Highlights
-
•
Lactobacillus, Streptococcus, and Lactococcus were majorly found in dairy products.
-
•
Major genera in fermented vegetables were Lactobacillus, Weissella, and Carnimonas.
-
•
Phages of Siphoviridae, Myoviridae, and Herelleviridae were found in dairy products.
-
•
Lp84-3 was significantly enriched in key enzymes of the butyrate metabolism pathway.
Keywords: Fermented foods, Microorganism, Bacteriophages, Flavor, CAZymes profiles, Lactiplantibacillus plantarum 84-3
Abstract
Microbes are critical for flavor formation in fermented foods; however, their mechanisms of action are not fully understood. The microbial composition of 51 dairy and 47 vegetable products was functionally annotated and the carbohydrate-active enzyme (CAZyme) profiles of Lactiplantibacillus plantarum 84-3 (Lp84-3), isolated from dairy samples, can promote resistant starch (RS) degradation, were analyzed. Lactobacillus, Streptococcus, and Lactococcus were the predominant genera in dairy products, whereas the major genera in vegetables were Lactobacillus, Weissella, and Carnimonas. Phages from Siphoviridae, Myoviridae, and Herelleviridae were also present in dairy products. Additionally, the glycosyl hydrolase (GHs) family members GH1 and GH13 and the glycosyltransferase (GTs) family members GT2 and GT4 were abundant in Lp84-3. Moreover, Lp84-3 was enriched in butanoate metabolism enzymes and butanoate metabolite compounds. Therefore, fermented food microbes, especially Lp84-3, have an abundant repertoire of enzymes that promote flavor production, as starter improving the flavor of fermented dairy and vegetable products.
Introduction
China has a variety of traditional fermented foods made from milk (Liang, Xie, Zhang, Ding, & Wu, 2021) and vegetables (Liang et al., 2022), such as the distinctive traditionally fermented dairy products (e.g., koumiss) and vegetables (e.g., Suancai), which are widely consumed due to their health functions, nutritional value, aroma, and taste (Xiao et al., 2020). These fermented foods include some probiotics that may also be involved in fermentation and affect the sensory and flavor properties of the final product (You, Yang, Jin, Kwok, Sun, & Zhang, 2022). Deep metagenomic sequencing can not only provide taxonomic resolution at the species and strain levels but also predict the functional potential of the metagenome (Niccum, Kastman, Kfoury, Robbat, & Wolfe, 2020). Previous reports have also indicated that the functional characteristics of fermented dairy and vegetable products regulate cholesterol levels in patients with hyperlipidemia (Hou et al., 2019). Therefore, metagenomic sequencing is necessary to understand the association between the functional potential of microbes in traditional fermented food production and their contributions to human health and nutrition.
The bacteriophage composition can be detected using metagenomic sequencing, which is crucial for the regulation of microbial composition. Phage predation is associated with the development of lactic acid bacteria (LAB) and important for the flavor and aroma of fermented foods (Z et al., 2012). However, because fermented dairy products contain a small number of strains, phage infection of the starter culture can have a negative effect on the fermentation process and final product performance (Spus et al., 2015), leading to production failure in the dairy fermentation industry. Among Lactobacillus phages, Lactococcus lactis, Lactococcus cremoris, and Streptococcus thermophilus have been extensively studied (Mahony et al., 2017). A previous study reported that clustered regularly interspaced short palindromic repeats (CRISPR) are widely distributed in bacteria and are used to resist phage infection (Horvath, Romero, Coûté, Richards, & Barrangou, 2008). However, there is a lack in our knowledge of the dynamics and mechanisms of bacteriophage composition that affect the fermentation process and flavor.
Lactiplantibacillus plantarum is a well-studied bacterium which can be isolated from a variety of sources, such as Tibetan kefir (Aziz et al., 2023a, Aziz et al., 2022, Aziz et al., 2023b), Inner Mongolian cheese (Zhao et al., 2023), and other fermented foods. Our previous findings showed that Lactiplantibacillus plantarum (Lp84-3) can affect resistant starch (RS) degradation to produce short-chain fatty acids (SCFAs) (Liang et al., 2023). Although metagenomic sequencing can further determine microbial composition during food fermentation, deep knowledge of microbial function is currently limited. Carbohydrate metabolism is an important energy source for the bacteria in fermented foods, with previous studies reporting that Paenibacillus peoriae, Bacillus stratosphericus, Bacillus toyonensi, and Bacillus cereus produce cellulases and pectinases (Zhang et al., 2018). Under the joint action of microorganisms and enzymes, sugars and proteins in raw materials are degraded into small molecular metabolites such as glucose, fructose, amino acids and organic acids, which were metabolized into various secondary metabolites including aroma-active compounds. Carbohydrate-active enzymes (CAZymes) participate in the synthesis of sugar complexes, oligosaccharides, and polysaccharides and the decomposition of complex carbohydrates, especially glycosylhydrolases (GHs) and glycosyltransferases (GTs), which have potential carbohydrate metabolic activity and play an important role in the flavor of fermented foods (You, Yang, Jin, Kwok, Sun, & Zhang, 2022). GHs are mainly used to hydrolyze glycosidic bonds or carbohydrates and glycosidic parts between carbohydrates (Tingley, Low, Xing, & Abbott, 2021); therefore, their involvement in the metabolism of complex carbohydrates is crucial (Renaud, Berlemont, Adam, & Martiny, 2015). At present, 135 GH subfamilies (GH1-GH135) participate in the degradation of complex carbohydrates, such as α-mannosidases, which are commonly found in GH families GH38 and GH47 (van et al., 2001). GH3 and GH5 mainly hydrolyze cellulose and hemicellulose, respectively (Zou et al., 2023); GH65 contains two classes of important enzymes, trehalases and maltose phosphorylases (Egloff, Uppenberg, Haalck, & Tilbeurgh, 2001). Maltose phosphorylase can be produced by Lactobacillus brevis and families GT8 and GT6 participate in the α-1,4-galactosyltransferase and α-1,3-galactosyltransferase from Neisseria meningitidis. While these findings suggest that GHs and GTs are crucial for flavor formation in fermented foods, there is limited information on microbes CAZymes in fermented foods and their impact on its flavor. Therefore, selecting optimal and appropriate bacterial strains is necessary to produce a large number of CAZymes that facilitate the formation of flavor substances in fermented foods.
In this study, 51 fermented dairy and 47 fermented vegetable samples were collected and the microbes of all samples were analyzed using high-throughput 16S rDNA sequencing combined with metagenomic sequencing. We conducted a comparative analysis of CRISPR activity and diversity of LAB species in fermented dairy and vegetable products, and the functional strain L. plantarum 84-3 (Lp84-3), which was isolated from the fermented dairy products (Liang et al., 2023). Furthermore, we analyzed CAZymes of microbes in fermented dairy and vegetable samples. Meanwhile, the CAZymes profiles and metabolites of genome and fermentation supernatant in Lp84-3 were determined, respectively. This information will help select the best and most suitable strains for degrading complex carbohydrates to produce excellent flavors from fermented dairy products and vegetable products.
Materials and methods
Sample collection
We obtained 47 fermented vegetables and 51 fermented dairy samples were from China, South Africa, and Sri Lanka based on the geographical origin and climate of the fermented products (Table S1). We collected 50 mL liquid samples and 0.5 g solid samples; packaged them into sterilized centrifuge tubes or bags, respectively; and transported them in a dry ice box to the Guangdong Provincial Key Laboratory of Microbial Safety and Health, Guangdong Institute of Microbiology (Guangzhou, China), after which the samples were stored at −80 ℃ until further experimentation.
DNA extraction and sequencing
The E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) was used to extract microbial DNA. Paired-end sequencing was performed using an Illumina NovaSeq/Hiseq Xten (Illumina Inc., San Diego, CA, USA) (Yang et al., 2021). The detailed protocol has been described previously (Liang et al., 2021).
16S rRNA gene data processing
Data were analyzed using the free online Majorbio Cloud platform (https://www.majorbio.com). The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered using FASTP version 0.20.0 (Chen, Zhou, Chen, & Gu, 2018) and merged using FLASH version 1.2.7 (Magoč, & Salzberg, 2011). Operational taxonomic units (OTUs) with a 97 % similarity cutoff were clustered. Alpha diversity and microbial taxa were determined using the Quantitative Insights into Microbial Ecology (QIIME) software (https://qiime.org/scripts/assign_taxonomy.html). Beta diversity analysis based on Unifrac was performed with non-metric multidimensional scaling analysis (NMDS), which was used to compare the differences among samples (https://UniFrac.colorado.edu/).
Metagenomic data processing
Prodigal (Hyatt, Chen, LoCascio, Land, Larimer, & Hauser, 2010)/MetaGene (https://metagene.cb.k.u-tokyo.ac.jp/) (Noguchi, Park, & Takagi, 2006) was used to predict the open reading frames (ORFs) for each assembly contig, and CD-HIT (https://www.bioinformatics.org/cd-hit/, version 4.6.1) was used to identify phage contigs. Representative sequences were compared to a non-redundant (NR) database for taxonomic annotation (https://www.diamondsearch.org/index.php, version 0.8.35). Diamond was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation (https://www.diamondsearch.org/index.php, version 0.8.35). CAZyme annotation was performed in the CAZy database using hmmscan (https://www.cazy.org/).
Complete genome sequencing and analysis of Lp84-3 CAZyme expression profiles
Lp84-3 was isolated from fermented dairy samples using traditional isolation and culture methods with De Man, Rogosa, and Sharpe (MRS) medium. The PacBio Sequel and Illumina HiSeq 4000 platforms (BGI, Shenzhen, China) were used to sequence the Lp84-3 genome. Cutadapt (v1.9.1) and single-molecule real-time (SMRT) sequencing were used for single-base correction to improve genome sequencing accuracy. Filtered Illumina reads were mapped to a bacterial plasmid database using SOAP to identify the plasmids. The best hits used for function annotation were conducted with the BLAST 9 (version 2.2.31+) alignment tool. The CAZy database was used for CAZyme functional annotation. CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/) was used to detect the CRISPR sequence and Cas gene in Lp84-3.
Metabolite analysis of Lp84-3
For metabolite extraction, 100 μL fermentation supernatant were suspended in a solvent (methanol: acetonitrile = 2:1, v:v, 300 μL), vortexed for 1 min, and incubated at −20 ℃ for 2 h before being centrifuged at 10 000 rpm for 20 min at 4 °C. The supernatant was evaporated in a cryo-vacuum concentrator and then 150 μL complex solution (methanol: H2O = 1:1, v:v) was added for resolution, vortexed for 1 min, and centrifuged at 10,000 rpm for 30 min at 4 °C. The supernatant was removed to a sample bottle for liquid chromatography tandem mass spectrometry (LC-MS) analysis. Subsequently, 10 μL of supernatant from each sample were mixed as quality control (QC) samples to evaluate the repeatability and stability of the LC-MS analysis process.
The raw data collected by LC-MS were imported into Compound Discoverer 3.1 (Thermo Fisher Scientific, USA) for data processing, which mainly included peak extraction, intra-and inter-group retention time correction, adjoint ion merging, missing value filling, background peak labeling, and metabolite identification. Finally, the molecular weights, retention times, peak areas, and identification results of the compounds were collected. Due to some metabolites was not identified using one database, the identification of metabolites was performed using several databases, including the BGI Library, mzCloud, ChemSpider, Human Metabolome Database (HMDB), KEGG, and LipidMaps.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). Spearman’s correlation of bacteria and phages, bacteria, and CAZymes were performed using Cytoscape (ver. 3.7.1). A t-test was used to evaluate differences between the fermented dairy and vegetable product groups, with p values of <0.05 indicating a significant difference.
Results and discussion
Characteristics of microbial community diversity
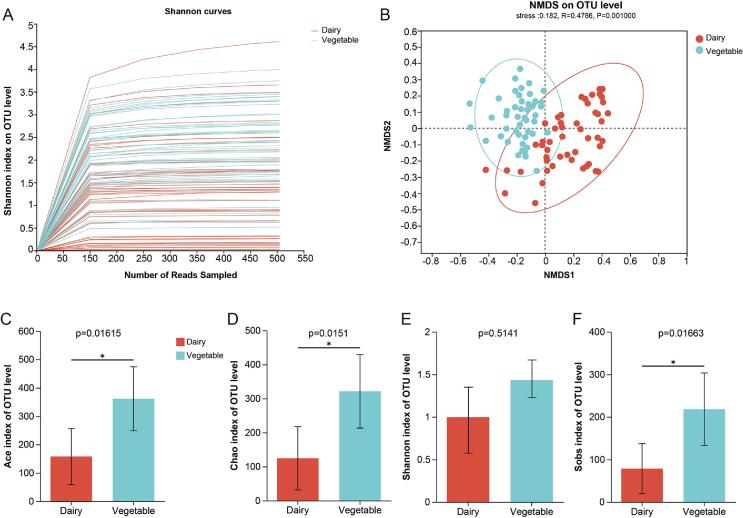
We obtained 4 932 916 reads from 98 fermented samples, which included 51 dairy and 47 vegetable samples. The Shannon curve indicated that the sequencing depth met the sequencing and analysis requirements (Fig. S1) and Good’s coverage estimators were 0.88–1.00 in all samples (Table S1), suggesting that the sequencing depth could predict the species richness of samples (Liu et al., 2019). The ACE and Chao1 indices represented microbial richness, whereas the Shannon and Simpson indices indicated microbial community diversity. Fig. S1C, D, E, F, and Table S1 show the differences in alpha diversity between dairy and vegetable products. The t-test results showed that ACE, Chao1, and Sobs indices of microbial composition in vegetable samples were significantly higher than those in dairy samples (p < 0.05), indicating a large abundance of microbial communities in fermented vegetable samples. A previous report suggested that the abundance of microbial communities of fermented dairy samples was low (Walsh, Crispie, O’Sullivan, Finnegan, Claesson, & Cotter, 2018), consistent with this and our other studies (Liang, Xie, Zhang, Ding, & Wu, 2021). However, no significant differences were observed between the dairy and vegetable samples with respect of the Shannon index (Fig. S1E, p < 0.05), which indicated that there was no significant difference in microbial community diversity between fermented dairy products and vegetable products. Beta-diversity analysis was used to explore the similarities or differences in bacterial community composition between the fermented dairy and vegetable samples. The results of NMDS analysis of the two groups were divided based on the microbiome at the OTU level, which showed that there were significant differences in the composition of microbes between the fermented vegetable and dairy products (Fig. S1B).
Microbial composition of fermented dairy and vegetables
The results from analyzing the microbial communities in fermented dairy and vegetables identified 27 phyla, 496 genera, and 784 species in all samples. At the phylum level (Fig. 1A), the four major phyla found in dairy and vegetable samples were Firmicutes (average mean of 84.2 % and 55.1 %, respectively), Proteobacteria (average mean of 10.8 % and 37.5 %, respectively), Actinobacteria (average mean of 2.2 % and 1.8 %, respectively), and Bacteroidetes (average mean of 2.0 % and 1.3 %, respectively), which were consistent with a study about the microbial community composition in fermented dairy (Liang, Xie, Zhang, Ding, & Wu, 2021) and vegetable products (Liang et al., 2022). Interestingly, our results demonstrated that the phyla Epsilonbacteraeota and Halanaerobiaeota were also present in the fermented vegetable samples, accounting for 3.1 % and 0.8 % of the composition, respectively (Fig. 1A), and were also observed in other traditional fermented products such as fermented stinky mandarin fish (Wu et al., 2022) and stinky eggs (Chen, Li, Gu, Tan, & Lu, 2020). In particular, Halanaerobiaeota was isolated from various hypersaline environments (Kushner, 2020) and were reported to accelerate fermentation and enhance the flavor of shrimp paste (Yu, Lu, Zi, Yang, Zheng, & Xie, 2022). We speculate that this is due to the extreme physical and chemical environments of fermented vegetable products that have very low water content and high salinity, which are conducive to the growth of complex microorganisms and the formation of flavors.
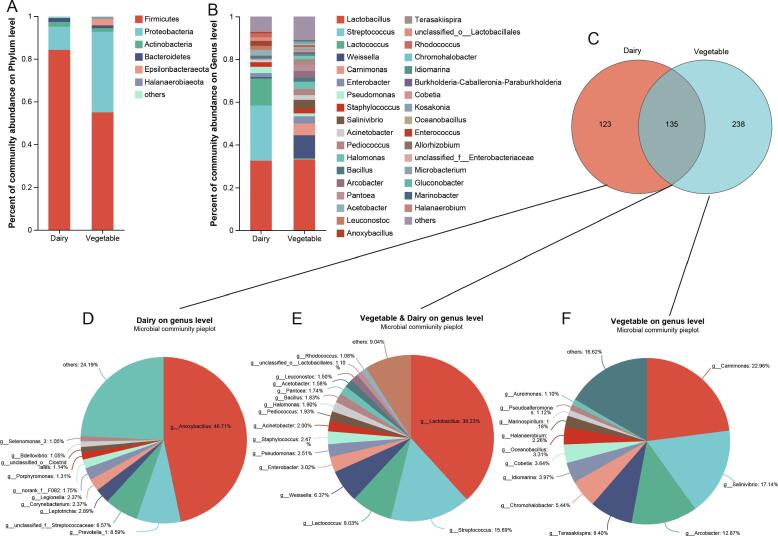
Fig. 1.
Microbial community structure of fermented dairy and vegetable products. (A) Relative abundance of the microbial community at the phylum level in fermented dairy and vegetable products. (B) Relative abundance of the microbial community at the genus level in fermented dairy and vegetable products. (C) Venn diagrams of the fermented dairy and vegetable products according to microbial biodiversity at the genus level. (D) The pieplot of unique microbial community in fermented dairy products at the genus level. (E) The pieplot of common microbial community in fermented dairy and vegetable products at the genus level. (F) The pieplot of unique microbial community in fermented vegetable products at the genus level.
At the genus level (Fig. 1B), the major genera in the fermented dairy products were Lactobacillus (32.6 %), Streptococcus (25.8 %), and Lactococcus (12.8 %), which was consistent with previous reports (Zhong et al., 2016). In fermented vegetable products, the following genus were majorly detected: Lactobacillus (33.1 %), Weissella (10.7 %), and Carnimonas (5.6 %), which was similar to studies on Paocai (Xiao, et al., 2020) as well as fermented green beans, bamboo shoots, and other fermented vegetables from Hainan (Peng et al., 2018). We identified 258 and 373 genera from fermented dairy and vegetable products, respectively; with 135 genera common between both samples (Fig. 1C). Lactobacillus, Streptococcus, Lactococcus, and Weissella were significantly enriched LAB genera in both fermented dairy and vegetable products (Fig. 1E), suggesting their importance in the production of both fermented dairy and vegetable products.
Several studies have reported that Lactobacillus is the most enriched genus in fermented food samples and can promote the production of organic acids, sugars, and other flavor metabolites as well as inhibit the growth of pathogenic bacteria (Chi, Tan, Gu, Yang, & Luo, 2021). For example, lactic acid bacterial fermentation can improve the flavor, bioactivity, and metabolic profiles of goji juice (Duan et al., 2023). Lactobacillus can also degrade polysaccharides in fermented dairy and vegetable products (Holzapfel & Wood, 2014); however, the possible mechanism of this property, such as whether it secretes enzymes, requires further study. Streptococcus and Lactococcus were more abundant in dairy samples than in vegetable samples in this study, which was consistent with a previous study that reported the major genera of bacteria in drinkable yogurt (Xu et al., 2015).
Notably, Lactobacillus, Streptococcus, Lactococcus, and Weissella were also enriched in fermented vegetable samples. The salted flowers of wild chive is a popular Asian fermented vegetable that has been reported to contain abundant Weissella (Liang et al., 2022). We found 238 unique genera, including Carnimonas, Salinivibrio, Arcobacter, Terasakiispira, and Chromohalobacter in the tested vegetable products (Fig. 1F). These bacteria were abundant in other traditionally fermented vegetables, such as pickled cucumber, pepper, and Brassica napobrassica (Liang et al., 2022).
We identified 123 unique genera in samples of the fermented dairy products, including Prevotella, Anoxybacillus, Leptotrichia, Corynebacterium, and Legionella (Fig. 1C, D). Prevotella bryantii 25A is a probiotic that reduces the risk of ruminal acidosis in dairy cows (Karaca, Karakaya, Ozcan, & Coleri, 2022); therefore, it may have originated from raw milk. Spore formation of Anoxybacillus occurs through their attachment to processed food surfaces, leading to a shortened shelf life of processed products (Karaca, Karakaya, Ozcan, & Coleri, 2022). In addition, Leptotrichia buccalis can cause bacteremia in patients (Ulstrup & Hartzen, 2006), whereas Corynebacterium pseudotuberculosis contamination resulted in severe mastitis in an Israeli dairy herd (Shpigel, Elad, Yeruham, Winkler, & Saran, 1993). Moreover, Legionella pneumophila infection in cattle can cause fatal pneumonia in calves (Fabbi, Pastoris, Scanziani, Magnino, & Di, 1998). Therefore, consumption of undercooked fermented dairy products may pose a safety risk to human consumers.
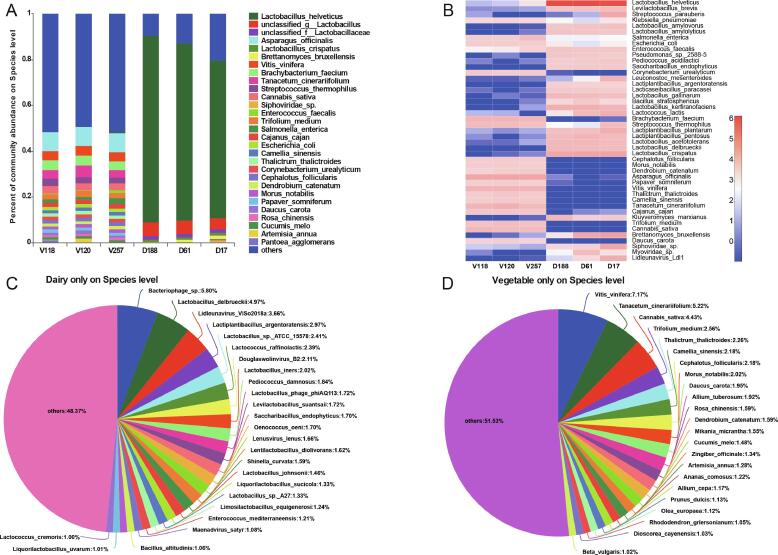
At the species level (Fig. 2A), the predominant species in dairy products were Lactobacillus helveticus (75.9 %) and Lactobacillus crispatus (1.1 %), which was similar to findings from a study that reported that the fermentation of dairy flavor profiles was significantly improved the LAB (Liang, Xie, Zhang, Ding, & Wu, 2021). However, other reports have suggested that L. delbrueckii is the most abundant LAB in yogurt samples (Suh & Kim, 2021), suggesting that the enriched species can differ depending on the types of fermented dairy sample. We also observed that Levilactobacillus brevis, Lactobacillus amylovorus, Lactobacillus amylolyticus, Lactiplantibacillus argentoratensis, Lacticaseibacillus paracasei, Lactobacillus gallinarum, Lactobacillus kerfiranofaciens, L. lactis, L. plantarum, Lactobacillus pentosus, and Leuconostoc mesenteroides were enriched in fermented dairy products (Fig. 2B, C), which can lead to high pH and low acidity. Therefore, isolating and identifying these gram-positive bacteria and other anaerobic species is necessary to explore the link between the microbial community structure and the flavor compounds of these fermented dairy products. However, the pathogenic bacterium Pseudomonas sp. 2588-5 was also present in these dairy samples, which has been widely reported in fresh dairy products due to food spoilage and can be transmitted to consumers (Quintieri, Fanelli, & Caputo, 2019). Therefore, appropriate measures must be taken to prevent contamination by opportunistic pathogens and other microorganisms.
Fig. 2.
Microbial community structure of fermented dairy and vegetable products at the species level. (A) Barplot of microbia communities in fermented dairy and vegetable products at the species level. (B) Heatmap of microbia communities in fermented dairy and vegetable products at the species level. (C) The pieplot of unique microbial community in fermented dairy products at the species level. (D) The pieplot of unique microbial community in fermented vegetable products at the species level.
Brachybacterium faecium (4.0 %) was the major species identified in vegetable products, with Klebsiella pneumoniae, Salmonella enterica, and Escherichia coli also abundant in the fermented vegetable samples (Fig. 2B, D), which are known as pathogenic bacteria. In general, simple washing of the vegetable surface is required before fermentation, suggesting that these fermented vegetable samples might be contaminated because of the lack of sterilization conditions.
Bacteriophage sequences
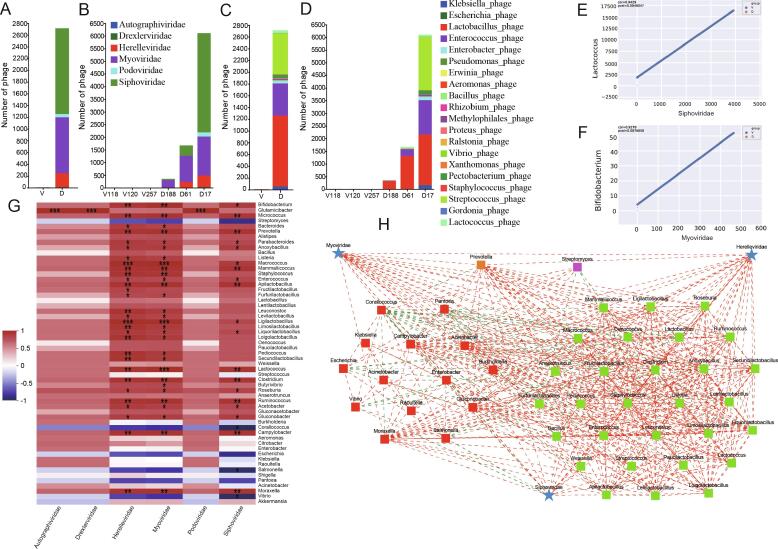
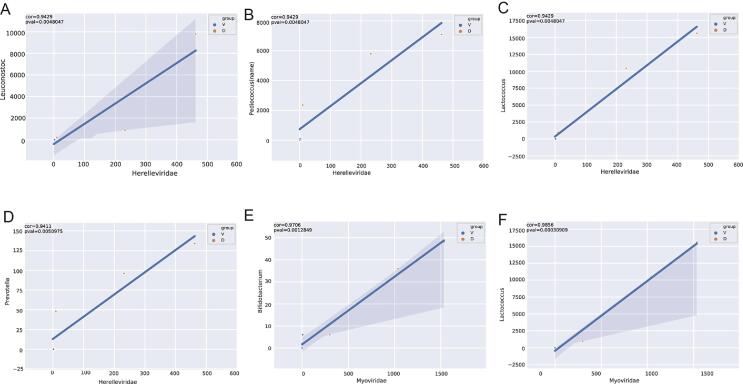
There is growing evidence that bacteriophages in fermented foods can directly affect the flavor and quality through their interactions with fermenting bacteria (White et al., 2022, Du et al., 2023). Previous studies have also indicated that the aroma from volatile acids such as acetic and propionic acids in starter cultures may be lost because of the destruction of aroma bacteria by specific phage strains (Ortiz, de, van, van, van, & Mahony, 2023). For example, Oenococcus oeni bacteriophages in wine may affect malolactic fermentation by interacting with the host bacteria (Costantini, Doria, Saiz, & Garcia-Moruno, 2017). The dominant phage families were Siphoviridae, Myoviridae, Herelleviridae, and Podoviridae in dairy products (Fig. 3A, B). A previous study also reported that dairy products like koumiss and yogurt were contaminated with these phages (You, Yang, Jin, Kwok, Sun, & Zhang, 2022). At the genus level, Streptococcus, Enterobacter, Enterococcus, Lactobacillus, and Klebsiella phages were detected only in dairy samples (Fig. 3C, D), with other studies reporting that L. lactis, Glutamicibacter, and Pseudoalteromonas genera phages are the most abundant in epoisse cheese (Dugat-Bony et al., 2020). Differences in geographical distribution, raw materials, and types of analytical technique may have contributed to the differences between the findings reported in these studies. Interestingly, we found linear correlations between Siphoviridae and Lactococcus and Myoviridae and Bifidobacterium (Fig. 3E, F; Fig. S2); whereas, nonlinear associations between Myoviridae and Lactobacillaceae and Sipoviridae and Lactobacillaceae have been previously reported (You, Yang, Jin, Kwok, Sun, & Zhang, 2022). These differences in results might be due to limited number of dairy samples included in this study and suggest that further research is needed to clarify the relationship between phage and bacterial populations in fermented dairy products.
Fig. 3.
Bacteriophage sequences in microbiome of fermented dairy and vegetable products. (A) Composition of the family-level phage metagenomes of the groups of fermented dairy and vegetable products. (B) Composition of the family-level phage metagenomes of the single samples of fermented dairy and vegetable products. (C) Composition of the genus-level phage metagenomes of the groups of fermented dairy and vegetable products. (D) Composition of the genus-level phage metagenomes of the single samples of fermented dairy and vegetable products. (E) Correlation between Lactococcus and Siphoviridae detected in the microbiome of fermented dairy and vegetable products. (F) Correlation between Bifidobacterium and Myoviridae detected in the microbiome of fermented dairy and vegetable products. (G) Heatmap analysis of correlation between bacteria and bacteriophages in fermented dairy and vegetable products. (H) Phage infection of fermented dairy and vegetable products-associated bacteria. *P < 0.05, **P < 0.01, ***P < 0.001.
A correlation heatmap and host-bacteriophage network were constructed to evaluate the relationship between bacteria and phages in fermented food products (Fig. 3G, H). The results indicated that the bacteriophage families Siphoviridae, Myoviridae, and Herelleviridae were associated with most of the abundant microbial populations, including 82 potential infection links. The most abundant bacteriophages in these three families were related to the phyla Firmicutes and Proteobacteria, suggesting that they are promiscuous. The Bacteroidetes genera, Prevotella, was linked with the families Myoviridae and Herelleviridae, which indicated that there were some phages related to more than one genus of bacteria in fermented dairy products. Together, these results showed that these bacteriophages may mainly infected the core bacteria involved in dairy-flavor fermentation (Du et al., 2023).
Generally, the bacterial CRISPR/CRISPR-associated proteins (Cas) system provides protection against phage re-infection in bacteria (Berg Miller et al., 2012). CRISPR elements were identified in LAB genomes using the microbial metagenome (Table S2), of which 17 strains of LAB had the Cas protein; L. helveticus, L. crispatus, Lactobacillus pasteurii, and Ligilactobacillus agilis had more than two Cas proteins, suggesting that these LAB were infected by different phages multiple times. There were seven types of Cas proteins in L. helveticus (Cas 1, 2, 3, 5, 7, 8b2, and 9), whereas L. crispatus (Cas 3, 5, 6, and 8b2) and L. pasteurii (Cas 1, 3, 5, and 6) had four types, and Ligilactobacillus agilis has two (Cas 3, 6e). The remaining 13 strains of LAB had only one Cas protein. These results suggested that LAB in fermented dairy samples have a defense system against phages and that LAB CRISPR/Cas elements play a role in regulating the community and food flavor (such as lactic acid).
Metagenomic function analysis
Mapping metagenomic ORFs to homologous gene clusters revealed an abundance of functional features through the use of KEGG pathway analysis and CAZymes annotation.
KEGG annotation profiles
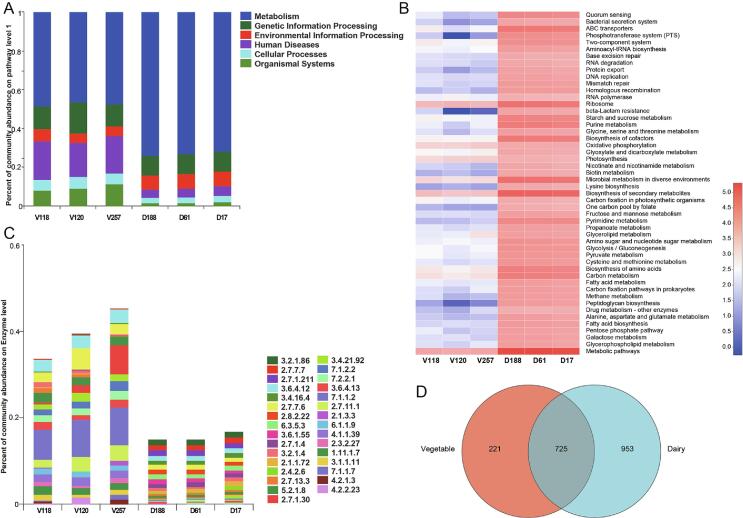
Our KEGG enrichment results for dairy and vegetable samples indicated that metabolism-related genes (73.1 % and 47.7 %, respectively) were the most enriched, followed by genes related to genetic information processing (10.2 % and 12.8 %, respectively), environmental information processing (7.4 % and 5.4 %, respectively), human diseases (4.6 % and 19.1 %, respectively), cellular processes (3.0 % and 5.6 %, respectively), and organismal systems (1.6 % and 9.4 %, respectively) (Fig. S3A). At level 2, carbohydrate metabolism was enriched in dairy and vegetable microbes (10.4 % and 5.5 %, respectively), suggesting that microbial communities, including both bacteria and bacteriophages, affected saccharification during dairy and vegetable fermentation (Du et al., 2023). This was followed by amino acid metabolism (4.4 % and 2.5 %, respectively), which are significant nutrients and flavor precursors in food fermentation that provide nitrogen sources for the growth of microbiota and contribute to the flavor of the food fermentation process (Cordente, Schmidt, Beltran, Torija, & Curtin, 2019). Further analysis indicated that glycolysis/gluconeogenesis (ko00010), citrate cycle (ko00020), pentose phosphate pathway (ko00030), pentose and glucuronate interconversions (ko00040), fructose and mannose metabolism (ko00051), galactose metabolism (ko00052), ascorbate and aldarate metabolism (ko00053), starch and sucrose metabolism (ko00500), as well as amino sugar and nucleotide sugar metabolism (ko00520) were abundant in dairy and vegetable microbe samples (Fig. S3B), suggesting that these metabolic pathways were the most important, which is consistent with the research results in fermented chili (Chen, Su, Mu, Jiang, & Qi, 2023). Moreover, 1678 and 946 enzymes were obtained from samples of dairy and vegetable products, respectively, with 725 enzymes common among dairy and vegetable products (Fig. S3D). The prevalent enzyme in the glycolysis/gluconeogenesis pathway was 6-phospho-beta-glucosidase (K01222, EC:3.2.1.86) and the prevalent enzyme in the starch and sucrose metabolism pathways was cellulase (K19357, EC:3.2.1.4) (Fig. S3C). A previous study emphasise that refractory carbohydrates (e.g. marine algal polysaccharides) can influence the fermentation process, which can promote the growth of beneficial microorganisms, ultimately leading to decreases in methane flavor emissions (Cheong et al., 2023). These results suggested that the genes involved in carbohydrate metabolism pathways were abundant in the fermented dairy and vegetable samples, and may contribute to the degradation of insoluble carbohydrates to produce flavor compounds and function in the final products.
CAZyme annotation profiles
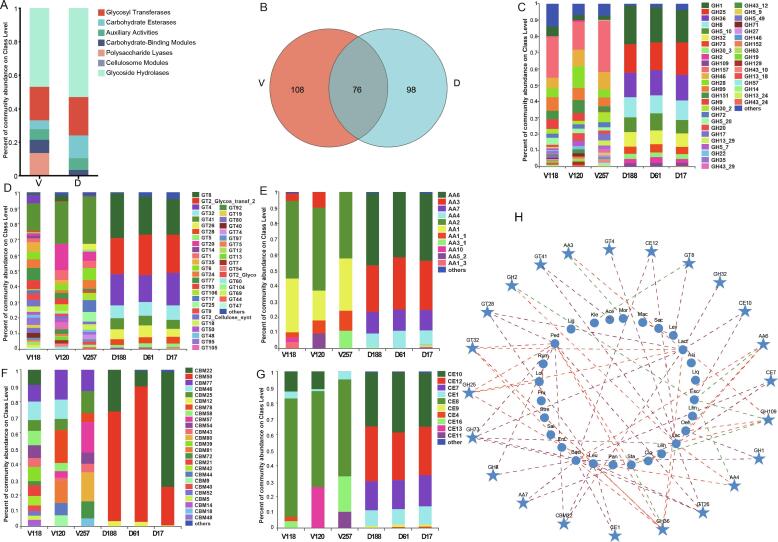
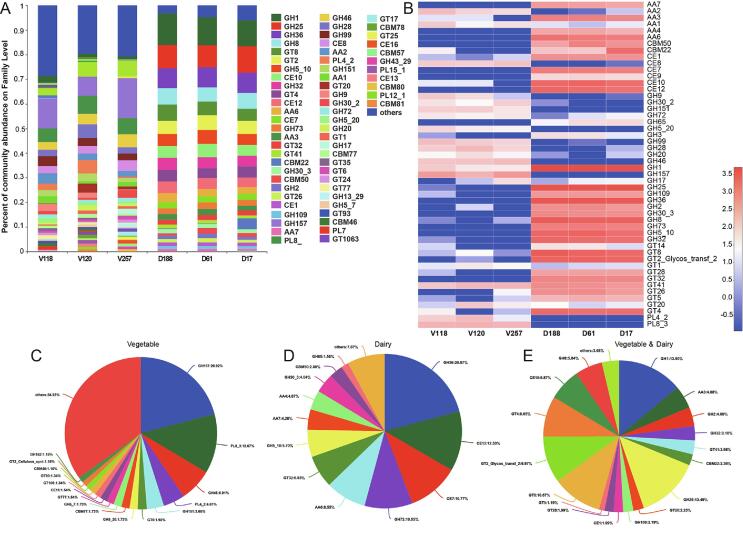
To investigate the microbial enzymes involved in carbohydrate metabolism in fermented dairy and vegetable products, scaffolds were searched based on gene families in the CAZy database (Fig. S4; Fig. 4). Venn analysis revealed that the fermented vegetable products contained unique CAZymes (Fig. 4B). The GHs (53.1 % and 47.0 %, respectively) and GTs (22.8 % and 20.1 %, respectively) were enriched in fermented dairy and vegetable product microbes, followed by carbohydrate esterases (CEs; 13.7 % and 5.1 %, respectively), auxiliary activities (AAs; 7.0 % and 6.5 %, respectively), and carbohydrate-binding modules (CBMs; 3.2 % and 7.8 %, respectively). Polysaccharide lyases were more abundant in fermented vegetable than in dairy microbe samples (Fig. 4A), which may be due to the high cellulose content that provides an abundant carbon source for the growth of microorganisms in fermented vegetable products.
Fig. 4.
CAZyme annotation profile of fermented dairy and vegetable products microbiome. (A) Overview of CAZyems classes in total fermented dairy and vegetable products microbes. (B) Venn diagrams of CAZymes subfamilies in the fermented dairy and vegetable products. (C) GH subfamily members in the fermented dairy and vegetable products. (D) GT subfamily members in the fermented dairy and vegetable products. (E) AA subfamily members in the fermented dairy and vegetable products. (F) CBM subfamily members in the fermented dairy and vegetable products. (G) CE subfamily members in the fermented dairy and vegetable products. (H) Correlation network of the dominant microbial and CAZymes subfamilies. CAZyme: carbohydrate-active enzyme; GH: Glycoside Hydrolases; GT: GlycosylTransferases; AA: Auxiliary Activities;CBM: Carbohydrate Binding Modules; CE: Carbohydrate Esterases. Mac: Macrococcus; Mor: Moraxella; Sec: Secundilactobacillus; Lev: Levilactobacillus; Lact: Lactococcus; Aci: Acinetobacter; Liq: Liquorilactobacillus; Esc: Escherichia; Lim: Limosilactobacillus; Oen: Oenococcus; Lac: Lactobacillus; Len: Lentilactobacillus; Clo: Clostridium; Sta: Staphylococcus; Pan: Pantoea; Leu: Leuconostoc; Baci: Bacillus; Ent: Enterococcus; Sal: Salmonella; Stre: Streptococcus; Fru: Fructilactobacillus; Loi: Loigolactobacillus; Rum: Ruminococcus; Ped: Pediococcus; Lig: Ligilactobacillus; Kle: Klebsiella; Ace: Acetobacter.
The top 10 most abundant GH subfamilies, including GH1, GH25, GH36, GH8, and GH510, were abundant in dairy products, whereas GH157, GH46, GH28, GH99, and GH151 were prevalent in vegetable products (Fig. 4C). Additionally, GT8, GT2, GT4, GT32, and GT41 were abundant in dairy products, whereas GT41, GT26, GT5, and GT20 were prevalent in vegetable products (Fig. 4D). The sequences that mapped to the CE10, CE12, CE7, and CE1 families were abundant in dairy products, whereas the CE10, CE1, CE8, CE16, CE13, and CE11 families were abundant in vegetable products (Fig. 4G). In addition, some metagenomic sequences were mapped to AA6, AA3, AA7, and AA4 in dairy products, and to AA3, AA2, AA1, AA1-1, AA3-1, AA10, AA5-2, and AA1-3 in vegetable products (Fig. 4E). Moreover, the families, CBM22, CBM50, and CBM12 families were enriched in dairy products, whereas the CBM77, CBM46, CBM25, CBM78, and CBM18 enzyme families were represented by ORFs in the metagenome of vegetable products (Fig. 4F). These results indicated that there were many CAZymes in fermented dairy and vegetable food microbes and might suggest that the microbial communities can degrade different polymers into glucose in fermented foods (Yi et al., 2019), and are likely associated with the final product flavor.
Notably, the GH1, GH2, GH5, GH8, GH13, GH30, and GH99 families are linked to pectin, cellulose, hemicellulose and starch, and promote polysaccharide degradation by β-glucosidase (EC 3.2.1.21), β-galactosidase (EC 3.2.1.23), β-1, 4-mannanase (EC 3.2.1.78), cellulase (EC 3.2.1.4), α-amylase (EC 3.2.1.1) and α-1,2 mannoside (Singh, Adsul, Vaishnav, Mathur, & Singhania, 2017). Moreover, GT8 is a well-known lipopolysaccharide α-1,3-galactosyltransferase (EC 2.4.1.44) that uses polysaccharides as substrates (Scott, Stéphane, Marie-France, Mandrell, Jarrell, Wakarchuk, et al. 2009). Therefore, we hypothesized that many GHs and GTs are secreted by fermented dairy and vegetable microorganisms, which reflects the importance CAZyme function in the flavor development of fermented foods.
In general, the formation of one flavor may be the result of synergistic interactions between microorganisms (Dzialo, Park, Steensels, Lievens, & Verstrepen, 2017). Our results showed that dairy and vegetable microorganisms, CAZymes, and Lactobacillus were positively correlated with GT28, GT32, GH25, GH73, GH36, GH109, CE10 and CBM22. Lactococcus was positively correlated with GH36, GT26, AA4, GH109, CE7, AA6, CE2, GT4, GT41, GH2, GH73, and AA7; Leuconostoc was positively correlated with CBM22, CE1, GH36, GT26, GT28, GT32, and GH25; and Pediococcus was positively correlated with GT28, GT32, GH25, GH73, GH8, CBM22, GH36, and GT26. However, Escherichia was positively correlated with CE10 and negatively correlated with AA6, GH109, AA4, and GH2; whereas Staphylococcus was negatively correlated with GH36 and GH109 expression (Fig. 4H). Previous studies have shown that Bacillus licheniformis is the main functional bacteria responsible for saccharification and flavor production during Baijiu fermentation (Du et al., 2023). Based on the our results, Lactobacillus and Lactococcus were closely related to the GHs and GTs subfamilies, suggesting that these fermented dairy and vegetable microbes may play an important role in the formation of unique flavor substances through the GHs- and GT-mediated degradation of polysaccharides and insoluble dietary fiber, especially those in the Lactobacillus and Lactococcus genera.
Analysis of KEGG and CAZymes profiles in Lp84-3
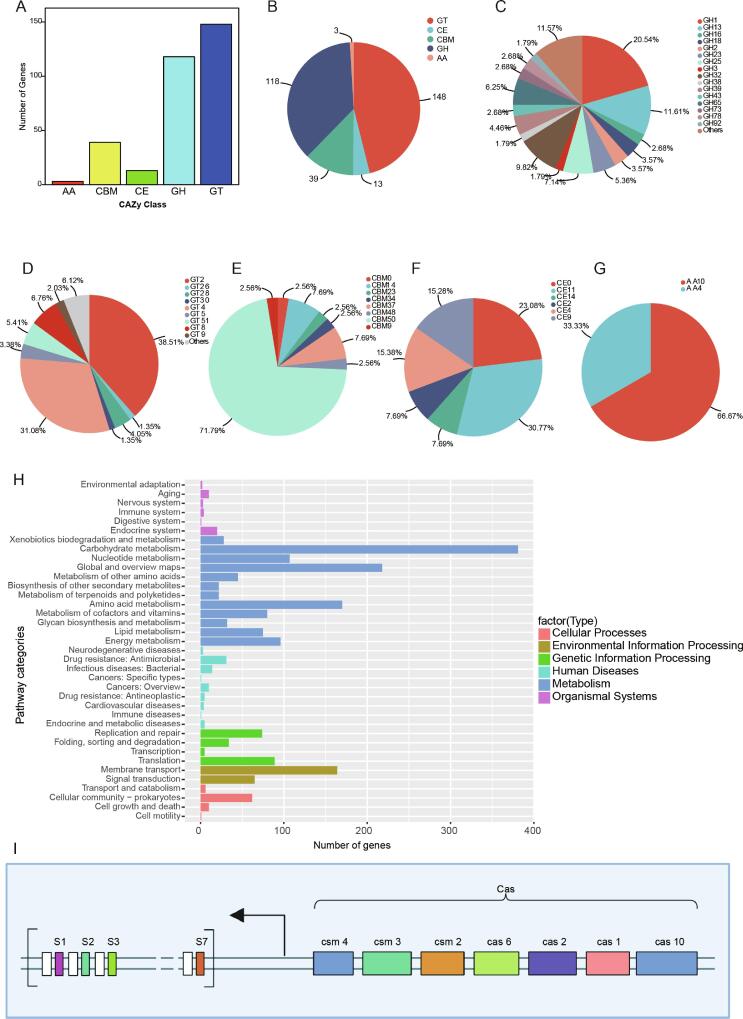
Studies have indicated that Lactipllantbacillus plantarum can convert carbohydrate into lactic acid via the Embden-Meyerhof pathway (EMP), promoting the formation of flavor (Zhang, Zeng, Hohn, & Vadlani, 2016). Lp84-3 isolated from fermented dairy products produced significantly more butyrate and enhanced RS degradation when compared with strains of L. plantarum 61-1, Limosilactobacillus fermentum 87-1, and Lactobacillus delbrueckii 42-1(Liang, et al., 2023). However, its mechanism is yet to be investigated. Using KEGG database annotation, 1576 functional genes were annotated in the Lp84-3 genome, of which 381 were mostly involved in carbohydrate metabolism (Fig. 5H). The CAZyme profile analysis of Lp84-3 identified 321 CAZymes, 148 GTs, 118 GHs, 39 CBMs, 13 CEs, and 3 AAs (Fig. 5A, B). Among these, GHs (GH1, GH13, GH23, GH25, GH32, and GH65), GTs (GT2 and GT4), CBMs (CBM14, CBM37, and CBM50), CEs (CE0, CE11, and CE4), and AAs (AA4 and AA10) were abundant in Lp84-3 (Fig. 5C, D, E, F, G). The GH, GT, and CE subfamilies of Lp84-3 were also investigated (Table S3) and numerous carbohydrate-degrading enzymes were identified that belonged to GH1, GH13, GH32, GH25, GH65, GT2, GT4, GT8, GT51, and CE11 families that function to degrade complex carbohydrates (e.g. RS) and give fermented food unique flavors (Table S3).
Fig. 5.
Basic information of CAZyme annotation profile in Lactiplantibacillus plantarum strain 84-3. (A) Barplot of CAZyems classes in Lp84-3. (B) Pieplot of CAZyems classes in Lp84-3. (C) GH subfamily members in Lp84-3. (D) GT subfamily members in Lp84-3. (E) CBM subfamily members in Lp84-3. (F) CE subfamily members in Lp84-3. (G) AA subfamily members in Lp84-3. (H) The functional pathways in Lp84-3 (Level 2). (I) Structure of the CRISPR Locus in Lp84-3.
Analysis of the key enzymes and genes associated with butyrate metabolism expressed in Lp84-3
Lactobacillus, as the dominant bacterial genus, can use carbohydrate decomposed sugars as energy sources to degrade them into flavor substances such as volatile fatty acids (Jang et al., 2015). In our previous study, Lp84-3 enhanced RS degradation to promote the production of SCFAs flavor compounds, especially butyric acid, which was significantly increased when compared with strains of L. plantarum 61-1, Limosilactobacillus fermentum 87-1, and Lactobacillus delbrueckii 42-1 (Liang, et al., 2023). However, the mechanism of this effect is yet to be elucidated. The key enzymes and genes related to butyrate metabolism were significantly enriched with enzymes in the Lp84-3 genome (Table S4), including the enzymes of EC:1.1.1. -, EC: 4.1.1.5, EC: 2.2.1.6, EC: 1.2.1.16, EC: 1.2.1.10, EC: 2.3.1.54, EC: 4.1.1.15, EC: 2.6.1.19, EC: 4.2.1. -, EC: 2.3.3.10, and EC: 2.8.3.8. Butyrate-producing bacteria mainly form butyric acid via the butyrate kinase and butyryl-CoA:acetate-CoA transfer (BUT) pathways (Vital et al., 2013). We found that the expression of enzymes related to the BUT pathway of butyric acid metabolism were enriched in the Lp84-3 genome, including EC:4.1.1.5, EC:2.2.1.6, EC:2.3.1.54, EC:2.3.3.10, and EC:2.8.3.8. In addition, the Lp84-3 genome was significantly enriched in other key enzymes involved in butyric acid metabolism, including gpr, alsD, budA, aldC, ilvB, ilvG, ilvI, gabD, adhE, pflD, gadB, gadA, GAD, puuE, hpaH, and citF (BUT). Therefore, Lp84-3 could promote butyric acid production to mediate RS via the BUT pathway, potentially through the expression of enzyme of EC:2.8.3.8 and the gene of citF.
Analysis of Lp84-3 carbohydrate and butyrate metabolites in the fermentation supernatant
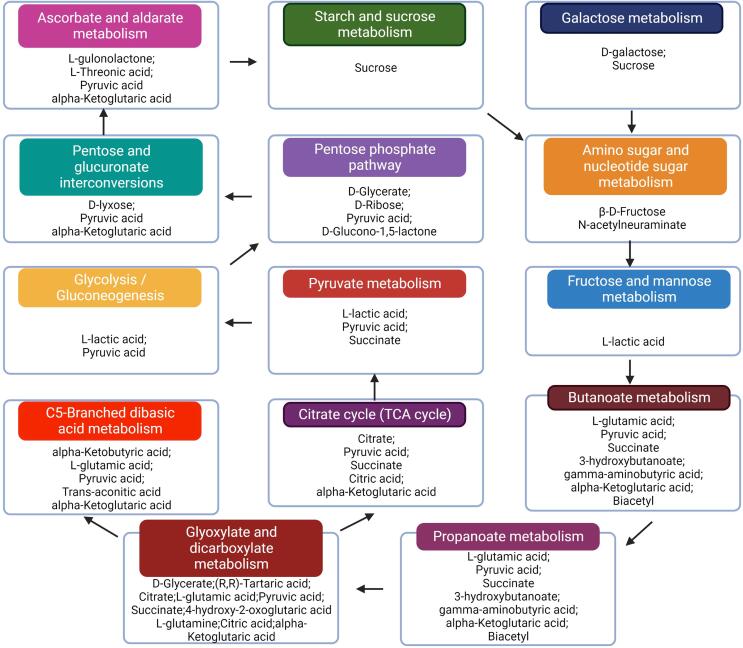
We identified 42 and 12 metabolites in pathways related to carbohydrate metabolism in the Electrospray ionization (ESI)− and ESI+ modes, respectively, that were involved in pentose phosphate pathway (4), amino sugar and nucleotide sugar metabolism (2), starch and sucrose metabolism (1), glyoxylate and dicarboxylate metabolism (10), glycolysis/gluconeogenesis (2), butanoate metabolism (7), ascorbate and aldarate metabolism (4), propanoate metabolism (5), citrate cycle (5), galactose metabolism (2), fructose and mannose metabolism (1), pentose and glucuronate interconversions (3), pyruvate metabolism (3), and C5-Branched dibasic acid metabolism (5) (Fig. 6, Table S5). Overall, Lp84-3 fermentation promoted the degradation of complex carbohydrates and the transformation of functional substances. The fermentation broth of Lp84-3 was significantly enriched in compounds related to the butyric acid metabolic pathway, including lactic, pyruvic, γ-aminobutyric, α-ketone glutaric, l-glutamic, and succinic acids (Table S6). We speculate that Lp84-3 mediates RS and regulates butyric acid production through lactic acid conversion; however, testing this hypothesis will require further research.
Fig. 6.
Metabolic network between the specific carbohydrate metabolism pathways in Lactiplantibacillus plantarum strain 84-3.
Analysis of the CRISPR-Cas system in Lp84-3
Two suspected CRISPR regions in the genome of Lp84-were identified using the CRISPRCasFinder software. Although these regions had simple repetitive intervals, they contained only one interval and did not contain Cas (Table S7). In addition, by analyzing the CRISPR system in Lp84-3, we found that seven Cas genes (Cas1, Cas2, Cas6, Cas10, Csm2, Csm3, and Csm4) were located on the side of the CRISPR sequence, which has a sequence length of 536 bp and contained seven spacer sequences (Fig. 5I). Previous studies have shown that the greater the number of spacer sequences, the higher the loci activity (Jackson, McKenzie, Fagerlund, Kieper, Fineran, & Brouns, 2017). Therefore, we speculated that Lp84-3 can specifically recognize foreign nucleic acid fragments (e.g., viral phages and plasmids), perhaps particularly those of antibacteriophages. Meanwhile, Lp84-3 can as a appropriate starter for the corresponding products to ensure the stability of product flavor.
Conclusions
In the present study, we characterized and compared the microbial diversity in and CAZyme profiles of fermented dairy and vegetable samples. Lactobacillus, Streptococcus, and Lactococcus constituted large proportions of the genera in dairy products, whereas the major genera present in the fermented vegetable products included Lactobacillus, Weissella, and Carnimonas. Many bacteriophage sequences were detected in fermented dairy products, including Siphoviridae, Myoviridae, Herelleviridae, and Podoviridae. Additionally, GHs, and GTs were abundant in fermented dairy and vegetable product microbes, and Lp84-3 isolated from dairy samples contained 148 GTs and 118 GHs being distributed, GHs families GH1, GH13, GTs families, GT2, and GT4 in high abundance, which might indicate that genes related to the degradation of complex carbohydrates produce the flavor. Additionally, Lp84-3 was significantly enriched in the key enzymes in the butyric acid metabolism pathway. Seven Cas genes were identified on the side of the CRISPR sequence in the Lp84-3 genome. These findings provide new insights into the mechanisms of flavor substance formation in fermented dairy and vegetable products, and enhance our understanding of how complex carbohydrate degradation contributes to the production of flavors during Lp84-3 fermentation, and Lp84-3 can be used as a starter for fermented foods production in the future.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This study was supported by research grants from the National Natural Science Foundation of China (81871734), Research foundation for advanced talents of Guandong Provincial People's Hospital (KJ012021097), Guangdong Province Basic and Applied Basic Research Fund Project (2022A1515110447).
Ethical approval
This study does not involve any human or animal testing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2023.101036.
Contributor Information
Tingting Liang, Email: 1320199507@qq.com.
Bo Dong, Email: dahe_78@163.com.
Qingping Wu, Email: wuqp203@163.com.
Bing Gu, Email: gubing@gdph.org.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
Supplementary figure 4.
Data availability
Data will be made available on request.
References
- Aziz T., Naveed M., Jabeen K., Shabbir M.A., Sarwar A., Zhennai Y., et al. Integrated genome based evaluation of safety and probiotic characteristics of Lactiplantibacillus plantarum YW11 isolated from Tibetan kefir. Frontiers in Microbiology. 2023;14:1157615. doi: 10.3389/fmicb.2023.1157615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz T., Naveed M., Makhdoom S.I., Ali U., Mughal M.S., Sarwar A., et al. Genome investigation and functional annotation of Lactiplantibacillus plantarum YW11 revealing streptin and Ruminococcin-A as potent nutritive bacteriocins against gut symbiotic pathogens. Molecules. 2023;28(2):491. doi: 10.3390/molecules28020491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz T., Naveed M., Sarwar A., Makhdoom S.I., Mughal M.S., Ali U., et al. Functional annotation of Lactiplantibacillus plantarum 13–3 as a potential starter probiotic involved in the food safety of fermented products. Molecules. 2022;27(17):5399. doi: 10.3390/molecules27175399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg Miller M.E., Yeoman C.J., Chia N., Tringe S.G., Angly F.E., Edwards R.A., et al. Phage–bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environmental Microbiology. 2012;14(1):207–227. doi: 10.1111/j.1462-2920.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Su W., Mu Y., Jiang L., Qi Q. Study on the quality formation mechanism of Zao chili with enhanced fermentation by Lactipllantbacillus plantarum 5–1. Food Chemistry: X. 2023;17 doi: 10.1016/j.fochx.2023.100626. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen W., Li Y., Gu X., Tan S., Lu H. Diversity of microbial community and key substances in naturally fermented stinky egg. Food science. 2020;41(22):158–165. [Google Scholar]
- Cheong K.L., Zhang Y., Li Z., Li T., Ou Y., Shen J., et al. Role of polysaccharides from marine seaweed as feed additives for methane mitigation in ruminants: A critical review. Polymers. 2023;15(15):3153. doi: 10.3390/polym15153153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi F., Tan Z., Gu X., Yang L., Luo Z. Bacterial community diversity of yak milk dreg collected from Nyingchi region of Tibet, China. LWT-Food Science and Technology-Food Science and Technology. 2021;145 [Google Scholar]
- Cordente A.G., Schmidt S., Beltran G., Torija M.J., Curtin C.D. Harnessing yeast metabolism of aromatic amino acids for fermented beverage bioflavouring and bioproduction. Applied Microbiology and Biotechnology. 2019;103:4325–4336. doi: 10.1007/s00253-019-09840-w. [DOI] [PubMed] [Google Scholar]
- Costantini A., Doria F., Saiz J.C., Garcia-Moruno E. Phage-host interactions analysis of newly characterized Oenococcus oeni bacteriophages: Implications for malolactic fermentation in wine. International Journal of Food Microbiology. 2017;246:12–19. doi: 10.1016/j.ijfoodmicro.2017.01.020. [DOI] [PubMed] [Google Scholar]
- Du H., Chen B., Fu W., Yang F., Lv X., Tan Y., et al. Composition and function of viruses in sauce-flavor baijiu fermentation. International Journal of Food Microbiology. 2023;387 doi: 10.1016/j.ijfoodmicro.2022.110055. [DOI] [PubMed] [Google Scholar]
- Duan W., Guan Q., Zhang H.L., Wang F.Z., Lu R., Li D.M., et al. Improving flavor, bioactivity, and changing metabolic profiles of goji juice by selected lactic acid bacteria fermentation. Food Chemistry. 2023;408 doi: 10.1016/j.foodchem.2022.135155. [DOI] [PubMed] [Google Scholar]
- Dugat-Bony E., Lossouarn J., De Paepe M., Sarthou A.-S., Fedala Y., Petit M.-A., et al. Viral metagenomic analysis of the cheese surface: A comparative study of rapid procedures for extracting viral particles. Food Microbiology. 2020;85 doi: 10.1016/j.fm.2019.103278. [DOI] [PubMed] [Google Scholar]
- Dzialo M.C., Park R., Steensels J., Lievens B., Verstrepen K.J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiology Reviews. 2017;41(Supp_1):S95–S128. doi: 10.1093/femsre/fux031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Uppenberg J., Haalck L., Tilbeurgh H.V. Crystal structure of maltose phosphorylase from Lactobacillus brevis: Unexpected evolutionary relationship with glucoamylases. Structure. 2001;9(8):689–697. doi: 10.1016/s0969-2126(01)00626-8. [DOI] [PubMed] [Google Scholar]
- Fabbi M., Pastoris M.C., Scanziani E., Magnino S., Di Matteo L. Epidemiological and environmental investigations of Legionella pneumophila infection in cattle and case report of fatal pneumonia in a calf. Journal of Clinical Microbiology. 1998;36(7):1942–1947. doi: 10.1128/jcm.36.7.1942-1947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel W.H., Wood B.J. John Wiley & Sons; 2014. Lactic acid bacteria: Biodiversity and taxonomy. [Google Scholar]
- Horvath P., Romero D.A., Coûté-Monvoisin A., Richards M., Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. Journal of Bacteriology. 2008;190(4):1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q., Li C., Liu Y., Li W., Chen Y., Siqinbateer B., et al. Koumiss consumption modulates gut microbiota, increases plasma high density cholesterol, decreases immunoglobulin G and albumin. Journal of Functional Foods. 2019;52:469–478. [Google Scholar]
- Hyatt D., Chen G.L., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:1–11. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J. CRISPR-Cas: Adapting to change. Science. 2017;356(6333):eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- Jang G.J., Kim D.W., Gu E.J., Song S.H., Lee J.I., Lee S.B., et al. GC/MS-based metabolomic analysis of the radish water kimchi, Dongchimi, with different salts. Food Science and Biotechnology. 2015;24:1967–1972. [Google Scholar]
- Karaca B., Karakaya A.B., Ozcan B., Coleri Cihan A. Rapid detection of Geobacillus and Anoxybacillus species by quantitative qPCR (qPCR) in commercial dairy products. Journal of Food Safety. 2022;42(2):e12964. [Google Scholar]
- Kushner D.J. The biology of halophilic bacteria. CRC Press; 2020. Growth and nutrition of halophilic bacteria; pp. 87–103. [Google Scholar]
- Liang T., Xie X., Ma J., Wu L., Xi Y., Zhao H., et al. Microbial communities and physicochemical characteristics of traditional Dajiang and Sufu in north China revealed by high-throughput sequencing of 16S rRNA. Frontiers in Microbiology. 2021;12 doi: 10.3389/fmicb.2021.665243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Xie X., Wu L., Li L., Li H., Xi Y., et al. Microbial communities and physiochemical properties of four distinctive traditionally fermented vegetables from north China and their influence on quality and Safety. Foods. 2022;11(1):21. doi: 10.3390/foods11010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Xie X., Wu L., Li L., Yang L., Jiang T., et al. Metabolism of resistant starch RS3 administered in combination with Lactiplantibacillus plantarum strain 84–3 by human gut microbiota in simulated fermentation experiments in vitro and in a rat model. Food Chemistry. 2023 doi: 10.1016/j.foodchem.2023.135412. [DOI] [PubMed] [Google Scholar]
- Liang T., Xie X., Zhang J., Ding Y., Wu Q. Bacterial community and composition of different traditional fermented dairy products in China, South Africa, and Sri Lanka by high-throughput sequencing of 16S rRNA genes. LWT- Food Science and Technology. 2021;144(7) [Google Scholar]
- Liu Z., Li J., Wei B., Huang T., Xiao Y., Peng Z., et al. Bacterial community and composition in Jiang-shui and Suan-cai revealed by high-throughput sequencing of 16S rRNA. International Journal of Food Microbiology. 2019;306 doi: 10.1016/j.ijfoodmicro.2019.108271. [DOI] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J., Moscarelli A., Kelleher P., Lugli G.A., Ventura M., Settanni L., et al. Phage biodiversity in artisanal cheese wheys reflects the complexity of the fermentation process. Viruses. 2017;9(3):45. doi: 10.3390/v9030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccum B.A., Kastman E.K., Kfoury N., Robbat A., Jr, Wolfe B.E. Strain-level diversity impacts cheese rind microbiome assembly and function. Msystems. 2020;5(3):e00149–e00220. doi: 10.1128/mSystems.00149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H., Park J., Takagi T. MetaGene: Prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Research. 2006;34(19):5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz Charneco G., de Waal P.P., van Rijswijck I.M., van Peij N.N., van Sinderen D., Mahony J. Bacteriophages in the dairy industry: A problem solved? Annual Review of Food Science and Technology. 2023;14:367–385. doi: 10.1146/annurev-food-060721-015928. [DOI] [PubMed] [Google Scholar]
- Peng Q., Jiang S., Chen J., Ma C., Huo D., Shao Y., et al. Unique microbial diversity and metabolic pathway features of fermented vegetables from Hainan, China. Frontiers in Microbiology. 2018;9:399. doi: 10.3389/fmicb.2018.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintieri L., Fanelli F., Caputo L. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: An underestimated risk and the control strategies. Foods. 2019;8(9):372. doi: 10.3390/foods8090372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud, Berlemont, Adam, Martiny Genomic potential for polysaccharide deconstruction in bacteria. Applied & Environmental Microbiology. 2015 doi: 10.1128/AEM.03718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott H.R., Stéphane B., Marie-France K., Mandrell R.E., Jarrell H.C., Wakarchuk W.W., et al. Complete chemoenzymatic synthesis of the Forssman antigen using novel glycosyltransferases identified in Campylobacter jejuni and Pasteurella multocida. Glycobiology. 2009;2:153–159. doi: 10.1093/glycob/cwn117. [DOI] [PubMed] [Google Scholar]
- Shpigel N., Elad D., Yeruham I., Winkler M., Saran A. An outbreak of Corynebacterium pseudotuberculosis infection in an Israeli dairy herd. The Veterinary Record. 1993;133(4):89–94. doi: 10.1136/vr.133.4.89. [DOI] [PubMed] [Google Scholar]
- Singh, A., Adsul, M., Vaishnav, N., Mathur, A., & Singhania, R. R. (2017). Improved cellulase production by Penicillium janthinellum mutant.
- Spus M., Li M., Alexeeva S., Wolkers-Rooijackers J.C.M., Zwietering M.H., Abee T., et al. Strain diversity and phage resistance in complex dairy starter cultures. Journal of Dairy Science. 2015;98(8):5173–5182. doi: 10.3168/jds.2015-9535. [DOI] [PubMed] [Google Scholar]
- Suh S.H., Kim M.K. Microbial communities related to sensory characteristics of commercial drinkable yogurt products in Korea. Innovative Food Science & Emerging Technologies. 2021;67 [Google Scholar]
- Tingley J.P., Low K.E., Xing X., Abbott D.W. Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues. Biotechnology for Biofuels. 2021;14(1):1–19. doi: 10.1186/s13068-020-01869-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulstrup A.K., Hartzen S.H. Leptotrichia buccalis: A rare cause of bacteraemia in non-neutropenic patients. Scandinavian Journal of Infectious Diseases. 2006;38(8):712–716. doi: 10.1080/00365540500452465. [DOI] [PubMed] [Google Scholar]
- van, den, Elsen, J., M., H., Kuntz, D., A., & Rose. (2001). Structure of Golgi alpha-mannosidase II: a target for inhibition of growth and metastasis of cancer cells. EMBO Journal. [DOI] [PMC free article] [PubMed]
- Vital M., Penton C.R., Wang Q., Young V.B., Antonopoulos D.A., Sogin M.L., et al. A gene-targeted approach to investigate the intestinal butyrate-producing bacterialcommunity. Microbiome. 2013;1:1–14. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh A.M., Crispie F., O’Sullivan O., Finnegan L., Claesson M.J., Cotter P.D. Species classifier choice is a key consideration when analysing low-complexity food microbiome data. Microbiome. 2018;6:1–15. doi: 10.1186/s40168-018-0437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Yu J.-H., Eraclio G., Dal Bello F., Nauta A., Mahony J., et al. Bacteriophage-host interactions as a platform to establish the role of phages in modulating the microbial composition of fermented foods. Microbiome Research Reports. 2022;1(1):3. doi: 10.20517/mrr.2021.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang Y., Yuan D., Zhang J., Chu L., Wang T., et al. Effects of ultra-high pressure treatment on bacterial community structure and quality of smelly Mandarin fish. Food Science. 2022;007 [Google Scholar]
- Xiao M., Huang T., Huang C., Hardie J., Peng Z., Xie M., et al. The microbial communities and flavour compounds of Jiangxi yancai, Sichuan paocai and Dongbei suancai: Three major types of traditional Chinese fermented vegetables. LWT-Food Science and Technology. 2020;121 [Google Scholar]
- Xu H., Liu W., Gesudu Q., Sun Z., Zhang J., Guo Z., et al. Assessment of the bacterial and fungal diversity in home-made yoghurts of Xinjiang, China by pyrosequencing. Journal of the Science of Food and Agriculture. 2015;95(10):2007–2015. doi: 10.1002/jsfa.6912. [DOI] [PubMed] [Google Scholar]
- Yang C., You L., Kwok L.Y., Jin H., Peng J., Zhao Z., et al. Strain-level multiomics analysis reveals significant variation in cheeses from different regions. LWT-Food Science and Technology. 2021;151 [Google Scholar]
- You L., Yang C., Jin H., Kwok L.-Y., Sun Z., Zhang H. Metagenomic features of traditional fermented milk products. LWT-Food Science and Technology. 2022;155 [Google Scholar]
- Yu J., Lu K., Zi J., Yang X., Zheng Z., Xie W. Halophilic bacteria as starter cultures: A new strategy to accelerate fermentation and enhance flavor of shrimp paste. Food Chemistry. 2022;393 doi: 10.1016/j.foodchem.2022.133393. [DOI] [PubMed] [Google Scholar]
- Z., Lu, I., M., Pérez-Díaz, J., S., Hayes, F., & Breidt. (2012). Bacteriophage ecology in a commercial cucumber fermentation. Applied & Environmental Microbiology. [DOI] [PMC free article] [PubMed]
- Zhao H., Ali U., Ren Q., Yao M., Lai T., Naz S., et al. Integrated metabolomic analysis of Lactiplantibacillus plantarum NMGL2 reveals its survival and response to combinational cold and acidic conditions during storage of fermented milk. Food Bioscience. 2023 [Google Scholar]
- Zhang F., Yao Y., Ren Y., Wan Q., He K., Chi Y., Yuanlong Microbial composition of spoiled industrial-scale Sichuan paocai and characteristics of the microorganisms responsible for paocai spoilage. International Journal of Food Microbiology. 2018;275 doi: 10.1016/j.ijfoodmicro.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zeng F., Hohn K., Vadlani P.V. Metabolic flux analysis of carbon balance in Lactobacillus strains. Biotechnology Progress. 2016;32(6):1397–1403. doi: 10.1002/btpr.2361. [DOI] [PubMed] [Google Scholar]
- Zhong Z., Hou Q., Kwok L., Yu Z., Zheng Y., Sun Z., et al. Bacterial microbiota compositions of naturally fermented milk are shaped by both geographic origin and sample type. Journal of Dairy Science. 2016;99(10):7832–7841. doi: 10.3168/jds.2015-10825. [DOI] [PubMed] [Google Scholar]
- Zou Y., Yuan Y., Liu M., Li X., Lai Y., Liu X., et al. Metagenomics reveal the role of microorganism and GH genes contribute to Sichuan South-road dark tea quality formation during pile fermentation. LWT-Food Science and Technology. 2023;178 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.