Abstract
Background
Acute myeloid leukemia (AML) is characterized by a high recurrence and mortality rate. Cuproptosis is involved in cell death regulation in in a variety of solid tumors. Long non-coding RNAs that regulate cuproptosis genes in the pathogenesis of acute leukemia have yet to be explored.
Methods
First, cuproptosis genes with distinct expression levels were discovered by contrasting AML with normal samples from the TCGA and GTEx cohorts. Pearson correlation and univariate Cox-regression analysis were performed to identify cuproptosis-associated lncRNAs with significant prognostic values. Then the least absolute shrinkage and selection operator (LASSO) Cox regression was utilized to establish a multi-gene signature to predict AML prognosis. Next, Kaplan-Meier estimator, receiver operating characteristic curve, and a nomogram were performed to evaluate the predictive capacity of the risk signature. Functional enrichment analyses were employed to assess their function. Moreover, qRT-PCR testing of lncRNA expression in AML samples was conducted. The competing endogenous RNA (ceRNA) network was constructed to find the target genes.
Results
A risk model based on the signature of three cuproptosis-associated lncRNAs was developed. The results showed that the model possessed excellent prognostic potential. The nomogram raised the accuracy in predicting AML survival. In addition, functional enrichment analyses demonstrated an enrichment of inflammatory and immune-related pathways. Moreover, correlations between the risk signature and clinicopathological variables, tumor mutational burden, RNA stemness score, immune profile, and drug sensitivity were observed. Furthermore, we discovered that TRAF3IP2-AS1 may function as a ceRNA to regulate cuproptosis and ferroptosis gene expression.
Conclusion
The risk signature established in this study could serve as a reliable biosignature for AML prognosis. And the findings presented here may facilitate research on cuproptosis in AML.
Keywords: Cuproptosis, Long noncoding RNA, Acute myeloid leukemia, Nomogram, Competing endogenous RNA
1. Introduction
Acute myeloid leukemia (AML) is known as an aggressive group of blood cell malignancies [1]. We have achieved significant advancement in understanding the pathogenesis and therapy strategies of AML in the last few decades. Standard treatments, such as chemotherapy or hematopoietic stem cell transplantation (HSCT) played an important role to improve the survival of eligible AML patients [2]. As a result, the survival rate for patients with AML has significantly increased. However, overall outcomes remain unsatisfactory, with most patients experiencing illness relapse or dying from leukemia after a few months of complete remission (CR) [1]. The long-term overall survival (OS) rate is less than 40 % in younger patients (<60 years) and only 15 % in elderly patients (≥60 years) [3]. While targeted therapies (IDH1/2 inhibitors, BCL-2 antagonists, targeting FLT3 mutation, and targeting TP53 mutation) and immunotherapy (antibody-based treatment, suppression of immune negative regulators, and possible CAR-T cells) further expand treatment options, AML can develop resistance to chemotherapy and targeted therapies over time, leading to treatment failure and disease relapse [[2], [3], [4]]. And targeted therapies are only applicable to patients with specific genetic mutations, leaving a significant proportion of AML patients without targeted treatment options [1]. The molecular complexity of AML relapse and therapy resistance remains a challenge [3]. The identification and validation of novel molecular biomarkers serve a vital role in predicting and evaluating the clinical prognosis of AML patients [1]. These biomarkers have the potential to provide valuable insights into the evolution of diseases, the response to treatment, the exploration of novel therapeutic approaches, and the improvement of the long-term prognosis for patients with AML [2,4].
Copper is an essential trace element for a variety of biochemical activities, including cellular respiration, peptide biosynthesis, connective tissue biosynthesis, and antioxidant defense [5,6] Recent research indicates that excessive copper could trigger cell death in an unprecedented manner, also known as "cuproptosis", which is a confirmed new type of programmed cell death (PCD) that may influence tumor growth [7]. However, few investigations [6,8] have explored the role of cuproptosis in acute leukemia. Copper levels have been found to be significantly altered in the serum and tumor tissue of patients suffering from hematologic tumors [9,10]. Copper levels beyond the normal range are linked to an elevated relapse possibility of hematologic malignancies, while normal concentrations of copper are associated with CR [9,10]. Moreover, an innovative approach to treating cancer, particularly for treatment-resistant cases, involves stimulating cancer cells to induce cuproptosis [8]. For instance, elesclomol exerts potent anti-cancer effects in the HL-60 leukemic cell line by increasing copper levels and mitochondrial oxidative stress, whereas has no effect on these levels in peripheral blood mononuclear cells [6]. In addition, elesclomol is capable of targeting cancer cells resistant to cisplatin and proteasome inhibitors. Furthermore, leukemic stem cells are dependent on the metabolism of oxidants; hence, disturbing the copper distribution in their mitochondria can prevent them from self-renewing [11].
Long non-coding RNA (lncRNA), a crucial regulator of gene expression, plays a vital role in regulating gene expression, chromatin structure, and various cellular processes [12]. Research revealed that lncRNAs have emerged as key players in AML disease pathogenesis, prognosis, and potential therapeutic targets. Several lncRNAs can act as oncogenes and promote leukemogenesis through dysregulation of gene expression networks. They could interact with chromatin modifiers, transcription factors, and signaling pathways to drive AML development [[13], [14], [15]]. Moreover, certain lncRNAs can modulate drug efflux pumps, DNA repair mechanisms, or anti-apoptotic pathways, thereby contributing to chemotherapy resistance. Recently, it was discovered that high- or low-expression of specific lncRNAs is associated with different prognoses, such as survival rates, AML recurrence, and response to AML therapy [15,16]. Their functions in AML make them attractive targets for further research and potential therapeutic interventions. On the impact of cuproptosis-associated lncRNAs on the development and prognosis of AML, however, relatively little research has been conducted.
In this study, we aimed to systematically evaluate the predictive value of lncRNAs linked to cuproptosis in AML patients. The prognostic signature comprised of three lncRNAs has been constructed, and the nomogram has significant clinical application potential. Furthermore, we also introduced a competing endogenous RNA (ceRNA) network that could contribute to providing new insight into identifying potential regulatory mechanisms for AML development.
2. Materials and methods
2.1. Data acquisition
Clinical information and RNA sequencing result of de novo AML adults were obtained from the TCGA-LAML database in the FPKM format (https://xenabrowser.net/datapages/). Due to the absence of normal samples in TCGA-LAML, normal bone marrow data from the Genotype-Tissue Expression (GTEx) (https://xenabrowser.net/datapages/) were used to fill in the gaps. The Toil method was used to uniformly process the data for both TCGA-LAML and GTEx [17]. In the TCGA-LAML data set, AML patients were treated in accordance with NCCN guidelines (www.nccn.org), with an emphasis on enrollment in therapeutic clinical trials wherever possible. Patients with unfavorable risk underwent allogeneic stem cell transplantation (allo-HSCT) if they were medically fit for the risks of transplantation and a suitably matched donor was available. Many patients with intermediate risk also underwent allo-HSCT at some point during the course of their disease [18]. Ten essential genes for cuproptosis have been identified by researchers, including seven positive regulators (FDX1, DLD, DLAT, LIPT1, LIAS, PDHB, and PDHA1) and three inhibitory regulators (GLS, MTF1, and CDKN2A) [7]. The expressions of cuproptosis regulators in TCGA-LAML were compared to those in GTEx, which served as a normal control. We used the limma package to screen out the cuproptosis-related lncRNAs (|Pearson R| >0.4 and P < 0.001).
2.2. LncRNA signature development and evaluation
In the first step, we screened for cuproptosis-associated lncRNAs linked with AML survival utilizing the univariate Cox regression analysis (P < 0.05). Samples lacking survival information were omitted. The lncRNAs obtained in the previous step were subsequently analyzed for expression differences between AML and normal samples. LncRNAs were determined to be substantially differentially expressed if they had a |log2FC| >1 and a false discovery rate (FDR) < 0.05. Next, all patients were randomly assigned 1:1 to a validation and training set (Supplementary Table S1) for integrated analysis utilizing the "caret" package. Through least absolute shrinkage and selection operator (LASSO) regression, the screened lncRNAs were ultimately chosen and verified for the training cohort. Finally, a novel three-lncRNA prognostic signature was established stepwise. Each AML patient has a risk score based on the following algorithm: Risk Score = expression of AC079466.1 × 0.818 + expression of TRAF3IP2-AS1 × (−0.886) + expression of NADK2-AS1 × (−0.511). Moreover, using the median risk value, AML individuals were separated into favorable and poor risk categories. Additionally, survival analyses were used to compare the outcomes of different subsets of AML adults according to their clinicopathological parameters and risk value. We used the R "ggpubr" package to characterize the variation in risk assessments regarding different clinical parameters. Pan-cancer expression analyses of the three lncRNAs across 33 cancers and their matched controls were conducted.The threshold values for |log2FC| and FDR were 1 and 0.01, respectively. The differential expression and prognostic performance of the model lncRNAs were validated using the Gene Expression Omnibus (GEO) datasets of adult AML cohorts (https://www.ncbi.nlm.nih.gov/geo/) and the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) AML database. The datasets GSE6891, GSE12662, and GSE13159 were downloaded in the form of a normalized expression matrix, while the Target AML data obtained from the TCGA TARGET GTEX cohort (https://xenabrowser.net/datapages/) were log2(norm_count+1) transformed. Among the datasets utilized in this study, GSE6891 (consisting of 537 AML samples) and the Target dataset (comprising 228 AML samples) were employed to assess the prognostic significance of model lncRANs by analyzing gene expression data alongside corresponding clinical information. Additionally, GSE12662 (comprising 91 AML samples and 15 normal samples) and GSE13159 (consisting of 542 AML samples and 75 normal samples) were utilized to compare the expression differences of risk lncRNAs in AML and normal tissues. Furthermore, AML specimens obtained from the TARGET database were juxtaposed with normal tissue samples acquired from the GTEX database for the purpose of performing differential gene expression analysis. During the process of conducting survival analysis, we excluded the samples that lacked survival time or exhibited a gene expression level of zero.
2.3. Nomogram construction and evaluation
To determine whether the model merited to be a predictive factor independent of other clinicopathological parameters, univariate and multivariate prognostic performances were evaluated. The AML prognosis was predicted using a nomogram, and the accuracy of the nomogram was evaluated using calibration curve, decision curve analysis (DCA), and ROC curve.
2.4. Functional enrichment analysis and tumor immune microenvironment landscape
The R "clusterProfiler" package was utilized according to risk score categorization, filtered by P < 0.05. The "ESTIMATE" R package was applied in AML samples for immune infiltration, stromal content, and the combined score. Estimating Relative Subsets of RNA Transcripts (CIBERSORT) was then used to determine the frequency of 22 distinct categories of immune cells in groups with favorable and poor risk [19]. The immune function score was calculated using the "GSVA" package [20]. The "limma", "ggpubr", and "ggplot2" packages were performed to analyze the data and provide visual representations in the form of box plots, vioplots, and heatmaps, respectively. Moreover, tumor stem cell-like characteristics based on RNA stemness score (RNAss) were examined using the PanCancer TCGA database of tumor stem cell transcriptome data [21]. The correlation between RNAss and risk score was identified using Spearman correlative analysis.
2.5. Tumor mutational burden (TMB) and principal component analysis (PCA)
Information on genetic mutations was collected from the TCGA-LAML database, while the R package "mafools" assessed genetic variance across risk groups. Our subsequent step was to compare favorable and poor risk groups with regards to TMB and to perform survival studies between four distinct TMB and risk categories. The separation pattern between the two risk categories was analyzed by means of PCA utilizing the "ggplot2" package.
2.6. CeRNA network and PPI network analysis
Based on lncRNA target prediction database LncBase v3.0 [22], we studied interactions between lncRNAs and miRNAs. Next, three miRNA target prediction databases were used to examine miRNA-mRNA interactions, including TargetScan, miRDB, and miRTarBase. Target differentially expressed mRNAs (DEmRNAs) were finally selected by "limma" Package, with the threshold set at |log2FC| ≥3 and FDR ≤0.05. The ceRNA network was represented visually using Cytoscape (version 3.7.2). Utilizing the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, a protein-protein interaction (PPI) network was constructed [23]. The PPI hub genes were found using CytoHubba. Enrichment analysis based on Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was then performed on DEmRNAs to identify biological processes and potential signaling pathways.
2.7. Drug sensitivity analysis
We also predicted differences in drug response rates between two risk groups using the pRRophetic algorithm based on the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancer-rxgene.org/) [24]. Wilcoxon signed-rank test was used to evaluate and compare the IC50 values between the two risk subgroups.
2.8. AML cell line culture
The THP-1 cells (human acute monocytic leukemia cell line) were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10 U/ml streptomycin, 100 U/ml penicillin, and 10 % heat-inactivated fetal bovine serum (Gibco, USA). THP-1 cells were cultured in a 37 °C, 5 % CO2 and 95 % humidity incubator.
2.9. Validation of risk lncRNAs expression by qRT-PCR
Bone marrow samples from 20 novel AML patients and peripheral blood samples from 17 healthy controls were collected from The Second Affiliated Hospital of Chongqing Medical University. This research was approved by the Ethics Committee of The Second Affiliated Hospital of Chongqing Medical University. Informed consent and approval were obtained from all participants. After isolating the mononuclear cells from the sample, total RNA from AML patients, healthy controls, and THP-1 cells were extracted using RNAiso Plus (TAKARA). RNA was then converted to cDNA using the TUREscript 1st Strand cDNA Synthesis Kit (Aidlab Biotechnologies). The genetic expression was normalized to that of GAPDH. SYBR Green qPCR Mix (Aidlab Biotechnologies) was used to quantify the real-time PCR assay. The primer sequences are presented in Supplementary Table S2. Gene expression was quantified using the 2−ΔΔCt method.
2.10. Statistical analysis
Statistical analyses were performed using R software (version 4.2.0) and GraphPad Prism 9.4.0. The Wilcoxon rank-sum test was used to assess differential functions between the two groups. P < 0.05 was considered statistically significant.
3. Results
3.1. Clinical factors of AML individuals
The clinical information of TCGA-LAML included age, gender, hemoglobin (HB), white blood cell (WBC) count, platelet (PLT) count, bone marrow blast cell (BMBC) percentage, peripheral blood blast cell (PBBC) percentage, cytogenetic risk category, French-American-British (FAB) classification, and genetic mutation (i.e., FLT3, NPM1, IDH1, and RAS) (Supplementary Table S1).
3.2. Identification of prognostic cuproptosis-associated lncRNAs
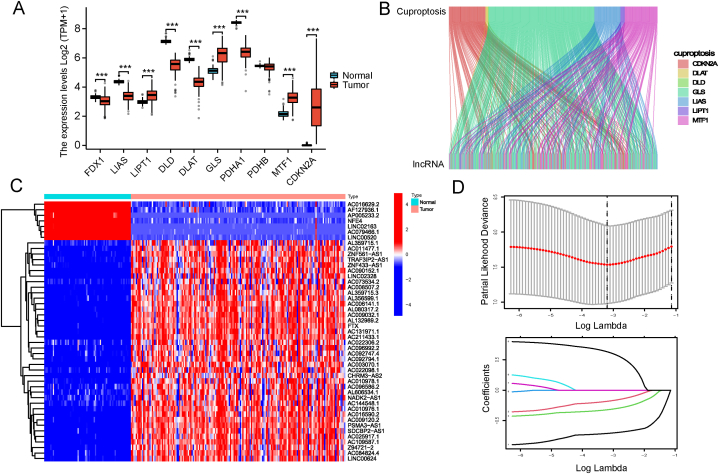
Supplementary Fig. S1 depicts the investigation's flowchart. 14 087 lncRNAs were initially obtained from 151 AML patients in TCGA-LAML datasets. In AML patients, the expression of PDHA1, LIAS, DLAT, FDX1, and DLD decreased, while the expression of LIPT1, GLS, CDKN2A and MTF1 increased (Fig. 1A). As PDHB expression did not differ significantly between AML patients and controls, only 9 cuproptosis genes were retained for further study. From the expression matrix of 9 cuproptosis regulators, 470 lncRNAs significantly related to cuproptosis were identified (Fig. 1B). Eleven AML patients lacking survival information were omitted. A total of 140 AML patients participated in the prognosis analysis, and 139 lncRNAs were found to be significantly associated with the clinical outcomes of the patients (P < 0.05). Upon comparing the expression levels of the 139 lncRNAs in healthy and AML samples, we discovered that 47 lncRNAs were abnormally expressed in AML (Fig. 1C).
Fig. 1.
Identifying prognostic associated cuproptosis-related lncRNA. (A) Cuproptosis regulators expression of AML patients in TCGA compared to normal control in GTEx. (B) Correlation between cuproptosis regulators and related lncRNAs in the Sankwy diagram. (C) Heatmap showed the 47 differentially expressed Cuproptosis-associated prognostic lncRNAs in AML versus normal samples (Wilcox test) (D) LASSO Cox algorithm was developed to generate a prognostic lncRNAs model. (***P < 0.001; **P < 0.01; *P < 0.05; ns, not statistically significant).
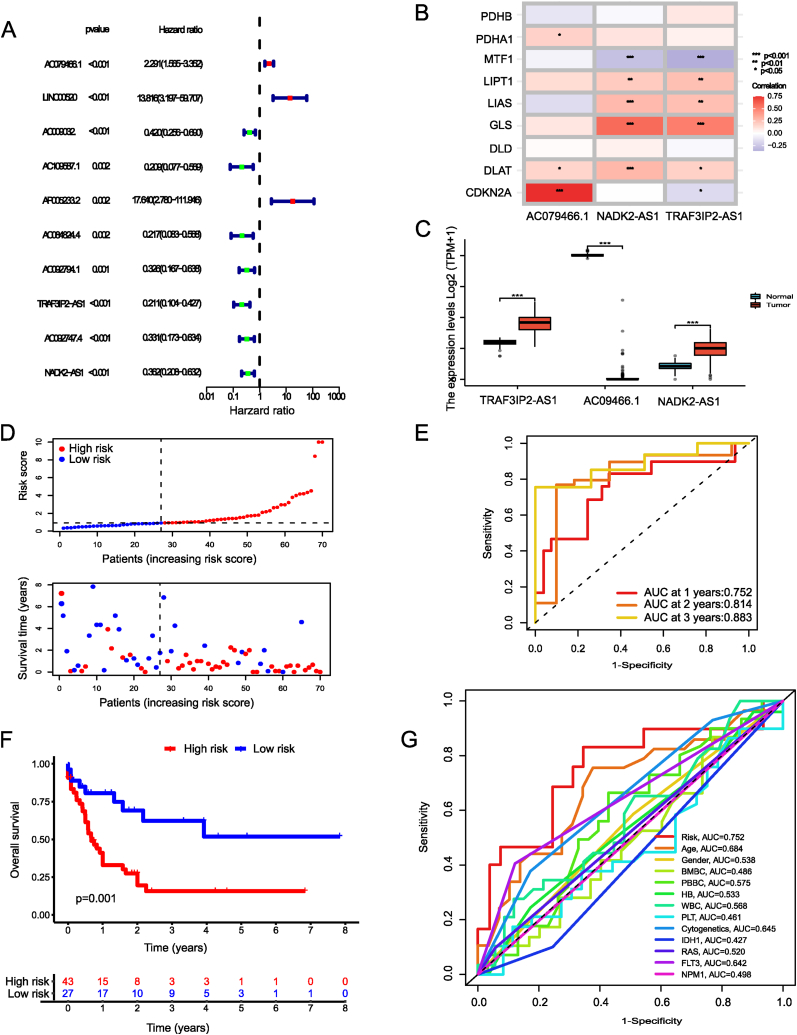
3.3. Prognostic signature construction
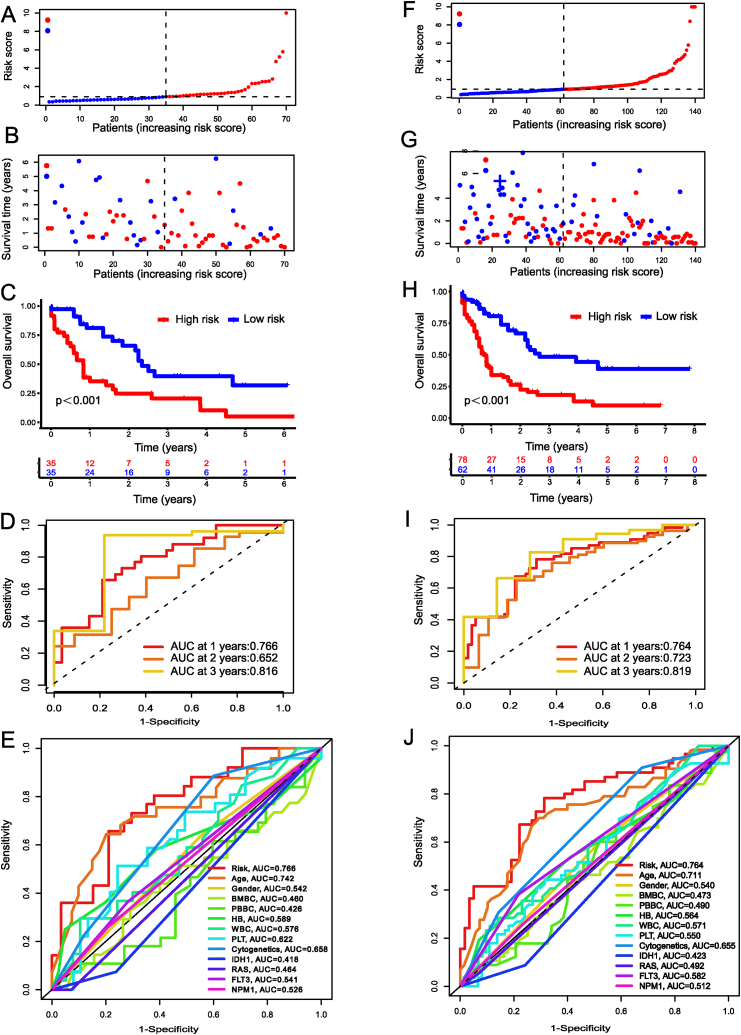
From the 47 lncRNAs found to be differentially expressed between AML cases and controls, we were able to identify 10 with relevance to AML prognosis using univariate Cox regression analysis (P < 0.005). Then, we conducted LASSO and Cox regression analyses with multiple variables to identify the cuproptosis-associated lncRNAs signature (Fig. 1D). Fig. 2Adepicts the hazard ratios of 10 lncRNAs related to cuproptosis. Fig. 2B illustrates the correlation between cuproptosis regulators and the three signature lncRNAs (TRAF3IP2-AS1, AC079466.1, and NADK2-AS1). The box plot showed the differences between AML samples and normal samples in terms of the three lncRNAs expression (Fig. 2C). Following this, 140 TCGA-LAML patients were split evenly between the testing (n = 70) and training (n = 70) cohorts. The clinicopathological features of AML patients were identical in both groups (Supplementary Table S1). Fig. 2D displayed the risk category and survival rate for each patient in the training cohort. The AUC of the risk signature for predicting 1-, 3-, and 5-year OS was 0.752, 0.814, and 0.883, respectively (Fig. 2E). And patients in the low-risk group had a prolonged OS, as shown by the survival curves in the training data (Fig. 2F). Compared to other clinicopathological features, the risk signature had the highest AUC value (Fig. 2G). The survival rates and time distributions of two risk categories in the testing (Fig. 3A–B) and entire (Fig. 3F–G) datasets were consistent with the training datasets. Results from both the training and entire databases revealed that patient in the low-risk category displayed a higher survival probability (Fig. 3C, H). The 1-, 3-, and 5-year AUC values in the testing set were 0.766, 0.652, and 0.816, while for the entire dataset were 0.764, 0.723, and 0.819 (Fig. 3D, I). Clinical ROC plots demonstrated that the risk model had the greatest prediction performance compared to other clinical characteristics (Fig. 3E, J).
Fig. 2.
Development of cuproptosis associated lncRNAs predictive signature. (A) Forrest plot showing 10 prognostic cuproptosis-related lncRNAs by Univariable Cox regression analyses. (B) Correlations between cuproptosis regulators and the 3 model lncRNAs. (C) Bar plot reflecting the expression of signature lncRNAs in TCGA and GTEx database. (D) Survival time and status in the low- and high-risk groups in the training set. (E) ROC curves at 1, 3 and 5 years are described for the training set. (F) Survival analysis shows that survival outcomes are significantly different between low- and high-risk sufferers in training set. (G) Clinical ROC curves showing the prediction reliability between the risk score and other single clinical feature at one year. ROC curve, receiver operating characteristics curve; AUC, area under the curve. (***P < 0.001; **P < 0.01; *P < 0.05; ns, not statistically significant).
Fig. 3.
Risk model performance in the testing and entire TCGA data. Distribution of survival status and survival times for individuals in testing set (A) and entire set (F). Risk score for each one in the testing dataset (B) and whole dataset (G). Kaplan-Meier survival curves between low- and high-risk group in the testing (C) and entire datasets (H). ROC evaluation of 1-, 3-, 5-year survival prediction in the testing (D) and entire datasets (I). Clinical ROC curve showing the prediction accuracy between the risk score and every clinical feature at one year in the testing (E) and whole datasets (J).
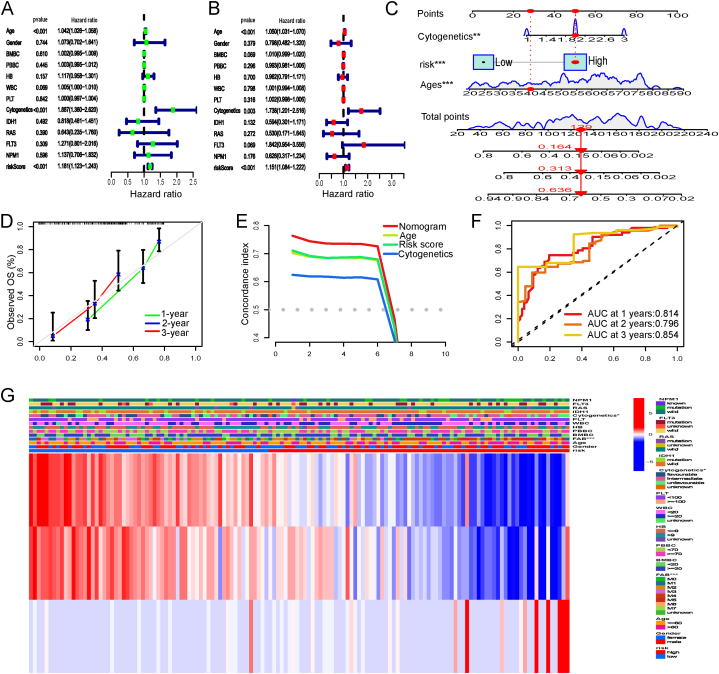
3.4. Constructing a predictive nomogram
The univariate and multivariate analyses revealed that risk score, age, and cytogenetic category were all significant predictors of AML (P < 0.05) (Fig. 4A–B). Next, we built a nomogram using clinical data (age and cytogenetic risk) and risk score to objectively estimate the probability of survival for each subject (Fig. 4C). To ensure the nomogram's accuracy, survival calibration curves for 1, 3, and 5 years were generated (Fig. 4D). Moreover, the concordance index (C-index) quantified the performance measure of different prediction factors (i.e., nomogram, risk score, age, and cytogenetic risk) in survival analysis, demonstrating the superiority of the nomogram in predicting outcomes (Fig. 4E). As seen by the ROC curves (Fig. 4F), the nomogram also performed exceptionally well in predicting 1-, 3-, and 5-year OS (AUC = 0.814, 0.796, and 0.854, respectively) as well. In addition, Fig. 4G depicts a heatmap of the distribution of patients stratified by clinical characteristics into two risk categories. Above all, the nomogram showed an optimal predictor of survival for adult AML patients.
Fig. 4.
Univariate (A) and multivariate (B) Cox regression analyses of clinical characteristics and risk score in the whole TCGA datasets. (C) Nomogram for 1-, 3-, 5-year survival prediction based on the clinical factors and risk score. (D) The calibration curves evaluating the consistency of nomogram. (E) The nomogram and other prognostic parameters are evaluated using the C-index curves. (F) Analysis of the nomogram's ROC curve at 1, 3, and 5 years. (G) Heatmap showing the distribution of clinicopathological features between low- and high-risk groups in the TCGA-LAML datasets.
3.5. Stratification analyses of the risk model with clinical traits
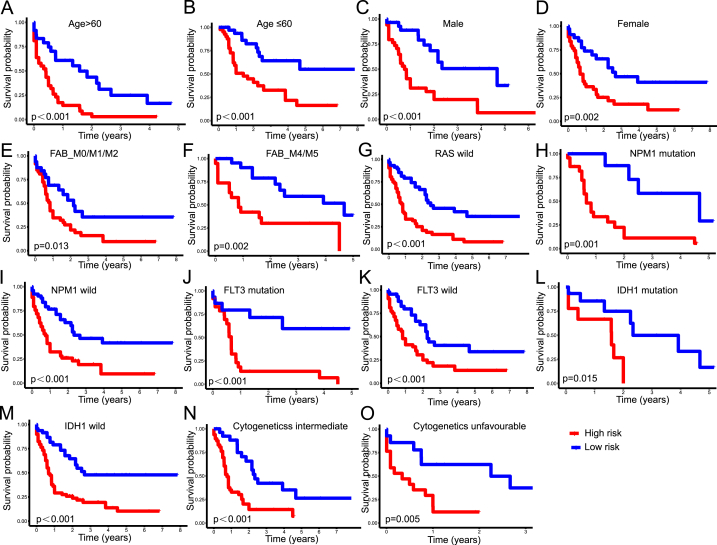
In this study, stratification analysis was performed based on clinicopathological parameters consisting of age (Fig. 5A and B), gender (Fig. 5C and D), FAB classification (Fig. 5E and F), cytogenetic risk category (Fig. 5N and O), and gene mutations of RAS (Fig. 5G), NPM1 (Fig. 5H and I), FLT3 (Fig. 5J and K), and IDH1 (Fig. 5L and M). Kaplan-Meier curves showed that the high-risk group exhibited worse survival rates versus the low-risk group when classified by various clinical parameters, apart from favorable chromosomal abnormalities, M3 types, and RAS mutation. In addition, we discovered that M3 patients had the lowest risk scores of all FAB subtypes, whereas M4/M5 patients had slightly higher risk scores than patients with M0/M1/M2 (Fig. S2A). Individuals with a favorable cytogenetic risk had lower risk scores than those with an intermediate or adverse cytogenetic risk (Fig. S2B). Moreover, AC079466.1 expression increased significantly in the group with a poor cytogenetic risk. TRAF3IP3-AS1 expression increased substantially in the group with a favorable cytogenetic risk (Supplementary Figs. S2C and D). Furthermore, NADK2-AS1 and TRAF3IP2-AS levels were elevated in AML patients with t [15,17] or NPM1 mutation. In contrast, AC079466.1 expression was elevated in patients with complex karyotypes or those who lacked the inv [16] or IDH1-R132/R140 mutation (Supplementary Figs. S2E–J).
Fig. 5.
Correlation between clinical traits and cuproptosis-related lncRNA signature. Kaplan-Meier survival curves depicting survival probability between the low and high-risk groups stratified by age (A–B), gender (C–D), FAB subtype (E–F), RAS mutation (G), NPM1 mutation (H–I), FLT3 mutation (J–K), IDH1 mutation (L–M), and cytogenetic risk categories (N–O).
3.6. PCA and TMB analysis
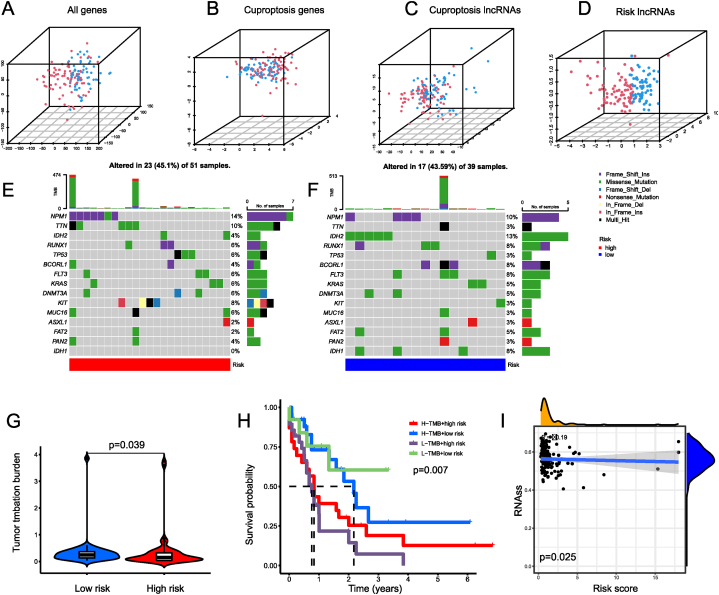
Using PCA analysis, an overview of the distributional trends of two prognostic groups was presented. Results from the entire gene set (Fig. 6A), genes involved in cuproptosis (Fig. 6B), and lncRNAs associated to cuproptosis (Fig. 6C) demonstrated highly dispersed distributions. Nevertheless, results obtained from the signature lncRNAs (Fig. 6D) revealed that two prognostic subgroups exhibited distinct distributions, indicating that the risk model could distinguish AML patients with various clinical characteristics. We subsequently compared mutation frequencies between two risk categories. A waterfall diagram was created using the somatic mutations of 90 individuals with AML (Fig. 6E and F). In the low-risk group, the TMB was more diverse (Fig. 6G). Furthermore, it may be possible to screen out a subset of high-risk, low-TMB individuals with the worst survival rates (Fig. 6H). From these results, it appears that the risk score in conjunction with TMB can be used to evaluate a patient's clinical prognosis. Furthermore, we observed a slightly negative correlation between risk scores and RNAss (Fig. 6I).
Fig. 6.
PCA and TMB Analysis. Data distribution shown via PCA scatterplots based on the expression of all genes (A), cuproptosis genes (B), cuproptosis related lncRNAs (C), and 3 risk lncRNAs in the proposed signature (D). Waterfall plot illustrating the frequently mutated genes in low- (E) and high-risk group (F). According to the frequency of their mutations, the top 15 genes were listed. The upper panel showing the frequencies of mutated genes, and the bottom panel showing the types of mutated genes. (G) TMB between the low- and high-risk group. (H) Analysis of patient survival for risk score and TMB groups. (I) correlation between the risk score and the RNAss. ssGSEA, computational single sample gene set enrichment analysis; RNAss, RNA stemness score; PCA, principal component analysis; TMB, tumor mutational burden.
3.7. Correlation of the risk signature and immune landscape
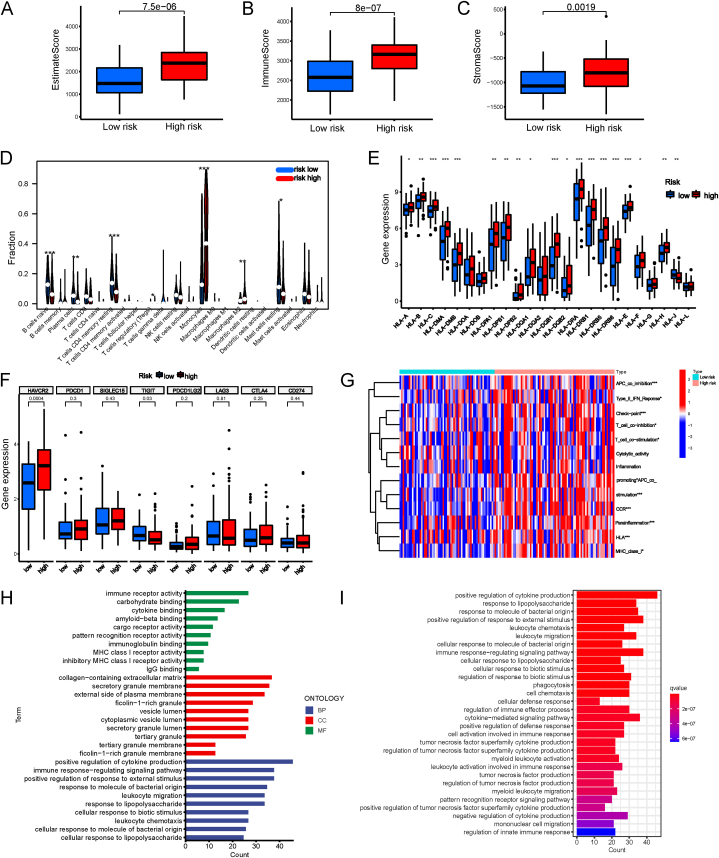
Compared to the favorable risk group, the ImmuneScores, StromalScores, and ESTIMATEScores of the high-risk group were all higher (Fig. 7A–C). And immune infiltration results showed that CD4 memory resting T cells, CD4 memory resting T cells, naive B cells, plasma cells, and resting mast cells were significantly enriched in the low-risk group; on the contrary, the high-risk group had more M2 macrophages, regulatory T (Treg) cells and monocytes (Fig. 7D). In addition, among the 24 HLA genes, 18 (75 %) were upregulated in the high-risk group (Fig. 7E). What's more, the difference between the two risk subtypes' immune checkpoints (HAVCR2, PDCD1, SIGLEC15, TIGIT, PDCD1LG2, LAG3, CTLA4, and CD274) was analyzed. The levels of HAVCR2 and TIGIT varied significantly between the two groups (Fig. 7F). These findings suggested that elevated risk scores may promote HLA expression in AML. Based on GSEA of immune-related function, we discovered that the following immune activities differed significantly between both risk groups: APC co-stimulation, Type II IFN Response, T cell co-stimulation, Checkpoint, Inflammation promoting, T cell co-inhibition, CCR (Chemokine receptors), Type I IFN Response, Parainflammation, HLA, and MHC class I (Fig. 7G).
Fig. 7.
Landscape of the tumour immune microenvironment. (A–C) Diferences in estimate, immune and stromal scores in two risk groups. (D) Types of immune cells that infiltrate tumors differ between high- and low-risk groups. (E) Comparison of the HLA gene expression levels in the low- and high-risk groups. (F) Immune checkpoint gene expression comparison between two risk categories. (G) The heatmap shows the distribution of ssGSEA scores of 13 immunological functions. (H) Analysis of functional enrichment between groups at low and high risk. Bar plot showing the biological process (BP), molecular function (MF), and cellular component (CC) by GO analysis. (I) Analysis of KEGG enrichment between two risk categories. (*P < 0.05, **P < 0.01, ***P < 0.001).
3.8. GO and KEGG enrichment analyses
For a molecular comprehension of the differences between the two risk categories, GO and KEGG analyses were conducted. Most enriched terms were related to inflammation and immunity, including immune response regulating signaling pathway, positive regulation of cytokine production, carbohydrate binding, positive regulation of response to external stimulus, and leukocyte migration (GO biological processes). The ten most significant GO terms and thirty KEGG pathways are shown in Fig. 7H–I.
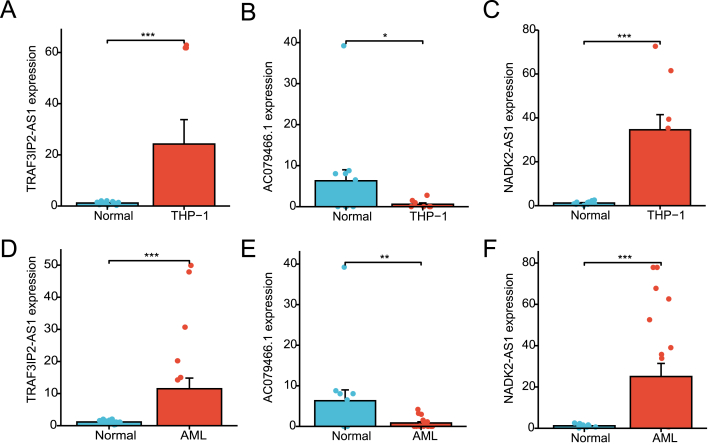
We subsequently performed qRT-PCR assay of three cuproptosis-related lncRNAs in THP-1 cells, 20 newly diagnosed AML patient samples, and 14 healthy adult samples. The three lncRNAs were identified to be significantly differentially expressed in AML samples (Fig. 8).
Fig. 8.
qRT-PCR assay of three cuproptosis related lncRNAs. (A–C) Three lncRNAs associated with cuproptosis were differentially expressed in blood samples from healthy persons and an AML cell line (THP-1). (D–F) Expression levels of 3 lncRNAs in 20 newly diagnosed AML patients and 14 healthy adult samples. (*P < 0.05, **P < 0.01, ***P < 0.001).
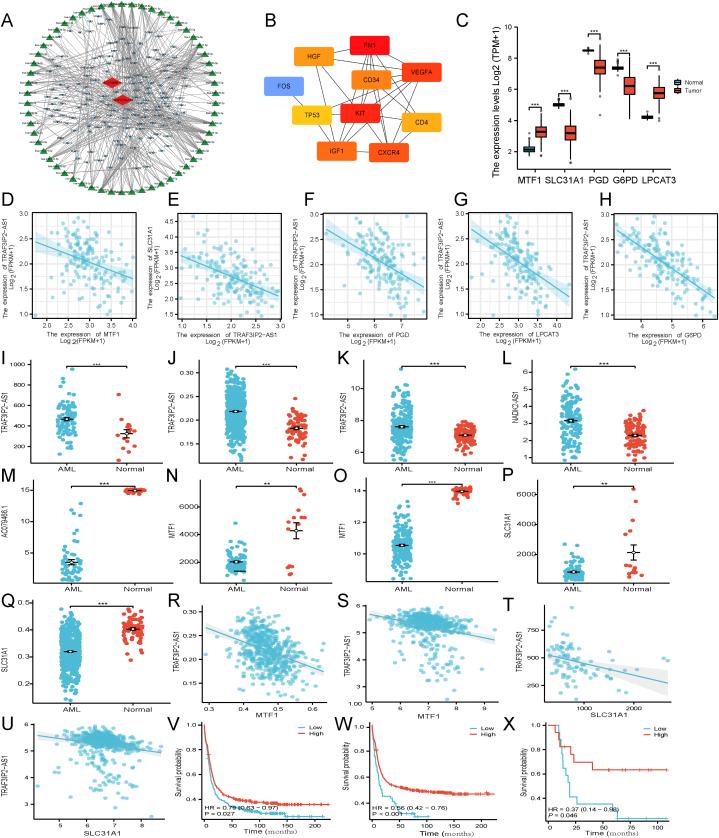
3.9. Construction of the ceRNA and PPI network
Numerous studies highlighted that lncRNAs, acting as ceRNAs, can bind to miRNA to influence the transcription of downstream mRNAs [15]. A lncRNA-miRNA-mRNA ceRNA network was constructed to investigate the potential roles of the cuproptosis-associated lncRNAs in AML. Target miRNAs for AC079466.1 and TRAF3IP2-AS1 were discovered using the LncBase database (no data available for NADK2-AS1). A total of two lncRNAs, 58 miRNA, and 144 mRNAs were identified to construct the ceRNA network (Fig. 9A). Using the PPI network (Supplementary Fig. S3A), we were able to identify ten hub genes (Fig. 9B) and two major modules dominated by FN1 and KIT nodes (Supplementary Fig. S3A). Moreover, the lncRNA-miRNA-mRNA regulatory aix analysis elucidated that TRAF3IP2-AS1 may regulate mRNA expression of MTF1 (cuproptosis gene) [7], SLC31A1 (copper importers) [25], PGD, G6PD and LPCAT3 (ferroptosis-associated genes) [25] through targeting miRNAs. MTF1 was suggested as the downstream target gene of five miRNAs (miR-93-5p, miR-106a-5p, miR-20b-5p, miR-17-5p, miR-20a-5p). Also, PGD and G6PD were probable target mRNA of miR-1-3p. In addition, candidate targets of miR-185-5p and miR-124-3p were LPCAT3 and SLC31A1, respectively. Due to the lack of corresponding miRNA expression data in the TCGA-LAML and GTEx datasets, we searched PubMed for studies on these miRNAs in AML. From previous literature, the expression of miR-93-5p [26,27], miR-124-3p [28,29], miR-185-5p [30,31], miR-20a-5p [32,33], and miR-17-5p [34,35] differs in AML patients compared to healthy controls. Meanwhile, miR-106a-5p was consistently overexpressed in settings where AML was resistant to therapy. MiR-1-3p [36] and miR-20b-5p [37], associated with poor prognosis in AML, were upregulated in leukemia with FLT3 or RUNX1 mutation, respectively.
Fig. 9.
CeRNA network construction and correlation analysis between the expression of the signature lncRNAs and cuproptosis gene. (A) CeRNA network of 2 model lncRNAs (AC079466.1 and TRAF3IP2-AS1; red diamonds), DEmRNAs (blue dot), and target miRNAs (green triangle). (B) PPI network and ten hub gene. (C) The expression of MTF1, SLC31A1, PGD, G6PD, and LPCAT3 in TCGA-LAML and normal tissues. (D–H) Correlation of cuproptosis (MTF1, SLC31A1) and ferroptosis (PGD, G6PD, and LPCAT3) genes with TRAF3IP2-AS1. (I–K) Compared to the normal blood samples obtained from GSE12662 (I), GSE13159 (J), and the GTEx database (K), the expression levels of TRAF3IP2-AS1 were significantly upregulated in AML samples from GSE12662 (I), GSE13159 (J), and the Target database (K). (L) The expression of NADK2-AS1 was significantly upregulated in AML samples from the Target cohort vs. normal samples from the GTEx dataset. (M) The scatter plot depicted the levels of AC079466.1 in AML samples sourced from the Target database, as well as normal samples obtained from the GTEx database. (N–O) The levels of MTF1 in AML samples from the GSE12662 (N) and Target database (O) were compared to normal samples from GSE12662 (N) and the GTEx database (O). (P–Q) In GSE12662 and GSE13159, SLC31A1 expression exhibited higher levels in normal blood samples compared to tumor tissue. (R–S) The expression level of MTF1 demonstrated a negative correlation with TRAF3IP2-AS1 expression in AML samples from GSE6891 and GSE13159. (T–U) The expression levels of SLC31A1 exhibited a negative correlation with the expression of TRAF3IP2-AS1 in AML samples obtained from GSE12662 and GSE6891. (V–W) In GSE6891, it was observed that AML patients with lower levels of TRAF3IP2-AS1 and NADK2-AS1 had a significantly worse prognosis compared to those with higher levels (P = 0.027, P < 0.001, respectively). (X) The expression levels of AC079466.1 were found to be capable of distinguishing AML patients into different prognostic groups in the Target database (p = 0.046). CeRNAs, competing endogenous RNAs. (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, compared to normal tissues in the GTEx, the expression of MTF1, SLC31A1, PGD, G6PD, and LPCAT3 was different in TCGA-LAML (Fig. 9C) and negatively correlated with the expression of TRAF3IP2-AS1 (Fig. 9D–H). Furthermore, we conducted a verification analysis to assess the differential expression levels of TRAF3IP2-AS1, NADK2-AS1, AC079466.1, MTF1, and SLC31A1 in AML samples compared to normal tissues using both GEO chip data and the Target dataset (Fig. 9I-Q). The expression patterns observed were consistent with those found in the TCGA-LAML dataset. Additionally, the GEO gene expression profile data sets revealed a negative correlation between TRAF3IP2-AS1 expression and both MTF1 and SLC31A1 (Fig. 9R–U). Notably, patients with low TRAF3IP2-AS1 or NADK2-AS1 expression exhibited a shorter survival time, which aligns with the findings from the TCGA databases (Fig. 9V-W). Following the application of the identical methodology to analyze the data from Target AML data, a notable disparity in prognosis was observed between the cohort exhibiting elevated AC079466.1 expression and the group with diminished expression levels (Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10X). According to TCGA's pan-cancer database, the expression levels of three signature lncRNAs were measured in 33 kinds of cancer (Supplementary Fig. S4). Moreover, the most enriched GO terms included regulation of mononuclear cell differentiation, myeloid leukocyte differentiation, and lymphocyte differentiation (GO biological processes, Fig. S3B). As determined by KEGG analysis, potential biological activities, such as the 'MAPK', 'PI3K-Akt', and 'Ras' signaling pathway, were linked to AML progression. Some other pathways such as 'Transcriptional misregulation in cancer', 'Th1 and Th2 cell differentiation', and 'Proteoglycans in cancer' were also involved in tumor development.
Fig. 10.
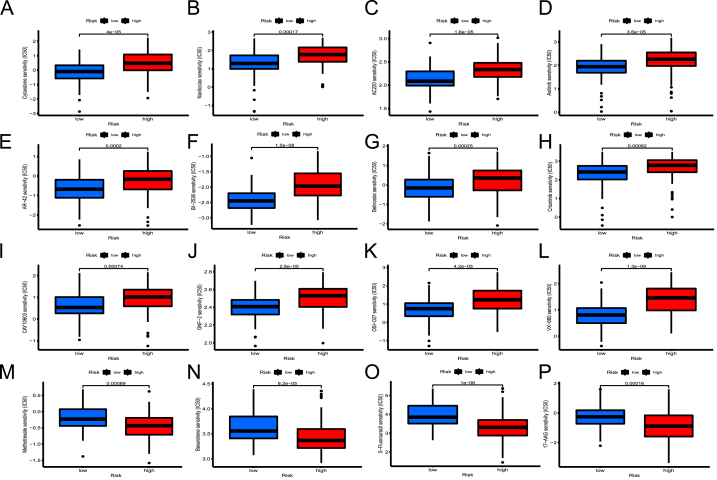
Patients in low- and high-risk categories differ in their susceptibility to chemotherapy drugs. (A–M) For cytarabine, n avitoclax, AC220, axitinib, AR-42, BI-2536, belinostat, crizotinib, CAY10603, GNF-2, OSI-027, and VX-680, IC50 values were greater in the high-risk category than in the low-risk category. (N–Q) Drug sensitivity of methotrexate, bexarotene, 5-fluorouracil, and 17-AGG were greater in the low-risk category than in the high-risk category.
3.10. Role of risk models in pharmacotherapy
Following that, there were obvious differences in drug sensitivity between the two risk populations. Several therapies have shown better results in the high-risk individuals than the favorable risk ones, including cytarabine, navitoclax (BCL-2/2 BCL-XL inhibitor), AC220 (FLT3 inhibitor), GNF2 (BCR-ABL inhibitor), AR-42, belinostat, CAY10603 (histone deacetylase inhibitor, HDACi), BI 2536 (polo-like kinase 1 inhibitor), crizotinib (ALK kinase inhibitor), VX-680 (aurora kinase inhibitor), OSI-027 (mTOR inhibitor) and axitinib (VEGFR and c-KIT inhibitor) (Fig. 10A-L). For methotrexate, bexarotene (retinoid X receptor agonist), 5-fluorouracil, and 17AGG (Hsp90 inhibitor), treatment response of low-risk patients was improved (Fig. 10M − P). These findings suggested that this risk model could aid in the optimization of personalized treatment strategies for AML patients.
4. Discussion
The pathophysiology of AML is complex, and our comprehension of its etiology and contributing factor is limited [2]. The prognostic stratification of AML is currently predominantly determined by its clinical characteristics, FAB typing, cytogenetic risk, and molecular abnormalities [2,4,9]. Nonetheless, the outcomes of AML patients in the same risk category who received similar treatment vary substantially, suggesting that the current staging system may not adequately guide treatment decisions and prognostic assessments [2]. Cuproptosis is a distinct form of PCD with promising potential as a cancer treatment. Growing evidence has linked cancer to disturbances in copper homeostasis [5,6]. There is an urgent need for cuproptosis detection indicators in cancer tissues. As previously indicated, lncRNA could be a reliable biomarker for AML prognosis and therapeutic efficacy [13,15]. However, research on the role of lncRNAs associated with cuproptosis in AML progression and prognosis is still in its infancy, and further studies are required.
Using univariate Cox regression, we initially identified 139 cuproptosis-associated lncRNAs related to prognostic value, and subsequently found that 47 of them expressed differently in AML samples and normal controls. Finally, utilizing multivariate Cox and LASSO regression analysis, we established a signature including three cuproptosis-associated lncRNAs to predict AML outcomes. NADK2-AS1 and TRAF3IP2-AS1 were protective factors, while AC079466.1 posed a threat. Furthermore, TRAF3IP2-AS1 and NADK2-AS1 were upregulated in patients with t(15,17) chromosomal translocation, whereas the expression of AC079466.1 was elevated in patients with complex karyotypes. In addition, qRT-PCR confirmed the differential expression of three lncRNAs between AML patients and healthy controls. In the meantime, those lncRNAs differential expression were also verified by the external validation cohort.
In previous studies, TRAF3IP2-AS1 has been revealed to serve as a tumor suppressor and may act as a ceRNA that competitively binds to miR-200a-3p/153-3p/141-3p and prevents them from reducing phosphatase and tensin homolog (PTEN) levels in renal cell carcinoma [38]. Also, TRAF3IP2-AS1 was shown to be a useful diagnostic marker for cervical cancer [39]. Besides, TRAF3IP2-AS1 functions as a negative regulator of IL-17 signaling in autoimmune diseases [40]. Moreover, TRAF3IP2-AS1 has been recognized as a probable prognostic indicators for pancreatic adenocarcinoma [41], glioblastoma [42], and AML [43]. Furthermore, AC079466.1 significantly correlated with the overall survival of gastric cancer and hepatocellular carcinoma [44,45]. Notably, NADK2-AS1 was disclosed for the first time. Although relatively few studies have described AC079466.1 and NADK2-AS1, these two lncRNAs are only differentially expressed in TCGA-LAML among 31 tumor types according to the pan-cancer analysis. The above findings implied a potential link between cuproptosis-associated lncRNAs and the development and prognosis of AML.
The Kaplan-Meier and ROC curves demonstrated the high predictability of the three-lncRNAs signature. In both univariate and multivariate Cox regression models, the risk score was shown to be a reliable predictive indicator. In addition, the proposed predictive model may be capable of identifying individuals with a worse OS based on survival studies of classified clinicopathologic variables. Moreover, the risk model exhibited the most accurate prediction performance when compared to other clinicopathological characteristics. Furthermore, the nomogram was assessed by ROC curve, calibration curve, DCA, and C-index. Finally, these findings demonstrated that our nomogram was highly reliable and could be used to assess AML prognosis. TMB was shown to be greater in the low-risk group compared to the high-risk group when stratified by risk score. In fact, the low TMB has been shown to cause immune therapy failure [46]. The Kaplan-Meier curves illustrated that TMB combined with risk signature could distinguish four different prognostic categories among patients. Apart from TMB, our results revealed a negative correlation between risk score and the stemness scores of AML samples. Recent investigations have proven the importance of non-coding RNA in regulating cancer stemness. However, the precise modulating mechanism remains poorly understood.
Further GO and KEGG analysis indicated that the pathways implicated are primarily focus on inflammation and immunity. It has been reported that copper can also promote cytokine secretion essential for tumor progression and metastasis, such as interleukin 1 (IL-1) and proangiogenic factors [6,47]. Several cuproptosis genes have been shown to play a crucial role in the immune-inflammatory pathology of rheumatoid arthritis [48]. For instance, MFT1 stimulates the release of inflammatory factors and facilitates the arrest of pro-inflammatory T-cells. FDX1 affects the activities of Treg cells, macrophages, dendritic cells, and monocytes. CDKN2A regulates adaptive immune function and inflammatory factor production, as well as promoting pro-inflammatory responses. GLS influences B cell activation and antibody production. LIAS is responsible for regulating inflammation and oxidative stress. Moreover, PDHA1 can affect the inflammatory response of macrophages [48].
Immune cell infiltration, immunological function, checkpoint, and HLA transcription were found to differ significantly between the two risk groups. Consequently, cuproptosis is probably essential for tumor immune response. Firstly, our immune function results were consistent with those obtained from a ferroptosis-related prognostic model for AML [25], indicating commonalities in anti-tumor immunity of cuproptosis and ferroptosis. Secondly, the results of immune cell abundance analysis demonstrated significantly higher levels of Treg cells, monocytes, and M2 macrophages in the high-risk subtype. Researchers have found that both Treg cells and M2 macrophages contribute to cancer development [49,50]. A decreased proportion of plasma cells, B cells, and CD4+ memory resting T cells indicated that adaptive immune system was impaired in the high-risk group. Thirdly, the immune score, HLA gene, and hepatitis A virus cellular receptor 2 (HAVCR2, also known as TIM-3) were upregulated in the high-risk group. Immune cell infiltration was found to be more prevalent in tumors with higher HLA gene expression, and the patient's response to the immune checkpoint inhibitor is related to the amount of HLA gene expression [51]. Our findings yielded similar results. Immunological response in AML relies on the immune-related checkpoint [52]. This means that targeting HAVCR2 strategies may be beneficial for high-risk patients. HAVCR2 has emerged as a therapeutic target for curing AML; blocking it can kill two birds with one stone by eradicating leukemia stem cells and activating anti-tumor immunity in AML [53].
The ceRNA network and functional enrichment analysis suggested that cuproptosis-associated lncRNAs play critical roles in myeloid cell differentiation, tyrosine autophosphorylation, and energy metabolism. The significantly enriched pathways primarily focused on cancer hallmark relevant pathways. These results indicated that cuproptosis-related lncRNAs may influence myeloid differentiation and cancer cell proliferation, shedding light on the potential functions of these lncRNAs in AML.
Interestingly, our research findings revealed that TRAF3IP2-AS1 may regulate the expression of MTF1 SLC31A1, PGD, G6PD, and LPCAT3 by targeting miRNAs associated with myeloid differentiation, drug resistance, treatment failure, proliferation, and invasion of AML [[26], [27], [28], [29], [30], [31], [32], [33]]. Moreover, TRAF3IP2-AS1 was negatively correlated with these mRNAs as a protective factor. MTF1, as a metal-binding transcription factor, is involved in copper homeostasis and has been reported as a negative regulator of cuproptosis [7]. SLC31A1 encodes a high-affinity copper transporter and plays a vital role in maintaining the intracellular copper concentration [42]. PGD, G6PD, and LPCAT3 have been discovered as ferroptosis-related genes with significance for AML survival [25]. LPCAT3, involved in lipid metabolism, was critical pro-ferroptosis gene [54]. PGD and G6PD are both associated with energy metabolism and have the ability to prevent erastin-induced ferroptosis [55]. In addition, TRA3IP2-AS1 has been identified as a ferroptosis-related lncRNA implicated in pancreatic cancer prognosis [41]. The evidence presented above suggested that TRAF3IP2-AS1 might function as a form of ceRNA to regulate the transcription of genes related to cuproptosis and ferroptosis.
Drug sensitivity analysis illustrated that each risk group was sensitive to its representative medications, which may contribute to better clinical outcomes. The high-risk group may benefit from FDA-approved chemotherapeutic agents, including methotrexate, BCL-2, HDACi, FLT3, and BCR-ABL inhibitors [2,4].
This study has several limitations. Firstly, our analysis relied upon retrospective cases available in public datasets. Because the TCGA database does not provide specific treatment project information for each patient, survival data regarding treatments could not be analyzed further. No external database was available for lncRNA expression and clinical data to validate the reliability of the signature. Additional verification using prospective, multicenter, real-world data is required for this risk model. Secondly, we only verified the differential expression of three lncRNAs in AML versus normal subjects with a small sample size. Thirdly, we did not conduct any further studies to explore the mechanisms and applications of the signature. Consequently, additional experiments are still required to deeply investigate the function of cuproptosis-associated lncRNAs in AML.
5. Conclusions
Our work established a signature of 3 cuproptosis-associated lncRNAs (AC079466.1, TRAF3IP2-AS1, and NADK2-AS1) to predict the clinical outcomes of AML patients with high accuracy. Besides, we established a related ceRNA network of those lncRNAs. TRAF3IP2-AS1 may act as a ceRNA to regulate the transcription of genes involved in AML cuproptosis and ferroptosis. Our findings contribute to current understanding of cuproptosis processes and their application to the prognosis and treatment of AML, despite the need for further functional investigation.
Human ethics
Ethical approval was obtained from the hospital's Ethics Committee and the Ethics Review Boards at Second Affiliated Hospital of Chongqing Medical University (Ethic code: 2020438).
Funding
This work was supported by the Chongqing Natural Science Foundation of China [grant number 2020FYYX227].
Data availability statement
The datasets utilized in this study were available to the public. All data can be download from TCGA (https://portal.gdc.cancer.gov/repository), GTEx (https://xenabrowser.net/datapages/), GEO (https://www.ncbi.nlm.nih.gov/geo/), and Target (https://ocg.cancer.gov/programs/target) datasets. Data will be made available on request.
CRediT authorship contribution statement
Chun Cao: Data curation, Investigation, Methodology, Project administration, Software, Validation, Writing - original draft. Teng Wang: Data curation, Investigation, Resources, Software, Validation. Yun Luo: Data curation, Methodology, Validation, Writing - original draft. Yin Zhang: Data curation, Resources, Software, Validation, Visualization. Yue-yu Dai: Formal analysis, Resources, Validation, Visualization. Yan Shen: Conceptualization, Funding acquisition, Project administration, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Yan Shen reports financial support was provided by Chongqing Natural Science Foundation of China. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the great technical assistance provided by the department of haematology staff at the Second Affiliated Hospital of Chongqing Medical University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22532.
Contributor Information
Chun Cao, Email: 303774@hospital.cqmu.edu.cn.
Teng Wang, Email: willwangteng@hospital.cqmu.edu.cn.
Yun Luo, Email: 300333@cqmu.edu.cn.
Yin Zhang, Email: 13996304411@163.com.
Yue-yu Dai, Email: 18782475758@163.com.
Yan Shen, Email: shenyan@hospital.cqmu.edu.cn, c1997@hospital.cqmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Diagram illustrating the study's design process.
Correlation between risk signature and clinicopathological characteristics in TCGA-LAML. (A-B): Distribution of risk score with reference to FAB and cytogenetic classification. (C-D) TRAF3IP2-AS1 and AC079466.1 expression across three cytogenetic risk groups. (E-J) Gene transcription of NADK2AS1, TRAF3IP2-AS1, and AC079466.1 in AML patients with or without t(15,17), inv(16), complex karyotype, IDH1 R132 mutation, IDH1R140 mutation, and NPM1 mutation. (***P < 0.001; **P < 0.01; *P < 0.05; ns, not statistically significant).
PPI network construction and functional enrichment analysis of DEmRNA. (A) The PPI network associated with model lncRNAs in AML. (B) The most enriched GO terms were regulation of mononuclear cell differentiation, myeloid leukocyte differentiation, and lymphocyte differentiation. DEmRNA, differentially expressed mRNA.
Expression of TRAF3IP2-AS1 (A) NADK2-AS1 (B), and AC079466.1 (C) , and TRAF3IP2-AS1 in 31 33 tumors and normal tissues. (*P≤ 0.01, |log2FC|≥1). Red box, lncRNAs expression levels of 31 tumors in TCGA; black box, lncRNAs expression levels of normal tissues in the TCGA and GTEx. (***P < 0.001; **P < 0.01; *P < 0.05)
References
- 1.Liu H. Emerging agents and regimens for AML. J. Hematol. Oncol. 2021;14:49. doi: 10.1186/s13045-021-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kantarjian H., Kadia T., DiNardo C., et al. Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 2021;11:41. doi: 10.1038/s41408-021-00425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koenig K., Mims A. Relapsed or primary refractory AML: moving past MEC and FLAG-ida. Curr. Opin. Hematol. 2020;27:108–114. doi: 10.1097/MOH.0000000000000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Döhner H., Wei A.H., Appelbaum F.R., et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–1377. doi: 10.1182/blood.2022016867. [DOI] [PubMed] [Google Scholar]
- 5.Michniewicz F., Saletta F., Rouaen J.R.C., et al. Copper: an intracellular Achilles' Heel Allowing the targeting of Epigenetics, kinase pathways, and cell metabolism in cancer therapeutics. ChemMedChem. 2021;16:2315–2329. doi: 10.1002/cmdc.202100172. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y., Huo Z., Qi X., et al. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine (London, England) 2022;17:303–324. doi: 10.2217/nnm-2021-0374. [DOI] [PubMed] [Google Scholar]
- 7.Tsvetkov P., Coy S., Petrova B., et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science (New York, NY) 2022;375:1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliveri V. Selective targeting of cancer cells by copper Ionophores: an overview. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.841814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadbeigi S., Javadian S., Ebrahimi-Rad M., et al. Assessment of trace elements in serum of acute lymphoblastic and myeloid leukemia patients. Exp. Oncol. 2019;41:69–71. [PubMed] [Google Scholar]
- 10.Kim S., Freeland-Graves J.H., Babaei M., et al. Quantifying the association between acute leukemia and serum zinc, copper, and selenium: a meta-analysis. Leuk. Lymphoma. 2019;60:1548–1556. doi: 10.1080/10428194.2018.1540043. [DOI] [PubMed] [Google Scholar]
- 11.Singh R.P., Jeyaraju D.V., Voisin V., et al. Disrupting mitochondrial copper distribution Inhibits leukemic stem cell self-renewal. Cell Stem Cell. 2020;26 doi: 10.1016/j.stem.2020.04.010. 926-37.e10. [DOI] [PubMed] [Google Scholar]
- 12.Grasedieck S., Rueß C., Pochert N., et al. Identification of novel Lncrnas that predict survival in AML patients and modulate leukemic cells. Blood. 2018;132:3909. [Google Scholar]
- 13.Li Z.J., Cheng J., Song Y., et al. LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Laboratory investigation; a journal of technical methods and pathology. 2021;101:341–352. doi: 10.1038/s41374-020-00519-9. [DOI] [PubMed] [Google Scholar]
- 14.Tao Y., Zhang J., Chen L., et al. LncRNA CD27-AS1 promotes acute myeloid leukemia progression through the miR-224-5p/PBX3 signaling circuit. Cell Death Dis. 2021;12:510. doi: 10.1038/s41419-021-03767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Cheng Z., Pang Y., et al. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019;12:51. doi: 10.1186/s13045-019-0734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J., Shi X., Dai R.Q. The prognostic impact of abnormally expressed, long noncoding RNAs in acute myeloid leukemia: a meta-analysis. Hematology (Amsterdam, Netherlands) 2020;25:219–228. doi: 10.1080/16078454.2020.1779480. [DOI] [PubMed] [Google Scholar]
- 17.Vivian J., Rao A.A., Nothaft F.A., et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017;35:314–316. doi: 10.1038/nbt.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genomic and Epigenomic Landscapes of adult de novo acute myeloid leukemia. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rooney M.S., Shukla S.A., Wu C.J., et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malta T.M., Sokolov A., Gentles A.J., et al. Machine Learning Identifies stemness features associated with oncogenic Dedifferentiation. Cell. 2018;173 doi: 10.1016/j.cell.2018.03.034. 338-54.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karagkouni D., Paraskevopoulou M.D., Tastsoglou S., et al. DIANA-LncBase v3: indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2020;48 doi: 10.1093/nar/gkz1036. D101-D110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D., Franceschini A., Wyder S., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y., Tian S., Zhang P., et al. Construction and validation of a novel ferroptosis-related prognostic model for acute myeloid leukemia. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.708699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim E.L., Trinh D.L., Ries R.E., et al. MicroRNA expression-based model indicates Event-Free survival in Pediatric acute myeloid leukemia. J. Clin. Oncol. : official journal of the American Society of Clinical Oncology. 2017;35:3964–3977. doi: 10.1200/JCO.2017.74.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Wu Y., Uddin M.N., et al. Identification of MiR-93-5p targeted pathogenic markers in acute myeloid leukemia through integrative bioinformatics analysis and clinical validation. Journal of oncology. 2021;2021 doi: 10.1155/2021/5531736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K., Dai J., Liu T., et al. Retracted Article: LncRNA ZEB2-AS1 regulates the drug resistance of acute myeloid leukemia via the miR-142-3p/INPP4B axis. RSC Adv. 2019;9:39495–39504. doi: 10.1039/c9ra07854a. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Li S., Ma Y., Tan Y., et al. Profiling and functional analysis of circular RNAs in acute promyelocytic leukemia and their dynamic regulation during all-trans retinoic acid treatment. Cell Death Dis. 2018;9:651. doi: 10.1038/s41419-018-0699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esa E., Hashim A.K., Mohamed E.H.M., et al. Construction of a microRNA-mRNA regulatory network in de novo Cytogenetically normal acute myeloid leukemia patients. Genet. Test. Mol. Biomarkers. 2021;25:199–210. doi: 10.1089/gtmb.2020.0182. [DOI] [PubMed] [Google Scholar]
- 31.Pang B., Mao H., Wang J., et al. MiR-185-5p suppresses acute myeloid leukemia by inhibiting GPX1. Microvasc. Res. 2022;140 doi: 10.1016/j.mvr.2021.104296. [DOI] [PubMed] [Google Scholar]
- 32.Bao F., Zhang L., Pei X., et al. MiR-20a-5p functions as a potent tumor suppressor by targeting PPP6C in acute myeloid leukemia. PLoS One. 2021;16 doi: 10.1371/journal.pone.0256995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J.M., Li M., Luo W., et al. Curcumin attenuates Adriamycin-resistance of acute myeloid leukemia by inhibiting the lncRNA HOTAIR/miR-20a-5p/WT1 axis. Laboratory investigation; a journal of technical methods and pathology. 2021;101:1308–1317. doi: 10.1038/s41374-021-00640-3. [DOI] [PubMed] [Google Scholar]
- 34.Hu L., Liu J., Meng Y., et al. Long non-coding RNA HOTAIR regulates myeloid differentiation through the upregulation of p21 via miR-17-5p in acute myeloid leukaemia. RNA Biol. 2021;18:1434–1444. doi: 10.1080/15476286.2020.1854520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Uddin M.N., Hao J.P., et al. Identification of potential novel prognosis-related genes through transcriptome sequencing, bioinformatics analysis, and clinical validation in acute myeloid leukemia. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.723001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S., Chen Y., Zhu Z., et al. Identification of the key genes and microRNAs in adult acute myeloid leukemia with FLT3 mutation by bioinformatics analysis. Int. J. Med. Sci. 2020;17:1269–1280. doi: 10.7150/ijms.46441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y., Chen S., Lu J., et al. MicroRNA-363-3p promote the development of acute myeloid leukemia with RUNX1 mutation by targeting SPRYD4 and FNDC3B. Medicine. 2021;100 doi: 10.1097/MD.0000000000025807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L., Chen Y., Liu N., et al. Low expression of TRAF3IP2-AS1 promotes progression of NONO-TFE3 translocation renal cell carcinoma by stimulating N(6)-methyladenosine of PARP1 mRNA and downregulating PTEN. J. Hematol. Oncol. 2021;14:46. doi: 10.1186/s13045-021-01059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo C., Pang X. IncRNA TRAF3IP2-AS1 restrains cervical cancer progression by Sponging miR-3677-3p and acts as a diagnostic marker in cervical cancer. Crit. Rev. Eukaryot. Gene Expr. 2022;33:43–51. doi: 10.1615/CritRevEukaryotGeneExpr.2022044035. [DOI] [PubMed] [Google Scholar]
- 40.He R., Wu S., Gao R., et al. Identification of a long noncoding RNA TRAF3IP2-AS1 as key regulator of IL-17 signaling through the SRSF10-IRF1-Act1 Axis in autoimmune diseases. J. Immunol. 2021;206:2353–2365. doi: 10.4049/jimmunol.2001223. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Zhang J., Tao S., et al. Prognostication of pancreatic cancer using the cancer Genome Atlas based ferroptosis-related long non-coding RNAs. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.838021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zan X.Y., Li L. Construction of lncRNA-mediated ceRNA network to reveal clinically relevant lncRNA biomarkers in glioblastomas. Oncol. Lett. 2019;17:4369–4374. doi: 10.3892/ol.2019.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong F., Yao F., Cheng Y., et al. m6A-related lncRNAs predict prognosis and indicate immune microenvironment in acute myeloid leukemia. Sci. Rep. 2022;12:1759. doi: 10.1038/s41598-022-05797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W., Chen Q.F., Huang T., et al. Identification and validation of a prognostic lncRNA signature for hepatocellular carcinoma. Front. Oncol. 2020;10:780. doi: 10.3389/fonc.2020.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng S., Chen Y., Yu S., et al. DISC1 as a prognostic biomarker correlated with immune infiltrates in gastric cancer. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strickler J.H., Hanks B.A., Khasraw M. Tumor mutational burden as a predictor of immunotherapy response: is more Always better? Clin. Cancer Res. 2021;27:1236–1241. doi: 10.1158/1078-0432.CCR-20-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L., Min J., Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Targeted Ther. 2022;7:378. doi: 10.1038/s41392-022-01229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J., Guo S., Schrodi S.J., et al. Cuproptosis and cuproptosis-related genes in rheumatoid arthritis: Implication, prospects, and perspectives. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.930278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouketsu A., Sato I., Oikawa M., et al. Regulatory T cells and M2-polarized tumour-associated macrophages are associated with the oncogenesis and progression of oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2019;48:1279–1288. doi: 10.1016/j.ijom.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Erlandsson A., Carlsson J., Lundholm M., et al. M2 macrophages and regulatory T cells in lethal prostate cancer. Prostate. 2019;79:363–369. doi: 10.1002/pros.23742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaafsma E., Fugle C.M., Wang X., et al. Pan-cancer association of HLA gene expression with cancer prognosis and immunotherapy efficacy. Br. J. Cancer. 2021;125:422–432. doi: 10.1038/s41416-021-01400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daver N. Immune checkpoint inhibitors in acute myeloid leukemia. Best Pract. Res. Clin. Haematol. 2021;34 doi: 10.1016/j.beha.2021.101247. [DOI] [PubMed] [Google Scholar]
- 53.Kikushige Y., Miyamoto T. TIM-3 as a novel therapeutic target for eradicating acute myelogenous leukemia stem cells. Int. J. Hematol. 2013;98:627–633. doi: 10.1007/s12185-013-1433-6. [DOI] [PubMed] [Google Scholar]
- 54.Kagan V.E., Mao G., Qu F., et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao M., Monian P., Quadri N., et al. Glutaminolysis and Transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagram illustrating the study's design process.
Correlation between risk signature and clinicopathological characteristics in TCGA-LAML. (A-B): Distribution of risk score with reference to FAB and cytogenetic classification. (C-D) TRAF3IP2-AS1 and AC079466.1 expression across three cytogenetic risk groups. (E-J) Gene transcription of NADK2AS1, TRAF3IP2-AS1, and AC079466.1 in AML patients with or without t(15,17), inv(16), complex karyotype, IDH1 R132 mutation, IDH1R140 mutation, and NPM1 mutation. (***P < 0.001; **P < 0.01; *P < 0.05; ns, not statistically significant).
PPI network construction and functional enrichment analysis of DEmRNA. (A) The PPI network associated with model lncRNAs in AML. (B) The most enriched GO terms were regulation of mononuclear cell differentiation, myeloid leukocyte differentiation, and lymphocyte differentiation. DEmRNA, differentially expressed mRNA.
Expression of TRAF3IP2-AS1 (A) NADK2-AS1 (B), and AC079466.1 (C) , and TRAF3IP2-AS1 in 31 33 tumors and normal tissues. (*P≤ 0.01, |log2FC|≥1). Red box, lncRNAs expression levels of 31 tumors in TCGA; black box, lncRNAs expression levels of normal tissues in the TCGA and GTEx. (***P < 0.001; **P < 0.01; *P < 0.05)
Data Availability Statement
The datasets utilized in this study were available to the public. All data can be download from TCGA (https://portal.gdc.cancer.gov/repository), GTEx (https://xenabrowser.net/datapages/), GEO (https://www.ncbi.nlm.nih.gov/geo/), and Target (https://ocg.cancer.gov/programs/target) datasets. Data will be made available on request.