Abstract
The pandemic of coronavirus disease 2019 (COVID-19) caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a significant impact on the economy and public health worldwide. Therapeutic options such as drugs and vaccines for this newly emerged disease are eagerly desired due to the high mortality. Using the U.S. Food and Drug Administration (FDA) approved drugs to treat a new disease or entirely different diseases, in terms of drug repurposing, minimizes the time and cost of drug development compared to the de novo design of a new drug. Drug repurposing also has some other advantages such as reducing safety evaluation to accelerate drug application on time. Carvedilol, a non-selective beta-adrenergic blocker originally designed to treat high blood pressure and manage heart disease, has been shown to impact SARS-CoV-2 infection in clinical observation and basic studies. Here, we applied computer-aided approaches to investigate the possibility of repurposing carvedilol to combat SARS-CoV-2 infection. The molecular mechanisms and potential molecular targets of carvedilol were identified by evaluating the interactions of carvedilol with viral proteins. Additionally, the binding affinities of in vivo metabolites of carvedilol with selected targets were evaluated. The docking scores for carvedilol and its metabolites with RdRp were − 10.0 kcal/mol, − 9.8 kcal/mol (1-hydroxyl carvedilol), − 9.7 kcal/mol (3-hydroxyl carvedilol), − 9.8 kcal/mol (4-hydroxyl carvedilol), − 9.7 kcal/mol (5-hydroxyl carvedilol), − 10.0 kcal/mol (8-hydroxyl carvedilol), and − 10.1 kcal/mol (O-desmethyl carvedilol), respectively. Using the molecular dynamics simulation (100 ns) method, we further confirmed the stability of formed complexes of RNA-dependent RNA polymerase (RdRp) and carvedilol or its metabolites. Finally, the drug-target interaction mechanisms that contribute to the complex were investigated. Overall, this study provides the molecular targets and mechanisms of carvedilol and its metabolites as repurposed drugs to fight against SARS-CoV-2 infection.
Subject terms: Bioinformatics, Antivirals, SARS-CoV-2
Introduction
The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection causes a global coronavirus disease 2019 (COVID-19) pandemic. According to the report of World Health Organization (WHO) (https://covid19.who.int/), there are 771,820,937 confirmed COVID-19 cases and 6,978,175 deaths among these cases worldwide by November 8, 2023. In May 2023, WHO announced the end of the COVID-19 emergency and started the transition to long-term management of COVID-19. There is an immeasurable economic cost related to COVID-19 during and post-pandemic periods. It is caused by an enveloped, single-stranded positive-sense RNA virus, belonging to the genus Betacoronavirus, one of the four coronaviral genera (Alphacoronavirus (α-), Betacoronavirus (β-), Gammacoronavirus (γ-), and Deltacoronavirus (δ-))1–4. Within the Betacoronavirus, SARS-CoV-2 shares 79% of sequence identity with SARS-CoV-1 and 50% of sequence similarity with MERS-CoV5. The genome of SARS-CoV-2 is 29.9 kb in length, which encodes structural proteins comprising spike protein (S), envelope protein (E), membrane protein (M), nucleocapsid protein (N), 16 non-structural proteins (NSP1-NSP16) in the open reading frame (ORF)1a and ORF1b regions, and 9 accessory proteins located in the region that contains the structural proteins6–8. S protein plays an essential role in the entry of SARS-CoV-2 into host cells, including subunits S1 and S2. Subunit S1 recognizes and binds with the human angiotensin-converting enzyme 2 (hACE2) receptor located on the surface of host epithelium cells in the lungs as well as blood vessels, then subunit S2 functions on the membrane fusion between the viral membrane and host cell membrane to finish the virus entry process9. Of note, there are two pathways of membrane fusion. One pathway is through the direct membrane fusion between the viral membrane and host cell membrane on the cell surface to release the viral genomic RNAs into the host cells, while the other pathway is processed by endocytosis to finish the membrane fusion inside the host cells and the release of viral RNAs10,11. After virus entry and membrane fusion, viral replication and proliferation proceed, then the newly made viruses are released from the infected host cells to infect other host cells.
Some important viral proteins can serve as therapeutic targets. Viral proteins including the structure and non-structure proteins play a pivotal role in the viral life cycle and pathogenesis7,12. The receptor binding domain (RBD) of the spike protein subunit S1 binds with human ACE2 to form the RBD-ACE2 complex facilitating the membrane fusion and virus entry into host cells. Based on this mechanism, a therapeutic agent that can inhibit or block the binding process by targeting the RBD-ACE2 complex is one of the therapeutic options to fight against SARS-CoV-2 infection13,14. The 3-chymotrypsin-like protease (3CLpro), also known as main protease (Mpro) or NSP5, is another favorable therapeutic target, which is associated with the release and maturation of NSP4-NSP16 and has the role in the cleavage of both virus and host proteins. 3CLpro also contributes to the interruption of host innate immunity15,16. Additionally, NSP12, well-known as RNA-dependent RNA polymerase (RdRp) that shares more than 95% sequence identified with SARS-CoV-1, can serve as a promising therapeutic target due to its close association with viral genome transcription and virus replication. This polymerase is the inhibitor target of the FDA-approved drug remdesivir (VEKLURY) for emergency application in SARS-CoV-2 treatment17–20. Papain-like protease (PLpro, a domain of Nsp3) is responsible for virus spread via the generation of replicase complex, a target for inhibiting viral transmission21,22. Furthermore, NSP13 helicase contributes the energy to facilitate the unwinding of RNA, which is critical to virus RNA replication and virus propagation23,24. NSP2 also plays an important role in facilitating virus replication by suppressing the production of immunomodulator IFN-β to impair host innate immunity to virus infection25. The above-mentioned proteins serve as ideal therapeutic drug targets for the treatment of COVID-1926,27.
A recent study discovered that the usage of carvedilol was associated with a lower rate of COVID-19-positive diagnosis. As a U.S. FDA-approved drug, carvedilol belongs to the third generation of beta-blockers, which has been used to treat hypertension due to its vasodilating properties28. In contrast to the traditional beta-blockers that can increase the expression of ACE2, carvedilol has the capability to reduce the expression of ACE. ACE serves as the binding receptor of SARS-CoV-2 on the membrane of host cells, facilitating virus entry into host cells. Therefore, the reduction of ACE expression level could limit virus binding and entry into host cells. Thus, carvedilol may inhibit SARS-CoV-2 infection by preventing the binding of viruses with host cells through inhibition of ACE expression29. In vitro assay also tested that carvedilol displayed strong anti-SARS-CoV-2 function in human lung epithelial cells. Metadata network analysis of virus protein-host protein interaction (PPI) revealed that carvedilol can serve as a potential drug for repurposing to treat COVID-19 due to its involvement in multiple important pathways, such as mRNA splicing and glucose metabolism30. In addition, carvedilol has the effect of reducing platelet aggregation31, which has been shown to be increased in COVID-19 patients in the intensive care unit (ICU)32. Thus, using carvedilol may contribute to the modulation of platelet levels in COVID-19 patients. Furthermore, it also has been suggested that carvedilol can regulate the expression of pro-inflammatory cytokines (e.g., IL-6) to reduce inflammation in COVID-19 patients33–36.
Although there are some beneficial effects of carvedilol against COVID-19, its anti-SARS-CoV-2 infection mechanism currently remains unclear. In this study, we first investigated the molecular mechanisms of carvedilol against SARS-CoV-2 infection using an in silico computational molecular docking approach. In addition, we further investigated the binding affinity of in vivo metabolites of carvedilol with SARS-CoV-2 virus, including 1-hydroxyl carvedilol, 3-hydroxyl carvedilol, 4-hydroxyl carvedilol, 5-hydroxyl carvedilol, 8-hydroxyl carvedilol, and O-desmethyl carvedilol. Finally, molecular dynamic simulation (MD) was employed to perform the in silico molecular simulation to study the stability of each drug-target complex and analyze the contact and interaction between small molecules and target proteins.
Results
In silico molecular docking of carvedilol with target proteins of SARS-CoV-2
To investigate the potential mechanisms of carvedilol against SARS-CoV-2 infection, we first applied computational-based approaches to test the binding affinities of carvedilol with potential target proteins of SARS-CoV-2, including RdRp, 3CL/Mpro, NSP13 helicase, NSP2, PLpro, RBD, ACE2, and the complex of RBD-ACE2 (Fig. 1). To find the best protein–ligand binding complex with the highest binding affinity, we performed blind molecular docking using RdRp crystal structure (PDB ID:6XQB, resolution of 3.40 Å, Chain A) to dock with carvedilol and its metabolites. The molecular docking method allowed to screen the best binding sites of candidate compounds with target proteins. In silico molecular docking results showed a higher docking score (− 10.0 kcal/mol) between carvedilol and viral protein RdRp compared to the scores of other tested proteins, including 3CL/Mpro (− 7.2 kcal/mol), NSP13 Helicase (− 7.4 kcal/mol), NSP2 (− 7.5 kcal/mol), PLpro (− 7.9 kcal/mol), RBD (− 6.8 kcal/mol), ACE2(− 8.0 kcal/mol), and the complex of RBD-ACE2 (− 8.1 kcal/mol). This suggests that RdRp could be an ideal potential drug target for carvedilol against SARS-CoV-2 infection.
Figure 1.
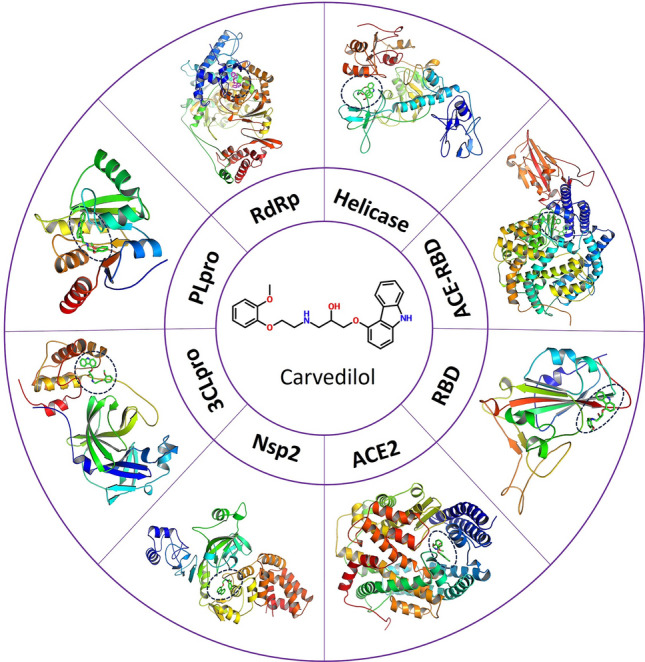
Schematic illustration of different SARS-CoV-2 proteins used to dock with carvedilol. Prepared protein structures used for docking are originally generated from the crystal structure obtained from the PDB database with details in the Methods. RdRp: RNA-directed RNA polymerase, Helicase: Non-structural protein 2 (NSP13), ACE2: angiotensin-converting enzyme 2, RBD: SARS-CoV-2 spike protein S1 receptor-binding domain, NSP2: Non-structural protein 2, 3CLpro/Mpro: main protease, PLpro: Papain-like protease (NSP3).
RdRp is also known as the drug target of remdesivir. Then, we used the same docking system to perform the molecular docking between remdesivir and RdRp. The molecular docking scores between remdesivir and carvedilol with RdRp were − 9.1 kcal/mol and − 10.1 kcal/mol, respectively, suggesting that carvedilol had a higher binding affinity with RdRp compared to remdesivir (Table 1).
Table 1.
Molecular docking results for the binding affinity scores of viral target proteins and carvedilol and its metabolites.
| Proteins targets/Molecules | RdRp (kcal/mol) | 3CL/Mpro (kcal/mol) | Helicase/NSP13 (kcal/mol) | NSP2 (kcal/mol) | PLpro (kcal/mol) | RBD (kcal/mol) | ACE2 (kcal/mol) | RBD-ACE2 complex (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| Carvedilol | − 10.0 | − 7.2 | − 7.4 | − 7.5 | − 7.9 | − 6.8 | − 8.0 | − 8.1 |
| 1-hydroxyl carvedilol | − 9.8 | − 7.5 | − 7.4 | − 8.1 | − 7.6 | − 6.9 | − 8.5 | − 8.1 |
| 3-hydroxyl carvedilol | − 9.9 | − 6.7 | − 7.9 | − 7.4 | − 7.2 | − 6.8 | − 8.0 | − 8.2 |
| 4-hydroxyl carvedilol | − 9.8 | − 7.1 | − 7.2 | − 8.1 | − 8.2 | − 6.9 | − 8.0 | − 8.4 |
| 5-hydroxyl carvedilol | − 9.7 | − 7.1 | − 7.8 | − 7.9 | − 7.7 | − 6.9 | − 8.2 | − 8.2 |
| 8-hydroxyl carvedilol | − 10.0 | − 7.3 | − 7.5 | − 8.3 | − 7.7 | − 7.1 | − 7.8 | − 8.1 |
| O-Desmethyl carvedilol | − 10.1 | − 7.3 | − 7.3 | − 7.6 | − 8.0 | − 6.9 | − 8.5 | − 8.3 |
| Remdesivir | − 9.1 | − 7.9 | − 8.7 | − 8 | − 8.7 | − 7.2 | − 8.3 | − 8.7 |
Molecular docking results of binding affinity between each target viral protein with carvedilol and its metabolites (1-hydroxyl carvedilol, 3-hydroxyl carvedilol, 4-hydroxyl carvedilol, 5-hydroxyl carvedilol, 8-hydroxyl carvedilol, and O-desmethyl carvedilol), as well as the control remdesivir, showed that different molecules had a higher binding affinity with viral RdRp compared to the other tested viral proteins (3CL/Mpro, helicase, NSP2, PLpro, RBD, ACE2, RBD-ACE2 complex).
In silico molecular docking of carvedilol metabolites with target viral proteins of SARS-CoV-2
Considering in vivo metabolites of carvedilol also could bind with viral proteins, we then performed the molecular docking between viral proteins of interest and 6 metabolites of carvedilol37, including 1-hydroxyl carvedilol, 3-hydroxyl carvedilol, 4-hydroxyl carvedilol, 5-hydroxyl carvedilol, 8-hydroxyl carvedilol, and O-desmethyl carvedilol (Fig. 2). The docking scores of each metabolite with protein RdRp of SARS-CoV-2 were separately analyzed and shown in a Table 1. The binding affinity scores of each carvedilol metabolite with RdRp were − 9.8 kcal/mol (1-hydroxyl carvedilol), − 9.7 kcal/mol (3-hydroxyl carvedilol), − 9.8 kcal/mol (4-hydroxyl carvedilol), − 9.7 kcal/mol (5-hydroxyl carvedilol), − 10.0 kcal/mol (8-hydroxyl carvedilol), and − 10.1 kcal/mol (O-desmethyl carvedilol), respectively.
Figure 2.
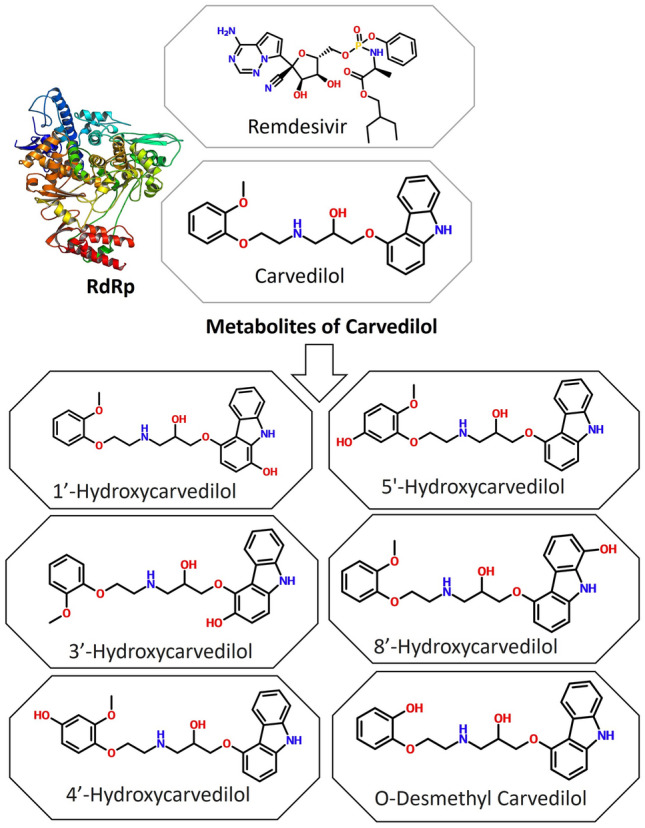
Secondary molecular structures of carvedilol and its metabolites and tertiary structure of RdRp of SARS-CoV-2. Remdesivir is used as a reference for binding affinity.
In addition to docking with the viral protein RdRp, other viral proteins of interest were also used for molecular docking with 6 metabolites of carvedilol. The results showed that all 6 metabolites of carvedilol displayed a higher binding affinity with the viral RdRp domain compared to their binding affinity with each of the other protein domains (Table 1). Overall, RdRp has the highest binding affinity with carvedilol and its metabolites, suggesting that RdRp is the most potent druggable target for carvedilol and its metabolites. In addition, the properties of carvedilol and its metabolites binding with multiple viral proteins suggest a possibility of exploring antiviral drugs using dual or multiple targets of interest.
Analysis of the key residues contributing to the interaction of carvedilol and its metabolites with drugable target protein RdRp
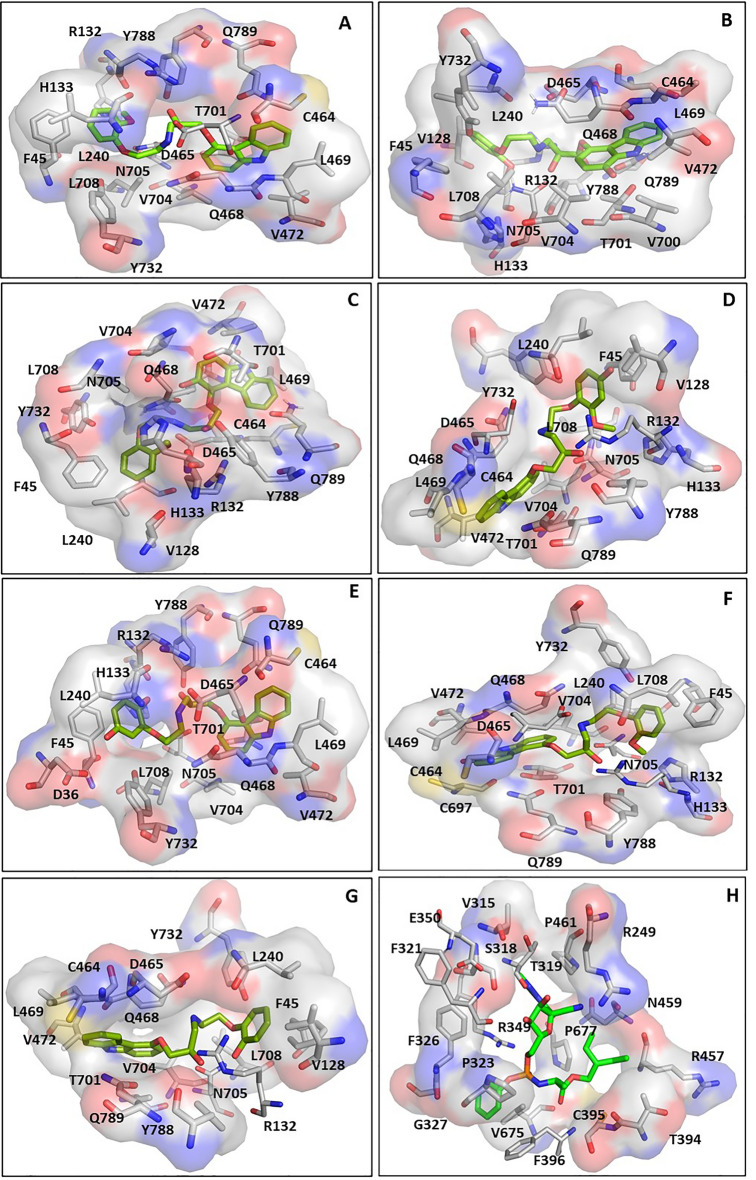
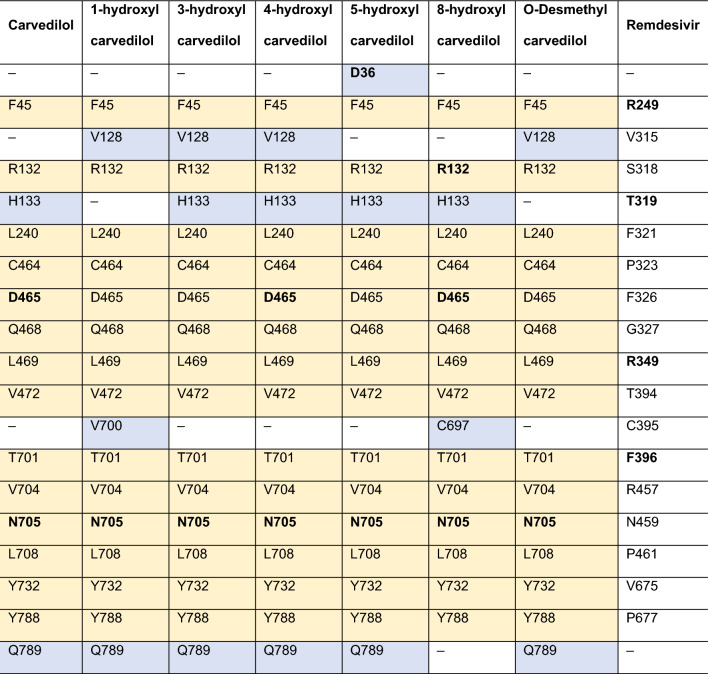
Given the higher binding affinity of carvedilol and its metabolites with RdRp compared to other virus proteins, we then focused on the analysis of the binding features or properties of carvedilol and its metabolites in complex with RdRp. The key amino acids contributing to the interaction of RdRp with carvedilol and its metabolites were analyzed and shown in a figure (Fig. 3) and listed in a table (Table 2). The results showed that the binding pocket position for carvedilol and its metabolites binding with RdRp was the same. Although RdRp interacted with different ligands, including carvedilol and its metabolites, most of these key residues that contribute to the binding complexes were shared. For example, the residues (F45, R132, L240, C464, D465, Q468, L469, V472 T701, V704, N705, L708, Y732, and Y788) contributed to the interaction of all the complexes, including hydrophobic interactions. In addition to the residues shared by different complexes, some residues such as D36, V128, H133, V700, C697, and Q789 served as the key residues only in some of the complexes. The binding pocket position of carvedilol and its metabolites with RdRp was different from the binding pocket position of remdesivir-RdRp. The key residues involved in the remdesivir-RdRp complex were different from the above-mentioned key residues involved in the complex of carvedilol/its metabolites-RdRp, listed in Table 2.
Figure 3.
The binding sites of RdRp in the complexes with carvedilol and its metabolites. Remdesivir was applied as a control. (A) The binding site of RdRp with carvedilol. (B) The binding site of RdRp with 1-hydroxyl carvedilol. (C) The binding site of RdRp with 3-hydroxyl carvedilol. (D) The binding site of RdRp with 4-hydroxyl carvedilol. (E) The binding site of RdRp with 5-hydroxyl carvedilol. (F) The binding site of RdRp with 8-hydroxyl carvedilol. (G) The binding site of RdRp with O-desmethyl carvedilol. (H) The binding site of RdRp with remdesivir.
Table 2.
Key residues involved in the interactions between viral protein RdRp and carvedilol and its metabolites.
The interactions between viral protein RdRp and carvedilol (column 1) and its metabolites (columns 2–7) were analyzed, and remdesivir was used as a control (column 8). Residues in bold text contribute to the formation of hydrogen bonds. The yellow highlighted residues stand for the same residues of RdRp that contribute to the ligand–protein interaction between different complexes.
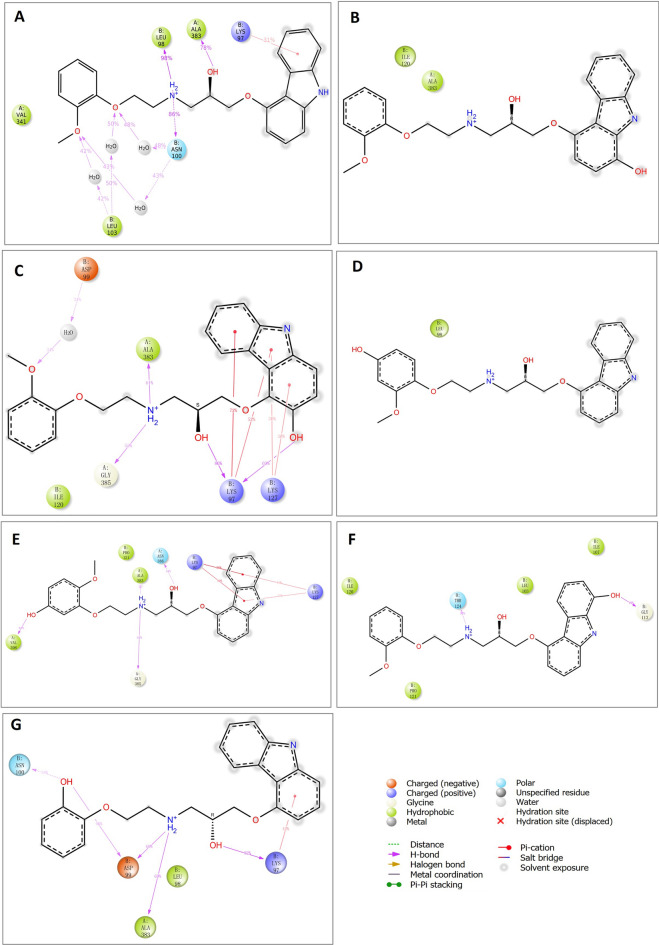
Analysis of the hydrogen bond and other interactions that contribute to the binding affinity of RdRp with carvedilol and its metabolites
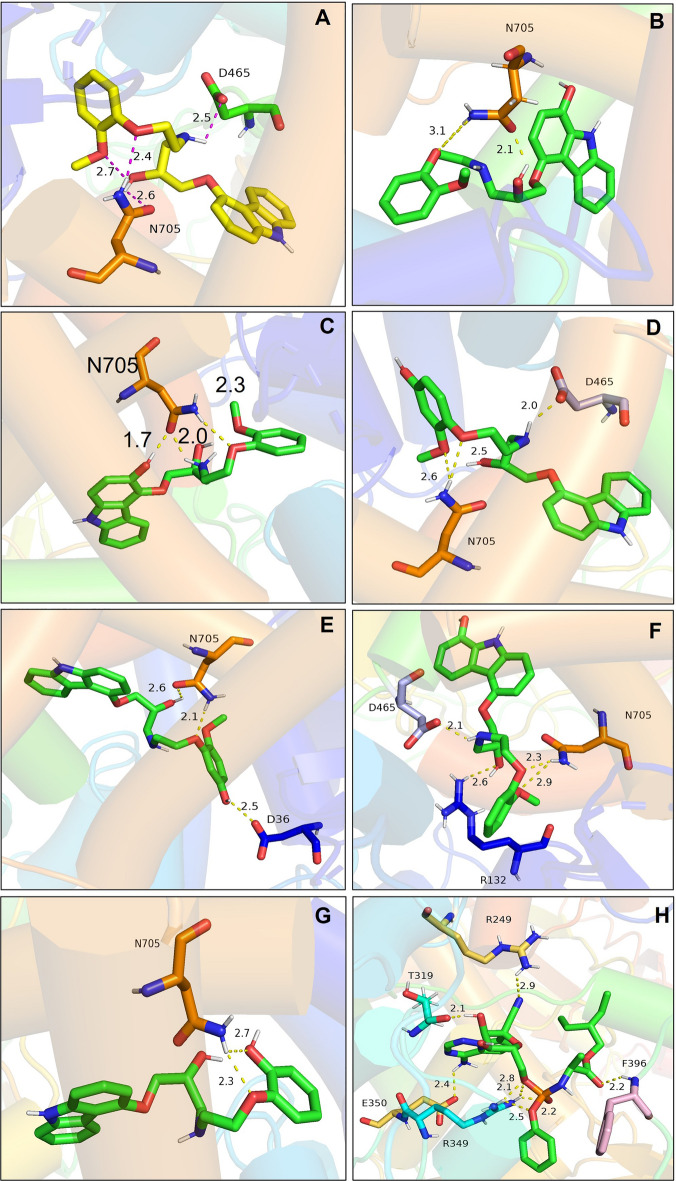
The hydrogen bonds contributing to the binding affinity between RdRp and carvedilol and its metabolites were demonstrated in a figure (Fig. 4A–G). In the binding complexes, two or three stable hydrogen bonds were formed between residue N705 of RdRp and carvedilol and its metabolites. In addition to N705, the residues D465 contributed to hydrogen bond formation in the complex of RdRp binding with carvedilol, 4-hydroxyl carvedilol, and 8-hydroxyl carvedilol. Residue D36 formed the hydrogen bond in the complex of RdRp binding with 5-hydroxyl carvedilol, and residue R132 contributed to the hydrogen bond formation in the complex of RdRp binding with 8-hydroxyl carvedilol. In contrast, the residues of RdRp forming hydrogen bonds with remdesivir were different from those in the complexes of RdRp with carvedilol and its metabolites, including R249, T319, R349, E350, and F396 (Fig. 4H). All these residues are highlighted in Table 2, contributing to the protein–ligand interaction.
Figure 4.
Hydrogen bonds in the binding complexes. The key residues of the RdRp that contribute to the formation of hydrogen bonds in different complexes of RdRp with carvedilol or its metabolites were analyzed. (A) carvedilol. (B) 1-hydroxyl carvedilol. (C) 3-hydroxyl carvedilol. (D) 4-hydroxyl carvedilol. (E) 5-hydroxyl carvedilol. (F) 8-hydroxyl carvedilol. (G) O-desmethyl carvedilol. (H) The interaction between RdRp and remdesivir was tested as a control. The labeled number is the distance between two atoms that form the hydrogen bond.
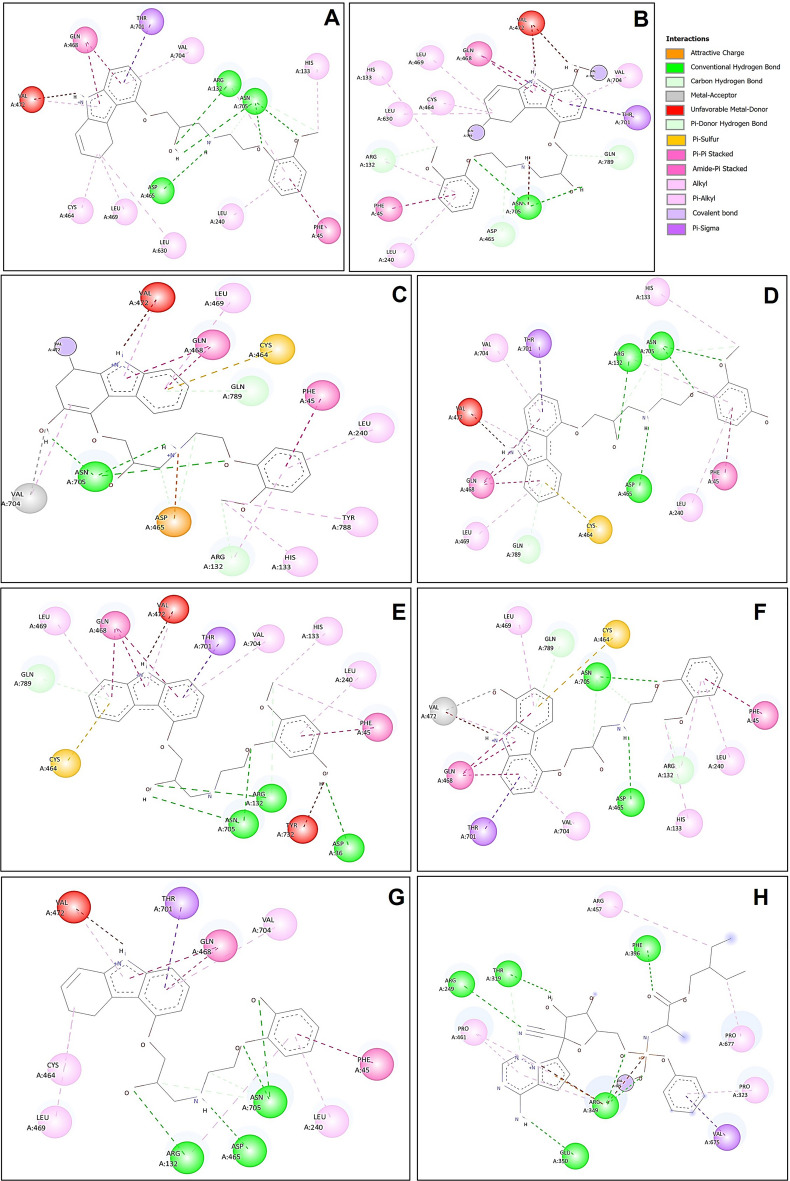
In addition to the hydrophobic interaction and hydrogen bonds, the interactions between the ligand and protein also include Van der Waals bonds, Pi–Sigma, Pi–Pi Stack, Pi–Alkyl Pi–Sulfur, Pi–Amide stack, and covalent bonds, contributing to the stability of the protein–ligand complexes (Fig. 5).
Figure 5.
2D graph of all the interactions in the binding sites of RdRp with carvedilolor and its metabolites. (A) carvedilol. (B) 1-hydroxyl carvedilol. (C) 3-hydroxyl carvedilol. (D) 4-hydroxyl carvedilol. (E) 5-hydroxyl carvedilol. (F) 8-hydroxyl carvedilol. (G) O-desmethyl carvedilol. (H) The interaction between RdRp and remdesivir was also tested and displayed.
Analysis of the binding pocket of viral RdRp and drugable properties of carvedilol and its metabolites
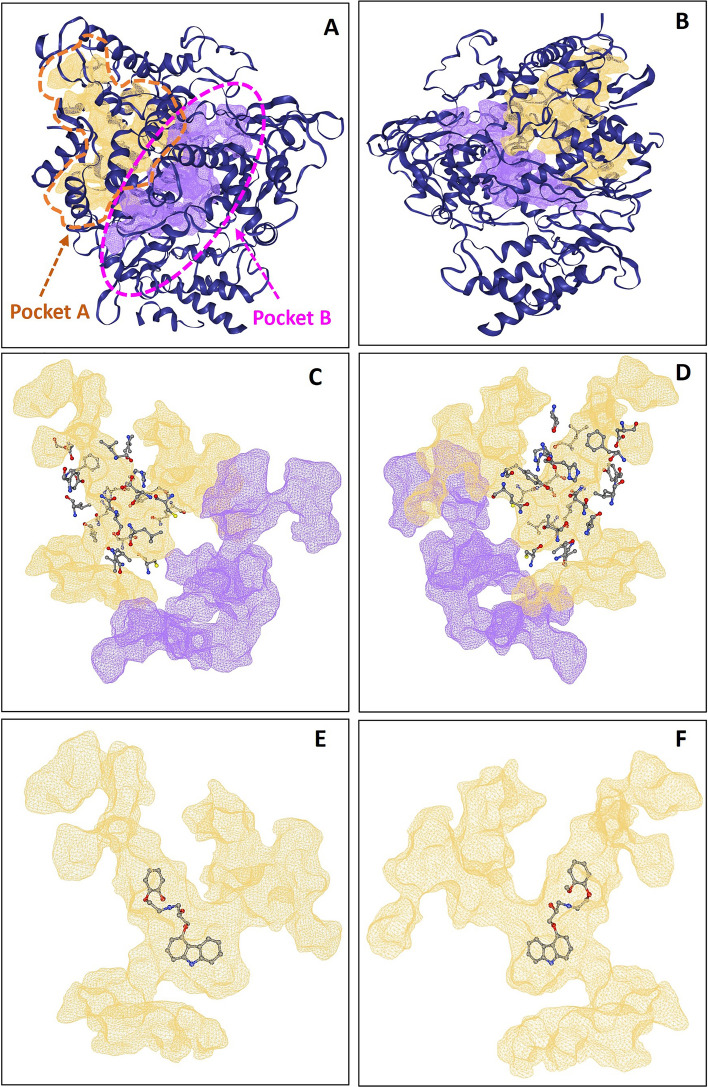
To further confirm the availability and druggability of the binding pocket, the computational-based prediction of druggable major pockets of viral RdRp protein was analyzed. Two major druggable pockets were identified with a higher score of druggability 0.81 for both pockets (pocket A and pocket B) (Score range: 0–1) (Fig. 6). The volume [Å3] of predicted binding pocket A was 2450.27 and the surface [Å2] of predicted binding pocket A was 2553.03. Pocket A was formed by 133 amino acids (Table 3) with functional groups, including 52 hydrogen bond donors, 185 hydrogen bond acceptors, and 53 hydrophobic interactions with a hydrophobicity ratio of 0.18. The amino acid composition of predicted pocket A showed that the apolar amino acid ratio was 0.59, the polar amino acid ratio was 0.31, the positive amino acid ratio was 0.07, and the negative amino acid ratio was 0.04.
Figure 6.
Major binding pockets of RdRp. (A) The front side of the two identified major binding pockets (Pocket A in yellow color and Pocket B in purple color). (B) The backside of the two identified major binding pockets (Pocket A in yellow color and Pocket B in purple color). The front side (C) and backside (D) of the two major identified binding pockets. Pocket A (yellow) was surrounded by the key amino acids that contribute to the interaction of the protein–ligand binding between RdRp and carvedilol or its metabolites. The front side (E) and backside (F) of the binding site of ligand (carvedilol)-RdRp complex used as an example.
Table 3.
List of the amino acids that form the identified major binding pocket A. 133 amino acids form one of the major pockets (Pocket A) of RdRp (columns 1–7) and the key residues of RdRp in complex with carvedilol and its metabolites (column 8).
| 133 amino acids that form one of the major pockets (Pocket 1) of RdRp | Key residues | ||||||
|---|---|---|---|---|---|---|---|
| ALA_34_A | VAL_166_A | TYR_217_A | THR_248_A | PRO_620_A | SER_709_A | LEU_775_A | ASP_36_A |
| PHE_35_A | PRO_169_A | ASP_218_A | ARG_249_A | LYS_621_A | VAL_720_A | VAL_776_A | PHE_45_A |
| ASP_36_A | ILE_171_A | PHE_219_A | TYR_455_A | ARG_624_A | LEU_723_A | ALA_777_A | VAL_128_A |
| TYR_38_A | LEU_172_A | GLY_220_A | ARG_457_A | ALA_625_A | GLN_724_A | PHE_782_A | ARG_132_A |
| PHE_45_A | VAL_174_A | ASP_221_A | TYR_458_A | PRO_627_A | LEU_727_A | SER_784_A | HIS_133_A |
| MET_124_A | TYR_175_A | PHE_222_A | ASN_459_A | LEU_630_A | TYR_728_A | VAL_785_A | LEU_240_A |
| ALA_125_A | ALA_176_A | PRO_232_A | LEU_460_A | MET_633_A | LEU_731_A | LEU_786_A | CYS_464_A |
| LEU_127_A | LEU_178_A | VAL_233_A | PRO_461_A | ASN_695_A | TYR_732_A | TYR_787_A | ASP_465_A |
| VAL_128_A | ARG_181_A | VAL_234_A | THR_462_A | ILE_696_A | ARG_733_A | TYR_788_A | GLN_468_A |
| TYR_129_A | VAL_182_A | SER_236_A | MET_463_A | CYS_697_A | PHE_741_A | GLN_789_A | LEU_469_A |
| ALA_130_A | THR_189_A | TYR_237_A | CYS_464_A | GLN_698_A | PHE_745_A | ASN_790_A | VAL_472_A |
| LEU_131_A | PHE_192_A | TYR_238_A | ASP_465_A | ALA_699_A | TYR_748_A | ASN_791_A | CYS_697_A |
| ARG_132_A | CYS_193_A | SER_239_A | GLN_468_A | VAL_700_A | LEU_749_A | VAL_792_A | VAL_700_A |
| HIS_133_A | VAL_204_A | LEU_240_A | LEU_469_A | THR_701_A | HIS_752_A | THR_701_A | |
| PHE_134_A | LEU_205_A | LEU_241_A | PHE_471_A | ALA_702_A | PHE_753_A | VAL_704_A | |
| THR_141_A | THR_206_A | PRO_243_A | VAL_472_A | ASN_703_A | MET_755_A | ASN_705_A | |
| LEU_142_A | LEU_207_A | ILE_244_A | VAL_475_A | VAL_704_A | ILE_757_A | LEU_708_A | |
| ILE_145_A | ASP_208_A | LEU_245_A | TYR_479_A | ASN_705_A | ALA_762_A | TYR_732_A | |
| LEU_146_A | ASN_209_A | THR_246_A | MET_615_A | LEU_707_A | VAL_764_A | TYR_788_A | |
| PHE_165_A | TRP_216_A | LEU_247_A | TRP_617_A | LEU_708_A | TYR_770_A | GLN_789_A | |
Residues in bold are the key residues that contribute to the formation of RdRp in complexes with carvedilol and its metabolites.
The results from our study showed the binding position of carvedilol and its metabolites were located within the major binding pocket A. All the key amino acids that contributed to the interaction of the binding affinity of the protein–ligand complex between the RdRp and carvedilol or its metabolites were included in the 133 amino acids that formed pocket A (Table 3). The size of binding pockets of carvedilol and its metabolites were smaller compared to pocket A, and the predicted druggable major binding pocket of these molecules was exactly located inside of the major binding pocket A. These results suggest that carvedilol and its metabolites have druggable properties against SARS-CoV-2 infection by targeting viral RdRp domain.
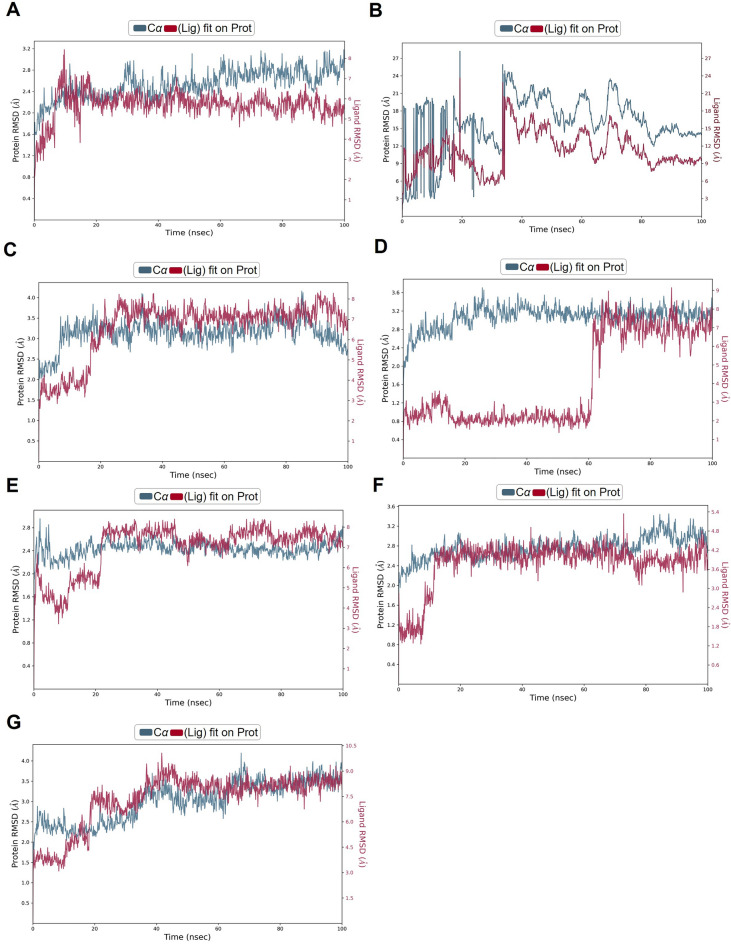
Molecular dynamic simulation to validate the binding of complexes
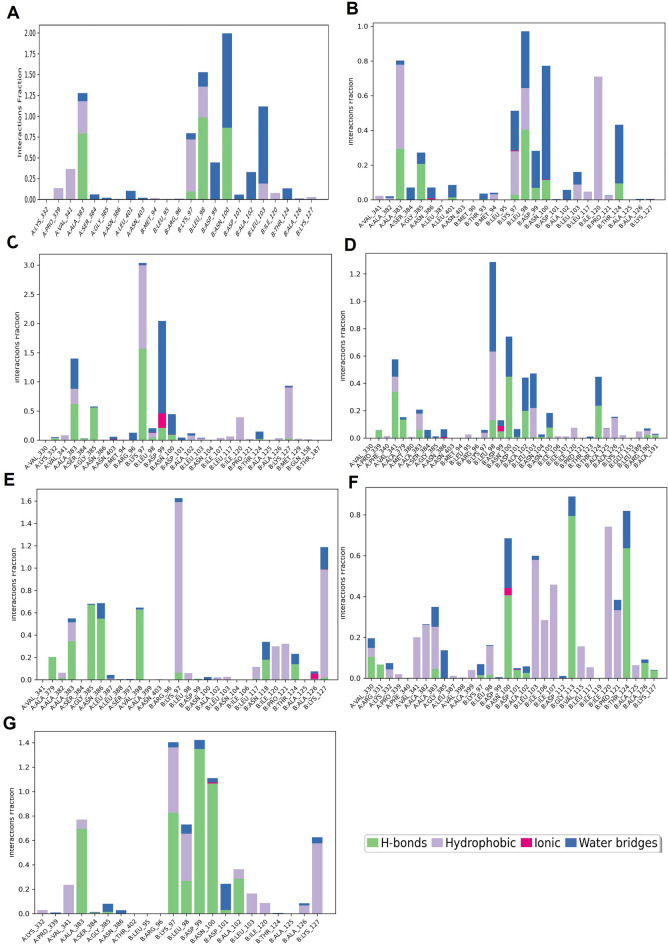
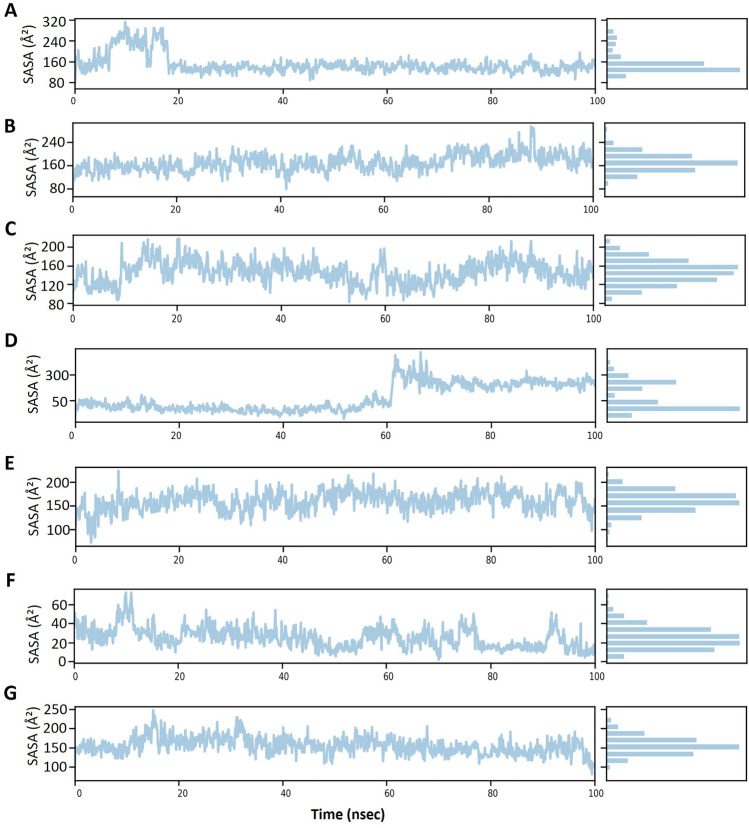
To further test the binding stability between carvedilol and target proteins, we chose the most conserved viral protein RdRp comprising a catalytic subunit NSP12 and two acessary subunits NSP7 and NSP8, also known as a target of FDA-approved drug remdesivir, to conduct the molecular simulation. Within the simulated time range of 100 ns, except for the RdRp-1-hydroxyl carvedilol complex (Fig. 7B), all the other tested complexes reached a relatively stable stage (Fig. 7A and C–G). The root mean square fluctuation (RMSF)38 results of proteins and ligands were further analyzed to test the fluctuation of atoms around the average position (Supplementary Fig. 1). These tested complexes include carvedilol-RdRp and carvedilol metabolite-RdRp complexes. The interacting features and properties of molecules with target proteins were characterized and analyzed for each complex, respectively. For instance, these interactions included hydrogen bound, hydrophobicity, charge distribution, polarization, Pi-cation, etc. (Fig. 8). The interacting residues and fractions that contributed to the interaction of hydrogen bonds, hydrophobic, ionic, and water bridges were analyzed (Fig. 9). These residues included the amino acids from RdRp NSP12 (displayed in chain A, Fig. 9) and amino acids from co-factors NSP8 (displayed in Chain B, Fig. 9). The solvent accessible surface area (SASA) measures the surface area of a molecule that is accessible to a solvent, which is a valuable metric for protein–ligand interactions. The fluctuation of SASA for carvedilol and its metabolites in complex with RdRp was displayed in the result of SASA profile generated within 100 ns of dynamic simulation (Fig. 10).
Figure 7.
Results of molecular dynamics simulation of RdRp in complex with carvedilol and its metabolites. (A) The complex of RdRp (PDB ID: 6XQB) and carvedilol. (B) The complex of RdRp and 1-hydroxyl carvedilol. (C) The complex of RdRp and 3-hydroxyl carvedilol. (D) The complex of RdRp and 4-hydroxyl carvedilol. (E) The complex of RdRp and 5-hydroxyl carvedilol. (F) The complex of RdRp and 8-hydroxyl carvedilol. (G) The complex of RdRp and O-Desmethyl carvedilol.
Figure 8.
2D schematic diagrams of the ligand atoms interacting with target proteins. The ligand atoms of carvedilol and its metabolites interacting with target protein RdRp (PDB ID: 6XQB) were analyzed, respectively. (A) The complex of RdRp and carvedilol. (B) The complex of RdRp and 1-hydroxyl carvedilol. (C) The complex of RdRp and 3-hydroxyl carvedilol. (D) The complex of RdRp and 4-hydroxyl carvedilol. (E) The complex of RdRp and 5-hydroxyl carvedilol. (F) The complex of RdRp and 8-hydroxyl carvedilol. (G) The complex of RdRp and O-desmethyl carvedilol. Labeled numbers stand for the percentages of contributions of atoms for the binding within 100 ns simulation time range.
Figure 9.
Stacked bar chart plot showing the protein-ligands interactions found during the 100 ns simulation run. The X-axis shows the residues of the protein that contribute to the protein–ligand interaction under the four categories including hydrogen bonds, hydrophobic, ionic, and water bridges. The residues from RdRp NSP12 (displayed as chain A) and amino acids from co-factors NSP8 (displayed as Chain B) (PDB ID: 6XQB). The Y-axis stands for the interaction fraction that describes how long interactions are maintained over the simulation run. An interaction fraction of 1.0 suggests that this interaction is maintained in the simulation 100% of the time. If the value is > 1.0, this means that the protein residue is involved in multiple interaction contacts.
Figure 10.
Solvent accessible surface area (SASA) results from the molecular dynamic simulation of the ligand–protein complexes. The interactions include carvedilol and its metabolites interacting with the target protein RdRp (PDB ID: 6XQB), respectively. (A) The complex of RdRp and carvedilol. (B) The complex of RdRp and 1-hydroxyl carvedilol. (C) The complex of RdRp and 3-hydroxyl carvedilol. (D) The complex of RdRp and 4-hydroxyl carvedilol. (E) The complex of RdRp and 5-hydroxyl carvedilol. (F) The complex of RdRp and 8-hydroxyl carvedilol. (G) The complex of RdRp and O-desmethyl carvedilol. The simulation time is 100 ns, SPC (simple point charge) water model as solvent for simulation was used.
AMDET analysis results showed that all the tested molecules meet the Lipinski drug-likeness criteria and were accepted by the criteria applied by Pfizer for drug discovery screening. The drug-likeness results also met the criteria of Egan (Pharmacia) filter and Muegge (Bayer) filter for drug screening (Table 4).
Table 4.
Pharmacokinetic properties ADMET analyses of absorption, distribution, excretion, metabolism, toxicity, and drug-likeness assessment.
| ADMET | Carvedilol | 1-hydroxyl carvedilol | 3-hydroxyl carvedilol | 4-hydroxyl carvedilol | 5-hydroxyl carvedilol | 8-hydroxyl carvedilol | O-desmethyl carvedilol |
|---|---|---|---|---|---|---|---|
| Physicochemical Properties | |||||||
| Molecular Weight | 406.47 | 422.47 | 422.47 | 422.47 | 422.47 | 422.47 | 392.45 |
| H-bond acceptors | 5 | 6 | 6 | 6 | 6 | 6 | 5 |
| H-bond donors | 3 | 4 | 4 | 4 | 4 | 4 | 4 |
| Number of rings | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Number of carbons | > 4 | > 4 | > 4 | > 4 | > 4 | > 4 | > 4 |
| Rotatable bonds | 10 | 10 | 10 | 10 | 10 | 10 | 9 |
| Heteroatoms | 6 | 7 | 7 | 7 | 7 | 7 | 6 |
| TPSA | 75.74 | 95.97 | 95.97 | 95.97 | 95.97 | 95.97 | 86.74 |
| Lipophilicity | |||||||
| iLOGP | 3.45 | 2.15 | 3.2 | 2.31 | 3.41 | 3.08 | 2.48 |
| XLOGP3 | 4.37 | 4.01 | 4.01 | 4.01 | 4.01 | 4.01 | 4.1 |
| WLOGP | 3.74 | 3.44 | 3.44 | 3.44 | 3.44 | 3.44 | 3.44 |
| MLOGP | 1.96 | 1.43 | 1.43 | 1.43 | 1.43 | 1.43 | 1.75 |
| Consensus Log P | 3.61 | 3.02 | 3.23 | 3.05 | 3.27 | 3.21 | 3.15 |
| Pharmacokinetics | |||||||
| GI absorption | High | High | High | High | High | High | High |
| BBB permeant | Yes | No | No | No | No | No | No |
| Pgp substrate | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP1A2 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP2C19 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP2C9 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP2D6 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| CYP3A4 inhibitor | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| log Kp (cm/s) | − 5.68 | − 6.03 | − 6.03 | − 6.03 | − 6.03 | − 6.03 | − 5.78 |
| Drug-likeness | |||||||
| Lipinski (Pfizer) | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Egan (Pharmacia) | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Muegge (Bayer) | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Bioavailability | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
Criteria for Lipinski (Pfizer) filter: molecular weight ≤ 500, H-bond acceptors ≤ 10, H-bond donors ≤ 5, MLOGP ≤ 4.15; Criteria for Egan (Pharmacia) filter: WLOGP ≤ 5.88, TPSA ≤ 131.6; Criteria for Muegge (Bayer) filter: 200 ≤ molecular weight ≤ 600, -2 ≤ XLOGP ≤ 5, TPSA ≤ 150, number of rings ≤ 7, number of carbon > 4, Rotatable bonds ≤ 15, Heteroatoms > 1, H-bond acceptors ≤ 10, H-bond donors ≤ 5.
TPSA: Topological polar surface area; iLOGP, XLOGP3, WLOGP, MLOGP, and Silicos-IT Log P are five methods used to calculate the n-octanol/water partition coefficient (log Po/w); Consensus Log P: coefficient average value calculated by the aforementioned five methods; GI absorption: Gastrointestinal absorption; BBB permeant: blood–brain barrier permeant; Pgp substrate: P-glycoprotein substrate; CYP1A2: Cytochrome P450 1A2; CYP2C19: Cytochrome P450 2C19; CYP2C9: Cytochrome P450 2C9; CYP2D6: Cytochrome P450 2D6; CYP3A4: Cytochrome P450 3A4; log Kp (cm/s): Skin permeability coefficient.
Discussion
Repurposing the FDA-approved drug for medical use to treat diseases can provide several benefits from different perspectives compared to de-novo drug discovery, such as the time-saving process, low cost for drug development, and reduced load of safety testing for applications39. For example, using molecular docking and dynamics simulation studies to screen chemical compounds from natural products against the receptor-binding domain of SARS-CoV-2 provides many drug candidates for further validation in wet labs40.
Molecular docking is an essential computational approach in drug discovery, allowing the exploration of potential leading drugs or compounds, identifying suitable therapeutic drugs, and accelerating drug design and development41–44. Computer-aided drug discovery is also a powerful tool to facilitate the mechanistic investigation of drugs and their targets, such as the binding affinity, binding mode, and essential residues that contribute to the binding interaction and stability of the complex between molecules and their target proteins45–50. Most importantly, the comprehensive and rational docking experiments applied in the computational approach should be based on the evidence of clinical observation and a deep understanding of the properties of the molecules and diseases, which is critical for harnessing the efficacy of drug discovery using computational methodologies and approaches51–53.
Results of clinical observation, network analysis, and in vitro studies have demonstrated that there are beneficial effects of the usage of carvedilol against SARS-CoV-2 infection. In this study, we explored the potential usage of carvedilol and its metabolites as re-purposed drugs to fight against SARS-CoV-2 infection using computer-aided mechanistic analysis. Our results have demonstrated that both carvedilol and its metabolites showed the properties to bind with multiple viral proteins (Table 1). The binding affinity of carvedilol/metabolites with RdRp showed the highest score compared to other tested viral proteins. RdRp can serve as the most promising druggable site for both carvedilol and its metabolites (Fig. 6).
Molecular dynamics simulation is a powerful computational method and invaluable tool in drug discovery, aiming to simulate the actual physical system and process54,55. Computing power also allows the characterization and analysis of ligand–protein interaction, which provides important information for understanding the biological and molecular mechanisms of small molecules and target proteins. The analysis results can also provide knowledge-based insight for further modification and optimization of druggable compounds. The results from this study suggest that there is a relatively stable binding complex formed between viral protein RdRp and carvedilol, as well as its metabolites with RdRp (Fig. 7). These results indicate repurposing carvedilol as a potential drug against COVID-19 infection exerts further investigation in research studies.
Therefore, the computational approach is a valuable and promising strategy to perform drug repurposing. Especially, in the pandemic area, computational-aided drug discovery could provide an unreplaceable method to accelerate the pace of drug discovery. During the COVID-19 pandemic, a lot of investigation and progress has been made using this approach56–58. Most importantly, the mutation and variant of interest could also be deeply investigated using the computational repurposing method59,60. In the current study, we incorporated the drug action mechanism of the FDA-approved drug carvedilol and clinical observation using a molecular docking approach to explore the potential of repurposing carvedilol as an anti-viral agent against SARS-CoV-2 infection. The molecular dynamic simulation was used to further confirm the findings. As demonstrated by the dynamic results, within the 100 ns dynamic simulation, the complexes could reach a relatively stable stage, except for the complex of RdRp and 1-hydroxyl carvedilol shown in Figure (Fig. 7B).The results of ADMET analysis results of the candidate molecules including carvedilol and its metabolites further demonstrated the feasibility of using those molecules as anti-viral compounds for future investigation.
For future perspectives, both in vitro and in vivo studies are needed to verify the treatment efficacy of those molecules. The exploration and optimization of the dosages are also required for the application in SARS-CoV-2 infection treatment. Meanwhile, reducing or minimizing the side effects is important given its functionality as a blood pressure modulator. The dual or multiple drug targets need to be further investigated to gain a synergistic effect against viral infection, considering the multiple druggable binding sites and several potential target proteins in the virus61. Moreover, given the modulatory properties of carvedilol on the levels of IL-6, the immune modulators may be applied as a synergistic treatment to improve the host immune system to result in better efficacy against viral infection62,63.
Drug repurposing is an alternative approach for developing new disease treatments, which has multiple benefits, such as low cost for drug development and risk assessment. However, the original effects of a drug should also be considered when applying it as a repositioning drug. Carvedilol can treat high blood pressure; therefore, it might not be applicable to use it as an anti-viral drug in subjects with low blood pressure or hypotension. In two conditions, repositioning drugs may be more applicable, including (1) incorporating the established function and repositioning function together to enhance treatment efficacy in subjects with both diseases and (2) shifting the original function to a newly proposed therapy for disease treatment. The side effects derived from the drug's original function and its application dosage of a repurposed drug for COVID-19 treatment should be evaluated and measured, while they are applied in patients with only SARS-CoV-2 infection. Newly merged studies suggest the possibility of SARS-CoV-2 infection can cause high blood pressure or hypertension64. Thus, the blood control agent carvedilol will confer a beneficial effect for controlling both blood pressure and viral infection. However, it may not be applicable for patients with only viral infections, as reducing the blood pressure function of carvedilol is a side effect. Overall, well-designed studies are needed to further assess the functions of the repurposed drugs. Modification and optimization of these agents may be necessary before the clinical trials.
In conclusion, based on computational analysis and molecular dynamics simulation, this study provides promising insight for further investigation of using carvedilol and its metabolites as repurposed drugs to fight against SARS-CoV-2 infection.
Materials and methods
Protein preparation
The crystal structure protein models used for docking in this study were originally obtained from the Protein Data Bank (www.rcsb.org): including the RNA-directed RNA polymerase, RdRp (PDB ID:6XQB) with a resolution of 3.40 Å, and the Chain A used for this study; the crystal structure of COVID-19 main protease 3CL/Mpro (PDB ID:6LU7, resolution of 2.16 Å, Chain A); was the crystal structure of NSP13 helicase (PDB ID:7CYQ, resolution of 2.83 Å, Chain E); non-structural protein 2 (NSP2) (PDB ID: 7MSX, resolution of 3.15 Å); papain-like protease NSP3/PLpro (PDB ID:7BF6, resolution 2.15 Å, chain A); SARS-CoV-2 spike protein S1 receptor-binding domain RBD (PDB ID: 6M0J, resolution of 2.45 Å, ligand removed); the crystal structure of human angiotensin-converting enzyme 2 (ACE2, PDB ID: 6M0J, resolution of 2.45 Å; and the crystal structure of the RBD-ACE2 complex (PDB ID: 6M0J, resolution of 2.45 Å). All these proteins were prepared for docking such as adding hydrogen, removal of water, elimination of all non-standard residues, adding Gasteiger charges, and removal of ligands if necessary.
Preparation of small molecules
Carvedilol (Compound CID: 2585), 1-hydroxyl carvedilol (Compound CID: 182523), 3-hydroxyl carvedilol (Compound CID: 127014), 4-hydroxyl carvedilol (Compound CID: 4572774), 5-hydroxyl carvedilol (Compound CID: 4181439), 8-hydroxyl carvedilol (Compound CID: 9979639), O-desmethyl carvedilol (Compound CID: 155763), remdesivir (Compound CID: 121304016) were obtained from the https://pubchem.ncbi.nlm.nih.gov/database. All the molecules were ensured with the hydrogen added and prepared for docking.
Molecular docking
Molecular modeling approach is a useful method for drug discovery65. Molecular docking was performed using an online server CB-Dock that adopted the docking and calculation algorithms based on the Auto Dock Vina (version 1.1.2). The blind docking was performed without defining the grid box to ensure that the best binding site with the highest binding affinity was identified. The parameter of identified best binding cavities between carvedilol and proteins of interest were listed in detail: 3CL/Mpro (Volume = 688; Center: x = − 23.771; y = 1.402; z = 56.178; Size: x = 26, y = 26, z = 26). NSP13 Helicase (Volume = 377; Center: x = 241.975; y = 170.098; z = 173.922; Size: x = 26, y = 26, z = 26). NSP2 (Volume = 561; Center: x = 126.469; y = 114.941; z = 117.846; Size: x = 21, y = 21, z = 21). PLpro (Volume = 570; Center: x = 22.645; y = − 3.141; z = 3.537; Size: x = 26, y = 26, z = 26). RBD (Volume = 94; Center: x = − 20.253; y = 26.578; z = 35.025; Size: x = 26, y = 26, z = 26). ACE2 (Volume = 377; Center: x = − 30.131; y = 32.105; z = − 21.789; Size: x = 26, y = 26, z = 26). RBD-ACE2 complex (Volume = 706; Center: x = − 30.503; y = 17.837; z = − 2.023; Size: x = 26, y = 26, z = 26). The best model with the highest binding affinity was identified for RdRp in complex with carvedilol and its metabolites (Volume = 783; Center: x = 101.643, y = 95.028, z = 80.186; Size: x = 26, y = 26, z = 26). Remdesivir Volume = 4738; Center: x = 106.442, y = 107.143, z = 103.191; Size: x = 25, y = 31, z = 31)66–68.
Docking complexes used for molecular dynamics simulation were prepared using crystal structure from PDB ID: 6XQB with all chains kept and containing RdRp (NSP12) in complex with its co-factors NSP7 and NSP8.
Binding pocket identification
RNA-directed RNA polymerase, RdRp, the major binding pockets identification was performed using prepared protein obtained from the crystal structure from Protein Data Bank (www.rcsb.org) and online server bioinformatics analysis tool https://proteins.plus/69–71.
Analysis of interaction on binding sites
The analysis and visualization of interaction on binding sites and 2D interaction graphs were generated using LigPlus (V.2.2.4)72, PyMol Molecular Graphics System (Version 2.0 Schrödinger, LLC) software, and Discover Studio Visualizer (2021) software.
Molecular dynamic simulation
To further confirm the stability of the molecules in complex with RdRp, the molecular dynamics (MD) simulation was carried out up to 100 ns by applying the Desmond package (the force field OPLS2005 (Schrödinger, LLC, New York, NY, 2015). The calculation of effective evaluation parameters was conducted, such as the root mean square deviation (RMSD) and the root mean square fluctuation (RMSF). RMSD referred to the overall spatial difference between the reference protein compared to the protein that was docked with ligand, which was used to measure the conformational stability of the complex, while the RMSF referred to the individual residue fluctuation (flexibility) during the simulation, which was also used to calculate and measure the stability of the docked complex. The intermolecular interactions of protein–ligand were also calculated to examine the stability and strength of the docked complex over the simulation period. The MD simulation was carried out under thermodynamic conditions (applied volume, density, pressure, and temperature) with the annealed and equilibrated complete system73,74.
Pharmacokinetic properties ADMET and druglikeness evaluation
The platform SWISSADME was used for systematic analysis of the absorption, distribution, excretion, metabolism, and toxicity (ADMET), as well as the evaluation of drug-likeness for the current study75.
Supplementary Information
Acknowledgements
The authors acknowledge the available software used for data analysis in this study.
Author contributions
Conceptualization, C.Z. and M.Y.; methodology, formal analysis, software, and investigation, C.Z., J.L., Y.S., S.L.; and M.Y.; writing—original draft preparation, C.Z., Y.S., S.L.; writing—review and editing, C.Z., Y.S., S.L., and M.Y. All authors have read and agreed to the published version of the manuscript.
Data availability
All the data supporting reported results can be found publicly and presented in the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-48398-6.
References
- 1.Wang W, Tang J, Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J. Med. Virol. 2020;92:441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaimes JA, André NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J. Mol. Biol. 2020;432:3309–3325. doi: 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu A, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JF, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan R, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yadav R, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021 doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Yang M. Newly emerged antiviral strategies for SARS-CoV-2: From deciphering viral protein structural function to the development of vaccines, antibodies, and small molecules. Int. J. Mol. Sci. 2022 doi: 10.3390/ijms23116083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020 doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021 doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss SR, Leibowitz JL. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:85–164. doi: 10.1016/b978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shamsi A, et al. Potential drug targets of SARS-CoV-2: From genomics to therapeutics. Int. J. Biol. Macromol. 2021;177:1–9. doi: 10.1016/j.ijbiomac.2021.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zahradník J, et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 15.Yan W, Zheng Y, Zeng X, He B, Cheng W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target Ther. 2022;7:26. doi: 10.1038/s41392-022-00884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sacco MD, et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci. Adv. 2020 doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra A, Rathore AS. RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2. J. Biomol. Struct. Dyn. 2022;40:6039–6051. doi: 10.1080/07391102.2021.1875886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb RL, et al. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N. Engl. J. Med. 2022;386:305–315. doi: 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alhammad YMO, et al. The SARS-CoV-2 conserved macrodomain Is a mono-ADP-ribosylhydrolase. J. Virol. 2021 doi: 10.1128/jvi.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman JA, et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12:4848. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng H, et al. SARS-CoV-2 helicase NSP13 hijacks the host protein EWSR1 to promote viral replication by enhancing RNA unwinding activity. Infect. Med. 2022;1(1):7–16. doi: 10.1016/j.imj.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, et al. SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation. Proc. Natl. Acad. Sci. U S A. 2022;119:e2204539119. doi: 10.1073/pnas.2204539119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eskandarzade N, et al. Network for network concept offers new insights into host- SARS-CoV-2 protein interactions and potential novel targets for developing antiviral drugs. Comput. Biol. Med. 2022;146:105575. doi: 10.1016/j.compbiomed.2022.105575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiNicolantonio JJ, Hackam DG. Carvedilol: A third-generation β-blocker should be a first-choice β-blocker. Expert Rev. Cardiovasc. Ther. 2012;10:13–25. doi: 10.1586/erc.11.166. [DOI] [PubMed] [Google Scholar]
- 29.Saijonmaa O, Nyman T, Fyhrquist F. Carvedilol inhibits basal and stimulated ACE production in human endothelial cells. J. Cardiovasc. Pharmacol. 2004;43:616–621. doi: 10.1097/00005344-200405000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, et al. A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets. Nat. Biotechnol. 2022 doi: 10.1038/s41587-022-01474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petríková M, Jancinová V, Nosál R, Májeková M, Danihelová E. Antiplatelet activity of carvedilol in comparison to propranolol. Platelets. 2002;13:479–485. doi: 10.1080/0953710021000057848. [DOI] [PubMed] [Google Scholar]
- 32.Battinelli EM. COVID-19 concerns aggregate around platelets. Blood. 2020;136:1221–1223. doi: 10.1182/blood.2020007805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skayem C, Ayoub N. Carvedilol and COVID-19: A potential role in reducing infectivity and infection severity of SARS-CoV-2. Am. J. Med. Sci. 2020;360:300. doi: 10.1016/j.amjms.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumura T, et al. Effects of carvedilol on plasma levels of interleukin-6 and tumor necrosis factor-alpha in nine patients with dilated cardiomyopathy. J. Cardiol. 2002;39:253–257. [PubMed] [Google Scholar]
- 35.Palanques-Pastor T, López-Briz E, Poveda Andrés JL. Involvement of interleukin 6 in SARS-CoV-2 infection: Siltuximab as a therapeutic option against COVID-19. Eur. J. Hosp. Pharm. 2020;27:297–298. doi: 10.1136/ejhpharm-2020-002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang CY, Liu S, Yang M. Crosstalk between gut microbiota and COVID-19 impacts pancreatic cancer progression. World J. Gastrointest. Oncol. 2022;14:1456–1468. doi: 10.4251/wjgo.v14.i8.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oldham HG, Clarke SE. In vitro identification of the human cytochrome P450 enzymes involved in the metabolism of R(+)- and S(-)-carvedilol. Drug Metab. Dispos. 1997;25:970–977. [PubMed] [Google Scholar]
- 38.Ahmad S, et al. Multitargeted molecular dynamic understanding of butoxypheser against SARS-CoV-2: An in silico study. Nat. Prod. Commun. 2022;17:1934578x221115499. doi: 10.1177/1934578x221115499. [DOI] [Google Scholar]
- 39.Pushpakom S, et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 40.Yadav MK, et al. Predictive modeling and therapeutic repurposing of natural compounds against the receptor-binding domain of SARS-CoV-2. J. Biomol. Struct. Dyn. 2023;41:1527–1539. doi: 10.1080/07391102.2021.2021993. [DOI] [PubMed] [Google Scholar]
- 41.Pinzi L, Rastelli G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crampon K, Giorkallos A, Deldossi M, Baud S, Steffenel LA. Machine-learning methods for ligand-protein molecular docking. Drug Discov. Today. 2022;27:151–164. doi: 10.1016/j.drudis.2021.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Choudhury C, Arul Murugan N, Priyakumar UD. Structure-based drug repurposing: Traditional and advanced AI/ML-aided methods. Drug Discov. Today. 2022;27:1847–1861. doi: 10.1016/j.drudis.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanzione F, Giangreco I, Cole JC. Use of molecular docking computational tools in drug discovery. Prog. Med. Chem. 2021;60:273–343. doi: 10.1016/bs.pmch.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug. Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McConkey B, Sobolev V, Edelman M. The performance of current methods in ligand-protein docking. Curr. Sci. 2002;83:845. [Google Scholar]
- 47.Sadybekov AV, Katritch V. Computational approaches streamlining drug discovery. Nature. 2023;616:673–685. doi: 10.1038/s41586-023-05905-z. [DOI] [PubMed] [Google Scholar]
- 48.de Ruyck J, Brysbaert G, Blossey R, Lensink MF. Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinform. Chem. 2016;9:1–11. doi: 10.2147/aabc.S105289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Sui Y, Liu S, Yang M. Anti-viral activity of bioactive molecules of silymarin against COVID-19 via in silico studies. Pharmaceuticals. 2023;16:1479. doi: 10.3390/ph16101479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang M, Liu S, Zhang C. Antimicrobial peptides with antiviral and anticancer properties and their modification and nanodelivery systems. Curr. Res. Biotechnol. 2023;5:100121. doi: 10.1016/j.crbiot.2023.100121. [DOI] [Google Scholar]
- 51.Saikia S, Bordoloi M. Molecular docking: Challenges, advances and its use in drug discovery perspective. Curr. Drug Targets. 2019;20:501–521. doi: 10.2174/1389450119666181022153016. [DOI] [PubMed] [Google Scholar]
- 52.Jazayeri A, Dias JM, Marshall FH. From G protein-coupled receptor structure resolution to rational drug design. J. Biol. Chem. 2015;290:19489–19495. doi: 10.1074/jbc.R115.668251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keretsu S, Bhujbal SP, Cho SJ. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020;10:17716. doi: 10.1038/s41598-020-74468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salo-Ahen OMH, et al. Molecular dynamics simulations in drug discovery and pharmaceutical development. Processes. 2021;9:71. doi: 10.3390/pr9010071. [DOI] [Google Scholar]
- 55.Fu Y, Zhao J, Chen Z. Insights into the molecular mechanisms of protein-ligand interactions by molecular docking and molecular dynamics simulation: A case of oligopeptide binding protein. Comput. Math. Methods Med. 2018;2018:3502514. doi: 10.1155/2018/3502514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain R, Mujwar S. Repurposing metocurine as main protease inhibitor to develop novel antiviral therapy for COVID-19. Struct. Chem. 2020;31:2487–2499. doi: 10.1007/s11224-020-01605-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mujwar S, Harwansh RK. In silico bioprospecting of taraxerol as a main protease inhibitor of SARS-CoV-2 to develop therapy against COVID-19. Struct. Chem. 2022;33:1517–1528. doi: 10.1007/s11224-022-01943-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kciuk M, et al. Computational bioprospecting guggulsterone against ADP ribose phosphatase of SARS-CoV-2. Molecules. 2022 doi: 10.3390/molecules27238287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fidan O, Mujwar S, Kciuk M. Discovery of adapalene and dihydrotachysterol as antiviral agents for the Omicron variant of SARS-CoV-2 through computational drug repurposing. Mol. Divers. 2023;27:463–475. doi: 10.1007/s11030-022-10440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mujwar S. Computational repurposing of tamibarotene against triple mutant variant of SARS-CoV-2. Comput. Biol. Med. 2021;136:104748. doi: 10.1016/j.compbiomed.2021.104748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murugan NA, Kumar S, Jeyakanthan J, Srivastava V. Searching for target-specific and multi-targeting organics for Covid-19 in the Drugbank database with a double scoring approach. Sci. Rep. 2020;10:19125. doi: 10.1038/s41598-020-75762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coomes EA, Haghbayan H. Interleukin-6 in Covid-19: A systematic review and meta-analysis. Rev. Med. Virol. 2020;30:1–9. doi: 10.1002/rmv.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Potere N, et al. The role of IL-6 and IL-6 blockade in COVID-19. Expert Rev. Clin. Immunol. 2021;17:601–618. doi: 10.1080/1744666x.2021.1919086. [DOI] [PubMed] [Google Scholar]
- 64.Zhang V, Fisher M, Hou W, Zhang L, Duong TQ. Incidence of new-onset hypertension post-COVID-19: Comparison with influenza. Hypertension. 2023;80:2135–2148. doi: 10.1161/hypertensionaha.123.21174. [DOI] [PubMed] [Google Scholar]
- 65.Tripathi, M. K., Ahmad, S., Tyagi, R., Dahiya, V. & Yadav, M. K. in Computer Aided Drug Design (CADD): From Ligand-Based Methods to Structure-Based Approaches (eds Mithun Rudrapal & Chukwuebuka Egbuna) 125–155 (Elsevier, 2022).
- 66.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021;61:3891–3898. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Y, et al. CB-Dock: A web server for cavity detection-guided protein-ligand blind docking. Acta Pharmacol. Sin. 2020;41:138–144. doi: 10.1038/s41401-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volkamer A, Griewel A, Grombacher T, Rarey M. Analyzing the topology of active sites: On the prediction of pockets and subpockets. J. Chem. Inf. Model. 2010;50:2041–2052. doi: 10.1021/ci100241y. [DOI] [PubMed] [Google Scholar]
- 70.Volkamer A, Kuhn D, Grombacher T, Rippmann F, Rarey M. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 71.Schöning-Stierand K, et al. ProteinsPlus: Interactive analysis of protein-ligand binding interfaces. Nucleic Acids Res. 2020;48:W48–w53. doi: 10.1093/nar/gkaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 73.Bowers, K. J. et al. in SC '06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. 43–43.
- 74.Desmond Molecular Dynamics System, D. E. Shaw Research. Maestro-Desmond Interoperability Tools, SchrödingerSchrödinger, New York, NY.
- 75.Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting reported results can be found publicly and presented in the current study.