Summary
Primary human lung organoid-derived air-liquid interface (ALI) cultures serve as a physiologically relevant model to study human airway epithelium in vitro. Here, we present a protocol for establishing these cultures from cryopreserved human lung tissue. We describe steps for lung tissue cryostorage, tissue dissociation, lung epithelial organoid generation, and ALI culture differentiation. We also include quality control steps and technical readouts for monitoring virus response. This protocol demonstrates severe acute respiratory syndrome coronavirus 2 infection in these cultures as an example of their utility.
For complete details on the use and execution of this protocol, please refer to Diana Cadena Castaneda et al. (2023).1
Subject areas: Immunology, Microscopy, Organoids
Graphical abstract
Highlights
-
•
Human lung tissue dissection and tissue cryopreservation
-
•
Lung epithelium organoid generation from cryopreserved human lung tissue
-
•
Studying viral infection in organoid-derived air-liquid interface cultures
-
•
Viral response assessment via RNA-seq, flow cytometry, viral titer, and imaging
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Primary human lung organoid-derived air-liquid interface (ALI) cultures serve as a physiologically relevant model to study human airway epithelium in vitro. Here, we present a protocol for establishing these cultures from cryopreserved human lung tissue. We describe steps for lung tissue cryostorage, tissue dissociation, lung epithelial organoid generation, and ALI culture differentiation. We also include quality control steps and technical readouts for monitoring virus response. This protocol demonstrates severe acute respiratory syndrome coronavirus 2 infection in these cultures as an example of their utility.
Before you begin
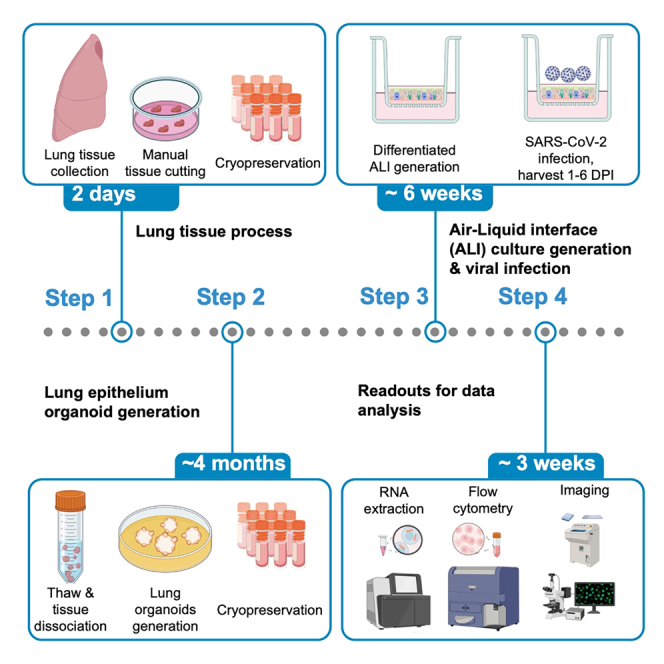
The protocol below describes the generation of primary human lung epithelial organoid-derived air-liquid-interface cultures from lung tissue and their use to investigate the impact and dynamics of infection with different SARS-CoV-2 variants. These cultures mimic in vivo airway epithelium architecture2,3,4,5,6,7 and find extensive application in studying responses to respiratory virus.8 We base the generation of lung epithelial organoids on the methodology from Sachs et al.9 And we will summarize it here. The study involves transcriptional analysis (Bulk RNA), flow cytometry (dissociated ALI), viral titer measurements (apical supernatant washes), and imaging to assess the outcomes. Perform all steps within a type II biological safety cabinet in a biological safety level 2 laboratory (BSL2). Conduct all work with SARS-CoV-2 variants under BSL3 conditions.
Institutional permissions
Obtention of de-identified human lung tissues from NDRI (Project: RPAK1 01) in compliance with relevant American institutional laws and NIH/NIAID guidelines. We conduct the study procedures in the context of the U19AI142733 grant at the Jackson Laboratory. Before performing these procedures, we obtain permission from the relevant institutions.
Human lung tissue processing & cryopreservation
Timing:2 days
-
1.On receipt of lung tissue, process immediately.Note: Throughout the process, we give attention to the lung tissue’s anatomical structure, facilitating dissection based on alveolar and bronchial areas.
-
a.Section a portion of the lung into approximately 3 cm × 3 cm pieces for embedding in OCT.
-
b.Snap-freeze in liquid nitrogen.
-
c.Mince the remaining tissue into smaller pieces.
-
d.Place around 25 of these small pieces in each cryovial.
-
e.Cryopreserve in FBS with 10% DMSO.
-
f.Store in liquid nitrogen before further processing.Note: This procedure ensures an appropriate preservation of human lung tissue for organoid generation or other applications (See Figure 1).
-
a.
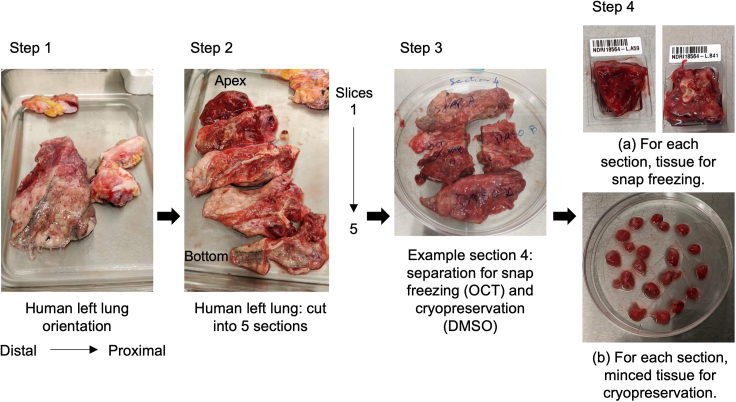
Figure 1.
Setup for human lung tissue processing and cryopreservation: Representative pictures for lung tissue processing
(Step 1) The left lung, including a partial portion of the trachea, is used for the processing. The tissue is oriented with the distal (alveolar) portion on the left and the proximal (bronchial) portion on the right. (Step 2) Cut the tissue into five sections from the apex (upper part) to the lower part. (Step 3–4) Cut each section into smaller pieces. Track the pieces from different areas of the lung as follows: for example, those from the alveolar region are named LA, and those from the bronchial region are named LB. From each region, either snap-freeze small pieces in OCT for histology or subject them to cryopreservation (10% FBS and DMSO).
Human lung tissue viable freeze-thawing & lung airway organoid generation
Timing: approximately 4 months
-
2.
Thaw cryopreserved lung fragments (from 2–3 cryovials).
-
3.
Proceed to tissue dissociation to yield a single cell suspension.
-
4.
Assess cell viability.
-
5.
Mix the single cell suspension with cold Cultrex growth factor reduced BME type 2 (Matrigel-like matrix).
-
6.
Dispense as droplets into a P24-well plate.
Note: Approximately 3 × 105 cells in 40 μL per well.
-
7.
Place the plates in a 37°C cell incubator 5% CO2 in a humidified atmosphere for 20 min.
Note: to allow gelation of droplets.
-
8.Fill the wells with warm complete media for organoids (AO).
-
a.Change the media every day after.
-
b.Pass the organoids every 2 weeks.
-
i.Disrupt the organoids into single cells.
-
ii.Replate the single cells for organoid amplification.
-
iii.Repeat steps 4–8.
-
iv.Cryopreserve the cells (approximately 1.5 × 106 cells/mL per cryovial) in FBS with 10% DMSO.
-
v.Store in liquid nitrogen before further processing.
-
i.
-
a.
Note: Passages allow to eliminate connective tissue, typically after 4–7 passages. This progression leads to well-defined, spherical organoids devoid of connective tissue remnants.
CRITICAL: During all organoid-related steps, pre-coat tips, pipettes, and tubes with 5% BSA to avoid organoid adherence to plasticware and thus prevent cell loss. Quality control screening for epithelial enrichment (e.g., cytokeratin markers) in airway organoids by immunofluorescence (IF) staining is essential after 4–7 passages, once organoids are clear of connective tissue (e.g., fibronectin marker), as outlined in the step-by-step method details section (See Figure 2).
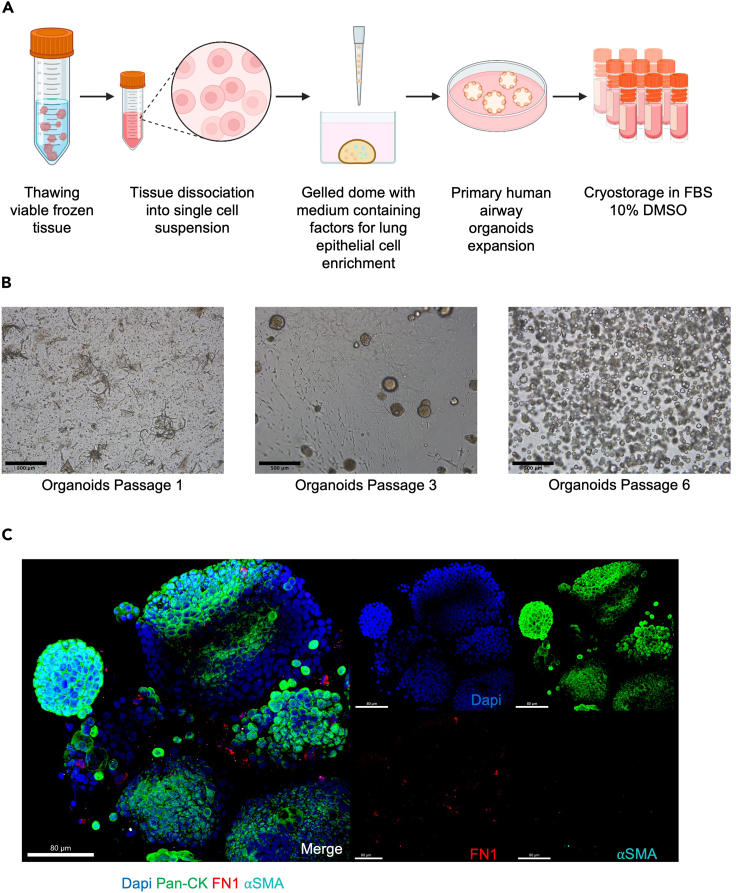
Figure 2.
Viable frozen tissue processing and generation of primary airway lung organoids
(A) Schematic experimental design for primary human lung organoid generation: viable frozen tissue is thawed and digested to obtain a single cell lung suspension containing airway cell progenitors. The cell lung suspension is resuspended in Cultrex, dispensed as a dome, and cultured in a medium containing factors allowing airway organoid expansion and lung epithelium enrichment. After each passage dissociated organoids are cryopreserved.
(B) Representative photomicrographs of primary lung organoids at different passages were captured using a bright-field microscope. Images were generated using ImageJ. Scale bars 500 μm, in black on the left corner.
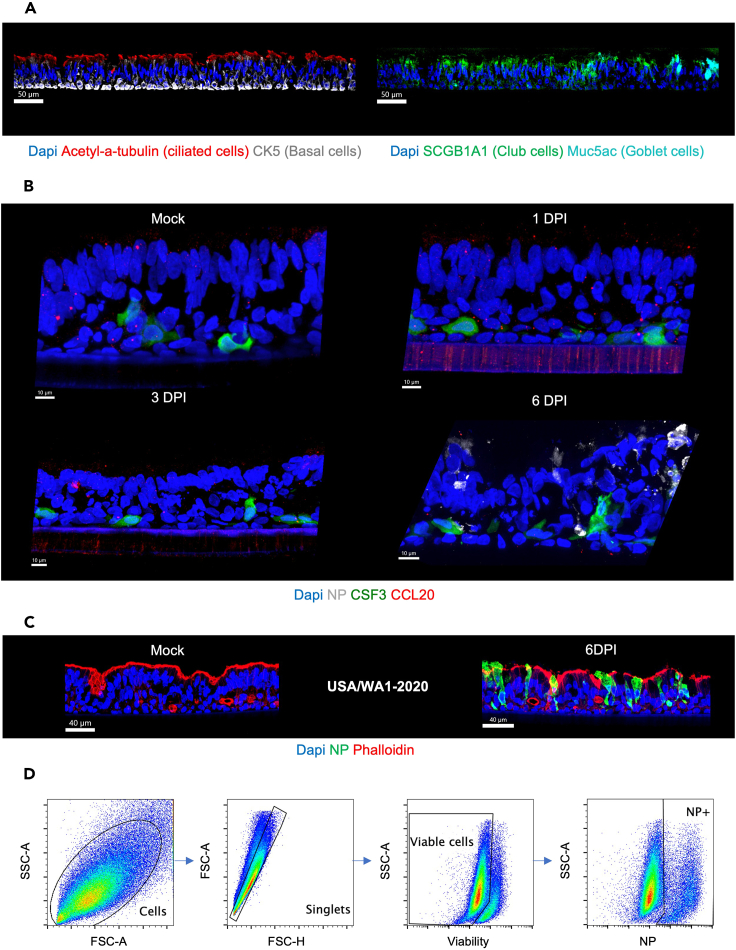
(C) Representative immunofluorescent (IF) images of whole mounted lung organoids showing markers for epithelial cells (PAN-CK, green), extracellular matrix (Fibronectin, red), and nuclei (DAPI, blue). Pan-CK is present exclusively in epithelial cells and fibronectin allows visualization of the presence of the remaining extracellular matrix from tissue dissociation. The fibronectin disappears after four passages and characterizes the enrichment of lung epithelial cells. Scale bar 80 μm, in white in the left corner. Note: Figure 2C reprinted with permission from Diana Cadena Castaneda et al. 20231 (Cell Press, Open Access).
Primary human lung organoid-derived air-liquid interface (ALI) culture generation, differentiation & viral exposure
Timing: approximately 6 weeks
-
9.
Thaw cryopreserved single cells derived from human lung organoids (from 1 cryovial).
-
10.Proceed to air-liquid interface (ALI) generation & differentiation.Note: One cryovial allows the generation of 48 ALI cultures (P24-well plate format; one ALI culture = one insert seed (0.33 cm2) with 3 × 104 cells per insert or approximately 9 × 104 cells/cm2). This process comprises three main steps, as detailed in the step-by-step method details section.
-
a.(1) Cell expansion in submerged culture to achieve confluence at 100%.
-
b.(2) Initial differentiation in submerged cultures.Note: To foster tight junctions and barrier integrity by monitoring TEER values (>500 Ω cm2).
-
c.(3) Final differentiation into pseudo-stratified epithelium.Note: Removing the apical media, “Airlift”, allows the ALI differentiation.
-
a.
-
11.
Monitor cilia beating and mucus production.
Note: Throughout approximately 4 weeks by using a bright-field microscope (See Figure 3).
-
12.Fully differentiated ALI cultures are ready for viral study protocols.Note: Our investigation comprises SARS-CoV-2 (USA/WA1-2020, Wuhan-like virus), Beta (B.1.351), Delta (B.1.617.2), and Omicron (B.1.1.529, BA.1) variants.
-
a.Perform an iterative wash of apical mucus.Note: Wash out the mucus before apical viral exposure.
-
i.Apply apically pre-warmed 1× PBS.
-
ii.Incubation for 15 min (37°C, 5% CO2 in a humidified atmosphere).
-
iii.Pipette up/down
-
iv.Eliminate the mucus.
-
i.
-
b.Optimal virus quantity determination (preliminary data).
-
i.Use two doses of Wuhan-like virus: 104 and 105 PFU, (data not shown).Note: A fully differentiated ALI contains approximately 3 × 106 cells/cm2, corresponding to MOIs (multiplicity of infection) of approximately 0.01 and 0.1 for 104 and 105 PFU, respectively.
-
ii.Add the virus or virus-free (control non-infected) media apically.
-
iii.Harvest ALI cultures daily from days 1 to 6 post-infection (DPI).Note: Each insert represents one culture.
-
iv.Assess the relative percentage of viral infection using histocytometry.Note: Histocytometry, utilizing immunofluorescence (IF) imaging targeting the viral nucleoprotein (NP) marker, reveals infection rates of approximately 2% for the 0.01 MOI and 34% for the 0.1 MOI at 6 DPI. Based on these results, we select the 0.1 MOI (equivalent to 105 PFU) for subsequent kinetic experiments.
 CRITICAL: TEER evaluation is critical during ALI generation to monitor the health of the cultures. For viral exposure, use at least 25 μL or a maximum of 100 μL of viral suspension for apical infection. Using larger volumes can induce tissue damage. Multiple readouts allow the assessment of the response to the virus (see sections related to “examples of readouts to assess response to viral exposure”).
CRITICAL: TEER evaluation is critical during ALI generation to monitor the health of the cultures. For viral exposure, use at least 25 μL or a maximum of 100 μL of viral suspension for apical infection. Using larger volumes can induce tissue damage. Multiple readouts allow the assessment of the response to the virus (see sections related to “examples of readouts to assess response to viral exposure”).
-
i.
-
a.
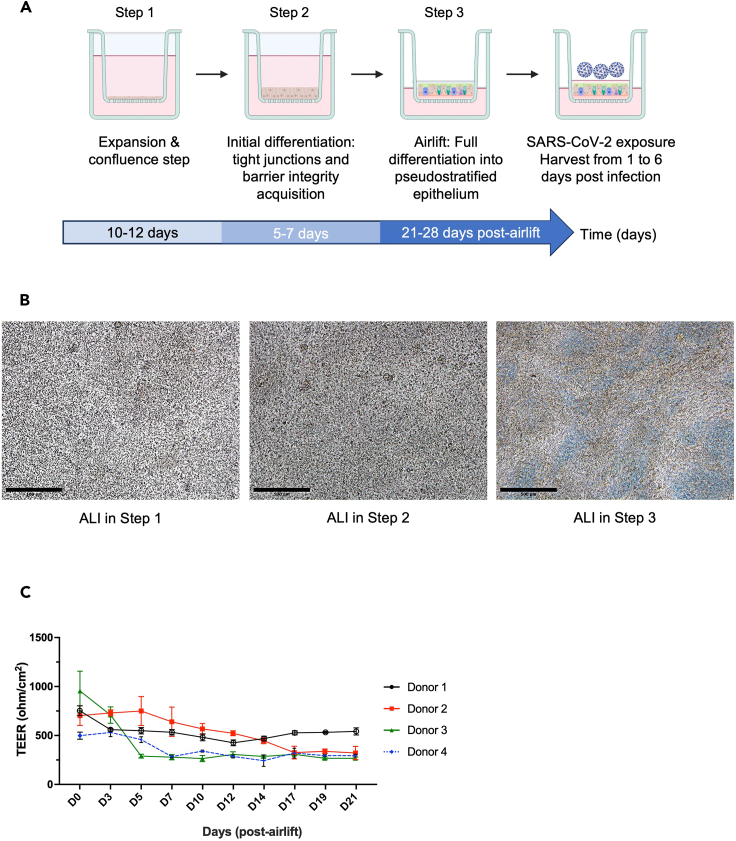
Figure 3.
Generation of primary human lung organoid-derived ALI cultures to study response to virus
(A) Schematic experimental design for primary human lung organoid-derived ALI culture generation: (1) cell expansion in submerged culture to obtain confluence at 100%; (2) initial differentiation in submerged cultures to foster tight junctions and barrier integrity, monitored by TEER values (>500 Ω cm2). (3) TEER goals are achieved, and cultures are transitioned to airlift by removal of apical media, which initiates final differentiation into pseudo-stratified epithelia. Cultures are monitored for a minimum of 4 weeks for the presence of beating cilia and mucus production.
(B) Representative photomicrographs of primary lung organoids-derived ALI cultures at Step 1 (3–4 days after seeding), Step 2 (at confluence approximately 12–14 days after seeding) and Step 3 (at 34 days post-airlift) captured using a bright-field microscope. Images were generated using ImageJ. Scale bars 500 μm, in black on the left corner.
(C) Measurement of trans-epithelial electrical resistance (TEER, Ω cm2) with error bars (mean ± SD), 3 measurements per time-point performed 3 times per week starting when the epithelium is confluent, one representative experiment per donor (four donors). Fully pseudo-stratified differentiated ALI cultures are obtained from primary lung organoid progenitors within 3–4 weeks (post-airlift). Note: Figure 3C reprinted with permission from Diana Cadena Castaneda et al. 20231 (Cell Press, Open Access).
Examples of readouts to assess response to viral exposure
Timing: approximately 3 weeks
We provide an overview of potential readouts to assess the response to viral exposure.
-
13.At each time-point, assess SARS-CoV-2 infection by using:
-
a.Flow cytometry on single cell suspensions of infected/non-infected ALI.Note: This step requires ALI dissociation, then, tracking the expression of the viral nuclear protein (NP), as outlined in the step-by-step method details section.
-
b.Viral titer on apical washes through plaque assays.Note: This step provides insight into viral particle release on the apical side.
-
c.Histocytometry, to evaluate infection through analysis of NP expression.Note: Additionally, the IF imaging method allows us to appreciate cellular ALI composition and relevant markers induced or elevated in response to the virus, such as CSF3 and CCL20.
-
a.
-
14.To appreciate the transcriptional response to the virus:
-
a.Dissociate ALI cultures at each time point.
-
b.Total RNA isolation using Direct-zol RNA MicroPrep kits.
-
c.Sequencing and subsequent bulk RNA analysis.
-
a.
-
15.
NanoString’s GeoMx Digital Spatial Profiler (DSP) on ALI culture frozen sections offers insights into in situ RNA data in response to the virus.
CRITICAL: Correct embedding of ALI cultures in OCT is crucial to obtaining high-quality frozen sections. During immunofluorescence staining, it is essential to carefully choose conjugated and unconjugated antibodies with the correct secondary antibodies with appropriate fluorophores to avoid misleading staining.
The materials and equipment section provides comprehensive recipes for all solutions, while the step-by-step method details section outlines the detailed procedures mentioned earlier. Prepare solutions beforehand, if possible, except for cell culture solutions. Please refer to the key resources table for a comprehensive list of materials and equipment.
Alternatives: Some of these readouts require a BSL3 setting, including flow cytometry, ALI dissociation & RNA extraction, and plaque assays for viral titers on the apical supernatant washes. However, other methods do not require BSL3, such as imaging (immunofluorescence, IF) or GeoMX DSP on ALI frozen sections (following PFA 4% fixation for 12 h at 20°C–25°C and OCT embedding for snap-frozen tissue in LN) or dissociated ALI after TRIzol treatment (for RNA preparation for sequencing). All work must comply with institutional guidelines.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-SCGBA1 (clone 394324, rat IgG) antibody dilution: 1/250 | R&D Systems | Cat#MAB4218; RRID: AB_2183286 |
| Anti-acetylated-alpha-tubulin (clone 6-11B-1, mouse IgG2b) antibody dilution: 1/250 | Thermo Fisher Scientific | Cat#32-2700; RRID: AB_2533073 |
| Anti-MUC5AC AF700 (clone 45M1, mouse IgG1) antibody dilution: 1/250 | Novus | Cat#NBP2-32732AF700; RRID: AB_2894883 |
| Anti-CCL20 (rabbit polyclonal) antibody dilution: 1/200 | Abcam | Cat#ab224188; RRID: AB_2894878 |
| Anti-CSF3 AF594 (clone CSF3/900, mouse IgG1) antibody dilution: 1/400 | Novus | Cat#NBP2-47934AF594; RRID: AB_2933966 |
| Anti-NP (SARS-CoV) AF488 or AF647 antibody dilution: 1/250 | Center for Therapeutic Antibody Development at Icahn School of Medicine at Mount Sinai | NP-1C7C7 |
| Anti-cytokeratin 5 AF594 (rabbit polyclonal) antibody dilution: 1/250 | Novus | Cat#NBP2-61931AF594; RRID: AB_2933967 |
| Anti-pan-cytokeratin (clone AE-1/AE-4, mouse IgG1) antibody dilution: 1/300 | BioLegend | Cat#914204; RRID: AB_2616960 |
| Anti-aSMA (clone 1A4, mouse IgG2a) antibody dilution: 1/200 | Abcam | Cat#Ab7817; RRID: AB_262054 |
| Anti-fibronectin (rabbit polyclonal) antibody dilution: 1/400 | Abcam | Cat#Ab32419; RRID: AB_732379 |
| Anti-mouse IgG1 AF488 antibody dilution: 1/2,000 | Thermo Fisher Scientific | Cat#A21121; RRID: AB_2535764 |
| Anti-mouse IgG2a AF647 antibody dilution: 1/2,000 | Thermo Fisher Scientific | Cat#A21241; RRID: AB_2535810 |
| Anti-rabbit IgG AF568 antibody dilution: 1/2,000 | Thermo Fisher Scientific | Cat#A11011; RRID: AB_143157 |
| Anti-mouse IgG2b AF555 antibody dilution: 1/2,000 | Thermo Fisher Scientific | Cat#A-21147; RRID: AB_2535783 |
| Anti-rabbit IgG AF555 antibody dilution: 1/200 | BioLegend | Cat#406412; RRID: AB_2563181 |
| Anti-rat IgG AF488 antibody dilution: 1/2,000 | Thermo Fisher Scientific | Cat#A-11006; RRID: AB_2534074 |
| Anti-NP (SARS-CoV-2; polyclonal) antibody dilution: 1/2,000 | Novus | Cat#NB100-56576A; RRID: AB_838838 |
| Anti-spike (SARS-CoV-2; polyclonal) antibody dilution: 1/50 | Novus | Cat#NBP2-24808AF647; RRID: AB_2933968 |
| Anti-cytokeratin 5 (clone EP1601Y) antibody dilution: 1/60 | Abcam | Cat#ab52635; RRID: AB_869890 |
| Anti-mouse IgG HRP antibody dilution: 1/5,000 | ProteinSimple | Cat# 042-205; RRID: AB_2860576 |
| Bacteria and virus strains | ||
| SARS-CoV-2 (USA/WA1-2020) | BEI Resources | NR-52281 |
| SARS-CoV-2 (Beta variant B.1.351) | Gift from Dr. Andy Pekosz | N/A |
| SARS-CoV-2 (Delta variant B.1.617.2) | Obtained from Dr. Viviana Simon | Mount Sinai Pathogen Surveillance program |
| SARS-CoV-2 (Omicron variant B.1.1.529, BA.1) | Obtained from Dr. Viviana Simon | Mount Sinai Pathogen Surveillance program |
| Biological samples | ||
| Human lung tissue | This study: NDRI | Project: RPAK1 01 |
| Chemicals, peptides, and recombinant proteins | ||
| Advanced DMEM/F12 | Thermo Fisher Scientific | Cat#12634-028 |
| MEM media 10× | Thermo Fisher Scientific | Cat#11430030 |
| GlutaMAX supplement | Thermo Fisher Scientific | Cat#35050-061 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat#50-753-2978 |
| Penicillin/streptomycin (PS) | Thermo Fisher Scientific | Cat#150-70-063 |
| Sodium bicarbonate 7.5% solution | Thermo Fisher Scientific | Cat#25080094 |
| HEPES (1 M) | Thermo Fisher Scientific | Cat#15-630-080 |
| Red blood cell lysis buffer | Roche | Cat#11814389001 |
| 1× PBS | Thermo Fisher Scientific | Cat#70011-044 |
| Cultrex growth factor reduced BME type 2 | Trevigen (R&D Systems) | Cat#3533-010-02 |
| Primocin | InvivoGen | Cat#Ant-pm-1 |
| Nicotinamide | Sigma | Cat#N0636 |
| N-acetylcysteine | Sigma | Cat#A9165 |
| B-27 supplement | Gibco | Cat#1750444 |
| SB202190 | Sigma | Cat#S7067 |
| A83-01 | Tocris | Cat#2939 |
| Noggin | PeproTech | Cat#120-10C |
| FGF 10 | PeproTech | Cat#100-26 |
| FGF 7 | PeproTech | Cat#100-19 |
| R-spondin 1 | PeproTech | Cat#120-38 |
| Y-27632 | AbMole | Cat#M1817 |
| Collagen I rat protein | Thermo Fisher Scientific | Cat#A1048301 |
| PneumaCult-Ex plus medium | STEMCELL | Cat#05040 |
| PneumaCult-ALI medium | STEMCELL | Cat#05001 |
| Hydrocortisone | STEMCELL | Cat#07926 |
| Heparin solution | STEMCELL | Cat#07980 |
| DNase I | Sigma | Cat#D4513 |
| Dispase I | Sigma | Cat#4942086001 |
| Collagenase I | Sigma | Cat#9C9407 |
| Paraformaldehyde 16% | Thermo Fisher Scientific | Cat#28906 |
| Triton X-100 | Thermo Fisher Scientific | Cat#HFH10 |
| Phalloidin ATTO647N | Sigma | Cat#65906 |
| Fluromount-G | Thermo Fisher Scientific | Cat#00-4958-02 |
| DAPI | Thermo Fisher Scientific | Cat#D1306 |
| Fc receptor blocker | Innovex | Cat#NB309 |
| Background buster | Innovex | Cat#NB306 |
| Mouse serum | Jackson ImmunoResearch | Cat#015-000-120 |
| BSA (IgG-free, Protease-free; for immunofluorescence, IF) | Jackson ImmunoResearch | Cat#001-000-161 |
| BSA (cell culture grade) | Sigma | Cat#A9418 |
| Dimethyl sulfoxide (DMSO) | Sigma | Cat#67-68-5 |
| TrypLE express enzyme | Thermo Fisher Scientific | Cat#12605010 |
| Trypsin-EDTA (0.05%) phenol red | Thermo Fisher Scientific | Cat#25-300-062 |
| Zombie Aqua Fixable Viability Kit | BioLegend | Cat#423101 |
| Zombie Green Fixable Viability Kit | BioLegend | Cat#423111 |
| Saponin | Sigma | Cat#S7900-100G |
| Oxoid purified agar | Thermo Fisher Scientific | Cat#LP0028 |
| DEAE-dextran | Thermo Fisher Scientific | Cat#J63781.14 |
| TPCK/trypsin | Thermo Fisher Scientific | Cat#20233 |
| TrueBlue peroxidase substrate | SeraCare | Cat#5510-0030 |
| Critical commercial assays | ||
| Cytofix/Cytoperm Fixation/ Permeabilization solution Kit | BD Biosciences | Cat#554714 |
| Direct-zol RNA MicroPrep Kits | Zymo Research | Cat#R2062 |
| RNase-free DNase set | QIAGEN | Cat#79254 |
| Kapa Stranded mRNA-seq Library Prep Kit | Kapa Biosystems | Cat#KK8401 |
| Viral stock protocol | ARTIC | https://artic.network/ncov-2019 |
| Viral-RNA Kit | Omega Bio-tek | Cat#R6874-02 |
| Native Barcoding Expansion Kit | ONT | Cat#EXP-NBD104 |
| Deposited data | ||
| Raw data and analyzed data | This study | GEO: GSE225603 |
| Experimental models:Cell lines | ||
| African green monkey (Chlorocebus sabaeus): VeroE6/TMPRSS2 cells | Cellosaurus R Matsuyama et al.10 |
Cat#JCRB1819; RRID: CVCL_YQ49 |
| African green monkey (Chlorocebus sabaeus): VeroE6 | ATCC | ATCC CRL-1586, clone E6; RRID: CVCL_0574 |
| Software and algorithms | ||
| Imaris 9.4 | Bitplane | https://imaris.oxinst.com |
| ImageJ | ImageJ | https://imagej.nih.gov/ij/ |
| Leica (LAS) X | Leica Microsystems | https://www.leica-microsystems.com/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| BD FACSuite software | BD Biosciences | https://www.bdbiosciences.com/ |
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com/ |
| FlowJo v10.3 | FlowJo LLC | https://www.flowjo.com/ |
| DNAstar | Lasergene | https://www.dnastar.com/software/lasergene/ |
| R (v4.2.0) | The R Foundation | https://www.r-project.org/ |
| ggplot2 (R package v3.3.6) | R package | https://rpkgs.datanovia.com/survminer/index.html |
| ComplexHeatmap package (v2.12.1) | R package | https://jokergoo.github.io/ComplexHeatmap/ |
| RSEM v1.3.3 | Li and Dewey11 | https://github.com/deweylab/RSEM/releases/tag/v1.3.3 |
| Bowtie | Langmead et al.12 | http://bowtie-bio.sourceforge.net/index.shtml |
| FastQC | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | N/A |
| MultiQC | Ewels et al.13 | N/A |
| BBDuk tool | BBMap download | SourceForge.net | https://sourceforge.net/projects/bbmap/ |
| DESeq2 | R package Love et al.14 |
N/A |
| SVAseq | R package | https://doi.org/10.1093/nar/gku864 |
| Reference manager | Mendeley | https://www.mendeley.com/search/ |
| Other | ||
| Greiner CELLSTAR multiwell culture plates (P24-well) | Sigma | Cat#M9312 |
| Corning Transwell polyester membrane cell culture inserts (P24-well, 0.4 μm) | Sigma | Cat#3470 |
| Corning sterile cell strainers (100 μm) | Fisher Scientific | Cat#431752 |
| Petri dishes 100 × 15 mm | Thermo Fisher Scientific | Cat#150350 |
| Petri dishes 60 × 15 mm | Thermo Fisher Scientific | Cat#174888 |
| 50 mL tubes | Fisher Scientific | Cat#339652 |
| 2 mL cryovial tubes | Fisher Scientific | Cat#431386 |
| Tissue-Tek OCT compound | Thermo Fisher Scientific | Cat#4583 |
| Tissue-Tek cryomold | VWR | Cat#4566 |
| Slides | Denville Scientific | Cat#M1021 |
| Cover glass | Thermo Scientific | Cat#152450 |
| Optimized freezing container | Thermo Fisher Scientific | Cat#5100-0001 |
| MACSmix tube rotator | Miltenyi Biotec | Cat#130-090-753 |
| EVOM2 ohm meter | WPI | Cat#EVOM2 |
| Piston from insulin syringe | BD Biosciences | Cat#324921 |
| Membrane filter, 0.22 μm pore size | Sigma | Cat#GSWP04700 |
| Beakers (1,000 mL) | VWR | Cat#13912-284 |
| Pyrex container | Cole-Parmer | Cat#3175-10 |
| Autoclaving foil | VWR | Cat#47734-154 |
| Saran wrap | VWR | Cat#46610-056 |
| Parafilm | VWR | Cat#52858-076 |
| Surgical blue wrap | VWR | Cat#56222-053 |
| Scalpel | VWR | Cat#76457-484 |
| Surgical forceps | VWR | Cat#10806-206 |
| Fine-point forceps | VWR | Cat#470005-440 |
| Surgical scissors | VWR | Cat#76457-366 |
| Pipet-Lite LTS pipette L-1000XLS+ | Rainin | Cat#17014382 |
| Pipettes (tips 1 mL) | Mettler Toledo | Cat#30389217 |
| Electric pipette controller Eppendorf Easypet 3 | Eppendorf | Cat#2231000955 |
| Serological pipettes (10 mL) | Thermo Fisher Scientific | Cat#170356N |
| Serological pipettes (25 mL) | Thermo Fisher Scientific | Cat#170357N |
| Schematic and graphical abstract images | BioRender | https://www.biorender.com |
Materials and equipment
Human lung tissue processing & cryopreservation
Equipment setup:
-
•
Sterile material (autoclave): beakers (1000 mL in size), large Pyrex container (24 cm × 36 cm x 6 cm) for tissue reception, saran wraps to seal beakers, surgical blue wrap to avoid blood spills. Sterile scalpel, surgical forceps, fine-point forceps, surgical scissors.
-
•
Pipette tips: 1 mL.
-
•
Serological pipettes: 10 mL, 25 mL.
-
•
Single channel pipette (1 mL), Pipet-Lite LTS Pipette L-1000XLS+.
-
•
Electric pipette controller Eppendorf Easypet 3.
-
•
Petri dishes 100 × 15 mm (Thermo Scientific, 150350).
-
•
Petri dishes 60 × 15 mm (Thermo Scientific, 174888).
-
•
50 mL centrifuge tubes (Fisher Scientific, 339652).
-
•
2 mL cryovial tubes (Fisher Scientific, 431386).
-
•
Tissue-Tek OCT Compound (Thermo Scientific, 4583).
-
•
Tissue-Tek OCT cryomold (VWR, 4566).
-
•
Optimized freezing container, Mr. Frosty (Thermo Scientific, 5100-0001)
-
•
Media for tissue preservation: 1× PBS (Thermo Scientific, 70011-044) supplemented with 50% FBS (Thermo Scientific, 50-753-2978). Store at 4°C for up to 2 months.
-
•
Cryopreservation media: cold 90% FBS with 10% DMSO (Sigma, 67-68-5). Store at 4°C for up to 2 months.
Human lung tissue viable freeze-thawing & lung airway organoid generation
Equipment setup:
-
•
Pipettes: 1 mL, 10 mL, 25 mL.
-
•
Petri dishes 60 × 15 mm.
-
•
50 mL centrifuge tubes.
-
•
Tube rotator (Miltenyi Biotec, MACSmix Tube Rotator, 130-090-753).
-
•
Sterile slides (Globe Scientific, #1324W).
-
•
Sterile Cell Strainers, 100 μm (Fisher Scientific, 431752).
Note: Pre-wet strainers before filtering tissue or cell suspensions with 1–2 mL 1× PBS.
-
•
Piston from insulin syringe (BD, 324921).
-
•
Cultrex growth factor reduced BME type 2 (R and D, 3533-010-02).
Note: Store at −20°C for up to 12 months. Melt the Matrigel-like matrix reagent 24 h prior to viable frozen tissue processing in the fridge at 4°C.
-
•
BSA solution (0.1%): dissolve 0.01 g of BSA (Cell culture grade, Thermo Scientific, A9418) in 10 mL 1× PBS, filter through 0.2 μm filter. Store at 4°C for up to 2 months.
-
•
Greiner CELLSTAR P24-well culture plates (Sigma, M9312).
-
•
TrypLE express enzyme (Thermo Fisher Scientific, 12605010)
Basal media for airway organoid (AdDF+)
| Reagent | Final concentration | Amount |
|---|---|---|
| Advanced DMEM/F12 | N/A | 485 mL |
| Penicillin/Streptomycin (100×) | 1× | 5 mL |
| HEPES (1 M) | 10 mM | 5 mL |
| GlutaMAX Supplement (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C for up to 2 months.
-
•
Media for tissue digestion: add 100–200 μL of collagenase I (Sigma-C9407, stock solution at 200 mg/mL in 1× PBS; store at −20°C for up to 1 year) for a final concentration of 1–2 mg/mL into 10 mL final of AdDF+ media (see above). Always prepare and use fresh.
Complete media for airway organoid generation (AO)
| Reagent | Final concentration | Amount |
|---|---|---|
| AdDF+ (See above) | N/A | 29.115 mL |
| R-Spondin 1 Reconstitute in 1× PBS 0.1% BSA at 200 μg/mL |
500 ng/mL | 75 μL |
| FGF-7 Reconstitute in 1× PBS 0.1% BSA at 20 μg/mL |
25 ng/mL | 37.5 μL |
| FGF-10 Reconstitute in 1× PBS 0.1% BSA at 100 μg/mL |
100 ng/mL | 30 μL |
| Noggin Reconstitute in 1× PBS 0.1% BSA at 40 μg/mL |
100 ng/mL | 75 μL |
| B-27 supplement (50×) | 1× | 600 μL |
| Primocin (50 mg/mL) | 50 μg/mL | 30 μL |
| A83-01 Reconstitute in DMSO at 10 M/mL |
500 nM/mL | 1.5 μL |
| Y-27632 (10 mM/mL) | 5 μM/mL | 15 μL |
| SB202190 Reconstitute in DMSO at 10 mM/mL |
5 μM/mL | 15 μL |
| N-Acetylcysteine Reconstitute in DMSO at 12.5 M/mL |
1.25 mM/mL | 3 μL |
| Nicotinamide Reconstitute in DMSO at 50 M/mL |
5 mM/mL | 3 μL |
| Total | N/A | 30 mL |
Always prepare and use fresh.
Note: Filter grows factors or chemical reagent solutions using a 0.22 μM filter. Aliquot and keep grow factors or chemical reagent at −20°C for at least 1 year, avoiding freeze and thaw cycles.
-
•
Media to stop tissue digestion reaction: prepare 1× PBS supplemented with 2% FBS. Store at 4°C for up to 2 months.
Primary human lung organoid-derived air-liquid interface (ALI) generation, differentiation & viral exposure
Equipment setup:
-
•
Pipettes: 1 mL, 10 mL, 25 mL.
-
•
50 mL centrifuge tubes.
-
•
EVOM2 ohm meter (WPI). For more details on how to use the device, refer directly to WPI supplier website.
-
•
Corning Transwell polyester membrane cell culture insert (P24-well, 0.4 μm; Sigma, 3470).
-
•
Collagen coating solution: add 50 μL of Collagen I Rat Protein (Thermo Fisher Scientific, A1048301; stock at 3 mg/mL, store at 4°C for up to 1 year) for a final concentration of 30 μg/mL into 5 mL of 1× PBS. Always prepare and use fresh.
-
•
Sterile forceps.
-
•
STEMCELL PneumaCult Ex-Plus (#05040), and STEMCELL PneumaCult ALI Maintenance media (05001).
Bellow, the preparation of all media for the ALI generation, comprises three main steps (1–3) and media for viral exposure.
Basal media for ALI culture expansion (step 1)
| Reagent | Final concentration | Amount |
|---|---|---|
| PneumaCult-Ex Plus Basal Medium | N/A | 442.5 mL |
| PneumaCult-Ex Plus 50× Supplement | 1× | 50 mL |
| Hydrocortisone Stock Solution (200×) | 1× | 2.5 mL |
| Penicillin/Streptomycin (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C for up to 2 months.
-
•
Complete media for ALI culture expansion (step 1): add 50 μL of Y-27632 (Stock solution at 10 mM/mL; store at −20°C for at least 1 year) for a final concentration of 0.01 mM/mL into 50 mL of basal media for ALI culture expansion (see above). Always prepare and use fresh.
Basal media for ALI culture differentiation and maintenance (steps 2–3)
| Reagent | Final concentration | Amount |
|---|---|---|
| PneumaCult-ALI Basal Medium | N/A | 445 mL |
| PneumaCult-ALI 10× Supplement | 1× | 50 mL |
| Penicillin/Streptomycin (100×) | 1× | 5 mL |
| Total | N/A | 500 mL |
Aliquot (e.g., 50 mL) and store at −20°C for at least 1 year, thaw when necessary.
Complete media for ALI culture differentiation and maintenance (steps 2–3)
| Reagent | Final concentration | Amount |
|---|---|---|
| Basal media for ALI culture differentiation and maintenance (See above) | N/A | 49.1 mL |
| Hydrocortisone Stock Solution (200×) | 1× | 250 μL |
| Heparin Solution (500×) | 1× | 100 μL |
| PneumaCult-ALI Maintenance Supplement (100×) | 1× | 500 μL |
| Y-27632 (10 mM/mL) Halt the addition after the ALI airlift. | 0.01 mM/mL | 50 μL |
| Total | N/A | 50 mL |
Always prepare and use fresh.
CRITICAL: Confluence and Initial Media Change (Step 2), (See Figure 3): After achieving full confluence of ALI cultures (around 10–12 days post-seeding), change the media on both the apical and basal sides. Use media designed for ALI culture differentiation and maintenance, enriched with Y-27632. This compound is essential for the initial phase of pseudo-stratified epithelium differentiation, spanning approximately 5–7 days (varies based on donor characteristics).
TEER Goal Achievement and Transition to Airlift (Step 3): When TEER values surpass 500 Ω cm2, commence the airlift phase. At this point, discontinue the addition of media to the apical side while maintaining media supply to the basal side and halt the supplementation with Y-27632. The airlift step marks the transition to the second differentiation phase, lasting approximately 4 weeks.
By supplementing with Y-27632 and altering media application according to the differentiation phases, you facilitate the progression of ALI cultures toward a pseudo-stratified epithelial state.
-
•
Media to apply on the basal side of ALI culture during viral exposure experiments: PneumaCult-ALI Basal Medium supplemented with PneumaCult-ALI 10× supplement (1× final). Store this media at 4°C for up to 2 weeks.
Note: Conduct all experiments involving live SARS-CoV-2 in a biosafety level 3 (BSL-3) facility. Follow all necessary safety measures during the viral exposure experiments.
CRITICAL: Sequence and confirm the genomic integrity of all SARS-CoV-2 viral stocks according to the ARTIC protocol (https://artic.network/ncov-2019).15 Store virus aliquots in a secured −80°C freezer until use.
Example of readouts to assess response to viral exposure
-
•
Preparation of material and reagents for plaque assays.
Collect viral supernatants at all time points by adding 150 μL of 1× PBS on the apical side and incubating for 15 min at 37°C, 5% CO2 in a humidified atmosphere. Then, store supernatants at −80°C until further processing.Note: Conduct all experiments involving live SARS-CoV-2 isolates in a biosafety level 3 (BSL-3) facility.Equipment setup:-
○Pipettes: 1 mL, 10 mL, 25 mL.
-
○1.5 mL Eppendorf tubes.
-
○BSA solution (3.5%): dissolve 3.5 g of BSA (Cell culture grade, Thermo Scientific, A9418) in 100 mL 1× PBS, sterile filter through 0.2 μm filter. Store at 4°C for up to 2 months.
-
○1% Bovine pre-seeded VeroE6 cell (for USA/WA1-2020) or VeroE6 TMPRSS2 (for Beta, Delta and Omicron) monolayers.
-
○2% Agar (w/v): dissolve 2 g of purified Oxoid (LP0028) agar in 100 mL diH2O.
-
○1% DEAE Dextran (w/v): dissolve 0.5 g of DEAE Dextran in 50 mL of diH2O, sterile filter through 0.2 μm filter.2× MEM solution
Reagent Final concentration Amount MEM media (10×) 2× 200 mL GlutaMAX supplement (100×) 2× 20 mL Sodium bicarbonate (7.5%) 0.24% 32 mL HEPES (1 M) 20 mM 20 mL Penicillin/Streptomycin (100×) 2× 20 mL BSA solution (3.5%) 0.04% 12 mL diH2O N/A 696 mL Total N/A 1000 mL Store at 4°C for up to 2 months.Note: Sterile filter using a Millipore (0.22 μM) vacuum-driven filter.Overlay mediaReagent Final concentration Amount 2× MEM solution (see above) 1× 12.5 mL 2% Oxoid agar 0.6% 8.5 mL 1% DEAE Dextran 0.01% 250 μL TPCK/trypsin (1 mg/mL) 1 μg/mL 25 μL diH2O N/A 4 mL Total N/A 25 mL Always prepare and use fresh. -
○PFA 4% fixation for immune staining: mix 1 mL PFA 16% (Thermo Fisher, 28906) with 3 mL 1× PBS.
-
○To reveal viral infection: Anti-SARS-CoV-2 NP antibody (1C7C7, 1/250) and HRP-conjugated secondary anti-mouse antibody (1/5000).
-
○TrueBlue peroxidase substrate (SeraCare).
-
○
-
•
Preparation of reagents for ALI dissociation, intracellular staining for infection (Flow cytometry), and RNA extraction.
Dissociate infected and non-infected ALI at all time-points, separately, one insert for flow cytometry and one insert for RNA extraction. See the step-by-step method details section for the detailed procedure.Note: Conduct all experiments involving live SARS-CoV-2 in a biosafety level 3 (BSL-3) facility.Equipment setup:-
○Pipettes: 1 mL, 10 mL, 25 mL.
-
○Trypsin-EDTA (0.05%) phenol red (Thermo Fisher Scientific, 25-300-062).
-
○Dispase I/DNase I solution: dissolve 1 vial of dispase I (Sigma, 4942086001; stock at 38 U; store at 4°C for up to 12 months) into 20 mL 1× PBS for a final concentration of 1.9 U/mL and add 20 μL of DNase I (Sigma, D4513; stock at 100 mg/mL in 1× PBS; store aliquots at −20°C for at least 1 year) for a final concentration of 0.1 mg/mL. Always prepare and use fresh.
-
○Media to stop tissue digestion reaction: 1× PBS supplemented with 2% FBS. Store at 4°C for up to 2 months.
-
○1.5 mL Eppendorf tubes.
-
○Flow cytometry:
-
-Viability assessment: staining in 1× PBS with Zombie aqua (#423101) or Zombie green (#423111); (BioLegend).
-
-Cytofix/Cytoperm Fixation/Permeabilization solution kit (BD, 554714).
-
-SARS-CoV-2 N antibody (1C7C7, AF488 or AF647)
-
-
-
○Buffer for cell suspension 1% BSA: dissolve 1 g of BSA (Cell culture grade, Thermo Scientific, A9418) in 100 mL 1× PBS, sterile filter through 0.2 μm filter. Store at 4°C for up to 2 months.
-
-BD FACSuite Software and FlowJo.
-
-
-
○RNA extraction:
 Pause point: Store extracted RNA at −80°C, until further processing.
Pause point: Store extracted RNA at −80°C, until further processing.-
-Direct-zol RNA MicroPrep kits (Zymo Research, R2062).
-
-RNase-free DNase set (QIAGEN, 79254).
-
-PolyA mRNA selection and Kapa Stranded mRNA-seq Library Prep kit (Kapa Biosystems, KK8401).
-
-DNAstar (Lasergene).
-
-
-
○
-
•
Preparation of ALI cultures for imaging and NanoString GeoMx whole transcriptome atlas (WTA).
Equipment setup:-
○Pipettes: 1 mL, 10 mL, 25 mL.
-
○Tissue-Tek OCT Compound (Thermo Scientific, 4583).
-
○Tissue-Tek OCT cryomold (VWR, 4566).
-
○Cryostat Leica.
-
○Immunofluorescence (IF):
-
-PFA 4% fixation for immune staining: mix 1 mL PFA 16% (Thermo Fisher Scientific, 28906) with 3 mL 1× PBS.
-
-Permeabilization buffer: add 10 μL of Triton X-100 into 10 mL 1× PBS for a final concentration of 0.1% Triton X-100. Store at 4°C for up to 1 month.Alternatives: For non PFA-fixed tissue, a one single permeabilization/fixation step with cold acetone (−30°C) is possible. For example, for organoids, submerge the tissue in cold Acetone, approximately 5 min. Use this reagent in a fume hood.
-
-Saturation (Innovex, NB309) and background reagents (Innovex, NB306).
-
-Saponin 10%: dissolve 1 g of saponin (Sigma, S7900-100G) in 10 mL 1× PBS and sterile filter through 0.2 μm filter. Store at 4°C for several months.
-
-Staining buffer: dissolve 0.5 g of BSA (IgG-Free, Protease-Free; Jackson ImmunoResearch, 001-000-161) in 10 mL of 1× PBS and add 50 μL of saponin 10% for a final concentration of 0.05% saponin. Sterile filter and always prepare and use fresh.
-
-Primary and secondary antibodies (see key resources table).
-
-Mouse serum, (Jackson ImmunoResearch, 015-000-120).
-
-Phalloidin ATTO647N, (Sigma, 65906).
-
-DAPI (Thermo Fisher Scientific, D1306)
-
-Hydrophobic pen (pap-pen, Daido Sangyo, N33)
-
-Mounting media (Thermo Fisher Scientific, 00-4958-02).
-
-
-
○
-
•Superfrost plus Slides (Denville Scientific, M1021) and cover-glass (Thermo Fisher, 152450)
-
○GeoMx WTA: Slides with 6 μm thickness sections of ALI cultures at different conditions. Store slides at −80°C until process.
-
-PFA 4% fixation for immune staining (see above).
-
-Prepare the tissue sections according to MAN-10131-03.Note: For more details, please refer to the NanoString website.
-
-Primary antibodies: Nuclei Syto83 (NanoString), CK5 (Abcam, ab52635, FITC), SARS-CoV-2 Spike (Novus, NBP2-24808AF647), and SARS-CoV-2 Nucleocapsid (Novus, NB100-56576).
-
-Secondary antibodies: Anti-rabbit IgG AF555 (BioLegend, 406412).
-
-Viral-RNA kit (Omega Bio-tek, R6874-02)
-
-Native Barcoding Expansion kit (ONT, EXP-NBD104).
-
-DNAstar (Lasergene).
 CRITICAL: Reagents or biological materials may be harmful and/or toxic; therefore, use appropriate laboratory equipment. For flow cytometry, IF, and GeoMx staining, make fresh reagents and antibody mix for each staining and keep them on ice in the dark.
CRITICAL: Reagents or biological materials may be harmful and/or toxic; therefore, use appropriate laboratory equipment. For flow cytometry, IF, and GeoMx staining, make fresh reagents and antibody mix for each staining and keep them on ice in the dark.
-
-
-
○
Step-by-step method details
Human lung tissue processing & cryopreservation
Timing:2 days
This step allows the selection of suitable tissues for cryopreservation and airway organoid generation based on tissue anatomy, elimination of necrotic or damaged areas, and cryopreservation of multiple vials from different regions of the lung.
-
1.Lung tissue processing (Figure 1): Receive human lung tissue under clean/sterile conditions.
-
a.Transfer the lung tissue to a sterile beaker (1000 mL) containing media for tissue preservation 1× PBS supplemented with 50% FBS.Note: Add approximately 400–500 mL to cover the tissue.
-
b.Seal the beaker with sterile aluminum foil and parafilm.
 Pause point: You can keep the lung tissue at 4°C under the conditions described above (a-b) for at least 8 h until tissue processing.
Pause point: You can keep the lung tissue at 4°C under the conditions described above (a-b) for at least 8 h until tissue processing. -
c.Transfer the lung into a large sterile Pyrex container (24 cm × 36 cm × 6 cm).
-
d.Anatomically orientate the lung tissue.Note: For example, for the left lung, place the lung facing the researcher in a way that the distal area (alveolar part) is on the left and the proximal part (bronchial part) is on the right close to the trachea (See Figure 1, Step 1).
-
e.Cut the lung tissue into slices (approximately 4–5 cm thick) with a sterile scalpel in function of anatomical orientation.Note: For example, start from the apex (upper part) to the bottom part of the lung, then from distal (more alveolar) to proximal (more bronchial), (See Figure 1, Step 2).
-
f.Cut each lung tissue slice into smaller pieces (approximately 3 cm × 3 cm in size), with a sterile scalpel (See Figure 1, Step 3).
-
g.Embed representative lung pieces from each region (approximately 3 cm × 3 cm in size) in OCT.
-
h.Snap freeze in liquid nitrogen.Note: Name each OCT tissue block based on anatomical region and donor code: for example, LA for “alveolar” and LB for “bronchial” (See Figure 1, Step 4(a)).
-
i.Mince the remaining tissue into smaller pieces, of approximately 3 mm × 3 mm in size.
-
j.Place around 25 small pieces into each cryovial (2 mL cryovial size).
-
k.Immediately add 1 mL of cold cryopreservation media (FBS 10% DMSO) to each cryovial.
-
l.Transfer to an optimized freezing container.
 Pause point: Store the cryovials at −80°C for at least 24 h before transferring them to liquid nitrogen for long-term storage. Similarly, label each cryovial based on anatomical region, donor code, and date: (See Figure 1, Step 4(b)).
Pause point: Store the cryovials at −80°C for at least 24 h before transferring them to liquid nitrogen for long-term storage. Similarly, label each cryovial based on anatomical region, donor code, and date: (See Figure 1, Step 4(b)). CRITICAL: Using this method, we process human lung tissue sections by anatomical region, allowing us to obtain organoids efficiently and fully differentiated ALI cultures from each area.
CRITICAL: Using this method, we process human lung tissue sections by anatomical region, allowing us to obtain organoids efficiently and fully differentiated ALI cultures from each area.
-
a.
Human lung tissue viable freeze-thawing & lung airway organoid generation
Timing: approximately 4 months
This section describes the workflow to culture primary airway organoids, including the isolation, expansion, and cryostorage of lung organoid-derived progenitor cells, along with quality control screening of organoids for epithelial enrichment (Figure 2).
-
2.Processing of viable frozen lung pieces and digestion for airway organoid generation.
-
a.Prepare AdDF+ media for washing steps.
-
b.Prepare AO media for organoid generation.
-
c.Prepare media for tissue digestion containing 1–2 mg/mL of collagenase I.
-
d.Prepare media to stop tissue digestion reaction (1× PBS supplemented with 2% FBS).
-
e.Thaw Cultrex growth factor reduced BME type 2 (Matrigel-like matrix).Note: Thaw in the fridge 24 h prior to viable frozen lung pieces processing.
-
f.Thaw the cryopreserved tissue by placing 2–3 cryovials into a dry bath at 37°C for 5 min.
-
g.Immediately, transfer the minced tissue into a 50 mL tube containing AdDF+ media (20–30 mL).Note: Pre-warm the media in a dry bath (37°C).
-
h.Centrifuge at 300g for 5 min (20°C–25°C)Note: This step allows to remove the DMSO.
-
i.Resuspend the minced tissue with 10 mL of media for tissue digestion.Note: Pre-warm the media in a dry bath (37°C).
-
j.Incubate on an orbital shaker at 37°C, 5% CO2 in a humidified atmosphere for 1 h. Troubleshooting 1.
-
k.Shear the digested tissue between sterile slides.
-
l.Strain the digested tissue through a 100 μm filter.Note: To improve the straining, use the piston of an insulin syringe to press the tissue through the filter. For tissue remaining in the filter, repeat the shearing and filtration steps to remove as much connective tissue as possible to obtain a single cell suspension.
-
m.Stop the digestion process by adding 1× PBS supplemented with 2% FBS (20°C–25°C).
-
n.Centrifuge at 300g for 5 min (20°C–25°C).
-
o.Resuspend the pellet in 1× PBS (approximately 5 mL, at 20°C–25°C).Alternatives: In case a visible red pellet is present, centrifuge for 5 min at 300g (20°C–25°C) and proceed to erythrocyte lysis using 2 mL red blood cell lysis buffer for 5 min at 20°C–25°C before the addition of 10 mL AdDF+ to stop the lysis.
-
p.Strain the cells through a 100 μm filter using a new insulin syringe piston.
-
q.Centrifuge at 300g for 5 min (20°C–25°C). Troubleshooting 2.
-
r.Resuspend the pellet in 1 mL of AO media (pre-warmed, 37°C in a dry bath).
-
s.Assess the cell viability.Note: Counting on a chamber Malassez. Dilute approximately, 10 μL of single cell suspension 5 times with 1× PBS. Then, mix one volume of the diluted cell suspension with one volume of trypan blue (0.4% in 1× PBS) to reveal blue dead cells. Apply approximately 10 μL of the cell mix to the counting chamber. Count the cells within 3–5 min of mixing with trypan blue, on the four corner squares plus the central big square of the chamber (5 big squares total). At this point, we expect to obtain 5 to 7.5 × 106 cells/mL with a viability of 80%–90%. To calculate the total cell concentration and then deduce cell viability, use the following formula:
-
a.
-
3.Primary human lung organoid generation is performed based on Sachs et al.9 and will be summarized in this protocol manuscript (Figure 2A).
-
a.Resuspend the lung cell suspension at 7.5 × 106 cells/mL final in a mix of ice-cold AO media and liquid Cultrex growth factor reduced BME type 2.Note: Use the liquid Cultrex growth factor reduced BME type 2, at 1 mg/mL final of basement membrane extract protein concentration for optimal obtention of organoids. To keep the Cultrex growth factor reduced BME type 2 liquid, place the mix on ice until you are ready to proceed to cell culturing. For more details refer to the Cultrex growth factor reduced BME type 2 manufacturer (R&D, Bio-Techne) methodology resource.
-
b.Mix the lung cell suspension.
-
c.Dispense approximately 300,000 cells/40 μL/well as a dome.Note: In this step pre-warm the CELLSTAR P24-well culture plate.
-
d.Gentle, flip the plate upside-down.
-
e.Incubate the plate in a humidified 37°C, 5% CO2 incubator at ambient O2 for 20 min to allow gellification. Troubleshooting 3.
-
f.Gentle, flip the plate right side up.
-
g.Add 400 μL of pre-warmed AO media (37°C in a dry bath).Note: AO media contains factors allowing the enrichment of lung epithelial cells.
-
h.Change media every day.
-
i.Passage organoids every 2 weeks as described below.
-
i.Remove media and add ice-cold 1× PBS.
-
ii.Incubate the plate on ice for 30 min.Note: the incubation on ice allows to melt the gel without full disruption of the organoids.
-
iii.Pipette (1 mL pipette) up/down to break the organoids.
-
iv.Combine the organoids of all the wells in a 50 mL tube.
-
v.Centrifuge at 300g for 5 min (7°C).
-
vi.For full organoid dissociation, add pre-warmed 1× TrypLE express.
-
vii.Incubate 5 min at 20°C–25°C.
-
viii.Mechanically, by pipetting up/down, dissociate the organoids.
-
ix.Stop the reaction by adding ice-cold 1× PBS supplemented with 2% FBS.
-
x.Centrifuge at 300g for 5 min (7°C).Note: During the process of organoids dissociation, monitor cell viability on chamber Malassez as described above in step (2. s), allowing also to verify that dissociation is complete. This leads to a distinct single cell suspension, free from intact organoids. If necessary, repeat the steps above from (vi) to (x).
-
i.
-
j.Resuspend the cells in a mix of ice-cold AO media and liquid Cultrex growth factor reduced BME type 2.
-
k.Replate 300 000 cells/40 μL as a dome.
-
l.Repeat the steps above from (b) to (i).
-
m.After each dissociation, count cells.
-
n.Cryopreserve at least 3–4 cryovials with 1.5 × 106 cells/mL/vial in FBS with 10% DMSO.Note: Having these backup stocks for later expansion is critical as bacterial or fungal contamination may occur during the expansion, which is a risk while working with primary tissues. Once the organoids are clear of the connective tissue, (after approximately 4–7 passages), generate a large frozen stock of dissociated organoids at 1.5 × 106 cells/cryovial. At this stage, there are usually 3–4, P24-well plates of organoids in culture from which you could expect to produce 20–25 cryovials.
 CRITICAL: During the initial 1–4 passages, there is a substantial amount of connective tissue, which gradually decreases with each passage (Figure 2B). Perform a quality control check on domes containing spherical organoids free of connective tissue (Figure 2C). Typically, after 4–7 passages, you can expect a high enrichment of lung epithelial progenitors. This outcome may vary depending on the donor.
CRITICAL: During the initial 1–4 passages, there is a substantial amount of connective tissue, which gradually decreases with each passage (Figure 2B). Perform a quality control check on domes containing spherical organoids free of connective tissue (Figure 2C). Typically, after 4–7 passages, you can expect a high enrichment of lung epithelial progenitors. This outcome may vary depending on the donor.
-
a.
-
4.Quality control screening for epithelial enrichment in airway organoids by immunofluorescence (Figure 2C).
-
a.Keep 10–20 μL of the organoid suspension.Note: After removing the gel, keep a sample of organoids, (See above step 3.i.ii).
-
b.Centrifuge at 300g for 5 min (4°C).Note: Keeping the centrifuge cold allows to remove the liquid Cultrex growth factor reduced BME type 2.
-
c.Gently, resuspend the intact organoids in 20 μL of cold 1× PBS.
-
d.Transfer the organoid suspension on a Superfrost plus slide.
-
e.Incubate at 20°C–25°C for 10 min in a fume hood.
-
f.Fix and permeabilize organoids by applying 20 μL of −30°C acetone.
-
g.Incubate approximately for 5 min in a fume hood until acetone evaporation.
-
h.Use a hydrophobic pen (pap-pen) to create a hydrophobic barrier around the organoids.Note: The hydrophobic barrier will help to define the area of interest for staining.
-
i.Gently rinse the organoids with 1× PBS.
-
j.Treat the organoids with Fc Receptor Block (one drop, 45 min, at 20°C–25°C).
-
k.Wash three times with 1× PBS (5 min at 20°C–25°C each time).
-
l.Treat the organoids with Background Buster treatment (one drop, 30 min, at 20°C–25°C).Note: One drop of each product is approximately 60 μL.
-
m.Repeat the wash step above in (L).
-
n.Prepare the mix of primary antibodies (unconjugated, approximately 100 μL per slide, see key resources table).Note: Dilute the antibodies anti-pan-Cytokeratin (pan-CK, to reveal enrichment of epithelial cells, 1.6 μg/mL final), anti-alpha-SMA (aSMA, to reveal presence of fibroblasts, 1 μg/mL final) and anti-Fibronectin (FN1, to reveal presence of connective tissue, 0.15 μg/mL final) in staining buffer 1× PBS/5% BSA/0.05% saponin.
-
o.Incubate in the dark for 1 h at 20°C–25°C.
-
p.Repeat the wash step above in (L).
-
q.Prepare the mix of secondary antibodies (conjugated, against primary antibodies, approximately 100 μL per slide, see key resources table).Note: Dilute the antibodies anti-mouse IgG1 (specie of the anti-pan-CK antibody), anti-mouse IgG2a (specie of the anti-aSMA antibody) and anti-rabbit IgG (specie of the anti-Fibronectin antibody) in staining buffer 1× PBS/5% BSA/0.05% saponin.
-
r.Incubate in the dark for 30 min at 20°C–25°C.
-
s.Repeat the wash step above in (L).
-
t.Counterstain sections with 1 μg/mL final concentration (in 1× PBS, 5 min, at 20°C–25°C) of 4′,6-diamidino-2-phenylindole (DAPI),
-
u.Repeat the wash step above in (l).
-
v.Whole mount organoids slide with Fluoromount-G (Thermo Fisher Scientific) and cover glass.
-
w.Image acquisition using a confocal microscope.Note: For example, Leica SP8 for high-resolution images.
-
x.Image analysis.Note: For example, using Imaris software (Bitplane).
-
a.
Primary human lung organoid-derived air-liquid interface (ALI) generation, differentiation & viral exposure
Timing: approximately 6 weeks
This part describes the workflow to culture primary human lung organoid-derived air-liquid interface (ALI) cultures, along with TEER measurement to monitor the quality of tight junction formation (Figure 3).
-
5.Prepare the Transwells (P24-well plate) for cell seeding.
-
a.Pre-coat the Transwells (2-well plate) apically with Collagen I (rat) (100 μL, final concentration of 30 μg/mL in 1× PBS) for 1 h at 37°C in a cell incubator.
-
b.Rinse the wells with 1× PBS.
-
c.Submerge the inserts in 1× PBS (at 37°C in a cell incubator) until cell seeding (approximately 30 min to 1 h).
-
a.
-
6.ALI culture generation includes 3 steps (Figures 3A and 3B).
-
a.Prepare fresh complete medial for ALI expansion supplemented with Y-27632.
-
b.Thaw lung organoid-derived cell suspension by placing 1 cryovial (containing 1.5 × 106 cells/mL) into a dry bath at 37°C for 5 min.
-
c.Transfer the cells into a conical 50 mL tube with fresh pre-warmed (37°C in a dry bath) ALI Expansion media (1 cryovial with 10 mL media).
-
d.Centrifuge at 300g for 5 min (20°C–25°C).
-
e.Resuspend in ALI Expansion media (final concentration: 300,000 cells/mL).
-
f.Plate the cells (30 000 cells/100 μL/well).Note: One cryovial can usually seed ∼48 inserts.
-
g.(1) Expansion step:
-
i.Use complete ALI expansion media supplemented with Y-27632 until reaching 100% confluence.
-
ii.Change the media every day after on the apical (100 μL) and on the basal side (500 μL).Note: ALI cultures reach 100% confluence in approximately 10–12 days, depending on the donor.
-
i.
-
h.(2) Differentiation step 1:
-
i.Use complete ALI differentiation and maintenance media supplemented with Y-27632.
-
ii.Change the media every day after on the apical (100 μL) and on the basal side (500 μL).Note: During this step, ALI cultures establish tight junctions. Perform TEER measurements using the EVOM2 ohm meter during days 13–19 after every media change (3 times per week). Once TEER values rise values > 500 Ω cm2, this indicates a healthy confluent layer (Figure 3C).
-
i.
-
i.(3) Differentiation step 2:
-
i.Airlift ALI cultures by removal of media from the apical side.
-
ii.Use complete ALI differentiation and maintenance media without Y-27632.
-
iii.Keep maintaining media supply to the basal side, (500 μL/well).Note: At this stage, ALI cultures initiate the differentiation into a pseudo-stratified epithelium. Typically, after airlift, the TEER values drop to approximately 250–500 Ω cm2 depending on the donor.
-
iv.Change the media on the basal side, 3 times per week.Note: Check ALI cultures under a bright-field microscope for evidence of beating cilia (Methods video S1) and mucus production. It takes approximately 4 weeks post-airlift until full differentiation.Alternatives: Assess mucus and cilia presence by immunofluorescence as outlined further in the step-by-step method details section related to “examples of readouts to assess response to viral exposure”.Methods video S1. Video illustrating movement of beating cilia in ALI cultures at D34 post-airlift, related to step 6 (iv) in the “step-by-step method details” sectionDownload video file (41.4MB, mp4)
-
i.
-
a.
-
7.ALI culture exposure to the SARS-CoV-2 virus. Troubleshooting 4.
-
a.Before viral exposure, remove mucus by applying pre-warmed 1× PBS apically.
-
b.Incubate for 15 min at 37°C, 5% CO2 in a humidified atmosphere.
-
c.Gently pipette up and down to remove the mucus.
-
d.Repeat the steps (a-c) above until mucus elimination.Note: Do not disrupt the cell layer during the wash step. Usually, the apical wash viscosity indicates the presence of mucus.
-
e.Change basal media.
-
f.Proceed to viral exposure by applying 105 PFU virus apically.Note: Approximately 25–100 μL at day 0 (D0). Usually, we do not remove the viral suspension.
-
g.Incubate the mock inserts (non-infected controls) with an infection medium containing no virus with a similar volume to the infected conditions.
-
h.Perform viral infection in a kinetic fashion.
-
i.Harvest inserts every day from 1 to 6 post-infection for analysis.Note: As summarized in this protocol, multiple readouts can help assess the response to a virus.
 CRITICAL: Note that during ALI generation, the TEER evaluation is critical. The cultures will collapse if you airlift ALI cultures without reaching the TEER peak. Additionally, using volumes larger than 100 μL for apical infection may result in tissue damage and collapse of the culture.
CRITICAL: Note that during ALI generation, the TEER evaluation is critical. The cultures will collapse if you airlift ALI cultures without reaching the TEER peak. Additionally, using volumes larger than 100 μL for apical infection may result in tissue damage and collapse of the culture.
-
a.
Examples of readouts to assess response to viral exposure
Timing: approximately 3 weeks
This part describes five methods to assess the response to SARS-CoV-2 virus: Plaque assays (Viral titers); Flow cytometry (virus detection on dissociated tissue); Immunofluorescence on non-dissociated tissue (quality controls steps during ALI culture generation, cell composition, virus detection, immune marker detection); RNA data (bulk RNA) and GeoMx analysis (probe-based), (Figure 4A). After viral exposure, harvest ALIs daily from 1 to 6 days post-infection. Keep at least 1 insert for each readout at each time point.
-
8.Plaque assays for viral titers on apical supernatants:
-
a.Add 150 μL of pre-warmed 1× PBS to the apical side of each ALI culture.
-
b.Incubate in a humidified 37°C, 5% CO2 incubator at ambient O2 for 15 min.
-
c.Collect supernatants by pipetting up/down.
-
d.Store at −80°C in a BSL3 facility until further use.
-
e.Serially dilute (10-fold) each apical supernatant from infected and non-infected ALI (mock) cultures in 1× PBS containing 1% BSA.Note: Adjust the volume for a P12-well plate.
-
f.Overlay each dilution per condition on pre-seeded confluent VeroE6 cell (for USA/WA1-2020) or Vero-E6 TMPRSS2 (for Beta, Delta, and Omicron) monolayers.
-
g.Incubate at 37°C for 1 h with gentle shaking every 5 min.Note: When performing in 12-well plates, infect each well with 200 μL of diluted samples.
-
h.Overlay a solution containing 2% Oxoid agarose mixed with 2× MEM supplemented with 0.3% FBS.
-
i.Incubate for 72 h at 37°C.
-
j.Fix with 4% formaldehyde solution (100 μL apically and 500 μL on the basal side, at least 12 h at 20°C–25°C).
-
k.Wash with 1× PBS, 3 times 5 min at 20°C–25°C.
-
l.Visualize viral infection by immune staining with SARS-CoV-2 NP antibody (1C7C7, 1/250) primary antibody for 1.5 h at 20°C–25°C with gentle shaking.
-
m.Repeat the wash step above in (k).
-
n.Incubate with an HRP-conjugated secondary anti-mouse antibody (1/5000) for 1 h at 20°C–25°C with gentle shaking.
-
o.Repeat the wash step above in (k).
-
p.Reveal plaques using TrueBlue peroxidase substrate (SeraCare, 250 μL for a P12-well plate for around 10 min).
-
q.Calculate titers as plaque-forming units per mL (PFU/mL) for every sample:
-
a.
-
9.Flow cytometry: for intracellular viral detection on dissociated tissue
-
a.Single cell generation: Wash ALI cultures with 1× PBS (150 μL on the apical and 500 μL on the basal side).
-
b.Treat with 0.05% trypsin on the apical (150 μL) and basal chamber (500 μL) for 15 min, 37°C.
-
c.Pipette up/down to dissociate the ALI tissue.
-
d.Transfer the cell suspension into a 15 mL tube.
-
e.Neutralize trypsin with an equal volume of 1× PBS containing 2% FBS (650 μL, at 20°C–25°C).
-
f.Centrifuge at 300g for 5 min (20°C–25°C).
-
g.Resuspend cells in a dispase I/DNase I solution (3 mL).Note: Final concentration in 1× PBS, 1.9 U/mL dispase I (Sigma) and 0.1 mg/mL DNase I (Sigma).
-
h.Incubate for 10–15 min at 37°C in a dry bath.
-
i.Centrifuge at 300g for 5 min (at 25°C–28°C).
-
j.Viability evaluation: resuspend cells in 1× PBS and stain with zombie live dead stain.Note: Use Zombie aqua or Zombie green (100 test size vial, BioLegend, concentration not available) according to manufacturer protocol, at 1/350 dilution (for approximately 3 × 106 cells) for 15 min at 25°C–28°C.
-
k.Centrifuge at 300g for 5 min (at 25°C–28°C).
-
l.Resuspend the cells in 4% methanol-free formaldehyde (100 μL/condition).
-
m.Fix over at least 12 h at 4°C.
-
n.Add 1 mL of 1× perm wash buffer (BD Biosciences) per tube containing fixed cells.
-
o.Centrifuge at 300g for 5 min (at 25°C–28°C).
-
p.Resuspend the pellet in 100 μL perm wash buffer with conjugated SARS-CoV-2 N antibody (1C7C7).Note: 1/100 or 1 μg/mL final dilution of Alexa Fluor 488-conjugated or 1/200 or 1 μg/mL final of Alexa Fluor 647-conjugated SARS-CoV-2 N antibody (1C7C7)
-
q.Incubate in the dark at 25°C–28°C for 45 min.
-
r.Add 1 mL perm wash buffer.
-
s.Centrifuge at 300g for 5 min (at 25°C–28°C).
-
t.Resuspend the pellet in 200 mL 1% BSA solution in 1× PBS.
-
u.Analyze by flow cytometry (BD, FlowJo) for live SARS-CoV-2 NP positive cells (Figure 4A).
-
a.
-
10.Immunofluorescence: for intracellular viral detection on non-dissociated tissue
-
a.Fix ALI cultures using 4% PFA (16% PFA methanol free diluted in 1× PBS), for 15 min at 4°C.
-
b.Wash with 1× PBS, then maintain in 1× PBS at 4°C until further processing (stable approximately for 36 months).
-
c.For each condition cut ALI mesh in half using a sterile scalpel.
-
d.Embed in OCT (25°C–28°C).
-
e.Snap freeze in liquid nitrogen, (Figure 4B). Troubleshooting 5.
 Pause point: You can store the OCT blocks at −80 for several months, until further processing.
Pause point: You can store the OCT blocks at −80 for several months, until further processing. -
f.Cut frozen sections at 8 μm, and air dry on Superfrost plus slides (Figure 4C).
-
g.Use a hydrophobic pen (pap-pen) to create a hydrophobic barrier around the tissue sections.Note: This will help to define the area of interest for staining.
-
h.Fix sections with 4% PFA (15 min, at 25°C–28°C).
-
i.Wash tissue sections three times with 1× PBS (5 min, at 25°C–28°C).
-
j.Permeabilize with 1× PBS/0.1% Triton X-100 (15 min, at 25°C–28°C).
-
k.Repeat the wash step above in (i).
-
l.Treat sections with Fc Receptor Block (45 min, at 25°C–28°C).
-
m.Repeat the wash step above in (i).
-
n.Background Buster treatment (30 min, at 25°C–28°C).Note: One drop of each product is approximately 60 μL.
-
o.Repeat the wash step above in (i).
-
p.Stain first with unconjugated antibodies, (1 h at 25°C–28°C),
-
q.Repeat the wash step above in (i).
-
r.Apply secondary antibodies (30 min, at 25°C–28°C).Note: Dilute antibodies in staining buffer 1× PBS/5% BSA/0.05% saponin.
-
s.Repeat the wash step above in (i).
-
t.Add mouse serum to saturate secondary antibodies.Note: Dilute normal mouse serum 1/20 in staining buffer 1× PBS/5% BSA/0.05% saponin.
-
u.Incubate for 15 min at 25°C–28°C.
-
v.Repeat the wash step above in (i).
-
w.Continue the staining with directly conjugated antibody mix for 1 h at 25°C–28°C.
-
x.Repeat the wash step above in (i).Alternatives: Depending on the staining panel (See below, Panel 3), stain for actin filaments with 10 nmol units/mL final concentration (in 1× PBS, 15 min, at 25°C–28°C) of Phalloidin ATTO647N (Sigma 65906).
-
y.Counterstain sections with 1 μg/mL final concentration (in 1×PBS, 5 min, at 25°C–28°C) of 4′,6-diamidino-2-phenylindole (DAPI).
-
z.Repeat the wash step above in (i).
-
aa.Mount ALI slides with Fluoromount-G (Thermo Fisher Scientific) and cover glass.Note: Image acquisition using a confocal microscope (for high-resolution images) or a wide-field microscope (for histocytometry). Perform image analysis by using Imaris software (Bitplane).In the context of our study, three panels of antibodies are detailed here as examples (See key resources table).
Panel 1 Panel 2 Panel 3 Cell composition: See Figure 5A CSF3 and CCL20 expression on infected ALI: See Figure 5B Infected and non-infected ALI: See Figure 5C Unconjugated Abs: Anti-SCGB1A1 Anti-Acetyl alpha-tubulin Unconjugated Abs: Anti-CCL20 Unconjugated Abs: NA Secondary Abs: Anti-rat IgG AF488 Anti-mouse IgG2b AF555 Secondary Abs: Anti-rabbit IgG AF555 Secondary Abs: NA Serum saturation: Yes Serum saturation: Yes Serum saturation: NA Conjugated Abs:
Anti-cytokeratin 5 AF594
Anti-MUC5AC AF700Conjugated Abs:
Anti-CSF3 AF594
Anti-NP SARS-CoV-2 AF488 (1C7C7)Conjugated Abs:
Anti-NP SARS-CoV-2 AF488 (1C7C7)Phalloidin ATTO647N: NA Phalloidin ATTO647N: NA Phalloidin ATTO647N: Yes Note: it is important that the ALI culture is correctly embedded in OCT (Figures 4B and 4C and Methods video S2). These steps will ensure the quality of sections for immunofluorescence and GeoMx WAT.Methods video S2. Video illustrating the embedding of ALI cultures in OCT, related to step 10 (aa) in the “step-by-step method details” sectionDownload video file (58.8MB, mp4)
-
a.
-
11.Bulk RNA sequencing:
-
a.First, dissociate ALI as described in steps (9.a.) to (9j.).Note: From 1 insert, you can recover approximately 1–2 x 106 cells.
-
b.Wash dissociated ALI cell suspension by centrifugation (300g, 5 min, at 25°C–28°C).
-
c.Isolate total RNA using Direct-zol RNA MicroPrep kits.Note: Around 200 μL of Direct-zol buffer, following Zymo Research manufacturer’s protocols.
-
d.Perform a DNase treatment using the RNase-free DNase set (QIAGEN).
-
e.Prepare cDNA libraries using polyA mRNA selection and Kapa Stranded mRNA-seq Library Prep kit (Kapa Biosystems).
-
f.Perform paired-end sequencing (i.e., 2 × 150 bp) of stranded total RNA libraries using Illumina NovaSeq or similar.
-
g.Use FastQC for quality control.16
-
h.Remove rRNA contamination with MultiQC.13
-
i.Trim reads with BBDuk tool.17
-
j.Map viral reads to the FDA-ARGOS SARS-CoV-2 reference sequences.Note: Use FDAARGOS_983, with bowtie,12,18 and map human reads to GRCh38 genome using bowtie2 (https://www.nature.com/articles/nmeth.1923.
-
k.Quantify gene expression using RSEM.11
-
l.Apply batch correction using R package SVAseq.19
-
m.Finally, use DESeq214 for read normalization and differential gene expression.Note: Adding time-matched non-infected conditions to the experimental design is important. This will ensure the quality and accuracy of RNA-seq data analysis. As an example of resulting data see Figure 6A, a Heatmap representing differentially expressed genes (DEGs) over time in response to SARS-CoV-2 variants. We conducted infection experiments on ALI from four donors with SARS-CoV-2 virus. Harvest samples for sequencing at 1, 2, 3, 4, 5, and 6 day-post infection (DPI) and similarly for mock-infected (control media without virus) samples.
-
a.
-
12.GeoMx whole Transcriptome Atlas: this assay is a probe-based method of reporting in situ RNA data analysis that can detect the expression of 18,676 genes. It includes a first step with immunofluorescence (3 targets + nuclei: basal cells (cytokeratin 5), virus NP, and Spike SARS-CoV-2 proteins) to define the regions of interest (ROI).
-
a.Cut 6 μm thick sections from ALI culture-OCT blocks and air dry on Superfrost plus slides.
 Pause point: Store the slides in a secured −80°C freezer until use, it can las for 1–2 weeks.
Pause point: Store the slides in a secured −80°C freezer until use, it can las for 1–2 weeks. -
b.Prepare PFA (4% in 1× PBS) fixed-frozen tissue slides.Note: Refer to the GeoMx NGS automated Leica Bond RNA Slide Preparation Manual (NanoString, MAN-10131-03). For more details, please refer to the NanoString website.
-
c.Load slides into the slide holder of the GeoMx digital spatial profiling (DSP) instrument.
-
d.Cover with 2 mL of manufacturer buffer S.
-
e.Stain each slide with the selected antibodies.Note: Respectively for the morphology markers nuclei Syto83 (Cy3/568 nm), Cytokeratin 5 (CK5, FITC/525 nm), SARS-CoV-2 Spike (Texas Red/615 nm), and SARS-CoV-2 Nucleocapsid (Cy5/666 nm).
-
f.Select regions of interest (ROIs) based on the antibody staining.Note: See Figure 6B, a representative image of ALI infected at 6 day-post-infection (DPI) with ROIs selection. The thick purple polygons represent the ROI strategy selection for the apical cytokeratin 5- cells (CK5-) vs basal side CK5+.
-
g.Expose selected ROIs to UV photocleavable barcode RNA probes.
-
h.Process DSP and PCR according to the manufacturer’s protocol.
-
i.Sequence purified libraries using an Illumina NovaSeq 6000 or similar machine.
-
j.GeoMX data processing and QCs according to NanoString protocols.
-
k.Perform data analysis in R (v4.2.0).
-
l.Produce graphics using ggplot2 unless otherwise stated (v3.3.6).
-
m.Differentially expressed genes (DEG):
-
i.Use a Wilcoxon rank-sum test to identify DEGs between different regions of interest.
-
ii.Adjust p values using Benjamini-Hochberg multiple test correction.
-
iii.Use p value cut-off of 0.05 and a logFC of 2.
-
iv.Identify gene variability across the whole dataset by a Kruskal-Wallis test (p < 0.05) to show the clustering of different ROI groups by heatmap.
-
i.
-
n.Gene expression heatmaps:
-
i.Produce heatmaps using the ComplexHeatmap package (v2.12.1).Note: Heatmaps use by-row scaling.
-
ii.Group ROIs first by infection type, order by Day, and then
-
iii.Cluster using the default hierarchical clustering algorithm.
 CRITICAL: GeoMx was initially optimized for FFPE tissue but has been validated on PFA-fixed frozen tissues.
CRITICAL: GeoMx was initially optimized for FFPE tissue but has been validated on PFA-fixed frozen tissues.
-
i.
-
a.
Figure 4.
Readouts to assess response to a virus, ALI culture OCT embedding, and OCT cutting
(A) Schematic workflow presenting five methods to assess response to virus. Flow cytometry and RNA extraction require tissue dissociation versus Immunofluorescence, plaque assay (viral titer on apical supernatant) and GeoMx WTA do not require tissue dissociation.
(B) Procedure of embedding ALIs in OCT and cryopreservation. First, turn the insert upside-down, and with a scalpel cut the mesh in the middle and partially around the edges, enough to hold the mesh and pull out from the insert. Be careful not to damage the cell layer. Place the ALI mesh on top of a layer of OCT and cover it with OCT to fully embed the ALI insert. Gently, with a pipette tip make sure the ALI is not curved or too close to the bottom or surface of the cryomold. Then, snap freeze in liquid nitrogen. To ensure no liquid nitrogen (LN) enters the tissue, the cryomold should be placed on top of a plastic lid resistant to LN.
(C) ALI section cutting and transfer to Superfrost plus slides. Make a mark with a Sharpie pen to indicate the location of the ALI culture on the cryomold then on the solidify OCT block. This tip will facilitate the OCT trimming until the mark and visualize the ALI (slightly in yellow). The diagram allows to picture the configuration of the ALI culture in the OCT block. Finally, ALI culture sections could be transferred into Superfrost plus slides. These steps will ensure good-quality sections for further experiments.
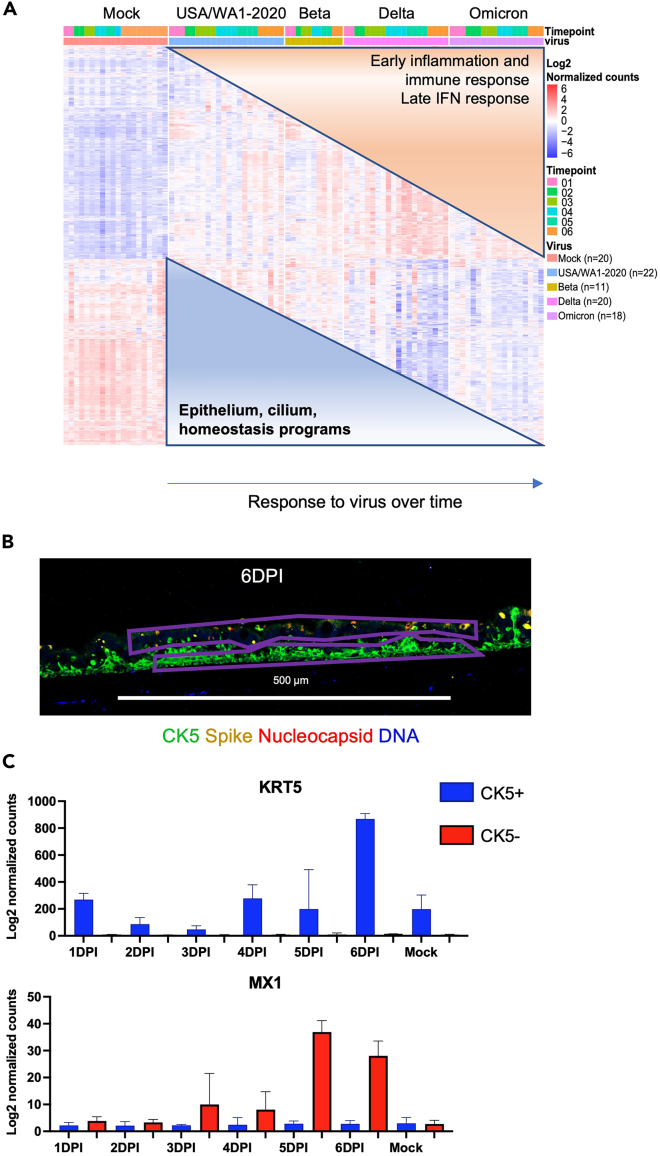
Figure 6.
Examples of expected outcomes of transcriptional response to SARS-CoV-2 variants and ROI selection for GeoMx data
(A) Heatmap representing differentially expressed genes over time in response to SARS-CoV-2 variants. ALI cultures from four donors were infected with SARS-CoV-2 and harvested for sequencing at 1, 2, 3, 4, 5, and 6 dpi, and mock-infected (control media without virus) samples were collected from days 1 through 6 days as well. The sequencing was performed in multiple batches with at least 2 independent experiments at each time-point, the cut-off used for defining differentially expressed genes: |logFC| > 1; < 0.01; normalized counts 10. Rows represent individual transcripts and columns represent individual biological replicates ordered by time-points and SARS-CoV-2 variants. Batch effect was removed using SVAseq R package. All variants induced expression of genes associated with the viral response at later time-points. This response to the virus from 1 to 6 DPI is depicted by the schematic covering the heatmap, with two “clusters”: one on the lower part “down-regulation” from 1 to 6 DPI, enriched for cilia and epithelium maintenance signatures whereas the upper part showed “up-regulation” of signatures enriched for inflammatory, immune and IFN response.
(B) ROI selection for GeoMx data: Representative image of infected ALI culture section with SARS-CoV-2 (105 PFU), at 6DPI stained for nuclei (DAPI), viral nucleoprotein (red), spike viral protein (yellow) and cytokeratin 5 (CK5). The thick purple polygons represent selected ROIs for the apical cytokeratin 5- cells (CK5-) vs. basal side CK5+. Scale bar 500 μm (white). (C) GeoMx data (one representative experiment): Bar graphs with error bars (mean ± SD) were generated using Graphpad (Prism 5) and illustrate the gene expression (log 2 normalized counts) over time within selected ROIs (at least 3 ROIs per condition) based on cytokeratin 5 protein expression (CK5 +) through KRT5 gene expression and based on cytokeratin 5 negative expression (CK5 -) of infected cells positive for SARS-CoV-2 Spike and NP, through MX1 as part of the anti-viral response which is mainly increased at later time-points. Note: Figure 6 reprinted with permission from Diana Cadena Castaneda et al. 20231 (Cell Press, Open Access).
Expected outcomes
The generation of primary human lung organoids has become an invaluable and widely utilized technique in biomedical research. Their use as progenitors for air-liquid interface (ALI) cultures has been particularly critical in advancing our understanding of the host response to respiratory viruses such as SARS-CoV-2. By using this protocol, researchers should be able to generate a large stock of organoid-derived lung epithelial progenitor cells for ALI culture production. Here we also describe some readouts that could assess ALI development and to measure the response to the virus. Notably, a simple control under a bright-field microscope allows for monitoring the proper development of ALI cultures by following mucus formation and cilia beating as illustrated in Methods video S1. Furthermore, Figures 5A–5C illustrate the results of immunofluorescence readout to monitor respectively ALI differentiation by following cell composition; viral infection with CSF3 localization on the basal side and CCL20 on the apical side, and a simple staining with anti-NP and phalloidin report levels of ALI culture infection. Flow cytometry, another protein-based method allows to measure viral infectivity as illustrated in Figure 5D. Finally, Figure 6A illustrates the transcriptional response to the virus across time-points and variants. Furthermore, Figures 6B and 6C illustrate the quality of the sections, and the precision of ROI selection resulting at RNA level (probe-based) with a “transcriptional” separation between cells expressing cytokeratin 5 (CK5+) and those infected not expressing CK5 (CK5-) based on MX1 expression as downstream of infection.
Figure 5.
Examples of expected outcomes of immunofluorescent images and flow cytometry gating strategy
(A) Representative immunofluorescent (IF) section (8 μm) of differentiated lung organoid-derived ALI cultures. The left panel, merged figures, shows markers for ciliated cells (acetylated a-tubulin, red) and goblet cells (MUC5AC, cyan). Right panel, merged figures, showing club cells (SCGB1A1, green), and basal cells (CK5, white). Nuclei (DAPI, blue). Scale bar 50 μm, in white on the left corner.
(B) Representative images of ALI cultures mock-infected at 6 days and ALI infected with SARS-CoV-2 USA/WA1-2020 (105 PFU) at 1, 3, and 6 days post-infection (DPI) stained for nuclei (DAPI, blue), viral NP (white) to reveal the effective viral replication, CSF3 (green) and CCL20 (red). Scale bars 10 μm, in white on the left corner.
(C) Representative images of donor 3 of mock-infected (control media without virus at 6 days) and infected ALI cultures with SARS-CoV-2 (105 PFU) at 6 days post-infection (DPI) stained for nuclei (DAPI), viral nucleoprotein (NP, green) to reveal the effective viral replication and phalloidin (Actin filament, red) to reveal tissue structure. Scale bar 40 μm, in white on the left corner.
(D) Gating strategy example for flow cytometry analysis. A single cell suspension was prepared from SARS-CoV-2 infected ALI cultures. Cell suspensions were stained for viability and viral infection using an anti-NP antibody specific to SARS-CoV-2. Plots from left to right show serial gating to identify percentages of infected (NP-positive) viable cells. Note: Figure 5 reprinted with permission from Diana Cadena Castaneda et al. 20231 (Cell Press, Open Access).
Limitations
This protocol involves the use of viable frozen lung tissue from whole lung donors to generate primary human lung organoids, which then serve to generate air-liquid-interface (ALI) cultures for studying the response to viruses. However, due to donor-to-donor variations in lung tissue, we may observe differences during organoid expansion and variations in ALI culture composition and response to the virus. Primary cell cultures can also be susceptible to microbial and fungal outgrowth during the organoid and ALI generation process. To prevent cross-contamination between wells or plates and to maintain culture integrity, we take the following measures: Prepare one tube of media per plate, and to minimize the risk of cross-contamination, change tips between each well to avoid transferring potential contaminants. If plates become contaminated, add immediately bleach and discarded them to prevent further contamination spread. By implementing these precautions, researchers can reduce the risk of contamination and preserve the quality and consistency of primary human lung organoid cultures, leading to more reliable and reproducible experimental results.
Troubleshooting
Problem 1
Incomplete or partial tissue digestion, related to the section “human lung tissue viable freeze-thawing & lung airway organoid generation” step (2 d) involving collagenase digestion.
Potential solution
-
•
Transfer the tissue to a Petri dish.
-
•
Mechanically shred the tissue with sterile slides to break the connective tissue.
-
•
Centrifuge at 300g for 5 min (25°C–27°C).
-
•
Repeat step (j) with new 10 mL of complete media for primary lung airway organoids (AO) containing 1–2 mg mL collagenase I on an orbital shaker at 37°C for an additional hour.
Problem 2
Incomplete red blood cell lysis, related to “Human lung tissue viable freeze thawing & lung airway organoid generation” step (2 f).
Potential solution
-
•
Use cell culture grade water to dilute the red blood cell lysis buffer. Do not use PBS.
-
•
Incubate samples on ice for 3–4 min.
-
•
Stop the reaction by the addition of 3 mL of 1× PBS.
Problem 3
Loss of cells during organoid passaging due to their high adherence to the plastic, related to the “human lung tissue viable freeze-thawing & lung airway organoid generation” step (3).
Potential solution
-
•
To avoid organoid adherence, pre-coat, tips, pipettes, and tubes with 1× PBS with 1% BSA for every step involving organoids.
Problem 4
Loss of cells during the viral infection, due to excess of apical buffer and/or mucus. Related to “primary human lung organoid-derived air-liquid interface (ALI) generation, differentiation & viral exposure” step (7).
Potential solution
-
•
Prevent mucus buildup on the apical side by washing every 48–72 h before infection.
-
•
For every infection, use between 25–100 μL virus infection medium. More than 100 μL will likely drown the cells if left for more than 24–48 h.
-
•
If using more than 100 μL, remove the infection medium after 3 h of exposure.
Problem 5
Difficulty locating the ALI tissue within the OCT block. Related to “Immunofluorescence: for intracellular viral detection on non-dissociated tissue” step (10 b).
Potential solution
-
•
During the embedding of ALI in the OCT, mark the tissue’s start on the plastic mold surface using a sharpie prior to the snap-freeze process (See Figure 4B).
-
•
Prepare the OCT block for sectioning, use the pre-marked guide to draw a sharpie line directly on the OCT block, Trim (10–30 μm) until nearing the mark and then precisely section (8 μm) until tissue becomes visible (See Figure 4C).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Karolina Palucka (Karolina.palucka@jax.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All sequencing data generated mentioned here has been deposited to GEO and is publicly available: GSE225603.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this working paper is available from the lead contact upon request.
Acknowledgments
We thank our tissue donors and the Microscopy, Single Cell Biology, Genome Technology, and CTRS Scientific Services of The Jackson Laboratory. We thank Randy Albrecht for support with the BSL3 facility and procedures at the Icahn School of Medicine at Mount Sinai (ISMMS). This work is partially supported by NIAID grants U19AI142733 (K.P.) and U19AI135972 (A.G.-S.); R01AI141609 (A.W.); R01AI160706 (MS); NIDDK grant R01DK130425 (M.S.); NCI grants P30 CA034196 (K.P.) and U54CA260560 (A.G.-S.); CRIPT (Center for Research on Influenza Pathogenesis and Transmission); and NIAID funded Center of Excellence for Influenza Research and Response (CEIRR, contract number 75N93021C00014) to A.G.-S.
Author contributions
Conceptualization, D.C.C., A.W., and K.P.; methodology, A.W., F.M., D.C.C., and K.P.; validation, D.C.C. and S.J.; formal analysis and data curation, M.Y.; investigation, D.C.C., S.J., M.S., J.M., M. Callender, M. Coxe, A.C., J.G.-B.D., T.-C.W., and F.M.; resources, A.G.-S. and M.S.; writing – original draft, D.C.C.; writing – review and editing, D.C.C., S.J., A.W., M.Y., M.S., A.G.-S., and K.P.; funding acquisition, A.W., A.G.-S., M.S., and K.P.; supervision, D.C., A.G.-S., M.S., A.W., and K.P.
Declaration of interests
The A.G.-S. laboratory has received research support from GSK, Pfizer, Senhwa Biosciences, Kenall Manufacturing, Blade Therapeutics, Avimex, Johnson & Johnson, Dynavax, 7 Hills Pharma, PharmaMar, ImmunityBio, Accurius, nanoComposix, Hexamer Therapeutics, N-fold LLC, Model Medicines, Atea Pharmaceuticals, Applied Biological Laboratories, and Merck, outside of the reported work. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: CastleVax, Amovir, Vivaldi Biosciences, ContraFect, 7 Hills Pharma, Avimex, Pagoda, Accurius, Esperovax, Farmak, Applied Biological Laboratories, PharmaMar, CureLab Oncology, CureLab Veterinary, Synairgen, Paratus, and Pfizer, outside of the reported work. A.G.-S. has been an invited speaker in meeting events organized by Seqirus, Janssen, Abbott, and AstraZeneca. A.G.-S. is an inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections and cancer, owned by the Icahn School of Medicine at Mount Sinai, New York, outside of the reported work. The M.S. laboratory has received unrelated funding support in sponsored research agreements from Phio Pharmaceuticals, 7 Hills Pharma, argenx, and Moderna. K.P. is a stockholder in Cue Biopharma and Guardian Bio, scientific advisor to Cue Biopharma and Guardian Bio, and co-founder of Guardian Bio. K.P. declares unrelated funding support from Guardian Bio (current) and Merck (past).
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102735.
Contributor Information
Adam Williams, Email: adam.williams@northwestern.edu.
Karolina Palucka, Email: karolina.palucka@jax.org.
References
- 1.Castaneda D.C., Jangra S., Yurieva M., Martinek J., Callender M., Coxe M., Choi A., García-Bernalt Diego J., Lin J., Wu T.-C., et al. Spatiotemporally organized immunomodulatory response to SARS-CoV-2 virus in primary human broncho-alveolar epithelia. iScience. 2023;26 doi: 10.1016/j.isci.2023.107374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fulcher M.L., Gabriel S., Burns K.A., Yankaskas J.R., Randell S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 3.Rayner R.E., Makena P., Prasad G.L., Cormet-Boyaka E. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for in vitro Lung Model Studies. Sci. Rep. 2019;9:500. doi: 10.1038/s41598-018-36735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh B., Park B., Bhowmik D., Nishida K., Lauver M., Putcha N., Gao P., Ramanathan M., Hansel N., Biswal S., Sidhaye V.K. Strong correlation between air-liquid interface cultures and in vivo transcriptomics of nasal brush biopsy. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;318:L1056–L1062. doi: 10.1152/ajplung.00050.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida K., Brune K.A., Putcha N., Mandke P., O’Neal W.K., Shade D., Srivastava V., Wang M., Lam H., An S.S., et al. Cigarette smoke disrupts monolayer integrity by altering epithelial cell-cell adhesion and cortical tension. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313:L581–L591. doi: 10.1152/ajplung.00074.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh B., Reyes-Caballero H., Akgün-Ölmez S.G., Nishida K., Chandrala L., Smirnova L., Biswal S., Sidhaye V.K. Effect of sub-chronic exposure to cigarette smoke, electronic cigarette and waterpipe on human lung epithelial barrier function. BMC Pulm. Med. 2020;20:216. doi: 10.1186/s12890-020-01255-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kliment C.R., Nguyen J.M.K., Kaltreider M.J., Lu Y., Claypool S.M., Radder J.E., Sciurba F.C., Zhang Y., Gregory A.D., Iglesias P.A., et al. Adenine nucleotide translocase regulates airway epithelial metabolism, surface hydration and ciliary function. J. Cell Sci. 2021;134 doi: 10.1242/jcs.257162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung-Luk B.H., Narayanan G.A., Ghosh B., Wally A., Lee E., Mokaya M., Wankhade E., Zhang R., Lee B., Park B., et al. SARS-CoV-2 infection alters mitochondrial and cytoskeletal function in human respiratory epithelial cells mediated by expression of spike protein. mBio. 2023;14 doi: 10.1128/mbio.00820-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Böttinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:1–20. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323–338. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550–570. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathnasinghe R., Jangra S., Ye C., Cupic A., Singh G., Martínez-Romero C., Mulder L.C.F., Kehrer T., Yildiz S., Choi A., et al. Characterization of SARS-CoV-2 Spike mutations important for infection of mice and escape from human immune sera. Nat. Commun. 2022;13:3921–3934. doi: 10.1038/s41467-022-30763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews S. 2010. FastQC A Quality Control Tool for High Throughput Sequence Data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 17.BBMap Download | SourceForge.Net. https://sourceforge.net/projects/bbmap/.
- 18.Bowtie: An Ultrafast, Memory-Efficient Short Read Aligner. http://bowtie-bio.sourceforge.net/index.shtml.
- 19.Zhang Y., Parmigiani G., Johnson W.E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020;2:lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All sequencing data generated mentioned here has been deposited to GEO and is publicly available: GSE225603.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this working paper is available from the lead contact upon request.