Abstract
A Tn10 insertion affecting SEF14 fimbrial synthesis in Salmonella enteritidis was located 13 bp upstream of a gene designated fimU. The 77-bp DNA sequence of fimU from S. enteritidis was identical to that of fimU encoding tRNAArg (UCU) from Salmonella typhimurium and 96% identical to that of the Escherichia coli argU homolog. Furthermore, the open reading frame adjacent to and overlapping the 3′ end of fimU was similar to the prophage DLP12 integrase gene. The fimU-encoded transcript comigrated with total cellular tRNA and was predicted to form a tRNA-like cloverleaf structure containing the arginine anticodon UCU. Thus, fimU encoded a tRNAArg specific for the rare codon AGA. fimU mapped to the SEF21 fim operon located 15 C’s from the sef14 gene cluster. Although fimU was located within the SEF21 fim gene cluster, the fimU Tn10 insertion mutant of S. enteritidis was found to be defective in SEF14 as well as SEF21 (type 1) fimbria production. SEF17 and SEF18 fimbria production was not affected. Complementation of this mutant with plasmid-borne fimU restored normal production of the fimbrins SefA and FimA as well as their respective fimbriae SEF14 and SEF21. This is the first description of tRNA simultaneously controlling the production of two distinct fimbriae.
Regulation of fimbria biosynthesis in bacteria is multifactorial and complex. In Escherichia coli, the expression of type 1 fimbriae is transcriptionally regulated in part by an inversion-dependent, phase-variable mechanism that involves two site-specific recombinases (17, 24, 27) and a tRNALeu molecule (32). tRNALeu, specific for the rare leucine codon UUG, stimulates type 1 fimbria synthesis by influencing the switch from phase off to phase on (35). Recently, type 1 fimbria expression in Salmonella typhimurium has been shown to be regulated by mechanisms that are different from those controlling type 1 fimbria expression in E. coli (41). However, a common regulatory theme does exist in that a tRNA, specific for the rare arginine codons AGA and AGG, is required (40). Swenson et al. (40) suggest that the amount of tRNAArg (UCU) available in S. typhimurium may influence the expression of three genes encoding regulatory proteins of the fim gene cluster, since in each of these genes there is a high frequency of rare AGA codons recognized by tRNAArg (UCU).
Salmonella enteritidis 27655-3b produces at least four fimbrial types: SEF17 (10), SEF18 (6), SEF21 (type 1 fimbriae) (30), and SEF14 (7, 14). Although little is known about how the expression of the operons is regulated, SEF21 and SEF14 fimbriae are produced under similar environmental conditions (5, 12). Thus, the question arises as to whether or not their expression is coregulated. In a previous study, a Tn10 insertion mutant, S. enteritidis 3b-122, was generated which no longer produced SEF14 fimbriae and carried the transposon outside of sefA, the structural gene for these fimbriae (14). Further characterization of 3b-122 in this study indicated that this mutant was also defective in type 1 fimbria (SEF21) production, suggesting that the Tn10 interrupted a gene whose product coregulated the expression of both SEF14 and SEF21 fimbriae. The results of this study show for the first time that the production of two fimbriae is coregulated by the same tRNA.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
S. enteritidis 27655-3b, originally isolated from human feces, was provided by T. Wadstrom (University of Lund, Lund, Sweden). S. enteritidis 27655-3b-122, a Tn10 insertion mutant of the parent strain, was constructed by Feutrier et al. (14). E. coli DH5α and S. enteritidis 3b-122 were used as hosts for pSFA (11), pLU/TA 4-1, and pGEM-T1. To create pLU/TA 4-1, PCR-amplified fimU was cloned into pGEM-T (Promega Corp.), a TA cloning vector containing 3′-terminal thymidines. To create pGEM-T1, the 3′-overhanging thymidines of pGEM-T were filled in with dATP and T4 DNA polymerase prior to ligation (38).
Bacteria were grown at 37°C with shaking in Luria-Bertani (LB) broth (36) supplemented with ampicillin to a final concentration of 250 μg/ml except where noted. To analyze the production of fimbriae by S. enteritidis, the cells were grown in various liquid media under different growth conditions (Table 1). Cultures grown in LB broth and terrific broth (TFB) (38) were transferred to ice 24 h after inoculation, whereas cultures grown in colonization factor antigen (CFA) medium (13) and T broth (10) were transferred to ice 48 h after inoculation. All the cultures were standardized to an optical density at 630 nm (OD630) of 1.
TABLE 1.
Production of SefA, FimA, AgfA, and SefD fimbrins by S. enteritidis 3b and S. enteritidis 3b-122
| Mediuma | °C | rpm | Fimbrin productionb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
S. enteritidis 3b
|
S. enteritidis 3b-122
|
|||||||||
| SefA | FimA | AgfA | SefD | SefA | FimA | AgfA | SefD | |||
| TFB | 28 | 0 | − | + | − | +++ | − | − | − | +++ |
| TFB | 250 | − | ++ | ± | +++ | − | − | − | +++ | |
| TFB | 37 | 0 | + | ++ | − | +++ | − | − | − | +++ |
| TFB | 250 | −c | ++ | ± | +++ | − | − | + | +++ | |
| LB | 28 | 0 | − | ++ | ++ | +++ | − | − | − | +++ |
| LB | 250 | − | ++ | +++ | +++ | − | − | +++ | +++ | |
| LB | 37 | 0 | +++ | +++ | ++ | +++ | − | − | − | +++ |
| LB | 250 | ++ | ++ | +++ | +++ | − | − | + | +++ | |
| CFA | 28 | 0 | − | ++ | +++ | +++ | − | ± | +++ | +++ |
| CFA | 250 | − | + | +++ | +++ | − | ± | +++ | +++ | |
| CFA | 37 | 0 | +++ | ++ | +++ | +++ | − | ± | +++ | +++ |
| CFA | 250 | −c | + | +++ | +++ | − | ± | +++ | +++ | |
| T | 28 | 0 | − | ++ | +++ | +++ | − | − | +++ | +++ |
| T | 250 | − | + | +++ | +++ | − | − | +++ | +++ | |
| T | 37 | 0 | + | ++ | +++ | +++ | − | − | +++ | +++ |
| T | 250 | −c | + | +++ | +++ | − | − | +++ | +++ | |
All cultures were grown in liquid medium for 24 (TFB and LB) or 48 h (T and CFA).
The intensities of fimbrin bands detected on Western blots are indicated as follows: −, none detected; ±, very faint; +, weak; ++, moderate; +++, strong.
SefA was not detected on four of six Western blots but a very faint SefA band was detected on two of six Western blots.
Subcloning Tn10 from S. enteritidis 27655 3b-122.
S. enteritidis 3b-122 chromosomal DNA was isolated by the method of Alm et al. (1), purified by CsCl centrifugation (38), and digested with HindIII. To subclone the Tn10-containing chromosomal DNA fragment, size fractionated HindIII fragments (2 to 3 and 3 to 5 kb) were purified from an agarose gel with Sephaglas (Pharmacia Biotech), ligated to HindIII-digested and -dephosphorylated cloning vector pTZ19R, and then introduced into E. coli DH5α by transformation (38). A total of 2,880 colonies grown on Hybond N+ membranes (Amersham) were screened by hybridization to the oligonucleotide probe Tn10 IS10L+R (5′ GCAGAATTGGTAAAGAGA 3′). This probe, complementary to the sequence located 134 bp inside the insertion sequence of Tn10, was used to identify Tn10-containing clones. The probe, end labelled with [γ-32P]ATP, was hybridized to the membranes at 45°C in prehybridization buffer (38) containing 200 μg of herring sperm DNA (Sigma)/ml. Following hybridization, the membranes were washed in 0.2× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate (SDS) at 45°C, and the results were recorded by autoradiography on Kodak BioMax film.
DNA sequencing and computer analyses.
The Tn10-positive clones and the three fimU PCR products amplified with primers located outside the fimU gene were sequenced with Sequenase version 2 (United States Biochemicals). The custom oligonucleotide primer Tn10IS10L+R was synthesized on a PCR-MATE EP model 391 DNA synthesizer (Applied Biosystems Inc.). The DNA sequences obtained were analyzed with DNA Strider 1.1 (26). Similarity searches of the National Center for Biotechnology Information (NCBI) databases were conducted with the program BLASTN (2).
PCR amplification of fimU.
Custom oligonucleotide primers fimULT (TAATAGCGATACGCAGAATTCAAAAATATCCTACACGGCAGG) and fimULB (CAGATATGCTCACCTAAGCTTTAATCATTTAACGGAACACGG) were designed based on the S. typhimurium chromosomal DNA sequence flanking fimU and were synthesized by Gibco BRL. fimU was PCR amplified from a previously prepared cosmid clone, pPB523 (12), with fimULT and fimULB. To facilitate the cloning of the amplified product, the primers were designed to contain an EcoRI site and a HindIII site, respectively (underlined). Amplification was carried out in a 100-μl reaction volume containing 10 μl of pPB523 (0.01 μg/ml), 25 pmol of each primer, the four deoxynucleotide triphosphates (Boehringer Mannheim) at 0.5 mM each, and 2 U of Taq DNA polymerase (Boehringer Mannheim) in reaction buffer consisting of 50 mM Tris-HCl (pH 8.5), 20 mM KCl, 2.5 mM MgCl2, and 0.5 mg of bovine serum albumin/ml. The Taq enzyme was added after an initial 3-min denaturation step at 95°C (4). Thermocycling was performed in a PTC-100TM Programmable Thermal Controller (MJ Research Inc.) as follows: 1 cycle of 75°C, 1 min; 50°C, 2 min; 74°C, 2 min and 30 cycles of 95°C, 1 min; 50°C, 1 min; 74°C, 2 min, followed by an 8-min elongation at 74°C.
Subcloning PCR-amplified fimU.
PCR-amplified fimU was purified from a 1% agarose gel with Sephaglas, ligated to pGEM-T according to the manufacturer’s instructions (Promega Corp.), and then transformed into E. coli DH5α (38).
Mapping of fimU on genomic restriction maps of Salmonella and E. coli strains.
The fimU gene was mapped as previously described for the four fimbrin genes sefA, agfA, fimA, and sefD (8). The fimU probe, prepared by running EcoRI- and HindIII-digested pLU/TA 4-1 on a 1% agarose gel and purifying the fragment with Sephaglas, was labelled with [α32P]dATP (Pharmacia Biotech) by nick translation. The radiolabelled fimU probe was hybridized to nitrocellulose blots containing XbaI- and BlnI-digested E. coli, S. typhimurium, and S. enteritidis genomic DNA separated by pulsed field gel electrophoresis (blots were provided by K. Sanderson and S.-L. Liu; see reference 8).
RNA extraction and Northern blot analysis.
Total RNA was prepared from S. enteritidis 3b, 3b-122, 3b-122 pGEM-T1, and 3b-122 pLU/TA 4-1 grown statically in LB or CFA broth at 37°C for 45 h by a modification of the procedure of McCormick et al. (28) as described in Clouthier et al. (7). For fimU transcript analysis, the RNA was separated on a 10% polyacrylamide gel containing 8 M urea and transferred onto Hybond N+ membranes (Amersham) with transfer buffer (0.025 M phosphate buffer [pH 6.5]) and an LKB Pharmacia semidry blotting apparatus. For sefA transcript analysis, the electrophoretic separation of total cellular RNA and its subsequent transfer to Hybond N+ membranes (Amersham) were performed as described in Fourney et al. (15). The fimU- and sefA-specific probes used for Northern blot analysis were gel purified from EcoRI and HindIII digests of pLU/TA 4-1 and pSFA, respectively, with Sephaglas. The probes were labelled with [α32P]dATP (Pharmacia Biotech) by nick translation and hybridized to the blots at 65°C for 18 h in the presence of 200 μg of herring sperm DNA (Sigma)/ml. The membranes were washed at high stringency (0.2× SSC buffer–0.1% SDS, 65°C), and the results were recorded on Kodak BioMax or X-Omat AR5 film.
SDS-PAGE and Western blot analysis.
Whole-cell lysates of S. enteritidis 3b or clones of this strain were screened for the presence of four fimbrial types. SEF14, 18, and 21 were solubilized from whole cells with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer supplemented with 0.2 M glycine (pH 2, 100°C, 10 min), whereas SEF17 fimbriae were solubilized from whole cells with formic acid according to the method of Collinson et al. (9, 10). A portion of each culture (1 OD630 unit) was resuspended in 200 μl of sample buffer, and 10 μl (0.01 OD630 unit) was loaded per lane. Proteins in these samples were separated by SDS-PAGE, electrophoretically transferred to nitrocellulose, and screened with rabbit polyclonal anti-SEF14 (7), SEF17 (10), SEF18 (6), or SEF21 immune serum (30). Immunoreactive proteins were detected with goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugates (Cedarlane) and visualized with 5-bromo-4-chloro-3-indolylphosphate and Nitro Blue Tetrazolium (Sigma).
Electron microscopy.
SEF14 and SEF21 fimbriae on S. enteritidis 3b, 3b-122, 3b-122 pGEM-T1, and 3b-122 pLU/TA 4-1 were immunogold labelled with SEF14- or SEF21-specific rabbit polyclonal immune sera followed by incubation with protein A–15-nm-diameter gold particles (Cedarlane). Negative staining was performed as described previously (10).
Nucleotide sequence accession number.
The nucleotide sequence reported herein for fimU has been submitted to GenBank and has been given the accession number AF013136.
RESULTS
Fimbria production in S. enteritidis 3b-122.
Production of SEF14, -17, -18, and -21 fimbriae by the S. enteritidis Tn10 mutant 3b-122 grown under various growth conditions was assessed by Western blotting with fimbria-specific antisera, and the results were compared to those obtained with the wild-type strain S. enteritidis 3b. The Tn10 mutation in 3b-122 had a pronounced effect on SEF14 and SEF21 production but little or no effect on SEF17 and SEF18 production (Table 1). As previously reported, SEF14 fimbriae were not expressed by 3b-122 grown in static CFA broth at 37°C (Table 1). Further characterization in this study, however, showed that 3b-122 lost SEF14 expression under all the growth conditions in which 3b was SEF14 positive (Table 1). In addition to the SEF14-negative phenotype, 3b-122 was also defective for type 1 fimbria (SEF21) production. The wild-type strain produced FimA under all growth conditions tested, whereas 3b-122 only produced FimA in CFA broth cultures. Thus, the result of the Tn10 insertion was the complete loss of SEF14 expression under all growth conditions and selective loss of SEF21 expression under certain growth conditions. The altered production of SEF14 and SEF21 fimbriae in the Tn10 insertion mutant relative to the wild-type expression patterns suggested that the transposon insertion interrupted a gene whose product was required for both SEF14 and SEF21 fimbria expression.
Identification of the Tn10 insertion site in S. enteritidis 3b-122.
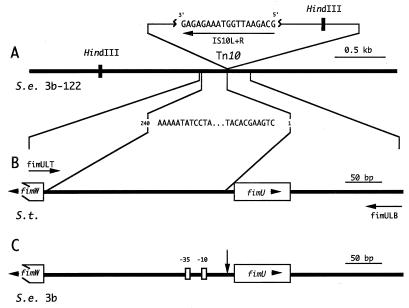
To determine the Tn10 insertion site, HindIII fragments of 3b-122 chromosomal DNA were subcloned into pTZ19. Clones containing Tn10 were identified with the probe IS10L+R, which hybridized within the insertion sequence located at either end of the transposon (Fig. 1A). Of the 17 Tn10-positive clones identified, 3 were subjected to DNA sequence analysis. Comparison of the 3b-122 DNA sequence flanking Tn10 to sequences listed in the NCBI databases revealed that the sequence was 99% identical to that of the region located between fimW and fimU of the S. typhimurium type 1 fimbrial gene cluster. Thus, on the basis of DNA sequence comparison, Tn10 was inserted 13 bp upstream of the predicted start site of the gene, which will hereafter be referred to as fimU (Fig. 1A and B).
FIG. 1.
Location of Tn10 on the S. enteritidis 3b-122 chromosome and identification of the genes flanking the Tn10 insertion in S. enteritidis 3b. (A) Schematic diagram of S. enteritidis (S.e.) 3b-122 chromosomal DNA (black line) showing the Tn10 insert and the strategy used to obtain the chromosomal DNA sequence adjacent to one side of this insert. A 3-kb HindIII fragment comprising 3b-122 chromosomal DNA fused to one end of Tn10 was identified by hybridization with the Tn10 oligonucleotide IS10L+R. IS10L+R was also used as a sequencing primer to obtain 240 bp of DNA sequence from the subcloned HindIII fragment. (B) Schematic diagram of the S. typhimurium (S.t.) chromosome (black line) between fimW and fimU of the type I fimbrial gene cluster that was homologous to the 240 bp of S. enteritidis 3b-122 DNA sequence. Two 42-bp oligonucleotide primers, fimULT and fimULB (horizontal arrows), were made based on the S. typhimurium sequence previously deposited in GenBank (L19338) by Swenson and Clegg (39). (C) Segment of the S. enteritidis 3b chromosome (black line) amplified by PCR with the primers fimULT and fimULB. This amplified DNA segment was subcloned and sequenced (Fig. 2) to identify the DNA flanking the Tn10 insert. The Tn10 insertion point (vertical arrow) was determined to be between the −10 region and the start of the fimU gene. The presence of fimW, fimU, and the −35 region on the 3b chromosome is also noted.
DNA sequence analysis of fimU subcloned from S. enteritidis 3b.
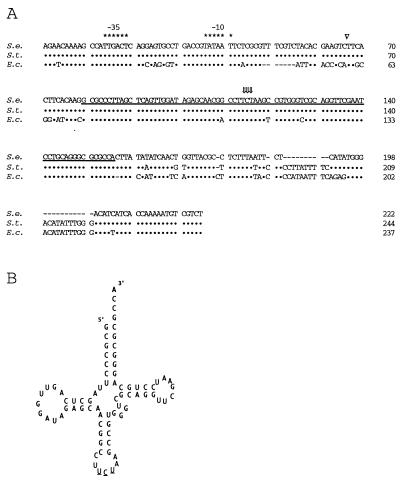
By using primers fimULT and fimULB designed from the sequence flanking the fimU gene in S. typhimurium, a 490-bp fragment was PCR amplified from the cosmid clone pPB523-G containing 35 kb of S. enteritidis 3b DNA (Fig. 1B). The fimU PCR product was subcloned into vector pGEM-T (Fig. 1B). Nucleotide sequence analysis of three clones revealed a potential promoter, but a putative translated protein could not be detected by open reading frame analysis. Comparison of the DNA sequence downstream of the potential promoter to sequences listed in the NCBI databases showed that the sequence was identical to that of fimU of S. typhimurium (Fig. 1C and 2B) and 96% similar to that of argU/dnaY of E. coli (Fig. 2A). These genes encode arginine-specific tRNAs that recognize the rare AGA codon. The nucleotide sequence of fimU from S. enteritidis 3b contained 4 inverted repeats, which were predicted to fold the sequence into the characteristic tRNA-like cloverleaf structure (Fig. 2B) with UCU in the expected tRNA anticodon position. Together, these data suggested that fimU from S. enteritidis 3b encoded an arginine-specific tRNA.
FIG. 2.
Sequence comparison of fimU from S. enteritidis 3b (S.e.) with fimU of S. typhimurium (S.t.) and argU of E. coli (E.c.) as well as the predicted fimU RNA secondary structure. (A) Alignment of S. enteritidis fimU DNA sequence with both the S. typhimurium fimU (39) and E. coli (31) argU gene sequences. Symbols: •, DNA sequence identity; −, gaps introduced to maximize homology; ∗, bases constituting the −35 and −10 boxes; ⇓, bases constituting the anticodon; ▿, position of the Tn10 insertion on the S. enteritidis 3b-122 chromosome. The DNA sequence corresponding to the proposed mature tRNAArg (UCU) is underlined. (B) Diagram of the proposed secondary structure for tRNAArg (UCU) from S. enteritidis 3b. The anticodon bases are underlined.
Analysis of the nucleotide sequence downstream of fimU revealed an open reading frame oriented in the opposite direction such that the 3′ ends of the two genes overlapped. The predicted amino acid sequence was 88% similar to that of the prophage DLP12 integrase of E. coli. The sequence further downstream of fimU displayed 60 to 88% similarities to those encoding transposases of the IS3 family of insertion elements.
Mapping fimU on the S. enteritidis 3b genome.
Like fimA, fimU was localized to chromosomal XbaI and BlnI fragments in the 98.5- to 13.0-C’s region of the chromosome in both Salmonella serovars. By using a series of S. typhimurium and S. enteritidis Tn10 mutants the fimU gene was more precisely mapped to between purE884::Tn10 at 12.6 C’s and the first XbaI restriction site at 13.6 C’s in S. enteritidis or 13.0 C’s in S. typhimurium. Thus, fimU mapped to the same region shown previously to contain the fimA gene in the fim gene cluster.
Analysis of fimU transcription.
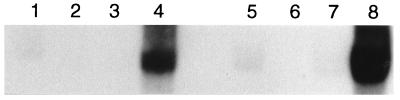
To determine if the fimU transcript was the same size as tRNA, a fimU-specific probe was hybridized to a blot containing total RNA isolated from S. enteritidis 3b, 3b-122, 3b-122 pGEM-T1, or 3b-122 pLU/TA 4-1 grown under conditions optimal for type 1 fimbria (SEF21) production in S. enteritidis 3b (static LB broth, 48 h, 37°C). The fimU-specific probe hybridized to a transcript that was present in total RNA from 3b and 3b-122 pLU/TA 4-1 (Fig. 3). The fimU transcript was consistently difficult to detect on Northern blots of 3b RNA even with excessive amounts of RNA loaded on the gels (25 μg [Fig. 3, lanes 1 to 4] and 50 μg [Fig. 3, lanes 5 to 8]) and extended exposure of the blots to X-ray film. Although the transcript was not found in RNA prepared from 3b-122, trace levels were evident in 3b-122 carrying the vector pGEM-T (Fig. 3), but the transcript was even more difficult to detect than its counterpart in 3b. The transcript detected with the fimU-specific probe comigrated with tRNA, suggesting that the product of fimU from S. enteritidis was indeed a tRNA molecule.
FIG. 3.
Northern blot analysis of tRNAArg (UCU) production in S. enteritidis 3b strains. A fimU-specific probe was hybridized to PAGE-separated total RNA from S. enteritidis 3b (lanes 1 and 5), 3b-122 (lanes 2 and 6), 3b-122 pGEM-T1 (lanes 3 and 7), or 3b-122 pLU/TA 4-1 (lanes 4 and 8). Lanes 1 to 4 contain 25 μg of RNA, and lanes 5 to 8 contain 50 μg of RNA.
Complementation of fimbrin expression and fimbria assembly in 3b-122.
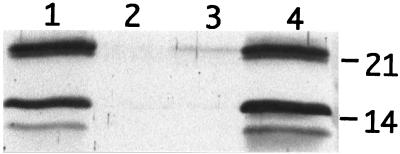
Fimbria expression was examined in S. enteritidis 3b, 3b-122, 3b-122 pGEM-T1, and 3b-122 LU/TA 4-1 grown under conditions that promoted production of both SEF14 and SEF21 by the wild-type strain (static CFA broth, 48 h, 37°C). Western blot analysis of whole-cell lysates using SEF14- or SEF21-specific antisera showed that complementation of the insertion mutation in 3b-122 with pLU/TA 4-1 restored SEF14 and SEF21 fimbria expression (Fig. 4). Thus fimU affected the production of two fimbrins encoded by genes located on two different gene clusters.
FIG. 4.
Complementation analysis of S. enteritidis 3b-122 Tn10 mutant with the fimU-containing recombinant plasmid pLU/TA 4-1. Whole-cell extracts were analyzed by Western blotting to determine the presence of SefA (21 kDa) and FimA (14 kDa) fimbrin proteins in S. enteritidis 3b (lane 1), 3b-122 (lane 2), 3b-122 pGEM-T1 (lane 3), and 3b-122 pLU/TA 4-1 (lane 4). Numbers at right indicate positions of SefA (21) and FimA (14).
Assembly of SEF14 and SEF21 fimbriae on the cell surface of S. enteritidis 3b, 3b-122, 3b-122 pGEM-T1, or 3b-122 pLU/TA 4-1 was determined by immunogold labelling and electron microscopy performed on cells grown in static CFA broth for 48 h at 37°C. The wild-type strain, S. enteritidis 3b, expressed both SefA and FimA and assembled the respective subunits into SEF14 and SEF21 fimbriae (Table 2). Similar analyses of 3b-122 and 3b-122 pGEM-T1 showed that SEF14 was not produced and that SEF21 fimbriae on the cell surfaces of these two strains were rarely detected (Table 2), a result consistent with the Western blot data (Fig. 4). In contrast, SEF14 and SEF21 fimbriae were evident on the surface of 3b-122 pLU/TA 4-1 (Fig. 5) at levels equal to or greater than that on 3b. Thus, expression of SefA and FimA fimbrins and assembly of their respective fimbriae were restored by complementation of the Tn10 fimU mutation in 3b-122 with a wild-type copy of the fimU gene on pLU/TA 4-1. Cells producing SEF17 were readily seen without immunolabelling on all the grids prepared for electron microscopy (Table 2).
TABLE 2.
Detection of assembled SEF14 and SEF21 fimbriae in various S. enteritidis strains by immunoelectron microscopy with specific antifimbrial sera
| S. enteritidis 3b strain | Fimbria productiona
|
||
|---|---|---|---|
| SEF14 | SEF21 | SEF17 | |
| Wild type | ++ | + | +++ |
| 3b-122 | − | ±b | +++ |
| 3b-122 pGEM-T1 | − | ±c | +++ |
| 3b-122 pLU/TA 4-1 | +++ | + | +++ |
The percentages of cells labelled by immunogold are indicated as follows: −, none; ±, less than 10%; +, 10 to 50%; ++, 50 to 75%; +++, 75 to 100%.
Two of approximately 400 cells were densely labeled.
Two of approximately 600 cells were sparsely labeled.
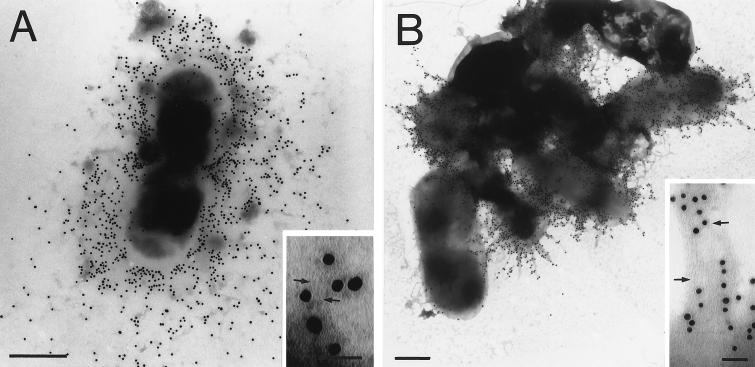
FIG. 5.
Analysis of SEF14 and SEF21 fimbria assembly in S. enteritidis 3b-122 pLU/TA 4-1 by immunogold electron microscopy. S. enteritidis 3b-122 pLU/TA 4-1 was labeled with protein A-gold and negatively stained following incubation with immune serum generated to SEF14 (A) or SEF21 (B). Arrows indicate individual immunogold-labeled SEF14 and SEF21 fimbriae in panel A and B insets, respectively. The average diameter of the gold particles was 15 nm. Bar, 0.5 μm (electron micrograph) or 50 nm (inset).
Analysis of sefA transcription.
The effect of tRNAArg (UCU) on sefA transcript production was analyzed by hybridizing a sefA-specific probe to a blot containing total cellular RNA isolated from S. enteritidis 3b, 3b-122, 3b-122 pGEM-T1, or 3b-122 LU/TA 4-1 grown under conditions optimal for SEF14 production in 3b (static CFA broth, 48 h, 37°C). The sefA-specific probe hybridized to a 660-base transcript that was present in RNA from 3b and 3b-122 pLU/TA 4-1 but absent from RNA from 3b-122 and 3b-122 pGEM-T1 (Fig. 6). The strains expressing the sefA transcript corresponded to those carrying a functional fimU gene, suggesting that fimU was required for expression of sefA.
FIG. 6.
Northern blot analysis of sefA transcription in S. enteritidis 3b strains. A sefA-specific probe was hybridized to 10 μg of total RNA from S. enteritidis 3b (lane 1), 3b-122 (lane 2), 3b-122 pGEM-T1 (lane 3), or 3b-122 LU/TA 4-1 (lane 4).
DISCUSSION
fimU, located in the fim (sef21) operon of S. enteritidis 3b, encodes an arginine-specific tRNA that is required for expression of not only SEF21 fimbriae (type 1) but also SEF14 fimbriae. The product of fimU in 3b is a tRNA, since Northern blot analysis of RNA from 3b and 3b-122 pLU/TA 4-1 demonstrates that the fimU transcript comigrates on polyacrylamide gels with tRNA. Furthermore, the 77-nucleotide sequence of the fimU gene of 3b is identical to that of fimU of S. typhimurium (39) and shares extensive homology with that of argU encoding tRNAArg (UCU) from E. coli (18). The fimU-encoded transcript from 3b can be folded into a typical tRNA cloverleaf structure containing the 3′-terminal sequence CCA as well as the invariant or semivariant nucleotides common to tRNA molecules (19, 34). Finally, the DNA sequence 5′ to the fimU gene contains features common to the promoters of tRNA operons including the consensus E. coli −10 and −35 promoter elements (16, 21) and a G+C-rich discriminator sequence (16, 42, 43). The regulatory mechanisms controlling fimU expression are unknown. Recently, however, the integration and excision of plasmids, phage, and pathogenicity islands into and out of the chromosomes at tRNA loci have been shown to affect tRNA gene expression (20, 33). As shown with the E. coli tRNA gene argU (25), the open reading frame adjacent to and overlapping fimU is a homolog of the integrase gene (int) from the defective lambdoid prophage DLP12. Integration of prophage DLP12 at this site prevents cotranscription of the int gene with fimU, which may contribute to the regulation of fimU expression in S. enteritidis 3b.
The influence of tRNAArg (UCU) encoded by fimU on SEF14 and SEF21 fimbria production is evident in the Tn10 insertion mutant S. enteritidis 3b-122. The transposon, inserted between the predicted promoter and the 5′ end of the mature fimU transcript, disrupts transcription of fimU and thus tRNAArg (UCU) production. This mutation results in the loss of SefA production and selective loss of FimA production, i.e., the subunits of SEF14 and SEF21 (type 1) fimbriae, respectively. Thus, in S. enteritidis 3b, tRNAArg (UCU) is required for SEF14 production and enhances type 1 fimbria (SEF21) production. In E. coli, cross-talk has also been reported to occur between adhesin gene clusters (29), and tRNA molecules have been shown to play a key role in global regulatory cascades (20). However, this is the first study to show that a tRNA-specific locus found on one fimbrial operon influences the production of two fimbrins whose operons are separated by 15 C’s on the chromosome.
tRNAArg (UCU) encoded by fimU is required for transcription of sefA, the gene encoding the subunit of SEF14 fimbriae in S. enteritidis 3b. The regulatory mechanism is unknown, but a direct correlation between the abundance of tRNAs and the occurrence of the respective codons in protein genes (22, 23) has been suggested to control the translation of genes containing rare codons (3, 37). Since AGA, the codon recognized by the tRNAArg (UCU) species encoded by fimU, is one of the least-used codons for arginine, then perhaps the limited availability of charged tRNAs for this minor codon controls the level of translation of the sefA transcript or of a transcript whose protein product is involved in the regulation of sefA transcription. sefA contains neither of the rare arginine codons AGA or AGG recognized by tRNAArg (UCU), indicating that fimU expression would not have a direct effect on the translation of sefA mRNA. However, sefE, the gene encoding the putative AraC-like transcriptional activator of the sef14 gene cluster, contains 13 arginine codons, including 9 AGA codons and 1 AGG codon (5). Perhaps the tRNAArg (UCU) encoded by fimU regulates translation of sefE, which would in turn affect transcription of sefA and the downstream genes.
With the exception of the gene fimA encoding the subunit of SEF21 fimbriae (12), the remainder of the sef21 gene cluster has not been characterized in S. enteritidis 3b. Thus, the mechanism for regulation of type 1 fimbria synthesis by fimU remains to be determined. However, type 1 fimbria (SEF21) production is optimal when S. enteritidis 3b is grown at 37°C in a static broth culture but suboptimal when the cells are grown at lower temperatures (21 to 30°C) in shaking broth culture or on solid medium (12). Similarly, expression of SEF14 fimbriae by S. enteritidis 3b is environmentally controlled by temperature, medium composition, and aeration, and is optimal at 37°C in static, aerobic CFA broth (5). Thus, the coregulation of SefA and FimA fimbrin production by fimU-encoded tRNAArg (UCU) results in the corresponding fimbriae being expressed under similar environmental conditions, which may give the bacteria a competitive advantage for survival.
ACKNOWLEDGMENTS
This project was supported by operating grants to W.W.K. provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Bacterial Diseases Network. S.C.C. was the recipient of an Industrial Postdoctoral Fellowship from NSERC, and A.P.W. was the recipient of a Graduate Research Engineering and Technology Award scholarship from the Science Council of British Columbia and a Graduate Research scholarship from NSERC.
We thank S. Lee for helpful discussions on tRNA.
REFERENCES
- 1.Alm R A, Guerry P, Trust T J. Distribution and polymorphism of the flagellin genes among isolates of Campylobacter coli and Campylobacter jejuni. J Bacteriol. 1993;175:3051–3057. doi: 10.1128/jb.175.10.3051-3057.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Chen G-F, Inouye M. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 1994;8:2641–2652. doi: 10.1101/gad.8.21.2641. [DOI] [PubMed] [Google Scholar]
- 4.Chou Q, Russell M, Birch D E, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplification. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clouthier S C. Ph.D. thesis. Victoria, Canada: University of Victoria; 1995. [Google Scholar]
- 6.Clouthier S C, Collinson S K, Kay W W. Unique fimbriae-like structures encoded by sefD of the SEF14 fimbrial gene cluster of Salmonella enteritidis. Mol Microbiol. 1994;12:893–903. doi: 10.1111/j.1365-2958.1994.tb01077.x. [DOI] [PubMed] [Google Scholar]
- 7.Clouthier S C, Müller K-H, Doran J L, Collinson S K, Kay W W. Characterization of three fimbrial genes, sefABC, of Salmonella enteritidis. J Bacteriol. 1993;175:2523–2533. doi: 10.1128/jb.175.9.2523-2533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collinson S K, Liu S-L, Clouthier S C, Banser P A, Doran J L, Sanderson K E, Kay W W. The location of four fimbrin-encoding genes, agfA, fimA, sefA, and sefD, on the Salmonella enteritidis and/or S. typhimurium XbaI-BlnI genomic restriction maps. Gene. 1996;169:75–80. doi: 10.1016/0378-1119(95)00763-6. [DOI] [PubMed] [Google Scholar]
- 9.Collinson S K, Doig P C, Doran J L, Clouthier S C, Trust T J, Kay W W. Thin aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinson S K, Emödy L, Müller K-H, Trust T J, Kay W W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doran J L, Collinson S K, Clouthier S C, Cebula T A, Koch W H, Burian J, Banser P A, Todd E C D, Kay W W. Diagnostic potential of sefA DNA probes to Salmonella enteritidis and certain other O-serogroup D1 Salmonella serovars. Mol Cell Probes. 1996;10:233–246. doi: 10.1006/mcpr.1996.0033. [DOI] [PubMed] [Google Scholar]
- 12.Doran J L, Collinson S K, Kay C M, Banser P A, Burian J, Munro C K, Lee S H, Somers J M, Todd E C D, Kay W W. fimA and tct based DNA diagnostics for Salmonella. Mol Cell Probes. 1994;8:291–310. doi: 10.1006/mcpr.1994.1041. [DOI] [PubMed] [Google Scholar]
- 13.Evans D G, Evans D J, Jr, Tjoa W. Hemagglutination of human group A erythrocytes by enterotoxigenic Escherichia coli isolated from adults with diarrhea: correlation with colonization factor. Infect Immun. 1977;18:330–337. doi: 10.1128/iai.18.2.330-337.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feutrier J, Kay W W, Trust T J. Purification and characterization of fimbriae from Salmonella enteritidis. J Bacteriol. 1986;168:221–227. doi: 10.1128/jb.168.1.221-227.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fourney R M, Miyakoshi J, Kay III R S, Paterson M C. Northern blotting: efficient RNA staining and transfer. Focus. 1992;10:5–7. [Google Scholar]
- 16.Fournier M J, Ozeki H. Structure and organization of the transfer ribonucleic acid genes of Escherichia coli K-12. Microbiol Rev. 1985;49:379–397. doi: 10.1128/mr.49.4.379-397.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 18.Garcia G M, Mar P K, Mullin D A, Walker J R, Prather N E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986;45:453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- 19.Gauss D H, Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1983;11:1–53. [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 21.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 23.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- 24.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsey D F, Mullin D A, Walker J R. Characterization of the cryptic lambdoid prophage DLP12 of Escherichia coli and overlap of the DLP12 integrase gene with the tRNA gene argU. J Bacteriol. 1989;171:6197–6205. doi: 10.1128/jb.171.11.6197-6205.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marck C. “DNA Strider”: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClain M S, Blomfield I C, Eisenstein B I. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick J R, Zengel J M, Lindahl L. Intermediates in the degradation of mRNA from the lactose operon of Escherichia coli. Nucleic Acids Res. 1991;19:2767–2776. doi: 10.1093/nar/19.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morschhauser J, Vetter V, Emody L, Hacker J. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol Microbiol. 1994;11:555–566. doi: 10.1111/j.1365-2958.1994.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 30.Müller K-H, Collinson S K, Trust T J, Kay W W. Type 1 fimbriae of Salmonella enteritidis. J Bacteriol. 1991;173:4765–4772. doi: 10.1128/jb.173.15.4765-4772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullin D A, Garcia G M, Walker J R. An E. coli DNA fragment 118 base pairs in length provides dnaY+ complementing activity. Cell. 1984;37:669–674. doi: 10.1016/0092-8674(84)90399-4. [DOI] [PubMed] [Google Scholar]
- 32.Newman J V, Burghoff R L, Pallesen L, Krogfelt K A, Kristensen C S, Laux D C, Cohen P S. Stimulation of Escherichia coli F-18Col− type-1 fimbriae synthesis by leuX. FEMS Microbiol Lett. 1994;122:281–287. doi: 10.1016/0378-1097(94)00337-8. [DOI] [PubMed] [Google Scholar]
- 33.Reiter W-D, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rich A, RajBhandary U L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- 35.Ritter A, Gally D L, Olsen P B, Dobrindt U, Friedrich A, Klemm P, Hacker J. The Pai-associated leuX specific tRNALeu affects type 1 fimbriation in pathogenic Escherichia coli by control of FimB recombinase expression. Mol Microbiol. 1997;25:871–882. doi: 10.1111/j.1365-2958.1997.mmi517.x. [DOI] [PubMed] [Google Scholar]
- 36.Rosner J L. Formation, induction and curing of bacteriophage P1 lysogens. Virology. 1972;48:679. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- 37.Saier M H., Jr Differential codon usage: a safeguard against inappropriate expression of specialized genes? FEBS Lett. 1995;362:1–4. doi: 10.1016/0014-5793(95)00185-c. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Swenson D L, Clegg S. Salmonella fimbriae. In: Klemm P, editor. Fimbriae: adhesion, biogenesis, genetics and vaccines. London, United Kingdom: CRC Press; 1994. pp. 105–113. [Google Scholar]
- 40.Swenson D L, Kim K-J, Six E W, Clegg S. The gene fimU affects expression of Salmonella typhimurium type 1 fimbriae and is related to the Escherichia coli tRNA gene argU. Mol Gen Genet. 1994;244:216–218. doi: 10.1007/BF00283525. [DOI] [PubMed] [Google Scholar]
- 41.Swenson D L, Clegg S. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J Bacteriol. 1992;174:7697–7704. doi: 10.1128/jb.174.23.7697-7704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Travers A A. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 1984;12:2605–2618. doi: 10.1093/nar/12.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Travers A A. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]