Abstract
The alc gene cluster of Bordetella pertussis includes three genes, alcA, alcB, and alcC, which are involved in alcaligin siderophore biosynthesis in response to iron starvation. The production of AlcA, AlcB, and AlcC in Bordetella cells and the transcriptional organization of alcA, alcB, and alcC were investigated by using a set of three alc′-′lacZ gene fusion constructs that were contiguous with the known promoter upstream of alcA and extended to fusion junctions within each alc cistron. All three alc′-′lacZ fusions exhibited iron-repressible reporter gene expression which was abolished by deletion of the 105-bp alcA promoter-operator region. In an immunoblot analysis using a monoclonal antibody specific for β-galactosidase, the AlcA-LacZ, AlcB-LacZ, and AlcC-LacZ hybrid proteins were detected in Bordetella cells grown under iron-depleted conditions. A B. pertussis mutant in which the 105-bp alcA promoter-operator region was deleted by allelic exchange was unable to produce detectable levels of siderophore. Hybridization analysis using gene-specific probes showed that alc-specific transcript levels in the mutant were negligible compared with those of the wild-type parent. These results confirm that alcA, alcB, and alcC are cotranscribed from an iron-regulated control region immediately upstream of alcA. Transcript analysis using hybridization probes representing regions downstream of alcC demonstrated that alc transcription extends approximately 3.6 kb further downstream from the alcC coding region, suggesting the cotranscription of additional, uncharacterized alcaligin system genes.
To establish infection, pathogenic bacteria must successfully compete for a limited iron pool (9, 32). As a defense mechanism to prevent bacterial growth, the mammalian host maintains extremely low levels of free extracellular iron through the action of iron-binding proteins, such as transferrin and lactoferrin (4). One bacterial iron retrieval strategy involves the secretion of high-affinity iron-chelating siderophores (18, 25). Siderophores are produced in response to iron limitation and are capable of removing iron from host sources such as transferrin and lactoferrin (4, 20, 32).
Bordetella pertussis and Bordetella bronchiseptica are gram-negative bacterial pathogens that cause respiratory diseases in mammals. The native siderophore of both B. pertussis and B. bronchiseptica is the macrocyclic dihydroxamate alcaligin (8, 22) which is expressed in low-iron growth conditions and is under the control of the ferric uptake regulator protein, Fur (2, 6). The phenotypes of previously isolated B. bronchiseptica siderophore-deficient mutants suggested that multiple genes were involved in alcaligin biosynthesis (1); these mutants were adopted as tools with which to identify the homologous alcaligin biosynthesis genes in B. pertussis. Analysis of one class of mutants led to the identification of the Bordetella odc gene, which encodes an ornithine decarboxylase catalyzing the conversion of ornithine to putrescine, an essential alcaligin precursor (7). In related studies, a 4.5-kb BamHI-SmaI B. pertussis genomic DNA fragment which corresponded to the mutated chromosomal regions of three B. bronchiseptica siderophore mutants was identified (16). Mutant complementation analysis using subclones of the 4.5-kb region, nucleotide sequence analysis, and protein expression studies suggested the existence of a putative iron-responsive promoter upstream of three alcaligin biosynthesis genes, alcA, alcB, and alcC, which appeared to be organized in an operon (16). The deduced AlcA proteins of B. pertussis (16) and B. bronchiseptica (15) and the deduced AlcB and AlcC proteins of B. pertussis (16) share strong primary amino acid sequence similarities with IucD, IucB, and IucC, respectively, involved in the biosynthesis of the Escherichia coli siderophore aerobactin (19, 24). The transcription initiation site of alcA was mapped to a position adjacent to a putative Fur repressor binding site (16).
In previous studies, we and others readily visualized the iron-regulated AlcC protein expressed in B. pertussis and B. bronchiseptica, while expression of AlcA and AlcB was not apparent in either Bordetella cells or in E. coli by use of a T7 RNA polymerase-promoter protein expression system (16). Although genetic complementation results showing polarity of transposon insertion mutations on downstream alc genes and nucleotide sequence data suggested the transcriptional linkage of alcA, alcB, and alcC, conclusive evidence for the cotranscription of these genes was still required, and the 3′ genetic limit of the operon remained unknown. Potentially, the operon may include additional, as-yet-undefined genes downstream of alcC. In this study, we examined the alc operon, using reporter gene fusion constructs and a B. pertussis alc promoter-operator region deletion mutant. We report the expression of AlcA, AlcB, and AlcC hybrid proteins in Bordetella cells and establish the existence of an iron-repressible operon transcribed from a promoter upstream of alcA.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. bronchiseptica B013N, a nalidixic acid-resistant derivative of wild-type strain B013 (1), and B. pertussis UT25Sm1, a streptomycin-resistant derivative of wild-type B. pertussis strain UT25 (12), have been described previously. E. coli DH5α [F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1] (Gibco BRL, Gaithersburg, Md.) was used as the host for general DNA manipulations. E. coli DH5α harboring pRK2013 (13) provided mobilization functions in triparental matings. Plasmid vectors pGEM3Z (Promega, Madison, Wis.), pBluescript SK+ (Stratagene, La Jolla, Calif.), and the broad-host-range plasmid vector pBBR1MCS (17) were used for the construction of recombinant plasmids, and suicide plasmid vector pSS1129 (30) was employed for allelic-exchange mutagenesis. β-Galactosidase translational fusions were constructed by using plasmids YIp356, YIp357, and YIp358R (23). Recombinant cosmid pCP1.11 (16) was the source of B. pertussis UT25 DNA for alcaligin system gene probes used in hybridization experiments; pBSK+4 contains a 4.5-kb BamHI-SmaI B. pertussis DNA subfragment of pCP1.11 encompassing alcA, alcB, and alcC (16).
Growth conditions.
E. coli was grown aerobically on Luria-Bertani (LB) medium (26); B. bronchiseptica and B. pertussis were cultured on LB agar and Bordet-Gengou agar (5), respectively. Iron-replete or iron-depleted modified Stainer-Scholte (SS) medium (28) was used for liquid culture as described previously (1). Growth was monitored with a Klett-Summerson colorimeter fitted with a no. 54 filter (Klett Manufacturing Co., Long Island City, N.Y.). Bordetella cells grown on agar plates were used to inoculate iron-replete SS broth seed cultures. Seed cultures were grown at 37°C with shaking, and the cells were harvested, washed twice with iron-depleted SS broth, and used to inoculate iron-replete or iron-depleted SS media to an initial density of 25 to 30 Klett units. For selection of plasmid-containing strains or selection of mutants constructed by allelic exchange, appropriate antibiotics were added to the culture media at the indicated concentrations (in micrograms per milliliter): ampicillin, 100 for E. coli and 50 for B. pertussis; chloramphenicol, 30; gentamicin, 10; kanamycin, 50; nalidixic acid, 35; streptomycin, 50; and tetracycline, 15.
General DNA manipulations.
Recombinant plasmid isolation, transformation of E. coli, restriction endonuclease analysis, and ligation of DNA fragments were performed as described previously (26). Transfer of plasmids from E. coli to Bordetella cells was carried out by triparental crosses as described by Brickman and Armstrong (7). Nucleotide sequencing using double-stranded plasmid templates was performed by the dideoxy chain termination method (27) as modified by DeShazer and coworkers (11), using [α-32P]dATP (ICN Radiochemicals, Irvine, Calif.) and a Sequenase version 2.0 kit (United States Biochemical Corp., Cleveland, Ohio). Southern and colony DNA hybridizations were performed under high-stringency conditions as described previously (26). Hybridization probes were labelled with [α-32P]dCTP (ICN Radiochemicals) by the random priming method using the Random Primers DNA Labeling System (Gibco BRL). B. pertussis chromosomal DNA was isolated by a method described previously (33).
Siderophore detection.
The chrome azurol S (CAS) universal siderophore detection assay (29) was performed to monitor siderophore production by Bordetella cells grown in iron-replete or iron-depleted SS medium by measuring the decrease in A630 of the CAS dye reaction as reported previously (1).
Construction of the alcA promoter-operator region deletion plasmid.
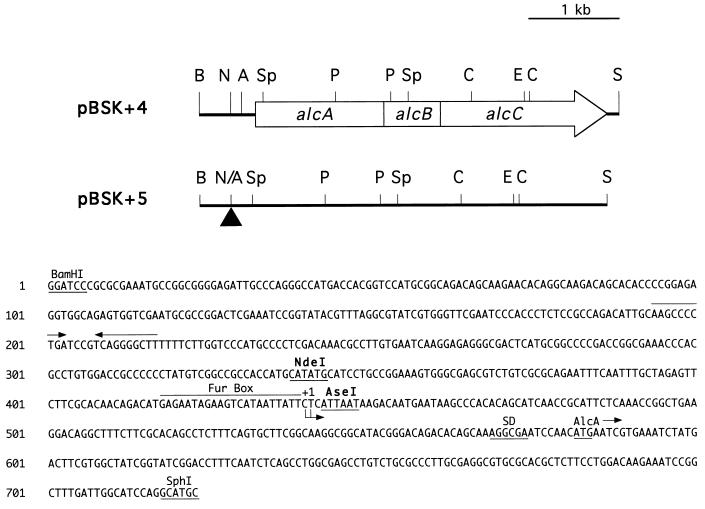
The 0.7-kb BamHI-SphI DNA fragment was isolated from pBSK+4 and digested with restriction endonucleases NdeI and AseI, which generated compatible cohesive ends. The 343-bp BamHI-NdeI and 280-bp AseI-SphI fragments were ligated with the vector pGEM3Z digested with BamHI and SphI, resulting in plasmid p3Z17. The resulting NdeI-AseI deletion removed the 105-bp alcA promoter-operator region containing the putative Fur binding site and transcription start site of alcA (16). The correct deletion and ligation were verified by nucleotide sequencing. The original 0.7-kb BamHI-SphI fragment of pBSK+4 was replaced with a 0.6-kb BamHI-SphI deletion derivative fragment isolated from p3Z17, generating plasmid pBSK+5 (see Fig. 1).
FIG. 1.
Schematic overview of the genetic organization of B. pertussis alcABC genes and the direction of transcription. The nucleotide sequence of the 723-bp BamHI-SphI DNA region used in the construction of the NdeI-AseI deletion of the alcA promoter-operator is shown below the diagram. The putative Fur repressor binding site (Fur Box), the alcA transcription initiation site determined previously (16) (+1), the position of a Shine-Dalgarno-like sequence (SD) upstream from the alcA open reading frame, and the upstream position of a putative transcription terminator (converging arrows) are indicated. Plasmid pBSK+5 carries the same insert DNA fragment as pBSK+4 but has the 105-bp NdeI-AseI region upstream of alcA deleted (triangle). Abbreviations for restriction endonuclease sites: A, AseI; B, BamHI; C, ClaI; E, EcoRI; N, NdeI; P, PvuII; S, SmaI; Sp, SphI; N/A, NdeI-AseI deletion junction.
Construction of protein fusions.
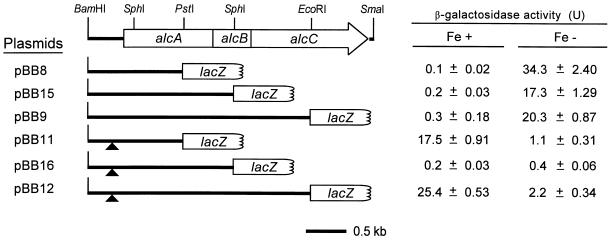
For the construction of alcA′-′lacZ, alcAB′-′lacZ, and alcABC′-′lacZ translational fusion plasmids, the 1.5-kb BamHI-PstI, 2.3-kb BamHI-SphI, and 3.6-kb BamHI-EcoRI alc DNA fragments isolated from pBSK+4 were ligated upstream of the promoterless ′lacZ genes of the vectors YIp357, YIp356, and YIp358R, respectively. Similarly, to construct the corresponding alcA promoter-operator region deletion derivatives, the 1.4-kb BamHI-PstI, 2.2-kb BamHI-SphI, and 3.5-kb BamHI-EcoRI DNA fragments isolated from deletion plasmid pBSK+5 were ligated in frame with the ′lacZ gene of the vectors YIp357, YIp356, and YIp358R, respectively. Because these vectors harbor the ColE1 origin of replication for maintenance in E. coli, the alc′-′lacZ fusions were subcloned as BamHI-ApaI fragments into the broad-host-range vector pBBR1MCS (17) for use in Bordetella species. The resultant plasmids were named pBB8, pBB15, pBB9, pBB11, pBB16, and pBB12 (see Fig. 2). In-frame fusion of each alc′-′lacZ construct was verified by nucleotide sequencing.
FIG. 2.
Iron-regulated expression and cotranscription of alcA, alcB, and alcC. Genetic maps of alcA′-′lacZ, alcAB′-′lacZ, and alcABC′-′lacZ translational fusions carried on the designated plasmids are shown. Deletions of the 105-bp NdeI-AseI fragment containing the alcA promoter-operator region shown in Fig. 1 are indicated (triangles). Levels of β-galactosidase expressed in wild-type B. bronchiseptica B013N containing alc′-′lacZ fusion plasmids in response to iron-replete (Fe+) and iron-depleted (Fe−) growth conditions are reported in Miller units (21) and are expressed as means of triplicate measurements (n = 3) ± standard deviations.
β-Galactosidase assays.
β-Galactosidase assays were performed by the method of Miller (21). B. bronchiseptica cells grown in iron-replete or iron-depleted SS medium were permeabilized with chloroform-sodium dodecyl sulfate (SDS). The enzyme activities were measured by cleavage of the chromogenic substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) and expressed in Miller units.
Immunoblot analysis.
Cultures grown under iron-replete and iron-depleted conditions were concentrated by centrifugation and each adjusted to an optical density at 600 nm equivalent to 5.0. A 50-μl volume of cell suspension was treated by being boiled for 5 min in digestion buffer consisting of 0.65% SDS, 6.26% glycerol, 6.25% 2-mercaptoethanol, 0.0025% bromophenol blue, 3% urea, and 0.125 M Tris (pH 6.8). Proteins were separated by SDS-polyacrylamide gel electrophoresis using 7.5% polyacrylamide gels containing 3% urea (28), transferred electrophoretically to nitrocellulose membranes as described by Towbin et al. (31), and processed as described previously (14). The membranes were blocked with 3% bovine serum albumin in 10 mM Tris–0.9% NaCl (pH 7.4) and incubated with a 1:5,000 dilution of mouse monoclonal antibody specific for β-galactosidase (Promega) and then a 1:2,000 dilution of peroxidase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.). The positive control was 1 μl of high-molecular-mass protein standards product (Bio-Rad Laboratories, Hercules, Calif.) containing β-galactosidase.
RNA preparation and analysis.
Total RNA of B. pertussis UT25Sm1 and mutant PM-4 was isolated by the acid guanidinium thiocyanate-phenol-chloroform extraction method (10) from cells grown under iron-replete or iron-depleted conditions as previously described (16).
For Northern hybridization, 50-μg samples of RNA were subjected to electrophoresis on 1.2% agarose-formaldehyde gels and transferred to nitrocellulose membranes as described elsewhere (26). For dot blot hybridizations, twofold dilutions of RNA samples (20 to 0.63 μg) were applied to a nitrocellulose membrane by using a 96-well vacuum manifold apparatus, and each well was rinsed twice with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membranes were baked at 80°C for 90 min, prehybridized, and incubated at 42°C with radiolabelled gene- or region-specific DNA probes in a solution containing 5× Denhardt’s solution, 50% formamide, 6× SSPE (1× SSPE is 0.15 M NaCl, 0.01 M NaH2PO4, and 1.25 mM EDTA), 0.5% SDS, and 100 μg of denatured sheared salmon sperm DNA per ml. Transcript levels were quantitated with a PhosphorImager (model 425E; Molecular Dynamics, Sunnyvale, Calif.). Alternatively, membranes were subjected to autoradiography, and quantitation of signal intensities was performed on a Macintosh PowerPC computer using the public domain NIH Image version 1.61 software package (developed at the National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
Construction of the B. pertussis alcA promoter-operator deletion mutant.
The 3.5-kb BamHI-EcoRI DNA fragment isolated from deletion plasmid pBSK+5 was subcloned to the suicide vector pSS1129 (30) and conjugally transferred to B. pertussis UT25Sm1 to transfer the mutation to the chromosome by homologous recombination. Presumptive mutants lacking the 105-bp alcA promoter-operator region were identified by colony hybridization using the 105-bp NdeI-AseI DNA fragment isolated from pBSK+4 as a probe. Correct allelic exchange in mutant PM-4 was verified by Southern hybridization analysis of chromosomal DNA using probes spanning the deletion junction.
RESULTS
Iron-regulated expression of alcA, alcB, and alcC.
Since expression of the alcA and alcB gene products was not detected in an earlier study (16), a set of β-galactosidase translational fusions was constructed. Each fusion carried DNA sequences contiguous with the known promoter upstream of alcA and extending to fusion junctions within each alc cistron. To investigate the potential cotranscription of alcA, alcB, and alcC directed by the promoter-operator located upstream of alcA, a 105-bp deletion (positions −100 to +5 relative to the alcA transcription start site) (Fig. 1) was introduced into each alc′-′lacZ fusion construct to produce the corresponding deletion set of fusions (Fig. 2). The 105-bp deletion encompasses the putative Fur binding and transcriptional start sites, yet does not impinge on the alcA coding region.
Wild-type B. bronchiseptica B013N harboring the fusion plasmid constructs was cultured in iron-replete or iron-depleted growth conditions to detect the expressed Alc-LacZ hybrid proteins by measurement of β-galactosidase fusion protein activity (Fig. 2). The iron starvation status of the cultures was monitored by measurement of siderophore activity in supernatants (data not shown). Bordetella cells containing pBB8, pBB15, and pBB9 expressed approximately 34-, 17-, and 20-fold increases in levels of β-galactosidase activity, respectively, under iron starvation growth conditions compared with iron-replete conditions. This iron-regulated expression of the Alc-LacZ hybrid proteins in Bordetella cells confirms the in vivo expression of the proteins encoded by alcA, alcB, and alcC. Moreover, deletion of the 105-bp alcA promoter-operator region in the alc fusion constructs (derivatives pBB11, pBB16, and pBB12) abolished the expression of β-galactosidase activities in B. bronchiseptica cells grown under iron-depleted conditions, indicating that the 105-bp region is required for iron-responsive transcription of not only alcA, but of alcB and alcC as well. This mutation also resulted in increased LacZ expression in cells carrying fusions pBB11 (encoding AlcA-LacZ) and pBB12 (AlcC-LacZ) under iron-replete, versus iron-depleted, growth conditions. However, cells carrying pBB16 (AlcB-LacZ), in which the same 105-bp DNA region was deleted, expressed negligible β-galactosidase activity, regardless of the iron status of the growth medium.
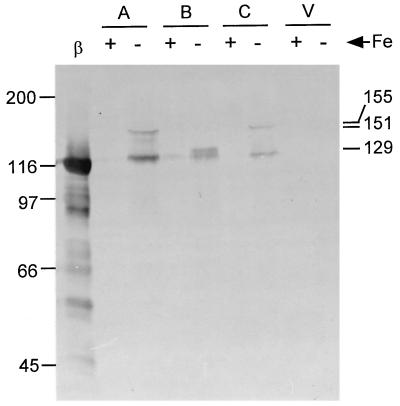
To visualize the AlcA-LacZ, AlcB-LacZ, and AlcC-LacZ hybrid proteins expressed in B. bronchiseptica, proteins from solubilized cells from the same cultures used for the β-galactosidase assays were separated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblot analysis using a monoclonal antibody specific for β-galactosidase. The immunoreactive AlcA-LacZ, AlcB-LacZ, and AlcC-LacZ hybrid proteins were detected in cells grown under iron-depleted conditions, and their apparent molecular masses were approximately 151, 129, and 155 kDa, respectively (Fig. 3). Little or no antibody reactivity was detected in cells grown in high-iron medium or in plasmid vector control samples. Densitometric analysis of the immunoblots showed that levels of fusion protein expression were proportional to the β-galactosidase enzyme activities measured in the cells (data not shown).
FIG. 3.
Immunoblot analysis of B. bronchiseptica B013N containing fusion plasmids. Cells grown in iron (Fe)-replete (+) and iron-depleted (−) conditions were subjected to immunoblot analysis as described in Materials and Methods. The Alc-LacZ hybrid proteins were visualized by reactivity with anti-β-galactosidase monoclonal antibody. Lanes: β, β-galactosidase positive control; A, cells carrying alcA′-′lacZ plasmid pBB8; B, alcAB′-′lacZ (pBB15); C, alcABC′-′lacZ (pBB9); V, vector plasmid pBBR1MCS. The relative migration positions of protein standards (left) and estimated molecular masses of Alc-LacZ hybrid proteins (right) are indicated in kilodaltons.
On the basis of the nucleotide sequences of the alc′-′lacZ fusions, the calculated molecular masses of AlcA-LacZ, AlcB-LacZ, and AlcC-LacZ are 152.1 kDa (1,332 amino acid residues), 128.8 kDa (1,133 amino acid residues), and 155.2 kDa (1,366 amino acid residues), respectively. The observed and calculated molecular masses determined from these studies are therefore consistent with the predicted translation start codons for the alcA, alcB, and alcC open reading frames identified in our previous studies (16). The immunoreactive species of approximately 120 kDa detected in cells containing the alc′-′lacZ constructs is hypothesized to be a degradation product and may correspond to the LacZ portion of the fusion proteins.
Construction and transcriptional analysis of a B. pertussis alcA promoter-operator region deletion mutant.
To further establish the role of the alcA promoter in directing cotranscription of alcABC and to evaluate transcription of the entire operon, the 105-bp alcA promoter-operator region deletion was introduced into the chromosome of B. pertussis by allelic exchange. The mutant, PM-4, was unable to produce detectable levels of alcaligin siderophore (data not shown), indicating that the chromosomal deletion of 105 bp upstream of alcA abrogated the expression of alcaligin biosynthesis genes, consistent with the results observed in the alc′-′lacZ reporter gene plasmid experiments. Supplying alcABC in trans as the 4.5-kb BamHI-SmaI fragment restored siderophore activity to PM-4 (data not shown).
Results from this study and previous work (16) for the alcaligin gene cluster were consistent with a polycistronic transcriptional organization for alcA, alcB, and alcC. To provide biochemical evidence for the proposed operonic structure of the alcABC region, transcript analysis was performed using total RNA isolated from both wild-type B. pertussis and B. pertussis alc promoter-operator mutant PM-4 grown in high- and low-iron medium.
(i) Northern blot analysis.
RNA samples from wild-type cells and PM-4 were subjected to Northern hybridization analysis using the following DNA probes (Fig. 4A) specific for each alc gene: alcA, 0.5-kb SphI-PstI fragment; alcB, 0.2-kb PstI-SphI fragment; and alcC, 0.6-kb internal ClaI fragment. While strong hybridization signals were detected for the RNA samples isolated from wild-type cells grown in low-iron conditions, no signal was observed for RNA samples from wild-type cells grown in high-iron conditions or from the promoter deletion mutant grown in either low- or high-iron conditions (data not shown). However, the sizes of the RNA transcripts from iron-starved wild-type cells could not be determined with confidence because of apparent rapid turnover of alc mRNA. Therefore, RNA dot hybridization using gene- and region-specific probes was employed as an alternative approach to quantitate alc-specific messages.
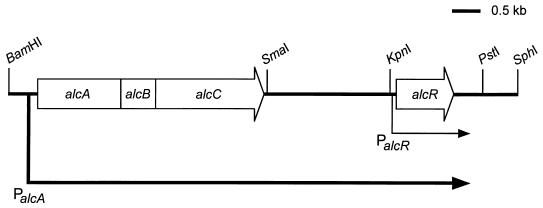
FIG. 4.
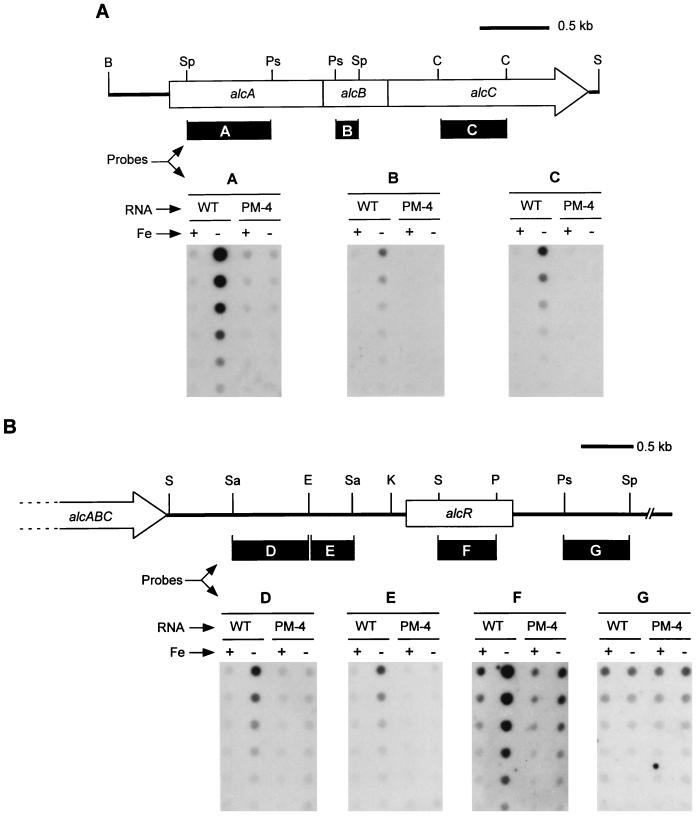
RNA dot hybridization showing transcriptional linkage of alcA, alcB, and alcC and the 3′ limit of the alc operon. RNA was isolated from wild-type B. pertussis UT25Sm1 (WT) and its isogenic alcA promoter deletion mutant (PM-4) cultured in parallel under high- and low-iron conditions (Fe+ or −, respectively). Twofold serial dilutions of denatured RNA samples (from 20 to 0.63 μg) were applied to the nitrocellulose membranes. (A) DNA fragments used as probes and derived from each alc gene are indicated (solid bars): A, 770-bp SphI-PstI fragment; B, 220-bp PstI-SphI fragment; and C, 620-bp ClaI fragment. (B) Physical map of the DNA region downstream of alcABC, including the alcR gene. DNA fragments representing subregions downstream from alcC which were used as probes are indicated (solid bars): D, 700-bp SacI-EcoRI fragment; E, 400-bp EcoRI-SacI fragment; F, 530-bp SmaI-PvuII fragment; G, 600-bp PstI-SphI fragment. Abbreviations: B, BamHI; C, ClaI; E, EcoRI; K, KpnI; P, PvuII; Ps, PstI; S, SmaI; Sa, SacI; Sp, SphI.
(ii) Cotranscription of alcA, alcB, and alcC from the alcA promoter.
RNA samples isolated from both wild-type cells and the promoter deletion mutant PM-4 grown in low- and high-iron media were hybridized with the DNA probes specific for alcA, alcB, and alcC. In the RNA samples from wild-type cells grown in low-iron conditions, alcA, alcB, and alcC transcripts were detected, whereas few or no alc-specific transcripts were detected in RNA preparations from these cells grown in high-iron conditions. RNA samples isolated from mutant PM-4 cultured under either low- or high-iron conditions hybridized weakly, if at all, with the alcA, alcB, and alcC probes (Fig. 4A). Densitometric analysis of the autoradiograms showed at least a 3- to 10-fold increase in the levels of alcA, alcB, and alcC transcripts from iron-starved wild-type cells versus those grown in high-iron conditions, confirming the iron-regulated transcription of alcA, alcB, and alcC noted in our previous studies (16). Further, these results establish that alcA, alcB, and alcC are cotranscribed from an iron-regulated promoter-operator region upstream of alcA. Deletion of this promoter region abrogates transcription of these three alc genes, which comprise all or part of the known alcaligin biosynthesis operon.
(iii) Determination of the 3′ genetic limit of the alcABC-containing operon.
To determine the 3′ limit of the alcABC-containing operon, RNA dot hybridization was performed using the RNA from wild-type B. pertussis and mutant PM-4 with DNA probes representing genetic regions downstream of alcC. Probes derived from a 0.7-kb SacI-EcoRI DNA region (0.6 kb downstream from alcC) and a 0.4-kb EcoRI-SacI DNA region (1.3 kb downstream of alcC) hybridized with the RNA samples in an iron-repressible pattern essentially the same as that observed in the dot blots using the alcA, alcB, and alcC probes. Iron-regulated transcripts were detected in wild-type cells but were negligible in B. pertussis mutant PM-4 RNA samples (Fig. 4B, probes D and E). Densitometric analysis of the autoradiograms also revealed at least a three- to fivefold increase in levels of alc region transcripts from wild-type cells grown in low-iron over high-iron conditions, similar to the patterns observed for alcA, alcB, and alcC transcripts.
We have obtained the nucleotide sequence of a 1.6-kb KpnI-PstI fragment located 2 kb downstream of alcC and identified a gene, alcR, which is involved in the regulation of alcaligin siderophore system genes (3). A DNA probe derived from a 0.5-kb SmaI-PvuII fragment internal to alcR hybridized strongly with RNA from wild-type cells grown in low-iron conditions compared with the results for transcripts from iron-starved mutant PM-4 (Fig. 4B, probe F). Quantitative analysis of multiple hybridization experiments (including the data set shown in Fig. 4B) consistently showed that the levels of transcripts detected in iron-starved wild-type cells were elevated approximately fourfold over transcripts detected in iron-starved mutant PM-4 (data not shown), indicating that transcription of alcR is also under control of the alcA promoter. Interestingly, although the level of alcR transcripts detected in mutant PM-4 was reduced due to deletion of the alcA promoter-operator region, significant residual iron-regulated alcR transcription was consistently observed. Approximately twofold-higher levels of alcR transcripts were observed in low-iron conditions than in high-iron conditions in the absence of a functional alcA promoter. These observations strongly suggest that alcR transcription is directed from the alcA promoter as well as from an iron-regulated secondary promoter unaffected by the deletion mutation in PM-4.
Downstream of alcR, a 0.6-kb PstI-SphI DNA fragment probe hybridized to all RNA samples isolated from both wild-type B. pertussis and mutant PM-4, regardless of iron status (Fig. 4B, probe G). This result indicates that alcR is most likely the last gene transcribed from the alcA promoter and is monocistronic with respect to the putative secondary promoter (Fig. 5).
FIG. 5.
Transcriptional organization of the alcaligin biosynthesis operon. Transcription from the alcA promoter (PalcA) extends 3.6 kb downstream from alcC. alcR is the last gene contained in the alc operon and is also transcribed from its own promoter (PalcR) (3).
DISCUSSION
Previous studies indicated that the three Bordetella alcaligin biosynthesis genes, alcA, alcB, and alcC, were carried on a 4.5-kb B. pertussis BamHI-SmaI DNA fragment and were likely organized as a polycistronic transcriptional unit (16). Because we were able to visualize only the AlcC protein in Bordetella cell preparations, in this study we constructed Alc-LacZ protein fusions to confirm the expression of the three Alc proteins. B. bronchiseptica cells harboring alc′-′lacZ translational fusions expressed iron-regulated hybrid proteins which were detected by β-galactosidase activity assays and immunoblot analysis using anti-β-galactosidase monoclonal antibody. Although these alc genes are cotranscribed, different levels of β-galactosidase activity and Alc-LacZ fusion proteins were observed. This result is likely due to differences in translation initiation efficiencies of each alc cistron or differential stabilities or enzymatic activities of the Alc-LacZ hybrid proteins. The observed molecular masses of the fusion proteins corresponded to the predicted masses of the native AlcA, AlcB, and AlcC proteins based on nucleotide sequence predictions (16).
The results of the present study unambiguously confirmed the transcriptional linkage of alcA, alcB, and alcC and localized the 3′ genetic limit of the alc operon. Deletion of the 105-bp DNA region encompassing the alcA promoter-operator abolished alcA′-′lacZ, alcAB′-′lacZ, and alcABC′-′lacZ reporter gene expression under iron-depleted growth conditions. However, this deletion had variable effects on Alc-LacZ hybrid protein activities under high-iron conditions. Deletion of the alcA promoter-operator region did not result in the apparent formation of a functional promoter from newly juxtaposed sequences at the deletion junction, and there are no apparent promoters located upstream. The variable β-galactosidase fusion expression levels observed under high-iron conditions may reflect differential stabilities of the three Alc-LacZ transcripts or hybrid proteins in the Bordetella host background. In the direct analysis of RNA transcripts, negligible levels of alcA, alcB, and alcC transcripts were observed in RNA isolated from the alcA promoter deletion mutant PM-4 grown in low-iron medium. Together, the results confirm that the three alcaligin biosynthesis genes, alcA, alcB, and alcC, are cotranscribed from the iron-regulated alcA promoter-operator region.
RNA analyses suggested that the regulatory gene alcR, located 2.1 kb downstream of alcC, is included in the alc operon, because deletion of the alcA promoter region resulted in significantly lower abundance of alcR transcripts. A 0.6-kb PstI-SphI DNA probe immediately downstream of alcR hybridized uniformly to all RNA samples isolated from both the wild type and PM-4, regardless of iron status, indicating that alcR is likely to represent the 3′-terminal gene of the alc operon. The fact that iron-regulated transcription of alcR was decreased in PM-4 but not abrogated (as was observed with the upstream alcABC genes) suggested that it has its own secondary promoter. Indeed, primer extension analysis of alcR revealed iron-regulated transcription from an initiation site immediately upstream of this gene and adjacent to potential Fur binding sequences (3). The 2.1-kb region between alcC and alcR has not yet been fully characterized. On the basis of the hypothetical alcaligin biosynthesis pathway, at least one other enzyme activity is predicted to be required for the complete synthesis of alcaligin (16). Procaryotic genes encoding activities which function in related cellular processes are most often organized in polycistronic operons where transcription is most efficiently regulated from a single control region. Therefore, it is likely that these predicted enzyme activities are encoded in the region downstream of alcC, making this iron-responsive operon dedicated to alcaligin biosynthesis and regulatory functions.
ACKNOWLEDGMENTS
We thank Timothy J. Brickman for helpful discussion, assistance with densitometry, and provision of plasmids for construction of the translational fusions. We also acknowledge Fiona Beaumont for sharing alcR sequence information.
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
ADDENDUM IN PROOF
After submission of this paper, a study reporting the transcriptional linkage of the B. bronchiseptica alcABC genes was published (P. C. Giardina, L.-A. Foster, S. I. Toth, B. A. Roe, and D. W. Dyer, Gene 194:19–24, 1997). Pradel and coworkers have also identified the Bordetella alcR gene and determined the nucleotide sequence of the alcC-alcR intergenic region (E. Pradel, N. Guiso, and C. Locht, J. Bacteriol. 180:871–880, 1998).
REFERENCES
- 1.Armstrong S K, Clements M O. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J Bacteriol. 1993;175:1144–1152. doi: 10.1128/jb.175.4.1144-1152.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall B, Sanden G N. Cloning and initial characterization of the Bordetella pertussis fur gene. Curr Microbiol. 1995;30:1–4. doi: 10.1007/BF00293637. [DOI] [PubMed] [Google Scholar]
- 3.Beaumont F C, Kang H Y, Brickman T J, Armstrong S K. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezkorovainy A. Iron proteins. In: Bullen J J, Griffiths E, editors. Iron and infection. New York, N.Y: John Wiley and Sons; 1987. pp. 27–67. [Google Scholar]
- 5.Bordet J, Gengou O. Le microbe de la coqueluche. Ann Inst Pasteur (Paris) 1906;20:731–741. [Google Scholar]
- 6.Brickman T J, Armstrong S K. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickman T J, Armstrong S K. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J Bacteriol. 1996;178:54–60. doi: 10.1128/jb.178.1.54-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman T J, Hansel J-G, Miller M J, Armstrong S K. Purification, spectroscopic analysis, and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. BioMetals. 1996;9:191–203. doi: 10.1007/BF00144625. [DOI] [PubMed] [Google Scholar]
- 9.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.DeShazer D, Wood G E, Friedman R L. Boiling eliminates artifact banding when sequencing double-stranded templates. BioTechniques. 1994;17:288–290. [PubMed] [Google Scholar]
- 12.Field L H, Parker C D. Differences observed between fresh isolates of Bordetella pertussis and their laboratory-passaged derivatives. In: Manclark C R, Hill J C, editors. International Symposium on Pertussis. U.S. Washington, D.C: Department of Health, Education, and Welfare; 1979. pp. 124–132. [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank D W, Parker C D. Interaction of monoclonal antibodies with pertussis toxin and its subunits. Infect Immun. 1984;46:195–201. doi: 10.1128/iai.46.1.195-201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giardina P C, Foster L-A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 16.Kang H Y, Brickman T J, Beaumont F C, Armstrong S K. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4884. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 18.Lankford C E. Bacterial assimilation of iron. Crit Rev Microbiol. 1973;2:273–331. [Google Scholar]
- 19.Martinez J L, Herrero M, de Lorenzo V. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J Mol Biol. 1994;238:288–293. doi: 10.1006/jmbi.1994.1290. [DOI] [PubMed] [Google Scholar]
- 20.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Moore C H, Foster L-A, Gerbig D G, Dyer D W, Gibson B W. Identification of alcaligin as the siderophore produced by Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 24.Neilands J B. Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-30) Can J Microbiol. 1992;38:728–733. doi: 10.1139/m92-119. [DOI] [PubMed] [Google Scholar]
- 25.Neilands J B. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider D R, Parker C D. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect Immun. 1982;38:548–553. doi: 10.1128/iai.38.2.548-553.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 30.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinberg E D. Acquisition of iron and other nutrients in vivo. In: Roth J A, Bolin C A, Brogden K A, Minion F C, Wannemuehler M J, editors. Virulence mechanisms of bacterial pathogens. 2nd ed. Washington, D.C: American Society for Microbiology; 1995. pp. 79–93. [Google Scholar]
- 33.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]