Abstract
Patients with germ line mutations in the VHL tumor suppressor gene are predisposed to the development of highly vascularized tumors within multiple tissues. Loss of pVHL results in constitutive activation of the transcription factors HIF-1 and HIF-2, whose relative contributions to the pathogenesis of the VHL phenotype have yet to be defined. In order to examine the role of HIF in von Hippel-Lindau (VHL)-associated vascular tumorigenesis, we utilized Cre-loxP-mediated recombination to inactivate hypoxia-inducible factor-1α (Hif-1α) and arylhydrocarbon receptor nuclear translocator (Arnt) genes in a VHL mouse model of cavernous liver hemangiomas and polycythemia. Deletion of Hif-1α did not affect the development of vascular tumors and polycythemia, nor did it suppress the increased expression of vascular endothelial growth factor (Vegf) and erythropoietin (Epo). In contrast, phosphoglycerokinase (Pgk) expression was substantially decreased, providing evidence for target gene-dependent functional redundancy between different Hif transcription factors. Inactivation of Arnt completely suppressed the development of hemangiomas, polycythemia, and Hif-induced gene expression. Here, we demonstrate genetically that the development of VHL-associated vascular tumors in the liver depends on functional ARNT. Furthermore, we provide evidence that individual HIF transcription factors may play distinct roles in the development of specific VHL disease manifestations.
Germ line mutations in the VHL tumor suppressor gene result in von Hippel-Lindau (VHL) disease, a familial tumor syndrome that predisposes affected patients to the development of highly vascularized neoplasms. These include hemangioblastomas of the retina and central nervous system (CNS), renal-cell carcinomas (RCC) of the clear-cell type, and endocrine and exocrine pancreatic tumors, as well as pheochromocytomas (34). In addition, VHL has also been found to be inactivated in the majority of sporadic RCC (8, 13).
VHL deficiency leads to constitutive activation of hypoxia-inducible factor (HIF) and increased expression of its target genes irrespective of the oxygen concentration (38). The VHL gene product (pVHL), together with elongins B and C (6, 25), Cullin-2 (43), and Rbx1 (23), forms an E3 ubiquitin ligase (19), which targets the hydroxylated, oxygen-sensitive α subunits of HIF-1, -2, and -3 for ubiquitination and subsequent degradation by the 26S proteasome (18, 20, 39). As a normal physiological response to hypoxia, HIF-1 and HIF-2 facilitate both oxygen delivery and adaptation to oxygen deprivation by regulating genes that are involved in glucose uptake and metabolism, angiogenesis, erythropoiesis, cell proliferation, and apoptosis (50, 62). HIFs belong to the PAS (Per-Arnt-Sim) family of basic helix-loop-helix (bHLH) transcription factors that bind to DNA as heterodimers composed of an oxygen-sensitive α subunit and a constitutively expressed β subunit, also known as the arylhydrocarbon receptor nuclear translocator (ARNT). ARNT is the general binding partner for the bHLH/PAS domain-containing proteins. In addition to forming heterocomplexes with HIF, ARNT also heterodimerizes with single minded (SIM), which is involved in neural development, and with the arylhydrocarbon receptor (AhR), which is involved in the xenobiotic response to environmental toxins (for a review, see reference 24).
The expression patterns of the HIF subunits differ within embryonic and adult tissues. In the adult, Hif-1α mRNA is ubiquitously expressed, and Hif-1α protein can be detected at baseline levels within multiple cell types in various tissues under normoxia and is significantly enhanced under conditions of hypoxia (55). In contrast, while Hif-2α mRNA expression has been detected within many tissues, Hif-2α protein has been found to be restricted to specific cell types within various tissues. In addition to being expressed in endothelial cells, Hif-2α is also expressed in glial cells of the brain, type II pneumocytes of the lung, cardiomyocytes, fibroblasts of the kidney, intersitial cells of the pancreas and duodenum, and hepatocytes (26, 63). Arnt expression is ubiquitous, and it seems to be the only Hif-β subunit present in the liver (21).
pVHL appears to have multiple functions besides regulating HIF, and its contributions to the development of VHL-associated tumors are presently subject to intense investigations. As a result of these efforts, it has been shown that pVHL plays an important role in fibronectin extracellular-matrix assembly (41) and microtubule stabilization (12). The importance of proper pVHL function during development and in the adult is illustrated by the fact that homozygous deletion of the murine Vhl gene (Vhlh) results in embryonic lethality. Mice homozygously deficient in Vhlh die in utero between embryonic days 10.5 and 12 due to a defect in embryonic vasculogenesis of the placenta (9). We and others have previously shown that mice with a heterozygous germ line deletion of Vhlh have a predisposition to develop cavernous hemangiomas of the liver (10, 32). These hepatic vascular tumors display some of the histological features observed in VHL hemangioblastomas, which can also be found at low frequency in the livers of patients with VHL disease (10). Tissue-specific deletion of floxed Vhlh via Albumin-Cre mediated recombination in hepatocytes recapitulated the vascular phenotype found in Vhlh heterozygotes and suggested that loss of pVhl function in hepatocytes was responsible for the development of vascular tumors (10).
In order to examine the role of HIF in the development of VHL-associated vascular tumors, we utilized Cre-loxP-mediated recombination to inactivate either Hif-1α or Arnt in Vhlh-deficient hepatocytes. In this report, we show that loss of Arnt is sufficient to suppress the development of liver hemangiomas and erythrocytosis in Vhlh mutant mice, while loss of Hif-1α is not. We also provide evidence that in regard to HIF-regulated gene expression, target gene-dependent functional redundancy exists between different HIF homologues. Based on our data, we propose that in patients, the development of VHL-associated hemangiomas is mediated by increased HIF transcriptional activity and that the different HIF homologues may play distinct roles in the development of certain clinical features associated with VHL disease.
MATERIALS AND METHODS
Generation and genotyping of mice.
The generation of mice carrying the Vhlh, Hif-1α 2-lox, and Arnt 3-lox alleles and the Albumin-Cre transgenes has been described (10, 45, 48). Phosphoenolpyruvate carboxykinase-Cre (PEPCK-Cre) transgenic mice were generated by targeted single-copy transgenesis in embryonic stem cells (Rankin and Haase, unpublished data). Mutant mice were in a mixed genetic background (BALB/c, 129Sv/J, and C57BL/6). All procedures involving mice were performed in accordance with the National Institutes of Health guidelines for the use and care of live animals and were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
The following primers were used to detect 2-lox, 1-lox, and wild-type alleles of Vhlh: FwdI (5′-CTGGTACCCACGAAACTGTC-3′), FwdII (5′-CTAGGCACCGAGCTTAGAGGTTTGCG-3′), and Rev (5′-CTGACTTCCACTGATGCTTGTCACAG-3′). The Vhlh 2-lox allele was identified by the 460-bp band, the 1-lox allele was identified by a 260-bp band, and the wild-type allele was identified as a 290-bp band. The Hif1-α 2-lox, 1-lox, and wild-type alleles were detected with the following primers: FwdI (5′-TTGGGGATGAAAACATCTGC-3′), FwdII (5′-GCAGTTAAGAGCACTAGTTG-3′), and Rev (5′-GGAGCTATCTCTCTAGACC-3′). The Hif-1α 1-lox allele was identified as a 270-bp band, the 2-lox allele was identified as a 260-bp band, and the wild-type allele was identified as a 240-bp band. The Arnt 3-lox, 1-lox, and wild-type alleles were identified with the following primers: Fwd (5′-GCAACTTTGACAAGGCAGCATTTA-3′), RevI (5′-GGCAGGGGGAATCTCTGAGTTCT-3′), and RevII (5′-ACACCCTTCTTTCACTTCACAG-3′). The 1-lox allele band was identified as a 400-bp band, the 3-lox allele was identified as a 174 bp, and the wild-type allele was identified as a 140-bp band.
DNA and RNA isolation.
Mouse tail DNA was isolated according to the method of Laird et al. (31) and was used for genomic PCR. DNA and RNA from mouse livers were isolated using Trizol reagent (Invitrogen) according to the manufacturer's guidelines.
Protein preparation and immunoblot analysis.
Cytoplasmic and nuclear protein fractions were isolated using the protocol described by Camenisch et al. (1). Mouse liver was homogenized and lysed in buffer A. Cytoplasmic protein fractions were collected, and nuclei were lysed in buffer C to obtain nuclear protein extracts. Nuclear proteins were then dialyzed twice for 2 h each time at 4°C in buffer D. Protein concentrations were determined using the Bio-Rad Protein Assay. Twenty micrograms of each protein extract was size separated by 3 to 8% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Invitrogen) and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech). The membranes were stained with Ponceau S solution (Sigma) to determine equal protein loading. After being blocked in 10% nonfat dry milk (Carnation)-Tris-buffered saline-Tween 20 solution, the blots were incubated with 5% nonfat dry milk-Tris-buffered saline-Tween 20 solutions containing rabbit anti-mouse Hif-2α (raised against amino acids 580 to 693) or monoclonal Hif-1α (Novus Biologicals). The blots were then washed and incubated with horseradish peroxidase-conjugated goat anti-rabbit (Bio-Rad Laboratories) or sheep anti-mouse (Amersham Pharmacia Biotech) secondary antibodies according to the manufacturers' instructions. Enhanced chemiluminescence reagents obtained from Amersham Pharmacia Biotech were used as a detection system according to the manufacturer's instructions. Blots were stripped in a 2× 7 M guanidine HCl-50 mM Tris-HCl (pH 7.4) solution for 30 min at room temperature, followed by a 1× solution for 30 min, before being blocked and reprobed.
Gel shift.
Nuclear proteins were isolated, and an electrophoretic mobility shift assay (EMSA) was performed using the erythropoietin (Epo) hypoxia response element (HRE) as previously described (1). The Epo sense oligonucleotide 5′-GCCCTACGTGCTGTCTCACACAGC-3′ was annealed to the antisense oligonucleotide 5′-GCTGTGTGAACAGCACGTA-3′ in 1× annealing buffer containing 10 mM Tris-Cl (pH 7.8) and 50 mM NaCl by heating them to 95°C for 3 min and slowly cooling them to room temperature. Similarly, the AP-1 competitor sense (5′-TTCCGGCTGACTCATCAAGCG-3′) and antisense (5′-CGCTTGATGAGTCAGCCGGAA-3′) oligonucleotides were annealed together. The annealed Epo oligonucleotides were then labeled with 50 μCi of α-dCTP and α-dGTP (Amersham) using Klenow (Promega). The labeled oligonucleotides were then run through a G-50 microcolumn (Amersham), counted on a scintillation counter, and diluted to 30,000 cpm/μl in Tris-EDTA buffer. Protein binding reactions and electrophoresis were performed as previously described (Camenisch et al. [1]). Supershift reactions were performed using either monoclonal Hif-1α (Novus Biologicals) or monoclonal Hif-2α (Novus Biologicals) antibody and were incubated with the protein AP-1 (activator protein 1) competitor oligonucleotide and probe for 2 h at 4°C prior to electrophoresis.
Blood and histological analysis.
For the determination of the blood hemoglobin (Hgb) concentration and red blood cell (RBC) numbers, blood was collected and analyzed with a Hemavet 1500 CBC analyzer (CDC Technologies). For histological analysis, tissues were fixed in 10% phosphate-buffered formalin overnight at 4°C and processed for routine embedding in paraffin. Sections of 6-μm thickness were stained with hematoxylin and eosin using standard staining procedures. LacZ staining was performed on tissue sections frozen in optical cutting temperature (OCT; Tissue-Tek) according to the method of MacGregor et al. (33).
Reverse transcription-PCR.
cDNA was synthesized from 4 μg of DNase (Invitrogen)-treated RNA isolated from mouse liver using the SuperScript first-strand synthesis system for reverse transcription-PCR (Invitrogen). One microliter of cDNA was subjected to PCR amplification using either SYBR GREEN PCR Master Mix or Taqman Universal PCR Master Mix (Applied Biosystems). The following primer and probe sets were used to amplify specific target genes: SYBR GREEN primers, Vhlh (Fwd, 5′-GCCTATTTTTGCCAACATCACA-3′; Rev, 5′-TCATTCTCTCTATGTGCTGGCTTT-3′), Hif-1α (Fwd, 5′-CAAGATCTCGGCGAAGCAA-3′; Rev, 5′-GGTGAGCCTCATAACAGAAGCTTT-3′), Hif-2α, Fwd (5′-CAACCTGCAGCCTCAGTGTATC-3′; Rev, 5′-CACCACGTCGTTCTTCTCGAT-3′), VegfA (Fwd, 5′-CCACGTCAGAGAGCAACATCA-3′; Rev, 5′-TCATTCTCTCTATGTGCTGGCTTT-3′), Epo (Fwd, 5′-CATCTGCGACAGTCGAGTTCTG-3′; Rev, 5′-CACAACCCATCGTGACATTTTC-3′), Bnip3 (Fwd, 5′-GACGAAGTAGCTCCAAGAGTTCTCA-3′; Rev, 5′-CTATTTCAGCTCTGTTGGTATCTTGTG-3′), Pgk (Fwd, 5′-GGAAGCGGGTCGTGATGA-3′; Rev, 5′-GCCTTGATCCTTTGGTTGTTTG-3′; Taqman primers and probes, 18S (Human 18S rRNA Taqman set [Applied Biosystems]), Arnt (Fwd, 5′-CGAAAACCAGACAAGCTAACCA-3′; Rev, 5′-TGTTGCCAGTTCCCCTCAAG-3′; Probe, 5′-CTTACGCATGGCCGTTTCTCACATGAA-3′). PCR amplification was performed on the Prism 7700 Sequence Detection System (Applied Biosystems). The thermal-cycling profile used was denaturation at 50°C for 2 min and 95°C for 10 min, followed by cycles of denaturation at 95°C for 15 s and 60°C for 1 min. 18S was used to normalize mRNA. Relative quantitation of mRNA expression levels was determined using the relative standard curve method according to the manufacturer's instructions (Applied Biosystems).
Statistical analysis.
For statistical analysis and comparison of numbers of liver hemangiomas in Vhlh and Vhlh/Hif-1α mutant mice, the chi-square test was performed. For statistical analysis of gene expression data, red blood cell numbers, and hemoglobin concentrations, analysis of variance, followed by the unpaired Student's t test, was performed. P values of <0.05 were considered statistically significant.
RESULTS
Efficient deletion of Hif-1α and Arnt in Vhlh-deficient hepatocytes.
We have previously reported that mice that are heterozygously deficient for Vhlh are predisposed to develop cavernous hemangiomas of the liver with high phenotypical penetrance at old age (10). Vascular lesions can be reproduced by Cre-mediated recombination of a Vhlh conditional allele (2-lox allele) in hepatocytes (10). To examine the requirement for HIF in VHL-associated vascular tumorigenesis, we used two Cre-transgenic lines, Albumin-Cre (45) and PEPCK-Cre, to inactivate both Vhlh and Hif-1α or Vhlh and Arnt in hepatocytes. In the Vhlh conditional allele, deletion of exon 1 and the promoter by Cre-mediated recombination results in a Vhlh null allele (1-lox allele) (Fig. 1A) (10). In the Hif-1α and Arnt conditional alleles, Hif-1α exon 2 and Arnt exon 6, which both encode the bHLH domain, undergo Cre-mediated out-of-frame deletion, resulting in the absence of Hif-1α or Arnt (Fig. 1A) (48, 60). Inactivation of Hif-1α results in the inability to form functional Hif-1, while it does not affect the formation of Hif-2 heterocomplexes (48, 53), whereas inactivation of Arnt prevents the formation of functional Hif-1 and Hif-2, but not nuclear translocation of Hif-α homologues (2).
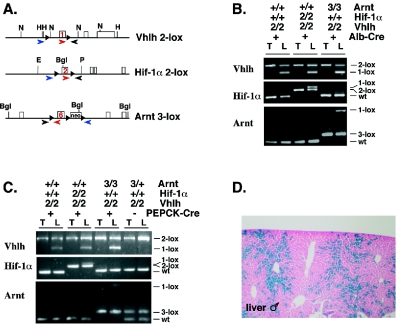
FIG. 1.
Generation of mice deficient for Vhlh, Vhlh/Hif-1α, and Vhlh/Arnt in hepatocytes. (A) Genomic maps for the targeted Vhlh 2-lox, Hif-1α 2-lox, and Arnt 3-lox alleles. Exons are depicted as rectangular boxes with the loxP floxed exon numbered. loxP sites are represented by black triangles. The locations of primers used for genomic PCR are shown with colored arrows. All three primers were used in a single PCR to specifically amplify the nonrecombined conditional allele (2-lox) or wild type with the red and black primers and the recombined allele (1-lox) with the blue and black primers. Genomic maps are not complete or drawn to scale. Abbreviations: neo, neomycin selection marker; H, HindIII; N, NcoI; E, EcoRI; P, PstI; Bgl, BglII. (B) Genotype analysis of Albumin-Cre mutant mice by genomic PCR. Abbreviations: 2, 2-lox; 3, 3-lox; +, wild type (wt) (+ for Alb-Cre indicates the presence of the Albumin-Cre transgene); T, tail; L, liver. (C) Genotype analysis of PEPCK-Cre mutant mice by genomic PCR. (D) Qualitative analysis of PEPCK-Cre expression in the liver. LacZ staining of a liver section collected from a male PEPCK-Cre mouse crossed with the ROSA26 LacZ reporter mouse (52). Blue staining indicates hepatocytes that have undergone Cre-mediated recombination.
It has been reported that Albumin-Cre is active in >80% of hepatocytes and recombines floxed alleles with high efficiency (45). High-efficiency gene deletion in Albumin-Cre mutants (Vhlh2lox/2lox, Hif-1α2lox/2lox; Albumin-Cre or Vhlh32lox/2lox, Arnt3lox/3lox; Albumin-Cre [referred to hereafter as Albumin-Vhlh/Hif-1α or Albumin-Vhlh/Arnt double mutants]) is shown in Fig. 1B and is consistent with previously reported findings (10). Since Albumin-Cre-mediated inactivation of Vhlh results in relatively complex liver pathology, which includes an increase in nonhepatocyte cell types, such as endothelial cells (10), genomic PCR of DNA isolated from whole livers most likely underestimates recombination efficiency in Albumin-Vhlh and Vhlh/Hif-1α livers. Recombination efficiency is therefore best assessed in Vhlh/Arnt double mutants, which have histologically normal livers (Fig. 1B; also see Fig. 3). Due to the high degree of Cre-mediated recombination in these mice, Albumin-Cre mutant mice were used for gene expression studies. Albumin-Cre-mediated targeting of Vhlh results in severe hepatic steatosis (neutral fat accumulation in hepatocytes) and angiectasis. Mutant mice died at an early age (10), thus precluding long-term studies of adult mice.
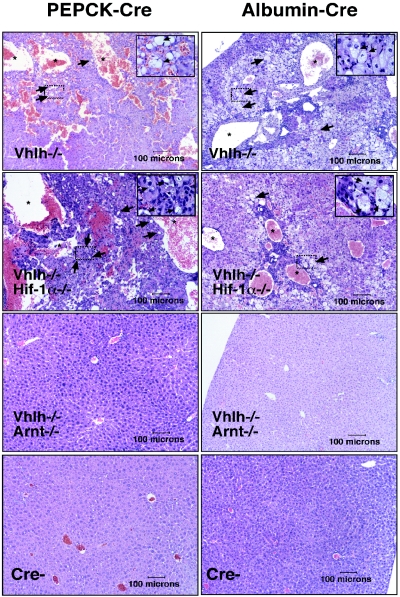
FIG. 3.
Inactivation of Arnt suppresses Vhlh-associated vascular lesions and steatosis. Hematoxylin and eosin staining of PEPCK-Cre (left; >6 months of age) and Albumin-Cre (right; 4 to 6 weeks of age) mutant liver sections (magnification, ×100). Note that large vascular spaces (stars) and steatosis (arrows) were observed in both Vhlh and Vhlh/Hif-1α mutant mice. The black boxes outline areas that are shown at higher magnification (×1,000) in the upper right corners.
In contrast to Albumin-Cre mutants, PEPCK-Cre Vhlh mutants lived to at least 15 months of age and developed cavernous liver hemangiomas (Fig. 2A). In the PEPCK-Cre transgene Cre-recombinase is under the control of the rat PEPCK promoter (42). The PEPCK-Cre transgene was generated by targeted single-copy transgenesis in embryonic stem cells and is located on the X chromosome upstream of the hypoxanthine-phosphoribosyltransferase (Hprt) gene (E. B. Rankin and V. H. Haase, unpublished data). Since PEPCK-Cre is located on the X chromosome, it is most likely subject to random X chromosome inactivation (for a review, see reference 4); we observed variable recombination efficiency in female mice that were heterozygous for the transgene (data not shown). In order to reduce experimental variability, the majority of mice used for analysis of the PEPCK mutant phenotype were male mice.
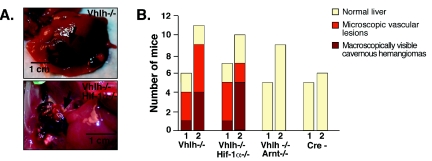
FIG. 2.
Inhibition of vascular-tumor development in Vhlh/Arnt mutant mice. (A) Photographs of gross cavernous liver hemangiomas observed in PEPCK-Vhlh and PEPCK-Vhlh/Hif-1α mutant mice. Hemangiomas are indicated by arrows. (B) Incidence of macroscopic liver hemangiomas and microscopic vascular lesions observed in PEPCK-Cre mutant mice. Mice from the indicated genotypes were divided into two age groups for analysis, mice 2 to 6 months of age (group 1) and mice >6 months of age (group 2). Each bar represents the number of mice from the indicated genotype with macroscopic hemangiomas, microscopic vascular lesions in the liver, and normal liver. A chi-square test revealed that there were no significant differences between the distributions of macroscopic and microscopic liver hemangiomas in the Vhlh- and Vhlh/Hif-1α-deficient mice.
To determine the requirement for Hif-1α and Arnt in the development of VHL-associated liver hemangiomas, we generated mice that were made double deficient for either Vhlh and Hif-1α or Vhlh and Arnt by PEPCK-Cre-mediated recombination. For this purpose, PEPCK-Cre transgenic mice were bred to mice that were homozygous for either Vhlh (10) and Hif-1α (48) or Vhlh and Arnt (60) conditional alleles to generate double-mutant mice: Vhlh2lox/2lox, Hif-1α2lox/2lox; PEPCK-Cre or Vhlh2lox/2lox, Arnt3lox/3lox; PEPCK-Cre (hereafter referred to as either PEPCK-Vhlh/Hif-1α or PEPCK-Vhlh/Arnt double mutants). PEPCK-Cre-mediated recombination efficiency was examined by genomic PCR and with the ROSA26 LacZ reporter (Fig. 1C and D) (52). Consistent with the expression pattern of the endogenous gene (42), PEPCK-Cre was found to be active in ∼20 to 30% of hepatocytes. Genomic PCR results were found to be comparable with the results from the ROSA26 Cre reporter study (Fig. 1C and D).
Inactivation of Arnt is required to suppress the development of liver hemangiomas in Vhlh mutants.
To determine the requirement for Hif-1α and Arnt in the development of liver hemangiomas, we examined the livers of PEPCK-Vhlh/Hif-1α and PEPCK-Vhlh/Arnt double-mutant mice macroscopically and histologically. Mice were divided into two age groups, 2 to 6 months of age (group 1) and >6 months of age (group 2). The oldest mice analyzed were 15 months of age. We observed that, similar to PEPCK-Vhlh mutants, 6 of 17 Vhlh/Hif-1α double mutants developed large cavernous hemangiomas compared to 5 of 17 PEPCK-Vhlh mutants, resulting in no significant difference in the number of hemangiomas that developed between the two mutants, as determined by the chi-square test (P = 0.71) (Fig. 2A and B). Although the number of large cavernous hemangiomas per individual liver was usually limited to <5, we observed focal microscopic changes throughout the liver that were similar to those previously reported for Vhlh heterozygotes and Albumin-Cre mutants (10). Typically, those microscopic vascular lesions consisted of hepatocellular steatosis, angiectasis, and endothelial cell proliferation (Fig. 3). The frequencies of mice that exhibited only focal microscopic lesions were similar for the Vhlh single mutant (8 of 17) and for the Vhlh/Hif-1α double mutant (6 of 17). Strikingly, we did not observe any of the same lesions (microscopic or macroscopic) in PEPCK-Vhlh/Arnt livers, which at an early age were histologically identical to control livers (Fig. 2B). Moderate steatosis without angiectasis or cellular proliferation was found in Vhlh/Arnt double mutants (three of nine) at older ages (>9 months), which may be a result of the inability to form Hif-2 heterodimers, as hepatic steatosis has been reported in Hif-2α germ line knockout mice (49).
Similar to PEPCK mutants, Albumin-Vhlh single mutants did not differ significantly from Albumin-Vhlh/Hif-1α double mutants and developed severe hepatic steatosis associated with angiectasis and endothelial cell proliferation, as previously reported (10). Albumin-Vhlh/Arnt double mutants appeared histologically normal at the age used for this study (4 to 6 weeks of age) (Fig. 3). We conclude from our data that the inability to generate transcriptionally active Hif-1 is not sufficient to prevent the development of VHL-associated vascular tumors and that inactivation of Arnt is sufficient to suppress the VHL phenotype in mice.
Inactivation of Hif-1α is not sufficient to suppress the development of VHL-associated polycythemia.
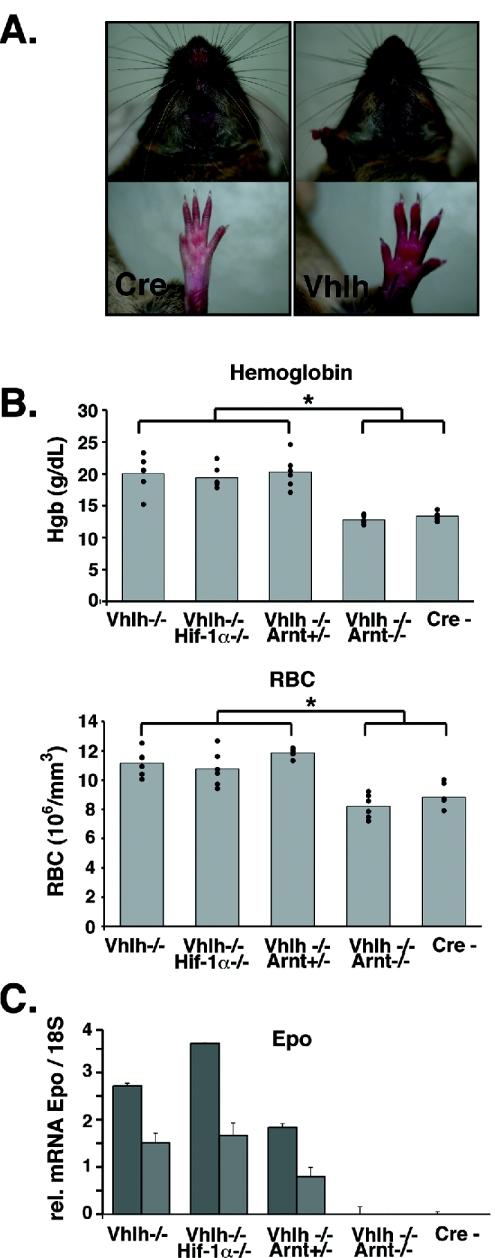
A subset of VHL patients with renal cell carcinoma and capillary hemangioblastomas develop polycythemia, which correlates with upregulation of Epo expression in tumors (5, 30). Epo is a classic hypoxia-inducible gene, and investigations aimed at the identification of transcription factors responsible for hypoxic induction of Epo mRNA eventually led to the purification of HIF-1 (for a review, see reference 51). We previously reported that inactivation of Vhlh in the mouse liver also results in polycythemia as a result of increased serum Epo levels (10). Similarly, we observed that PEPCK-Vhlh mutant mice developed erythrocytosis that clinically manifested as redness of the paws and muzzle (Fig. 4A). To determine whether the development of VHL-associated erythrocytosis was exclusively dependent on increased Hif-1 transcriptional activity, we compared RBC numbers and total Hgb concentrations in PEPCK-Vhlh mutants to those in PEPCK-Vhlh/Hif-1α and PEPCK-Vhlh/Arnt mice. We found that PEPCK-Vhlh- and Vhlh/Hif-1α-deficient mice developed similar degrees of erythrocytosis, with average RBC counts and Hgb values of 11.17 × 106 ± 0.9 × 106/mm3 and 20.05 ± 2.81 g/dl for Vhlh mutants and 10.77 × 106 ± 1.22 × 106/mm3 and 19.38 ± 1.76 g/dl for Vhlh/Hif-1α double mutants (n = 6 in all groups). In contrast, Vhlh/Arnt double-mutant mice had normal RBC counts, with average RBC numbers and Hgb values of 8.21 × 106 ± 0.82 × 106/mm3 and 12.8 ± 0.635 g/dl, respectively, compared to values of 9.3 × 106 ± 0.62 × 106/mm3 and 13.24 ± 0.48 g/dl in control mice (Fig. 4B).
FIG. 4.
Inactivation of Vhlh and Vhlh/Hif-1α in hepatocytes induces erythrocytosis. (A) Control (Cre−) and PEPCK-Vhlh muzzles and paws. Note the increased red coloration of the skin in the PEPCK-Vhlh mutant mouse. (B) Elevated hemoglobin and red blood cell numbers in PEPCK-Vhlh- and PEPCK-Vhlh/Hif-1α-deficient mutant mice. The results shown are average hemoglobin concentrations and RBC numbers in blood collected from PEPCK-Cre mutant mice determined by a CBC analyzer. Note that Vhlh/Arnt mutant mice and control mice have similar hemoglobin concentrations and red blood cell numbers (no statistical differences as determined by t test). Vhlh-, Vhlh−/−/Hif-1α−/−, and Vhlh−/−/Arnt+/− mice exhibited increased levels of hemoglobin and red blood cells compared to control and Vhlh−/−/Arnt−/− mice (*, P < 0.05). Six mice from each genotype were analyzed and are represented by individual dots. (C) Liver erythropoietin expression correlates with erythrocytosis. Shown are Epo mRNA expression levels from PEPCK-Cre mutant livers determined by real-timePCR. Each bar represents the average of three values obtained for an individual mouse. The error bars represent standard deviations. 18S was used to normalize mRNA. Two representative mice from an n of 3 are shown for each genotype.
To determine if erythrocytosis correlated with increased Epo expression, we measured Epo mRNA levels in whole-liver homogenates. PEPCK-Vhlh and PEPCK-Vhlh/Hif-1α mice exhibited increased Epo mRNA expression, whereas PEPCK-Vhlh/Arnt and control mice did not express detectable levels of Epo mRNA transcripts, as determined by real-time PCR (Fig. 4C). We conclude that increased production of Epo in Vhlh-deficient livers is not dependent on Hif-1, suggesting that loss of Hif-1 can be compensated for by other Hif heterocomplexes.
Differential suppression of Hif target genes in Vhlh/Hif-1α double-mutant mice.
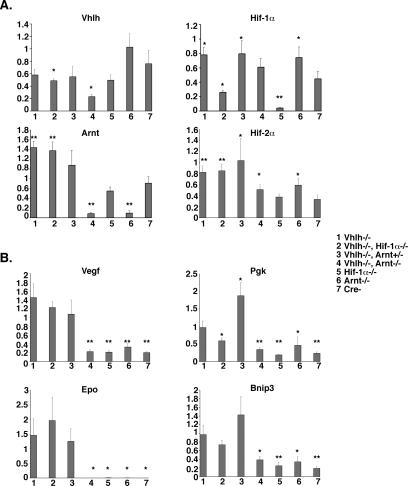
The liver expresses at least two Hif homologues that have been shown to function as hypoxia-responsive transcriptional activators, namely, Hif-1 and Hif-2 (26, 55, 63). In order to determine to what degree Hif target gene expression in the liver is Hif-1 dependent, we examined Vegf, Epo, phosphoglycerokinase (Pgk), and Bnip3 mRNA levels in wild-type and Vhlh-, Vhlh/Hif-1α-, and Vhlh/Arnt-deficient livers. For these studies, we analyzed Albumin-Cre mutants due to the limited expression of PEPCK-Cre in the liver. We first determined mRNA expression levels for the loxP targeted genes in each of the mice analyzed. Real-time PCR analysis revealed a robust reduction of Hif-1α and Arnt mRNA expression in all Albumin-Cre mutant mice homozygous for the Hif-1α (groups 2 and 5) or Arnt (groups 4 and 6) floxed alleles (Fig. 5A). Reduction of Vhlh mRNA in whole-liver homogenates from Vhlh mutants (Fig. 5A, group 1) was less pronounced, probably due to the presence of nonrecombined cells, such as proliferating endothelial and other nonhepatocyte cell types (Fig. 3), which also may have resulted in a relative increase in Hif-1α and Arnt mRNA levels. Consistent with the increased presence of the Vhlh 1-lox allele (Fig. 1B) is the reduction of Vhlh mRNA levels by ∼75% in Vhlh/Arnt double mutants (Fig. 5A, group 4). In addition, we determined the relative expression levels of Hif-2α in the Albumin-Cre mutant livers. We observed that, similar to Arnt, expression of Hif-2α was significantly increased in Vhlh and Vhlh/Hif-1α mutants (P < 0.001; groups 1 and 2). This may be either a result of Vhlh deletion, as induction of HIF-2α mRNA expression in VHL-deficient renal cell carcinoma cells has been described (29), or the result of the increased presence of nonrecombined cells.
FIG. 5.
Inactivation of Arnt is sufficient to suppress Hif target gene induction in Vhlh-deficient livers. (A) Suppression of targeted gene (Vhlh, Hif-1α, and Arnt) expression in Albumin-Cre mutant livers. Shown are relative mRNA transcript levels normalized to 18S levels determined by real-time PCR. Note that Albumin-Cre-mediated inactivation of Vhlh is best represented in the Vhlh/Arnt double-mutant mice. Each bar represents the average mRNA expression level of four individual mice for each genotype. The error bars represent the standard deviations. Asterisks indicate a significant increase or decrease in target gene expression compared to control mice (Cre−) (*, P < 0.05; **, P < 0.001) as determined by the t test. (B) Relative mRNA expression levels for HIF target genes (Vegf, Epo, Pgk, and Bnip3) determined by real-time PCR. Each bar represents the average mRNA expression level of four individual mice for each genotype. The error bars represent standard deviations. Asterisks indicate a significant increase or decrease in target gene expression compared to Vhlh−/− mice (*, P < 0.05; **, P < 0.001) as determined by the t test. 18S was used to normalize mRNA.
Real-time PCR analysis of HIF target genes revealed that mRNA transcripts for Vegf and Epo were increased in both Vhlh- and Vhlh/Hif-1α-deficient livers, whereas Vhlh/Arnt double-mutant livers expressed transcript levels comparable to those in control mice (Fig. 5B). Similarly, Pgk and Bnip3 mRNA transcripts were increased in Vhlh- and Vhlh/Hif-1α-deficient livers and were expressed at baseline levels in the Vhlh/Arnt mutant livers. However, expression levels of Pgk in Vhlh/Hif-1α double mutants were significantly reduced compared to those in Vhlh mutants (P < 0.01), while Bnip3 mRNA levels were less affected (Fig. 5B). These results suggest that the functional redundancy between different Hif transcription factors is target gene dependent. The inability to form Hif-1 heterodimers had little effect on Vegf and Epo gene expression, which is consistent with the persistence of excessive erythrocytosis in Vhlh/Hif-1α double mutants and their predisposition to the development of vascular tumors. Our data demonstrate that inactivation of Arnt is sufficient to suppress Hif target gene induction in Vhlh-deficient livers and suggest that another Arnt binding partner is responsible for Vegf and Epo target gene induction in Vhlh-deficient livers.
Hif target gene expression data from Vhlh/Hif-1α double-deficient mice suggest that Hif-2 is transcriptionally active in hepatocytes. To examine Hif-2α protein levels and nuclear localization, we performed immunoblot analysis on nuclear protein fractions isolated from Albumin-Cre mutants. Nuclear Hif-2α, as well as Hif-1α, was easily detectable in livers from Vhlh and Vhlh/Arnt mutants, while only Hif-2α was easily detectable in Vhlh/Hif-1α mutants (Fig. 6A). In addition, we found that both Hif-1 and Hif-2 complexes bound to the Epo HRE in Vhlh-deficient livers, whereas only Hif-2 heterocomplexes were detected in Vhlh/Hif-1α-deficient livers (Fig. 6B). Neither Hif-1α- nor Hif-2α-containing complexes were bound to the Epo HRE in the Vhlh/Arnt and control livers. Taken together, our data suggest that in Vhlh-deficient livers, Hif-2α is able to translocate to the nucleus, where it is transcriptionally active only when Arnt is present. These data are consistent with the observation that Arnt is the only Hif-β subunit expressed in the liver at significant levels (21).
FIG. 6.
Hif-2α protein stabilization and HRE binding in Vhlh-deficient livers. (A) Nuclear protein (N) extracts were isolated from livers of Albumin-Vhlh, Albumin-Vhlh/Hif-1α, and Albumin-Vhlh/Arnt mutant and control mice (Cre−). Immunoblot analysis for Hif-2α revealed that Hif-2α protein was stable and nuclear in all Vhlh-deficient livers. Hif-1α protein was also detectable above baseline levels in Vhlh- and Vhlh/Arnt-deficient livers. Ponceau S staining is shown to demonstrate equal protein loading. Abbreviations: 2, 2-lox; 3, 3-lox; +, wild type (+ or − for Alb-Cre indicates the presence or absence of the Albumin-Cre transgene). (B) Hif-1 and Hif-2 HRE binding activity by EMSA. Nuclear protein extracts isolated from Albumin-Vhlh, Vhlh/Hif-1α, Vhlh/Arnt, and Cre− livers were tested for Hif binding to the Epo HRE in vitro by EMSA. Supershifts with Hif-1α and Hif-2α antibodies revealed that both Hif-1 and Hif-2 form heterocomplexes at the Epo HRE in Vhlh-deficient livers. Note that Hif-2 binding to the Epo HRE was still observed in Albumin-Vhlh/Hif-1α nuclear extracts despite a decrease in total Hif HRE binding. Abbreviations: n.s., nonspecific; s.s., supershift; CREB, cAMP response element binding protein.
DISCUSSION
In this report, we have used a conditional knockout mouse model of VHL-associated vascular tumors and erythrocytosis to investigate the contributions of HIF hetercomplexes to the development of the VHL phenotype. Typically, patients with VHL disease suffer from a variety of highly vascularized tumors, which include retinal and CNS hemangioblastomas, as well as RCC of the clear-cell type (for a review, see reference 34). While the biological behaviors of these tumors are very different—hemangioblastomas are usually benign and do not metastasize, whereas RCC are malignant—they share several molecular features. In both cases, mutant pVHL lacks the ability to target HIF for proteasomal degradation, resulting in constitutively active HIF (3, 15). VHL tumors express high levels of HIF target genes that regulate angiogenic growth factors, such as VEGF; glucose uptake and metabolism; and erythropoiesis (17, 30, 64). Besides regulating HIF, pVHL is involved in microtubule stabilization and extracellular-matrix fibronectin assembly and other non-HIF-related cellular processes (for a review, see reference 22). While the biology of VHL-associated hemangioblastomas can more easily be attributed to dysregulated HIF, VHL-associated renal carcinogenesis is more complex. Although HIF can modulate RCC metastatic potential through regulation of the chemokine receptor CXCR4 (54) and tumor growth is HIF2 dependent in nude-mouse xenograft models of RCC (27, 28, 65), RCC tumorigenesis most likely requires genetic events in addition to loss of pVHL function (36, 44).
In mice with germ line deficiency for one copy of the murine VHL homologue Vhlh, the liver seems to be the preferred organ site for vascular-tumor development. In contrast to mice, hepatic vascular tumors are rare manifestations of VHL disease in humans (11, 40, 46). Despite several microscopic features shared with VHL-associated CNS hemangioblastomas, VHL-associated vascular tumors in the murine liver are histogenetically distinct (57). Nevertheless, our model provides a genetic tool to study the role of HIF transcription factors and their individual contributions to the development of VHL-associated vascular tumorigenesis and alterations of gene expression.
Our data suggest that in regard to HIF-regulated gene expression, the level of redundancy among different HIF heterocomplexes is target gene dependent. While the inability to form Hif-1 heterodimers does not affect VHL-mediated angiogenesis or polycythemia, Pgk, a gene involved in glycolysis, was significantly reduced with the loss of Hif-1 heterodimers. Consistent with this finding is a report by Hu et al. that demonstrates that in genetically modified 786-0 cells, glycolytic gene expression is preferentially regulated by HIF-1 and not HIF-2 (16). Hepatocytes express the Hif-α homologues Hif-1α, Hif-2α, and Hif-3α (26, 55, 63). Both Hif-1α and Hif-2α form transcriptionally active heterocomplexes in hepatocytes, while the role of Hif-3α, another target for pVHL-mediated proteolysis (39), in hypoxic signaling of hepatocytes is unclear. Based on our studies, Arnt appears to be the only functional Hif-β subunit expressed in hepatocytes, which is consistent with previously published observations (21, 37).
Inactivation of Vhlh in hepatocytes results in excessive erythrocytosis from increased Epo production. We have previously shown that serum Epo levels are increased up to 40-fold over normal values (10). Despite the very high hematocrits, we have not observed that PEPCK mutants are prone to thromboembolic complications. Although PEPCK mutants are able to live for at least 15 months, several mice have died at a younger age when exposed to stressful situations. The cause of death in these situations is unclear. Similar observations were made in transgenic mice that express a human EPO transgene under the control of the PDGF-B chain promoter (59). EPO transgenic mice develop similar levels of erythrocytosis and are able to adapt to high hematocrits by increasing eNOS activity, resulting in vasodilatation. Higher blood viscosity from increased erythrocyte flexibility at physiological shear rates appears to be an additional mechanism to prevent cardiovascular complications in these mice (47, 56). Although it has not been formally examined, it is very likely that PEPCK-Cre mutants adapt to erythrocytosis by similar mechanisms. EPO transgenics do not develop liver angiectasis or cavernous hemangiomas (M. Gassmann, personal communication), which makes the involvement of high systemic Epo levels and erythrocytosis in the development of vascular tumors in PEPCK-Cre mutants unlikely. This notion is supported by the fact that Vhlh heterozygotes in which polycythemia is not a clinical feature are still prone to develop vascular tumors in the liver.
It is surprising that the development of erythrocytosis and Epo expression in Vhlh mutants is not affected by the inability to form functional Hif-1 heterodimers in Vhlh/Hif-1α double-knockout animals. This finding suggests that Hif-2 is able to fully compensate for the loss of Hif-1 in regard to Epo expression in the liver. Based on recently published studies, it has been speculated that the HIF-2 heterodimer may be the more relevant HIF in the transcriptional regulation of EPO in experimental retinopathy of prematurity and in human hepatoma and neuroblastoma cell lines (14, 61). By EMSA, we found significant binding of both Hif-1α- and Hif-2α-containing heterocomplexes to the Epo HRE in Vhlh-deficient liver protein extracts, suggesting that qualitative rather than quantitative differences between Hif-1 and Hif-2 may affect Epo expression in the liver. Efforts in our laboratory are under way to determine whether inactivation of Hif-2α in contrast to Hif-1α deletion will lead to a more significant attenuation of VHL-associated erythrocytosis.
The pharmacological disruption of HIF signaling may be an important therapeutic adjunct for the successful treatment and prevention of VHL-associated vascular tumors that are difficult to manage surgically, such as multifocal CNS hemagioblastomas (for a recent review of clinical management issues, see reference 14). To what degree the different HIF isoforms (HIF-1 versus HIF-2) contribute to the development of specific VHL-associated tumors is still under investigation. Both HIF-1α and HIF-2α are constitutively expressed in RCC (29, 64), where HIF-2 in particular seems to have a tumor-promoting effect even in the presence of wild-type pVHL (27, 28, 35, 65). High levels of HIF-2α have been found in stromal cells (the neoplastic component of VHL hemangioblastomas) (58) and correlate well with VEGF mRNA levels, while correlation of HIF-1α expression with tumor VEGF levels was less obvious (7). Taken together, these findings and our data suggest that therapeutic intervention strategies aimed at HIF signaling must be designed to efficiently target both HIF homologues. We have shown genetically in mice that inactivation of Arnt in a Vhlh-deficient background is sufficient to suppress the development of VHL-associated vascular tumors in the liver. Our results should stimulate future investigations into targeting either ARNT directly or its ability to dimerize with HIF-α subunits as a therapeutic strategy in the treatment of VHL vascular tumors.
In summary, our data suggest that in a mouse model of VHL-associated liver hemangiomas, vascular tumorigenesis is mediated by dysregulation of HIF transcription factors. We further propose that different HIF heterocomplexes may play distinct roles in the development of certain clinical features linked to VHL disease.
Acknowledgments
This work was supported by the Center for Molecular Studies in Digestive and Liver Disease (P30-DK50306) and in part by NIH grants R03-DK062060 and R01-CA100787, as well as seed money from the Department of Medicine, University of Pennsylvania (all to V.H.H.).
We are grateful to the members of the Morphology Core, Center for Molecular Studies in Digestive and Liver Disease (in particular, Gary Swain), for help with the preparation of tissues for histology and image analysis and to Nikita Shrimanker for technical assistance.
REFERENCES
- 1.Camenisch, G., R. H. Wenger, and M. Gassmann. 2002. DNA-binding activity of hypoxia-inducible factors (HIFs). Methods Mol. Biol. 196:117-129. [DOI] [PubMed] [Google Scholar]
- 2.Chilov, D., G. Camenisch, I. Kvietikova, U. Ziegler, M. Gassmann, and R. H. Wenger. 1999. Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF-1α. J. Cell Sci. 112:1203-1212. [DOI] [PubMed] [Google Scholar]
- 3.Clifford, S. C., M. E. Cockman, A. C. Smallwood, D. R. Mole, E. R. Woodward, P. H. Maxwell, P. J. Ratcliffe, and E. R. Maher. 2001. Contrasting effects on HIF-1α regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum. Mol. Genet. 10:1029-1038. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, D. E., and J. T. Lee. 2002. X-chromosome inactivation and the search for chromosome-wide silencers. Curr. Opin. Genet. Dev. 12:219-224. [DOI] [PubMed] [Google Scholar]
- 5.Da Silva, J. L., C. Lacombe, P. Bruneval, N. Casadevall, M. Leporrier, J. P. Camilleri, J. Bariety, P. Tambourin, and B. Varet. 1990. Tumor cells are the site of erythropoietin synthesis in human renal cancers associated with polycythemia. Blood 75:577-582. [PubMed] [Google Scholar]
- 6.Duan, D. R., A. Pause, W. H. Burgess, T. Aso, D. Y. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 7.Flamme, I., M. Krieg, and K. H. Plate. 1998. Up-regulation of vascular endothelial growth factor in stromal cells of hemangioblastomas is correlated with up-regulation of the transcription factor HRF/HIF-2α. Am. J. Pathol. 153:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnarra, J. R., K. Tory, Y. Weng, L. Schmidt, M. H. Wei, H. Li, F. Latif, S. Liu, F. Chen, F. M. Duh, et al. 1994. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 7:85-90. [DOI] [PubMed] [Google Scholar]
- 9.Gnarra, J. R., J. M. Ward, F. D. Porter, J. R. Wagner, D. E. Devor, A. Grinberg, M. R. Emmert-Buck, H. Westphal, R. D. Klausner, and W. M. Linehan. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 94:9102-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haase, V. H., J. N. Glickman, M. Socolovsky, and R. Jaenisch. 2001. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl. Acad. Sci. USA 98:1583-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayasaka, K., Y. Tanaka, T. Satoh, and H. Mutoh. 1999. Hepatic hemangioblastoma: an unusual presentation of von Hippel-Lindau disease. J. Comput. Assist. Tomogr. 23:565-566. [DOI] [PubMed] [Google Scholar]
- 12.Hergovich, A., J. Lisztwan, R. Barry, P. Ballschmieter, and W. Krek. 2003. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 5:64-70. [DOI] [PubMed] [Google Scholar]
- 13.Herman, J. G., F. Latif, Y. Weng, M. I. Lerman, B. Zbar, S. Liu, D. Samid, D. S. Duan, J. R. Gnarra, W. M. Linehan, et al. 1994. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. USA 91:9700-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hes, F. J., R. B. van der Luijt, and C. J. Lips. 2001. Clinical management of Von Hippel-Lindau (VHL) disease. Neth. J. Med. 59:225-234. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman, M. A., M. Ohh, H. Yang, J. M. Klco, M. Ivan, and W. G. Kaelin, Jr. 2001. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum. Mol. Genet. 10:1019-1027. [DOI] [PubMed] [Google Scholar]
- 16.Hu, C. J., L. Y. Wang, L. A. Chodosh, B. Keith, and M. C. Simon. 2003. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 23:9361-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliopoulos, O., A. P. Levy, C. Jiang, W. G. Kaelin, Jr., and M. A. Goldberg. 1996. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93:10595-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 19.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 21.Jain, S., E. Maltepe, M. M. Lu, C. Simon, and C. A. Bradfield. 1998. Expression of ARNT, ARNT2, HIF1 alpha, HIF2 alpha and Ah receptor mRNAs in the developing mouse. Mech. Dev. 73:117-123. [DOI] [PubMed] [Google Scholar]
- 22.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 23.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 24.Kewley, R. J., M. L. Whitelaw, and A. Chapman-Smith. 2004. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 36:189-204. [DOI] [PubMed] [Google Scholar]
- 25.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 26.Kietzmann, T., Y. Cornesse, K. Brechtel, S. Modaressi, and K. Jungermann. 2001. Perivenous expression of the mRNA of the three hypoxia-inducible factor alpha-subunits, HIF1α, HIF2α and HIF3α, in rat liver. Biochem. J. 354:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo, K., W. Y. Kim, M. Lechpammer, and W. G. Kaelin, Jr. 2003. Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol 1:E83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo, K., J. M. Klco, E. Nakamura, M. Lechpammer, and W. G. Kaelin. 2002. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1:237-246. [DOI] [PubMed] [Google Scholar]
- 29.Krieg, M., R. Haas, H. Brauch, T. Acker, I. Flamme, and K. H. Plate. 2000. Up-regulation of hypoxia-inducible factors HIF-1α and HIF-2α under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene 19:5435-5443. [DOI] [PubMed] [Google Scholar]
- 30.Krieg, M., H. H. Marti, and K. H. Plate. 1998. Coexpression of erythropoietin and vascular endothelial growth factor in nervous system tumors associated with von Hippel-Lindau tumor suppressor gene loss of function. Blood 92:3388-3393. [PubMed] [Google Scholar]
- 31.Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, W., L. Tessarollo, S. B. Hong, M. Baba, E. Southon, T. C. Back, S. Spence, C. G. Lobe, N. Sharma, G. W. Maher, S. Pack, A. O. Vortmeyer, C. Guo, B. Zbar, and L. S. Schmidt. 2003. Hepatic vascular tumors, angiectasis in multiple organs, and impaired spermatogenesis in mice with conditional inactivation of the VHL gene. Cancer Res. 63:5320-5328. [PubMed] [Google Scholar]
- 33.MacGregor, G. R., B. P. Zambrowicz, and P. Soriano. 1995. Tissue non-specific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development 121:1487-1496. [DOI] [PubMed] [Google Scholar]
- 34.Maher, E. R., and W. G. Kaelin, Jr. 1997. von Hippel-Lindau disease. Medicine (Baltimore) 76:381-391. [DOI] [PubMed] [Google Scholar]
- 35.Maranchi, J. K., J. R. Vasselli, J. Riss, J. S. Bonifacio, W. M. Linehan, and R. D. Klausner. 2002. The contribution of VHL substrate binding and HIF-1α to the phenotype of vhl loss in renal cell carcinoma. Cancer Cell 1:247-253. [DOI] [PubMed] [Google Scholar]
- 36.Martinez, A., P. Fullwood, K. Kondo, T. Kishida, M. Yao, E. R. Maher, and F. Latif. 2000. Role of chromosome 3p12-p21 tumour suppressor genes in clear cell renal cell carcinoma: analysis of VHL dependent and VHL independent pathways of tumorigenesis. Mol. Pathol. 53:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maxwell, P. H., G. U. Dachs, J. M. Gleadle, L. G. Nicholls, A. L. Harris, I. J. Stratford, O. Hankinson, C. W. Pugh, and P. J. Ratcliffe. 1997. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. USA 94:8104-8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 39.Maynard, M. A., H. Qi, J. Chung, E. H. Lee, Y. Kondo, S. Hara, R. C. Conaway, J. W. Conaway, and M. Ohh. 2003. Multiple splice variants of the human HIF-3α locus are targets of the VHL E3 ubiquitin ligase complex. J. Biol. Chem. 21:21. [DOI] [PubMed] [Google Scholar]
- 40.McGrath, F. P., R. G. Gibney, D. C. Morris, D. A. Owen, and S. R. Erb. 1992. Case report: multiple hepatic and pulmonary haemangioblastomas—a new manifestation of von Hippel-Lindau disease. Clin. Radiol. 45:37-39. [DOI] [PubMed] [Google Scholar]
- 41.Ohh, M., R. L. Yauch, K. M. Lonergan, J. M. Whaley, A. O. Stemmer-Rachamimov, D. N. Louis, B. J. Gavin, N. Kley, W. G. Kaelin, Jr., and O. Iliopoulos. 1998. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol. Cell 1:959-968. [DOI] [PubMed] [Google Scholar]
- 42.Patel, Y. M., J. S. Yun, J. Liu, M. M. McGrane, and R. W. Hanson. 1994. An analysis of regulatory elements in the phosphoenolpyruvate carboxykinase (GTP) gene which are responsible for its tissue-specific expression and metabolic control in transgenic mice. J. Biol. Chem. 269:5619-5628. [PubMed] [Google Scholar]
- 43.Pause, A., S. Lee, R. A. Worrell, D. Y. Chen, W. H. Burgess, W. M. Linehan, and R. D. Klausner. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 94:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pavlovich, C. P., and L. S. Schmidt. 2004. Searching for the hereditary causes of renal-cell carcinoma. Nat. Rev. Cancer 4:381-393. [DOI] [PubMed] [Google Scholar]
- 45.Postic, C., M. Shiota, K. D. Niswender, T. L. Jetton, Y. Chen, J. M. Moates, K. D. Shelton, J. Lindner, A. D. Cherrington, and M. A. Magnuson. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305-315. [DOI] [PubMed] [Google Scholar]
- 46.Rojiani, A. M., D. A. Owen, K. Berry, B. Woodhurst, F. H. Anderson, C. H. Scudamore, and S. Erb. 1991. Hepatic hemangioblastoma. An unusual presentation in a patient with von Hippel-Lindau disease. Am. J. Surg. Pathol. 15:81-86. [PubMed] [Google Scholar]
- 47.Ruschitzka, F. T., R. H. Wenger, T. Stallmach, T. Quaschning, C. de Wit, K. Wagner, R. Labugger, M. Kelm, G. Noll, T. Rulicke, S. Shaw, R. L. Lindberg, B. Rodenwaldt, H. Lutz, C. Bauer, T. F. Luscher, and M. Gassmann. 2000. Nitric oxide prevents cardiovascular disease and determines survival in polyglobulic mice overexpressing erythropoietin. Proc. Natl. Acad. Sci. USA 97:11609-11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 49.Scortegagna, M., K. Ding, Y. Oktay, A. Gaur, F. Thurmond, L. J. Yan, B. T. Marck, A. M. Matsumoto, J. M. Shelton, J. A. Richardson, M. J. Bennett, and J. A. Garcia. 2003. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat. Genet. 35:331-340. [DOI] [PubMed] [Google Scholar]
- 50.Semenza, G. L. 2001. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 13:167-171. [DOI] [PubMed] [Google Scholar]
- 51.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 52.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 53.Sowter, H. M., R. R. Raval, J. W. Moore, P. J. Ratcliffe, and A. L. Harris. 2003. Predominant role of hypoxia-inducible transcription factor (Hif)-1α versus Hif-2α in regulation of the transcriptional response to hypoxia. Cancer Res. 63:6130-6134. [PubMed] [Google Scholar]
- 54.Staller, P., J. Sulitkova, J. Lisztwan, H. Moch, E. J. Oakeley, and W. Krek. 2003. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature 425:307-311. [DOI] [PubMed] [Google Scholar]
- 55.Stroka, D. M., T. Burkhardt, I. Desbaillets, R. H. Wenger, D. A. Neil, C. Bauer, M. Gassmann, and D. Candinas. 2001. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 15:2445-2453. [DOI] [PubMed] [Google Scholar]
- 56.Vogel, J., I. Kiessling, K. Heinicke, T. Stallmach, P. Ossent, O. Vogel, M. Aulmann, T. Frietsch, H. Schmid-Schonbein, W. Kuschinsky, and M. Gassmann. 2003. Transgenic mice overexpressing erythropoietin adapt to excessive erythrocytosis by regulating blood viscosity. Blood 102:2278-2284. [DOI] [PubMed] [Google Scholar]
- 57.Vortmeyer, A. O., S. Frank, S. Y. Jeong, K. Yuan, B. Ikejiri, Y. S. Lee, D. Bhowmick, R. R. Lonser, R. Smith, G. Rodgers, E. H. Oldfield, and Z. Zhuang. 2003. Developmental arrest of angioblastic lineage initiates tumorigenesis in von Hippel-Lindau disease. Cancer Res. 63:7051-7055. [PubMed] [Google Scholar]
- 58.Vortmeyer, A. O., J. R. Gnarra, M. R. Emmert-Buck, D. Katz, W. M. Linehan, E. H. Oldfield, and Z. Zhuang. 1997. von Hippel-Lindau gene deletion detected in the stromal cell component of a cerebellar hemangioblastoma associated with von Hippel-Lindau disease. Hum. Pathol. 28:540-543. [DOI] [PubMed] [Google Scholar]
- 59.Wagner, K. F., D. M. Katschinski, J. Hasegawa, D. Schumacher, B. Meller, U. Gembruch, U. Schramm, W. Jelkmann, M. Gassmann, and J. Fandrey. 2001. Chronic inborn erythrocytosis leads to cardiac dysfunction and premature death in mice overexpressing erythropoietin. Blood 97:536-542. [DOI] [PubMed] [Google Scholar]
- 60.Walisser, J. A., M. K. Bunger, E. Glover, E. B. Harstad, and C. A. Bradfield. 2004. Patent ductus venosus and dioxin resistance in mice harboring a hypomorphic Arnt allele. J. Biol. Chem. 279:16326-16331. [DOI] [PubMed] [Google Scholar]
- 61.Warnecke, C., Z. Zaborowska, J. Kurreck, V. A. Erdmann, U. Frei, M. Wiesener, and K. U. Eckardt. 2004. Differentiating the functional role of hypoxia-inducible factor (HIF)-1α and HIF-2α (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2α target gene in Hep3B and Kelly cells. FASEB J. 18:1462-1464. [DOI] [PubMed] [Google Scholar]
- 62.Wenger, R. H. 2002. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 16:1151-1162. [DOI] [PubMed] [Google Scholar]
- 63.Wiesener, M. S., J. S. Jurgensen, C. Rosenberger, C. K. Scholze, J. H. Horstrup, C. Warnecke, S. Mandriota, I. Bechmann, U. A. Frei, C. W. Pugh, P. J. Ratcliffe, S. Bachmann, P. H. Maxwell, and K. U. Eckardt. 2003. Widespread hypoxia-inducible expression of HIF-2α in distinct cell populations of different organs. FASEB J. 17:271-273. [DOI] [PubMed] [Google Scholar]
- 64.Wiesener, M. S., P. M. Munchenhagen, I. Berger, N. V. Morgan, J. Roigas, A. Schwiertz, J. S. Jurgensen, G. Gruber, P. H. Maxwell, S. A. Loning, U. Frei, E. R. Maher, H. J. Grone, and K. U. Eckardt. 2001. Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1α in clear cell renal carcinomas. Cancer Res. 61:5215-5222. [PubMed] [Google Scholar]
- 65.Zimmer, M., D. Doucette, N. Siddiqui, and O. Iliopoulos. 2004. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol. Cancer Res. 2:89-95. [PubMed] [Google Scholar]