Abstract
The “MEG-MASC” dataset provides a curated set of raw magnetoencephalography (MEG) recordings of 27 English speakers who listened to two hours of naturalistic stories. Each participant performed two identical sessions, involving listening to four fictional stories from the Manually Annotated Sub-Corpus (MASC) intermixed with random word lists and comprehension questions. We time-stamp the onset and offset of each word and phoneme in the metadata of the recording, and organize the dataset according to the ‘Brain Imaging Data Structure’ (BIDS). This data collection provides a suitable benchmark to large-scale encoding and decoding analyses of temporally-resolved brain responses to speech. We provide the Python code to replicate several validations analyses of the MEG evoked responses such as the temporal decoding of phonetic features and word frequency. All code and MEG, audio and text data are publicly available to keep with best practices in transparent and reproducible research.
Subject terms: Language, Cortex
Background & Summary
Humans have the unique ability to produce and comprehend an infinite number of novel utterances. This capacity of the human brain has been the subject of vigorous studies for decades. Yet, the core computational mechanisms upholding this feat remain largely unknown1–3.
To tackle this issue, a common experimental approach has been to decompose language processing into elementary computations using highly controlled factorial designs. This approach allows experimenters to compare average brain responses to carefully chosen stimuli and make inferences based on the select ways that those stimuli were designed to differ. The field has learnt a lot about the neurobiology of language by taking this approach; however, factorial designs also face several key challenges4. First, this method has led the community to study language processing in atypical scenarios (e.g. using unusual text fonts5, meaningless syntactic constructs6,7, or words and phrases isolated from context8,9). Presenting language in this unconventional manner runs the risk of studying phenomena that are not representative of how language is naturally processed. Second, high-level cognitive functions can be difficult to fully orthogonalize in a factorial design. For instance, comparing brain responses to words and sentences matched in length, syntactic structure, plausibility and pronunciation is often close to impossible. In the best case, experimenters will be forced to make concessions on how well the critical contrasts are controlled. In the worst case, unidentified confounds may drive differences associated with experimental contrasts, leading to incorrect conclusions.
During the past decade, several studies have complemented the factorial paradigm with more natural experiments. In these studies, participants listen to continuous speech10–12, read continuous prose13,14 or watch videos that include verbal communication15. This approach is more likely to recruit neural computations that are representative of day-to-day language processing. Complications arising from correlated language features can be overcome by explicitly modeling properties of interest, in tandem with potential confounds. This allows variance belonging to either source to be appropriately distinguished.
To analyze the brain responses to the complex stimulation that natural language provides, a variety of encoding and decoding methods have proved remarkably effective10,16–21. Consequently, language studies based on naturalistic designs have since flourished11,22. The popularity of this approach has some of its roots in the rise of natural language processing (NLP) algorithms, which map remarkably onto brain responses to written and spoken sentences23–30. Such tools also allow experimenters to annotate the language stimuli for features of interest, without relying on time-consuming annotations done by hand. These data have allowed researchers to identify the main semantic components10, recover the hierarchy of integration constants in the language network31, distinguish syntax and semantics hubs32 and to track the hierarchy of predictions elicited during speech processing28,33,34. More generally, brain responses to natural stories have proved useful in keeping participants engaged, while studying the neural representations of phonemes, word surprisal and entropy22,35,36.
While large and high-quality functional Magnetic Resonance Imaging (fMRI) datasets related to language processing have recently been released37,38, there is currently little publicly available high-quality temporally-resolved brain recordings acquired during story listening. The most extensive databases of such data include:
van Essen et al.39: 72 subjects recorded with fMRI and MEG as part of the Human Connectome Project, listening to 10 minutes of short stories, no repeated session39
Brennan and Hale40: 33 subjects recorded with EEG, listening to 12 min of a book chapter, no repeated session40
Broderick et al.11: 9–33 subjects recorded with EEG, conducting different speech tasks, no repeated sessions11
Schoffelen et al.38: 100 subjects recorded with fMRI and MEG, listening to de-contextualised sentences and word lists, no repeated session38
Armeni et al.41: 3 subjects recorded with MEG, listening to 10 h of Sherlock Holmes, no repeated session41
Until the present release of our dataset, there existed no public magneto-encephalography (MEG) with (1) several hours of story listening (2) multiple sessions (3) a systematic audio, phonetic and word annotations (4) a standardized data structure. Thus, our dataset offers a powerful resource to the scientific community.
In the present study, 27 English-speaking subjects performed ~two hours of story listening, punctuated by random word lists and comprehension questions in the MEG scanner. Except if stated otherwise, each subject listened to four distinct fictional stories twice.
Methods
Participants
Twenty-seven English-speaking adults were recruited from the subject pool of NYU Abu Dhabi (15 females; age: M = 24.8, SD = 6.4). All participants provided a written informed consent and were compensated for their time. Participants reported having normal hearing and no history of neurological disorders. All participants were right-handed, as evaluated using the Edinburgh Handedness Inventory questionnaire42. All but one participant (S21) were native English speakers - this person was a native speaker of Hindi, and learned English at 10 years old. All but five participants (S3, S12, S16, S20, S21) performed two identical one-hour-long sessions. These two recording sessions were separated by at least one day and at most two months depending on the availability of the experimenters and of the participants. The study was approved by the Institutional Review Board (IRB) ethics committee of New York University Abu Dhabi.
Procedure
Within each ∼1 h recording session, participants were recorded with a 208 axial-gradiometer MEG scanner built by the Kanazawa Institute of Technology (KIT), and sampled at 1,000 Hz, and online band-pass filtered between 0.01 and 200 Hz while they listened to four distinct stories through binaural tube earphones (Aero Technologies), at a mean level of 70 dB sound pressure level.
Before the experiment, participants were exposed to 20 sec of each of the distinct speaker voices used in the study to (i) clarify the structure of the session and (ii) familiarize the participants with these voices. The sound files and scripts are available in (‘/stimuli/exp_intro/’).
The order in which the four stories were presented was assigned pseudo-randomly, thanks to a “Latin-square design” across participants. The story order for each participant can be found in ‘participants.tsv’. This participant-specific order was used for both recording sessions. Our motivation for running two identical sessions was to (i) give researchers the ability to average the data across the two recordings to boost signal-to-noise; (ii) provide a like-for-like data reliability measure; (iii) give the opportunity for matched train and test datasets if attempting to run cross validated analyses.
To ensure that the participants were attentive to the stories, they responded, every ∼3 min and with a button press, to a two-alternative forced-choice question relative to the story content (e.g. ‘What precious material had Chuck found? Diamonds or Gold’). Participants performed this task with an average accuracy of 98%, confirming their engagement with and comprehension of the stories. The questions and answers are provided in (‘stimuli/task/question_dict.py’).
Participants who did not already have a T1-weighted anatomical scan usable for the present study were scanned in a 3 T Magnetic-Resonance-Imaging (MRI) scanner after the MEG recording to avoid magnetic artefacts. Twelve participants returned for their T1 scan.
Before each MEG session, the head shape of each participant was digitized with a hand-held FastSCAN laser scanner (Polhemus), and co-registered with five head-position coils. The positions of these coils with regard to the MEG sensors were collected before and after each recording and stored in the ‘marker’ file, following the KIT’s system. The experimenter continuously monitored head position during the acquisition to ensure that the participants did not move.
Stimuli
Four English fictional stories were selected from the Manually Annotated Sub-Corpus (MASC) which is part of the larger Open American National Corpus43. MASC is distributed without license or other restrictions (https://anc.org/data/masc/corpus/577-2/):
‘LW1’: a 861-word story narrating an alien spaceship trying to find its way home (5 min, 20 sec)
‘Cable Spool Boy’: a 1,948-word story narrating two young brothers playing in the woods (11 min)
‘Easy Money’: a 3,541-word fiction narrating two friends using a magical trick to make money (12 min, 10 sec)
‘The Black Willow’: a 4,652-word story narrating the difficulties an author encounters during writing (25 min, 50 sec)
An audio track corresponding to each of these stories was synthesized using Mac OS Mojave © version 10.14 text-to-speech. To help decorrelate language features from acoustic representations, we varied both voices and speech rate every 5–20 sentences. Specifically, we used three distinct synthetic voices:‘Ava’, ‘Samantha’ and ‘Allison’ speaking between 145 and 205 words per minute. Additionally, we varied the silence between sentences between 0 and 1,000 ms. Both speech rate and silence duration were sampled from a uniform distribution between the min and max values.
Each story was divided into ~3 min sound files. In between these sounds – approximately every 30 s – we played a random word list generated from the unique content words (nouns, proper nouns, verbs, adverbs and adjectives) selected from the preceding 5 min segment presented in random order. We decided to include word lists to allow data users to compare brain responses to content words within and outside of context, following experimental paradigms of previous studies38,44. In addition, a very small fraction (<1%) of non-words were inserted into the natural sentences, on average every 30 words. We decided to include non-words to allow comparisons between phonetic sequences that do and do not have an associated meaning.
Hereafter, and following the BIDS labeling45, each “task” corresponds to the concatenation of these sentences and word lists. Each subject listened to the exact same set of four tasks, in a different block order.
Preprocessing
MEG
The MEG dataset and its annotations are shared raw (i.e. not preprocessed) organized according to the Brain Imaging Data Structure45 MNE-BIDS46.
MRI
Structural MRIs were collected with separate averages of the T1w images using 3D MPRAGE sequence with 0.8 mm isotropic resolution (FOV = 256 mm, matrix = 320,208 sagittal slices in a single slab), TR = 2400 ms, TE = 2.22 ms, TI = 1000 ms, FA = 8 degrees, Bandwidth (BW) = 220 Hz per pixel, Echo Spacing (ES) = 7.5 ms, phase encoding undersampling factor GRAPPA = 2, no phase encoding oversampling.
To avoid subject identification, the T1-weighted MRI anatomical scan was defaced using PyDeface47 (https://github.com/poldracklab/pydeface) and manually checked.
For four subjects (02, 06, 07, 19) we were unable to record structural MRIs, and so instead we provide the scaled FreeSurfer average MRIs in their place.
The alignment between the spaces of (1) the head-position coils, (2) the MEG sensors and (3) the T1 MRI was co-registered manually with MNE-Python48.
Stimuli
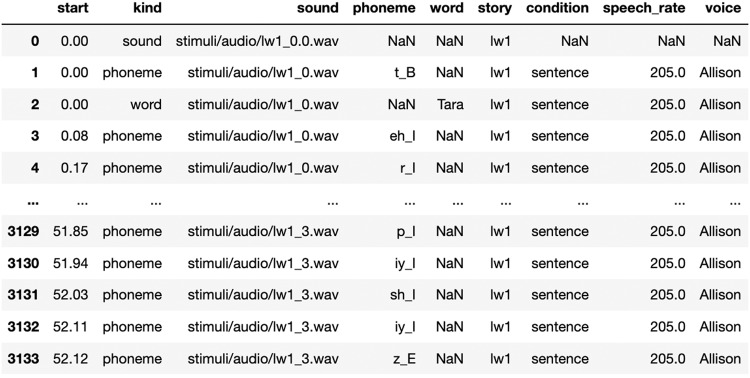
We include in the dataset: the original stories (‘stimuli/text’), the stories intertwined with the word lists (‘stimuli/text_with_wordlists’) and their corresponding audio tracks (‘stimuli/audio’).he alignment between the MEG data and the words and phonemes is provided for each participant separately (e.g., /sub-01/ses-0/meg/sub-01_ses-0_task-1_events.tsv’).
Both sentences and word lists were annotated for phoneme boundaries and labels (107 phoneme labels, detailing phoneme category and its location in the word (Beginning; Internal; End) using the ‘Gentle aligner’ from the Python module lowerquality https://lowerquality.com/gentle/. However, the inclusion of the original audio leaves the possibility for future research to develop more advanced alignment technique and recover additional features.
For each phoneme and word, we indicate the corresponding voice, speech rate, wav file, story, word position within the sequence, and sequence position within the story, and whether the sequence is a word list or a sentence.
Computing environment
In addition to the packages mentioned in this manuscript, the processing of the present data is based on the free and open-source ecosystem of the neuroimaging community. In particular, we used:
MNE BIDS46 (https://mne.tools/mne-bids)
Bids-Validator (https://github.com/bids-standard/bids-validator)
Nibabel49 (https://nipy.org/nibabel/)
Scikit-Learn50 (https://scikit-learn.org/)
Pandas51 (https://pandas.pydata.org/)
Data Records
The dataset is organized according to Brain Imaging Data Structure (BIDS) 1.2.145 and publicly available on the Open Science Framework data repository52 10.17605/OSF.IO/AG3KJ under a Creative Common Licence 0. An image of the folder structure is provided in Fig. 1. The detailed description of the BIDS file system is available at http://bids.neuroimaging.io/. In summary,
Fig. 1.
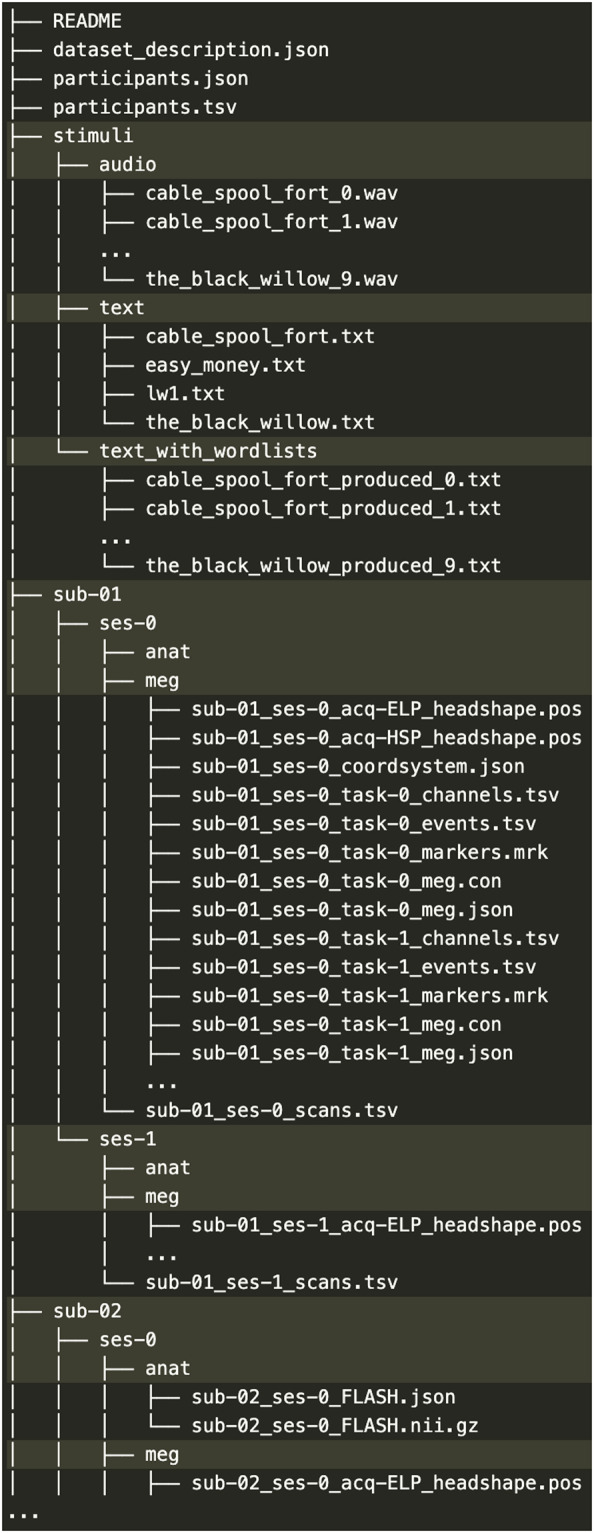
Dataset file structure.
• ‘./dataset_description.json’ describes the dataset
• ‘./participants.tsv’ indicates the age and gender of each participant, the order in which they heard the stories, whether they have an anatomical MRI scan, and how many recording sessions they completed
• ‘./stimuli/’ contains the original texts, the modified texts (i.e. with word lists), the synthesized audio tracks.
Each’./sub-SXXX’ contains the brain recordings of a unique participant divided by session (e.g.’ses-0’ and’ses-1’)
In each session folder lies the anatomical and the meg data, and the timestamp annotations (see Fig. 4).
Sessions are numbered by temporal order (s0 is first; s1 is second).
Tasks are numbered by a unique integer common across participants.
The dataset can be read directly with MNE-BIDS46.
Fig. 4.
MEG data annotations: Pandas DataFrame of sound, phoneme and word time-stamps.
Technical Validation
We checked that the present dataset complies with the standardized brain imaging data structure by using the Bids-Validator (https://github.com/bids-standard/bids-validator).
MEG recordings are notoriously noisy and thus challenging to validate empirically. In particular, MEG can be corrupted by environmental noise (nearby electronic systems) and physiological noise (eye movement, heart activity, facial movements)53. To address this issue, several labs have proposed a myriad of preprocessing techniques based on temporal and spatial filtering54 and trial and channel rejection55. However, there is currently no accepted standard for the selection and ordering these preprocessing steps. Consequently, we here opted for (1) a minimalist preprocessing pipeline derived from MNE-Python’s default pipeline48 followed by (2) median evoked responses and (3) standard single-trial linear decoding analyses.
Minimal preprocessing
For each subject separately, and using the default parameters of MNE-Python, we:
bandpass filtered the MEG data between 0.5 and 30.0 Hz with raw.load_data().filter(0.5, 30.0, n_jobs = 1),
temporally-decimate the data 10x, segment these continuous signals between −200 ms and 600 ms after word and phoneme onset, and apply a baseline correction between −200 ms to 0 ms with mne.Epochs(tmin = −0.2, tmax = 0.6, decim = 10, baseline = (−0.2, 0.0)),
and clip the MEG data between fifth and ninety-fifth percentile of the data across channels.
Evoked
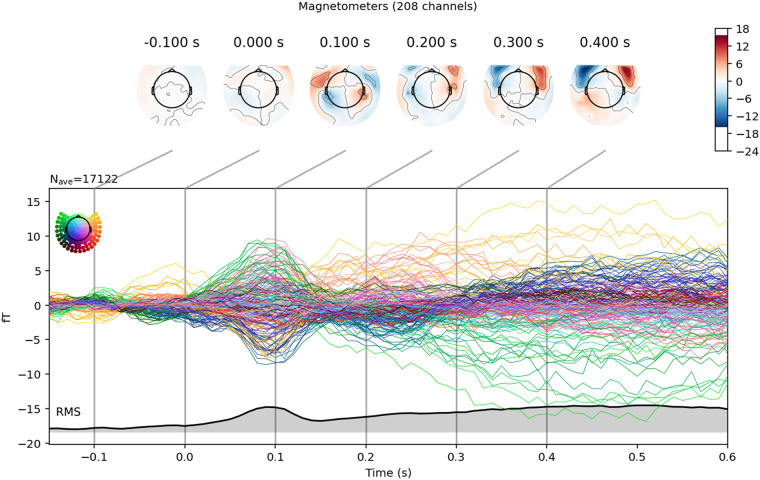
Figure 2 displays the median evoked responses across participants and words onset and after phoneme onsets, respectively. Both of these topographies are typical of auditory activity in MEG36.
Fig. 2.
Median (across subjects) evoked response to all words. The gray area indicates the global field power (GFP).
Decoding
For each recording independently, our objective was to verify the alignment between the word annotations and the MEG recordings. To this end, we trained a linear classifier W ∈ Rd across all d = 208 magnetometers (X ∈ Rn × d), for each time sample relative to word (or phoneme) onset independently, and for each subject separately. The classifier consisted of a standard scaler, followed by a linear discriminant regression implemented by scikit-learn50 using model = make_pipeline(StandardScaler(), LinearDiscriminantAnalysis())
decode high versus low median zipf-frequency of each word, as defined by the WordFreq package56.
decode whether the phoneme is voiced or not.
The decoding pipeline was trained and evaluated using a five-split cross-validation scheme (with shuffling) using cv = KFold(5, shuffle = True, random_state = 0) The scoring metric reported is Pearson R correlation between the continuous probabilistic output of the classifier on each trial, and the ground truth label (high vs. low for word frequency; voiced vs. voiceless for voicing). The full decoding pipeline can be found in the script check_decoding.py.
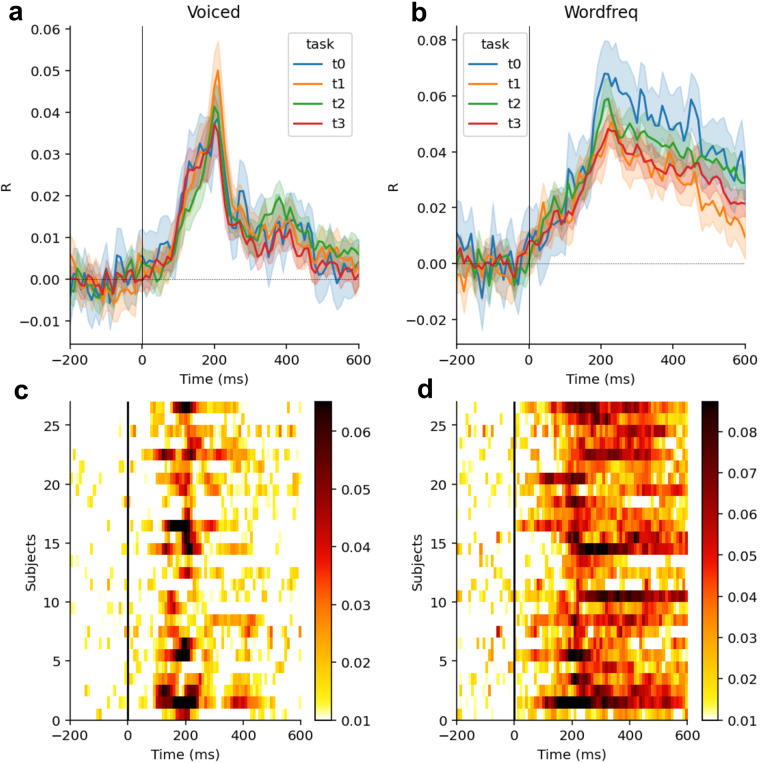
The results displayed in Fig. 3 show a reliable decoding at the phoneme and at the word level, across both subjects and tasks (i.e. stories).
Fig. 3.
(a) Average (mean) decoding of whether the phoneme is voiced or not as a function of time following phoneme onset. The four colors refer to the four tasks (stories + word lists). Error bar are SEM across subjects. (b) Same as A for the decoding of words’ zipf frequency as a function of word onset. (c) Decoding of voicing (average across all tasks) for each participant, as a function of time following phoneme onset. (d) Same as C for decoding of word frequency (average across all tasks) for each participant, as a function of time following word onset.
The success of our decoding analysis demonstrates: (i) the data have been correctly time-stamped relative to phoneme and word onset, in order to elicit a zero-aligned decoding timecourse; (ii) the data contain reliable signals that contain speech-related properties, suitable for further investigation; (iii) information at multiple levels (phonetic and lexical) are present in the data, allowing users to test hypotheses at different linguistic levels of description. We anticipate that encoding models would provide equally compelling results. Note that a decoding performance of Pearson R = 0.08 is typical for single-trial MEG data of continuous listening, and is of the same magnitude that has been reported in previous studies38. We have a large number of events (tens of thousands of phonemes; thousands of words), and this dataset has been demonstrated to provide sufficient statistical power to yield significant results, despite small effect sizes36,57.
Usage Notes
import pandas as pdimport mne bidsbids_path = mne_bids.BIDSPATH( subject = ’01’, session = ’0’, task = ’0’, datatype = ”meg”, root = ’my/data/path’)raw = mne_bids.read_raw_bids(bids_path)raw.load_data().data # channels X timesdf = raw.annotations.to_data_frame()
Accessing all sound, word and phoneme annotation is directly readable in a Pandas58 DataFrame format:
df = pd.DataFrame(df.description.apply(eval).to_list())
Acknowledgements
This work was funded in part by FrontCog grant ANR-17-EURE-0017 (JRK); Abu Dhabi Research Institute (G1001) (AM; LP); Dingwall Foundation (LG).
Author contributions
L.G. and J.-R.K. conceived the experiment. L.G. conducted the experiment. G.F. assisted with MEG and MRI data collection. L.G. and J-R.K. analyzed the results. A.M., L.P., D.P. and J.-R.K. contributed to the funding of the study. All authors reviewed the manuscript.
Code availability
The code is available on https://github.com/kingjr/meg-masc/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hickok G, Poeppel D. The cortical organization of speech processing. Nat. reviews neuroscience. 2007;8:393–402. doi: 10.1038/nrn2113. [DOI] [PubMed] [Google Scholar]
- 2.Berwick RC, Friederici AD, Chomsky N, Bolhuis JJ. Evolution, brain, and the nature of language. Trends cognitive sciences. 2013;17:89–98. doi: 10.1016/j.tics.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Dehaene S, Meyniel F, Wacongne C, Wang L, Pallier C. The neural representation of sequences: from transition probabilities to algebraic patterns and linguistic trees. Neuron. 2015;88:2–19. doi: 10.1016/j.neuron.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Hamilton LS, Huth AG. The revolution will not be controlled: natural stimuli in speech neuroscience. Lang. cognition neuroscience. 2020;35:573–582. doi: 10.1080/23273798.2018.1499946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gwilliams L, King J-R. Recurrent processes support a cascade of hierarchical decisions. ELife. 2020;9:e56603. doi: 10.7554/eLife.56603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pallier C, Devauchelle A-D, Dehaene S. Cortical representation of the constituent structure of sentences. Proc. Natl. Acad. Sci. 2011;108:2522–2527. doi: 10.1073/pnas.1018711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersson K-M, Folia V, Hagoort P. What artificial grammar learning reveals about the neurobiology of syntax. Brain language. 2012;120:83–95. doi: 10.1016/j.bandl.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Gwilliams L, Linzen T, Poeppel D, Marantz A. In spoken word recognition, the future predicts the past. J. Neurosci. 2018;38:7585–7599. doi: 10.1523/JNEUROSCI.0065-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bemis DK, Pylkkänen L. Simple composition: A magnetoencephalography investigation into the comprehension of minimal linguistic phrases. J. Neurosci. 2011;31:2801–2814. doi: 10.1523/JNEUROSCI.5003-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huth AG, De Heer WA, Griffiths TL, Theunissen FE, Gallant JL. Natural speech reveals the semantic maps that tile human cerebral cortex. Nature. 2016;532:453–458. doi: 10.1038/nature17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broderick MP, Anderson AJ, Di Liberto GM, Crosse MJ, Lalor EC. Electrophysiological correlates of semantic dissimilarity reflect the comprehension of natural, narrative speech. Curr. Biol. 2018;28:803–809. doi: 10.1016/j.cub.2018.01.080. [DOI] [PubMed] [Google Scholar]
- 12.Brodbeck C, Simon JZ. Continuous speech processing. Curr. Opin. Physiol. 2020;18:25–31. doi: 10.1016/j.cophys.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuster S, Hawelka S, Hutzler F, Kronbichler M, Richlan F. Words in context: The effects of length, frequency, and predictability on brain responses during natural reading. Cereb. Cortex. 2016;26:3889–3904. doi: 10.1093/cercor/bhw184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wehbe L, et al. Simultaneously uncovering the patterns of brain regions involved in different story reading subprocesses. PloS one. 2014;9:e112575. doi: 10.1371/journal.pone.0112575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haxby JV, Connolly AC, Guntupalli JS. Decoding neural representational spaces using multivariate pattern analysis. Annu. review neuroscience. 2014;37:435–456. doi: 10.1146/annurev-neuro-062012-170325. [DOI] [PubMed] [Google Scholar]
- 16.Naselaris T, Kay KN, Nishimoto S, Gallant JL. Encoding and decoding in fmri. Neuroimage. 2011;56:400–410. doi: 10.1016/j.neuroimage.2010.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriegeskorte N, Mur M, Bandettini PA. Representational similarity analysis-connecting the branches of systems neuroscience. Front. systems neuroscience. 2008;2:4. doi: 10.3389/neuro.06.004.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King J-R, Dehaene S. Characterizing the dynamics of mental representations: the temporal generalization method. Trends cognitive sciences. 2014;18:203–210. doi: 10.1016/j.tics.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, J.-R. et al. Encoding and decoding neuronal dynamics: Methodological framework to uncover the algorithms of cognition (2018).
- 20.King J-R, Charton F, Lopez-Paz D, Oquab M. Back-to-back regression: Disentangling the influence of correlated factors from multivariate observations. NeuroImage. 2020;220:117028. doi: 10.1016/j.neuroimage.2020.117028. [DOI] [PubMed] [Google Scholar]
- 21.Chehab, O., Defossez, A., Loiseau, J.-C., Gramfort, A. & King, J.-R. Deep recurrent encoder: A scalable end-to-end network to model brain signals. arXiv preprint arXiv:2103.02339 (2021).
- 22.Mesgarani N, Cheung C, Johnson K, Chang EF. Phonetic feature encoding in human superior temporal gyrus. Science. 2014;343:1006–1010. doi: 10.1126/science.1245994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian, P., Qiu, X. & Huang, X. Bridging lstm architecture and the neural dynamics during reading. arXiv preprint arXiv:1604.06635 (2016).
- 24.Jain, S. & Huth, A. Incorporating context into language encoding models for fmri. Adv. neural information processing systems31 (2018). [PMC free article] [PubMed]
- 25.Toneva, M. & Wehbe, L. Interpreting and improving natural-language processing (in machines) with natural language-processing (in the brain). Adv. Neural Inf. Process. Syst. 32 (2019).
- 26.Millet, J. & King, J.-R. Inductive biases, pretraining and fine-tuning jointly account for brain responses to speech. arXiv preprint arXiv:2103.01032 (2021).
- 27.Caucheteux C, King J-R. Brains and algorithms partially converge in natural language processing. Commun. Biology. 2022;5:1–10. doi: 10.1038/s42003-022-03036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein A, et al. Shared computational principles for language processing in humans and deep language models. Nat. neuroscience. 2022;25:369–380. doi: 10.1038/s41593-022-01026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrimpf, M. et al. The neural architecture of language: Integrative modeling converges on predictive processing. Proc. Natl. Acad. Sci. 118 (2021). [DOI] [PMC free article] [PubMed]
- 30.Caucheteux, C., Gramfort, A. & King, J.-R. Gpt-2’s activations predict the degree of semantic comprehension in the human brain (2021).
- 31.Caucheteux, C., Gramfort, A. & King, J.-R. Model-based analysis of brain activity reveals the hierarchy of language in 305 subjects. arXiv preprint arXiv:2110.06078, (2021).
- 32.Caucheteux, C., Gramfort, A. & King, J.-R. Disentangling syntax and semantics in the brain with deep networks. In International Conference on Machine Learning, 1336–1348 (PMLR, 2021).
- 33.Caucheteux, C., Gramfort, A. & King, J.-R. Long-range and hierarchical language predictions in brains and algorithms. arXiv preprint arXiv:2111.14232 (2021).
- 34.Heilbron, M., Ehinger, B., Hagoort, P. & De Lange, F. P. Tracking naturalistic linguistic predictions with deep neural language models. arXiv preprint arXiv:1909.04400, (2019).
- 35.Gillis M, Vanthornhout J, Simon JZ, Francart T, Brodbeck C. Neural markers of speech comprehension: measuring eeg tracking of linguistic speech representations, controlling the speech acoustics. J. Neurosci. 2021;41:10316–10329. doi: 10.1523/JNEUROSCI.0812-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gwilliams L, King JR, Marantz A, Poeppel D. Neural dynamics of phoneme sequences reveal position-invariant code for content and order. Nature Communications. 2022;13(1):1–14. doi: 10.1038/s41467-022-34326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nastase SA, et al. The “narratives” fmri dataset for evaluating models of naturalistic language comprehension. Sci. data. 2021;8:1–22. doi: 10.1038/s41597-021-01033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoffelen, J. et al. Mother of unification studies, a 204-subject multimodal neuroimaging dataset to study language processing (2019). [DOI] [PMC free article] [PubMed]
- 39.Van Essen DC, et al. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennan JR, Hale JT. Hierarchical structure guides rapid linguistic predictions during naturalistic listening. PloS one. 2019;14(1):e0207741. doi: 10.1371/journal.pone.0207741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armeni K, et al. A 10-hour within-participant magnetoencephalography narrative dataset to test models of language comprehension. Sci Data. 2022;9:278. doi: 10.1038/s41597-022-01382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veale JF. Edinburgh handedness inventory–short form: a revised version based on confirmatory factor analysis. Laterality: Asymmetries of Body, Brain and Cognition. 2014;19(2):164–177. doi: 10.1080/1357650X.2013.783045. [DOI] [PubMed] [Google Scholar]
- 43.Ide, N. & Macleod, C. The american national corpus: A standardized resource of american english. In Proceedings of corpus linguistics (Vol. 3, pp. 1–7). Lancaster, UK: Lancaster University Centre for Computer Corpus Research on Language (2001).
- 44.Fedorenko, E. et al Neural correlate of the construction of sentence meaning. Proceedings of the National Academy of Sciences, 113(41), E6256-E6262. Chicago (2016). [DOI] [PMC free article] [PubMed]
- 45.Gorgolewski KJ, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. data. 2016;3:1–9. doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Appelhoff, S. et al. Mne-bids: Organizing electrophysiological data into the bids format and facilitating their analysis. The J. Open Source Software. 4 (2019). [DOI] [PMC free article] [PubMed]
- 47.Gulban OF, 2019. poldracklab/pydeface: v2. 0.0. Zenodo. [DOI]
- 48.Gramfort, A. et al. Meg and eeg data analysis with mne-python. Front. neuroscience267 (2013). [DOI] [PMC free article] [PubMed]
- 49.Brett M, 2020. nipy/nibabel: 3.2.1. Zenodo. [DOI]
- 50.Pedregosa F, et al. Scikit-learn: Machine learning in python. J. machine Learn. research. 2011;12:2825–2830. [Google Scholar]
- 51.W McKinney. Data Structures for Statistical Computing in Python. In van der Walt, S. & Millman, J. (eds.) Proceedings of the 9th Python in Science Conference, 56–61, 10.25080/Majora-92bf1922-00a (2010).
- 52.King JR, Gwilliams L. 2022. MASC-MEG. OSF. [DOI]
- 53.Hämäläinen M, Hari R, Ilmoniemi RJ, Knuutila J, Lounasmaa OV. Magnetoencephalography—theory, instrumentation, and applications to noninvasive studies of the working human brain. Rev. modern Phys. 1993;65:413. doi: 10.1103/RevModPhys.65.413. [DOI] [Google Scholar]
- 54.de Cheveigné A, Nelken I. Filters: when, why, and how (not) to use them. Neuron. 2019;102:280–293. doi: 10.1016/j.neuron.2019.02.039. [DOI] [PubMed] [Google Scholar]
- 55.Jas M, Engemann DA, Bekhti Y, Raimondo F, Gramfort A. Autoreject: Automated artifact rejection for meg and eeg data. NeuroImage. 2017;159:417–429. doi: 10.1016/j.neuroimage.2017.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speer R, Chin J, Lin A, Jewett S, Nathan L. 2018. Luminosoinsight/wordfreq: v2.2. Zenodo. [DOI]
- 57.Gwilliams, L., Marantz, A., Poeppel, D. & King, J. R. Top-down information shapes lexical processing when listening to continuous speech. Language, Cognition and Neuroscience, 1–14 (2023).
- 58.pandas development team T. 2020. pandas-dev/pandas: Pandas. Zenodo. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Gulban OF, 2019. poldracklab/pydeface: v2. 0.0. Zenodo. [DOI]
- Brett M, 2020. nipy/nibabel: 3.2.1. Zenodo. [DOI]
- King JR, Gwilliams L. 2022. MASC-MEG. OSF. [DOI]
- Speer R, Chin J, Lin A, Jewett S, Nathan L. 2018. Luminosoinsight/wordfreq: v2.2. Zenodo. [DOI]
- pandas development team T. 2020. pandas-dev/pandas: Pandas. Zenodo. [DOI]
Data Availability Statement
The code is available on https://github.com/kingjr/meg-masc/.