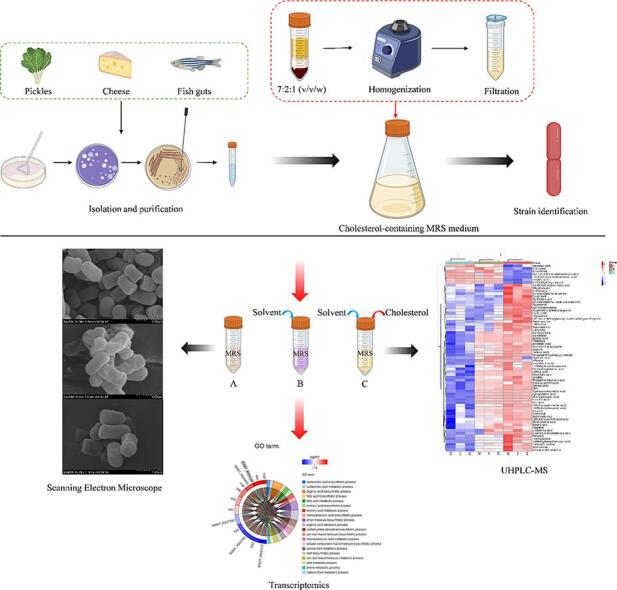
Graphical abstract
Keywords: Lactic acid bacteria, Cholesterol, Transcriptomics, Metabolomics, Molecular docking, Ultrasound
Highlights
-
•
A novel practical MRS medium for screening cholesterol-lowering LAB was developed.
-
•
L. plantarum 54–1 was found to have the highest ability to eliminate cholesterol.
-
•
PTS, ABC transporter and insulin signaling pathways were the main reason.
-
•
Transcriptomics found 5 genes that may be related with cholesterol-lowering in LAB.
Abstract
In this study, an efficient cholesterol-lowering strain of Lactiplantibacillus plantarum 54–1 was screened and its degradation molecular mechanism was investigated. Furthermore, a novel practical MRS medium for screening cholesterol-lowering lactic acid bacteria (LAB) was developed based on ultrasound treatment. L. plantarum 54–1 was found to have the highest ability to eliminate cholesterol (340.69 ± 5.87 µg/mL). According to SEM and the count of viable LAB results, the morphology of LAB in the cholesterol-containing medium developed in this experiment was close to the normal (full and smooth), and it can grow normally. Metabolomics revealed that L. plantarum 54–1 initially converted a portion of cholesterol to 7α-hydroxy-cholesterol and then to the key metabolite taurine, via the phosphotransferase system. These metabolites were further transformed into L-alanine, L-lysine, N6-Acetyl-L-lysine, (R)-b-aminoisobutyric acid, and 2-oxoarginine, through glycine, serine, and threonine metabolism, citrate cycle, D-arginine and D-ornithine metabolism, lysine degradation, and pyruvate metabolism pathways. Prokaryotic reference transcriptomics found that this may be mainly regulated by the bsh, phnE, ptsP, B0667_RS04545, and B0667_RSRS12300 genes, which was further validated by qPCR. Furthermore, molecular docking results demonstrated that 8 differential metabolites might bind to another portion of cholesterol via PI-PI conjugation and hydrophobic interactions and lower cholesterol via co-sedimentation. This study has strategic implications for developing probiotic powder food that lowers cholesterol.
1. Introduction
According to clinical research, humans typically have between 2.8 and 6.0 mmol/L, serum cholesterol levels, and greater levels are linked to a higher risk of illness [1]. A 1 % decrease in serum cholesterol levels results in a 2–3 % reduction in the prevalence of coronary heart disease [2]. The primary methods for controlling blood cholesterol levels can be divided into 3 categories. First, the intake of foods high in cholesterol was reduced. For example, monkeys served a meal containing 0.19 % cholesterol saw a considerable increase in LDL levels after 7 d [3]. However, it is challenging to sustain this strategy over time. Second, the autologous synthesis of cholesterol was blocked by drugs (monacolin, lovastatin, etc.), promoting the conversion of cholesterol to bile acids and inhibiting the activity of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase, reducing the synthesis of cholesterol [4]. Nevertheless, these drugs have strong side effects and can damage the liver with long-term use. Third, physical, chemical, and biotransformation techniques were employed to eliminate cholesterol from food. However, these techniques may have affected the meal's flavor and nutritional value [5]. Consequently, developing foods that lower cholesterol while being safe and healthy is crucial.
In the post-epidemic era, probiotic powder food with probiotic functions is widely accepted by consumers [6]. Lactic acid bacteria (LAB), as a kind of probiotic, has many beneficial functions for the human body, such as anti-aging, preventing depression, regulating intestinal flora, etc [7], [8]. Identifying LAB that can efficiently degrade cholesterol has been a hot research topic. For example, a strain of LAB named Lacticaseibacillus paracasei M5 was screened from breast milk with a cholesterol degradation rate of 84.75 % [9]. Lye et al. in 2012 found that ultrasound enhanced the growth and cholesterol removal of Limosilactobacillus fermentum FTDC 1311 mother cells, but had no effect on subsequent passaged cells [10]. When cholesterol removal was used as an evaluation criterion, a strain named Lacticaseibacillus casei extracted from ewe milk could remove up to 35.41 µg/mL cholesterol [11]. However, it is difficult to compare the performance of strains obtained by different scholars due to differences in experimental conditions such as cholesterol solvent (cholesterol solutions are cumbersome to prepare and water-soluble cholesterol is expensive), cholesterol starting concentration in the medium. Therefore, the preparation of simple and easy-to-use media for cholesterol-lowering LAB screening to standardize the measurements, and screen an efficient cholesterol-lowering LAB strain, are crucial for developing probiotic powder foods. Furthermore, the molecular mechanism of cholesterol-lowering by LAB has always been controversial [12]. We speculate that the highly efficient gene expression in regulating bile salt hydrolase expression in LAB may be the determining factor.
Untargeted metabolomics is the qualitative and quantitative analysis of all low molecular weight metabolites (MW < 1000) of an organism or cell in a specific physiological period, based on cohort index analysis, high throughput detection, and data processing [13]. For example, in our previous study, metabolomics revealed a high GABA content and its metabolic pathway in yogurt fermented by Levilactobacillus brevis CGMCC1.5954 [14]. In addition, by utilizing untargeted metabolomics, Shi et al. unraveled the mechanism behind nitrite degradation in Limosilactobacillus fermentum RC4 [15]. Prokaryotic reference transcriptomics is the comprehensive study of gene transcription and transcriptional control in cells, examining RNA-level gene expression [16]. For example, Lee et al. investigated the transcriptome reactions of Latilactobacillus curvatus WiKim38, Companilactobacillus allii WiKim39, and Lactococcus lactis WiKim0124, which were isolated from kimchi, upon exposure to hydrogen peroxide [17]. Therefore, we aim to investigate the molecular mechanism of cholesterol-lowering by LAB using metabolomics and transcriptomics in a combined multi-omics analysis.
First, a novel, stable, and convenient cholesterol-containing LAB culture medium was created. In addition, a strain of cholesterol-degrading LAB was screened and identified from raw materials such as kimchi, yogurt, and fish sausage with high efficiency. Finally, a multi-omics combination of metabolomics and transcriptomics was used to analyze the molecular mechanism of cholesterol-lowering by this strain of LAB. It has strategic implications for the screening methods of cholesterol-lowering LAB, the investigation of molecular mechanisms, and the development of cholesterol-lowering probiotic powder food.
2. Materials and methods
2.1. Materials
The deep-sea fish sausages, cheese, and kimchi were bought at the Ningbo Farmers' Market (Zhejiang, China). Tween-80, ether, and cholesterol were supplied by China Pharmaceutical Group Co (Beijing, China). In addition, the total cholesterol content test kit, methanol (99 % purity), 2-chloro-L-phenylalanine (internal standard), and 0.22 µm filter membrane were produced by Nanjing Jiancheng Institute of Biological Engineering (Nanjing, China), Fisher Scientific (Loughborough, UK), Aladdin (Shanghai, China), and TCI (Shanghai, China) respectively.
2.2. Preparation of cholesterol-containing media
2.2.1. Preparation of cholesterol solution
First, 4 methods of preparing cholesterol solutions were selected for comparison (cholesterol solutions should be homogeneous in texture, free of precipitation, non-reactive with solvents, stable, and easy to prepare and use). In control group A: 1 g of cholesterol was dissolved in 9 mL of anhydrous ethanol and vortexed for 30 min. Control group B consists of 0.8 g of cholesterol, 2 g of bovine bile salt, 1 g of sucrose fatty acid ester, 1 mL of Tween-80, 5 mL of anhydrous ethanol, water bath at 80 °C for 10 min, sonication at 80 W, and passage through a 0.45 µm filter membrane [18]. Control group C experiment contained 8 mL of ether, 1 mL of Tween-80, and 1 g of cholesterol, which were vortexed for 30 min before being passed through a 0.22 µm filter membrane. Finally, group D contained 7 mL of ether, 2 mL of Tween-80, and 1 g of cholesterol and was subjected to a 100 W ultrasound for 10 min, a 20 min vortex, and passed through a 0.22 µm membrane.
2.2.2. Preparation of cholesterol-containing MRS liquid medium
First, 500 mL of MRS liquid medium was sterilized for 30 min at 121 °C before cooling to room temperature. Next, 3 mL of sterile cholesterol solution was aspirated from experimental group D, added to 500 mL of MRS medium, shaken, and then mixed for 5 min to homogenize. The Total Cholesterol Content Assay Kit was used to calculate the cholesterol content. After 7 d of standing at room temperature, the cultures were checked for precipitation in the medium. A blank MRS liquid medium was used as a control.
2.2.3. Cholesterol-containing medium-quality confirmation
LAB (an efficient cholesterol-degrading strain screened in section 2.3) was inoculated at 2 % (v/v) into MRS medium containing cholesterol, as described in section 2.2.2, and incubated for 24 h at 37 °C. Blank MRS medium and blank cholesterol-containing medium were also included as controls. After incubation, the cultures were centrifuged at 8000 × g and 4 °C for 10 min, with the cholesterol-containing MRS liquid medium left standing for 7 d.
2.2.4. Scanning electron microscope (SEM)
To compare the effect of the cholesterol-containing MRS medium developed in this experiment with that of MRS medium prepared from water-soluble cholesterol (Sigma-Aldrich, St. Louis MO, USA) solution on the morphology of the bacteria at high cholesterol concentrations, SEM was carried out. The LAB (an efficient cholesterol-degrading strain screened in section 2.3) was inoculated at 2 % (v/v) in the blank group (blank MRS medium), control group (MRS medium with added water-soluble cholesterol solution), and experimental group (MRS medium with added cholesterol solution developed in this study) MRS liquid medium and incubated at 37 °C for 16 h. The suspension was centrifuged at 3000 × g for 5 min at 4 °C. The supernatant was removed and washed twice with PBS buffer. Then, the bacteria were resuspended with 2.5 % (v/v) glutaraldehyde, mixed thoroughly, and fixed at 4 °C for 4 h. The organisms were washed 3 times with PBS buffer, once with a gradient of ethanol (50 %, 705 %, 90 %, 95 %, v/v) for 10 min, and finally 3 times with anhydrous ethanol for 10 min. Finally, the samples were centrifuged at 3000 × g for 5 min at 4 °C and dried in an oven at 37 °C for 12 h. After gold spraying, the bacterium’s surface was observed using SEM [18].
2.3. Screening and identification of cholesterol-lowering LAB
2.3.1. Primary screening of LAB
The ingredients, such as kimchi, sauerkraut, and fish sausage, were diluted in sterile saline and ground. Then, 100 µL was pipetted and coated in MRS solid medium containing 1.2 % (w/v, mg/mL) bromocresol violet. The plates were incubated at 37 °C for 24 h, and the single bacteria with color change on the plates were picked and purified by successive scribing for 3 generations. Finally, they were transferred to a liquid medium, DNA was extracted, and their 16S rRNA sequences were determined.
2.3.2. Rescreening of cholesterol-lowering LAB
The LAB from the primary sieve and laboratory conserved were inoculated at 2 % (v/v) in a liquid medium with a cholesterol content of 942.88 µg/mL and then incubated at 37 °C for 24 h. Subsequently, the culture bacterial fluid was centrifuged at 5000 rpm for 10 min at 4 °C. The supernatant was extracted and the cholesterol-lowering ability of each LAB was determined using the Total Cholesterol Content Kit. The strain identification of the screened LAB with the highest cholesterol-lowering ability was performed based on Gram staining, physiological and biochemical indices, and genotype [19].
2.4. Number of viable bacteria of L. Plantarum 54–1 in solvent and cholesterol
In order to investigate the reason for the cholesterol-lowering of L. plantarum 54–1 and to exclude the interference caused by solvents, its effect in the blank MRS (group A), in the MRS group to which solvents were added (group B, containing ether and Tween-80), and in the MRS group to which cholesterol solutions were added (group C, containing ether, Tween-80, and cholesterol) were therefore determined. L. plantarum 54–1 was inoculated into the above three MRS liquid media at a ratio of 2 % (v/v), and then incubated at 37 °C for 16 h. Then, 100 µL of bacterial solution was aspirated onto a blank solid MRS plate using the 10-fold gradient dilution smear plate method and incubated at 37 °C for 24 h. Finally, the number of viable bacteria on the plate was determined using the icount 22 automatic colony counter (Hangzhou Xunxun Counting Ltd., Hangzhou, China).
2.5. UHPLC-MS combined with metabolomics at the metabolic level to investigate the cholesterol-lowering mechanism of L. Plantarum 54–1
2.5.1. Sample preparation and UHPLC-MS detection
The target strains were inoculated at 2 % (v/v) into the blank (group A), solvent (containing Tween-80 and ether, group B), and cholesterol MRS liquid medium (containing Tween-80, ether, and cholesterol, group C) and incubated at 37 °C for 16 h. After fermentation, they were vortexed for 1 min and thoroughly mix. After concentrating and drying 1.5 mL of suspension in a 2 mL centrifuge tube, 500 µL of methanol was added, vortexed for 1 min, and then centrifuged at 12000 × g for 10 min at 4 °C. The supernatant was extracted and concentrated. Then 150 µL of 2-chloro-L-phenylalanine (4 ppm) in 80 % methanol–water was added and filtered through a 0.22 μm membrane for Ultra Performance Liquid Chromatography - Mass Spectrometry (UHPLC-MS) [15]. An ACQUITY UPLC® HSS T3 column (2.1 x 150 mm, 1.8 µm) was used with a Thermo Vanquish UPLC system (Thermo Fisher Scientific, USA) at a flow rate, column temperature, and injection volume of 0.25 mL/min, 40 °C, and 2 μL, respectively. Up-sampling assays were performed according to the method of Zelena et al [20].
2.5.2. Metabolomics data processing and analysis
Initially, the MSConvert utility in the Proteowizard package (v3.0.8789) was used to convert the raw mass spectrometry downlink file to the mzXML file format. Afterwards, peak detection, peak filtering, and peak alignment processes were performed to obtain a quantitative list of substances. Compounds were identified through the public databases HMDB (https://www.hmdb.ca), massbank (https://www.massbank.jp/), LipidMaps (https://www.lipidmaps.org), mzclound (https://www.mzcloud.org), and KEGG (https://www.genome.jp/kegg/) with parameters set to ppm < 30 ppm [8], [13], [15].
2.6. Molecular docking
The 3D molecular structure of the screened differential compounds with cholesterol was downloaded from the PubChem database. The small molecules were then imported into AutoDockTools (v1.5.6) for hydrogenation, charge calculation, charge assignment, rotation key setting and finally saved in “pdbqt” format. Molecular docking was performed using AutoDock Vina1.1.2 software and the conformation was optimised using a genetic algorithm based on the principle of minimum docking energy. The docking results were then analysed for interaction patterns using PyMOL 2.3.0 and Discovery Studio.
2.7. Transcriptomics-based investigation of the molecular mechanism of cholesterol-lowering at the transcriptional level of L. Plantarum 54–1
Firstly, L. plantarum 54–1 was grouped and cultured under the same conditions as in section 2.5.1. Total RNA was then extracted from the LAB samples, and the concentration and purity of the extracted RNA were examined by Nanodrop2000. The TruSeqTM Stranded Total RNA Library Prep Kit was used to build the library. Then, the second strand of cDNA was created by replacing dTTP with dUTP in the dNTPs reagent, resulting in bases in the second strand of cDNA that are A/U/C/G. Next, the second strand of the cDNA was digested with the UNG enzyme before PCR amplification so that the library only contained the first strand. Afterward, total RNA was obtained, and rRNA was eliminated. mRNA was then fragmented and reversed transcribed into cDNA by reverse transcriptase. cDNA structure of the adaptor duplex was then joined with sticky ends, which were patched to flat ends by adding End Repair Mix, followed by adding an A base at the 3′ end for joining the Y junction. Finally, the cDNA duplex was digested by the UNG enzyme and sequenced using the Illumina Hiseq sequencing platform [21].
2.8. Quantitative real-time PCR (qPCR) validation
L. plantarum 54–1 was grouped and cultured under the same conditions as in section 2.7. An RNA extraction kit was used to extract the RNA from the L. plantarum 54–1, and then cDNA was synthesized using a reverse transcription kit. Primers were designed using Primer Premier 6 and validated by comparison using the BLAST database in NCBI. Table 1 shows the designed primers, with 16S rRNA serving as the internal reference gene. The qPCR reaction for the differential genes was performed on a 20 μL reaction system using a Roche LightCycler® 96 instrument, following the method of Fan et al. with slight modifications [22].
Table 1.
Primer sequences for qPCR.
| Genes name | Sequence |
|---|---|
| bsh | f: 5′-CCGCTACATTGGTTGGTT-3′ |
| r: 5′-TAAGGCACGATAGTTGTTCA-3′ | |
| phnE | f: 5′-GACGCTAGGCTATCTAACA-3′ |
| r: 5′-CACAGAAGACACATTGGAA-3′ | |
| ptsP | f: 5′-ACATTGGTGGTCGGACTA-3′ |
| r: 5′-GTAATCAGCAACTTCAGCAT-3′ | |
| B0667-RS04545 | f:5′-TGAACTCGTTATCGTTGACT-3′ |
| r: 5′-CACCAGCCGTAATGACAA-3′ | |
| B0667-RS12300 | f: 5′-AGGCACCGATGGATTACT-3′ |
| r: 5′-CGAGAATGACGCTTGGAT-3′ | |
| 16S rRNA | f: 5′-GACAATTACGCCGAGTGA-3′ |
| r: 5′-GCACAAGCATCCAAGACT-3′ |
2.9. Statistical analysis
All experiments were repeated at least 3 times for data analysis. SPSS 22.0 was used to analyze data statistics for ANOVA with a significance level of difference of P < 0.05. Origin 8.0 was used to generate graphs. Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) dimensionality reduction analysis were performed using the R software package Ropls. The P value was calculated according to the statistical test, the variable projected importance (VIP) was calculated by the OPLS-DA dimensionality reduction method, and the multiplicity of difference of the components was calculated by fold change, which measured the strength of the influence of the content of each metabolite component on the classification discrimination of the samples and the explanatory ability, and assisted in the screening of marker metabolites.
3. Results and discussion
3.1. Preparation of cholesterol-containing media
3.1.1. Preparation of cholesterol solution
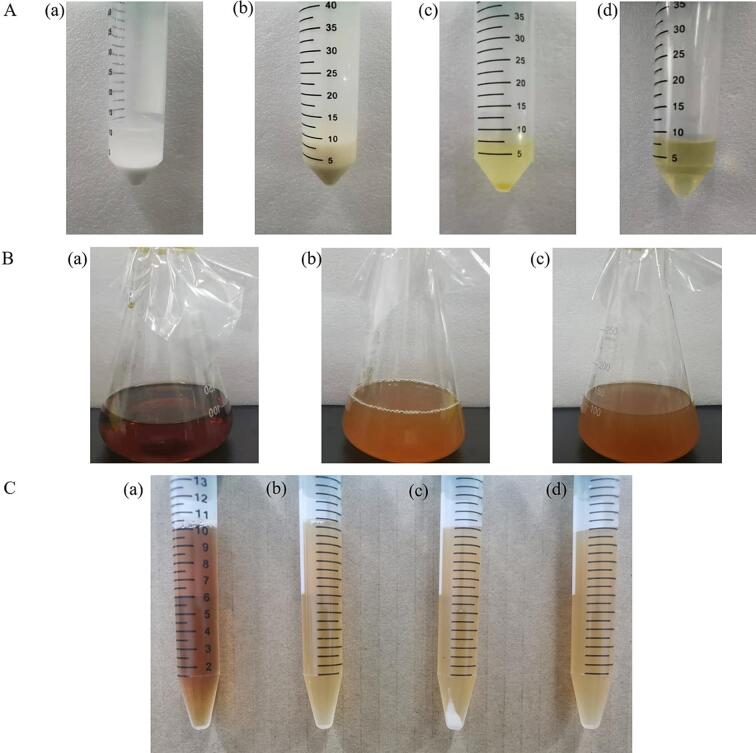
Cholesterol is comparable to fat in terms of solubility. It is insoluble in water and marginally soluble in anhydrous ethanol. When screening for LAB capable of degrading cholesterol, it is challenging to formulate a liquid medium containing a specific cholesterol concentration. Fig. 1A shows that cholesterol was almost insoluble in anhydrous ethanol (a). In contrast, it appears as a homogeneous emulsion in control B (b). However, it is very viscous and not easily removable. The solution in control group C showed a clear stratification (c). In contrast, the solution in the experimental group was homogeneous and clear (d), and did not precipitate when left at room temperature for 7 d. It was demonstrated that cholesterol solutions could be prepared in 40 min using 7 mL of ether, 2 mL of Tween-80, 1 g of cholesterol, and 100 W ultrasound 10 min after vortexing for 20 min, and the quality of the solutions was stable.
Fig. 1.
Preparation of cholesterol-containing MRS medium. (A): Preparation of cholesterol solution. Figure a-d shows the cholesterol dissolved in anhydrous ethanol, control 1 (0.8 g cholesterol, 2 g bovine bile salt, 1 g sucrose fatty acid ester, 1 mLTween-80, 5 mL anhydrous ethanol, water bath at 80 °C for 10 min, after 80w sonication over 0.45 µm filter membrane), control 2 (ether: Tween-80: cholesterol of 8:1:1, v/v/w), and experimental group (ether: Tween-80: cholesterol of 7:2:1, v/v/w). (B): Cholesterol-containing MRS liquid medium. Figures a-c show the normal MRS liquid medium, MRS medium after adding cholesterol solution and homogenization, and MRS medium after adding cholesterol solution and resting for 7 d, respectively. (C): Cholesterol-containing MRS medium after 37 °C fermenting 16 h and 4 °C, 8000 × g centrifugating 10 min. Figure a-d shows the blank MRS medium, MRS medium after homogenization with cholesterol solution, MRS medium inoculated with 2 % (v/v) LAB containing cholesterol solution, and MRS medium after 7 d of resting, respectively.
3.1.2. Preparation of cholesterol-containing MRS liquid medium
Compared with the blank control group (Fig. 1B-a), the medium with newly added cholesterol solution had a uniform texture and no precipitation (Fig. 1B-b). In addition, the cholesterol-containing medium remained intact at room temperature for 7 d without cholesterol precipitation (Fig. 1B-c). It demonstrated that the cholesterol solution prepared in this experiment was stable when used in an MRS medium. Fig. 1C shows that precipitates were produced in the tubes containing LAB after centrifugation (c). However, no precipitates were produced in the blank MRS control (a), the blank MRS control with cholesterol solution (b), and the MRS blank control with cholesterol solution (d) after 7 d of resting. It indicated that all the precipitates were LAB and that centrifugation, resting, or incubation at 37 °C did not affect the stability of the MRS medium containing cholesterol solution. This indicated that the experiment was unaffected and met the requirements for the experiment.
3.1.3. SEM to observe the morphology of the LAB under different cholesterol medium conditions
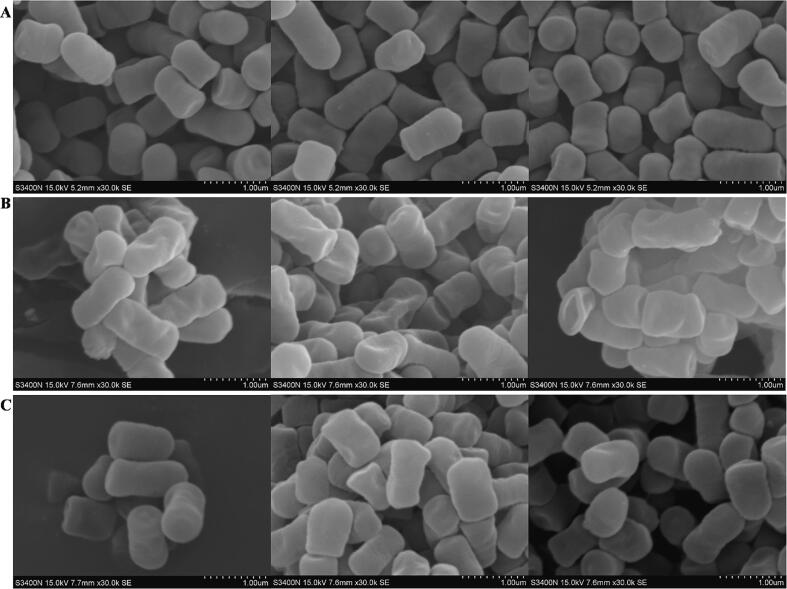
The LAB screened for efficient cholesterol degradation in section 2.3 is used at this point. As shown in Fig. 2A, when L. plantarum 54–1 was cultured in the blank group MRS, the bacterium was short rod-shaped, full, and stout, about 0.7–1.2 μm long and 0.5–0.6 μm wide, with wrinkled surface and arranged singly. In addition, when high concentrations of cholesterol were present (MRS medium prepared from water-soluble cholesterol), the morphology of the LAB changes to a slender, long rod-like shape, about 0.8 ∼ 1.4 μm long and 0.3–0.5 μm wide, with a concave surface and varying length (Fig. 2B). However, the morphology of the bacteria in the experimental group (the cholesterol solvent developed in this experiment) was close to the normal (group A), fuller, and smoother (about 0.7–1.2 μm long and 0.4–0.6 μm wide) (Fig. 2C). Furthermore, Kimoto [23] reported that Tween 80 enhances the bile tolerance of Lactococcus lactis. Therefore, it may be due to the protective effect of tween-80 as a pro-solvent, stabilizer, and protector on LAB cell membranes, which might allow LAB to maintain normal metabolism in a high cholesterol concentration medium [24], [25].
Fig. 2.
SEM. (A): SEM of LAB in blank MRS. (B): SEM of LAB in the control group (MRS medium with added water-soluble cholesterol solution). (C): SEM of LAB in the MRS medium containing cholesterol solution developed in this study. Notes: The total length of the scale in the lower right corner of the figure is 1 µm.
3.2. Screening and identification of cholesterol-lowering LAB
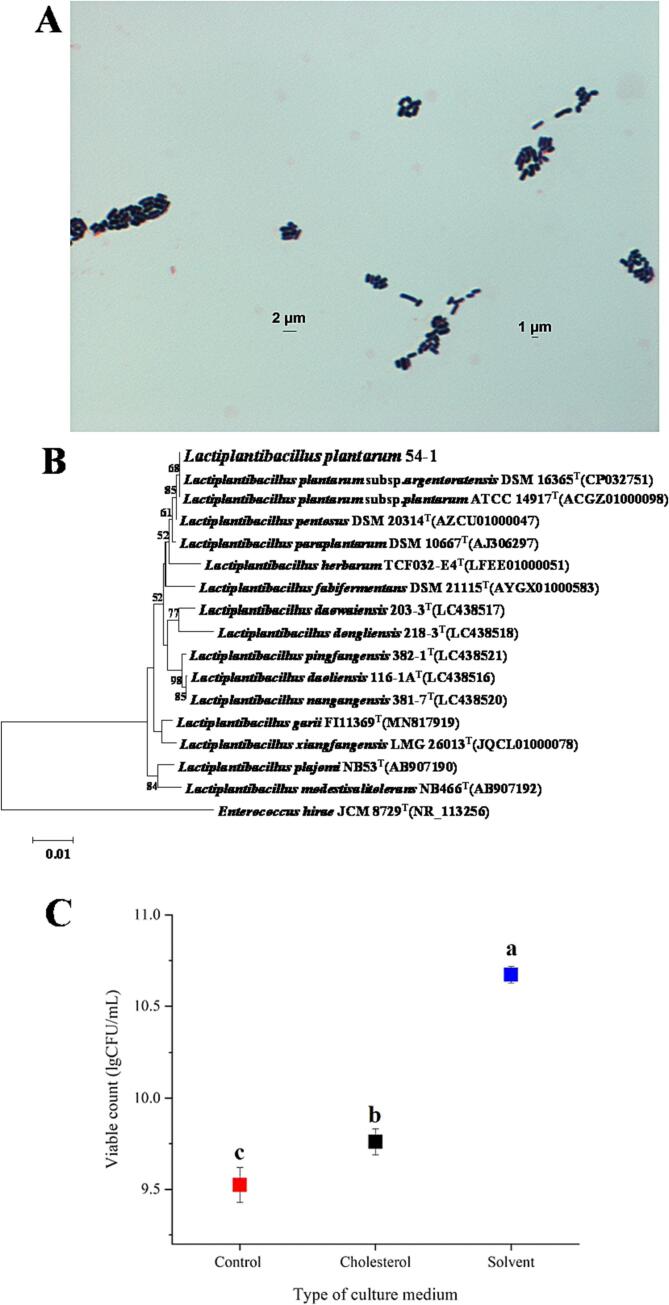
After the initial screening, 62 strains of LAB were screened cumulatively, and 35 strains of LAB with different genotypes were identified by 16S rRNA sequencing (16S sequencing results were available in the Supplementary Material). The high-efficiency cholesterol-lowering LAB was screened alongside 13 strains of LAB from the laboratory collection. The top 50 % cholesterol-lowering capacity ranking is shown in Table 2. One strain, L. plantarum 54–1, had the highest ability to remove cholesterol from MRS at 340.69 ± 5.87 µg/mL, with a clearance rate of 36.13 ± 0.62 %. Le & Yang [26] screened 191 strains of LAB from traditional salted shrimp foods with cholesterol-lowering abilities ranging from 12.5 to 43.1 μg/mL (total 100 μg/mL), with L. rhamnosus GG (LGG) and P. pentosaceus KACC 12311 showing the highest clearance rates of 38.2 % and 26.1 %, respectively. In terms of total amount cholesterol removal, L. plantarum 54–1 screened in this experiment was much higher. The bacterium was identified as L. plantarum subsp. plantarum by Gram staining (Fig. 3A), API 50CH (Table 3), and constructing a phylogeny (Fig. 3B). In addition, it was conserved in the China General Microbiological Culture Collection Center (CGMCC) No. 1.5953.
Table 2.
Screening of cholesterol-lowering LAB.
| Strain name | Cholesterol clearance (µg/mL) | Clearance rate (%) |
|---|---|---|
| Lactobacillus acidophilus NCFM | 258.75 ± 2.34f | 27.44 ± 0.25f |
| Lactococcus lactis | 255.41 ± 1.62 fg | 27.09 ± 0.17 fg |
| Lactiplantibacillus plantarum 7XCJ-1 | 217.53 ± 3.23i | 23.07 ± 0.34i |
| Lactobacillus acidophilus 6074 | 189.80 ± 6.98jk | 20.13 ± 0.74jk |
| Lactobacillus delbrueckii subsp. bulgaricus | 199.20 ± 8.36j | 21.13 ± 0.88j |
| Lacticaseibacillus casei | 325.45 ± 2.09b | 34.52 ± 0.22b |
| Lactiplantibacillus plantarum 52–1 | 191.30 ± 5.27jk | 20.29 ± 0.56jk |
| Levilactobacillus brevis 55 | 193.21 ± 5.08jk | 20.49 ± 0.54jk |
| Limosilactobacillus fermentum | 229.93 ± 6.42 h | 24.38 ± 0.68 h |
| Lactiplantibacillus plantarum 22–1 | 252.04 ± 3.1 g | 26.73 ± 0.33 g |
| Leuconostoc lactis 60–7 | 184.18 ± 7.35jk | 19.53 ± 0.78jk |
| Limosilactobacillus reuteri WQY | 272.99 ± 2.0e | 28.95 ± 0.21e |
| Limosilactobacillus reuteri WX | 174.06 ± 10.3 k | 18.46 ± 1.09 k |
| Lacticaseibacillus paracasei SH-14–7 | 290.97 ± 4.82d | 30.86 ± 0.51d |
| Lactiplantibacillus plantarum 51–1 | 281.64 ± 7.38de | 29.87 ± 0.78de |
| Leuconostoc mesenteroides 12–1 | 273.73 ± 6.93e | 29.03 ± 0.73e |
| Limosilactobacillus reuteri SH-23 | 153.07 ± 15.82 k | 16.23 ± 1.68 k |
| Pediococcus pentosaceus 13–16 | 99.48 ± 20.64 l | 10.55 ± 2.17 l |
| Lactiplantibacillus plantarum PDD-2 | 266.62 ± 2.03e | 28.27 ± 0.21e |
| Lactiplantibacillus plantarum F8 | 253.09 ± 4.84 fg | 26.84 ± 0.51 fg |
| Lactobacillus helveticus | 267.74 ± 3.97e | 28.39 ± 0.42e |
| Weissella cibaria 12–18 | 268.08 ± 6.01e | 28.43 ± 0.64e |
| Weissella paramesenteroides 60–8 | 319.45 ± 2.17c | 33.88 ± 0.23c |
| Lactiplantibacillus plantarum 54–1 | 340.69 ± 5.87a | 36.13 ± 0.62a |
Fig. 3.
Identification of strains and their viable counts. (A): Gram staining of the target strain. (B): Phylogenetic tree of the target strain. (C): Effect of solvent (containing ether and Tween-80) and cholesterol solution (containing ether, Tween-80, and cholesterol) on the growth of LAB.
Table 3.
Acid production experiments with assimilated carbon sources (API 50CH).
| Detection | Result | Detection | Result |
|---|---|---|---|
| control | – | esculin ferric citrate | + |
| glycerol | – | salicin | + |
| erythritol | – | D-cellobiose | + |
| D-arabinose | – | D-maltose | + |
| L-arabinose | – | D-lactose | + |
| D-ribose | + | D-melibiose | + |
| D-xylose | – | D-saccharose(sucrose) | + |
| L-xylose | – | D-trehalose | + |
| D-ardonitol | – | inulin | – |
| Mehyl-βD-xylopyranoside | – | D-melezitose | + |
| D-galactose | + | D-raffinose | + |
| D-glucose | + | amidon(starch) | – |
| D-fructose | + | glycogen | – |
| D-mannose | + | xylitol | – |
| L-sorbose | – | gentiobiose | + |
| L-rhamnose | – | D-turanose | + |
| dulcitol | – | D-lyxose | – |
| inositol | – | D-tagatose | – |
| D-mannitol | + | D-fucose | – |
| D-sorbitol | + | L-fucose | – |
| Mehyl-αD-mannopyranoside | – | D-arabitol | – |
| Mehyl-αD-glucopyranoside | – | L-arabitol | – |
| N-acetylglucosamine | + | potassium gluconate | + |
| amygdalin | + | potassium 2-ketogluconate | – |
| arbutin | + | potassium 5-ketogluconate | – |
3.3. Number of viable bacteria of L. Plantarum 54–1 in solvent and cholesterol
As shown in Fig. 3C, the addition of solvents (containing ether and Tween-80) significantly increased the viable number of L. plantarum 54–1 (10.67 lg (CFU/mL)) compared to the blank control group (9.52 lg (CFU/mL)). In contrast, adding cholesterol solution (containing ether, Tween-80, and cholesterol) significantly decreased the viable L. plantarum 54–1 (9.76 lg (CFU/mL)) compared to the solvent group, consistent with the previous findings that high cholesterol concentration inhibits the growth of L. plantarum 54–1. In addition, compared with the blank group, there was a slight increase in the number of live bacteria after the addition of cholesterol solution, which could be seen by controlling with the solvent-added group (containing ether and Tween-80) and combining with the SEM results, which was due to the protective effect of the Tween-80 in the solvent on the L. plantarum 54–1, so that they could grow and metabolize normally in the cholesterol solution, and the pro-growth might have been induced by the ether[23].
3.4. UHPLC-MS combined with metabolomics analysis
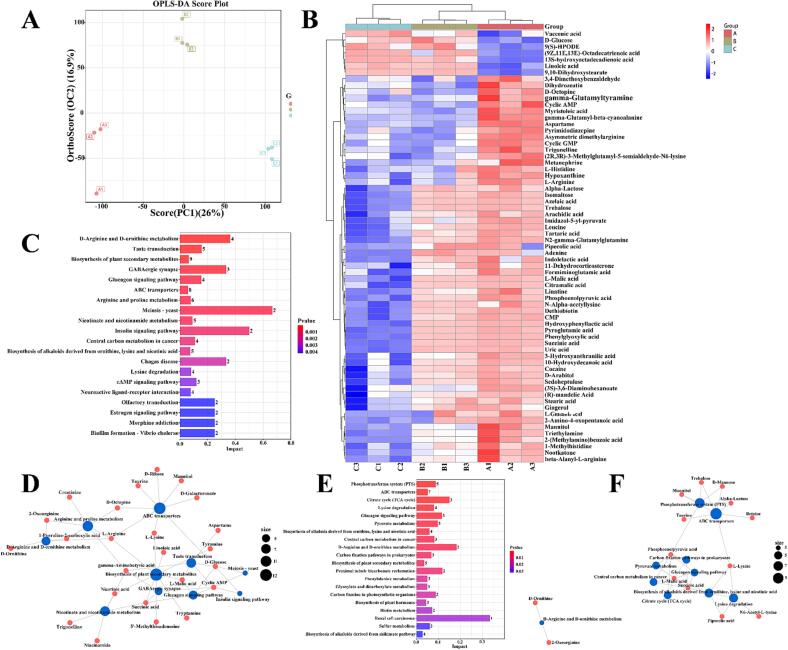
The cholesterol-lowering mechanism of LAB is still controversial, but numerous studies have shown that LAB lower cholesterol concerning the bile salt hydrolase produced during their growth [27]. Therefore, we performed metabolomic analysis to investigate the degradation mechanism. As shown in Fig. 4A, the same group of samples was strictly clustered together, indicating excellent experimental reproducibility. In contrast, samples from different groups were far apart, indicating more significant metabolite variability between groups (a plot of PCA scores between the two groups was shown in Supplementary Fig. 1) [28].
Fig. 4.
Untargeted metabolomics analysis. (A): PCA score chart. (B): Heat map. (C): Differential metabolic pathways between groups A and B. (D): Network diagram of differential metabolic pathways between groups A and B. (E): Differential metabolic pathways between groups B and C. (F): Network diagram of differential metabolic pathways between groups B and C. Group A is the blank group, containing only MRS medium. Group B is the control group (the MRS medium contains only ether and Tween-80). Group C is the experimental group and the MRS medium contains the cholesterol solution (ether, Tween-80, and cholesterol) prepared for this experiment.
A Heat-map was drawn to distinguish between the three groups of sample metabolites (Fig. 4B). P-value ≤ 0.05, VIP ≥ 1, and fold change ≥ 1.5 or ≤ 0.667 were generally considered statistically significant for the screening of differential metabolites [7]. Comparing group B (solvent, containing Tween-80 and ether) and group A (blank control), there were 78 differential metabolites, of which 69 were significantly down-regulated and 9 were significantly up-regulated. This is mainly due to the presence of Tween-80 and ether in the solvent affecting the metabolic pathways such as ABC Transporters and Biosynthesis of plant secondary metabolites in L. plantarum 54–1 (Fig. 4C, D). In addition, Biofilm formation-vibrio cholera was significantly affected in the differential metabolic pathway, demonstrating that Tween-80 did have a protective effect on L. plantarum 54–1 [24].
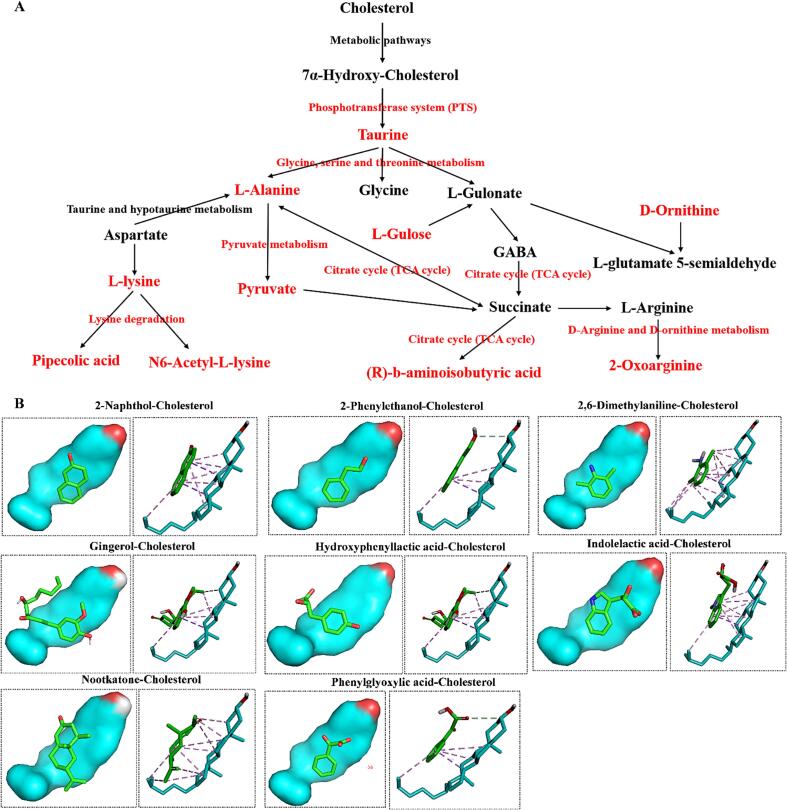
Comparing group C (containing ether, Tween-80, and cholesterol) with group B (containing Tween-80 and ether), 52 differential metabolites were identified, of which 33 were significantly down-regulated and 19 were significantly up-regulated. The analysis of their differential metabolic pathways revealed that this was mainly related to the Phosphotransferase system (PTS), ABC Transporters, Citrate cycle, and Glucagon signaling pathway (Fig. 4E, F). Koh et al. [29] reported that L. rhamnosus strain GG enhances their attachment to HT-29 intestinal epithelial cells and their cholesterol-lowering activity. The active cholesterol efflux pathway is mainly mediated by the ATP-binding cassette (ABC) transport proteins ABCA1 and ABCG1. ABCA1 is essential for cholesterol and phospholipids to apolipoprotein AI and high-density lipoprotein (HDL) biogenesis [30]. Therefore, ABCG1 mainly promotes cholesterol efflux to HDL particles. Studies have shown that the ATP-binding cassette (ABC) transport proteins ABCA1 and ABCG1 are essential in reducing macrophage foam cell formation, inflammation, and atherosclerosis [31]. Two ABCG1 (ABC subfamily G member 1) variants regulate cellular cholesterol [32]. In addition, ATP-citrate lyase is a key lipogenic enzyme that converts cytoplasmic citrate derived from the TCA cycle into acetyl coenzyme A in the cytoplasm. Studies have reported that the CoA-conjugated form 326E -CoA inhibits ATP-citrate lyase activity in vitro to treat cholesterolemia [33]. Furthermore, treatment with AAV8-mediated shRNA silenced the hepatic glucagon receptor (GcgR) in obese mice, effectively lowering blood glucose in obese mice and demonstrating the functional importance of glucagon on cholesterol metabolism [34]. The summarized metabolic mechanism was shown in Fig. 5A. Cholesterol was first converted to 7α-hydroxy-cholesterol and then to the key metabolite taurine via the phosphotransferase system (PTS) which were further converted to L-alanine, L-lysine, N6-Acetyl-L-lysine, (R)-b-aminoisobutyric acid and 2-oxoarginine through glycine, serine and threonine metabolism, citrate cycle (TCA cycle), D-arginine and D-ornithine metabolism, Lysine degradation and pyruvate metabolism.
Fig. 5.
Metabolic pathways and molecular docking. (A): Cholesterol-lowering pathway of L. plantarum 54–1. (B): Molecular docking.
3.5. Molecular docking
The 52 differential compounds screened in group B and group C were individually molecularly docked to cholesterol, and eight of them showed good docking with cholesterol molecules (Fig. 5B). Among them, 2-naphthol, 2-phenylethanol, 2,6-dimethylaniline, gingerol, hydroxyphenyllactic acid, indolelactic acid, and phenylglyoxylic acid can form PI-PI conjugate interactions with cholesterol and the carbon atoms of the compounds form hydrophobic interactions with cholesterol, while the carbon atoms of nootkatone are able to form hydrophobic interactions directly with cholesterol. This indicated that the metabolites of L. plantarum 54–1 may bind to cholesterol and reduce the cholesterol content of the solution by co-sedimentation.
3.6. Transcriptomic analysis
3.6.1. Transcriptome sequence alignment analysis
The high-quality sequences obtained after QC were compared with the reference genome of L. plantarum 54–1 by the Burrows-Wheeler (mapping) method. Table 4 displays the total number of sequences that matched the reference genome and the number of sequences that did not match the reference genome after sequencing for each sample. The sequences that can match the reference genome in the transcripts of all samples were more than 90 %, with the lowest being 91.61 % and the highest being 92.09 %, indicating that the high-quality sequences after quality control can map to the reference genome.
Table 4.
Statistics of transcriptome sequence of L. plantarum 54–1 compared with the reference genome.
| Sample Name | Total Reads | Genome Mapped Reads | Genome Mapped Ratio(%) | Unmapped Reads | Unmapped Reads Ratio(%) | Uniq Mapped Reads | Uniq Mapped Reads Ratio(%) |
|---|---|---|---|---|---|---|---|
| A1 | 27,116,868 | 24,881,845 | 91.76 | 2,235,023 | 8.24 | 24,738,078 | 91.23 |
| A2 | 32,227,162 | 29,594,771 | 91.83 | 2,632,391 | 8.17 | 29,432,794 | 91.33 |
| A3 | 27,781,414 | 25,466,287 | 91.67 | 2,315,127 | 8.33 | 25,322,513 | 91.15 |
| B1 | 28,004,946 | 25,674,533 | 91.68 | 2,330,413 | 8.32 | 25,534,912 | 91.18 |
| B2 | 28,210,626 | 25,978,407 | 92.09 | 2,232,219 | 7.91 | 25,854,522 | 91.65 |
| B3 | 28,975,162 | 26,561,450 | 91.67 | 2,413,712 | 8.33 | 26,391,048 | 91.08 |
| C1 | 29,375,078 | 26,909,945 | 91.61 | 2,465,133 | 8.39 | 26,772,998 | 91.14 |
| C2 | 27,693,254 | 25,553,876 | 92.27 | 2,139,378 | 7.73 | 25,430,408 | 91.83 |
| C3 | 25,217,778 | 23,213,563 | 92.05 | 2,004,215 | 7.95 | 23,094,345 | 91.58 |
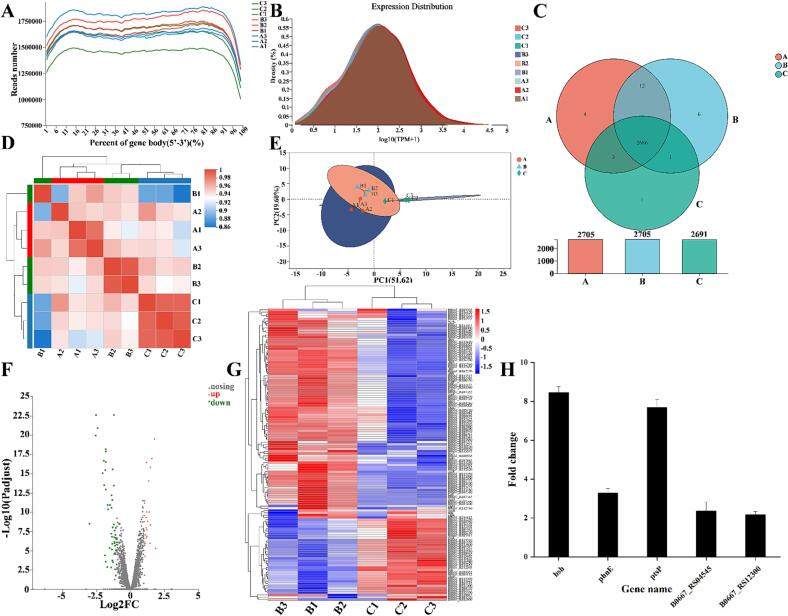
Fig. 6A shows the distribution of L. plantarum 54–1 transcriptome sequencing coverage, in which the horizontal coordinates indicate from the 5′ end to the 3′ end of a gene, and the vertical coordinates indicate the total number of sequences matched to all genes in the genome at the corresponding positions of the horizontal coordinates. All the gene sequences on the L. plantarum 54–1 genome have at least 100,000 sequences matched at the relative proportional positions and a maximum of 200,000 sequences, indicating a high coverage of the transcriptome sequencing. It can also be seen that the shapes of the curves of the three samples from each group of blank, solvent (containing Tween-80 and ether), and experimental (containing ether, Tween-80, and cholesterol) were the same, indicating that the sequences obtained from the transcriptome sequencing of each group are evenly distributed on the genes without bias.
Fig. 6.
Prokaryotic reference transcriptomics. (A): Sequencing coverage analysis. (B): Expression distribution density map. (C): Expression Venn diagram. (D): Inter-sample correlation heat map. (E): PCA analysis. (F): Expression difference volcano map. (G): Heat map of gene set clustering analysis. (H): Differential gene screening. Group A is the blank group, containing only MRS medium. Group B is the control group (the MRS medium contains only ether and Tween-80). Group C is the experimental group and the MRS medium contains the cholesterol solution (ether, Tween-80, and cholesterol) prepared for this experiment.
3.6.2. Gene expression analysis
The distribution of gene expression in the expression of the transcriptome of L. plantarum 54–1 is shown in Fig. 6B. The gene expression distribution in the transcriptome of the blank group A, the solvent group B (containing Tween-80 and ether), and the experimental group (containing ether, Tween-80, and cholesterol) reached the peak gene density at log10(TPM + 1) of 2. The distribution of gene expression was more consistent between the blank group and the control group (solvent group), while the distribution of gene expression in the experimental group (cholesterol group) was slightly different.
Fig. 6C shows the Venn diagram of gene co-expression and specific expression between samples. In total, 2686 genes were co-expressed in the 3 samples, and only 1 gene was explicitly expressed in the experimental group. The lower bar graph in the figure shows the total number of genes expressed in each sample, with groups A (blank group) and B (solvent group) consistently expressing 2705 genes, but group C only 2691. The results indicated that the solvent (Tween-80 and ether) has little effect on expressed genes in terms of co-expression and specific expression of genes, and the cholesterol treatment is more effective. Based on the expression matrix, inter-sample correlation analysis was performed. Fig. 6D shows the correlation heat map between the transcriptome samples. Groups A and B were well clustered together, while the difference with group C was significant. In the PCA score plot, A, B, and C were distinguished along PCA1 51.62 % and PCA2 19.60 %, and samples of the same group were strictly clustered together, indicating good reproducibility of the experiment (Fig. 6E). In contrast, the closer distance between groups A and B indicated smaller differences, while the farther distance between group C and groups A and B indicated larger differences between the samples. The results showed that the solvent has little effect on expressed genes in terms of co-expression and specific gene expression and that the cholesterol treatment is more effective. Therefore, subsequent comparisons of differential gene expression between groups B and C were performed to elucidate the genes that play key regulatory roles in the cholesterol-lowering process of L. plantarum 54–1.
3.6.3. Gene differential expression analysis
Fig. 6F is a volcano plot of transcriptome expression differences, showing the differential expression of genes in group B compared to group C. The presence of cholesterol resulted in many significantly up-or down-regulated genes in the transcriptome and a small number of genes with no significant differences. As shown in the heat map of Fig. 6G, there were 84 differentially expressed genes between groups B and C, of which 27 were significantly up-regulated and 56 were significantly down-regulated. Five differential genes that might be associated with cholesterol-lowering in L. plantarum 54–1 were screened, namely bsh, phnE, ptsP, B0667_RS04545, and B0667_RSRS12300.
The gene bsh can regulate the expression of choloylglycine hydrolase. It was shown that L. plantarum Lp91 and Lp21 reportedly produced 99.29 nmol/mL and 88.63 nmol/mL glycine per minute, respectively, and exhibited higher choloylglycine hydrolase activity compared to other probiotic bacteria [35]. Conjugated bile salts are formed when primary bile acids bind to glycine or taurine through a N⁃acyl amide bond (C-24) to form glycine or taurine bile acids, which are readily absorbed and utilized by the hepatic and intestinal cycles, thus causing cholesterol accumulation in the body [36]. In contrast, choloylglycine hydrolase activity leads to uncoupling bound bile salts, i.e., choloylglycine hydrolase hydrolyzes the N⁃acyl amide bond (C-24) of glycine or taurine bound bile salts, and the unbound bile salts are poorly soluble and the free bile acids produced are excreted in the feces. To compensate for the loss of bile acids, cholesterol, a precursor substance of bile acids, is partially converted to bile acids, thus delaying the rise of cholesterol [37]. Therefore, the highly expressed bsh gene may be one of the reasons for the high cholesterol-lowering ability of L. plantarum 54–1.
The ptsP gene controls phosphoenolpyruvate, a protein phosphotransferase, and it has been demonstrated that tagatose affects the viability and enhances the cholesterol-lowering activity of L. rhamnosus strain GG via the phosphotransferase system (PTS) [29]. In addition, the PTS enhances the stress resistance of bacteria in vivo and in vitro [38]. The gene phnE can encode a phosphate ABC transporter %2C permease protein. The genes phnE and B0667_12300 can encode the phosphonate ABC transporter%2C permease protein PhnE and ABC transporter ATP-binding protein, respectively. The ATP-binding box (ABC) protein is essential for sterol homeostasis, and several ABC subfamily members function as cholesterol transporters across cell membranes [39]. Gene B0667_RS04545 regulates the expression of L-lactate dehydrogenase, mainly associated with propionate metabolism, pyruvate metabolism, glycolysis/gluconeogenesis, and glucagon signaling pathway. The first step in L-lactate catabolism is the conversion of L-lactate to pyruvate by lactate dehydrogenase, which is used in glycolysis and is closely related to cholesterol metabolism [40], [41]. These were consistent with what we found in metabolomics. The qPCR result revealed that cholesterol significantly increased the expression of bsh (8 ± 1 fold), phnE (3 ± 1 fold), ptsP (7 ± 1 fold), B0667_RS04545(4 ± 1 fold), and B0667_RS12300 (2 ± 1 fold) genes in L. plantarum 54–1 (Fig. 6H), which were further validated the results in transcriptomics.
4. Conclusion
An efficient cholesterol-lowering strain of L. plantarum 54–1 (total amount cholesterol removal 340.69 ± 5.87 µg/mL) was screened and its degradation molecular mechanism was investigated, in this study. Moreover, a novel practical MRS medium for screening cholesterol-lowering LAB was developed based on ultrasound treatment. The formula of cholesterol solution is ether: tween-80: cholesterol 7:2:1 (v/v/w), 100 W ultrasound for 10 min, vortex for 20 min, and pass through 0.22 µm filter membrane. Its quality is stable within 7 d after adding it to the MRS medium. SEM and the count of viable LAB results demonstrated that the morphology of LAB in the cholesterol-containing medium developed in this experiment was close to the normal, and it can grow normally. Metabolomics revealed that L. plantarum 54–1 initially converted a portion of cholesterol to 7α-hydroxy-cholesterol and then to the key metabolite taurine, via the phosphotransferase system. These metabolites were further transformed into L-alanine, L-lysine, N6-Acetyl-L-lysine, (R)-b-aminoisobutyric acid, and 2-oxoarginine, through glycine, serine, and threonine metabolism, citrate cycle, D-arginine and D-ornithine metabolism, lysine degradation, and pyruvate metabolism pathways. Prokaryotic reference transcriptomics found that this may be mainly regulated by the bsh, phnE, ptsP, B0667_RS04545, and B0667_RSRS12300 genes, which was further validated by qPCR. Furthermore, molecular docking results demonstrated that 8 differential metabolites might bind to another portion of cholesterol via PI-PI conjugation and hydrophobic interactions and lower cholesterol via co-sedimentation. This is extremely important for screening cholesterol-lowering LAB, developing cholesterol-lowering functional foods fermented by LAB, and exploring the cholesterol-lowering mechanism of LAB.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Natural Science Funding of China (31972048, 32272339, 32072195), the Zhejiang Provincial Natural Science Foundation of China (LR23C200001), National Key R&D Program of China (2021YFD2100104, 2022YFD2100603), and Ningbo University Postgraduate Research Innovation Fund (IF2023053).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ultsonch.2023.106698.
Contributor Information
Xiaoqun Zeng, Email: zengxiaoqun@nbu.edu.cn.
Daodong Pan, Email: daodongpan@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yadav N., Sharma V., Kapila S., Malik R.K., Arora S. Hypocholesterolaemic and prebiotic effect of partially hydrolysed psyllium husk supplemented yoghurt. J. Funct. Foods. 2016;24:351–358. [Google Scholar]
- 2.Raveschot C., Coutte F., Frémont M., Vaeremans M., Dugersuren J., Demberel S., Cudennec B. Probiotic Lactobacillus strains from Mongolia improve calcium transport and uptake by intestinal cells in vitro. Food Res. Int. 2020;133 doi: 10.1016/j.foodres.2020.109201. [DOI] [PubMed] [Google Scholar]
- 3.Wang J., Zheng W., Zheng S., Yuan Y., Wen W., Cui W., Zhang H. Targeting ANGPTL3 by GalNAc-conjugated siRNA ANGsiR10 lowers blood lipids with long-lasting and potent efficacy in mice and monkeys. Molecular Therapy-Nucleic Acids. 2023;31:68–77. doi: 10.1016/j.omtn.2022.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel K.K., Kashfi K. Lipoproteins and cancer: The role of HDL-C, LDL-C, and cholesterol-lowering drugs. Biochem. Pharmacol. 2022;196 doi: 10.1016/j.bcp.2021.114654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miremadi F., Sherkat F., Stojanovska L. Hypocholesterolaemic effect and anti-hypertensive properties of probiotics and prebiotics: A review. J. Funct. Foods. 2016;25:497–510. [Google Scholar]
- 6.Sharma A., Kaur J., Lee S., Park Y.-S. Tracking of intentionally inoculated lactic acid bacteria strains in yogurt and probiotic powder. Microorganisms. 2019;8(1):5. doi: 10.3390/microorganisms8010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, X., Du, L., Xu, J., Shi, Z., Zhang, T., Jiang, X., Pan, D. (2022). Effect of single probiotics Lacticaseibacillus casei CGMCC1. 5956 and Levilactobacillus brevis CGMCC1. 5954 and their combination on the quality of yogurt as fermented milk. LWT, 163, 113530.
- 8.Fan X., Li X., Zhang T., Guo Y., Shi Z., Wu Z., Pan D. Novel millet-based flavored yogurt enriched with superoxide dismutase. Front. Nutr. 2022;8:1179. doi: 10.3389/fnut.2021.791886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhat B., Bajaj B.K. Multifarious cholesterol lowering potential of lactic acid bacteria equipped with desired probiotic functional attributes. 3. Biotech. 2020;10(5):200. doi: 10.1007/s13205-020-02183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lye H.S., Khoo B.Y., Karim A.A., Rusul G., Liong M.T. Ultrasound enhanced growth and cholesterol removal of Lactobacillus fermentum FTDC 1311 in the parent cells but not the subsequent passages. Ultrason. Sonochem. 2012;19(4):901–908. doi: 10.1016/j.ultsonch.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 11.Iranmanesh M., Ezzatpanah H., Mojgani N. Antibacterial activity and cholesterol assimilation of lactic acid bacteria isolated from traditional Iranian dairy products. LWT-Food Science and Technology. 2014;58(2):355–359. [Google Scholar]
- 12.Ding W., Shi C., Chen M., Zhou J., Long R., Guo X. Screening for lactic acid bacteria in traditional fermented Tibetan yak milk and evaluating their probiotic and cholesterol-lowering potentials in rats fed a high-cholesterol diet. J. Funct. Foods. 2017;32:324–332. [Google Scholar]
- 13.Damiani C., Gaglio D., Sacco E., Alberghina L., Vanoni M. Systems metabolomics: From metabolomic snapshots to design principles. Curr. Opin. Biotechnol. 2020;63:190–199. doi: 10.1016/j.copbio.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Fan X., Yu L., Shi Z., Li C., Zeng X., Wu Z., Pan D. Characterization of a novel flavored yogurt enriched in γ-aminobutyric acid fermented by Levilactobacillus brevis CGMCC1. 5954. J. Dairy Sci. 2023;106(2):852–867. doi: 10.3168/jds.2022-22590. [DOI] [PubMed] [Google Scholar]
- 15.Shi J., Xia C., Tian Q., Zeng X., Wu Z., Guo Y., Pan D. Untargeted metabolomics based on LC–MS to elucidate the mechanism underlying nitrite degradation by Limosilactobacillus fermentum RC4. LWT. 2022;163 [Google Scholar]
- 16.Breschi A., Gingeras T.R., Guigó R. Comparative transcriptomics in human and mouse. Nat. Rev. Genet. 2017;18(7):425–440. doi: 10.1038/nrg.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee M., Kim D., Choi E.J., Song J.H., Kang J.Y., Lee K.W., Chang J.Y. Transcriptome responses of lactic acid bacteria isolated from kimchi under hydrogen peroxide exposure. Food Res. Int. 2023;168 doi: 10.1016/j.foodres.2023.112681. [DOI] [PubMed] [Google Scholar]
- 18.Lin D., Long X., Xiao L., Wu Z., Chen H., Zhang Q., Xing B. Study on the functional properties and structural characteristics of soybean soluble polysaccharides by mixed bacteria fermentation and microwave treatment. Int. J. Biol. Macromol. 2020;157:561–568. doi: 10.1016/j.ijbiomac.2020.04.133. [DOI] [PubMed] [Google Scholar]
- 19.Kingkaew E., Konno H., Hosaka Y., Phongsopitanun W., Tanasupawat S. Characterization of lactic acid bacteria from fermented fish (pla-paeng-daeng) and their cholesterol-lowering and immunomodulatory effects. Microbes Environ. 2023;38(1):ME22044. doi: 10.1264/jsme2.ME22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zelena, E., Dunn, W. B., Broadhurst, D., Francis-McIntyre, S., Carroll, K. M., Begley, P., HUSERMET Consortium Development of a robust and repeatable UPLC− MS method for the long-term metabolomic study of human serum. Anal. Chem. 2009;81(4):1357–1364. doi: 10.1021/ac8019366. [DOI] [PubMed] [Google Scholar]
- 21.Mou Z.-L., Wang L., Zeng Z.-X., Su X.-G., Ji S.-J., Shan W., Zhao Y.-T. Metabolomics integrated with transcriptomics unveil the regulatory pathways of modified atmosphere packaging–maintained leaf quality of Chinese flowering cabbage. Food Chem. 2023;405 [Google Scholar]
- 22.Fan X., Li X., Zhang T., Xu J., Shi Z., Wu Z., Du L. A novel qPCR method for the detection of lactic acid bacteria in fermented milk. Foods. 2021;10(12):3066. doi: 10.3390/foods10123066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimoto H., Ohmomo S., Okamoto T. Enhancement of bile tolerance in lactococci by Tween 80. J. Appl. Microbiol. 2002;92(1):41–46. doi: 10.1046/j.1365-2672.2002.01505.x. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg I., Eschar L. Stability of lactic acid bacteria to freezing as related to their fatty acid composition. Appl. Environ. Microbiol. 1977;33(3):489–496. doi: 10.1128/aem.33.3.489-496.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reitermayer D., Kafka T.A., Lenz C.A., Vogel R.F. Interrelation between Tween and the membrane properties and high pressure tolerance of Lactobacillus plantarum. BMC Microbiol. 2018;18:1–14. doi: 10.1186/s12866-018-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le B., Yang S.-H. Identification of a novel potential probiotic Lactobacillus plantarum FB003 isolated from salted-fermented shrimp and its effect on cholesterol absorption by regulation of NPC1L1 and PPARα. Probiotics Antimicrob. Proteins. 2019;11:785–793. doi: 10.1007/s12602-018-9469-9. [DOI] [PubMed] [Google Scholar]
- 27.Sui Y., Liu J., Liu Y., Wang Y., Xiao Y., Gao B., Zhu D. In vitro probiotic characterization of Lactobacillus strains from fermented tangerine vinegar and their cholesterol degradation activity. Food Biosci. 2021;39 [Google Scholar]
- 28.Fan X., Shi Z., Xu J., Li C., Li X., Jiang X., Wu Z. Characterization of the effects of binary probiotics and wolfberry dietary fiber on the quality of yogurt. Food Chem. 2023;406 doi: 10.1016/j.foodchem.2022.135020. [DOI] [PubMed] [Google Scholar]
- 29.Koh J.H., Choi S.H., Park S.W., Choi N.-J., Kim Y., Kim S.H. Synbiotic impact of tagatose on viability of Lactobacillus rhamnosus strain GG mediated by the phosphotransferase system (PTS) Food Microbiol. 2013;36(1):7–13. doi: 10.1016/j.fm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Tall A.R., Yvan-Charvet L., Terasaka N., Pagler T., Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7(5):365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Dunn P.J., Salm E.J., Tomita S. ABC transporters control ATP release through cholesterol-dependent volume-regulated anion channel activity. J. Biol. Chem. 2020;295(16):5192–5203. doi: 10.1074/jbc.RA119.010699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small D.M. Role of ABC transporters in secretion of cholesterol from liver into bile. Proc. Natl. Acad. Sci. 2003;100(1):4–6. doi: 10.1073/pnas.0237205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Z., Zhang M., Song Q., Cheng L., Zhang X., Song G., Zhang Y. Development of the novel ACLY inhibitor 326E as a promising treatment for hypercholesterolemia. Acta Pharm. Sin. B. 2023;13(2):739–753. doi: 10.1016/j.apsb.2022.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spolitu S., Ozcan L. Glucagon receptor signaling-mediated regulation of PCSK9 and cholesterol metabolism. Arterioscler. Thromb. Vasc. Biol. 2018;38(Suppl_1):A177–A. [Google Scholar]
- 35.Kumar R., Grover S., Batish V.K. Hypocholesterolaemic effect of dietary inclusion of two putative probiotic bile salt hydrolase-producing Lactobacillus plantarum strains in Sprague-Dawley rats. Br. J. Nutr. 2011;105(4):561–573. doi: 10.1017/S0007114510003740. [DOI] [PubMed] [Google Scholar]
- 36.Reis S., Conceição L., Rosa D., Siqueira N., Peluzio M. Mechanisms responsible for the hypocholesterolaemic effect of regular consumption of probiotics. Nutr. Res. Rev. 2017;30(1):36–49. doi: 10.1017/S0954422416000226. [DOI] [PubMed] [Google Scholar]
- 37.Tsai C.-C., Lin P.-P., Hsieh Y.-M., Zhang Z.-Y., Wu H.-C., Huang C.-C. Cholesterol-lowering potentials of lactic acid bacteria based on bile-salt hydrolase activity and effect of potent strains on cholesterol metabolism in vitro and in vivo. Scientific World Journal. 2014;2014 doi: 10.1155/2014/690752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horng Y.-T., Wang C.-J., Chung W.-T., Chao H.-J., Chen Y.-Y., Soo P.-C. Phosphoenolpyruvate phosphotransferase system components positively regulate Klebsiella biofilm formation. J. Microbiol. Immunol. Infect. 2018;51(2):174–183. doi: 10.1016/j.jmii.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 39.Xavier B.M., Jennings W.J., Zein A.A., Wang J., Lee J.-Y. Structural snapshot of the cholesterol-transport ATP-binding cassette proteins. Biochem. Cell Biol. 2019;97(3):224–233. doi: 10.1139/bcb-2018-0151. [DOI] [PubMed] [Google Scholar]
- 40.Oburoglu L., Mansell E., Canals I., Sigurdsson V., Guibentif C., Soneji S., Woods N.B. Pyruvate metabolism guides definitive lineage specification during hematopoietic emergence. EMBO Rep. 2022;23(2):e54384. doi: 10.15252/embr.202154384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanko R.T., Reynolds H.R., Lonchar K.D., Arch J.E. Plasma lipid concentrations in hyperlipidemic patients consuming a high-fat diet supplemented with pyruvate for 6 wk. Am. J. Clin. Nutr. 1992;56(5):950–954. doi: 10.1093/ajcn/56.5.950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.