Abstract
In Saccharomyces cerevisiae, the maturation of both pre-rRNA and pre-small nucleolar RNAs (pre-snoRNAs) involves common factors, thereby providing a potential mechanism for the coregulation of snoRNA and rRNA synthesis. In this study, we examined the global impact of the double-stranded-RNA-specific RNase Rnt1p, which is required for pre-rRNA processing, on the maturation of all known snoRNAs. In silico searches for Rnt1p cleavage signals, and genome-wide analysis of the Rnt1p-dependent expression profile, identified seven new Rnt1p substrates. Interestingly, two of the newly identified Rnt1p-dependent snoRNAs, snR39 and snR59, are located in the introns of the ribosomal protein genes RPL7A and RPL7B. In vitro and in vivo experiments indicated that snR39 is normally processed from the lariat of RPL7A, suggesting that the expressions of RPL7A and snR39 are linked. In contrast, snR59 is produced by a direct cleavage of the RPL7B pre-mRNA, indicating that a single pre-mRNA transcript cannot be spliced to produce a mature RPL7B mRNA and processed by Rnt1p to produce a mature snR59 simultaneously. The results presented here reveal a new role of yeast RNase III in the processing of intron-encoded snoRNAs that permits independent regulation of the host mRNA and its associated snoRNA.
Bacterial pre-rRNA processing is carried out by a defined set of nucleases (3-5, 43, 52). Key among this set is RNase III, initially isolated by its ability to bind and cleave duplex RNA (47, 48). RNase III generates the immediate precursors to the mature 16S and 23S rRNAs from the primary transcripts by cleaving within two extended RNA duplexes formed by long-range interactions that pair the termini of each rRNA (7, 63). These long-range interactions provide a simple method of coordinating the processing events at both ends of the transcript. In eukaryotes, pre-rRNA processing is more complex and requires many more small nucleolar RNAs (snoRNAs) and protein components with overlapping functions (13, 15, 16, 41, 46). For example, the removal of the 5′ external-transcribed spacer requires 4 snoRNAs (U3, snR30, U14, and snR10) and about 64 snoRNAs are required for rRNA modifications (24, 57). snoRNAs are divided into two major subclasses: the first includes box C/D snoRNAs that function mostly as a guide for the methylation of rRNA (6, 20, 21, 55), while the second includes H/ACA snoRNAs that guide RNA pseudouridine formation (25, 39, 59). Most mammalian snoRNAs are encoded within intron sequences and are processed from either unspliced precursors or lariat species (18, 19, 64). In Saccharomyces cerevisiae, most snoRNAs are transcribed either as independent units or as a part of polycistronic transcript, while only 7 of the 66 known snoRNAs are located in the introns of mRNAs (14, 44, 53). Several polycistronic snoRNAs, and a few monocistronic ones, are processed by Rnt1p, the orthologue of the bacterial RNase III (30), which is also required for the processing of the pre-rRNA's 3′ end (2, 10, 11, 23, 33). Following processing by Rnt1p, the RNAs are trimmed by exonucleases producing the mature ends (22, 56).
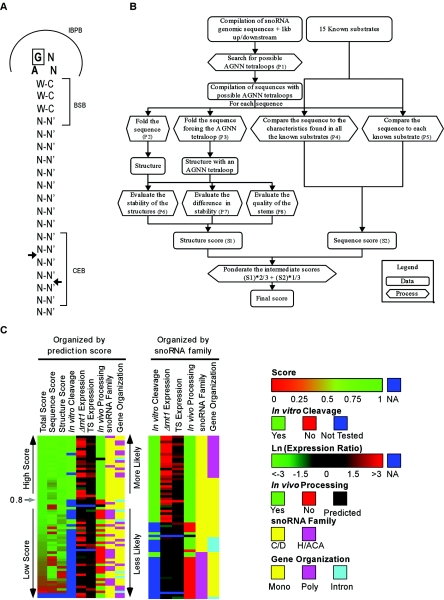
Unlike other RNase IIIs, Rnt1p recognizes substrates with conserved stem-loop structures and has a low affinity for generic RNA duplexes (27). Most Rnt1p substrates exhibit a conserved AGNN tetraloop structure (9, 28, 32, 61). Rnt1p cleaves at a fixed distance from the conserved loop, generating a product with staggered ends (28). Mutations (28), chemical protection assays (27), chemical interference (9), and nuclear magnetic resonance analysis (28) indicate that Rnt1p binding and cleavage are regulated by reactivity epitopes grouped into three boxes (see Fig. 1A). These are the initial binding and positioning box (IBPB), located at the tetraloop; the binding stability box (BSB), located adjacent to the tetraloop; and the cleavage efficiency box (CEB), located near the cleavage site. Alteration of the sequences of both the IBPB and the BSB inhibits cleavage and reduces binding, while alteration of the CEB sequence inhibits cleavage without affecting the binding efficiency. Thus, despite the lack of universally conserved residues, the nucleotide composition of the reactivity epitopes contributes to substrate selectivity. The second nucleotide of the IBPB is believed to be universally conserved, and changing it to any nucleotide other than G reduces binding to known substrates and blocks cleavage (9, 27, 28, 38). The solution structure of the Rnt1p/substrate complex indicates that the enzyme interacts with the minor groove adjacent to the 3′ end of the tetraloop, suggesting that substrate recognition depends on the shape of the groove (60). However, accurate identification of the universal features of Rnt1p substrates requires the identification of a large set of substrates that allows statistical analysis of the cleavage signals.
FIG. 1.
A combined in silico and in vitro approach identifies Rnt1p cleavage signals near known snoRNAs. (A) Schematic representation of a model RNA substrate illustrating the features used for the selection of new Rnt1p substrates. Arrows indicate the sites of cleavage. N represents any nucleotide, and N′ is its counterpart. W-C indicates a position where base pairing is predominant and is required for optimal activity (28). The IBPB indicates nucleotides that position the enzyme for cleavage. The BSB indicates nucleotides that stabilize the binding of Rnt1p and enhance cleavage. The CEB indicates nucleotides that directly contribute to the Rnt1p cleavage efficiency without affecting substrate binding. (B) Methods used for the selection of Rnt1p substrates in silico. The illustrated procedure takes as input two independent compilations: one contains snoRNAsequences, and the other contains known Rnt1p substrates. P1 indicates the search performed using RNAMotif (34). P2 and P3 indicate RNA folding obtained with the Vienna RNA package (51) and the minimum free-energy folding algorithm (65) and using the dynamic programming algorithm (36). P4 and P5 indicate comparisons performed using an algorithm developed during this study. P6 and P7 indicate an evaluation performed with the Vienna RNA package (51). P8 indicates the evaluation performed using an algorithm developed in the course of this study. S1 indicates a score for which the highest value 1 is for structures with a long and stable stem having few bulges and an internal loop. The lowest value of S1 is given to a sequence that, when unconstrained, does not fold into an AGNN tetraloop and has a high stability difference compared to the constrained structure. S2 indicates a score for which the highest value of 1 is given to primary sequences that shares the characteristics common to all known substrates and is highly similar to at least one of the known substrates. The lowest value of S2 (0) is given to a sequence that does not resemble any of the known substrates. (C) Summary of the data obtained from the prediction algorithm, the microarray data, and both in vitro and in vivo validation. Details of the experimental data and the references of previously published results are indicated in Table S1 in the supplemental material. A score was given to each of the potential Rnt1p cleavage sites within 1 kb of all known snoRNAs. The score was assigned as described for panel B. In vitro cleavage was tested by the incubation of total RNA with purified Rnt1p as described in the Material and Methods section. Expression level detected upon the inactivation of the Rnt1p temperature-sensitive mutant (TS Expression) was recorded after a 4-h shift to the nonpermissive temperature. In vivo processing was assessed by Northern blot analysis of RNA extracted from Δrnt1 cells, and a defect in processing was scored by the accumulation of a snoRNA precursor in the absence of Rnt1p. Information about the snoRNA families and gene organization was obtained from the snoRNA database (49). The snoRNAs were organized either according to the prediction score (left) or according to the snoRNA gene family (right). Notice that most C/D-box snoRNAs are processed by Rnt1p, while only a few H/ACA-box snoRNAs reside near Rnt1p cleavage signals.
In this study, we searched for new Rnt1p substrates by examining the expression profiles of all known snoRNAs in the presence and absence of Rnt1p. In parallel, we developed a program that identifies potential Rnt1p cleavage signals near known snoRNA sequences. All newly identified substrates were tested for cleavage in vitro, and their contributions to snoRNA processing were verified in vivo. Our combined in silico and in vitro approach identified all known substrates of Rnt1p and revealed seven new snoRNA substrates. In general, monitoring the expression of snoRNAs was most effective when Rnt1p cleavage was not redundant with other processing events that could lead to the maturation of the snoRNA in question. In contrast, the in silico screen was most effective in identifying snoRNAs that harbored conserved processing signals, regardless of their processing pathway in vivo.
MATERIALS AND METHODS
Strains and plasmids.
Yeast cells were grown and manipulated using standard procedures (1, 31). The effect of Rnt1p depletion was studied using the strains W303-1A and Δrnt1 (12). The Δrnt1Δdbr1 strain (40) was constructed by crossing Δrnt1 cells with Δdbr1 cells kindly provided by J. D. Boeke, Johns Hopkins University. The temperature-sensitive strain was a recreation of the rnt1-ts strains described previously (38). The temperature-sensitive strain prp2ts was a kind gift from Ren-Jang Lin, City of Hope.
Microarray-based analysis of snoRNA expression.
The microarray experiment was conducted and analyzed at the Genome Quebec Innovation Center (Montreal, Quebec, Canada). The RNA was extracted from W303, Δrnt1, and RNT1-TS cells (31, 38) grown at either 26 or 37°C in either yeast complete (YC) medium or YC medium without leucine (54).
In silico screening of Rnt1p substrates.
The sequence homology score was calculated using two methods. One looked for homology to the sequences conserved in all known substrates, and the other used an algorithm that searched for the best sequence homology to any single known substrate. For both methods, a nucleotide probability matrix was generated from the alignment of known substrates by using their tetraloops as anchor points. In the second method, a score was given to each substrate based on its sequence homology to all known substrates and the sum of the probability of its nucleotides in relation to the distance to the tetraloop. Higher significance was given to the nucleotides near the tetraloop. In order to identify the best sequence homology to a known substrate, an intermediate score was weighted for each known substrate. Only the highest intermediate score was kept. The intermediate scores were calculated by comparing the nucleotides of potential substrates to those of known substrates. For each nucleotide comparison, when the 2 nucleotides (nt) were identical, the intermediate score was raised by a distance-weighted factor. When the 2 nt were not identical, the intermediate score was raised only according to the probability matrix multiplied by the weight factor. In addition to thermostability, an evaluation of the secondary structure of the potential substrates was based on the quality of its stem. In this study, this was calculated by giving a positive score to nucleotides downstream of the tetraloop that were paired to upstream nucleotides and vice versa. This score was weighted according to the distance from the nucleotide to the tetraloop, with higher significance being given to the nucleotides close to the tetraloop. For any potential substrate, the sum of its nucleotide scores represented the quality of its stem.
In vitro RNA cleavage.
Cleavage reactions were performed essentially as described previously (28) using either 0.2 pmol of purified Rnt1p (26) or total cell extracts (29). For the in vitro cleavage assay, 3 fmol of internally labeled RNA was incubated in the presence of 0.2 pmol of Rnt1p for 20 min at 30°C in 20 μl of reaction buffer (30 mM Tris [pH 7.5], 5 mM spermidine, 10 mM MgCl2, 0.1 mM dithiothreitol, 0.1 mM EDTA [pH 7.5]). Yeast extracts were prepared using 3 liters of yeast culture (W303 or ΔRNT1 strain) grown to an optical density at 600 nm of 0.8 at 26°C in yeast extract-peptone-dextrose. Cells were harvested by centrifugation, washed, and resuspended in 0.4 times the cell pellet's volumes of AGK buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 200 mM KCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 μg of leupeptin/ml, 1 μg of aprotinin/ml, 1 μg of pepstatin A/ml, 1 μg of antipain/ml). Following cell lysis in liquid nitrogen, the frozen powder was transferred to a centrifuge tube and centrifuged at 18,900 × . for 30 min. The supernatant was then centrifuged at 94,000 × . for 30 min and dialyzed for 3 h against 2 liters of dialysis buffer (20 mM HEPES [pH 7.0], 0.2 mM EDTA, 0.5 mM dithiothreitol, 50 mM KCl, 20% glycerol). Finally, the extract was centrifuged at 18,900 × . for 20 min and the supernatant was stored at −80°C. The model snR55 substrate was generated by T7 RNA polymerase (1). 5′-End-labeled RNA was produced as described previously (27). The snR55, snR56, and snR48 templates were prepared by in vitro transcription using PCR products as templates. Each PCR product was obtained from genomic DNA by using a forward primer carrying a T7 promoter located 200 to 300 nt upstream of the mature 5′ end of snoRNA and a reverse primer located 200 to 300 nt downstream of the mature 3′ end of the snoRNA. The oligonucleotides are listed in Oligonucleotide List S1 in the supplemental material. Cleavage of total RNA extracted from both wild-type and Δrnt1 cells was conducted as described previously (8) using total RNA (50 μg) incubated with purified Rnt1p (10 pmol) in the reaction buffer described above.
Northern blot analyses.
Northern blot analyses were performed with total RNA (10 to 15 μg) run on 4 to 8% denaturing polyacrylamide gels as described previously (1). The RNA was visualized using either randomly labeled probes or 5′-end-labeled oligonucleotides. The oligonucleotides used are listed in Oligonucleotide List S2 in the supplemental material.
Primer extension.
Primer extension reactions were performed as described previously (2). The oligonucleotides used for the primer extensions of snR50, snR52, snR54, snR56, snR57, snR58, snR59 snR60, snR62, snR64, snR67, snR68, snR69, and snR71 are listed in Oligonucleotide List S2 in the supplementary material. Oligonucleotides specific to snR67, snR55, and snR39 are listed in Oligonucleotide List S3 in the supplemental material.
RESULTS
In silico and in vivo search for Rnt1p-dependent snoRNA.
In order to identify snoRNAs that require Rnt1p cleavage for their maturation, we screened the sequences within 1 kb upstream and downstream of all known yeast snoRNAs for Rnt1p cleavage signals (Fig. 1A). As indicated in Fig. 1B, the sequences were scanned for the presence of an AGNN tetraloop followed by three Watson and Crick base pairs. Scores were given for each substrate based on sequence homology, secondary structure stability, and secondary structure similarity to known substrates. Based on the scores of known substrates, we set the score cutoff to 0.8. Above this cutoff, the identified RNA structures were expected to be cleaved by Rnt1p. A total of 64 snoRNAs with potential stem-loops were identified, but only 26 obtained a score above 0.8 and 2 did not associate with any predicted structure (Fig. 1C). The other 38 snoRNAs were found near structures with a score lower than 0.8. As expected, all tested structures with scores higher than 0.8 were cleaved by Rnt1p in vitro, validating the efficiency of the selection scheme (see Table S1 in the supplemental material). Interestingly, most of Rnt1p substrates with scores higher than 0.8 were found near C/D-box snoRNAs and only three were found near H/ACA-box snoRNAs (Fig. 1C; see also Table S1 in the supplemental material). The 38 snoRNA-associated structures with scores lower than 0.8 include two cleaved by Rnt1p in a loop-independent but Nop1-dependent manner and five associated with snoRNAs expressed as part of polycistronic units and processed using a stem associated with an adjacent snoRNA within the polycistronic unit. On the other hand, 22 other snoRNAs associated with structures that were not cleaved by Rnt1p and their processing were not affected by RNT1 deletion in vivo (see Table S1 in the supplemental material and references therein). The snoRNA snR17b (U3) carried a cleavage signal identical to that of snR17a and thus was presumed to be processed by Rnt1p (22) but was not directly tested. Finally, four snoRNAs were processed by Rnt1p by using structures that differ from the canonical AGNN tetraloop motif that was used in the search for Rnt1p cleavage sites (Fig. 1). Only four AGNN stem-loop structures with scores between 0.675 and 0.8 were cleaved by Rnt1p in vitro. Therefore, by using a score cutoff of 0.8, we missed four substrates with the canonical AGNN tetraloop, resulting in a false-negative rate of 11%. In silico search using NGNN as the starting motif did not identify new snoRNA-associated motifs that are affected by the deletion of RNT1 in vivo (data not shown).
In order to assess the efficiency of the AGNN tetraloop as indicator for Rnt1p-dependent snoRNAs, we examined the expression profile of snoRNAs in the presence and absence of Rnt1p. The microarray-based expression profile of wild-type cells was compared to that of cells carrying a complete deletion of Rnt1p. In parallel, the snoRNA expression profile of cells carrying a temperature-sensitive allele of Rnt1p grown at the permissive temperature was compared to that of cells grown at the restrictive temperature. It is important to note that the microarray analysis will not differentiate between precursor and mature snoRNA, and therefore, the increase in the expression level of any snoRNA could reflect the accumulation of a pre-snoRNA, a mature snoRNA, or both. The expression level of most snoRNAs located near stem-loops that were cleaved by Rnt1p in vitro was increased more than 1.5-fold upon deletion of Rnt1p (Fig. 1C). The expression level of only five snoRNAs near the cleavable Rnt1p processing signal did not increase in the absence of Rnt1p, probably due to rapid degradation of the unprocessed RNA transcript. All but nine of the snoRNAs that were cleaved in vitro were affected 4 h after a shift to the restrictive temperature. Most of the in vitro substrates that were not overexpressed in Δrnt1 cells were associated with monocistronic or intron-encoded snoRNAs. The most sensitive substrates to Rnt1p deletion or inactivation were those expressed as polycistronic units. All independently transcribed snoRNAs and intron-encoded snoRNAs except snR42 with scores inferior to 0.8 did not exhibit Rnt1p-dependent expression. The result of the in silico and in vivo screens identified 7 new substrates and indicated that all but 22 snoRNAs, mostly H/ACA snoRNAs, are processed by Rnt1p.
Identification of Rnt1p substrates that form through long-range base pairing.
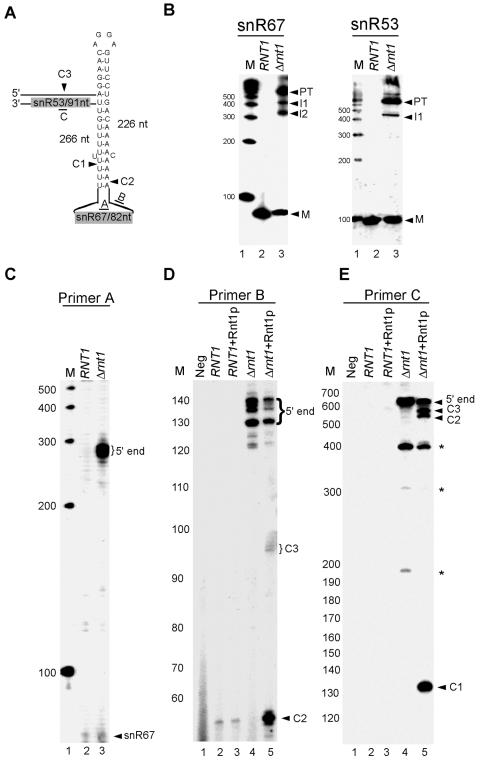
The ideal Rnt1p substrate (Fig. 1A) is a perfect uninterrupted A-U-rich stem capped with an AGNN tetraloop, a feature which is very easy to identify by searching for a stable structural motif. However, most Rnt1p substrates are interrupted stems that in many cases are not structurally stable when taken out of their native RNA context. Furthermore, it has been previously shown that some Rnt1p substrates can form through long-range interactions that are very difficult to identify using conventional motif-based searches or folding programs like mfold (65-67). In contrast, the in silico screen we have developed is capable of identifying AGNN tetraloops with 3-bp stems regardless of either the context or the global folding of the targeted RNA. Consequently, we were able to identify a hidden 3-bp stem capped with an AGGA tetraloop that could not form a stable local stem within the polycistronic unit of snR67/snR53 (Fig. 2A). Northern blot analysis of RNA extracted from either wild-type or Δrnt1 cells hybridized to a probe corresponding to the mature sequence of snR67 revealed an accumulation of a large RNA precursor in Δrnt1 cells and a decrease in the level of the mature snR67 (Fig. 2B). Hybridization to a probe corresponding to snR53 also showed an accumulation of a precursor corresponding to the size of the unprocessed polycistronic transcript. However, the level of the mature snR53 was less affected than that of snR67, suggesting that the localization of snR53 near the 3′ end of the primary transcript makes it less sensitive to Rnt1p deletion. Extension of a primer hybridized to the mature snR67 sequence confirmed the accumulation of an RNA species that extends to the predicted 5′ end of the polycistronic subunit in the absence of Rnt1p (Fig. 2C). The capacity of Rnt1p to directly cleave the primary transcript of snR67/snR53 was tested in vitro using purified Rnt1p and total RNA extracted from Δrnt1 cells. Northern blot analysis of total RNA cleaved by Rnt1p in vitro confirmed that the primary transcript of snR67/snR53 is a direct substrate of Rnt1p (data not shown). Primer extension of total RNA incubated with purified Rnt1p revealed three cleavages, two near the 5′ end of snR67 and one between snR53 and snR67 (Fig. 2D and E). This indicates the presence of two redundant cleavages at the 5′ end of snR67. We concluded that the snR67/snR53 polycistronic transcript is a direct substrate of Rnt1p in vitro and that Rnt1p is required for the efficient processing of both snR53 and snR67 in vivo.
FIG. 2.
Rnt1p is required for the maturation of the polycistronic snR67/snR53 unit. (A) Schematic representation of the predicted stem-loop structure that forms through long-range interactions between the sequences surrounding snR67 and snR53. The arrowheads indicate the positions of the cleavage sites identified in vitro. (B) Northern blot analysis of the snR67 and snR53 expression patterns both in the presence and in the absence of Rnt1p. RNA was extracted from either wild-type or Δrnt1 cells, separated on a 6% acrylamide gel, and hybridized to radioactive probes corresponding to the mature sequence of either snR67 or snR53. The positions of the mature snoRNAs (M), the processing intermediates (I1 and I2), and the primary transcripts (PT) are indicated on the right. (C) Primer extension mapping of the mature and extended termini of snR67. RNA extracted from both wild-type and Δrnt1 cells was subjected to primer extension using primer A, which was complementary to the coding sequence of snR67. Positions of the mature RNA (snR67) and the extended forms detected in the absence of Rnt1p (5′ end) are indicated on the right. (D) Mapping Rnt1p cleavages upstream of snR67. RNA extracted from either wild-type or Δrnt1 cells was incubated with purified Rnt1p enzyme. The cleaved RNA was subjected to primer extension using primer B that hybridized to the sequence upstream of snR67. The positions of the cleavage sites (C2 and C3) and of the 5′ ends are indicated on the right. (E) Mapping of the Rnt1p cleavage sites downstream of snR67. RNA was treated as described for panel D, but the primer extension was performed using a primer (primer C) that hybridized downstream of snR67.
A single stem-loop structure directs cleavage in two independent stems.
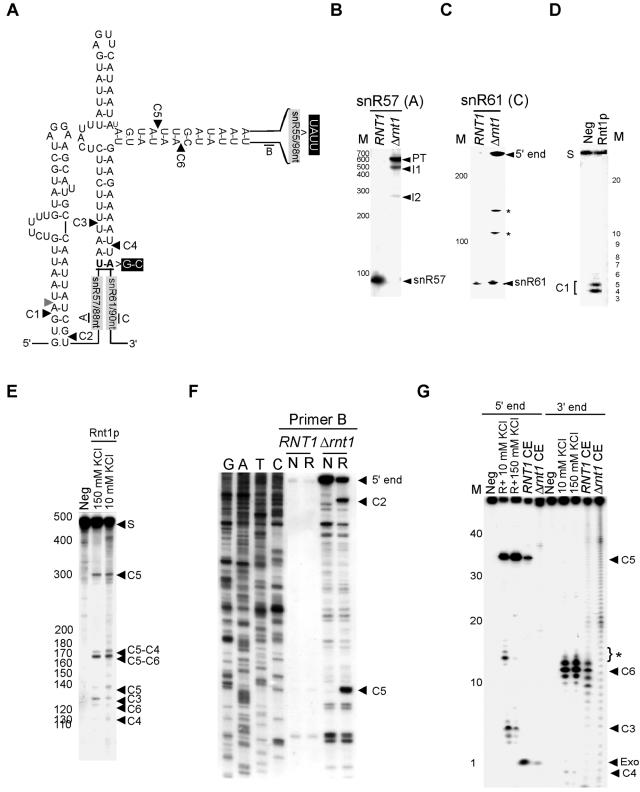
The in silico prediction assay identified a short stem capped with an AGUU tetraloop located in the middle of the snR57/snR55/snR61 polycistronic unit. This single tetraloop was predicted to direct cleavages in two independent stems; one released snR61, and the other released both snR55 and snR57. An additional canonical stem was also identified at the 5′ end of snR57 (Fig. 3A). Very little signal of the mature snR57, snR61, and snR55 was detected upon deletion of RNT1 (Fig. 3B and C and data not shown). In contrast, an RNA species corresponding to the size of the unprocessed precursor was detected, indicating that Rnt1p is required for the processing of this transcript (Fig. 3B). Extension of a primer corresponding to the sequence at the 5′ end of snR57 (primer A) confirmed that in the absence of Rnt1p, the processing of the stem at the 5′ end does not occur (data not shown). Incubation of purified Rnt1p with a T7-transcribed model substrate corresponding to the predicted stem-loop structure upstream of snR57 resulted in a specific cleavage at the predicted distance from the AGGA loop (Fig. 3D). This indicated that the predicted stem is a direct substrate of Rnt1p. In order to test whether or not the predicted structure located between snR55 and snR57/snR61 is cleaved by Rnt1p, and to map the cleavage site, we produced a T7 transcript that corresponded to the entire region located between snR57 and snR61 and incubated it with purified Rnt1p. Incubation of the T7 transcript with Rnt1p revealed six different cleavage products consistent with four cleavage events in two independent stems at the right distance from the tetraloop (Fig. 3E). Two cleavages released snR55, one released the 3′ end of snR57, and one liberated the 5′ end of snR61 (Fig. 3E and data not shown). Mutation of the AGUU tetraloop to GAAA blocked all cleavages in both stems (data not shown). These data clearly demonstrate that the predicted AGUU tetraloop is required for cleavage events that occur in the sequence that separate snR57/snR55 from snR61.
FIG. 3.
Rnt1p uses a single tetraloop to release three different snoRNAs. (A) Illustration of the predicted cleavage sites associated with the snR57/snR55/snR61 cluster is given. Arrowheads indicate the cleavage sites identified in vitro. The positions of the primers used are indicated by letters. The positions of the mutations used in G are indicated in black boxes. (B) Northern blot analysis of snR57 is shown. Northern analysis was performed as described in the legend to Fig. 2B. (C) Northern blot analysis of snR61 processing intermediates is shown. The hybridization was carried out using probe C, which corresponds to the mature sequence of snR61 (indicated in panel A). The asterisk indicates truncated RNA that accumulated in the absence of Rnt1p. (D) Cleavage of a model substrate representing the stem-loop structure found upstream of snR57 is shown. The T7-transcribed RNA representing the 45-nt-long stem-loop structure near the 5′ end of snR57 was 5′ end labeled and incubated with purifiedRnt1p. The cleavage product corresponding to a cleavage in position C1 is indicated on the left. (E) An internally labeled T7-transcribed substrate corresponding to the entire snR57/snR55/snR61 cluster was incubated in the presence of Rnt1p. The cleaved RNA was separated using a 4% polyacrylamide electrophoresis gel and directly visualized by autoradiography. The bands corresponding to the different cleavage sites are indicated on the right. (F) Primer extension mapping of total RNA cleaved by Rnt1p in vitro is shown. The RNA was cleaved to completion and then incubated with primer B that corresponded to the sequence near the 3′ end of snR55 (indicated in panel A). Unrelated DNA sequence was used as a marker. (G) In vitro cleavage of an artificial substrate confirmed Rnt1p's capacity to direct four cleavage events by using a single binding site. A T7 transcript of an RNA harboring the AGUU stem-loop structure found in the snR57/snR55/snR61 cluster was tested for cleavage in vitro. The snR55 sequence of this RNA was replaced by a UAUU tetraloop, and the snR57 and snR61 sequences were replaced by terminating the RNA with a G-C base pair. The RNA was labeled either at the 3′ or the 5′ end and was incubated in the presence of either purified Rnt1p, wild-type cell extracts, or Δrnt1 cell extracts. The cleavage products are indicated on the right. Exo indicates products produced by exonucleases found in cell extracts. The asterisks indicate unspecific cleavages that occurred under low-salt conditions.
The predicted cleavage sites at positions C4 and C6 were confirmed by reverse transcription using a primer corresponding to the mature sequence of snR55 (data not shown). The cleavage sites predicted 5′ to snR55 were determined by extending a primer corresponding to the sequence at the 3′ end of snR55 (primer B) after incubation of total RNA with purified Rnt1p (Fig. 3F). In the context of the native RNA subunit that accumulates in the absence of Rnt1p (Fig. 3B and C), we detected in vitro cleavage at the 5′ end of snR55 (C5) and at the 3′ end of snR57 (C2). These same cleavage sites were also detected using a primer corresponding to the mature sequence of snR61 (data not shown). Failure to detect cleavage at C3 by using a primer at the 3′ end of C5 (Fig. 3A and F) indicated that under this cleavage condition, all substrates that were cleaved at C3 were also cleaved at C5. In contrast, cleavage at C3 was detected using internally labeled and 5′-end-labeled RNA transcripts (Fig. 3E and G and data not shown). Together, these data suggest that a single tetraloop may direct Rnt1p cleavages within two separate stems. In order to directly examine this possibility, we produced a short T7 transcript representing a model substrate that maintains the two stems linked to the AGUU tetraloop binding site. The first stem ends with the 2 nt located below the C4 cleavage site, and the other terminates with a UAUU tetraloop that replaces the naturally occurring snR55 sequence. The UAUU tetraloop cannot direct cleavage by itself, and no tetraloop other than the AGUU was found within this RNA transcript. Thus, any cleavage detected in the adjacent stem would be directed by the AGUU tetraloop. The model T7 substrate was labeled at either the 3′ or the 5′ ends and incubated with purified Rnt1p either at a low, monovalent salt concentration or at physiological salt concentration. As shown in Fig. 3G, all four cleavages were detectable at both salt concentrations. The cleavage sites were mapped based on the sizes of the released fragments (Fig. 3G). The simultaneous detection of cleavages at C3/C5 and C4/C6 within a single-end-labeled RNA species suggested that the four cleavages are not produced from a single binding event. Incubation of the model substrate in either a wild-type or a Δrnt1 cell extract confirmed that the native Rnt1p could also cleave substrates with bifurcated stems. Consequently, kinetic analysis using similar bifurcated RNA substrate showed that each stem is cleaved by a distinct binding event (B. Lamontagne and S. Abou Elela, unpublished results).
The AGNN tetraloop is not conserved near all Rnt1p-dependent snoRNAs.
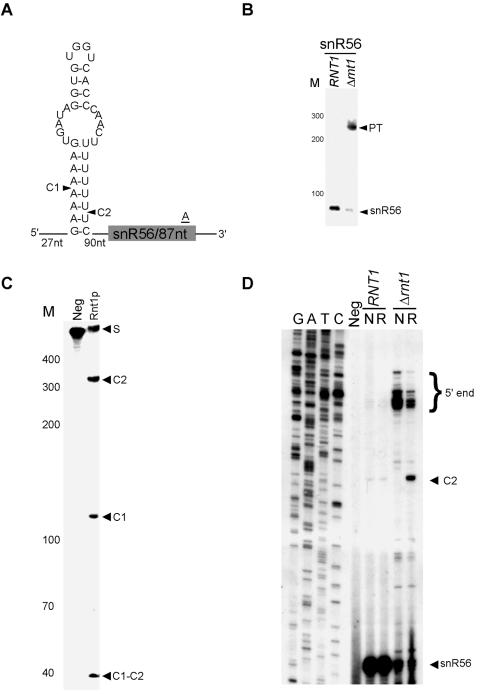
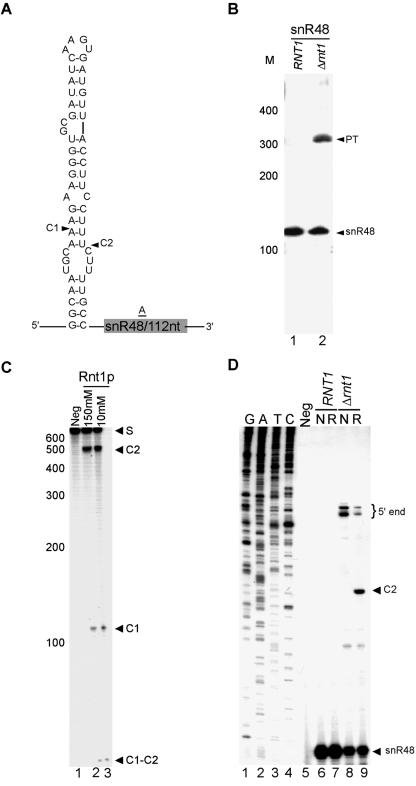
Analysis of the expression profiles of snoRNAs suggested that the monocistronic snR56 and snR48 snoRNAs are Rnt1p dependent. However, we failed to identify AGNN tetraloops near the termini of these snoRNAs. Northern blot analysis confirmed that the deletion of Rnt1p impairs the processing of these two snoRNAs (Fig. 4B and 5B). In vitro cleavage of a T7-transcribed RNA corresponding to the 5′ end of snR56 indicated that Rnt1p could directly cleave the RNA near snR56 despite the absence of an AGNN tetraloop (Fig. 4C). Extension of a primer corresponding to a portion of the snR56 mature sequence confirmed cleavage at a stem capped with UGGU that occurs at the predicted distance from the loop (Fig. 4D). While this study was in progress, it was also reported that Rnt1p could cleave a stem in the intron of RPL18A that is capped with UGGU (12). Thus, Rnt1p does not require A in the first position of the tetraloop for cleavage. Similarly, in vitro cleavage of a model substrate near the 5′ end of snR48 confirmed that Rnt1p could cleave this substrate. However, no canonical stem capped with an NGNN tetraloop was found in the vicinity of the cleavage. Primer extension analysis indicated cleavage near an AAGU-terminal tetraloop (Fig. 5D). Since all known substrates to date contain a G in the second position of the tetraloop and it has previously been shown that changing this G to any other nucleotide blocks cleavage in vitro (9), we presume that a special feature of either this RNA stem or a combination of the loop and stem sequence permits Rnt1p cleavage in this case. We concluded that the AGNN tetraloop is not essential for the Rnt1p-dependent maturation of snoRNA.
FIG. 4.
Rnt1p does not require the presence of an AGNN tetraloop for the maturation of snR56. (A) A schematic representation of the predicted cleavage sites associated with snR56. The cleavage sites identified in vitro are indicated by arrowheads. (B) Northern blot analysis of snR56. RNA extraction and Northern analysis were performed as described in the legend to Fig. 2B. (C) In vitro cleavage of a model substrate representing the cleavage signals found near snR56. Internally labeled T7-transcribed RNA possessing the stem-loop structure indicated for panel A was incubated with purified Rnt1p. The cleavage products were separated by polyacrylamide gel electrophoresis and visualized by autoradiography. (D) Mapping of the Rnt1p cleavage site by using primer extension. The primer extension was performed as described in the legend to Fig. 2C. The mature RNA, extended ends, and cleavage site are indicated on the right.
FIG. 5.
Rnt1p is required for the processing of snR48 in the absence of any detectable NGNN tetraloop. (A) A schematic representation of the predicted stem near the 5′ end of snR48. The cleavage sites identified in vitro are indicated by arrowheads. (B) Northern blot analysis of snR48. RNA extraction and Northern analysis were performed as described in the legend to Fig. 2B. (C) In vitro cleavage of a model substrate representing the cleavage signals found near snR48. Internally labeled T7-transcribed RNA exhibiting the stem-loop structure indicated for panel A was incubated with purified Rnt1p, and the cleavage products were separated by polyacrylamide gel electrophoresis and visualized by autoradiography. (D) Mapping of the Rnt1p cleavage site by using primer extension. The primer extension was conducted as described in the legend to Fig. 2C. The mature RNA, extended ends, and cleavage site are indicated on the right.
Direct processing of intron-embedded snoRNAs by Rnt1p.
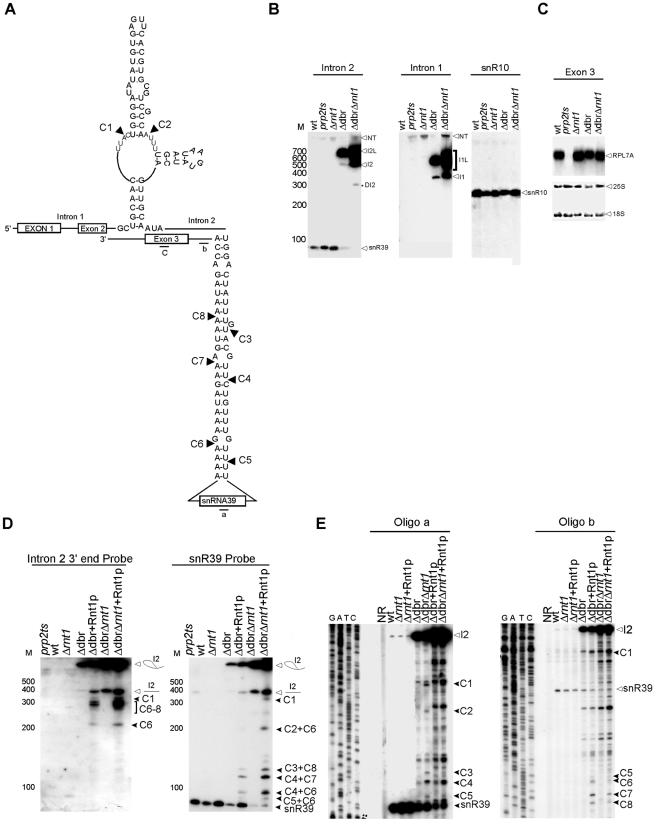
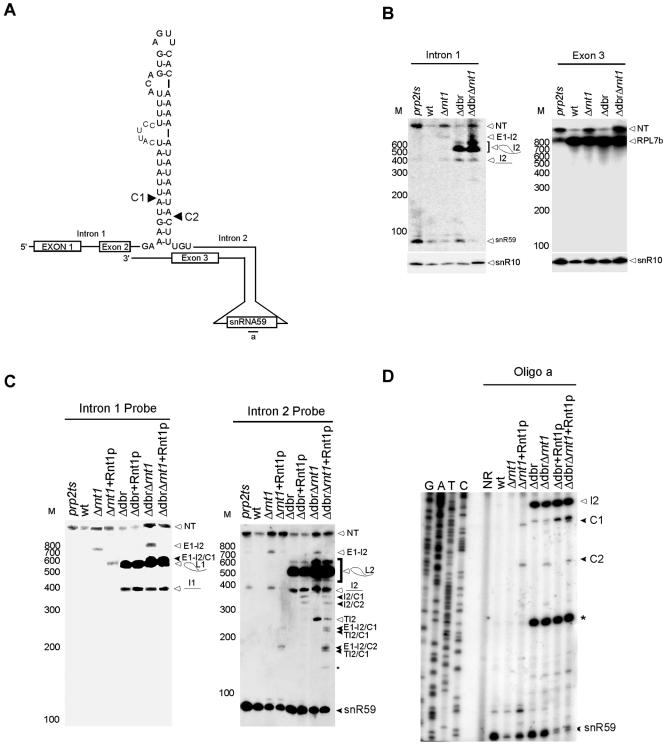
Two of the predicted Rnt1p cleavage signals were located in the introns of the pre-mRNA of the ribosomal protein (r-protein) RPL7A and that of its nearly identical isoform RPL7B near snR39 and snR59, respectively. Consistently, the microarray expression profile indicated that the expression of both snoRNAs was induced upon the deletion of RNT1 (see Table S1 in the supplemental material). In order to examine the impact of Rnt1p on the processing of snR39 and snR59, we monitored the RNA profiles of these two RNAs in both the absence and the presence of Rnt1p. As shown in Fig. 6B, the deletion of Rnt1p did not inhibit the accumulation of mature snR39 or of the mature RPL7A mRNA (Fig. 6C) but caused mild accumulation of the unspliced pre-mRNA precursor. The Northern blot analysis suggests that the increase in the expression of snR39 observed by microarray analysis (see Table S1 in the supplemental material) is due to either an accumulation of the unspliced mRNA or a partially degraded snoRNA and not to the accumulation of snR39 or its immediate precursor. In contrast, deletion of Rnt1p causes the accumulation of a precursor of snR59, a significant accumulation of the pre-mRNA, and a reduction in the level of mature snR59 (Fig. 7B). The accumulation of RPL7B pre-mRNA did not result in a decrease in the level of the mature mRNA, suggesting that splicing is not impaired, as is observed upon inactivation of the essential splicing factor Prp2p (Fig. 7B).
FIG. 6.
Rnt1p assists in releasing the intron-encoded snR39 from the lariat of RPL7A pre-mRNA. (A) Schematic representation of the structure found near snR39 within the intron of RPL7A. The cleavage sites identified in vitro are indicated by arrowheads. (B) Northern blot analysis of snR39 and the associated mRNA sequence. RNA extraction and Northern analysis were performed as described in the legend to Fig. 2B. Probes corresponding to the snR39 mature sequence (intron 2), intron 1, or exon 3 were used. A probe corresponding to snR10 that was notaffected by Rnt1p deletion is shown as a control for both loading and RNA quality. NT indicates the nascent transcript. wt indicates the wild type. (C) Northern blot analysis using a probe specific to exon 3 of RPL7A. The RNA used was extracted and manipulated as described for panel B except that it was fractionated on 1% agarose gel. (D) In vitro cleavage of total RNA. RNA extracted from either wild-type cells or Δrnt1 cells was incubated with purified Rnt1p. Northern blot analysis using probes specific to the sequence near either the 3′ end of snR39 or the mature sequence was used to display the cleavage products. The positions of the different cleavages are indicated on the right. L designates the position of the lariat sequence. (E) Primer extension mapping of Rnt1p cleavage in vitro. The experiment was conducted as described in the legend to Fig. 2C.
FIG. 7.
Rnt1p is required for the processing of the intron-encoded snR59 from the pre-mRNA of RPL7B. (A) Schematic representation of the structure found near snR59 within the intron of RPL7B. The cleavage sites identified in vitro are indicated by arrowheads. (B) Northern blot analysis of snR59 and the associated mRNA sequence. Probes corresponding to either intron 1 or exon 3 were used. A probe corresponding to snR10 that was not affected by Rnt1p deletion is shown as a control for both loading and RNA quality. E1 and E2 indicate exons 1 and 2, respectively, I1 indicates intron 1, and I2 indicates intron 2. L indicates the splice lariat. NT indicates the nascent transcript. C indicates the site of cleavage. TI2 indicates truncated fragments of intron 2. (C) In vitro cleavage of total RNA. RNA was extracted from either wild-type or Δrnt1 cells incubated with purified Rnt1p. Northern blot analysis using probes specific to the sequences near either the 3′ end of snR59 or the mature sequence were used to display the cleavage products. The positions of the different cleavages are indicated on the right. L designates the position of the lariat sequence. (D) Primer extension mapping of Rnt1p cleavage in vitro. The experiment was conducted as described in the legend to Fig. 2C.
In order to examine the possibility of a lariat-dependent processing pathway of snR39 and snR59, we studied the accumulation of these two snoRNAs in cells lacking the debranching enzyme Dbr1p and in cells containing deletions in the DBR1 and RNT1 genes. In the case of snR39, the deletion of DBR1 resulted in the accumulation of a lariat containing snR39 and in a severe reduction in the mature form of snR39. Deletion of both DBR1 and RNT1 completely abolished the production of mature snR39 (Fig. 6B). These data indicate that Rnt1p plays a minor role in releasing snR39 from the lariat, while the major processing pathway of snR39 is through the trimming of the debranched lariat. In vitro cleavage assays and primer extension mapping of the cleavage site revealed that Rnt1p could cleave the lariat containing the snR39 but not the primary mRNA transcript. Eight different cleavage sites were mapped near both the 5′ and 3′ ends of snR39 (Fig. 6D and E). Two of these were located at the predicted distance from the identified AGUU tetraloop, while the six others were not near any recognizable tetraloop motif. It is possible that an alternative fold brings these cleavage sites close to the identified terminal tetraloop. In contrast, in vitro cleavage indicated that Rnt1p targets RPL7B pre-mRNA and not the produced lariat for cleavage (Fig. 7C and D). The cleavage of Rnt1p occurred at the predicted distance from the AGUU tetraloop (Fig. 7A and D). These data indicate that Rnt1p is required for the maturation of snR59. We concluded that, while the processing of snR39 is dependent on the splicing of RPL7A pre-mRNA, the splicing of RPL7B pre-mRNA is inhibited when Rnt1p-dependent snR59 RNA processing occurs.
DISCUSSION
In this study, we presented a combined in silico, in vitro, and in vivo approach for the detection of Rnt1p substrates and demonstrated its utility for the identification of key processing events. Most Rnt1p processing signals were found near box C/D snoRNAs, while only three were found near box H/ACA snoRNAs, indicating either a distinct evolutionary origin or a distinct regulatory pathway for each class of snoRNA. The location and organization of Rnt1p cleavage signals were found to vary from one snoRNA transcript to another. Rnt1p-cleaved substrates formed through base pairing between distantly located RNA sequences, thereby ensuring the maturation of both ends of the targeted snoRNA (Fig. 2 and 3), as previously suggested for the bacterial RNase III (63). In other cases, a single NGNN tetraloop directed cleavages at two distinct cleavage sites, thereby relating the processing of two adjacent snoRNAs (Fig. 3). The cleavage signals of all monocistronic snoRNAs that are processed by Rnt1p were found near the 5′ end of the precursor except the two isoforms of U3 snoRNA that contain introns, which were instead matured through splicing (22). Finally, depending on the nature and context of the Rnt1p cleavage signal, the processing of intron-encoded snoRNAs could be either linked to or separated from the splicing of the host pre-mRNA, allowing a flexible control of the snoRNA-associated redundant r-protein isoforms (Fig. 6 and 7). The data presented here indicate the capacity of Rnt1p processing signals to provide a flexible tool to relate rRNA, snoRNA, and r-protein production.
In yeast, most intron-containing pre-mRNAs encode r-proteins and are redundant (45). It has been suggested that this organization is important for fine-tuning the expression of r-proteins and linking it to rRNA production (58). Indeed, in several cases, the expression of one r-protein isoform regulates the splicing or the mRNA transport of another. However, it is not clear how the production of two r-protein isoforms might be regulated if they harbor a functionally distinct snoRNA, as is the case for RPL7A and RPL7B. The two proteins are nearly identical and have similarity to the Escherichia coli L30 and rat L7 r-proteins (35, 62). Deletion of RPL7A that harbors snR39 within its introns moderately impairs growth and affects budding (37); however, deletion of RPL7B that harbors snR59 in its intron has no effect on growth (37). Deletion of both genes is lethal, reflecting their housekeeping function as part of the ribosome. Like other r-proteins, the expression of these two isoforms needs to be regulated in order to achieve an equimolar production of both the protein and the rRNA it binds. For example, the expression of these two proteins is shut down along with that of all other r-proteins when no rRNA is produced (58). However, controlling the transcriptional level will also affect the snoRNAs encoded within the introns of these proteins, which are required for the production of normal, mature rRNA. Although the snoRNAs embedded in the introns of both RPL7 isoforms are not essential like most snoRNAs, they are conserved among fungi (49, 50) and their expression is expected to be controlled like most components of the ribosome biogenesis machinery (42). In this study, we show that in the case of RPL7B, cleavage by Rnt1p could release snR59 while preventing the production of RPL7B. In contrast, snR39 production appears to be linked to RPL7A since Rnt1p could cleave only the splicing by-product of RPL7A and not the mature RNA. Therefore, the cells could control the overall level by slowing the splicing of RPL7B, which would lead to an increase in cleavage by Rnt1p, producing snR59 and reducing the amount of RPL7B. This is consistent with the fact that the deletion of RPL7A has a greater effect on growth than the deletion of RPL7B. Introns of r-protein mRNA were previously searched for Rnt1p cleavage sites, and no cleavage was detected other than that identified within the introns of RPS22B and RPL18 pre-mRNAs (12). We have searched all other mRNAs containing introns in yeast (17) for potential Rnt1p cleavage sites, and we did not find mRNAs with intronic stem-loops above 0.8 that are significantly overexpressed upon Rnt1p deletion other than those previously identified (J. Gagnon and S. Abou Elela, unpublished results). This indicates that Rnt1p cleavage within pre-mRNA introns might be restricted to r-proteins, possibly due to the specific need to regulate ribosome biogenesis. However, we cannot exclude a more general but redundant role of Rnt1p in the regulation of intron-containing mRNAs that cannot be easily detected by the deletion of RNT1.
Supplementary Material
Acknowledgments
We thank J. D. Boeke for the Δdbr1 strain and Ren-Jang Lin for the prp2 temperature-sensitive strain. We also thank Stéphanie Larose for preparing the yeast cell extract used in this study. We are indebted to all members of the Abou Elela lab for stimulating discussions and critical reading of the manuscript.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada. S.A. is a Chercheur-Boursier Junior II of the Fonds de la Recherche en Santé du Québec.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela, S., H. Igel, and M. Ares, Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85:115-124. [DOI] [PubMed] [Google Scholar]
- 3.Apirion, D. 1983. RNA processing in a unicellular microorganism: implications for eukaryotic cells. Prog. Nucleic Acid Res. Mol. Biol. 30:1-40. [DOI] [PubMed] [Google Scholar]
- 4.Apirion, D., and P. Gegenheimer. 1981. Processing of bacterial RNA. FEBS Lett. 125:1-9. [DOI] [PubMed] [Google Scholar]
- 5.Apirion, D., and A. Miczak. 1993. RNA processing in prokaryotic cells. Bioessays 15:113-120. [DOI] [PubMed] [Google Scholar]
- 6.Bachellerie, J. P., and J. Cavaille. 1997. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22:257-261. [DOI] [PubMed] [Google Scholar]
- 7.Bram, R. J., R. A. Young, and J. A. Steitz. 1980. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell 19:393-401. [DOI] [PubMed] [Google Scholar]
- 8.Catala, M., B. Lamontagne, S. Larose, G. Ghazal, and S. Abou Elela. 2004. Cell cycle-dependent nuclear localization of yeast RNase III is required for efficient cell division. Mol. Biol. Cell 15:3015-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chanfreau, G., M. Buckle, and A. Jacquier. 2000. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. USA 97:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanfreau, G., P. Legrain, and A. Jacquier. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975-988. [DOI] [PubMed] [Google Scholar]
- 11.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danin-Kreiselman, M., C. Y. Lee, and G. Chanfreau. 2003. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11:1279-1289. [DOI] [PubMed] [Google Scholar]
- 13.Eichler, D. C., and N. Craig. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49:197-239. [DOI] [PubMed] [Google Scholar]
- 14.Filipowicz, W., P. Pelczar, V. Pogacic, and F. Dragon. 1999. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 46:377-389. [PubMed] [Google Scholar]
- 15.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 16.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 17.Grate, L., and M. Ares, Jr. 2002. Searching yeast intron data at Ares lab Web site. Methods Enzymol. 350:380-392. [DOI] [PubMed] [Google Scholar]
- 18.Hirose, T., M. D. Shu, and J. A. Steitz. 2003. Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol. Cell 12:113-123. [DOI] [PubMed] [Google Scholar]
- 19.Hirose, T., and J. A. Steitz. 2001. Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 98:12914-12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss-Laszlo, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 21.Kiss-Laszlo, Z., Y. Henry, and T. Kiss. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kufel, J., C. Allmang, G. Chanfreau, E. Petfalski, D. L. J. Lafontaine, and D. Tollervey. 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20:5415-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kufel, J., B. Dichtl, and D. Tollervey. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lafontaine, D., and D. Tollervey. 1995. Trans-acting factors in yeast pre-rRNA and pre-snoRNA processing. Biochem. Cell Biol. 73:803-812. [DOI] [PubMed] [Google Scholar]
- 25.Lafontaine, D. L. J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamontagne, B., and S. Abou Elela. 2001. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 342:159-167. [DOI] [PubMed] [Google Scholar]
- 27.Lamontagne, B., and S. Abou Elela. 2004. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 279:2231-2241. [DOI] [PubMed] [Google Scholar]
- 28.Lamontagne, B., G. Ghazal, I. Lebars, S. Yoshizawa, D. Fourmy, and S. Abou Elela. 2003. Sequence dependence of substrate recognition and cleavage by yeast RNase III. J. Mol. Biol. 327:985-1000. [DOI] [PubMed] [Google Scholar]
- 29.Lamontagne, B., R. N. Hannoush, M. J. Damha, and S. Abou Elela. 2004. Molecular requirements for duplex recognition and cleavage by eukaryotic RNase III: discovery of an RNA-dependent DNA cleavage activity of yeast Rnt1p. J. Mol. Biol. 338:401-418. [DOI] [PubMed] [Google Scholar]
- 30.Lamontagne, B., S. Larose, J. Boulanger, and S. Abou Elela. 2001. The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 3:71-78. [PubMed] [Google Scholar]
- 31.Lamontagne, B., A. Tremblay, and S. Abou Elela. 2000. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 20:1104-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebars, I., B. Lamontagne, S. Yoshizawa, S. Aboul-Elela, and D. Fourmy. 2001. Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing. EMBO J. 20:7250-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, C. Y., A. Lee, and G. Chanfreau. 2003. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. RNA 9:1362-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macke, T. J., D. J. Ecker, R. R. Gutell, D. Gautheret, D. A. Case, and R. Sampath. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 29:4724-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchfelder, A., D. A. Clayton, and A. Brennicke. 1998. The gene for ribosomal protein L7a-1 in Schizosaccharomyces pombe contains an intron after the initiation codon. Biochim. Biophys. Acta 1397:146-150. [DOI] [PubMed] [Google Scholar]
- 36.McCaskill, J. S. 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29:1105-1119. [DOI] [PubMed] [Google Scholar]
- 37.Mizuta, K., T. Hashimoto, and E. Otaka. 1995. The evolutionary relationships between homologs of ribosomal YL8 protein and YL8-like proteins. Curr. Genet. 28:19-25. [DOI] [PubMed] [Google Scholar]
- 38.Nagel, R., and M. Ares, Jr. 2000. Substrate recognition by a eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA 6:1142-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 40.Ooi, S. L., D. A. Samarsky, M. J. Fournier, and J. D. Boeke. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 4:1096-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pederson, T. 1998. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 43.Perry, R. P. 1976. Processing of RNA. Annu. Rev. Biochem. 45:605-629. [DOI] [PubMed] [Google Scholar]
- 44.Petfalski, E., T. Dandekar, Y. Henry, and D. Tollervey. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Planta, R. J., and H. A. Raue. 1988. Control of ribosome biogenesis in yeast. Trends Genet. 4:64-68. [DOI] [PubMed] [Google Scholar]
- 46.Reeder, R. H. 1990. rRNA synthesis in the nucleolus. Trends Genet. 6:390-395. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, H. D. 1967. A nuclease specific for double-stranded RNA. Virology 12:718. [DOI] [PubMed] [Google Scholar]
- 48.Robertson, H. D., R. E. Webster, and N. D. Zinder. 1968. Purification and properties of ribonuclease III from Escherichia coli. J. Biol. Chem. 243:82-91. [PubMed] [Google Scholar]
- 49.Samarsky, D. A., and M. J. Fournier. 1999. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 27:161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samarsky, D. A., M. J. Fournier, R. H. Singer, and E. Bertrand. 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuster, P., W. Fontana, P. F. Stadler, and I. L. Hofacker. 1994. From sequences to shapes and back: a case study in RNA secondary structures. Proc. R. Soc. Lond. B Biol. Sci. 255:279-284. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava, A. K., and D. Schlessinger. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44:105-129. [DOI] [PubMed] [Google Scholar]
- 53.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 54.Tremblay, A., B. Lamontagne, M. Catala, Y. Yam, S. Larose, L. Good, and S. Abou Elela. 2002. A physical interaction between Gar1p and Rnt1p is required for the nuclear import of H/ACA small nucleolar RNA-associated proteins. Mol. Cell. Biol. 22:4792-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tycowski, K. T., C. M. Smith, M. D. Shu, and J. A. Steitz. 1996. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. USA 93:14480-14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venema, J., and D. Tollervey. 1995. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast 11:1629-1650. [DOI] [PubMed] [Google Scholar]
- 58.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 59.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, H., A. Henras, G. Chanfreau, and J. Feigon. 2004. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. USA 101:8307-8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, H., P. K. Yang, S. E. Butcher, S. Kang, G. Chanfreau, and J. Feigon. 2001. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 20:7240-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yon, J., A. Giallongo, and M. Fried. 1991. The organization and expression of the Saccharomyces cerevisiae L4 ribosomal protein genes and their identification as the homologues of the mammalian ribosomal protein gene L7a. Mol. Gen. Genet. 227:72-80. [DOI] [PubMed] [Google Scholar]
- 63.Young, R. A., and J. A. Steitz. 1978. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc. Natl. Acad. Sci. USA 75:3593-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, H., J. Zhao, C. H. Yu, Q. J. Luo, Y. Q. Chen, Y. Xiao, and L. H. Qu. 2004. Identification of a novel box C/D snoRNA from mouse nucleolar cDNA library. Gene 327:99-105. [DOI] [PubMed] [Google Scholar]
- 65.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuker, M., and A. B. Jacobson. 1998. Using reliability information to annotate RNA secondary structures. RNA 4:669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuker, M., and A. B. Jacobson. 1995. “Well-determined” regions in RNA secondary structure prediction: analysis of small subunit ribosomal RNA. Nucleic Acids Res. 23:2791-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.