Abstract
The human γ-globin genes form part of a 5-kb tandem duplication within the β-globin gene cluster on chromosome 11. Despite a high degree of identity between the two genes, we show that while the upstream Gγ-globin gene terminates transcription efficiently, termination in the Aγ gene is inefficient. This is primarily due to the different strengths of the poly(A) signals of the two genes; Gγ-globin has a functionally stronger poly(A) signal than the Aγ gene. The probable cause of this difference in poly(A) efficiency characteristics lies with a number of base changes which reduce the G/U content of the GU/U-rich region of the Aγ poly(A) signal relative to that of Gγ. The 3′ flanking regions of the two γ-globin genes have similar abilities to promote transcription termination. We found no evidence to suggest a cotranscriptional cleavage event, such as that seen in the human β-globin gene, occurs in either γ-globin 3′ flank. Instead we find evidence that the 3′ flank of the Gγ-globin gene contains multiple weak pause elements which, combined with the strong poly(A) signal the gene possesses, are likely to cause gradual termination across the 3′ flank.
Transcription termination is the process whereby the transcription complex and nascent RNA are both released from the template DNA (23), and it has several important consequences. The efficient release of the transcription complex facilitates either its return to the cellular pool or its recycling to promote further rounds of initiation from the same promoter (10a). Transcription termination is also necessary to prevent occlusion of nearby, downstream genes. Transcriptional interference caused by inefficient termination occurs in a number of genes in the closely spaced Saccharomyces cerevisiae genome and can result in a significant down-regulation of the downstream gene (15). Finally, transcription termination serves as the mechanism by which the nascent RNA is released. In the case of most eukaryotic RNA polymerase II (Pol II) genes, the event which releases the functional part of the transcript is not termination but, rather, a cotranscriptional 3′ processing event.
The 3′ end of all eukaryotic mRNAs, with the exception of that of the replication-dependent histone-encoding mRNAs, is defined by a tail of adenosine residues (approximately 200 in human mRNAs). These are not encoded but rather are added by the template-independent poly(A) polymerase following cleavage at a site specified in the primary transcript. The poly(A) site lies between a highly conserved hexanucleotide (AAUAAA) and a more poorly defined U- or GU-rich region. The polyadenylation machinery consists of a number of proteins which together recognize the poly(A) site, cleave the transcript, and add the adenosine residues. Cleavage and polyadenylation specificity factor interacts with the AAUAAA hexanucleotide, while cleavage stimulatory factor (CstF) binds to the GU/U-rich region; together, these complexes define the cleavage site. Two additional proteins, cleavage factors I and II, are needed for endonucleolytic cleavage to occur. Following the generation of the 3′ end by cleavage, poly(A) polymerase adds approximately 200 A residues to generate the mature mRNA (for a review of the biochemistry of polyadenylation, see reference 30).
It has been appreciated for a number of years that 3′ processing is an intimate component of the transcription termination mechanism for protein-coding genes; in the absence of a functional poly(A) signal, transcription does not terminate (11, 15). More-specific experiments using yeast have shown that cleavage, rather than polyadenylation factors, is necessary (8). Termination sites and signals have been mapped for a number of genes and fall into several categories. Pausing at AU-rich sequences has been implicated in termination in the human α-globin (14) and mouse β-major globin (27) genes. Binding sites for transcription factors, for example, a MAZ binding site downstream of the human complement C2 gene (6), also have pausing effects which lead to termination, although this is by no means ubiquitous among all protein binding sites. Data such as these have led to a number of different transcription termination models, including the antiterminator and the torpedo models. The former proposes that recognition of the poly(A) site causes a conformational change in the transcription complex which renders it competent to terminate (17). The demonstration that components of the poly(A) machinery interact with the carboxyl terminal domain of the largest Pol II (6a, 16a) suggested a biochemical mechanism for the antiterminator model; the departure of poly(A) factors from the transcription complex following the generation of the 3′ processing signals would be the change that renders the complex termination competent. The torpedo model suggests a rather different role for the poly(A) signal during termination; following cleavage at the poly(A) site, the downstream, nascent RNA fragment is left with an uncapped 5′ end, which renders it susceptible to exonucleases that degrade the molecule, culminating in termination when the degradation reaches the transcription complex (9, 25).
More-recent work has revealed additional aspects of the transcription termination mechanism. With the human β-globin gene it was shown that a cotranscriptional RNA cleavage (CoTC) event occurs in the 3′ flank and is an essential component of transcription termination (12). Meanwhile, in yeast it has been shown that chromatin structure can play a role in termination (2). Models which incorporate these new discoveries are presently being developed (for reviews, see references 24 and 26).
We have now studied transcription termination in the human γ-globin genes. Since these genes form part of a recent tandem duplication and are closely related, any similarities or differences in the ways they terminate may provide an insight into termination mechanisms. We have shown that in the Gγ-globin gene the combination of multiple weak pause sites and a strong poly(A) signal promotes efficient transcription termination. By contrast, termination in the Aγ-globin gene is less efficient, largely due to its weak poly(A) signal.
MATERIALS AND METHODS
Clones.
All of the γ-globin minigene clones used in this work were transcribed from the human immunodeficiency virus (HIV) long terminal repeat (LTR) promoter (10) from nucleotides (nt) −161 to +80 relative to the transcription start site. To this were fused exons 1 and 2 and the first 17 bp of exon 3 derived from the Aγ gene (position +35 to a BstEII site at +1373 relative to the transcription start site), since the two γ-globin genes are almost identical over this region. The remainder of exon 3 and various regions of flanking DNA from the two γ-globin genes were generated by PCR using PfG DNA polymerase (Stratagene) and primers which introduce unique restriction sites for subsequent cloning into this basic minigene as described below.
The HγA clone contains the region +1374 to +4347 from the Aγ-globin gene fused to the above-described minigene, while in the HγG (HγG14) clone the equivalent sequence from +1394 (BstEII) to +4730 relative to the Gγ-globin transcription start site was added. Coordinates of the Gγ-globin fragments added to the basic clones were as follows: HγG9, +1394 to +3529; HγG7, +1394 to +2722; HγG4, +1394 to +2003; and HγG3, +1394 to +1711. Coordinates of the internal deletions generated in HγG7 were as follows: Δ6, 2394 to 2620; Δ5, 2004 to 2409; and Δ4, 1714 to 2004. Clone HγA7 contained the region +35 to +2557 from the Aγ gene. The flank exchange clones were generated from HγA7, replacing various regions with the equivalent region of the Gγ-globin gene, as follows: for HγGpA/A7, +1374 to +1611 of Aγ was replaced with the equivalent region of Gγ; for HγApA/G7, +1611 to +2557 of Aγ was replaced with the same region of Gγ; for HγG4/A7, +1374 to +1966 of Aγ was replaced with Gγ; and for HγA4/G7, +1966 to +2557 of Aγ was replaced with Gγ. Poly(A) competition clones were as follows: HγApA/SPA contains the region of Aγ-globin from +35 to +1611 fused to the strong synthetic polyadenylation signal (SPA) (16) via the pBluescript polylinker; HγGpA/SPA was a similar clone in which the region of Aγ-globin from +1374 to +1630 was replaced with the equivalent region of Gγ-globin. Fragments of the Gγ 3′ flank were cloned into HγApA/SPA at a blunt-ended SalI site between the two poly(A) sites (fragment A, 1709 to 1982; fragment B, 1982 to 2198; fragment C, 2198 to 2372; fragment D, 2372 to 2722) (all locations relative to the Gγ-globin transcription start site). The α-globin/SPA clone with inactive method poly(A) signal has been described previously (19); fragments of the Aγ- and Gγ-globin genes from BstEII (positions +1373 and +1393, respectively) to DraI (+1611 and +1630) were inserted into a PvuII site between the α-globin gene and SPA. pVa is a pUC18 plasmid containing the VA1 gene and was used as a cotransfection control. The Tat plasmid has been described previously (1).
Cell culture and transfection.
HeLa cells were maintained in D-MEM supplemented with 10% bovine serum and 2 mM l-glutamine. Cells were transfected with plasmids as indicated in the figure legends by the use of calcium phosphate precipitation (11) for steady-state analysis or of Effectene according to the instructions of the manufacturer [QIAGEN]) for nuclear run-on (NRO) analysis. At 24 h after transfection cells were harvested by washing and scraping into cold phosphate-buffered saline (PBS).
Steady-state RNA isolation and analysis.
Transfected HeLa cells were fractionated into nuclear and cytoplasmic components as described previously (13). Cytoplasmic RNA was purified by phenol-chloroform extraction and precipitated with ethanol. RNA was extracted from isolated nuclei by the hot-phenol method: nuclei were resuspended in PBS and added to a 1:1 mixture of acid phenol and 100 mM NaCl-10 mM Tris-HCl (pH 7.5)-1 mM EDTA-1% sodium dodecyl sulfate. Following centrifugation the aqueous phase was reextracted with phenol-chloroform and chloroform, precipitated with ethanol, and resuspended in R-loop (80% formamide-40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid] [pH 6.8]-1 mM EDTA-400 mM NaCl).
A labeled riboprobe spanning the HIV transcription start site (−161 to +123) was generated, and RNase protection assays were carried out as previously described (5). The poly(A) site competition assay (using S1 nuclease analysis) has been described previously (14).
NRO analysis.
NRO analysis was carried out as described previously (4), with some modifications. Nuclei were isolated at 4°C by lysing the cells with 0.5% NP-40 in 10 mM Tris-HCl (pH 7.5)-10 mM NaCl-2.5 mM MgCl2, followed by centrifugation through a 10% sucrose cushion made in the same buffer. Transcription reactions were carried out in 40 mM Tris-HCl (pH 7.9)-300 mM KCl-10 mM MaCl2-40% glycerol-2 mM dithiothreitol supplemented with 250 μM concentrations each of ATP, CTP, and GTP plus 1.1 MBq of [α-32P]UTP. After 15 min at 30°C, the nuclei were pelleted briefly (30 s) and resuspended in PBS, and RNA was extracted using the hot-phenol approach. The precipitated RNA was treated with RNase-free DNase I, extracted with phenol-chloroform, and reprecipitated as previously described (4). RNA was then hydrolyzed with 0.2 M NaOH, neutralized with 0.5 M Tris-0.5 M HCl, and hybridized to filters containing single-stranded DNA probes as described in Table 1. Selection of RNAs contiguous with a biotinylated riboprobe containing the reverse complement of the region 1424 to 1711 [spanning the Gγ poly(A) site] was carried out as described for the β-globin gene (12). Briefly, run-on RNAs were hybridized to the biotinylated probe in R-loop buffer overnight and were selected using streptavidin-coated magnetic beads (Promega). After two washes in 300 mM NaCl-10 mM Tris-HCl (pH 7.5)-5 mM EDTA to reduce nonspecific binding, selected RNAs were released by hydrolysis with 0.2 M NaOH and neutralized with 0.5 M Tris-0.5 M HCl as described for the standard run-on technique.
TABLE 1.
Nuclear run-on probes
| Probe | Position | No. of Us |
|---|---|---|
| Gγ-globina | ||
| G3 | 1395-1629 | 75 |
| G4 | 1630-1999 | 94 |
| G5 | 1999-2394 | 104 |
| G6 | 2394-2632 | 75 |
| G7 | 2632-2863 | 54 |
| G8 | 2863-3200 | 132 |
| G9 | 3200-3506 | 121 |
| Aγ-globina | ||
| A3 | 1375-1609 | 76 |
| A4 | 1610-1961 | 105 |
| A5 | 1961-2232 | 74 |
| A6 | 2232-2469 | 79 |
| A7 | 2469-2705 | 61 |
| A8 | 2705-3042 | 129 |
| A9 | 3042-3346 | 121 |
| A18 | 3346-3562 | 9 |
| pUCb | ||
| A | 237-2622 | 78 |
| B | 2622-2351 | 86 |
| C | 2351-2060 | 71 |
| D | 1223-907 | 57 |
| E | 907-449 | 83 |
| Miscellaneous | ||
| M | Empty ssM13 | |
| His | Mouse histone H4 | NAc |
| 5S | Xenopus 5S RNA | NA |
| U3 | HIV LTR promoter (−161 to −20)a | 27 |
| PH | HIV LTR promoter (−20 to +123)a | 28 |
| pG7 | Gγ 2632-2722 plus pUC18 418-237 | 81 |
Position relative to the transcription start site.
Position numbering according to accession number L09136.
NA, not assessed.
RESULTS
The human γ-globin genes (Gγ and Aγ) are found in tandem within the β-globin gene cluster on chromosome 11, with a distance of only 3.3 kb between the poly(A) site of the upstream Gγ gene and the transcription start site of the Aγ gene. The γ-globins are predominantly expressed within fetal erythroid cells, where the upstream Gγ gene represents the more highly expressed (70:30) of the pair (21).
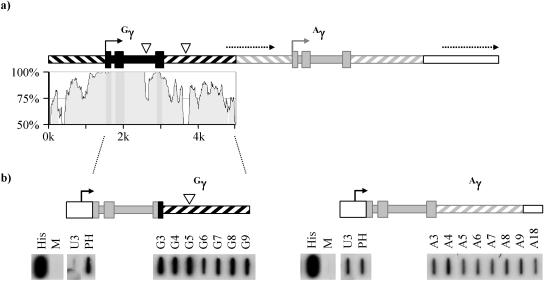
The two γ-globin genes are very closely related, with >95% identity across the proximal promoter and three exons. The two introns show similar degrees of conservation, with a 20-nt insertion in intron 2 of Gγ relative to Aγ marking the only major difference between them (Fig. 1a). The DNA flanking the γ-globin genes is also well conserved, with approximately 70 to 80% identity across the 1 kb upstream of the proximal promoter and the 2 kb downstream of the poly(A) site (Fig. 1a). Thus, as illustrated in Fig. 1a, the γ-globins lie within a 5-kb direct tandem repeat.
FIG. 1.
Structure of the human γ-globin repeat unit. (a) A tandem repeat of ∼5 kb in the human β-globin gene cluster encompasses the Gγ-globin (black boxes) and Aγ-globin (grey boxes) genes. Wide filled boxes indicate exons, and narrow filled boxes indicate introns. Transcription start sites are indicated by solid arrows. The repeat includes ∼1.5 kb of DNA upstream of the γ-globin transcription start site (diagonal lines sloping downward from left to right) and ∼1.9 kb downstream of the terminal exon (diagonal lines sloping upward). The approximate locations of intergenic transcripts downstream of the Gγ and Aγ genes are shown by dashed arrows. Inserts of 20 nt in intron 2 and 120 nt in the 3′ flank of Gγ are shown as white triangles. The degree of conservation between the two γ-globin genes is indicated in the VISTA plot (18) below. (b) Transcription profiles of the Gγ and Aγ genes. In each case, exons 1 and 2 and the first 17 bp of exon 3 were derived from the Aγ gene (position +35 to +1373 relative to the transcription start site) and were cloned downstream of the HIV LTR promoter (open box). The remainder of exon 3 and the 3′ flank of each of the γ-globin genes (positions +1374 to +4347 for Aγ and +1394 to +4730 for Gγ) were then fused to this to generate the two clones shown. Following cotransfection into HeLa cells with the HIV trans-activator Tat, NRO analysis was carried out and the resulting labeled RNA was hybridized to the series of probes shown. Probes A3 and G3 span the poly(A) site of the γ-globin genes and are 93% identical between the two genes. Probes G4 to G9 and A4 to A9 occupy equivalent positions within the 3′ flanks of the two γ-globin genes, while A18 is immediately adjacent to A9 (see Table 1). The His probe measures histone H4 expression and is a run-on control, while M represents the empty M13 vector and is a negative control. Probes U3 and PH lie upstream and downstream of the HIV LTR transcription start site, respectively. The efficiency of transcription termination was assessed by the amount of read round into the HIV promoter, as measured using the probe U3.
Previous attempts to define the sites of transcription termination in the γ-globin genes have been complicated by their repetitive nature and by the existence within the β-globin gene cluster of additional, so-called intergenic transcripts. These unstable transcripts initiate between the globin transcription units and within the locus control region of the β-globin gene cluster. The extent of intergenic transcription downstream of the two γ-globin genes is indicated in Fig. 1a. Previous work (4) has suggested that transcription initiating at the Aγ and Gγ promoters gradually reduces over their 3′ flanks but does not completely cease prior to the start of intergenic transcription.
The β-globin cluster intergenic transcripts are erythroid specific (4); however, they can be induced in nonerythroid cells by a variety of methods, including the provision of erythroid transcription factors, disruption of higher-order chromatin structure with histone deacetylase inhibitors (22), and the process of transinduction (4). This latter phenomenon occurs when plasmids which express parts of the β-globin cluster are transfected into nonerythroid cells; transcribing plasmids localize with the chromosomal β-globin cluster and trigger the transcription of the intergenic, but not genic, regions of the cluster by an undefined mechanism (4). This further complicates the analysis of transcription termination in the γ-globin genes, since termination within the γ-globin flank contained within a transfected plasmid can be masked by the stimulation of intergenic transcripts from the chromosome.
Comparison of the efficiencies of transcription termination in the Aγ- and Gγ-globin genes.
The β-globin gene and its 3′ flank were cloned downstream of the HIV LTR promoter, and, following cotransfection into HeLa cells with the trans-activator Tat (10), high levels of transcription were detected by NRO. Crucially, the efficiency of termination could be assessed using a probe (U3) which lies immediately upstream of the HIV LTR transcription start site, thus providing a measure of read-through transcription which is uncontaminated by signals derived from the trans-induced chromosomal β-globin cluster (11).
We have employed this assay to compare the efficiency characteristics of termination in the Gγ and Aγ-globin 3′ flanks. Since the two genes are almost identical (98%) over the coding region and the introns (with no mismatches in exons or sequences involved in splicing), the first two exons and the 5′ end of exon 3 (from position +35 to +1373) derived from the same (Aγ) gene were cloned downstream of the HIV LTR promoter. Exon 3 and the 3′ flank from the Gγ-globin gene (from position +1394 to +4730, 3.1 kb downstream of the polyadenylation site) were fused into this minigene, which was then assayed as described above. As shown in Fig. 1b, there is very little read round into probe U3, suggesting that efficient termination occurs in this clone even though NRO signals are detected across the entire Gγ 3′ flank (probes G4 to G9). By comparison, a similar region of the Aγ gene (to position +4347, 2.8 kb downstream of the polyadenylation site) shows significant amounts of read round into probe U3, indicating that transcription termination in the Aγ gene is much less efficient than in Gγ (Fig. 1b). Interestingly, the overall level of NRO signals derived from the Aγ clone was much lower than those derived from the Gγ clone (compare probe PH from the two panels of Fig. 1b). This is probably due to occlusion of the HIV LTR promoter by transcription complexes which read through the promoter and is a further indication of the inefficiency of termination in the Aγ gene.
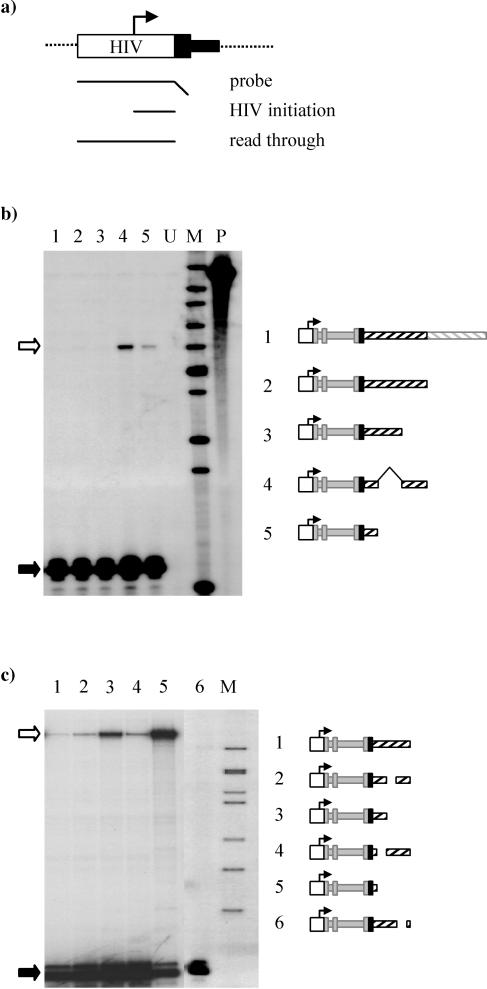
A 1.1-kb 3′ flanking region of the Gγ-globin gene is sufficient for efficient termination.
To facilitate the rapid analysis of large numbers of deletion clones we have developed a new transient transfection transcription termination assay based on read round into the HIV LTR promoter. As with the previous assay, transcripts which do not terminate read around the transfected plasmid and into the promoter from which they were initiated. These read-through transcripts are unstable and are not exported from the nucleus (data not shown); however, they can be detected in the nuclear RNA fraction (e.g., Fig. 2b, lanes 4 and 5). Thus, following transfection and 24 h of expression, transcripts which initiate at the HIV promoter can be differentiated from those which read round the plasmid by an RNase protection assay using a probe which spans the HIV promoter (Fig. 2a).
FIG. 2.
Identification of the minimal requirements for efficient termination in the Gγ gene. (a) The efficiency of termination was assessed by the amount of read round into the HIV promoter, as measured by analysis of nuclear RNA extracted from transfected cells. A riboprobe spanning the HIV promoter is protected by transcripts which initiate at the HIV promoter (80-nt product) and also by transcripts which fail to terminate and read round the transfected plasmid and across the HIV promoter (241-nt protected fragment). (b) Analysis of the efficiency of termination in Gγ deletion clones. The final exon of the Gγ-globin gene from +1394 relative to the transcription start site and various amounts of 3′ flank were cloned downstream of the HIV LTR promoter (−161 to +80) and exons 1 to 3 (position +35 to +1373) of the Aγ-globin gene. The resultant clones were cotransfected into HeLa cells with the Tat expression plasmid by calcium phosphate precipitation. After 24 h nuclear RNA was extracted and analyzed using the riboprobe described for panel a. Clones examined were as follows: lane 1, HγG14 (3.1-kb Gγ 3′ flank); lane 2, HγG9 (1.9 kb); lane 3, HγG7 (1.1 kb); lane 4, HγG9Δ5-7 [contains the 3′ flank regions up to 0.4 kb and 1.1 to 1.9 kb downstream of the poly(A) site]; lane 5, HγG4 (0.4-kb 3′ flank).Lane U represents the untransfected control. Lane M contains markers (sizes from top to bottom: 517, 394, 344, 285, 244, 210, 190, 148, 130, and 75 nt), while lane P contains the undigested riboprobe. The protected fragments corresponding to HIV initiation (filled arrow) and read round (open arrow) are indicated. (c) Fine deletions in the Gγ 3′ flank were generated and analyzed as described above. The clone HγG7 showing no read round (lane 1) was used as a base to delete the internal fragments. Deletions (relative to the Gγ transcription start site) were of nt +2004 to +2409 (HγG7Δ5; lane 2), +2004 to +2722 (HγG4; lane 3), +1714 to +2004 (HγG7Δ4; lane 4), +1714 to +2722 (HγG3; lane 5), and +2394 to +2620 (HγG7Δ6; lane 6). The Gγ poly(A) site is at position +1592. Deletion clones were cotransfected with the Tat plasmid and assayed as described above. Markers (lane M) of 244, 210, 190, 179, 148, 130, and 106 nt (top to bottom) are shown. Each clone was analyzed on at least two separate occasions, giving the same result each time.
To determine how much of the Gγ 3′ flank is necessary for efficient termination, a series of deletion clones were generated. As with the minigenes shown in Fig. 1, exons 1 and 2 and the first 17 bp of exon 3 derived from the Aγ gene were cloned downstream of the HIV LTR promoter and the remainder of exon 3 and various pieces of 3′ flank derived from the Gγ-globin gene were fused into this clone to give the minigenes shown in Fig. 2b. Three clones containing large regions of 3′ flank (HγG14, 3.1 kb; HγG9, 1.9 kb; HγG7, 1.1 kb) all gave no detectible read-through transcript, indicative of efficient termination (Fig. 2b, lanes 1 to 3). However, when the 3′ flanking region was reduced to 0.4 kb (HγG4), read-through transcripts were readily apparent (lane 5). Interestingly, when the region between 0.4 and 1.1 kb downstream of the Gγ poly(A) site was replaced with the adjacent region (1.1 to 1.9 kb), generating a clone of a similar size (HγG9Δ5-7), termination was inefficient. This suggests that sequences important for termination are located within the region 0.4 to 1.1 kb downstream of the Gγ poly(A) site.
Existence of multiple termination elements within the Gγ-globin 3′ flank.
To more precisely define the sequences needed for efficient termination in the Gγ gene, we generated a set of clones with deletions within 1.1 kb downstream of the poly(A) site. These were transiently transfected, and the amount of read-through transcript generated by each clone was compared to that of the HγG7 clone, which contains the entire 1.1-kb flank (Fig. 2c, lane 1). When the 3′ flank was deleted to within 120 nt of the poly(A) signal (HγG3), termination was very inefficient, with noticeable occlusion of the HIV promoter (Fig. 2c, lane 5; compare the amount of HIV-initiated transcripts to the results seen with larger clones). Smaller deletions of 290, 380, and 226 nt across the 3′ flank (lanes 2, 4, and 6) all had a minor effect on the amount of read round detected, and no promoter occlusion was apparent. Thus, no single region appears to be entirely responsible for efficient termination in the Gγ-globin gene, suggesting there may be multiple, weak elements within 1.1 kb downstream of the poly(A) site which are together sufficient for efficient termination to occur.
Inefficient termination in the Aγ-globin gene.
In comparison to Gγ results, it is apparent that termination in the Aγ-globin gene is less efficient (Fig. 1b). Although the 3′ flanks of the γ-globin genes are similar (averaging 75% identity over the first 1.1 kb) (Fig. 1a), there is considerable scope for sequences important in termination to differ between these two genes. In particular, the Gγ-globin gene possesses an insertion of 120 nt at a position 0.5 kb downstream of the poly(A) site, within the probe G5 (Fig. 1).
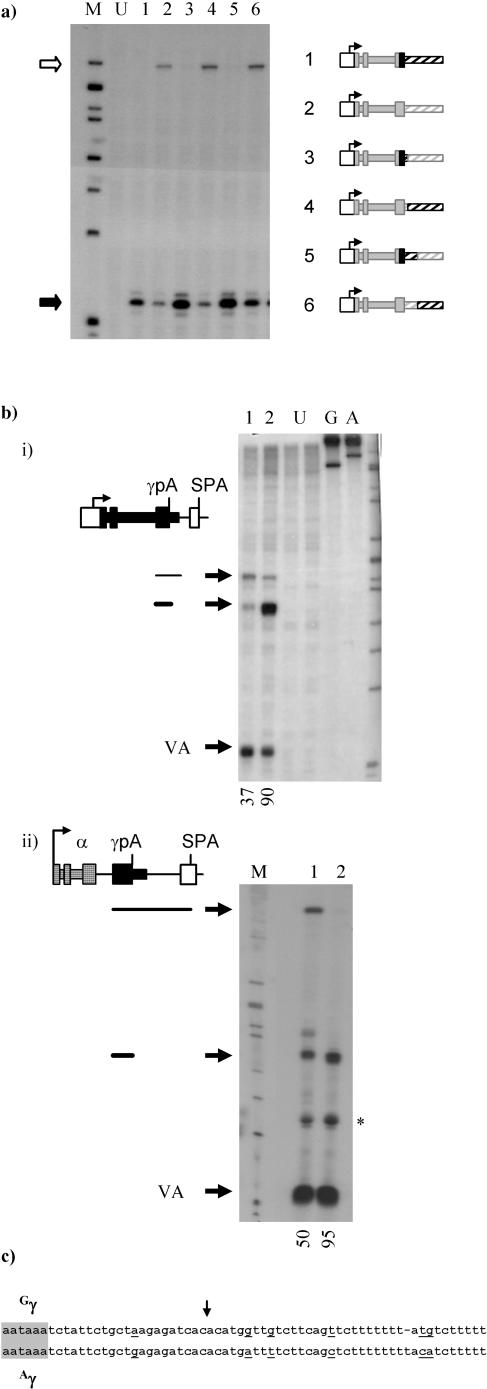
We have generated a set of clones with hybrid 3′ flanks to determine which differences in the 3′ flank of the γ-globin genes account for their different characteristics with respect to the ability to terminate. Figure 3a, lanes 1 and 2, emphasizes that while 1.1 kb of Gγ 3′ flank is sufficient for termination, the equivalent region of Aγ is not. Surprisingly, when various regions of the 3′ flanks of the two γ-globin genes were swapped (lanes 3 to 6) it became apparent that the major determinant of the efficiency of termination was the poly(A) signal utilized. Thus, when the entire Aγ 3′ flank was linked to the poly(A) signal of the Gγ gene (lane 3; HγGpA/A7) termination was efficient, while when the Gγ 3′ flank was coupled to the Aγ poly(A) signal significant read round was observed (lane 4; HγApA/G7). This was the case for every region tested (not all data shown); for example, when the regions from 0.4 to 1.1 kb downstream of the γ-globin poly(A) sites were swapped efficient termination was observed in the clone containing the Gγ poly(A) signal (lane 5; HγG4/A7) but in not that containing the Aγ poly(A) signal (lane 6; HγA4/G7).
FIG. 3.
Comparison of the transcription termination efficiencies of the Gγ- and Aγ-globin genes. (a) Gγ/Aγ hybrid clones were generated and cotransfected into HeLa cells with the trans-activator Tat. After 24 h nuclear RNA was isolated and termination efficiency was assessed using the amount of read round into the HIV LTR promoter (open arrow) compared to the amount of HIV initiation (black arrow) as described for Fig. 2. The source of DNA fragments present in the transfected clones is shown schematically in the accompanying diagrams according to the scheme defined in Fig. 1; black boxes (solid and hatched) indicate sequences derived from the Gγ-globin gene, while grey boxes were derived from the Aγ gene. HγG7 (lane 1) has been described for Fig. 2, and HγA7 (lane 2) contains the equivalent region (+35 to +2557) from the Aγ-globin gene cloned downstream of the HIV promoter. Flank swap clones were generated by replacing parts of HγA7 with the equivalent region of Gγ DNA specifically as follows: in HγGpA/A7 (lane 3), +1374 to +1611 of Aγ was replaced with the equivalent region of Gγ; in HγApA/G7 (lane 4), +1611 to +2557 of Aγ was replaced with Gγ; in HγG4/A7 (lane 5), +1374 to +1966 of Aγ was replaced with the same region of Gγ; and in HγA4/G7 (lane 6), +1966 to +2557 of Aγ was replaced with Gγ. Lane M contains markers (244, 210, 190, 179, 148, 130, 106, and 75 nt), and lane U is an untransfected control. This analysis has been repeated three times, with the same result on each occasion. (b) Comparison of poly(A) signal strength in the Gγ- and Aγ-globin genes. (i) The strong synthetic polyadenylation signal (SPA; white box) was cloned downstream of each of the γ-globin poly(A) signals at a position equivalent to 37 nt downstream of the cleavage site of each. Equal amounts of extraneous polylinker DNA were present in each of the two clones generated. At 48 h after cotransfection into HeLa cells with the Tat plasmid and the VA transfection control, total RNA was extracted. The amount of RNA corresponding to the use of each poly(A) signal was detected using S1 nuclease probes which span each of the γ-globin poly(A) cleavage sites. RNA that is cleaved at the γ-globin poly(A) site protects a 165-nt fragment, while RNA that is processed at the SPA protects a 202-nt fragment of the probe. The poly(A) signal from the Aγ genes (lane 1) and the Gγ gene (lane 2) were competed against the SPA, and the results are shown numerically below as percentages of use of the globin poly(A) site. Untransfected controls (lanes U) and undigested probes (lanes G and A) are also shown, along with the markers (from top, 244, 210, 190, 179, 148, 130, 106 nt and 75 nt). (ii) A similar experiment in which the two γ-globin poly(A) sites (+1373 to +1611 for Aγ and +1393 to +1630 for Gγ) were inserted into the α-globin/SPA expression clone (19) upstream of the SPA. α-globin sequences are shown as checkered boxes. Lane 1 shows competition with the Aγ-globin poly(A) site, while lane 2 shows the competition with Gγ-globin. *, partial digestion product generated from the VA probe. (c) Comparison of the Gγ and Aγ sequences surrounding the poly(A) cleavage site (downward arrow). Differences in Aγ relative to Gγ are underlined.
The read-round assay used in the experiment described above is not purely transcriptional; since it utilizes steady-state RNA, its outcome is also influenced by RNA stability. However, it should be noted that the read-round RNAs detected in this assay were unpolyadenylated nuclear RNAs and therefore, we believe, all unstable. It seems probable that transcription, and not RNA stability, is the primary determinant of the appearance of a read-round signal in this assay. Thus, although the amount of read round measured in this assay is not strictly quantitative, and indeed we have been careful to avoid treating it as such, it provides a rapid assessment of whether transcription reads round into the HIV promoter at all. The flank swap experiments described above provide strong evidence for the validity of these assays; two almost identical clones which differ only in their poly(A) signals (for example, HγG7 and HγApA/G7) gave opposite results in this assay (Fig. 3a, lanes 2 and 3), strongly suggesting that RNA stability is not a major determinant. In addition, the principle findings of the assay have been verified in later run-on experiments; therefore, we believe this read-round assay to be a useful initial analytical tool.
To investigate the difference between the poly(A) signals of the two γ-globin genes we carried out a poly(A) competition assay (Fig. 3b). In this assay the upstream γ poly(A) signal competes with a stronger downstream synthetic poly(A) signal, SPA (16). The balance of poly(A) site usage is influenced by the relative strengths of the two poly(A) signals, and so this assay can be used to rank poly(A) sites according to their strengths by comparing each to the SPA in turn (16).
The poly(A) site of each of the γ-globin genes was competed against the SPA by cloning the latter at a position 37 nt downstream of the cleavage site of each γ-globin gene in the HIV promoter minigene setting used in previous experiments (Fig. 3bi). In this context, the Aγ-globin poly(A) signal was able to compete at roughly 40% with the SPA (lane 1). By comparison the Gγ-globin poly(A) signal was much stronger, contributing over 90% of the signal (lane 2). We note the difference in the overall signal obtained from the two competition clones (lanes 1 and 2), and although we cannot presently explain this difference, the result was reproducible (data not shown) and all the usual checks on plasmid integrity revealed no inconsistencies. We have carried out the poly(A) competition assay in a different minigene setting in which transcription of the three exons of α-globin was driven by its own promoter under the additional regulation of the simian virus 40 enhancer. In this case we were able to measure the difference in the strengths of the two poly(A) signals without a change in the overall signal observed (Fig. 3bii).
These experiments suggest an obvious and simple difference between the two γ-globin genes. Gγ has a strong poly(A) signal which enables it, in the presence of its 3′ flank, to terminate transcription efficiently. By comparison, Aγ has a much weaker poly(A) signal, and therefore, despite having a termination competent 3′ flank (as demonstrated in Fig. 3a, lanes 3 and 5), this gene does not terminate efficiently. A sequence comparison between the two poly(A) sites (Fig. 3c) shows a number of differences in the downstream sequence element which have the net effect of reducing the G and U content and which would be predicted to reduce the efficiency of cleavage and polyadenylation (39).
Mechanism of termination in the Gγ-globin gene.
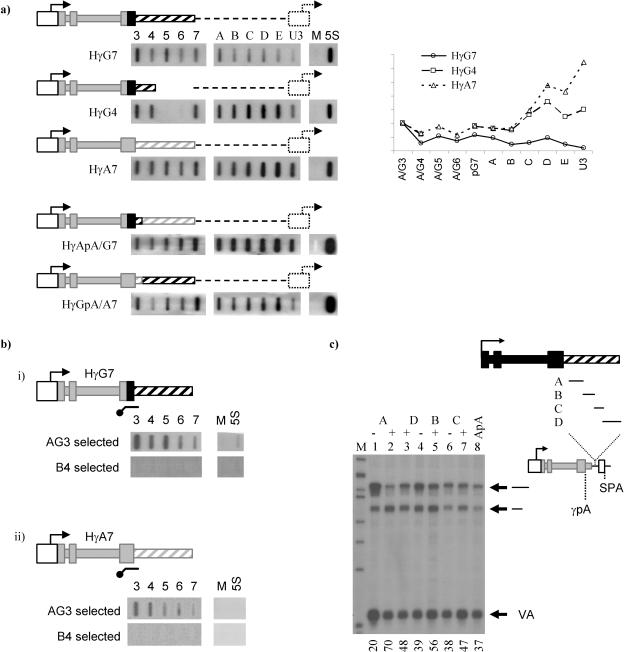
(i) Transcription does not stop within 1.1 kb of Gγ-globin. For a number of genes (in particular, yeast genes) it has been possible to use nuclear run on to show directly where transcription terminates. We have carried out NRO analysis on a number of the γ-globin minigenes we generated, using a panel of probes across the 3′ flank (probes A and G3 to G6) and into the plasmid backbone (probe pG7 and A to E) along with probe U3, upstream of the HIV transcription start site. It should be noted that the Tat expression plasmid contains related vector sequences, although transcription in this plasmid runs in the opposite orientation. Transfection of the Tat plasmid alone suggested that it makes a minor contribution to run-on signals across probes A to E (data not shown).
In the HγG7 minigene, transcription continues across the 3′ flank and into the plasmid backbone, where it gradually reduces before reaching the HIV promoter (Fig. 4a, top panel). By comparison, the Gγ deletion clone HγG4 terminates inefficiently, with higher run-on signals across the plasmid and into the HIV promoter (second panel from top). Termination in the Aγ-globin minigene HγA7 is less efficient still (third panel from top), showing high signals across the 3′ flank and into the plasmid as well as over the HIV promoter. The large effect that the poly(A) signal strength has on termination is reiterated in the bottom two panels; the HγApA/G7 minigene, containing the weak Aγ-globin poly(A) signal and the Gγ 3′ flank, fails to terminate transcription and reads into the plasmid and HIV promoter at high levels, while in the HγGpA/A7 clone the majority of transcription has ceased prior to the HIV promoter.
FIG. 4.
Mechanism of termination in the Gγ-globin gene. (a) HeLa cells were transfected with 3 μg of the HIV/γ-globin minigene clones indicated along with 1 μg of the Tat expression plasmid by the use of Effectene transfection reagent (QIAGEN). Following overnight incubation, NRO was carried out as described in Materials and Methods. Radiolabeled RNA was hybridized to a panel of probes (G3-pG7 or A3-pG7) appropriate to the transfected plasmid. Five vector probes (probes A to E; see Table 1 for locations) were also used, and read-round transcription was detected using probe U3 (HIV promoter). Probes M and 5S are an M13 background control and 5S ribosomal run-on control, respectively. NROs were quantified using a Molecular Dynamics PhosphorImager, and signals were corrected for U content and normalized for transfection efficiency to probes A3 and G3 and are shown graphically. (b) Hybrid selection run on. Cells were transfected with plasmids HγG7 or HγA7 plus the Tat expression plasmid as described above. Following NRO, isolated radiolabeled RNA was hybridized to a biotinylated riboprobe complementary to region 1424 to 1711 of Gγ-globin. This region is 93% identical between the two globin genes and therefore could be used for selection of transcripts from both genes. A control selection using the β-globin probe B4 (12) is shown below the diagram. Following hybridization to the biotinylated selection probe, contiguous transcripts were isolated using streptavidin-coated magnetic beads (Promega). Selected transcripts were hydrolyzed, neutralized, and then hybridized to the probe panels shown per the normal run-on technique. (c) Fragments of the Gγ-globin 3′ flank were cloned into the plasmid HγApA/SPA (as described for Fig. 2b) between the Aγ-globin poly(A) site and the SPA. Fragments corresponding to regions of the Gγ-globin gene from position +1709 to +1982 (fragment A; 273 nt), +1982 to +2198 (fragment B; 216 nt), +2198 to +2372 (fragment C; 174 nt), and +2372 to +2722 (fragment D; 350 nt) were cloned in positive and negative (fragments A, C, and D) orientations. Clones were transfected, and RNA was extracted and analyzed as described for Fig. 2b. Values at the bottom of the panel represent the percentages of transcripts processed at the Aγ-globin poly(A) site. The gel shown is one of two repeats, giving similar results each time.
It may be noted that the signal observed over probe U3 appears somewhat low in all cases, which may suggest a strong termination event upstream of this probe. However, the region corresponding to probe U3 is short and has a proportionally low U content (see Table 1) which reduces the amount of radioactivity incorporated into transcripts hybridizing to this region which in turn lowers the signal observed on the filter. When U content is taken into account it can be seen that there is not a strong termination event immediately prior to the HIV promoter (see graph in Fig. 4a, right panel).
This analysis suggests that there is not a single, strong termination site within the HγG7 minigene but rather that termination occurs in a gradual manner across both the Gγ-globin 3′ flank and the adjacent vector. This is in agreement with our previous finding that multiple regions across the Gγ-globin 3′ flank have weak termination-promoting activities.
(ii) The Gγ-globin gene shows no evidence of pretermination cleavage.
A novel transcription termination mechanism has been reported for the human β-globin gene (12), as suggested by hybrid selection nuclear run-on analysis. In this procedure, transcripts are hybridized to a complementary biotinylated RNA probe. Contiguous transcripts are then isolated with streptavidin-coated magnetic beads and, following their release, are detected by hybridization to a panel of single-stranded probes. In the case of the human β-globin gene, it was shown that there is a region 0.9 to 1.6 kb downstream of the poly(A) signal through which selection does not occur, suggesting that the transcripts in this region are not contiguous. Further work showed that the RNA generated in this region of the β-globin gene undergoes cleavage and that this event is an important precursor to transcription termination (12). These data were interpreted into a two-step model for termination in which a reiterative cotranscription cleavage (CoTC) event mediates the ultimate release of RNA polymerase following polyadenylation and the release of the mature mRNA (reviewed in reference 26).
We have used hybrid selection nuclear run-on analysis to look for CoTC in the γ-globin genes (Fig. 4b). A series of five nuclear run-on probes (G3 to pG7) provide complete coverage of the 1.1-kb region of the Gγ-globin 3′ flank which is required for transcription termination (Fig. 4bi). Using an antisense hybrid selection probe corresponding to region G3, which straddles the Gγ poly(A) site, we carried out hybrid selection NRO as outlined above. Contiguous transcripts were selected throughout the entire 1.1-kb 3′ flanking region (Fig. 4bi, top panel). By the use of the same selection probe, similar results were obtained for the Aγ-globin gene; contiguous transcripts were selected across 1 kb of the Aγ-globin 3′ flank (Fig. 4bii, top panel). A control selection performed using a probe generated from the β-globin gene (bottom panels) was carried out to ensure that selection was specific.
These data therefore rule out CoTC as a component of the termination mechanism in the γ-globin genes. Thus, the human γ-globin genes differ from the β-globin gene, where strong CoTC elements mediate transcription termination, and also from the human ɛ-globin gene, for which there is evidence that weak CoTC elements have a similar role.
(iii) Transcription pausing in the Gγ-globin 3′ flank.
It has previously been demonstrated for a number of yeast and mammalian genes that transcriptional pausing downstream of the poly(A) signal may contribute towards the ability of Pol II to terminate (3, 7, 27). For example, in the human α-globin gene, a strong pause site 350 nt downstream of the poly(A) signal brings about transcription termination (14). We have analyzed the 3′ flank of the Gγ-globin gene to determine whether a similar mechanism is utilized.
The poly(A) competition assay was developed as a means of detecting transcriptional pausing (14). As described above, when a weak upstream poly(A) signal is placed in close proximity to a stronger downstream poly(A) signal the resulting competition will tend to favor the stronger signal and will reflect their relative strengths. By inserting fragments of DNA between the two poly(A) signals the balance of competition can be pushed in favor of the upstream site, since as larger fragments are inserted it will begin to gain a temporal advantage by virtue of being transcribed first. A transcription pause site effectively has a much larger effect on the balance of poly(A) competition; by slowing the rate of transcription, it has a greater temporal effect. With this assay it is also possible to identify polyadenylation enhancers; indeed, transcription pause sites and terminators in general form one class of poly(A) enhancer.
We have inserted fragments of the Gγ 3′ flank, ranging from 180 to 350 nt, between the weak Aγ poly(A) signal and the strong SPA (Fig. 4c). The fragments were inserted in both orientations to allow us to distinguish pausing effects over and above the distance effects one might expect to see. As shown in Fig. 4c, in the absence of any intervening sequences the upstream Aγ poly(A) signal contributes 37% of the mRNA produced from this clone (lane 8). Each of the four fragments A to D increased this contribution when inserted in the positive orientation (lanes 2, 5, 7, and 3, respectively), with the largest increase observed when region A [117 to 390 nt downstream of the Gγ poly(A) site] was analyzed (lane 2). Importantly, the effect of the increase in distance between the two poly(A) signals, assessed by analyzing the largest and smallest fragments (fragments D and C, respectively), is not significant in this assay (lanes 4 and 6).
The data presented above show enhanced use of the weak upstream poly(A) site in the assay, which may be interpreted in three ways. Firstly, polyadenylation enhancement as a result of the binding of positive regulatory factors to the flanking RNA is possible, although since the effect was shown to spread over 1.1 kb of the 3′ flank this seems unlikely. Secondly, it is possible that the Gγ-globin 3′ flank forms an RNA secondary structure which enhances polyadenylation, although since the four separate regions of the flank each have an individual effect of poly(A) site usage this again seems unlikely. In addition, since we have shown that the Gγ-globin poly(A) signal is extremely efficient even in the absence of its 3′ flank (Fig. 3b) it seems unlikely that poly(A) enhancers such as these would affect its usage in the whole-gene setting. We therefore favor a third explanation, according to which transcriptional pausing in the Gγ-globin 3′ flank accounts for the enhanced use of the upstream poly(A) site in our assay. This, in combination with the strong Gγ-poly(A) site, would be expected to contribute toward transcription termination in the Gγ gene.
DISCUSSION
The two human γ-globin genes form part of a 5-kb tandem repeat within the β-globin gene cluster and show a high degree of identity across their exons and introns (average > 95%, as shown in Fig. 1a) and also across both 5′ and 3′ flanks (average, 75 to 80%). In the 5′ flank and coding region, this similarity reflects the conserved regulation and function of the two genes. In the 3′ flank of the γ-globin genes, the similarity may merely be indicative of a recent evolutionary origin; however, it is also possible that here too it reflects conserved regulation, potentially in the termination stage of transcription.
We have compared the transcription profiles of the Gγ- and Aγ-globin genes by the use of a transient transfection system. Plasmid minigenes under the control of the HIV LTR promoter were generated for both human γ-globins, and these were transfected into HeLa cells and nascent transcription was measured using NRO. In both cases nascent transcripts were detected across the entire flank. However, when a probe immediately upstream of the HIV LTR transcription start site (probe U3) was used it was seen that while transcription of the Gγ-globin gene terminates prior to the HIV promoter, presumably within the backbone of the plasmid, in the Aγ-gene termination is inefficient and transcripts across probe U3 are readily apparent (Fig. 1b). This resulted in a noticeable promoter occlusion effect in the Aγ clone relative to the Gγ clone results. We predict that the more-efficient termination process for the Gγ-globin gene may be required to minimize transcriptional interference with the downstream, coexpressed Aγ-globin gene.
We investigated the difference in the termination efficiency characteristics of the two γ-globin genes further by generating a series of heterologous minigenes containing various combinations of the two γ-globin genes. These minigenes revealed that the primary difference in the termination efficiency characteristics of the two γ-globin genes is due to the different strengths of their poly(A) signals. Thus, while the strong poly(A) signal of the Gγ-globin gene correlates with efficient transcription termination, the weaker poly(A) signal of the Aγ gene results in inefficient termination, even in the context of the Gγ 3′ flank (Fig. 3). Within the region tested, only seven nucleotide differences occur between the two poly(A) sites; the majority of these lower the GU content in the downstream GU/U-rich region of the Aγ signal and would therefore be predicted to reduce its strength. The correlation between poly(A) signal strength and transcription termination is in agreement with the results seen with other studies (15b, 27a). Furthermore, in our previous studies using a similar β-globin minigene, mutation of the poly(A) signal significantly increased the amount of transcriptional read through into the HIV promoter, as assessed by NRO (11). In an alternative approach, plasmid minigenes containing weak, intermediate, or strong poly(A) signals were injected into Xenopus oocytes and it was shown using electron microscopy that poly(A) efficiency correlates both with the percentage of terminating plasmids and with the site of termination (20).
We also attempted to dissect the Gγ-globin 3′ flank to define precisely the sequences required to promote termination and the mechanism by which termination in the Gγ-globin gene occurs. Sequences up to 1.1 kb downstream of the poly(A) site were sufficient to cause efficient termination in combination with the strong Gγ poly(A) site (Fig. 2b and 4a). However, our efforts to map more precisely which sequences are necessary for termination suggested that there is no single region responsible but rather that multiple sequences spread across the 3′ flank each weakly contribute to the combined termination efficiency of the Gγ-globin gene (Fig. 2c). These results are supported by our efforts to define the mechanism of termination in the Gγ-globin gene; using the poly(A) competition assay we have shown that a number of weak pause elements exist across the Gγ 3′ flank (Fig. 4c).
Transcriptional pausing has been shown to be an important component of the termination mechanism for a number of mammalian and yeast genes. In the fission yeast Schizosaccharomyces pombe, pause sites in the 3′ flanks of the ura4 and nmt2 genes are required for transcription termination (3, 7). Similar pause sites have been identified in the human α2-globin (14), C2 complement (6), and mouse immunoglobulin M (21a) genes. Pausing downstream of the poly(A) site is an integral part of the Torpedo model of termination; transcription complexes which are slowed by pausing are more readily overtaken by the RNA-degrading activity, which eventually causes template release (25). Recent evidence from both yeast and mammalian systems has confirmed that the major nuclear 5′-3 exonuclease (Rat1p in yeast and Xrn2 in mammals) is indeed required for efficient Pol II termination (15a, 27b). Analysis of the C2 pause site in an in vitro-coupled transcription-polyadenylation system showed that pausing can also stimulate polyadenylation (28, 29). In addition the latter studies showed that not all pause sites are able to stimulate polyadenylation; however, they do suggest an additional mechanism by which pausing promotes transcription termination (26).
An alternative type of terminator for the human β-globin gene has recently been described (12). A termination region was identified 900 to 1,600 nt downstream of the poly(A) site of the β-globin gene, and hybrid selection analysis of the nascent transcripts showed that they were not contiguous across this region, instead being cleaved in a cotranscriptional event. Although cotranscriptional cleavage (CoTC) is not dependent on polyadenylation, both events are necessary for termination to occur downstream of the site of CoTC (12). We have looked for cotranscriptional cleavage in both the Gγ- and Aγ-globin 3′ flanks but have found no evidence that it occurs in either gene (Fig. 4b). We therefore predict that while some genes employ CoTC to mediate efficient Pol II termination, other genes utilize pausing mechanisms instead.
The emerging view of transcription termination is one in which events integrated through the carboxy-terminal domain influence the conformation of the transcription complex, RNA processing, and the structure of the chromatin through which transcription occurs (24). Ultimately, this results in transcription termination, not in some random fashion but in a way which is regulated by activities occurring across the genes. Polyadenylation is at the heart of the events leading to transcription termination, and the work on the duplicated γ-globin genes in humans presented here highlights this role by directly comparing two poly(A) signals which have different strengths, ultimately leading to markedly different profiles of transcriptional termination.
Acknowledgments
We thank the members of the lab of N.J.P. for advice during these studies.
This work was supported by the Wellcome Trust (program grant to N.J.P.).
REFERENCES
- 1.Adams, S. E., I. D. Johnson, M. Braddock, A. J. Kingsman, S. M. Kingsman, and R. M. Edwards. 1988. Synthesis of a gene for the HIV transactivator protein Tat by a novel single stranded approach involving in vivo gap repair. Nucleic Acids Res. 16:4287-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alén, C., N. A. Kent, H. S. Jones, O'Sullivan, J., A. Aranda, and N. J. Proudfoot. 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10:1441-1452. [DOI] [PubMed] [Google Scholar]
- 3.Aranda, A. and N. J. Proudfoot. 1999. Definition of transcriptional pause elements in fission yeast. Mol. Cell. Biol. 19:1251-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashe, H. L., J. Monks, M. Wijgerde, P. Fraser, and N. J. Proudfoot. 1997. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 11:2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashe, M. P., P. Griffin, W. James, and N. J. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008-3025. [DOI] [PubMed] [Google Scholar]
- 6.Ashfield, R., A. J. Patel, S. A. Bossone, H. Brown, R. D. Campbell, K. B. Marcu, and N. J. Proudfoot. 1994. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 13:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birse, C. E., B. A. Lee, K. Hansen, and N. J. Proudfoot. 1997. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 16:3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birse, C. E., L. Minvielle Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. [DOI] [PubMed] [Google Scholar]
- 9.Connelly, S. and J. L. Manley. 1988. A functional messenger-RNA polyadenylation signal is required for transcription termination by RNA polymerase-ii. Genes Dev. 2:440-452. [DOI] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 1993. Does HIV-1 Tat induce a change in viral initiation rights? Cell 73:417-420. [DOI] [PubMed] [Google Scholar]
- 10a.Dieci, G., and A. Sentenac. 2002. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 28:202-209. [DOI] [PubMed] [Google Scholar]
- 11.Dye, M. J., and N. J. Proudfoot. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3:371-378. [DOI] [PubMed] [Google Scholar]
- 12.Dye, M. J., and N. J. Proudfoot. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105:669-681. [DOI] [PubMed] [Google Scholar]
- 13.Eggermont, J., and N. J. Proudfoot. 1993. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 12:2539-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enriquez Harris, P., N. Levitt, D. Briggs, and N. J. Proudfoot. 1991. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 10:1833-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greger, I. H., and N. J. Proudfoot. 1998. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Kim, M., N. J. Krogan, L. Vasiljeva, O. J. Rando,E. Nedea, J. F. Greenblatt, and S. Buratowski. 2004. The Rat1 exonuclease promotes transcriptional termination by RNA polymerase II. Nature 432:517-522. [DOI] [PubMed] [Google Scholar]
- 15b.Kim, S. I., and H. G. Martinson. 2003. Poly(A) dependent transcription termination. J. Biol. Chem. 278:41691-41701. [DOI] [PubMed] [Google Scholar]
- 16.Levitt, N., D. Briggs, A. Gil, and N. J. Proudfoot. 1989. Definition of an efficient synthetic poly(A) site. Genes Dev. 3:1019-1025. [DOI] [PubMed] [Google Scholar]
- 16a.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 17.Logan, J., E. Falck Pedersen, J. E. Darnell, and T. Shenk. 1987. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl. Acad. Sci. USA 84:8306-8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayor, C., M. Brudno, J. R. Schwartz, A. Poliakov, E. M. Rubin, K. A. Frazer, L. S. Pachter, and I. Dubchak. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046-1047. [DOI] [PubMed] [Google Scholar]
- 19.Moreira, A., M. Wollerton, J. Monks, and N. J. Proudfoot. 1995. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 14:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osheim, Y. N., N. J. Proudfoot, and A. L. Beyer. 1999. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell 3:379-387. [DOI] [PubMed] [Google Scholar]
- 21.Papassotiriou, I., R. Ducrocq, C. Prehu, J. Bardakdjian-Michau, and H. Wajcman. 1998. Gamma chain heterogeneity: determination of Hb F composition by perfusion chromatography. Hemoglobin 22:469-481. [DOI] [PubMed] [Google Scholar]
- 21a.Peterson, M. L., S. Bertolino, and F. Davis. 2002. An RNA polymerase pause site is associated with the immunoglobulin μs poly(A) site. Mol. Cell. Biol. 22:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plant, K. E., S. J. E. Routledge, and N. J. Proudfoot. 2001. Intergenic transcription in the human β-globin gene cluster. Mol. Cell. Biol. 21:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt, T. 1998. RNA structure in transcription elongation, termination, and antitermination, p. 541-574. In R. Simons and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16:272-278. [DOI] [PubMed] [Google Scholar]
- 25.Proudfoot, N. J. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14:105-110. [DOI] [PubMed] [Google Scholar]
- 26.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating rnRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 27.Tantravahi, J., M. Alvira, and E. Falck Pedersen. 1993. Characterization of the mouse βmaj globin transcription termination region: a spacing sequence is required between the poly(A) signal sequence and multiple downstream termination elements. Mol. Cell. Biol. 13:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Tran, D. P., S. J. Kim, N. J. Park, T. M. Jew, and H. G. Martinson. 2001. Mechanism of poly(A) signal transduction of RNA polymerase II in vitro. Mol. Cell. Biol. 21:7495-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27b.West, S., N. Gromak, and N. J. Proudfoot. 2004. Human 5′->3′ exonuclease Xrn2 promotes transcription termination from sites of co-transcriptional cleavage. Nature 432:522-525. [DOI] [PubMed] [Google Scholar]
- 28.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3:593-600. [DOI] [PubMed] [Google Scholar]
- 29.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]