei-Gurske IP, Polgar WE, Brandt SR, Adapa ID, Rodriguez L, et al. Standard binding and functional assays related to medications development division testing for potential cocaine and opiate narcotic treatment medications. NIDA Res Monogr. 1998;178:440–466. [PubMed] [Google Scholar]
- 327.Zhu J, Luo LY, Li JG, Chen C, Liu-Chen LY. Activation of the cloned human kappa opioid receptor by agonists enhances [35S]GTPgammaS binding to membranes: determination of potencies and efficacies of ligands. J Pharmacol Exp Ther. 1997;282(2):676–684. [PubMed] [Google Scholar]
- 328.Cynthia J. Vaughn. Drugs and lactation database: LactMed. 2012;9(4):272–7. 10.1080/15424065.2012.735134.
- 329.Yoshie O. CCR4 as a therapeutic target for Cancer Immunotherapy. Cancers (Basel) 2021 doi: 10.3390/cancers13215542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Peters GL. Migraine overview and summary of current and emerging treatment options. Am J Manag Care. 2019;25(2 Suppl):23–S34. [PubMed] [Google Scholar]
- 331.Latuszek A, Liu Y, Olsen O, Foster R, Cao M, Lovric I, et al. Inhibition of complement pathway activation with Pozelimab, a fully human antibody to complement component C5. PLoS ONE. 2020;15(5):e0231892. doi: 10.1371/journal.pone.0231892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Goadsby PJ, Reuter U, Hallstrom Y, Broessner G, Bonner JH, Zhang F, et al. A controlled trial of Erenumab for episodic migraine. N Engl J Med. 2017;377(22):2123–2132. doi: 10.1056/NEJMoa1705848. [DOI] [PubMed] [Google Scholar]
- 333.Duvic M, Evans M, Wang C. Mogamulizumab for the treatment of cutaneous T-cell Lymphoma: recent advances and clinical potential. Ther Adv Hematol. 2016;7(3):171–174. doi: 10.1177/2040620716636541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kanazawa T, Hiramatsu Y, Iwata S, Siddiquey M, Sato Y, Suzuki M, et al. Anti-CCR4 monoclonal antibody mogamulizumab for the treatment of EBV-associated T- and NK-cell lymphoproliferative Diseases. Clin Cancer Res. 2014;20(19):5075–5084. doi: 10.1158/1078-0432CCR-14-0580. [DOI] [PubMed] [Google Scholar]
- 335.Yamauchi J, Coler-Reilly A, Sato T, Araya N, Yagishita N, Ando H, et al. Mogamulizumab, an anti-CCR4 antibody, targets human T-lymphotropic virus type 1-infected CD8 + and CD4 + T cells to treat associated myelopathy. J Infect Dis. 2015;211(2):238–248. doi: 10.1093/infdis/jiu438. [DOI] [PubMed] [Google Scholar]
- 336.Maeda S, Murakami K, Inoue A, Yonezawa T, Matsuki N. CCR4 Blockade Depletes Regulatory T Cells and prolongs survival in a canine model of Bladder Cancer. Cancer Immunol Res. 2019;7(7):1175–1187. doi: 10.1158/2326-6066CIR-18-0751. [DOI] [PubMed] [Google Scholar]
- 337.Jiao X, Wang M, Zhang Z, Li Z, Ni D, Ashton AW, et al. Leronlimab, a humanized monoclonal antibody to CCR5, blocks Breast cancer cellular Metastasis and enhances cell death induced by DNA damaging chemotherapy. Breast Cancer Res. 2021;23(1):11. doi: 10.1186/s13058-021-01391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Verkleij CPM, Broekmans MEC, van Duin M, Frerichs KA, Kuiper R, de Jonge AV, et al. Preclinical activity and determinants of response of the GPRC5DxCD3 bispecific antibody talquetamab in Multiple Myeloma. Blood Adv. 2021;5(8):2196–2215. doi: 10.1182/bloodadvances.2020003805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pillarisetti K, Edavettal S, Mendonca M, Li Y, Tornetta M, Babich A, et al. A T-cell-redirecting bispecific G-protein-coupled receptor class 5 member D x CD3 antibody to treat Multiple Myeloma. Blood. 2020;135(15):1232–1243. doi: 10.1182/blood2019003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Pettus J, Boeder SC, Christiansen MP, Denham DS, Bailey TS, Akturk HK, et al. Glucagon receptor antagonist volagidemab in type 1 Diabetes: a 12-week, randomized, double-blind, phase 2 trial. Nat Med. 2022;28(10):2092. doi: 10.1038/s41591-022-02011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Wang MY, Yan H, Shi Z, Evans MR, Yu X, Lee Y, et al. Glucagon receptor antibody completely suppresses type 1 Diabetes phenotype without insulin by disrupting a novel diabetogenic pathway. Proc Natl Acad Sci U S A. 2015;112(8):2503–2508. doi: 10.1073/pnas1424934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Jahchan NS, Mujal AM, Pollack JL, Binnewies M, Sriram V, Reyno L, et al. Tuning the Tumor Myeloid Microenvironment to Fight Cancer. Front Immunol. 2019;10:1611. doi: 10.3389/fimmu.2019.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Tran TTN, Tran QH, Nguyen QT, Le MT, Trinh DTT, Tran VH, et al. LY3041658/ interleukin-8 complex structure as targets for IL-8 small molecule inhibitors discovery using a combination of in silico methods. SAR QSAR Environ Res. 2022;33(10):753–778. doi: 10.1080/1062936X.2022.2132536. [DOI] [PubMed] [Google Scholar]
- 344.Boyles JS, Beidler CB, Strifler BA, Girard DS, Druzina Z, Durbin JD, et al. Discovery and characterization of a neutralizing pan-ELR + CXC chemokine monoclonal antibody. MAbs. 2020;12(1):1831880. doi: 10.1080/19420862.2020.1831880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Carvelli J, Demaria O, Vely F, Batista L, Chouaki Benmansour N, Fares J, et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588(7836):146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Rubio-Beltran E, Correnti E, Deen M, Kamm K, Kelderman T, Papetti L, et al. PACAP38 and PAC(1) receptor blockade: a new target for headache? J Headache Pain. 2018;19(1):64. doi: 10.1186/s10194-018-0893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zhang C, Wang X, Zhang H, Yao C, Pan H, Guo Y, et al. Therapeutic monoclonal antibody antagonizing endothelin receptor A for pulmonary arterial Hypertension. J Pharmacol Exp Ther. 2019;370(1):54–61. doi: 10.1124/jpet.118.252700. [DOI] [PubMed] [Google Scholar]
- 348.Hye T, Hossain MR, Saha D, Foyez T, Ahsan F. Emerging biologics for the treatment of pulmonary arterial Hypertension. J Drug Target. 2023;31(5):1–15. doi: 10.1080/1061186X.2023.2199351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Lee SH, Chu SY, Rashid R, Phung S, Leung IW, Muchhal US, et al. Abstract 3633: Anti-SSTR2 × anti-CD3 bispecific antibody induces potent killing of human tumor cells in vitro and in mice, and stimulates target-dependent T cell activation in monkeys: A potential immunotherapy for neuroendocrine tumors. Cancer Res. 2017;77(13_Supplement):3633. doi: 10.1158/1538-7445.AM2017-3633%. [DOI] [Google Scholar]
- 350.Dao M, Francois H. Cannabinoid receptor 1 inhibition in chronic Kidney Disease: a New Therapeutic Toolbox. Front Endocrinol (Lausanne) 2021;12:720734. doi: 10.3389/fendo.2021.720734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Kuhne MR, Mulvey T, Belanger B, Chen S, Pan C, Chong C, et al. BMS-936564/MDX-1338: a fully human anti-CXCR4 antibody induces apoptosis in vitro and shows antitumor activity in vivo in hematologic malignancies. Clin Cancer Res. 2013;19(2):357–366. doi: 10.1158/1078-0432.CCR-12-2333. [DOI] [PubMed] [Google Scholar]
- 352.Kashyap MK, Kumar D, Jones H, Amaya-Chanaga CI, Choi MY, Melo-Cardenas J, et al. Ulocuplumab (BMS-936564 / MDX1338): a fully human anti-CXCR4 antibody induces cell death in chronic lymphocytic Leukemia mediated through a reactive oxygen species-dependent pathway. Oncotarget. 2016;7(3):2809–2822. doi: 10.18632/oncotarget6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Huang W, Luo J, Li Y, Fei D, Qin X, Li R. Abstract 6020: Preclinical activity of LM-305 targeting G-protein-coupled receptor class 5 member D (GPRC5D) antibody drug conjugate for the treatment of Multiple Myeloma. Cancer Res. 2022;6020(12_Supplement):6020. doi: 10.1158/1538-7445.AM2022-6020%. [DOI] [Google Scholar]
- 354.Diamond JR, Henry JT, Falchook GS, Olszanski AJ, Singh H, Leonard EJ, et al. Phase 1a/1b study design of the novel STING agonist, immune-stimulating antibody-conjugate (ISAC) TAK-500, with or without pembrolizumab in patients with advanced solid tumors. 2022;40(16_suppl):TPS2690. 10.1200/JCO.2022.40.16_suppl.TPS2690.
- 355.Mateu-Albero T, Juarez-Sanchez R, Loscertales J, Mol W, Terron F, Munoz-Calleja C, et al. Effect of ibrutinib on CCR7 expression and functionality in chronic lymphocytic Leukemia and its implication for the activity of CAP-100, a novel therapeutic anti-CCR7 antibody. Cancer Immunol Immunother. 2022;71(3):627–636. doi: 10.1007/s00262-021-03014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Cuesta-Mateos C, Fuentes P, Schrader A, Juarez-Sanchez R, Loscertales J, Mateu-Albero T, et al. CCR7 as a novel therapeutic target in t-cell PROLYMPHOCYTIC Leukemia. Biomark Res. 2020;8:54. doi: 10.1186/s40364-020-00234-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Broussas M, Boute N, Akla B, Berger S, Beau-Larvor C, Champion T, et al. A new Anti-CXCR4 antibody that blocks the CXCR4/SDF-1 Axis and mobilizes Effector cells. Mol Cancer Ther. 2016;15(8):1890–1899. doi: 10.1158/1535-7163MCT-16-0041. [DOI] [PubMed] [Google Scholar]
- 358.Fouquet G, Guidez S, Richez V, Stoppa AM, Le Tourneau C, Macro M, et al. Phase I dose-escalation study of F50067, a humanized anti-CXCR4 monoclonal antibody alone and in combination with lenalidomide and low-dose dexamethasone, in relapsed or refractory Multiple Myeloma. Oncotarget. 2018;9(35):23890–23899. doi: 10.18632/oncotarget.25156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Behin A, Le Panse R. New pathways and therapeutic targets in Autoimmune Myasthenia Gravis. J Neuromuscul Dis. 2018;5(3):265–277. doi: 10.3233/JND-170294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Gurevich VV, Gurevich EV. Biased GPCR signaling: possible mechanisms and inherent limitations. Pharmacol Ther. 2020 doi: 10.1016/j.pharmthera.2020.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Mullard A. Setting GPCRs free. Nat Rev Drug Discovery. 2023;22(5):347–348. doi: 10.1038/d41573-023-00064-2. [DOI] [PubMed] [Google Scholar]
- 362.Nguyen ATN, Nguyen DTN, Koh HY, Toskov J, MacLean W, Xu A, et al. The application of artificial intelligence to accelerate G protein-coupled receptor drug discovery. Br J Pharmacol. 2023 doi: 10.1111/bph.16140. [DOI] [PubMed] [Google Scholar]
- 363.Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80–93. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.