Abstract
Background
and Purpose: Fuzitang decoction (FZT), a classic prescription of traditional Chinese medicine (TCM), has excellent efficacy in treating gouty arthritis (GA). However, the underlying molecular mechanism remains obscure. In the present study, we aimed to explore the underlying mechanisms of FZT in treating GA by virtual screening combined with experimental verification.
Methods
In this study, the active components of FZT and their corresponding targets were screened from the TCMSP database and TargetNet database. Then, the potential targets of FZT against GA were retrieved from multiple databases to generate a network. Protein-protein interaction, herbal-component-target, Gene Ontology (GO) enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were applied to identify potential targets and related signaling pathways. Furthermore, molecular docking simulation was applied to identify the interactions between the drug and targets. Finally, in vitro experiments were conducted to validate the potential targets and signaling pathways.
Results
In the present study, several crucial components, including kaempferol, luteolin, catechin, deoxyandrographolide, and perlolyrine in FZT, were obtained through network pharmacology, and several potential targets to treat GA were developed, such as PPARG, CYP3A4, PTGS2 (known as COX2), VEGFA, and CYP1A1. Experimental validation suggested that deoxyandrographolide significantly suppressed the expression of IL-1β, COX2, NLRP3 and IL-6 in inflammatory monocyte cells.
Conclusions
Our results identified a novel anti-inflammatory compound, deoxyandrographolide, which helps to explain the potential mechanism of FZT in treating GA and provides evidence to support FZT's clinical use.
Keywords: Fuzitang decoction, Gouty arthritis, Virtual screening, Experimental verification, Molecular docking
1. Introduction
Gouty Arthritis (GA) is a kind of hyperuricemia-related disease caused by the deposition of monosodium urate (MSU) crystals in the joint [1], which shows characteristics of recurrent gout flares and tophus [2]. GA negatively impacts the long-term prognosis and quality of life, as it is usually accompanied by many comorbidities, such as kidney disease, hypertension, obesity, diabetes, and cardiovascular diseases [3]. For example, a Taiwanese study showed that the periodicity of gout flares was associated with an odds ratio of 1.18 for myocardial infarction [4]. Banishing gout flares and preventing tophus are considered two central focuses of gout treatment [3]. However, the inflammatory reaction of the deposition of MSU in the joint is the key pathogenesis of gout flares and joint injury [5]. Therefore, anti-inflammatory therapy is a key strategy in the treatment of GA. Currently, colchicine, nonsteroidal anti-inflammatory drugs (NSAIDs), and corticosteroids have been clinically used as first-line drugs for GA [6]. However, anti-inflammation for GA treatment remains challenging, since many patients have incomplete responses or contraindications to one or more of the drugs. In addition, gastrointestinal and cardiovascular adverse reactions are often observed in patients with long-term anti-inflammatory treatment [7]. Therefore, optimized treatments need to be explored to overcome these limitations.
Traditional Chinese medicine (TCM) has a long history and unique curative effect on the treatment of GA. Fuzitang decoction (FZT) is a TCM formula (classical prescription) that was first described by Zhongjing Zhang in “Treatise on Febrile Disease Caused by Cold (Shanghan Lun in Chinese)”, which consists of five herbs: Fuzi (Aconitum carmichaeli Debeaux), Baishao (Paeonia lactiflora Pall), Baizhu (Atractylodes macrocephala Koidz), Dangshen (Codonopsis pilosula) and Fuling (Poria cocos). FZT was used to treat joint pain from 200 CE. In recent years, it has also been reported that the prescription has a good clinical effect in the treatment for GA. Studies have shown that FZT significantly relieves joint pain and swelling of GA, reduces white blood cell count (WBC) and serum uric acid (uric acid), and mediates the level of triglycerides (TG) [8]. Meanwhile, the adverse reaction was significantly lower than that of the anti-inflammatory treatment group. Some animal studies have demonstrated that FZT can reduce joint swelling and inflammation in rats with collagen-induced arthritis by modulating the PPAR- γ/NF-κB signaling pathway and decreasing the expression of pro-inflammatory cytokines [9]. Aconitine in Fuzi has anti-inflammatory activity and can significantly inhibit xylene-induced mouse auricle swelling and capillary permeability increasement, exerting an anti-inflammatory effect [10]. Baishao is the dried root of the buttercup plant Paeonia lactiflora, which exhibits analgesic, anti-inflammatory, and immunoregulatory effects [11]. Baizhu can regulate multiple signaling pathways, such as NF-κB, MAPK, ERK1/2 and p38, to exert anti-inflammatory effects [12]. Dangshen can inhibit pro-inflammatory factors secretion by regulating the miR-361–5p/TLR4/NF-κB pathway [13]. Extracts of Fuling have been reported to have a variety of biological activities, including anti-inflammatory effects [14]. This evidence indicates that FZT reduces inflammatory reactions and ameliorates the symptom of GA. However, FZT contains toxic ingredient-aconitine, which causes some side-effects and damages its clinical promotion. Thus, extreme caution is required when the usage of FZT, specific anti-inflammatory omponents and potential mechanism of FZT in the treatment of GA remain unclear. Therefore, it is particularly significant to virtually screen the effective components with good anti-inflammatory effects and verify them experimentally to provide a basis for follow-up animal experiments.
Virtual screening has been successfully used in the research of active ingredients and potential targets of TCM following the construction of the TCM database and biological database. Network pharmacology and molecular docking are important tools for virtual screening. Network pharmacology can construct a network consisting of multiple drugs, multiple components, multiple targets, and multiple channels. It has become the most suitable tool to explore the active components of herbal medicine at the molecular level. Molecular docking, a computer virtual docking method, can evaluate the docking effect between active components and targets and visualize the docking sites, free energy, molecules and proteins. In the present study, we aimed to explore the main bioactive ingredients of FZT for GA and clarify the molecular mechanisms. The workflow of this study is illustrated in Fig. 1.
Fig. 1.
Graphic abstract of this study. In this study, virtual screening combined with experimental verification were used to identify anti-inflammation compounds and to reveal the potential mechanism of Fuzitang Decoction against Gouty Arthritis.
2. Materials & methods
2.1. Active ingredient screening of FZT
The chemical ingredients of FZT were collected from the Traditional Chinese Medicine System Pharmacology (http://tcmspw.com/tcmsp.php, TCMSP), a unique pharmacology platform for Chinese herbal medicines [15]. The Chinese herbal names of “Fuzi”, “Baishao”, “Baizhu”, “Dangshen”, and “Fuling” were retrieved. According to the ADME (adsorption, distribution, metabolism, excretion) parameters, the active compounds of FZT were selected using oral bioavailability (OB ≥ 30 %) and drug-likeness (DL ≥ 0.18) as the filtration criteria [16]. The two-dimensional (2D) structure, canonical smiles, and PubChem ID of the ingredients were found by the PubChem (https://pubchem.ncbi.nlm.nih.gov/) database.
2.2. Screening of related targets corresponding to active ingredients
The related targets corresponding to the active ingredients of FZT were collected from the TSMSP database. In addition, the targets of the active ingredients of FZT were obtained from the TargetNet (http://targetnet.scbdd.com/calcnet/index/) database by submitting the canonical smiles of the active ingredients (prob ≥1 as the filtration criteria). The Perl Programming Language (version 5.34.0) was used to remove the duplicated target names. Gene symbols of each target were obtained from the Uniprot database (https://www.uniprot.org/).
2.3. Identifying the potential targets of FZT in the context of GA
The therapeutic target genes of GA were searched in the GeneCards database (https://www.genecards.org/) [17], Online Mendelian Inheritance in Man database (OMIM, https://omim.org/) [18], Pharmgkb database (https://www.pharmgkb.org/) [19], Therapeutic target database (TTD, http://db.idrblab.net/ttd/) [20]and DrugBank database (https://go.drugbank.com/) [21]. The keywords “gout”, “gouts”, and “gouty arthritis” were retrieved. The putative target genes were selected by setting an elevance score ≥1 as the filtration criteria in the GeneCards database. The Perl Programming anguage (version 5.34.0) was used to remove the duplicated genes from these 5 databases. To obtain the target genes at the intersection of FZT and GA (F&G), the Venn diagram in the ebsite (https://bioinfogp.cnb.csic.es/tools/venny/) was applied. The F&G targets were regarded as potential targets through which FZT exerts anti-GA effects.
2.4. Protein-protein interaction (PPI) network construction
The PPI network of F&G targets was constructed by STRING database (version 11.5, https://www.string-db.org/) [22]. The gene symbols of F&G were submitted to STRING with the following settings: “Homo sapiens”, the confidence score ≥0.4, and K-means clustering (number of clusters set as 3) [23,24]. PPI network information was downloaded from the STRING website. Based on this information, we constructed the PPI network using Cytoscape (version 3.7.2, Agilent Technologies Company, USA) and obtained the core network by cytoNCA [25].
2.5. Herb-component-target network construction
The F&G targets and their corresponding ingredients were included in the herb-component-target network. Cytoscape (v3.7.2) was used to construct the network [25]. Each node in the network represents a herb, ingredient, or target. If an interaction occurred between nodes, they were connected by a line.
2.6. GO and KEGG pathway enrichment
GO and KEGG enrichment of the associated proteins was performed using the Metascape (http://metascape.org/). The GO analysis includes biological process (BP), molecular function (MF), and cellular component (CC) categories [26]. The results of GO and KEGG enrichment were considered significant when the P value < 0.05. Then, we used the herbs, ingredients, F&G targets, and enriched KEGG pathways to construct the herb-ingredient-target-pathway network by Cytoscape 3.7.2 [25,27].
2.7. Molecular docking
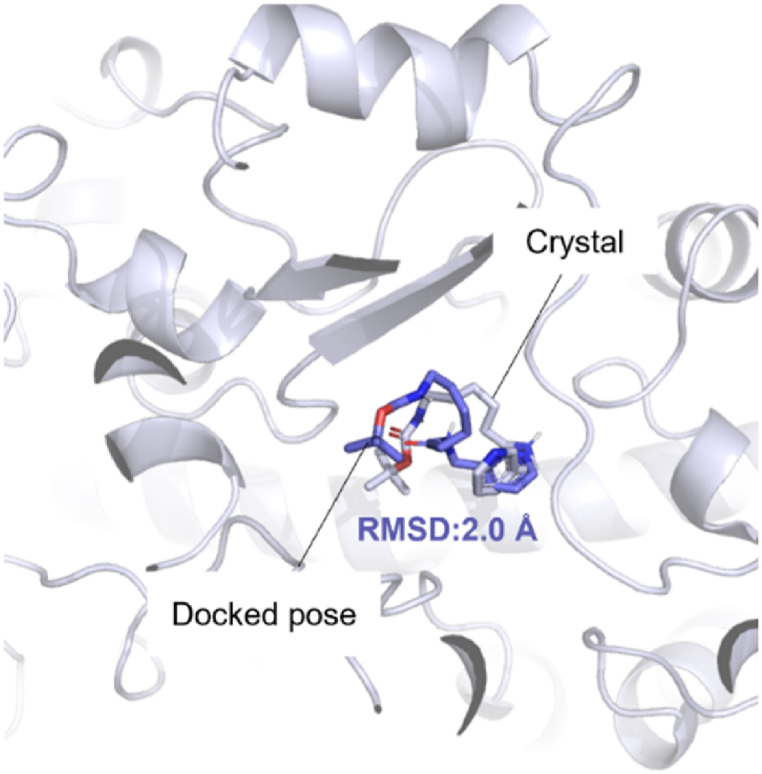
Firstly, the identifier of putative targets were chosen using the X-ray of method and high resolution as the filtration criteria on the UniProt database (https://www.uniprot.org/). Then, the crystal structure of the corresponding protein (PDB IDs:4I8V, 4D6Z, 3U9Q, 5F19, and 1MKK) was taken from PDB database (https://www.rcsb.org/). In these structures, the ligands and water motifs were removed by PyMOL v2. X software (Fig. 2). The selected proteins were added with Kollman atom charges, solvation parameters, and polar hydrogens, and the potential binding site was confirmed and detected by AutoDock Tools- 1.5.7 and CavityPlus web server (http://www.pkumdl.cn/cavityplus). Secondly, the three-dimensional (3D) structures of candidate ingredients were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). Then, the energy of the compound was minimized through ChemBio3D and converted into mol2 file format. After adding an atomic charge and assigning an atomic type, each compound was imported into AutoDock Tools- 1.5.7 [28] and save as pdbqt format for docking. Third, molecular docking was performed by AutoDock Vina software to compute the Gibbs free energy based on the hydrogen bonding, van der Waal forces, desolvation electrostatic, and rotatable bonds in the ligand [29]. The docking grid was set to 45 × 45 × 45 xyz points with a grid spacing of 0.375 Å based on the predicted active site of each protein. The reasonable docked poses of compounds binding in the corresponding protein were selected according to the docking energy as well as the binding mode in the protein‒ligand complex. The 3D presentations of binding modes were depicted by PyMOL v2. X software. To validate the accuracy of molecular docking process, the re-docking protocol by deleting the cocrystal ligand PK9 from the protein structure (PDB ID 4D6Z) was conducted for re-examination. As shown in Fig. 2, RMSD value of 2.0 Å between the docked conformation of the ligand and X-ray crystal conformation show the accuracy of the docking program. An acceptable RMSD value of 2.0 Å and reproducibility of important interactions indicated that a valid docking protocol [30].
Fig. 2.
Validation of docking protocol by redocking the co-crystal ligand PK9 in protein (PDB ID:4D6Z).
2.8. Cell culture
Human-derived THP- 1 monocyte cell lines (procell, Wuhan, China) were grown in 1640 medium (procell, Wuhan, China) supplemented with 0.05 mM β-mercaptoethanol (procell, Wuhan, China), 10 % fetal bovine serum (FBS), and 1%–2% penicillin/streptomycin (Gibicol, USA) in 5 % CO2 at 37 °C.
2.9. Inflammation model establishment in vitro
THP- 1 cells were seeded in 6-well plates (Corning, USA) at a density of 1 × 105/well and cultured until confluence. The cells were then treated with PMA (phorbol 12-myristate 13-acetate, 50 ng/mL) (InvivoGen, USA) for 3 h and subsequently stimulated with 200 μg/mL MSU crystals, which led to the formation of an inflammatory reaction. After treatment with MSU for 2 h, the cells were exposed to deoxyandrographolide at concentrations of 5, 25, and 50 μM for 12 h and then harvested for gene expression analysis or Western blot detection.
2.10. Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated by TRIzol Reagent (Solarbio, China). Total RNA (500 ng) of each sample was reverse transcribed into cDNA and amplified using a PrimeScripttm RT reagent kit with gDNA Eraser (Takara, Japan) according to the manufacturer's instructions. PCR was then conducted using the UltraSYBR Mixture (CWBIO, China). The primer sequences were as follows: GAPDH, F: 5′- ACAACTTT GGTATCGTGGAAGG -3′, R: 5′- GCCATCACGCCACAGTTTC-3’; IL 1β, F: 5′- AAAGAGAGCACACCAGTCAAT-3’; R: 5′- GGGTAAGAAGTAGCAGTGTCTGT -3’; COX2, F: 5′- GTTCCACCCGCAG TACAGAA -3′, R: 5′- AGGGCTTCAGCAT AAAGCGT -3’; IL6, F: 5′- CCACCGGG AACGAAAGAGAA -3′, R: 5′- TCTCCTG GGGGTATTGTGGA -3’; NLRP3, F: 5′- CAACCTCACGTCACACTGCT -3′, R: 5′- TTTCAGACAACCCCAGGTTC’.
2.11. Western blot
Stimulated cells were lysed in ice-cold RIPA buffer, supplemented with 1 mM phenylmethylsulfonyl fluorid (Solarbio, China). Proteins were obtained by centrifugation at 12,000×g for 20 min at 4 °C. Equal amounts of proteins (10 μg) were loaded onto a 10 % SDS‒PAGE gel unit and transferred to a nitrocellulose (NC) membrane (Millipore, USA) by electroblotting. The NC membranes were blocked with 5 % nonfat-dried milk in Tris-buffered saline/Tween-20, stained with primary antibodies against GADPH, COX2, and NLRP3 (Beyotime, China) at dilutions of 1:1000 and then incubated overnight at 4 °C. Membranes were then probed with peroxidase-conjugated secondary antibody at a dilution of 1:10,000 (CST, USA) for 1 h at room temperature. The antigen-antibody complexes were then detected with enhanced chemiluminescence reagent (Beyotime, China), visualized using an AMERSHAmersham Image Quant 800 system (GE, USA), and analyzed using ImageJ Software.
2.12. Immunofluorescence staining
THP- 1 cells were seeded on glass coverslips placed in 24-well plates. Following different treatments, cells were fixed with 2 % paraformaldehyde for 10 min, followed by permeabilization with 0.2 % Triton X- 100 in PBS for 20 min. Next, the cells were blocked in 0.3 M glycine in PBS for 1 h and incubated overnight with primary antibodies (COX2 and NLRP3) at 4 °C. Appropriate secondary antibodies were added and incubated for 3 h at room temperature. The nuclei were stained with DAPI for 1 h. Images were captured using a fluorescence microscope (Leica, Germany).
3. Results
3.1. Identifying the active ingredients and related targets of FZT
Given that the herbal formula is characterized by multiple components and multiple targets, we first screened the active ingredients with satisfactory pharmacokinetic properties from the herbal formula. The TCMSP database and TargetNet database were applied, and 77 active compounds and 417 related targets of each herb were selected after retrieval in the TCMSP database and setting the requirements of both OB ≥ 30 % and DL ≥ 0.18 (Table 1). Similarly, the related targets were also obtained from the TargetNet database by searching the canonical smiles of the active ingredients (Table 2). We set the prob ≥1 as the filtration criteria. After the duplicated target names were removed, we obtained 239 targets in total, which were then converted into gene symbols by the UniProt database for further study.
Table 1.
Active ingredients and related targets of FZT in the TCMSP database.
| Herb | The cout of ingredients | The cout of target |
|---|---|---|
| Paeonia lactiflora Pall (Bai Shao) | 13 | 123 |
| Atractylodes macrocephala Koidz (Bai Zhu) | 7 | 23 |
| Codonopsis pilosula (DangShen) | 21 | 211 |
| Poria Cocos (FuLing) | 15 | 30 |
| Aconitum carmichaeli Debeaux (Fu Zi) | 21 | 30 |
Table 2.
Active ingredients and related targets of FZT in the TargetNet database.
| Herb | The cout of ingredients | The cout of SMILES | The cout of target (prob≥ 1) |
|---|---|---|---|
| Paeonia lactiflora Pall (Bai Shao) | 13 | 8 | 125 |
| Atractylodes macrocephala Koidz (Bai Zhu) | 7 | 1 | 12 |
| Codonopsis pilosula (DangShen) | 21 | 15 | 223 |
| Poria Cocos (FuLing) | 15 | 11 | 44 |
| Aconitum carmichaeli Debeaux (Fu Zi) | 21 | 14 | 127 |
3.2. Identifying the potential targets of FZT against GA
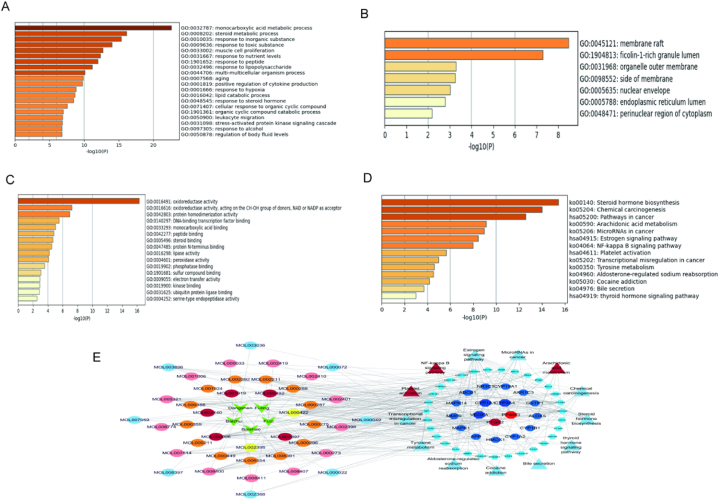
To further predict the potential targets of FZT against GA, we obtained 536 GA-related targets by using the GeneCards, OMIM, Pharmgkb, TTD, and DrugBank databases (n = 174 in DrugBank, n = 19 in TTD, n = 5 in Pharmgkb, n = 16 in OMIN, n = 322 in GeneCards). A Venn diagram between these 5 databases was shown in Fig. 3A. In total, we obtained 498 therapeutic targets of GA after removing the duplicated targets. Then, we selected the target genes at the intersection of FZT and GA (F&G). As shown in Fig. 3B, the 49 overlapping F&G targets were regarded as potential targets involved in the anti-GA effect of FZT.
Fig. 3.
Construction of the PPI network and herb-component-target network. (A) Venn diagram displaying the overlap of GA-related targets between 5 databases. (B) Venn diagram displaying the overlap between the GA-related targets and the potential targets of FZT. (C) PPI network of 49 F&G targets. (D) Screening of core targets in the PPI network. (E) The construction of the herb-component-target network.
3.3. The construction of the PPI network and herbal-component-target network
The PPI network of F&G targets was established by the STRING database. A total of 49 F&G targets contained 48 nodes and 232 edges, representing the interaction between F&G targets (Fig. 3C). Based on the frequency of each node and the combined score between two nodes, the core targets in the PPI network were screened by cytoNCA, including PPARG (Peroxisome proliferator-activated receptor gamma), CYP3A4, PTGS2, VEGFA, and CYP1A1 (Fig. 3D), which may play central roles in the anti-GA effect of FZT. Then, the 49 F&G targets and their corresponding 41 active ingredients were collected to establish the herb-component-target network by Cytoscape (Fig. 3E). The correspondence table of herbal ingredient codes is shown in Table 3.
Table 3.
Chinese medicine ingredient code correspondence table.
| MOLID | MOLName |
|---|---|
| MOL001919 | (3S,5R,8R,9R,10S,14S)-3,17-dihydroxy-4,4,8,10,14-pentamethyl-2,3,5,6,7,9-hexahydro-1H-cyclopenta [a]phenanthrene-15,16-dione |
| MOL001924 | paeoniflorin |
| MOL000211 | Mairin |
| MOL000358 | beta-sitosterol |
| MOL000359 | sitosterol |
| MOL000422 | kaempferol |
| MOL000492 | (+)-catechin |
| MOL000022 | 14-acetyl-12-senecioyl-2E,8Z,10E-atractylentriol |
| MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta [a]phenanthren-3-ol |
| MOL000049 | 3β-acetoxyatractylone |
| MOL000072 | 8β-ethoxy atractylenolide III |
| MOL001006 | poriferasta-7, 22E-dien-3 beta-ol |
| MOL002140 | Perlolyrine |
| MOL003036 | ZINC03978781 |
| MOL000449 | Stigmasterol |
| MOL003896 | 7-Methoxy-2-methyl isoflavone |
| MOL005321 | Frutinone A |
| MOL000006 | luteolin |
| MOL006774 | stigmast-7-enol |
| MOL007059 | 3-beta-Hydroxymethyllenetanshiquinone |
| MOL007514 | methyl icosa-11,14-dienoate |
| MOL008397 | Daturilin |
| MOL008400 | glycitein |
| MOL008407 | (8S,9S,10R,13R,14S,17R)-17-[(E,2R,5S)-5-ethyl-6-methylhept-3-en-2-yl]-10,13-dimethyl-1,2,4,7,8,9,11,12,14,15,16,17-dodecahydrocyclopenta [a]phenanthren-3-one |
| MOL008411 | 11-Hydroxyrankinidine |
| MOL006554 | Taraxerol |
| MOL008391 | 5alpha-Stigmastan-3,6-dione |
| MOL000273 | (2 R)-2-[(3S,5R,10S,13R,14R,16R,17R)-3,16-dihydroxy-4,4,10,13,14-pentamethyl-2,3,5,6,12,15,16,17-octahydro-1H-cyclopenta [a]phenanthren-17-yl]-6-methylhept-5-enoic acid |
| MOL000275 | trametenolic acid |
| MOL000296 | hederagenin |
| MOL000287 | 3 beta-Hydroxy-24-methylene-8-lanostene-21-oic acid |
| MOL000289 | pachymic acid |
| MOL002211 | 11,14-eicosadienoic acid |
| MOL002388 | Delphin_qt |
| MOL002392 | Deltoin |
| MOL002395 | Deoxyandrographolide |
| MOL002398 | Karanjin |
| MOL002397 | karakoline |
| MOL002401 | Neokadsuranic acid B |
| MOL002410 | 1-benzoylnapelline |
| MOL002419 | (R)-Norcoclaurine |
3.4. GO and KEGG enrichment analysis
To clarify the anti-GA mechanism of FZT, GO and KEGG enrichment analyses were conducted based on 49 F&G targets. For GO analysis, BP, CC, and MF were included, and 20 items were obtained via BP enrichment analysis (including monocarboxylic acid metabolic process, steroid metabolic process, etc.); 7 items were obtained via CC enrichment analysis (including membrane raft, ficolin-1-rich granule lumen, etc.); 16 items were generated via MF nrichment analysis (including oxidoreductase activity, monocarboxylic acid binding, etc.). The results of Fig. 4A–C displays the GO enrichment analysis in the BP, CC, and MF categories. To further elucidate the pathways regulated by the therapeutic target genes, we performed a KEGG pathway analysis. We selected the top enriched genes to construct a histogram, and KEGG pathway enrichment analysis is shown in Fig. 4D. To visualize the interactions among herbs, active ingredients, targets, and signaling pathways, the herb-ingredient-target-pathway network was constructed by Cytoscape (Fig. 4E).
Fig. 4.
GO and KEGG enrichment of 49 F&G targets. (A) The BP category of GO enrichment. (B) The CC category of GO enrichment. (C) The MF category of GO enrichment. (D) KEGG pathway enrichment in a bar plot. (E) The construction of herb-ingredient-target-pathway network.
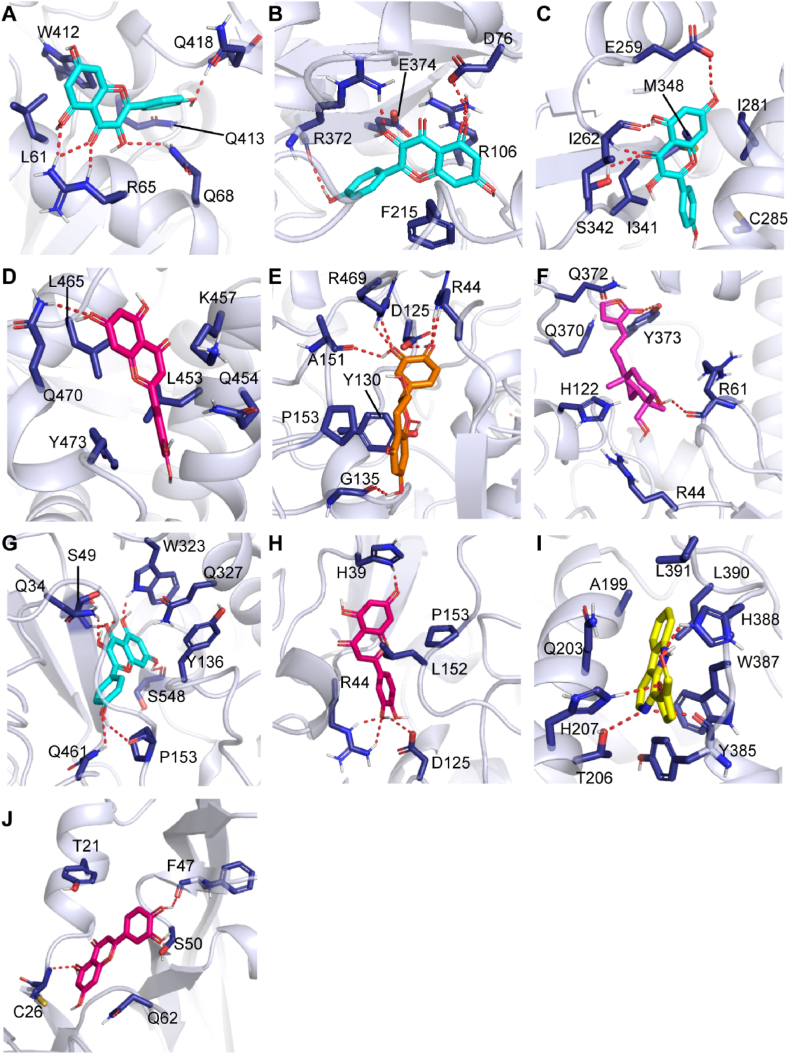
3.5. Molecular docking between the targets and compounds
To reveal the potential protein-ligand interactions between active compounds and F&G targets, we selected the active compounds that were moderate by ADMET to perform molecular docking with the core target genes by AutoDock Tools. The Gibbs free energies between active compounds and potential targets are displayed in Table 4. The results shows that the free energy of selected active compounds ranging from −7.0 to −9.8 kcal/mol, indicating a strong binding affinity between the ligand and the target protein [31]. The 3D presentations of binding modes in each protein-ligand system are depicted in Fig. 5. Three residues (R65, Q68 and Q418) in CYP1A1, two residues (R372 and D76) in CYP3A4, and three residues (I262, E259, and S342) in PPARG were involved in the hydrogen bond interaction with kaempferol (Fig. 5A–C). In the luteolin- PPARG complex, residue Q470 formed a hydrogen bond interaction with the ligand (Fig. 5D). In addition, residue L465, L453, and K457 engaged in hydrophobic interactions, and Y473 formed π-π interaction with the ligand. For the five PTGS2-bound complexes, five residues (G135, A151, R469, D125, and R44) in the catechin-bound complex, residue R61 in the deoxyandrographolide-bound complex, six residues (Q461, P153, Q34, S49, W323, and S548) in the kaempferol-bound complex, three residues (H39, R34 and D125) in the luteolin-bound complex, and four residues (T206, H207, Y385, and H388) in the Perlolyrine-bound complex participated in hydrogen bond interactions with PTGS2 (Fig. 5E–I). Meanwhile, the non-polar residues such as residues A151, L152, P153, A199, L390 and L391 of PTGS2 mainly interacts with active compounds via hydrophobic interactions. In the luteolin-VEGFA bound complex, residue C26 and F47 simultaneously formed hydrogen bond interactions with the ligand (Fig. 5J). By analyzing the binding mode of these active compounds with corresponding protein, the important residues predominantly formed hydrogen bond interactions or hydrophobic interactions with the active compounds.
Table 4.
Gibbs free energy results from molecular docking calculation.
| Target genes | PDB ID | Active ingredients ADMET: (Moderate) | Drug | Affinity (kcal/mol) |
|---|---|---|---|---|
| CYP1A1 | 4i8v | kaempferol | Baishao | −7.0 |
| CYP3A4 | 4d6z | kaempferol | Baishao | −8.7 |
| PPARG | 3u9q | kaempferol | Baishao | −7.7 |
| PPARG | 3u9q | luteolin | Dangshen | −7.9 |
| PTGS2 | 5f19 | luteolin | Dangshen | −9.8 |
| PTGS2 | 5f19 | (+)_catechin | Baishao | −9.2 |
| PTGS2 | 5f19 | Deoxyandrographolide | Fuzi | −8.2 |
| PTGS2 | 5f19 | kaempferol | Baishao | −9.2 |
| PTGS2 | 5f19 | Perlolyrine | Dangshen | −9.2 |
| VEGFA | 1mkk | luteolin | Dangshen | −7.8 |
Fig. 5.
Binding modes of active compounds with potential targets derived from molecular docking calculations. (A–C) The ligand kaempferol (cyan) in the CYP1A1, CYP3A4 and PPARG. (D–J) The compounds luteolin (red), catechin (orange), Deoxyandrographolide (purple), kaempferol (cyan), luteolin (red), Perlolyrine (yellow) and luteolin (red) in the PPARG, PTGS2, PTGS2, PTGS2, PTGS2, PTGS2, and VEGFA, respectively. The ligands in the binding site are shown a stick model, and the protein structures are represented as cartoon. The activate residues involve in the interaction with ligand are shown as blue stick and the red dashed line denotes hydrogen bonding. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
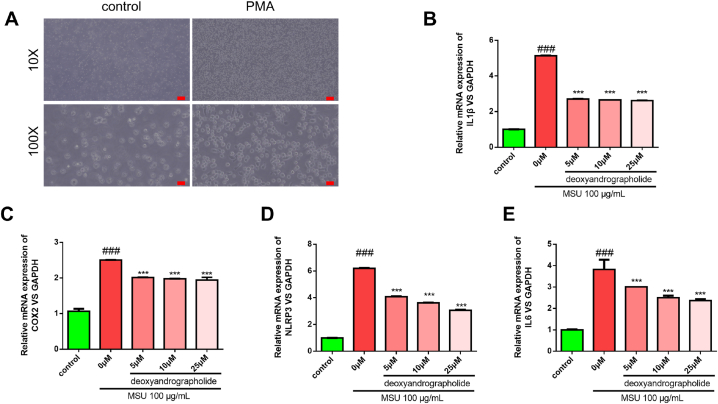
3.6. Experimental verification
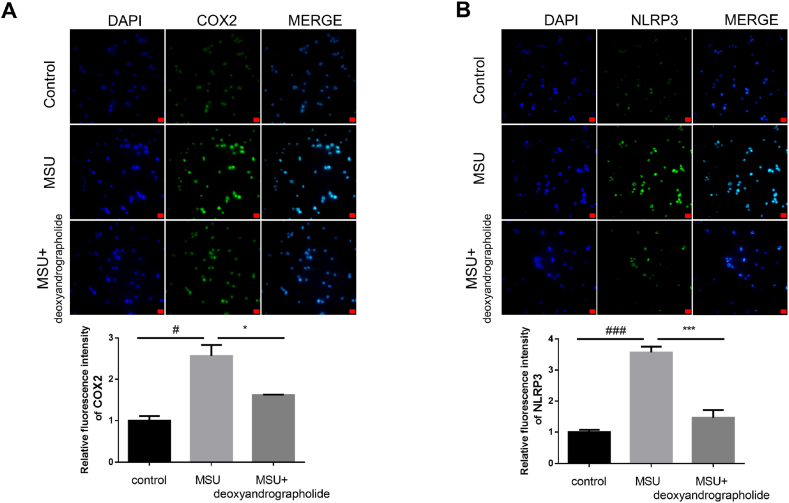
To elucidate the anti-GA effects of FZT in vitro, experimental verification was employed. The THP- 1 monocytes transformed into macrophages after treatment with PMA (Fig. 6A). Relative expression of inflammation biomarkers (IL- 1β, COX2, NLRP3, and IL-6) were detected by Q-PCR or Western blotting. The Q-PCR results showed that the inflammation biomarkers were significantly increased when treated with MSU. However, the groups treated with deoxyandrographolide (5, 10, and 25 μM) exhibited a decreasing trend in IL- 1β, COX2, NLRP3, and IL-6 expression compared with the MSU groups (Fig. 6B–E). We next detected the protein expression of COX2 and NLRP3 by western blotting. The results showed that their expression was significantly increased with MSU treatment while they were decreased with deoxyandrographolide treatment (25 μM) (Fig. 7A–B). The immunofluorescence results further confirmed that the deoxyandrographolide treatment had a good anti-inflammatory effect. As shown in Fig. 8A–B, the COX2 and NLRP3 expression was suppressed with deoxyandrographolide treatment compared with MSU treatment alone. In summary, these results indicated that deoxyandrographolide effectively reduced the inflammatory response of monocyte cells by regulating the expression of inflammation-related factors, which in turn verified the results of the network pharmacology analysis.
Fig. 6.
Deoxyandrographolide alleviated the MSU-induced inflammation by influencing the mRNA expression of IL-1β, COX2, NLRP3, and IL-6. (A) Representative images of cell adherence after PMA treatment (the original magnification was 100 px). (B–E) Monocyte cells were exposed to 100μ g/mL MSU for 2 h to induce inflammation. Then, monocyte cells were treated with different concentrations of deoxyandrographolide (5, 20, and 50 μM) for 12 h. Relative mRNA expression levels of IL- 1β, COX2, NLRP3, and IL-6 were detected by Q-PCR. The quantitative data are shown as the mean ± SD (n = 3). ***r < 0.001 versus the MSU group. ###r < 0.001 versus the control group.
Fig. 7.
Deoxyandrographolide alleviated the MSU-induced inflammation by influencing the protein expression of COX2 and NLRP3. (A–B) Monocyte cells were exposed to 100 μg/mL MSU for 2 h to induce inflammation. Then, monocyte cells were treated with 25 μM deoxyandrographolide for 12 h. Relative protein expression levels of COX2 and NLRP3 were detected by western blotting assay. GAPDH was used as the loading control. The quantitative data are shown as the mean ± SD (n = 3). *P < 0.05, versus the MSU group. #P < 0.05, ##P < 0.01 versus the control group.
Fig. 8.
The anti-inflammatory effect of deoxyandrographolide detected by immunofluorescence assay. (A–B) Monocyte cells were exposed to 100 μg/mL MSU for 2 h to induce inflammation. Then, monocyte cells were treated with 25 μM deoxyandrographolide for 12 h. The immunofluorescence of COX2 and NLRP3 was observed by a fluorescence microscope and the representative images are shown (scale bar as 20 μm). Quantification of fluorescence intensity was calculated by Image-Pro software. Each column represents the mean ± SD (n = 5). *P < 0.05, ***P < 0.001 versus the MSU group. #P < 0.05, ###P < 0.001 versus the control group.
4. Discussion
At present, the pathogenesis of GA is mainly related to metabolism, inflammation, and immunity [32]. The final metabolic product of purines is uric acid, which forms urate and is deposited in joints and tissues, causing GA. Second, mono-urate crystals are deposited in the articular cavity, cartilage, and kidney, which mediate the inflammatory response of GA by activating phagocytes, inflammatory bodies and Toll-like receptors [33]. As a multifactorial inflammatory disorder, the prevalence of GA worldwide is 2–4%, and the disability rate is increasing yearly [34]. FZT is a classical TCM formula and has been widely used to treat joint pain diseases for thousands of years. Previous studies have also confirmed that FZT is positive and effective in the treatment of GA [35]. However, the underlying mechanism of its anti-GA effects is elusive. Thus, we explored the potential mechanisms of the therapeutic action of FZT against GA by a network pharmacology approach and molecular docking analysis.
In this study, we identified 49 important targets and 41 active ingredients for herb-ingredient-target-disease network construction. The complex network demonstrated that most components of FZT adjusted multiple targets. The results of this study showed that five key ingredients of FZT, kaempferol, luteolin, catechin, perlolyrine, and deoxyandrographolide, had strong binding effects on the target proteins based on the Gibbs free energy of binding score. The Gibbs free energy of binding, which was reported to be greater than 6.0 kcal/mol (the absolute value of the docking score), indicates the possibly active drug, and a Gibbs free energy less than 6.0 kcal/mol means an inactive drug [31]. Kaempferol is considered to possess significant anti-inflammatory properties through regulation of pro-inflammatory enzyme activity and inflammation-related gene expression [36]. Increasing evidence have demonstrated that kaempferol, luteolin, and catechin inhibited the activity of xanthine oxidase to reduce excessive production of uric acid in the liver and regulate the expression of uric acid transporters [[37], [38], [39]]. Luteolin, a plant-derived flavonoid, has been verified to possess anti-inflammatory properties by activating the NF-κB signaling pathway and reducing pro-inflammatory cytokines in macrophages [40]. Catechin can inhibit the inflammatory factor IL-6, thus displaying anti-inflammatory effects and protecting articular cartilage [41]. Deoxyandrographolide exhibits anti-inflammatory functions, which is possibly connected with the inhibition of NF-κb [42]. Perlolyrine is an alkaloid compound with pain-relieving, circulation-boosting and anti-thrombotic properties [43]. According to these results, we considered that the mechanism of activated ingredients of FZT mainly focused on anti-inflammatory, analgesic, antioxidative stress and uric acid-lowering effects,explaining the efficacy in the treatment of GA.
By means of PPI network analysis, we finally identified five central targets, including PPARG, CYP3A4, PTGS2, VEGFA, and CYP1A1 that may play core roles in the anti-GA effects of FZT. PPARG regulates the expression of a large number of genes, participates in the physiological and pathological regulation of the body, and inhibits the production of inflammatory signaling pathways and inflammatory mediators [44,45]. Previous studies have shown that PPARG plays a key role in anti-inflammatory and analgestic effects [46,47]. CYP3A4 exhibits good anti-inflammatory activity by catalyzing two major metabolites in humans, M − 2 and M − 5 [48]. PTGS2, also known as COX2, is a key rate-limiting enzyme in arachidonic acid metabolism. It widely exists in immune and inflammatory processes and plays an important role in inflammation and pain [49,50]. VEGF is a highly specific vascular endothelial growth factor that can enhance vascular permeability and promote the exudation of vascular substances, thus stimulating the formation and development of inflammation [51]. In 1999, Maeno found that the level of serum VEGF was positively correlated with joint inflammation, and it was considered that the level of VEGF in peripheral blood could be used as an important index to judge the severity and prognosis of GA [52]. CYP1A1 could contribute to relieving damage caused by inflammatory overreaction [53]. Therefore, the mechanisms of FZT in the treatment of GA may be related to the inhibition of the immune inflammatory response and analgesia. The results of network pharmacology and molecular docking showed that FZT may play a therapeutic role in GA by regulating multiple targets, such as PPARG, CYP3A4, PTGS2, VEGFA, and CYP1A1, as well as regulating multiple biological functions or pathways, such as the immune inflammatory response, oxidative stress reaction and uric acid production.
According to KEGG enrichment analysis, the pharmacological effect of FZT on GA mainly includes arachidonic acid metabolism and the NF-κB signaling pathway. Arachidonic acid is a kind of unsaturated fatty acid that produces a variety of metabolites through three pathways cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP450), causing different inflammatory reactions [54]. The activation of NF- κB signaling is the core medium of the inflammatory response, which increases the expression of inflammatory factors such as IL- 1β, IL-6, and TNF-α, thus leading to the inflammatory response of GA [55,56]. The results demonstrated that FZT may relieve GA by inhibiting arachidonic acid metabolism and NF-κB signaling and reducing the expression of inflammatory factors.
In this study, molecular docking simulations were used to identify the potential protein binding sites and decipher the potential binding mechanism of active compounds binding to proteins at the molecular level. By analyzing the molecular mechanism of active compounds of FZT, such as the binding site residues involved in hydrogen bonding, hydrophobic interactions with ligand, etc., these important residues may impact the function of corresponding proteins, such as inflammasome signal transduction. This enables us to better understand the polypharmacology of their effects. Based on the above analysis, we noticed that deoxyandrographolide has never been reported in treating GA. Then, THP- 1 monocyte cells with an inflammatory response induced by MSU were used as a cell model. Through Q-PCR and Western blot detection, it was found that the expression of inflammatory biomarkers in the MSU-treatment group was significantly increased, suggesting that MSU induced an inflammatory response in THP- 1 monocytes, and that the cell model was reliable. The pathogenesis of GA is closely linked to the production of large amounts of inflammatory factors such as IL-1β. The NLRP3 inflammasome signaling pathway indirectly induces the maturation and secretion of IL- 1β through the activation of caspase- 1, resulting in an inflammatory response [57]. The expression of IL-1β, COX2, NLRP3, and IL-6 were decreased in the deoxyandrographolide-treatment group, indicating that deoxyandrographolide could reduce the inflammatory reactionin GA model in vitro. These results further provided theoretical basis for explaining FZT used in treating GA.
However, our study still has some limitations. First, all of the active ingredients were screened from the databases, but the core components were not verified by mass spectrometry due to the limitation of research conditions. Then, we proved in vitro that deoxyandrographolide resisted MSU-induced inflammasome release from macrophages but failed to carry out the experiment in vivo and observe its effect on uric acid metabolism.
5. Conclusions
Chinese medicine plays a crucial role in treating GA. In this study, five key ingredients and five central targets were screened based on network pharmacological analysis. Molecular docking simulation identified the important residues that involved the key interactions with active compounds of FZT, which may influence the immune inflammatory response, oxidative stress reaction and uric acid production by regulating the expression of PPARG, CYP3A4, PTGS2, VEGFA, and CYP1A1, thus playing a role in the treatment of GA. The results of in vitro experiments also showed that the active ingredient (deoxyandrographolide) of FZT could reduce the level of inflammation biomarkers and reduce the inflammatory response to GA. In summary, our research not only provides a comprehensive understanding of the active components and molecular mechanism of FZT in the treatment of GA, but also identifies a novel anti-inflammatory compound in treating GA. At last, our paper provides new idea for further development and clinical application of FZT in the treatment of GA.
Funding
This work was financially supported by the key specialty construction project of traditional Chinese medicine in Shenzhen, the scientific research project of Guangdong Provincial Bureau of traditional Chinese Medicine (Nos. 20211338, 20232148), and the health public welfare project in Futian District, Shenzhen (Nos. FTWS2019043, FTWS2022076).
Data availability
All data generated or analyzed during this study are included in this published article.
CRediT authorship contribution statement
Yufeng Xie: Writing – original draft, Conceptualization. Zhongxiao Lin: Writing – review & editing, Conceptualization. Jianmei Zhang: Writing – original draft. Yun Chen: Writing – review & editing, Supervision. Jianhao Huang: Data curation. Hong Tang: Data curation. Jieting Chen: Writing – review & editing. Yuhe Lei: Writing – review & editing. Ziliang Qian: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Yuhe Lei, Email: 448598296@qq.com.
Ziliang Qian, Email: 2283373364@qq.com.
References
- 1.Lin Y., Luo T., Weng A., et al. Gallic acid alleviates gouty arthritis by inhibiting NLRP3 inflammasome activation and pyroptosis through enhancing Nrf 2 signaling. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.580593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang Z.H., Waizy H. Current concepts in the treatment of gouty arthritis. Orthop. Surg. 2013;5(1) doi: 10.1111/os.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X., Wu M., Xie Y., et al. Identification of Glycine receptor α3 as a colchicine-binding protein. Front. Pharmacol. 2018;9:1238. doi: 10.3389/fphar.2018.01238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J.C., Lin C.L., Chen M.C., et al. Gout, not hyperuricemia alone, impairs left ventricular diastolic function. Arthritis Res. Ther. 2015;17:323. doi: 10.1186/s13075-015-0842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwabl C., Taljanovic M., Widmann G., et al. Ultrasonography and dual-energy computed tomography: impact for the detection of gouty deposits. Ultrasonography (Seoul, Korea) 2021;40(2):197–206. doi: 10.14366/usg.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Y.Y., Jean Y.H., Lin S.C., et al. Etoricoxib prevents progression of osteolysis in repeated intra-articular monosodium urate-induced gouty arthritis in rats. J. Adv. Res. 2020;24:109–120. doi: 10.1016/j.jare.2020.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emanuela R., Cecilia C., Yew T.S., et al. Cyclooxygenase-2, asymmetric dimethylarginine and the cardiovascular hazard from NSAIDs. Circulation. 2018;138(21):2367–2378. doi: 10.1161/CIRCULATIONAHA.118.033540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan W.X. Fujian University of Traditional Chinese medicine; 2021. Clinical Study on Treatment of Acute Gouty Arthritis of Taiyin Shaoyin Syndrome with Modified Fuzi Decoction Combined with Pricking and Cupping. [D]; [Fuzhou (Fujian)] [Google Scholar]
- 9.Dong L.L., Feng C.R., Zhao Y.Y., et al. Effect and mechanism of Shaogan Fuzi decoction on rats with collagen-induced arthritis based on PPAR- γ/NF- κ B signal pathway. Central South Pharm. 2022;20(1):45–51. [Google Scholar]
- 10.Nesterova Y.V., Povet’yeva T.N. Analgesic activity of diterpene alkaloids from Aconitum baikalensis. Bull. Exp. Biol. Med. 2014;157(4):488–491. doi: 10.1007/s10517-014-2598-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q., He X.Y., Zhou M.J., et al. Research progress on chemical components, pharmacological effects and clinical application of radix paeoniae alba. Clinic. Res. Practice. 2021;6(11):187–189. [Google Scholar]
- 12.Zhang W.X., Su P., Zhao A. J. Research progress on processing methods and pharmacological effects of baizhu(atractylodes macrocephala) Guiding J. Tradit. Chin. Med. Pharm. 2022;28(5):110–115. [Google Scholar]
- 13.Yang Z.H., Zhou H.L. Effects of Codonopsis pilosum polysaccharide on the proliferation, apoptosis and expression of inflammatory factors in gastric cancer cells AGS by regulating miR-361-5p/TLR4/NF-κB pathway. Immunol. J. 2022;38(4):347–353. [Google Scholar]
- 14.Chao C.L., Wang C.J., Huang H.W., et al. Poria cocos modulates Th1/Th2 response and attenuates airway inflammation in an ovalbumin-sensitized mouse allergic asthma model. Life. 2021;11(5) doi: 10.3390/life11050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Wang Z.Y., Zheng J.H., et al. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin. J. Nat. Med. 2021;19(1):1–11. doi: 10.1016/S1875-5364(21)60001-8. [DOI] [PubMed] [Google Scholar]
- 17.Stelzer G., Rosen N., Plaschkes I., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protocols Bioinf. 2016;54 doi: 10.1002/cpbi.5. 1.30.1-1..3. [DOI] [PubMed] [Google Scholar]
- 18.Amberger J.S., Bocchini C.A., Schiettecatte F., et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic acids Res. 2015;43(Database issue):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whirl-Carrillo M., Mcdonagh E.M., Hebert J.M., et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Therapeut. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y.H., Yu C.Y., Li X.X., et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic acids Res. 2018;46(D1) doi: 10.1093/nar/gkx1076. D1121-d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart D.S., Feunang Y.D., Guo A.C., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic acids Res. 2018;46(D1) doi: 10.1093/nar/gkx1037. D1074-d82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Gable A.L., Nastou K.C., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic acids Res. 2021;49(D1) doi: 10.1093/nar/gkaa1074. D605-d12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Chen L., Ju L., et al. Novel biomarkers associated with progression and prognosis of bladder cancer identified by Co-expression analysis. Front. Oncol. 2019;9:1030. doi: 10.3389/fonc.2019.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D., Morris J.H., Cook H., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids Res. 2017;45(D1) doi: 10.1093/nar/gkw937. D362-d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The Gene Ontology Resource 20 years and still GOing strong. Nucleic acids Res. 2019;47(D1) doi: 10.1093/nar/gky1055. D330-d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M., Furumichi M., Sato Y., et al. KEGG: integrating viruses and cellular organisms. Nucleic acids Res. 2021;49(D1) doi: 10.1093/nar/gkaa970. D545-d51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodsell D.S., Sanner M.F., Olson A.J., et al. The AutoDock suite at 30. Protein Sci. : Publ. Protein Soc. 2021;30(1):31–43. doi: 10.1002/pro.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen N.T., Nguyen T.H., Pham T.N.H., et al. Autodock Vina adopts more accurate binding poses but Autodock 4 forms better binding affinity. J. Chem. Inf. Model. 2020;60(1):204–211. doi: 10.1021/acs.jcim.9b00778. [DOI] [PubMed] [Google Scholar]
- 30.Castro-Alvarez A., Costa A.M., Vilarrasa J. The performance of several docking programs at reproducing protein-macrolide-like crystal structures. Molecules. 2017;22(1) doi: 10.3390/molecules22010136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shityakov S., Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Comput. Biol. Chem. Adv. Appl. : AABC. 2014;7:23–36. doi: 10.2147/AABC.S63749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akahoshi T. [Pathological mechanisms of gouty arthritis] Nihon Rinsho. 2008;66(4):705–710. [PubMed] [Google Scholar]
- 33.Martinon F. Mechanisms of uric acid crystal-mediated autoinflammation. Immunol. Rev. 2010;233:218–232. doi: 10.1111/j.0105-2896.2009.00860.x. [DOI] [PubMed] [Google Scholar]
- 34.Galozzi P., Bindoli S., Doria A., et al. Autoinflammatory features in gouty arthritis. J. Clin. Med. 2021;10(9):1880. doi: 10.3390/jcm10091880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu B., Xu Q.Q., Wang C. 44 cases of acute gouty arthritis with wind-cold-dampness arthralgia syndrome by warming needle moxibustion combined with modified Fuzi decoction. Zhejiang J. Tradit. Chin. Med. 2021;56(12):912. [Google Scholar]
- 36.Devi K.P., Malar D.S., Nabavi S.F., et al. Kaempferol and inflammation: from chemistry to medicine. Pharmacol. Res. 2015;99:1–10. doi: 10.1016/j.phrs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Yu H.F., Huang L.S., Han J.X., et al. Study on the effect of luteolin on reducing uric acid in hyperuricemia mice and its mechanism. Lishizhen Med. Mater. Med. Res. 2021;32(5):1071–1074. [Google Scholar]
- 38.Wu D., Zhang W., Lai X., et al. Regulation of catechins on uric acid metabolism disorder related human diseases. Mini Rev. Med. Chem. 2020:20. doi: 10.2174/1389557520666200719015919. [DOI] [PubMed] [Google Scholar]
- 39.Mehmood A., Rehman A.U., Ishaq M., et al. In vitro and in silico xanthine oxidase inhibitory activity of selected phytochemicals widely present in various edible plants. Comb. Chem. High Throughput Screen. 2020;23(9):917–930. doi: 10.2174/1386207323666200428075224. [DOI] [PubMed] [Google Scholar]
- 40.Seelinger G., Merfort I., Schempp C.M. Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. 2008;74(14):1667–1677. doi: 10.1055/s-0028-1088314. [DOI] [PubMed] [Google Scholar]
- 41.Sun D.D., Cheng H.B., Shen W.X., et al. In vitro anti-inflammatory activity of five flavonoids from bushen huoxue recipe based. Chin. J. Exp. Tradit. Med. Formulae. 2015;21(7):137–141. [Google Scholar]
- 42.Nguyen V.S., Lon, Yi X., et al. Specificity and inhibitory mechanism of andrographolide and its analogues as antiasthma agents on NF-kappa B p50. J. Nat. Prod. 2015;78(2) doi: 10.1021/np5007179. [DOI] [PubMed] [Google Scholar]
- 43.Liu X.M. Dalian University of Technology; 2019. Systems Pharmacology Study on the Mechanism of Action of Chuanxiong-Shengdi Herb Pair [D]; [Dalian (Liaoning)] [Google Scholar]
- 44.Ren L.J., Zhao H., Bi B. Research progress of peroxisome proliferator-activated receptor gamma in inflammatory-related diseases. Lab. Med. 2017;32(2):153–157. [Google Scholar]
- 45.Ahmadian M., Suh J.M., Hah N., et al. PPARγ signaling and metabolism: the good, the bad and the future. Nat. Med. 2013;19(5):557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeda T., Kishioka S. PPAR and pain. Int. Rev. Neurobiol. 2009;85(85):165–177. doi: 10.1016/S0074-7742(09)85013-7. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-Boyanapalli R., Frasch S.C., Riches D.W., et al. PPARγ activation normalizes resolution of acute sterile inflammation in murine chronic granulomatous disease. Blood. 2010;116(22):4512–4522. doi: 10.1182/blood-2010-02-272005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui Y.L. Dalian Medical University; 2017. Study on the in Vitro Metabolism of AKBA and KBA of Boswellia Serrata [D]; [Dalian (Liaoning)] [Google Scholar]
- 49.Menon V.P., Sudheer A.R. Antioxidant and anti-inflammatory properties of curcumin. Oxygen Transport Tissue XXXIII. 2007;595(1):105. doi: 10.1007/978-0-387-46401-5_3. [DOI] [PubMed] [Google Scholar]
- 50.Gan K.L., Li H.C., Peng X.P. Effect of optimization of wuhua tea formula on febrile model rats and its effect on liver and kidney. World Chin. Med. 2019;14(8):1999–2003. [Google Scholar]
- 51.Liang Q.H., He J.H., Li X.L., et al. Effect of bizhongxiao decoction on the expression of VEGF in the synovial membrane of CⅡ-induced rheumatoid arthritis in rats [J] J. Cent. S. Univ. 2002;(6):491–494. [PubMed] [Google Scholar]
- 52.Maeno N., Takei S., Imanaka H., et al. Increased circulating vascular endothelial growth factor is correlated with disease activity in polyarticular juvenile rheumatoid arthritis. J. Rheumatol. 1999;26(10):2244–2248. [PubMed] [Google Scholar]
- 53.Zhang W.Y., Wang H., Qi S., et al. CYP1A1 relieves lipopolysaccharide-induced inflammatory responses in bovine mammary epithelial cells. Mediat. Inflamm. 2018;2018:1–10. doi: 10.1155/2018/4093285. 2018-2-28), 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T., Fu X., Chen Q., et al. Arachidonic acid metabolism and kidney inflammation. Int. J. Mol. Sci. 2019;20(15) doi: 10.3390/ijms20153683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Q., Bing W., Di Y., et al. Kinsenoside screening with a microfluidic chip attenuates gouty arthritis through inactivating NF-κB signaling in macrophages and protecting endothelial cells. Cell Death Dis. 2016;7(9) doi: 10.1038/cddis.2016.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q., Lin B., Li Z., et al. Cichoric acid ameliorates monosodium urate-induced inflammatory response by reducing NLRP3 inflammasome activation via inhibition of NF-kB signaling pathway. Evid. base Compl. Alternative Med. : eCAM. 2021;2021 doi: 10.1155/2021/8868527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.