Abstract
The CpxA/R two-component signal transduction system of Escherichia coli can combat a variety of extracytoplasmic protein-mediated toxicities. The Cpx system performs this function, in part, by increasing the synthesis of the periplasmic protease, DegP. However, other factors are also employed by the Cpx system for this stress-combative function. In an effort to identify these remaining factors, we screened a collection of random lacZ operon fusions for those fusions whose transcription is regulated by CpxA/R. Through this approach, we have identified a new locus, cpxP, whose transcription is stimulated by activation of the Cpx pathway. cpxP specifies a periplasmic protein that can combat the lethal phenotype associated with the synthesis of a toxic envelope protein. In addition, we show that cpxP transcription is strongly induced by alkaline pH in a CpxA-dependent manner and that cpxP and cpx mutant strains display hypersensitivity to growth in alkaline conditions.
The Cpx signal transduction system of Escherichia coli consists of a two-component inner membrane sensor (CpxA) and a cognate response regulator (CpxR) (1, 8). The activity of this signal transduction system has been linked to the physiology of the bacterial envelope. For example, the activity of CpxA is stimulated by overproduction of various extracytoplasmic proteins (6, 15, 22, 35). In response to such stimuli, the Cpx system increases the transcription of various envelope stress-combative proteins, including DegP, DsbA, and RotA (5, 6, 26, 27).
Our laboratory has previously shown that the Cpx pathway can combat various extracytoplasmic protein-mediated stresses (4, 35). For example, when highly produced, the LamB-LacZ-PhoA fusion protein forms a disulfide-bonded aggregate in the bacterial envelope, ultimately causing cell lysis. Activation of the Cpx pathway suppresses this lethal phenotype (4, 35). High-level synthesis of the processing-defective maltoporin, LamBA23D, is also toxic. Specifically, LamBA23D confers upon E. coli a hypersensitivity to detergents such as sodium dodecyl sulfate (SDS), implying that this protein perturbs the structure of the outer membrane. Activation of the Cpx pathway also suppresses this SDS-hypersensitive phenotype (4).
In both cases, the Cpx-mediated suppression is due in part to the increased synthesis of the periplasmic protease, DegP. However, tests of epistasis indicate that the activated Cpx system can partially ameliorate these extracytoplasmic toxicities even in the absence of DegP and DsbA (4, 34, 35). Thus, it seemed possible that the Cpx pathway would control other factors that could also combat extracytoplasmic protein-mediated stresses. Here, we describe a screen for genes whose transcription is stimulated by overproduction of the outer membrane lipoprotein, NlpE (which activates the Cpx signal transduction pathway). With this screen, we have identified a new Cpx-regulated gene, cpxP. cpxP is a pH-regulated locus that encodes a periplasmic protein that aids in combating extracytoplasmic protein-mediated toxicity.
MATERIALS AND METHODS
Media, reagents, and enzymes.
Media were prepared as described elsewhere (30). Liquid cultures were grown either in Luria broth or in M63 minimal medium supplemented with thiamine (50 μg/ml) and 0.4% carbon source. The final concentrations of antibiotics used in the growth media were as follows: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 20 μg/ml; spectinomycin, 50 μg/ml; and chloramphenicol, 20 μg/ml. Standard microbiological techniques were used for strain construction and bacterial growth (30). 5-Bromo-4-chloro-3-indolyl-d-galactoside (X-Gal) was purchased from Fischer.
Strains and phage.
λRS88, λNK1324, and λplacMu53 have been described elsewhere (3, 16, 31). Lysogenization of λRS88[cpxP-lacZ] was performed as described previously (31). λRS88 operon fusions were shown to be located in single copy at the λatt locus by P1 transduction (6). Strain PND541 (MC4100 ara+ nadA::Tn10 Δ[gal-att-bio]) was used in the initial screen for Cpx-regulatable loci. Strains harboring the ara-74::cam mutation were used to generate data shown in Fig. 5 and 6. The ara-74::cam mutation renders MC4100 (which is normally sensitive to growth in the presence of arabinose) arabinose resistant and ara mutant.
FIG. 5.
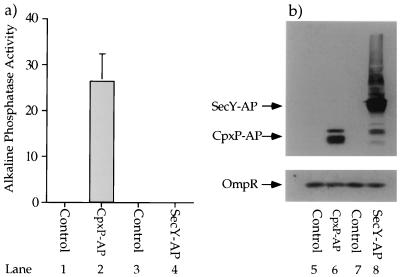
CpxP-AP possesses relatively high alkaline phosphatase activity. Strain SP627a (MC4100, ara74::cam) was transformed with pBR322 (control for pND24) (lanes 1 and 5), pND24 (produces CpxP-AP) (lanes 2 and 6), pBAD18 (control for pCH215) (lanes 3 and 7), and pCH215 (produces SecY-AP) (lanes 4 and 8). The transformants were grown in Luria broth containing 50 μg of ampicillin per ml supplemented with 0.4% l-arabinose to induce the synthesis of SecY-AP from pCH215. (a) The CpxP-AP fusion protein displays relatively high alkaline phosphatase activity. The alkaline phosphatase activities of each of these four transformant strains were determined in the presence of 5 mM IAA (lanes 1 to 4). (b) The amounts of alkaline phosphatase-cross-reacting species generated by the CpxP-AP and SecY-AP fusion proteins are comparable. Immunoblot analysis was performed on whole-cell protein extracts generated from SP627a transformed with pBR322, pND24, pBAD18, and pCH215 (lanes 5 to 8). The whole-cell extracts were separated by SDS-polyacrylamide gel electrophoresis (equal OD600 units were loaded in each lane) and subjected to immunoblot analysis with anti-alkaline phosphatase and anti-OmpR antisera. OmpR serves as an additional loading control.
FIG. 6.
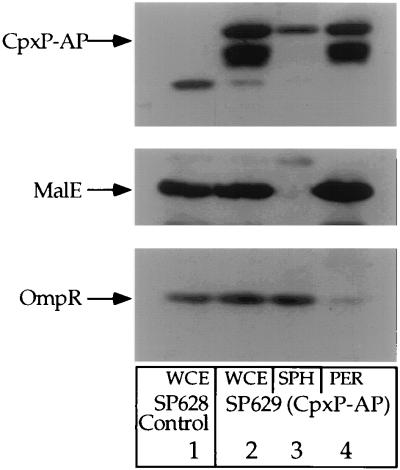
Subcellular fractionation of CpxP-AP. Whole-cell protein extracts were prepared from strain SP627a (MC4100 ara74::cam) transformed with plasmid pBR322 (control for pND24) (lane 1). Whole-cell, spheroplast, and periplasmic extracts were prepared from SP627a transformed with pND24 (lanes 2 to 4). The protein extracts were separated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot analysis with anti-alkaline phosphatase, anti-MalE, and anti-OmpR antisera. Abbreviations: WCE, whole-cell extract; SPH, spheroplast extract; PER, periplasmic extract. MalE and OmpR serve as model periplasmic and cytoplasmic proteins, respectively. Both strains were grown to late-log phase in M63 minimal medium supplemented with 0.4% maltose and 50 μg of ampicillin per ml. Protein extracts were then generated as described in Materials and Methods.
Plasmid construction and primer information.
The following plasmids have been described elsewhere. pND18 overexpresses nlpE (6). pBAD18 is the parent of pND18 (11). pLD404 overexpresses nlpE (35). pBR322 is the parent of pLD404 (2). pPR272 overexpresses ompF (20). pLG338 is the parent of pPR272 (36). pRAM1006 overexpresses ompC. pRAM1005 is the parent of pRAM1006 (21).
To construct the plasmid producing the CpxP-alkaline phosphatase (CpxP-AP) fusion protein, the 5′ region of the cpxP open reading frame, along with 1,031 nucleotides of upstream sequence, was amplified by PCR using the Cpx2828 (5′ CTG GTA AGC TTT GAT GGT TTC G 3′) and Finpho (5′ CAT TAA CAG GAT CCT GTT CGT GCC 3′) primers. The amplified DNA was digested with HindIII and BamHI and was subcloned into the corresponding restriction sites of pBR322, generating pND23. pND23 contains the first 71 codons of the cpxP open reading frame. A 2.6-kb BamHI fragment containing the alkaline phosphatase open reading frame, but lacking the signal sequence coding region, was removed from plasmid pPHO7 (10) and subcloned into the BamHI site of pND23. The proper insertion and orientation of this phoA coding sequence were confirmed by restriction analysis. In this way, pND24 was generated. pND24 fuses the first 71 codons of the cpxP open reading frame with the coding sequence for the mature portion of alkaline phosphatase.
Construction of lacZ fusions.
All lacZ fusions derived from pRS415 were recombined onto λRS88 as previously described (31) and the resulting recombinant phage were introduced at the λatt site of MC4100.
To construct λRS88[cpxP-lacZ], the Finlac (5′ CTC AAG GCC GAG AAT TCG ATC AAG 3′) and Finpho primers were used to amplify the promoter region and a portion of the cpxP open reading frame from the chromosome of MC4100. This amplified DNA was digested with EcoRI and BamHI and subcloned into the corresponding restriction sites of pRS415, generating pND22. This amplified DNA includes nucleotides from positions −410 to +214 with respect to the cpxP translation start site.
Maltose sensitivity disc assays.
Maltose sensitivity disc assays were performed as follows. Each strain was grown to saturation overnight at 37°C in 5 ml of M63 minimal medium supplemented with 0.4% glycerol, Luria broth (final concentration, 1%), and ampicillin. Then 3 ml of molten F top agar (55°C) was mixed with 100 μl of each overnight culture and immediately spread onto M63 minimal agar supplemented with 0.4% glycerol and ampicillin (warmed to 23°C). The top agar was allowed to solidify for 2 min. A Schleicher & Schuell analytical paper filter disc (7-mm diameter) was then placed in the middle of the M63 glycerol plate; 10 μl of 40% (wt/vol) maltose was placed on the filter disc, and the plates were incubated overnight at 37°C. The zone of clearing, which is defined as the diameter of inhibited growth minus the diameter of the filter disc, was measured 18 h after the inception of incubation. Each value shown in Fig. 7 is the average of four replicate experiments. The error bars represent the standard deviation from each average.
FIG. 7.
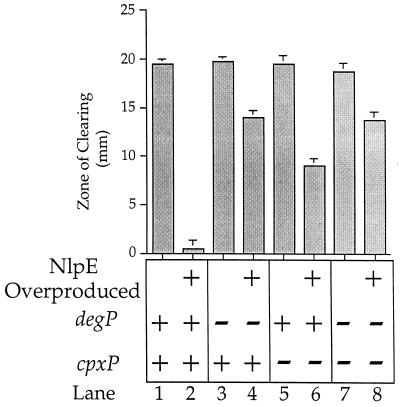
CpxP combats extracytoplasmic stress. Ten microliters of 40% maltose was added to filter discs that had been placed on lawns of strains WBS164 (MC4100 Φ(lamB-lacZX90) Hyb42-1[λp1(209)]) (lanes 1 and 2), SP9 (WBS164 degP::Tn10) (lanes 3 and 4), SP10 (WBS164 λplacMu53[cpxP-lacZ]) (lanes 5 and 6), and SP24 (WBS164 degP::Tn10 λplacMu53[cpxP-lacZ]) (lanes 7 and 8). Strains in odd-numbered lanes were transformed with pBR322 (control for pLD404); strains in even-numbered lanes were transformed with pLD404 (overproduces NlpE and activates the Cpx signal transduction pathway). The values displayed along the y axis (zone of clearing) represent the amount of growth inhibition caused by the addition of maltose. The zone of clearing value is defined as the diameter of growth inhibition around the maltose-saturated filter disc minus the diameter of the filter disc itself (7 mm). The maltose disc assays were performed on M63 minimal agar containing 50 μg of ampicillin per ml and 0.2% glycerol as a carbon source.
Enzyme assays. (i) Alkaline phosphatase assays.
Alkaline phosphatase assays were performed as described previously (34). All assays were performed in the presence of 5 mM iodoacetamide (IAA).
(ii) β-Galactosidase assays.
Cells were grown overnight in Luria broth, supplemented (when necessary) with 0.4% carbon source (see figure legends for details). Cells were then subcultured (1:40) into 2 ml of the same medium and grown to mid-log phase. β-Galactosidase activities were determined by a microtiter plate assay (32). β-Galactosidase activities are expressed as (units/A600) × 103, where units are defined as micromoles of product formed per minute. A minimum of four independent isolates from each strain were used to determine the β-galactosidase activities, and the results were averaged to obtain the indicated activities. Error bars indicate the standard deviation. The absence of error bars indicates that the standard deviation fell below the resolution limit of the graphing program.
Protein analysis. (i) Preparation of whole-cell, periplasmic, and spheroplast protein extracts.
All procedures were performed on ice, and all solutions were chilled on ice. Whole-cell extracts were prepared by pelleting 1 ml of cells, resuspending the pellet in loading buffer (30), and boiling it for 10 min. Periplasmic and spheroplast protein extracts were prepared as previously described (5).
(ii) Immunoblot analysis.
Protein samples were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The membranes were blocked in MTS buffer (0.9% NaCl, 0.01 M Tris-HCl [pH 7.5], 2.5% powdered milk) at 4°C for approximately 12 h and then incubated with the primary antibody (either anti-OmpR, anti-alkaline phosphatase, or anti-MalE, diluted 1:5,000 in MTS) at 4°C overnight. Membranes were washed with wash buffer (0.2% Tween 20, 0.9% NaCl, 0.01 M Tris-HCl [pH 7.5]) for 2 h with at least four changes of buffer and then incubated as before with horseradish peroxidase-linked secondary antibody (anti-rabbit diluted 1:10,000 in MTS). Membranes were washed as before, and antibody was detected with ECL detection reagents as described by the supplier (Amersham).
Nucleotide sequence accession number.
The GenBank accession number for the sequence shown in Fig. 3 is L19201.
FIG. 3.
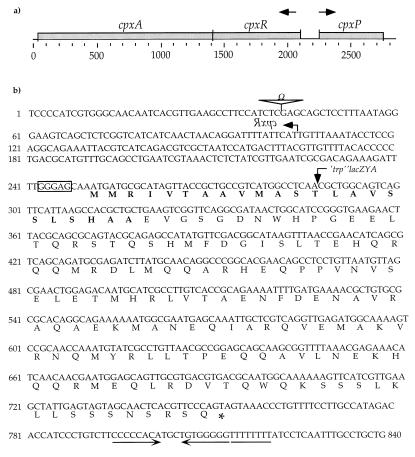
The cpxP locus. (a) The cpxP open reading frame shown in relation to the cpx operon. The cpx operon and cpxP are divergently transcribed, as shown by the arrows. The size of this genomic region is also shown in nucleotides. (b) Nucleotide and deduced amino acid sequences of the cpxP open reading frame. The site of the cpxR::Ω insertion is marked with Ω. The start codon of cpxR (which is shown in reversed typeface) is depicted by a leftward-pointing arrow. The deduced primary amino acid sequence of cpxP is shown below the nucleotide sequence. A putative Shine-Dalgarno sequence (GGGAG) is enclosed within a box. The residues comprising the putative CpxP signal sequence are shown in boldface. The position of the λplacMu53[cpxP-lacZ] fusion joint is marked by a downward-pointing arrow within the 13th codon of the cpxP open reading frame. An asterisk marks the stop codon of the cpxP open reading frame. A putative rho-independent transcriptional terminator stem loop is underlined with inverted arrows. The adjacent sequence of eight consecutive thymine nucleotides in this putative rho-independent transcriptional terminator is underlined. The nucleotide sequence shown in panel b corresponds to positions 67200 to 68039 of the published DNA sequence for the E. coli chromosomal region from 87.2 to 89.2 min.
RESULTS
The screen for Cpx-regulated loci.
Since the CpxA and CpxR proteins control the expression of degP at the transcriptional level, and since CpxR is homologous to the OmpR transcription factor, we reasoned that the Cpx signal transduction system would likely control its other regulatory targets at the transcriptional level as well. Accordingly, we screened a collection of lacZ operon fusions for those fusions whose transcription could be induced upon activation of the Cpx pathway. Specifically, we generated lacZ operon fusions throughout the chromosome of strain PND541 (MC4100 ara+ nadA::Tn10 Δ[gal-att-bio]). PND541 also harbors plasmid pND18, which expresses the nlpE locus from the arabinose promoter, pBAD (6). Since overproduction of NlpE activates the Cpx pathway (6, 35), this signal transduction system can be conditionally activated in PND541 by growing the strain in the presence of arabinose.
We used λplacMu53 to generate lacZ operon fusions throughout the chromosome of PND541. λplacMu53 carries a kanamycin resistance determinant, allowing a direct selection for the creation of lacZ fusions (3). Twelve independent pools of PND541 were infected with λplacMu53. From these pools, a total of 13,213 colonies harboring stable integrants of λplacMu53 were individually streaked onto two types of medium: (i) Luria agar containing 1.6 μg of X-Gal per ml and (ii) Luria agar containing 1.6 μg of X-Gal per ml supplemented with 0.4% l-arabinose. Individual streaking of these colonies provided the most reproducible visual assay for comparing Lac activity between strains grown with and without arabinose. In addition, the X-Gal concentration of 1.6 μg/ml was chosen because this concentration was optimal for distinguishing between degP-lacZ expression with and without overproduction of NlpE.
Of these 13,213 colonies, 107 displayed a qualitative increase in Lac activity when grown on Luria agar in the presence of l-arabinose. In general, colonies displaying increased Lac activity under these conditions should fall into two classes: the colonies could harbor a lacZ fusion that is transcriptionally regulated by an arabinose-inducible promoter; alternatively, the lacZ fusion could be under the control of an NlpE-inducible promoter.
To distinguish between these possibilities, we transferred each λplacMu53-generated lacZ fusion from the 107 strains described above to strain PND900 (MC4100, ara+). These 107 strains were then transformed with either pBR322 (control for pLD404) or pLD404. pLD404 constitutively overproduces NlpE and thus provides an arabinose-independent means of activating the Cpx pathway (35). Of the 107 strains transformed with pLD404, one harbored a lacZ fusion whose transcription was induced by this plasmid. The lacZ fusion carried by this strain was named cpxP-lacZ. We believe that a large proportion of the 106 remaining strains contained fusions that were arabinose inducible as a consequence of having integrated into plasmid pND18.
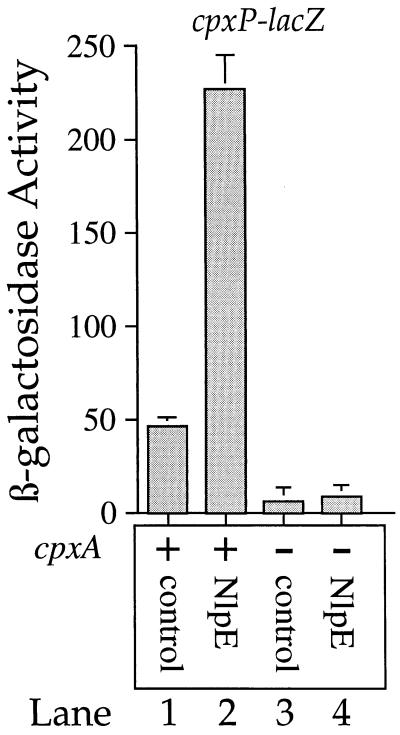
Figure 1 shows that overproduction of NlpE stimulates cpxP-lacZ transcription approximately fivefold (compare lanes 1 and 2) and also illustrates two other aspects of cpxP-lacZ transcription: first, the Cpx system is the major contributor to cpxP-lacZ transcription, because the cpxA null mutation nearly abolishes transcription of this fusion (compare lanes 1 and 3); second, in the absence of CpxA, overproduction of NlpE does not stimulate cpxP-lacZ transcription (compare lanes 1 and 2 with lanes 3 and 4). Thus, cpxP-lacZ transcription is activated by overproduction of NlpE in a CpxA-dependent fashion.
FIG. 1.
cpxP-lacZ transcription is induced in a CpxA-dependent fashion by overproduction of the outer membrane lipoprotein NlpE. β-Galactosidase activities were determined for SP1 (MC4100 ara+ λplacMu53[cpxP-lacZ]) (lanes 1 and 2) and SP7 (SP1 cpxA::cam) (lanes 3 and 4). The strains whose β-galactosidase activities are depicted in lanes 1 and 3 were transformed with plasmid pBR322 (control for pLD404). The strains whose β-galactosidase activities are depicted in lanes 2 and 4 were transformed with pLD404 (overproduces NlpE). All strains were grown in Luria broth containing 50 μg of ampicillin per ml as described in Materials and Methods.
Ac∼P can mediate the transcriptional induction of cpxP-lacZ in the absence of CpxA.
We have previously (6) shown that degP transcription can be stimulated in the absence of CpxA when acetyl-phosphate (Ac∼P) levels are increased by growth in the presence of glucose. This effect is most likely mediated by hyperphosphorylation of CpxR via Ac∼P. This phenomenon is also observed with cpxP.
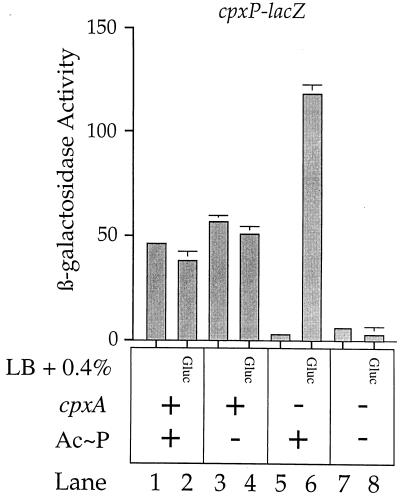
Specifically, Fig. 2 shows that growth in the presence of glucose has little effect on cpxP transcription in a wild-type background (compare lanes 1 and 2). In addition, growth in the presence of glucose has little effect on cpxP transcription in a cpxA+ background that is deleted for the genes responsible for Ac∼P synthesis (compare lanes 3 and 4). Lane 5 again shows that the cpxA mutation drastically reduces cpxP-lacZ transcription. However, when this same cpxA strain is grown in the presence of glucose, cpxP-lacZ transcription is stimulated more than 100-fold (compare lanes 5 and 6). This induction is completely eliminated by a mutation that abolishes Ac∼P synthesis (compare lanes 5 and 6 with lanes 7 and 8). Thus, Ac∼P can mediate the transcriptional induction of cpxP-lacZ in the absence of CpxA. This result has two implications. First, it suggests that the transcriptional activation of degP and that of cpxP by the Cpx system proceed via the same mechanism. Second, it supports the hypothesis that CpxR-phosphate (CpxR-P) is the activating species for the Cpx signal transduction system (6).
FIG. 2.
Ac∼P can mediate the transcriptional induction of cpxP-lacZ in the absence of CpxA. β-Galactosidase activities were determined for strains SP34 (MC4100 ara+ λplacMu53[cpxP-lacZ] zej::Tn10) (lanes 1 and 2), SP35 (SP34 Δ[pta ackA hisQ hisP]) (lanes 3 and 4), SP36 (SP34 cpxA::cam) (lanes 5 and 6), and SP37 (SP34 cpxA::cam Δ[pta ackA hisQ hisP]) (lanes 7 and 8). Strains whose β-galactosidase activities are depicted in odd-numbered lanes were grown in Luria broth (LB); strains whose β-galactosidase activities are depicted in even-numbered lanes were grown in Luria broth supplemented with 0.4% glucose to stimulate Ac∼P production.
Our laboratory has also recently obtained biochemical evidence that supports the view that CpxR-P is the activating species of the CpxA/R system. Specifically, in vitro studies indicate that the wild-type CpxA protein possesses CpxA autokinase, CpxR kinase, and CpxR-P phosphatase activities. In contrast to the wild-type protein, gain-of-function CpxA* mutant proteins (which stimulate cpxP transcription) are devoid of CpxR-P phosphatase activity (28). Thus, transcription of Cpx-regulated loci is stimulated when the kinase/phosphatase ratio of CpxA is increased. cpxA null strains lack both kinase and phosphatase activities. Consequently, low-level phosphorylation of CpxR mediated by Ac∼P results in significant accumulation of CpxR-P, stimulating cpxP transcription as shown in Fig. 2.
ςE does not regulate cpxP transcription.
Previous studies have indicated that both the Cpx and ςE regulatory systems control transcription of the degP locus (6, 27). In contrast to degP, transcription of the dsbA locus is controlled only by the Cpx system and not by ςE (5). Based on these results, we were interested in determining if cpxP transcription was coregulated by CpxA/R and ςE (as for degP) or whether its transcription was controlled only by CpxA/R (as for dsbA). To address this issue, we transformed SP1 (MC4100 ara+ λplacMu53[cpxP-lacZ]) with plasmids that overproduce the outer membrane porin proteins, OmpF and OmpC. Mecsas and colleagues (19) have shown that overproduction of these outer membrane proteins will stimulate ςE activity. However, these plasmids have no effect on cpxP transcription compared with their respective control plasmids (data not shown).
In a complementary analysis, we also assayed cpxP-lacZ transcription in a strain lacking a functional rpoE gene (which encodes ςE) (14, 27, 29). This strain did not decrease cpxP transcription compared to an isogenic control strain (data not shown), which indicates that cpxP transcription is not regulated by ςE. Thus, the cpxP locus falls into the same class as dsbA, as both genes are controlled by CpxA/R and not ςE.
Identification of the cpxP locus.
We mapped the location of the cpxP-lacZ fusion to the 88- to 89-min region of the E. coli chromosome. Surprisingly, we found that the cpxP-lacZ fusion was tightly linked to the cpxR::Ω insertion (>99.5% linkage by P1 transduction).
Because of the tight linkage between the cpxP-lacZ fusion and the cpx operon, we directly sequenced the region surrounding cpxR and found that the cpxP-lacZ fusion is inserted within the open reading frame of the gene immediately upstream of cpxR. This open reading frame was previously referred to as ORF_o167 (25).
Figure 3 shows the cpxP DNA sequence, the corresponding predicted amino acid sequence, and other salient features of this locus. For example, the fusion joint that creates cpxP-lacZ is positioned within the 13th codon of the cpxP open reading frame (Fig. 3b), disrupting the final 93% of this open reading frame. Hence, strains containing this λplacMu53-generated fusion are also cpxP.
Moreover, the first 23 codons of the cpxP open reading frame appear to specify a signal sequence. Specifically, the predicted amino acid sequence of this region begins with a basic residue (R), followed by a hydrophobic stretch of 20 residues ending in an alanine, which is immediately followed by an acidic residue (E). These features are the hallmark of a signal sequence, and they imply that cpxP encodes an exported protein (23).
cpxP transcription is regulated in trans by the Cpx proteins.
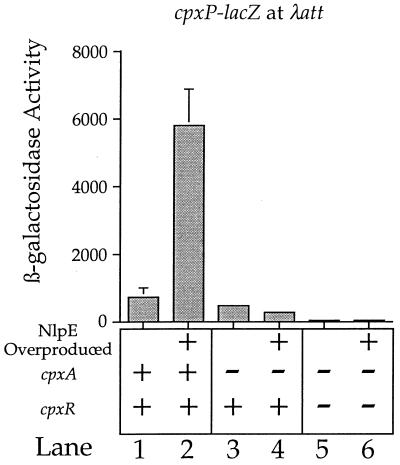
Since the cpxP locus was situated immediately upstream of the cpxRA operon, we were interested in determining if the cpxP-lacZ fusion was regulated by the Cpx system in trans. To address this issue, we generated a cpxP-lacZ fusion de novo (see Materials and Methods) and situated it at the λatt site, away from the cpx operon. This fusion behaves in a qualitatively similar manner to the λplacMu53-generated cpxP-lacZ fusion (Fig. 4).
FIG. 4.
The Cpx signal transduction system regulates transcription of a cpxP-lacZ fusion situated at the λatt site on the E. coli chromosome. Lanes 1, 3, and 5 show β-galactosidase activities of strains transformed with pBAD18 (control for pND18); lanes 2, 4, and 6 show β-galactosidase activities of strains transformed with pND18 (overexpresses nlpE). Lanes 1 and 2, SP702 (MC4100 ara+ zej::Tn10 Δ[pta ackA hisQ hisP] λRS88[cpxP-lacZ]); lanes 3 and 4, SP704 (SP702 cpxA::cam); lanes 5 and 6, SP706 (SP702 cpxR::Ω). All strains were grown in Luria broth containing 0.4% l-arabinose and 50 μg of ampicillin per ml (see Materials and Methods for details). These experiments were performed with strains deleted for pta and ackA. Since NlpE synthesis is driven from the araB promoter (11) in these experiments, full transcriptional induction requires growth in arabinose. Hence, Ac∼P synthesis must be eliminated to prevent hyperphosphorylation of CpxR in the cpxR+ cpxA background.
For example, Fig. 4 shows that the cpxR::Ω mutation abolishes transcription of the cpxP-lacZ fusion situated at the λatt site (compare lanes 1 and 5). Note that because of the tight linkage between cpxP and cpxR, we were unable to create a strain harboring both λplacMu53[cpxP-lacZ] and the cpxR::Ω mutation. The Ω cassette is inserted within the 21st codon of the cpxR open reading frame, leaving only 246 nucleotides between the Ω cassette and the cpxP-lacZ fusion (Fig. 3b). Figure 4 also shows that overproduction of NlpE stimulates transcription of the cpxP-lacZ fusion situated at λatt in a CpxA-dependent fashion (compare lanes 1 and 2 with lanes 3 and 4). Thus, the cpxP-lacZ fusion is controlled by the Cpx system in trans.
There is one quantitative difference between the cpxP-lacZ fusion situated within the cpxP open reading frame (λplacMu53[cpxP-lacZ]) and the cpxP-lacZ fusion at λatt (λRS88[cpxP-lacZ]). Specifically, the absolute level of cpxP-lacZ transcription is significantly higher when situated at λatt than when situated at the cpxP chromosomal locus. There are two possible explanations for this difference in absolute levels of transcription. First, λplacMu-generated fusions contain more than 2 kb of linker DNA between their target sequence and the lacZ gene (3). λRS88[cpxP-lacZ], which is positioned at λatt, does not contain this large amount of intervening sequence. Thus, it is possible that this intervening sequence reduces the absolute amount of transcription proceeding to lacZ in the λplacMu53[cpxP-lacZ] fusion compared to λRS88[cpxP-lacZ]. Second, since the λplacMu53[cpxP-lacZ] fusion is inserted within the cpxP open reading frame, and since this fusion is adjacent to the cpxRA operon, the fusion may affect the entire Cpx regulon, either by reducing cpxR/A activity in cis or by abolishing CpxP synthesis. The results presented here do not distinguish between these possibilities, and in fact, the difference in transcription between the two cpxP-lacZ fusions may result from a combination of the possibilities described above. Nevertheless, the results shown in Fig. 1 and 4 show that cpxP is regulated in trans by CpxRA.
CpxP is exported to the periplasm.
Since the deduced amino acid sequence suggested that the CpxP protein contained a signal sequence, we wanted to determine the subcellular location of this protein. To this end, we fused the first 71 codons of the cpxP open reading frame to the alkaline phosphatase coding sequence (lacking a signal sequence coding region), thus generating a CpxP-AP protein fusion (see Materials and Methods). We then determined (i) the amount of alkaline phosphatase activity generated by this fusion and (ii) the subcellular location of the CpxP-AP protein fusion. Since alkaline phosphatase is active only in relatively oxidizing subcellular compartments like the periplasm, the alkaline phosphatase activity generated by the CpxP-AP fusion provides an indirect assessment of the subcellular location of CpxP.
Figure 5 shows the results of alkaline phosphatase assays performed on SP627a (MC4100 ara-74::cam) harboring (i) control plasmids that do not produce alkaline phosphatase fusion proteins, (ii) plasmid pND24, which produces the CpxP-AP fusion, and (iii) pCH215. pCH215 produces a SecY-AP fusion protein, in which the alkaline phosphatase moiety is fused to cytoplasmic loop 5 of the inner membrane protein, SecY (13a). The SecY-AP fusion serves as an example of an alkaline phosphatase protein fusion that is not situated in an oxidizing compartment. Accordingly, this fusion should display little to no alkaline phosphatase activity.
Figure 5a shows that the only strain displaying an appreciable amount of alkaline phosphatase activity is the CpxP-AP fusion (lane 2). Note that the alkaline phosphatase assays used to generate the data illustrated in Fig. 5 were performed in the presence of 5 mM IAA. IAA covalently bonds to free sulfhydryl groups and as a consequence prevents the spontaneous activation of reduced (cytoplasmic) alkaline phosphatase molecules after cell lysis (7). Thus, all of the alkaline phosphatase activity observed in this experiment results from exported alkaline phosphatase molecules.
The data in Fig. 5a imply that the CpxP-AP fusion protein is exported to an oxidizing compartment. However, when performing such alkaline phosphatase assays, it is important to correct the observed amount of alkaline phosphatase activity for the amount of fusion protein that is responsible for generating this activity. Figure 5b shows the results of an immunoblot of whole-cell protein extracts from the four strains whose alkaline phosphatase activities are shown in Fig. 5a. The whole-cell extracts were probed with anti-alkaline phosphatase and anti-OmpR antisera (the OmpR protein serves as an internal loading control). Figure 5b shows that the amount of alkaline phosphatase-cross-reacting material generated in the CpxP-AP-producing strain is less than that generated in the SecY-AP-producing strain (compare lanes 6 and 8). This result substantiates the hypothesis that unlike the SecY-AP fusion protein, CpxP-AP is exported to an extracytoplasmic oxidizing compartment.
To determine the precise subcellular location of CpxP-AP, we fractionated proteins from a strain producing this fusion protein into whole-cell, spheroplast, and periplasmic fractions. Figure 6 shows an immunoblot of protein fractions prepared from SP627a transformed with pBR322 (control for pND24; lane 1) and pND24 (produces CpxP-AP; lanes 2 through 4). The immunoblot was probed with anti-alkaline phosphatase, anti-MalE, and anti-OmpR antisera. MalE and OmpR serve as model periplasmic and cytoplasmic proteins, respectively.
Lane 1 of Fig. 6 shows the endogenously encoded MalE and OmpR proteins from whole-cell protein extracts of SP627a transformed with pBR322 (control for pND24). Lane 2 shows the CpxP-AP fusion, MalE, and OmpR proteins from whole-cell protein extracts. Note that there are two bands generated by the CpxP-AP fusion. Either these two bands represent precursor (signal sequence unprocessed) and mature forms of the fusion protein or the lower band is a degradation product of the full-length CpxP-AP protein fusion. Nevertheless, the entire portion of the lower band is found in the periplasmic protein fraction, indicating that CpxP-AP is predominantly a periplasmic protein (compare lanes 3 and 4). As expected, MalE is found within the periplasmic protein fraction, while OmpR is predominantly found in spheroplast protein fractions (compare lanes 3 and 4).
The periplasmic location of CpxP is consistent with our proposed function for the Cpx signal transduction system. Specifically, if the Cpx regulon is involved in combating protein-mediated toxicities in the bacterial envelope, we would expect the members of the Cpx regulon to be found within the envelope. This prediction is clearly supported by the subcellular location of DegP, DsbA, and RotA and now CpxP as well.
CpxP is a stress-combative member of the Cpx regulon.
Since cpxP encodes a periplasmic protein, and since cpxP transcription is controlled by the Cpx system, we were interested in determining if CpxP was an extracytoplasmic stress-combative factor. To address this issue, we created three derivatives of WBS164 (MC4100 Φ(lamB-lacZX90) Hyb42-1[λp1(209)]): SP9 (WBS164 degP::Tn10), SP10 (WBS164 λplacMu53[cpxP-lacZ]), and SP24 (WBS164 degP::Tn10 λplacMu53[cpxP-lacZ]). Recall that λplacMu53[cpxP-lacZ] disrupts the cpxP open reading frame, and thus strains carrying this fusion are also cpxP mutant.
LamB-LacZX90 is a derivative of the LamB-LacZ hybrid fusion that contains a late nonsense mutation near its carboxy terminus (34). Much like the LamB-LacZ-PhoA tribrid fusion protein, LamB-LacZX90 forms a disulfide-bonded aggregate in the bacterial envelope and causes cell lysis as a result of its export to the envelope. Activation of the Cpx pathway suppresses this lethal phenotype (34).
To determine if CpxP functions to ameliorate the toxicity associated with LamB-LacZX90, we transformed WBS164, SP9, SP10, and SP24 with either pBR322 (control for pLD404) or pLD404 (overproduces the envelope lipoprotein NlpE). Previous studies have shown that overproduction of NlpE alleviates the toxicity associated with LamB-LacZX90 by activating the Cpx pathway (33).
We then quantified the susceptibility of each of the eight transformed strains to growth in the presence of maltose, which induces synthesis of LamB-LacZX90. Figure 7 shows the zone of growth inhibition caused by the addition of 10 μl of 40% maltose to a filter disc that was placed on a lawn of each of the eight transformed strains. The larger the zone of inhibition, the more susceptible the strain is to the synthesis of LamB-LacZX90.
Lane 1 of Fig. 7 shows that the parent strain, WBS164, transformed with the control vector, pBR322, is very maltose sensitive. Lane 2 shows that WBS164 transformed with the NlpE-overproducing plasmid (pLD404) is protected from the synthesis of LamB-LacZX90. Lane 4 shows that SP9 (WBS164 degP::Tn10) transformed with the NlpE-overproducing plasmid is more maltose sensitive than the isogenic degP+ strain (compare lanes 2 and 4). However, this strain is still less maltose sensitive than the parent strain (WBS164) transformed with the pBR322 control vector. Thus, as previously described (4, 35), the Cpx signal transduction system utilizes DegP (as well as other factors) to combat the extracytoplasmic toxicity conferred by LamB-LacZX90.
Lane 6 of Fig. 7 shows that SP10 (WBS164 λplacMu53[cpxP-lacZ]) transformed with the NlpE-overproducing plasmid is also more maltose sensitive than WBS164 transformed with the same NlpE-overproducing plasmid (compare lanes 2 and 6). Thus, cpxP is also utilized by the Cpx signal transduction system to combat the toxicity associated with LamB-LacZX90. Finally, when SP24 (WBS164 degP::Tn10 λplacMu53[cpxP-lacZ[) is transformed with the NlpE-overproducing plasmid, the strain is no more maltose sensitive than SP9 (WBS164 degP::Tn10) transformed with the same plasmid (compare lanes 4, 6, and 8). This final result has two implications. First, it suggests that CpxP and DegP may function within the same pathway to combat the toxicity associated with LamB-LacZX90. Second, since the degP cpxP double mutation does not completely abolish the ability of NlpE overproduction to ameliorate the LamB-LacZX90-associated toxicity, the Cpx signal transduction system must control still other factors that can combat this toxicity.
cpxP transcription is stimulated by extracellular alkaline pH.
A previous study (24) has demonstrated that the cpx locus affects the pH regulation of the Shigella sonnei virulence locus, virF. When introduced into E. coli, virF transcription is repressed at pH 6.0 and induced at pH 7.4. However, in a cpxA null mutant strain, virF transcription is derepressed at pH 6.0. Interestingly, a cpxR null mutation abolishes virF transcription altogether at both pH 6.0 and 7.4. Considered together, these results suggest that the activity of the Cpx signal transduction system may be affected (either directly or indirectly) by extracellular pH.
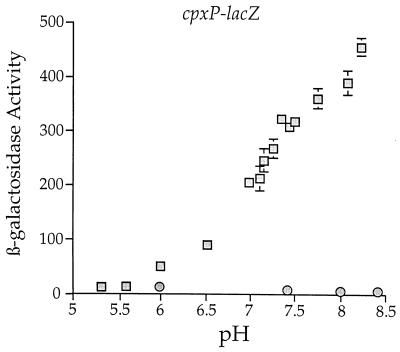
This study prompted us to examine the transcriptional regulation of cpxP as a function of pH. To this end, we determined the β-galactosidase activity of SP1 (MC4100 ara+ λplacMu53[cpxP-lacZ]) and SP7 (SP1 cpxA::cam) as a function of pH. Specifically, SP1 and SP7 were grown in Luria broth buffered with 100 mM sodium phosphate ranging in pH from 5.3 to 8.4, and the resulting β-galactosidase activities of these strains were determined. From this analysis, we found that the amount of cpxP transcription rises almost 50-fold from pH 5.3 to 8.4 (Fig. 8). This transcriptional induction is not observed in the cpxA::cam background (Fig. 8), indicating that this phenomenon is dependent on CpxA. Note that qualitatively similar increases in cpxP-lacZ transcription were also observed as a function of pH when the cpxP-lacZ fusion was positioned at the λatt site (data not shown).
FIG. 8.
Transcription of the cpxP-lacZ fusion is induced by alkaline pH. The β-galactosidase activities of SP1 (MC4100 ara+ λplacMu53[cpxP-lacZ]) (squares) and SP7 (SP1 cpxA::cam) (circles) were determined after strains had been grown at the indicated pH values. Strains were grown in buffered Luria broth ranging from pH 5.3 to 8.4. Luria broth was buffered with 100 mM sodium phosphate as described in Materials and Methods.
The buffering of Luria broth with 100 mM sodium phosphate necessarily changes sodium ion concentration as well as the pH of the medium. Thus, it was important to determine if alterations in sodium ion concentration affected transcription of cpxP-lacZ. Alterations in sodium ion concentration do not alter transcription from either cpxP-lacZ operon fusion (data not shown). The observed changes in cpxP-lacZ transcription shown in Fig. 8 are due to changes in pH.
Note that in the process of generating data for Fig. 8, bacterial growth in the buffered Luria broth did not alter extracellular pH by more than 0.1 pH unit. In addition, the pH of the β-galactosidase assay buffer (Z buffer) was not affected by the residual buffered Luria broth associated with the cell pellets. All β-galactosidase assays were performed at pH 7.1.
The cytoplasmic pH of E. coli is maintained at an approximate value of 7.7 irrespective of the external pH (13). As a consequence of this homeostatic phenomenon, the ΔpH component of the proton motive force (PMF) diminishes as the external pH rises. Because of this effect, we considered the possibility that the alkaline-pH-mediated induction of cpxP transcription was due to a collapse of the ΔpH gradient across the inner membrane. This model predicts that cpxP transcription should also be induced by protonophore uncouplers, such as carboxyl cyanide m-chlorophenylhydrazone (CCCP) (18). To test this model, we determined the amount of cpxP transcription from SP569 (MC4100 ara+ ilv::Tn10ΔuncBC λplacMu53[cpxP-lacZ]) before and after growth in the presence of CCCP, an uncoupler of the PMF. The ΔuncBC mutation in SP569 is required to prevent the strain from depleting its ATP reserves in an effort to buttress a collapsing PMF in the presence of CCCP (9).
Samples of SP569 were grown in Luria broth (buffered at pH 6.1 with 100 mM sodium phosphate) and then incubated in the presence of 50 μM CCCP for a period of time ranging from 0 to 135 min (this was also done with CCCP concentrations of 25, 100, and 125 μM). The β-galactosidase activities of each of these samples were then determined. cpxP-lacZ transcription does not rise under these conditions, indicating that cpxP transcription is not stimulated by a collapse of the ΔpH gradient across the inner membrane (data not shown). Rather, cpxP transcription appears to be induced by the alkalinity of the external environment. Note that the optical density at 600 nm (OD600) of SP569 continued to rise after CCCP treatment, indicating that this strain was subjected to sublethal doses of CCCP.
cpxP and cpx null mutations confer a hypersensitivity upon E. coli to alkaline pH.
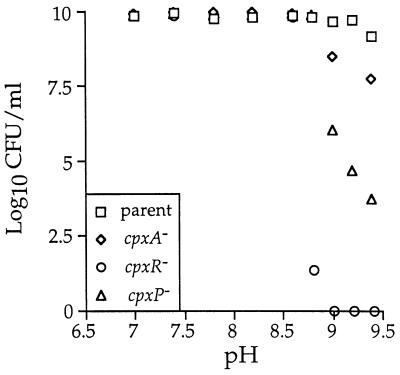
In light of the alkaline pH regulation of cpxP, we wanted to determine if the cpxP or cpx null mutation conferred upon E. coli any growth defects under conditions of extreme alkaline pH. Strains SP744 (MC4100), SP754 (SP744 cpxA::cam), SP762 (SP744 cpxR::Ω), and SP774 (SP744 λplacMu53[cpxP-lacZ]) were grown to saturation in Luria broth. Serial dilutions of each strain were then plated on Luria broth containing 100 mM Tris-HCl (ranging from pH 7.0 to 9.4), and plates were incubated at 37°C for 48 h. After this incubation period, the number of CFU/milliliter was determined for each strain at each pH value. Figure 9 shows the log10 of the CFU/milliliter value of each of these strains grown at pH 7.0 to 9.4. In contrast to the parental strain, the cpxA, cpxR, and cpxP strains each display an inability to form colonies in the pH range from 8.8 to 9.4. The cpxR strain is the most alkaline-hypersensitive strain, as it does not form any colonies at pH 9.0 and above. The cpxP strain is the second-most alkaline-hypersensitive strain, while the cpxA strain is least hypersensitive to alkaline pH (Fig. 9). The cpxR null mutation confers a higher degree of alkaline hypersensitivity than the cpxP null mutation, which implies that CpxR controls another factor(s), in addition to CpxP, that is also needed for growth under alkaline conditions.
FIG. 9.
cpxP and cpx null strains are hypersensitive to alkaline pH. Strain SP744 (MC4100 (squares), SP754 (SP744 cpxA::cam) (diamonds), SP762 (SP744 cpxR::Ω) (circles), and SP774 (SP744 λplacMu53[cpxP-lacZ]) (triangles) were grown to saturation in Luria broth. Serial dilutions of each culture were then plated on Luria broth buffered with 100 mM Tris-HCl (ranging from pH 7.0 to 9.5). Strains were incubated at 37°C for 48 h, and the number of CFU/milliliter of each culture at each pH value was determined as described in Materials and Methods. The results are plotted as the log10 of the CFU/milliliter values.
DISCUSSION
Studies by Cosma et al. (4) and Snyder et al. (35) have shown that activation of the Cpx signal transduction system is capable of combating extracytoplasmic protein-mediated toxicities. Activation of the Cpx pathway can combat toxicities conferred by at least two different types of envelope proteins, which argues that the members of the Cpx regulon most likely ameliorate these toxicities by performing general and fundamental functions within the envelope. Indeed, the identification of the DegP protease and the DsbA disulfide bond catalyst as regulatory targets of the Cpx system supports this notion (5, 6, 26, 27).
However, our laboratory has shown DegP is not the only factor that is regulated by the Cpx system to combat extracytoplasmic stresses (4, 35). This information provided the impetus to search for novel regulatory targets of the Cpx signal transduction system. Through this search, we have identified CpxP, a periplasmic protein whose synthesis is increased at the transcriptional level by the Cpx system. Intriguingly, the cpxP locus is adjacent to the cpx operon, intimating a fundamental link between the functions of these two loci.
The Cpx and ςE regulons.
Previous studies have shown that transcription of the degP locus is coregulated by CpxA/R and the heat shock-inducible ς factor, ςE (6, 27). In the case of degP, the Cpx pathway functions in concert with ςE to stimulate transcription from this locus (6, 26). Interestingly, the activities of ςE and Cpx are each attuned to the physiology of the bacterial envelope. For example, ςE activity rises in response to the overproduction of various outer membrane proteins, while Cpx activity rises when the outer membrane lipoprotein NlpE is overproduced (6, 19, 35). In contrast to the coregulation of degP transcription, other members of the Cpx regulon (e.g., dsbA and rotA) (5, 26) are not coregulated by ςE. Thus, there are at least two classes of Cpx-regulated loci: those that are coregulated by ςE and CpxA/R (degP) and those that are regulated only by CpxA/R (dsbA and rotA). cpxP falls into the latter class, since its transcription is not affected by alterations in ςE activity.
The extracytoplasmic stress-combative function of CpxP.
Tests of epistasis indicate that both CpxP and DegP are utilized by the activated Cpx pathway to suppress the toxic effects associated with the exported LamB-LacZX90 protein fusion. Interestingly, when the Cpx pathway is activated in the cpxP degP double mutant strain (Fig. 7, lane 8), the strain is no more sensitive to the effects of LamB-LacZX90 than the cpxP+ degP strain (Fig. 7, lane 4).
This result implies that CpxP and DegP function within the same pathway to suppress the toxic effects of LamB-LacZX90. For example, CpxP may function upstream of DegP, perhaps preparing substrates for this protease, or CpxP may function downstream of DegP, perhaps further processing DegP’s proteolytic products. Alternatively, CpxP may directly alter the activity of DegP. However, since the precise biochemical function of CpxP is not known, we cannot at present distinguish among such possibilities.
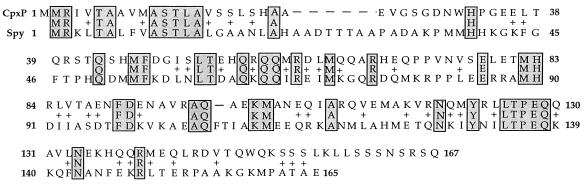
Interestingly, the predicted CpxP amino acid sequence displays limited similarity (29% identity over 101 amino acids) with Spy (spheroplast protein y), a periplasmic protein whose synthesis is increased when the outer membrane is stripped away from E. coli cells (Fig. 10) (12). Since Spy synthesis is induced by the extracytoplasmic stress of spheroplast formation, the homology between CpxP and Spy may intimate a functional relationship between these two proteins.
FIG. 10.
Homology between the CpxP and Spy proteins. Identical amino acids are shaded; similar amino acids are connected with plus signs.
Activation of the Cpx pathway by alkaline pH.
In addition to the stress-combative function of cpxP, we have found that the transcription of this locus is strongly induced by alkaline conditions in a CpxA-dependent fashion. It is unclear whether CpxA directly senses extracellular pH or whether it senses an indirect consequence that high pH imposes upon E. coli.
We favor the latter model for the following reason. The Cpx pathway can be activated by overproduction of various envelope proteins, including NlpE and the P-pilus subunits PapG and PapE (6, 15, 35). It seems unlikely that overproduction of these proteins activates the Cpx pathway by radically altering the pH of the bacterial envelope.
We note that other stress-combative envelope proteins are also required to protect E. coli from alkaline conditions. For example, surA null strains, which are defective for the efficient assembly of outer membrane porins, are also hypersensitive to alkaline pH (17). Similarly, strains that do not produce the stress-inducible envelope protein PspA are hypersensitive to alkaline pH in stationary phase (37). Taken together, these results suggest that structural damage may be inflicted upon the bacterial envelope by alkaline pH, and E. coli may utilize proteins such as SurA, PspA, and the regulatory targets of CpxR to weather such onslaughts.
ACKNOWLEDGMENTS
We thank Joe Pogliano and members of the Silhavy Lab, especially Christine Cosma, Leslie Pratt, and Bill Snyder, for helpful discussions. We also thank Chris Harris for providing pCH215.
P.N.D. gratefully acknowledges support from NIGMS training grant GM07312. T.J.S. was supported by NIGMS grant GM34821.
REFERENCES
- 1.Albin R, Weber R, Silverman P M. The Cpx proteins of Escherichia coli K12. Immunologic detection of the chromosomal cpxA gene product. J Biol Chem. 1986;261:4698–4705. [PubMed] [Google Scholar]
- 2.Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene. 1978;4:121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- 3.Bremer E, Silhavy T J, Weinstock G M. Transposable λplacMu bacteriophages for creating lacZ operon fusions and kanamycin resistance insertions in Escherichia coli. J Bacteriol. 1985;162:1092–1099. doi: 10.1128/jb.162.3.1092-1099.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosma C L, Danese P N, Carlson J H, Silhavy T J, Snyder W B. Activation of the Cpx two-component signal transduction pathway in Escherichia coli suppresses envelope associated stresses. Mol Microbiol. 1995;18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- 5.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 6.Danese P N, Snyder W B, Cosma C L, Davis L J B, Silhavy T J. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 7.Derman A I, Beckwith J. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J Bacteriol. 1991;173:7719–7722. doi: 10.1128/jb.173.23.7719-7722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong J S, Iuchi S, Kwan H S, Lu Z, Lin E C C. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene. 1993;136:227–230. doi: 10.1016/0378-1119(93)90469-j. [DOI] [PubMed] [Google Scholar]
- 9.Fraimow H S, Greenman J B, Leviton I M, Dougherty T J, Miller M H. Tobramycin uptake in Escherichia coli is driven by either electrical potential or ATP. J Bacteriol. 1991;173:2800–2808. doi: 10.1128/jb.173.9.2800-2808.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez C, Devedjian J C. A plasmid facilitating in vitro construction of phoA gene fusions in Escherichia coli. Nucleic Acids Res. 1989;17:3999. doi: 10.1093/nar/17.10.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagenmaier S, Stierhof Y-D, Henning U. A new periplasmic protein of Escherichia coli which is synthesized in spheroplasts but not in intact cells. J Bacteriol. 1997;179:2073–2076. doi: 10.1128/jb.179.6.2073-2076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harold F M, Maloney P C. Energy transduction by ion currents. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 283–306. [Google Scholar]
- 13a.Harris, C. Unpublished data.
- 14.Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 17.Lazar S W, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis K, Naroditskaya V, Ferrante A, Fokina I. Bacterial resistance to uncouplers. J Bioenerg Biomembr. 1994;26:639–646. doi: 10.1007/BF00831539. [DOI] [PubMed] [Google Scholar]
- 19.Mecsas J, Rouvière P E, Erickson J W, Donohue T J, Gross C A. The activity of ςE, an Escherichia coli heat-inducible ς-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 20.Misra R, Reeves P R. Role of micF in the tolC-mediated regulation of OmpF, a major outer membrane protein of Escherichia coli K-12. J Bacteriol. 1987;169:4722–4730. doi: 10.1128/jb.169.10.4722-4730.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misra R, Benson S A. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J Bacteriol. 1988;170:3611–3617. doi: 10.1128/jb.170.8.3611-3617.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missiakas D, Raina S. Signal transduction pathways in response to protein misfolding in the extracytoplasmic compartments of E. coli: role of two new phosphoprotein phosphatases PrpA and PrpB. EMBO J. 1997;16:1670–1685. doi: 10.1093/emboj/16.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy C K, Beckwith J. Export of proteins to the cell envelope. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 967–978. [Google Scholar]
- 24.Nakayama S-I, Watanabe H. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol. 1995;177:5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plunkett G, III, Burland V, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993;21:3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogliano J, Lynch A S, Belin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 27.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raivio T L, Silhavy T J. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouvière P E, de las Peñas A, Mecsas J, Lu C Z, Rudd K E, Gross C A. rpoE, the gene encoding the second heat-shock sigma factor, ςE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 31.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusion. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 32.Slauch J M, Silhavy T J. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder W B. The secretion related toxicities of LacZ. Ph.D. thesis. Princeton, N.J: Princeton University; 1993. [Google Scholar]
- 34.Snyder W B, Silhavy T J. β-Galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J Bacteriol. 1995;177:953–963. doi: 10.1128/jb.177.4.953-963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder W B, Davis L J B, Danese P N, Cosma C L, Silhavy T J. Overproduction of NlpE, a new outer-membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol. 1995;177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 37.Weiner L, Model P. Role of an Escherichia coli stress-response operon in stationary-phase survival. Proc Natl Acad Sci USA. 1994;91:2191–2195. doi: 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]