Abstract
Aims
M2 macrophage is believed to play an important role in the development of endometriosis. This study aimed to identify several key genes related to the M2 macrophage in endometriosis.
Method
Differential expressed genes between endometriosis and non-endometriosis were identified based on three microarray datasets from the Gene Expression Omnibus database. Gene modules significantly associated with M2 macrophage were identified from the weighted gene co-expression network analysis. Furthermore, by intersecting the differential expressed genes and M2 macrophage-associated module genes, M2 macrophage-related genes in endometriosis were identified. Functional analyses of the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes for these genes were then performed. Following, the least absolute shrinkage and selection operator, random forest, and receiver operating characteristic curves were further conducted to identify the key M2 macrophage-related genes in endometriosis. Finally, the expressions of key genes in endometriosis, as well as their correlations with M2 macrophages were verified in an independent validation cohort.
Results
Totally, 185 M2 macrophage-related genes were identified, and they were mainly enriched in functions associated with the cell cycle, oocyte maturation, and immune response. Following machine learning algorithms, eight key genes were selected in the endometriosis: PGR, OLFM4, PIP5K1B, CCNA1, BRIP1, CADM1, PRAME, and GCNT1. The eight key genes were confirmed to be negative with M2 macrophage infiltration levels. Furthermore, the expression levels of these genes were significantly lower in the middle secretory stage while relevantly higher in the proliferative stage. The validation analysis also showed similar outcomes with the above results.
Conclusion
Eight M2 macrophage-related genes were identified as potential biomarkers of endometriosis, providing novel understanding of immune cells in the endometriosis.
Keywords: Endometriosis, Macrophage, Biomarker, Immune cell, Menstrual cycles
Highlights
-
•
Eight key M2 macrophage-related genes were identified in endometriosis.
-
•
The eight key genes were negatively related to the M2-macrophage.
-
•
The eight key genes were significantly down-expressed in the middle secretory stage.
-
•
The eight key genes were relevantly up-expressed in the proliferative stage.
1. Introduction
Endometriosis is a chronic estrogen-dependent inflammatory disease defined as lesions of endometrial-like tissue outside the uterus [1,2]. It is estimated that endometriosis affects 10% of women of reproductive age [3], equating to 190 million women worldwide [4]. The diagnosis of endometriosis is mainly through laparoscopy and the common treatment method is complete laparoscopic surgical resection of ectopic endometrial tissues [5]. Combined hormonal contraceptives with or without nonsteroidal anti-inflammatory drugs are first-line options in therapy of endometriosis and have tolerable adverse effect profile [6]. However, it is still unsatisfying that the progression of endometriosis is relatively slow. Usually, it takes 7–10 years before the onset of the endometriosis symptoms, which delays the diagnosis and optimal treatment timing [5]. Moreover, the pathogenesis of endometriosis is poorly understood. Therefore, it is urgent to find the possible diagnostic biomarkers and molecular mechanisms for endometriosis to improve the early diagnosis and treatment of endometriosis.
Recently, the disturbance of the immune microenvironment is demonstrated to play a pivotal role in the pathophysiology and development of endometriosis [7]. Macrophages are ubiquitous immune cells and play important roles in both innate and acquired immunity. The macrophages are abundant throughout female reproductive tissues like ovary, uterus, oviduct and mammary gland [8], and the dysregulation in the endometriosis is common [9]. Increasing of macrophages stimulate cytotoxic T helper cells to release excessive inflammatory cytokines, resulting in promoting the shape of a pro-inflammatory environment in the endometrium and eventually developing an endometriosis [7]. Alternatively activated or M2 macrophage, polarized by Th2 cytokines such as IL-4 and IL-13 and producing anti-inflammatory cytokines such as IL-10 and TGF-β, are anti-inflammatory and immunoregulatory in the diseases [10]. Moreover, aberrant increase and activation of anti-inflammatory M2 macrophage stimulate the abnormal gene expression that is associated with the ectopic endometrium [7]. Moreover, in a deeper investigation, M2 macrophage is elevated in III-IV endometriosis [11]. In addition, specific phenotypes and functions of macrophages also correspond to each phase of the menstrual cycle [12]. While short menstrual cycle is one of the features of high risk of endometriosis [3], and endometrial diseases have a menstrual cycle-dependent epigenetic profile [13]. All these evidences demonstrate that macrophages may play important role in endometriosis progression. Considering the contribution of M2 macrophages in the immunopathogenesis of endometriosis, the identification of M2 macrophage-associated genes may help in the search for potential diagnostic markers of the diseases.
Thanks to the rapid development of bioinformatics, it is convenient to recognize and explore the role of specific genes in diseases. A relevant bioinformatics analysis has reported five M2 macrophage-related genes [14]. Distinguishing from this, the present study aimed to identify key genes related to M2 macrophage through machine learning algorithms. These genes are novel and initially proposed by bioinformatics analyses. In addition to confirming the aberrant expression of these genes in endometriosis and their relationships with M2 macrophages, this study also explored the expression changes of these genes during menstrual cycles in endometriosis. These findings may provide a novel insight for the diagnosis and therapy for patients with endometriosis at the molecular level.
2. Method
2.1. Data source
Based on the Gene Expression Omnibus (GEO) database [15] (http://www.ncbi.nlm.nih.gov/geo/), this study screened microarray datasets with the following criteria: 1) endometriosis is used as keyword; 2) organisms are homo sapiens and samples are endometrium tissues; 3) the sample size is greater than 20. Finally, four datasets (GSE51981, GSE6364, GSE7305 and GSE135485) were obtained. GSE51981 contained 71 non-endometriosis and 77 endometriosis. GSE6364 contained 16 non-endometriosis and 21 endometriosis. GSE7305 contained 10 non-endometriosis and 10 endometriosis. GSE135485 contained 4 non-endometriosis and 54 endometriosis. Samples in the first three datasets were sequenced by GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array and samples in the last dataset were sequenced with Illumina HiSeq 3000.
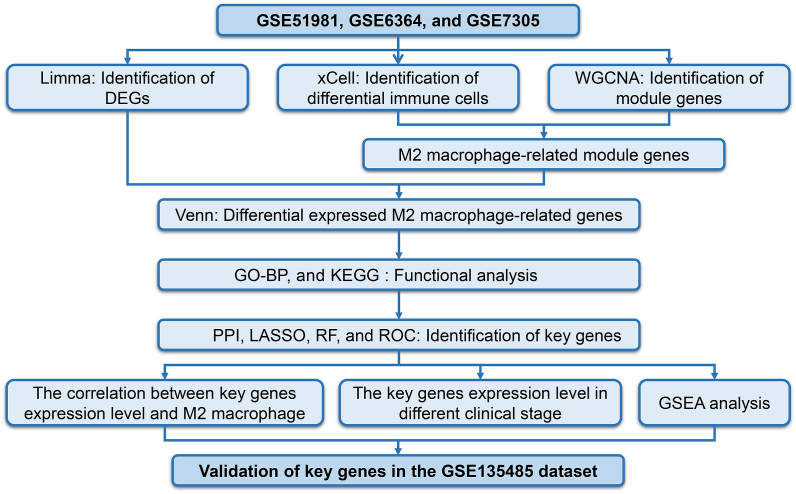
The study design was shown in Fig. 1.
Fig. 1.
The study design. DEGs, differential expressed genes; WGCNA, Weighed Gene Co-expression Network Analysis; GO-BP, Gene Ontology – Biological Process; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, Protein-Protein Interaction Networks; RF, Random Forest; LASSO, Least Absolute Shrinkage and Selection Operator; ROC, Receiver Operator Characteristic; GSEA, Gene Set Enrichment Analysis.
2.2. Data processing
The probe expression matrix of GSE51981, GSE6364, and GSE7305 after pretreatment, standardization, and log2 transformation were downloaded. Platform annotation file was also downloaded and was utilized to convert the probe into gene expression matrix. For the gene with numerous probes, the average value of probes was defined as the gene expression level. Data in GSE51981, GSE6364, and GSE7305 were all detected from GPL570, and this study performed a batch effect removing analysis by sva package [16] (version 3.38.0) in R 3.6.1. The combined gene expression data after removing the batch effects was named combat edata, which was utilized for the following analysis.
The GSE135485 dataset sequenced by Illumina HiSeq 3000 was standardized by TMM algorithm in edgeR package (version 3.4) [17,18]. The data were further converted into logCPM value for the following validation analysis.
2.3. Analysis of differential expression
This study performed a Classical Bayesian method using limma package [19] (version 3.10.3) in the combat edata obtained previously. Differential genes expression between endometriosis and non-endometriosis were analysed and relevant p.value and logfold change (FC) were obtained. In addition, multiple testing corrections were carried out to get adjusted p value namely adj.p.value using the Benjamini & Hochberg (BH) method [20]. This study selected differentially expressed thresholds of adj.p.value < 0.05 and |logFC| > 0.5 as the differentially expressed genes (DEGs).
2.4. Analysis of immune cell composition
In order to identify the significantly differential immune cells between disease group and the control group, this study estimated the relative abundance of immune cells and stromal cells in each sample using xCell package [21] in R. Furthermore, the significance p.value was calculated by Wilcox test, and the p value was corrected by BH to obtain p.adjust. Immune cells with significant differences between the disease group and the control group were found for the subsequent analysis with p.adjust <0.05. Then, the CIBERSORT algorithm [22,23] was further used to verify the difference in the proportion of M2 macrophages between two groups.
2.5. Analysis of macrophage M2-related genes
Top 3000 genes with large variation in the combat edata was selected to screen the highly coordinated variation gene modules using weighted correlation network analysis (WGCNA) [24]. In the WGCNA algorithm, the elements in the gene co-expression matrix are the weighted values of gene correlation coefficients, that is, the soft-threshold power. By setting a series of powers, this study calculated the square value of correlation coefficient of connectivity k and p(k) and the average connectivity under each power value, and then selected the appropriate power to allow the connections between genes in the network conform to the scale-free network distribution. Then, genes with similar expression pattern (similarity is greater than 0.3) are combined into a module based on cluster analysis and dynamic pruning method. By calculating the correlation between modules and phenotypes (immune cell infiltration abundance), the modules highly correlated with immune cells were selected as the immune cell-related module and genes in module were selected as immune cell-related genes. Moreover, this study intersected the immune cell-related genes with the DEGs as the differential immune cell-related genes. According to the results, macrophage M2 showed most significant positive and negative correlation with purple and brown modules, respectively. So the purple and brown modules were defined as the key module, and genes in the two modules were defined as a key immune cell (macrophage M2)-related module gene.
2.6. Functional analysis of macrophage M2-related genes and construction of Protein-Protein Interaction (PPI) Networks
Gene Ontology [25] biological process (GO-BP) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) [26] pathway were conducted in the differential macrophage M2-related genes using clusterProfiler package (version 3.8.1) [27] in R. BH was used for p value correction, after correction, P.adjust <0.05 was considered as a significant enrichment result. Due to the large number of GO-BP results, there was some redundancy. Therefore, simplifyEnrichment (Version: 1.4.0) [28] was further adopted for the further processing. The package used binary cut to divide the obtained GO similarity matrix into several classes. Through the annotation, the corresponding function of each class can be captured.
At the same time, differential macrophage M2-related genes were input into STRING (version 10.0) [29], defined species as homo, PPI parameter as 0.4 (medium confidence) to get the PPI relationship pairs. Cytoscape [30] was utilized to construct a PPI network.
2.7. Selection of key genes
In order to screen which genes in the above macrophage M2-related genes played important role in the disease, two algorithms were carried out. Least Absolute Shrinkage and Selection Operator (LASSO) was used to select the genes using glmnet package (version 4.0-2) [31]. The parameter is set to nfold = 20, that is, 20-fold cross-validation is performed. Random Forest (RF) algorithm [32] were subsequently used to select the Top30 genes. The parameter is set as: ntree = 1000, mtry = 6. The intersecting genes from LASSO and RF algorithms were selected to draw the Receiver Operator Characteristic (ROC) curve and screen genes with areas under the curve (AUC) ≥ 0.65 as the final key genes.
2.8. Analysis of expression features for key genes
Pearson correlations were conducted to validate the correlation between key genes and M2 macrophage infiltration level. The relationships between the key genes and clinical features were also analysed. Box plots of the severity of endometriosis (Minimal/Mild and Moderate/Severe) and the expression levels of key genes at different times (endometrial proliferative stage, early secretory stage, and middle secretory stage) were plotted.
2.9. Enrichment pathway analysis of key genes
In order to observe the mechanism of key genes in the process of endometriosis occurrence, Gene Set Enrichment Analysis (GSEA) was conducted for each key gene using GSEA software (version 3.0) [33]. Parameters are set as: Gene sets database: c2.cp.kegg.v7.4.symbols.gmt; Phenopyte labels: Use a gene as the phenotype; Metric for ranking genes: Pearson; Gene list sorting mode: real; Gene list ordering mode: descending; threshold: NOM p-val <0.01.
2.10. Validation of the key genes
The validation data set GSE135485 was used to extract the expression values of key genes in each sample. The differences of the gene expression levels between the disease group and the control group were analysed. xCell algorithm was also used to estimate the macrophage M2 infiltration level of each sample, and Pearson correlation coefficient between key genes and macrophages M2 was calculated.
3. Results
3.1. Identification of DEGs
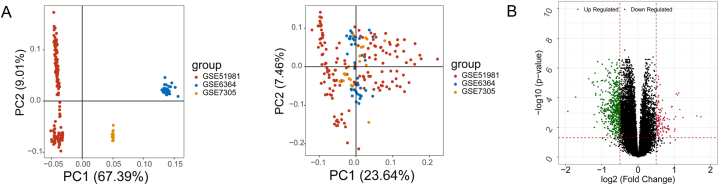
Data in GSE51981, GSE6364, and GSE7305 were processed to remove batch effects and all samples were generated into combat edata. There was no significant separation between samples after batch effects calibration (Fig. 2A), suggesting the data were able to use for the further analysis. According to the threshold setting in the method, 116 up-regulated and 519 down-regulated DEGs were identified (Fig. 2B).
Fig. 2.
Identification of DEGs. (A) PCA shows gene distributions before (left) and after (right) batch effect removal. (B) The volcano plot of DEGs. The red triangle represents the up-regulated genes, the green square represents the down-regulated genes, and the black indicates that the gene difference is not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Identification of differential expressed M2 macrophage-related genes
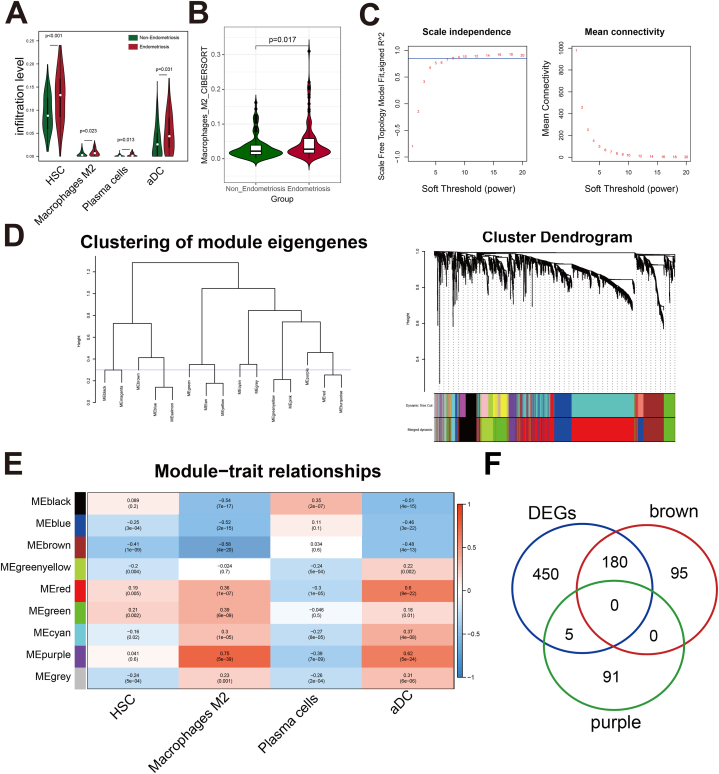
The immune infiltration levels of 64 stromal and immune cells were calculated using xCell package in R. There are significant differential infiltration levels between the non-endometriosis and endometriosis groups in four cells: hematopoietic stem cell (HSC), M2 macrophage, plasma cells, and activated dendritic cell (aDC) (p < 0.05). The infiltration level of HSC, M2 macrophage, plasma cells, and aDC in the endometriosis group were significantly higher than in the Non-endometriosis group (Fig. 3A). The infiltration level of macrophage M2 in xCell was provided in Table S1. To further verity the infiltration difference of M2 macrophage between two groups, the CIBERSORT algorithm was implemented. The results also suggested an increased level of M2 macrophage infiltration in the endometriosis group than that in the non-endometriosis group (Fig. 3B), which is consistent with the above findings.
Fig. 3.
Identification of differential expressed M2 macrophage-related genes. (A) Screening of four immune cells with significant infiltration differences between endometriosis and non-endometriosis groups based on the xCell algorithm. (B) The CIBERSORT algorithm was applied to confirm the infiltration difference of M2 macrophages between two groups. (C) Selection of the soft-threshold powers in WGCNA analysis. (D) Dendrogram of all genes clustered based on the measurement of dissimilarity. (E) Heat plot of correlation between different modules and immune cells. (F) Venn plot of intersecting of DEGs and M2 macrophage-related module genes.
The Top3000 genes in the combat edata were selected to conduct a WGCNA analysis. Finally, the genes were integrated into nine modules (Fig. 3C and D). Following, the correlations between the gene modules and the immune infiltration level were calculated (Fig. 3E). The brown (275 genes, r = −0.58, p < 0.001) and the purple (96 genes, r = 0.75, p < 0.001) modules were most significantly negative and positive with the M2 macrophage (Fig. 3E). Therefore, the brown and the purple modules were selected to be the key M2-related gene modules for the further analysis. Intersecting the key M2-related gene modules with the DEGs, 180 differential M2-related brown module genes and 5 differential M2-related purple module genes were obtained (Fig. 3F).
3.3. The function of M2-related genes and construction of the PPI network
GO-BP and KEGG pathway enrichment analyses were conducted to explore the function of the M2-related genes. As there were too many GO-BP items and it was redundancy, semantic similarity enrichment analysis was further conducted for GO-BP. Among them, 27 GO-BP clusters and eight GO-BP clusters were obtained from the brown and purple module genes, respectively. The most significant items in each cluster were visualized in Fig. 4A. Genes in the module brown were more likely to involve in chromosome segregation and nuclear division, while the purple module genes were mainly enriched in the regulation of syncytium formation and immunity. The KEGG pathway of the M2-related genes were enriched in the cell cycle, oocyte meiosis, DNA replication, progesterone-mediated oocyte maturation, p53 signaling pathway, human T-cell leukemia virus 1 infection, antifolate resistance, base excision repair, natural killer cell mediated cytotoxicity, and IL-17 signaling pathway (Fig. 4B).
Fig. 4.
The function analysis of the differential expressed M2 macrophage-related genes. (A) The most significant item in each GO BP clusters. (B) The KEGG pathway significantly enriched in differential module genes. (C) The PPI network constructed using differential expressed module genes.
Furthermore, a PPI network was constructed in the 185 differential M2-related module genes (Fig. 4C). A total of 3030 interaction pairs composed of 140 gene proteins were obtained. The list of these 140 genes and their connections was provided in Table S2. Of these, CDK1, CCNB1, and MAD2L1 may contribute more to the progression of endometriosis, since they have more degrees of connections in this network.
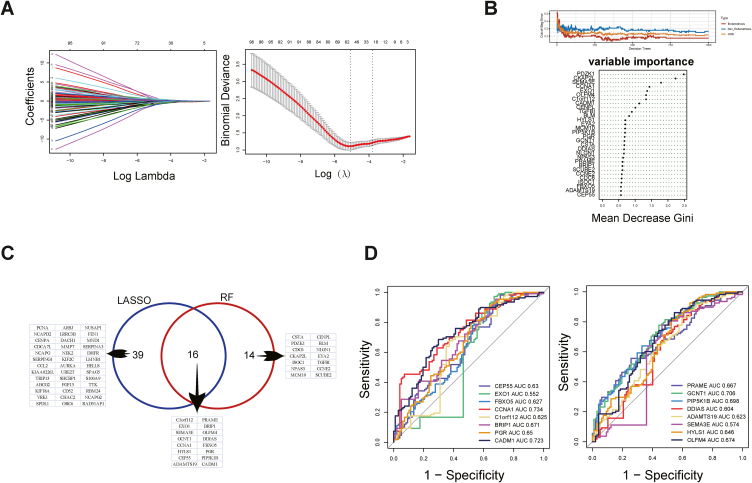
3.4. Screening of key genes
LASSO regression analysis identified 55 genes from 140 PPI genes (Fig. 5A). RF algorithm was used to select the Top30 genes (Fig. 5B). Following, intersecting the 55 genes from LASSO and 30 genes from RF, 16 genes were finally obtained (Fig. 5C). Finally, ROC analysis was performed in the intersecting genes and eight genes with AUC ≥0.65 were selected as the key genes (Fig. 5D). The eight key genes were PGR, OLFM4, PIP5K1B, CCNA1, BRIP1, CADM1, PRAME, and GCNT1.
Fig. 5.
Screening of the key genes. (A) LASSO coefficient distribution (left) and the likelihood bias of LASSO coefficient distribution (right). (B) The Top30 genes, ranked by importance, were selected using the RF algorithm. (C) Intersecting Venn plot of genes from LASSO and RF. (D) ROC curves of 16 intersecting genes, and eight genes with AUC >0.65 were screened as the key genes.
3.5. The analysis for key genes
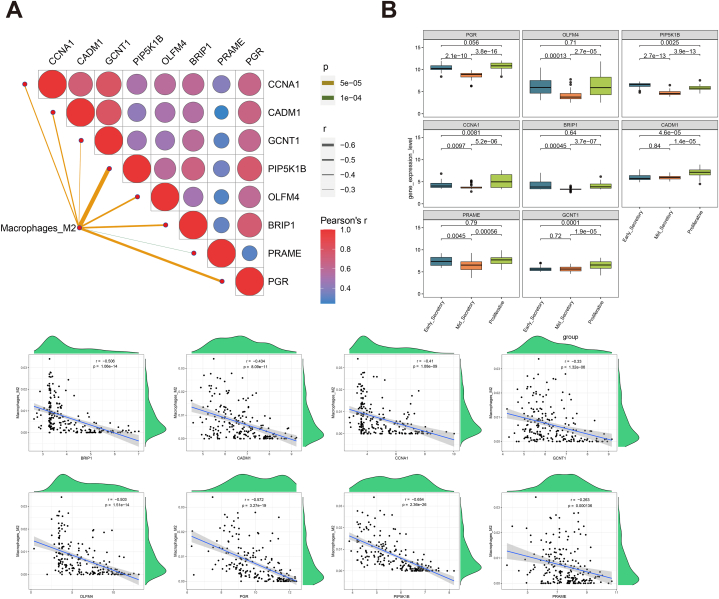
The correlations of each key gene and the M2 macrophage were evaluated, and the expressions of eight key genes were all significantly negative with the infiltration levels of M2 macrophage (Fig. 6A). In addition, the box plots of the gene expression level in different stage were plotted (Fig. 6B). The expression levels of the eight key genes were significantly lower in the middle secretory stage (p < 0.05) while relevantly higher in the proliferative stage. This study also compared the expression difference of the eight genes between different severity degree of endometriosis group in proliferative, early, and mid secretory phases of menstrual cycle, respectively (Fig. S1). The results showed that only PRAME in moderate/severe group were significantly lower than in the minimal/mild group in the early secretory stage. However, the expression difference of these genes between different severity degree of endometriosis group was not found in proliferative and mid secretory phases.
Fig. 6.
The analysis for key genes. (A) The correlation of each key gene expression and the M2 macrophage infiltration level. (B) Box plots of the gene expression level in different stage.
3.6. The GSEA enrichment pathway of the key genes
The GSEA enrichment of KEGG pathway analysis for eight genes was performed. The results showed that these genes were significantly enriched in pathways related to genomic instability, such as base excision repair, DNA replication, mismatch repair, nucleotide excision repair, RNA degradation spliceosome etc. (Fig. 7A–H).
Fig. 7.
The GSEA enrichment pathways of eight key genes.
The enrichment pathways for BRIP1, CADM1, CCNA1, GCNT1, PIP5K1B, PRAME, PGR, and OLFM4 are shown in panel A–H, respectively.
3.7. Validation of the key genes
GSE135485 was utilized as a validation dataset. The eight gene expression levels in the samples of non-endometriosis and endometriosis groups were analysed (Fig. 8A). All the eight genes showed downregulated trend in the endometriosis groups. Except for the OLFM4 and CADM1, the other 6 genes were all significant differences between the non-endometriosis and endometriosis groups (p < 0.05). The ROC curves showed five genes (PGR AUC = 0.796, PIP5K1B AUC = 0.759, CCNA1 AUC = 0.912, BRIP1 AUC = 0.773, CADM1 AUC = 0.694, and GCNT1 AUC = 0.829) with AUCs >0.65 (Fig. 8B). These results further demonstrated that the eight key genes were significantly associated with the endometriosis. Moreover, the Pearson coefficients were calculated to evaluate the correlation between the key genes and the M2 macrophage (Fig. 8C). The seven key genes were significantly negatively related to the M2 macrophage.
Fig. 8.
Validation of the key genes in the GSE135485. (A) The eight gene expression level in the non-endometriosis and endometriosis groups. (B) The ROC curves for the eight key genes. (C) The correlation between the key genes and the M2 macrophage.
4. Discussion
Endometriosis, affects around 10 % of reproductive women, with growth or deposition of endometrial tissue at extra uterine sites. M2 macrophages play important roles in the endometriosis which may stem from the regulation of M2 macrophage-related genes [7]. In the present study, by analyzing the data downloaded from the public GEO database, this study identified 185 M2 macrophage-related genes through a series bioinformatics method. These M2 macrophage-related genes were mainly enriched in the oocyte-related function, cell cycle, and immune cell cytokines-related function (Fig. 4A–B). The PPI network suggested that these genes had close interaction relationships and might play an important role in endometriosis progression. To obtain the optimal genes related to M2-macrophage, further LASSO, RF, and ROC analysis were conducted to screen the optimal key genes in the endometriosis. Finally, eight key genes were obtained: PGR, OLFM4, PIP5K1B, CCNA1, BRIP1, CADM1, PRAME, and GCNT1.
The eight key genes were all demonstrated to be negatively related to M2 macrophage, further confirming their closely correlations with the M2 macrophage. PGR, progesterone receptor, has been demonstrated to be important in regulating female reproduction [34]. A study has confirmed that the increased methylation of PGR isoforms may be associated with the reduced gene expression level, thereby impairing endometrial receptivity in patients with endometriosis [35]. The loss of PGR nuclear positivity in the proliferative endometrium of patients with endometriosis also suggests a decrease in the expression level of PGR [36]. These findings are consistent with our study, suggesting that PGR expression levels are regulated by methylation of PGR isoforms, which may further affect the endometrial receptivity. In addition, this study also found the elevated level of M2 macrophages and their significant negative correlations with PGR levels in patients with endometriosis. Jeong et al. revealed that a large number of CD163+ (M2) macrophages was associated with PGR negativity in tumor [37]. These evidences demonstrate that patients with endometriosis may be in a state of immunosuppression, leading to the increased infiltration of M2 macrophages, which can cause damage to the endometrium. OLFM4, olfactomedin-4, an extracellular matrix protein, is one of the top downregulated genes in patients with endometriosis compared with non-endometriosis controls [38]. However, it is highly expressed in human endometrium and has the highest expression level in proliferative-phase endometrium [39]. These conclusions are consistent with the present study, as this study found that OLFM4 expression in endometriosis is menstrual cycle-dependent, with decreased expression in the midsecretory phase and relatively high expression in the proliferative phase (Fig. 6B). In ulcerative colitis, the expression level of OLFM4 is also significantly negatively correlated with the infiltration of M2 macrophages [40]. Therefore, this study further hypothesized that OLFM4 may play an anti-inflammatory and endometrial stabilizing role by negatively regulating M2 macrophages. PIP5K1B, CCNA1, BRIP1, CADM1, PRAME, and GCNT1 were demonstrated to involve in various diseases and cancers, but their roles in endometriosis and their relationships with M2 macrophages remain unreported. Therefore, subsequent functional experiments are needed to investigate their immunoregulatory mechanisms.
In a menstrual cycle, endometrium undergoes transition from estrogen-dominant proliferative (follicular) phases, progesterone-dominant secretory (luteal) phases (early-secretory, mid-secretory, and late-secretory), to menstrual phase. In each endometrial cycle phase, the transcriptional profile exhibited distinct differences [41]. The transcriptomic profiles of uterine linings in patients with endometriosis will alter [38], demonstrating that the molecular feature of endometriosis may be a marker of onset of endometriosis. In the present study, the results of testing set and validation set indicated that the expressions of eight key genes were decreased in endometriosis. Meanwhile, in patients with endometriosis, the expression of these eight key genes shows a similar pattern, that is, the expression is decreased in the mid-secretion stage, but significantly up-regulated in the proliferative stage. Therefore, the expression of these eight key genes is considered to be menstrual cycle-dependent, and their aberrant down-regulation in endometriosis may be associated with the reduced expression during the mid-secretory phase. It is known that the short menstrual cycle is one of the risk factors for endometriosis [3], so increasing the expression levels of these genes in the mid-secretory phase may be beneficial in reducing the risk of endometriosis. However, these speculations still need to be validated by the clinical sample-based sequencing analyses.
There are still several limitations in the present study. Firstly, the down-regulation of these eight genes in endometriosis needs to be verified by in vitro experiments. Secondly, the relationships between these eight genes and M2 macrophages, as well as their regulatory mechanisms in endometriosis still need to be excavated by conducting in vivo and in vitro experiments. Finally, the lack of clinical information such as regional and ethnic limit the generalisability of this study. Therefore, how the eight genes affect the disease progression of endometriosis through the menstrual cycle still needs to be confirmed by experiments based on a large number of clinical samples.
5. Conclusions
Using machine learning algorithms, this study identified eight M2 macrophage-related genes (PGR, OLFM4, PIP5K1B, CCNA1, BRIP1, CADM1, PRAME, and GCNT1) that may be potential biomarkers of endometriosis. These genes were confirmed to be significantly under-expressed in endometriosis and were significantly negatively correlated with the infiltration of M2 macrophages. Furthermore, the expression pattern of these genes may be menstrual cycle-dependent. However, these conclusions need to be further explored by in vitro and in vivo experiments, as well as clinical sample-based analyses.
Funding
No funding was used in this study.
Data Availability statement
All data generated or analysed during this study are included in this article.
CRediT authorship contribution statement
Hongyan Ding: Writing – original draft, Conceptualization. Hongge Xu: Formal analysis, Data curation. Ting Zhang: Validation, Methodology, Investigation. Can Shi: Writing – review & editing, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22258.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Chapron C., Marcellin L., Borghese B., Santulli P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat. Rev. Endocrinol. 2019;15:666–682. doi: 10.1038/s41574-019-0245-z. [DOI] [PubMed] [Google Scholar]
- 2.Mińko A., Turoń-Skrzypińska A., Rył A., Bargiel P., Hilicka Z., Michalczyk K., Łukowska P., Rotter I., Cymbaluk-Płoska A. Endometriosis-A multifaceted problem of a modern woman. Int. J. Environ. Res. Publ. Health. 2021;18 doi: 10.3390/ijerph18158177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafrir A.L., Farland L.V., Shah D.K., Harris H.R., Kvaskoff M., Zondervan K., Missmer S.A. Risk for and consequences of endometriosis: a critical epidemiologic review. Best Pract. Res. Clin. Obstet. Gynaecol. 2018;51:1–15. doi: 10.1016/j.bpobgyn.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Kvaskoff M., Mahamat-Saleh Y., Farland L.V., Shigesi N., Terry K.L., Harris H.R., Roman H., Becker C.M., As-Sanie S., Zondervan K.T., Horne A.W., Missmer S.A. Endometriosis and cancer: a systematic review and meta-analysis. Hum. Reprod. Update. 2021;27:393–420. doi: 10.1093/humupd/dmaa045. [DOI] [PubMed] [Google Scholar]
- 5.Falcone T., Flyckt R. Clinical management of endometriosis. Obstet. Gynecol. 2018;131:557–571. doi: 10.1097/AOG.0000000000002469. [DOI] [PubMed] [Google Scholar]
- 6.Edi R., Cheng T. Endometriosis: evaluation and treatment. Am. Fam. Physician. 2022;106:397–404. [PubMed] [Google Scholar]
- 7.Vallvé-Juanico J., Houshdaran S., Giudice L.C. The endometrial immune environment of women with endometriosis. Hum. Reprod. Update. 2019;25:564–591. doi: 10.1093/humupd/dmz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu R., Van der Hoek K.H., Ryan N.K., Norman R.J., Robker R.L. Macrophage contributions to ovarian function. Hum. Reprod. Update. 2004;10:119–133. doi: 10.1093/humupd/dmh011. [DOI] [PubMed] [Google Scholar]
- 9.Wu J., Xie H., Yao S., Liang Y. Macrophage and nerve interaction in endometriosis. J. Neuroinflammation. 2017;14:53. doi: 10.1186/s12974-017-0828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S.A., Mardani F., Seifi B., Mohammadi A., Afshari J.T., Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 11.Poli-Neto O.B., Meola J., Rosa E.S.J.C., Tiezzi D. Transcriptome meta-analysis reveals differences of immune profile between eutopic endometrium from stage I-II and III-IV endometriosis independently of hormonal milieu. Sci. Rep. 2020;10:313. doi: 10.1038/s41598-019-57207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L.L., Li Z.H., Wang H., Kwak-Kim J., Liao A.H. Cutting edge: the regulatory mechanisms of macrophage polarization and function during pregnancy. J. Reprod. Immunol. 2022;151 doi: 10.1016/j.jri.2022.103627. [DOI] [PubMed] [Google Scholar]
- 13.Saare M., Modhukur V., Suhorutshenko M., Rajashekar B., Rekker K., Sõritsa D., Karro H., Soplepmann P., Sõritsa A., Lindgren C.M., Rahmioglu N., Drong A., Becker C.M., Zondervan K.T., Salumets A., Peters M. The influence of menstrual cycle and endometriosis on endometrial methylome. Clin. Epigenet. 2016;8:2. doi: 10.1186/s13148-015-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Z., Bhandari R., Lei Q., Lu M., Zhang L., Zhang M., Sun F., Feng L., Zhao S. Identification and exploration of novel macrophage M2-related biomarkers and potential therapeutic agents in endometriosis. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.656145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.-C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolayeva O., Robinson M.D. edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol. Biol. 2014;1150:45–79. doi: 10.1007/978-1-4939-0512-6_3. [DOI] [PubMed] [Google Scholar]
- 18.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smyth G.K. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Vol. Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Springer New York; New York, NY: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 20.Ghosh D. Wavelet-based Benjamini-Hochberg procedures for multiple testing under dependence. Math. Biosci. Eng. 2019;17:56–72. doi: 10.3934/mbe.2020003. [DOI] [PubMed] [Google Scholar]
- 21.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z., Lei K., Li H., He J., Shi E. Transcriptome-based network analysis related to M2-like tumor-associated macrophage infiltration identified VARS1 as a potential target for improving melanoma immunotherapy efficacy. J. Transl. Med. 2022;20:489. doi: 10.1186/s12967-022-03686-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao K., Zhao S., Yuan J., Pan Y., Song Y., Tang L. Construction of molecular subtypes and related prognostic and immune response models based on M2 macrophages in glioblastoma. Int. J. Gen. Med. 2022;15:913–926. doi: 10.2147/IJGM.S343152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu Z., Hübschmann D. simplifyEnrichment: an R/bioconductor package for clustering and visualizing functional enrichment results. bioRxiv. 2023;21:190–202. doi: 10.1016/j.gpb.2022.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucl. Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman J., Hastie T., Tibshirani R. Glmnet: lasso and elastic-net regularized generalized linear models. R Package Version. 2009;1 [Google Scholar]
- 32.Liaw A., Wiener M. Package 'randomForest': breiman and Cutler's random forests for classification and regression. R Development Core Team. 2014;4:6–10. [Google Scholar]
- 33.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Zhang B.Y., Feng G.D., Xiang W., Ma Y.X., Chen H., Chu M.X., Wang P.Q. The mechanism of miRNA-mediated PGR signaling pathway in regulating female reproduction. Yi Chuan. 2016;38:40–51. doi: 10.16288/j.yczz.15-293. [DOI] [PubMed] [Google Scholar]
- 35.Rocha-Junior C.V., Da Broi M.G., Miranda-Furtado C.L., Navarro P.A., Ferriani R.A., Meola J. Progesterone receptor B (PGR-B) is partially methylated in eutopic endometrium from infertile women with endometriosis. Reprod. Sci. 2019;26:1568–1574. doi: 10.1177/1933719119828078. [DOI] [PubMed] [Google Scholar]
- 36.Colón-Caraballo M., García M., Mendoza A., Flores I. Human endometriosis tissue microarray reveals site-specific expression of estrogen receptors, progesterone receptor, and Ki67. Appl. Immunohistochem. Mol. Morphol. 2019;27:491–500. doi: 10.1097/PAI.0000000000000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong H., Hwang I., Kang S.H., Shin H.C., Kwon S.Y. Tumor-associated macrophages as potential prognostic biomarkers of invasive breast cancer. J Breast Cancer. 2019;22:38–51. doi: 10.4048/jbc.2019.22.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirza Z., Abdel-Dayem U.A. Uncovering potential roles of differentially expressed genes, upstream regulators, and canonical pathways in endometriosis using an in silico genomics approach. Diagnostics. 2020;10 doi: 10.3390/diagnostics10060416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dassen H., Punyadeera C., Delvoux B., Schulkens I., Marchetti C., Kamps R., Klomp J., Dijcks F., de Goeij A., D'Hooghe T., Kyama C., Ederveen A., Dunselman G., Groothuis P., Romano A. Olfactomedin-4 regulation by estrogen in the human endometrium requires epidermal growth factor signaling. Am. J. Pathol. 2010;177:2495–2508. doi: 10.2353/ajpath.2010.100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He T., Wang K., Zhao P., Zhu G., Yin X., Zhang Y., Zhang Z., Zhao K., Wang Z., Wang K. Integrative computational approach identifies immune-relevant biomarkers in ulcerative colitis. FEBS Open Bio. 2022;12:500–515. doi: 10.1002/2211-5463.13357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponnampalam A.P., Weston G.C., Trajstman A.C., Susil B., Rogers P.A. Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol. Hum. Reprod. 2004;10:879–893. doi: 10.1093/molehr/gah121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article.