Abstract
Mcl-1 is one Bcl-2 family member that plays a pivotal role in animal development. The extremely labile nature of the Mcl-1 protein itself and the fact that the Mcl-1 level is a critical determinant in various cell survival pathways suggest that cellular processes that regulate Mcl-1 stability are as important as those that regulate Mcl-1 synthesis. Although transcriptional stimulation of Mcl-1 synthesis in response to various stimuli has been well documented, regulation of Mcl-1 stability has been hardly explored. In this study, we identified that the translationally controlled tumor protein (TCTP) was one cellular factor that interacted with Mcl-1 and modulated Mcl-1 stability. While overexpression of TCTP augmented the protein stability of Mcl-1, knockdown expression of TCTP by RNA interference destabilized Mcl-1. Furthermore, TCTP stabilized Mcl-1 through interfering with Mcl-1's degradation by the ubiquitin-dependent proteasome degradation pathway, and the TCTP binding-defective mutant of Mcl-1 (K257V) was much more susceptible to degradation and manifested a compromised antiapoptotic activity. Taken together, these results suggest that TCTP modulates Mcl-1's antiapoptotic activity by modulating its protein stability. The possible mechanism(s) involved in TCTP's modulation process is discussed.
The mcl-1 gene was originally identified as an early gene induced during differentiation of ML-1 myeloid leukemia cells (13), and its encoded gene product contains structural features (i.e., the Bcl-2 homologous domains, BH1, BH2, and BH3) that categorize it as a member of the Bcl-2 family.
Mcl-1 appears to function at an apical step in many regulatory programs that control cell survival and death. Its synthesis is quickly stimulated after treatment of cells with many cytokines or other signals that lead to cell survival and/or differentiation (2, 6, 10, 11, 14, 25, 33). On the other hand, when cells are deprived of survival factors, stimulated by genotoxic stress (e.g., UV irradiation), or infected by certain viruses, Mcl-1 synthesis is rapidly blocked, and the existing pool of Mcl-1 protein is quickly degraded (6, 8, 24). Overexpression of Mcl-1 delays but does not prevent cells from undergoing apoptosis (6, 26, 38), indicating that Mcl-1 enhances short-term cell survival. In the immune system, Mcl-1 protein levels are rapidly degraded in the extremely short-lived neutrophils that constitutively undergo apoptosis for the resolution of inflammation (1, 22). Moreover, Mcl-1 levels were reported to decrease markedly in peripheral blood B lymphocytes when they undergo spontaneous apoptosis in vitro (19), and the macrophage survival is strictly dependent on the expression of Mcl-1 (18). A recent study by conditional ablation of Mcl-1 in the lymphoid system further indicates that Mcl-1 is required for the development and maintenance of B and T lymphocytes (25). Lastly, Mcl-1 appears to be extremely important for early embryo development, since Mcl-1-null embryos die at the peri-implantation stage (27). Taken together, these studies indicate that proper regulation of Mcl-1 levels is pivotal to the success of many developmental processes, as well as of the various defense responses of the host system.
Cytokine stimulation of Mcl-1 expression is rapid but transient (6). This is mainly because Mcl-1 proteins and mRNA are both extremely labile and are quickly degraded after the initial synthesis step (6, 34). The extremely labile nature of the Mcl-1 protein itself and the fact that the Mcl-1 level is a critical determinant in various cell survival pathways suggest that cellular processes that regulate Mcl-1 stability are as important as those that regulate Mcl-1 synthesis. Although transcriptional stimulation of Mcl-1 synthesis in response to various stimuli has been well documented (6, 30, 31; see also reference 7 for a recent review), the regulation of Mcl-1 stability has been hardly explored.
Translationally controlled tumor protein (TCTP) has been identified in a wide range of eukaryotes, including fungus, yeast, insects, plants, and mammals (3, 20, 28, 36). The extremely high degree of sequence conservation during evolution suggests that TCTP plays an essential role in the development of various organisms (28). Although TCTP was implicated in the human allergic response (20), apoptosis regulation (15), and various other growth-related functions (9, 29, 35), the biological roles of this protein remain elusive.
In the present study, by using a yeast two-hybrid approach, we identified that TCTP was one cellular factor that interacted with Mcl-1, a result consistent with that recently reported by Zhang et al. (37). However, in contrast to their conclusion that Mcl-1 acts as a chaperone of TCTP (fortilin) (37), we found that TCTP bound to Mcl-1 and modulated the stability and antiapoptotic activity of Mcl-1. Furthermore, we demonstrated that TCTP stabilized Mcl-1 via interfering with Mcl-1's degradation by the ubiquitin-dependent proteasome degradation pathway. The role of TCTP in modulating Mcl-1's stability and antiapoptotic activity is discussed.
MATERIALS AND METHODS
Yeast two-hybrid screen.
PBTM116mMcl-1ΔC31 and PBTM116hMcl-1ΔC27 are two yeast expression vectors that could direct the synthesis of a fusion protein containing the LexA-binding domain and the mouse or human Mcl-1 protein without the C-terminal transmembrane domain (mMcl-1ΔC31 and hMcl-1ΔC27). These two vectors were used as bait to screen human lymphocyte and mouse embryonic day 7 cDNA libraries according to the manufacturer's instructions (Clontech). All positive clones were processed to recover the plasmid containing the library cDNAs, and the recovered plasmids were reintroduced into the yeast strain L40 transformed with the bait vector to confirm their positivity before their sequence was analyzed by the standard method.
Antibodies.
Mouse Mcl-1-specific polyclonal antibodies were generated in rabbits by using bacterially produced, histidine-tagged mMcl-1ΔC31 protein. TCTP-specific polyclonal antibodies were generated in guinea pigs by using recombinant TCTPΔN10. For most immunoprecipitation and immunoblotting analyses, partially purified rabbit or guinea pig sera were used. For immunocolocalization experiments, both TCTP and mMcl-1 antibodies were first affinity purified by using specific antigens cross-linked to CNBr-activated Sepharose (Amersham). Other antibodies used in the present study include those specific to hMcl-1, Bcl-2, Bax, ubiquitin, and green fluorescent protein (GFP; all from Santa Cruz Biotechnology); p53 (a gift from Young-Sun Lin, IBMS, Academia Sinica, Taipei, Taiwan); and α-tubulin (Sigma).
Cells.
NIH 3T3 is a mouse fibroblast cell line and was purchased from the American Type Culture Collection. CHOP is a Chinese hamster ovary cell line stably transfected with the polyomavirus large T antigen (9a) and was kindly provided by James W. Dennis (Mt. Sinai Hospital, Toronto, Ontario, Canada). Ba/F3 is a murine interleukin-3 (IL-3)-dependent pro-B-call line, and Ba/F3/hMcl-1 cells are Ba/F3 derivatives stably overexpressing the human Mcl-1 protein as previously described (30). Ba/F3hMcl-1/TCTP cells (clones T-20 and T-23) are Ba/F3/hMcl-1 (clone #23H) derivatives stably overexpressing FLAG-TCTP, whereas the Ba/F3hMcl-1/vector cells (clones V-13 and V-14) are the parental Ba/F3/hMcl-1 cells (clone #23H) stably transfected with an empty vector. Ba/F3/mMcl #Wt-34 and #Wt-38 are two representative clones of Ba/F3 derivatives stably overexpressing the hemagglutinin (HA)-tagged wild-type mMcl-1 protein. Ba/F3/E206R, Ba/F3/K257V and Ba/F3/K260V are all Ba/F3 derivatives stably overexpressing the HA-tagged mMcl-1 mutants with a single amino acid mutation at the position as indicated in their names. All Ba/F3 derivatives were generated by transfection of cells with respective expression vectors by electroporation as previously described (6). To analyze the viability of the Ba/F3 derivatives after deprivation of IL-3, cells were seeded at a density of 105/ml in RPMI 1640 medium supplemented with 10% fetal bovine serum and 10 U of recombinant IL-3/ml. At 2 days after seeding, cells were washed three times with phosphate-buffered saline (PBS) and seeded in low-serum (0.5%) medium without IL-3. Cell viability was determined by the trypan blue exclusion analysis. In some cases, the annexin V staining kit (BioVision) was used to quantify the relative number of cells that had undergone apoptosis.
Expression vectors.
pCDNA3-HA-mMcl-1, pCDNA3-HA-hMcl-1, pCDNA3-BID, pCDNA3-Bad, pCDNA3-Bcl-2, pCDNA3-A1, pCDNA3-HA-AKT, pCDNA3-HA-PU.1, pCDNA-FLAG-Bim, and pCDNA3-p53 are mammalian expression vectors for expressing various proteins as indicated in the construct names. pFLAG-CMV2-TCTP was generated by PCR amplification of the coding region of the mouse TCTP cDNA with two primers (sense, 5′-CCCAAGCTTATCATCTACCGGGACCTCATCAGCCATGACGAGCTGTTCTCCGAC-3′;antisense, 5′-GATCGGATCCTTAACATTTCTCCATCTCTAAGCC-3′). The amplified fragments were restricted with HindIII and BamH1 and cloned into the corresponding sites of the pFLAG-CMV2 vector (Kodak). pGEX4T-1-TCTPΔN10 is a bacterial expression vector which directs the synthesis of a TCTP mutant (without the first 10 amino acids) C-terminally fused to the glutathione S-transferase (GST) protein. This vector was constructed by subcloning the EcoRI fragment of the positive clone obtained from the yeast two-hybrid screen (Fig. 1A, clone 1) into the corresponding sites of the pGEX4T-1 vector (Amersham-Pharmacia). pGEX 4T-1-mMcl-1 was constructed by subcloning the EcoRI fragment of the mouse Mcl-1 cDNA into the corresponding sites of the pGEX 4T-1 vector. The expression vectors encoding the HA-tagged, mMcl-1 mutants (E206R, T240V, K257V, and K260V) were constructed by standard PCR-assisted mutagenesis-coupled cloning methods. The template used for PCR amplification was pCDNA3-HA-mMcl-1. The first PCR was performed with the antisense primer (5′-ATCGGGCCCCTATCTTATTAGATATGCCAGACC-3′), together with one of the following primers (underlined nucleotides differ from the wild-type sequence): E206R (5′-GGCGTGCAGCGCAACCACCGGACGGCCTTCCAGGGCATG-3′), T240V (5′-GTTTTCAAAGATGGCGTAGTAAACTGGGGCAGGATTGTG 3′), K257V (5′-TTCGGTGCCTTTGTGGCCGTACACTTAAAGAGCGTAAAC-3′), and K260V (5′-TTTGTGGCCAAACACTTAGTGAGCGTAAACCAAGAAAGC-3′). The second PCR was performed with the sense primer (5′-TACCGCCAGTCGCTGGAG-3′) and the PCR fragment obtained from the first PCR. The resultant Eco47III/ApaI fragment was used to replace the corresponding region of the wild-type mMcl-1 coding sequence within the pCDNA3-HA-mMcl-1 vector. All mutated nucleotides were confirmed by sequencing.
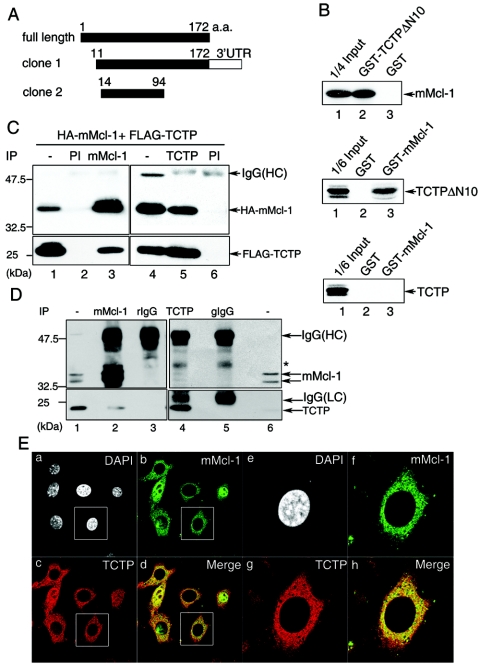
FIG. 1.
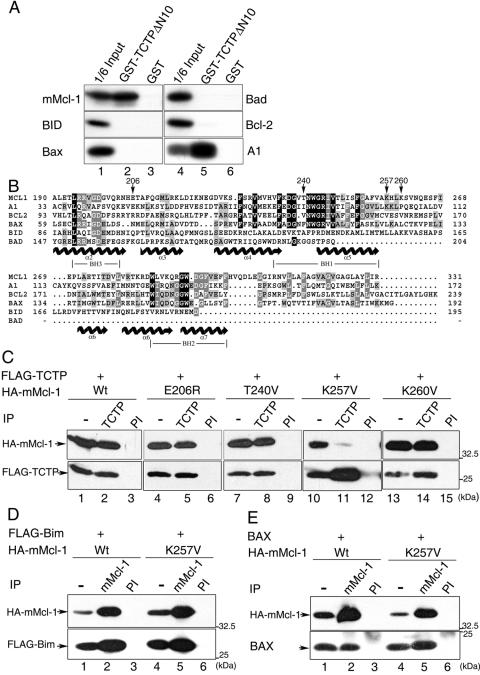
Mcl-1 interacts with TCTP. (A) Schematic representation of two TCTP cDNA-containing yeast clones fished out by the yeast two-hybrid screen with mMcl-1 ΔC31 as bait. (B) GST pull-down assays. Bacterially produced GST, GST-TCTPΔN10, or GST-mMcl-1 immobilized on glutathione beads were incubated with in vitro-synthesized [35S]methionine-labeled mMcl-1 (upper panel), TCTPΔN10 (middle panel), or TCTP (lower panel). Bound proteins and 1/6 or 1/4 of those added to the binding assay as indicated (input) were resolved by SDS-PAGE and specific signals revealed by fluorography. (C) Co-IP of TCTP and mMcl-1. Cell lysates from CHOP cells transiently overexpressing HA-Mcl-1 and FLAG-TCTP were immunoprecipitated with anti-mMcl-1 (lane 3), anti-TCTP (lane 5), or preimmune sera (PI, lanes 2 and 6). The cell lysates (lanes 1 and 4, 1/20 of those used in co-IP) and the immune complexes were then analyzed by immunoblotting with anti-HA (upper panel) or anti-FLAG (lower panel) antibodies. (D) In vivo interaction of mMcl-1 and TCTP. The co-IP experiment was carried out with NIH 3T3 cell lysates and affinity-purified anti-mMcl-1 (lane 2), anti-TCTP (lane 4), or control rabbit IgG (rIgG, lane 3) or guinea pig IgG (gIgG, lane 5). The immunoblotting following the co-IP step was sequentially probed with anti-mMcl-1 (upper panel) and anti-TCTP (lower panel) antibody. One five-hundredth of cell lysates used in co-IP was loaded in lanes 1 and 6 for direct immunoblotting analysis. A band marked by an asterisk indicates a nonspecific protein detected in this immunoprecipitation-Western blotting experiment. (E) Partial colocalization of Mcl-1 and TCTP. NIH 3T3 cells were stained simultaneously with antibodies specific to mMcl-1 (green) and TCTP (red), and the specific signals were examined by indirect immunofluorescence and confocal microscopy. Nucleus was counterstained by DAPI. Images of higher magnification of the cell marked in (a to d) are shown on the right (e to h). Control staining with primary antibodies preblocked with specific antigens did not show any signals (data not shown).
In vitro GST pull-down assay.
Bacterially produced GST, GST-TCTPΔN10, GST-TCTP, or GST-mMcl-1 fusion proteins were immobilized on glutathione-Sepharose 4B beads (Pharmacia) and then incubated for 2 h at 4°C with in vitro-synthesized, [35S]methionine-labeled proteins as indicated in Fig. 1B. After three extensive washes with the bead binding buffer (50 mM potassium phosphate [pH 7.5], 150 mM KCl, 10 mM MgCl2, 10% [vol/vol] glycerol, 1% [vol/vol] Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, 10 μg of aprotinin/ml), the bound proteins were resolved on a sodium dodecyl sulfate (SDS)-containing 10% polyacrylamide gel and visualized by fluorography.
Coimmunoprecipitation (co-IP) of TCTP and mMcl-1.
CHOP cells were transiently transfected with wild-type or mutant mMcl-1 mammalian expression vectors, together with pFLAG-CMV2-TCTP, by the liposome-mediated gene transfer method as previously described (17). At 18 h after transfection, cells were lysed with an ice-cold lysis buffer (10 mM HEPES [pH 7.2], 142.5 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.2%NP-40, 1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, and 10 μg of aprotinin/ml). Precleared cell lysates were then immunoprecipitated with partially purified antiserum specific to TCTP, mMcl-1, or the corresponding preimmune serum. The immunoprecipitated protein complexes were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to the polyvinylidene difluoride membrane (Millipore), and the coimmunoprecipitated proteins were analyzed by immunoblotting with anti-HA (Boehringer Mannheim), anti-FLAG M2 (Sigma), or other antibodies as indicated in the figures. After probing with an appropriate horseradish peroxidase-conjugated secondary antibody, specific signals on the membrane were visualized by using an enhanced chemiluminescence system (Amersham) according to the manufacturer's instructions. To analyze the interaction between endogenous TCTP and Mcl-1, the procedure described above was used except that the NIH 3T3 cell lysates were used in the assay.
Indirect immunofluorescence and confocal microscopy.
To determine the cellular localization of Mcl-1 and TCTP, NIH 3T3 cells, cultured on coverslips, were fixed in 2% paraformaldehyde in PBS for 15 min. After a brief rinse with PBS at room temperature, cells were blocked and permeabilized in PBS containing 2% goat serum, 0.1% saponin, and 0.02% NaN3 for 15 min. The cells were then incubated at room temperature with affinity-purified rabbit anti-mouse Mcl-1 and guinea pig anti-mouse TCTP antibodies (5 μg/ml each in PBS containing 3% bovine serum albumin). Two hours after incubation, cells were washed with PBS and then incubated with fluorescein-conjugated goat anti-rabbit and Alexa Fluor 568-conjugated goat anti-guinea pig immunoglobulin G (IgG; Molecular Probes) for an additional 1 h at room temperature. After three washes with PBS, stained cells were analyzed with confocal fluorescence microscopy (Zeiss type LSM-5). To show the specificity of the stained signals, cells to be analyzed were processed in an identical way except that the primary antibody used in the staining procedure was preblocked with specific antigen produced in bacterial cells.
Knockdown of TCTP expression with small interfering RNA (siRNA).
To knock down TCTP expression in the human cell lines, two TCTP siRNA-expressing vectors (siTCTP#1 and siTCTP#2) were constructed by using the pSUPER-neo+gfp vector (OligoEngine, Inc.). These two constructs were designed according to the manufacturer's protocol in such a way that a duplex siRNA corresponding to the following two regions of the human TCTP gene 5′-AAGGTACCGAAAGCACAGT-3′(siTCTP#1) or 5′-AACCATCACCTGCAGGAAA-3′ (siTCTP#2) could be synthesized inside mammalian cells transfected with these vectors (4).
RESULTS
Interaction between Mcl-1 and TCTP.
We sought here initially to identify cellular factors that might bind to Mcl-1 and modulate Mcl-1's activity. By using the yeast two-hybrid approach with Mcl-1 as bait (see Materials and Methods), several candidate molecules were identified. Two of these candidates turned out to be Bcl-2 family members (Bax and Bik) that were already reported to be able to interact with Mcl-1 in either mammalian or yeast cells (9b, 32). We first characterized, among many other candidates that were identified in our two-hybrid screen, the clone that encodes the mouse homolog of the human TCTP protein. Sequence analysis revealed that we had selected out two independent groups of TCTP cDNA clones. One would encode a truncated protein from amino acids 11 to 172 and the other from amino acids 14 to 94 (Fig. 1A).
We next carried out the following experiments to validate the specific interaction between Mcl-1 and TCTP. First, the in vitro GST pull-down assay was performed. As shown in Fig. 1B, the [35S]methionine-labeled Mcl-1 protein synthesized by the in vitro transcription-translation system was efficiently and specifically pulled down by the GST-TCTPΔN10 fusion protein (Fig. 1B, upper panel, lane 2). The observed in vitro binding activity of this fusion protein to Mcl-1 was specifically mediated by the TCTP moiety, since Mcl-1 was not pulled down by the GST protein alone. (Fig. 1B, upper panel, lane 3). The specific in vitro interaction between Mcl-1 and TCTP was also observed with the GST-Mcl-1 fusion protein and [35S]methionine-labeled, in vitro-synthesized TCTPΔN10 (Fig. 1B, middle panel, lane 3). Interestingly, in this in vitro binding assay, unlike TCTPΔN10, the full-length TCTP protein did not manifest any significant binding to Mcl-1 (compare the middle and the bottom panels of Fig. 1B, lanes 3).
Next, the in vivo interaction between Mcl-1 and TCTP was examined by a co-IP assay. In this assay, CHOP cells were transiently transfected with expression vectors encoding HA-tagged mMcl-1 (HA-mMcl-1) and FLAG-tagged full-length TCTP (FLAG-TCTP). As shown in Fig. 1C, immunoprecipitation of cell lysates containing HA-mMcl-1 and FLAG-TCTP (see Fig. 1C, lanes 1 and 4, for protein expression) with anti-mMcl-1, but not the preimmune serum, coimmunoprecipitated FLAG-TCTP as revealed by immunoblotting with anti-FLAG antibody (Fig. 1C, compare lanes 2 and 3). In a reciprocal experiment, the TCTP antibody also specifically coimmunoprecipitated HA-mMcl-1 (Fig. 1C, lane 5). Next, we examined whether the interaction between Mcl-1 and TCTP could be observed at endogenous levels of both proteins. For this experiment, co-IP assays with lysates from NIH 3T3 cells were performed. As shown in Fig. 1D, the purified mMcl-1 antibody, but not control rabbit IgG, coimmunoprecipitated endogenous TCTP (Fig. 1D, compare lanes 2 and 3). Also, in a reciprocal co-IP experiment, the mMcl-1 protein was found to be present in the immune complexes pulled down by the TCTP but not by the control guinea pig IgG (Fig. 1D, compare lanes 4 and 5). Lastly, the in vivo interaction between endogenous mMcl-1 and TCTP was further supported by an immunocolocalization experiment with affinity-purified antibodies specific to mMcl-1 and TCTP. As shown in Fig. 1E, the mMcl-1 protein is predominantly localized in the cytoplasm with a punctate staining structure. However, in some cells it is also found localized in the nucleus (Fig. 1Eb). Interestingly, no matter where mMcl-1 resides, the TCTP protein is only partially colocalized with mMcl-1, with some cells more prominent than others (Fig. 1Eb to d and f to h). This partial colocalization result is consistent with the low co-IP efficiency of the endogenous proteins as observed in Fig. 1D (compare Mcl-1 bands, shown in lanes 1 and 6, to those shown in lanes 2 and 4). Taken together, these results demonstrate that, in contrast to the negative interaction as observed in the GST pull-down assay (Fig. 1B, the bottom panel), full-length TCTP and mMcl-1 did interact with each other in vivo (Fig. 1C and D), albeit probably in a cell state-specific manner (see Discussion). Our results also suggest that additional cellular protein(s) might potentially be involved in mediating the TCTP and Mcl-1 interaction inside the cells.
TCTP enhances the protein stability of Mcl-1 but not vice versa.
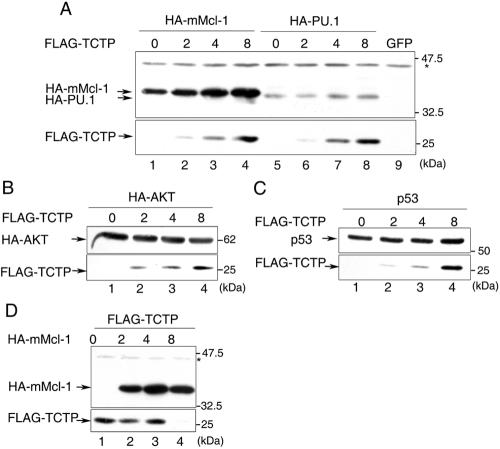
While performing the transient cotransfection experiments for analysis of the in vivo interaction between Mcl-1 and TCTP (Fig. 1C), we noticed that as we increased the amounts of TCTP-encoding plasmids in the transfection experiment, the protein levels of coexpressed HA-Mcl-1 elevated in a dose-dependent manner, despite the fact that equal amounts of HA-Mcl-1-expressing plasmids were used (Fig. 2A, compare lanes 1 to 4). Such TCTP-mediated enhancing effect toward intracellular Mcl-1 levels was specific, since levels of other examined proteins, such as HA-PU.1 (Fig. 2A, lanes 5 to 8), HA-AKT (Fig. 2B), and p53 (Fig. 2C), remained unchanged or marginally affected under identical experimental conditions. Conversely, in a reciprocal experiment with increasing amounts of the HA-mMcl-1 expression vector, we failed to observe any enhanced expression of coexpressed FLAG-TCTP (Fig. 2D). At higher dosages, overexpression of HA-Mcl-1 even inhibited the expression of coexpressed FLAG-TCTP (Fig. 2D, lane 4). Hence, this modulatory effect on protein levels seems to be unidirectional.
FIG. 2.
Coexpression of TCTP specifically augments the expression levels of Mcl-1 but not vice versa. (A to C) 293T cells were transiently transfected with expression vectors encoding HA-mMcl-1, HA-PU.1, HA-AKT, or P53 along with increasing amounts (0, 2, 4, or 8 μg) of the FLAG-TCTP expression vector as indicated on the top of each panel. After transfection, cell lysates were analyzed by immunoblotting with HA (upper parts of panels A and B), FLAG (lower parts of panels A to C), or p53 antibodies (upper part of panel C). (D) Same as in panels A to C, except that cells were transfected with the FLAG-TCTP expression vector, along with increasing amounts (0, 2, 4, and 8 μg) of vectors encoding HA-mMcl-1. The asterisk points to an unknown protein cross-reacted with the anti-HA antibody. This band served as a loading control for each sample.
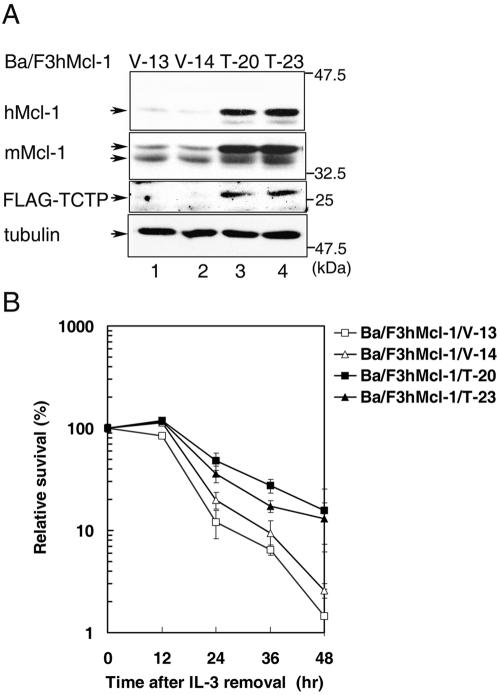
Next, we examined whether overexpression of TCTP would also affect the expression of endogenous Mcl-1 in a stable cell line system. To address this issue, Ba/F3 derivatives stably overexpressing human Mcl-1 (hMcl-1) was further engineered to overexpress FLAG-TCTP (see Materials and Methods). As shown in Fig. 3A, in comparison to cells transfected with a control vector (V-13 and V-14), overexpression of FLAG-TCTP in two independent subclones (Ba/F3hMcl-1/T-20 and Ba/F3hMcl-1/T-23) led to elevated steady-state levels of both endogenous mouse Mcl-1 and that of the ectopically overexpressed hMcl-1 proteins (compare lanes 3 and 4 to lanes 1 and 2). We next evaluated the functional consequence of the TCTP-mediated accumulation of intracellular Mcl-1 pool. As illustrated in Fig. 3B, cells of the two control clones (V-13 and V-14) were significantly more susceptible to IL-3-deprivation-induced cell death than those of the TCTP-overexpressing lines (T-20 and T-23). We have previously demonstrated that overexpression of Mcl-1 could significantly delay IL-3-removal-induced cell death (6, 30). Our present results therefore suggest that ectopic overexpression of TCTP in Ba/F3 cell lines results in the elevation of the steady-state levels of Mcl-1 and consequently the augmentation of Mcl-1's antiapoptotic effect.
FIG. 3.
(A) Overexpression of TCTP elevated Mcl-1 protein levels. Control (clones V-13 and V-14) or TCTP overexpressing Ba/F3hMcl-1 cells (clones T-20 and T-23) under normal growth conditions were lysed, and cell lysates were analyzed by immunoblotting with anti-FLAG, anti-hMcl-1, or anti-mMcl-1 antibodies to detect the ectopically overexpressed proteins (hMcl-1 and FLAG-TCTP) or endogenous mMcl-1, respectively. The same blot was also probed with anti-α-tubulin antibody to monitor the amount of proteins loaded for each sample. (B) TCTP delays IL-3-removal-induced cell death. IL-3 was removed from the culture of the indicated cell clones for various times, and the numbers of viable cells at each time points were determined by trypan blue staining. The percent survival relative to those seeded is plotted against hours after IL-3 removal. The results shown are means (± the standard deviation) of three independent experiments with very similar results.
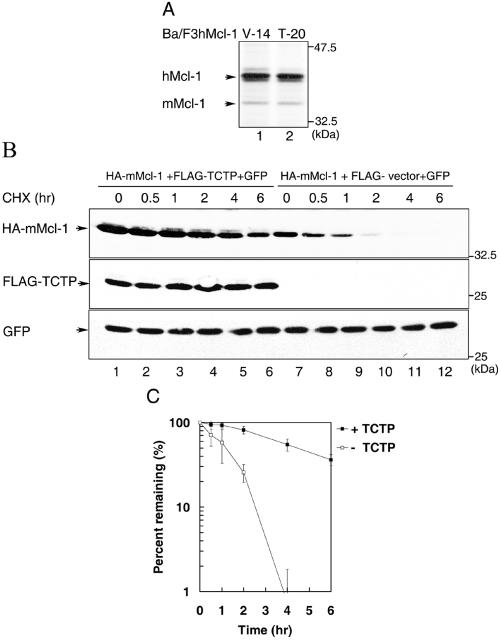
TCTP-mediated elevation of Mcl-1 protein levels observed in Fig. 2A and 3A could be either due to enhanced protein synthesis or inhibition of protein degradation or both. To distinguish these possibilities, we first carried out a pulse-labeling experiment to examine whether TCTP would enhance synthesis of Mcl-1. In this experiment, control Ba/F3hMcl-1 cells (clone V-14) or those stably overexpressing TCTP (clone T-20) were pulse-labeled with [35S]methionine for 30 min before the cell lysates were analyzed by immunoprecipitation with anti-mouse Mcl-1 antibody, which also cross-reacts with the human homolog of Mcl-1. As shown in Fig. 4A, although the steady-state levels of Mcl-1 (mouse or human) was elevated in the TCTP-overexpressing lines (Fig. 3A), the synthesis rate of Mcl-1 in these cells as reflected from the amounts of 35S-labeled band was not significantly different from those cells transfected with the empty expression vector (Fig. 4A, compare lanes 1 and 2). These results indicate that TCTP does not increase Mcl-1's steady-state levels via enhancing Mcl-1's protein synthesis rate. We next examined whether this modulation effect was at the protein stability level. To address this issue, CHOP cells were transiently transfected with HA-mMcl-1 in combination with FLAG-TCTP (Fig. 4B, lanes 1 to 6) or control expression vectors (Fig. 4B, lanes 7 to 12). A GFP expression vector was also included in this assay to monitor the transfection efficiency among samples. At 18 h after transfection, transfected cells were treated with cycloheximide to inhibit any further protein synthesis. At different time points after cycloheximide treatment, cell lysates were prepared and analyzed by immunoblotting with antibodies specific to the HA-tag (for detection of HA-mMcl-1), the FLAG-tag (for detection of FLAG-TCTP), or GFP. As shown in Fig. 4B and C, under such experimental conditions, the Mcl-1 protein in cells overexpressing TCTP had a half-life of ca. 4.5 h, which was much longer than those in cells transfected with an empty expression vector (∼1.5 h). Taken together, these results indicate that TCTP enhances the accumulation of Mcl-1 protein levels through increasing the protein stability of Mcl-1.
FIG. 4.
TCTP-enhanced expression of Mcl-1 is mediated through an increase in protein stability. (A) TCTP does not augment Mcl-1's protein synthesis rate. Control (clone V-14) or Ba/F3hMcl-1 cells stably overexpressing FLAG-TCTP (clone T-20) were pulse-labeled with [35S]methionine for 30 min before the cell lysates were harvested and immunoprecipitated with anti-mouse Mcl-1 antibody (which also immunoprecipitates the human homolog of Mcl-1). The immune complexes were resolved by SDS-PAGE, and specific bands visualized by fluorography. (B) CHOP cells were transiently transfected with the HA-mMcl-1 expression vector along with a vector expressing FLAG-TCTP (lanes 1 to 6) or an empty vector (FLAG-vector, lanes 7 to 12). To monitor the transfection efficiency among different samples, a GFP expression vector was also included in each transfection mixture. At 18 h after transfection, cells were treated with cycloheximide for various times as indicated before cell lysates were prepared and analyzed by sequential immunoblotting with HA, FLAG, or GFP antibodies. Specific bands are as indicated. Shown here is one representative result from experiments that were repeated three times with very similar results. The Mcl-1-specific bands were quantified with a luminescent image analyzer (LAS-1000plus) and the level of Mcl-1 at each time point was converted to the percentage of Mcl-1 level at time zero and plotted to determine the protein turnover rate. The Mcl-1 degradation kinetics determined from three independent experiments was plotted in panel C.
Knockdown expression of TCTP destabilizes Mcl-1.
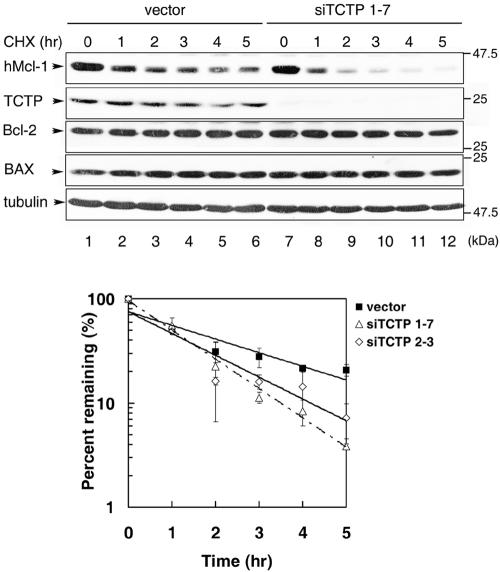
Ectopic overexpression of TCTP enhanced the protein stability of Mcl-1. We next examined whether decreased expression of TCTP would destabilize Mcl-1. To address this issue, we established a few stable HeLa cell lines whose TCTP levels were markedly downregulated by siRNA specific to the human TCTP gene (see Materials and Methods). The stability of Mcl-1 in all established TCTP knockdown lines were very similar, and the results of two representative clones are demonstrated in Fig. 5. As shown in this figure, after treatment of the cells with cycloheximide, the endogenous hMcl-1 levels decreased with a half-life of ca. 45 min in both TCTP knockdown lines, siTCTP1-7 and siTCTP2-3, which was significantly shorter than the half-life observed for the control cells (∼1.5 h). In contrast, under the same experimental conditions, the protein levels of two other TCTP noninteracting Bcl-2 family proteins (Bcl-2 and Bax, see Fig. 6A), and α-tubulin remained rather constant both in the control and the TCTP knockdown lines. Taken together, these results indicate that the stability of Mcl-1 is indeed specifically influenced by the cellular levels of TCTP.
FIG. 5.
Knockdown of TCTP destabilizes Mcl-1. TCTP knockdown (lines siTCTP1-7 and siTCTP2-3) or control cells (vector) were treated with cycloheximide (30 μg/ml) for various times as indicated before total cell lysates were prepared. Levels of Mcl-1, as well as other Bcl-2 family members, TCTP and α-tubulin (served as a protein loading control) in the cell lysates were then analyzed by immunoblotting with specific antibodies as indicated. The Mcl-1 specific signals at each time point were quantified and plotted against hours of treatment as that described in the legend to Fig. 4. Very similar results were observed for four TCTP knockdown lines. The top panel shows one representative result of the immunoblotting analysis. The bottom panel shows the turnover kinetics from three independent experiments with cell clones as indicated.
FIG. 6.
The Lys257 residue is critical for Mcl-1 to interact with TCTP. (A) The in vitro-synthesized, [35S]methionine-labeled Bcl-2 family members as indicated were subjected to GST pull-down assays with immobilized GST-TCTPΔN10 or GST. One-sixth of the input proteins added to the binding assay (lanes 1 and 4) were also analyzed by SDS-PAGE and visualized by fluorography. (B) Sequence alignment of selected Bcl-2 family members. Locations of the BH1, BH2, and BH3 domains are indicated. Conserved amino acid residues are highlighted in black or gray blocks, depending on their degree of conservation. Secondary structure (α-helix) assignments predicted from the Bcl-xL crystal structure (23) are shown below the aligned sequences with helical symbols. Residues subjected to mutagenesis in the present study are indicated by arrows. (C) Co-IP of TCTP and various mMcl-1 mutants. Cell lysates from CHOP cells transiently overexpressing FLAG-TCTP and wild-type or mutant mMcl-1 (all are HA tagged) was immunoprecipitated with TCTP or preimmune (PI) serum. The immune complexes were then analyzed by immunoblotting with anti-HA or anti-FLAG antibody. (D and E) Co-IP of FLAG-Bim (or Bax) and the wild-type or K257V mutant forms of mMcl-1. The experiment shown here is the same as in panel C except the CHOP cells were transfected with vectors overexpressing FLAG-Bim (or Bax) and the wild-type or K257V mutant of mMcl-1 and the cell lysates were immunoprecipitated with preimmune (lanes 3 and 6) or anti-mMcl-1 antibody (lanes 2 and 5). The subsequent immunoblotting analysis was with HA, FLAG, or Bax antibodies. Specific bands are as indicated.
The TCTP binding-defective Mcl-1 mutant is much less stable and manifests a compromised antiapoptotic activity.
We next sought to map the TCTP-interacting domain of Mcl-1. For this purpose, we initially constructed a few mammalian expression vectors that would direct the synthesis of Mcl-1 mutants with deletion of various N, C, or internal domains. However, most of these mutants, except for the two with deletions of ca. 30 amino acids at the N or C terminus, were expressed at a level that was too low to be detected in our co-IP experiment (data not shown). We therefore sought a different approach. We first examined whether TCTP also interacted with other Bcl-2 family members. Using GST pull-down assays, we demonstrated that among the Bcl-2 family proteins tested as indicated in Fig. 6A only A1 interacted with TCTP. The inability of BID, Bax, Bad, and Bcl-2 to associate with TCTP indicated that the interaction between TCTP and Mcl-1 (as well as A1) was unlikely to be mediated through the highly conserved Bcl-2 homology (BH) domains present in Mcl-1 (or A1). We therefore reasoned that there must be unique sequence information shared by Mcl-1 and A1 that served as a critical structural determinant in mediating the interaction with TCTP. To help identify TCTP-interacting domain in Mcl-1, we first made a sequence alignment analysis on Mcl-1, A1, and other TCTP noninteracting Bcl-2 members (Fig. 6B). We then mutated two candidate sites (i.e., E206 and K257 in Mcl-1) that met the following two criteria. First, the primary amino acid sequence was distinctly conserved between Mcl-1 and A1 but not among other Bcl-2 family proteins analyzed here. Second, the position of the candidate site was within a loop region, a rather relaxed structure that was more likely to be accessible for protein-protein interaction. As controls, we also mutated a few other neighboring sites that did not meet either or both of the aforementioned criteria (e.g., T240 and K260). Although mutation of E206 and the two sites selected as controls (i.e., T240 and K260) did not affect Mcl-1's binding affinity to TCTP (Fig. 6C), mutation of K257 to a valine residue (the same residue found in Bcl-2) markedly decreased the TCTP binding affinity of Mcl-1 (Fig. 6C, lane 11). Notably, the K257V mutant still retained the ability to interact with Bim and Bax, two known Mcl-1 interacting proteins (25, 32) (Fig. 6D and E, compare lanes 2 and 5). This result further suggests that mutation of K257 into a valine residue does not exert a global conformational change on the Mcl-1 protein itself, and therefore this residue is likely to reside in an important TCTP-interacting domain.
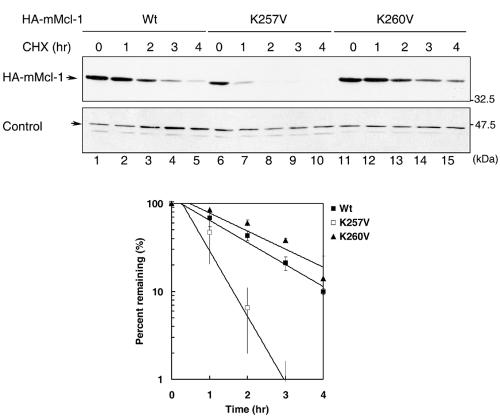
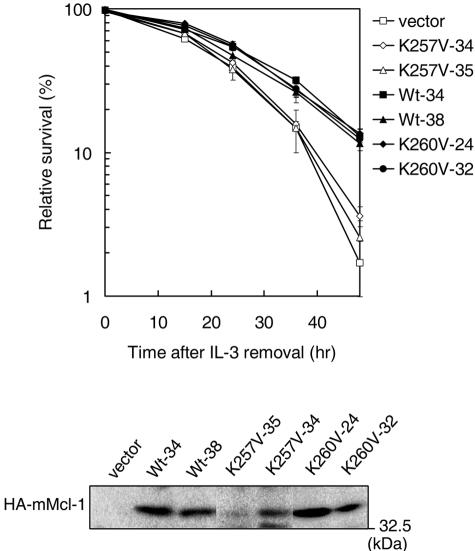
We next compared the protein stability of the K257V mutant to that of the wild-type or other Mcl-1 mutants whose TCTP binding activity was not significantly compromised. We noticed that the turnover rate of all TCTP binding-competent Mcl-1 mutants (only analysis of K260V was shown in Fig. 7) was comparable to that of the wild-type protein (t1/2, ∼1.5 h), whereas the TCTP binding-defective mutant K257V manifested a much shorter half-life (∼40 min). Interestingly, only the antiapoptotic activity of the TCTP binding-defective mutant (i.e., K257V) and not that of other TCTP binding-competent mutants was significantly compromised (Fig. 8 and data not shown for the T240V and E206R mutants). Taken together, these results indicate that the stability and the antiapoptotic activity of Mcl-1 strongly correlate with Mcl-1's ability to interact with TCTP.
FIG. 7.
The TCTP-binding defective mutant K257V is much less stable than the wild-type or the K260V mutant. 293T cells were transfected with plasmids encoding the wild-type, K257V, or K260V mutant forms of mMcl-1. Transfected cells were then treated with cycloheximide for 0 to 4 h before cell lysates were harvested and analyzed by immunoblotting with anti-HA antibody. The band labeled with “control” is an unknown protein cross-reacted with the anti-HA antibody. This band served as a loading control for cell lysates analyzed. The bottom panel is the quantification analysis of three independent experiments as that described in the legend to Fig. 4.
FIG. 8.
The antiapoptotic activity of the TCTP-binding defective mutant K257V is compromised. Ba/F3 stable clones overexpressing wild-type or mutant Mcl-1 (two independent clones of each) were examined for cell survival after IL-3 removal. Cells undergoing cell death was quantified by annexin V staining. The top panel shows the relative cell survival of various clones as indicated after IL-3 removal. The immunoblot on the bottom shows the protein levels of ectopically expressed wild type or mutant Mcl-1 (K257V or K260V).
TCTP blocks ubiquitination of Mcl-1.
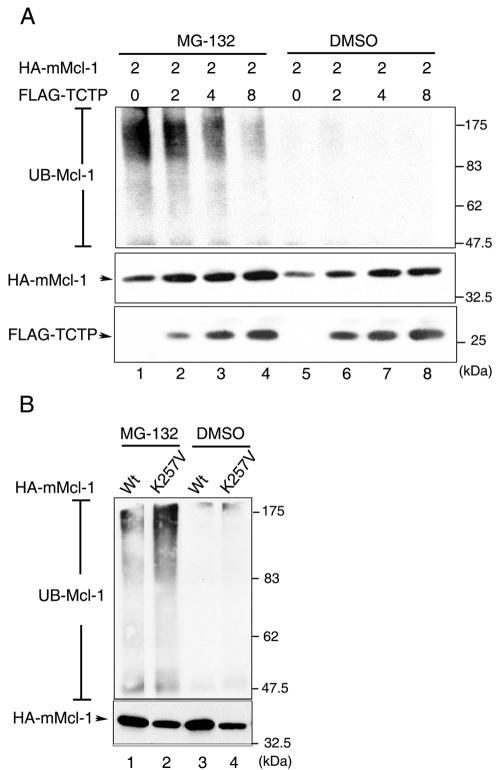
Mcl-1 is eliminated by the ubiquitin-dependent proteasome degradation pathway (24). We next examined whether TCTP plays any antagonistic role in Mcl-1's degradation process. As shown in Fig. 9, in the presence of the proteasome inhibitor MG132 the ubiquitinated form of Mcl-1 (UB-Mcl-1) could be detected in cells transiently transfected with the HA-mMcl-1 expression vector (Fig. 9A, lane 1). Notably, when cells were cotransfected with increasing amounts of the TCTP expression vector the extent of ubiquination of Mcl-1 diminished in a dose-dependent manner (Fig. 9A, compare lanes 1 to 4). Furthermore, we noticed that the TCTP binding-defective mutant K257V was much more susceptible to ubiquination than the wild-type Mcl-1 protein (Fig. 9B). Taken together, these results indicate that the susceptibility of Mcl-1 to undergo proteasome-dependent degradation is in a reverse relationship to its ability to interact with TCTP.
FIG. 9.
Overexpression of TCTP blocks Mcl-1 from undergoing ubiquitination. (A) CHOP cells were transiently transfected with the HA-mMcl-1 expression vector along with increasing amounts of the vector encoding FLAG-TCTP. At 16 h after transfection, cells were treated with MG-132 (2 μM, lanes 1 to 4) or dimethyl sulfoxide (vehicle control) (lanes 5 to 8) for 4 h before cell lysates were prepared and immunoprecipitated with anti-mMcl-1 antibody. The immune complex was then analyzed by immunoblotting with anti-ubiquitin (UB) antibody (top panel). The ubiquitinated form of mMcl-1 is as indicated (UB-Mcl-1). Direct immunoblotting analysis of the corresponding cell lysates (1/20 of those used in the top panel) with anti-HA and anti-FLAG antibodies are shown on the middle and the bottom panels, respectively. (B) The K257V mutant is more susceptible to undergoing ubiquitination. CHOP cells transiently overexpressing the HA-tagged wild-type (lanes 1 and 3) or K257V mutant (lanes 2 and 4) forms of mMcl-1 were treated and analyzed as described in panel A. The ubiquitinated form of mMcl-1 (wild-type or mutant) is as indicated. The lower panel is a direct immunoblotting analysis of cell lysates (1/20 of that used in the upper panel) with anti-HA antibody.
DISCUSSION
Mcl-1 is a labile protein with a half-life of ca. 30 to 40 min when measured by the pulse-chase analysis (6, 24) and ca. 1.5 h when measured by the cycloheximide blocking method (Fig. 4B, 5, and 7). The discrepancy between these two measurements suggests that either Mcl-1's degradation involves another labile protein whose synthesis is sensitive to cycloheximide treatment or Mcl-1 undergoes posttranslational modification(s), and the rate of disappearance of the newly synthesized protein (revealed by the pulse-chase analysis) is different from the actual half-life of the mature protein (revealed by the cycloheximide-blocking method). Although the involvement of another labile protein in the Mcl-1's degradation pathway has not been excluded, the identification of three forms of Mcl-1 with different protein half-lives (1 to 3 h for the upper two bands and undetermined for the lower one; see reference 34) favors the possibility that the newly synthesized and the mature forms of Mcl-1 have different turnover rates. In the results presented here, most endogenous Mcl-1 protein bands were not properly resolved to reveal the three bands (in most cases, we only observe two bands as that shown in Fig. 1D and 3A). Although the identity of these multiple forms of Mcl-1 remains to be determined, our preliminary observation seems to indicate that TCTP exerts a differential effect on the turnover rates of these different forms of Mcl-1. More experiments are required to investigate this issue.
Given the fact that overexpression of TCTP interferes with Mcl-1's degradation at the ubiquitination step and that the TCTP-binding defective mutant of Mcl-1 (K257V) is much more susceptible to ubiquitination and manifests a compromised antiapoptotic activity, we propose the following two models. First, TCTP may serve as a molecular chaperone of Mcl-1. Without TCTP, the newly synthesized Mcl-1 somehow cannot be properly processed to fold into a mature and functional protein and therefore gets degraded faster. While our work was in progress, we noticed that Zhang et al. reported an interaction between Mcl-1 and TCTP (fortilin) (37). In that study, the authors demonstrated that TCTP was destabilized when the Mcl-1 protein levels were knocked down by RNAi. The authors further suggested that Mcl-1 worked as a chaperone of TCTP and stabilized the latter protein, a result that was seemingly opposite to our observations. Under our experimental conditions, we noticed that TCTP was much more stable (t1/2 > 6 h) than Mcl-1. If Mcl-1 indeed functions to stabilize TCTP as suggested by Zhang et al. (37), it means that the cells would have to constantly stimulate the synthesis of the labile Mcl-1 protein so as to maintain the stability of a much more stable protein, such as TCTP, a cellular condition that does not seem to be logically favorable. Based on this argument and the recent structure study where a chaperone function was proposed for the yeast homolog of TCTP (28), TCTP is more likely to serve as a molecular chaperone of Mcl-1, if a chaperone function is indeed involved in the interaction between Mcl-1 and TCTP.
Alternatively, the functional outcome of Mcl-1 and TCTP interaction can be interpreted by a second model, in which TCTP may be a cofactor of Mcl-1, i.e., Mcl-1 carries out its biological activity only when it forms a functional complex with TCTP. In this model, the highly stable nature of the TCTP protein suggests that its interaction with Mcl-1 is probably dependent on a posttranslational modification of TCTP and possibly of Mcl-1 as well. TCTP was shown to be phosphorylated by the mitotic polo-like kinase Plk (35). Interestingly, overexpression of the Plk phosphorylation site-deficient mutant of TCTP (AA-TCTP) triggered cells to undergo multinucleation or cell death (35). It would be interesting to test whether the AA-TCTP mutant retains the ability to interact with Mcl-1 and modulates Mcl-1's antiapoptotic activity. If the second model is correct, the observed protection of Mcl-1 by TCTP from ubiquitination (Fig. 9A) is probably due to masking of the ubiquitination sites on Mcl-1 when TCTP forms a functional complex with Mcl-1. Under such conditions, this complex formation may also protect TCTP from degradation, a model that would be consistent with Zhang et al.'s finding that the Mcl-1 binding-defective mutant of TCTP is much more susceptible to degradation than the wild-type molecule (37). More experiments are certainly required to test these two models.
In addition to Mcl-1, we also identified a novel interaction between TCTP and another Bcl-2 family member, A1 (Fig. 6A). Although the functional significance of the latter interaction is not clear, we noticed that Mcl-1 and A1 share some molecular characteristics that are unique within the Bcl-2 family. First, they both are immediate-early genes activated by cytokines (6, 12, 16). Second, both mRNAs are extremely unstable (6, 21). It would be interesting to determine whether A1 is also a labile protein and whether TCTP also affects its stability and its antiapoptotic activity.
Growth-related functions have been reported for TCTP. However, the underlying mechanisms still remain largely uncharacterized (9). Recent studies have suggested that the cellular role of TCTP may be partly mediated by its association with factors involved in translation elongation or mitosis/cytokinesis (5, 35). The newly identified ability of TCTP in modulating the activity of the antiapoptotic protein Mcl-1 (and potentially other related factors) further strengthens TCTP's role in cell growth control. We noticed that, consistent with an indispensable role of TCTP in cell growth, some transfected cells (i.e., GFP-positive cells) underwent autophagy-like morphological changes and subsequently died in the course of selecting stable clones that overexpress the TCTP siRNA (data not shown). For stable clones that we were able to establish for subsequent analysis, we noticed that their TCTP levels were not completely knocked down (at least 10 to 20% of the original levels remained). The growth and morphology of these established clones are apparently not too much different from those of their parental cells (data not shown). This observation suggests that whereas the presence of TCTP is critical for optimal proliferation, residual levels of TCTP may be sufficient to carry out most of its biological activities. Under such conditions, the effect of TCTP on the Mcl-1 stability and function in the established TCTP knockdown lines may be underestimated, as residual amounts of TCTP are still present in these cells. An ultimate proof of the modulatory roles of TCTP on the Mcl-1 protein would require the generation of cells whose TCTP expression is completely inhibited, for example, from cells generated by the gene knockout approach.
The Mcl-1 protein was found localized in both the mitochondrial and the nonmitochondrial membrane fractions (34). However, despite the absence of a common nuclear localization sequence, one group did report a predominant nuclear localization of Mcl-1 (15, 37). In our immunofluorescence analysis, we observed that Mcl-1 was predominantly localized in mitochondria when cells were freshly seeded and analyzed on the next day (data not shown). However, if the immunostaining analysis was carried out with cells that had been cultured for a longer time (e.g., at least 2 days after seeding), Mcl-1 was found localized both in nucleus and cytoplasm. In either cellular compartment, we noticed that, whereas Mcl-1 and TCTP were only partially colocalized, such spatial association was more prominent in some cell states than others, suggesting that the interaction of these two proteins is potentially regulated in a cell-state-specific manner. The physiological consequence, as well as the responsible regulatory signaling events, of such cell state-dependent, subcellular translocation of these two proteins is currently not clear and awaits further investigation.
Acknowledgments
We thank Shiu-Ming Shih for his technical advice on the yeast two-hybrid screen, Yi-Ping Hsueh for the pSUPER-neo+gfp plasmid, and Cheng-Ting Chien for critical comments on the manuscript.
This study was supported in part by an intramural fund from Academia Sinica and by grants NHR1-EX91-9119BN, NSC90-2318-B-001-001-M51, NSC91-3112-B-001-004-M51, and NSC 92-3112-B-001-016 from National Health Research Institutes and the National Science Council of Taiwan to H.-F.Y.-Y.
REFERENCES
- 1.Akgul, C., D. A. Moulding, and S. W. Edwards. 2001. Molecular control of neutrophil apoptosis. FEBS Lett. 487:318-322. [DOI] [PubMed] [Google Scholar]
- 2.Altmeyer, A., R. C. Simmons, S. Krajewski, J. C. Reed, G. W. Bornkamm, and S. Chen-Kiang. 1997. Reversal of EBV immortalization precedes apoptosis in IL-6-induced human B-cell terminal differentiation. Immunity 5:667-677. [DOI] [PubMed] [Google Scholar]
- 3.Bohm, H., R. Benndorf, M. Gaestel, B. Gross, P. Nurnberg, R. Kraft, A. Otto, and H. Bielka. 1989. The growth-related protein P23 of the Ehrlich ascites tumor: translational control, cloning and primary structure. Biochem. Int. 19:277-286. [PubMed] [Google Scholar]
- 4.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 5.Cans, C., B. J. Passer, V. Shalak, V. Nancy-Portebois, V. Crible, N. Amzallag, D. Allanic, R. Tufino, M. Argentini, D. Moras, G. Fiucci, B. Goud, M. Mirande, R. Amson, and A. Telerman. 2003. Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. Proc. Natl. Acad. Sci. USA 100:13892-13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, J. R., J. M. Wang, S. F. Lee, H. W. Peng, Y. H. Lin, C. H. Chou, J. C. Li, H. M. Huang, C. K. Chou, M. L. Kuo, J. J. Yen, and H. F. Yang-Yen. 1998. mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol. Cell. Biol. 18:4883-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig, R. W. 2002. MCL1 provides a window on the role of the BCL2 family in cell proliferation, differentiation, and tumorigenesis. Leukemia 16:444-454. [DOI] [PubMed] [Google Scholar]
- 8.Cuconati, A., C. Mukherjee, D. Perez, and E. White. 2003. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 17:2922-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gachet, Y., S. Tournier, M. Lee, A. Lazaris-Karatzas, T. Poulton, and U. A. Bommer. 1999. The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J. Cell Sci. 8:1257-1271. [DOI] [PubMed] [Google Scholar]
- 9a.Heffernan, M., and J. W. Dennis. 1991. Polyoma and hamster papovavirus large T antigen-mediated replication of expression shuttle vectors in Chinese hamster ovary cells. Nucleic Acids Res. 19:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9b.Hsu, S. Y., A. Kaipia, E. McGee, M. Lomeli, and Hsueh, A. J. 1997. Bok is a proapoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective antiapoptotic Bcl-2 family members. Proc. Natl. Acad. Sci. USA 94:12401-12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, H. M., C. J. Huang, and J. J. Yen. 2000. Mcl-1 is a common target of stem cell factor and interleukin-5 for apoptosis prevention activity via MEK/MAPK and PI-3K/Akt pathways. Blood 96:1764-1771. [PubMed] [Google Scholar]
- 11.Jourdan, M., J. De Vos, N. Mechti, and B. Klein. 2000. Regulation of Bcl-2-family proteins in myeloma cells by three myeloma survival factors: interleukin-6, interferon-alpha and insulin-like growth factor 1. Cell Death Differ. 7:1244-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karsan, A., E. Yee, K. Kaushansky, and J. M. Harlan. 1996. Cloning of human Bcl-2 homologue: inflammatory cytokines induce human A1 in cultured endothelial cells. Blood 87:3089-3096. [PubMed] [Google Scholar]
- 13.Kozopas, K. M., T. Yang, H. L. Buchan, P. Zhou, and R. W. Craig. 1993. Mcl-1, a gene expressed in programmed myeloid cell differentiation has sequence similarity to Bcl2. Proc. Natl. Acad. Sci. USA 90:3516-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leu, C. M., C. Chang, and C. Hu. 2000. Epidermal growth factor (EGF) suppresses staurosporine-induced apoptosis by inducing mcl-1 via the mitogen-activated protein kinase pathway. Oncogene 19:1665-1675. [DOI] [PubMed] [Google Scholar]
- 15.Li, F., D. Zhang, and K. Fujise. 2001. Characterization of fortilin, a novel antiapoptotic protein. J. Biol. Chem. 276:47542-47549. [DOI] [PubMed] [Google Scholar]
- 16.Lin, E. Y., A. Orlofsky, M. S. Berger, and M. B. Prystowsky. 1993. Characterization of A1, a novel hemopoietic-specific early-response gene with sequence similarity to bcl-2. J. Immunol. 151:1979-1988. [PubMed] [Google Scholar]
- 17.Lin, Y. H., C. J. Huang, J. R. Chao, S. T. Chen, S. F. Lee, J. J. Y. Yen, and H. F. Yang-Yen. 2000. Coupling of osteopontin and its cell surface receptor CD44 to the cell survival response elicited by IL-3 or GM-CSF. Mol. Cell. Biol. 20:2734-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H., H. Perlman, L. J. Pagliari, and R. M. Pope. 2001. Constitutively activated Akt-1 is vital for the survival of human monocyte-differentiated macrophages. Role of Mcl-1, independent of nuclear factor (NF)-κB, Bad, or caspase activation J. Exp. Med. 194:113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomo, J., E. B. Smeland, S. Krajewski, J. C. Reed, and H. K. Blomhoff. 1996. Expression of the Bcl-2 homologue Mcl-1 correlates with survival of peripheral blood B lymphocytes. Cancer Res. 56:40-43. [PubMed] [Google Scholar]
- 20.MacDonald, S. M., T. Rafnar, J. Langdon, and L. M. Lichtenstein. 1995. Molecular identification of an IgE-dependent histamine-releasing factor. Science 269:688-690. [DOI] [PubMed] [Google Scholar]
- 21.Moulding, D. A., C. Akgul, M. Derouet, M. R. White, and S. W. Edwards. 2001. BCL-2 family expression in human neutrophils during delayed and accelerated apoptosis. J. Leukoc. Biol. 70:783-792. [PubMed] [Google Scholar]
- 22.Moulding, D. A., J. A. Quayle, C. A. Hart, and S. W. Edwards. 1998. Mcl-1 expression in human neutrophils: regulation by cytokines and correlation with cell survival. Blood 92:2495-2502. [PubMed] [Google Scholar]
- 23.Muchmore, S. W., M. Sattler, H. Liang, R. P. Meadows, J. E. Harlan, H. S. Yoon, D. Nettesheim, B. S. Chang, C. B. Thompson, S. L. Wong, S. L. Ng, and S. W. Fesik. 1996. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381:335-341. [DOI] [PubMed] [Google Scholar]
- 24.Nijhawan, D., M. Fang, E. Traer, Q. Zhong, W. Gao, F. Du, and X. Wang. 2003. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 17:1475-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opferman, J. T., A. Letai, C. Beard, M. D. Sorcinelli, C. C. Ong, and S. J. Korsmeyer. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426:671-676. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds, J. E., T. Yang, L. Qian, J. D. Jenkinson, P. Zhou, A. Eastman, and R. W. Craig. 1994. Mcl-1, a member of the Bcl-2 family, delays apoptosis induced by c-Myc overexpression in Chinese hamster ovary cells. Cancer Res. 54:6348-6352. [PubMed] [Google Scholar]
- 27.Rinkenberger, J. L., S. Horning, B. Klocke, K. Roth, and S. J. Korsmeyer. 2000. Mcl-1 deficiency results in peri-implantation embryonic lethality. Genes Dev. 14:23-27. [PMC free article] [PubMed] [Google Scholar]
- 28.Thaw, P., N. J. Baxter, A. M. Hounslow, C. Price, J. P. Waltho, and C. J. Craven. 2001. Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat. Struct. Biol. 8:701-704. [DOI] [PubMed] [Google Scholar]
- 29.Tuynder, M., L. Susini, S. Prieur, S. Besse, G. Fiucci, R. Amson, and A. Telerman. 2002. Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc. Natl. Acad. Sci. USA 99:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, J. M., J. R. Chao, W. Chen, M. L. Kuo, J. J. Y. Yen, and H. F. Yang-Yen. 1999. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol. Cell. Biol. 19:6195-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, J. M., M. J. Lai, and H. F. Yang-Yen. 2003. Interleukin-3 stimulation of mcl-1 gene transcription involves activation of the PU. 1 transcription factor through a p38 mitogen-activated protein kinase-dependent pathway. Mol. Cell. Biol. 23:1896-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, K., A. Gross, G. Waksman, and S. J. Korsmeyer. 1998. Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing. Mol. Cell. Biol. 18:6083-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, T., H. L. Buchan, K. J. Townsend, and R. W. Craig. 1996. MCL-1, a member of the BLC-2 family, is induced rapidly in response to signals for cell differentiation or death, but not to signals for cell proliferation. J. Cell Physiol. 166:523-536. [DOI] [PubMed] [Google Scholar]
- 34.Yang, T., K. M. Kozopas, and R. W. Craig. 1995. The intracellular distribution and pattern of expression of Mcl-1 overlap with, but are not identical to, those of Bcl-2. J. Cell Biol. 128:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarm, F. R. 2002. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell. Biol. 22:6209-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yenofsky, R., S. Cereghini, A. Krowczynska, and G. Brawerman. 1983. Regulation of mRNA utilization in mouse erythroleukemia cells induced to differentiate by exposure to dimethyl sulfoxide. Mol. Cell. Biol. 3:1197-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, D., F. Li, D. Weidner, Z. H. Mnjoyan, and K. Fujise. 2002. Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and fortilin: the potential role of MCL1 as a fortilin chaperone. J. Biol. Chem. 277:37430-37438. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, P., L. Qian, K. M. Kozopas, and R. W. Craig. 1997. Mcl-1, a Bcl-2 family member, delays the death of hematopoietic cells under a variety of apoptosis-inducing conditions. Blood 89:630-643. [PubMed] [Google Scholar]