Abstract
Histone methylation and the enzymes that mediate it are important regulators of chromatin structure and gene transcription. In particular, the histone H3 lysine 36 (K36) methyltransferase Set2 has recently been shown to associate with the phosphorylated C-terminal domain (CTD) of RNA polymerase II (RNAPII), implying that this enzyme has an important role in the transcription elongation process. Here we show that a novel domain in the C terminus of Set2 is responsible for interaction between Set2 and RNAPII. This domain, termed the Set2 Rpb1 interacting (SRI) domain, is encompassed by amino acid residues 619 to 718 in Set2 and is found to occur in a number of putative Set2 homologs from Schizosaccharomyces pombe to humans. Unexpectedly, BIACORE analysis reveals that the SRI domain binds specifically, and with high affinity, to CTD repeats that are doubly modified (serine 2 and serine 5 phosphorylated), indicating that Set2 association across the body of genes requires a specific pattern of phosphorylated RNAPII. Deletion of the SRI domain not only abolishes Set2-RNAPII interaction but also abolishes K36 methylation in vivo, indicating that this interaction is required for establishing K36 methylation on chromatin. Using 6-azauracil (6AU) as an indicator of transcription elongation defects, we found that deletion of the SRI domain conferred a strong resistance to this compound, which was identical to that observed with set2 deletion mutants. Furthermore, yeast strains carrying set2 alleles that are catalytically inactive or yeast strains bearing point mutations at K36 were also found to be resistant to 6AU. These data suggest that it is the methylation by Set2 that affects transcription elongation. In agreement with this, we have determined that deletion of SET2, its SRI domain, or amino acid substitutions at K36 result in an alteration of RNAPII occupancy levels over transcribing genes. Taken together, these data indicate K36 methylation, established by the SRI domain-mediated association of Set2 with RNAPII, plays an important role in the transcription elongation process.
Successful synthesis of mRNA by RNA polymerase II (RNAPII) requires tight regulation of the initiation, elongation, and termination processes of transcription. The process of transcription elongation is affected in part by the binding of regulatory factors to the phosphorylated C-terminal domain (CTD) of the RNAPII subunit Rpb1. Recent studies have highlighted an important role for histone methylation in the process of transcription elongation. In particular, studies have shown that the Set1 and Set2 methyltransferases, which catalyze methylation of histone H3 lysines 4 (K4) and 36 (K36), respectively, are associated with RNAPII at different stages of the transcription elongation cycle. While Set1 associates with RNAPII via the Paf1 transcription elongation complex in a manner that is dependent on the serine 5 (Ser5)-phosphorylated CTD, Set2 is recruited to RNAPII in a manner that is dependent on the CTD and the Ctk1 kinase (CTDK-I) that effects serine 2 CTD phosphorylation (10, 11, 18, 19, 21, 29, 42). Importantly, while studies show that Set1 preferentially associates with the 5′ end of genes, Set2 is found throughout the coding regions of genes (19, 29, 33, 42). These observations imply that K4 and K36 methylation have differing roles in the transcription elongation process.
Several lines of evidence indicate that Set2 is a phospho-CTD binding protein (10, 11, 19, 21, 22, 29, 42). For example, truncations of the RNAPII CTD severely reduce global K36 methylation levels in vivo (21, 42). In addition, deletion of CTDK-I results in a total abolition of K36 methylation (19, 42). These data have led to the model that Set2 preferentially binds to RNAPII that is phosphorylated at the Ser2 position of the CTD, which is supported by findings that Set2 binds to Ser2-phosphorylated CTD repeats in vitro (21, 22). Thus, a function for the CTDK-I-generated phospho-CTD in either the recruitment of Set2 and/or the control of K36 methylation activity has been proposed (36, 37).
While the association of Set2 with RNAPII is well established, the precise region(s) in Set2 required for this interaction, as well as the functional significance of this interaction on K36 methylation and transcription elongation, is poorly defined. In this report, we uncover a region in Set2 that mediates RNAPII interaction. We show that deletion of this Set2-Rpb1 interaction (SRI) domain abolishes K36 methylation on chromatin in vivo and leads to a transcription elongation defect, as assayed with 6-azauracil (6AU). In support of a direct role for Set2 methylation in the transcription elongation process, we find that set2 mutants or H3 K36 point mutations that prevent K36 methylation result in 6AU phenotypes similar to those of a complete SET2 deletion, which we in turn correlate with altered distribution of RNAPII along genes. These results define a novel domain in Set2 responsible for functional interaction with RNAPII and strongly suggest that the K36 methylation mediated by Set2 influences transcription elongation.
MATERIALS AND METHODS
Yeast strains.
The p3Flag-KanMX plasmid was used as a PCR template for genomic tagging of Saccharomyces cerevisiae Set2 (9). This provided for the generation of either full-length Set2 (Set2-3Flag) or a form with the SRI domain deleted [Set2(1-618)-3Flag] by homologous recombination. Primers used to generate Set2-3Flag in the BY4742 background were constructed previously (42). The primers for Set2(1-618)-3Flag were 5′-CAAAAGGAAGAGTCCAAAAAACTAGTGGAAGCAAAAGAGGCTAAGCGGTTGAAAAGGGAACAAAAGCTGGAG-3′ (forward) and 5′-AAAGAATTTATTCCAGTTGTGCTCTAGTCTTTGGGACTGGGAGACCGTTTTTCTTTACTATAGGGCGAATTGGGT-3′ (reverse). Bases which anneal to the p3Flag-KanMX plasmid are underlined, while the remaining sequence corresponds to the SET2 locus insertion position. The set2Δ and wild-type (WT) strains of the BY4742 background were obtained from Research Genetics, while the YCB652 strain, carrying an integrated URA3 gene, was obtained from Jeff Smith, University of Virginia School of Medicine (38). The SET2 gene was deleted in the W303 and YCB652 backgrounds by using a PCR product amplified from genomic DNA obtained from the BY4742 set2Δ strain, in which the SET2 gene had already been replaced by the KanMX gene (Research Genetics).
The H3/H4 shuffle strain WZY42 (in the S288C background) was used in 6AU analyses of H3 point mutants, and replacement of WT H3 with H3 mutants was accomplished as described previously (5, 44). Plasmids coding for the H3 S10A and K4R mutants have been described previously (5, 13). All other H3 point mutations were prepared by standard PCR-based site-directed mutagenesis using materials and methods previously described (5, 44).
Yeast WCE and nuclei preparation.
Yeast whole-cell extracts (WCEs) were prepared as described previously, but differed in the extraction buffer (42). The extraction buffer used consisted of 50 mM Tris-HCl at pH 8.0, 300 mM NaCl, 1 mM Mg(C2H3O2)2, 1 mM imidazole, 0.1% NP-40, 0.5 mM EDTA, and 10% glycerol. In addition, this buffer contained 0.5% phosphatase inhibitor cocktail I (Sigma), phenylmethylsulfonyl fluoride (2 mM), and leupeptin-pepstatin-aprotinin mix (each at 2 μg/ml). Nuclear extracts were prepared as previously described from strains grown in 200 ml of yeast extract-peptone-dextrose medium to an optical density at 600 nm (OD600) of 1.5 (7).
Electrophoresis and immunoblotting.
Western blotting and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses were performed according to procedures and reagents obtained from Amersham Life Sciences. The ECL Plus Western blotting detection kit (Amersham Pharmacia Biotech) was used for specific antibody detection. The rabbit anti-Me2(K36) antibody was obtained from Upstate Biotechnology Inc. and used at a dilution of 1:3,000. The antibody targeted against the C terminus of H3 was obtained from Abcam Inc. (AB1791) and used at a dilution of 1:5,000. The anti-phospho-CTD antibodies H5 and H14 were obtained from Covance, Inc.
Generation of SET2 expression constructs.
The Set2 constructs containing a C-terminal Flag epitope [Set2(1-618), Set2(262-475), Set2(445-538), Set2(528-638), Set2(619-733), Set2(634-733), Set2(619-718), and Set2(619-703)] were generated by PCR amplification using Vent DNA polymerase (New England BioLabs) and the Set2-Flag PN823 expression construct as the template. Full-length Set2, Set2(1-261), and Set2R195G constructs were prepared previously (39, 42). The PCR products were cloned into the PN823 yeast expression plasmid and sequenced for accuracy. Primer sequences are available upon request. For in vitro phospho-CTD binding experiments, the Set2(1-618) and Set2(619-733) constructs were subcloned into the pMAL-c2G vector (New England BioLabs), and proteins were purified according to the manufacturer's protocol.
Immunoprecipitations.
Coimmunoprecipitation (co-IP) experiments involving the various mutant Set2-Flag constructs or Set2-3Flag strains were performed essentially as previously described (42). In brief, a set2Δ strain (in the BY4742 background) was transformed with the indicated series of Set2 expression constructs and grown to an OD600 of ∼1.0 in synthetic complete medium lacking uracil, and WCEs were prepared using the extraction buffer described above. Co-IPs were performed in a final volume of 0.9 ml, equalized with extraction buffer containing 1.5 mg of WCE protein (or 2.0 mg for genomically tagged strains). The extracts were incubated with 12.5 μl of preequilibrated anti-Flag affinity beads (M2; Sigma) for 2 h at 4°C, after which extracts were washed three times for 2 min in extraction buffer. The beads were eluted in SDS-PAGE loading buffer with incubation at 100°C for 5 min, and bead-bound proteins were analyzed by immunoblotting with antibodies targeted against the phospho-CTD.
ChIP assays.
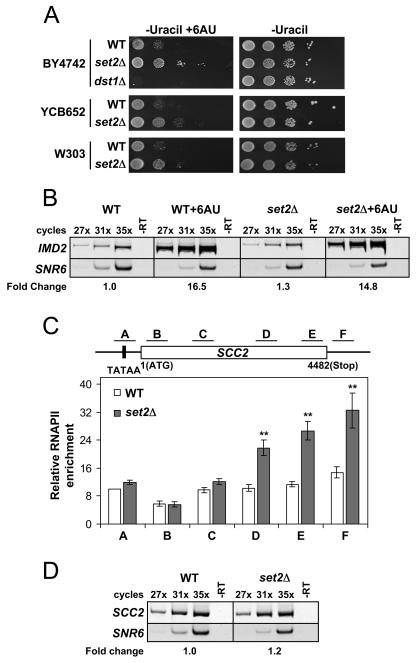
The chromatin immunoprecipitation (ChIP) assay using the H3 K36 dimethyl antibody was performed as described previously (42). Primers were used to amplify regions of SCC2 in the following ranges relative to the ATG start site: −277 to −27, 2 to 238, 984 to 1222, 3044 to 3276, 3981 to 4222, and 4489 to 4679. Intergenic chromosome V primers were used as a reference and loading control, as previously reported (17). The previously characterized general RNAPII-CTD antibody (not specific to any CTD modification state) was used in ChIP assays for RNAPII detection (34).
Set2-3Flag and Set2(1-618)-3Flag purification.
Set2-3Flag and Set2(1-618)-3Flag proteins were purified as previously described (42). A Coomassie-stained gel was used to visualize the associated proteins, and mass spectrometry analysis confirmed the presence or absence of Rpb1 and Rpb2 from excised gel slices.
In vitro HMT assays.
The Escherichia coli strain BL21 was transformed with pMAL vectors expressing the constructs maltose binding protein (MBP)-Set2 and MBP-Set2(1-618), which also contained the C-terminally tagged Flag epitope. Protein expression was induced in 100 μM isopropyl-β-d-thiogalactopyranoside for 3 h at 30°C, cells were lysed by sonication, and 20-μl histone methyltransferase (HMT) reaction mixtures were prepared as described previously (39). In brief, lysate volumes were used that resulted in equal amounts of each MBP-Set2 fusion per reaction mixture, as analyzed by Western blotting with the Flag antibody. HMT reaction mixtures contained 1 μCi of S-adenosyl-l-[methyl-3H]methionine (Amersham Biosciences), with or without 6 μg of chicken nucleosomes. Reaction mixtures were incubated at 30°C for 20 min and spotted on Whatman P-81 for liquid scintillation counting or analyzed by SDS-PAGE followed by fluorography.
Far Western analysis of Set2 fragments using phospho-CTD probes.
Far Western analysis using a phospho-CTD probe (generated by CTDK-I) was performed essentially as described previously (26, 27). In brief, picomole quantities of recombinant MBP-Set2(1-618) and MBP-Set2(619-733) were resolved on a 4-to-15% Tris-HCl criterion gel (Bio-Rad) and transferred to nitrocellulose (Hybond C Extra; Amersham Pharmacia Biotech). The nitrocellulose was stained with Ponceau S to visually ensure protein transfer from the gel. The membrane was blocked at 4°C for 24 h and probed with 2.5 μg of glutathione S-transferase (GST)-[32P]CTD in blocking buffer for 3.5 h at 4°C. The membrane was washed, air dried, and exposed to film. Reverse far Western analysis was performed according to published methods by resolving recombinant unphosphorylated or phosphorylated GST-CTD fusions on a 4-to-15% Tris-HCl gel (31). The gel was transferred to a nitrocellulose membrane and probed with MBP-Set2 or MBP-Set2(619-733), followed by detection using an antibody against MBP.
Determination of SRI-CTD specificity by using BIACORE.
The BIACORE sensor chip carrying three-repeat CTD peptides phosphorylated at either Ser2 (2-phospho), Ser5 (5-phospho), or both (2 + 5-phospho) or a control Ser-phosphorylated peptide that mimics the charge state of the 2 + 5-phospho peptide (6PC scrambled control) was generated as previously recorded (16, 31). Purified MBP-Set2(619-733) was interacted with the peptides, and association and dissociation were monitored. The response curves were normalized to that for the 6PC control peptide.
RT-PCR.
Reverse transcription (RT)-PCR analysis was performed as previously described (42, 43). Primer sequences are available upon request.
6AU growth assays.
Yeast strains used in this assay, except for YCB652, were transformed with the URA3+ CEN plasmid pRS316 and grown in synthetic defined medium lacking uracil (SD-Ura). Overnight cultures were diluted 1:20 and grown to an OD600 of 1.0, and 10-fold serial dilutions were plated on SD-Ura medium with or without 6AU (Aldrich) or mycophenolic acid (Sigma), each at 100 μg/ml. Plates were photographed after 30°C incubation for 2 to 3 days. Liquid cultures used for RT-PCR analysis were grown with 6AU at 50 μg/ml for 120 min. This time point was selected based on a recent study in which the IMD2 steady-state mRNA levels of a large-scale 6AU screening of yeast deletion mutants were analyzed (32).
RESULTS
A novel domain in Set2 mediates RNAPII interaction.
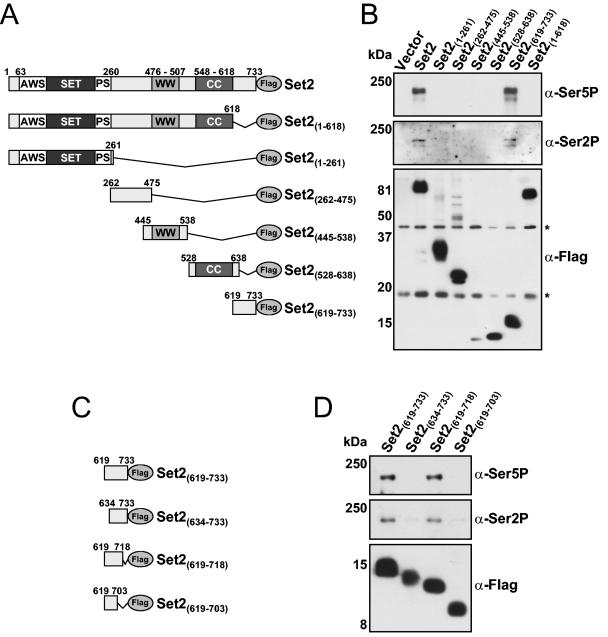
Although Set2 is known to bind to the phosphorylated CTD of RNAPII, the region(s) within Set2 responsible for this association is poorly defined (19, 21, 22, 33, 42). Thus, we generated a series of Set2 mutant yeast expression constructs that contained a C-terminal Flag epitope tag (Fig. 1A) and used them in co-IP studies with antibodies generated against RNAPII. Either full-length SET2, vector-only control, or the indicated SET2 mutant was expressed in a set2 deletion mutant (set2Δ), and WCEs were prepared. As expected, immunoprecipitation of full-length Set2-Flag resulted in coprecipitation of RNAPII as detected by immunoblotting with anti-phospho-CTD antibodies (Fig. 1B). We previously reported that specific deletion of the WW domain in Set2 does not disrupt Set2-RNAPII coprecipitation, and so we hypothesized that the coiled-coil region of Set2 may be responsible (42). Unexpectedly, we found that a precise deletion of the coiled-coil region in Set2 (amino acids 548 to 618) did not disrupt this interaction (data not shown), indicating that a previously undefined region in Set2 mediates this association. As shown in Fig. 1B, we found through further Set2 truncations that a region at the C terminus of Set2, encompassing amino acid residues 619 to 733, is both necessary and sufficient to mediate the interaction of Set2 with RNAPII (compare last two lanes). We therefore termed this region the SRI domain.
FIG. 1.
Identification of a novel region in Set2 required for RNAPII binding. (A) Schematic representation of the Set2 constructs used to probe for RNAPII interaction. The SET domain along with the AWS domain, postSET domain (PS), WW domain, and coiled-coil motif (CC) are shown. All constructs contained a C-terminal Flag epitope. (B) set2Δ cells were transformed with either vector only or plasmids coding for the indicated Set2-Flag construct, and WCE were prepared. WCE were immunoprecipitated with anti-Flag beads followed by immunoblotting with antibodies directed against serine 5-phosphorylated CTD (H14; α-Ser5P), serine 2-phosphorylated CTD (H5; α-Ser2P), or the Flag epitope. Significant to mention is that the H5 antibody may also recognize serine 5 CTD phosphorylation in addition to serine 2 phosphorylation (16). Asterisks indicate the location of nonspecific Flag antibody-reactive species. (C) Schematic representation of the Set2-SRI domain constructs used to determine the boundaries of the functional SRI domain. N- and C-terminal truncations of the SRI domain were made in 15-amino-acid increments as shown. All constructs contained a C-terminal Flag epitope. (D) set2Δ cells were transformed with the indicated plasmids, WCE were prepared, and co-IPs were performed using the antibodies indicated in panel B. Sizes of the molecular mass markers are shown.
We next sought to determine the precise boundaries of the SRI domain. To accomplish this, we generated additional Set2 constructs containing N- and C-terminal truncations of the SRI region (Fig. 1C) and used them in co-IP analyses as described for Fig. 1B. Results revealed that N-terminal truncation of the SRI domain beyond Set2 amino acid 619 abolished RNAPII binding. However, binding was still possible with a C-terminal truncation up to amino acid 718 of Set2 (Fig. 1D), thereby identifying the boundaries of the SRI domain as amino acids 619 to 718.
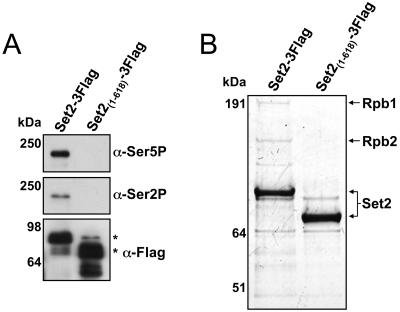
Due to the possibility that the observed interaction between the SRI domain and RNAPII shown in Fig. 1 might have been influenced by the high levels of recombinant Set2 protein produced (as these constructs are expressed from a plasmid using the highly active ADH1 promoter), we genomically tagged Set2 at amino acid 733 or 618 with a triple Flag sequence and reexamined its association with RNAPII. As shown in Fig. 2A, full-length Set2 (Set2-3Flag) again coimmunoprecipitated RNAPII as analyzed by immunoblot analysis with the anti-phospho-CTD antibodies. In contrast, a form of Set2 with the SRI domain deleted [Set2(1-618)-3Flag] resulted in the abolition of RNAPII interaction. We also confirmed these results by examining the protein associations of Set2 by affinity purification, which revealed that the readily detectable subunits of RNAPII (Rpb1 and Rpb2) were only observed in purifications involving full-length Set2 (Fig. 2B). Additionally, while Rpb1 and Rpb2 were detected by mass spectrometry in gel excised bands from the full-length Set2 purification, these proteins were not detected in a parallel gel region excised from the form of Set2 with the SRI domain deleted (data not shown). Collectively, these data confirm the importance of the SRI domain in mediating the Set2-RNAPII interaction.
FIG. 2.
The SRI domain is required for interaction of Set2 with RNAPII. (A) Yeast strains containing full-length Set2 (Set2-3Flag) or a form of Set2 without the SRI domain [Set2(1-618)-3Flag] were made via genomic tagging with the 3xFlag epitope. WCEs of these strains were immunoprecipitated with anti-Flag beads followed by immunoblotting with antibodies directed against serine 2-phosphorylated CTD (H5; α-Ser2P), serine 5-phosphorylated CTD (H14; α-Ser5P), or the Flag epitope. Sizes of the molecular mass markers are shown, and asterisks indicate the location of expected Set2-Flag products. (B) WCEs from the strains in panel A were incubated with anti-Flag resin, and the resulting bound proteins were eluted with 3xFlag peptide. Eluted proteins were resolved by a 4-to-12% NuPAGE gel and examined by Coomassie staining. Arrows indicate the protein identity of bands in the Set2-3Flag lane that were examined by mass spectrometry, while analysis of parallel regions in the Set2(1-618)-3Flag lane was negative for the presence of Rpb1 or Rpb2. Sizes of the molecular mass markers are shown.
The SRI domain of Set2 is conserved and interacts with the phosphorylated CTD in vitro.
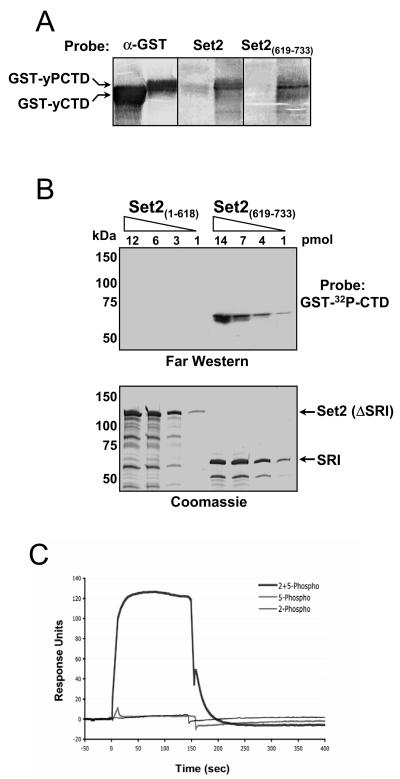
Previous studies have suggested that Set2 association with RNAPII is dependent, in part, on the Paf1 transcription elongation complex (12, 19). Thus, it was a formal possibility that the SRI domain linked Set2 to the polymerase by indirect protein association. To test whether the SRI domain of Set2 is responsible for direct association with the CTD of RNAPII, we generated a variety of MBP fusions of Set2 and examined their ability to associate with a recombinant GST-CTD fusion protein that was either unmodified (GST-yCTD) or phosphorylated by CTDK-I (GST-yPCTD). Using a reverse far Western approach (see reference 31), the GST-CTD fusions were resolved by SDS-PAGE, transferred to nitrocellulose, and then probed with MBP fusion proteins carrying full-length Set2 or only the SRI domain of Set2 [Set2(619-733)]. Results revealed that both the full-length form of Set2 and the SRI domain of Set2 preferentially bound to the phosphorylated CTD (Fig. 3A). To independently confirm this interaction and further address whether other regions of Set2 may bind to the phosphorylated CTD in vitro, we transferred increasing amounts of MBP fusions of Set2 lacking the SRI or containing only the SRI domain [Set2(1-618) and Set2(619-733), respectively] to nitrocellulose and probed with a CTDK-I-phosphorylated GST-[32P]CTD fusion. As shown in Fig. 3B, this far Western approach revealed that while the SRI domain of Set2 bound efficiently to the GST-[32P]CTD fusion, Set2 lacking the SRI domain did not. While Fig. 3B shows a 3.5-h exposure, it is noteworthy that a 90-h exposure revealed a potential weak interaction of Set2(1-618) to phosphorylated CTD (data not shown); however, it is unclear whether such interaction is physiologically relevant (Fig. 2 to 4) (31). In summary, our results show that the SRI domain in Set2 binds directly to the phospho-CTD of RNAPII. Thus, the ability of the Paf1 complex to modulate Set2 activity is likely an indirect consequence of the fact that this complex can regulate CTD phosphorylation (28).
FIG. 3.
The SRI domain of Set2 binds synergistically to doubly modified CTD repeats. (A) Reverse far Western analysis. GST-yCTD and CTDK-I-phosphorylated GST-CTD (GST-yPCTD) fusion proteins were subjected to SDS-PAGE and transferred to nitrocellulose. Membranes were probed individually with purified recombinant full-length MBP-Set2 (Set2) or with MBP-SRI [Set2(619-733)], and the bound MBP fusions were detected with an anti-MBP antibody. As a control, a duplicate membrane was probed with an anti-GST antibody (α-GST) to demonstrate the presence of both GST-CTD fusion proteins. (B) Increasing amounts of two MBP fusion proteins [Set2(1-618) and Set2(619-733)] were resolved in two SDS-polyacrylamide gels; one gel was subjected to far Western analysis with GST-[32P]CTD as a probe, and the other was stained with Coomassie. (C) BIACORE analysis of the SRI domain. The MBP-SRI fusion protein [MBP-Set2(619-733)] was analyzed by surface plasmon resonance (BIACORE) for binding to distinct phosphorylated synthetic three-repeat CTD peptides. These peptides were either Ser5 phosphorylated (5-phospho), Ser2 phosphorylated (2-phospho), or both (2 + 5-phospho) in each repeat (see Materials and Methods). Response units, on the y axis, represent binding to the peptides. The binding response to a scrambled control peptide carrying six SerPO4residues (see Materials and Methods) has been subtracted from each of the three response curves. Only the peptide carrying both Ser2PO4 and Ser5PO4 in each repeat showed binding above control levels, and we estimate the affinity of this interaction (after subtraction of background binding to the control peptide) to be 6 μM.
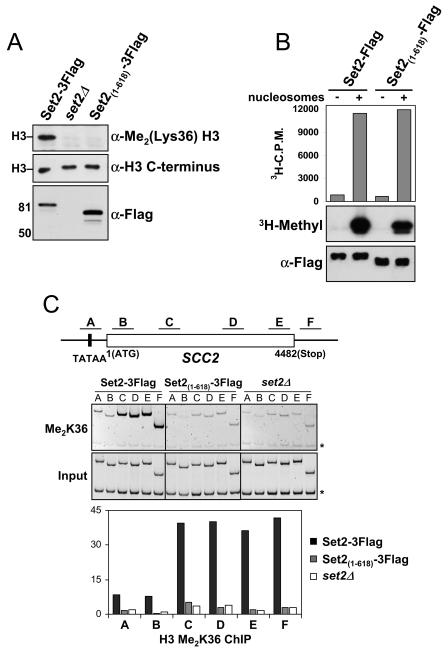
FIG. 4.
Deletion of the SRI domain in Set2 abolishes H3 K36 dimethylation. (A) Yeast nuclear extracts prepared from set2Δ cells or the indicated genomically tagged strains in the BY4742 background were probed with antibodies against dimethylated lysine 36 at H3 [α-Me2(Lys36) H3] to monitor the role of the SRI domain in global K36 methylation levels. An antibody directed against the C terminus of H3 (α-H3 C terminus) was used as a loading control. Nuclear levels of Set2 in these strains were monitored using the anti-Flag antibody. A similar loss of K36 methylation was observed when the SRI domain was genomically deleted in the W303 background (data not shown). (B) Full-length (Set2-Flag) or SRI domain-truncated [Set2(1-618)-Flag] recombinant forms of Set2, which also contained an N-terminal MBP epitope, were prepared and analyzed for their HMT activity in vitro. HMT reactions were prepared with bacterial lysates containing the indicated Set2 constructs with or without nucleosomes. Identical samples were analyzed by the filter-binding assay (upper) and fluorography (middle). Immunoblotting with the Flag antibody (lower) was performed to ensure equal amounts of protein were present in each reaction mixture. (C) WT or SRI-deleted yeast strains were analyzed by ChIP for K36 dimethyl levels on genes. DNA isolated from the K36 methylation IPs was used in PCRs with primer pairs for the indicated regions of the SCC2 gene (top). PCR products of the input DNA (input) and ChIP DNA (Me2K36) are shown (middle). The asterisks indicate the location of a PCR product pertaining to an intergenic region at chromosome V (ChV), which was used as a loading control in all PCRs. The histogram displays the relative enrichment values for K36 dimethylation (bottom). The values were calculated by dividing the ratio of band intensities for IP DNA/ChV with the ratio of intensities for the input DNA/ChV. Identical results were found for the PMA1, ENO1, and ADH1 genes (data not shown).
Next, we examined the specificity of the SRI domain for binding phospho-epitopes by BIACORE analysis with three-repeat synthetic CTD peptides that were phosphorylated in each repeat at either Ser2 (2-phospho), Ser5 (5-phospho), or both (2 + 5-phospho). As a control, a Ser-phosphorylated peptide that mimics the charge state of the 2 + 5-phospho peptide (6PC) was included. Sensor chips containing these CTD peptides were reacted with the SRI domain of Set2 [Set2(619-733)], and binding was monitored. Surprisingly, we found that the Set2 SRI domain bound preferentially to CTD repeats that were doubly phosphorylated (Fig. 3C, compare 2 + 5-phospho curve with 2-phospho and 5-phospho curves). Moreover, because these response curves were obtained after subtracting the contribution of the nonspecific control peptide (6PC), this binding depends on the presence of both Ser2P and Ser5P in the context of the CTD heptad repeat sequence. Based on additional BIACORE experiments involving titrated amounts of the Set2 SRI domain (not shown), we determined that the Set2 SRI domain binds to the 2 + 5-phospho peptide (relative to the control) with an apparent dissociation constant of about 6 μM. It is important to note that the ability of the SRI domain to bind to the nonspecific control peptide was nearly equivalent to that found for binding to the individually phosphorylated peptides (data not shown). We take this result to suggest that the SRI domain of Set2 has a specific requirement for Ser2- and Ser5-phosphorylated CTD epitopes. Collectively, these results reveal a novel and selective requirement for a specific CTD phosphorylation pattern in Set2 binding to RNAPII.
Given these findings, we next asked whether the SRI domain of Set2 is a conserved phospho-CTD-binding motif found in other proteins in budding yeast and beyond. By performing a PSI-BLAST search, we determined that the SRI domain of Set2 was unique to this enzyme alone in budding yeast (data not shown). However, the SRI domain showed significant homology to the C-terminal regions of proteins in other species that also displayed domain organizations similar to that of Set2 (AWS, SET, postSET, and WW), suggesting that these proteins may be the functional homologs of budding yeast Set2 and function with RNAPII (Table 1). Interestingly, the proteins identified in Table 1 represent only a subset of proteins that the SMART database revealed to contain AWS, SET, and postSET domains (>70), suggesting that not every putative histone methyltransferase that contains an AWS, SET, and postSET is by default a functional homolog of Set2. Indeed, recent evidence shows that the Drosophila melanogaster Ash1 protein, which falls into the Set2 family of HMTs (by way of having an AWS domain rather than an archetypal PreSET domain), is an H3 lysine 4 methyltransferase (4, 6). These results suggest that the SRI domain is a probable indicator of RNAPII-interacting enzymes that catalyze K36 methylation. To determine whether any of the putative SRI domains we identified by our PSI-BLAST search would actively bind to the phospho-CTD, we expressed and purified from bacteria a GST fusion protein carrying the C-terminal 178 residues of the human Huntington interacting protein B (HYPB) that includes the region of homology to Set2's SRI domain (Table 1). Using the far Western approach as described for Fig. 3B, we found that similar to Set2, the SRI-containing region in HYPB interacts efficiently with a CTDK-I-phosphorylated GST-[32P]CTD fusion (data not shown). Furthermore, additional BIACORE analyses (as described for Fig. 3C) revealed that the human SRI domain displays binding properties nearly identical to those of the budding yeast domain (H. P. Phatnani, A. L. Greenleaf, and P. Zhou, unpublished results). Taken together, these results suggest that the SRI domain is a highly conserved and novel phospho-CTD-interacting domain.
TABLE 1.
Putative Set2 homologs identified by PSI-BLAST searching with the SRI domain
| Species | Gene namea | GI accession | Sequence position of putative SRI domain | % Identity/similarity to SRI domainb |
|---|---|---|---|---|
| S. cerevisiae | Set2 | 6322293 | 619-718 | NA |
| Candida albicans | Ca019.9324 | 46435920 | 723-820 | 35/55 |
| Homo sapiens | HYPB | 30410779 | 1956-2056 | 23/37 |
| Mus musculus | XP_135176 | 38090181 | 2393-2493 | 23/37 |
| D. melanogaster | CG1716 | 24641786 | 2262-2356 | 21/39 |
| Neurospora crassa | XP_322355 | 32403484 | 569-653 | 19/37 |
| S. pombe | SPAC29B12 | 2408044 | 703-778 | 17/42 |
All proteins identified contain a domain architecture similar to yeast Set2, including the AWS, SET, postSET and, in some cases, WW domains. It is notable that the putative SRI domains, as with Set2, are found in the C termini of these proteins.
Identity refers to the percentage of identical amino acids present between the yeast Set2 SRI domain and the putative SRI domain of the indicated homolog, while similarity indicates amino acid replacements that exhibit similar charge or hydrophobicity.
The SRI-RNAPII interaction is required for H3 K36 methylation.
Given the conserved nature and potential importance of the SRI domain to Set2's cellular function, we next investigated the consequences of deleting this domain. Although studies suggest that the CTD and its proper phosphorylation are necessary for K36 methylation, it has not been formally excluded that the phospho-CTD might regulate the enzymatic activity of Set2 (19, 21, 33, 42). To determine if the loss of the SRI domain would result in a loss of genome-wide K36 methylation, we measured the K36 dimethylation levels in strains containing WT Set2 or Set2 with the SRI domain genomically deleted. Nuclei were prepared from these strains and then resolved on an SDS-PAGE gel, followed by Western blotting with an antibody specific to dimethylated K36. Results revealed that deletion of the SRI domain in Set2 abolishes global H3-K36 dimethylation (Fig. 4A). As a control, we examined the levels of H3 in parallel gels with an antibody specific to the C terminus of H3, which revealed that the levels of histones were similar in both nuclei preparations. Importantly, the nuclei of both strains showed the presence of Set2 by Western blot analysis using an anti-Flag antibody (Fig. 4A). This result indicates that the deletion of the SRI domain does not influence the nuclear localization of Set2 or significantly affect its stability. The requirement of the SRI domain for K36 methylation was independently confirmed in parallel studies in which a different strain background (W303) was genomically tagged either at the C terminus or at the beginning of the SRI domain at residue 618 (data not shown). In addition, we used the ChIP assay to analyze chromatin modifications at a gene-specific resolution and also observed a loss of K36 methylation at the SCC2 gene when the SRI domain of Set2 is removed (Fig. 4C). Analysis of the PMA1, ENO1, and ADH1 genes yielded similar results (data not shown).
To test the possibility that the SRI domain itself might regulate Set2's catalytic activity, we analyzed recombinant full-length Set2 or SRI-deleted MBP-Set2 fusion proteins in HMT assays with chicken oligo-nucleosomes. Results showed that both forms of the enzyme were equally active for K36 methylation, indicating that the SRI domain is not required for the catalytic activity of Set2 in vitro (Fig. 4B). In fact, we found that a region of Set2 encompassing the AWS, SET, and postSET domains (amino acid residues 1 to 261 in Set2) is fully active for histone methylation in vitro, indicating that the C terminus of Set2 does not intrinsically regulate its HMT activity (data not shown).
Set2 methylation influences transcription elongation and RNAPII occupancy on genes.
Growth phenotypes observed in the presence of the drug 6AU are frequently used as indicators of defects in transcription elongation (8, 23, 25, 41). We therefore asked whether deletion of SET2, or prevention of the Set2-RNAPII association by deletion of the SRI domain, would exhibit 6AU-dependent phenotypes. We began by examining the 6AU phenotypes caused by Set2 deletion in several strain backgrounds. Various WT (W303, BY4742/SC288C, and YCB652) and matched set2Δ strains were grown on control medium (no drug) or medium containing 6AU. The parent strain YCB652 contained the integrated URA3 gene, which is required for the 6AU assay, while others were transformed with the URA3 plasmid pRS316 (38). The survival and colony sizes of each strain were monitored after several days of growth and compared to those on control plates. As shown in Fig. 5A, we found that deletion of SET2 in these strain backgrounds resulted in a significant resistance phenotype to 6AU. Similar results were also observed when we used medium containing mycophenolic acid, another drug that reveals elongation defects but through a mechanism unique from that of 6AU (data not shown). Furthermore, we analyzed a dst1 null strain of the BY4742 background and observed the characteristic 6AU sensitivity known to exist for this mutant (Fig. 5A) (3, 30, 41).
FIG. 5.
Deletion of SET2 results in an elongation phenotype and an alteration of RNAPII occupancy on genes. (A) Various strains containing either WT, set2Δ, or dst1Δ alleles were plated on synthetic dextrose-uracil medium with or without 6AU (100 μg/ml) and grown at 30°C for 2 to 3 days to monitor for transcription elongation phenotypes. All strains contained the plasmid pRS316 containing the URA3 gene, except yeast strain YCB652, which contains an integrated URA3 gene. Results using mycophenolic acid (100 μg/ml) were found to yield identical results (data not shown). (B) The loss of Set2 does not aberrantly affect the increased expression of IMD2 in 6AU. Semiquantitative RT-PCR was used to monitor the expression of IMD2 and SNR6 (a polymerase III-transcribed gene used as a control) in WT or set2Δ strains in the absence or presence of 6AU (50 μg/ml). The results of RT-PCRs with (PCR amplification cycles indicated above) or without (-RT) reverse transcriptase are shown. The fold change in IMD2 expression under each condition is indicated, based on averages of the three cycling parameters for each strain, with WT set to 1.0 as reference. (C) WT or set2Δ strains were analyzed by ChIP for RNAPII levels. Isolated DNA from the RNAPII IPs were used in PCRs with primer pairs for regions of SCC2 as indicated in the schematic. The data shown represent the average of 13 individual ChIP assays from separate cell pellets. The standard error of the mean is indicated. Asterisks indicate the relative set2Δ RNAPII enrichment values that were statistically significant compared to their WT counterparts (P < 0.01 for primer set D and P < 0.001 for primer sets E and F). ChIP analyses using other RNAPII antibodies (8GW16 from Covance or Rpb3 from NeoClone) or the N terminus of Rpb1 (y-80; Santa Cruz) revealed the same pattern of RNAPII distribution as displayed in the figure (results not shown). (D) SCC2 expression was not changed in set2Δ despite the increase in RNAPII density detected by ChIP. The expression levels of SCC2 and SNR6 (control) were monitored by semiquantitative RT-PCR as described for panel B. The fold change of SCC2 expression in set2Δ cells compared to that in WT is displayed as described for panel C.
Given that previous studies have demonstrated that a proper response to 6AU is the induced expression of the IMD2 gene, which is a result of the elongation machinery's response to depleted nucleotide pools (35), we sought to verify that the resistance phenotype observed in the set2Δ deletion mutant was not due to an aberrant effect on the cellular metabolism of 6AU. Using semiquantitative RT-PCR, we monitored the expression levels of IMD2 and SNR6 (an RNAPIII-transcribed gene used as a control) in the presence or absence of 6AU. As shown in Fig. 5B, we found that the expression of the IMD2 gene was increased to equal degrees in both WT and set2Δ strains in the presence of the drug, confirming that the loss of Set2 results in a bona fide transcription elongation defect. Importantly, the IMD2 gene was not induced in the absence of 6AU for either the WT or set2Δ strains, indicating that Set2 does not act to repress the basal expression of this gene (Fig. 5B). In addition to our results with Set2, 6AU resistance has also been observed from the deletion or mutation of a variety of other elongation factors, including Chd1, Bye1, Isw1, and forkhead factor 1 (2, 40, 41).
Since transcription elongation defects are typically correlated with changes in the occupancy and distribution of RNAPII along genes, we therefore asked whether the loss of Set2 would result in an alteration in RNAPII levels on actively transcribed genes. Using an antibody that recognizes the general levels of RNAPII irrespective of its phosphorylation status (34), we examined RNAPII levels on the promoters and coding regions of active genes in WT and set2Δ strains by ChIP. As shown in Fig. 5C, we found that RNAPII levels in the set2 deletion mutant were significantly increased in the middle to late coding region of the actively transcribing SCC2 gene compared to the WT control strain. Interestingly, the gene locations that showed an RNAPII increase were also the same locations determined to be highly methylated by Set2 (Fig. 4C), suggesting the possibility that a relationship may exist between regions of chromatin highly methylated at K36 and RNAPII occupancy potential. We next examined a variety of other active genes in the set2 deletion mutant to determine how general this RNAPII defect would be. We examined the promoter and coding regions of TOM1, MDN1, PMA1, ENO1, and FIR1 for the presence of RNAPII as described above and found a similar pattern of RNAPII increase in the set2 deletion mutant as was observed for SCC2 (data not shown). We addressed the possibility that the observed increases in RNAPII might be a result of a general increase in transcript formation for these genes in the absence of Set2. Indeed, Set2 has been shown to play a role in the basal repression of GAL4 (20). We therefore examined the expression of the genes indicated above by semiquantitative RT-PCR and observed that the increased density of RNAPII did not correlate with any change in the steady-state mRNA levels (Fig. 5D and data not shown). These mRNA results were also confirmed independently by examining the gene expression microarray profiles found in WT and set2Δ cells (N. Krogan and J. Greenblatt, personal communication). Furthermore, we also examined TBP levels by ChIP at the promoters of several genes listed above (SCC2 and TOM1) and found no significant increases in TBP occupancy in the set2 deletion mutant (data not shown). Our data indicate that Set2 does not function as a basal repressor of the genes analyzed, but rather it affects the precise levels of RNAPII on genes, further supporting the 6AU results suggesting that Set2 can influence RNAPII elongation.
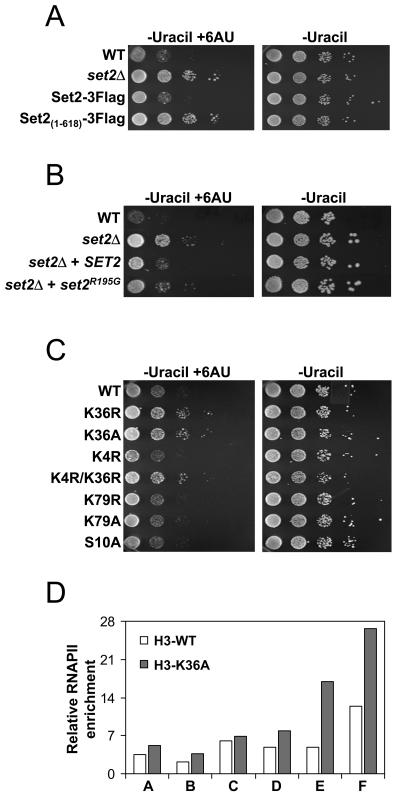
The above results suggest Set2 is important for transcription elongation, but they do not reveal whether this function of Set2 is dependent on its association with RNAPII and/or K36 methylation. To test if loss of the interaction between Set2 and RNAPII is responsible for the elongation defect, we assayed the growth of strains with SRI deleted [Set2(1-618)-3Flag], SET2 deleted, and the Set2-3Flag strains by using 6AU. We observed that deletion of the SRI domain resulted in a resistance to 6AU that was similar to that of the set2 deletion mutant, indicating that the interaction between Set2 and RNAPII is necessary for the normal transcription elongation functions of Set2 (Fig. 6A).
FIG. 6.
K36 methylation influences transcription elongation. (A) Genomically tagged strains containing either full-length Set2 (Set2-3Flag) or Set2 with the SRI domain deleted [Set2(1-618)-3Flag] were generated and assayed, as in Fig. 5A, for growth on 6AU and compared to WT and set2Δ strains. (B) set2Δ strains were transformed with a plasmid expressing WT Set2 (SET2) or a mutant form of Set2 which abolishes its catalytic activity (set2R195G) and assayed for growth on 6AU as before. (C) Yeast strains (WZY42 derived) bearing various point mutations on histone H3 were assayed for growth on 6AU as in Fig. 5A. (D) WT or a K36 mutated strain was analyzed by ChIP for RNAPII levels for the SCC2 gene, as in Fig. 5C. The histogram data are representative of two independent experiments which showed similar results.
We next asked whether K36 methylation per se is important for the activity of Set2 in this process. In one case, we transformed set2Δ cells with a plasmid coding for either full-length Set2 (SET2) or a form of Set2 containing a point mutation (set2R195G) that has been shown to abolish K36 methylation activity in vitro and in vivo (39). We found that expression of SET2 in the set2Δ strain nearly restored WT levels of 6AU sensitivity (Fig. 6B). However, set2Δ cells expressing set2R195G showed resistance to the drug (Fig. 6B), consistent with a role for K36 methylation in the elongation process. In the second case, we asked whether amino acid substitutions at K36 that prevent methylation (K36A and K36R) would result in resistance to 6AU and RNAPII density increases. As shown in Fig. 6C, the K36A and K36R strains were significantly resistant to 6AU compared to the WT H3 strain, whereas strains with mutations at other sites of methylation (K4 and K79) or sites of phosphorylation (serine 10) were not. In addition, the 6AU resistance caused by the K36A or K36R mutations was not suppressed by mutation of lysine 4 (Fig. 6C, K4R/K36R). Significantly, we also found the same pattern of increased RNAPII density for the SCC2 gene in the K36A strain as with the set2Δ strain (Fig. 6D), suggesting that the specific lack of K36 methylation is the primary cause of the 6AU phenotype and RNAPII defect. These data strongly implicate the methylation by Set2 as being functionally important in the elongation process.
DISCUSSION
Recent work in S. cerevisiae has revealed an unexpected role for the Set1 and Set2 histone methyltransferases in transcription elongation. Although Set2 associates with the elongating polymerase, very little is known regarding the influence of Set2 or K36 methylation on transcription elongation. Our results presented in this study reveal that (i) a novel region of Set2, which we have termed the SRI domain, is necessary and sufficient for the functional interaction between Set2 and RNAPII; (ii) the Set2-RNAPII interaction, established through the SRI domain, is required for H3 K36 methylation; and (iii) the K36 methylation mediated by Set2, in particular, influences RNAPII elongation.
A novel phospho-CTD binding motif in Set2.
Based on recent observations of other RNAPII CTD-interacting proteins, we expected that the WW and/or coiled-coil regions in Set2 would mediate its association with RNAPII. Surprisingly, neither of these domains was found to be involved in RNAPII binding. Instead, a region from 619 to 733 in Set2, which we have termed the SRI domain, was found to be both necessary and sufficient for the Set2-RNAPII interaction (Fig. 1 to 3). This is in conflict with an earlier report which stated that the WW domain of Set2 is required for the association of this enzyme with RNAPII (22). However, the technical approach used in this earlier study to analyze the involvement of the WW domain in RNAPII interaction employed a genomic insertion of the TAP tag in front of the WW domain of Set2, which removed its entire C terminus, including the SRI domain. Even though the WW and/or coiled-coil regions in Set2 are not likely to be involved in its association with RNAPII, they may play important roles in Set2 function by mediating the association of Set2 with other phosphorylated proteins and/or itself since Set2 is thought to exist as a homodimer in the cell (33, 39).
Through further examination of the binding requirements of the SRI domain, we uncovered that this domain binds preferentially to the CTD phosphorylated at both serines 2 and 5, compared to the singly phosphorylated form or a charge control peptide (Fig. 3C). This discovery strongly suggests that a synergistic relationship between CTD phosphorylation and Set2 binding exists. Interestingly, Ser2 phosphorylation is predominantly found in the body of genes, while it has been thought that Ser5 phosphorylation is generally restricted to the promoters and 5′ region of these same genes. Recent studies, however, reveal that Ser5 phosphorylation is indeed found at significant levels throughout the transcribed regions of genes (1). Furthermore, a recent paper has noted that CTDK-I preferentially phosphorylates serine 2 of the heptad repeat only when the serine 5 position is already phosphorylated (16). Thus, it seems likely that doubly phosphorylated CTD epitopes exist in yeast that may serve to direct the interaction of Set2 (and perhaps other phospho-CTD-interacting proteins).
Based on our BLAST search analysis, the SRI domain appears to be conserved in other organisms only in proteins that have a domain architecture nearly identical to that of Set2 (those containing AWS or SAC, SET, Post-SET, and WW) (Table 1). We take these data to suggest that the SRI domain is a probable indicator of proteins that are bona fide functional homologs of Set2. Consistent with this, we have determined that a construct containing the SRI region of HYPB (Table 1) binds directly to the phosphorylated CTD. Furthermore, our studies also reveal the putative S. pombe homolog of Set2 (Table 1) to be a robust K36 HMT that also interacts with RNAPII (S. A. Morris and B. D. Strahl, unpublished data). Based on these observations, it is likely that Set2 and K36 methylation have a conserved function in transcription elongation.
Role for H3 K36 methylation in transcription elongation.
Previous studies have implicated Set2 in the transcription elongation process. This has been established not only by the numerous biochemical and genetic analyses that have been performed on Set2, but also by the use of 6AU, a drug commonly used as an indicator for elongation defects. The compound 6AU depletes the available nucleotide pool by inhibition of the enzyme IMP dehydrogenase, which is responsible for catalyzing the rate-limiting step in de novo synthesis of GTP (8, 14, 15). Inhibition of IMP dehydrogenase results in a challenge to the transcription elongation machinery, which is continually in need of free nucleotides. Thus, strains with altered transcription elongation function display a sensitivity to the drug different from their WT parent strain. Our 6AU assays revealed that set2 deletion mutants or deletion of the SRI domain itself results in resistance to the drug, whether the URA3 gene is genomically inserted or carried ectopically on a plasmid (Fig. 5A). This is in contrast to some previous studies that showed that set2 mutants are modestly sensitive to 6AU (19, 22, 33). While it is unclear why varied 6AU results have been reported, it is noteworthy that others have also seen a 6AU resistance phenotype with set2 deletion mutants (21, 43; D. Stillman, personal communication), indicating that the differences may lie in context-specific differences in the genetic backgrounds used.
In addition to set2 deletion, resistance to 6AU has also been observed in strains mutated for a number of other factors, including Ess1, Bye1, Chd1, Isw1, and Fkh1 (2, 40, 41). Many of these factors have been shown to influence transcription elongation via regulation of specific phases in the transcription elongation cycle, such as the transition between elongation and initiation or termination (24, 25, 41). While 6AU phenotypes alone are not sufficient to allow conclusions regarding the mechanistic details of how a protein affects the elongation phase of transcription, 6AU resistance can be interpreted as a consequence of increased elongation efficiency resulting from the deletion of a factor which negatively regulates elongation. Indeed, it was found that strains with the Ess1 suppressor, Bye1, deleted displayed resistance to 6AU, and this protein may negatively regulate transcription elongation by inducing RNAPII pausing at elongation arrest sites (41). In addition, Chd1 null strains were found to exhibit 6AU resistance, which may be due to an indirect effect on elongation by its influence on termination (2, 40).
In an effort to better understand the nature of the 6AU phenotype of the set2 deletion mutant, we examined the distribution of RNAPII along genes. Interestingly, we found that RNAPII levels significantly increased at the 3′ ends of genes in the absence of Set2 or when methylation was inhibited (Fig. 5C and 6D). Furthermore, this increase generally paralleled K36 methylation levels, indicating the possibility of a direct mechanistic link between the levels of K36 methylation and RNAPII occupancy (Fig. 4C and 5C). While our results strongly implicate K36 methylation as having a direct role in RNAPII elongation, the precise role is currently unclear. We propose three possibilities: (i) K36 methylation generates a chromatin structure that is more permissive for RNAPII passage. In this scenario, the loss of Set2/K36 methylation would result in a chromatin structure that is more difficult for RNAPII to pass, which might appear as an increase in RNAPII occupancy along the gene through increased pausing. (ii) K36 methylation generates chromatin structure that is less permissive for RNAPII passage. In this case, K36 methylation acts as a negative regulator of RNAPII elongation, and the loss of this “mark” permits increased RNAPII passage. However, since the termination event at the 3′ end would be under tight checkpoint control and would be rate limiting, the loss of K36 methylation would result in a buildup or backlog of RNAPII across the gene, while overall transcript production remained consistent. Such a buildup of RNAPII might be viewed as advantageous to cells when presented with 6AU stress. Finally, (iii) termination efficiency is diminished in the absence of K36 methylation. Thus, similar to chd1 mutants, the reduced efficiency of termination would result in a backlog of RNAPII across the gene. Among these possibilities, we suggest scenario ii is most likely, given the 6AU resistance of set2Δ strains and the fact that other factors whose deletion results in 6AU resistance have been shown to be negative regulators of RNAPII elongation. In summary, our results provide strong evidence that K36 methylation, mediated by the SRI-dependent association of Set2 with RNAPII, plays a role in the elongation phase of transcription. Whether Set2's elongation role is mediated through the association of factors that bind to K36 methylation, or through the ability of this modification to control chromatin structure by regulating nucleosome-nucleosome or nucleosome-DNA interactions, is unknown but will be of keen interest for future studies.
Acknowledgments
This work is supported by NIH grants GM068088 (B.D.S.) and GM40505 (A.G.) and a grant from the American Heart Association (0465506U) to B.D.S. B.D.S. is a Pew Scholar in the Biomedical Sciences.
We thank C. D. Allis for providing the H3 S10A mutant plasmid, D. Bentley for providing the CTD antibody used for ChIP, S. Dent for providing the H3/H4 shuffle strain WZY42 and plasmids, M. Green for the TBP antiserum, M. MacDonald for the original HYPB clone, and T. Tsukiyama for the p3Flag-KanMX plasmid. We are grateful to J. Greenblatt, N. Krogan, and D. Stillman for sharing unpublished results as well as B. Temple for assistance in BLAST database searches. We also thank J. Lieb, M. Hall, G. Sancar, and Strahl lab members for helpful discussions and comments on the manuscript, as well as M. Hall for mass spectrometry analysis.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Alen, C., N. A. Kent, H. S. Jones, J. O′Sullivan, A. Aranda, and N. J. Proudfoot. 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10:1441-1452. [DOI] [PubMed] [Google Scholar]
- 3.Archambault, J., F. Lacroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857-862. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrd, K. N., and A. Shearn. 2003. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc. Natl. Acad. Sci. USA 100:11535-11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmondson, D. G., M. M. Smith, and S. Y. Roth. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 10:1247-1259. [DOI] [PubMed] [Google Scholar]
- 8.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22:9-11. [DOI] [PubMed] [Google Scholar]
- 9.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerber, M., and A. Shilatifard. 2003. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 278:26303-26306. [DOI] [PubMed] [Google Scholar]
- 11.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 12.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 14.Hyle, J. W., R. J. Shaw, and D. Reines. 2003. Functional distinctions between IMP dehydrogenase genes in providing mycophenolate resistance and guanine prototrophy to yeast. J. Biol. Chem. 278:28470-28478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, R. C., G. Weber, and H. P. Morris. 1975. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature 256:331-333. [DOI] [PubMed] [Google Scholar]
- 16.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 279:24957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 19.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landry, J., A. Sutton, T. Hesman, J. Min, R. M. Xu, M. Johnston, and R. Sternglanz. 2003. Set2-catalyzed methylation of histone H3 represses basal expression of GAL4 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:5972-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 23.Mandal, S. S., H. Cho, S. Kim, K. Cabane, and D. Reinberg. 2002. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 22:7543-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morillon, A., N. Karabetsou, J. O'Sullivan, N. Kent, N. Proudfoot, and J. Mellor. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115:425-435. [DOI] [PubMed] [Google Scholar]
- 25.Morillon, A., J. O′Sullivan, A. Azad, N. Proudfoot, and J. Mellor. 2003. Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science 300:492-495. [DOI] [PubMed] [Google Scholar]
- 26.Morris, D. P., J. M. Lee, D. E. Sterner, W. J. Brickey, and A. L. Greenleaf. 1997. Assaying CTD kinases in vitro and phosphorylation-modulated properties of RNA polymerase II in vivo. Methods 12:264-275. [DOI] [PubMed] [Google Scholar]
- 27.Morris, D. P., H. P. Phatnani, and A. L. Greenleaf. 1999. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-end formation. J. Biol. Chem. 274:31583-31587. [DOI] [PubMed] [Google Scholar]
- 28.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447-456. [DOI] [PubMed] [Google Scholar]
- 29.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 30.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 31.Phatnani, H. P., J. C. Jones, and A. L. Greenleaf. 2004. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry 43:15702-15719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riles, L., R. J. Shaw, M. Johnston, and D. Reines. 2004. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaft, D., A. Roguev, K. M. Kotovic, A. Shevchenko, M. Sarov, A. Shevchenko, K. M. Neugebauer, and A. F. Stewart. 2003. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, R. J., J. L. Wilson, K. T. Smith, and D. Reines. 2001. Regulation of an IMP dehydrogenase gene and its overexpression in drug-sensitive transcription elongation mutants of yeast. J. Biol. Chem. 276:32905-32916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shilatifard, A. 2004. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim. Biophys. Acta 1677:79-86. [DOI] [PubMed] [Google Scholar]
- 37.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 38.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl, B. D., P. A. Grant, S. D. Briggs, Z. W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodage, T., M. A. Basrai, A. D. Baxevanis, P. Hieter, and F. S. Collins. 1997. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 94:11472-11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, X., A. Rossettini, and S. D. Hanes. 2003. The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics 165:1687-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25:637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]