Abstract
The proopiomelanocortin (POMC) gene is expressed in the pituitary and arcuate neurons of the hypothalamus. POMC arcuate neurons play a central role in the control of energy homeostasis, and rare loss-of-function mutations in POMC cause obesity. Moreover, POMC is the prime candidate gene within a highly significant quantitative trait locus on chromosome 2 associated with obesity traits in several human populations. Here, we identify two phylogenetically conserved neuronal POMC enhancers designated nPE1 (600 bp) and nPE2 (150 bp) located approximately 10 to 12 kb upstream of mammalian POMC transcriptional units. We show that mouse or human genomic regions containing these enhancers are able to direct reporter gene expression to POMC hypothalamic neurons, but not the pituitary of transgenic mice. Conversely, deletion of nPE1 and nPE2 in the context of the entire transcriptional unit of POMC abolishes transgene expression in the hypothalamus without affecting pituitary expression. Our results indicate that the nPEs are necessary and sufficient for hypothalamic POMC expression and that POMC expression in the brain and pituitary is controlled by independent sets of enhancers. Our study advances the understanding of the molecular nature of hypothalamic POMC neurons and will be useful to determine whether polymorphisms in POMC regulatory regions play a role in the predisposition to obesity.
The proopiomelanocortin (POMC) gene encodes a prohormone expressed at significant levels in three discrete groups of cells of ectodermal neural plate origin that develop into either pituitary endocrine cells or brain neurons (54). Cell-specific posttranslational processing of the POMC prohormone generates a variety of biologically active peptides that mediate the mammalian stress response (46). In the pituitary gland, anterior lobe corticotrophs release adenocorticotropic hormone (ACTH) whereas intermediate lobe melanotrophs further process POMC to produce .-acetylated forms of α-melanocyte stimulating hormone (α-MSH) and β-endorphin. In the brain, a specialized population of neurons concentrated in the hypothalamic arcuate nucleus also express Pomc, giving rise to α-, β-, and γ-melanocortins and the potent analgesic opioid peptide β-endorphin (46, 50).
During the last few years, POMC hypothalamic neurons have received a great deal of attention because they express receptors for the adipostatic hormone leptin and play a central role in the control of energy homeostasis and body weight regulation (10, 52, 56). Once leptin crosses the blood-brain barrier, it stimulates both the transcription of the POMC gene and the activity of POMC arcuate neurons, leading to the release of melanocortins which, in turn, stimulate central melanocortin receptors to decrease food intake and increase metabolic rate (4, 52). The importance of the central melanocortin pathway in feeding behavior is clearly observed in mice and humans with null Pomc homozygous mutations, which display hyperphagia and early-onset obesity (8, 29, 30, 65). Even though total POMC deficiency is very rare in humans, POMC is a strong candidate gene to predispose to familial obesity. Several independent genome-wide scans for quantitative trait loci (QTL) have found a highly significant genetic linkage between a relatively narrow region in chromosome 2 containing the POMC locus and obesity-related traits (11, 15, 47). However, polymorphisms in the coding sequences of POMC that alter the structure or function of POMC peptides apparently do not account for this correlation (25), suggesting the alternative possibility that mutations in noncoding regulatory sequences may alter the level of POMC RNA transcripts and consequently the concentration of POMC peptides in the brain.
Transcriptional regulation of Pomc has been studied primarily in pituitary corticotrophs and melanotrophs because it has been possible to combine in vitro transfection in immortalized pituitary cell lines (40) with in vivo expression in transgenic mice (33, 34). Cell-specific expression of the rat or mouse Pomc genes in the pituitary is controlled by the combinatorial presence of cis-acting elements localized within approximately 400 nucleotides adjacent to the transcriptional start site (33, 34). Several transcription factors including Nurr77, NeuroD1/BETA2 heterodimers, Pitx1, SP1, and Tpit have been implicated in the pituitary regulation of Pomc expression or the determination of corticotroph and melanotroph lineages (31, 34, 43, 44, 45, 60). In contrast, little is known about enhancer regions and transcription factors that regulate Pomc expression in arcuate neurons. In previous studies, we demonstrated that the 400-bp proximal Pomc promoter region is incapable of directing reporter gene expression to the POMC arcuate neurons of transgenic mice (49). However, the inclusion of additional genomic sequences suggested that cis-acting regions controlling Pomc expression in the brain are localized within 11 kb of distal 5′ flanking sequences upstream of the Pomc promoter (13, 41, 66). Combining a functional expression analysis in transgenic mice together with in silico phylogenetic footprinting, we report here the identification of two conserved enhancers that are necessary and sufficient for transgene expression in POMC arcuate neurons. Our results also indicate that the Pomc promoter has a modular architecture that can allow independent transcriptional control of Pomc expression in the brain and the pituitary by a distal and proximal set of enhancers, respectively. These results advance the understanding of the molecular identity of POMC neurons and might contribute to a further determination of the role that POMC plays in feeding centers of the brain and the genetics of obesity.
MATERIALS AND METHODS
Transgenes.
Transgenes were constructed using standard molecular cloning techniques and the pBluescript SK (+/−) vector (Stratagene). To construct parental transgene 1, the coding region of enhanced green fluorescent protein (pEGFP-1, Clontech) followed by the polyadenylation sites from simian virus 40 were inserted into a StuI site located before the translational start codon in exon 2 of a mouse Pomc gene fragment, and subsequent subcloning extended the flanking regions to −13 and +8 kb from the Pomc gene (13). Based on this parental construct, transgenes 2, 3, 4, and 6 were constructed by performing deletions with restriction enzymes SmaI, BamHI-SmaI, NotI-BamHI, and NotI-EcoRI, respectively. Transgenes 7, 8, and 9 carry deletions of neuronal POMC enhancer 1 (nPE1) and nPE2, which were introduced using enzymes ApaI-HindIII (585-bp deletion) and NsiI-ApaI (172-bp deletion), respectively. To construct transgenes 5, 10, 11, and 12, a 2.3-kb fragment containing the minimal promoter of the thymidine kinase (TK) gene from the Herpes simplex virus upstream of the human growth hormone (GH) gene was excised from plasmid pTKGH (Nichols Institute) and cloned downstream of −13- and −9-kb POMC segments from previously constructed plasmids. To construct transgene 13, two human DNA fragments containing nPE1 and nPE2 were PCR amplified using primers HP1 (5′-TTTGAATTCCTGACCTCAAGCGATCCACCC-3′) and HP2 (5′-CTAGGCTCAGATCTAGAGTCAAGCTCTGTG-3′) that yield a 1.6-kb product and HP3 (5′-GACTCTAGATCTGAGCCTAGAATCATAGAA-3′) and HP4 (5′-CCCGATATCTCTGCAGGCATCTGGACCTC-3′) that yield a 1.2-kb product. Both fragments were ligated together via a natural XbaI site (underlined in the primer sequences) and subcloned into the pTrap vector (42) upstream of the minimal promoter of the chicken β-globin gene and the lacZ reporter gene. Sequence quality was assessed by automated PCR sequencing. Prior to microinjection, all transgenes were digested with appropriate restriction enzymes to discard vector sequences, size fractionated on agarose gels, and purified using Elutip columns (Schleicher and Schuell). Different reporter cassettes were used in this study according to methodological or practical reasons. Enhanced green fluorescent protein (EGFP) has been previously used by our group for the identification of live POMC neurons (13, 24), whereas the lacZ and human growth hormone cassettes were chosen because they contain expressionless minimal promoters and convenient upstream polylinkers and encode transgenic products that facilitate colocalization experiments with antibodies against Pomc peptides.
Production of transgenic mice.
Transgenic mice were generated by pronuclear microinjection of B6CBF2 or B6D2F2 zygotes as described previously (66). Microinjected zygotes were transferred to the oviduct of NIH or B6CB pseudopregnant females. Transgenic pups were identified by tail genomic DNA PCR as described previously (66). For transgenes 1 to 4 and 6 to 9, primers M329 (5′-GAAGTACGTCATGGGTCACT-3′) and M330 (5′-AGCTCCCTCTTGAACTCTAG-3′), which identify a heterologous insertion present in exon 3 of the transgenes (66), were used. For transgenes 5 and 10 to 12, primers TK (5′-CAGCGTCTTGTCATTGGCG-3′) and GH (5′-AGTGGTTCGGGGAGTTGGG-3′), which amplify a 170-bp product from the TKGH sequence, were used, and for transgene 13, primers HP3 and PPH9 (5′-TCAAGGGCAAAGAGGAATCA-3′), which amplify an 800-bp, human-specific product, were used. The numbers of independent transgenic lines (in parentheses) analyzed for each transgene were as follows: transgene 1 (1), transgene 2 (2), transgene 3 (2), transgene 4 (1), transgene 5 (3), transgene 6 (1), transgene 7 (2), transgene 8 (2), transgene 9 (5), transgene 10 (2), transgene 11 (1), transgene 12 (5), transgene 13 (3). All animal procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals by the U.S. Public Health Service.
Immunohistochemistry.
Mice were perfused with 4% paraformaldehyde (PFA) in KPBS (0.9% NaCl, 16 mM K2HPO4, 3.6 mM KH2PO4), and brains were excised, postfixed in 4% PFA-KPBS overnight at 4°C, and sectioned (50 μm) with Vibratome 1000 (Ted Pella, Redding, Calif.). Brain slices were treated with 1% H2O2 in KPBS for 20 min, washed twice with KPBS, and incubated overnight at 4°C with the primary antibodies diluted in KPBS-0.3% Triton X-100 and 2% normal goat serum. The primary antibodies were rabbit polyclonal anti-ACTH-IC-1 (1:1,000; National Hormone and Pituitary Program, National Institutes of Health [NIH]), anti-EGFP ab290 (1:10,000; AbCam), and anti-hGH-IC-3 (1:1,000; National Hormone and Pituitary Program, NIH) antibodies. The next day, slices were washed in KPBS and incubated with biotinylated anti-rabbit immunoglobulin G antibody (Vector) diluted 1:200 in KPBS-0.3% Triton X-100 for 2 h at room temperature (RT). After washing in KPBS, slices were incubated with avidin/biotin-horseradish peroxidase complex (Vectastain Elite ABC kit; Vector) for 1 h at RT, washed in KPBS, and developed with 25 mg of diaminobenzidine (DAB; Sigma)/ml and 0.05% H2O2 in TBS (150 mM NaCl, 50 mM Tris-HCl, pH 7.5). Alternatively, reactions were developed with a blue chromogen (SG kit; Vector). Stained slices were then mounted onto gelatin-coated slides.
Immunofluorescence.
To check for colocalization of EGFP and POMC, brain slices containing EGFP-expressing cells were subjected to an anti-ACTH immunohistochemistry as described above up to the second antibody incubation and then slices were incubated in streptavidin conjugated with Cy3 (1:800; Jackson Immunoresearch) in KPBS for 1 h at RT. Subsequently slices were washed in KPBS and mounted onto slides with FluorSave reagent (Calbiochem) and photographed with a microscope (Leica, Wetzlar, Germany) under UV light by using different filters to check for colocalization of green (EGFP) and red (ACTH-Cy3) fluorescence. The protocol for pituitaries was similar except that, prior to immunohistochemistry, perfused pituitaries were dehydrated in an ascending series of sucrose concentrations (10, 20, and 30% in PBS), frozen in embedding medium (Tissue-Tek), and sectioned with a cryostat (Microm HM 505N; Micron, Heidelberg, Germany) and then postfixed in 4% PFA.
Double in situ hybridization and immunohistochemistry.
To colocalize endogenous Pomc mRNA and the human GH (hGH) transgene product, nonradioactive POMC in situ hybridization was performed and followed by anti-hGH immunohistochemistry in 12-well plastic plates. A 600-bp mouse POMC exon 3 riboprobe was synthesized using a DIG-labeling kit (Roche) according to the instructions of the manufacturer. PFA-fixed, Vibratome-sectioned brain slices were treated with 0.1 M triethanolamine (TEA), pH 8.0, for 3 min, followed by an incubation in 0.0025% acetic anhydride in 0.1 M TEA for 10 min to block positive charges, and washed twice in 2× SSC (0.3 M NaCl, 0.03 M sodium citrate, pH 7.2). Brain slices were incubated in hybridization solution (50% formamide, 200 mM NaCl, 1× Denhardt's solution, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0], 10% dextran sulfate) for 2 h at 57°C. The riboprobe was heated at 65°C for 5 min (in 100 μl of water with 5 μg of tRNA) and added to the hybridization solution of each well. Hybridization followed overnight at 57°C. The next day, slices were washed for 10 s in 2× SSC and subjected to RNase A digestion (20 μg of RNase A/ml, 50 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) at 37°C for 30 min, de-salted in 1× and 0.25× SSC for 30 min at 60°C. Slices were washed in buffer 1 (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl) and incubated with anti-DIG mouse monoclonal antibody (1:5,000; Jackson ImmunoResearch) in buffer 1 containing 0.2% normal goat serum for 2 h at 37°C. Slides were then washed once in buffer 1 and once in KPBS and incubated with goat biotinylated anti-mouse immunoglobulin G antibody (1:200, Vector) for 2 h at RT and then developed with DAB as described above. After that, slides were washed in KPBS and then incubated with anti-hGH (1:1,000) and finally developed with benzidine hydrochloride (Sigma) as described in reference 18.
Radioactive in situ hybridization of pituitaries.
Pituitaries were fresh frozen in embedding medium (Tissue-Tek), and 12-μm-thick sections were collected on gelatinized glass slides with a cryostat microtome (Microm HM 505N; Micron). A 176-nucleotide riboprobe labeled with [α-35S]UTP was synthesized from a plasmid containing part of exon 5 of the hGH gene by using the MAXIscript kit (Ambion). The in situ hybridization was performed as described previously (66).
X-Gal staining.
The 50-μm-thick Vibratome brain slices of transgenic mice carrying construct 11 were stained with 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gal)/ml in staining solution (PBS [pH 7.3] containing 2.12 mg of potassium ferrocyanide/ml, 1.64 mg of potassium ferricyanide/ml, 2 mM MgCl2, 0.01% sodium deoxycolate, and 0.02% NP-40) for 4 h at 37°C. Stained slices were then subjected to anti-ACTH immunohistochemistry as described above.
Isolation and partial mapping of bovine nPE sequences.
A genomic clone containing bovine nPE sequences was isolated from a bovine genomic phage library (number BL1015, average insert size of 40 kb; Clontech) by a PCR-based strategy (27) using degenerate primers for a conserved segment of nPE1: PPH7 (5′-GACTGAGCTGAGTGCCTGT-3′) and PPH3 (5′-ANGAATGCNGAGTTCTCCA-3′). From this clone, partial sequences of nPE1 and nPE2 were amplified by PCR using primers PPH7 and PPH3 for nPE1 and PPH8 (5′-GGATAAAAGCNGTCTCAAGG-3′) and PPH9 for nPE2, subcloned into pGEM-T (Promega), and sequenced. The same clone also contained POMC exon 1 as confirmed by performing PCR using primers specific for the exon 1 of the bovine POMC gene (5-′ GGAAGTCCACTCAACGTC-3′ and 5′-CTGCGCCCTTACCTGTCA-3′), which were designed based on a published sequence (GenBank accession number J00016).
Sequences and programs.
POMC loci sequences from human, mouse, rat, chimpanzee, chick, and zebrafish were retrieved from the Ensembl website (http://www.ensembl.org). Partial dog sequences were retrieved from the National Center for Biotechnology Information (NCBI) Trace database (http://www.ncbi.nih.gov/Traces/trace.cgi). Takifugu rubripes and Xenopus tropicalis sequences were retrieved from the Joint Genome Institute website (http://www.jgi.doe.gov). Human and mouse sequences were compared using the DOTTER program (55) downloaded from http://www.cgb.ki.se/cgb/groups/sonnhammer/Dotter.html, and multiple sequences were compared using the PipMaker program (available at http://bio.cse.psu.edu/pipmaker). Multiple alignments were done using the program Clustal W (58) at http://www.ebi.ac.uk/clustalw. Identification of conserved and aligned transcription factor binding sites was performed using the program rVista2.0 (37) at http://www.dcode.org.
RESULTS
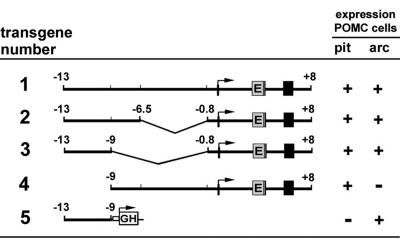
The region located between −13 and −9 kb of the mouse POMC gene directs expression to hypothalamic neurons. Our previous results showed that the region between −13 and −2 kb upstream of the mouse POMC transcriptional start site contains enhancers capable of directing proper cell-specific transgene expression to POMC neurons of the arcuate nucleus of the hypothalamus (66). To facilitate the analysis of transgene expression, we inserted the coding region of EGFP into mouse Pomc exon 2 immediately before the translational start codon of the POMC prohormone (Fig. 1, transgene 1). This 22-kb transgene containing the entire transcriptional unit of mouse Pomc, together with 13 kb of 5′ flanking regions and 2 kb of 3′ flanking regions, drives accurate EGFP expression to nearly 100% of that of POMC-expressing neurons in the arcuate nucleus and has allowed for electrophysiological studies of living POMC neurons (3, 13, 14, 24). To further narrow the flanking regions necessary for Pomc expression in the brain, we constructed two large deletions of transgene 1 by removing the region between −6.5 and −0.8 kb or between −9 and −0.8 kb (Fig. 1, transgenes 2 and 3). The expression pattern of EGFP was compared to that of endogenous Pomc in the serial brain sections of transgenic mice carrying constructs 2 and 3 by using EGFP and ACTH polyclonal antibodies. Colocalization of EGFP in POMC neurons was assessed by red-fluorescent immunohistochemistry coupled to the ACTH antibody on Vibratome-sectioned brain slices of several independent transgenic mouse pedigrees. The analysis of deletion of transgenes 2 and 3 demonstrated that EGFP was coexpressed with Pomc in neurons of the arcuate nucleus (Fig. 2A and B), as well as in pituitary corticotrophs and melanotrophs (data not shown), suggesting that the region between −9 and −0.8 kb is dispensable for neuron-specific expression of Pomc in the hypothalamus. In addition, complete deletion of the fragment spanning from −13 to −9 (Fig. 1, transgene 4) prevented EGFP expression in the arcuate nucleus, although EGFP was properly expressed in pituitary corticotrophs and melanotrophs (data not shown). Transgenes 1 and 4 showed a consistent ectopic expression of EGFP in the subgranular layer of the dentate gyrus of the hippocampus probably due to the presence of a cryptic enhancer located in the −2- to −0.8-kb region of the Pomc promoter, as recently reported (41). These results suggest that the 4-kb region located between −13 and −9 kb upstream of the murine Pomc gene contains one or more neuronal enhancers necessary to direct Pomc expression to arcuate neurons.
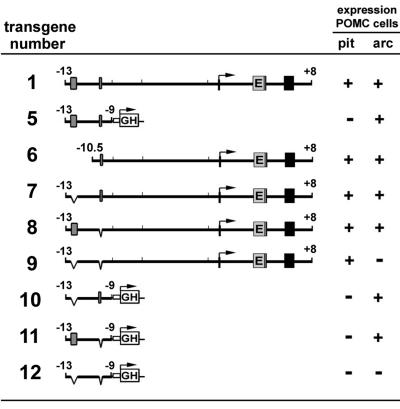
FIG. 1.
First set of transgenes used in this study. Transgenes 1 through 4 carry the whole transcriptional unit of mouse Pomc with the coding region of EGFP (E) inserted into exon 2 before the starting translational codon. Pomc exons are indicated by black boxes. Transgene 5 carries the distal enhancer region (kb −13 to −9) of mouse Pomc upstream of the viral TK minimal promoter followed by the hGH gene as reporter. Positive reporter expression in pituitary melanotrophs and corticotrophs (pit) and hypothalamic arcuate neurons (arc) are indicated (+).
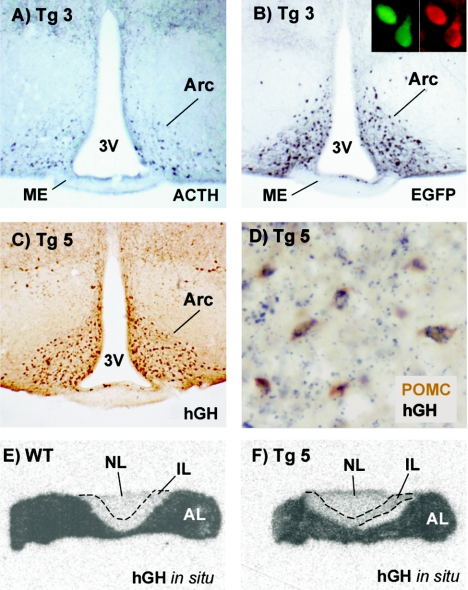
FIG. 2.
Reporter gene expression in the arcuate nuclei and pituitary glands of transgenic mice. (A and B) Coronal brain sections of a transgene 3 mouse immunostained for ACTH (panel A) and EGFP (panel B). The inset in panel B shows an example of colocalization of EGFP fluorescence (green) and ACTH immunofluorescence (red) in the arcuate of a transgene 3 mouse. (C) Coronal section of a transgene 4 mouse brain immunostained for hGH. (D) Colocalization of Pomc mRNA and hGH in the arcuate of a transgene 5 mouse. Brain slices were subjected to nonradioactive Pomc in situ hybridization (brown) followed by hGH immunostaining (dark blue granules). (E and F) Autoradiography of wild-type (panel E) and transgene 5 (panel F) pituitary coronal sections hybridized with a 35S-radiolabeled hGH antisense riboprobe. The probe detected endogenous GH mRNA in the anterior lobe (AL) of both pituitaries but not in the intermediate (IL) or neural (NL) lobes. Arc, arcuate nucleus; ME, median eminence; 3V, third ventricle.
We next sought to determine whether this 4-kb DNA region was sufficient to direct transgene expression to POMC neurons in the absence of the proximal Pomc promoter, which is known to be essential for expression in pituitary melanotrophs and corticotrophs. To this end, we constructed a transgene in which the region from kb −13 to −9 from the mouse Pomc gene was ligated upstream of a minimal heterologous promoter obtained from the Herpes simplex virus TK gene, followed by the hGH gene, which served as a reporter (Fig. 1, transgene 5). Transgenic mouse brains were analyzed for colocalization by in situ hybridization against endogenous Pomc mRNA by using a DIG-labeled riboprobe (developed with the brown chromogen diaminobenzidine) followed by immunohistochemistry against hGH (developed with the blue chromogen benzidine dihydrochloride). Interestingly, the fragment from positions −13 to −9 was able to target reporter gene expression to arcuate POMC neurons (Fig. 2C and D) in the total absence of sequences from the proximal promoter, exons, introns, or 3′ flanking regions of mouse Pomc, demonstrating that this distal 4-kb region contains neuronal-specific enhancer(s) sufficient to drive transcription in POMC arcuate neurons. This result was confirmed in three independent transgenic pedigrees that showed an average of 75% of POMC-positive arcuate neurons coexpressing the reporter hGH. Reporter gene expression was not detected in pituitary melanotrophs (Fig. 2E and F). The hGH riboprobe intensively labeled mouse somatotrophs in the pituitary anterior lobe due to cross-hybridization with mouse GH mRNA, precluding transgene expression analysis in corticotrophs. Variable patterns of ectopic transgene expression were also detected in the brains of these mouse lines in areas that included the supraoptic nuclei of the hypothalamus, amygdala, habenula, and cerebral cortex. Together, these results demonstrate that distal neuronal enhancers located within the −13- to −9-kb region of the mouse Pomc 5′ flanking region are sufficient to drive reporter expression to POMC neurons of the arcuate nucleus.
The Pomc neuron-specific region contains two highly conserved elements in mouse and human.
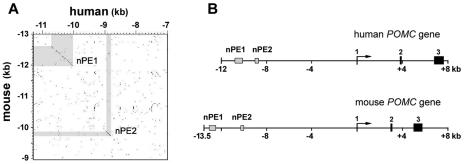
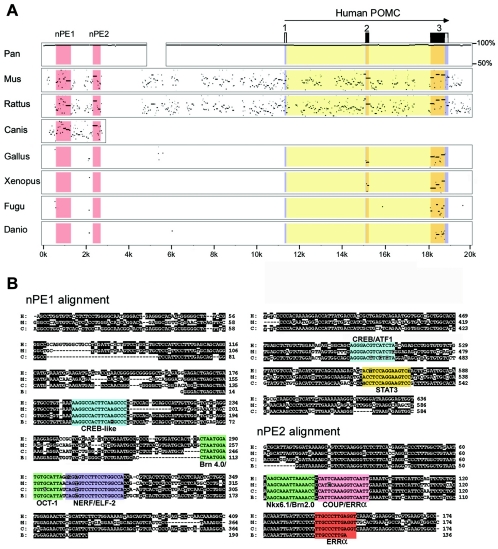
Among vertebrate genomes, regulatory elements are often more conserved than average intergenic regions (16, 57). Therefore, in order to identify candidate enhancers within the −13- to −9-kb mouse Pomc fragment, we sequenced this 4-kb region and compared it to the publicly available human POMC locus sequence. Figure 3A shows a global visualization of local sequence alignments (DOTTER program) (55) between the regions from kb −13 to −9 and kb −11 to −7 from the mouse and human POMC genes, respectively. Two regions of approximately 600 and 150 bp with high identities between the two species were readily identifiable. We named these two putative neuronal POMC enhancers nPE1 and nPE2. In addition to the conservation of nucleotide sequence, the distances from each element to the transcriptional start were also similar, indicating that the overall genomic organization at the POMC locus is conserved between these two species (Fig. 3B).
FIG. 3.
Identification of nPE enhancers. (A) Global visualization of local sequence alignments between regions from kb −13 to −9 of the mouse (vertical) and kb −11 to −7 of the human (horizontal) POMC genes by using the DOTTER program. Sequence conservation is shown as dark dots in the graph, and colinear conserved stretches appear as diagonals. Two regions of high sequence identity are highlighted: nPE1 (600 bp) and nPE2 (150 bp). (B) Scheme of mouse and human POMC genes showing the relative positions of nPE regions. The three POMC exons are numbered.
Conservation of neuronal enhancer function in the human POMC gene.
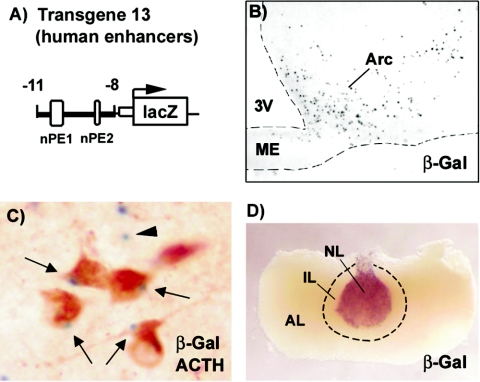
To determine whether the homologous human POMC region containing nPE1 and nPE2 sequences also has the functional capability to target transgenic expression to arcuate POMC neurons, we constructed a transgene in which the region between −11 and −8 kb of the human POMC gene (containing both nPE regions [Fig. 3]) was ligated upstream of the minimal promoter of the chicken β-globin gene followed by the lacZ reporter gene (transgene 13 [Fig. 4A ]). β-Galactosidase (β-Gal) activity was visualized in situ followed by immunohistochemistry against ACTH in coronal brain sections of transgenic mice from three independent pedigrees. Interestingly, X-Gal blue staining was evident in POMC-positive arcuate neurons (Fig. 4B and C), although the percentage of colocalization was highly variable among the three pedigrees analyzed (10, 68, and 95%). No reporter expression was detected in pituitary melanotrophs or corticotrophs (Fig. 4D). Different levels of ectopic β-Gal expression were observed in the brain of each transgenic pedigree in regions that included the amygdala, the habenula, the hippocampus, the cerebral cortex; the lateral, ventromedial, and arcuate nuclei of the hypothalamus; and some cells of the neural lobe of the pituitary that do not express Pomc (Fig. 4D). These results, together with those obtained from transgene 5 (see above), indicate that neuronal POMC and pituitary POMC gene expression are controlled by topographically distinct and functionally independent regulatory regions. In addition, the ability of the human genomic fragment containing nPE1 and nPE2 to target reporter expression to mouse POMC arcuate neurons suggests that these conserved sequences play a role in the expression of POMC in the human hypothalamus.
FIG. 4.
Conservation of human POMC enhancer function. (A) Scheme of transgene 13. The region between kb −11 and −8 of the human POMC gene was ligated upstream of the minimal promoter of chick β-globin and the lacZ transgene. nPE regions are indicated. (B) X-Gal staining of the hypothalamus of a transgene 13 mouse showing expression in the arcuate nucleus (Arc). (C) Colocalization of X-Gal staining (blue) and ACTH immunoreactivity (brown) in arcuate neurons. Arrows point to stained β-Gal inclusion bodies in POMC neurons; the arrowhead shows an example of a β-Gal inclusion body that does not colocalize with POMC. Abbreviations are as defined in the legend to Fig. 2.
nPE1 and nPE2 are necessary to direct transgene expression to POMC arcuate neurons.
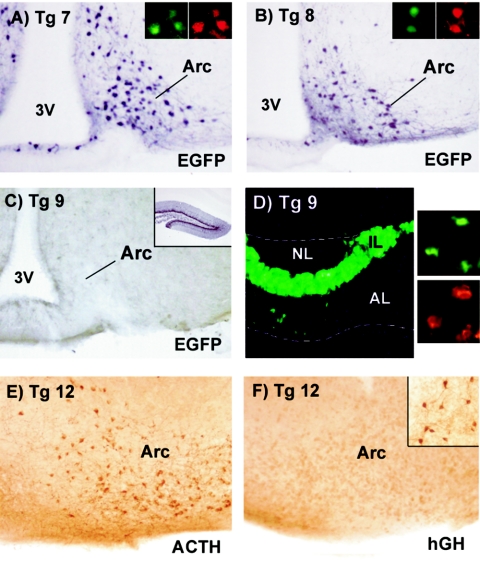
Within the region spanning kb −13 to −9 of the mouse Pomc promoter, nPE1 and nPE2 are the only nonrepetitive sequences that have been highly conserved during mammalian evolution. This conservation suggests that these sites may carry the necessary information to assure neuronal-specific expression of the POMC gene. To challenge this hypothesis, we constructed transgenic mice carrying deleted versions of the original POMC-EGFP transgene from kb −13 to +8 in which nPE1, nPE2, or both enhancers were deleted (transgenes 7, 8, and 9) (Fig. 5). EGFP expression and colocalization with POMC were determined in brain and pituitary slices of transgenic mice. Transgenic mice lacking either nPE1 (construct 7) or nPE2 (construct 8) reproducibly targeted EGFP expression to over 90% of POMC-expressing neurons in the arcuate nucleus (Fig. 6A and B), demonstrating that the absence of either one of these elements was not sufficient to disrupt neuronal-specific expression. A truncation from kb −13 to −10.5 that removed nPE1, together with neighboring sequences (transgene 6) (Fig. 5), also directed eutopic expression of EGFP in POMC arcuate neurons (data not shown), confirming that nPE1 is not absolutely necessary to guarantee neuronal Pomc gene expression. Nevertheless, the simultaneous deletion of nPE1 and nPE2 completely eliminated EGFP expression in POMC neurons of the arcuate in five independent transgenic lines (Fig. 6C). All transgenic lines carrying constructs 6, 7, 8, or 9 displayed EGFP expression in pituitary melanotrophs and corticotrophs (Fig. 6D), confirming that Pomc expression in the brain and pituitary gland is controlled by independent enhancers. Together, these results indicate that nPE1 and nPE2 function as neuronal transcriptional enhancers that are necessary for Pomc expression in arcuate neurons. In the absence of either one of these enhancers, the remaining element is still able to assure transgene expression in POMC neurons of the arcuate nucleus, indicating a functional redundancy between nPE1 and nPE2.
FIG. 5.
Second set of transgenes used in this study. nPE enhancers (gray boxes) were deleted from parental transgenes 1 and 5. Details are as described for Fig. 1.
FIG. 6.
nPE enhancers are necessary for POMC expression in the hypothalamus. (A through C) EGFP immunostainings performed on coronal hypothalamic sections of transgene 7 (panel A), 8 (panel B), and 9 (panel C) mice. Insets in panels A and B show colocalization of EGFP (green) and ACTH immunofluorescence (red) in arcuate neurons. Inset in panel C shows ectopic expression of transgene 9 in the hippocampal dentate gyrus. Abbreviations as in Fig. 2. (D) Image of a transgene 9 pituitary under UV light showing green fluorescence in melanotrophs in the intermediate lobe (IL) and corticotrophs in the anterior lobe (AL). NL, neural lobe. Panels on the right show colocalization of EGFP (green) and ACTH immunofluorescence (red) in corticotrophs of a transgene 9 pituitary. (E and F) Immunostainings of serial sagittal cuts of transgene 12 mouse hypothalamus. The arcuate nucleus expresses ACTH (panel E) but not the hGH reporter (panel F). The inset in panel F shows ectopic hGH expression surrounding the anterior commissure in the same brain section.
To confirm the importance of these enhancers in directing reporter expression to POMC arcuate neurons, we introduced deletions of nPE1, nPE2, or both elements simultaneously into the original transgene 5 in which a minimal viral TK promoter was fused to the reporter gene hGH (transgenes 10, 11, and 12 [Fig. 5]). As observed with the previous set of larger transgenes, deletion of either nPE1 or nPE2 alone did not prevent expression of reporter hGH in the arcuate POMC neurons (results not shown). However, the simultaneous deletion of both nPE1 and nPE2 completely abolished hGH expression in the POMC arcuate neurons of five independent lines of transgenic mice as assessed by anti-hGH immunohistochemistry (Fig. 6E and F). Transgenic mice carrying constructs 10, 11, and 12 did not show reporter gene expression in pituitary melanotrophs as predicted, but there was variable ectopic expression in the habenula, cerebral cortex, and hippocampus in all lines, showing that the transgenes were not inserted into transcriptionally silent regions (Fig. 6F, inset). These results further demonstrate that nPE1 and nPE2 are responsible for the neuronal enhancer activity that we identified in the fragment from kb −13 to −9 of the mouse Pomc 5′ flanking region.
nPE1 and nPE2 are conserved in mammals but not in other vertebrates.
Since the POMC gene is expressed in the ventromedial hypothalamic neurons of all vertebrate species studied to date, we investigated whether the neuronal regulatory elements nPE1 and nPE2 are conserved in different vertebrate orders. We searched the sequenced POMC loci from the completed genome projects of rat, chimpanzee, and chicken and the teleost fishes pufferfish (T. rubripes) and zebrafish (Danio rerio), as well as the ongoing genome projects of the dog and the frog X. tropicalis (see Materials and Methods). Figure 7A shows a multiple percentage-identity plot (PIP) (53) comparing the human POMC locus to the complete mouse, rat, chimpanzee, chicken, frog, pufferfish and zebra fish POMC loci, as well as to a partial dog sequence. All mammalian sequences showed extensive identity to the human POMC locus, including several conserved noncoding blocks. Among these, nPE1 and nPE2 displayed a particularly greater degree of conservation, evidenced by long stretches and a high percentage of sequence identity relative to those of other less conserved intergenic or intronic regions. The chimpanzee POMC locus was almost 100% identical to the human locus in coding and noncoding regions, as expected given the close evolutionary distance between these two primate species. In contrast, nonmammalian POMC genes did not show significant blocks of sequence identity to the human POMC locus, except for exons 2 and 3 (chicken and Xenopus) and exon 3 (fishes). These data show that both nPE1 and nPE2 are strongly conserved among mammals but cannot be recognized in birds, amphibians, or teleost fishes. Figure 7B shows a multiple alignment of nPE1 and nPE2 of four different mammals, including partial nPE sequences that we amplified by PCR from a bovine genomic phage clone containing the POMC gene. The alignment shows several blocks of high sequence identity, which might indicate possible binding sites for transcription factors. nPE1 had an overall mouse and human similarity of 76%, with a 40-bp core of 100% identity, whereas mouse and human nPE2 had 90% similarity (138 bp out of 153 bp are identical). Interestingly, the similarities between mouse and human exons 1, 2, and 3 of 64, 87, and 82%, respectively, were not higher than the interspecies identity for nPE1 and nPE2.
FIG. 7.
Conservation of POMC neuronal enhancers. (A) Comparison of the human POMC gene to several species by using the program PipMaker. The human POMC gene including 12 kb of the 5′ sequence was compared to equivalent regions of the mouse (Mus musculus), rat (Rattus norvegicus), chimpanzee (Pan troglodytes), chicken (Gallus gallus), frog (X. tropicalis), zebra fish (D. rerio), and pufferfish (T. rubripes), as well as to a 4-kb region obtained from the Dog (Canis familiaris) Genome Project. The exons of human POMC as well as nPE regions are indicated. For each species, alignments to human sequence are displayed by plotting the length and percent identity (between 50 and 100%) of aligned segments. In the plot, short noncoding segments that appear conserved between mammals and nonmammalian species are usually simple repeats, e.g., (AT)n, (GT)n. (B) ClustalW alignments of nPE1 and nPE2 enhancer regions from several mammalian species. H, human; M, mouse; C, dog; B, cow (Bos taurus). Sequences from cows are incomplete. Identical residues are highlighted black. Colored boxes denote conserved and aligned putative transcription factor binding sites.
A further comparison of human, mouse, dog, and cow nPE1 and nPE2 sequences by using the recently developed rVista version 2.0 bioinformatics tool (37) allowed the detection of a limited number of conserved and aligned transcription factor binding sites that could determine the transcriptional regulation of Pomc in arcuate neurons (Fig. 7B). Within nPE1, we identified one site that carried two adjacent consensus sequences for the POU domain transcription factors Brn 4.0 and OCT-1. Immediately downstream of this site was a consensus sequence for Elf-2, a member of the Ets transcription factor family. Interestingly, a sequence present at the 3′ end of nPE1 highly matched a consensus binding site for signal transducer and activator of transcription 3 (STAT3), a downstream effector of leptin known to upregulate Pomc transcription in hypothalamic neurons. In addition, two conserved cAMP-responsive element binding (CREB-like) sites were present within nPE1.
Within nPE2, we identified a sequence that showed high similarity to the consensus binding site for the homeobox gene Nkx6.1 and the POU domain gene Brn2.0. Contiguous to this site, there is a sequence that carries overlapping matches for chicken ovoalbumin upstream promoter (COUP) and estrogen-related receptor alpha (ERRα), two transcription factors that belong to the orphan nuclear receptor family. Another canonical ERRα site was located further downstream.
DISCUSSION
Identification of two neuronal enhancers that control POMC gene expression in the central nervous system.
The aim of the present study was to identify functional cis-acting elements that control neuronal POMC expression in the hypothalamus. In the absence of POMC-expressing neuronal cell lines, we performed a deletion and truncation analysis in transgenic mice that provided a highly faithful and efficient expression system in which to test the ability of different genomic regions to target reporter gene expression to POMC hypothalamic neurons. Here, we characterize two novel distal enhancers, nPE1 and nPE2, that play an essential role in the activation of POMC gene expression in a selected population of hypothalamic arcuate neurons. Our transgenic mouse and phylogenetic analyses show that (i) a distal genomic region containing nPE1 and nPE2 is necessary and sufficient to direct authentic neuron-specific expression of reporter genes to POMC arcuate neurons; (ii) either nPE1 or nPE2 assures proper reporter expression in POMC arcuate neurons, whereas simultaneous deletion of these two enhancers completely eliminates expression in POMC neurons; (iii) nPE1 and nPE2 nucleotide sequences and genomic organization are both highly conserved among mammals but not between mammals and birds, amphibians, or fishes; (iv) the enhancer activity of mouse and human genomic fragments containing nPE1 and nPE2 is functionally conserved; (v) POMC expression in the brain and pituitary gland is controlled by different and independent sets of enhancers; and (vi) a limited number of conserved and aligned putative transcription factor binding sites are present in these enhancers, including canonical DNA sequences for STAT3 and for the POU-domain proteins Brn 4.0 and OCT-1 in nPE1 and the homeodomain factors Nkx6.1 and Brn2.0 in nPE2.
In the arcuate nucleus of the mouse hypothalamus, approximately 3,000 POMC-expressing neurons are intermingled with neurons that express other neuropeptides or monoamines, including NPY/AGRP, ghrelin, somatostatin, or dopamine (13, 14, 51). The specific set of neuropeptides expressed in each of these neuronal subpopulations is presumably determined by particular combinations of transcription factors acting during ontogenesis and subsequently in the maintenance of established phenotypes. nPE enhancers probably play an important role in these processes, acting as recognition sites for the assembly of transcriptional complexes that participate in the remodeling of chromatin structure at the POMC locus and the expression of POMC in arcuate neurons. Although the mouse and human regions containing nPE1 and nPE2 proved to be necessary and sufficient to drive reporter gene expression to POMC arcuate neurons of transgenic mice, they were not able to isolate transgenes from the transcriptional influences present at the insertion sites. In consequence, most transgenic pedigrees that showed reporter expression in arcuate POMC neurons also displayed variable patterns of ectopic transgene expression.
Interestingly, our transgenic mouse analysis demonstrated that nPE1 and nPE2 do not need to be simultaneously present to direct transgene expression to POMC arcuate neurons, suggesting some level of functional redundancy between these two enhancers. Nevertheless, it is possible that each nPE enhancer plays a unique role in POMC transcriptional regulation. For example, leptin receptors localized in POMC arcuate neurons signal via phosphorylated STAT3 and deficits in this signal transduction pathway within these neurons correlate with very low levels of POMC mRNA (2, 17). A recent study performed in vitro using a heterologous cell line showed that STAT3 might increase POMC transcription by interacting with a noncanonical STAT3 binding site located immediately upstream of the TATAA box (39). Although it is still not known whether phosphorylation of STAT3 activates Pomc expression directly, nPE1 contains a STAT3 canonical site that is conserved and aligned across mammals and could participate in the leptin regulation of Pomc expression. Further studies are needed to address this possibility.
Modular architecture of the POMC gene.
Our results showed that genomic mouse and human fragments containing nPE1 and nPE2 fused to heterologous minimal promoters direct the expression of transgenes to POMC arcuate neurons in the absence of detectable reporter expression in pituitary melanotrophs or corticotrophs (transgenes 5 and 13). On the other hand, simultaneous deletion of nPE1 and nPE2 in the context of the entire POMC transcriptional unit and flanking regions abolishes reporter gene expression in the arcuate nucleus while maintaining authentic transgenic expression in pituitary POMC cells (transgene 9). Together, these results indicate that two independent sets of enhancers control the cell-specific expression of POMC: a distal module acting in the brain and a proximal module acting in the pituitary. Whether these two modular regions cross talk under particular physiological circumstances to regulate POMC expression in the pituitary or the brain remains to be investigated. Our previous studies demonstrated that all necessary elements to provide cell-specific and hormonally regulated expression of transgenes in pituitary POMC cells are located in approximately 300 bp upstream of the rat or mouse TATAA box (33, 34). Moreover, several transcription factors involved in pituitary POMC expression have been identified, including the homeobox protein Pitx1, the T-box factor Tpit/Tbx19, and basic helix-loop-helix proteins of the NeuroD1 subfamily (31, 32, 44). In contrast, the transcription factors controlling POMC expression in the brain are completely unknown. In this regard, nPE1 and nPE2 could provide a valuable platform for the discovery of such factors. Predictive analysis of binding sites based solely on computer algorithms that identify canonical sequences generally yields a high number of false positives (63) because transcription factors bind to short sequence motifs (6 to 12 bp) that occur very frequently in a genome and different functional DNA elements for the same transcription factor admit a considerable level of degeneracy. However, novel bioinformatics tools have been recently developed that select for canonical binding sites that are aligned within highly conserved sequences of two or more species. Using one of these tools, rVista2.0 (37), we were able to identify five potential transcription factor binding sites along the 600 bp of nPE1 and three sites along the 150 bp of nPE2 that complied with such stringent criteria. The high selective pressure that these sequences withstood during more than 65 million years of evolutionary distance existing between mice and men suggests that these sites play a key role in the commitment of the POMC hypothalamic lineage and the cell-specific and hormone-regulated expression of Pomc in the brain. In fact, nPE1 contains a conserved and aligned site carrying two consecutive canonical sequences for Brn 4.0 and OCT-1, two POU domain transcription factors that participate in the final differentiation of glucagon-expressing pancreatic α cells (26), and GnRH-expressing hypothalamic neurons, respectively (9). This site is adjacent to a consensus sequence for NERF/Elf-2, a member of the Ets transcription factor family. The close proximity of these two sites suggests the existence of a unique combination of a POU domain protein present in POMC neurons that would heterodimerize with an Ets transcription factor to control POMC expression. Both POU domain and Ets factors have been shown to form heterodimers to activate gene expression. In fact, physical interaction of the pituitary-specific POU homeodomain protein Pit-1 with Ets-1 has been shown to be critical to establish the lactotroph pituitary lineage and to induce prolactin gene expression (5). nPE1 also contains a perfect canonical site for STAT3 whose potential significance in the leptin regulation of POMC expression and the central control of satiety mechanisms have been addressed above. In addition, two separate conserved CREB-like sites are present in nPE1, suggesting that transcription factors from the leucine zipper family could also participate in the neuronal regulation of POMC expression.
Within nPE2, there is a conserved and aligned homeodomain binding site carrying a canonical sequence for Nkx6.1 and Brn2.0, two homeodomain transcription factors that participate in the final differentiation of pancreatic β cells (28) or hypothalamic neuropeptide neurons expressing oxytocin, vasopressin, and CRH (21), respectively. Binding of a homeobox or POU domain factor to this site may be critical to induce neuron-specific expression of POMC. Adjacent to this site, there are overlapping canonical sequences for COUP and ERRα, two transcription factors that belong to the orphan nuclear receptor family. The close proximity of these two binding sites suggests that an orphan nuclear receptor may physically interact with a homeodomain factor to repress POMC expression. Such a type of transcriptional repression has been observed between the homeodomain protein Pit-1 and different members of the orphan nuclear receptor factor family (20). An additional ERRα site is located further downstream at the 3′ end of nPE2. ERRα participates in the control of energy balance by regulating the mitochondrial β-oxidation of fatty acids (62) as well as the development of brown and white adipocytes (38, 64). ERRα-induced transcriptional regulation of hypothalamic POMC could provide another level for the control of energy homeostasis by regulating food intake in concert with leptin and NPY activation of POMC neurons. Taken together, it is tempting to speculate that some, if not all, of these highly conserved and aligned binding sites are the cis-acting platforms where unique clusters of transcription factors present in hypothalamic POMC neurons are recruited to specifically transactivate POMC. Additional functional and biochemical assays will have to be performed in order to finally characterize the cis- and trans-acting partners that control neuronal POMC expression.
The independent modular architecture of the POMC gene may also have important implications in the study of genetic predisposition to familial obesity. Total POMC deficiency causes a complicated syndrome that includes adrenal insufficiency, skin and hair pigmentation deficits, and early-onset obesity probably due to the lack of ACTH and peripheral and central melanocortins, respectively (8, 29, 65). Although homozygous null mutations of POMC are extremely rare in humans, genome-wide QTL scans have demonstrated a genetic linkage between obesity-related traits and a region in human chromosome 2 containing the POMC locus with a reported logarithm of odds score of 7.5, the highest ever found in a human genetic study of obesity-related traits (reviewed in reference 12). This linkage first determined in a population of Mexican-Americans has been reproduced in other studies that analyzed individuals of Caucasian French and African-American origins (23, 25, 47). Although the strongest candidate gene for this chromosome 2 QTL is POMC, no loss-of-function mutations were found within the POMC transcriptional unit (25). Therefore, it is tempting to speculate about the possibility that polymorphisms or mutations in nPE1 and nPE2 could decrease the transcription of POMC and thereby the neural content of POMC peptides, impairing central energy homeostatic mechanisms and predisposing to excessive weight gain without altering the levels or physiological actions of pituitary-released POMC products. Thus, nPE regions are natural candidates to screen for sequence polymorphisms that influence human obesity.
Phylogenetic footprinting to identify neuronal POMC enhancers.
Until recently, it was difficult to predict the localization of transcriptional regulatory elements based solely on nucleotide sequence analysis. However, the recent availability of genomic sequences from an increasing number of vertebrate species has allowed the identification of regulatory regions by comparing homologous intergenic regions from different species, a technique termed phylogenetic footprinting (16, 36, 57). This strategy is based on the idea that, as a result of selective pressure, mutations accumulate faster in nonfunctional nucleotide residues whereas functional residues remain more conserved. Therefore, multiple alignment comparisons between orthologous gene loci allow the discovery of potential regulatory sequences embedded in large intergenic regions.
In the study reported here, we combined phylogenetic footprinting with expression analysis in transgenic mice to identify two POMC neuronal enhancers. We found nPE1 and nPE2 in representative species of the mammalian orders Rodentia, Primates, Carnivora, and Artiodactyla, and it is reasonable to predict that nPEs will be found in all mammals. Conversely, we failed to identify conserved nPE enhancers outside Mammalia in spite of the fact that POMC is expressed in the ventromedial hypothalamus of the chicken and teleost fishes like trout, goldfish, and zebrafish (7, 19, 35, 59). Even though there are several examples of enhancers conserved among mammals, birds, and fishes (1, 6, 22, 48, 61), the only conserved regions we observed after comparing the human POMC locus with orthologous genomic sequences from other nonmammalian vertebrates were coding exons 2 and 3. Nevertheless, given that hypothalamic expression of POMC is a conserved vertebrate feature and that core recognition sites for transcription factors are usually very short (6 to 10 bp), it is likely that a similar set of transcription factors control POMC expression in all vertebrates but that the accumulation of neutral mutations throughout evolution prevented us from identifying neuronal POMC enhancers by global alignment programs. The example of POMC illustrates the fact that for each gene, a different set of species might be necessary to obtain useful results by phylogenetic footprinting. In this case, the human X mouse comparison provided an optimal degree of sequence conservation for the identification of functional enhancers while the human X chimpanzee pair was too similar and the sequences of birds, amphibians, or fishes was too different.
Acknowledgments
Our thanks go to B. Wyss, M. Ricca, N. Malarini, and L. Hayes for excellent technical assistance and to the OHSU transgenic core lab for microinjection of selected transgenes.
This work was supported in part by a Fogarty International Research Collaborative Award TW01233 (M.J.L. and M.R.), NIH grant DK68400 (M.J.L. and M.R.), International Research Scholar Grant of the Howard Hughes Medical Institute (M.R.), Agencia Nacional de Promoción Científica y Tecnológica (M.R.), JS Guggenheim Foundation (M.R.) and Universidad de Buenos Aires (M.R.). F. S. J. de Souza, A. M. Santangelo, V. F. Bumaschny, and M. E. Avale are recipients of fellowships from CONICET, Argentina.
M.J.L. declares a financial interest in transgenic models utilizing the POMC neural regulatory regions, and this potential conflict of interest has been disclosed to and managed by the OHSU Office for Integrity in Research.
REFERENCES
- 1.Bagheri-Fam, S., C. Ferraz, J. Demaille, G. Scherer, and D. Pfeifer. 2001. Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics 78:73-82. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S. H., W. H. Stearns, T. A. Dundon, M. Schubert, A. W. Tso, Y. Wang, A. S. Banks, H. J. Lavery, A. K. Haq, E. Maratos-Flier, B. G. Neel, M. W. Schwartz, and M. G. Myers, Jr. 2003. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature 421:856-859. [DOI] [PubMed] [Google Scholar]
- 3.Batterham, R. L., M. A. Cowley, C. J. Small, H. Herzog, M. A. Cohen, C. L. Dakin, A. M. Wren, A. E. Brynes, M. J. Low, M. A. Ghatei, R. D. Cone, and S. R. Bloom. 2002. Gut hormone PYY(3-36) physiologically inhibits food intake. Nature 418:650-654. [DOI] [PubMed] [Google Scholar]
- 4.Bjorbaek, C., and B. B. Kahn. 2004. Leptin signaling in the central nervous system and the periphery. Recent Prog. Horm. Res. 59:305-331. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, A. P., K. S. Brodsky, S. E. Diamond, L. C. Kuhn, Y. Liu, and A. Gutierrez-Hartmann. 2000. The Pit-1 homeodomain and beta-domain interact with Ets-1 and modulate synergistic activation of the rat prolactin promoter. J. Biol. Chem. 275:3100-3106. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, S., B. Venkatesh, W. H. Yap, C. F. Chou, A. Tay, S. Ponniah, Y. Wang, and Y. H. Tan. 2002. Conserved regulation of the lymphocyte-specific expression of lck in the Fugu and mammals. Proc. Natl. Acad. Sci. USA 99:2936-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerda-Reverter, J. M., H. B. Schioth, and R. E. Peter. 2003. The central melanocortin system regulates food intake in goldfish. Regul. Pept. 115:101-113. [DOI] [PubMed] [Google Scholar]
- 8.Challis, B. G., A. P. Coll, G. S. Yeo, S. B. Pinnock, S. L. Dickson, R. R. Thresher, J. Dixon, D. Zahn, J. J. Rochford, A. White, R. L. Oliver, G. Millington, S. A. Aparicio, W. H. Colledge, A. P. Russ, M. B. Carlton, and S. O'Rahilly. 2004. Mice lacking pro-opiomelanocortin are sensitive to high-fat feeding but respond normally to the acute anorectic effects of peptide-YY(3-36). Proc. Natl. Acad. Sci. USA 101:4695-4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, M. E., and P. L. Mellon. 1995. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol. Cell. Biol. 15:6169-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coll, A. P., I. S. Farooqi, B. G. Challis, G. S. Yeo, and S. O'Rahilly. 2004. Proopiomelanocortin and energy balance: insights from human and murine genetics. J. Clin. Endocrinol. Metab. 89:2557-2562. [DOI] [PubMed] [Google Scholar]
- 11.Comuzzie, A. G., J. E. Hixson, L. Almasy, B. D. Mitchell, M. C. Mahaney, T. D. Dyer, M. P. Stern, J. W. MacCluer, and J. Blangero. 1997. A major quantitative trait locus determining serum leptin levels and fat mass is located on human chromosome 2. Nat. Genet. 15:273-276. [DOI] [PubMed] [Google Scholar]
- 12.Comuzzie, A. G. 2002. The emerging pattern of the genetic contribution to human obesity. Best. Pract. Res. Clin. Endocrinol. Metab. 16:611-621. [DOI] [PubMed] [Google Scholar]
- 13.Cowley, M. A., J. L. Smart, M. Rubinstein, M. G. Cerdan, S. Diano, T. L. Horvath, R. D. Cone, and M. J. Low. 2001. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411:480-484. [DOI] [PubMed] [Google Scholar]
- 14.Cowley, M. A., R. G. Smith, S. Diano, M. Tschop, N. Pronchuk, K. L. Grove, C. J. Strasburger, M. Bidlingmaier, M. Esterman, M. L. Heiman, L. M. Garcia-Segura, E. A. Nillni, P. Mendez, M. J. Low, P. Sotonyi, J. M. Friedman, H. Liu, S. Pinto, W. F. Colmers, R. D. Cone, and T. L. Horvath. 2003. The distribution and mechanism of action of ghrelin in the CNS demonstrates a novel hypothalamic circuit regulating energy homeostasis. Neuron 37:649-661. [DOI] [PubMed] [Google Scholar]
- 15.Delplanque, J., M. Barat-Houari, C. Dina, P. Gallina, K. Clement, B. Guy-Grand, F. Vasseur, P. Boutin, and P. Froguel. 2000. Linkage and association studies between the proopiomelanocortin (POMC) gene and obesity in caucasian families. Diabetologia 43:1554-1557. [DOI] [PubMed] [Google Scholar]
- 16.Fickett, J. W., and W. W. Wasserman. 2000. Discovery and modeling of transcriptional regulatory regions. Curr. Opin. Biotechnol. 11:19-24. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Q., M. J. Wolfgang, S. Neschen, K. Morino, T. L. Horvath, G. I. Shulman, and X. Y. Fu. 2004. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc. Natl. Acad. Sci. USA 101:4661-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelman, D. M., D. Noain, M. E. Avale, V. Otero, M. J. Low, and M. Rubinstein. 2003. Transgenic mice engineered to target Cre/loxP-mediated DNA recombination into catecholaminergic neurons. Genesis 36:196-202. [DOI] [PubMed] [Google Scholar]
- 19.Gerets, H. H., K. Peeters, L. Arckens, F. Vandesande, and L. R. Berghman. 2000. Sequence and distribution of pro-opiomelanocortin in the pituitary and the brain of the chicken (Gallus gallus). J. Comp. Neurol. 417:250-262. [PubMed] [Google Scholar]
- 20.Gonzalez, M. M., and C. Carlberg. 2002. Cross-repression, a functional consequence of the physical interaction of non-liganded nuclear receptors and POU domain transcription factors. J. Biol. Chem. 277:18501-18509. [DOI] [PubMed] [Google Scholar]
- 21.Goshu, E., H. Jin, J. Lovejoy, J. F. Marion, J. L. Michaud, and C. M. Fan. 2004. Sim2 contributes to neuroendocrine hormone gene expression in the anterior hypothalamus. Mol. Endocrinol. 18:1251-1262. [DOI] [PubMed] [Google Scholar]
- 22.Göttgens, B., L. M. Barton, J. G. Gilbert, A. J. Bench, M. J. Sanchez, S. Bahn, S. Mistry, D. Grafham, A. McMurray, M. Vaudin, E. Amaya, D. R. Bentley, A. R. Green, and A. M. Sinclair. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18:181-186. [DOI] [PubMed] [Google Scholar]
- 23.Hager, J., C. Dina, S. Francke, S. Dubois, M. Houari, V. Vatin, E. Vaillant, N. Lorentz, A. Basdevant, K. Clement, B. Guy-Grand, and P. Froguel. 1998. A genome-wide scan for human obesity genes reveals a major susceptibility locus on chromosome 10. Nat. Genet. 20:304-308. [DOI] [PubMed] [Google Scholar]
- 24.Heisler, L. K., M. A. Cowley, L. H. Tecott, W. Fan, M. J. Low, J. L. Smart, M. Rubinstein, J. B. Tatro, J. N. Marcus, H. Holstege, C. E. Lee, R. D. Cone, and J. K. Elmquist. 2002. Activation of central melanocortin pathways by fenfluramine. Science 297:609-611. [DOI] [PubMed] [Google Scholar]
- 25.Hixson, J. E., L. Almasy, S. Cole, S. Birnbaum, B. D. Mitchell, M. C. Mahaney, M. P. Stern, J. W. MacCluer, J. Blangero, and A. G. Comuzzie. 1999. Normal variation in leptin levels is associated with polymorphisms in the proopiomelanocortin gene, POMC. J. Clin. Endocrinol. Metab. 84:3187-3191. [DOI] [PubMed] [Google Scholar]
- 26.Hussain, M. A., C. P. Miller, and J. F. Habener. 2002. Brn-4 transcription factor expression targeted to the early developing mouse pancreas induces ectopic glucagon gene expression in insulin-producing beta cells. J. Biol. Chem. 277:16028-16032. [DOI] [PubMed] [Google Scholar]
- 27.Israel, D. I. 1993. A PCR-based method for high stringency screening of DNA libraries. Nucleic Acids Res. 21:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iype, T., D. G. Taylor, S. M. Ziesmann, J. C. Garmey, H. Watada, and R. G. Mirmira. 2004. The transcriptional repressor Nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic beta-cell differentiation: evidence for feedback activation of the nkx6.1 gene by Nkx6.1. Mol. Endocrinol. 18:1363-1375. [DOI] [PubMed] [Google Scholar]
- 29.Krude, H., H. Biebermann, W. Luck, R. Horn, G. Brabant, and A. Gruters. 1998. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Nat. Genet. 19:155-157. [DOI] [PubMed] [Google Scholar]
- 30.Krude, H., H. Biebermann, D. Schnabel, M. Z. Tansek, P. Theunissen, P. E. Mullis, and A. Gruters. 2003. Obesity due to proopiomelanocortin deficiency: three new cases and treatment trials with thyroid hormone and ACTH4-10. J. Clin. Endocrinol. Metab. 88:4633-4640. [DOI] [PubMed] [Google Scholar]
- 31.Lamolet, B., A. M. Pulichino, T. Lamonerie, Y. Gauthier, T. Brue, A. Enjalbert, and J. Drouin. 2001. A pituitary cell-restricted T box factor, Tpit, activates POMC transcription in cooperation with Pitx homeoproteins. Cell 104:849-859. [DOI] [PubMed] [Google Scholar]
- 32.Lamonerie, T., J. J. Tremblay, C. Lanctot, M. Therrien, Y. Gauthier, and J. Drouin. 1996. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 10:1284-1295. [DOI] [PubMed] [Google Scholar]
- 33.Liu, B., G. D. Hammer, M. Rubinstein, M. Mortrud, and M. J. Low. 1992. Identification of DNA elements cooperatively activating proopiomelanocortin gene expression in the pituitary glands of transgenic mice. Mol. Cell. Biol. 12:3978-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, B., M. Mortrud, and M. J. Low. 1995. DNA elements with AT-rich core sequences direct pituitary cell-specific expression of the pro-opiomelanocortin gene in transgenic mice. Biochem. J. 312:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, N. A., H. Huang, Z. Yang, W. Herzog, M. Hammerschmidt, S. Lin, and S. Melmed. 2003. Pituitary corticotroph ontogeny and regulation in transgenic zebrafish. Mol. Endocrinol. 17:959-966. [DOI] [PubMed] [Google Scholar]
- 36.Loots, G. G., R. M. Locksley, C. M. Blankespoor, Z. E. Wang, W. Miller, E. M. Rubin, and K. A. Frazer. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136-140. [DOI] [PubMed] [Google Scholar]
- 37.Loots, G. G., and I. Ovcharenko. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217-W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, J., R. Sladek, J. Carrier, J.-A. Bader, D. Richard, and V. Giguère. 2003. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor α. Mol. Cell. Biol. 23:7947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Münzberg, H., L. Huo, E. A. Nillni, A. N. Hollenberg, and C. Bjorbaek. 2003. Role of signal transducer and activator of transcription 3 in regulation of hypothalamic proopiomelanocortin gene expression by leptin. Endocrinology 144:2121-2131. [DOI] [PubMed] [Google Scholar]
- 40.Newell-Price, J. 2003. Proopiomelanocortin gene expression and DNA methylation: implications for Cushing's syndrome and beyond. J. Endocrinol. 177:365-372. [DOI] [PubMed] [Google Scholar]
- 41.Overstreet, L. S., S. T. Hentges, V. F. Bumaschny, F. S. de Souza, J. L. Smart, A. M. Santangelo, M. J. Low, G. L. Westbrook, and M. Rubinstein. 2004. A transgenic marker for newly born granule cells in dentate gyrus. J. Neurosci. 24:3251-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeffer, P. L., M. Bouchard, and M. Busslinger. 2000. Pax2 and homeodomain proteins cooperatively regulate a 435 bp enhancer of the mouse Pax5 gene at the midbrain-hindbrain boundary. Development 127:1017-1028. [DOI] [PubMed] [Google Scholar]
- 43.Philips, A., M. Maira, A. Mullick, M. Chamberland, S. Lesage, P. Hugo, and J. Drouin. 1997. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol. Cell. Biol. 17:5952-5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poulin, G., B. Turgeon, and J. Drouin. 1997. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell. Biol. 17:6673-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulichino, A. M., S. Vallette-Kasic, J. P. Tsai, C. Couture, Y. Gauthier, and J. Drouin. 2003. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 17:738-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raffin-Sanson, M. L., Y. de Keyzer, and X. Bertagna. 2003. Proopiomelanocortin, a polypeptide precursor with multiple functions: from physiology to pathological conditions. Eur. J. Endocrinol. 149:79-90. [DOI] [PubMed] [Google Scholar]
- 47.Rotimi, C. N., A. G. Comuzzie, W. L. Lowe, A. Luke, J. Blangero, and R. S. Cooper. 1999. The quantitative trait locus on chromosome 2 for serum leptin levels is confirmed in African-Americans. Diabetes 48:643-644. [DOI] [PubMed] [Google Scholar]
- 48.Rowitch, D. H., Y. Echelard, P. S. Danielian, K. Gellner, S. Brenner, and A. P. McMahon. 1998. Identification of an evolutionarily conserved 110 base-pair cis-acting regulatory sequence that governs Wnt-1 expression in the murine neural plate. Development 125:2735-2746. [DOI] [PubMed] [Google Scholar]
- 49.Rubinstein, M., M. Mortrud, B. Liu, and M. J. Low. 1993. Rat and mouse proopiomelanocortin gene sequences target tissue-specific expression to the pituitary gland but not to the hypothalamus of transgenic mice. Neuroendocrinology 58:373-380. [DOI] [PubMed] [Google Scholar]
- 50.Rubinstein, M., J. S. Mogil, M. Japon, E. C. Chan, R. G. Allen, and M. J. Low. 1996. Absence of opioid stress-induced analgesia in mice lacking beta-endorphin by site-directed mutagenesis. Proc. Natl. Acad. Sci. USA 93:3995-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saper, C. B., T. C. Chou, and J. K. Elmquist. 2002. The need to feed: homeostatic and hedonic control of eating. Neuron 36:199-211. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, M. W., S. C. Woods, D. Porte, Jr., R. J. Seeley, and D. G. Baskin. 2000. Central nervous system control of food intake. Nature 404:661-671. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer, J. Bouck, R. Gibbs, R. Hardison, and W. Miller. 2000. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 10:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scully, K. M., and M. G. Rosenfeld. 2002. Pituitary development: regulatory codes in mammalian organogenesis. Science 295:2231-2235. [DOI] [PubMed] [Google Scholar]
- 55.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1-GC10. [DOI] [PubMed] [Google Scholar]
- 56.Spiegelman, B. M., and J. S. Flier. 2001. Obesity and the regulation of energy balance. Cell 104:531-543. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, J. W., and J. W. Touchman. 2002. Vertebrate genome sequencing: building a backbone for comparative genomics. Trends Genet. 18:104-108. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tollemer, H., C. A. Teitsma, J. Leprince, T. Bailhache, F. Vandesande, O. Kah, M. C. Tonon, and H. Vaudry. 1999. Immunohistochemical localization and biochemical characterization of two novel decapeptides derived from POMC-A in the trout hypothalamus. Cell Tissue Res. 295:409-417. [DOI] [PubMed] [Google Scholar]
- 60.Tremblay, J. J., C. Lanctot, and J. Drouin. 1998. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol. Endocrinol. 12:428-441. [DOI] [PubMed] [Google Scholar]
- 61.Uchikawa, M., Y. Ishida, T. Takemoto, Y. Kamachi, and H. Kondoh. 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4:509-519. [DOI] [PubMed] [Google Scholar]
- 62.Vega, R. B., and D. P. Kelly. 1997. A role for estrogen-related receptor alpha in the control of mitochondrial fatty acid beta-oxidation during brown adipocyte differentiation. J. Biol. Chem. 272:31693-31699. [DOI] [PubMed] [Google Scholar]
- 63.Wasserman, W. W., and A. Sandelin. 2004. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5:276-287. [DOI] [PubMed] [Google Scholar]
- 64.Willy, P. J., I. R. Murray, J. Qian, B. B. Busch, W. C. Stevens, Jr., R. Martin, R. Mohan, S. Zhou, P. Ordentlich, P. Wei, D. W. Sapp, R. A. Horlick, R. A. Heyman, and I. G. Schulman. 2004. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc. Natl. Acad. Sci. USA 101:8912-8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yaswen, L., N. Diehl, M. B. Brennan, and U. Hochgeschwender. 1999. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat. Med. 5:1066-1070. [DOI] [PubMed] [Google Scholar]
- 66.Young, J. I., V. Otero, M. G. Cerdan, T. L. Falzone, E. C. Chan, M. J. Low, and M. Rubinstein. 1998. Authentic cell-specific and developmentally regulated expression of pro-opiomelanocortin genomic fragments in hypothalamic and hindbrain neurons of transgenic mice. J. Neurosci. 18:6631-6640. [DOI] [PMC free article] [PubMed] [Google Scholar]