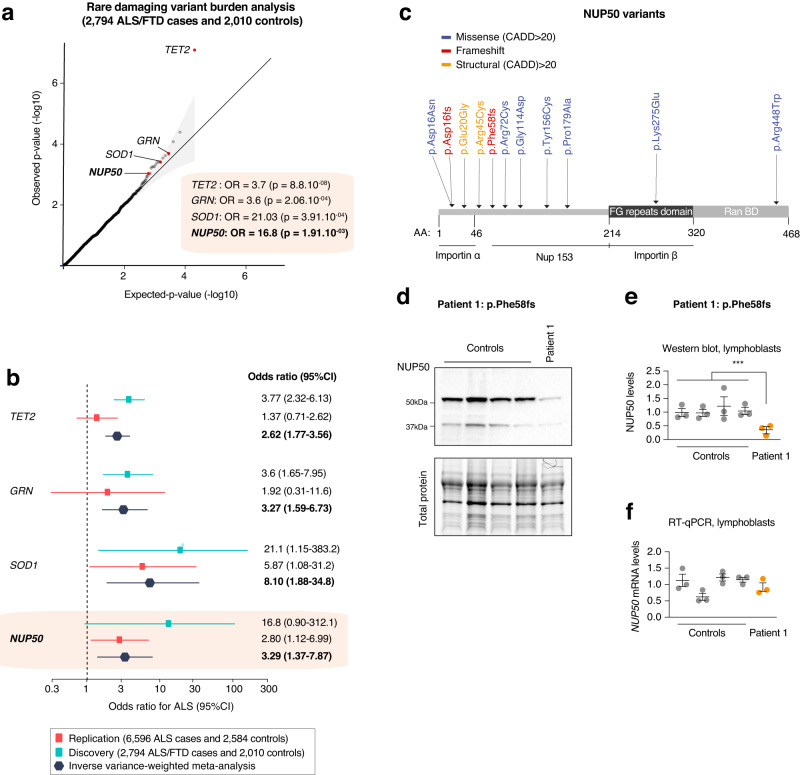
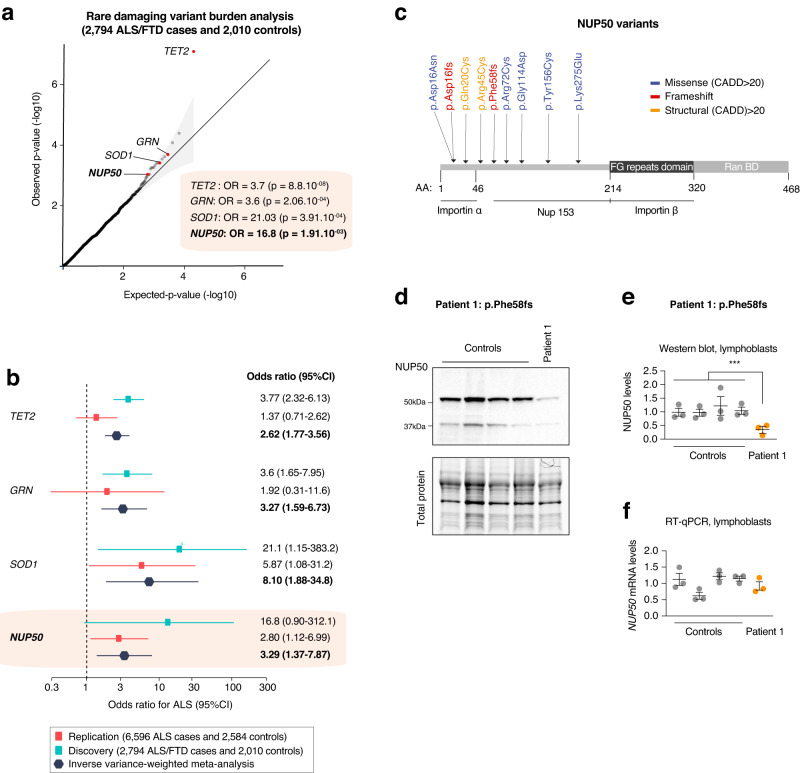
Salim Megat
Salim Megat
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,✉,
Natalia Mora
Natalia Mora
2Department of Molecular Neurobiology, Donders Institute for Brain, Cognition and Behaviour and Faculty of Science, Radboud University, Nijmegen, Netherlands
2,
Jason Sanogo
Jason Sanogo
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,
Olga Roman
Olga Roman
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,
Alberto Catanese
Alberto Catanese
3Institute of Anatomy and Cell Biology, Ulm University, Ulm, Germany
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
3,4,
Najwa Ouali Alami
Najwa Ouali Alami
5Clinical Neuroanatomy, Department of Neurology, Ulm University, Ulm, Germany
5,
Axel Freischmidt
Axel Freischmidt
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
6Department of Neurology, Ulm University, Ulm, Germany
4,6,
Xhuljana Mingaj
Xhuljana Mingaj
7Laboratory of Translational Research for Neurological Disorders, Imagine Institute, Université de Paris, INSERM UMR 1163, 75015 Paris, France
7,
Hortense De Calbiac
Hortense De Calbiac
7Laboratory of Translational Research for Neurological Disorders, Imagine Institute, Université de Paris, INSERM UMR 1163, 75015 Paris, France
7,
François Muratet
François Muratet
8Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, Paris, France
8,
Sylvie Dirrig-Grosch
Sylvie Dirrig-Grosch
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,
Stéphane Dieterle
Stéphane Dieterle
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,
Nick Van Bakel
Nick Van Bakel
2Department of Molecular Neurobiology, Donders Institute for Brain, Cognition and Behaviour and Faculty of Science, Radboud University, Nijmegen, Netherlands
2,
Kathrin Müller
Kathrin Müller
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
9Institute of Human Genetics, Ulm University, Ulm, Germany
4,9,
Kirsten Sieverding
Kirsten Sieverding
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
4,
Jochen Weishaupt
Jochen Weishaupt
10Division for Neurodegenerative Diseases, Neurology Department, University Medicine Mannheim, Heidelberg University, Mannheim, Germany
10,
Peter Munch Andersen
Peter Munch Andersen
11Department of Clinical Science, Neurosciences, Umea University, Umea, Sweden
11,
Markus Weber
Markus Weber
12Neuromuscular Disease Unit/ALS Clinic, Kantonsspital St. Gallen, St. Gallen, Switzerland
12,
Christoph Neuwirth
Christoph Neuwirth
12Neuromuscular Disease Unit/ALS Clinic, Kantonsspital St. Gallen, St. Gallen, Switzerland
12,
Markus Margelisch
Markus Margelisch
13Institute for Pathology, Kanstonsspital St. Gallen, St. Gallen, Switzerland
13,
Andreas Sommacal
Andreas Sommacal
13Institute for Pathology, Kanstonsspital St. Gallen, St. Gallen, Switzerland
13,
Kristel R Van Eijk
Kristel R Van Eijk
14Department of Neurology, University Medical Center Utrecht Brain Center, Utrecht University, Utrecht, The Netherlands
14,
Jan H Veldink
Jan H Veldink
14Department of Neurology, University Medical Center Utrecht Brain Center, Utrecht University, Utrecht, The Netherlands
14;
Project Mine Als Sequencing Consortium,
Géraldine Lautrette
Géraldine Lautrette
15Service de Neurologie, Centre de Référence SLA et autres maladies du neurone moteur, CHU Dupuytren 1, Limoges, France
15,
Philippe Couratier
Philippe Couratier
15Service de Neurologie, Centre de Référence SLA et autres maladies du neurone moteur, CHU Dupuytren 1, Limoges, France
15,
Agnès Camuzat
Agnès Camuzat
8Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, Paris, France
8,
Isabelle Le Ber
Isabelle Le Ber
8Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, Paris, France
8,
Maurizio Grassano
Maurizio Grassano
16ALS Center “Rita Levi Montalcini” Department of Neuroscience, University of Turin, Turin, Italy
16,
Adriano Chio
Adriano Chio
16ALS Center “Rita Levi Montalcini” Department of Neuroscience, University of Turin, Turin, Italy
16,
Tobias Boeckers
Tobias Boeckers
3Institute of Anatomy and Cell Biology, Ulm University, Ulm, Germany
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
3,4,
Albert C Ludolph
Albert C Ludolph
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
6Department of Neurology, Ulm University, Ulm, Germany
4,6,
Francesco Roselli
Francesco Roselli
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
6Department of Neurology, Ulm University, Ulm, Germany
4,6,
Deniz Yilmazer-Hanke
Deniz Yilmazer-Hanke
5Clinical Neuroanatomy, Department of Neurology, Ulm University, Ulm, Germany
5,
Stéphanie Millecamps
Stéphanie Millecamps
8Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, Paris, France
8,#,
Edor Kabashi
Edor Kabashi
7Laboratory of Translational Research for Neurological Disorders, Imagine Institute, Université de Paris, INSERM UMR 1163, 75015 Paris, France
7,#,
Erik Storkebaum
Erik Storkebaum
2Department of Molecular Neurobiology, Donders Institute for Brain, Cognition and Behaviour and Faculty of Science, Radboud University, Nijmegen, Netherlands
2,#,
Chantal Sellier
Chantal Sellier
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,#,
Luc Dupuis
Luc Dupuis
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
1,✉,#
1Université de Strasbourg, Inserm, Mécanismes Centraux et Périphériques de la Neurodégénérescence, UMR-S1118, Centre de Recherches en Biomédecine, Strasbourg, France
2Department of Molecular Neurobiology, Donders Institute for Brain, Cognition and Behaviour and Faculty of Science, Radboud University, Nijmegen, Netherlands
3Institute of Anatomy and Cell Biology, Ulm University, Ulm, Germany
4German Center for Neurodegenerative Diseases (DZNE) Ulm, Ulm, Germany
5Clinical Neuroanatomy, Department of Neurology, Ulm University, Ulm, Germany
6Department of Neurology, Ulm University, Ulm, Germany
7Laboratory of Translational Research for Neurological Disorders, Imagine Institute, Université de Paris, INSERM UMR 1163, 75015 Paris, France
8Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, APHP, Hôpital de la Pitié Salpêtrière, Paris, France
9Institute of Human Genetics, Ulm University, Ulm, Germany
10Division for Neurodegenerative Diseases, Neurology Department, University Medicine Mannheim, Heidelberg University, Mannheim, Germany
11Department of Clinical Science, Neurosciences, Umea University, Umea, Sweden
12Neuromuscular Disease Unit/ALS Clinic, Kantonsspital St. Gallen, St. Gallen, Switzerland
13Institute for Pathology, Kanstonsspital St. Gallen, St. Gallen, Switzerland
14Department of Neurology, University Medical Center Utrecht Brain Center, Utrecht University, Utrecht, The Netherlands
15Service de Neurologie, Centre de Référence SLA et autres maladies du neurone moteur, CHU Dupuytren 1, Limoges, France
16ALS Center “Rita Levi Montalcini” Department of Neuroscience, University of Turin, Turin, Italy
Subject terms: Quantitative trait, Amyotrophic lateral sclerosis, Genetics research
© The Author(s) 2023
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.