Abstract
Myocardin and the myocardin-related transcription factors (MRTFs) MRTF-A and MRTF-B are coactivators for serum response factor (SRF), which regulates genes involved in cell proliferation, migration, cytoskeletal dynamics, and myogenesis. MRTF-A has been shown to translocate to the nucleus and activate SRF in response to Rho signaling and actin polymerization. Previously, we described a muscle-specific actin-binding protein named striated muscle activator of Rho signaling (STARS) that also activates SRF through a Rho-dependent mechanism. Here we show that STARS activates SRF by inducing the nuclear translocation of MRTFs. The STARS-dependent nuclear import of MRTFs requires RhoA and actin polymerization, and the actin-binding domain of STARS is necessary and sufficient for this activity. A knockdown of endogenous STARS expression by using small interfering RNA significantly reduced SRF activity in differentiated C2C12 skeletal muscle cells and cardiac myocytes. The ability of STARS to promote the nuclear localization of MRTFs and SRF-mediated transcription provides a potential muscle-specific mechanism for linking changes in actin dynamics and sarcomere structure with striated muscle gene expression.
Serum response factor (SRF) is a MADS-box transcription factor that regulates genes involved in cell proliferation, migration, cytoskeletal dynamics, and myogenesis by binding a conserved DNA sequence [CC(A/T)6GG], known as a CArG box or serum response element (20). The ability of SRF to distinguish between different target genes depends on the presence of binding sites for other transcription factors and the number of CArG boxes, as well as the association of SRF with a plethora of cofactors, many of which are cell type specific and signal responsive (4, 7, 8; reviewed in reference 31).
At least two signaling pathways are known to modulate SRF activity, one involving the phosphorylation of ternary complex factors (TCFs) of the Ets domain family and another controlled by Rho family small GTPases and actin dynamics (9, 11, 13, 14, 22, 29). Phosphorylation of TCFs by mitogen-activated protein kinase signaling facilitates their association with SRF on target genes that contain TCF-binding sites adjacent to the CArG box (14). Association of TCFs with SRF can stimulate or repress SRF activity, depending on the promoter (14, 22, 35). In contrast, the activation of SRF target genes in response to Rho signaling is dependent on actin polymerization and occurs through a TCF-independent mechanism (11, 13, 29).
Myocardin and the myocardin-related transcription factors (MRTFs) MRTF-A (MAL/MKL1) and MRTF-B (MKL2) comprise a family of transcriptional coactivators that associate with SRF and stimulate SRF-dependent transcription (5, 12, 21, 32, 33; reviewed in references 6 and 34). Myocardin is expressed specifically in cardiac and smooth muscle cells and is sufficient and necessary for the activation of smooth muscle genes that are SRF dependent (10, 16, 32, 36). In contrast, MRTFs are expressed in a broad range of cell types and regulate numerous genes involved in cell proliferation and migration (5, 12, 27, 28, 33).
SRF activity is exquisitely sensitive to the state of actin polymerization. G-actin monomers inhibit SRF activity, whereas polymerization of actin in response to serum stimulation and RhoA signaling stimulates SRF activity by depleting the cellular pool of inhibitory G-actin (9, 13, 25, 29). Recently, MRTF-A/MAL was shown to associate with G-actin, resulting in its sequestration in the cytoplasm of NIH 3T3 cells. Serum stimulation and Rho signaling result in the translocation of MRTF-A to the nucleus and the activation of SRF target genes (21).
RhoA signaling has been implicated in the differentiation of smooth, cardiac, and skeletal muscle cells, as well as in cardiomyocyte hypertrophy (1, 2, 10, 17, 18, 26, 30, 37, 38). Dominant-negative RhoA can reduce myogenic differentiation, whereas activated RhoA can enhance myocyte differentiation and growth (30, 38). The Rho effector p160 rho kinase/ROCK and actin treadmilling are necessary for smooth muscle cell differentiation (17, 18), and RhoA signaling has been shown to induce the nuclear import of MRTF-A in smooth muscle cells, thereby triggering smooth muscle gene activation (10).
We have described an actin-binding protein, named striated muscle activator of Rho signaling (STARS), which is specifically expressed in cardiac and skeletal muscle (3). STARS stimulates SRF-dependent transcription through a mechanism involving actin polymerization and Rho activation, but the precise molecular mechanisms by which STARS activates SRF-dependent transcription have not been previously defined. In light of recent evidence that MRTF-A can translocate to the nucleus in response to Rho signaling, we investigated whether MRTF-A or -B might serve as a link between STARS and SRF. Here we show that STARS induces the nuclear accumulation of MRTF-A and -B and stimulates SRF-dependent transcription through a mechanism requiring Rho-actin signaling. These findings reveal a muscle-specific signaling mechanism that links actin dynamics with SRF-dependent transcription and have implications for understanding the regulation of muscle gene expression during development and disease.
MATERIALS AND METHODS
Plasmids.
Expression vectors encoding STARS, MRTF-A and -B, dominant-negative myocardin, and C3 transferase were described previously (3, 32, 33). N-terminal deletion mutants of MRTF-A and -B lack the RPEL domains and begin at amino acids 92 and 152, respectively. The luciferase reporter plasmids containing the SM22 promoter and tandem copies of the SM22 CArG (4× SM22-CArG-luciferase) were reported previously (15). The expression vectors for wild-type β-actin (pEF-FLAG-actin) and its mutant derivatives, actins R62D and S14C, kindly provided by R. Treisman, were described previously (25).
Cell culture and transfection.
NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% calf serum, and COS1, 293T, and HeLa cells were grown in DMEM with 10% fetal bovine serum. Transfections were performed with Fugene (Roche) according to the manufacturer's instructions. For immunocytochemistry, cells plated on glass coverslips in six-well dishes were shifted to DMEM without serum 12 h after transfection. Twenty-four hours later, cells were stimulated with 20% fetal bovine serum and immunocytochemistry was performed. Unless indicated otherwise, 500 ng of each plasmid was used. For luciferase assays, cells grown in 12-well dishes were shifted to DMEM without serum 12 h after transfection. Two days later, cells were lysed and reporter gene expression was assayed. Unless indicated otherwise, 200 ng of luciferase reporter plasmid and 100 ng of each expression plasmid were used. The total amount of DNA per well was kept constant by adding expression vectors without a cDNA insert. An RSV promoter-driven lacZ expression plasmid was included for all transfections as an internal control. Cytochalasin D (CD; 1 μM) (Sigma-Aldrich) and latrunculin B (LB; 0.5 μM) (Calbiochem) were added 12 h after transfection, and 10% ethanol (final concentration, 0.01%) was added to control wells.
The C2C12 mouse myoblast line was maintained with DMEM with 20% fetal bovine serum. Differentiation was induced in C2C12 cells grown in 12-well dishes at about 90% confluence by replacing medium with DMEM containing 2% horse serum. Primary neonatal rat ventricular myocyte culture was performed as described previously (3).
Luciferase assays.
Cells were harvested and luciferase and control β-galactosidase activities were measured according to the manufacturer's instructions (Promega). All assays were performed at least twice with triplicates for each assay.
Immunocytochemistry.
Cells were rinsed with phosphate-buffered saline (PBS), fixed with 4% formaldehyde for 10 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min. Cells were blocked for 30 min at room temperature in 3% bovine serum albumin in PBS. Cells were then incubated with primary antibody diluted in 3% bovine serum albumin in PBS for 60 min at room temperature. Anti-Myc polyclonal antibody (1:200; Santa Cruz) and anti-FLAG M2 monoclonal antibody (1:1,000; Sigma-Aldrich) were used. Secondary antibodies were rabbit anti-mouse immunoglobulin G fluorescein isothiocyanate or Texas Red (Vector Lab) used at a 1:200 dilution. Vectashield medium with DAPI (4′,6′-diamidino-2-phenylindole) (Vector) was used for mounting.
Immunoprecipitations and immunoblots.
For immunoprecipitations, 293T cells were grown in 6-cm-diameter dishes and transfected with several expression vectors by using Fugene. Twenty-four hours after transfection, the medium was replaced with DMEM without serum and further maintained for 24 h. Cells were then rinsed once with ice-cold phosphate-buffered saline and lysed in 1 ml of immunoprecipitation buffer (PBS containing 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail purchased from Sigma). The lysates were sonicated and cleared by centrifugation. FLAG-MRTF-A, FLAG-MRTF-B, or FLAG-β-actin was immunoprecipitated with 1 μl of anti-FLAG antibody (M2; Sigma) and protein G-Sepharose (Zymed). Beads were washed three times with immunoprecipitation buffer, and the proteins were resolved on a sodium dodecyl sulfate-polyacrylamide gel. The proteins were immunoblotted with anti-Myc (1:2,000 dilution; Santa Cruz), anti-actin (1:500 dilution; Chemicon), or anti-FLAG antibodies (1:5,000 dilution) and horseradish-conjugated secondary antibodies at a 1:5,000 dilution. The signals were visualized with Western blotting luminol reagent (Santa Cruz).
For STARS expression analysis in HeLa cells, cells were lysed in 100 μl of immunoprecipitation buffer. Twenty microliters of the lysates were immunoblotted by anti-Myc or anti-α-tubulin (Sigma) antibodies.
RNA interference.
For RNA interference analysis of STARS, a 21-nucleotide small interfering RNA (siRNA) duplex with two deoxythymidine bases as 3′ overhangs corresponding to STARS-specific sequence 5′-CCA GGT TAC TTT CGG AGA A-3′ was synthesized. BLOCK-iT fluorescent oligo (Invitrogen) was used as a nonspecific control. For luciferase assays, the transfection of 100 pmol of siRNA and 500 ng of 4× SM22-CArG-luciferase reporter plasmid was performed in undifferentiated C2C12 cells in DMEM with 20% fetal bovine serum at 60% confluence, differentiating C2C12 cells 2 days after the induction of differentiation or primary neonatal rat ventricular myocytes by using Fugene. An RSV promoter-driven lacZ expression plasmid was included for all transfections as an internal control. Cells were harvested for luciferase assay 60 h after transfection. To verify the efficiency of siRNA-mediated knockdown of STARS expression, HeLa cells maintained in six-well dishes with DMEM containing 10% fetal bovine serum were transfected with 100 ng of myc-STARS expression vector and 200 pmol of siRNA. Cells were harvested 48 h after transfection for Western blot analysis.
RESULTS
STARS activates SRF via MRTFs.
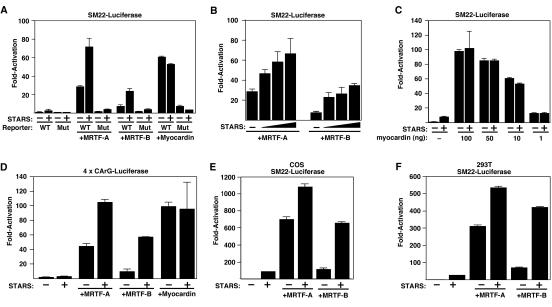
In light of the role of actin dynamics in the control of SRF activity and recent studies implicating MRTF-A as a transducer of Rho-actin signals to SRF, we investigated whether STARS, a muscle-specific actin-binding protein and activator of RhoA and SRF (3), might stimulate SRF activity via MRTFs. As shown in Fig. 1A and B, STARS and MRTF-A or -B synergistically activated a luciferase reporter controlled by the SM22 promoter, which contains a pair of SRF-binding sites (15), in serum-starved NIH 3T3 cells. In contrast, STARS did not enhance the transcriptional activities of various amounts of myocardin (Fig. 1A and C). The responsiveness of the SM22 promoter to STARS and MRTFs was abolished when the two SRF-binding sites were mutated (Fig. 1A). STARS also enhanced the transcriptional activities of MRTF-A and -B on a reporter plasmid containing four tandem copies of a CArG box from the SM22 promoter and the Elb minimal promoter (Fig. 1D). Thus, the CArG box is necessary and sufficient for the synergistic transcriptional activity of STARS and MRTFs. The synergistic activation by STARS and MRTFs was also observed in other cell types, including COS1 and 293T cells (Fig. 1E and F).
FIG. 1.
Stimulation of MRTF activity by STARS. nsrsid4287733\delrsid4287733 NIH 3T3 cells were transiently transfected with expression vectors encoding STARS, MRTF-A (20 ng), MRTF-B (100 ng), or myocardin (10 ng) and SM22-luciferase (A, B, and C) or 4× CArG-luciferase (D) as described in Materials and Methods. (B) Cells were transfected with expression vectors encoding STARS (10, 50, and 100 ng) and MRTF-A (20 ng) or MRTF-B (100 ng). (C) Cells were transfected with the indicated doses of expression vectors encoding myocardin with STARS (100 ng). Values are expressed as the levels of activation (n-fold) of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars). COS1 (E) and 293T (F) cells were transiently transfected with expression vectors encoding STARS, MRTF-A, or MRTF-B and SM22-luciferase as described above. Values are expressed as the levels of activation (n-fold) of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars).
STARS simulates nuclear translocation of MRTFs.
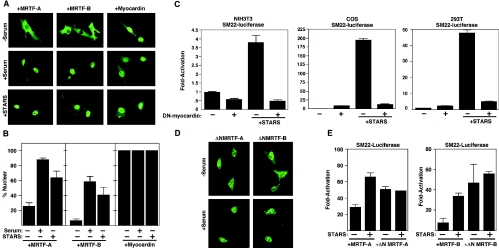
Consistent with previous reports (10, 21), MRTF-A was predominantly localized to the cytoplasm of NIH 3T3 cells in the absence of serum and accumulated in the nucleus in response to serum stimulation (Fig. 2A and B). MRTF-B also underwent nuclear translocation in response to serum stimulation, although it showed less responsiveness to serum than MRTF-A (Fig. 2A and B). Myocardin was localized predominately to the nucleus of NIH 3T3 cells irrespective of the presence or absence of serum (Fig. 2A).
FIG. 2.
Nuclear translocation of MRTFs in response to STARS. (A) NIH 3T3 cells were transiently transfected with expression vectors encoding FLAG-tagged MRTFs or myocardin in the presence or absence of 20% serum as indicated. In the bottom panels, cells maintained in the absence of serum were transfected with expression vectors encoding MRTFs, myocardin, and STARS as indicated. The subcellular distributions of MRTFs and myocardin were determined by immunostaining for the FLAG epitope. (B) The percentage of transfected cells from panel A that contained MRTFs or myocardin in the nucleus in the presence or absence of 20% serum or STARS was determined. At least 150 transfected cells were counted under each condition. Values are means ± standard deviations (error bars). (C) NIH 3T3, COS1, and 293T cells were transiently transfected with expression vectors encoding STARS or dominant-negative (DN) myocardin and SM22-luciferase as described in Materials and Methods. Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars). (D) NIH 3T3 cells were transiently transfected with expression vectors encoding FLAG-tagged MRTF mutants lacking the N-terminal RPEL domains in the presence or absence of 20% serum as indicated. The subcellular distribution of ΔN-MRTFs was determined by immunostaining for the FLAG epitope. (E) NIH 3T3 cells were transiently transfected with expression vectors encoding MRTFs or ΔN-MRTFs with or without STARS and SM22-luciferase as indicated. Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars).
In NIH 3T3 cells cotransfected with expression plasmids encoding MRTFs and STARS, the MRTFs were translocated to the nucleus in the absence of serum (Fig. 2A and B). The nuclear localization of myocardin was unchanged in the absence or presence of STARS. Thus, STARS can substitute for serum stimulation and promote the nuclear translocation of MRTFs with consequent activation of SRF-dependent transcription.
To confirm that MRTFs mediate STARS activity to induce SRF-dependent transcription, we next coexpressed STARS with a dominant-negative myocardin mutant, which can inhibit the transcriptional activities of myocardin and MRTF-A and -B, in several cell types. As shown in Fig. 2C, dominant-negative myocardin completely blocked the ability of STARS to induce SRF-dependent transcription in NIH 3T3, COS1, and 293T cells (Fig. 2C). Dominant-negative myocardin also inhibited the synergistic activity of STARS and MRTFs in NIH 3T3 cells (data not shown). STARS did not alter the level of expression of MRTFs, suggesting that STARS stimulates SRF-dependent transcription solely by promoting the nuclear translocation of MRTF-A and -B.
The N-terminal region of MRTF-A has been reported to sequester MRTF-A in the cytoplasm of serum-starved NIH 3T3 cells (21). Accordingly, deletion mutants of MRTF-A or -B lacking the conserved N-terminal regions (ΔN-MRTF-A or ΔN-MRTF-B) were constitutively localized to the nuclei of NIH 3T3 cells in the absence of serum or STARS (Fig. 2D). These N-terminal deletion mutants activated SRF-dependent reporters more effectively than the wild-type proteins in the absence of STARS, and STARS did not further enhance their transcriptional activities (Fig. 2E). Together, these results strongly support the notion that the nuclear translocation of MRTFs mediates the stimulatory effect of STARS on SRF activity.
Actin-binding activity of STARS is necessary and sufficient for nuclear accumulation of MRTF-A and -B.
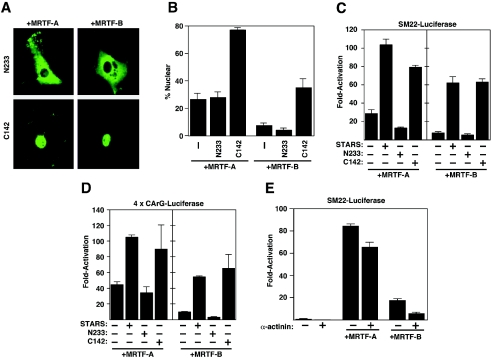
To further explore the mechanism through which STARS stimulates SRF activity, we tested the effects of a series of STARS mutants on MRTF nuclear translocation and activation of SRF-dependent transcription. The STARS protein contains 375 amino acids with the conserved actin-binding domain contained within the C-terminal 142 residues (3). The STARS C-terminal deletion mutant N233, which cannot bind actin or activate SRF, failed to induce the nuclear accumulation of MRTF-A and -B (Fig. 3A and B). In contrast, the C-terminal 142 amino acids of STARS (C142), which bind actin and stimulate SRF (3), induced the nuclear accumulation of MRTFs as efficiently as full-length STARS (Fig. 3A and B).
FIG. 3.
Actin-binding activity of STARS is necessary and sufficient for stimulation of MRTF activity. (A) NIH 3T3 cells were transiently transfected with expression vectors encoding FLAG-tagged MRTFs and the N233 or C142 STARS mutants as indicated. The subcellular distribution of MRTFs was determined by immunostaining for the FLAG epitope. (B) The percentages of transfected cells from panel A that contained MRTFs in the nucleus in the presence or absence of the N233 and C142 STARS mutants were determined. At least 150 transfected cells were counted under each condition. Values are means ± standard deviations (error bars). NIH 3T3 cells were transiently transfected with expression vectors encoding FLAG-tagged MRTFs and wild-type STARS or N233 and C142 STARS mutants and SM22-luciferase (C) or 4× CArG-luciferase (D) in the absence of serum as indicated. Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars). (E) NIH 3T3 cells were transiently transfected with expression vectors encoding α-actinin and STARS and SM22-luciferase in the absence of serum as indicated. Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars).
There was a precise correlation between the abilities of STARS mutants to induce MRTF nuclear localization and SRF-dependent transcription. STARS N233 failed to enhance MRTF-dependent activation of SRF-dependent reporters, whereas STARS C142 synergistically enhanced MRTF-mediated transcription to the same level as full-length STARS (Fig. 3C and D). These results demonstrate that the actin-binding domain of STARS is necessary and sufficient for the nuclear accumulation and transcriptional activation of MRTFs by STARS. To rule out the possibility that the activity of STARS is a general nonspecific response to the overexpression of an actin-binding protein, we tested whether α-actinin, an unrelated actin-binding protein, affected MRTF-mediated transcription. As shown in Fig. 3E, the overexpression of α-actinin failed to stimulate SRF-dependent transcription either alone or in combination with MRTFs. Thus, the effects of STARS appear to be specific.
Involvement of actin dynamics and Rho in the activity of STARS.
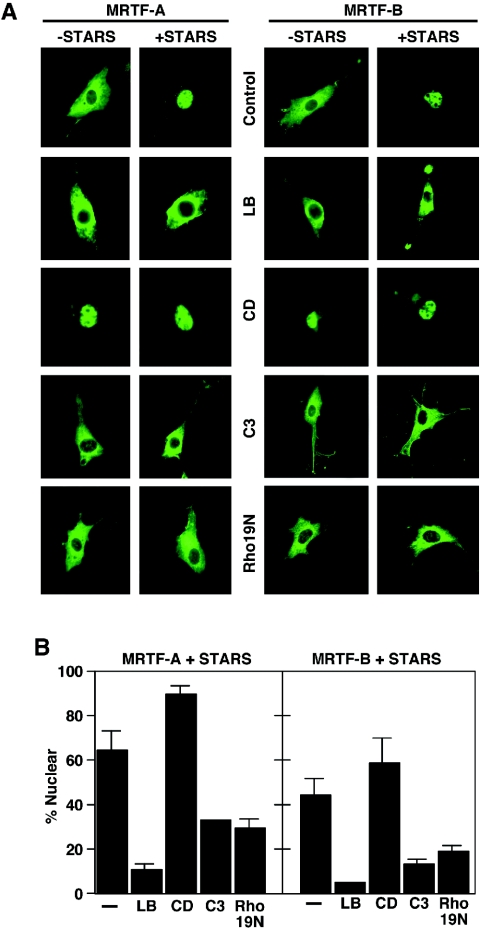
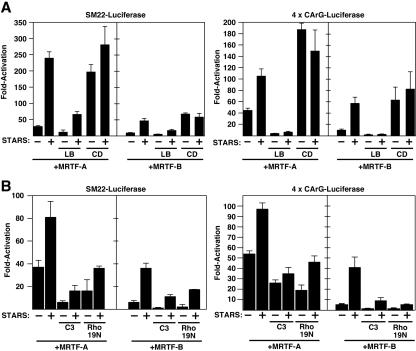
To assess the potential involvement of actin dynamics and Rho signaling in STARS-induced nuclear translocation and transcriptional activation of MRTFs, we examined the effects of a series of agents that affect actin polymerization on STARS activity. Treatment of NIH 3T3 cells with latrunculin B, which sequesters actin monomers and prevents Rho-dependent nuclear accumulation of MRTF-A and SRF activation (21), blocked nuclear accumulation of MRTF-A and -B in the presence of STARS (Fig. 4A and B). Conversely, cytochalasin D, which dimerizes actin but prevents actin polymerization and activates SRF, strongly induced the nuclear translocation of MRTFs, even in the absence of STARS (Fig. 4A and B). Consistent with their effects on MRTF nuclear import, LB significantly blocked the stimulatory effect of STARS on MRTF-dependent transcription and CD enhanced the activity of MRTFs alone (Fig. 5A). These results support the notion that actin dynamics are involved in the STARS-induced nuclear accumulation of MRTFs and transcriptional activation of SRF via MRTFs.
FIG. 4.
Effects of actin-modifying agents on the subcellular localization of MRTFs in the presence and absence of STARS. (A) NIH 3T3 cells transiently transfected with expression vectors encoding FLAG-tagged MRTFs with and without STARS were treated with LB (0.5 μM) or CD (1 μM) or cotransfected with vectors encoding C3 transferase (200 ng) or Rho19N (1 μg) in the absence of serum as indicated. The subcellular distribution of MRTFs was determined by immunostaining for the FLAG epitope. (B) The percentage of transfected cells from panel A that contained MRTFs in the nucleus in the presence of STARS was determined. At least 150 transfected cells were counted under each condition. Values are means ± standard deviations (error bars).
FIG. 5.
Effects of actin-modifying agents on the transcriptional activity of MRTFs in the presence and absence of STARS. NIH 3T3 cells were transiently transfected with expression vectors encoding STARS and MRTFs and SM22-luciferase or 4× CArG-luciferase as indicated. (A) Cells were treated with LB or CD as indicated. (B) Cells were cotransfected with expression vectors encoding C3 transferase (50 ng) or Rho19N (1 μg) as indicated. Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars).
Because Rho activity is crucial for actin dynamics, we next examined the potential involvement of RhoA in STARS-induced nuclear accumulation and enhancement of transcriptional activity of MRTFs. Expression of C3 transferase, which ADP-ribosylates and inactivates Rho, inhibited the nuclear accumulation of MRTFs and the stimulatory effect of STARS on the transcriptional activity of MRTFs, as did the dominant-negative RhoA mutant Rho19N (Fig. 4A and B and 5B). Although STARS requires Rho activity to induce actin treadmilling and MRTF nuclear translocation and the inhibition of Rho activity blocks STARS activity, assays of RhoA activity in STARS-transfected cells showed no difference from that in untransfected cells (data not shown). Thus, STARS does not appear to function as an upstream activator of Rho but requires Rho-actin signaling and changes in actin dynamics to evoke its stimulatory effects on MRTFs and SRF activity.
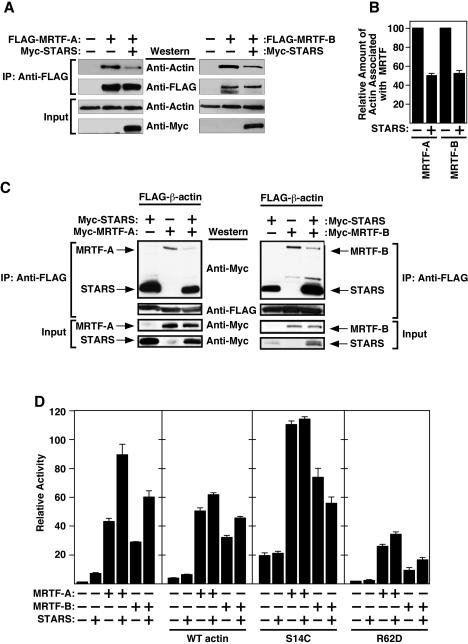
STARS promotes the dissociation of MRTFs from actin.
MRTF-A was previously shown to associate with actin (24). Activation of Rho signaling and actin treadmilling dissociate actin from MRTF-A and induce the nuclear accumulation of MRTF-A (21). Given the actin-binding and actin-polymerizing activities of STARS, we examined whether STARS might promote the nuclear import of MRTFs by dissociating MRTFs from actin. As shown in Fig. 6A and B, endogenous actin was immunoprecipitated together with FLAG-MRTF-A and FLAG-MRTF-B in extracts from 293T cells. The presence of STARS caused a decrease in the amount of endogenous actin coimmunoprecipitated with MRTFs, suggesting that STARS leads to dissociation of actin-MRTF complexes. Indeed, the amount of Myc-MRTFs coimmunoprecipitated with FLAG-β-actin was also reduced in the presence of STARS (Fig. 6C). No direct physical interaction between MRTFs and STARS was observed in coimmunoprecipitation experiments (data not shown).
FIG. 6.
Modulation of MRTF activity by STARS and actin. (A) STARS dissociates MRTFs from endogenous actin. 293T cells were transfected with 1 μg of expression vectors encoding FLAG-MRTF with or without Myc-STARS. Cell extracts were immunoprecipitated (IP) with anti-FLAG, and proteins were analyzed by Western blotting with the indicated antibodies. (B) The relative amounts of actin immunoprecipitation with MRTFs in panel A were determined by densitometry. Values are expressed as means ± standard deviations (error bars). (C) STARS leads to dissociation of the MRTF-actin complex. 293T cells were transfected with expression vectors encoding FLAG-β-actin (1 μg) with myc-MRTFs (4 μg) and/or myc-STARS (100 ng). Cell extracts were immunoprecipitated (IP) with anti-FLAG, and proteins were analyzed by Western blotting with the indicated antibodies. (D) Effects of various actin mutants on STARS-induced activation of MRTFs-mediated transcription. NIH 3T3 cells were transiently transfected with the expression vectors encoding STARS, MRTFs, and wild-type (WT) actin or various mutants of actin (S14C or R62D) and the SM22-luciferase reporter. Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene alone ± standard deviations (error bars).
As it was shown that unpolymerized G-actin controls MRTF activity (21) and we have previously shown that STARS induces actin polymerization (3), we hypothesized that STARS activates MRTFs by depleting the G-actin pool. We therefore tested the effects of the various actin mutants on STARS-induced activation of MRTF-mediated transcription. Expression of wild-type actin, which increases the amount of G-actin but does not alter the F-actin/G-actin ratio, reduced the ability of STARS to activate MRTF-dependent transcription, though it did not significantly alter the activity of MRTF in the absence of STARS. The expression of an actin S14C mutant, which favors F-actin formation and increases the F-actin/G-actin ratio (25), stimulated MRTF activity even in the absence of STARS and abolished further activation of MRTFs by STARS. In contrast, the actin R62D mutant, which is unable to polymerize and decreases the F-actin/G-actin ratio, inhibited MRTF activity and also reduced the ability of STARS to enhance MRTF activity (Fig. 6D). Together, these results support the notion that STARS stimulates MRTF activity by inducing the dissociation of MRTFs from actin by depleting the G-actin pool.
Inhibition of STARS reduces SRF activity in muscle cells.
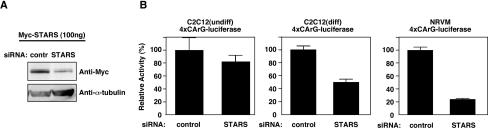
The experiments described above showed that forced expression of STARS, a muscle-specific protein, could stimulate MRTF/SRF activity in nonmuscle cells. To determine the potential role of STARS in SRF-mediated transcription in muscle cells, we knocked down endogenous STARS expression in C2C12 skeletal muscle cells and in rat cardiac myocytes by using siRNA. The effectiveness of STARS siRNA, which consisted of conserved sequences of mouse and rat STARS mRNA, was tested by overexpressing STARS in transfected HeLa cells in the presence of STARS siRNA and a nonspecific control siRNA. As shown in Fig. 7A, STARS siRNA effectively knocked down STARS protein expression.
FIG. 7.
STARS siRNA reduced SRF activity in muscle cells. (A) HeLa cells were transiently transfected with an expression vector encoding myc-STARS and STARS siRNA or control nonspecific siRNA. Cell lysates were analyzed by Western blotting with anti-myc or anti-α-tubulin antibodies. (B) STARS siRNA was cotransfected with 4× CArG-luciferase in undifferentiated (undiff) C2C12 cells, differentiated (diff) C2C12 cells (2 to 5 days after induction of differentiation), or primary neonatal rat ventricular myocytes (NRVM). Values are expressed as the levels of activation of luciferase expression above the level with the reporter gene with control siRNA ± standard deviations (error bars).
In C2C12 cells, STARS expression is induced 2 days after serum deprivation as myoblasts differentiate into myotubes (3). STARS siRNA efficiently reduced the expression of a luciferase reporter gene controlled by tandem SRF-binding sites in differentiated C2C12 cells 2 to 5 days after the induction of differentiation but not in undifferentiated C2C12 cells that do not express STARS (Fig. 7B). STARS siRNA also significantly reduced the expression of this reporter in primary neonatal rat ventricular myocytes, which express STARS (Fig. 7B). These results support the conclusion that endogenous STARS participates in activation of SRF in muscle cells.
DISCUSSION
STARS is a muscle-specific actin-binding protein capable of stimulating SRF-dependent transcription through a mechanism involving RhoA and actin polymerization (3). We speculated previously that STARS activates SRF by depleting the G-actin pool and that the activity of STARS may be mediated by an unidentified intermediary protein. The results of the present study demonstrate that STARS activates SRF-dependent transcription by inducing the nuclear accumulation of MRTF-A and -B through a mechanism dependent on RhoA and actin polymerization. These findings identify MRTF-A and -B as a link between STARS and SRF.
STARS induces nuclear translocation of MRTF-A and -B.
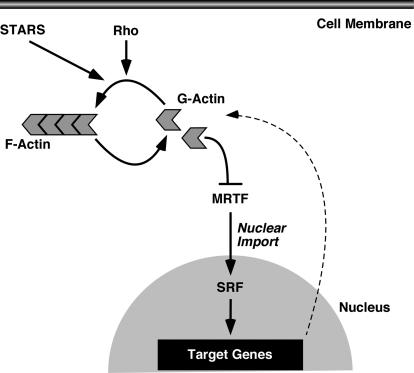
Consistent with recent reports that serum induces SRF-mediated transcription through the RhoA-dependent nuclear accumulation of MRTF-A (10, 21), our results show that STARS can substitute for serum signals and promote the nuclear translocation of MRTF-A and -B with consequent activation of SRF-dependent transcription. Stimulation of MRTF nuclear translocation by STARS requires only the actin-binding domain of STARS and is inhibited by LB, which sequesters G-actin monomers. Similarly, the inhibition of RhoA activity with C3 transferase or the expression of dominant-negative RhoA prevents the nuclear accumulation of MRTFs and their stimulation of SRF. Rho-actin signaling appears to promote the nuclear accumulation of MRTFs, at least in part, by enhancing actin polymerization and the depletion of monomeric G-actin with consequent dissociation of G-actin from MRTF-A and nuclear accumulation of MRTF-A (21). Indeed, MRTF-A/MAL was recently reported to interact directly with G-actin (24). As STARS directly associates with actin and appears to enhance actin polymerization, we favor the notion that STARS promotes the nuclear accumulation of MRTFs by depletion of the G-actin pool (Fig. 8). The ability of LB, which sequesters G-actin monomers, to block nuclear localization of MRTFs in response to STARS and the ability of STARS to disrupt the association of actin with MRTFs are consistent with this notion.
FIG. 8.
A model to explain the stimulation of MRTF activity by STARS. STARS binds to actin and promotes actin polymerization in the presence of basal Rho activity, which releases MRTFs from the inhibitory influence of G-actin, allowing the nuclear import of MRTFs and stimulation of SRF activity. SRF activates actin genes, which allows a negative-feedback loop.
The N-terminal regions of MRTFs contain a series of conserved amino acids referred to as RPEL motifs, which have been shown to sequester the MRTFs in the cytoplasm by association with actin (21, 25). Consistent with the possibility that STARS promotes the nuclear import of MRTFs by displacing them from monomeric G-actin, the RPEL motifs are required for the effects of STARS on MRTFs. The RPEL region of myocardin is divergent from those of MRTF-A and -B and does not retain myocardin in the cytoplasm. Hence, myocardin is localized constitutively to the nucleus and its activity is not enhanced by STARS.
Implications for muscle gene regulation during development and disease.
The cardiac and skeletal muscle specificities of STARS expression and the finding that STARS siRNA reduced SRF-mediated transcription in these cell types suggest that STARS provides a muscle-specific mechanism for SRF activation. This mechanism can be deployed in nonmuscle cells by the forced expression of STARS, as shown by its ability to stimulate MRTF nuclear import and SRF activity in a variety of nonmuscle cell types.
It is noteworthy that SRF regulates the expression of cytoskeletal actin genes and is itself a target of regulation by actin dynamics. Thus, the regulation of SRF activity by actin dynamics provides a regulatory loop whereby the cell can modulate the expression of genes encoding components of the actin cytoskeleton in response to changes in cytoskeletal dynamics. STARS may modulate this regulatory loop for muscle cells. Indeed, not only β-actin but skeletal muscle α-actin, which is encoded by an SRF target gene, also can bind to MRTF-A (24). In muscle cells, STARS may promote polymerization of muscle-specific actin and prevent unpolymerized actin from inhibiting MRTF activity, thereby maintaining SRF-induced transcription of muscle-specific actin genes. Because myocardin is insensitive to the effects of STARS, its target genes would be expected to be highly active irrespective of the polymerization state of actin, although STARS would be expected to further augment the expression of these genes through its actions on MRTF-A and -B, which are also expressed in cardiac muscle, and which form heterodimers with myocardin.
The ability of STARS to modulate SRF activity via MRTFs also has several interesting implications for striated muscle development and disease. For example, Rho signaling stimulates differentiation of cardiac and skeletal muscle cells, and MRTF-B has been shown to be necessary for skeletal muscle differentiation (28, 30, 37, 38). Thus, the up-regulation of STARS during myogenesis may provide a feed-forward mechanism for enhancing the activation of MRTF/SRF target genes and reinforcing the differentiation process. It is also well established that perturbation of the sarcomere can lead to pathological hypertrophy of cardiac myocytes with activation of fetal cardiac genes, many of which are regulated by SRF (reviewed in reference 23). The mechanisms whereby changes in sarcomere structure and function trigger changes in gene expression in the nucleus are unclear. The ability of STARS to interconnect actin dynamics with transcription via MRTFs provides a potential mechanism to account for these observations. It is notable in this regard that STARS has recently been reported to be up-regulated in the heart during pathological hypertrophy (19). Thus, the perturbation of the STARS-MRTF/SRF regulatory loop has potentially interesting therapeutic implications.
Acknowledgments
We thank A. Tizenor for assistance with graphics, J. Page for editorial assistance, and Richard Treisman for actin expression plasmids.
This research was supported by grants from the National Institutes of Health, the Donald W. Reynolds Cardiovascular Clinical Research Center, the Robert A. Welch Foundation, the Muscular Dystrophy Association, and the Texas Advanced Technology Program to E.N.O. G.C.T.P. was supported by a postdoctoral fellowship from the National Institutes of Health. K.K. was supported by a research fellowship from the Uehara Memorial Foundation.
REFERENCES
- 1.Aikawa, R., I. Komuro, T. Yamazaki, Y. Zou, S. Kudoh, W. Zhu, T. Kadowaki, and Y. Yazaki. 1999. Rho family small G proteins play critical roles in mechanical stress-induced hypertrophic responses in cardiac myocytes. Circ. Res. 84:458-466. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., S. Izumo, and J. Sadoshima. 1998. Angiotensin II activates RhoA in cardiac myocytes: a critical role of RhoA in angiotensin II-induced premyofibril formation. Circ. Res. 82:666-676. [DOI] [PubMed] [Google Scholar]
- 3.Arai, A., J. A. Spencer, and E. N. Olson. 2002. STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J. Biol. Chem. 277:24453-24459. [DOI] [PubMed] [Google Scholar]
- 4.Belaguli, N. S., J. L. Sepulveda, V. Nigam, F. Charron, M. Nemer, and R. J. Schwartz. 2000. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol. 20:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cen, B., A. Selvaraj, R. C. Burgess, J. K. Hitzler, Z. Ma, S. W. Morris, and R. Prywes. 2003. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 23:6597-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cen, B., A. Selvaraj, and R. Prywes. 2004. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J. Cell. Biochem. 93:74-82. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell 4:107-118. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and R. J. Schwartz. 1996. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell. Biol. 16:6372-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland, J. W., and R. Treisman. 2002. The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol. Biol. Cell 13:4088-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du, K. L., M. Chen, J. Li, J. J. Lepore, P. Mericko, and M. S. Parmacek. 2004. Megakaryoblastic leukemia factor-1 transduces cytoskeletal signals and induces smooth muscle cell differentiation from undifferentiated embryonic stem cells. J. Biol. Chem. 279:17578-17586. [DOI] [PubMed] [Google Scholar]
- 11.Gineitis, D., and R. Treisman. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 12.Han, Z., X. Li, J. Wu, and E. N. Olson. 2004. A myocardin-related transcription factor regulates activity of serum response factor in Drosophila. Proc. Natl. Acad. Sci. USA 101:12567-12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, C. S., J. Wynne, and R. Treisman. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159-1170. [DOI] [PubMed] [Google Scholar]
- 14.Janknecht, R., W. H. Ernst, V. Pingoud, and A. Nordheim. 1993. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 12:5097-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, L., Z. Liu, B. Mercer, P. Overbeek, and E. N. Olson. 1997. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev. Biol. 187:311-321. [DOI] [PubMed] [Google Scholar]
- 16.Li, S., D. Wang, Z. Wang, J. Richardson, and E. N. Olson. 2003. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. USA 100:9366-9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, J., T. E. Landerholm, J. S. Wei, X. R. Dong, S. P. Wu, X. Liu, K. Nagata, M. Inagaki, and M. W. Majesky. 2001. Coronary smooth muscle differentiation from proepicardial cells requires rhoA-mediated actin reorganization and p160 rho-kinase activity. Dev. Biol. 240:404-418. [DOI] [PubMed] [Google Scholar]
- 18.Mack, C. P., A. V. Somlyo, M. Hautmann, A. P. Somlyo, and G. K. Owens. 2001. Smooth muscle differentiation marker gene expression is regulated by RhoA-mediated actin polymerization. J. Biol. Chem. 276:341-347. [DOI] [PubMed] [Google Scholar]
- 19.Mahadeva, H., G. Brooks, D. Lodwick, N. W. Chong, and N. J. Samani. 2002. ms1, a novel stress-responsive, muscle-specific gene that is up-regulated in the early stages of pressure overload-induced left ventricular hypertrophy. FEBS Lett. 521:100-104. [DOI] [PubMed] [Google Scholar]
- 20.Miano, J. M. 2003. Serum response factor: toggling between disparate programs of gene expression. J. Mol. Cell. Cardiol. 35:577-593. [DOI] [PubMed] [Google Scholar]
- 21.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329-342. [DOI] [PubMed] [Google Scholar]
- 22.Murai, K., and R. Treisman. 2002. Interaction of serum response factor (SRF) with the Elk-1 B box inhibits RhoA-actin signaling to SRF and potentiates transcriptional activation by Elk-1. Mol. Cell. Biol. 22:7083-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olson, E. N., and, M. D., Schneider. 2003. Sizing up the heart: development redux in disease. Genes Dev. 17:1937-1956. [DOI] [PubMed] [Google Scholar]
- 24.Posern, G., F. Miralles, S. Guettler, and R. Treisman. 2004. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 23:3973-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posern, G., A. Sotiropoulos, and R. Treisman. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 13:4167-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sah, V. P., M. Hoshijima, K. R. Chien, and J. H. Brown. 1996. Rho is required for Galphaq and alpha1-adrenergic receptor signaling in cardiomyocytes. Dissociation of Ras and Rho pathways. J. Biol. Chem. 271:31185-31190. [DOI] [PubMed] [Google Scholar]
- 27.Selvaraj, A., and R. Prywes. 2004. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaraj, A., and R. Prywes. 2003. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J. Biol. Chem. 278:41977-41987. [DOI] [PubMed] [Google Scholar]
- 29.Sotiropoulos, A., D. Gineitis, J. Copeland, and R. Treisman. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98:159-169. [DOI] [PubMed] [Google Scholar]
- 30.Takano, H., I. Komuro, T. Oka, I. Shiojima, Y. Hiroi, T. Mizuno, and Y. Yazaki. 1998. The Rho family G proteins play a critical role in muscle differentiation. Mol. Cell. Biol. 18:1580-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 32.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 33.Wang, D. Z., S. Li, D. Hockemeyer, L. Sutherland, Z. Wang, G. Schratt, J. A. Richardson, A. Nordheim, and E. N. Olson. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99:14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, D. Z., and E. N. Olson. 2004. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr. Opin. Genet. Dev. 14:558-566. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Z., D. Z. Wang, D. Hockemeyer, J. McAnally, A. Nordheim, and E. N. Olson. 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428:185-189. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z., D. Z. Wang, G. C. Pipes, and E. N. Olson. 2003. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. USA 100:7129-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei, L., L. Wang, J. A. Carson, J. E. Agan, K. Imanaka-Yoshida, and R. J. Schwartz. 2001. Beta1 integrin and organized actin filaments facilitate cardiomyocyte-specific RhoA-dependent activation of the skeletal alpha-actin promoter. FASEB J. 15:785-796. [DOI] [PubMed] [Google Scholar]
- 38.Wei, L., W. Zhou, J. D. Croissant, F. E. Johansen, R. Prywes, A. Balasubramanyam, and R. J. Schwartz. 1998. RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J. Biol. Chem. 273:30287-30294. [DOI] [PubMed] [Google Scholar]