Abstract
Understanding the role of the mitogen-activated protein kinases (MAPKs) signalling pathway is essential in advancing treatments for neurodegenerative disorders like Alzheimer's. In this study, we investigate in-silico techniques involving computer-based methods to extract the MAPK1 sequence. Our applied methods enable us to analyze the protein's structure, evaluate its properties, establish its evolutionary relationships, and assess its prevalence in populations. We also predict epitopes, assess their ability to trigger immune responses, and check for allergenicity using advanced computational tools to understand their immunological properties comprehensively. We apply virtual screening, docking, and structure modelling to identify promising drug candidates, analyze their interactions, and enhance drug design processes. We identified a total of 30 cell-targeting molecules against the MAPK1 protein, where we selected top 10 CTL epitopes (PAGGGPNPG, GGGPNPGSG, SAPAGGGPN, AVSAPAGGG, AGGGPNPGS, ATAAVSAPA, TAAVSAPAG, ENIIGINDI, INDIIRTPT, and NDIIRTPTI) for further evaluation to determine their potential efficacy, safety, and suitability for vaccine design based on strong binding potential. The potential to cover a large portion of the world's population with these vaccines is substantial—88.5 % for one type and 99.99 % for another. In exploring the molecular docking analyses, we examined a library of compounds from the ZINC database. Among them, we identified twelve compounds with the lowest binding energy. Critical residues in the MAPK1 protein, such as VAL48, LYS63, CYS175, ASP176, LYS160, ALA61, LEU165, TYR45, SER162, ARG33, PRO365, PHE363, ILE40, ASN163, and GLU42, are pivotal for interactions with these compounds. Our result suggests that these compounds could influence the protein's behaviour. Moreover, our docking analyses revealed that the predicted peptides have a strong affinity for the MAPK1 protein. These peptides form stable complexes, indicating their potential as potent inhibitors. This study contributes to the identification of new drug compounds and the screening of their desired properties. These compounds could potentially help reduce the excessive activity of MAPK1, which is linked to Alzheimer's disease.
Keywords: Pharmacophore, Virtual screening, Molecular docking, Multiepitope, Vaccine design, MAPK1, Alzheimer, Zebrafish
Graphical abstract
1. Introduction
Alzheimerˈs disease (AD) is a chronic neurodegenerative disorder due to the overproduction or accumulation of Aꞵ (Amyloid ꞵ) plaques, which causes inhibition of phosphorylation of proteins that regulate signalling pathways [1]. The prevalence of AD increases with age. 5 % of the world's population with 65–74 age, 13.1 % with 75–85 age, and 33.2 % with 85 are diagnosed with Alzheimer's disease [2]. This disease is characterized by relentless progression of memory loss, such as motor ability, hallucinations, speech impairment, depression, aggressive behaviour, delusions, poor quality of life, and ultimately, death [3]. Despite of intensive understanding of the pathophysiology of this disorder, Risk gene identification biomarkers are still unknown and challenging since various susceptibility genes control this polygenic disorder due to the linkage of different types of AD on four different loci, particularly AD1, AD2, AD3, and AD4 [4]. Great efforts are needed to prevent disease formation, improve available treatments, and elucidate these neurodegenerative disorders' economic and social burden. Recent studies on the pathophysiology of this disease reveal the contribution of the MAPK1 Protein in AD [5]. MAPK1 is a protein coding gene and has a role in the MAPK cascade. MAPK1ise ERK2 (extracellular signals regulatory kinase 2) that regulate various physiological processes such as cell growth, differentiation, embryogenesis, proliferation, survival, death, and different signalling cascades [3].
The signalling pathway of MAPK is activated by diverse extracellular or intracellular stimuli such as hormones, peptides, cytokines, growth factors, and several cellular stressors (endoplasmic reticulum and oxidative stress). At the same time, its activation requires phosphorylation, which activates transcription factors that initiate the production of responded protein. Increased function of MAPK1 causes different neurodevelopmental disorders like AD. The deposition of extracellular Aꞵ plaques usually causes AD. Damaged neurons in AD are caused by Induced toxicity of Aꞵ plaques due to increased regulation of MAPK1 by inducing glutamate excitotoxicity, disruption of synaptic plasticity, and NF-kB activation, so the MAPK1 Protein should be a potential target to break down the Aꞵ plaques and to cure people with AD. In short, inhibition of MAPK1 Protein could be an effective and promising strategy to manage such neurodegenerative disorders.
Supportive efforts are ongoing to develop & and identify potent compounds to target the MAPK1 Protein to prevent disease formation and to improve the available treatments for this neurodegenerative disorder [6]. These efforts can be augmented by utilizing zebrafish as model organisms, as their genes are orthologous to human mutated genes for Alzheimer's disease. The genome of zebrafish has been fully characterized. It has shown a more straightforward nervous system than human and other rodent animal models for modelling or screening bioactive lead drug compounds, behavioural testing, and potential therapies for various neurodegenerative diseases. The CNS of zebrafish is traditionally arranged and separated into the forebrain, hindbrain, midbrain, motor spinal cord, cranial nerves, sensory nerves, and ascending and descending spinal cord. Transcriptome studies of Zebrafish have shown 87 % significant structural similarity to humans, highly conserved regions, accessible and e & malleable models for processing vertebrate genomes & and neural structures, neurodegenerative research, and facilitating the discovery of exceptional characteristics of genes that were difficult to observe by other model organism [7]. So, zebrafish is a viable animal model to target MAPK1 protein through peptides, and novel compound inhibitors should be utilized as potent drug candidates designed through different In-silico immunoinformatic approaches.
As in-vivo drug discovery is a lengthy process that takes 10–15 years and costs up to 2.448 billion USD for a drug to reach the market. It is a multi-step process, including identification, validation of drug targets, optimization, and preclinical and clinical trials, which is a long process. Therefore, the In-silico approach is utilized for targeted drug design to ease the in-vivo process in terms of time, validation, efficacy, and approval of a drug to reach the market [8].
The progression of computational drug designing and personalized medicine offers a way to understand different neurodegenerative diseases. Different structural and bioinformatics approaches were utilized to identify and design novel targeted active compounds for drug designing and peptides for epitope-based vaccine designing to treat several neurodegenerative diseases. Different immunoinformatic software was used for In Silico structure prediction by comparative modelling followed by identifying inhibitors, virtual screening based on physiochemical properties, and peptide design against MAPK1 in zebrafish. MAPK1 inhibitors could effectively mitigate the patient suffering and pathophysiology of Alzheimer's disease [5].
The in-silico design of a multiepitope-based-peptide (MBP) vaccine involves the computational identification of potential epitopes within a specific MAPK protein associated with Alzheimer's disease, followed by the selection of immunogenic epitopes, and the incorporation of these epitopes into a single peptide sequence. This peptide is optimized for immunogenicity and safety using computational tools, simulating its interaction with the immune system components and predicting potential immune responses. This approach leverages zebrafish as a model organism for Alzheimer's research, aiming to stimulate a broad and effective immune response against the targeted MAPK protein. However, experimental validation would be needed to confirm the vaccine's efficacy and safety.
This research's primary objectives encompass identifying and evaluating new drug compounds possessing specific physiochemical attributes to act as inhibitors targeting the overexpression of MAPK1, with a focus on potential applications for treating neurodegenerative disorders such as Alzheimer's disease. The study aims to understand the intricate MAPK signalling pathway's contribution to neurological disorders, providing a comprehensive context for diseases like Alzheimer's. Employing immunoinformatic analysis, the research aims to design epitope-based vaccines to precisely target the MAPK1 pathway, predicting β-cell and T-cell epitopes as potential drug targets and vaccine candidates. Leveraging the zebrafish model, the research investigates the effects of developed drug compounds and vaccines on neurological disorders. A crucial aspect involves identifying top-ranked peptides within the MAPK1 proteome capable of binding to major histocompatibility complex (MHC) molecules, facilitating vaccine development. Ultimately, this research strives to contribute to novel therapeutic strategies, offering insights for tailored drug design to address Alzheimer's disease and other neurodegenerative conditions through MAPK1 inhibition.
2. Materials and methods
We performed computational analysis to predict the structure of the target protein (MAPK1) as per the mothed of earlier researchers [9,10]. The amino acid sequence of this protein in FASTA format is retrieved from the UniProt database. We find appropriate template for the structure prediction of the target protein, the FASTA sequence of MAPK1 is subjected to BlastP, a protein-protein blast, against the Protein Data Bank (PDB) database as per the method of Ali, Ali, et al. [11]. We identify a suitable homologous protein structure that can serve as a guide for predicting the structure of the target protein. We set selection process for scrutinizing based on different factors such as query coverage and sequence identity, with the objective of identifying the homologous protein structure that best aligns with the target protein's sequence.
We utilized various methods for structure prediction, including homology/comparative modelling, ab initio modelling, and threading approach. Among these, the homology modeling method, executed through the MODELLER software, emerges as the most successful in generating accurate structure predictions. Homology modelling leverages the existing structural information of a related protein as a foundation for predicting the structure of the target protein. This methodology necessitates the utilization of multiple files and a command-based approach. Around 10 different model numbers are generated to construct the model, each involving a unique configuration. The criterion for selecting the optimal predicted model revolves around the concept of mol-pdf value, with the model possessing the lowest mol-pdf value being chosen as the most accurate representation.
After model generation, the energy of the predicted structure is refined and minimized through the use of UCSF Chimera and ChemDraw software, both of which enhance the overall stability of the model. Assessment of the predicted structure's reliability entails subjecting it to various structure evaluation tools. These tools encompass MolProbity, ERRAT, ENOLEA, Ramachandran plot analysis, ProCheck, and Verify 3D. This comprehensive evaluation aids in ascertaining the quality, accuracy, and overall credibility of the three-dimensional structure of the MAPK1 protein. An intricate depiction of the methodology employed in this process presented in Fig. 1, which outlines the step-by-step journey from sequence retrieval to reliable three-dimensional structure prediction.
Fig. 1.
Applied methodology for epitope-based vaccine designing and identification of novel drug compounds against MAPK1 protein.
2.1. Sequence retrieval
The amino acid sequence of mitogen-activated protein kinase (Uniprot ID: Q9DGR5) was retrieved from the Uniprot Knowledge base (https://www.uniprot.org) in FASTA format to determine the 3-Dimensional structure of the target protein (369 residues). Protpam was utilized to access the biochemical properties of MAPK1 Protein [12].
2.2. Physiochemical properties and antigenicity prediction of protein
Vxijen v2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was utilized to predict the protein's antigenicity by submitting the FASTA sequence of Protein with 0.4 thresholds [13]. The ExPAsy-Portparam tool (https://web.expasy.org/protparam/) was utilized to predict the physiochemical properties of MAPK1 Protein.
2.3. Template selection & structure prediction
The FASTA sequence of MAPK1 Protein was subjected to a BLASTp search against the RCSB Protein Data Bank (PDB: https://www.rcsb.org/) [14]to find an appropriate template structure for structure prediction through a comparative modelling approach. The Crystallographic structure of ERK2 in complex with compound 22 having a resolution of 1.76 Å [PDB ID; 5K4I] was chosen as an appropriate template having 94.54 % identity, 94 % query coverage, accession length of 352 residues, and E value 0.0 for reliable structure prediction. The 3-dimensional structure of MAPK1 was predicted by MODELLER 9.20 (https://salilab.org/modeller/) by satisfying all the spatial restraints [15]. The modeller generated 10 models. Computational Threading and ab initio approach (SWISS-MODEL (https://swissmodel.expasy.org/), I-TASSER (https://zhanggroup.org/I-TASSER/) Phyre2 (https://bio.tools/phyre), RaptorX (http://raptorx6.uchicago.edu/), ROBETTA (https://robetta.bakerlab.org/), M4t (http://manaslu.fiserlab.org/M4T/) were also utilized forMAPK1 Protein structure prediction. The energy minimization followed by geometrical optimization of the 3D predicted structure of MAPK1 was performed by UCSF chimaera (https://www.cgl.ucsf.edu/chimera/).
2.4. Structure evaluation/validation
The 3D predicted structure of MAPK1 Protein was evaluated using different evaluation tools. MolProbity (http://molprobity.biochem.duke.edu/) was used to assess the expected structure by the MODELLER (https://salilab.org/modeller/). At the same time, Rampage http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php [16], ProCheck https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/ [17], Anolea (http://leandro.iqm.unicamp.br/anolea/), and ERRAT (http://servicesn.mbi.ucla.edu/ERRAT/) were utilized to access the overall quality factor of predicted structure. The Ramachandran plot (https://molprobity.biochem.duke.edu/images/ramachandran.png) was generated to evaluate the forecast models. It showed the distribution of residues like Φ and Ψ distributions of non-proline and non-glycine residues. The plotted Φ and Ψ angles were used to distinguish the favourable and unfavourable regions. These angles were utilized to evaluate the quality of areas. Generally, a quality factor above 95 % was generated for high-resolution structures. UCSF Chimera 1.14 (https://www.cgl.ucsf.edu/chimera/) was used to visualize the 3D structure of MAPK1 protein. WinCoot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) was employed to correct all the outlier residues and poor rotamers from the final selected MAPK1 Protein structure.
2.5. Phylogenetic analysis
Six strains of MAPK1 were retrieved in FASTA format through the NCBI database (https://www.ncbi.nlm.nih.gov/). MSA (Multiple Sequence Alignment) was carried out to determine ancestral relationships; for this purpose, the well-known software MEGA-X (https://www.megasoftware.net/) [18] was employed for phylogenetic analysis of different strains of MAPK1 Protein. The phylogenetic tree of homologous sequences was built using a neighbour-joining distance-based approach, considering a bootstrap value of 500 replications [19]. Weblogo3 was utilized to find the conserved residues among different strains of MAPK1 [20].
2.6. World population coverage studies
IEBD (https://www.iedb.org/) was utilized to evaluate the world population coverage studies of predicted epitopes. Epitopes associated with specific HLA alleles were accessed to analyze MHC class I epitopes [21]. CTL epitopes were utilized against sets of particular alleles for selected population coverage studies. Different countries with higher affected rates of Alzheimer's disease were selected for whole world population coverage studies. Different epitopes with region-country-ethnicity combinations were utilized to analyze population coverage in the global population survey.
2.7. Prediction of linear and conformational B-cell epitopes
Plasma (antibody) cells and memory cells derived from B-lymphocytes because of interaction between anti-ꞵ-cells and epitopes of ꞵ-cells. The hydrophobic nature, flexibility, antigenicity, linear epitope forecast, and surface accessibility of ꞵ-cells are crucial for recognizing epitopes of ꞵ-cells [22]. ꞵ-cells epitopes were utilized by the free online IEBD abbreviated as Immune Epitope Database and Analysis Resource. The features of IEBD were set to 75 % specificity with 14 residual lengths of epitopes that were shown satisfactory to influence defensive immune response. The epitopes on the outer surface were selected, and those on the inner surface were rejected. The antigenicity, hydro and physiochemical properties, toxicity, digestibility, mutation, and allergenicity, were of selected epitopes were predicted. Hydrophilicity, linear epitopes forecast, flexibility, and surface acceptability studies were performed through Kolaskar & Tongaonkar antigenicity scale method [23], Karplus and Schulz for Flexibility Prediction [24], PHP (Parker's method for Hydrophilicity Prediction), Eminifor surface accessibility prediction [23]. Three different approaches were utilized in combination with Ellipro to forecast IEBD conformational ꞵ-cells epitopes (amino acid sequence having antigens interact with receptor protein of immune system), linear epitopes, along with adjacent clustering residues depending proximate 3D protein structure, PI (protrusion index) [25] and the criteria followed was 0.5 for possible least score and 6 for maximum distance [26].
2.8. Prediction of CTL epitopes and peptide designing
T cells epitopes are fragments of peptides that can provoke specific immune reactions and are immunodominant required for epitope-based peptide vaccine designing. CTL epitopes play an essential role in vaccine design by reducing wet lab experimentations' costs and time. The epitopes' cytotoxic effects on T lymphocytes were predicted. The predicted CTL epitopes were evaluated through NetCTL (https://services.healthtech.dtu.dk/services/NetCTL-1.2/) [27] for MHC class-I. For this purpose, the NetCTL features were set to super type A1 with TAP transport efficiency with 0.05 wt, thresholds for epitope prediction with 0.75, and C-terminal cleavage with 0.15. CTLs were activated through the antigenic surface of the MHC molecule. NetCTL server combined C-terminal proteasome cleavage, MHC Class 1 prediction, and transport efficiency TAP transporters into a single analytical system. The amino acid sequence of MAPK1 was used to analyze HLA (human leucocyte antigens) alleles and the length polypeptide chain. Artificial neural networks were used to estimate proteosome MHC class 1 binding and C-terminal cleavage, while T-epitome forecasts and weight matrix were utilized to evaluate transport efficiency TAP. Predicted epitopes for the Responded Protein, including antigenic residues, were analyzed through molecular docking studies. CTL epitopes of MAPK1Proteinrotein were predicted. The top 30 peptide sequences, along with the peptide's C-terminal proteasome cleavage affinity, Rescale-binding affinity, prediction score, MHC-binding affinity, and TAP transport efficiency, were generated by uploading the FASTA sequence of MAPK1 in the Net CTL server. The epitopes for MHC class II were predicted through the IEDB database. The N-align method was used to predict MHC class II Allele to be utilized as the best epitope candidate. The peptide's structure for respective Alleles was predicted by uploading the amino acid sequence of the peptide in the PEP-FOLD server, and a simulation of 100 runs was performed in several conformations' changes. The energy rating of sOPEP was utilized to examine compliance models in complex with the PEP-FOLD3. Only model 1 from all 10 Pepfold results was used for molecular docking [27].
2.9. Antigenicity and epitope conservancy prediction
The Vxijen 2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) and the conservancy prediction tool of IEBD (http://tools.iedb.org/conservancy/) were utilized to predict antigenicity/immunogenicity and conservancy of predicted peptides. More antigenic peptides were preferred to less antigenic ones for efficient T cell epitopes. So, the higher immunogenic epitopes were chosen for further analysis [28]. The conservancy implies a specific part of a protein sequence restrains the epitopes and shows availability with identity level [29].
2.10. Allergenicity and toxicity identification of predicted peptides
AllergenFP 1.0 (https://ddg-pharmfac.net/AllergenFP/) and AllerTOP v. 2.0 (https://www.ddg-pha/rmfac.net/AllerTOP/) were utilized to predict the Allergenicity of the predicted peptide as a candidate for the vaccine. AllergenFP 1.0 server indicates results through a non-alignment descriptor-based identification approach and can predict allergenicity and non-allergenicity with an accuracy of 87.9 % [30]. In contrast, a robust AllerTOP v. 2.0 server predicts results through a kNN (k nearest neighbours) based complementary approach to categorize allergens and non-allergens with an accuracy of 87.7 % [21]. The ToxinPred web server (http://crdd.osdd.net/raghava/toxinpred/) was employed to analyze the toxicity of predicted peptides. Toxinpred expects results using different peptide properties based on a quantitative matrix and machine learning approaches [31].
2.11. Molecular docking and structure modelling
2.11.1. Protein-peptide docking
For protein-peptide docking studies, the PatchDock server (http://bioinfo3d.cs.tau.ac.il/PatchDock/) was used to determine functionally interacting residues, the binding conformation, and the best structural information and to investigate the binding pattern of ligands to the respective protein [32]. The docked complexes with the least binding energy were chosen to analyze the receptor-ligand interaction and for further post-docking analysis using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/). The amino acid residues placed within 4 Å of the ligand and 10 interactions between receptor and ligand were identified. The docked complexes were categorized through geometric complementary, and all the redundant receptor atoms were removed from the docked complexes for reliable results. Fire-Dock (http://bioinfo3d.cs.tau.ac.il/FireDock/) software was used for flexible refinement and scoring of protein-ligand/protein-protein interactions. In the Firedock, the flexibility of docking experiments was set to increase while the scoring errors of docked complexes were fixed to decrease. UCSF Chimera 1.14, PyMOL, and Discovery Studio were employed for post-docking and interactional analysis to evaluate the H-bonding interaction and binding affinity of docked complexes.
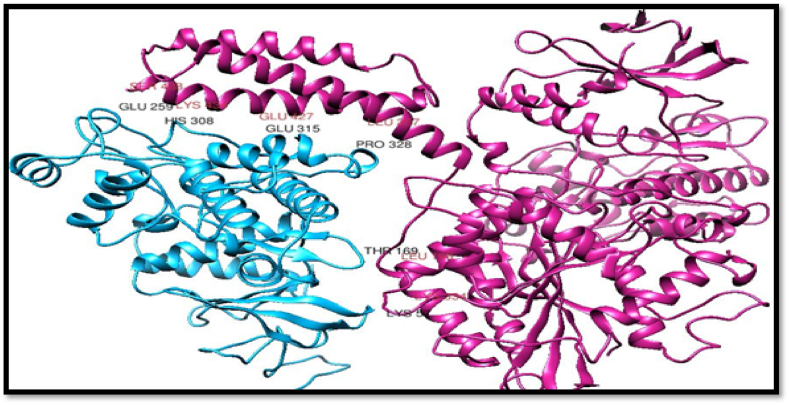
2.11.2. Protein-protein docking
The STRING database was employed to find out the interactions of MAPK1 Protein. It is an online database of known and predicted protein interactions, including direct (physical) and indirect (functional) relationships [19]. Docking of MAPK1 Protein with its interactive protein Atlastin GTPase 1(PDB ID: 3Q5D) was simulated using ClusPro and Patch Dock. The UCSF Chimera, Discovery Studio, and PyMol visualization software were utilized for post-docking studies of protein-protein interactions.
2.11.3. Protein-ligand docking
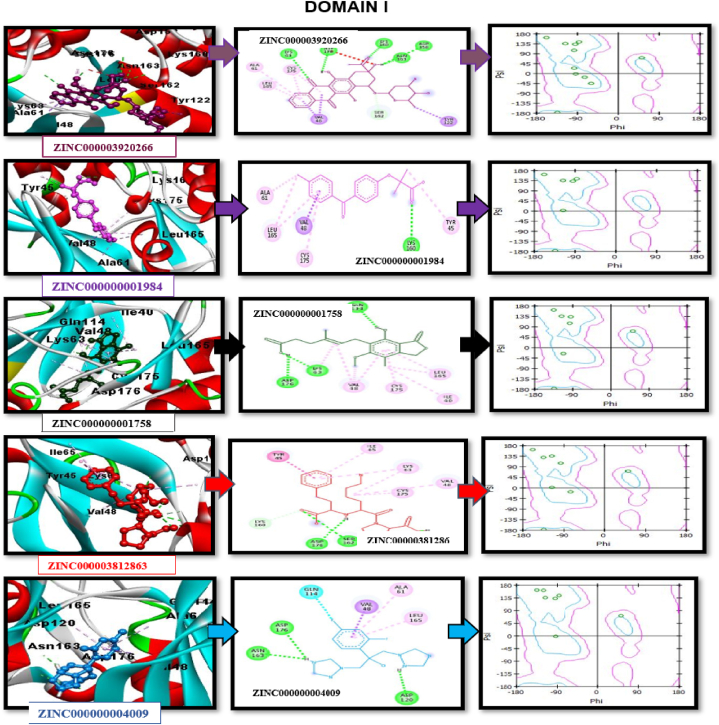
For molecular docking analysis and simulated scanning, the FDA-approved library of drug compounds was selected for the virtual screening of potential drug compounds. ZINC database (https://zinc.docking.org/) was utilized to download the library of 1800 FDA-approved drug compounds, which was minimized for further high throughput screening. The compound selection was based on pharmacophore [33,34]. This pharmacophore was screened against FDA FDA-approved zinc database library. Ligand Scout 4.4.8 software was used to build a library and to screen compounds. AutoDock 4 (https://autodock.scripps.eu/), PyRx (https://sourceforge.net/projects/pyrx/) [35], and AutoDock vina (https://vina.scripps.edu/) [36] were utilized to perform molecular docking studies [36]. The specific binding site of receptor-ligand interaction was identified through blind docking. PubChem database was utilized for getting the molecular coordinates of ligands. The ligandˈs SDF format was changed into.pdb format through ChemDraw [37]. The input files for ligand and receptor molecules should be in PDB format to execute AutoDock-vina. The polar H-atoms were introduced to the receptor Protein for docking studies, and thousands of docking runs were performed for each docking experiment. To examine the binding affinity between the selected ligand and MAPK1, an automated, flexible docking program was executed to the binding site of the ligand-protein through AutoDock Vina. The grid map of AutoDock facilitates the actual docking process. The gridˈs dimensions for molecular docking analysis were 56 × 56 × 56 Å for the X, Y, and Z axis with 0.648 Å spacing for the ligand to cover up the whole receptor protein between the grid points. The lowest binding energy and RMSD value of docking results were utilized to evaluate the best-docketed complex. The Druglike chemical and physical properties were assessed through ADMETlab (https://admetmesh.scbdd.com/) and admetSAR. UCSF Chimera 1.14, PyMOL(https://pymol.org/2/), Discovery studio (https://discover.3ds.com/discovery-studio-visualizer-download), and Liplot (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/) software's were employed to visualize and investigate the interacting residues.
The best structure of MAPK1 protein was selected with poor rotamers and lower Ramachandran outliers’ residues, which WinCoot further accessed to decrease the errors. The UCSF chimaera was utilized to visualize the predicted structure. It was accessed for different In-silico analyses of MAPK1 Protein followed by peptide designing and novel drug compounds used as effective inhibitors against MAPK1 protein. After the structure prediction and its evaluation, the peptides were designed against the target protein based on epitopes. The Net CTL version 1.2 server was utilized for CTL epitope prediction, providing multiple possible peptide sequences from the query protein found on the presence of CTL and MHC 1 binding epitopes. The peptide sequence obtained from the Net CTL was placed in PEPFOLD version 1.3, and the 30-peptide model was predicted for each peptide sequence. The epitope against the target protein and the 3D structure of the proposed peptide were analyzed using molecular docking by Patchdock with a threshold value of 4.0. Different docking solutions were obtained from Patchdock. The docked complexes with the least binding energy were chosen to analyze the receptor-ligand interaction and for further post-docking analysis using UCSF Chimera by refining patchdock results. Firedock gives the 10 best-refined docking solutions of protein and peptide based on global energy. protein and peptide docking results from Firdock were visualized by UCSF chimaera. The results show that the predicted 3D structure has two binding domains. The String database observed protein-protein interactions and their docking studies by the Cluspro web server, and UCSF Chimera and Discovery Studio visualized their docking interactions. The computation drug designing ZINC database was accessed for high throughput FDA-approved drug compound library screening for molecular docking studies. The FDA-approved drug library was screened through ligand Scoutt, and the best 50 compounds based on interactions were selected for further analysis. Different docking tools such as Autodock4, PyRx, and Autodock-vina were utilized for molecular docking studies, and 12 drug compounds with the lowest binding energy and RMSD difference of less than 2 were selected as innovative and effective inhibitors against MAPK1 protein. Most drug compounds interacted at the same binding pocket or active site of the target protein.
3. Results
3.1. 3D structure prediction and evaluation
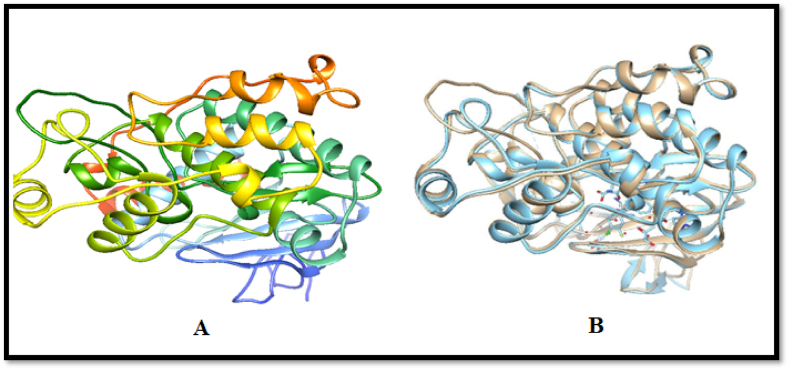
X-ray crystallography and nuclear magnetic resonance have not yet reported the 3D structures of MAPK1. 3D structures of Mitogen-activated protein kinase were predicted using threading and comparative homology modelling [19,38]. The MAPK1 amino acid sequence was subjected to BLASTp against the PDB database to find an appropriate template. A template was chosen for homology modelling comparison based on its rank as having the highest possible score regarding query coverage, maximal similarity, E value, and overall score. Protein sequence alignment revealed that conserved regions share similar activities [18,34,39]. MAPK1 3D structures were generated from the analyzed templates, as shown in Fig. 2 (A & B).
Fig. 2.
(A) MAPK1 structures predicted using 5K4I template with 0 E-value and 94 % query coverage. (B) Superimposition of 5K4I and predicted MAPK1 structures, demonstrating high identity and structural similarity.
In general, homology modelling revealed query coverage and similarities of>94 % between all the used templates and MAPK1, which is deemed adequate for robust structures. The threading method was used to fix the mistakes and improve the 3D model's structure. Different software (I-TASSER, SWISS-MODEL, M4t, Robetta, Raptor X Phyre2, 3D-jigsaw, and MODELLER) were employed to predict numerous models of MAPK1 by satisfying the sequence. Modeller 9.20 made a total of 10 models. The second one was the best because it had the lowest molpdf value. Favored and allowed regions, outliers, the overall quality factor, and binding regions were used to assess each of the created models. Different evaluation tools like Rampage and ERRAT were used to investigate the precision and accuracy of the forecast model. According to the ERRAT forecast, Model 2 was the best-predicted structure because it has an 84.225 % overall quality factor, as shown in Fig. 3.
Fig. 3.
Quality Assessment of Predicted MAPK1 Structure (Overall Quality Factor: 84.225) by Errat. Interactions visualized: white (bonded), red (non-bonded), and yellow (allowed residues).
Model 2 seemed to have the highest quality by ERRAT, with an overall quality factor of 84.225 %. Consistently, ERRAT was used to evaluate all the models' overall quality. After thoroughly analysing the evaluation factors that decided which structures would move forward, UCSF Chimera 1.14 was used to examine the secondary structures, side chains, ions, and ligands of proteins. Table 1 indicates that of 20 top-ranked 3D predicted structures of MAPK1 by MODELLER, the overall quality factor of predicted structure, Pro-check gives the %value of amino acid residue presents in favored, allowed, and outlier regions of a protein, and z score values for predicted 3D structures of MAPK 1.
Table 1.
Total of 20 top-ranked 3D predicted structures of MAPK1 base on In-silco investigations.
| MODELS | SOFTWARE | ERRAT % | VERIFY 3D (RESIDUE VALUE %) | PROCHECK |
PROSA |
||
|---|---|---|---|---|---|---|---|
| FAVORED REGION | ALLOWED REGION | OUTLIERS | Z SCORE VALUE % | ||||
| Model 1 | MODELLER | 74.5098 | 98.64 % | 90.10 % | 9.00 % | 0.30 % | −8.77 |
| Model 2 | MODELLER | 84.225 | 98.10 % | 91.00 % | 7.40 % | 0.60 % | −8.6 |
| Model 3 | MODELLER | 70.028 | 98.37 % | 89.50 % | 9.60 % | 0.00 % | −8.58 |
| Model 4 | MODELLER | 68.0556 | 96.75 % | 89.80 % | 8.70 % | 0.30 % | −8.72 |
| Model 5 | MODELLER | 70.9141 | 96.48 | 85.80 % | 12.70 % | 0.30 % | −9.15 |
| Model 6 | MODELLER | 68.6275 | 100 % | 87.30 % | 9.90 % | 0.90 % | −9.01 |
| Model 7 | MODELLER | 69.8324 | 96.75 % | 88.20 % | 9.90 % | 1.20 % | −8.29 |
| Model 8 | MODELLER | 70.0831 | 97.83 % | 89.50 % | 9.90 % | 0.00 % | −8.66 |
| Model 9 | MODELLER | 69.3593 | 98.37 % | 87.30 % | 10.50 % | 0.30 % | −8.96 |
| Model 10 | MODELLER | 72.6257 | 98.10 % | 88.50 % | 10.20 % | 0.30 % | −9.09 |
| Model 11 | MODELLER | 72.0339 | 98.92 % | 88.90 % | 9.30 % | 1.20 % | −8.72 |
| Model 12 | MODELLER | 67.22 | 96.56 % | 87.90 % | 9.60 % | 0.60 % | −8.59 |
| Model 13 | MODELLER | 68.7151 | 97.56 % | 88.50 % | 10.50 % | 0.00 % | −8.61 |
| Model 14 | MODELLER | 72.0222 | 100 % | 88.20 % | 10.50 % | 0.60 % | −8.8 |
| Model 15 | MODELLER | 71.3889 | 100 % | 89.80 % | 9.00 % | 0.30 % | −8.69 |
| Model 16 | MODELLER | 70.5556 | 100 % | 87.90 % | 10.20 % | 0.60 % | −8.59 |
| Model 17 | MODELLER | 61.9444 | 97.29 % | 87.00 % | 10.20 % | 0.60 % | −8.82 |
| Model 18 | MODELLER | 74.3733 | 98.64 % | 89.80 % | 9.00 % | 0.30 % | −8.6 |
| Model 19 | MODELLER | 72.1127 | 99.19 % | 87.90 % | 10.80 % | 0.30 % | −8.98 |
| Model 20 | MODELLER | 70.7042 | 98.37 % | 88.90 % | 9.60 % | 0.30 % | −9.04 |
3.2. Phylogenetic analysis
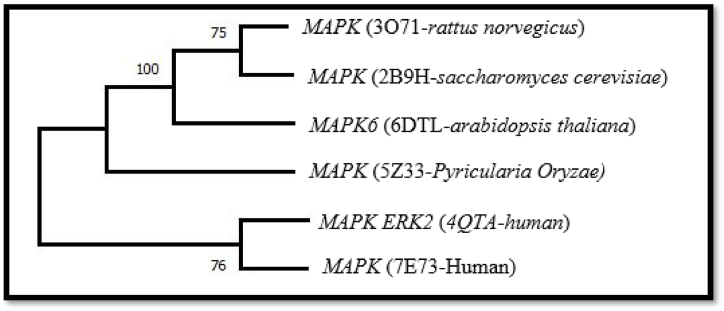
A phylogenetic tool called MEGA X was used to make the nearest tree of a chosen candidate gene to determine how it changed over time in humans, mammals, and Rattus norvegicus. The Molecular Evolutionary Algorithm Based (MEGA X), the most widely used software for phylogenetic studies, was used on MAPK1 to find the relationships between its ancestors. MSA of protein sequences represents the conserved part of MAPK1 strains having similar functions, as mentioned in Fig. 4.
Fig. 4.
Conserved residues in six MAPK1 strains highlighted by color-coded diagram, demonstrating high similarity across orthologous species. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
These templates were also used to create 3D structures of MAPK1. The distance-based method was used with neighbour-joining and a bootstrap valuation of 500 replications to make the phylogenetic tree, as mentioned in Fig. 5.
Fig. 5.
MAPK1 strain phylogeny by neighbour-joining method.
For making a phylogenetic tree or to find out the evolutionary relationship of MAPK strains, different strains in humans were downloaded through BlastP search against a database (PDB). Six top-ranked optimally aligned proteins FASTA sequences with maximum E-value, query coverage, and % identity were selected in Table 2.
Table 2.
Maximum score, query coverage, % identity, and E-value of MAPK1 strains in humans, Rattus norvegicus, saccharomyces cerevisiae, Arabidopsis thaliana, Pyricularia oryzae.
| Accession ID | Max. score | Total score | Query coverage | Max. identity | E-value |
|---|---|---|---|---|---|
| AQTA | 690 | 690 | 94 % | 94.54 % | 0.0 |
| 3O71 | 690 | 690 | 94 % | 94.54 % | 0.0 |
| 2B9H | 353 | 353 | 91 % | 50.14 % | 0.0 |
| 6DTL | 338 | 338 | 92 % | 51.16 % | 0.0 |
| 5Z33 | 337 | 337 | 92 % | 50.72 % | 0.0 |
| 7E73 | 688 | 688 | 94 % | 94.25 % | 0.0 |
3.3. Structure-based prediction of MPAK1 epitopes
Surface acceptability, antigenicity, flexibility, and predicted epitopes of 3D structures were evaluated to reduce the errors [40]. The interaction between antibody and protein was examined for selected epitopes, and the best leading conformational epitopes of MAPK1 having greater than 0.7 scores were further investigated. As per earlier research, conformational ꞵ-cells epitopes, linear epitopes, and adjacent clustering residues were determined for the target 3D protein structure through PI value [41]. The parameter was set at maximum distance and minimum length to predict 3D conformational epitopes, residual position, size, and name. The score range was observed between 0.685 and 0.714 for predicted peptides; a graphical representation of all applied parameters is mentioned in Fig. 6 (A).
Fig. 6.
6(A) we represent the detail of Structure-based prediction of MPAK1 epitopes, 6(B) Surface acceptability, 6(C) Surface flexibility 6(D) hydrophilicity 6(E) Antigenicity and 6(F) represent the World population Coverage Analysis.
3.4. Surface acceptability analysis of MAPK1
The surface accessibility >1.0 was observed for predicted peptides, indicating the presence of selected peptides on the protein surface. The forecast peptides were pragmatic based on the Y-axis, and reliably predicted peptides against MAPK1 were chosen for sequence position and surface probability analysis. Graphically representation of all applied parameters mentioned in Fig. 6 (B), such as the highest value observed for predicted peptides was 0.998with sequence PTIDQM ranging from 102 To 107 amino acids, while the lowest value observed for predicted peptides was 0.051 having sequence VGCILA ranging from 223 to 228 amino acids.
3.5. Surface flexibility analysis of MAPK1
Schulz and Karplus's flexibility method was employed to analyze and calculate different atomic vibration motions of 3D protein structures determined by temperature and B-factor [24]. The structure's organization, quality, and stability were identified through values of the B-factor, which determine the quality of the structure. The highest value B-factor suggests poorly ordered and less ordered structures, while the lower B-factor values depict reliable and highly organized and ordered structures [23]. The highest and lowest flexibility values were determined as 1.125 and 0.896 at locations 141–147 and 254-260for heptapeptide sequences [23]. The graphical representation of all applied parameters is mentioned in Fig. 6 (C).
3.6. Hydrophilicity analysis of MAPK1
The hydrophilicity of peptides was analyzed through Parker's method by utilizing Reverse-phase HPLC on column C18 to determine the retention duration of the predicted peptide. Parker's hydrophilicity predictions have the highest hydrophilicity values. The graphical representation of all applied parameters is mentioned in Fig. 6 (D), such as interactions between hydrophilic regions and antigenic sites were determined because of immunological studies. The hydrophilic graph was utilized to predict the peptides' hydrophilicity, in which the graph's X-axis represents the residual position. At the same time, Y-axes are shown measuring hydrophobicity to analyze the hydrophilicity of MAPK1-predicted peptides.
3.7. Antigenicity analysis of MAPK1 by Kolaskar and Tongaokar method
The antigenicity of MPAK1 was predicted by uploading the amino acid FASTA sequence of MAPK1 protein to the Vaxijen v2.0 server. The highest antigenicity was observed as 0.675 for protein sequence GGGPNPG at position 17. In contrast, the least antigenicity was observed as 0.242 for protein sequence CYFLYQI at positions 136 and 142, and a graphical representation of all applied parameters is mentioned in Fig. 6 (E).
3.7.1. World population coverage and allergenicity analysis
IEBD was utilized to evaluate the world population coverage studies of predicted epitopes, especially MHC HLA binding alleles. Epitopes associated with specific HLA alleles were accessed to analyze MHC class I of T cells. MHC HLA binding allele distribution varies among diverse ethics groups and geographic areas. Therefore, population coverage is crucial for effective vaccine design. The present study studied MHC-I-binding alleles of 30 epitopes for population coverage analysis. Significantly higher population coverage was observed for selected epitopes in different regions of the globe. The highest population Epitope coverage analysis for MHC class I was observed at 90 % worldwide and higher in Finland and UK populations. The graphical representation of all applied parameters is mentioned in Fig. 6 (F).
Allergenicity and toxicity prediction is vital during vaccine designing because many vaccines show allergic reactions because of immune response by initiating T helper cells type II and immunoglobulins. Therefore, the allergenicity and toxicity of predicted peptides were determined through AllergenFP. 1.0, ToxinPred server, and AllenTOP v. 2.0. The 3D structure of MHC class I and MHC class II of Tcells epitopes and 3D structure linear epitopes of B cells were predicted by the PEPFOLD server. The peptides PAGGGPNPG, GGPNPGSGA, EMVRGQAFD, RYSNLSYIG, MATAAVSAP, DNKVRVAIK, TAAVSAPAG, AVSAPAGGG, GINDIIRTP, INDIIRTPT, IIRTPTIDQ were determined as potential allergens to human. In contrast, the rest of the peptides are probable non-allergens to humans. The 3D structure of B cell, MHC class-1, and class-II of T cell epitopes was determined by submitting their peptide sequence into the PEPFOLD server.
3.8. Peptide designing and structure prediction
The Net CTL 1.2 webserver was utilized to predict CTL epitopes in a protein sequence for 12 supertypes. The amino acid sequence of MAPK1 was uploaded to the Net CTL server. A table was generated for the top 30 peptides having peptide Rescale binding affinity, TAP transport efficiency, MHC binding affinity, Prediction score, and C- terminal cleavage affinity through utilizing weigh matrix and artificial neural network, MHC class 1 epitopes respectively, as shown in Table 3.
Table 3.
Top-ranked 30 T-Cell epitopes with interacting MHC-1 alleles, epitope antigenicity, conservancy, score, allergenicity, mutation, hydrophobicity, hydrophilicity, and toxicity.
| Sr. No. | Peptide Sequence | Peptide MHC binding affinity | Rescale binding affinity | C-terminal cleavage affinity | TAP transport efficiency | Prediction score of MHC binding affinity | Conservancy | Antigenicity score | Toxicity | Allergenicity |
Mol wt. | Hydrophobicity | Hydrophilicity | Mutation prediction | Charge | SVM Score |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AllerTOP 2.0 |
AllergenFP 1.0 |
||||||||||||||||
| 1. | PAGGGPNPG | 0.0501 | 0.2127 | 0.046 | −1.832 | 0.1281 | 100 % | 1.5096 | NO | NO | YES | 722.88 | 0.00 | −0.03 | NO | 0.00 | −0.66 |
| 2. | ATAAVSAPA | 0.0827 | 03510 | 0.9067 | −0.3600 | 04690 | 100 % | 1.4078 | NO | NO | NO | 757.94 | 0.14 | −0.46 | NO | 0.00 | −0.47 |
| 3 | GGGPNPGSG | 0.0481 | 0.2042 | 0.0444 | −1.850 | 0.1184 | 14.9 % | 1.2459 | NO | NO | NO | 698.82 | −0.03 | 0.06 | NO | 0.00 | −0.75 |
| 4. | GGPNPGSGA | 0.0497 | 0.2108 | 0.3556 | −1.026 | 0.2129 | 33.3 % | 0.8650 | NO | NO | YES | 712.84 | −0.02 | 0.00 | NO | 0.00 | −0.91 |
| 5. | NPGSGAEMV | 0.0599 | 0.2542 | 0.7530 | −0.261 | 0.3541 | 100 % | 0.5841 | NO | NO | NO | 861.06 | −0.02 | 0.02 | NO | −1.00 | −0.56 |
| 6. | PGSGAEMVR | 0.0433 | 0.1840 | 0.4734 | 0.8750 | 0.2988 | 100 % | 0.8645 | NO | NO | NO | 903.14 | −0.15 | 0.33 | NO | 0.00 | −0.75 |
| 7. | EMVRGQAFD | 0.0555 | 0.2358 | 0.0281 | −1.666 | 0.1567 | 100 % | 1.2480 | NO | NO | YES | 1052.29 | −0.22 | 0.38 | NO | −1.00 | −0.92 |
| 8. | APAGGGPNP | 0.0524 | 0.2127 | 0.4669 | −1.8320 | 0.1281 | 100 % | 1.2377 | NO | NO | NO | 736.90 | 0.01 | −0.19 | NO | 0.00 | −0.59 |
| 9. | PRYSNLSYI | 0.0510 | 0.2163 | 0.7480 | 0.4180 | 0.3494 | 33 % | 1.5095 | NO | NO | NO | 1112.37 | −0.19 | −0.49 | NO | 1.00 | −90 |
| 10. | RYSNLSYIG | 0.0520 | 0.2207 | 0.1556 | −1.036 | 0.1922 | 33 % | 1.3891 | NO | NO | YES | 1072.31 | −0.16 | −0.49 | NO | 1.00 | −0.60 |
| 11. | MATAAVSAP | 0.0664 | 0.2820 | 0.0742 | −0.1660 | 0.3014 | 100 % | 0.9822 | NO | NO | YES | 923.12 | 0.04 | −0.32 | NO | −1.00 | −0.72 |
| 12. | SAYDRDNKV | 0.0713 | 0.3027 | 0.8990 | 0.6000 | 0.4675 | 100 % | 0.7874 | NO | NO | NO | 1067.23 | −0.49 | 0.91 | NO | 0.00 | −0.73 |
| 13. | DRDNKVRA | 0.0493 | 0.2095 | 0.2071 | −0.873 | 0.1969 | 100 % | 1.9579 | NO | NO | NO | 973.15 | −0.74 | 1.65 | NO | 1.00 | −0.78 |
| 14. | RDNKVRVAI | 0.0576 | 0.2447 | 0.1973 | 0.2380 | 0.2862 | 85.4 % | 0.7266 | NO | NO | NO | 1070.38 | −0.44 | 0.77 | NO | 2.00 | −0.84 |
| 15. | DNKVRVAIK | 0.0474 | 0.2013 | 0.7292 | 0.1260 | 0.3170 | 74.6 % | 0.5797 | NO | NO | YES | 1042.37 | −0.36 | 0.77 | NO | 2.00 | −0.90 |
| 16. | AIKKISPFE | 0.0504 | 0.2141 | 0.0247 | −1.248 | 0.1554 | 33.3 % | 0.8488 | NO | NO | NO | 1032.37 | −0.09 | 0.30 | NO | 1.00 | −1.26 |
| 17. | SAPAGGGPN | 0.0656 | 0.2659 | 0.0342 | −1.1780 | 0.2121 | 14.9 % | 1.1791 | NO | NO | NO | 726.86 | −0.06 | −0.01 | NO | 1.50 | −0.85 |
| 18. | TAAVSAPAG | 0.0580 | 0.2461 | 0.0348 | −1.1930 | 0.1917 | 72.4 % | 0.5891 | NO | NO | YES | 743.92 | 0.13 | −0.40 | NO | 1.50 | −1.41 |
| 19. | AVSAPAGGG | 0.0586 | 0.2488 | 0.0286 | −0.9630 | 0.2050 | 100 % | 1.0560 | YES | NO | NO | 685.85 | 0.16 | −0.30 | NO | 0.05 | −1.28 |
| 20. | ISPFEHQTY | 0.2829 | 1.2012 | 0.9750 | 3.0740 | 1.5011 | 24.4 % | 0.5730 | NO | NO | NO | 1121.34 | −0.10 | −0.44 | NO | −0.50 | −0.80 |
| 21. | ENIIGINDI | 0.0532 | 0.2259 | 0.5385 | 0.2240 | 0.3178 | 94.2 % | 0.7984 | NO | NO | NO | 1000.26 | 0.05 | −0.09 | NO | −2.00 | −0.78 |
| 22. | IIGINDIIR | 0.0569 | 0.2416 | 0.0822 | 1.6140 | 0.3346 | 86.6 % | 1.0368 | NO | NO | NO | 1026.39 | 0.08 | −0.31 | NO | 0.00 | −0.41 |
| 23. | IGINDIIRT | 0.0581 | 0.2467 | 0.0302 | −0.871 | 0.2077 | 100 % | 0.6975 | NO | NO | NO | 1014.33 | −0.02 | −0.16 | NO | 0.00 | −0.555 |
| 24. | GINDIIRTP | 0.0533 | 0.2264 | 0.2396 | −0.031 | 0.2608 | 100 % | 1.1470 | NO | YES | NO | 998.28 | −0.11 | 0.04 | NO | 0.00 | −0.64 |
| 25. | INDIIRTPT | 0.0541 | 0.2295 | 0.0301 | −1.041 | 0.1820 | 82.9 % | 1.0889 | NO | YES | NO | 1042.33 | −0.15 | 0.00 | NO | 0.00 | −0.74 |
| 26. | NDIIRTPTI | 0.0506 | 0.2148 | 0.4440 | 0.3420 | 0.2985 | 23.4 % | 0.6587 | NO | NO | NO | 1042.33 | −0.15 | 0.00 | NO | 0.00 | −0.52 |
| 27. | DIIRTPTID | 0.0535 | 0.2273 | 0.0282 | −1.793 | 0.1419 | 100 % | 0.7061 | NO | NO | NO | 1043.31 | −0.16 | 0.31 | −1.00 | NO | −0.49 |
| 28. | IIRTPTIDQ | 0.0537 | 0.2281 | 0.0335 | −0.044 | 0.2310 | 42.3 % | 0.5511 | NO | YES | NO | 1056.36 | −0.16 | 0.00 | NO | 0.00 | −0.45 |
| 29. | RTPTIDQMK | 0.0909 | 0.3858 | 0.4122 | 0.6810 | 0.4817 | 14.29 % | 1.3169 | NO | NO | NO | 1089.40 | −0.41 | 0.59 | NO | 1.00 | −0.86 |
| 30. | DQMKDVYIV | 0.0586 | 0.2487 | 0.5812 | 0.1480 | 0.3433 | 33.3 % | 0.8014 | NO | NO | NO | 1110.42 | −0.13 | −0.09 | NO | −1.00 | −0.82 |
We explore that T-cell epitopes are immunodominant fragments of peptides that can provoke an immune response and are integral for epitope-based peptide vaccine design (see Table 4). For this purpose, The NetCTL server was utilized for CTL (cytotoxic T-lymphocytes) epitope prediction from an amino acid sequence of the respective protein. It predicts human CD8+ T-cell epitopes for twelve major supertypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, and B62) considering TAP transport efficiency, MHC class I binding, binding affinity, proteasomal C terminal cleavage through weight matrix and neural network method. In this study, we predicted epitopes for all twelve supertypes considering threshold value 1.25, which has 0.993 specificity and 0.54 sensitivity, respectively. Initially, 150 epitopes considering the highest combined score were selected, which were further evaluated through Vaxijen v2.0 software to analyze the immunogenicity, toxicity, and antigenicity of predicted peptides for final selection and toxicity prediction is a vital step during vaccine designing because many vaccines show allergic reactions because of immune response by initiating T helper cells type II and immunoglobulins. Therefore, the allergenicity and toxicity of predicted peptides were determined through AllergenFP. 1.0, ToxinPred server, and AllenTOP v. 2.0.
Table 4.
Prediction of top-ranked 30 MHC-II alleles for desired epitopes.
|
MHC-II Allele |
Position | Core Sequence | Peptide Sequence | IC50 value | Rank | Antigenicity Score | Toxicity | Hydroph0bicity | Hydrophilicity | Mutation Prediction | Charge | SVM score | Molecular weight | Allergenicity |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AllerTOP 2.0 |
AllergenFP 1.0 |
||||||||||||||
| HLA-DRB1*04:05 | 144–158 | YIHSANVLH | RGLKYIHSANVLHRD | 6.70 | 0.01 | 0.4749 | No | −0.29 | 0.12 | No | 3.00 | −0.98 | 1779.26 | Yes | No |
| HLA-DRB1*04:05 | 145–159 | GLKYIHSANVLHRDL | 6.80 | 0.01 | 0.2509 | No | −0.13 | −0.20 | No | 2.00 | −0.98 | 1736 | No | No | |
| HLA-DRB1*04:05 | 146–160 | LKYIHSANVLHRDLK | 7.60 | 0.01 | 0.5898 | No | −0.22 | 0.00 | No | 3.00 | −1.08 | 1807.36 | No | No | |
| HLA-DRB3*02:02 | 269–283 | YLLSLPLRS | ARNYLLSLPLRSKVP | 9.00 | 0.45 | 0.4488 | No | −0.20 | −0.11 | No | 3.00 | −1.29 | 1727.30 | No | No |
| HLA-DRB5*01:01 | 268–282 | KARNYLLSLPLRSKV | 8.10 | 0.77 | 0.6047 | No | −0.27 | −0.09 | No | 4.00 | −1.30 | 1758.36 | No | No | |
| HLA-DPA1*03:01/DPB1*04:02 | 344–358 | TLKELIFEX | LDDLPKETLKELIFE | 959.3 | 31.0 | 0.3343 | No | −0.15 | −0.61 | No | −3.00 | −1.77 | 1803.31 | No | No |
| HLA-DPA1*02:01/DPB1*01:01 | 299–313 | MLTFNPHK | LDKMLTFNPHKRIEV | 1069 | 31.0 | 0.8176 | No | −0.23 | 0.24 | No | 1.50 | −1.43 | 1841.43 | Yes | No |
| HLA-DQA1*01:02/DQB1*06:02 | 225–339 | AEMLSNRPI | CILAEMLSNRPIFPG | 962.7 | 31.0 | 0.3552 | No | 0.03 | −0.40 | No | 0.00 | −0.90 | 1661.23 | Yes | No |
| HLA-DRB1*11:01 | 56–70 | VRVAIKKIS | NKVRVAIKKISPFEH | 3.40 | 0.02 | 0.4654 | No | −0.24 | 0.36 | No | 3.50 | −1.16 | 1766.36 | No | No |
| HLA-DRB1*04:05 | 147–161 | YIHSANVLH | KYIHSANVLHRDLKP | 13.20 | 0.07 | 0.4161 | No | −0.26 | 0.12 | No | 3.00 | −0.99 | 1791.31 | No | No |
| HLA-DRB1*12:01 | 260–274 | IINIKARNY | DLNCIINIKARNYLL | 15.70 | 0.08 | 1.0557 | No | −0.09 | −0.33 | No | 1.00 | −0.58 | 1776.36 | No | No |
| HLA-DRB1*12:01 | 261–275 | LNCIINIKARNYLLS | 16.50 | 0.1 | 0.9209 | No | −0.06 | −0.51 | No | 2.00 | −0.61 | 1748.35 | No | No | |
| HLA-DRB1*12:01 | 269–273 | EDLNCIINIKARNYL | 18.20 | 0.01 | 1.0401 | No | −0.17 | −0.01 | No | 0.00 | −0.45 | 1792.31 | No | No | |
| HLA-DRB1*12:01 | 262–276 | NCIINIKARNYLLSL | 19.30 | 0.11 | 0.8983 | No | −0.06 | −0.51 | No | 2.00 | −0.52 | 1748.35 | No | No | |
| HLA-DRB1*12:01 | 258–272 | QEDLNCIINIKARNY | 22.90 | 0.13 | 1.0882 | No | −0.25 | 0.12 | No | 0.00 | −0.03 | 1807.28 | Yes | No | |
| HLA-DRB1*12:01 | 263–277 | CIINIKARNYLLSLP | 24.90 | 0.15 | 0.7558 | No | −0.03 | −0.53 | No | 2.00 | −0.83 | 1731.36 | No | No | |
| HLA-DRB4*01:01 | 120–134 | LLKTQHLSN | DLYKLLKTQHLSNDH | 14.00 | 0.25 | 0.9376 | No | −0.27 | 0.12 | No | 1.00 | −0.90 | 1825.75 | No | No |
| HLA-DRB4*01:01 | 21–35 | LYKLLKTQHLSNDHI | 13.50 | 0.23 | −0.6296 | No | −0.17 | −0.20 | No | 2.00 | −0.93 | 1823.37 | No | No | |
| HLA-DRB1*11:01 | 57–71 | VRVAIKKIS | KVRVAIKKISPFEHQ | 5.80 | 0.25 | 0.2936 | No | −0.25 | 0.36 | No | 3.50 | −1.31 | 780.37 | No | Yes |
| HLA-DRB1*11:01 | 53–67 | DRDNKVRVAIKKISP | 4.90 | 0.18 | 0.6534 | No | −0.43 | 0.96 | No | 3.00 | −1.34 | 1739.26 | Yes | No | |
| HLA-DRB1*11:01 | 54–68 | RDNKVRVAIKKISPF | 3.90 | 0.11 | 0.6600 | No | −0.34 | 0.59 | No | 4.00 | −1.30 | 1771.35 | No | No | |
| HLA-DRB1*11:01 | 55–69 | DNKVRVAIKKISPFE | 3.80 | 0.09 | 0.5399 | No | −0.26 | 0.59 | No | 2.00 | −1.28 | 1744.28 | No | No | |
| HLA-DRB1*01:01 | 268–282 | YLLSLPLRS | KARNYLLSLPLRSKV | 3.30 | 0.09 | 0.6047 | No | −0.27 | 0.09 | No | 4.00 | −1.30 | 1758.36 | No | Yes |
| HLA-DRB1*01:01 | 269–283 | ARNYLLSLPLRSKVP | 3.50 | 0.11 | 0.4485 | No | −0.20 | −0.11 | No | 3.00 | −1.29 | 1727.30 | No | No | |
| HLA-DRB1*01:01 | 270–284 | RNYLLSLPLRSKVPW | 3.80 | 0.14 | 0.1028 | No | −0.19 | −0.31 | No | 3.00 | −.12 | 1842.44 | No | No | |
| HLA-DRB1*04:05 | 118–132 | YKLLKTQH | ETDLYKLLKTQHLSN | 16.20 | 0.13 | 1.0167 | No | −0.25 | 0.13 | No | 0.50 | −1.05 | 1803.23 | No | No |
| HLA-DRB1*04:05 | 119–133 | TDLYKLLKTQHLSND | 16.90 | 0.14 | 0.9217 | No | −0.26 | 0.13 | No | 0.50 | −0.88 | 1789.25 | No | No | |
| HLA-DRB1*04:05 | 117–131 | METDLYKLLKTQHLS | 17.40 | 0.15 | 0.9272 | No | −0.19 | 0.03 | No | 0.50 | −1.15 | 1820.37 | No | No | |
| HLA-DRB1*12:01 | 137–151 | YQILRGLKY | YFLYQILRGLKYIHS | 25.30 | 0.16 | 0.5531 | No | −0.02 | −0.83 | No | 2.50 | −0.66 | 1914.52 | No | No |
| HLA-DRB1*12:01 | 136–150 | CYFLYQILRGLKYIH | 2.70 | 0.17 | 0.6318 | No | 0.00 | −0.91 | No | 2.50 | −0.42 | 1930.58 | Yes | No | |
The 3D structure of MHC class I and MHC class II of Tcells epitopes and the 3D structure of linear epitopes of B cells were predicted by the PEPFOLD server. The peptides PAGGGPNPG, GGPNPGSGA, EMVRGQAFD, RYSNLSYIG, MATAAVSAP, DNKVRVAIK, TAAVSAPAG, AVSAPAGGG, GINDIIRTP, INDIIRTPT, IIRTPTIDQ were determined as potential allergens to human. In contrast, the rest of the peptides are probable non-allergens to humans. The 3D structure of B cell, MHC class-1, and class II of T cell epitopes was determined by submitting their peptide sequence into the PEPFOLD server.
Among 150 primarily designed epitopes for T-cells, 79 epitopes were observed under 0.00 threshold value on the Vaxijen webserver, while 41 peptides were observed under 0.00 immunogenicity value. This observation shows that all the predicted epitopes with the highest combined score were unreliable and couldn't be considered best. Thus, 30 epitopes with positive immunogenicity and antigenicity scores were selected as highly immunogenic epitopes that can interact with MHC alleles with high binding affinity to generate an effective and strong immune response. The IEBD tools were utilized to study immunogenicity and conservancy analysis of predicted peptides. Conservancy refers to a specific protein sequence with a particular level of identity that shows availability and restrains an epitope. Higher immunogenic peptides are preferable to less immunogenic peptides to be considered reliable candidates for vaccine development. The conservancy analysis of 30 epitopes resulted in 13 epitopes with 100 % conservancy scores and 19 epitopes having greater than 70 % conservancy scores, which were reliable for effective vaccine development. The higher conservancy score of predicted epitopes ensures the best target among different variants of respective proteins for improved, effective, and reliable vaccine design. Among 30 peptides, 13 epitopes show greater than 1 Vaxijen score which reveals the antigenic nature of the corresponding peptides which could be a good criterion for reliable epitope selection as the higher score increases the probability to provoke an effective and reliable immune response. While the rest of the 11 epitopes were considered less immunogenic for a vaccine candidate.
We also predict the most frequently occurring MHC binding alleles to predict potent peptides. The predicted peptides with lower IC50 values were chosen as potent candidates because this shows the higher affinity of peptides for respective epitopes. We also utilized these epitopes to predict MHC II alleles and CD4+ T-cell epitopes. The epitopes, MHC class II alleles, and individual peptides are presented in Table 4. The Accessibility, flexibility, epitope prediction, and antigenicity of the predicted 3D structure were evaluated to decrease the errors. The antibody-protein interaction for the selected epitope and top-ranked conformational epitope with more than 0.7score of MAPK1 were analyzed further by isoelectric point. The linear conformational and discontinuous epitopes were predicted through Bepipred 2.0 as B cells are categorized into these recognized through humoral immunity through antibodies or B cell receptors.
Therefore, predicting these B cell peptides is essential for peptide-based vaccine designing. 11 linear B cell peptides with different lengths were predicted from antigenic MAPK1 protein through the BepiPred 2.0 method. The peptide structure was predicted using the Pepfold server to study various post-docking analyses. The antigenicity, toxicity, hydrophobicity, hydrophilicity, IC50 value, and mutations were also predicted for respective peptides. Epitope 9 was shown to be toxic among all predicted epitopes for MHC-1, while different allergenicity results were observed for different software. This finding indicates that these epitopes were reliable for peptide-based vaccine design against MAPK1.
In this computational in-silico analysis, the epitope AVSAPAGGG was observed as toxic for vaccine design. Allergenicity of predicted peptides was anticipated through AllergenFP 1.0 and AllerTOP v.2.0, and toxicity was expected through the ToxinPred webserver. Allergenicity analysis is vital before vaccine development as many vaccines reverse immune response to hypersensitivity and allergic reaction through IgE and Type II helper T cells. The predicted allergenicity was different for different software's like AllerTOP shows allergenicity for 3 epitopes, while AllergenFP shows allergenicity for 7 epitopes. Each software offers allergenicity even for 100 % conserved epitopes. Successful recognition of predicted epitopes through HLA molecules with high binding affinity is not enough for an effective and reliable immune response rather, it also depends on considerable immunogenicity and antigenicity scores. So, the potential epitopes with the highest antigenicity, immunogenicity, non-allergens, and non-toxins recognized by many HLA molecules were selected to induce a strong and effective immune response.
Identification of B cell epitopes is a critical step, especially for epitope-based peptide vaccine designing as B cells are antigenic sequences that were recognized either by antibodies or specific receptors of B cells because of the humoral response. The epitopes of B cells are categorized into two main types: linear or continuous epitopes of B cells and conformational and discontinuous epitopes of B cells. Most research reveals that above 90 % of B cell epitopes are conformational. Therefore, B cell epitopes from highly antigenic MAPK1 protein were identified through Insilico analysis using online Bepipred2.0 software which is the best-known epitope prediction method for B cell epitopes and shows reliable results for linear epitopes data predicted through the IEDB database and crystallographic approach. The discontinuous and confirmation epitopes of B cells were predicted through IEDB B-cell conformational epitope prediction tools ElliPro by using default parameters of a minimum score of 0.8 and a maximum distance of 7 Å. B-cell conformational epitopes are antigenic amino acid sequences interacting with immune system receptors.
In contrast, Ellipro predicts both conformational and linear epitopes based on the 3D structure of protein and indicates score as protrusion index PI value. The bepiPred 2.0 server predicted 11 linear epitopes of B cells with different variations in length. The linear epitopes of B cells with different variations in length, as presented in Table 5. Among these 11 predicted peptides, 4 B-cell epitopes were observed as nonantigenic through Vaxijen v 2.0 forecasts, and these epitopes couldn't be considered potential vaccine candidates. In contrast, the remaining 7 epitopes could be regarded as potential vaccine candidates due to their high antigenicity score. After that, allergenicity and toxicity were also predicted for these epitopes. The toxicity, antigenicity, allergenicity, and conservancy analysis reveals that epitopes 5 and 11 could be considered potential and reliable B-cell epitope for effective vaccine designing. In this computational analysis total of 5 conformational and discontinuous epitopes of B-cells were predicted by utilizing the IEDB tool Elllipro. The epitopes having PI scores greater than 0.6 were considered. The protrusion index value of 8 predicted epitopes was analyzed between 0.658 and 0.747. The higher score of residues reveals greater solvent accessibility. The Conformation epitopes of B-cells, their length, individual residues, score, and residue position are presented in Table 6.
Table 5.
Prediction of linear B-cell epitopes of MAPK1 protein through Bepipred-2.0
| Sr. No. | Peptide Sequence | Position | Peptide MHC pair | Global energy (Kcal/Mol) | Attractive energy (Kcal/Mol) | H-bond energy (Kcal/Mol) | Conserved Residues | Antigenicity score | Toxicity | Allergenicity |
Mol wt. | Hydrophobicity | Hydrophilicity | Mutation prediction | Charge | SVM Score |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AllerTOP 2.0 |
AllergenFP 1.0 |
||||||||||||||||
| 1. | NHILGILGSPSQEDLNCIINIKARNYLLSLPLRSKVPWNRLFPNADPKALD | 247–297 | PRO102 CG-VAL7 CG1, PRO365 CG-AR6 CG3, ILE99 CG2-IL9 C, ALA10 N-VAL5 CG2, ALA4 CA-VAL5 CG1, VAL30 C-ILE9 CB, PRO32 O-ASP2 OD2, ASN15 N-LYS4 NZ, ALA4 CA-VAL5 CG1, GLY13 O-LYS4 CE, PRO14 CA-LYS4 NZ, ALA4 N-ASN3 O, THR3 CA-ASN3 O. | −31.36 | −36.51 | −4.71 | ARG33, ILE99, ARG100, PRO102, THR101, GLN106, GLY12, GLY11, ALA10. | 0.5251 | NO | NO | NO | 5625.38 | −0.12 | −0.17 | NO | 3.50 | −0.77 |
| 2. | MATAAVSAPAGGGPNPGSGAEMVRGQAFDVGPRYSNLS | 1–38 | GLY12 N-SER2 CB, ASP290-PRO5 CA, ASP290 OD2-LEU1 C. | −31.24 | −32.94 | −4.84 | GLY12, ASP290. | 0.9304 | NO | NO | YES | 3649.57 | −0.06 | −0.10 | NO | 0.00 | −1.14 |
| 3 | ARFQPGNRP | 361–369 | MET22 CE-ARG2 NH2 | −23.29 | −23.56 | −2.51 | MET22 | 0.0448 | NO | YES | YES | 1042.28 | −0.44 | −1.67 | NO | 2.00 | −1.24 |
| 4. | ADPDHDHTGFL | 183–193 | PRO9 CG-LYS6 CA, PRO9 CG-VAL7 CG2, ALA1 O-ARG3 CD. | −29.46 | −29.34 | −1.03 | ARG100, GLN364. | −0.248 | NO | NO | YES | 1224.40 | −0.15 | 0.25 | NO | −2.00 | −0.57 |
| 5. | YDRDNKVR | 52–59 | PRO365 O-ARG8 O2, PRO365 CB-ASN5 CB, ASN5 C-LYS6 N, | −35.15 | −35.20 | −4.58 | PRO365, PRO9, ALA10, PRO102 | 1.1874 | NO | NO | NO | 1065.25 | −0.77 | 1.43 | NO | 1.00 | −0.70 |
| 6. | EEALAHPYLEQYYDPTDEPVAEAPFKFDMELDDLPKETLKELIFE | 314–358 | PRO365 CB-ILE2 CD, VAL6 O-ILE3 CG2, ALA4 CB-ILE8 CG1, ARG33 CG-THR7 CB, ASP55 N-THR7 CG2, ARG54 CA-THR7 CG2, ARG54 CB-PROR6 O2. | −30.72 | −34.41 | −4.67 | PRO365, VAL6, ALA4, ARG33, ASP55, ARG54. | 0.2551 | NO | NO | YES | 5331.56 | −0.15 | 0.40 | NO | −10.5 | −1.28 |
| 7. | RTPTIDQMKD | 100–109 | PRO102 CG-ARG8 CG, GLN364 E-ARG8 CB, PRO9 CG-ARG3 C. | −14.29 | −29.88 | 0.00 | ASN5, ARG8, LYS6, VAL7, ARG3. | 1.4968 | NO | YES | YES | 1204.50 | −0.44 | 0.83 | NO | 0.00 | −0.94 |
| 8. | KTQHLSNDHICY | 126–137 | THR199 CG2-GLN9 C, ASP158 OD1-GLN9 O1, LYS160 NZ-ASP8 C, TYR122 CD1-ILE7 CG1, THR45 CE1-ARG3 NH2, ALA44 CA-ARG3 NE, GLU42 O-ILE2 C, GLY41 CA-ILE2 C, GLY41 CA-ILE1 O. | −27.51 | −30.54 | −2.44 | CYS160, ASP158, SER162, THR122, ALA44, GLY43, TYR45, GLU42. | 0.1609 | NO | YES | NO | 1458.79 | −0.26 | −0.13 | NO | 1.00 | −0.51 |
| 9. | GKHYLD | 239–244 | GLN364 OE1-TYR4 CD1, PRO102 CG-GLYI C, PRO102 CG-LEU5 CG, GLN106 OE1-LEU5 CD, PRO9 CB-LYS2 CG, ALA10 CA-LYS52 CE, GLY11 N-LYS2 NZ, GLY11 CA-ASP6 OD1, ALA10 C-ASP6 OD2. | −42.53 | −19.78 | −1.52 | PRO365, GLN364, PRO102, GLN106, GLY11, ALA10, PRO9. | −1.416 | NO | YES | YES | 731.89 | −0.25 | 0.23 | NO | 0.50 | −0.93 |
| 10. | NTTCD | 167–171 | ALA8 CB-ILE9 CD, ALA10 C-OLE3 CD, GLY11 N-ILE3 CD, PRO14 CD-ASN1 ND2, ASP29 N-ARG5 NH2, GLY12 CA-ASP2 OD2, VAL30 C-ARG5 CD, GLY12 N-ASN1 N, ALA5 N-ILE9 O1, PRO32 CG-TRY6 OG1, ALA4 CB-THR8 CA, PRO9 C-ILE3 CG1, PRO9 O-ILE9 CG1, PRO9 CG-THR8 N, ALA6 O-ILE9 CA | −24.26 | −20.54 | −1.51 | ALA4, PRO32, VAL30, PRO14, GLY12, GLY11, ASP29, ALA10, PRO9, ALA8, ALA5. | __ | NO | NO | YES | 552.61 | −0.34 | 0.28 | NO | −1.00 | 0.74 |
| 11. | LSNRP | 231–235 | PRO102 CG-ARG44 NH2, ARG100 C-PRO5 CG, ILE99 CG2-PRO5 CB, ILE99 CG2-ARG4 C. | −32.34 | −14.77 | −1.20 | LEU1, PRO5, ARG4, ASN3, SER2. | __ | NO | NO | NO | 585.72 | −0.44 | 0.34 | NO | 1.00 | −0.66 |
Table 6.
Prediction of discontinuous or conformational B-cell epitopes of MAPK1 protein.
| Sr. no. | Residues | Number of residues | score |
|---|---|---|---|
| 1. | A:M1, A:A2, A:T3, A:A4, A:A5, A:V6, A:S7, A:A8, A:P9, A:A10, A:G11, A:G12, A:13, A:P14, A:N15, A:P16, A:G17, A:S18, A:G19, A:A20, A:E21, A:M22, A:V23, A:R24, A:G25, A:Q26, A:A27, A:F28, A:D29, A:V30, A:G31, A:P32, A:R33, A:S35, A:N36, A:L37, A:S38, A:Y52, A:D53, A:R54, A:D55, A:N56, A:K57, A:V58 | 44 | 0.747 |
| 2. | A:H187, A:D188, A:H189, A:T190, A:G191, A:F192, A:L193, A:T194, A:N210, A:S21, A:K212, A:F237, A:P238, A:G239, A:K240, A:H241, A:Y242, A:L243, A:D244, A:N247, A:H248, A:L250, A:G251, A:I252, A:G254, A:S255, A:P256, A:S257, A:Q258, A:E259, A:D260, A:L261, A:N262, A:C263, A:I264, A:I265, A:N266, A:I267, A:K268, A:A269, A:R270, A:N271, A:Y272, A:L274, A:S275, A:L276, A:P277, A:L278, A:R279, A:S280, A:K281, A:V282, A:P283, A:N285, A:R286, A:T304, A:F305, A:N306, A:P307, A:H308, A:K309. | 62 | 0.699 |
| 3. | A:F68, A:R100, A:T101, A:P102, A:T103, A:I104, A:D105, A:Q106, A:M107, A:K108, A:D1009, A:F338, A:K339, A:F340, A:D341, A:M342, A:E343, A:L344, A:D345, A:D345, A:D346, A:L347, A:P348, A:K349, A:E350, A:T351, A:L352, A:K353, AA:E354, A:L355, A:F357, A:E358, A:A361, A:Q364, A:P365, A:G366, A:N367 | 36 | 0.672 |
| 4. | A: K126, A: T127, A: Q128 | 3 | 0.661 |
| 5. | A:E118, A:H129, A:L130, A:S131, A:N132, A:D133, A:H134, A:C136, A:Y137, A:A152, A:N167, A:T168, A:T169, A:C170, A:D171, A:L231, A:S232, A:N233, A:R234, A:P235 A:L287, A:F288, A:P289, A:N290, A:A291, A:D292, A:P293, A:K294, A:A295, A:L296, A:I311, A:E312, A:E314, A:E315, A:L317, A:A318, A:H319, A:P320, A:Y321, A:L322, A:E323, A:Q324, A:Y325, A:Y326, A:D327, A:P328, A:T329, A:D330, A:E331, A:P332, A:V333, A:A334, A:E335, A:A336, A:P337 | 55 | 0.658 |
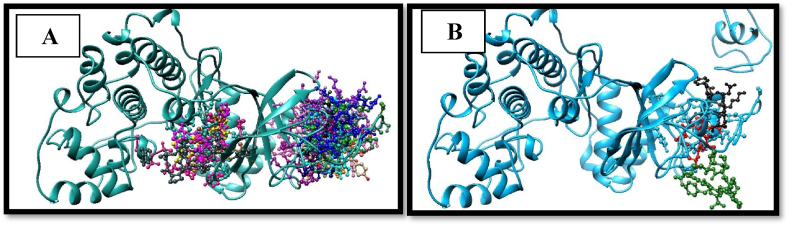
3.9. Protein-peptide docking
The PatchDock software was utilized to examine the docking conformation, functionally interacting residues and optimal structural information of the peptide with its target protein (see Table 7). The binding energy of each complex was calculated after docking, and the one with the lowest energy was chosen for further study using UCSF Chimera to examine receptor-peptide interactions. For this study, we looked at how likely amino acid residues within 4 Å of the peptide would interact with each other (Sehgal, 2017). Three hundred thirty interactions were detected between Peptide and receptor Protein (MAPK1). Fire Dock docking software was used for flexible refinement and scoring protein-peptide interactions. UCSF chimaera was used to visualize protein-peptide interacting residues.
Table 7.
Peptide-MAPK1 docking: Assessing interactions and energy factors.
| Sr. No. | Peptide Sequence | Position | Global energy (Kcal/Mol) | Attractive energy (Kcal/Mol) | H-bond energy (Kcal/Mol) | Peptidase MHC-pair | Bond Distance Å | Interactive Residues of Peptide | Interactive Residues of Protein | Conserved Residues |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PAGGGPNPG | 9–17 | −52.26 | −24.88 | −2.57 | ASP120 OD2-PRO6 CD, LYS123 CE-PRO6 CD. CYS75 SG-PRO 4 CG, SER162 O-ASN5CB, LYS63 CD-GLY30, GLU42O-PRO6 O, ASO176 OD2-GLY9 O1, LEU176 OD1-GLY9 O2, GLY46 CA-GLY10, TRY45 N-GLY GLY1 CA, ALA44 N-GLY1 CA, GLY43 C-GLY1 CA | 2.395 Å, 3.068 Å, 2.148 Å, 2.206 Å, 2.313 Å, 2.1175 Å, 1.806 Å, 2.179 Å, 2.133 Å, 2.849 Å, 2.449 Å, 2.358 Å | PRO4, PRO6, GLY3, SER8, GLY1, GLY7, ASN5, GLY9, | LYS63, LEU40, GLU42, GLY43, SER162, LYS123, THR122, ASP176, THR45, ALA44, LEU179, ASP120, CYS175, | LYS63, GLU42, GLY43, SER162, LYS123, THR122, ASP176, THR45, ALA44, LEU179, ASP120, CYS175, |
| 2 | ATAAVSAPA | 2–10 | −52.43 | −26.95 | −2.21 | PRO32 CG-THR 2 N, ASP29 O-ALA4 CB, ASP29 OD-ALA7 CB, ARG 100 CA-ALA9 O2, GLN106 OE1-ALA7 O. | 2.553 Å, 2.421 Å, 2.374 Å, 2.345 Å, 2.027 Å | ALA1, GLY3, GLY4, ASN6, PRO7, SER9 | GLY43, ASP176, SER162, ASP120, LYS160, GLY42 | GLY43, ASP176, SER162, ASP120, LYS160. |
| 3 | GGGPNPGSG | 11–19 | −37.75 | −21.85 | −1.60 | ASP120 OD2-VAL9 CG4, MET8 CE-ASP176 CG, MET8 CA-VAL48 CG2, LYS63 CD-MET8 CG, ASN163-GLY5 CA, LYS160 NZ-GLY3 O, VAL9 CG1-LUE 165 CD2, VAL9 ASP120-CG, SER162-ALA6 CA, GLY5 CA-ASP176 OD2, ASN1 N-THR199 CG2, MET8 CA-LYS63 NZ, GLY 43CA-GLU7 CG, GLU42 O-GLU7 CB | 2.452 Å, 2.604 Å, 2.855 Å, 2.862 Å, 2.439 Å, 1.798 Å, 2.670 Å, 3.006 Å, 2.505 Å, 2.426 Å, 2.866 Å, 2.766 Å, 2.36 Å, 2.70 Å | ANS1, VAL3, ALA6, GLY5, GLY3, GLU7, MET8 | ASP120, LEU165, VAL48, GLU42, SER162, ASN 163, LYS 160, LYS63, GLY43, ASP176, THR199, ARG203. | ASP120, LEU165, VAL48, SER162, ASN 163, LYS 160, LYS63, GLY43, ASP176, ARG203 |
| 4 | GGPNPGSGA | 12–20 | −40.36 | −27.92 | −3.44 | LEU165 CD2-VAL9 CG1, ASP 120 CB-VAL9 CG1, SER162 O-VAL9 CG2, ASN63 OD1-GLY5 CA, ARG203 NH2-ASN1 ND2, ASP176 OD2-GLY5 CA, ASP176 CG-MET8 CE, LYS63 NZ-MET8 CE, THR199 CG2-ASN21 N, LYS160 NZ-GLY30, GLY43 CA-GLU7 CG, GLU42 O-GLU7 CB. | 2.670 Å, 2.730 Å, 2.414 Å, 2.414 Å, 2.439 Å, 2.505 Å, 2.587 Å, 2.426 Å, 2.604 Å, 2.766 Å, 2.826 Å, 2.866 Å, 1.798 Å, 2.366 Å, 2.270 Å | VAL9, MET8, LY5, ALA6, GLU7, GLY3, ASN1 | LYS63, ALA48, VAL48, ASP120, ASN163, LYS160, ARG203, THR199, GLU92, GLY43, SER62, LEU165, VAL48. | LYS63, VAL48, ASP120, ASN163, LYS160, ARG203, THR199, GLY43, LEU165, VAL48. |
| 5 | NPGSGAEMV | 15–23 | −10.23 | −8.94 | 0.00 | ALA10 CB-GLN6 CB, ALA8 O-GLN6 O, ALA5 CB-ALA7 CB, ALA8 CB-ASP9 O1 | 2.962 Å, 2.031 Å, 2.85 Å, 2.55 Å | ASP9, GLN8, ALA7 | ALA10, ALA8, ALA5 | ALA10, ALA8, ALA5 |
| 6 | PGSGAEMVR | 16–24 | −17.71 | −27.64 | −1.77 | THR103 CG2-VAL6 CG1, PRO102 CD-GLY1 N, PRO365 CG-PRO8 CG, RO365 CG-GLN2 CG, ALA8 CA-AR9 O2, PRO9 CD-ARG9 C, PRO9 CB-ALA3 CA, ALA10-ALA3 CB. | 2.685 Å, 2651 Å, 2.933 Å, 2.730 Å, 2.545 Å, 2.860 Å, 2.705 Å, 2.354 Å. | PRO8, GLN2, ARG9, VAL6, ALA3, GLY1. | PRO365, THR103, ALA3, PRO9, ALA10, PRO102 | PRO36, PRO9, ALA10, PRO102 |
| 7 | EMVRGQAFD | 21–29 | −52.29 | −26.93 | −1.89 | ASN367 O-SER4 O, PRO369 C-SER4 O, PRO369 CD-ALA5 CA. | 2.035 Å, 2.612 Å, 3.143 Å. | ALA5, SER4 | ASN367, PRO369. | ASN367, PRO369 |
| 8 | APAGGGPNP | 25–33 | −40.72 | −19.49 | −2.17 | GLN364 OE1-PRO7 CB, PRO102 CG-ASN8 N, PRO102 CG-PRO7 CA, ILE99 CG2-GLY6 CA, ALA10 O-PRO2 CB, PRO32 CG-PRO2 CA, PRO9 CB-ALA3 CB. | 2.45 Å, 2.525 Å, 2.943 Å, 3.100 Å, 2.374 Å, 2.723 Å, 2.575 Å, 2.602 Å. | PRO9, ASN8, PRO102, GLY6, ALA3, PRO5 | PRO9, ALA10, ALA4, PRO32, PRO102, ILE99, GLN364 | PRO9, ALA10, ALA4, PRO32, PRO102, ILE99, GLN364 |
| 9 | PRYSNLSYI | 32–40 | −38.32 | −24.80 | −1.68 | ASP55 CG-ASN ND2 | 2.743 Å | ASN1 | ASP55 | NO |
| 10 | RYSNLSYIG | 33–41 | −14.20 | −5.06 | −0.99 | PRO365 CA-TYR2 CE1, PRO365 CB-SER6 OG, VAL6 O-ILE8 CA, ALA5-O-ILE8 O, ALA4 CB-GLY9 CA. | 2.99 Å, 2.528 Å, 2.50 Å, 2.50 Å, 1.827 Å, 2.656 Å | TYR2, SER6, ILE8, GLY9 | ASN367, GLY366, PRO365, PRO9, VAL6, ALA5, ALA4. | ASN367, PRO365, PRO9, VAL6, ALA5, ALA4 |
| 11 | NLSYIGEGA | 36–44 | −45.02 | −20.54 | −2.16 | LYS160 CD-MET1 CG, SER162 CB-MET1 CE, SER162 OG-ALA4 CB, GLU42 O-ALA5 CB, ILE40 C-ALA5 O, GLY43 CA-VAL6 CG2, ASP176 OD2-THR3 C, LEU179 CD1-THR3 CG2, ASP176 CG-SER7 CB, LYS63 NZ-SER7 CB, LYS63 CD-VAL 6 O, GLY46 N-VAL6 CG1. | 2.847 Å, 2.849 Å, 2.433 Å, 2.571 Å, 2.463 Å, 2.505 Å, 2.227 Å, 2.949 Å, 2.411 Å, 2.791 Å, 2.939 Å, 2.662 Å, 2.390 Å, 2.836 Å. | VAL6, SER7, ALA4, MET1, ALA5, PRO9, THR3. | GLY46, GLY43, LYS63, GLU42, LEU179, ASP176, ILE40, LYS160, SER162. | GLY46, GLY43, LYS63, GLU42, LEU179, ASP176, LYS160, SER162. |
| 12 | SAYDRDNKV | 50–58 | −23.24 | −26.81 | −2.81 | VAL6 O-VAL9 CG1, PRO32 CB-LYS8, GLY12 N-ARG5 NH, GLY11 CA-ARG5 NH1, ALA10 O-AR 5 CD, PRO9 CG-ASP6 CA, PRO365 CG-ASP6 CB, PRO365 CD-TYR3 CA, PHE363 O-TYR3 CD1. | 2.545 Å, 2.488 Å, 2.227 Å,2.456 Å, 2.628 Å, 3.089 Å, 2.925 Å, 2.749 Å, 2.368 Å. | TYR3, ASP6, VAL9, LYS8, ARG5. | PHE363, PRO365, VAL6, GLY12, PRO9, PRO32, ALA10, GLY11. | PHE363, PRO365, VAL6, GLY12, PRO9, PRO32, ALA10, GLY11. |
| 13 | DRDNKVRA | ____ | −27.91 | −29.21 | −2.79 | ARG33 CB-ARG7 NH2, PRO9 CD-VAL8 CG1, ALA10 O-ASN4 O, ALA10 L-LYS5 CA, GLY11 CA-LYS5 CB, GLY11 CA-ASP1 CG, GLY12 N-ASP1 CB, GLY106 OE1-ARG2 CG2, PRO102 CG-VAL6 CG2, ILE99 CG2-ASP3 OD1, THR101 CG2-ARG2 CD, ARG100 CA-ARG2 NH2. | 2.428 Å, 3.116 Å, 1.859 Å, 2.679 Å, 3.148 Å, 3.069 Å, 2.101 Å, 2.587 Å, 2.398 Å, 2.418 Å, 2.917 Å, 2.822 Å. | VAL6, ARG7, VAL6, ASP3, ARG2, LYS5, ASP1, ASN4. | ARG33, ILE99, ARG100, PRO102, THR101, GLN106, GLY12, GLY11, ALA10, PRO9. | ARG33, ILE99, ARG100, PRO102, THR101, GLN106, GLY12, GLY11, ALA10, PRO9. |
| 14 | RDNKVRVAI | _____ | −31.14 | −23.34 | −3.51 | PRO102 CG-VAL7 CG1, PRO365 CG-AR6 CG3, ILE99 CG2-IL9 C, PRO9 CB-VAL5 CB, ALA10 N-VAL5 CG2, ALA4 CA-VAL5 CG1, PRO32 CD-ILE9 CG2, VAL30 C-ILE9 CB, PRO32 O-ASP2 OD2, ASN15 N-LYS4 NZ, VAL30 O-ILE9 CD, GLY31 CA-ILE9 CG, ALA4 CA-VAL5 CG1, GLY13 O-LYS4 CE, PRO14 CA-LYS4 NZ, ALA4 N-ASN3 O, THR3 CA-ASN3 O. | 2.898 Å, 3.103 Å, 2.692 Å, 3.046 Å, 2.513 Å, 2.940 Å, 2.809 Å, 1.974 Å, 2.604 Å, 1.989 Å, 2.243 Å, 2.549 Å, 1.924 Å, 2.940 Å, 2.592 Å, 2.920 Å, 2.010 Å, 1.803 Å. | ILE9, ARG6, VAL5, ASN3, LYS4, VAL7, ARG2. | PRO369, PRO102, ALA10, PRO9, ILE99, VAL30, ALA4, GLY13, ASN15, THR3, ALA2, PRO14, GLY31, PRO32, VAL30. | PRO369, PRO102, ALA10, PRO9, ILE99, VAL30, ALA4, THR3, ALA2, GLY31, PRO32, VAL30. |
| 15 | DNKVRVAIK | ____ | −7.77 | −22.40 | −5.20 | PRO365 CD-ARG6 CD, GLN364 OE1-ARG6 CD, ARG33 CD-ILE9 CA, ARG33 CD-ALA8 C, PRO32 CD-VAL7 CG1, GLY31 N-VAL7 CG1, PRO9 CG-VAL5 CG1. | 2.453 Å, 2.363 Å, 2.920 Å, 2.872 Å, 2.629 Å, 2.907 Å, 2.575 Å. | ILE9, VAL5, VAL7, ALA8, ARG6. | ILE365, GLN364, PRO9, GLY31, VAL30, PRO32, ARG33. | GLN364, PRO9, GLY31, VAL30, PRO32, ARG33. |
| 16 | AIKKISPFE | 1–9 | −23.99 | −23.85 | −0.55 | GLY366 CA-PRO7 CD, ALA8 CB-ILE2 CG1, ALA8 CB-ALA1 N, PRO9 CD-ILE5 CD. | 2.906 Å, 2.805 Å, 2.848 Å, 3.033 Å. | ILE5, ILE2, ALA1, PRO7. | ALA8, PRO9, GLY363. | ALA8, PRO9, GLY36 |
| 17 | SAPAGGGPN | 7–13 | −42.08 | −25.39 | −3.27 | GLY46 O-GLY5 CA, GLY43 CA-GLY5 CA, GLY43 CA-GLY6 N, GLU42 O-PRO8 CB, CYS175 SG-PRO3 CD, LYS123 CE-ASN9 D2, SER162 O-PRO3 CG, ASP120 OD2-ASN9 O2, LEU165 CD2-SER1. | 2.679 Å, 2.581 Å, 2.784 Å, 2.343 Å, 2.681 Å, 2.571 Å, 2.432 Å, 2.163 Å, 2.532 Å. | SER1, PRO3, ALA4, ASN9, PRO8, GLY6, GLY5. | LEU165, CYS175, VAL48, GLY46, ASP120, LYS123, SER162, GLU42, GLY43. | LEU165, CYS175, VAL48, GLY46, ASP120, LYS123, SER162, GLU42, GLY43. |
| 18 | TAAVSAPAG | 3–11 | −42.15 | −26.07 | −1.24 | ASP29 O-ALA8 O, ASP29 OD1-PRO7 O2, ARG33 CB-VAL4 CG1, PRO9 CB-ALA6 CB, PRO9 CB-ALA3 CA, PRO9 CG-ALA2 C, PRO365CB-ALA2 N, PRO365 CB-THR1 CB. | 2.124 Å, 2.681 Å, 3.082 Å, 2.719 Å, 2.966 Å, 2.797 Å, 2.833 Å, 3.005 Å. | THR1, ALA2, VAL4, ALA3, ALA6, ALA8, PRO7. | PRO365, ARG33, ASP29, PRO9. | PRO365, ARG33, PRO9. |
| 19 | AVSAPAGGG | 4–12 | −44.41 | −25.19 | −0.97 | TYR45 CZ-ALA1 CB, LYS64 O-GLY9 O1, LYS63 CG-GLY9 O2, GLY46 CA-GLY9 O1, VAL48 CG2-GLY8 CA, LYS123 CE-PRO5 O2, ASP120 OD2-PRO5 O2, ASP120 OD2-ALA6 CB, ASN163 OD1-VAL2 CG1, LYS160CE-VAL2 CG2, LEU165 OD2-PRO5 CD, LEU165 OD2-ALA4 CA, SER162 O-ALA4 CB. | 2.854 Å, 1.919 Å, 2.206 Å, 2.121 Å, 3.093 Å, 2.120 Å, 2.159 Å, 2.166 Å, 2.200 Å, 2.812 Å, 2.518 Å, 3.060 Å, 2.663 Å, 2.270 Å | ALA1, GLY9, GLY8, GLY7, VAL2, ALA6, ALA4, PRO5. | VAL48, LYS63, GLY46, GLU42, TYR45, ASP176, CYS175, LEU165, ASP120, LYS123, ASN163, SER162, LYS160. | VAL48, LYS63, GLY46, GLU42, TYR45, ASP176, CYS175, LEU165, ASP120, LYS123, ASN163, SER162, LYS160. |
| 20 | ISPFEHQTY | 5–13 | −45.03 | −36.51 | −3.30 | PRO365 CB-PHE4 CE1, GLN364 CB-GLN7 OE1, PRO9 CG-PHE4 CE2, PRO102 CB-HIS6 C, ARG100 O-PRO3 CG, GLN106 NE2-HIS6 CE1, GLN11 CA-ILE1 CG2, GLY11 CA-GLU5 OE1, ALA10 O-GLU5 CD, PRO9 CB-THR8 OG1, PRO9 CG-PHE4 CE2. | 3.007 Å, 2.499 Å, 2.9003 Å, 3.117 Å, 2.879 Å, 2.670 Å, 2.287 Å, 2.445 Å, 2.639 Å, 2.381 Å, 2.476 Å, 2.903 Å. | PHE4, THR8, GLN7, PRO3, HIS6, GLU5, ILE1 | GLN364, ARG100, PRO102, GLN106, GLY11, ALA10, PRO9, ILE90 | GLN364, ARG100, PRO10, GLY11, ALA10, PRO9, ILE90 |
| 21 | ENIIGINDI | 30–38 | −24.92 | −21.34 | −0.85 | ARG33 CD-ASP8 O, ARG33 CB-ILE9 CA, VAL30 O-ILE9 CD, PRO32 CA-ILE9 CG2, PRO32 CB-GLU1 N, PRO32 CB-ILE3 CG1, ALA4 CB-ASN2 O, ALA10 O-ILE3 O, PRO9 CG-ASN2 CG, GLY5 C-ILE6 N, GLN364 OE1-ILE6 CD, PRO365 CD-ILE6 CG2, PHE363 O-ASP8 OD2, PRO365 CG-ASN7 N. | 2.055 Å, 3.142 Å, 2.826 Å, 3.672 Å, 2.622 Å, 1.667 Å, 2.668 Å, 1.326 Å, 2.248 Å, 2.62 Å, 2.124 Å, 2.150 Å | ASN7, ASP8, ILE6, GLN2, ILE3, GLU1, ILE9, ASN2. | PRO365, PHE363, GLN364, PRO9, ALA10, VAL30, ALA4, ARG33, PRPO32. | PRO365, PHE363, GLN364, PRO9, ALA10, VAL30, ALA4, ARG33, PRPO32. |
| 22 | IIGINDIIR | 32–40 | −29.61 | −30.79 | −0.68 | ALA10 CA-ARG9 NH2, PRO9 O-ARG9 NH2, PRO9 O-ARG9 CB, ALA8 O-ARG9 O2. | 2.669 Å, 2.258 Å, 2.121 Å. | ARG4. | PRO9, ALA8, ALA10. | PRO9, ALA8, ALA10 |
| 23 | IGINDIIRT | 33–41 | −45.59 | −27.75 | −3.37 | GLU358 CA-ILE7 CD, PHE357 OD2-ILE7 CD, ALA361 N-ILE3 CG2, PHE363 N-ILE3 CD, ALA361 N-ILE3 CG2, PHE363 N-ILE3 CD, PRO369 OXT-ASP5 CG, ARG368 O-GLY2 CA, ARG368 N-ILE1 C, ASN367 N-ILE1 CG2, GLY366 CA-ILE1 CD, GLN365 CD-ILE1 CD. | 3.050 Å, 2.679 Å, 2.519 Å, 2.880 Å, 2.675 Å, 1.791 Å, 2.127 Å, 2.591 Å, 2.390 Å, 2.964 Å, 2.689 Å, 3.092 Å. | ASP5, GLY2, ILE3, ILE7, ILLE1. | PRO369, ARG368, ASN367, GLY377, PRO365, GLN364, PHE363, ARG362, PHE357, ALA361, GLU358. | PRO369, ASN367, PRO365, GLN364, PHE363. |
| 24 | GINDIIRTP | 34–42 | −35.8 | −27.63 | −1.39 | ALA8 CB-ARG7 NH2, ALA10 CB-ARG7 CB | 2.890 Å, 3.038 Å | ARG7 | ALA8, ALA10 | ALA8, ALA10 |
| 25 | INDIIRTPT | 35–43 | −17.39 | −28.72 | −2.57 | ASP29 CG-PRO8 CA, ILE99 CG2-ILE5 O, GLY31 CA-ILE4 CG2, GLY12 N-THR9 C, GLY11 C-THR9 O2, PRO32 CG-ILE1 CG1, ALA4 CB-ILE1 CG1, ARG100 C-ARG6 O, THR101 CA-ARG60, GLN106 OE1-THR7 CB, THR101 CG2THR7 CG2, PRO102 CG-THR7 CG2, GLN364 OE1-ARG6 CG, PRO9 CG-ASN2 ND2, PRO9 CB-ASP3 OD1, ALA10 CA-ASP3 CG. | 3.128 Å, 2.09 Å, 2.897 Å, 2.543 Å, 2.179 Å, 3.063 Å, 2.513 Å, 2.402 Å, 1.889 Å, 2.353 Å, 2.769 Å, 2.897 Å, 2.656 Å, 2.306 Å, 2.146 Å, 2.971 Å. | PRO8, THR9, ILE4, ILE1, ILE5, ASP3, ASN2, THR7, ARG6. | PRO102, ARG100, THR101, GLN106, PRO9, ALA10, ALA4, GLY11, GLY31, PRO32, GLY12, ASP29, GLN364, ASP29. | PRO102, ARG100, THR101, GLN106, PRO9, ALA10, ALA4, GLY11, GLY31, PRO32, GLY12, ASP29, GLN364, ASP29. |
| 26 | NDIIRTPTI | 36–44 | −30.71 | −25.00 | −1.58 | ALA8 CB-ILE9 CD, ALA10 CB-ILE9 CG2, ALA10 C-OLE3 CD, GLY11 N-ILE3 CD, PRO14 CD-ASN1 ND2, ASP29 N-ARG5 NH2, VAL30 CA-ARG5 NE, GLY12 CA-ASP2 OD2, GLY31 N-ARG5 NE, VAL30 C-ARG5 CD, GLY12 N-ASN1 N, GLY11 C-ASN1 N, ALA4 C-ILE9 O1, ALA5 N-ILE9 O1, PRO32 CG-TRY6 OG1, ALA4 CB-THR8 CA, PRO9 C-ILE3 CG1, PRO9 O-ILE9 CG1, PRO9 CG-PRO7 CA, PRO9 CG-THR8 N, PRO9 CA-ILE9 N, ALA6 O-ILE9 CA. | 2.185 Å, 2.436 Å, 2.449 Å, 2.483 Å, 2.483 Å, 2.746 Å, 2.583 Å, 2.632 Å, 1.997 Å, 2.025 Å, 2.279 Å, 1.935 Å, 2.587 Å, 2.370 Å, 1.830 Å, 2.457 Å, 3.128 Å, 2.678 Å, 2.787 Å, 2.602 Å, 2.255 Å, 2.168 Å. | ILE4, ILE3, ASN1, ARG5, ASP2, THR6, THR8, PRO7, ILE2. | ALA4, PRO32, GLY31, VAL30, PRO14, GLY12, GLY11, ASP29, ALA10, PRO9, ALA8, ALA5. | ALA4, PRO32, GLY31, VAL30, GLY12, GLY11, ASP29, ALA10, PRO9, ALA8, ALA5. |
| 27 | DIIRTPTID | 37–45 | −25.24 | −30.03 | −2.30 | PRO365 CB-ILE2 CD, VAL6 O-ILE3 CG2, ALA4 CB-ILE8 CG1, ARG33 CG-THR7 CB, ASP55 N-THR7 CG2, ARG54 CA-THR7 CG2, ARG54 CB-PROR6 O2. | 2.938 Å, 2.160 Å, 3.159 Å, 2.698 Å, 2.497 Å, 3.003 Å, 2.603 Å. | ILE3, THR7, PRO6, ILE2, ILE8. | PRO365, VAL6, ALA4, ARG33, ASP55, ARG54. | PRO365, VAL6, ALA4, ARG33. |
| 28 | IIRTPTIDQ | 38–46 | −28.22 | −22.26 | −0.53 | PRO102 CB-THR6 CG2, ALA8 CB-GLN9 CG, SER7 CB-GLN9 NE, PRO9 CD-ILE7 CG2, PRO9 O-ILE1 CG2, ALA10 CB-ILE CA. | 3.137 Å, 2.823 Å, 2.907 Å, 2.907 Å, 2.980 Å, 2.453 Å., 3.078 Å. | THR6, ILE7, GLN9, ILE1. | ALA10, PRO02, PRO9, ALA8, SER7. | ALA10, PRO02, PRO9, ALA8. |
| 29 | RTPTIDQMK | 40–48 | −16.23 | −36.39 | −5.34 | THR199 CG2-GLN9 C, ASP158 OD1-GLN9 O1, LYS160 NZ-ASP8 C, TYR122 CD1-ILE7 CG1, THR45 CE1-ARG3 NH2, ALA44 CA-ARG3 NE, GLU42 O-ILE2 C, GLY41 CA-ILE2 C, GLY41 CA-ILE1 O. | 2.679 Å, 1.976, 2.342 Å, 1.746 Å, 2.486 Å, 2.581 Å, 2.782 Å, 2.403 Å, 1.887 Å, 2.355 Å, 2.415, Å, 2.54 Å. | ILE2, ARG3, ILE1, ILE7, GLN9, ASP8. | CYS160, ASP158, SER162, THR122, ALA44, GLY43, TYR45, GLY41, GLU42. | SER162, THR122, ALA44, GLY43, TYR45, GLU42. |
| 30 | DQMKDVYIV | 45–53 | −34.00 | −23.16 | −1.22 | PRO365 CB-ASP1 N, PRO365 CG-G1N 2 N, PRO102 CB-GLN2 NE2, PRO102 CD-VAL6 CG2, ILE99 CG2-ASP5 CB, GLN364 OE1-GLN2 CG, ASP29 O-ASP5 O, PHE28 O-ILE8 CD, ASP29 CA-ILE8 CD, GLY31 N-ILE8 CD, ALA2 O-VAL9 CA, ASP29 OD2-VAL6 O. | 2.377 Å, 2.830 Å, 2.512 Å, 2.849 Å, 2.318 Å, 1.946 Å, 1.842 Å, 2.644 Å, 2.517 Å, 2.724 Å, 2.848 Å, 2.295 Å, 3.037 Å, I.835 Å. | GLN2, ASP1, MET3, TYR7, VAL6, LYS4, ASP5, ILE8, VAL9. | PRO365, GLN364, PRO102, PRO9, ALA10, GLY11, ILE99, THR3, PRO32, ALA2, VAL30, ASP29, GLY31. | PRO365, GLN364, PRO102, PRO9, ALA10, GLY11, ILE9, PRO32, VAL30, ASP29, GLY31. |
3.10. Fire dock results in evaluation by UCSF chimera
30 CTL epitope and their sequences were identified against MAPK1. It was also evaluated that from all Protein-peptide docking solutions, six peptides (1,3,11,17,19,29) were present in the same binding pocket A, and 16 peptides were present in the other same binding pocket B. In comparison, eight peptides (5,7,9,10, 22,24,27,28) were present on different active sites/binding pocket of MAPK1 protein as shown in Fig. 7(A). Only the 4 top-ranked linear B cell epitopes show interaction at the secondary binding domain among 11 as shown in Fig. 7(B).
Fig. 7.
(A) Docking of 30 CTL predicted epitopes with MAPK1, 6 CTL epitopes were bound on the primary binding site of the protein. In contrast, 16 epitopes were present on the secondary active site of the protein. (B) shows the interaction of top-ranked 4 linear B cell epitopes at the secondary binding domain.
The Individual docked complexes along their binding residues are presented in Fig. 8.
Fig. 8.
Docking of CTL epitopes on primary A and secondary B binding/active site pocket interacting residues were labelled in different colors. A represents docking of 5 peptides MHC class1 HLA-B interacting residues of MAPK1 were presented in different colors. B represents the docking of 3 peptides MHC class1 HLA-B interacting residues of MAPK1 were shown in different colors. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Different docking interactions of predicted peptides of MHC class-1 and class-II against MAPK1, along with attractive energy, global energy, hydrogen bonding, Bond distance, Bond distance, interacting residues, and conserved residues of docking complexes are presented in Table [7 & 8].
The 3D representation of linear B-cell epitopes of MAPK1 protein is presented in Fig. 9. However, the position of confirmational epitopes on 3D protein structure is presented in Fig. 10.
Fig. 9.
The position of Linear epitopes on 3D protein structure was presented.
Fig. 10.
The 3D representation displays B-cell epitopes of the antigenic MAPK1 protein. The docking complex is depicted, with the protein in grey sticks and epitopes in yellow surface. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.11. Protein-ligand docking
Various bioinformatics tools such as ChemDraw, AutoDock Tools, PyRx, PyMol [42], UCSF Chimera, and Discovery Studio were used for the In-silico study ofMAPK1 [43]. The receptor molecule (Protein) was prepared by adding polar H-atoms. Colman's charges are added in preparation for ligand molecule. The minimization of the predicted structure was performed with a 1000 bootstrap value. FDA-approved library from the zinc database was screened for the best-interacting compound having the potential to inhibit target protein. FDA-approved compound library of 11600 compounds was selected for making Ligand-based pharmacophores. After screening, the 50 best-interacting compounds were selected based on high binding affinity and the difference between upper and lower RMS values. MAPK1 Protein has two main domains, amino acid sequence from 1 to 33 has domain I, and from 34 to 322 amino acid sequence contains Domain II. In-depth Analysis shows that Domain II with amino acid sequence from 34 to 322 is functionally active. The two domains (Domain I & Domain II) of MAPK1 protein are presented with two different colors along with the Top-ranked potent compound as shown in Fig. 11 (see Table 8).
Fig. 11.
The figure displays MAPK protein's binding domains: Domain I (green) and Domain II (deep sky-blue). Compounds interact at Domain II, highlighting its functional importance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 8.
Firedock evaluation of docked complexes of antigenic protein and MHC II alleles for desired epitopes.
| Sr. No. | Peptide Sequence | Global energy (Kcal/Mol) | Attractive energy (Kcal/Mol) | H-bond energy (Kcal/Mol) | MHC-II Alleles |
|---|---|---|---|---|---|
| 1 | RGLKYIHSANVLHRD | −18.17 | −44.22 | −5.04 | HLA-DRB1*04:05 |
| 2 | GLKYIHSANVLHRDL | −53.69 | −31.42 | −3.95 | HLA-DRB1*04:05 |
| 3 | LKYIHSANVLHRDLK | −4.02 | −8.08 | −0.89 | HLA-DRB1*04:05 |
| 4 | ARNYLLSLPLRSKVP | −10.12 | −26.81 | −5.31 | HLA-DRB3*02:02 |
| 5 | KARNYLLSLPLRSKV | −34.13 | −30.48 | −4.72 | HLA-DRB5*01:01 |
| 6 | LDDLPKETLKELIFE | −19.24 | −20.85 | −1.49 | HLA-DPA1*03:01/DPB1*04:02 |
| 7 | LDKMLTFNPHKRIEV | −3.43 | −3.66 | −1.60 | HLA-DPA1*02:01/DPB1*01:01 |
| 8 | CILAEMLSNRPIFPG | −23.04 | −29.01 | −5.22 | HLA-DQA1*01:02/DQB1*06:02 |
| 9 | NKVRVAIKKISPFEH | −21.78 | −30.90 | −3.02 | HLA-DRB1*11:01 |
| 10 | KYIHSANVLHRDLKP | −7.82 | −27.65 | −6.87 | HLA-DRB1*04:05 |
| 11 | DLNCIINIKARNYLL | −22.43 | −37.30 | −5.48 | HLA-DRB1*12:01 |
| 12 | LNCIINIKARNYLLS | −30.76 | −23.23 | −3.97 | HLA-DRB1*12:01 |
| 13 | EDLNCIINIKARNYL | −21.43 | −34.38 | −3.81 | HLA-DRB1*12:01 |
| 14 | NCIINIKARNYLLSL | −42.75 | −23.50 | −1.46 | HLA-DRB1*12:01 |
| 15 | QEDLNCIINIKARNY | −18.31 | −23.39 | −2.85 | HLA-DRB1*12:01 |
| 16 | CIINIKARNYLLSLP | −18.04 | −25.48 | −1.50 | HLA-DRB1*12:01 |
| 17 | DLYKLLKTQHLSNDH | −19.19 | −36.85 | −3.18 | HLA-DRB4*01:01 |
| 18 | LYKLLKTQHLSNDHI | −13.74 | −26.41 | −3.27 | HLA-DRB4*01:01 |
| 19 | KVRVAIKKISPFEHQ | −34.33 | −32.43 | −4.59 | HLA-DRB1*11:01 |
| 20 | DRDNKVRVAIKKISP | −6.58 | −21.47 | −0.63 | HLA-DRB1*11:01 |
| 21 | RDNKVRVAIKKISPF | −52.64 | −39.49 | −2.70 | HLA-DRB1*11:01 |
| 22 | DNKVRVAIKKISPFE | −6.99 | −26.27 | −0.96 | HLA-DRB1*11:01 |
| 23 | KARNYLLSLPLRSKV | 0.43 | −22.98 | −3.85 | HLA-DRB1*01:01 |
| 24 | ARNYLLSLPLRSKVP | −15.37 | −26.04 | −2.66 | HLA-DRB1*01:01 |
| 25 | RNYLLSLPLRSKVPW | −43.91 | −25.96 | −3.15 | HLA-DRB1*12:01 |
| 26 | ETDLYKLLKTQHLSN | −23.95 | −23.20 | −1.83 | HLA-DRB1*04:05 |
| 27 | TDLYKLLKTQHLSND | −14.62 | −7.40 | −0.95 | HLA-DRB1*04:05 |
| 28 | METDLYKLLKTQHLS | −29.39 | −24.37 | −0.64 | HLA-DRB1*04:05 |
| 29 | YFLYQILRGLKYIHS | −20.64 | −9.91 | −0.54 | HLA-DRB1*12:01 |
| 30 | CYFLYQILRGLKYIH | −29.67 | −22.08 | −1.47 | HLA-DRB1*12:01 |
Fig. 12, Fig. 13 shows the interaction of the 12 best drug compounds on protein domains I and II with the potential to inhibit MAPK1 Protein expression according to ADMET analysis, as presented in Table 9. Extensive In-silico studies show that these compounds can inhibit MAPK1 (target protein) by satisfying the desired drug properties, as presented in Fig. 14.
Fig. 12.
Interactions of docked protein-drug complexes in Domain I are shown. Top-ranked drug compound's interacting residues, 2D pharmacophore diagram, and Ramachandran plot illustrate complex interactions.
Fig. 13.
Interaction of docked complexes of protein and drug compounds for Domain II, Interacting residues of the top-ranked selected drug compound, 2D diagram pharmacophore-based drug compound, and Ramachandran plot showing interaction of docked complexes.
Table 9.
Virtual screening and comparative docking: Identifying novel drug compounds with ADMET analysis.
| Ligands | Binding Affinity | RMSD value | Molecular weight | Interacting Residues | No. of heavy atoms | No. of rotatable bonds | No. of H acceptor | No. of H donors | Density | Consensus Logp | Pharmacokinetics |
Medicinal Chemistry |
Drug Likeliness |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GI absorption | BBB (blood-brain barrier) | P-gp substrate. | P-gp Inhibitor | PAINS | Synthetic Accessibility | Lipinski’ Rules | Bioavailability score | Water solubility | Acute Oral Toxicity (kg/Mol) | GSK | Pfizer Rule | |||||||||||
| ZINC000003920266 | −9.7 | 1.5 | 497.5 | LYS160, ASP158, SER162, VAL48, LEU165, ALA61, LYS63, ARG176, TYR122. | 36 | 3 | 10 | 5 | 1.032 | 2.801 | Low | No | Yes | No | No | 5.63 | Accepted | 0.94 | −3.41 | 4.45 | Rejected | Accepted |
| ZINC000000001984 | −7.6 | 0.591 | 318.756 | ALA61, VAL48, LEU165, LYS160, TYR45, CYS175. | 12 | 5 | 4 | 1 | 1.011 | 3.34 | High | No | No | No | No | 2.14 | Accepted | 0.85 | −4.62 | 2.145 | Accepted | Rejected |
| ZINC000000001758 | −7.5 | 1.2 | 320.341 | GLN114, LYS63, ASP176, VAL48, CY175, LEU165, ILE40. | 23 | 6 | 6 | 2 | 0.993 | 2.72 | High | Yes | No | No | No | 3.02 | Accepted | 0.56 | −4.19 | 2.821 | Accepted | Accepted |
| ZINC000003812863 | −7.4 | 1.746 | 405.496 | TYR45, ILE65, LYS63, VAL48, CYS175, LYS160, ASP176, SER162. | 29 | 13 | 7 | 4 | 0.975 | 0.13 | High | Yes | No | No | No | 3.67 | Accepted | 0.84 | −2.18 | 2.003 | Rejected | Accepted |
| ZINC000000004009 | −7.2 | 0.304 | 306.276 | ASN163, ASP176, ASP120, LEU165, VAL48, GLN114, ALA61. | 22 | 5 | 7 | 1 | 1.108 | 0.88 | High | Yes | No | No | No | 2.45 | Accepted | 0.55 | −1.86 | 1.781 | Accepted | Accepted |
| ZINC000000599734 | −9.7 | 0.559 | 369.368 | GLY46, LYS64, TYR45, ILE65, CYS63, VAL48, LEU165, ALA61. | 25 | 6 | 7 | 1 | 1.124 | 3.13 | High | Yes | Yes | No | No | 3.40 | Accepted | 0.928 | −4.22 | 2.041 | Accepted | Accepted |
| ZINC000028467879 | −7.9 | 1.72 | 554.552 | TRP261, LYS160, GLY43, ASP176, LYS63, GLN114, CYS175, VAL48. | 36 | 9 | 11 | 4 | 1.193 | −0.67 | Low | No | Yes | yes | No | 5.08 | Rejected | 0.64 | −3.09 | 2.017 | Rejected | Accepted |
| ZINC000035653007 | −7.2 | 1.904 | 373.449 | LYS63, ILE65, TYR45, GLY46, GLY43, ASP176, ALA4. | 26 | 6 | 5 | 0 | 1.026 | 3.78 | High | Yes | Yes | Yes | No | 3.76 | Accepted | 0.55 | −3.74 | 3.246 | Accepted | Rejected |
| ZINC000003927870 | −7.2 | 1.62 | 365.214 | GLU42, ASP158, LYS160, SER162, ASN163, ASN163, ASP176, LYS63, CYS175, VAL48, GLN14. | 24 | 9 | 11 | 5 | 1.269 | −1.86 | Low | No | No | No | No | 4.43 | Accepted | 0.68 | −3.08 | 1.254 | Accepted | Accepted |
| ZINC000008551177 | −7.6 | 11.50 | 477.603 | ILE40, CYS175, SER62, TYR202, LYS160, GLU42. | 33 | 7 | 12 | 8 | 1.032 | −2.22 | Low | No | No | No | No | 6.51 | Rejected | 0.88 | −2.13 | 1.858 | Rejected | Accepted |
| ZINC000004468780 | −7.4 | 1.96 | 455.474 | PRO32, AR33, ILE99, ASP97, GLY13, ASP29, GLN364, PRO365, PHE363, ILE998. | 30 | 9 | 9 | 3 | 1.161 | −0.27 | Low | No | No | Yes | No | 4.79 | Accepted | 0.75 | −2.81 | 1.284 | Rejected | Accepted |
| ZINC000003913937 | −8.2 | 1.92 | 517.564 | PRO32, PRO365, ASP29, LYS08, ASP109, THR101, ARG100, PRO102. | 36 | 9 | 7 | 3 | 1.070 | −0.24 | Low | No | Yes | No | No | 4.91 | Rejected | 0.91 | −3.24 | 2.027 | Rejected | Accepted |
Fig. 14.
2D structures of FDA-approved selected drug library from Zinc database (a) Idarubicidin, (b) Finofibric acid (c) Myfortic (d) Lisinopril (e) Diflucan (f) Lansoprazole, (g) Ceftriaxone (h) prasugrel (i) fludarabine (j) gentamincin mixture (l) Cefotaxime (k) gentamincin mixture (m) piperacillin.
3.12. Protein-protein docking
Different Interactions of MAPK1 protein with functionally and structurally partner proteins were observed through the STRING database by integrating other interactions by predicting various physical and structural associations between proteins. The results of STRIG are mentioned in Fig. 15.
Fig. 15.
Predicted Interaction: MAPK1 and dusp1 (Uniprot ID: Q7T2L8) through STRING Server.
MAPK1 protein may directly or indirectly link or interact with other proteins to perform a specific function. Best interactions of MAPK1 protein were observed dusp6 (Uniprot ID: Q7T2L8) Protein. The docking of MAPK1 Protein with its best functionally interacting partner protein dusp6 (Uniprot ID: Q7T2L8), along with their interacting residues, are mentioned in Fig. 16. The 3D structure of dusp6 was downloaded for protein-protein Docking analysis through Cluspro and Ppatchdock. UCSF Chimera was utilized to find interacting residues and other post-docking analyses.
Fig. 16.
Docking of MAPK1 with dusp6 (Uniprot ID: Q7T2L8) and Their Interacting Residues.
4. Discussion
Research on neurodegenerative disorders is imperative to facilitate the discovery of effective treatments for Alzheimer's disease [44]. The contribution of the MAPK signaling pathway is a critical element of various neurological disorders case neurodegenerative diseases. Scientific efforts are needed to prevent disease formation and improve the current treatment options. An immunoinformatic analysis is borderline for targeting epitope-based vaccine designs to control MAPK1 pathways, as per the investigation of earlier researchers [45]. In our study investigations, the zebrafish allow us to explore various neurological-related disorders. We used the Uniprot database to retrieve the MAPK1 sequence for identifications of details analysis related to MSA for Structure prediction, evaluation, phylogeny, and population Coverage, and the results were matched [46]. We also predict epitopes and their Antigenicity and Allergenicity using high-throughput bioinformatics approaches.
Further, we carry out Pharmacophore-Based Virtual Screening Molecular Docking and structure modelling where we explore tool-based Protein-peptide docking, Protein-Protein Docking, and Protein-ligand docking as per the analysis of earlier researchers [10,11]. The relation of the MAPK pathway with Alzheimer's disease was the idea leading to this present study. Identifying MAPK1 inhibitors effectively eradicates the patient's suffering and pathophysiology of Alzheimer's disease [5]. Extensive In-silico analysis was executed to identify, evaluate, and design innovative compounds by targeting MAPK1 [47]. The 3D structure of MAPK1 has not yet been documented in PDB or determined by NMR or X-ray crystallography.
The six leading optimally aligned appropriate templates with maximum query coverage, identity, score, and E-value were selected. Among the chosen templates, the 5K4I [PDB ID] template showed the best evaluation for reliable structure prediction for MAPK1 homology or comparative modelling using MODELLER 9.20. The target sequence (MAPK1) query coverage and template sequence query coverage were satisfactory for the 3D structure prediction of MAPK1. Threading and ab initio approaches were also utilized to decrease the chance of errors and to predict the reliable 3D structure of MAPK1 [48]. Different structure Evaluation software was used to find the most reliable 3D predicted structure of MAPK1, followed by protein-ligand, protein-peptide, and protein-protein docking analysis. The Ramachandran Plot showed that 91.00 % of residues of the predicted structure were present in the favored region, 7.40 % were in the allowed region, and 0.30 % were outliers. The overall quality of the predicted structure was 97.44 %, as observed by ERRAT.
Multiple sequence alignment was performed to trace the conserved region of the target candidate gene through the time of evolution in humans. For this purpose, the genome of six MAPK1 strains was retrieved to build a neighbour-joining tree using an evolutionary tool (MEGAX) as per the method of earlier researchers [49,50]. The observation shows the binding domain or active site was conserved among all MAPK1 strains. Different molecular Docking analyses were performed for ligand-based drug designing [51] and peptide-based vaccine designing [52]against MAPK1 in zebrafish. The protein-ligand docking analysis was performed using Pyrx, protein-protein docking utilizing ClusPro, and protein-peptide docking using Patch Dock. UCSF Chimera, Discovery Studio, and Pymol then visualize the docking analysis. The energy of docking complexes was minimized to fix & and remove the steric hinderances.
PatchDock server was used to perform protein-peptide docking, followed by peptide-based vaccine designing. Receptor-peptide docking reagents were sourced from various databases, most frequently NetCTL 1.2 and PEP-FOL. NetCTL webserver was utilized for CTL prediction from protein sequence by submitting FASTA sequence of Protein on NetCTL 1.2 server, which utilizes MHC supertype A1 and 0.75000. Top-ranked 30 CTL epitopes of MHC class I of T cells and their sequence were identified against MAPK1. These antigenic determinants associated with specific HLA alleles were assessed to analyze MHC class 1 & and MHC class ll epitopes present in 90 % of the world population. The epitopes of MHC class II of T cells and linear and discontinuous epitopes of B cells were predicted using the IEDB server. Comparative molecular-docking Analysis of predicted epitope followed by comparative and structural genomic analysis strategies to identify effective drug and novel inhibitors for Alzheimer's disease in humans by targeting MAPK1 isoforms. PEPFOLD 2.0 server was then utilized to predict 3D models of responding epitopes for protein-peptide docking analysis.
Top-ranked docked complexes were evaluated for similar residues [53]. The binding energy of each complex was calculated after docking [54], and the one with the lowest energy was chosen for further study using UCSF Chimera to examine peptide-receptor interactions [55]. It was also evaluated that from all 30 peptides, 6 CTL epitopes were present in the primary binding pocket and 16 CTL epitopes were present in the secondary binding pocket. Protein-peptide docking analysis reveals that VAL48, LYS63, GLU42, GLY43, CYS175, ASP176, LYS 160, ALA 61, LEU165, ALA61, ASP120, SYS175, LYS160, VAL48, ASN163, ALA4, ALA5, ALA8, ALA10, TYR45, SER162, GLY11, GLY12, VAL6, GLN364, PRO9, PRO32, PRO102, ILE99, ASN367, ARG33, PRO365, PHE363, ILE40, ASN 163, ASP29, GLY31, VAL30, GLN106, ARG100, VAL6 and GLU42 are critical residues for protein-peptide interaction followed by peptide-based vaccine designing.
The FDA-approved ZINC database library of 1600 compounds was selected and screened by post-Docking Analysis [56], and 50 potential top-ranked drug compounds were selected for further analysis. For this purpose, PyRx (virtual screening) software, Auto Dock Vina & and Auto Dock 4 are Utilized for receptor-ligand Docking Studies [57], and ChemDraw& Open Babel for energy minimization, removing salts bridges and importing SDF files [58]. These findings would be helpful in the novel therapeutic discovery of targeted drug designing to cure Alzheimer's disease. Receptor-ligand decking was simulated on Auto Dock software [59]. The docked complexes with the least binding energy were selected to analyze protein-ligand interaction using VMD [60]. Protein-ligand docking analysis reveals that VAL48, LYS63, CYS175, ASP176, LYS 160, ALA 61, LEU165, ALA61, TYR45, SER162, ARG33, PRO365, PHE363, ILE40, ASN 163 and GLU42 are critical residues for protein-drug interaction. The interaction was observed at a similar site among all docked complexes. Extensive In-silico studies show that these compounds can inhibit MAPK1 (target protein) by satisfying the desired drug properties [61]. Protein-protein docking analysis reveals that THR169, PRO328, GLU315, HIS 308, LYS 5, and GLU259 residues of MAPK1 interact with its functional partner protein dusp6 (Uniprot ID: Q7T2L8). Protein-protein docking [62]was performed on Patch Dock and ClusPro followed by post-docking analysis on UCSF chimaera [63].
An emergent need for effective AD treatment [64]. Developing a vaccine is of substantial curiosity to the peptide inhibitors [65]. The predicted peptide target has the most negligible side effect, lower toxicity, and is faster in action than other traditional ligand-based medicinal products [66]. CTL epitopes of MAPK protein were predicted against Alzheimer's disease. In-silico approaches facilitate scientists to minimize laboratory work burden, cost-effective and less expedient [67]. Thirty peptides for B cells, MHC class I and II of T cells were selected with the most remarkable binding affinities and antigenicity; among these peptides (PAGGGPNPG, GGGPNPGSG, SAPAGGGPN, AVSAPAGGG, AGGGPNPGS, ATAAVSAPA, TAAVSAPAG, ENIIGINDI, INDIIRTPT, and NDIIRTPTI showed effective and maximum interaction depending on global energies. We identified a total of 30 CTL against MAPK1 Protein, where 10 top is further evaluated and found potent peptides binding to MHC class I and MHC class II that are 80–90 % identical with experimentally determined epitopes and this will likely be beneficial for quick progression and promising candidates for vaccine design with adequate world population coverage of 88.5 % and 99.99 %, respectively. The molecular docking analyses were conducted on the ZINC database library, and ten top selected from 30 compounds having the least binding energy were scrutinized. Protein-ligand docking analysis reveals that VAL48, LYS63, CYS175, ASP176, LYS 160, ALA 61, LEU165, ALA61, TYR45, SER162, ARG33, PRO365, PHE363, ILE40, ASN 163 and GLU42 are critical residues for protein-drug interaction. Furthermore, docking analyses revealed that the predicted peptides engaged in strong bonding with the MAPK1 Protein, where docked complexes have maximum interaction as a potent inhibitor.
The in-silico design of a multiepitope-based-peptide (MBP) vaccine against the MAPK protein for Alzheimer's disease in zebrafish presents promising implications. Through a rational epitope selection process, epitopes were chosen based on their potential to induce an immune response and alignment with the MAPK protein. These approaches can potentially modulate the pathways associated with Alzheimer's disease progression, offering a novel strategy for therapeutic intervention. The multiepitope-based process demonstrates advantages in targeting diverse aspects of the MAPK protein, increasing the likelihood of an effective immune response. While limitations of in silico methods and assumptions exist, this research lays a foundation for future studies encompassing in vitro and in vivo validation, epitope prediction improvements, and the exploration of cross-species applicability. The ethical considerations associated with animal experimentation and responsible research practices further underscore the significance of these findings. In conclusion, this study contributes to understanding Alzheimer's disease treatment strategies by proposing a computational approach that warrants further investigation and experimental validation.
5. Conclusion
Our computer-based efforts to create a vaccine (a way to fight diseases) using multiple epitopes, focused on the MAPK protein connected to Alzheimer's disease in zebrafish, have shown promising results. Our analysis of the MAPK1 protein was conducted based on multiple steps, including sequence retrieval from Uniprot, physiochemical property prediction via ExPAsy-ProtParam, and antigenicity assessment using VaxiJen. The 3D structure was predicted through a comparative modelling approach with ERK2 template (PDB ID: 5K4I) via MODELLER and validated using various tools. Phylogenetic analysis was performed using MEGA-X. Epitope prediction encompassed world population coverage, B-cell epitopes, and CTL epitopes through IEDB. Allergenicity and toxicity were evaluated with AllergenFP, AllerTOP, and ToxinPred. Molecular docking involved protein-peptide (PatchDock, FireDock), protein-protein (STRING, ClusPro), and drug compound interactions from ZINC database.
Findings were integrated for potential vaccine and drug development insights. The study's main results encompass diverse aspects of MAPK1 analysis. Notably, the absence of its 3D structure reported by X-ray crystallography and NMR prompted threading and homology modelling. BLASTp aided template selection based on coverage, similarity, E value, and score, resulting in robust 3D models. Phylogenetic analysis via MEGA X and Molecular Evolutionary Algorithm revealed conserved regions in protein sequences. Epitope prediction involved assessing surface attributes and interactions, unveiling conformational β-cells and linear epitopes, while hydrophilicity analysis showcased distinctive regions. Antigenicity evaluation with Vaxijen indicated world population coverage preference in MHC class I of T cells. Peptide design produced reliable CTL and B cell epitopes, while protein-peptide docking highlighted interactions and residues. Structural visualization depicted epitope positions and B cell epitopes' 3D representation. Additionally, in-silico protein-ligand interactions yielded potential inhibitors from an FDA-approved compound library, identifying key binding domains and interactions. Protein-protein docking demonstrated MAPK1's interaction with dusp1, emphasizing its functional associations. We picked specific CTL epitopes (PAGGGPNPG, GGGPNPGSG, SAPAGGGPN, AVSAPAGGG, AGGGPNPGS, ATAAVSAPA, TAAVSAPAG, ENIIGINDI, INDIIRTPT, and NDIIRTPTI) that could trigger a strong defense response and match well with the MAPK protein. Protein-ligand docking analysis reveals that VAL48, LYS63, CYS175, ASP176, LYS 160, ALA 61, LEU165, ALA61, TYR45, SER162, ARG33, PRO365, PHE363, ILE40, ASN 163 and GLU42 are critical residues for protein-drug interaction. This extensive In-silico analysis proposed that reported compounds are effective against Alzheimer's disease by targeting and inhibiting MAPK1. This approach seems capable of influencing the processes linked to Alzheimer's disease. Using many epitopes also seems smart as it can deal with the MAPK protein's complexity. The study provided comprehensive insights into MAPK1's structure, epitopes, interactions, and potential therapeutic applications. By offering this In-silicon study, our work lays a foundation for future researchers to test and develop new ways to fight Alzheimer's disease. This study could inspire new ideas among scientists, showing a way forward to find a solution for Alzheimer's by creating a special vaccine.
Funding
Not applicable.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Data availability statement
All data are available in this manuscript.
Conflicts of authors
The authors declare no conflicts of interest.
Consent for publication
All the authors have read and approved the manuscript and all are aware of its submission to the journal.
Code availability
Not applicable.
Ethics approval
Not applicable.
CRediT authorship contribution statement
Yasir Arfat: Writing – original draft, Software, Methodology, Investigation, Data curation, Conceptualization. Imran Zafar: Writing – original draft, Visualization, Project administration, Investigation, Formal analysis, Data curation, Conceptualization. Sheikh Arslan Sehgal: Writing – original draft, Validation, Project administration, Investigation, Funding acquisition, Data curation, Conceptualization. Mazhar Ayaz: Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Formal analysis. Muhammad Sajid: Writing – original draft, Validation, Project administration, Methodology, Investigation, Formal analysis. Jamal Muhammad Khan: Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation. Muhammad Ahsan: Writing – review & editing, Visualization, Validation, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation. Mohd Ashraf Rather: Writing – review & editing, Visualization, Validation, Project administration, Investigation, Data curation. Azmat Ali Khan: Writing – review & editing, Validation, Supervision, Software, Resources, Methodology, Formal analysis, Data curation. Jamilah M. Alshehri: Writing – review & editing, Visualization, Project administration, Methodology, Investigation, Funding acquisition. Shopnil Akash: Writing – review & editing, Validation, Software, Methodology, Investigation, Formal analysis. Eugenie Nepovimova: Writing – review & editing, Project administration, Investigation, Data curation. Kamil Kuca: Writing – review & editing, Methodology, Formal analysis, Data curation. Rohit Sharma: Writing – review & editing, Supervision, Software, Resources.
Declaration of competing interest
The authors declare no conflict of interest, financial or otherwise.
Acknowledgement
This work was funded by the Researchers Supporting Project Number (RSP2023R339) at King Saud University, Riyadh 11451, Saudi Arabia. This study was supported by the Excellence project PrF UHK 2208/2023–2024 and by the Ministry of Health project - MH CZ - DRO (UHHK, 00179906).
Contributor Information
Yasir Arfat, Email: yasirarfat886.pk@gmail.com.
Imran Zafar, Email: bioinfo.pk@gmail.com.
Sheikh Arslan Sehgal, Email: arslansehgal@yahoo.com.
Mazhar Ayaz, Email: mmazharayaz@cuvas.edu.pk.
Muhammad Sajid, Email: sajid@uo.edu.pk.
Jamal Muhammad Khan, Email: jamalmkhan@cuvas.edu.pk.
Muhammad Ahsan, Email: ahsan_m@outlook.com.
Mohd Ashraf Rather, Email: biotechashraf786@gmail.com.
Azmat Ali Khan, Email: azkhan@ksu.edu.sa.
Jamilah M. Alshehri, Email: jalshehri@ksu.edu.sa.
Shopnil Akash, Email: shopnil.ph@gmail.com.
Eugenie Nepovimova, Email: eugenie.nepovimova@uhk.cz.
Kamil Kuca, Email: Kamil.kuca@uhk.cz.
Rohit Sharma, Email: rohitsharma@bhu.ac.in.
References
- 1.Cheignon C.m., Tomas M., Bonnefont-Rousselot D., Faller P., Hureau C., Collin F. Oxidative stress and the amyloid beta peptide in Alzheimer's disease. Redox Biol. 2018;14:450–464. doi: 10.1016/j.redox.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton S., Matthews F.E., Barnes D.E., Yaffe K., Brayne C. Potential for primary prevention of Alzheimer's disease: an analysis of population-based data. Lancet Neurol. 2014;13(8):788–794. doi: 10.1016/S1474-4422(14)70136-X. [DOI] [PubMed] [Google Scholar]
- 3.Kim E.K., Choi E.-J. Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2010;1802(4):396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe D.J. Translating cell biology into therapeutic advances in Alzheimer's disease. Nature. 1999;399(6738):A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 5.Munoz L., Ammit A.J. Targeting p38 MAPK pathway for the treatment of Alzheimer's disease. Neuropharmacology. 2010;58(3):561–568. doi: 10.1016/j.neuropharm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 6.English J.M., Cobb M.H. Pharmacological inhibitors of MAPK pathways. Trends Pharmacol. Sci. 2002;23(1):40–45. doi: 10.1016/s0165-6147(00)01865-4. [DOI] [PubMed] [Google Scholar]
- 7.Newman M., Verdile G., Martins R.N., Lardelli M. Zebrafish as a tool in Alzheimer's disease research. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2011;1812(3):346–352. doi: 10.1016/j.bbadis.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Eiben A.E., Smith J. From evolutionary computation to the evolution of things. Nature. 2015;521(7553):476–482. doi: 10.1038/nature14544. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad H.M., Abrar M., Izhar O., Zafar I., Rather M.A., Alanazi A.M.…Wani T.A. Characterization of fenugreek and its natural compounds targeting AKT-1 protein in cancer: pharmacophore, virtual screening, and MD simulation techniques. J. King Saud Univ. Sci. 2022;34(6) [Google Scholar]
- 10.Ali S., Noreen A., Qamar A., Zafar I., Ain Q., Nafidi H.-A.…Sharma R. Amomum subulatum: a treasure trove of anti-cancer compounds targeting TP53 protein using in vitro and in silico techniques. J. F. i. C. 2023;11 doi: 10.3389/fchem.2023.1174363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali S., Ali U., Qamar A., Zafar I., Yaqoob M., ul Ain Q.…Jardan Y.A.B. Predicting the effects of rare genetic variants on oncogenic signaling pathways: a computational analysis of HRAS protein function. J. F. i. C. 2023:11. doi: 10.3389/fchem.2023.1173624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. 2005:571–607. [Google Scholar]
- 13.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8(1):1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L.…Dutta S. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eswar N., Eramian D., Webb B., Shen M.-Y., Sali A. Structural Proteomics. Springer; 2008. Protein structure modeling with MODELLER; pp. 145–159. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y.H., Specht T. 2000 IEEE Aerospace Conference. Proceedings (Cat. No. 00TH8484) 2000. Remote access multi-mission processing and analysis ground environment (RAMPAGE) [Google Scholar]
- 17.Laskowski R., MacArthur M., Thornton J. 2006. PROCHECK: Validation of Protein-Structure Coordinates. [Google Scholar]
- 18.Sajid M., Marriam S., Mukhtar H., Sohail S., Sajid M., Sehgal S.A. Epitope-based peptide vaccine design and elucidation of novel compounds against 3C like protein of SARS-CoV-2. PLoS One. 2022;17(3) doi: 10.1371/journal.pone.0264700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sehgal S.A., Khattak N.A., Mir A. Structural, phylogenetic and docking studies of D-amino acid oxidase activator (DAOA), a candidate schizophrenia gene. Theor. Biol. Med. Model. 2013;10(1):1–13. doi: 10.1186/1742-4682-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayati A., Falahati M., Irannejad H., Emami S. Synthesis, in vitro antifungal evaluation and in silico study of 3-azolyl-4-chromanone phenylhydrazones. Daru. 2012;20(1):1–7. doi: 10.1186/2008-2231-20-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP v. 2—a server for in silico prediction of allergens. J. Mol. Model. 2014;20(6):1–6. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- 22.Blois S., Tometten M., Kandil J., Hagen E., Klapp B.F., Margni R.A., Arck P.C. Intercellular adhesion molecule-1/LFA-1 cross talk is a proximate mediator capable of disrupting immune integration and tolerance mechanism at the feto-maternal interface in murine pregnancies. J. Immunol. 2005;174(4):1820–1829. doi: 10.4049/jimmunol.174.4.1820. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., Wang G., Zhang D., Yin H., Wang M. Identification of novel B cell epitopes within Toxoplasma gondii GRA1. Exp. Parasitol. 2013;135(3):606–610. doi: 10.1016/j.exppara.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Parker J., Guo D., Hodges R. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25(19):5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 25.Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9(1):1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrescu I., Lamotte-Brasseur J., Chessa J.-P., Ntarima P., Claeyssens M., Devreese B.…Gerday C. Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles. 2000;4(3):137–144. doi: 10.1007/s007920070028. [DOI] [PubMed] [Google Scholar]
- 27.Waqas M., Haider A., Rehman A., Qasim M., Umar A., Sufyan M.…Rasool D. Immunoinformatics and molecular docking studies predicted potential multiepitope-based peptide vaccine and novel compounds against novel SARS-CoV-2 through Virtual screening. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/1596834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calis J.J., Maybeno M., Greenbaum J.A., Weiskopf D., De Silva A.D., Sette A.…Peters B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013;9(10) doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinf. 2007;8(1):1–12. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- 31.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R., Consortium O.S.D.D., Raghava G.P. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hassan S., Javid J., Sadiqua A., Mushtaq S., Fatima N., Mohsin A., Akram H. Thyroid Cancer TC-1: an insight from 3D structure prediction to virtual screening for computational drug design. Biomed. Lett. 2020;6(1):17–22. [Google Scholar]
- 33.Khan T., Lawrence A.J., Azad I., Raza S., Joshi S., Khan A.R. Computational drug designing and prediction of important parameters using in silico methods-A review. Curr. Comput. Aided Drug Des. 2019;15(5):384–397. doi: 10.2174/1573399815666190326120006. [DOI] [PubMed] [Google Scholar]
- 34.Tahir R.A., Hassan F., Kareem A., Iftikhar U., Sehgal S.A. Ligand-based pharmacophore modeling and virtual screening to discover novel CYP1A1 inhibitors. Curr. Top. Med. Chem. 2019;19(30):2782–2794. doi: 10.2174/1568026619666191112104217. [DOI] [PubMed] [Google Scholar]
- 35.Vilar S., Cozza G., Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008;8(18):1555–1572. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 36.Dallakyan S., Olson A.J. Chemical Biology. Springer; 2015. Small-molecule library screening by docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 37.Mendelsohn L.D. ChemDraw 8 ultra, windows and macintosh versions. J. Chem. Inf. Comput. Sci. 2004;44(6):2225–2226. [Google Scholar]
- 38.Tahir R.A., Sehgal S.A., Khattak N.A., Khan Khattak J.Z., Mir A. Tumor necrosis factor receptor superfamily 10B (TNFRSF10B): an insight from structure modeling to virtual screening for designing drug against head and neck cancer. Theor. Biol. Med. Model. 2013;10(1):1–14. doi: 10.1186/1742-4682-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh D., Ghosh Dastidar D., Roy K., Ghosh A., Mukhopadhyay D., Sikdar N.…Das A. Computational prediction of the molecular mechanism of statin group of drugs against SARS-CoV-2 pathogenesis. Sci. Rep. 2022;12(1):1–26. doi: 10.1038/s41598-022-09845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martina B.E., Haagmans B.L., Kuiken T., Fouchier R.A., Rimmelzwaan G.F., Van Amerongen G.…Osterhaus A.D. SARS virus infection of cats and ferrets. Nature. 2003;425(6961) doi: 10.1038/425915a. 915-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adhikari U.K., Tayebi M., Rahman M.M. Immunoinformatics approach for epitope-based peptide vaccine design and active site prediction against polyprotein of emerging oropouche virus. Journal of immunology research. 2018;2018 doi: 10.1155/2018/6718083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan S., Chan H.S., Hu Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017;7(2) [Google Scholar]
- 43.Tahir R.A., Sehgal S.A., Ijaz A. In silico comparative modeling of PapA1 and PapA2 proteins involved in Mycobacterium tuberculosis sulfolipid-1 biosynthesis pathway. International Journal Bioautomation. 2012;16(3):155. [Google Scholar]
- 44.Abd El Aty A.A., Alshammari S.O., Alharbi R.M., Soliman A.A. Astragalus sarcocolla gum-mediated a novel green-synthesis of biologically active silver-nanoparticles with effective anticancer and antimicrobial activities. Jordan J. Biol. Sci. 2023;16(1) [Google Scholar]
- 45.Marriam S., Afghan M.S., Nadeem M., Sajid M., Ahsan M., Basit A.…Microbiology I. Elucidation of novel compounds and epitope-based peptide vaccine design against C30 endopeptidase regions of SARS-CoV-2 using immunoinformatics approaches. 2023;13:568. doi: 10.3389/fcimb.2023.1134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rafique Q., Rehman A., Afghan M.S., Ahmad H.M., Zafar I., Fayyaz K. Reviewing methods of deep learning for diagnosing COVID-19, its variants and synergistic medicine combinations. Medicine. 2023 doi: 10.1016/j.compbiomed.2023.107191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagic Z., Ruzic D., Djokovic N., Djikic T., Nikolic K. In silico methods for design of kinase inhibitors as anticancer drugs. Front. Chem. 2020;7:873. doi: 10.3389/fchem.2019.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orengo C., Bray J., Hubbard T., LoConte L., Sillitoe I. Analysis and assessment of ab initio three‐dimensional prediction, secondary structure, and contacts prediction. Proteins: Struct., Funct., Bioinf. 1999;37(S3):149–170. doi: 10.1002/(sici)1097-0134(1999)37:3+<149::aid-prot20>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- 49.Zafar I., Pervez M.T., Rather M.A., Babar M.E., Raza M.A., Iftikhar R. Genome-Wide identification and expression analysis of PPOs and POX gene families in the selected plant species. Biosciences Biotechnology Research Asia. 2020;17(2):301–318. [Google Scholar]
- 50.Zafar I., Rubab A., Aslam M., Ahmad S.U., Liyaqat I., Malik A.…Khan A.A. Genome-wide identification and analysis of GRF (growth-regulating factor) gene family in Camila sativa through in silico approaches. J. King Saud Univ. Sci. 2022;34(4) [Google Scholar]
- 51.Proschak E., Stark H., Merk D. Polypharmacology by design: a medicinal chemist's perspective on multitargeting compounds. J. Med. Chem. 2018;62(2):420–444. doi: 10.1021/acs.jmedchem.8b00760. [DOI] [PubMed] [Google Scholar]
- 52.Singh A., Thakur M., Sharma L.K., Chandra K. Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-73371-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozakov D., Beglov D., Bohnuud T., Mottarella S.E., Xia B., Hall D.R., Vajda S. How good is automated protein docking? Proteins: Struct., Funct., Bioinf. 2013;81(12):2159–2166. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choudhary M.I., Shaikh M., tul-Wahab A.-., ur-Rahman A.-. In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation. PLoS One. 2020;15(7) doi: 10.1371/journal.pone.0235030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grebner C., Niebling S., Schmuck C., Schlücker S., Engels B. Force field-based conformational searches: efficiency and performance for peptide receptor complexes. Mol. Phys. 2013;111(16–17):2489–2500. [Google Scholar]
- 56.Pant S., Singh M., Ravichandiran V., Murty U.S.N., Srivastava H.K. Peptide-like and small-molecule inhibitors against Covid-19. J. Biomol. Struct. Dyn. 2021;39(8):2904–2913. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruz J.V., Serafim R.B., da Silva G.M., Giuliatti S., Rosa J., Araújo Neto M.F.…Santos C.B. Computational design of new protein kinase 2 inhibitors for the treatment of inflammatory diseases using QSAR, pharmacophore-structure-based virtual screening, and molecular dynamics. J. Mol. Model. 2018;24(9):1–16. doi: 10.1007/s00894-018-3756-y. [DOI] [PubMed] [Google Scholar]
- 58.Ogunyemi O.M., Gyebi G.A., Ibrahim I.M., Olaiya C.O., Ocheje J.O., Fabusiwa M.M., Adebayo J.O. Dietary stigmastane-type saponins as promising dual-target directed inhibitors of SARS-CoV-2 proteases: a structure-based screening. RSC Adv. 2021;11(53):33380–33398. doi: 10.1039/d1ra05976a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grieco L., Calzone L., Bernard-Pierrot I., Radvanyi F., Kahn-Perles B., Thieffry D. Integrative modelling of the influence of MAPK network on cancer cell fate decision. PLoS Comput. Biol. 2013;9(10) doi: 10.1371/journal.pcbi.1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y., Zhao J., Chen Z. Insights into the molecular mechanisms of protein-ligand interactions by molecular docking and molecular dynamics simulation: a case of oligopeptide binding protein. Comput. Math. Methods Med. 2018;2018 doi: 10.1155/2018/3502514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hillisch A., Pineda L.F., Hilgenfeld R. Utility of homology models in the drug discovery process. Drug Discov. Today. 2004;9(15):659–669. doi: 10.1016/S1359-6446(04)03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonvin A.M. Flexible protein–protein docking. Curr. Opin. Struct. Biol. 2006;16(2):194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Morris C.J., Corte D.D. Using molecular docking and molecular dynamics to investigate protein-ligand interactions. Mod. Phys. Lett. B. 2021;35(8) [Google Scholar]
- 64.Reiman E.M., Langbaum J., Fleisher A.S., Caselli R.J., Chen K., Ayutyanont N.…Tariot P.N. Alzheimer's Prevention Initiative: a plan to accelerate the evaluation of presymptomatic treatments. J. Alzheim. Dis. 2011;26(s3):321–329. doi: 10.3233/JAD-2011-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen R.C., Popat R., Diggle S.P., Brown S.P. Targeting virulence: can we make evolution-proof drugs? Nat. Rev. Microbiol. 2014;12(4):300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 66.Craik D.J., Fairlie D.P., Liras S., Price D. The future of peptide‐based drugs. Chem. Biol. Drug Des. 2013;81(1):136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 67.Bal-Price A.K., Coecke S., Costa L., Crofton K.M., Fritsche E., Goldberg A.…Lucchini R. Altex; 2012. Conference Report: Advancing the Science of Developmental Neurotoxicity (DNT): Testing for Better Safety Evaluation. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available in this manuscript.