Abstract
The role of cell adhesion molecules in mediating interactions with neighboring cells and the extracellular matrix has long been appreciated. More recently, these molecules have been shown to modulate intracellular signal transduction cascades critical for cell growth and proliferation. Expression of adhesion molecule on glia (AMOG) is downregulated in human and mouse gliomas, suggesting that AMOG may be important for growth regulation in the brain. In this report, we examined the role of AMOG expression on cell growth and intracellular signal transduction. We show that AMOG does not negatively regulate cell growth in vitro or in vivo. Instead, expression of AMOG in AMOG-deficient cells results in a dramatic increase in cell size associated with protein kinase B/Akt hyperactivation, which occurs independent of phosphatidylinositol 3-kinase activation. AMOG-mediated Akt phosphorylation specifically activates the mTOR/p70S6 kinase pathway previously implicated in cell size regulation, but it does not depend on tuberous sclerosis complex/Ras homolog enriched in brain (Rheb) signaling. These data support a novel role for a glial adhesion molecule in cell size regulation through selective activation of the Akt/mTOR/S6K signal transduction pathway.
Cues received from the extracellular environment by membrane receptors influence diverse intracellular signaling pathways that regulate cell survival, differentiation, and growth. Cell adhesion molecules have been primarily implicated in maintaining cell-cell and cell-matrix interactions important for maintaining tissue integrity. However, recent evidence indicates that these adhesion molecules, like other membrane-localized receptors, can influence intracellular signal transduction (34, 61). Numerous adhesion molecules, including cadherins, integrins, and immunoglobulin-like adhesion molecules, modulate these signaling pathways' effects on cell growth and proliferation.
In the central nervous system (CNS), altered expression of a number of cellular adhesion molecules has been associated with brain tumor formation, including neural cell adhesion molecule (NCAM), the L1 adhesion molecule, and multiple members of the cadherin family. Increased expression of NCAM, a member of the immunoglobulin superfamily, has been implicated in invasion of glioma cells (47). Upon clustering of the 140-kDa NCAM protein by homophilic binding or interactions with heparan sulfate proteoglycans, the NCAM cytoplasmic tail activates the Ras/mitogen-activated protein (MAP) kinase (MAPK) signaling cascade (56), which likely contributes to increased tumor proliferation. In addition, overexpression of the L1 adhesion molecule in high-grade gliomas promotes cell-matrix and intercellular interactions and facilitates glioma cell migration (33, 59). Similarly, numerous members of the cadherin family have been implicated in brain tumor formation. N-cadherin promotes oligodendrocyte migration and adhesion to astrocytes (57), and E-cadherin expression in WC5 rat astrocyte-like cells results in increased cell adhesion and decreased cell motility (14). Expression of another cadherin protein, cadherin 11, was shown to be decreased in gliomas, where it has been implicated in tumor invasion (79). In this regard, our laboratory has shown that T-cadherin, a novel cadherin protein lacking the catenin intracellular binding domain, functions as a glioma growth regulator (30). In these studies, T-cadherin was reduced in mouse and human gliomas, and its re-expression in T-cadherin-deficient glioma cells resulted in a p21-dependent G2 growth arrest.
Our laboratory has employed a transgenic mouse glioma model in which activated H-Ras is expressed in astrocytes to identify novel genetic changes associated with astrocytoma formation (25). Gene expression profiling of neoplastic and nonneoplastic astrocytes from these mice revealed that another adhesion molecule expressed in the brain, adhesion molecule on glia (AMOG), is downregulated in neoplastic cells (25). Similarly, Senner et al. (60) showed that AMOG expression was decreased in neoplastic cells in human glioma specimens relative to normal astrocytes, and that this decrease in expression correlated with increasing tumor grade. These observations suggested that AMOG may play a role in regulating glioma growth and proliferation.
AMOG was first described as a unique membrane glycoprotein mediating neuron and astrocyte adhesion in the central nervous system, where it has been implicated in neurite outgrowth and neuronal migration (4, 5, 6, 39, 45, 46). AMOG is first expressed in the brain shortly before granule cell migration, and its expression increases during early postnatal development to reach its highest levels in adult glial cells (48). While phenotypically normal at birth, Amog-deficient mice develop motor incoordination and paralysis in early postnatal life and die 17 to 18 days after birth (44).
Analysis of the predicted amino acid sequence revealed sequence similarity between AMOG and the β1 subunit of the Na+/K+ ATPase (23). This Na+/K+ ATPase enzyme consists of a catalytic α subunit and a regulatory β subunit (74), and it is required for the maintenance of ionic homeostasis in most mammalian cells. AMOG associates with the catalytic α subunit to form a functional ion pump and, therefore, was also identified as the β2 subunit of the Na+/K+ ATPase (23). In this regard, this unique molecule serves both cell-cell adhesion and ion exchange functions.
The role of the Na+/K+ ATPase in cell growth control has been most extensively studied in cardiac myocytes, where inhibition of the ion pump with the cardiac glycoside ouabain results in myocyte hypertrophy (26, 27, 75). In myocytes exposed to nontoxic concentrations of ouabain, myocyte protein synthesis increases while the DNA content of the cells remains unchanged, suggesting that pump inhibition primarily influences cell growth rather than cell proliferation without affecting intracellular ion concentrations (43). This growth phenotype was the result of the Na+/K+ ATPase-dependent alterations in intracellular signaling pathway activation, which directly contributed to myocyte hypertrophy (26, 27, 36, 75). Collectively, these data support a role for the Na+/K+ ATPase or its subunits in regulating cell growth; however, the mechanisms underlying its signaling have not been fully elucidated.
Because AMOG is downregulated in mouse and human tumors and the Na+/K+ ATPase pump has been implicated in cell growth control, we sought to define the biological consequence of AMOG re-expression in AMOG-deficient cells and to characterize the mechanism underlying AMOG-mediated intracellular signaling. Consistent with its role as an adhesion molecule, AMOG expression resulted in increased cell aggregation and attachment. Furthermore, our studies show that AMOG expression in glioma cells results in a dramatic increase in cell soma size, which is associated with hyperactivation of the mTOR/p70S6 kinase (S6K) signal transduction pathway. In addition, AMOG increases Akt activation independent of phosphatidylinositol 3-kinase (PI3K), and it stimulates ribosomal S6 phosphorylation by a mechanism that requires mTOR but is independent of the tuberin/hamartin complex (THC) and the Rheb GTPase protein. These data provide a link between cell adhesion and cell size regulation and demonstrate that Akt regulation of the mTOR pathway can be selectively modulated.
MATERIALS AND METHODS
AMOG cloning and cell culture.
RNA was extracted from normal mouse brain using TRIzol Reagent (Invitrogen, Carlsbad, Calif.), and 3 μg of RNA was subjected to first-strand synthesis using random polyhexamer primers and Superscript II reverse transcriptase (Invitrogen) at 42°C. Two microliters of first-strand cDNA was PCR amplified using the forward AMOG primer corresponding to bases 574 to 597 (National Center for Biotechnology Information accession number BC034586; 5′GCGGATCCAGTCATCCAGAAAGAGAAGAAGAGC-3′) and the reverse primer corresponding to bases 2224 to 2244 (5′TCAGGTTTTGTTGATCCGGAG-3′). The AMOG cDNA was cloned into pCR2.1 using the TA cloning system (Invitrogen) to generate pCR2.1.AMOG. To generate a tagged AMOG protein, a second forward primer containing a myc tag at the 5′ end (myc tag, bases 3 to 36; 5′-GCCATGGAACAAAAACTCATCTCAGAAGAGGATCTGGTCATCCAGAAAGAGAAGAAGAG-3′)was used to amplify myc.AMOG from pCR2.1.AMOG. The myc-tagged AMOG was cloned into pCR2.1 using the TA cloning system and was subcloned into pcDNA3 at the EcoRI site to generate pcDNA3.myc.AMOG. Gene sequence was confirmed by direct sequencing using the ABI PRISM dGTP BigDye Terminator ready reaction cycle sequencing kit (version 3.0; Applied Biosytems, Foster City, Calif.). The protein product of pcDNA3.myc.AMOG plasmid (65 kDa) was identified using the coupled in vitro transcription/translation kit (Promega, Madison, Wis.) using the anti-AMOG polyclonal antibody (BD Transduction Laboratories, Lexington, Ky.).
AMOG-expressing and control cell lines were generated by transfecting U87-MG human glioma cells with either pcDNA3.myc.AMOG or pcDNA3.myc vector. Stably transfected cells were selected in 500 μg of Geneticin (G418)/ml. Twenty-one clones were screened for AMOG expression by Western blotting. Two cell lines transfected with the vector (V8 and V9) and three cell lines expressing AMOG (A4, A13, and A16) were selected for further study. Transfected cells were maintained in Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS; Invitrogen) supplemented with nonessential amino acids (Mediatech, Herndon, Va.) and 10 mM sodium pyruvate (Mediatech). For immunofluorescence images, the cytoskeleton was stained with BODIPY-conjugated phalloidin (0.2 U in 50 μl; Molecular Probes, Eugene, Oreg.) and the nuclei were stained with Hoechst (Molecular Probes).
LY294002 and the farnesyltransferase inhibitor FTI-277 were purchased from Calbiochem (La Jolla, Calif.). Rapamycin was purchased from Sigma (St. Louis, Mo.). Cultures containing equal numbers of cells were treated with 10 μM FTI-277, 2 ng of rapamycin/ml, or 20 μM LY294002 in DMEM containing 10% fetal bovine serum for 24 h. Cells were harvested, lysed, and analyzed by Western blotting as described below. Each experiment was performed at least three times with comparable results.
Western blotting and antibodies.
Cells were washed with cold phosphate-buffered saline (PBS), scraped, transferred to an Eppendorf tube, and lysed with cold lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Na-pyrophosphate, 1 mM β-glycerol phosphate, 1 mM Na3VO4, 1 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Lysates were centrifuged at 16,000 relative centrifugal forces for 10 min at 4°C. Protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, Ill.). Equal total protein was loaded on sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, Mass.). Membranes were blocked in Tris-buffered saline-Tween 20 (TBS-T) with 5% nonfat dry milk for 1 h at room temperature. Membranes were then incubated with primary antibodies in TBS-T with 5% nonfat dry milk or 5% bovine serum albumin (for phospho antibodies), washed, and incubated with the appropriate secondary antibody in TBS-T with 5% nonfat dry milk. Chemiluminescence detection was performed using the ECL reagent (Amersham Biosciences, Arlington Heights, Ill.). Each experiment was performed at least three times with identical results.
All antibodies are from Cell Signaling Technology (Beverly, Mass.) unless otherwise indicated. The following commercial primary antibodies were used: anti-PDK1 (3062), anti-Akt (9272), anti-S6K (9202), anti-S6 (2212), anti-MAPK (9102), anti-Rheb (4935), anti-P-S6K (9205), anti-P-S6 (2215), anti-P-MAPK (9101), anti-P-Akt (9271), anti-Gab1 (3232), anti-Na+/K+ ATPase β2 subunit (AMOG; 610914; BD Transduction Laboratories), anti-Na+/K+ ATPase β3 subunit (N74120; BD Transduction Laboratories), anti-Myc (sc-40; Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-cleaved caspase 3 (9661), anti-P-src (2101), anti-P-insulin-like growth factor receptor (IGFR) (Tyr1131/1426; 3021), anti-P-Tyr (PY20; BD Transduction Laboratories), anti-fibroblast growth factor receptor-1 (FGFR-1; (34720), anti-epidermal growth factor receptor (EGFR) (2232), anti-platelet-derived growth factor receptor (PDGFR) (3162), anti-P-tuberin Ser939, Thr1462, and Tyr1571 (3611, 3614, 3615), anti-P-tyr p85 PI3K binding motif (3821), and anti-α-tubulin (T-9026; Sigma). The Na+/K+ ATPase α1 and α3 subunits were identified with polyclonal antibodies to synthetic peptides from the N termini of the subunits. The α2 subunit was identified with an antibody (McB2) provided by K. Sweadner (Massachusetts General Hospital), and the Na+/K+ ATPase β1 subunit was identified with an antibody provided by A. Askari (Medical College of Ohio). Secondary antibodies used were anti-rabbit horseradish peroxidase (HRP) conjugate (7074) and anti-mouse HRP conjugate (7076).
Rubidium-86 uptake.
Cells were grown to 70% confluency in 12-well dishes and were washed with 1.5 ml of medium containing 145 mM NaCl, 2.5 mM KCl, 1.5 mM MgSO4, 2 mM Na2HPO4, 1 mM CaCl2, 25 mM HEPES, pH 7.40 (buffer A). Cells were resuspended in buffer A containing 0.1 mM bumetanide with 0.1 mM ouabain as indicated. After 5 min, the solution was aspirated and replaced with 250 μl of the corresponding solution containing 86Rb (1.5 × 105 cpm). At various time points (0 to 10 min), the flux medium was aspirated and the cells were washed three times with 2.5 ml of 116 mM MgCl2 at 4°C. Cells were dissolved by the addition of 300 μl of 0.1 M NaOH-0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate. Radioactivity in 200-μl samples was determined by liquid scintillation. The protein concentration of each well was determined using bovine serum albumin as standard and the Bio-Rad Protein Assay reagent as described by the supplier (Hercules, Calif.).
Growth assays.
To assess log-phase growth, 105 cells were plated in 60-mm-diameter culture dishes, and cell numbers for each clone were determined by direct counting on a hemacytometer at days 3, 6, and 9.
To measure DNA synthesis, thymidine incorporation assays were performed. For these experiments, 105 vector-transfected or AMOG-expressing cells were plated in 6 wells of a 24-well plate. After 24 h, the medium was changed to serum-free DMEM and was incubated at 37°C for 24 h. The medium was aspirated and replaced with serum-free DMEM containing 1 mCi of [3H]thymidine (Amersham Biosciences, Piscataway, N.J.)/ml and incubated at 37°C for 4 h. Cells were washed twice with PBS and solubilized in 200 mM NaOH. Disintegrations per minute (dpm) were determined in a Liquid Scintillation Beta Analyzer (Packard Instruments Co., Meridan, Conn.), and the mean and standard deviation was determined for each cell line. Each experiment was repeated three times with similar results.
To measure cell growth in vivo, AMOG-expressing clones (A4 and A16) and vector-transfected (V8 and V9) cells were trypsinized and resuspended in Matrigel at a final concentration of 107 cells/ml. One-hundred microliters was injected into the flanks of 6- to 8-week old male immunocompromised athymic nu/nu mice. Four mice were injected with each clone. All procedures followed the Interdisciplinary Principles and Guidelines for the Use of Animals in Research, Marketing, and Education, issued by the New York Academy of Sciences' Ad Hoc Committee on Animal Research. The tumor volumes were measured with calipers for 3 weeks after injection. Tumor volume was calculated according to the formula tumor volume (in cubed millimeters) = (L × W2)/2, where L represents the longest dimension and W the shortest dimension of the tumor. Half of each tumor was homogenized and lysed in MAPK lysis buffer. One-hundred micrograms of total protein from each tumor was analyzed by Western blotting to confirm AMOG expression. The experiment was repeated twice with identical results.
Aggregation assays.
Cells were washed with Ca2+- and Mg2+-free PBS twice and digested with 0.05% trypsin containing 0.5 mM Ca2+. The cells were washed with HCMF (10 mM HEPES [pH 7.4], 0.137 M NaCl, 5.4 mM KCl, 0.34 mM Na2HPO4 · 12H2O, 0.1% glucose) while on ice, as previously described. Cells were resuspended at 105 cells per 0.5 ml of HCMF. In quadruplicate, 0.5 ml of the cell suspension was seeded in the wells of a 24-well plate that was precoated with 1% bovine serum albumin (BSA). Ca2+ was added to a final concentration of 1 mM. The plates were placed in a 37°C shaker and rotated at 80 rpm for 1 h. Cells were fixed with 0.5 ml of 8% paraformaldehyde on ice for 15 min. The wells were gently stirred to prevent cells from settling. The number of aggregates (≥3 cells/cluster) and single cells were counted with a hemacytometer. To calculate the aggregation index, the total number of particles (aggregates and single cells) was divided by the total number of cells in the suspension. The assay was repeated three times with identical results.
Cell adhesion.
Cell adhesion was analyzed as described previously (30). A 96-well plate was precoated with fibronectin (10 μg/ml; Sigma) in sterile PBS at 4°C overnight. The wells were washed twice with PBS and incubated with 2% heat-inactivated BSA for 2 h at 37°C. The cells were digested with 0.05% trypsin containing 0.5 mM CaCl2, resuspended at a density of 106 cells/ml in serum-free DMEM, and incubated for 2 h at 37°C. Cells (100 μl) were added to each well and allowed to adhere for 1 or 4 h. At the end of the incubation period, cells were washed three times with PBS and stained with 0.5% crystal violet for 30 min at room temperature. After being washed, 50 μl of 1% sodium dodecyl sulfate was added to each well overnight at room temperature. The number of adherent cells was quantitated by absorbance at 540 nm. Each experiment was repeated at least three times with identical results.
Flow cytometry.
Cultured cells were trypsinized, fixed in 2% formaldehyde-0.2% glutaraldehyde, RNase A treated, and stained with propidium iodide according to established protocols. Astrocyte cell size was analyzed using a FACSCalibur flow cytometer (Becton-Dickinson, San Jose, Calif.). Events were gated according to forward scatter and side scatter to exclude debris and aggregates. A second gating by FL2-A was then used to select cells in G0/G1, and the gated events were plotted as a forward-scatter histogram to evaluate cell size. Each experiment was performed at least three times with identical results.
PI3K activity assay.
Vector-transfected (V8) or AMOG-expressing (A16) cells were cultured to 80% confluency in 100-mm-diameter tissue culture plates and serum starved overnight. As negative controls, one dish of each cell line was treated with serum-free DMEM containing 0.5% NP-40 (Sigma) to inhibit PI3K activity. Culture dishes were washed three times in ice-cold buffer A (137 mM NaCl, 20 mM Tris-HCl [pH 7.4], 1 mM CaCl2, 1 mM MgCl2, 0.1 mM sodium orthovanadate). Cells were lysed in 1 ml of buffer A containing 1% NP-40 and 1 mM PMSF at 4°C for 20 min. Cells were scraped, and cellular debris was removed by high-speed centrifugation at 4°C. Supernatants were incubated with anti-PI3K antibody (Upstate Biotechnology, Lake Placid, N.Y.) and precipitated with protein A-agarose beads. Cells were washed sequentially in buffer A containing 1% NP-40, wash buffer (100 mM Tris-HCl [pH 7.4], 5 mM LiCl, and 0.1 mM Na3VO4), and TNE buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, and 0.1 mM sodium Na3VO4). To test PI3K activity, the precipitated enzyme-antibody complex was incubated with 30 μCi of [γ-32P]dATP (Amersham Biosciences) containing 20 mM MgCl2 in the presence of 20 μg of phosphatidylinositol (Avanti Polar Lipids, Alabaster, Ala.) in a 37°C shaker (75 rpm) for 10 min. The reaction was terminated with the addition of 20 μl of 6 N HCl. Lipids were extracted in 160 μl of CHCl3:methanol (1:1). Samples were centrifuged at 16,100 × g for 10 min. Silicon thin-layer chromatography (TLC) plates were pretreated with 1 mM EDTA-1% potassium oxalate diluted in methanol-H2O (40:60) and activated at 100°C for 1 h. One microliter of the lower organic phase from each sample was spotted on the TLC plate and developed by chromatography in CHCl3-methanol-H2O-NH4OH (60:47:11.3:2). Plates were allowed to dry, and radiolabeled lipids were visualized by autoradiography and quantitated by liquid scintillation counting. Each experiment was performed at least three times with identical results.
siRNA-mediated Rheb inhibition.
Rheb siRNA constructs are described elsewhere (68). The plasmids p5RαIRESGFP (murine stem cell virus [MSCV] vector) and pΦ2 helper plasmid were a kind gift from Jason Weber (Washington University, St. Louis, Mo.). Empty MSCV vector, MSCV-H1non (control), and MSCV-Rheb653 were cotransfected with pM2 helper vector into the 293T packaging cell line using Lipofectamine 2000 (Invitrogen). The medium was changed 6 h later. Vector-only (V8) or AMOG-expressing (A16) cells were transduced with the filtered supernatant of the transfected 293T cells in the presence of 10 μg of polybrene (Specialty Media, Phillipsburg, N.J.)/ml five times over the ensuing 60 h. Six hours after the last application of filtered media, the medium was exchanged for DMEM containing 10% FBS. Twenty-four hours later, the cells were harvested and lysed, and Western blotting was performed as described above. Each experiment was performed at least two times with identical results.
RESULTS
AMOG expression in glioma cells does not alter Na+/K+ ATPase expression or function.
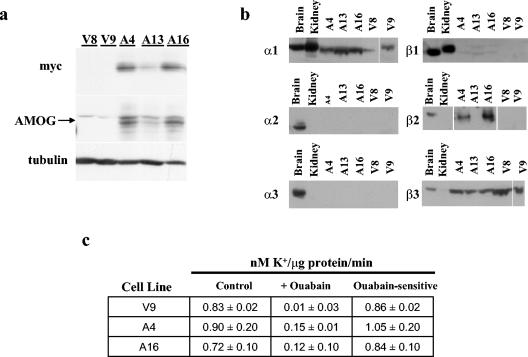
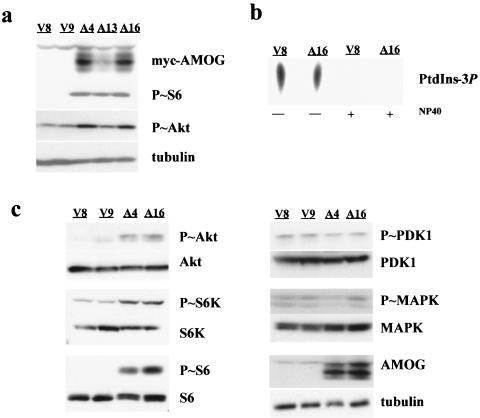
To examine the potential role of AMOG in regulating cell growth and proliferation independent of its role as an ion pump subunit, we re-expressed AMOG in U87-MG glioma cells that do not express endogenous AMOG or have apparent deficits in ionic homeostasis. We generated several cell lines expressing myc-tagged AMOG and chose three for further analysis (Fig. 1a). Clones A4 and A16 expressed high levels of AMOG, while expression was lower in clone A13. Two vector-transfected cell lines (V8 and V9) were used as controls. Re-expression of AMOG did not interfere with protein expression of the other Na+/K+ ATPase subunits expressed in the brain (Fig. 1b).
FIG. 1.
Stable expression of AMOG does not affect cell proliferation in vitro and in vivo. (a) U87 glioma cells were transfected with pcDNA3 or pcDNA3.myc.AMOG and selected in Geneticin to generate stable cell lines. Three cell lines with various levels of AMOG expression (arrow) were selected for further characterization: A4 (high expression), A13 (low expression), and A16 (high expression). Cells that incorporated the pcDNA3 vector only (V8 and V9) were used as controls. (b) All three α and β subunits are expressed in mouse brain while the α1 and β1 subunits are expressed in kidney. Stable expression of AMOG (Na+/K+ ATPase β2 subunit) in clones A4, A13, and A16 did not disrupt expression of the other Na+/K+ ATPase subunits. The A13 clone expresses low levels of β2 that were detectable with longer film exposures. (c) Function of the Na+/K+ ATPase was assessed by uptake of the K+ analog rubidium-86. K+ transport was similar in AMOG-expressing (A4 and A16) and control cells (V9; control). This flux was sensitive to ouabain (+ ouabain), indicating that Na+/K+ ATPase mediates K+ transport in these cell lines (ouabain sensitive). Similar results were obtained for A13 and V8 cells (data not shown). Each experiment was repeated at least three times with similar results. Error bars indicate standard deviations.
While most studies have used ouabain to study the effects of the Na+/K+ ATPase on cell growth, we evaluated AMOG-dependent effects directly by expressing AMOG in cells that lack endogenous AMOG. Present evidence suggests that while the beta subunit is required for the formation of active enzyme and the targeting of the alpha subunit to the plasma membrane, it is not directly involved in ion exchange (22). To ensure that Na+/K+ ATPase pump function was not disrupted in AMOG-expressing cells, we measured uptake of the K+ analog rubidium-86. Rubidium-86 uptake was similar in AMOG-expressing and control cells (Fig. 1c). This transport was sensitive to ouabain, indicating that ion flux was mediated by the Na+/K+ ATPase. These data show that expression of AMOG (β2) in β2-deficient cells did not disrupt the expression of other Na+/K+ ATPase subunits (α1, α2, α3, β1, and β3) or alter the function of the Na+/K+ ATPase ion pump.
AMOG expression does not alter cellular proliferation in vitro or in vivo.
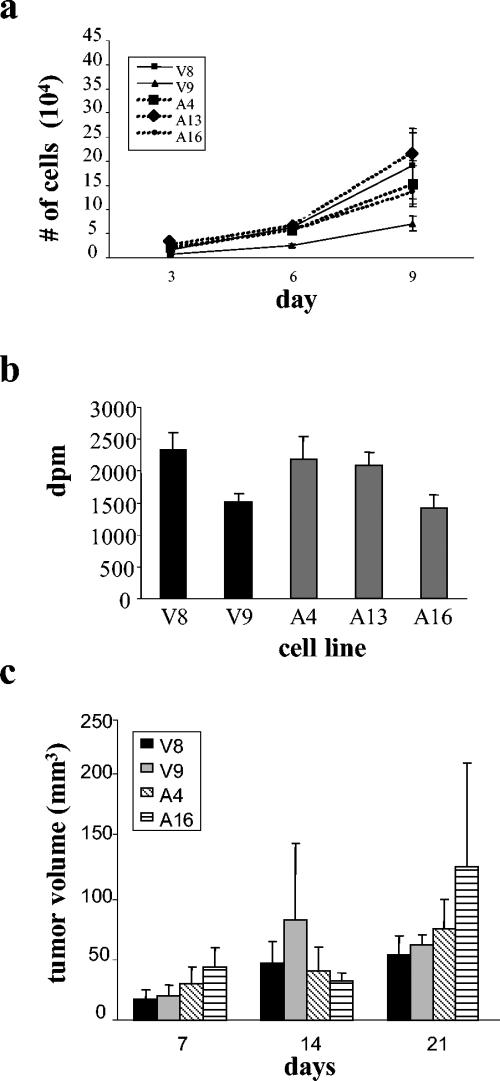
To determine if AMOG expression altered cell proliferation, we analyzed cultures during log-phase growth. When equal numbers of cells were plated and counted after 3, 6, and 9 days in culture, no differences in cell number were observed (Fig. 2a). Similarly, we observed no differences in saturation density when cells were grown for 6 days past confluence (data not shown). In addition, [3H]thymidine incorporation assays showed no significant differences in DNA synthesis between vector control cell lines and those expressing AMOG (Fig. 2b). Lastly, we observed no differences in DNA content by flow cytometry (data not shown).
FIG. 2.
AMOG expression does not alter cell proliferation. (a) Equal numbers of cells were plated and cells were counted during log-phase growth (days 3, 6, and 9 after plating). There were no significant differences in cell number between the vector-transfected (V8 and V9) or AMOG expressing (A4, A13, and A16) stable cell lines at any time point. (b) Thymidine incorporation assays revealed no significant differences in DNA synthesis between the cell lines. (c) Vector-only or AMOG-expressing cells were injected subcutaneously in the flanks of 6- to 8-week-old nude mice. Tumors volumes were determined weekly for 3 weeks. At all time points, there was no significant difference in tumor volume produced from the vector-transfected or the AMOG-expressing cell lines. Error bars indicate standard deviations.
Because the in vivo environment may influence cell proliferation, we tested the ability of these cells to proliferate when grown as subcutaneous tumor implants in the flanks of immunocompromised athymic (nu/nu) mice. As observed with the in vitro growth assays, we observed no differences between the two clones with the highest AMOG expression (A4 and A16) and the two vector control cell lines (V8 and V9) after 3 weeks in vivo (Fig. 2c). At the conclusion of the experiment, we verified AMOG expression in the A4 and A16 lines (data not shown). These results demonstrate that, both in vitro and in vivo, AMOG expression does not alter cellular proliferation.
To determine if cell death was altered in AMOG-expressing glioma cells, we stained cultured cells for cleaved (activated) caspase 3, a critical mediator of both the extrinsic and intrinsic apoptotic pathways (9). We observed no consistent differences in the number of caspase 3-expressing cells (data not shown). In addition, when equal cell numbers were plated and the number of live, Trypan blue-excluding cells counted, we observed no differences in the number of surviving cells (data not shown). Taken together, these data suggest that AMOG does not regulate cell proliferation or apoptosis.
AMOG expression increases cellular aggregation and attachment.
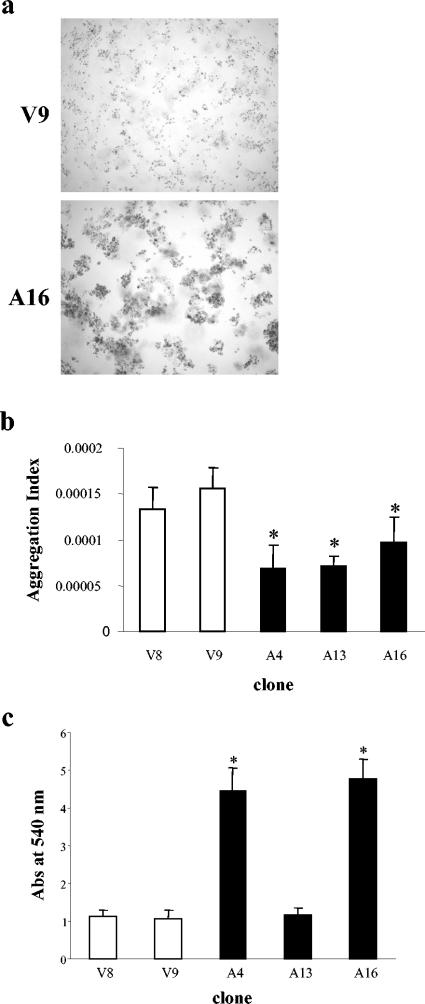
Because AMOG was originally identified as a glial adhesion molecule, we next analyzed the ability of AMOG-expressing cells to promote cell aggregation in vitro. When cells were allowed to aggregate in culture plates pretreated with BSA to prevent cell attachment, AMOG-expressing cells formed large cellular aggregates containing ≥3 cells (Fig. 3a). To quantitate this observation, the aggregation index was calculated by determining the total particle number (single cells and aggregates) in suspension cultures. Fewer particles were present in AMOG-expressing cultures, indicating a higher tendency of these cells to form clusters (Fig. 3b).
FIG. 3.
AMOG expression increases cellular aggregation and adhesion. (a) When grown in suspension in precoated plates and visualized by phase-contrast microscopy, AMOG-expressing cells exhibit increased cell-cell aggregation compared to that of vector-transfected control cells. (b) The aggregation index was calculated for AMOG-expressing and vector-transfected cells. A decrease in aggregation index is indicative of an increased tendency to aggregate. The asterisk denotes a statistically signification decrease in this index. Error bars indicate standard deviations. (c) Cell lines expressing high levels of AMOG (clones A4 and A16) exhibit increased cell adhesion to fibronectin-coated plates 1 h after seeding. The single asterisk denotes a statistically significant increase in absorbance (Abs) at 540 nm. Error bars indicate standard deviations.
To further assess AMOG's role in adhesion, we analyzed the ability of these cells to attach to fibronectin-coated plates. At 1 h, attachment to fibronectin was increased in those cells that expressed high levels of AMOG (A4 and A16; Fig. 3c). These results confirm AMOG's role as an adhesion molecule and demonstrate that AMOG mediates both cell-cell and cell-substrate interactions.
AMOG-expressing cells exhibit an increased cell size.
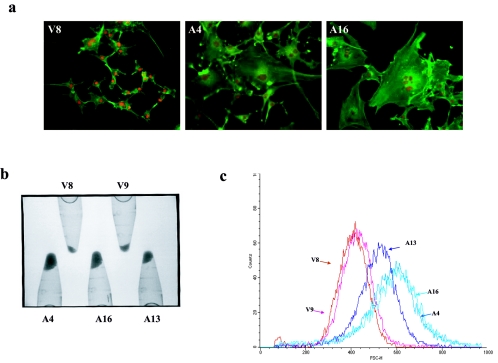
While no effects of AMOG expression on cell proliferation, apoptosis, or motility (data not shown) were observed, we noted a striking increase in the cell size of the AMOG-expressing cell lines. Phalloidin staining of the actin cytoskeleton revealed an increased cell soma size (Fig. 4a). Many of these cells were multinucleated, as determined by Hoechst staining. In contrast, vector-transfected cells were small and singly nucleated. To provide support for these qualitative observations, equal numbers of cells were pelleted by centrifugation. The pellets from the AMOG-expressing cell lines were significantly larger than those produced from the vector-only stable cell lines (Fig. 4b). In addition, we determined the size distribution of these cells using forward scatter analysis by flow cytometry. The cell size histogram demonstrated a right shift compared to control cells, indicative of an increase in cell size in the AMOG-expressing cells (Fig. 4c). These results show, using three independent methods, that AMOG expression results in an increased cell soma size.
FIG. 4.
AMOG-expressing cells exhibit increased cell soma size. (a) Fluorescence images of phalloidin-stained actin (green) demonstrate the increased cellular size of the AMOG-expressing cells (A4 and A16) compared to that of vector-expressing cells (V8). Several of the AMOG-expressing cells are multinucleated, as shown by Hoescht staining (red). (b) The overall cellular volume was increased in AMOG-expressing cells, as determined by centrifugation of 2.5 million cells from each cell line and directly visualizing pellet size. (c) Flow cytometry events were collected for vector (V8 and V9) and AMOG-expressing cells (A4, A13, and A16) and counts were plotted according to forward scatter. The cell size distribution histogram demonstrates the increased cell size of the AMOG-expressing cells.
Activation of Akt and ribosomal S6 in AMOG-expressing cells occurs independently of PI3K.
Previous work from our laboratory and others (7, 38, 68) has implicated hyperactivation of the Akt/mTOR/S6K pathway in cell size regulation in the brain. To determine whether AMOG expression resulted in increased Akt/mTOR/S6K pathway activation, we monitored phosphorylation of ribosomal S6 as a surrogate marker of mTOR pathway hyperactivation. In AMOG-expressing cells, ribosomal S6 was hyperphosphorylated (Fig. 5a). This effect was dose dependent, as cells expressing high levels of AMOG (A4 and A16) expressed more phosphorylated S6 protein than cells with an intermediate level of AMOG expression (A13). In contrast, vector-transfected cells have only low levels of phosphorylated S6 when grown in medium containing 10% serum or under conditions of serum starvation (Fig. 5a; data not shown).
FIG. 5.
AMOG-expressing cells exhibit increased Akt/mTOR/S6 pathway signaling. (a) The increase in cell size was associated with increased levels of phosphorylated S6 (Ser240/244), as determined by Western blotting, in AMOG-expressing cells (A4 and A16) compared to that of vector-only controls (V8 and V9). Activation of Akt (Ser473) was assessed using a phosphorylation-specific antibody. Western blotting for tubulin demonstrates equal protein loading. (b) The enzymatic activity of PI3K was determined in vitro by monitoring the conversion of PtdIns to PtdIns-3P by lipid extraction and separation by thin-layer chromatography. When vector-transfected or AMOG-expressing cells were serum starved overnight, there was no difference in PI3K activity. NP-40 eliminates PI3K activity in both cell lines. (c) Activation of the insulin-like growth factor receptor signaling pathway, as determined by Western blotting with phosphospecific antibodies, revealed that AMOG expression is associated with an increase in phosphorylated Akt (Ser473), S6K (Thr389), and S6 (Ser240/244). Upstream targets of this pathway, including PDK1 (Ser241) and MAPK (Thr202/Tyr204), were not activated in the AMOG-expressing cells. Expression of these proteins was not altered in AMOG expressing cells, as demonstrated by immunoblotting with antibodies that detect both phosphorylated and nonphosphorylated forms of these proteins. Immunoblotting for tubulin serves as a loading control. All experiments were repeated at least three times with comparable results.
The signal transduction cascade downstream of the insulin-like growth factor-1 receptor (IGF1-R) is known to regulate S6 phosphorylation in both Drosophila melanogaster and mammalian cells (18, 51). This signaling pathway is dependent on activation of phosphatidylinositol 3-kinase and its downstream target, Akt. We sought to better understand how AMOG regulates this pathway by examining activation of the signaling intermediates. Previous work using ouabain-treated cells has shown that the Na+/K+ ATPase pump regulates Src activation (26, 27, 74). Src activation and translocation to the membrane can, in turn, activate receptor tyrosine kinases (RTKs). Using phosphospecific antibodies, we determined that there was no increase in the expression of phosphorylated Src (data not shown). These results suggest that AMOG regulates this pathway downstream of Src.
We also examined the phosphorylation status of the IGF1-R using a phosphospecific antibody and found no differences in AMOG-expressing cells compared to controls. In addition, immunoprecipitation of cell lysates with a phosphotyrosine antibody revealed no differences in the phosphorylation status of other RTKs, including platelet-derived growth factor receptor, epidermal growth factor receptor, and fibroblast growth factor receptor-1 (FGFR-1; data not shown). IGF1-R transduces signals by recruiting and phosphorylating the docking protein Grb2-associated binder 1 (Gab1) (28). Phosphorylated Gab1 associates with a variety of downstream effector molecules, including PI3K, to regulate several different biological processes that include cell proliferation and survival (37). Western blotting using phospho-specific antibodies specific for activated Gab1 similarly revealed no differences in activation between AMOG-expressing and vector control cells (data not shown). These results demonstrate that AMOG expression does not alter activation of these growth factor receptors or the docking protein Gab1.
To establish the level at which AMOG influences this signaling pathway, we examined the activation status of molecules downstream of RTKs. To determine whether PI3K was constitutively activated in AMOG-expressing cells, we used an in vitro assay to monitor the ability of immunoprecipitated PI3K to phosphorylate phosphatidylinositol (PtdIns) to produce the radioactive lipid product PtdIns3P. We found no difference in PI3K activity in the AMOG-expressing cells compared to that of control cells under any of the conditions tested (Fig. 5b). Similarly, we observed no increase in phosphorylation of PI3K targets using an antibody that specifically recognizes a PI3K phosphorylation motif (data not shown). Finally, to determine whether AMOG expression resulted in activation of the PI3K target, PDK1, we used phospho-specific PDK1 antibodies and found no differences in PDK1 activation (Fig. 5c).
However, we did observe a robust increase in the levels of activated (phosphorylated) Akt in AMOG-expressing cells, suggesting that AMOG-mediated activation of this pathway occurs at the level of Akt activation (Fig. 5c). This activation was dose dependent, as cells expressing intermediate levels of AMOG (clone A13) exhibited an intermediate level of Akt activation (Fig. 5a). Consistent with this observation, other downstream effectors, including S6 kinase and ribosomal S6, were also hyperactivated in AMOG-expressing cells. Collectively, these results indicate that AMOG regulates this signaling pathway at the level of Akt and that this activation occurs independently of PI3K or PDK1 activation. This finding suggests a new mechanism for Akt regulation, similar to that reported for the phosphatidylinositol kinase enhancer A (PIKE-A) protein, which binds to and activates Akt independent of PI3K activity (2). To determine if AMOG-mediated Akt activation influenced other Akt-regulated signaling pathways, we examined MAPK activation by immunoblotting with an antibody specific for phosphorylated MAPK. We observed no differences in MAPK activation in AMOG-expressing cells (data not shown). These data support a model in which AMOG-mediated Akt activation preferentially activates the mTOR/S6K signaling cascade to modulate cell size.
Akt-mediated activation of ribosomal S6 is mTOR-dependent but occurs independently of signaling by the THC complex and Rheb.
Recent evidence suggests that Akt regulates ribosomal S6 activation through the tuberin/hamartin complex (THC) (16, 52, 64), and that tuberin directly inhibits activation of the mTOR/S6K pathway by functioning as a GTPase-activating protein (GAP) for a Ras-like molecule, Ras-homolog enriched in brain (Rheb), to prevent mTOR activation (16, 31, 55, 62, 72). Present models suggest that Akt phosphorylates tuberin to inactivate the THC, resulting in hyperactivation of the mTOR/S6K pathway. We examined the phosphorylation status of tuberin, in light of reports that Akt activation results in tuberin phosphorylation on residues Ser939, Thr1463, and Tyr1572 (16, 32, 52, 65). No hyperphosphorylation (inactivation) of tuberin on any of these amino acid residues was observed in AMOG-expressing cells (data not shown), suggesting that AMOG-induced Akt and S6 activation do not involve inactivation of the THC.
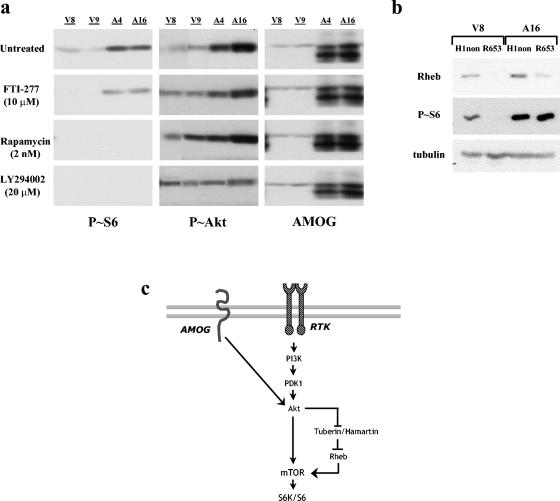
To determine whether AMOG-mediated regulation of this pathway was dependent on known downstream effector molecules, we inhibited Rheb, mTOR, and S6K using both pharmacologic and genetic inhibition. We used the farnesyltransferase inhibitor FTI-277 to inhibit Rheb. Previous work has shown that Rheb is a farnesylated protein and that this modification is critical for its function (11, 15). When AMOG-expressing cells were treated with 10 μM FTI-277 for 24 h, we observed no decrease in S6 activation by Western blotting (Fig. 6a). This concentration of drug has been shown to completely ameliorate S6 activation in hamartin-deficient astrocytes in which the THC is disrupted (68). In contrast, rapamycin treatment (2 ng/ml) inhibits mTOR activation and results in complete amelioration of S6 hyperactivation in AMOG-expressing cells. Similarly, S6 activation in control cells was sensitive to rapamycin, supporting the role of mTOR in growth-factor-mediated activation of this pathway. Finally, we used low doses of LY294002 (20 μM) to selectively inhibit the function of S6K but not PI3K (1), and we found complete blockade of S6, but not Akt, activation in AMOG-expressing and control cells (Fig. 6a). Phosphorylation of MAPK or Akt was not altered in cells treated with FTI-277, rapamycin, or LY294002 (Fig. 6a and data not shown). These data suggest that S6 activation in these cells operates through a pathway that is dependent on Akt and mTOR.
FIG. 6.
Pharmacologic and genetic inhibition of the mTOR/S6K pathway reveals Rheb-independent regulation in AMOG-expressing cells. (a) Vector-only (V8 and V9) and AMOG-expressing (A4 and A16) cells were treated with the following inhibitors for 24 h: FTI-277 (10 nM), rapamycin (2 nM), and low-dose LY294002 (20 μM). Activation of ribosomal S6 by phosphorylation (Ser240/244) was used as a marker of pathway activation. Untreated cells show high levels of activated S6. Inhibition of Rheb with the farnesyltransferase inhibitor FTI-277 did not alter P-S6 levels. Inhibition of mTOR with rapamycin or inhibition of S6K with low-dose LY294002 completely blocked activation of S6. Activation of Akt (Ser473) was assessed using a phosphorylation-specific antibody. Western blotting for AMOG is shown as a control. Blotting for tubulin demonstrated equal protein loading (data not shown). (b) Genetic inactivation of Rheb by siRNA (R653) did not alter S6 activation in AMOG-expressing cells but did eliminate S6 activation in vector-only cells. As a negative control, cells were transduced with a construct containing the H1 promoter alone (H1). Tubulin immunoblots are shown as loading controls. (c) AMOG regulates Akt by a mechanism that occurs independently of PI3K. AMOG-mediated Akt activation preferentially influences mTOR/S6K signaling that regulates cell size. AMOG-dependent activation of this pathway requires mTOR but occurs independently of THC and Rheb signaling, suggesting that the molecular input (i.e., AMOG) to Akt differentially influences activation of specific downstream pathways.
Farnesyltransferase inhibitors can influence other signaling pathways, including those regulated by other Ras family members (58). To directly determine whether S6 activation in these cells was Rheb independent, we inhibited Rheb expression using a previously generated siRNA construct. We used murine stem cell virus to deliver Rheb-specific siRNA (Rheb653) or a control construct (H1) to AMOG-expressing and vector-only control cells. This genetic approach has been shown to efficiently inhibit S6 activation in Tsc1-deficient astrocytes (68). Transduction with the Rheb siRNA inhibited Rheb expression and S6 phosphorylation in the control cells but did not reverse S6 hyperactivation in the AMOG-expressing cells (Fig. 6b). Moreover, this Rheb siRNA specifically targets Rheb1 and not Rheb2. While both Rheb1 and Rheb2 mRNA are expressed in U87 cells by quantitative reverse transcription-PCR (RT-PCR) (data not shown), these results suggest that Rheb1 is the primary Rheb molecule involved in THC signaling in glioma cells. Coupled with the pharmacologic data, these results support a Rheb-independent mechanism of S6 activation in AMOG-expressing cells (Fig. 6c).
DISCUSSION
Cellular adhesion molecules play numerous important roles both in maintaining tissue integrity via extracellular interactions and by modulating intracellular signal transduction pathways important for cellular homeostasis. Altered expression of these adhesion molecules can disrupt intracellular signals important for cell growth regulation and contribute to tumor formation (12). Although adhesion molecules have been implicated in brain tumor formation, little is known about the specific intracellular signals that these molecules modulate. We have examined the role of the nervous system astrocyte adhesion molecule AMOG in cell growth control and intracellular signal transduction in light of recent observations that this molecule is downregulated in both human and mouse brain tumors.
We examined the effect of AMOG re-expression on several cellular properties important for brain tumor formation and progression, including cell proliferation, cell adhesion, cell migration, and cell size. Previous studies using the cardiac glycoside ouabain to partially inhibit the Na+/K+ ATPase support a role for this ion pump in regulating cell growth rather than cell proliferation (29, 36, 49, 75). In these studies, ouabain-induced inhibition increased the expression of a number of growth-related genes and stimulated nonproliferative cell growth (hypertrophy). Similarly, Senner et al. (60) showed that AMOG expression in AMOG-deficient C6 glioma cells did not reduce cell proliferation. Instead, AMOG expression increased cell adhesion and decreased cell migration on Matrigel. These results are consistent with our results with U87 human glioma cells, in which we did not observe any effects of AMOG re-expression on glioma proliferation or apoptosis in vitro and in vivo. Collectively, these data show that AMOG does not modulate cell proliferation in vitro or in vivo and raise the possibility that AMOG modulates other tumor-associated properties, such as cell adhesion and motility.
Adhesion molecules mediate homophilic and heterophilic cellular interactions with neighboring cells and with the extracellular matrix (ECM). Consistent with the role of AMOG as an adhesion molecule, our data show that AMOG expression promotes homophilic aggregation. Furthermore, AMOG-expressing cells exhibit increased cell-substrate interactions, such as attachment to fibronectin. The mechanism underlying these cell-cell and cell-substrate interactions remains unknown, because the putative AMOG receptor has not yet been identified. Previous studies have shown that AMOG has a unique extracellular domain and does not bind to NCAM and L1. In this regard, AMOG-mediated adhesion is not inhibited by antibodies to either NCAM or L1 (4, 5).
The study by Senner and colleagues (60) using a single AMOG-expressing glioma cell line showed that AMOG expression in glioma cells decreases cell motility. We assessed the migration of several cell lines with various levels of AMOG expression by using a Boyden chamber assay (data not shown). However, the large size of the AMOG-expressing cells impeded their movement through the 8-μm-pore-size membrane barriers and through Matrigel. Although we cannot rule out a direct effect on cell motility, the most striking phenotype of AMOG-expressing cells in our experiments was a dramatic increase in cell size. Because cell size was not specifically addressed by Senner et al., it is possible that changes in cell size also contributed to observed decreases in cell motility in their study.
The increase in cell soma size observed in our AMOG-expressing cells was reminiscent of cellular phenotypes observed in Drosophila mutants in which various components of the insulin signaling pathway, including the insulin receptor (dinr) (13), insulin receptor substrate 1 (chico) (8, 24), and PI3K (Dp110) (40, 72), are disrupted (71). Insulin receptor-mediated PI3K activation generates inositol lipids that stimulate a variety of downstream effectors important for cell growth and proliferation. One of these targets, the kinase PDK1, recruits Akt to the cell membrane and phosphorylates Akt on threonine-308 (3, 53, 70). In addition to insulin receptor-mediated PI3K activation, other growth factor receptor tyrosine kinases (RTKs) and their associated adaptor proteins, including Src and Gab1, activate PI3K and result in Akt phosphorylation (9). In addition, focal adhesion kinase (FAK) relays integrin-mediated signals to PI3K/Akt in response to the ECM (10, 73). Importantly, numerous adhesion molecules have been shown to regulate PI3K/Akt signaling. E-cadherin, an integral membrane glycoprotein important for homophilic interactions at adherens junctions, activates this pathway by recruiting PI3K to E-cadherin protein complexes (14). Similarly, intracellular adhesion molecule 2 (ICAM-2)-dependent ezrin phosphorylation in immune cells recruits PI3K to the cell membrane, promoting production of inositol lipids and activating PDK1 and Akt (50). Expression of the melanoma-associated adhesion molecule (MelCAM) recruits FAK to a focal adhesion complex, resulting in PI3K activation (41).
Insights into alternative mechanisms of Akt activation have derived from recent studies on a unique molecule termed PI3K enhancer (PIKE). PIKE is a nuclear GTPase that exists as three isoforms: PIKE-S (short form), PIKE-L (long form), and PIKE-A, as a result of alternative splicing (PIKE-S and -L) or the use of an alternative transcription initiation site (PIKE-A). PIKE-S and PIKE-L bind to PI3K via a proline-rich N-terminal domain and stimulate its lipid kinase activity, resulting in Akt activation (77). PIKE-A, however, lacks this N-terminal domain and instead binds directly to and activates Akt (2). These results indicate that, while most known activators rely on PI3K-dependent mechanisms of Akt phosphorylation, Akt activation can occur independently of PI3K. In this regard, heat shock, oxidative stress, and cytosolic Ca2+ have been shown to activate Akt and are insensitive to pharmacologic PI3K inhibitors (70).
In this study, we identify AMOG as an adhesion molecule that regulates Akt (Fig. 6c). In contrast to signal transduction mediated by other adhesion molecules, AMOG-mediated regulation of Akt signaling does not involve activation of RTKs or RTK-associated proteins. Similar to PIKE-A-mediated signaling, AMOG activates Akt independently of PI3K. It is possible that AMOG directly activates Akt; however, we were unable to demonstrate any biochemical interaction between AMOG and Akt, as had been reported for PIKE-A. Alternative routes to Akt activation might involve other signaling intermediates that bind AMOG and subsequently activate Akt. Further studies will be required to define the molecular interactions that facilitate AMOG activation of Akt.
While many inputs influence Akt activation via PI3K-dependent and -independent mechanisms, the downstream targets of Akt signaling are equally diverse. Akt activation modulates numerous cellular processes, including apoptosis, proliferation, differentiation, and nutrient metabolism (18, 70). Central to its role in regulating cell growth, Akt has been shown to phosphorylate tuberin to inactivate the THC, resulting in increased activation of the Rheb/mTOR/S6K pathway (11, 16, 21, 31, 32, 62). Loss of Tsc1 or Tsc2 in both Drosophila and mammalian cells results in an increase in cell size but does not influence cell number (20, 32, 51, 63, 68). However, recent studies suggest that Akt growth regulation does not require Tsc expression in Drosophila, suggesting that Akt signaling includes both THC-dependent and -independent pathways (17). Activation of S6K and its target, ribosomal S6, results in increased translation of key components of the translational machinery, including ribosomal proteins and elongation factors (9). Hyperactivation of this pathway, therefore, increases protein production, resulting in increased cell soma size. In this regard, pharmacologic inhibition of mTOR signaling results in reduced protein translation and decreased cell soma size (38). The importance of mTOR signaling to cell size regulation is underscored by our finding that the AMOG-induced increase in S6 phosphorylation is inhibited by treatment with rapamycin and low-dose LY294002 for 24 h. While prolonged rapamycin treatment has been shown to partially rescue the cell size defects in Tsc1-deficient astrocytes (68), this lengthy exposure was associated with significant cellular toxicity in glioma cells, precluding detailed analysis of this cellular phenotype in the presence of these pharmacologic agents.
Using two complementary approaches, our studies demonstrate that AMOG-mediated activation of the mTOR/S6K signaling pathway occurs independently of Rheb GTPase activation. Previous studies examining the effect of farnesylation on Rheb function have shown that farnesylation-defective Rheb mutants exhibit compromised ability to activate S6K (11, 42, 66) and that Rheb farnesylation is required for cell cycle progression and amino acid uptake in yeast (69, 76). In our experiments, cells were treated with FTI-277 for 24 h. This treatment was sufficient to completely block S6 activation in control cells but not in AMOG-expressing glioma cells. However, it is possible that previously synthesized, farnesylated protein persisted in these cells to permit S6 activation. To address this, we used a genetic approach to inhibit Rheb. Again, S6 activation was completely blocked in control cells treated with Rheb siRNA but not in AMOG-expressing cells. While it remains formally possible that the low level of persistent Rheb expression following siRNA-mediated inhibition is responsible for S6 activation, our studies both in control U87-MG glioma cells and primary astrocytes (Fig. 6) (68) indicate that the level of Rheb inhibition achieved using this siRNA approach is sufficient to completely block S6 activation in these cells. These observations, coupled with the use of two independent experimental approaches to assess the Rheb dependence of mTOR/S6K activation in glioma cells, support our conclusion that stimulation of this signaling pathway by AMOG occurs primarily by a Rheb-independent mechanism.
In addition to modulating the mTOR/S6K pathway, Akt regulates expression of the cyclin-dependent kinase inhibitor p27Kip1, a negative regulator of cell proliferation and size. p27Kip1 inhibits formation of the cyclin E/cyclin-dependent kinase 2 complex that is required for the G1-to-S cell cycle transition. p27Kip1 expression is decreased in cells that overexpress Akt or lack Tsc1 expression. In Tsc1-deficient astrocytes, our laboratory has found that decreased expression of p27Kip1 is associated with a decrease in contact inhibition growth arrest (67). Moreover, mice lacking p27Kip1 expression exhibit enhanced growth and multiple organ enlargement (19, 35, 47). The enhanced organ size was shown to be due to an increase in cell number in those tissues studied, with no significant increase in cell size, suggesting Akt modulates cell soma size through mTOR pathway activation and not p27Kip1. In AMOG-expressing cells, we did not find any reproducible changes in p27Kip1 expression (data not shown), suggesting that mTOR/S6K pathway activation is the primary mechanism involved in AMOG-induced cell size regulation. Similarly, we found no differences in the rates of programmed cell death, as determined by monitoring activation of the apoptotic marker caspase 3, in AMOG-expressing cells (data not shown), despite the role for Akt in regulating apoptosis (9). These data suggest that the input to Akt specifically regulates the activation of downstream effectors. Similar pathway specificity has been observed in cells in which Akt is activated by PIKE-A overexpression. While Akt is hyperphosphorylated by PIKE-A and the rates of apoptosis are robustly decreased, PIKE-A does not appear to influence cell size regulation (2, 54).
Our data suggest that AMOG-mediated activation of mTOR and its downstream effectors occurs independently of tuberin/hamartin signaling (Fig. 6C). This is consistent with previous observations in Drosophila showing that the effects of Pten and Tsc1 deletion on cell size control are synergistic (63) and further supports tuberin/hamartin-independent roles of Akt in cell size control. Unlike our observations of hamartin-deficient cells (68), S6 hyperactivation in AMOG-expressing cells occurs independently of Rheb activation. Collectively, these data support tuberin/hamartin-dependent and -independent mechanisms of Akt-mediated regulation of mTOR signaling and suggest that the molecular input regulating Akt phosphorylation differentially influences Akt-dependent pathways, contributing to unique cellular phenotypes.
Acknowledgments
We are grateful for flow cytometry assistance from Erik Uhlmann.
This work was supported by grants from the U.S. Army (DAMD17-03-1-0073) and NINDS (NS41097) to D.H.G. D.K.S. is supported by a National Research Service Award (Medical Scientist 5 T32 GM07200).
REFERENCES
- 1.Adi, S., N.-Y. Wu, and S. M. Rosenthal. 2001. Growth factor-stimulated phosphorylation of Akt and p70S6K is differentially inhibited by LY294002 and wortmannin. Endocrinology 142:498-501. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J.-Y., R. Rong, T. G. Kroll, E. G. Van Meir, S. H. Snyder, and K. Ye. 2004. PIKE (phosphatidylinositol 3-kinase enhancer)-A GTPase stimulates Akt activity and mediates cellular invasion. J. Biol. Chem. 279:16441-16451. [DOI] [PubMed] [Google Scholar]
- 3.Alessi, D. R., M. Deak, A. Casamayor, F. B. Caudwell, N. Morrice, D. G. Norman, P. Gaffney, C. N. MacDougall, D. Harbison, A. Ashworth, and M. Bownes. 1997. 3-Phosphoinositide-dependent protein kinases-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol. 7:776-789. [DOI] [PubMed] [Google Scholar]
- 4.Antonicek, H., and M. Schachner. 1988. The adhesion molecule on glia (AMOG) incorporated into lipid vesicles binds to subpopulations of neurons. J. Neurosci. 8:2961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonicek, H., E. Persohn, and M. Schachner. 1987. Biochemical and functional characterization of a novel neuron-glia adhesion molecule that is involved in neuronal migration. J. Cell Biol. 104:1587-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avila, J., D. Alvarez de la Rosa, L. M. Gonzalez-Martinez, E. Lecuona, and P. Martin-Vasallo. 1998. Structure and expression of the human Na,K ATPase beta 2-subunit gene. Gene 208:221-227. [DOI] [PubMed] [Google Scholar]
- 7.Backman, S. A., V. Stambolic, A. Suzuki, J. Haight, A. Elia, J. Pretorius, M.-S. Tsao, P. Shannon, B. Bolon, G. O. Ivy, and T. W. Mak. 2001. Deletion of Pten in mouse brain causes seizures, ataxia, and defects in soma size resembling Lhermitte-Duclos disease. Nat. Genet. 29:396-403. [DOI] [PubMed] [Google Scholar]
- 8.Bohni, R., J. Riesgo-Escovar, S. Oldham, W. Brogiolo, H. Stocker, B. F. Andruss, K. Buckingham, and E. Hafen. 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell 37:865-875. [DOI] [PubMed] [Google Scholar]
- 9.Brazil, D. P., Z-Z. Yang, and B. A. Hemmings. 2004. Advances in protein kinase B signaling: AKTion on multiple fronts. Trends Biochem. Sci. 29:233-242. [DOI] [PubMed] [Google Scholar]
- 10.Carloni, V., R. M. Defranco, A. Caligiuri, A. Gentilini, S. C. Sciammetta, E. Baldi, B. Lottini, P. Gentilini, and M. Pinzani. 2002. Cell adhesion regulates platelet-derived growth factor-induced MAP kinase and PI-3 kinase activation in stellate cells. Hepatology 36:582-591. [DOI] [PubMed] [Google Scholar]
- 11.Castro, A. F., J. F. Rebhun, G. J. Clark, and L. A. Quilliam. 2003. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J. Biol. Chem. 278:32493-32496. [DOI] [PubMed] [Google Scholar]
- 12.Cavallaro, U., and G. Christofori. 2004. Multitasking in tumor progression: signaling functions of cell adhesion molecules. Ann. N. Y. Acad. Sci. 1014:58-66. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C., J. Jack, and R. S. Garofalo. 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137:846-856. [DOI] [PubMed] [Google Scholar]
- 14.Chen, H., N. E. Paradies, M. Fedor-Chaiken, and R. Brackenbury. 1997. E-cadherin mediates adhesion and suppresses cell motility via distinct mechanisms. J. Cell Sci. 110:345-356. [DOI] [PubMed] [Google Scholar]
- 15.Clark, G. J., M. S. Kinch, K. Rogers-Graham, S. M. Sebti, A. D. Hamilton, and C. J. Der. 1997. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J. Biol. Chem. 272:10608-10615. [DOI] [PubMed] [Google Scholar]
- 16.Dan, H. C., M. Sun, L. Yang, R. I. Feldman, X.-M. Sui, C. C. Ou, M. Nellist, R. S. Yeung, D. J. J. Halley, S. V. Nicosia, W. J. Pledger, and J. Q. Cheng. 2002. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J. Biol. Chem. 277:35364-35370. [DOI] [PubMed] [Google Scholar]
- 17.Dong, J., and D. Pan. 2004. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 18:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edinger, A. L., and C. B. Thompson. 2002. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell 13:2276-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fero, M. L., M. Rivkin, M. Tasch, P. Porter, C. E. Carow, E. Firpo, K. Polyak, L. H. Tsai, V. Broudy, R. M. Perlmutter, K. Kaushansky, and J. M. Roberts. 1996. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 85:733-744. [DOI] [PubMed] [Google Scholar]
- 20.Gao, X., Y. Zhang, P. Arrazola, O. Hino, T. Kobayashi, R. S. Yeung, B. Ru, and D. Pan. 2002. Tsc tumor suppressor proteins antagnozie amino-acid-TOR signaling. Nat. Cell Biol. 4:669-704. [DOI] [PubMed] [Google Scholar]
- 21.Garami, A., F. J. T. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11:1457-1466. [DOI] [PubMed] [Google Scholar]
- 22.Geering, K., I. Theulaz, F. Verrey, M. T. Hauptle, and B. T. Rossier. 1989. A role for the beta-subunit in the expression of functional Na,K-ATPase in Xenopus oocytes. Am. J. Physiol. 257:C851-C858. [DOI] [PubMed] [Google Scholar]
- 23.Gloor, S., H. Antonicek, K. Sweadner, S. Pagliusi, R. Frank, M. Moos, and M. Schachner. 1990. The adhesion molecule on glia (AMOG) is a homologue of the beta subunit of the Na,K-ATPase. J. Cell Biol. 110:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goberdhan, D. C., N. Paricio, E. C. Goodman, M. Mlodzik, and C. Wilson. 1999. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 13:3244-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutmann, D. H., Z.-Y. Huang, N. M. Hedrick, H. Ding, A. Guha, and M. A. Watson. 2002. Mouse glioma gene expression profiling identifies novel human glioma-associated genes. Ann. Neurol. 51:393-405. [DOI] [PubMed] [Google Scholar]
- 26.Haas, M., H. Wang, J. Tian, and Z. Xie. 2002. Src-mediated inter-receptor cross-talk between the Na+/K+-ATPase and the epidermal growth factor receptor relays the signal from ouabain to mitogen-activated protein kinases. J. Biol. Chem. 277:18694-18702. [DOI] [PubMed] [Google Scholar]
- 27.Haas, M., A. Askari, and Z. Xie. 2000. Involvement of Src and epidermal growth factor receptor in the signal-transducing function of Na+/K+-ATPase. J. Biol. Chem. 275:27832-27837. [DOI] [PubMed] [Google Scholar]
- 28.Holgado-Madruga, M., D. R. Emlet, D. K. Moscatello, A. K. Godwin, and A. J. Wong. 1996. A Grb2-associated docking protein in EGF- and insulin-receptor signaling. Nature 379:560-564. [DOI] [PubMed] [Google Scholar]
- 29.Huang, L., P. Kometiani, and Z. Xie. 1997. Differential regulation of Na/K ATPase alpha-subunit isoform gene expression in cardiac myocytes by ouabain and other hypertrophic stimuli. J. Mol. Cell. Cardiol. 29:3157-3167. [DOI] [PubMed] [Google Scholar]
- 30.Huang, Z.-Y., Y. Wu, N. Hedrick, and D. H. Gutmann. 2001. T-cadherin-mediated cell growth regulation involves G2 phase arrest and requires p21CIP1/WAF1 expression. Mol. Cell. Biol. 23:566-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoki, K., Y. Li, T. Xu, and K.-L. Guan. 2003. Rheb GTPase is a direct target of Tsc2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoki, K., Y. Li, T. Zhu, J. Wu, and K.-L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signaling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 33.Izumoto, S., T. Ohnishi, N. Arita, S. Hiraga, T. Taki, and T. Hayakawa. 1996. Gene expression of neural cell adhesion molecule L1 in malignant gliomas and biological significance of L1 in glioma invasion. Cancer Res. 15:1440-1444. [PubMed] [Google Scholar]
- 34.Juliano, R. L. 2002. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu. Rev. Pharm. Toxicol. 42:283-323. [DOI] [PubMed] [Google Scholar]
- 35.Kiyokawa, H., R. D. Kineman, K. O. Manova-Todorova, V. C. Soares, E. S. Hoffman, M. Ono, D. Khanam, A. C. Hayday, L. A. Frohman, and A. Koff. 1996. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 85:721-732. [DOI] [PubMed] [Google Scholar]
- 36.Kometiani, P., J. Li, L. Gnudi, B. B. Kahn, A. Askari, and Z. Xie. 1998. Multiple signal transduction pathways link Na+/K+-ATPase to growth-related genes in cardiac myocytes. J. Biol. Chem. 273:15249-15256. [DOI] [PubMed] [Google Scholar]
- 37.Korhonen, J. M., F. A. Said, A. J. Wong, and D. R. Kaplan. 1999. Gab1 mediates neurite outgrowth, DNA synthesis, and survival in PC12 cells. J. Biol. Chem. 274:37307-37314. [DOI] [PubMed] [Google Scholar]
- 38.Kwon, C.-H., X. Zhu, J. Zhang, and S. J. Baker. 2003. mTor is required for hypertrophy of Pten-deficient neuronal soma in vivo. Proc. Natl. Acad. Sci. USA 100:12923-12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecuona, E., S. Luquin, J. Avila, L. M. Garcia-Segura, and P. Martin-Vasallo. 1996. Expression of the beta 1 and beta 2 (AMOG) subunits of the Na,K ATPase in neural tissues: cellular and developmental expression patterns. Brain Res. Bull. 40:167-174. [DOI] [PubMed] [Google Scholar]
- 40.Leevers, S. J., D. Weinkove, L. K. MacDougall, E. Hafen, and M. D. Waterfield,. 1996. The Drosophila phosphoinositide 3-kinases Dp110 promotes cell growth. EMBO J. 15:6584-6594. [PMC free article] [PubMed] [Google Scholar]
- 41.Li, G., J. Kalabis, X. Xu, F. Meier, M. Oka, T. Bogenrieder, and M. Herlyn. 2003. Reciprocal regulation of MelCAM and AKT in human melanoma. Oncogene 22:6891-6899. [DOI] [PubMed] [Google Scholar]
- 42.Li, Y., K. Inoki, and K. L. Guan. 2004. Biochemical and functional characterizations of the small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24:7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, J., J. Tian, M. Haas, J. I. Shapiro, A. Askari, and Z. Xie. 2000. Ouabain interaction with cardiac Na+/K+-ATPase initiates signal cascades independent of changes in intracellular Na+ and Ca+2 concentrations. J. Biol. Chem. 275:27838-27844. [DOI] [PubMed] [Google Scholar]
- 44.Magyar, J., U. Bartsch, Z. Wang, N. Howells, A. Aguzzi, E. Wagner, and M. Schachner. 1994. Degeneration of neural cells in the central nervous system of mice deficient in the gene for the adhesion molecule on glia, the beta 2 subunit of murine Na,K-ATPase. J. Cell Biol. 127:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller-Husmann, G., S. Gloor, and M. Schachner. 1993. Functional characterization of beta isoforms of murine Na,K-ATPase. The adhesion molecule on glia (AMOG/beta 2), but not beta 1, promotes neurite outgrowth. J. Biol. Chem. 268:26260-26267. [PubMed] [Google Scholar]
- 46.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D. Y. Loh, and K. Nakayama. 1996. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707-720. [DOI] [PubMed] [Google Scholar]
- 47.Owens, G. C., E. A. Orr, B. K. DeMasters, R. J. Muschel, M. E. Berens, and C. Kruse. 1998. Overexpression of the transmembrane isoform of neural cell adhesion molecule alters the invasiveness of rat CNS-1 glioma. Cancer Res. 58:2020-2028. [PubMed] [Google Scholar]
- 48.Pagliusi, S. R., M. Schachner, P. H. Seeburg, and B. D. Shivers. 1990. The adhesion molecule on glia (AMOG) is widely expressed by astrocytes in developing and adult mouse brain. Eur. J. Neurosci. 2:471-480. [DOI] [PubMed] [Google Scholar]
- 49.Peng, M., L. Huang, Z. Xie, W. H. Huang, and A. Askari. 1996. Partial inhibition of Na+/K+ ATPase by ouabain induces the Ca+2-dependent expressions of early-response genes in cardiac myocytes. J. Biol. Chem. 271:10372-10378. [DOI] [PubMed] [Google Scholar]
- 50.Perez, O. D., S. Kinoshita, Y. Hitoshi, D. G. Payan, T. Kitamura, G. P. Nolan, and J. B. Lorens. 2002. Activation of the PKB/AKT pathway by ICAM-2. Immunity 16:51-65. [DOI] [PubMed] [Google Scholar]
- 51.Potter, C. J., H. Huang, and T. Xu. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105:357-368. [DOI] [PubMed] [Google Scholar]
- 52.Potter, C. J., L. G. Pedraza, and T. Xu. 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4:658-665. [DOI] [PubMed] [Google Scholar]
- 53.Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma, B. A. Hemmings, and G. Thomas. 1998. Phosphorylation and activation of p70s6k by PDK1. Science 279:707-710. [DOI] [PubMed] [Google Scholar]
- 54.Rong, R., J. Y. Ahn, H. Huang, E. Nagata, D. Kalman, J. A. Kapp, J. Tu, P. F. Worley, S. H. Snyder, and K. Ye. 2003. PI3 kinase enhancer-Homer complex couples mGluRI to PI3 kinase, preventing neuronal apoptosis. Nat. Neurosci. 6:1153-1161. [DOI] [PubMed] [Google Scholar]
- 55.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signaling network. Nat. Cell Biol. 5:566-571. [DOI] [PubMed] [Google Scholar]
- 56.Schmid, R. S., M. D. Schaller, S. Chen, M. Schachner, J. J. Hemperly, and P. F. Maness. 1999. NCAM stimulates the Ras-MAPK pathway and CREB phosphorylation in neuronal cells. J. Neurobiol. 38:542-548. [PubMed] [Google Scholar]
- 57.Schnadelbach, O., O. W. Blaschuk, M. Symonds, B. J. Gour, P. Doherty, and J. W. Fawcett. 2000. N-cadherin influences migration of oligodendrocytes on astrocyte monolayers. Mol. Cell Neurosci. 15:288-302. [DOI] [PubMed] [Google Scholar]
- 58.Sebti, S. M., and C. J. Der. 2003. Searching for the elusive targets of the farnesyltransferase inhibitors. Nat. Rev. Cancer 3:945-951. [DOI] [PubMed] [Google Scholar]
- 59.Senner, V., E. Kismann, S. Puttmann, N. Hoess, I. Baur, and W. Paulus. 2002. L1 expressed by glioma cells promotes adhesion but not migration. Glia 38:146-154. [DOI] [PubMed] [Google Scholar]
- 60.Senner, V., S. Schmidtpeter, S. Braune, S. Puttmann, S. Thanos, U. Bartsch, M. Schachner, and W. Paulus. 2003. AMOG/β2 and glioma invasion: does loss of AMOG make tumour cells run amok? Neuropathol. Appl. Neurobiol. 29:370-377. [DOI] [PubMed] [Google Scholar]
- 61.Sporns, O., G. Edelman, and K. Crossin. 1995. The neural cell adhesion molecule inhibits proliferation in primary cultures of rat astrocytes. Proc. Natl. Acad. Sci. USA 92:542-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stocker, H., T. Radimerski, B. Schindelholz, F. Witterwer, P. Belawat, P. Daram, S. Breuer, G. Thomas, and E. Hafen. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559-565. [DOI] [PubMed] [Google Scholar]
- 63.Tapon, N., N. Ito, B. J. Dickson, J. E. Treisman, and I. K. Hariharan. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105:345-355. [DOI] [PubMed] [Google Scholar]
- 64.Tee, A. R., D. C. Fingar, B. D. Manning, D. J. Kwiatkowski, L. C. Cantley, and J. Blenis. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 99:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tee, A. R., R. Anjum, and J. Blenis. 2003. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J. Biol. Chem. 278:37288-37296. [DOI] [PubMed] [Google Scholar]
- 66.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase activating protein complex toward Rheb. Curr. Biol. 13:1259-1268. [DOI] [PubMed] [Google Scholar]
- 67.Uhlmann, E. J., A. J. Apicelli, R. L. Baldwin, S. P. Burke, M. L. Bajenaru, H. Onda, D. Kwiatkowski, and D. H. Gutmann. 2002. Heterozygosity for the tuberous sclerosis complex (TSC) gene products results in increased astrocyte numbers and decreased p27-Kip1 expression in TSC2+/− cells. Oncogene 21:4050-4059. [DOI] [PubMed] [Google Scholar]
- 68.Uhlmann, E. J., W. Li, D. K. Scheidenhelm, C. Gau, F. Tamanoi, and D. H. Gutmann. 2004. Loss of tuberous sclerosis complex 1 (Tsc1) expression results in increased Rheb/S6K signaling important for astrocyte cell size regulation. Glia 47:180-188. [DOI] [PubMed] [Google Scholar]
- 69.Vanhaesebroeck, B., and D. R. Alessi. 2000. The PI3K-PDK1 connection: more than just a road to PKB. Biochem. J. 346:561-576. [PMC free article] [PubMed] [Google Scholar]
- 70.Urano, J., C. Ellis, G. J. Clark, and F. Tamanoi. 2001. Characterization of Rheb functions using yeast and mammalian systems. Methods Enzymol. 333:217-231. [DOI] [PubMed] [Google Scholar]
- 71.Verdu, J., M. A. Buratovich, E. L. Wilder, and M. J. Birnbaum. 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1:500-506. [DOI] [PubMed] [Google Scholar]
- 72.Weinkove, D., T. P. Neufeld, T. Twardzik, M. D. Waterfield, and S. J. Leevers. 1999. Regulation of imaginal disc cell size, cell number, and organ size by Drosophila class I(A) phosphoinositide 3-kinases and its adaptor. Curr. Biol. 9:1019-1029. [DOI] [PubMed] [Google Scholar]
- 73.Xia, H., R. S. Nho, J. Kahm, J. Kleidon, and C. A. Henke. 2004. Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a beta 1 integrin viability signaling pathway. J. Biol. Chem. 279:33027-33034. [DOI] [PubMed] [Google Scholar]
- 74.Xie, Z. 2003. Molecular mechanisms of Na/K-ATPase-mediated signal transduction. Ann. N. Y. Acad. Sci. 986:497-503. [DOI] [PubMed] [Google Scholar]
- 75.Xie, Z., P. Kometiani, J. Liu, J. Li, J. I. Shapiro, and A. Askari. 1999. Intracellular reactive oxygen species mediate the linkage of Na+/K+-ATPase to hypertrophy and its marker genes in cardiac myocytes. J. Biol. Chem. 274:19323-19328. [DOI] [PubMed] [Google Scholar]
- 76.Yang, W., A. P. Tabancay, Jr., J. Urano, and F. Tamanoi. 2001. Failure to farnesylate Rheb protein contributes to the enrichment of G0/G1 phase cells in the Schizosaccharomyces pombe farnesyltransferase mutant. Mol. Microbiol. 41:1339-1347. [DOI] [PubMed] [Google Scholar]
- 77.Ye, K., K. J. Hurt, F. Y. Wu, M. Fang, H. R. Luo, J. J. Hong, S. Blackshaw, C. D. Ferris, and S. H. Snyder. 2000. Pike A nuclear GTPase that enhances PI3 kinase activity and is regulated by protein 4.1N. Cell 103:919-930. [DOI] [PubMed] [Google Scholar]
- 78.Zhang, Y., X. Gao, L. J. Saucedo, R. Binggen, B. A. Edgar, and D. Pan. 2003. Rheb is a direct target of the tuberous sclerosis tumor suppressor proteins. Nat. Cell Biol. 5:578-581. [DOI] [PubMed] [Google Scholar]
- 79.Zhou, R., and O. Skalli. 2000. Identification of cadherin-11 down-regulation as a common response of astrocytoma cells to transforming growth factor-alpha. Differentiation 66:165-172. [DOI] [PubMed] [Google Scholar]