Abstract
The function of putative regulatory sequences identified in cell transfection experiments can be elucidated only through in vivo experimentation. However, studies of gene regulation in transgenic mice (TgM) are often compromised by the position effects, in which independent transgene insertions differ in expression depending on their location in the genome. In order to overcome such a dilemma, a method called transgene coplacement has been developed in Drosophila melanogaster. In this method, any two sequences can be positioned at exactly the same genomic site by making use of Cre/loxP recombination. Here we applied this method to mouse genetics to characterize the function of direct repeat (DR) sequences in the promoter of the human angiotensinogen (hAGT) gene, the precursor of the vasoactive octapeptide angiotensin II. We modified a hAGT bacterial artificial chromosome to use Cre/loxP recombination in utero to generate TgM lines bearing a wild-type or a mutant promoter-driven hAGT locus integrated at a single chromosomal position. The expression analyses revealed that DR sequences contribute 50 or >95% to hAGT transcription in the liver and kidneys, respectively, whereas same sequences are not required in the heart and brain. This is the first in vivo dissection of DNA cis elements that are demonstrably indispensable for regulating both the level and cell type specificity of hAGT gene transcription.
The renin-angiotensin system (RAS) plays a key role in the regulation of blood pressure (BP) and electrolyte homeostasis. Angiotensinogen (AGT), a plasma protein synthesized predominantly in the liver, is catalyzed by renin, an aspartyl protease synthesized mainly in the juxtaglomerular cells of the kidney, to produce decapeptide angiotensin I (AI). The latter is further processed by angiotensin-converting enzyme to an octapeptide, angiotensin II (AII), which mediates vasoconstriction and aldosterone secretion through specific cell surface receptors present throughout the cardiovascular system. Aldosterone then causes sodium reabsorption in the collecting ducts of the kidney to increase body fluid, leading to increased BP.
Since the reaction catalyzed by renin is the rate-limiting step of this enzymatic cascade and AGT concentrations in plasma are close to the Km value for renin (12), the level of AGT is directly reflected by BP phenotype. In accord with this notion, transgenic overexpression (8, 18) or targeted gene inhibition of AGT in the mouse (39) has demonstrated that this gene is a key genetic determinant of BP. It has been also shown in experimental animal models that as little as a 20% change in AGT expression was reflected as a BP phenotype (18, 19). Furthermore, in humans, genetic linkage between human AGT (hAGT) and essential hypertension has been reported (4, 16). Given these correlations, it may be of fundamental importance to elucidate the molecular mechanisms underlying hAGT transcriptional regulation in order to define the origins of BP physiology and pathology. To this end, we have identified a number of cis-acting DNA elements and trans-acting factors that may be critical for determining the level and cell type specificity of hAGT gene expression (25, 26, 43-45) by using a human hepatoma (HepG2) cell line. Although these cell culture experiments have been instructive, it is nonetheless imperative to evaluate the functional significance of these putative regulatory elements in vivo to reflect a normal physiological environment.
Among the candidate cis-regulatory elements identified thus far, Yang and Sigmund (47) evaluated the function of two sequences in the hAGT locus through analysis of a 14-kbp promoter-proximal construct in transgenic mice (TgM). Two sequences, AGE2 (37) and d61-2 (25), had powerful enhancer activities when assayed in transiently transfected HepG2 cells, conferring 10- and 50-fold activation to a minimal hAGT promoter, respectively. Surprisingly, however, disruption of these elements did not produce a significant phenotype in vivo, i.e., neither the level nor the cell type specificity of hAGT expression was altered by the mutations in TgM. This result argues that these elements are redundant and therefore dispensable for the hAGT expression in TgM but does not necessarily mean that they are nonfunctional elements in the locus.
A series of 5′ deletion mutants of the hAGT promoter (from −1222 to +44 bp) identified two regions that were responsible for robust cell-type-specific expression of the reporter gene in HepG2 cells (43); these regions were designated “C” (from −431 to −380 bp) and “J” (from −281 to −252, Fig. 1). Subsequent electrophoretic mobility shift assay experiments with HepG2 nuclear extracts identified hepatocyte nuclear factor 4 (HNF4) as the predominant binding activity to regions C and J. The liver-enriched transcription factor HNF4 is an orphan member of the nuclear receptor superfamily that has been implicated in modulating the onset and progression of various diseases (13) and interacts with the direct-repeat (DR) AGGTCA motif (33). Three or two copies of this motif are found in regions C and J, respectively (Fig. 1B). Further transactivation experiments in HepG2 cells confirmed that regions C and J are in fact responsive to force-expressed HNF4 protein (43).
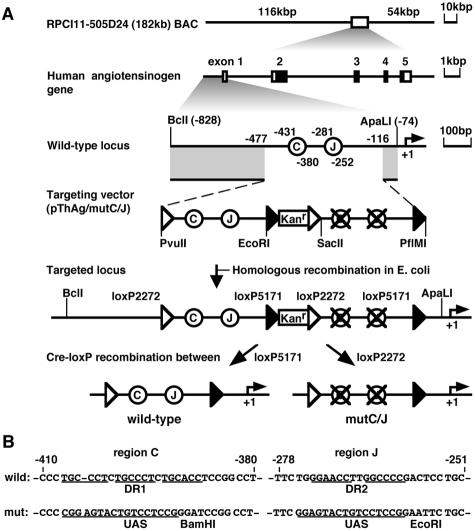
FIG. 1.
Schematic outline of transgenic coplacement strategy used to generate wild-type and mutant hAGT TgM. (A) Structures of the hAGT BAC (top) and the gene (second line) were depicted. The BAC clone contains 116 kb of upstream and 54 kb of downstream regions, in addition to the hAGT coding sequences. Coding and noncoding exons are shown as solid and open boxes, respectively. An enlarged map of the hAGT promoter region is shown (third line), together with the structure of the targeting vector (pThAg/mutC/J, fourth line). Regions C and J were represented as open circles relative to the transcriptional start site (+1). Positions of loxP2272 and loxP5171 are indicated as open and solid triangles, respectively. A BclI-ApaLI 2.5-kb DNA fragment was used for targeting in E. coli. Expected structure of the targeted region in the BAC DNA was shown (fifth line; parental) and was used for establishing TgM lines. After the intercrossing of “parental” TgM with cre-expressing TgM, selective excision in utero of the DNA segment between a pair of loxP5171 or loxP2272 generated either wild-type or mutant promoters, respectively (sixth line). The two resulting sister lines thus contained either wild-type or mutant hAGT transgene at identical genomic positions. Kanr, Kanr gene. (B) Sequence comparison of wild-type (top) and mutant (bottom) hAGT promoter regions. DR motifs in the regions C and J were underlined. DR1 (in region C) and DR2 (in region J) motifs were replaced with UAS sequences and restriction enzyme sites (BamHI and EcoRI) were introduced so as to facilitate subsequent structural analysis of transgene and to maintain the overall length of the promoter region.
To evaluate the functional significance of the DR motifs in the hAGT gene promoter in vivo, we generated TgM lines harboring a 182-kbp bacterial artificial chromosome (BAC), containing either wild-type or mutant hAGT promoter sequences, integrated into the same chromosomal site (Fig. 1A). To accomplish this, we used a modified cre/loxP-mediated coplacement strategy (2, 29, 31, 32) to eliminate potential position effects that might complicate experimental interpretations that commonly arise as a consequence of examining transgenes at different positions in the mouse genome. Northern blot and real-time reverse transcription-PCR (RT-PCR) analyses of transgenic hAGT expression revealed that DR sequences only modestly contribute to hAGT gene expression in the liver. Although differing in magnitude, this in vivo result confirmed our previous transfection studies (43). Surprisingly, however, the same mutation completely abolished hAGT transcription in the kidney, another major site of hAGT synthesis in TgM (34, 46). Thus, this in vivo strategy clearly revealed a cell-type-dependent and DR sequence-dependent activity within the hAGT promoter through which different trans-acting factor environments differentially regulate the level of hAGT transcription.
MATERIALS AND METHODS
BAC targeting vector.
To construct phAGT13(-C-J-), a 1.3-kbp (from −1222 to +44 bp) DNA fragment containing the hAGT gene promoter was cloned into the pGV-B luciferase (Luc) reporter plasmid (Wako). To generate phAGT13(-M-M-), in which DR motifs in regions C and J of phAGT13(-C-J-) are disrupted by substituting with upstream activating sequences (UAS) (Fig. 1), three fragments were independently amplified with the following primer sets, digested, and ligated together in order to replace the corresponding portion in the phAGT13(-C-J-) (the UAS [42] are italicized): M1-5′, 5′-TGTCTATTAGAGGCCTTTGCAC-3′; M1-3′, 5′-GCCGGATCCCGGAGGACAGTACTCCGGGGGAGAGTCTTGCTTAGGCAAC-3′ (BamHI); M2-5′, 5′-ATGGGATCCGGCCTGCATGTCCCTG-3′ (BamHI); M2-3′, 5′-AGAATTCCGGAGGACAGTACTCCGAAACGGGAGCATCTCCCTAGGTG-3′ (EcoRI); M3-5′, 5′-GAATTCTGCAAACTTCGGTAAATGTG-3′ (EcoRI); and M3-3′, 5′-GCTGGAGAGCAACTGCATAA-3′.
Next, LCJL and LMML DNA fragments were PCR amplified by using the following primer sets and phAGT13(-C-J-) or phAGT13(-M-M-), respectively, as templates and digested with appropriate enzymes (loxP sequences were italicized in each of the following oligonucleotides): 5′-LoxP2272, 5′-GGAGCAGCTGATAACTTCGTATAGGATACTTTATACGAAGTTATCCGCGGAAGGTCACACATCCCATGAG-3′ (PvuII and SacII); and 3′-loxP5171, 5′-CAGACCACAGGCTGGATAACTTCGTATAATGTGTACTATACGAAGTTATGAATCCCAGAAGGACAGATGCCAGA-3′ (PflMI and EcoRI).
The kanamycin resistance (Kanr) gene cassette was amplified with the following primer sets by using pIGCN21 (21) as the template and digested with EcoRI and SacII: E5171Kan-5S, 5′- GGAATTCATAACTTCGTATAGTACACATTATACGAAGTTATCTGCAAGGCGATTAAGTTGG-3′ (EcoRI); and SII2272Kan-3A, 5′-GTCCCCGCGGATAACTTCGTATAAAGTATCCTATACGAAGTTATAAGCTCGCGGATCCGAACAA-3′ (SacII).
The LCJL (PvuII-EcoRI), Kanr (EcoRI-SacII), and LMML (SacII-PflMI) fragments were simultaneously cloned into the PvuII/PflMI sites of phAGT13(-C-J-) to generate pThAg/mutC/J (Fig. 1A).
BAC mutagenesis.
The BAC clone containing hAGT gene (RPCI11-505D24) was purchased from Children's Hospital Oakland Research Institute (CHORI), and its DNA was used to transform EL350 Escherichia coli strains (a gift from N. A. Jenkins) by electroporation. We used a prophage-based recombination system (21) for BAC mutagenesis.
TgM.
Modified BAC DNA was purified by using a NucleoBond BAC 100 kit (BD Biosciences Clontech). The DNA insert was released by NotI digestion of BAC DNA, followed by separation on pulsed-field gels as described elsewhere (38). Purified DNA was microinjected into the pronuclei of fertilized eggs from ICR mice (Charles Liver). Transgenic founders were screened by PCR with hAGT-specific primers 5′-CAGGGATGTGCTAGTGTAAG-3′ and 5′-CACGTCCTCAGCACTCAAAG-3′ and subsequently confirmed by Southern blot analysis.
Structural analysis of the transgene.
For a long-range structural analysis, high-molecular-weight DNA was prepared from the thymi of each line, embedded in agarose plugs and digested with SfiI. After pulsed-field gel electrophoresis, DNA was transferred onto a nylon membrane. Blots were hybridized at 65°C with α-32P-labeled DNA probes spanning the hAGT locus. Probes I to VI (unpublished data) were amplified by PCR and subcloned into plasmids, and the sequence fidelity was confirmed by DNA sequencing. Nucleotide positions of each human probe sequence in GenBank are as follows: I (positions 36578 to 37066; AL591291), II (positions 48188 to 48803; AL512328.15), III (positions 42035 to 42647; AL512328.15), IV (positions 6744 to 7239; AL512328.15), V (positions 758 to 1239; AL512328.15), and VI (positions 138972 to 139512; AL158214). All of these probes do not cross-hybridize with mAGT sequences.
In order to discriminate wild-type and mutant transgenic loci, PCR was performed with the a primer set specific for hAGT promoter region (5′-GGAGCAGCTGATAACTTCGTATAGGATACTTTATACGAAG TTATCCGCGGAAGGTCACACATCCCATGAG-3′ and 5′-CAGACCACAGGCTGGATAACTTCGTATAATGTGTACTATACGAAGTTATGAATTCCAGAAGGACAGATGCCAGA-3′) using tail DNA from TgM. PCR products were then subjected to RsaI enzyme digestion, followed by electrophoresis on an 8% polyacrylamide gel.
Northern blot analysis.
Total RNA was denatured with glyoxal, separated on 1.2% agarose gel, and transferred onto nylon membranes by capillary blotting. Blots were hybridized at 60°C with α-32P-labeled DNA probes. A 293-bp DNA fragment (ApaI/EcoRI) excised from exon 5 of the hAGT gene was used as the hAGT-specific probe.
Real-time RT-PCR.
Total RNA was reverse transcribed into cDNA by using random hexamers and reverse transcriptase (ReverTraAce; Toyobo). Real-time RT-PCR was performed by using the LightCycler System and the FastStart DNA Master SYBR Green I kit (Roche Diagnostics). The PCR profile was as follows: DNA denaturation (95°C for 10 min) and amplification (95°C for 10 s, 60°C for 6 s, and 72°C for 10 s) for up to 40 cycles. A sample value was extrapolated from the standard curve, and levels of transgenic hAGT mRNA were normalized to the endogenous mouse angiotensinogen (mAGT) mRNA level. The primers used were as follows: hAGT, 5′-CTTCACAGAACTGGATG-3′ (forward) and 5′-GAACTCCTGGGGCTCG-3′ (reverse) (product size, 241 bp); and mAGT, 5′-CTTTTGGGTGCGGAGGC-3′ (forward) and 5′-CAGGCTGCTGGACAGACGTG-3′ (reverse) (product size, 133 bp).
In situ hybridization.
A kidney of 2-month-old hAGT BAC TgM was fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 3-μm serial sections. Sections were deparaffinized and hybridized for 15 h at 50°C with sense or antisense RNA probes. A 298-bp DNA fragment (AatI-EcoRI) from the hAGT cDNA (35) was used for making digoxigenin (DIG)-labeled probes (Roche Diagnostics). The hybridization signal was visualized by using a DIG nucleic acid detection kit (Roche Diagnostics).
RESULTS
Transgenic coplacement strategy for direct comparison of gene expression in vivo.
Studies of gene regulation in TgM are often obscured by “position effects” in which qualitative and quantitative traits of transgene expression can differ significantly among independent TgM lines bearing identical constructs. Depending on the chromatin environment in the vicinity of a randomly integrated transgene, expression can be positively or negatively affected, and the magnitude of this effect is far more evident in smaller transgene constructs. In such circumstances, it becomes difficult to compare the levels of transgene expression from wild-type and mutant loci. Hence, the analysis of several TgM lines is mandatory, especially when differences are expected to be subtle. In order to circumvent this problem, we coupled two new technologies.
First, we used BAC DNA as the substrate for genetic manipulation and transgenesis. BAC inserts carry more genetic information than plasmid constructs and often include enhancers, silencers, and chromatin modulatory elements, which are required for proper temporal and spatial control over the gene under study. In addition, genes borne on BACs appear to be more resistant to position effects than are plasmids, possibly because genomic DNA flanking both sides of the gene can “buffer” such effects. However, even when very large transgenes (e.g., a 150-kbp locus control region-containing yeast artificial chromosome) are examined, the level of expression can differ by as much as 50% between lines bearing an identical construct (10). We therefore incorporated a second strategy, transgene coplacement (2, 29, 31, 32), in which two genes can be introduced into the same position in the genome, thus subjecting both genes to the identical chromatin environment. This strategy has been successfully applied to the analysis of gene regulation in Drosophila melanogaster (2, 5).
To pursue this strategy, we first identified a hAGT BAC clone in silico. Clone RPCI11-505D24 was derived from a human BAC library and was used as a sequencing template in the human genome project (28). Thus, the sequence of the entire 182-kbp insert, including 116 kbp of 5′ and 54 kbp of 3′ information (Fig. 1A, top), was available and facilitated the structural analysis of the hAGT transgene in mice. We then constructed a BAC targeting vector (Fig. 1A, third and 4th lines), in which a bacterial selectable marker encoding the Kanr gene, and both the wild-type and mutant promoter sequences were flanked by 351 and 42 bp of hAGT genomic sequences at the 5′ and 3′ ends, respectively. The two promoter sequences were identical except for 17- and 11-bp mutations within regions C and J, respectively, and were flanked by a set of loxP sequence variants, loxP2272 and loxP5171. These two different loxP elements recombine efficiently between themselves but not with one another (22). To target this mutation into the hAGT locus within the BAC (Fig. 1A, 5th line), we used the prophage system for homologous recombination in E. coli (21). Successful recombination was confirmed by Southern blots, followed by direct sequencing of the modified BAC (data not shown). We refer below to this hAGT promoter configuration (containing both wild-type and mutant promoters) as “parental.”
Generation of TgM bearing the hAGT BAC.
We purified the parental BAC DNA insert from pulsed-field gels and injected it into fertilized oocytes to generate TgM. Tail DNA was prepared from founder offspring and screened with hAGT-specific PCR primer sets, followed by detailed Southern blot hybridizations (data not shown). To determine the transgene copy number in the several different BAC transgenic lines, we compared the signal intensity of a hAGT internal fragment to that from the endogenous mAGT locus in genomic Southern blots. Three of the lines (lines 97, 87, and 106) carried a single copy of the hAGT BAC (data not shown). To determine the integrity of the BACs in these lines, we conducted a long-range structural analysis of each of the transgenes (unpublished data). Based on these results, we concluded that TgM lines 97, 87, and 106 harbored a single copy of the parental hAGT transgene that was, at a minimum, 182, 91.3, or 46.5 kbp in size, respectively.
Generation of TgM lines in which wild-type and mutant hAGT loci are present in the same genomic location.
As shown in Fig. 1A (bottom two lines), recombination between the pairs of loxP2272 or loxP5171 sites mediated by cre recombinase should occur randomly, resulting in the excision of the intervening wild-type or mutant hAGT promoter sequences, as well as the Kanr gene, to generate the mutant (mutC/J) or wild-type hAGT loci, respectively. In order to initiate this reaction in vivo, we bred the “parental” hAGT lines with TgM ubiquitously expressing cre recombinase. Pups carrying both transgenes (Cre-F0) were confirmed to have undergone the desired recombination event by Southern blot analysis (data not shown). These individuals as adults were intercrossed with wild-type animals to remove the cre locus so as to prevent any potential disturbance on expression. The progeny from the latter intercross (Cre-F1) were genotyped by Southern blot analysis; the data showed that in each sibling TgM subline recombination occurred as anticipated (unpublished data). To further confirm the recombination events, we conducted a fine structural analysis of the wild-type and mutant promoters. Since the UAS sequence contained an RsaI restriction enzyme site (see Materials and Methods), PCR amplification of promoter sequences, followed by RsaI enzyme digestion, could be used to clearly discriminate between the wild-type and mutant loci. The PCR amplicons generated from tail DNA were either sequenced (data not shown) or digested with RsaI and then size fractionated on polyacrylamide gels. The results were consistent with those obtained by Southern blot analysis (unpublished data), with only one exception. Interestingly, some of the animals from line 97 underwent an unpredicted recombination event, which resulted in the disruption of region C but left region J intact (mutC). Although the mechanistic basis for this unexpected recombination event is currently unresolved, this rare, fortuitous event allowed us to assess the function of region C alone on hAGT transcription.
The wild-type BAC recapitulates normal hAGT expression in TgM.
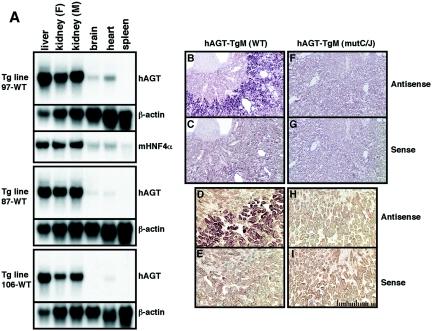
Expression of the hAGT gene in the wild-type TgM lines was analyzed by Northern blot analysis with a hAGT-specific probe. Total RNA was prepared from several tissues of BAC TgM, and representative results are shown in Fig. 2A. In all three lines, transgenic hAGT expression was robust in the liver and kidney, low in the brain and heart, and not detectable in the spleen, thus accurately reflecting tissue-specific AGT gene expression in mice (36) and humans (9), with the exception of relatively high level expression in the kidney of both species. We and others have previously observed an unexpectedly high level of hAGT expression in the mouse kidney when a smaller hAGT gene (14 kbp) construct was analyzed in transgenic studies (34, 46). In situ hybridization analysis of kidney sections from hAGT BAC TgM, however, revealed that transgene expression was specifically restricted to the proximal convoluted tubule cells of the renal cortex (Fig. 2B and D). No expression was detected in the renal medulla (Fig. 2B and D) or in the kidney of non-TgM (data not shown). Similarly, no hybridization signal was detected when a control sense RNA probe was hybridized to serial kidney sections from TgM (Fig. 2C and E). These observations indicated that the cell type specificity of hAGT gene expression was not altered in the kidneys of TgM.
FIG. 2.
Tissue-specific and uniform expression of the hAGT gene in three independent TgM lines. (A) Northern blot analysis of hAGT mRNA isolated from several tissues obtained from 1-month-old female (F) animals of three wild-type TgM lines. The kidney was isolated from a male (M) animal as well. The RNA blot was hybridized to hAGT, mouse HNF4α, or β-actin (41) probes. (B to I) In situ hybridization analysis of the hAGT expression in the kidney of wild-type (WT; B to E) and mutant (mutC/J; F to I) hAGT TgM. Kidney sections were hybridized with the hAGT antisense (B, F, D, and H) and sense (C, G, E, and I) probes. The scale divisions are 10 μm.
Although TgM lines 87 and 106s (with smaller, truncated transgenes) exhibited slightly lower hAGT abundance (in the brain and heart, for example [Fig. 2A]), the overall expression pattern (level and cell type specificity) did not differ significantly among the different lines, confirming the validity and reproducibility of this approach with BAC DNA.
Transgenic hAGT expression in the liver.
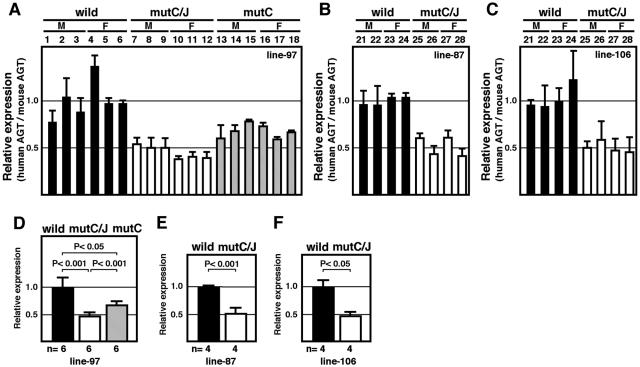
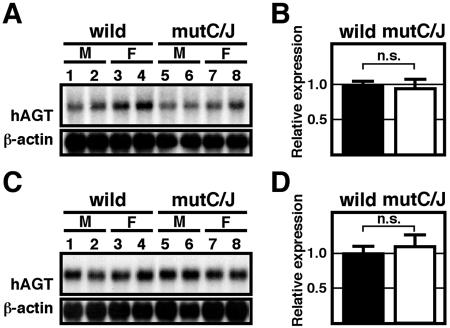
To determine whether the previously implicated DR elements contributed to hAGT gene expression in the liver, we first analyzed RNAs from wild-type and mutant hAGT TgM by Northern blot analysis (unpublished data). Although mutC/J animals expressed significant levels of hAGT mRNA, their expression tended to be lower than that observed in the wild-type animals (wild) when normalized to β-actin expression. There was no obvious difference in transgenic expression between males (M) and females (F). Quantitative analysis of the hybridization signals (unpublished data) demonstrated a significant difference in the expression of hAGT between wild and mutC/J TgM. However, the effect of individual differences was relatively large in this analysis. To accurately assess the in vivo function of the DR sequences, we performed quantitative RT-PCR analysis with endogenous mAGT mRNA expression as the internal control (Fig. 3A to C) and statistically analyzed the data (Fig. 3D to F). As shown in Fig. 3D to F, the level of hAGT expression in mutC/J animals was significantly lower (50% reduction, P < 0.001) than in wild-type animals. This phenotype was also observed in the liver of mutC TgM (30% reduction, P < 0.05, Fig. 3D), indicating that the remaining DR motif in region J of the hAGT promoter could not fully compensate for the loss of the DR motif in region C.
FIG. 3.
Mutation in DR motifs leads to moderate loss of hepatic hAGT expression. (A to C) Liver RNA was isolated from TgM and expression level of human and mouse AGT mRNA was quantitatively analyzed by real-time RT-PCR. Each value represented the ratio of hAGT to mAGT gene expression, which served as an internal control. Relative hAGT/mAGT value for individual mouse was shown after normalization to the average value of wild-type group (see panels D to F), which was arbitrarily set at 1.0. Each sample was analyzed at least three times and the mean ± standard deviation are shown. (D to F) The means ± the standard deviations of individuals in each group in panels A to C were summarized. P values were determined by using the Student t test. A and D, line 97; B and E, line 87; C and F, line 106.
Transgenic hAGT expression in the kidney.
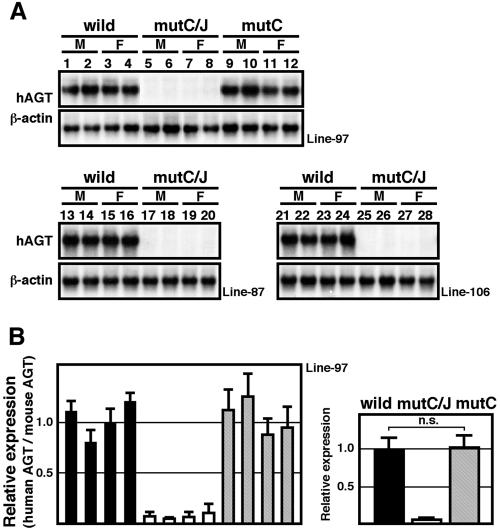
We next analyzed the expression of hAGT in the kidney, one of the major sites for hAGT synthesis, in TgM. Surprisingly, Northern blot (Fig. 4A) and RT-PCR (Fig. 4B) analyses demonstrated that renal hAGT expression was almost completely absent in mutC/J TgM (lines 97, 106, and 87). In contrast to the case in the liver, this phenotype was fully rescued by the DR sequences contained in region J (mutC, Fig. 4), conclusively demonstrating a fundamental difference between liver and kidney cells in their functional requirement for intact DR motifs in regions C and J.
FIG. 4.
Mutation in DR motifs leads to severely attenuated hAGT expression in the kidney. Renal RNA from TgM (line 97, 87, and 106) was analyzed by Northern blotting (A) and real-time RT-PCR (B) assays. See the legend to Fig. 3 for details. n.s., not significant.
It is well established that renal expression of both endogenous mAGT (15) and transgenic hAGT (Fig. 2B) (46) is strictly restricted to the proximal tubule cells of the renal cortex, where mouse HNF4 is also abundantly expressed (17). In situ hybridization analysis of kidney sections revealed that hAGT gene expression was lost in mutC/J TgM (Fig. 2F and H). No hybridization was detected when a control sense RNA probe was hybridized to serial kidney sections (Fig. 2G and I). These results clearly demonstrate that the DR sequences are indispensable for appropriate cell-specific expression of hAGT in the kidney.
Transgenic hAGT expression in the brain and the heart.
We also analyzed hAGT expression in other organs by Northern blot (Fig. 5A and C) and real-time RT-PCR analyses (Fig. 5B and D) to further confirm the cell-type-dependent activity of the DR sequences. In both the brain (Fig. 5A and B) and the heart (Fig. 5C and D), no significant difference in the hAGT expression was detected between wild-type and mutC/J TgM, indicating that both DR elements are dispensable for hAGT expression in those organs.
FIG. 5.
Mutation in DR motifs do not alter hAGT expression in the brain and heart of TgM. RNA from brain (A) or heart (B) of TgM (line 97) was analyzed by Northern blotting (A and C) and real-time RT-PCR (B and D) assays. See the legend to Fig. 3 for details. n.s., not significant.
DISCUSSION
Small DNA constructs (<20 kbp) have been widely used to investigate the regulation of gene activity in TgM, and are generally favored because of their ease of manipulation and purification. Although this approach is powerful, there are potential problems associated with such transgenes. Smaller transgenes carry less potential regulatory information that might be required for proper temporal and spatial gene expression and therefore may not accurately reproduce gene expression. Equally importantly, small transgenes are susceptible to position-of-integration site effects from the surrounding chromatin environment, which generates often significant line-to-line heterogeneity in the level of transgene expression. Finally, smaller transgenes usually integrate in a multicopy tandem arrangement that does not accurately reflect the situation for the endogenous gene. These drawbacks in examining the activities of small DNA constructs make it extremely difficult to sensitively detect possible subtle phenotypic differences that may exist between wild-type and mutant TgM lines.
In the systemic RAS, renin is secreted into the circulation from juxtaglomerular cells of the kidney and catalyzes cleavage of AGT produced mainly by the liver. In this report, we present definitive in vivo evidence that DR motifs within the hAGT gene promoter are functionally required for setting its expression level in the liver, which in turn may largely determine BP. In addition to systemic RAS, an important role for local RAS in the regulation of BP has also been proposed. For example, animal models have suggested a role for brain RAS in BP regulation (reviewed in reference 1). In the kidney, RAS has been postulated to modulate renal function, including aspects such as blood flow, sodium-hydrogen exchange, and tubular-glomerular feedback (reviewed in reference 3). AGT is expressed in proximal tubule kidney cells, transported through an apical membrane, and secreted into the tubular lumen, where it is subsequently cleaved to angiotensin I (AI) and AII to stimulate local AII receptors. It is therefore conceivable that inappropriately activated AGT gene expression may cause excessive salt and water retention, leading to pathogenesis exhibited as hypertension or renal injury (24). It has been suggested that the hypertension associated with autosomal dominant polycystic kidney disease may be related to an increase in the intratubular AII concentration (23). In accord with this notion, recent reports examining cell-type-specific TgM demonstrated a possible role for renal RAS in the pathogenesis of hypertension that is independent of systemic RAS (6, 20). Although the physiological relevance of intrarenal RAS secretion is debated, it seems increasingly relevant to elucidate the local functions of RAS and to determine how the tissue-restricted gene regulation of each RAS component is achieved.
We previously discovered a pair of DR motifs in the hAGT promoter that preferentially activate its expression in HepG2 cells, and we proposed that nuclear receptor HNF4 was the activator of these effector sites (43). In the present study, using a reliable genetic manipulation system in mice, we evaluated the function of these sequences in the regulation of hAGT gene expression. In the livers of BAC TgM (Fig. 3), the levels of hAGT expression in the mutant line (mutC/J) were significantly lower (a 50% reduction) compared to those seen in the corresponding control lines (wild type), confirming the results of the cell transfection studies (43). However, the magnitude of transcriptional diminution from the hAGT promoter was less pronounced in vivo, which may be attributable to a difference in the regulatory information carried in the BAC (182-kbp locus) versus the reporter plasmid (1.3-kbp promoter sequence). One can easily imagine that the contribution of a single cis-regulatory element to overall transcriptional activity might be magnified when a smaller promoter DNA fragment is assayed.
As noted earlier, we suggested HNF4 as the candidate trans-acting factor of DR motifs based on compiled in vitro evidence (43). In the present in vivo study, which clearly highlights the significance of the DR motifs in the regulation of hAGT, we still do not know whether HNF4 is the bona fide factor binding to DR sequences in the hAGT promoter. We discovered an even more prominent role for the DR motifs in the proximal tubule cells of the kidney than in the liver (Fig. 2 and 4), and immunohistochemical analysis of renal sections revealed that HNF4 localized in nuclei of proximal tubule cells of murine (11) and rat (14, 30) kidneys, coincident with hAGT gene expression (Fig. 1 and 4). In addition, we found that the DR sequences were dispensable for hAGT gene expression in the brain and heart (Fig. 5), where the endogenous mouse HNF4α expression level is negligible (40). These correlative observations thus strengthen the notion that HNF4 may be the authentic activator of hAGT transcription through the DR elements in vivo. Recently, Odom et al. conducted a genome-scale location analysis of HNF4 binding sites by using chromatin immunoprecipitation, and they concluded that the hAGT promoter an in vivo target for HNF4 (27).
AGT is widely expressed in a variety of tissues, such as liver, kidney, brain, heart, and adipose tissues. It is not clear which trans-acting factors or their combinations are controlling hAGT expression in these tissues. Detailed analyses of our mice would help us unveil the identities of such transcription factor complexes. It is generally established that AGT synthesis is modulated by steroid hormones, such as glucocorticoids, thyroid hormones, and estrogens, all of whose receptors bind to DR motifs (7). It will therefore be interesting to see whether such hormones can regulate mutC/J transgene in vivo. Finally, transgenic coplacement strategy is a powerful genetic tool that can be used to investigate with precision the change in transcriptional activity in vivo attributed to a minor change in DNA cis-elements, which can potentially lead to the pathogenesis of genetic diseases, such as hypertension.
Acknowledgments
We thank Y. Tanimoto for technical assistance in preparing TgM. We also thank Kim-Chew Lim for help with the manuscript.
This study was supported by grants from the Yamanouchi Foundation for Research on Metabolic Disorders (K.T.); the Japan Heart Foundation (K.T.); the Uehara Memorial Foundation (K.T.); the Tokyo Biochemical Research Foundation (K.T.); the 21st Century COE Program (A.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); a grant for the Support of Young Researchers (MEXT [K.T.]); and a Grant-in-Aid for Scientific Research (MEXT [A] to A.F. and [B] to K.T.).
REFERENCES
- 1.Bader, M., and D. Ganten. 2002. It's renin in the brain: transgenic animals elucidate the brain renin angiotensin system. Circ. Res. 90:8-10. [PubMed] [Google Scholar]
- 2.Butler, J. E., and J. T. Kadonaga. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15:2515-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey, R. M., and H. M. Siragy. 2003. The intrarenal renin-angiotensin system and diabetic nephropathy. Trends Endocrinol. Metab. 14:274-281. [DOI] [PubMed] [Google Scholar]
- 4.Caulfield, M., P. Lavender, M. Farrall, P. Munroe, M. Lawson, P. Turner, and A. J. Clark. 1994. Linkage of the angiotensinogen gene to essential hypertension. N. Engl. J. Med. 330:1629-1633. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. L., K. L. Huisinga, M. M. Viering, S. A. Ou, C. T. Wu, and P. K. Geyer. 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davisson, R. L., Y. Ding, D. E. Stec, J. F. Catterall, and C. D. Sigmund. 1999. Novel mechanism of hypertension revealed by cell-specific targeting of human angiotensinogen in transgenic mice. Physiol. Genomics 1:3-9. [DOI] [PubMed] [Google Scholar]
- 7.Feldmer, M., M. Kaling, S. Takahashi, J. J. Mullins, and D. Ganten. 1991. Glucocorticoid- and estrogen-responsive elements in the 5′-flanking region of the rat angiotensinogen gene. J. Hypertension 9:1005-1012. [DOI] [PubMed] [Google Scholar]
- 8.Fukamizu, A., K. Sugimura, E. Takimoto, F. Sugiyama, M. S. Seo, S. Takahashi, T. Hatae, N. Kajiwara, K. Yagami, and K. Murakami. 1993. Chimeric renin-angiotensin system demonstrates sustained increase in blood pressure of transgenic mice carrying both human renin and human angiotensinogen genes. J. Biol. Chem. 268:11617-11621. [PubMed] [Google Scholar]
- 9.Fukamizu, A., S. Takahashi, and K. Murakami. 1990. Expression of the human angiotensinogen gene in human cell lines. J. Cardiovasc. Pharmacol. 16(Suppl. 4):S11-S13. [DOI] [PubMed] [Google Scholar]
- 10.Gaensler, K. M., M. Kitamura, and Y. W. Kan. 1993. Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human beta-globin locus in transgenic mice. Proc. Natl. Acad. Sci. USA 90:11381-11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gociman, B., A. Rohrwasser, P. Lantelme, T. Cheng, G. Hunter, S. Monson, J. Hunter, E. Hillas, P. Lott, T. Ishigami, and J. M. Lalouel. 2004. Expression of angiotensinogen in proximal tubule as a function of glomerular filtration rate. Kidney Int. 65:2153-2160. [DOI] [PubMed] [Google Scholar]
- 12.Gould, A. B., and D. Green. 1971. Kinetics of the human renin and human substrate reaction. Cardiovasc. Res. 5:86-89. [DOI] [PubMed] [Google Scholar]
- 13.Hertz, R., J. Magenheim, I. Berman, and J. Bar-Tana. 1998. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4α. Nature 392:512-516. [DOI] [PubMed] [Google Scholar]
- 14.Ingelfinger, J. R., R. E. Pratt, K. Ellison, and V. J. Dzau. 1986. Sodium regulation of angiotensinogen mRNA expression in rat kidney cortex and medulla. J. Clin. Investig. 78:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingelfinger, J. R., W. M. Zuo, E. A. Fon, K. E. Ellison, and V. J. Dzau. 1990. In situ hybridization evidence for angiotensinogen messenger RNA in the rat proximal tubule: an hypothesis for the intrarenal renin angiotensin system. J. Clin. Investig. 85:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeunemaitre, X., F. Soubrier, Y. V. Kotelevtsev, R. P. Lifton, C. S. Williams, A. Charru, S. C. Hunt, P. N. Hopkins, R. R. Williams, J. M. Lalouel, et al. 1992. Molecular basis of human hypertension: role of angiotensinogen. Cell 71:169-180. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, S., T. Tanaka, H. Iwanari, H. Hotta, H. Yamashita, J. Kumakura, Y. Watanabe, Y. Uchiyama, H. Aburatani, T. Hamakubo, T. Kodama, and M. Naito. 2003. Expression and localization of P1 promoter-driven hepatocyte nuclear factor-4α (HNF4α) isoforms in human and rats. Nucl. Recept. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. S., J. H. Krege, K. D. Kluckman, J. R. Hagaman, J. B. Hodgin, C. F. Best, J. C. Jennette, T. M. Coffman, N. Maeda, and O. Smithies. 1995. Genetic control of blood pressure and the angiotensinogen locus. Proc. Natl. Acad. Sci. USA 92:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, H. S., G. Lee, S. W. John, N. Maeda, and O. Smithies. 2002. Molecular phenotyping for analyzing subtle genetic effects in mice: application to an angiotensinogen gene titration. Proc. Natl. Acad. Sci. USA 99:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavoie, J. L., K. D. Lake-Bruse, and C. D. Sigmund. 2004. Increased blood pressure in transgenic mice expressing both human renin and angiotensinogen in the renal proximal tubule. Am. J. Physiol. Renal Physiol. 286:F965-F788. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 22.Lee, G., and I. Saito. 1998. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216:55-65. [DOI] [PubMed] [Google Scholar]
- 23.Loghman-Adham, M., C. E. Soto, T. Inagami, and L. Cassis. 2004. An intrarenal renin-angiotensin system in autosomal dominant polycystic kidney disease. Am. J. Physiol. Renal Physiol. 287:F775-F788. [DOI] [PubMed] [Google Scholar]
- 24.Navar, L. G., and A. Nishiyama. 2004. Why are angiotensin concentrations so high in the kidney? Curr. Opin. Nephrol. Hypertens. 13:107-115. [DOI] [PubMed] [Google Scholar]
- 25.Nibu, Y., S. Takahashi, K. Tanimoto, K. Murakami, and A. Fukamizu. 1994. Identification of cell type-dependent enhancer core element located in the 3′-downstream region of the human angiotensinogen gene. J. Biol. Chem. 269:28598-28605. [PubMed] [Google Scholar]
- 26.Nibu, Y., K. Tanimoto, S. Takahashi, H. Ono, K. Murakami, and A. Fukamizu. 1994. A cell type-dependent enhancer core element is located in exon 5 of the human angiotensinogen gene. Biochem. Biophys. Res. Commun. 205:1102-1108. [DOI] [PubMed] [Google Scholar]
- 27.Odom, D. T., N. Zizlsperger, D. B. Gordon, G. W. Bell, N. J. Rinaldi, H. L. Murray, T. L. Volkert, J. Schreiber, P. A. Rolfe, D. K. Gifford, E. Fraenkel, G. I. Bell, and R. A. Young. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osoegawa, K., A. G. Mammoser, C. Wu, E. Frengen, C. Zeng, J. J. Catanese, and P. J. de Jong. 2001. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 11:483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsch, J. 2004. Functional analysis of Drosophila melanogaster gene regulatory sequences by transgene coplacement. Genetics 168:559-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richoux, J. P., J. L. Cordonnier, J. Bouhnik, E. Clauser, P. Corvol, J. Menard, and G. Grignon. 1983. Immunocytochemical localization of angiotensinogen in rat liver and kidney. Cell Tissue Res. 233:439-451. [DOI] [PubMed] [Google Scholar]
- 31.Siegal, M. L., and D. L. Hartl. 1998. An experimental test for lineage-specific position effects on alcohol dehydrogenase (Adh) genes in Drosophila. Proc. Natl. Acad. Sci. USA 95:15513-15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegal, M. L., and D. L. Hartl. 1996. Transgene Coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics 144:715-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sladek, F. M., W. M. Zhong, E. Lai, and J. E. Darnell, Jr. 1990. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 4:2353-2365. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, S., A. Fukamizu, T. Hasegawa, M. Yokoyama, T. Nomura, M. Katsuki, and K. Murakami. 1991. Expression of the human angiotensinogen gene in transgenic mice and transfected cells. Biochem. Biophys. Res. Commun. 180:1103-1109. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi, S., A. Fukamizu, T. Hatae, Y. Yamada, F. Sugiyama, N. Kajiwara, K. Yagami, and K. Murakami. 1992. Species-specific kinetics of mouse renin contribute to maintenance of normal blood pressure in transgenic mice with overexpressed human angiotensinogen. J. Vet. Med. Sci. 54:1191-1193. [DOI] [PubMed] [Google Scholar]
- 36.Tamura, K., K. Tanimoto, S. Takahashi, M. Sagara, A. Fukamizu, and K. Murakami. 1992. Structure and expression of the mouse angiotensinogen gene. Jpn. Heart J. 33:113-124. [DOI] [PubMed] [Google Scholar]
- 37.Tamura, K., S. Umemura, M. Ishii, K. Tanimoto, K. Murakami, and A. Fukamizu. 1994. Molecular mechanism of transcriptional activation of angiotensinogen gene by proximal promoter. J. Clin. Investig. 93:1370-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanimoto, K., Q. Liu, F. Grosveld, J. Bungert, and J. D. Engel. 2000. Context-dependent EKLF responsiveness defines the developmental specificity of the human epsilon-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 14:2778-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanimoto, K., F. Sugiyama, Y. Goto, J. Ishida, E. Takimoto, K. Yagami, A. Fukamizu, and K. Murakami. 1994. Angiotensinogen-deficient mice with hypotension. J. Biol. Chem. 269:31334-31337. [PubMed] [Google Scholar]
- 40.Taraviras, S., A. P. Monaghan, G. Schutz, and G. Kelsey. 1994. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech. Dev. 48:67-79. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga, K., H. Taniguchi, K. Yoda, M. Shimizu, and S. Sakiyama. 1986. Nucleotide sequence of a full-length cDNA for mouse cytoskeletal beta-actin mRNA. Nucleic Acids Res. 14:2829.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster, N., J. R. Jin, S. Green, M. Hollis, and P. Chambon. 1988. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 transactivator. Cell 52:169-178. [DOI] [PubMed] [Google Scholar]
- 43.Yanai, K., K. Hirota, K. Taniguchi-Yanai, Y. Shigematsu, Y. Shimamoto, T. Saito, S. Chowdhury, M. Takiguchi, M. Arakawa, Y. Nibu, F. Sugiyama, K. Yagami, and A. Fukamizu. 1999. Regulated expression of human angiotensinogen gene by hepatocyte nuclear factor 4 and chicken ovalbumin upstream promoter-transcription factor. J. Biol. Chem. 274:34605-34612. [DOI] [PubMed] [Google Scholar]
- 44.Yanai, K., Y. Nibu, K. Murakami, and A. Fukamizu. 1996. A cis-acting DNA element located between TATA box and transcription initiation site is critical in response to regulatory sequences in human angiotensinogen gene. J. Biol. Chem. 271:15981-15986. [DOI] [PubMed] [Google Scholar]
- 45.Yanai, K., T. Saito, K. Hirota, H. Kobayashi, K. Murakami, and A. Fukamizu. 1997. Molecular variation of the human angiotensinogen core promoter element located between the TATA box and transcription initiation site affects its transcriptional activity. J. Biol. Chem. 272:30558-30562. [DOI] [PubMed] [Google Scholar]
- 46.Yang, G., D. C. Merrill, M. W. Thompson, J. E. Robillard, and C. D. Sigmund. 1994. Functional expression of the human angiotensinogen gene in transgenic mice. J. Biol. Chem. 269:32497-32502. [PubMed] [Google Scholar]
- 47.Yang, G., and C. D. Sigmund. 1998. Regulatory elements required for human angiotensinogen expression in HepG2 cells are dispensable in transgenic mice. Hypertension 31:734-740. [DOI] [PubMed] [Google Scholar]