Abstract
Genetically modified macrophage infusion has been proven to be a novel treatment for cancer. One of the most important processes in macrophage-based therapy is the efficient transfer of genes. HIV-1-derived lentiviruses were widely used as delivery vectors in chimeric antigen receptor T and NK cell construction. While macrophages are relatively refractory to this lentiviral vector transduction as a result of the myeloid-specific restriction factor SAMHD1, which inhibited the virion cycle through exhausting the dNTPs pool and degradating RNAs. An efficient macrophage transduction strategy has been developed via packaging the HIV-2 accessory protein Vpx into the virion. Vpx counteracts SAMHD1 through CRL4 (DCAF1) E3 ubiquitin ligase mediated SAMHD1 degradation, yet the influence by the introduction of Vpx on macrophage has not been fully evaluated. Here, we constructed the chimeric lentiviral vector HIV-1-Vpx and systematically analyzed the infection efficiency of this vector in time-dependent manner. Our results showed that the simplified chimeric virus exhibited dramatically enhanced infection in human macrophages compared to normal lentivirus. Moreover, transcriptome sequencing was performed to evaluate the cellular status after chimeric virus infection. The sequencing results indicated that Vpx introduction promoted macrophage remodeling towards a proinflammatory phenotype, without affecting classic M1/M2 cell surface markers. Our results suggest that the Vpx-containing lentivirus could be used as an ideal tool for the generation of genetically engineered macrophages with high gene transfer efficiency and poised proinflammatory gene sets, especially for solid tumor treatment.
Keywords: Vpx, lentivirus, macrophage, hMDMs, Transcriptome
1. Introduction
Lentiviral vectors (LVs) are the newest vector system that establishes permanent integration into the genome of both dividing and nondividing cells. Unlike other retroviruses, LVs preintegration complex can get through the intact membrane of the nucleus of the target cell. Lentivector particles are normally generated by coexpressing the virion packaging elements with the vector genome embedded. Currently, lentiviral vectors (LVs) are available from several species, such as human immunodeficiency virus type 1 (HIV-1) and type 2 (HIV-2), simian immunodeficiency virus (SIV), equine infectious anemia virus (EIAV), bovine immunodeficiency virus (BIV), caprine arthritis-encephalitis virus (CAEV), and porcine immunodeficiency virus (FW). LVs are becoming popular tools for gene and cell therapies due to their ability to integrate the genome in dividing and nondividing cells. LVs can carry various exogenous genes or small guide RNAs to integrate into the host chromosome to achieve stable expression, gene knockout, or knockdown [1]. LVs showed high transduction efficiency in nerve cells, liver cells, brain cells, and other tissue cells. However, their ability to infect human primary myeloid cells is extremely poor.
Lentivirus genome encodes for three major structural proteins, gag, pol, and env [2]. Gag encodes core structural proteins, pol encodes enzymes required in the process of genome integration into host cells, and env encodes envelope proteins. However, lentiviruses such as HIV and SIV have substantially more complicated genomes, as they concede several regulatory and accessory proteins that aid in efficient viral replication [3]. The main accessory proteins of lentivirus are Vpr, Vpx, Vpu, Nef, and Vif, each with a unique function in viral replication and virus packaging [4,5]. Vpu allows virus release by counteracting BST-2/tetherin. Nef inhibits the presentation of antigens by downregulating the expression of major histocompatibility complex proteins. Vif safeguards the virus genome by downregulating the host viral response. Vpx and Vpr are all localized to the nucleus and involved in the nuclear transport of preintegration complex [6,7]. While Vpr is encoded by every known lentivirus, Vpx is only available for HIV-2 and SIV and is absent from HIV-1. The carboxy-terminal Gag protein, p6, interacts in a unique way with both the Vpr and Vpx proteins to package them into the assembling virion. Virus packaging is blocked when removing the whole p6 region or specific part of motifs within p6 [8].
Vpx is an HIV-2/SIV accessory viral protein that is the non-essential factor for virus infection in cell lines [[9], [10], [11]]. However, Vpx was found to be indispensable for HIV-2/SIV infection in non-dividing primary target cells such as human monocyte-derived macrophages (hMDMs) [10,[12], [13], [14]]. Vpx counteracts the sterile alpha motif and HD domain-containing protein 1 (SAMHD1) in hMDMs [15,16]. SAMHD1 is a myeloid-specific restriction factor, belonging to a group of closely related DNA hydrolases and nucleases with SAM domains and HD domains that prevents viral infection at an early stage of reverse transcription [17,18]. SAMHD1 is a dNTPase in cells, which restricts infection by HIV-1 by decreasing dNTPs below the amount necessary for reverse transcription [19,20]. Besides, SAMHD1 functions as RNase which causes cellular RNA degradation [21]. Virion-packaged Vpx loads SAMHD1 to CRL4DCAF1 E3 ubiquitin ligase, inducing its ubiquitin-proteasome degradation [17,18,21]. Then the reverse transcription block can be relieved by the replenishment of intracellular dNTP pool [20].
Macrophages are an important class of phagocytic cells that engulf and digest dying/dead cells as well as invasive bacteria and viruses. Macrophages play an essential role in tissue growth, homeostasis, and repair, as well as immunity [22]. In mammals, macrophages are mainly generated from the bone marrow after birth where hematopoietic stem cells (HSCs) give rise to monocytes. These monocytes then differentiate into tissue-resident macrophages after migrating from the blood into the specific tissue [23,24]. Macrophages are heterogeneous and plastic cells that acquire various phenotypes and functions due to local stimuli [24]. Macrophages can be divided into two types in response to different microenvironments: activated pro-inflammatory M1 macrophages and activated anti-inflammatory M2 macrophages [25]. M1 macrophages mediate antimicrobial and antitumor responses by expressing pro-inflammatory cytokines such as IFNγ and TNFα, but when overactivated, they can also cause inflammation and tissue damage. M2 macrophages, on the other hand, mediate tissue repair by expressing anti-inflammatory cytokines such as IL-10, TGFβ, and arginase, but can also mediate fibrosis if dysregulated. Macrophages have become important seed cells in cell therapy, and genetic modification of macrophages can improve the efficiency of cell therapy. In recent years, researchers have paid a lot interest in chimeric antigen receptor macrophages (CAR-M) for solid tumor immunotherapy, after CAR-T cell therapy shown a low success rate due to the complex tumor microenvironment in solid tumors [26]. Until October 2022, three clinical trials based on the CAR-M strategy have been registered at https://clinicaltrials.gov. However, human-derived macrophages are hardly infected by the HIV-1-based lentivirus. Therefore, it is essential to modify the structure of lentiviral vectors to enhance the infection efficiency on human-derived macrophages.
Here, we generated Vpx-containing HIV-1-based virions, and systematically analyzed the infection efficiency in time-dependent manner on human primary macrophages. To explore the influence of Vpx on host gene expression, we carried out RNA sequencing and found that Vpx introduction promoted macrophage remodeling towards proinflammatory phenotype, indicating Vpx-containing lentivirus could be used as an ideal tool for macrophages carrying exogenous genes utilized in solid tumor treatment.
2. Materials and methods
2.1. Plasmids
The packaging vector psPAX2-Vpx and envelop vector pMD2.G-Vpx were generated by PCR. A SphI-to-BstZ17I subclone containing the Vpx packaging motif was subcloned into psPAX2. Vpx packaging motif from SIV (amino acids 17–26) was placed in the p6 region of the Gag/Pol expression psPAX2 vector, the mutated fragment was cloned into psPAX2 at SphI and BstZ17I sites. SIVmac Vpx was amplificated by overlapping PCR and cloned into pMD2.G at PmlI site, placing a Flag epitope tag at the N-terminus.
2.2. Cells and cell culture
293T were maintained in Dulbecco's modified Eagle medium (DMEM, Gibco), while RAW264.7 were seeded in RPMI 1640 (Corning). The 293T and RAW264.7 were obtained from the Applied Biology Laboratory, Shenyang University of Chemical Technology. The medium is all containing 10 % FBS (LONSERA), 100 μg/ml Penicillin-Streptomycin (Sangon), and 100 μg/ml Glutamine (Sangon). Peripheral blood mononuclear cells (PBMC) were isolated from buffy coats of normal human donor blood, provided by Chinese PLA General Hospital. The blood was transferred into anticoagulant EDTA tube and PBMC were isolated by gradient density centrifugation with density reagent (tbdscience). For this procedure, 2 ml of blood was mixed with 2 ml of PBS (tbdscience), slowly added into 5 ml of density reagent and centrifuged for 20 min at 600 g. The cell layer direct on the second of the density reagent contains PBMC. This layer was transferred into a new 15 ml tube, resuspended in 10 ml of PBS, and centrifuged (700 g, 10 min). Subsequently, cells were counted in a haematocytometer, aliquoted, frozen and stored in liquid nitrogen. CD14+ monocytes were isolated from PBMC by positive selection using magnetic beads (Miltenyi Biotech), seeded in RPMI 1640 (Corning) supplemented with 10 % FBS (Excell), 100 μg/ml Penicillin-Streptomycin, and 100 ng/mL M-CSF (PeproTech). Then human monocyte-derived macrophages (hMDMs) subsequently polarized into M1 or M2 macrophages by adding to the medium with 20 ng/mL interferon gamma (IFNγ, PeproTech) and 50 ng/mL LPS (PeproTech), or 20 ng/mL interleukin 4 (IL-4, PeproTech), respectively. All of these cells were cultured at 37 °C and 5 % CO2.
2.3. Lentivirus preparation and infection
Virus was produced by standard transfection of 293T cells with Lipofectamine 2000 transfection reagent (Invitrogen), the mass ratio of psPAX2, pMD2.G, pcDNA-GFP is 3:1:4, followed by media change. Supernatants were harvested at 24 h, 48 h, and 72 h post-transfection, passed through a 0.45 μm-pore-size filter, and centrifugation with an Amicon Ultra-15 Centrifugal Filter (Millipore Sigma) to concentrate virus, which was aliquoted and frozen at −80 °C. The groups of lentivirus were shown as Fig. 2B. The virus titer was measured using Lenti-X™ p24 Rapid Titer Kit (Takara). Then the sample was infected by 4.0 × 107 purified virus particles with 1.0 × 105 hMDMs.
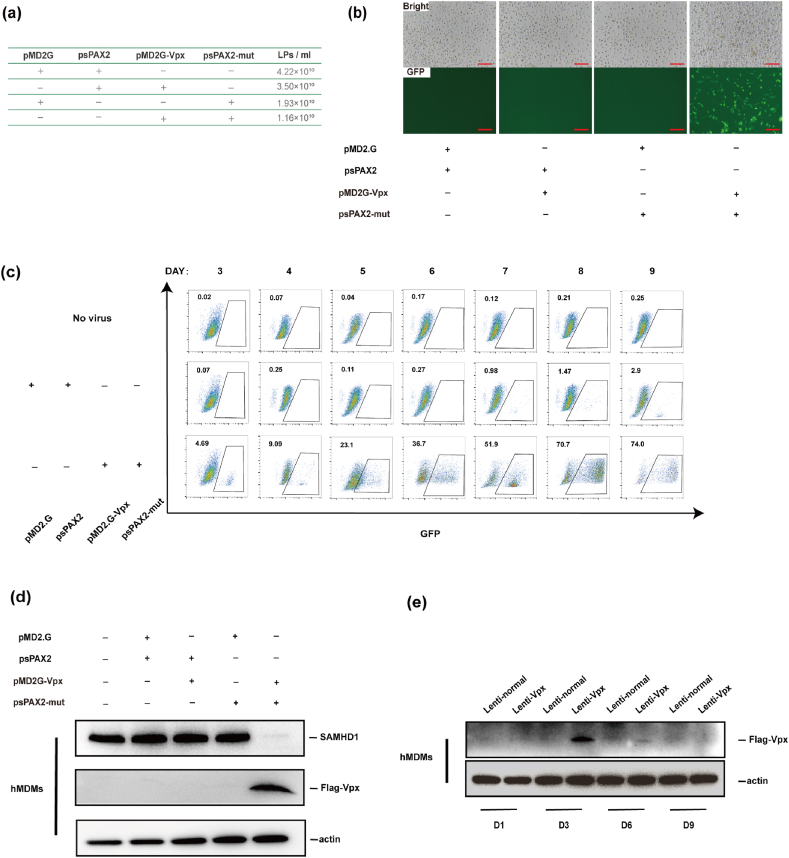
Fig. 2.
Vpx dramatically enhances the ability of lentivirus to infect primary human macrophages. (a) Virus titers of various lentivirus. (b) Fluorescence of human primary macrophages infected with various lentivirus. Scale bars 100 μm. (c) Analysis of the infection efficiency of Vpx-containing and control lentivirus on human primary macrophages by flow cytometry on the day indicated. (d) Analysis of the SAMHD1 and Vpx expression of different types of lentiviruses on human primary macrophages on day 5. (e) Analysis of Vpx protein levels of Vpx-containing and non-Vpx-containing lentivirus infecting human primary macrophages on the day indicated.
2.4. Immunoblotting
Cell lysis and virus were supplemented with 4x loading buffer and ran on denaturing SDS-PAGE, transferred to PVDF membrane, incubated first with blocking solution (Biofroxx), then with primary antibodies (1:5000), washed with PBST, incubated with secondary antibody (Beyotime), washed, and scanned using ChemiDoc Touch Imaging System (BIO-RAD). The primary antibodies used were: mouse anti-Flag (Cell Signaling Technology), mouse anti-Actin (Beyotime), and rabbit anti-p24 (Abcam).
2.5. Flow cytometry analysis
Following lentiviral transduction, adherent cells hMDMs were washed twice with PBS, incubated with Trypsin-EDTA (Gbico) at 37 °C for 5 min, centrifuged for 5 min at 750 g, and resuspended in PBS to be analyzed for green fluorescence light emission at 530 nm (FITC) using flow cytometry (NovoCyte, ACEA Biosciences).
For detection of CD11b surface expression, 1.0 × 105 cells were washed twice with PBS supplemented with 2 % FACS and incubated in 100 μl antibody dilution (1:100 in PBS + 2 % FACS; APC-conjugated CD11b antibody, BD) for 30 min at 4 °C in the dark. Prior to cytometric detection (APC: 660 nm), cells were washed twice and diluted in 100 μl PBS + 2 % FACS. Further analyses were performed in FlowJo.
2.6. RT-qPCR
To detect hMDMs polarization and proliferation, hMDMs were seeded in six-well plates and infected with wt lentivirus, or Vpx-containing lentivirus, respectively. Cells chosen to be positive control should polarize into M1 or M2 macrophages by adding 20 ng/mL IFNγ and 50 ng/mL LPS, or 20 ng/mL IL-4 24h before harvesting. RNA was isolated using Eastep Super Total RNA Extraction Kit (Promega). CDNA was generated using Transcriptor reverse transcriptase (Vazyme) primed with oligo(dT). The real-time PCR was performed on a CFX96 Real Time PCR detection system (Bio-Rad). The 25 μL PCR including TBGreen™ Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa), 0.4 μM forward and reverse primer and the RT product was diluted at least 10 × (Table 1).
Table 1.
List of sequences of primers used to detect inflammatory and proliferation relative genes of macrophages with qRT-PCR.
| Target | Sequence (5′-3′) |
|---|---|
| hIL-6 F | CCCCTGACCCAACCACAAAT |
| hIL-6 R | ATTTGCCGAAGAGCCCTCAG |
| hIL-1β F | GCTCGCCAGTGAAATGATGG |
| hIL-1β R | GGTGGTCGGAGATTCGTAGC |
| hTNF F | GCTGCACTTTGGAGTGATCG |
| hTNF R | TCACTCGGGGTTCGAGAAGA |
| hPIK3AP1 F | ATGAGGCTGTGCTCCACTTC |
| hPIK3AP1 R | CTGTCTTCGGGTGATGCTGT |
| hCCL2 F | AATCAATGCCCCAGTCACCT |
| hCCL2 R | CTTCTTTGGGACACTTGCTGC |
| hCXCL5 F | ACCACGCAAGGAGTTCATCC |
| hCXCL5 R | TCCTTCCCGTTCTTCAGGGA |
| hBCL2 F | TTTGAGTTCGGTGGGGTCAT |
| hBCL2 R | GGAGAAATCAAACAGAGGCCG |
| hMYC F | GCGAACACACAACGTCTTGG |
| hMYC R | ACTACCTTGGGGGCCTTTTC |
| hCSF1R F | ATCCGGCTGAAAGTGCAGAA |
| hCSF1R R | TGTTGAGGGATTGCGAGCTT |
| hPIK3R5 F | CATCACTGGGGAGAAGACGAC |
| hPIK3R5 R | GGACCTGACGCCTTGATGG |
| hLIF F | GCTGTACCGCATAGTCGTGT |
| hLIF R | CTAAGGAGGCCTCGCAGGAT |
2.7. RNA sequencing of human macrophage
RNA was isolated from human macrophages from volunteer donors, treated each with wt-lentivirus and Vpx-containing lentivirus and lytic by TRIzol. RNA-seq libraries were sequenced on 150-bp paired-end reads using a DNBSEQ2000 (BGI).
After sequencing, the raw reads were filtered. Data filtering includes removing adaptor sequences, contamination and low-quality reads from raw reads. First, raw data with adapter sequences or low-quality sequences were filtered. We went through a series of data processing to remove contamination and obtain valid data. This step was completed by SOAPnuke software developed by BGI. Then cleaned reads were mapped to human genome (GRCh38, Ensembl) using STAR. Gene count was calculated using featureCounts. EdgeR package with log fold change of 2 and adjusted P value of 0.05 was used to identify differentially expressed genes. As well as Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were conducted by R package clusterProfile.
2.8. MTT
HMDMs were seeded into 12 well plates, and then transfected by normal lentivirus and Vpx-containing lentivirus. Prepare the MTT solution (0.5 mg/ml), then add 500 μl of MTT solution to each well, and incubate the plate at 37 °Cfor additional 1 h. Remove the medium solution from each well, then add 1 ml of DMSO (Sangon) to each well to dissolve formazan. Shake the plate for 5 min until homogeneousness is reached. Take 100 μl the above solution into 96 well plate, Measure the absorbance at 490 nm by a microplate reader spectrophotometer.
2.9. Phagocytosis assay
Isolated CD14+ monocytes were seeded in a 6-well culture plate at 5 × 105 cells per well. SKOV3 cells were seeded in a 6-well cultural plate at 1.5 × 106 cells per well. Then, CD14+ monocytes were infected with Vpx-containing lentivirus and differentiated. Before coculture, SKOV3 cells were stained with Deep red celltracker (Invitrogen) at 37 °C for 30 min avoid light, cells then washed with PBS. SKOV3-red cells and hMDMs then co-incubated with 6 h, cells were washed, followed by flow cytometry analysis.
3. Results
3.1. Construction of lentiviruses carrying vpx component
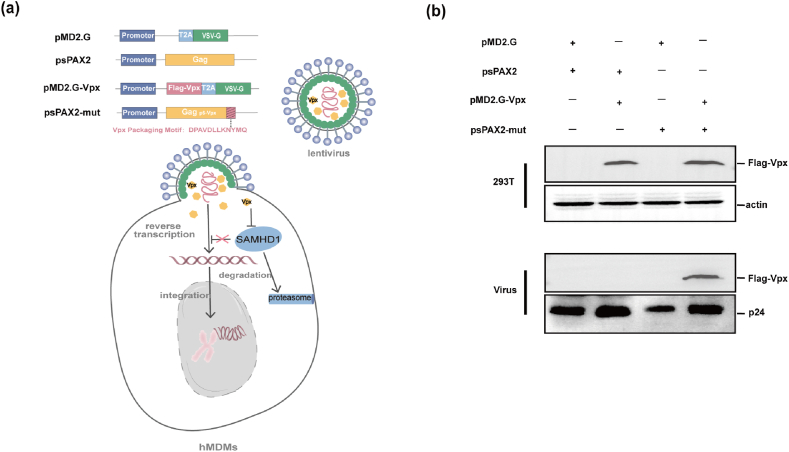
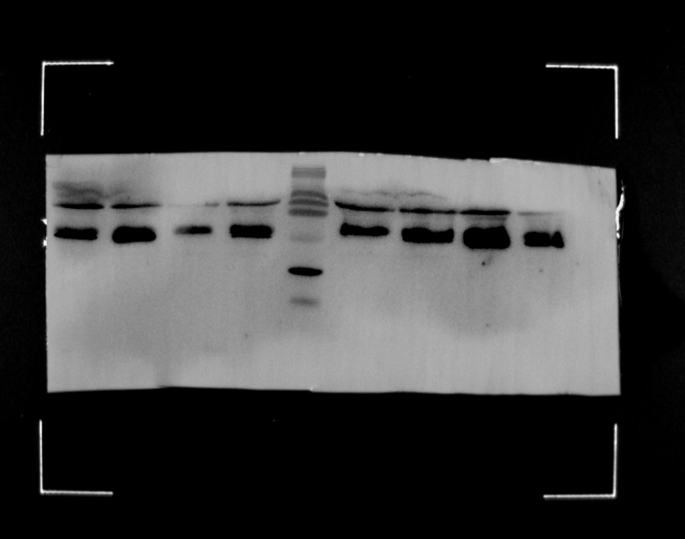
To generate lentivirus that contained Vpx, we constructed a simplified expression system consisting of two vectors. One of the vectors derived from the normally used HIV-1 psPAX vector, but harbored a Vpx-binding motif sequence by inserting the SIVmac p6 17–26 aa (DPAVDLLKNYMQ) sequence into the C-terminal of Gag, named psPAX-mut. Meanwhile, a codon-optimized SIVmac vpx open reading frame was inserted into pMD2.G vector to allow for Vpx expression along with VSV-g envelope protein, referred to as pMD2.G-Vpx (Fig. 1a, Supplementary Fig. 1). Immunoblot analysis of virions produced from various vector groups showed that only in the group that vector psPAX-mut and pMD2G-Vpx were both included, Vpx protein could be integrated into the virions (Fig. 1b). This result suggests that our two modified vectors system successfully packaged Vpx protein into HIV-1 virions.
Fig. 1.
Expression of Vpx protein in Virion. (a) Schematic diagram of Vpx construction of vectors; (b) Cell lysates and supernatant containing virions were analyzed by Western-blot with an antibody against Flag-tagged Vpx, HIV-1 p24 CA, or actin.
3.2. Vpx dramatically enhanced lentivirus infection efficiency in macrophages
To investigate whether modified lentivirus could enhance infection efficiency in macrophages, we generated chimeric and control green fluorescent protein reporter viruses from 293T cells. The infectious titers of the viruses were measured and normalized, and then used to infect hMDMs prepared from healthy donors at a multiplicity of infection (MOI) of 400 lentivirus particles (LPs) per cell (Fig. 2a). Through fluorescence microscopy, we found that only cells infected with the virus containing Vpx protein showed GFP positive signal (Fig. 2b). To determine the ratios of infected cells, GFP-positive cells were quantified by flow cytometry. We found that the Vpx-containing lentivirus infected more than 74% of hMDMs compared to nearly 3% infection efficiency by the control lentivirus (Fig. 2c). Additionally, the WB analysis shows that SAMHD1 expression is also reduced in hMDMs which infected with Vpx-containing lentiviruses (Fig. 2d and e). These results suggest that the Vpx protein and p6 motif mutant are both indispensable for the highly infection.
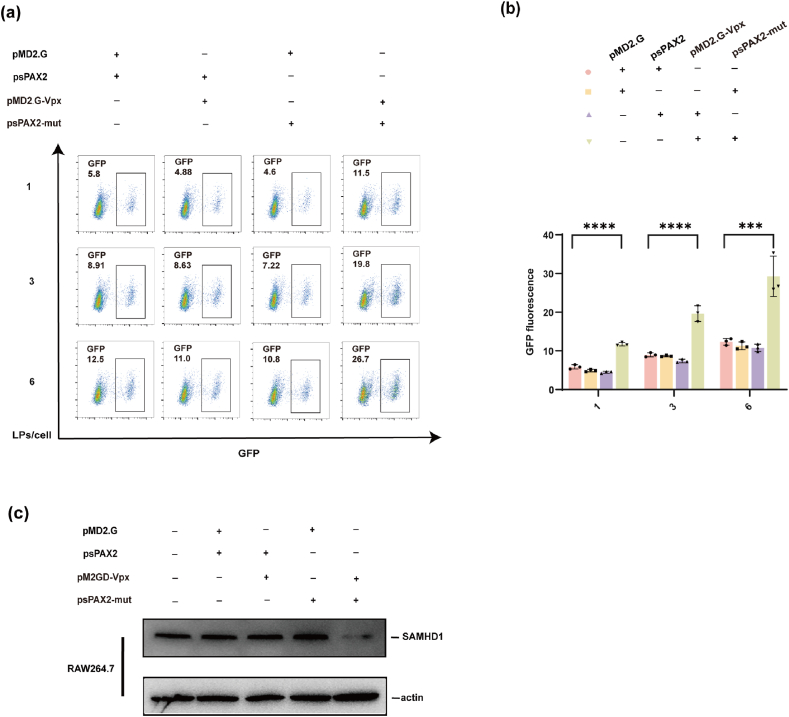
Then to ensure the infectivity ability of the modified virus on murine macrophages which were considered can be infected by normal lentivirus, we infected RAW264.7 cells with four kinds of virus mentioned above at different viral volumes. The flow cytometry results indicated that although non-Vpx-containing virus infection resulted in less than 20 % infection efficiency, the infection efficiency of Vpx-containing virus was about 2-fold as high at all three virus titers (Fig. 3a and b). We also detect a decrease of SAMHD1 expression in RAW264.7 cells (Fig. 3c). These results suggest that the Vpx protein dramatically elevates lentivirus infection efficiency in murine macrophages cell line RAW264.7.
Fig. 3.
Vpx dramatically enhances the ability of lentivirus to infect murine macrophages cell line RAW264.7. (a and b) Analysis of the infection efficiency of different types of lentiviruses on RAW264.7 cells by flow cytometry on day 3 after infection with different titers of virus. (c) Analysis of the SAMHD1 expression in different types of lentiviruses infected RAW264.7 cells on day 3.
3.3. Lentiviruses carrying vpx manipulate the expression of inflammatory genes in macrophages
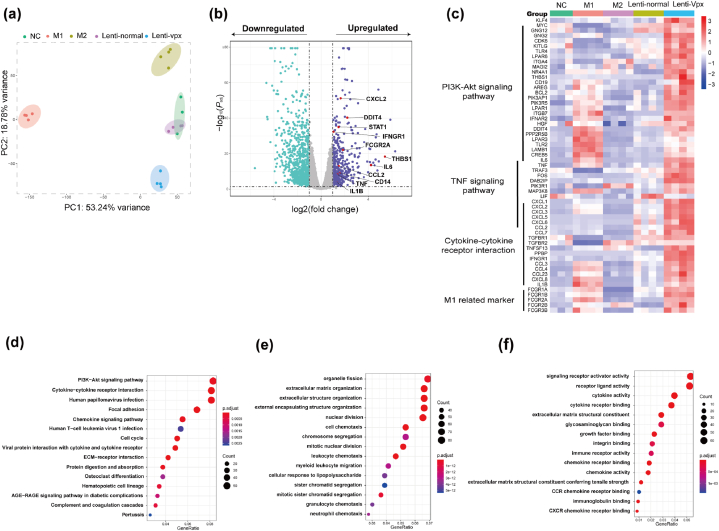
To determine the impact of Vpx-lentivirus infection on hMDMs, we profiled the transcriptomic landscape of the untransduced (NC), Vpx-containing lentivirus transduced (lenti-Vpx), normal lentivirus transduced (lenti-normal), classically activated (M1) and alternatively activated (M2) macrophage. When NC, lenti-Vpx, lenti-normal, M1, and M2 macrophage transcriptomes were subjected to nonbiased principal component analysis (PCA), Vpx-containing lentivirus transduced macrophages clustered not towards to classically activated macrophages nor alternatively activated macrophages (Fig. 4a). Then interestingly, we found that the macrophages infected by Vpx-containing lentivirus had a high expression of inflammatory genes, such as interleukin 6 (IL6), interleukin 1β (IL1B), tumor necrosis factor (TNF), and C-X-C motif chemokine (Fig. 4b and c). Pathway analysis confirmed the inflammatory-associated pathways and HIV-infected associated pathways, such as the P13K-Akt signaling pathway, cytokine-cytokine receptor interaction, and human papillomavirus infection pathway (Fig. 4d). We also found that macrophages infected by Vpx-containing lentivirus clustered on organelle fission, extracellular matrix organization, signaling receptor activator activity, and receptor ligand activity (Fig. 4e and f).
Fig. 4.
Transcriptome analysis of macrophages infected by Vpx-containing lentivirus on day 5. (a) Gene expression principal component analysis clustering from NC, Vpx-containing lentivirus transduced, normal lentivirus transduced, classically activated M1, and alternatively activated M2 human primary macrophages. (b) Volcano plot of differentially expressed genes in normal lentivirus versus Vpx-containing lentivirus. Purple indicates Padj < 0.05 and log2 fold change >1 or < −1. Red dots indicate significantly upregulated inflammatory genes. Statistical significance was calculated using the Wald test for edgeR data. (c) Gene expression heatmap about pro-inflammatory and proliferation genes from NC, Vpx-containing lentivirus transduced, normal lentivirus transduced, classically activated M1 and alternatively activated M2 human macrophage. (d) KEGG pathways analysis about normal lentivirus versus Vpx-containing lentivirus. (e) Biological process of differentially expressed genes in Vpx-containing lentivirus versus normal lentivirus. (f) Molecular function of differentially expressed genes in Vpx-containing lentivirus versus normal lentivirus. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
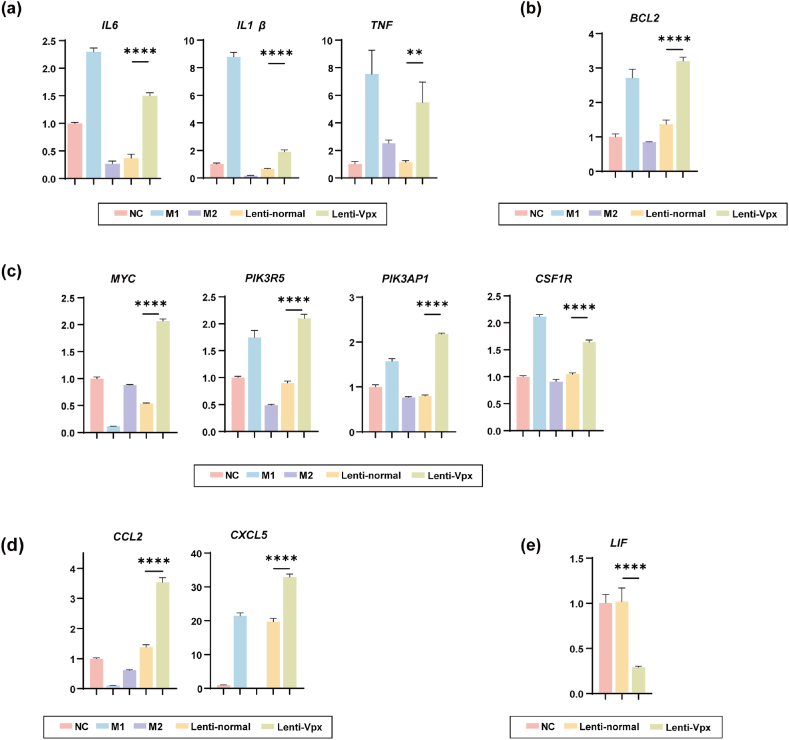
Then, qPCR was performed to confirm the transcriptomic results. Besides inflammation-related factors IL6, IL1B, and TNF (Fig. 5a, Supplementary Fig. 2), anti-apoptotic factor Bcl2 also got an up-regulation (Fig. 5b). Myc, lysophosphatidic acid receptor 1 (LPAR1), phosphoinositide-3-kinase regulatory subunit 5 (PIK3R5), phosphoinositide-3-kinase adaptor protein 1 (PIK3AP1), and colony stimulating factor 1 receptor (CSF1R) which promote macrophage proliferation (Fig. 4, Fig. 5c, Supplementary Fig. 3) were upregulated too. Other up-regulated factors CCL2 and CXCL5 have been reported to enhance macrophage recruitment (Fig. 5d). LIF can generate a local immunosuppressive microenvironment and some tumors express aberrantly high levels of LIF [27]. Vpx-containing lentivirus transduced could suppress the expression of LIF (Fig. 5e).
Fig. 5.
RT-qPCR analysis of inflammatory related genes. (a) IL6, IL1β, and TNF mRNA expression levels determined by RT-qPCR on human primary macrophages. (b) Expression of anti-apoptosis gene BCL2 in human primary macrophages. (c) Proliferation genes MYC, PIK3R5, PIK3AP1, and CSF1R expression levels on human primary macrophages. (d) CCL2 and CXCL5 mRNA expression levels on human primary macrophages. (e) LIF mRNA expression levels determined by RT-qPCR on human primary macrophages treated by normal lentivirus and Vpx-containing lentivirus. Error bars reflect the standard deviation of fold change. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.001. ns, non-significant (P > 0.05).
3.4. Enhanced phagocytosis upon vpx mediated high chimeric antigen receptor lentivirus infection efficiency
We generated the CAR-HER2 Vpx-lentivirus with the CAR structure as described, with the replacement of intracellular domain CD3 to CD32a, which is endogenous expressed in monocytes and macrophages (Fig. 6 a). As shown in Fig. 6 b, the Vpx-lentivirus significantly increased the infection efficiency of CAR-HER2, up to 70 % after 7 days of infection. We next analysized the phagocytosis by using human ovarian adenocarcinoma SKOV3 HER2 positive cells. After co-incubation of 6 h, Vpx-lenti-CAR-HER2 infeccted macrophages and SKOV3 cells were harvested and SKOV3 were stained with Deep red celltracker. FACS showed the CAR-HER2 infected macrophages had higher phagocytic activity compared to none CAR Vpx-lenti infected cells (Fig. 6c).
Fig. 6.
(A) Diagram of CAR-Her2 structure. (b) GFP expression on hMDMs 7days after lentivirus transduction. (c) Phagocytosis rates of CAR-Her2-Vpx-lenti or non-CAR Vpx-lenti infeccted hMDMs and SKOV3 cells.
4. Discussion
In our study, we conduct a Vpx-containing HIV-1 based lenti-virion, which shows robust infection efficiency to human PBMC derived macrophages. Compared to normal HIV-1 virion, the infection efficiency raised from an extremely low level to 74 % in a time-dependent manner. Notably, Vpx protein regulates human macrophage transcriptome towards proinflammatory phenotype, which brings broad prospects in the cancer therapy field.
Immune-cell therapies, such as CAR-T, NK, TCR-T, TIL, and CAR-M therapy, offer promising applications for the treatment of tumors and aging-related diseases. Exogenous gene delivery to immune cells is critical in immune cell preparation. Gene delivery tools include viruses, transposons, and circular RNAs, among which, the most commonly used viruses are adenovirus, adeno-associated virus, and lentivirus. Adenovirus has a high transfection rate and does not integrate into the genome; however, it has the disadvantage of losing exogenous genes easily. On the other hand, lentiviruses can integrate into the genome, resulting in the stable presence of exogenous genes in target cells.
Lentivirus vectors are designed from HIV-1, which lacks Vpx accessory protein. Vpx is an important lentivirus accessory protein that exists in SIV in sooty mangabeys and rhesus macaques/HIV-2, which degrades SAMHD1 by recruiting CUL4A-DCAF1 E3 ubiquitin ligase to improve dNTPs pools for viral RNA genome reverse transcription in myeloid cells, resting T cells, and dendritic cells [18,[28], [29], [30]]. The p6 carboxy-terminal domain of gag interacts with Vpx to produce Vpx-containing virions. The minimal Vpx packaging motif of p6 gag, which contains 10 amino acids 17DPAVDLLKNY26 at the carboxy-terminal, was identified from simian immunodeficiency virus SIVmac239 [31]. In our study, the minimal Vpx packaging domain was cloned into psPAX2 vector, and Vpx sequence was cloned into pMD2.G vector. This combination promoted Vpx to be packaged into lentivirus successfully. The infection efficiency increased drastically in a time-dependent manner. On day 6, the ratio of transfected-positive cells elevated up to 36.7 %, and the percentage was up to 74 % after 9 days post-infection as shown in Fig. 2a. As previously reported, the knockdown of SAMHD1 in murine monocytic cell line RAW264.7 improved HIV-1 infection efficiency [32]. In our study, RAW264.7 cell lines showed elevated susceptibility towards Vpx-containing lentiviruses compared to wt lentiviruses. All the data indicate that Vpx-containing lentivirus can be a promising tool for enhancing both primary human macrophage and mouse RAW264.7 macrophage susceptibility to HIV-1 lentivirus transfection. However, the Vpx-containing lentivirus vectors here also show some parts of unsatisfactory. As other lentivirus, they have the potential of intergrating into some unwanted loci, as the random integration events. Also due to the Vpx was deliverd as a protein. the limited expression is sufficient for lentivirus infection, but might not be enough for lasting the proinflammation phenotype.
The immunosuppressive state of the tumor microenvironment (TME) results in tumor progression and resistance to therapy [33]. Oncotherapy involves reprogramming the TME from immunosuppressive to proinflammatory state. Tumor associated macrophages (TAMs) are heterogeneous, including M1-like and M2-like TAMs. The predominant phenotype in the TME is the M2-like phenotype, which is immunosuppressive and promote cancer aggravation [34]. Turning the immunosuppressive phenotype into proinflammatory and maintaining M1 can control tumor growth. Ad5f35 adenoviral vectors were used for gene delivery to generate chimeric antigen receptor macrophages. Ad5f35-engineered CAR-M maintained an M1 phenotype, produced numerous proinflammatory cytokines, converted bystander M2 macrophages to M1 in the TME, and stimulated T cell antigen presentation. Meanwhile, the adenovirus Ad5f35 had no effect on macrophage markers [26]. Similarly, in our study, transcriptome analysis indicated that Vpx protein in lentivirus increased gene expression related to proinflammatory molecules and signals. Similar to the M1 phenotype, IL-6, IL-1β, and TNF-α expression levels and some chemokines including CCL2 and CXCL5 increased when Vpx was incorporated into lentivirus, which help maintain the pro-inflammatory state of the TME and recruit more circulating immune cells to the tumor site. The RNA-sequencing analysis and RT-qPCR data revealed that CSF1R was elevated in Vpx group compared to wild type lentivirus group. CSF1R is a kind of tyrosine-protein kinase that functions as a cell-surface receptor for CSF1. CSF1/CSF1R promotes macrophages and monocytes survival, proliferation, and differentiation. Especially, CSF1/CSF1R signaling enhances proinflammatory chemokines and cytokines expression of macrophages [35,36]. Even if growth factors are present, terminal differentiation of myeloid precursors limits cell proliferation. Proliferation challenges limit large scale macrophage generation in vitro. The transcription factors Oct-4, Sox-2, KLF4, and c-Myc are capable of remodeling differentiated cells into pluripotent stem cells (iPSC) [37]. c-Myc, a proto-oncogene, promoted the differentiation of monocytes into macrophages and increased macrophage expansion [38,39]. According to the transcriptome profiling, c-Myc was significantly increased due to Vpx modification. Besides, Kruppel-like factor 4 (Klf4) promotes cell survival and inhibits cell apoptosis [[40], [41], [42], [43], [44], [45]]. In our study, the Klf4 expression level is much higher in Vpx-containing lentivirus infection group than wild type lentivirus. Moreover, Vpx-containing lentivirus dramatically increased primary hMDMs cell viability. Our data highlighted the potential for Vpx to promote cell growth. In addition, the PI3K-Akt signaling pathway was strengthened. BCL2, a member of the PI3K-Akt signaling pathway, was discovered in a type of B-cell lymphoma at the t (14; 18) chromosomal breakpoint, which was elevated in our study. BCL2 potentiates cell survival and inhibits various types of apoptosis [31], suggesting that the Vpx protein promotes macrophage survival. All the transcriptome data indicate that Vpx-containing lentivirus may promote primary human macrophages maintaining in proinflammatory state, keeping proliferation, and reducing apoptosis.
5. Conclusion
In this study, we constructed a simplified Vpx-containing lentivirus packaging system. This system dramatically increased lentivirus infection efficiency for both primary human macrophages and murine RAW264.7 macrophages. Moreover, Vpx-containing lentivirus reprogrammed hMDMs towards proliferative and proinflammatory phenotype, resulting in primary hMDMs cell growth promotion. Finally, we found that Vpx mediated a high chimeric antigen receptor lentivirus infection efficiency which augmented the phagocytic ability of human macrophages. All the data indicate that our lentivirus packaging system can can be a promising tool for antitumor therapy.
Funding
This work was supported by Liaoning Revitalization Talents Program (XLYC2002027), construction of Liaoning technological innovation center (1,590,826,279,052), Central government funds for guiding local scientific and Technological Development (2021JH6/10,500,225), 2021 Shenyang Science and technology plan (21-103-0-20 and 21-102-0-10), National Key Research and Development Program [2019YFA0110200, 2019YFA0110201], Programs from the Department of Education of Liaoning Province (LJKZ0464 and LQ2020022).
Data Availability statement
The RNA-seq data presented in this study are available on request from https://www.ncbi.nlm.nih.gov/sra/PRJNA893383.
Declarations
Collection and analysis of human donor blood were approved by Roc Rock Biotechnology ethics committee. The ethics approval number is 2,022,002. The experiment using human blood was agreed under the medial and rules for each participating individual and the written informed consent was waived.
CRediT authorship contribution statement
Yun Gao: Data curation, Formal analysis, Methodology, Writing - original draft. Yue Ju: Data curation. Xiaomeng Ren: Writing - review & editing. Luo Zhang: Conceptualization, Writing - review & editing. Xiushan Yin: Conceptualization, Resources, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Fenghua Guo and Yue Jing for their help about the analysis of transcriptome.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21886.
Contributor Information
Xiaomeng Ren, Email: renxm@syuct.edu.cn.
Luo Zhang, Email: marbleluo@126.com.
Xiushan Yin, Email: xiushanyin@gmail.com.
Appendix A. Supplementary data
The following are the supplementary data to this article.
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
figs8.
figs9.
figs10.
figs11.
References
- 1.Naldini L., Trono D., Verma I.M. Lentiviral vectors, two decades later. Science. 2016;353(6304):1101–1102. doi: 10.1126/science.aah6192. [DOI] [PubMed] [Google Scholar]
- 2.Varmus H. Retroviruses, Science. 1988;240(4858):1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- 3.Collins D.R., Collins K.L. HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathog. 2014;10(1) doi: 10.1371/journal.ppat.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malim M.H., Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3(6):388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Greenwood E.J.D., Williamson J.C., Sienkiewicz A., et al. Promiscuous targeting of cellular proteins by Vpr drives systems-level proteomic remodeling in HIV-1 infection. Cell Rep. 2019;27(5):1579–1596.e7. doi: 10.1016/j.celrep.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belshan M., Mahnke L.A., Ratner L. Conserved amino acids of the human immunodeficiency virus type 2 Vpx nuclear localization signal are critical for nuclear targeting of the viral preintegration complex in non-dividing cells. Virology. 2006;346(1):118–126. doi: 10.1016/j.virol.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Yurkovetskiy L., Guney M.H., Kim K., et al. Primate immunodeficiency virus proteins Vpx and Vpr counteract transcriptional repression of proviruses by the HUSH complex. Nat Microbiol. 2018;3(12):1354–1361. doi: 10.1038/s41564-018-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunseri N., O'Brien M., Bhardwaj N., et al. Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J. Virol. 2011;85(13):6263–6274. doi: 10.1128/jvi.00346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guyader M., Emerman M., Montagnier L., et al. VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. Embo j. 1989;8(4):1169–1175. doi: 10.1002/j.1460-2075.1989.tb03488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueno F., Shiota H., Miyaura M., et al. Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microbes Infect. 2003;5(5):387–395. doi: 10.1016/s1286-4579(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 11.Yu X.F., Yu Q.C., Essex M., et al. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 1991;65(9):5088–5091. doi: 10.1128/jvi.65.9.5088-5091.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher T.M., 3rd, Brichacek B., Sharova N., et al. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM) Embo j. 1996;15(22):6155–6165. [PMC free article] [PubMed] [Google Scholar]
- 13.Pancio H.A., Vander Heyden N., Ratner L. The C-terminal proline-rich tail of human immunodeficiency virus type 2 Vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J. Virol. 2000;74(13):6162–6167. doi: 10.1128/jvi.74.13.6162-6167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita M., Otsuka M., Miyoshi M., et al. Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 2008;82(15):7752–7756. doi: 10.1128/jvi.01003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bobadilla S., Sunseri N., Landau N.R. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 2013;20(5):514–520. doi: 10.1038/gt.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H., Guo H., Su J., et al. Inhibition of vpx-mediated SAMHD1 and vpr-mediated host helicase transcription factor degradation by selective disruption of viral CRL4 (DCAF1) E3 ubiquitin ligase assembly. J. Virol. 2017;91(9) doi: 10.1128/jvi.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hrecka K., Hao C., Gierszewska M., et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laguette N., Sobhian B., Casartelli N., et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstone D.C., Ennis-Adeniran V., Hedden J.J., et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 20.Lahouassa H., Daddacha W., Hofmann H., et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 2012;13(3):223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maharana S., Kretschmer S., Hunger S., et al. SAMHD1 controls innate immunity by regulating condensation of immunogenic self RNA. Mol Cell. 2022;82(19):3712–3728.e10. doi: 10.1016/j.molcel.2022.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Wynn T.A., Chawla A., Pollard J.W. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginhoux F., Schultze J.L., Murray P.J., et al. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2016;17(1):34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 24.Okabe Y., Medzhitov R. Tissue biology perspective on macrophages. Nat. Immunol. 2016;17(1):9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- 25.Mosser D.M., Edwards J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klichinsky M., Ruella M., Shestova O., et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020;38(8):947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pascual-García M., Bonfill-Teixidor E., Planas-Rigol E., et al. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8(+) T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun. 2019;10(1):2416. doi: 10.1038/s41467-019-10369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romani B., Cohen E.A. Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr Opin Virol. 2012;2(6):755–763. doi: 10.1016/j.coviro.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhard C., Bottinelli D., Kim B., et al. Vpx rescue of HIV-1 from the antiviral state in mature dendritic cells is independent of the intracellular deoxynucleotide concentration. Retrovirology. 2014;11:12. doi: 10.1186/1742-4690-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldauf H.M., Stegmann L., Schwarz S.M., et al. Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc Natl Acad Sci U S A. 2017;114(10):2729–2734. doi: 10.1073/pnas.1613635114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin X.M., Oltvai Z.N., Korsmeyer S.J. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature. 1994;369(6478):321–323. doi: 10.1038/369321a0. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R., Bloch N., Nguyen L.A., et al. SAMHD1 restricts HIV-1 replication and regulates interferon production in mouse myeloid cells. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0089558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitale I., Manic G., Coussens L.M., et al. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30(1):36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Han S., Wang W., Wang S., et al. Tumor microenvironment remodeling and tumor therapy based on M2-like tumor associated macrophage-targeting nano-complexes. Theranostics. 2021;11(6):2892–2916. doi: 10.7150/thno.50928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee A.W., States D.J. Colony-stimulating factor-1 requires PI3-kinase-mediated metabolism for proliferation and survival in myeloid cells. Cell Death Differ. 2006;13(11):1900–1914. doi: 10.1038/sj.cdd.4401884. [DOI] [PubMed] [Google Scholar]
- 36.Otero K., Turnbull I.R., Poliani P.L., et al. Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and beta-catenin. Nat. Immunol. 2009;10(7):734–743. doi: 10.1038/ni.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi K., Tanabe K., Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Blasi E., Mathieson B.J., Varesio L., et al. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature. 1985;318(6047):667–670. doi: 10.1038/318667a0. [DOI] [PubMed] [Google Scholar]
- 39.Aziz A., Soucie E., Sarrazin S., et al. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science. 2009;326(5954):867–871. doi: 10.1126/science.1176056. [DOI] [PubMed] [Google Scholar]
- 40.Rowland B.D., Bernards R., Peeper D.S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005;7(11):1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., Goldstein B.G., Chao H.H., et al. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol. Ther. 2005;4(11):1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 42.Ghaleb A.M., Katz J.P., Kaestner K.H., et al. Krüppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26(16):2365–2373. doi: 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Talmasov D., Xinjun Z., Yu B., et al. Krüppel-like factor 4 is a radioprotective factor for the intestine following γ-radiation-induced gut injury in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308(2):G121–G138. doi: 10.1152/ajpgi.00080.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C., DeRoo E.P., Stecyk C., et al. Impaired autophagy in mouse embryonic fibroblasts null for Krüppel-like Factor 4 promotes DNA damage and increases apoptosis upon serum starvation. Mol. Cancer. 2015;14:101. doi: 10.1186/s12943-015-0373-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuruvilla J.G., Kim C.K., Ghaleb A.M., et al. Krüppel-like factor 4 modulates development of BMI1(+) intestinal stem cell-derived lineage following γ-radiation-induced gut injury in mice. Stem Cell Rep. 2016;6(6):815–824. doi: 10.1016/j.stemcr.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data presented in this study are available on request from https://www.ncbi.nlm.nih.gov/sra/PRJNA893383.