Abstract
Several models have been proposed for the mechanism of chromatin remodelling across the promoters of inducible genes in mammalian cells. The most commonly held model is one of cooccupation where histone proteins are modified by acetylation or phosphorylation and nucleosomes are remodelled, allowing the assembly of transcription factor complexes. Using chromatin immunoprecipitation, we observed an apparent decrease of histone acetylation and phosphorylation signals at the proximal promoter region of the inducible interleukin-2 and granulocyte-macrophage colony-stimulating factor genes in response to T-cell activation. We showed that this apparent decrease was due to a loss of histone H3 and H4 proteins corresponding to a decrease in nucleosome occupation of the promoter. This histone loss is reversible; it is dependent on the continual presence of appropriate activating signals and transcription factors and is not dependent on the acetylation status of the histone proteins. These data show for the first time that histone proteins are lost from a mammalian promoter upon activation of transcription and support a model of activation-dependent disassembly and reassembly of nucleosomes.
The chromatin environment of a gene has a major influence on its transcription efficiency. In general, genes that are packaged into highly compacted chromatin structures are maintained in a silent state, whereas highly expressed genes have a more loosely packaged chromatin conformation. The activation and repression of genes during differentiation and development have been associated with alterations in chromatin structure and nuclear location (3, 16, 20, 40). Several general mechanisms for altering chromatin structure have been elucidated in recent years. A group of enzymes with ATPase activity, the Swi/Snf- and iSwi-type proteins, remodel chromatin structure by altering the interaction between nucleosomes and DNA, allowing movement of nucleosomes along the DNA in some cases (14, 19, 24, 41). The N-terminal tails of the histone proteins are highly modified through the addition of acetyl or methyl groups to lysine residues and the addition of phosphate groups to serine residues (19, 42, 45). Specific modifications have been associated with active or silent genes; for example, acetylation of histone H3 on lysine 9 (H3K9) is associated with gene activation, whereas methylation of the same residue is associated with gene silencing (19, 42, 45). Phosphorylation of serine 10 on H3 is also associated with active transcription (8, 27). There is extensive cross talk between the different modifications so that synergistic acetylation of K14 and phosphorylation of S10 occurs but S10 phosphorylation antagonizes methylation at K9 (8, 27). Modification of the histones is performed by proteins that are also coactivators or corepressors of gene transcription, such as the CBP/p300 proteins (42, 45).
Rapid and transient induction of gene transcription is a feature of the response of cells of the immune system to infection. Transient changes in DNase I, micrococcal nuclease (MNase), or restriction enzyme digestion patterns have been documented for promoters and enhancers of many inducible genes in immune cells, indicating that changes in chromatin structure occur in response to cell activation (10, 44). For most genes, the biochemical events underlying these changes have not been well described. However, several general mechanisms of inducible promoter chromatin remodelling have been proposed. The majority of reports in the literature describe an increase in specific histone acetylation and phosphorylation across gene promoter regions in response to activation, concurrent with recruitment of transcription factors (reviewed in reference 11). These data imply a model of cooccupation of promoter regions by modified nucleosomes and transcription factor complexes. A detailed study of the promoter of the beta interferon gene (IFN-β) has shown that a nucleosome positioned across the transcription start site slides along the DNA in response to the binding of transcription complexes and coactivators to a nucleosome-free region immediately upstream (23). Recently, several studies on the inducible Saccharomyces cerevisiae PHO5 gene have shown that histones are lost from the promoter region of this gene leading to a model of nucleosome disassembly across the region of transcription factor recruitment (1, 6, 7, 32).
The mammalian cytokine interleukin-2 (IL-2) gene (IL-2) is rapidly induced after T-cell activation (26). The proximal promoter region has been defined in transfection assays as a region from positions +1 to −300 upstream from the transcription start site (12, 39), although further upstream regions are required for correct transgene function in transgenic animal experiments (46). This promoter region contains defined binding sites for many inducible transcription factors, such as NF-κB, NFAT, and AP-1 (18, 28, 36, 38). The proximal promoter region shows inducible DNase I and restriction enzyme accessibility in response to T-cell activation (26, 44). We have previously reported that the promoter is completely insensitive to MNase digestion in resting T cells and that within 1.5 to 2 h of activation, the entire region (positions +1 to −300) becomes accessible to MNase (30), implying an alteration or removal of nucleosomes. Transcription from another cytokine gene, the granulocyte-macrophage colony-stimulating factor gene (GM-CSF), is also induced after T-cell activation (10, 38), and the promoter, which is incorporated in the first 120 bp from the transcription start site, becomes accessible to DNase I and MNase (17). The changes in chromatin accessibility are reversible and are dependent on new protein synthesis and on the presence of the NF-κB protein, c-Rel (29, 30). We have previously provided evidence for a positioned nucleosome across the promoter region of IL-2 located from positions −60 to −200, and the assembly of this nucleosome in vitro blocks the binding of recombinant transcription factors, such as c-Rel, AP-1, and NFAT, implying a role for this positioned nucleosome in controlling IL-2 gene transcription (4, 17). The role of histone modification in IL-2 gene activation has not been investigated, although many of the transcription factors that activate the IL-2 gene, such as NF-κB and NFAT, have been shown to interact with histone acetyltransferase proteins, such as p300/CBP, and to cooperate with these coactivators for transcription activation (15, 47, 48).
We have now examined some of the biochemical events underlying chromatin remodelling on the IL-2 and GM-CSF gene promoters. We provide evidence that histone proteins are lost from the promoter regions in an activation-dependent manner and suggest a model of nucleosome disassembly from inducible promoters to allow the assembly of transcription complexes.
MATERIALS AND METHODS
Cell culture.
EL-4 thymoma cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, gentamicin, and penicillin. Cells in suspension were stimulated for various times at 106 cells/ml with both phorbol 12-myristate 13-acetate and Ca2+ ionophore (P/I) at final concentrations of 10 ng/ml and 1 μM, respectively. For the stimulus withdrawal experiment, stimulated cells were washed after 4 h of stimulation in 50 ml of fresh medium three to five times prior to incubation for the designated times. In some cases, cells were pretreated with trichostatin A (TsA) (Upstate Biotechnology) for 5 h or cyclosporin A (CsA) for 30 min prior to activation with P/I.
RNA isolation and PCR amplification.
Total RNA was isolated from 5 × 106 cells using Tri reagent (Sigma-Aldrich). cDNA was prepared, and SYBR green PCR was performed with 50 ng of cDNA in a total volume of 25 μl. The primer sets used for IL-2, GM-CSF, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification are shown in Table 1. The cycle threshold (CT) values generated for each time point were quantified using a standard curve generated with cDNA from cells stimulated for 4 h and normalized to GAPDH and expressed as changes relative to GAPDH.
TABLE 1.
Sequences of primers used for chromatin accessibility assays, ChIP, or mRNA measurements by quantitative PCR
| Primer | Sequence of primer
|
Amplified region (bp) | |
|---|---|---|---|
| Sense (5′→3′) | Antisense (5′→3′) | ||
| IL-2 A | CCTGCAGGCATGTACAGCAT | CGCTGTTGACAAGGAGCACA | +38 to +107 |
| IL-2 B | CACAGGTAGACTCTTTGAAAATATGTGTAA | CATGGGAGGCAATTTATACTGTTAATG | −110 to −16 |
| IL-2 C | TCACCTAAATCCATTCAGTCAGTGTA | GTGGCAGAAAGCATTACCTTTG | −201 to −111 |
| IL-2 D | CTTTTGTGTCTCCACCCCAAA | CACACTTAGGTGGCAGTTTTAATTCAT | −309 to −225 |
| IL-2 D1 | CCATCTTGAAACAGGAAACCA | TCCTGAACCTGTAAGCAGCC | −459 to −342 |
| IL-2 E | TGTCCTTAGCTTACTATTTCTCTGGCT | GGTTTTACCTTCTTATCTACACAATGAGC | −652 to −460 |
| IL-2 F | CATGCAGAGTTTTTTGTTGTTTTCTAG | GCCTAAAGTCTCTCACAAAGAACAGA | −1982 to −1890 |
| IL-2 cDNA | CCTGAGCAGGATGGAGAATTACA | TCCAGAACATGCCGCAGAG | |
| GAPDH | CATGGAGAAGGCTGGGGCTC | AACGGATACATTGGGGGTAG | |
| GM-CSF I | AAGGTCCTGAGGAGGATGTG | GAGGTTCAGGGCTTCTTTGA | +17 to +157 |
| GM-CSF A | GCCTGACAACCTGGGGGAAG | TGATTAATGGTGACCACAGAACTC | −162 to −47 |
| GM-CSF B | AAAAGGAGAGGCTAGCCAGA | TAAGCCCTTCCAAGAACTGG | −281 to −181 |
| GM-CSF F | TGGAATGAGCCACCAGAGTA | GGCTCTTGCTTCCATAGCAC | −429 to −355 |
| GM-CSF H | TCATTCTCACTGCTCCCAAG | ATAAGGTCCAGCCCAATGAC | −630 to −507 |
Primary T-cell preparation.
All mice were maintained in a pathogen-free environment in barrier facilities. Spleens were isolated from C57BL6 mice (5 to 6 weeks old). The CD4+ cells were purified using the automated magnetic cell sorting (MACS) or autoMACS columns (Miltenyi Biotec) after incubation with MACS CD4+ (LT34) beads according to the manufacturer's guidelines. The cells were subsequently stained and analyzed by flow cytometry with T-cell populations shown to be more than 90% pure using antibodies against CD4+ T cells and less than 5% were dead using propidium iodide or 7-amino-actinomycin D (BD Biosciences). Cells were maintained in MLC medium (0.999% DMEM, 0.4% d-glucose, 0.006 g of folic acid per liter, 0.0036 g of l-asparagine per liter, 0.116 g of l-arginine per liter, 0.2% NaHCO3) supplemented with 10% fetal calf serum, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, gentamicin, and penicillin.
ChIP assay.
Chromatin immunoprecipitation (ChIP) analysis was performed following the instructions recommended by the supplier of acetyl-H3 and acetyl-H4 antibodies (Upstate Biotechnology) with some modifications. In brief, 2.0 × 106 cells were used in each ChIP assay. Cells were harvested, and proteins were cross-linked to DNA by adding formaldehyde to a final concentration of 1.0% for 10 min at room temperature followed by the addition of 0.25 M glycine. Cells were washed twice with phosphate-buffered saline and resuspended in sodium dodecyl sulfate lysis buffer (Upstate Biotechnology) with the addition of complete EDTA-free protease inhibitor cocktail tablets (Roche Molecular Biochemicals). Formaldehyde-fixed cells were incubated on ice for 10 min and sonicated to shear chromatin by using an Utrasonic processor (Cole/Parmer) under optimized conditions (65% of maximum output, eight 20-s pulses) that gave a range of DNA fragments from 100 to 1,000 bp, as determined by 1.5% agarose gel electrophoresis. Before the addition of antibody, samples were precleared with 60 μl of salmon sperm DNA-protein A-agarose for 30 min at 4°C with rotation. The soluble chromatin fraction was incubated with either 4 μg of anti-acetyl-H3 (raised against residues 1 to 20), anti-acetyl-H4 (raised against residues 2 to 19), anti-acetyl-H3K9, anti-acetyl-H3K18, anti-phosphorylated-H3, anti-acetyl-H3K14-phospho-H3S10 (Upstate Biotechnology), anti-trimethyl-H3K9 (Abcam), or 1.5 μg of anti-histone H3 and anti-histone H4 (Abcam) antibodies or without antibodies as a control, overnight with rotation at 4°C. Immunocomplexes were collected with salmon sperm DNA-protein A-agarose, washed with low-salt buffer, high-salt buffer, LiCl wash buffer (2×), and Tris-EDTA buffer (2×) (Upstate Biotechnology) and eluted with elution buffer (Upstate Biotechnology). Protein-DNA cross-links were reversed at 65°C overnight. Samples were recovered by phenol-chloroform extraction twice and ethanol precipitated. DNA pellets were washed in 70% ethanol, resuspended in 200 μl of Millipore purified water, and stored at −20°C for real-time PCR amplification.
Real-time PCR analysis.
The IL-2 and GM-CSF primers were designed using the computer software Primer Express (Perkin-Elmer/PE Applied Biosystems) for SYBR green PCR analysis with the ABI PRISM 7700 sequence detector (Perkin-Elmer/PE Applied Biosystems) (Table 1). For all primer sets, identical thermocycler conditions were used: stage 1, 2 min at 50°C; stage 2, 10 min at 95°C; and stage 3, 40 cycles, with 1 cycle consisting of 15 s at 95°C and 1 min at 60°C. Each PCR was performed in duplicate. Controls were included for all PCRs to exclude PCR amplification of contaminating genomic DNA and to ensure that amplification was not due to contamination of other components within the PCR mixture. Since PCR amplification was being monitored by SYBR green, the absence of nonspecific amplification was determined by analyzing the dissociation curve of the PCR amplification products. Data collected were analyzed and plotted using Microsoft Excel. The copy number of a given target sequence precipitated by a specific antibody was determined from a standard curve generated using genomic DNA for each primer set. The amount of precipitated target sequence was calculated by normalization with the total input after subtraction of the no-antibody background.
Real-time PCR analysis of mononucleosomal DNA.
Nuclei were isolated from stimulated and unstimulated EL-4 cells as described previously (30). Briefly, cells were washed once in ice-cold phosphate-buffered saline. Pelleted cells were resuspended in lysis buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40, 0.15 mM spermine, 0.5 mM spermidine). After 5 min of incubation on ice, the lysate was spun at 3,000 rpm (Eppendorf 5415R) for 5 min. The nuclei were resuspended in MNase digestion buffer and incubated with 200 U of MNase for 30 min at 37°C. Mononucleosome fragments (∼150 bp) were purified from an agarose gel. Five nanograms of mononucleosomal DNA was used to perform SYBR green real-time PCR using primer sets and thermocycler conditions described above (Perkin-Elmer/PE Applied Biosystems).
Southern blotting.
Nuclei were prepared from EL-4 cells as described above. The nuclei were incubated with increasing amounts of MNase (10, 50, and 150 U) in 100 μl per 105 cells at 37°C for 30 min. The digestion reaction was stopped by the addition of 0.5 mM EDTA. Genomic DNA was purified using a QIA amp blood kit (QIAGEN) and separated in a 1.5% agarose gel and transferred to a HybondN+ membrane (Amersham Biosciences). Probes (approximately 100 bp) were generated by PCR amplification of genomic DNA with the primer sets described. Probe labeling and hybridization were performed using Gene Images Random Prime Labeling and Gene Images CDP-star detection kits (Amersham Biosciences) according to the manufacturer's instructions.
RESULTS
Histone H3 and H4 acetylation decreases at the IL-2 promoter after T-cell activation.
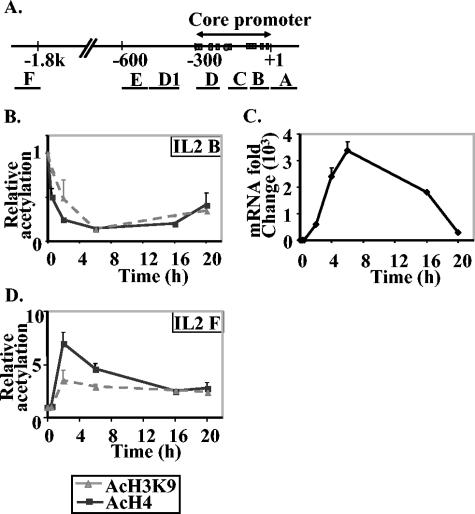
We have previously shown that the proximal promoter region of the IL-2 gene but not a region 2 kb upstream from the transcription start site (−2 kb) becomes accessible to MNase digestion after T-cell activation (29, 30). To determine the relationship between MNase accessibility and histone modification, we examined H4 and H3 acetylation after P/I activation of EL-4 T cells using ChIP and measured the resultant immunoprecipitated DNA by quantitative PCR amplification of 100-bp amplicons at these regions (Fig. 1A and Table 1). Surprisingly, we found that the levels of both H4 acetylation and H3K9 acetylation decreased in a time-dependent manner at the proximal promoter (primer set IL-2 B; Fig. 1B). This decrease was detected as early as 30 min and was greatest for both antibodies at 6 h (Fig. 1B), which correlates with the time at which maximum levels of IL-2 mRNA were detected (Fig. 1C). The level of acetylation was slightly increased again at 20 h when IL-2 mRNA levels had returned to near-baseline levels (Fig. 1B and C). In contrast, the −2-kb region (primer set IL-2 F; Fig. 1A) showed a transient increase in both H4 and H3K9 acetylation, which peaked at 2 h of stimulation and decreased again by 6 h (Fig. 1D).
FIG. 1.
Time course of acetylation of histone H3K9 and histone H4 on the IL-2 gene during transcription activation. (A) Diagram of the IL-2 gene promoter and upstream regions. The regions amplified by IL-2 primer sets designed for real-time PCR (Table 1) are indicated by the lines labeled A to F under the map. (B and D) Time course of acetylation of histone H3K9 (AcH3K9) and histone H4 (AcH4) at the B and F regions (shown in panel A) on the IL-2 gene during gene activation. Enrichment of specific DNA sequences in chromatin immunoprecipitates was detected by real-time PCR. The data are presented as the ratios of immunoprecipitated DNA to total input DNA and normalized to the unstimulated cell sample, which was set at a value of 1. The data presented are the combined means ± standard errors (error bars) of replicate PCR values from three independent experiments. (C) Time course of IL-2 mRNA expression in EL-4 T cells. SYBR green real-time PCR was performed on cDNA from unstimulated and P/I-stimulated EL-4 T cells for the time points indicated. The CT values generated for each time point were quantified using a standard curve generated with cDNA and normalizing to GAPDH.
To determine whether the H3 phosphorylation status was also altered at the IL-2 gene by T-cell activation, we examined the time-dependent changes in H3 S10 phosphorylation (H3-phosphoS10 antibody) and H3 S10 phosphorylation and K14 acetylation (H3-phosphoS10-acetylK14 antibody) in EL-4 cells in response to P/I activation. In agreement with the results above, there was a time-dependent decrease in H3-phosphoS10 and H3-phosphoS10-acetylK14 at the proximal promoter region but not at upstream regions (data not shown). We also examined the H3K9 trimethylation status across the IL-2 gene and found barely detectable levels of methylation in both nonstimulated and stimulated cells (data not shown).
Surprisingly, these results showed that a decrease in both histone acetylation and phosphorylation occurred at the promoter region of the IL-2 gene where we and others have previously described an increase in chromatin accessibility. Conversely, the increase in histone acetylation observed at −2 kb did not correspond to a change in chromatin accessibility.
The decrease in histone acetylation is localized to the proximal promoter region of IL-2 in a T-cell line and in primary T cells.
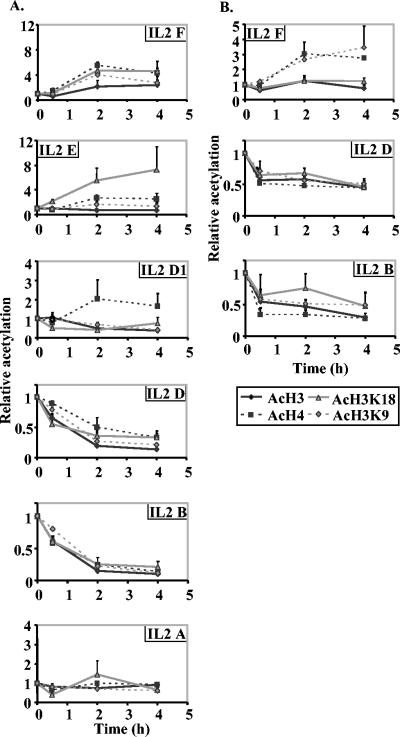
To map the pattern of histone modification at other regions of the IL-2 gene, specific primer sets were used to amplify DNA fragments spread across the gene with antibodies to detect general H3 or H4 acetylation as well as specific acetylation of lysines K9 and K18 on H3 (Fig. 1A and Table 1). In unstimulated EL-4 cells, the level of histone H3 and H4 acetylation appeared to be relatively constant across the regions assayed, although the −600 region had a lower baseline H4 modification than other regions (data not shown). This was not due to unequal amplification by different primer sets, as each of these primer sets has previously been shown to amplify genomic DNA with equal efficiency (29). There was a consistent time-dependent decrease in acetylated histone H3 and H4 with two primer sets (primer sets IL-2 B and IL-2 D) that amplified regions within the proximal promoter (Fig. 2A). A smaller decrease for acetylated H3 but not acetylated H4 was observed using a primer set located at approximately −350 to −450 (primer set IL-2 D1) (Fig. 2A). In fact, H4 acetylation levels increased slightly (ca. twofold) at this region. In contrast, the −2-kb region showed large increases in acetylation with all antibodies (primer set IL-2 F) (Fig. 2A), whereas a region at −600 showed an increase in H3K18 acetylation and a smaller increase for H4 acetylation (primer set IL-2 E) (Fig. 2A). A region immediately downstream from the transcription start site (primer set IL-2 A) showed little or no change in acetylation patterns across the time frame examined (Fig. 2A).
FIG. 2.
Histone acetylation on the IL-2 gene during transcription activation in EL-4 cells and CD4+ primary T cells. (A) ChIP assays were performed with anti-acetylated histone H3 (AcH3), anti-acetylated histone H4 (AcH4), anti-acetylated histone H3K9 (AcH3K9), and anti-acetylated histone H3K18 (ACH3K18) antibodies using unstimulated EL-4 cells or cells stimulated for 30 min, 2 h, and 4 h. Real-time PCR analysis was performed on the immunoprecipitated DNA with the IL-2 primer sets amplifying different regions along the IL-2 gene (Fig. 1A and Table 1). The data are presented as the ratios of immunoprecipitated DNA to total input DNA and normalized to the unstimulated sample, which was set at a value of 1. The combined means ± standard errors (error bars) of replicate CT values from three independent experiments are shown. (B) ChIP experiments were performed on CD4+ primary T cells with the same antibodies as in panel A. Immunoprecipitated samples were amplified with primer sets B, D, and F and the data presented as in panel A.
Next we asked whether the results described above were also seen in primary T cells by performing ChIP assays on CD4+ T cells purified from mouse spleens. A time-dependent decrease was observed across the proximal promoter region of IL-2 for all four antibodies, although the amount of decrease was not as large as for the EL-4 T cells (primer sets IL-2 B and IL-2 D) (Fig. 2B). Once again, the major decreases were limited to the proximal promoter regions with only minor alterations seen downstream of the transcription start site or upstream at −600 (data not shown). The increases in acetylation at the −2-kb region were more variable in the CD4+ T cells than in the EL-4 T cells, with an increase in H3K9 and H4 acetylation, but not in general H3 or H3K18 acetylation (primer set IL-2 F) (Fig. 2B). The less dramatic changes seen in primary CD4+ cells compared with EL-4 cells probably reflects the fact that only 20 to 25% of the primary CD4+ cell population produce detectable IL-2 in response to P/I activation (I. Elsum, T, Juelich, and M. F. Shannon, unpublished observations)
These data show that decreases in histone acetylation occur at the proximal promoter region of the IL-2 gene in a T-cell line and in primary T cells.
Histone proteins are lost from the IL-2 promoter after T-cell activation.
There are at least two possible explanations for the reduction in acetylated and phosphorylated histones at the proximal promoter of IL-2. Either histone modification levels are specifically reduced or histones are removed from the DNA in these regions, leading to an apparent loss of modification.
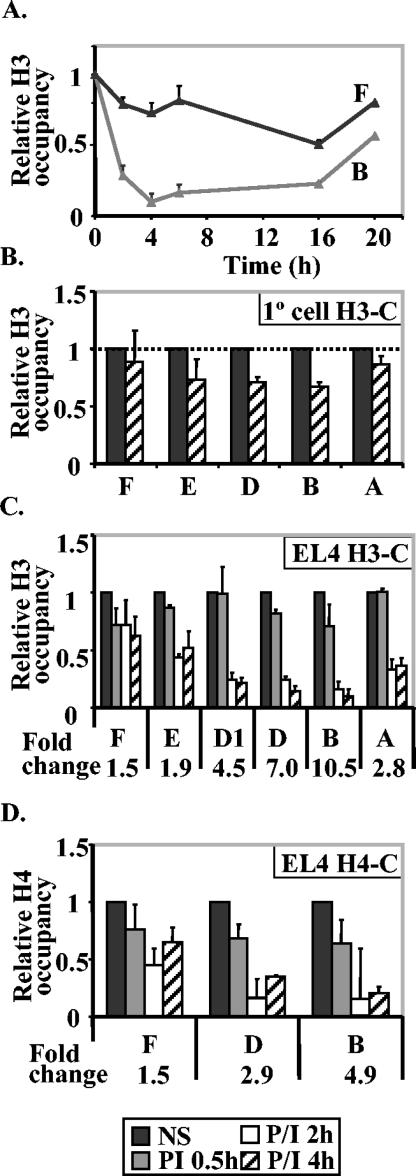
To investigate whether histone loss occurred at the IL-2 promoter, we used an antibody against the C terminus of histone H3 in ChIP assays to determine the total amount of histone H3 that was associated with the gene promoter after activation. We observed a time-dependent decrease in H3 levels at the proximal promoter after activation (Fig. 3A). The time course of this decrease closely parallelled the time course of the decrease in acetylated H3 at the same region (compare Fig. 3A and Fig. 1B). By 20 h, the histone level was returning to the baseline level (Fig. 3A). In contrast, there was a slower and less significant decrease in histone H3 levels at the −2-kb region that did not correlate with the time frame of acetylation changes at that region (compare Fig. 3A and Fig. 1D). We also observed a decrease in histone H3 in primary CD4+ cells at the promoter region but not at the −2-kb region, although the amount of the decrease was once again not as great as for the EL-4 T-cell line, presumably because only 20 to 25% of the CD4+ cells actually activate the IL-2 gene (Fig. 3B).
FIG. 3.
Histones are lost from the IL-2 promoter after T-cell activation. (A) ChIP assay was performed with an antibody against the C terminus of histone H3 on EL-4 cells stimulated with P/I for the times shown. Real-time PCR was performed on the proximal promoter (IL-2 primer set B) and on the −2-kb upstream region (IL-2 primer set F). The data are plotted as the ratio of immunoprecipitated DNA to total input DNA and normalized to the unstimulated cell value, which was set at 1. (B) ChIP assay was performed on primary (1°) CD4+ T cells either left unstimulated or stimulated with P/I for 4 h and DNA amplified with the IL-2 primer sets shown. Data are presented as in panel A. (C and D) ChIP assays were performed with an antibody against the C terminus of histone H3 (H3-C) (C) and an antibody against the C terminus of histone H4 (H4-C) (D) on EL-4 cells unstimulated or stimulated for 30 min, 2 h, and 4 h. Real-time PCR analysis was performed with the immunoprecipitated DNA with IL-2 primers amplifying the regions indicated along the IL-2 gene (Fig. 1A). The CT value was converted to the relative difference (fold) in copy number and then normalized against the value for nonstimulated (NS) cells, which was set at 1 for each primer set. The change (fold) from the nonstimulated cell sample to the cell sample stimulated for 4 h is shown below the graph for each gene region.
Next we determined the extent of the spread of this decrease in H3 levels across the gene. Prior to activation, histone H3 levels were very constant across the genes (Fig. 3C). There was a significant decrease in histone H3 levels at the proximal promoter region (10.5- and 7-fold decreases with primer sets IL-2 B and IL-2 D, respectively) with the decrease spreading as far as −500 (4.5-fold decrease with primer set IL-2 D1) and downstream (2.8-fold decrease with primer set IL-2 A) (Fig. 3C). Relatively small changes occurred at regions further upstream at bp −600 and kb −2 (decreases of less than twofold with primer sets IL-2 E and IL-2 F) (Fig. 3C).
To determine whether other histones were also lost from the promoter, we used an antibody to the C terminus of H4 in ChIP assays. The level of H4 also decreased at the proximal promoter of IL-2 but not at the −2-kb region (Fig. 3D).
These data show that histones are lost from the promoter region of the IL-2 gene after T-cell activation and imply that the apparent decrease in histone modification at the IL-2 promoter may be explained by a loss of histone proteins.
Histone loss correlates with regions of increased MNase accessibility and lower nucleosome density.
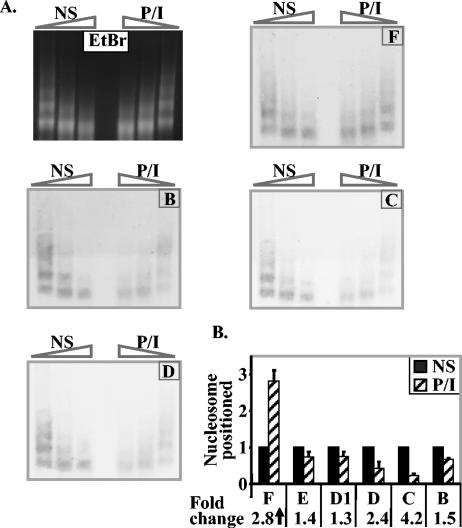
In order to correlate chromatin accessibility (as measured by MNase accessibility) with histone loss, we performed limited MNase digestion followed by Southern blotting with probes from the proximal promoter region and the −2-kb region of the IL-2 gene in both nonstimulated and P/I-stimulated EL-4 T cells. In agreement with the results of our previous PCR-based analysis (29), we found that the proximal promoter region was represented at lower levels in the MNase-digested DNA after stimulation compared to nonstimulated cells (Fig. 4A, regions B, C, and D). In contrast, the −2-kb region was equally represented in nonstimulated and stimulated samples (Fig. 4A, region F).
FIG. 4.
Increased MNase accessibility and decreased mononucleosome density on the IL-2 promoter after T-cell activation. (A) EL-4 T cells were stimulated with P/I for 4 h, and genomic DNA from nonstimulated (NS) and stimulated cells (P/I) was subjected to limited digestion with increasing amounts of MNase to generate a nucleosome ladder visualized by ethidium bromide (EtBr) staining. Southern blots of this DNA were probed with labeled probes representing each of the IL-2 gene regions shown on the diagram. Bands were visualized with enhanced chemiluminescence. (B) Genomic DNA from nonstimulated or P/I-stimulated EL-4 cells was digested to mononucleosome-sized fragments with MNase, and the mononucleosome fraction was purified and subjected to quantitative PCR analysis with the IL-2 primer sets shown. Data are presented for each IL-2 primer set relative to the amount of product in the unstimulated samples. The data are the means ± standard errors (error bars) of three independent experiments, and the change (fold) from the value for nonstimulated cells is shown below the graph.
To determine whether nucleosomes were lost from the proximal promoter region, genomic DNA from nonstimulated or P/I-stimulated EL-4 cells was digested to generate mainly mononucleosomal DNA, and the mononucleosomal fraction was gel purified and subjected to PCR analysis with the primer sets described above. Once again the proximal promoter region had reduced representation in the mononucleosomal DNA in activated T cells compared to nonstimulated cells (4.2- and 2.4-fold for primer sets IL-2 C and IL-2 D, respectively [Fig. 4B]), whereas there was little change upstream of −450 bp (primer sets IL-2 E and IL-2 D1 [Fig. 4B]) and an increase for the −2-kb region (2.8-fold for primer set IL-2 F)(Fig. 4B).
These results show that the region of greatest histone loss corresponds to the region that shows reduced mononucleosome density and becomes more accessible to MNase digestion after activation.
Histone loss also occurs on the GM-CSF promoter after T-cell activation.
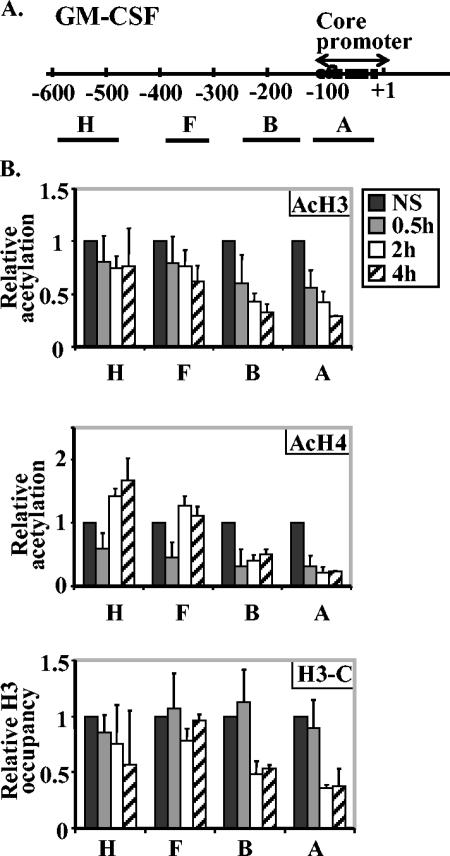
To determine whether the histone loss that we observed on the IL-2 promoter is a specific phenomenon of this gene or occurs on other T-cell-induced genes, we examined histone protein and modification levels on the promoter of the GM-CSF gene, another cytokine gene that responds to T-cell activation. Previously we showed that the proximal promoter region of the GM-CSF gene also becomes more accessible to MNase after T-cell activation (17). We used primer sets for the promoter and upstream regions of the GM-CSF gene (Fig. 5A and Table 1) for quantitative PCR on DNA obtained from total histone H3, acetylated histone H3, or acetylated histone H4 ChIP assays. The levels of acetylated histones H3 and H4 and total H3 decreased significantly across the GM-CSF promoter (Fig. 5B, primer sets GM-CSF A and B, P < 0.005 for the sample stimulated for 4 h compared with the nonstimulated sample for all antibodies, except acetylated H3, which had a P value of <0.02, but the decrease was not significant at regions further from the promoter region after T-cell activation [primer sets F and H]). However, we did find a decrease in total H3 at the −600 region outside the defined promoter (Fig. 5B), but its significance is unclear at this stage.
FIG. 5.
Histone proteins are lost across the GM-CSF promoter after EL-4 T-cell activation. (A) Diagram of the GM-CSF gene promoter showing the transcription start site (+1) and the defined core promoter region. The regions amplified by GM-CSF PCR primer sets are shown by the lines labeled with capital letters under the map. The sequences of the GM-CSF PCR primer sets are shown in Table 1. (B) EL-4 cells were left nonstimulated (NS) or stimulated with P/I for 30 min, 2 h, or 4 h, and ChIP assays were performed with antibodies directed against acetylated H3 (AcH3), acetylated-H4 (AcH4), and the C terminus of H3 (H3-C). The immunoprecipitated DNA was amplified with the primer sets shown. The data are shown relative to total input DNA and normalized to the value for nonstimulated cells for each GM-CSF primer set. The values represented the combined means ± standard errors (error bars) of three independent experiments.
These results extend our findings on the IL-2 promoter to a second inducible promoter and suggest that histone loss may be a general phenomenon of highly inducible gene promoters.
Histone acetylation does not determine histone loss.
For many genes, increased histone acetylation has been associated with increased chromatin accessibility and gene transcription (11). Although we observed a decrease in histone H3 levels as early as 30 min after activation, it was possible that there was a transient increase in histone acetylation prior to histone loss. Therefore, we examined H3K9 and H4 acetylation during the first 30 min of activation on the IL-2 proximal promoter and the −2-kb region. There was no increase in H4 or H3K9 acetylation at early time points after stimulation at either the promoter or the −2-kb region (Fig. 6A). In fact, we observed a transient dip in acetylation 5 to 10 min after P/I activation at both regions, which was especially notable for acetylated H4 (Fig. 6A).
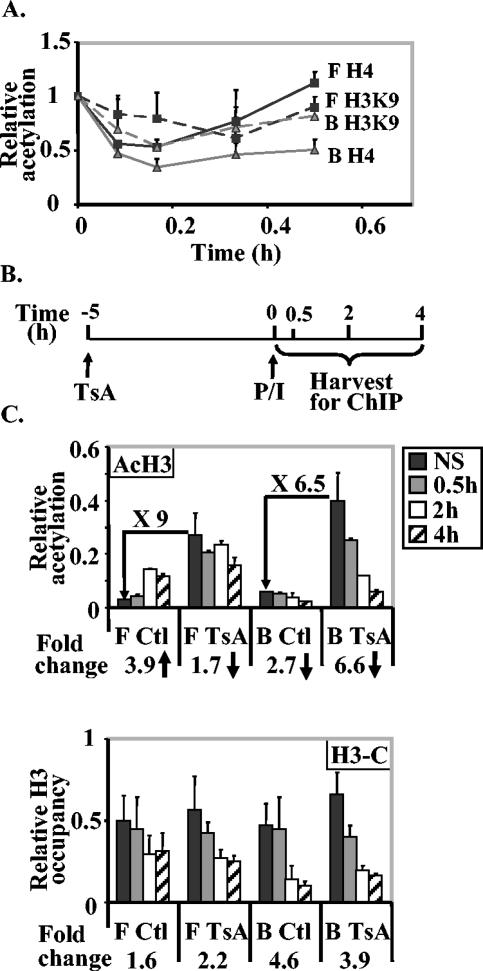
FIG. 6.
Histone acetylation is not directly related to histone loss at the IL-2 gene promoter. (A) Time course of histone acetylation during the first 30 min of P/I activation of EL-4 T cells. Cells were stimulated with P/I for 5, 10, 20, and 30 min, and ChIP assays were performed with antibodies against acetylated H4 (solid lines) and acetylated H3K9 (stippled lines). Immunoprecipitated DNA was amplified with IL-2 primer sets B (grey) and F (black). The data are presented as the ratios of immunoprecipitated DNA to total input DNA and normalized to the value for the unstimulated cell sample, which was set at 1. The data presented are the combined means ± standard errors (error bars) of replicate PCR values from three independent repeats. (B) Schematic showing the time frame of treatment of cells with TsA followed by activation with P/I and the times of harvesting for ChIP assays. (C) Histone hyperacetylation does not promote histone loss at the IL-2 promoter. EL-4 cells were treated with TsA for 5 h and then not stimulated (NS) or stimulated with P/I for 30 min, 2 h, or 4 h. Cells were also activated without prior treatment with TsA (control [Ctl]). ChIP assays were performed with antibodies against acetylated H3 (AcH3) and the C terminus of H3 (H3-C), and the immunoprecipitated DNA was amplified with IL-2 primer sets B and F. Data are plotted as ratios of immunoprecipitated sample to total input sample. The increase (fold) in acetylation levels after TsA treatment and the fold decrease (fold) in total H3 after activation are also shown on or below the graph.
These data imply that histones can be lost from the promoter without the need for prior hyperacetylation, although it is still possible that the time between modification and loss is so short that we cannot detect it without the ability to inhibit histone loss.
Next we asked whether treatment of cells with the histone deacetylase inhibitor TsA could lead to histone loss at the IL-2 promoter or could affect histone loss after T-cell activation (Fig. 6B). EL-4 T cells were pretreated with TsA prior to activation by P/I for various times up to 5 h with no significant impact on IL-2 gene mRNA levels (data not shown). As expected, there was a general increase in histone H3 acetylation (approximately 5- to 10-fold) across the gene after TsA treatment as illustrated here for the promoter and −2-kb regions (Fig. 6C, primer sets IL-2 B and F). However, underlying total H3 levels were not significantly altered across these regions (Fig. 6C), showing that acetylation per se does not alter histone density across the gene. Upon stimulation with P/I after pretreatment with TsA, we observed decreases in both acetylated and total H3 across the promoter region but not at the −2-kb region as previously described (Fig. 6C). The kinetics and degree of decrease were not significantly affected by prior TsA treatment, implying that the loss of histone H3 after activation was not closely linked with its acetylation status. Of note, we also found that activation after TsA treatment led to no further increase in H3 acetylation at the −2-kb region (Fig. 6C).
Therefore, it appears that histone hyperacetylation is neither sufficient nor necessary for the histone loss that accompanies activation of the IL-2 promoter.
Histone loss requires appropriate and continuous cell stimulation.
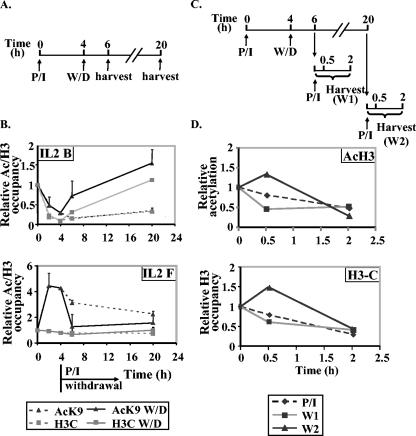
The time course experiments presented above suggest that histone loss is relatively stable and does not revert to a prestimulation state during the first 20 h of activation. To determine whether this stability is dependent on the continual presence of the stimulus, EL-4 cells were treated with P/I for 4 h, stimulus was removed for either 2 h or 16 h, and histone levels were measured on the promoter region and the −2-kb region (Fig. 7A). The level of acetylated H3K9 or total H3 decreased after 4 h of stimulation as described above, but after 2 h of stimulus withdrawal, the amount of histone protein detected had increased, and by 16 h of stimulus withdrawal, the total H3 levels had reverted to nonstimulated levels, while H3K9 levels were somewhat higher than baseline levels (Fig. 7B). At −2 kb, the transient increase in acetylation dropped rapidly to baseline levels once the stimulus was removed. These results show that the presence of the stimulus is necessary for the continued removal of histones from the promoter and that histones reassociate with the promoter relatively rapidly in the absence of stimulus. The maintenance of increased acetylation at −2 kb is also dependent on the presence of the stimulus.
FIG. 7.
Histone removal is reversible and requires the continuous presence of the stimulus. (A) Schematic of the experiment to test the effect of stimulus withdrawal on histone loss from the IL-2 promoter. The times of stimulation (P/I), withdrawal (W/D), and harvest are shown. (B) EL-4 T cells were stimulated with P/I (stippled lines) for the times indicated. In parallel samples, the stimulus was removed after 4 h, and cells were harvested 2 h and 20 h later (solid lines). These samples were all subjected to ChIP assays with antibodies against acetylated H3K9 (AcK9) and the C terminus of H3 (H3C), and the immunoprecipitated DNA was amplified with IL-2 primer sets B and F. Data are presented as ratios of immunoprecipitated sample to total input normalized to unstimulated cell samples, which were given a value of 1. (C) Schematic of the experiment to test the effect of stimulus withdrawal and restimulation on histone loss at the IL-2 promoter. The times of stimulation (P/I), stimulus withdrawal (W/D), restimulation (P/I), and harvest are shown. (D) EL-4 cells were stimulated with P/I for 4 h, and the stimulus was withdrawn for a further 2 or 16 h and then restimulated with P/I for 30 min and 2 h. In parallel, untreated cells were stimulated for 30 min and 2 h with P/I. The amount of acetylated H3 (AcH3) and total H3 were measured by ChIP assays using IL-2 primer set B. Data are presented as ratios of immunoprecipitated sample to total input normalized to unstimulated cell samples, which were given a value of 1.
Next we asked whether histones could be removed from and reassembled onto the promoter in a cyclical fashion. EL-4 cells were initially stimulated for 4 h with P/I and then the stimulus was withdrawn for 2 or 16 h before restimulating for 30 min or 2 h (Fig. 7C). Acetylated H3 and total histone H3 were lost and then reassembled upon primary stimulation and stimulus withdrawal as described above (Fig. 7B). Upon secondary stimulation, histone H3 and acetylated H3 were again lost from the promoter with a similar degree of loss observed by 2 h, irrespective of whether it was the primary or secondary stimulus (Fig. 7D). The loss of histone H3 does not appear to proceed at a faster rate upon restimulation.
Therefore, histone loss is dependent on the continuous presence of the appropriate signal and can operate in a cyclical fashion.
The presence of specific transcription factors is required for histone loss.
It has previously been shown that in the absence of specific transcription factors known to drive IL-2 gene transcription, the IL-2 promoter is no longer accessible to DNase I or MNase in activated T cells (26, 29, 30). Therefore, we speculated that if chromatin accessibility equated with histone loss, then the absence of specific transcription factors should block histone loss. Treatment of EL-4 T cells with CsA is known to inhibit the activation-dependent translocation of NFAT transcription factors to the nucleus (9), so we asked whether CsA pretreatment for 30 min prior to P/I activation affected histone loss. As expected, CsA pretreatment dramatically inhibited IL-2 mRNA accumulation (data not shown) and had no obvious affect on acetylated or total histone measured across the IL-2 gene in unstimulated cells (data not shown). However, CsA pretreatment completely inhibited the loss of both acetylated H3 and total H3 from the IL-2 promoter (Fig. 8A). Surprisingly, CsA pretreatment also blocked the increase in H3 acetylation levels normally observed at the −2-kb region (Fig. 8A), suggesting that there is a connection between NFAT-mediated events at the promoter and upstream histone acetylation.
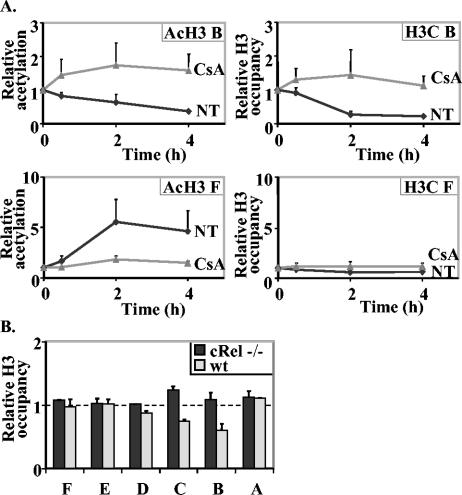
FIG. 8.
Transcription factor requirement for histone loss. (A) CsA inhibits histone loss from the IL-2 promoter. EL-4 cells were pretreated with CsA for 30 min prior to stimulation with P/I or stimulated with P/I alone (NT) for the times indicated. Cells were then subjected to ChIP assays with antibodies against acetylated H3 (AcH3) and C terminus of H3 (H3C), and the immunoprecipitated DNA was amplified with IL-2 primer sets B and F. The data are presented as the ratios of immunoprecipitated DNA to total input DNA and normalized to the unstimulated cell sample, which set at a value of 1. The data are the combined means ± standard errors (error bars) of replicate PCRs in two independent experiments. (B) The transcription factor c-Rel is required for histone loss at the IL-2 promoter. CD4+ T cells were purified from the spleens of wild-type (wt) mice and c-Rel−/− mice stimulated with P/I for 4 h and then subjected to ChIP assays with an antibody against the C terminus of H3. The immunoprecipitated DNA was amplified in PCRs with primer sets spanning the IL-2 gene. Data are presented as described above and show the means ± standard errors (error bars) of four replicate PCRs from a representative experiment.
Because CsA treatment is not totally specific for the NFAT transcription factors and may have unknown effects on chromatin, we also examined histone loss in primary T cells lacking the c-Rel transcription factor. Previously we showed that the loss of c-Rel affects MNase and restriction enzyme accessibility across the IL-2 promoter (29). We found that in contrast to the reduction in total histone H3 in wild-type primary CD4+ T cells at the promoter region (primer sets IL-2 B to D), we did not detect a loss of total histone H3 across the IL-2 gene promoter in c-Rel−/− CD4+ T cells (Fig. 8B).
These results suggest that the presence of appropriate transcription factors, particularly c-Rel, is required for histone loss at the inducible IL-2 promoter.
DISCUSSION
The major finding presented in this paper is that histone proteins are lost from a defined region of both the IL-2 and GM-CSF promoters after T-cell activation. This histone loss is (i) limited to the regions close to the proximal promoters, (ii) reversible, and (iii) dependent on the continual presence of an appropriate stimulus. Mechanistically, the histone loss is not correlated with changes in histone acetylation or phosphorylation but is dependent on the presence of specific transcription factors.
Several models of chromatin remodelling on inducible promoters have been presented in the literature. The vast majority of published reports show an increase in histone acetylation associated with inducible gene transcription (2, 11, 25, 34, 35). For example, an increase in acetylation of promoter histones has been shown for the IFN-β gene (2) and for a number of genes, such as the MCP-1, Rantes, and IL-6 genes, in response to lipopolysaccharide stimulation of dendritic cells (34, 35). These changes are accompanied by binding of transcription factors to these promoter regions, thus suggesting a model of cooccupation of active promoters by transcription complexes and modified nucleosomes. Our data on the IL-2 and GM-CSF genes do not support a cooccupation model. Rather, all of our data support a model of histone and nucleosome loss from the IL-2 and GM-CSF promoters after T-cell activation. However, increases in histone acetylation patterns were observed at upstream regions that may be associated with gene activation.
Although the ChIP assay does not allow a precise definition of the boundaries of histone loss, the core region of histone loss corresponds approximately to the active promoter regions that have been defined in transient- or stable-transfection assays in T cells (reviewed in references 18, 36, 37, and 38). The proximal promoter regions of these genes have long been known to become DNase I hypersensitive after T-cell activation, and recently, we showed that they also become MNase hypersensitive after T-cell activation (17, 30). However, the physical or biochemical events involved have not been defined. While DNase I hypersensitivity was initially assumed to represent regions of nucleosome-free DNA and MNase digests only nucleosome-free DNA at least in vitro, there has been much recent debate about the possibility that MNase or DNase I accessibility could represent DNA assembled into persistently altered nucleosome structures (31). We show here in limited digestion experiments that IL-2 promoter DNA is much more easily digested by MNase in activated cells than in nonstimulated cells and that the IL-2 promoter is found in lower levels in mononucleosomes prepared from activated T cells than in nonstimulated T cells. These data agree with our previous results showing increased MNase, DNase I, and restriction accessibility, measured by real-time PCR, in activated T cells across both the GM-CSF and IL-2 promoters (17, 29, 30). From all our data, MNase accessibility appears highly correlated with the loss of histone proteins and not with their modification. By way of example, the promoter regions of both IL-2 and GM-CSF genes become MNase accessible after T-cell activation (17, 29, 30) and concomitantly lose contact with histone proteins (data shown here). On the other hand, histones at the −2-kb region of the IL-2 gene become more highly acetylated upon activation, but there is no change in MNase accessibility. Thus, we would propose that promoters with increased MNase accessibility are likely to represent regions of histone loss, although we cannot completely rule out the possibility of highly altered nucleosome structures where the histone proteins cannot be cross-linked to DNA.
Two methods of nucleosome movement have been proposed from studies of other inducible genes in mammalian and yeast cells. Detailed mapping of the IFN-β promoter has shown that the promoter region is nucleosome free in the repressed state with a positioned nucleosome located across the transcription start site (23). Upon activation, transcription complexes assemble on the nucleosome-free promoter region and recruit various chromatin remodelling complexes, which then leads to sliding of the positioned nucleosome approximately 36 bp along the DNA, allowing transcription initiation (23). Studies of the PHO5 gene in S. cerevisiae have shown evidence of nucleosome disassembly rather than nucleosome sliding for an inducible promoter (7). This disassembly is dependent upon the histone chaperone Asf-1 (1). Our findings for the mammalian IL-2 and GM-CSF genes have many of the same characteristics as those observed for the PHO5 gene and suggest that a disassembly mechanism may also be operating on these genes. First, the IL-2, GM-CSF, and PHO5 promoters, in contrast to the IFN-β promoter, appear to be assembled into nucleosome arrays prior to activation (4, 17, 30). Second, histone loss is centered on the region defined as the functional promoter in each gene. This region is relatively large and probably represents the removal of several nucleosomes in all cases. Third, the loss of histones is reversible suggesting a dynamic cycle of disassembly and reassembly. Recent biophysical studies on the PHO5 promoter strongly support a disassembly model (7) as does the recent finding that the histone chaperone Asf1p is required (1). While it is possible that several nucleosomes could coordinately slide along the DNA to create the nucleosome-free regions observed on the IL-2 and GM-CSF genes, sliding on this scale has not yet been described and might be expected to result in long-range perturbation of chromatin structure. We observed no changes in nucleosome structure at least in the immediate vicinity of the remodelled promoters (data not shown), but the ultimate proof of a disassembly model versus a sliding model requires further experimentation.
In the presence of the activating signals, histone loss is relatively stable and is not reversed even 20 h after activation, when IL-2 mRNA levels have returned to near resting levels. However, histones were detected on the promoter as soon as 2 h after stimulus withdrawal and once again could be removed by readding the stimulus. These results suggest a dynamic process of histone removal and reassembly that may be linked with the presence of transcription factors. Early work on the IL-2 promoter showed an all-or-nothing phenomenon where the absence of any signal or factor required for IL-2 gene transcription led to a loss of all DNase I accessibility across the promoter (33). Our own data showing that the lack of a single transcription factor (c-Rel) could prevent MNase access across the entire promoter after T-cell activation also argues for a dynamic interplay between the transcription factors required for activation of the IL-2 gene and the chromatin status of the promoter (29). It is possible to propose a model whereby the presence of the appropriate transcription factor complexes are required to lead to or maintain histone removal from an activated promoter. Our data presented here showing that CsA inhibits histone removal and that the NF-κB protein c-Rel is required for histone removal also supports such a model. Whether these transcription factors help to recruit chromatin remodelling complexes or compete with histones for promoter occupation remains to be tested. It has recently been shown in a genome-wide survey of promoter occupancy by nucleosomes in yeast that the transcription factor Rap1 was associated with nucleosome-depleted promoters but that it was unlikely to be the only determinant (5, 13). Another determinant appeared to be the presence of poly(dA-dT) elements which have been associated with nucleosome destabilization (5), and it is interesting that the IL-2 promoter sequence is highly A/T rich (4). It should be noted, however, that A/T-rich stretches have also been associated with the ability of DNA to position nucleosomes and that the IL-2 promoter has a strong ability to position a nucleosome in vitro (4). Both functions of A/T stretches may not be mutually exclusive with positioned nucleosomes on A/T stretches playing a repressive role in transcription and being easily displaced under the correct activation conditions.
Direct signaling to chromatin remodelling complexes or to chromatin, promoting histone loss, cannot be completely ruled out at this stage. We find no evidence for changes in histone acetylation or phosphorylation at the proximal promoter of the IL-2 or GM-CSF gene after T-cell activation, although we did observe a transient increase in acetylation at a region upstream of the IL-2 gene. In addition, inhibition of histone deacetylases by TsA did not trigger nor affect the kinetics of histone dynamics on the IL-2 promoter. The simplest explanation for these data is that acetylation is not required for histone loss. It is possible, however, that changes in histone acetylation are masked by the binding of proteins with bromo-domains, as on the Swi/Snf complexes, or that histones are lost from the chromatin very rapidly after acetylation. For the PHO5 gene, histone hyperacetylation preceded histone loss, but this was observed only by slowing down histone loss with the deletion of chromatin-modifying complexes from the cell (32). In a recent study in yeast, histone loss from the coding region of the GAL1 gene was shown to be dependent on active transcription but independent of histone acetylation (21). In contrast, other genes (GCN5 and ELP3) do not lose histones but do become acetylated as a result of transcription (21). In this study, it was suggested that histones may be lost from a gene with a very high transcription rate and density of polymerase (such as GAL1) but not from genes with lower rates of transcription (21). It has recently been shown that there is a relationship between gene transcription rate and promoter occupancy in a global study of yeast chromatin structure (5, 22). The IL-2 and GM-CSF genes are very highly induced after T-cell activation, and histone loss on the promoter may also be associated with transcription rate and the level of promoter occupancy by the transcriptional machinery. It should be possible to test this hypothesis for mammalian cells by studying histone dynamics and transcription from an array of genes with distinct levels of response to T-cell activation. We have used expression profiling on CD4+ T cells to identify genes with different rates of transcription (S. Rao, K. Bunting, T. Juelich, G. S. Denyer, D. Woltring, S. Gerondakis, and M. F. Shannon, unpublished results), and this set of genes would be useful in testing this hypothesis.
Thus, it is possible that there may be several mechanisms of promoter chromatin remodelling ranging from histone modification through to partial or complete loss of histones depending on the rate of transcription from the gene. Indeed, a recent study of the mouse mammary tumor virus promoter showed that histone H2A/H2B dimers (but not H3/H4) were lost from the active promoter (43). All of these mechanisms may also involve increased movement of nucleosomes along the DNA which can be viewed as sliding and may be an intermediate step in the process of histone loss.
It should be noted that most studies of histone modification in mammalian cells do not take into account the underlying level of histone occupancy and so may generate misleading results. Our studies here clearly show that decreases in histone modification are solely due to depletion of histones and not to any changes in modification.
This study demonstrates for the first time that core histones H3 and H4 lose contact with the DNA on inducible promoters of genes in mammalian cells and presents a mechanism of chromatin remodelling that appears to be conserved across eukaryotic evolution from yeast cells to mammalian cells.
Acknowledgments
We thank Assam El Osta for help with the ChIP technique and Peter Milburn, Biomolecular Resource Facility, Australian National University for advice on real-time PCR.
The work was funded in part by a National Health and Medical Research Council project grant to M.F.S. X. Chen is funded by an Australian National University postgraduate scholarship.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 3.Arney, K. L., and A. G. Fisher. 2004. Epigenetic aspects of differentiation. J. Cell Sci. 117:4355-4363. [DOI] [PubMed] [Google Scholar]
- 4.Attema, J. L., R. Reeves, V. Murray, I. Levichkin, M. D. Temple, D. J. Tremethick, and M. F. Shannon. 2002. The human IL-2 gene promoter can assemble a positioned nucleosome that becomes remodeled upon T cell activation. J. Immunol. 169:2466-2476. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 7.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 8.Clayton, A. L., and L. C. Mahadevan. 2003. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 546:51-58. [DOI] [PubMed] [Google Scholar]
- 9.Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357:695-697. [DOI] [PubMed] [Google Scholar]
- 10.Cockerill, P. N., M. F. Shannon, A. G. Bert, G. R. Ryan, and M. A. Vadas. 1993. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc. Natl. Acad. Sci. USA 90:2466-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 12.Durand, D. B., J. P. Shaw, M. R. Bush, R. E. Replogle, R. Belagaje, and G. R. Crabtree. 1988. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol. Cell. Biol. 8:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ercan, S., M. J. Carrozza, and J. L. Workman. 2004. Global nucleosome distribution and the regulation of transcription in yeast. Genome Biol. 5:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaus, A., and T. Owen-Hughes. 2001. Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev. 11:148-154. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Rodriguez, C., and A. Rao. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgopoulos, K. 2002. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat. Rev. Immunol. 2:162-174. [DOI] [PubMed] [Google Scholar]
- 17.Holloway, A. F., S. Rao, X. Chen, and M. F. Shannon. 2003. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor κB proteins. J. Exp. Med. 197:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain, J., C. Loh, and A. Rao. 1995. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7:333-342. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Kioussis, D., and W. Ellmeier. 2002. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2:909-919. [DOI] [PubMed] [Google Scholar]
- 21.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 23.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261-271. [DOI] [PubMed] [Google Scholar]
- 24.Lusser, A., and J. T. Kadonaga. 2003. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25:1192-1200. [DOI] [PubMed] [Google Scholar]
- 25.Martens, J. H., M. Verlaan, E. Kalkhoven, and A. Zantema. 2003. Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 23:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novak, T. J., P. M. White, and E. V. Rothenberg. 1990. Regulatory anatomy of the murine interleukin-2 gene. Nucleic Acids Res. 18:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20:214-220. [DOI] [PubMed] [Google Scholar]
- 28.Powell, J. D., J. A. Ragheb, S. Kitagawa-Sakakida, and R. H. Schwartz. 1998. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol. Rev. 165:287-300. [DOI] [PubMed] [Google Scholar]
- 29.Rao, S., S. Gerondakis, D. Woltring, and M. F. Shannon. 2003. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J. Immunol. 170:3724-3731. [DOI] [PubMed] [Google Scholar]
- 30.Rao, S., E. Procko, and M. F. Shannon. 2001. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 167:4494-4503. [DOI] [PubMed] [Google Scholar]
- 31.Reinke, H., and W. Horz. 2004. Anatomy of a hypersensitive site. Biochim. Biophys. Acta 1677:24-29. [DOI] [PubMed] [Google Scholar]
- 32.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 33.Rothenberg, E. V., and S. B. Ward. 1996. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc. Natl. Acad. Sci. USA 93:9358-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saccani, S., and G. Natoli. 2002. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 16:2219-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saccani, S., S. Pantano, and G. Natoli. 2001. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 193:1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serfling, E., A. Avots, and M. Neumann. 1995. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim. Biophys. Acta 1263:181-200. [DOI] [PubMed] [Google Scholar]
- 37.Shannon, M. F., L. S. Coles, M. A. Vadas, and P. N. Cockerill. 1997. Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit. Rev. Immunol. 17:301-323. [DOI] [PubMed] [Google Scholar]
- 38.Shannon, M. F., S. R. Himes, and L. S. Coles. 1995. GM-CSF and IL-2 share common control mechanisms in response to costimulatory signals in T cells. J. Leukoc. Biol. 57:767-773. [DOI] [PubMed] [Google Scholar]
- 39.Siebenlist, U., D. B. Durand, P. Bressler, N. J. Holbrook, C. A. Norris, M. Kamoun, J. A. Kant, and G. R. Crabtree. 1986. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol. Cell. Biol. 6:3042-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smale, S. T., and A. G. Fisher. 2002. Chromatin structure and gene regulation in the immune system. Annu. Rev. Immunol. 20:427-462. [DOI] [PubMed] [Google Scholar]
- 41.Tsukiyama, T. 2002. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nat. Rev. Mol. Cell Biol. 3:422-429. [DOI] [PubMed] [Google Scholar]
- 42.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 43.Vicent, G. P., A. S. Nacht, C. L. Smith, C. L. Peterson, S. Dimitrov, and M. Beato. 2004. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol. Cell 16:439-452. [DOI] [PubMed] [Google Scholar]
- 44.Ward, S. B., G. Hernandez-Hoyos, F. Chen, M. Waterman, R. Reeves, and E. V. Rothenberg. 1998. Chromatin remodeling of the interleukin-2 gene: distinct alterations in the proximal versus distal enhancer regions. Nucleic Acids Res. 26:2923-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weissmann, F., and F. Lyko. 2003. Cooperative interactions between epigenetic modifications and their function in the regulation of chromosome architecture. Bioessays 25:792-797. [DOI] [PubMed] [Google Scholar]
- 46.Yui, M. A., G. Hernandez-Hoyos, and E. V. Rothenberg. 2001. A new regulatory region of the IL-2 locus that confers position-independent transgene expression. J. Immunol. 166:1730-1739. [DOI] [PubMed] [Google Scholar]
- 47.Zhong, H., M. J. May, E. Jimi, and S. Ghosh. 2002. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell 9:625-636. [DOI] [PubMed] [Google Scholar]
- 48.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]