Abstract
Phase separation compartmentalizes many cellular pathways. Given that the same interactions that drive phase separation mediate the formation of soluble complexes below the saturation concentration, the contribution of condensates versus complexes to function is sometimes unclear. Here, we characterized several new cancer-associated mutations of the tumor suppressor speckle-type POZ protein (SPOP), a substrate recognition subunit of the Cullin3-RING ubiquitin ligase. This pointed to a strategy for generating separation-of-function mutations. SPOP self-associates into linear oligomers and interacts with multivalent substrates, and this mediates the formation of condensates. These condensates bear the hallmarks of enzymatic ubiquitination activity. We characterized the effect of mutations in the dimerization domains of SPOP on its linear oligomerization, binding to its substrate DAXX, and phase separation with DAXX. We showed that the mutations reduce SPOP oligomerization and shift the size distribution of SPOP oligomers to smaller sizes. The mutations therefore reduce the binding affinity to DAXX but unexpectedly enhance the poly-ubiquitination activity of SPOP toward DAXX. Enhanced activity may be explained by enhanced phase separation of DAXX with the SPOP mutants. Our results provide a comparative assessment of the functional role of complexes versus condensates and support a model in which phase separation is an important factor in SPOP function. Our findings also suggest that tuning of linear SPOP self-association could be used by the cell to modulate activity and provide insights into the mechanisms underlying hypermorphic SPOP mutations. The characteristics of cancer-associated SPOP mutations suggest a route for designing separation-of-function mutations in other phase-separating systems.
Keywords: biomolecular condensate, phase transition, soluble complex, ubiquitin ligase, endometrial cancer
Phase separation orchestrates spatial and temporal compartmentalization of the cell (1, 2) and influences fundamental processes such as chromatin compartmentalization (3, 4, 5), membrane receptor signaling (6, 7), localized translation (8), and the stress response (9, 10, 11). Dysregulation of phase separation can cause neurodegenerative diseases and cancer (12, 13). Hence, phase separation is a fundamentally important process used ubiquitously in cells. Yet, important questions remain regarding the extent of the functional role phase separation plays in several processes. Phase separation is mediated by multivalent interactions and leads to the formation of condensates above the saturation concentration, but these same multivalent interactions can also mediate the formation of higher-order oligomers, or so-called “clusters”, in subsaturated solutions (14, 15). If clusters and condensates are formed via the same interactions, how can their relative functional contributions be determined?
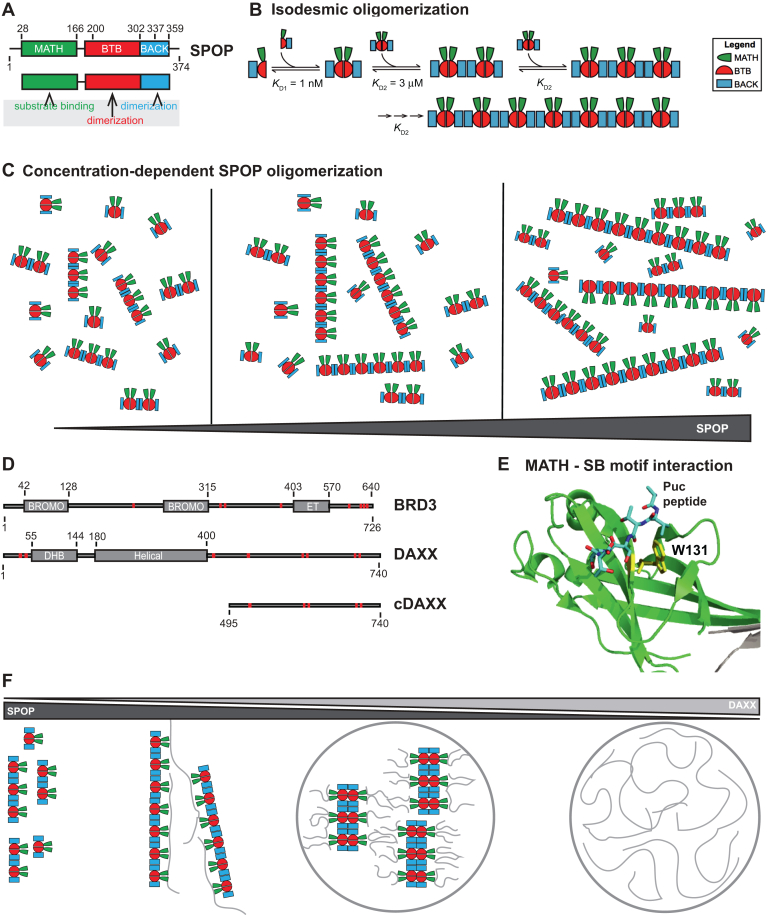
We address this question using the tumor suppressor speckle-type POZ protein (SPOP), a substrate recognition subunit of the Cullin3-RING ubiquitin ligase (CRL3). SPOP recruits substrates to the CRL3, and these substrates are subsequently polyubiquitinated and degraded (16, 17, 18). We previously demonstrated that SPOP forms linear higher-order oligomers via its two dimerization domains, the broad-complex, tramtrack, and bric-a-brac (BTB) and BTB and C-terminal kelch (BACK) domains (19) (Fig. 1, A and B). Concentration-dependent oligomerization of SPOP results in an exponential distribution of oligomer sizes, wherein increasing protein concentrations promote the formation of longer and longer oligomers, but long oligomers are always also in equilibrium with short oligomers (Fig. 1C). Given that each SPOP monomer contains a meprin and TRAF homology (MATH) domain that mediates substrate binding, these SPOP oligomers are inherently multivalent for substrates. Substrates themselves can also contain multiple SPOP-binding (SB) motifs (20, 21, 22) (Fig. 1D), each of which can bind in the substrate-binding cleft of the MATH domain (Fig. 1E). The resulting multivalent interactions encoded by SPOP and its substrates mediate phase separation and the formation of SPOP/substrate condensates that coexist with a dilute phase (23).
Figure 1.
The substrate adaptor SPOP forms higher-order oligomers that are multivalent for substrates.A, SPOP has three functional domains; the MATH domain (green) with the substrate-binding site, and the BTB and BACK domains (red and blue, respectively) that dimerize. B, SPOP undergoes isodesmic oligomerization via its tandem self-association domains: KD1 and KD2 are the self-association affinities for the BTB and BACK domains, respectively. KD2 remains the same independent of oligomer size (19). C, increasing SPOP concentrations shift the size distribution to longer oligomers, while short and long oligomers are always in equilibrium with each other. D, schema of the SPOP substrates BRD3 and DAXX. BRD3 has two bromodomains and one extra-terminal (ET) domain. DAXX contains a DAXX helical bundle (DHB) domain and a helical domain. SPOP-binding motifs are depicted as red bars. E, cartoon model of the MATH domain structure with the canonical SPOP-binding motif from the substrate Puc shown in stick representation. W131 (shown in yellow stick representation) is a residue in the substrate-binding cleft important for substrate binding. F, schematic representation of possible SPOP, DAXX, and SPOP/DAXX assemblies: SPOP alone forms oligomers (left) (19). At low molar ratios of DAXX:SPOP, large clusters of SPOP/DAXX are established by stabilization of SPOP oligomers through multivalent DAXX. DAXX can also crosslink oligomers (middle left). At higher molar ratios of DAXX:SPOP, DAXX cannot bind along SPOP oligomers, and SPOP-DAXX brushes are formed, which crosslink via intermolecular DAXX-DAXX interactions and lead to the formation of SPOP/DAXX condensates (middle right). DAXX alone can form condensates (right) (23, 24). BACK, BTB and C-terminal kelch; BTB, broad-complex, tramtrack, and bric-a-brac; MATH, meprin and TRAF homology; SPOP, speckle-type POZ protein.
The ability of SPOP to undergo linear oligomerization is required for its subcellular localization to nuclear speckles and activity as a substrate recognition subunit of the CRL3 (19). Mutations that disrupt one or both SPOP dimerization interfaces result in constitutive SPOP monomers or dimers and substantial functional defects; the SPOP mutants localize diffusely in the nucleoplasm instead of in nuclear speckles or other nuclear bodies, and they have strongly reduced activity toward substrate polyubiquitination (19, 23). But is SPOP oligomerization important because it drives phase separation with substrates (Fig. 1F, middle right panel), or are the higher-order SPOP/substrate oligomers, that is, the clusters that form below the saturation concentration, the functional entities (Fig. 1F, middle left panel)? In other words, are phase-separated condensates uniquely suited for function or can clusters perform similar functions? The condensates formed by SPOP and its substrate DAXX have the hallmarks of compartments that are active for SPOP-mediated ubiquitination (23). In vitro reconstituted SPOP/DAXX condensates are also active, but so are the clusters that form below the saturation concentration (23). Given that both types of structures are formed by multivalent interactions between SPOP and DAXX, separation-of-function mutations are difficult to envision. This is the case not only for the SPOP system but in general for multivalent systems that both undergo phase separation and form clusters that may also mediate activity.
We previously characterized the network structure of clusters and condensates in the SPOP/DAXX system (24) (Fig. 1F). We demonstrated that these clusters are large SPOP oligomers that are stabilized and cross-linked by the binding of multivalent DAXX. In this system, the resulting clusters are large enough to be light microscopically observable. At higher DAXX/SPOP molar ratios, multivalent DAXX cannot stabilize SPOP oligomers and instead leads to the formation of SPOP-DAXX brushes; in these structures, multivalent DAXX molecules hang off SPOP oligomers (Fig. 1F, middle right). DAXX-DAXX interactions between SPOP-DAXX brushes mediate the formation of condensates above the saturation concentration. Therefore, the network structures (and their underlying interactions) differ between clusters and condensates. These conclusions are also in agreement with theoretical considerations and computational studies of associative polymers, in which cohesive interactions (by so-called stickers, which can make noncovalent physical crosslinks) mediate networking (25, 26), but accompanying density transitions are mediated by the relative insolubility (i.e., poor solvation) of other parts of the molecules (which are typically called spacers) (27, 28). Our recent work also shows that for prion-like low-complexity domains, the solubility of the protein molecules (typically encoded in the spacers) as well as their networking ability (via the stickers) together determine the driving force for phase separation (29). Hence, generation of mutations that produce opposite effects on clusters versus condensates should be attainable (15) and would allow for the interrogation of their respective functional contributions.
Here, we assess the molecular mechanism of action of a previously uncharacterized set of cancer-associated SPOP mutations and find that they have opposing effects on the stability of SPOP/substrate clusters and condensates; that is, they increase the formation of condensates and reduce the formation of clusters. The mutations target conserved residues in the BTB and BACK domain interfaces and do not appear in healthy individuals but are found in patients with endometrial, skin, and other cancers. We show that the mutations result in a shift in the SPOP oligomer size distribution to smaller sizes relative to WT and reduce binding affinity to multivalent substrates in vitro. Paradoxically, the mutations enhance substrate ubiquitination in cells, a surprising observation given the importance of oligomerization for function. We provide a possible molecular explanation for this phenotype by demonstrating that the SPOP interface mutants have a stronger driving force for phase separation together with substrates in vitro. Mutations that differentially affect clusters and phase-separated condensates therefore enable the comparative assessment of these functions. Our results point to the importance of phase separation for SPOP function and suggest a strategy for disentangling the role of clusters versus condensates in other biological processes.
Results
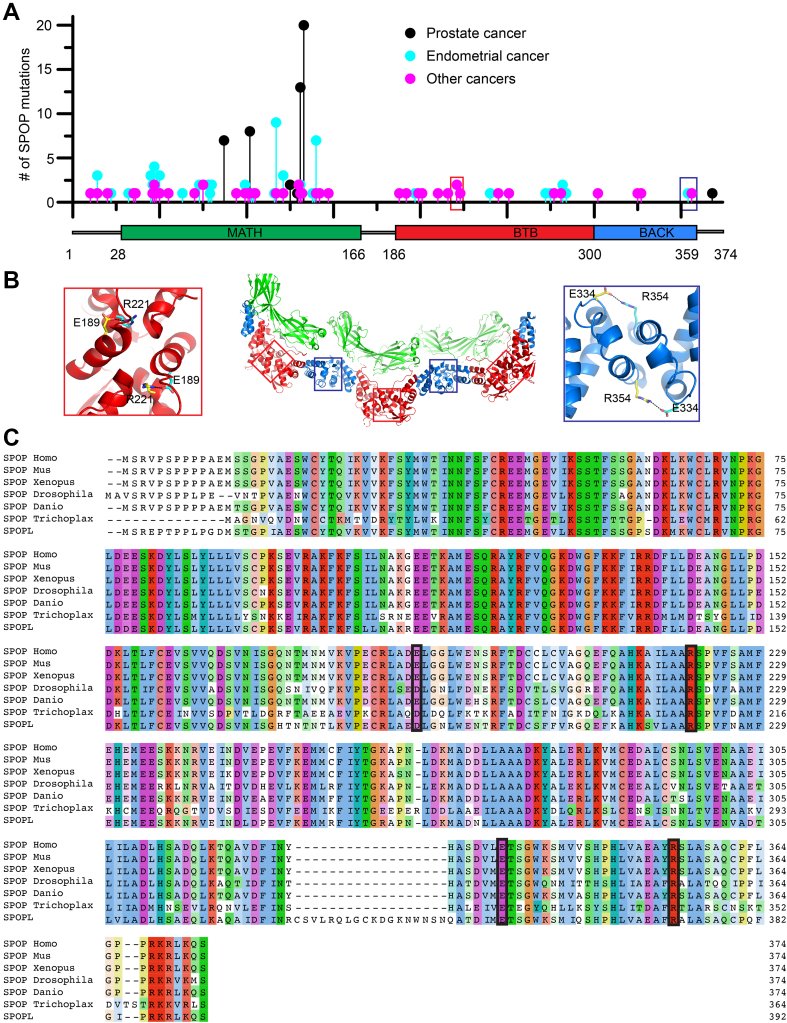
We and others previously established the importance of higher-order SPOP oligomerization for its function (19, 23, 30); mutations that completely abrogate dimerization of either or both dimerization interfaces result in altered subcellular localization and loss of ubiquitination activity (19, 23). To probe the impact of more subtle changes to oligomerization on SPOP function, we reviewed cancer genomes for SPOP mutations that may modulate oligomerization. The most prevalent SPOP mutations found in cancer patients are those in the substrate-binding cleft of the MATH domain (Fig. 2A); they reduce substrate binding and turnover of proto-oncogenic substrates and result in prostate cancer oncogenesis (31). By contrast, mutations on the periphery of the MATH domain are present in patients with endometrial cancer (Fig. 2A) and have counterintuitive effects on substrate ubiquitination; some substrates undergo enhanced ubiquitination in their presence while others undergo reduced ubiquitination (32). Additional mutations of unknown significance occur with low prevalence across most of the sequence. However, two uncharacterized mutations in the dimerization domains, R221C and R354H, stood out due to their somewhat higher prevalence as well as their locations (Fig. 2A). The mutated residues are located in the BTB and BACK domain interfaces, respectively, where we expect them to interfere with ion pair bonding across the interface; R221 usually forms a salt bridge with E189, and R354 forms a bridge with E334 (Fig. 2B). R221C was found in four patients with skin, mouth, bladder, or large intestine cancers and one additional patient had the mutation R221H; R354H was found in two patients with endometrial or large intestine cancer, and three additional patients had the mutation R354C. R221 and R354 are conserved across animals (Fig. 2C), and mutation of R221 does not appear as a normal SNP in the population according to the gnomAD database; R354H was seen in one individual. Based on these results, we predicted that the interface mutations reduce linear SPOP oligomerization and that they are potentially pathogenic.
Figure 2.
Conserved residues that form contacts across the dimerization interfaces are mutated in cancer patients.A, the lollipop plot shows mutations identified in cancer patients in all three domains of SPOP. Prostate cancer–associated mutations (black) are prevalent in the substrate-binding cleft of the MATH domain. These mutations impair SPOP–substrate interactions, resulting in the stabilization of oncoproteins in prostate cancer patients. Endometrial cancer mutations (blue), which have intermediate frequency, also cluster in the MATH domain. However, these surprisingly promote ubiquitination of some substrates while inhibiting the ubiquitination of others. Among the other SPOP mutations identified (magenta), R221C (red box) and R354H (blue box) are located in the BTB and BACK domains, respectively. Mutation data were collated from cBioPortal (47, 48). B, model of a SPOP oligomer generated by superimposing the SPOP crystal structures for the BTB dimer (36) and BACK dimer (35). Red and blue boxes show the locations of R221 and R354 in the BTB/BTB and BACK/BACK interfaces, respectively. Both residues form salt bridges with glutamic acid residues across their respective dimerization interfaces. C, sequence alignment of human SPOP with SPOP homologs in other species and with human SPOP-like protein (SPOPL). The alignment shows evolutionary conservation of R221 and R354 and their corresponding salt bridge partners E189 and E334. Homo, Homo sapiens; Mus, Mus musculus; Xenopus, Xenopus laevis; Drosophila, Drosophila melanogaster; Danio, Danio rerio; and Trichoplax, Trichoplax adhaeren. BACK, BTB and C-terminal kelch; BTB, broad-complex, tramtrack, and bric-a-brac; MATH, meprin and TRAF homology; SPOP, speckle-type POZ protein.
SPOP interface mutants reduce higher-order oligomerization
We first assessed the effects of the mutations on the overall structure of SPOP using CD spectropolarimetry. Both mutants had similar CD spectra relative to the WT protein (Fig. S1A), indicating that the mutations did not affect the protein structure. Next, we used composite-gradient multiangle light scattering (CG-MALS) experiments to characterize the linear oligomerization of SPOPWT and the SPOP mutants (Fig. 3A). We previously characterized SPOP self-association using CG-MALS and found that the SPOP dimers formed via BTB domain dimerization are the building blocks in an isodesmic self-association through the BACK domains, in which addition of each dimer is governed by the same KD value (19). Recent characterization using a series of small-angle X-ray scattering experiments, as a function of SPOP concentration, in combination with molecular dynamics simulations showed good agreement with this mechanism (33).
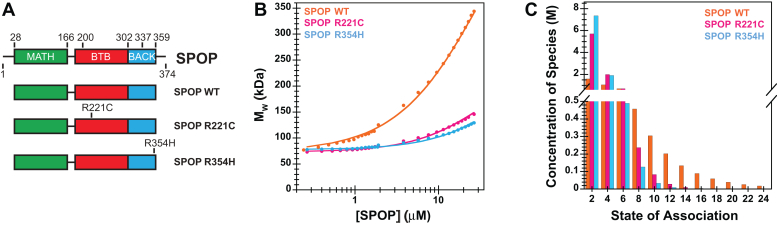
Figure 3.
Interface mutations reduce SPOP self-association and oligomer size.A, cartoon schematics of SPOPWT, SPOPR221C, and SPOPR354H. The experiments with purified protein used constructs comprising residues 28 to 359; experiments in cells used the full-length protein. B, experimental weight-average molar mass (Mw) from CG-MALS (shown in circles) for SPOPWT and the interface mutants were fitted to an isodesmic self-association model in which SPOP dimers are the self-associating unit (solid lines) [as in (19)]. The largest SPOP oligomer taken into account was an undecamer of SPOP dimers [(SPOP2)11]. C, graphical representation of the SPOP concentration of each oligomeric species in a 10 μM solution of SPOPWT or the interface mutants. CG-MALS, composite-gradient multiangle light scattering; SPOP, speckle-type POZ protein.
We generated a dilution series of SPOP solutions and determined their static light scattering intensities. As expected, SPOPR221C and SPOPR354H showed reduced self-association relative to SPOPWT as evidenced by lower scattering intensities at given protein concentrations (Fig. 3B). To interpret the data, we used an isodesmic oligomerization model in which SPOP dimers (i.e., BTB dimers) self-associate via BACK domain interactions, and the addition of each successive BTB dimer occurs with the same affinity (Fig. 1B). We previously determined the KD of BTB dimerization to be ∼1 nM (19). The KD for the isodesmic step increased from 2.7 μM for SPOPWT to 14.8 μM for SPOPR221C and 26.9 μM for SPOPR354H, respectively (Table 1). The mutation R221C is expected to alter the BTB dimerization affinity, but we can capture the shift to smaller oligomers with a fit to the same isodesmic model.
Table 1.
Affinities for SPOP self-association and substrate binding
| SPOP variant (residues 28–359) | Isodesmic KDa [μM] | KD for peptide [μM]b | KD for cDAXX [μM]b |
|---|---|---|---|
| WT | 2.7 ± 0.4 | 9.8 ± 0.7 | 1.2 ± 0.1 |
| R221C | 14.8 ± 2.0 | 5.9 ± 0.4 | 3.2 ± 0.6 |
| R354H | 26.9 ± 2.5 | 5.1 ± 0.6 | 3.0 ± 0.4 |
| W131G | N.D. | No binding | N.D. |
| mutBACK | N.D. | 4.3 ± 0.3 | N.D. |
| mutBTB | N.D. | 5.7 ± 0.4 | N.D. |
| mutBTB-BACK | N.D. | 7.4 ± 0.4 | N.D. |
BACK, BTB and C-terminal kelch; BTB, broad-complex, tramtrack, and bric-a-brac; ND, not determined.
From CG-MAS; mean from at least three replicates ± S.D.
From fluorescence anisotropy assay; mean from at least three replicates ± S.D.
These in vitro assays were performed using truncated constructs encompassing residues 28 to 359 of SPOP; this construct lacks the extreme termini which were previously thought to be disordered. We recently determined the cryo-EM structure of full-length SPOP (34), which revealed linear oligomers in agreement with models previously generated by superposition of available crystal structures of individual domains (19). However, the SPOP termini formed additional contacts in the structure not previously appreciated, and we therefore tested whether these might enhance the enzymatic activity of SPOP. In vitro ubiquitination assays revealed that full-length SPOP was less active than SPOP28–359 toward ubiquitination of BRD3 (Fig. S1B), potentially because additional contacts in the full-length protein restrict the conformational freedom of the MATH domains and reduce its ability to engage multivalent substrates. We expect that the mutations R221C and R354H lead to similar reductions in the self-association of full-length SPOP that are observed for SPOP28–359. In conclusion, SPOP interface mutants can form higher-order oligomers, but their size distributions are shifted to smaller sizes (Fig. 3C). The effect of the R221C mutant was more pronounced than that of the R354H mutant. The interface mutants thus do not affect SPOP folding but reduce linear SPOP oligomerization to different extents.
SPOP interface mutants have enhanced ubiquitination activity
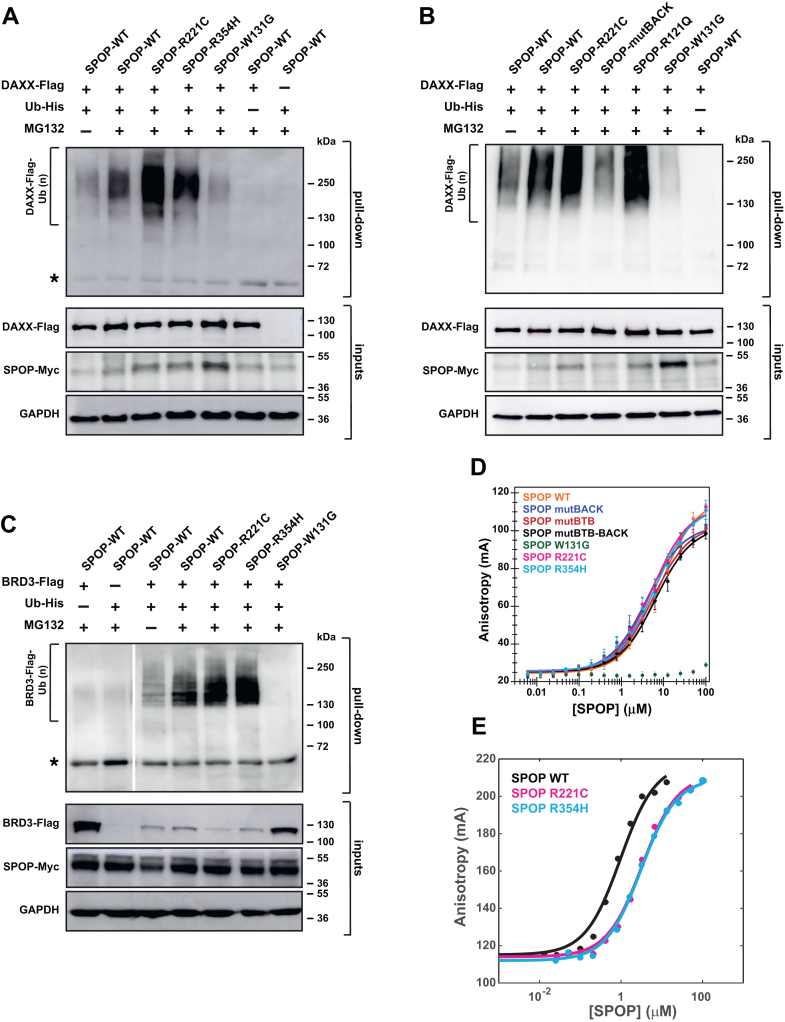
To test whether the SPOP interface mutants had functional defects, we established inducible expression systems for full-length SPOP WT and the SPOP mutants in T-REx cells wherein SPOP was expressed at relatively low levels (Fig. S2), and we performed in-cell ubiquitination assays with DAXX as the substrate. While the prostate cancer mutant SPOPW131G had only weak activity as expected, the interface mutants SPOPR221C and SPOPR354H polyubiquitinated DAXX more strongly than SPOPWT (Fig. 4A and S3A). As a control, we examined the effect of a SPOP mutant harboring the mutation Y353E (resulting in SPOPmutBACK) (35), a mutation we previously used to completely abrogate BACK domain dimerization (19, 23). We confirmed that SPOPmutBACK has low ubiquitination activity toward DAXX, which is in agreement with our previous observations that linear higher-order SPOP oligomerization is required for full function (19, 23) (Fig. 4B). The opposite effects observed for the mutations Y353E and R354H on substrate ubiquitination highlighted that complete loss of BACK domain dimerization strongly suppresses activity (and also leads to mislocalization of the protein), while a mild reduction of BACK domain dimerization, and the accompanied reduced SPOP oligomer size, increases substrate ubiquitination.
Figure 4.
SPOP interface mutations enhance ubiquitination but reduce substrate binding.A, SPOPR221C and SPOPR354H enhance DAXX polyubiquitination. Representative immunoblot showing ubiquitination in T-REx cells transfected to express SPOP-Myc, DAXX-Flag, and His-tagged ubiquitin. Twenty-four hours posttransfection, cells were incubated with DMSO or 20 μM MG132 for 4 to 5 h. The cells were lysed, and the resulting lysates were used in His-tag pull-down assays, using nickel-NTA beads under denaturing conditions. The samples were then run on SDS–PAGE gels, and the gels were immunoblotted with an anti-Flag antibody. Protein input was verified using antibodies for Myc, Flag, and GAPDH (loading control). For a replicate experiment, Fig. S3A. B, similar to the interface mutant R221C, the endometrial cancer mutant R121Q promotes SPOP polyubiquitination activity. The opposite effect is observed for mutBACK, the SPOP mutant that completely disrupts BACK domain dimerization. Experimental conditions as in (A). C, SPOPR221C and SPOPR354H enhance BRD3 polyubiquitination, indicating that gain-of-function for the interface mutants is not limited to the substrate DAXX. Experimental conditions as in (A), except that BRD3-Flag is expressed as the substrate instead of DAXX-Flag. Asterisks in panels A and C indicate a cross-reacting band. Slightly higher SPOPW131G levels are typical (23, 34) and seem to stem from loss of autoregulatory processes. For a replicate experiment, see Fig. S3B. D, SPOP affinity for its canonical binding motif is not altered for the SPOPR221C and SPOPR354H mutants relative to SPOPWT, but it is reduced in SPOPW131G, a prostate cancer mutant that abrogates substrate binding. The affinities were measured using a fluorescence anisotropy binding assay with a fluorescently labeled peptide that was derived from a SPOP-binding motif from the substrate Puckered. Markers show average values from three replicate experiments, the error bars represent the standard deviation, and solid lines show a fit to a general binding model (49). E, the binding affinity for SPOPR221C and SPOPR354H to the multivalent substrate is slightly decreased. Affinities were measured using a fluorescence anisotropy binding assay with fluorescently labeled cDAXX. Markers show experimentally determined values, and solid lines show a fit to a general binding model (49). Three replicate measurements were performed. BACK, BTB and C-terminal kelch; BTC, broad-complex, tramtrack, and bric-a-brac; DMSO, dimethylsulfoxide; NTA, nitriloacetic acid; SPOP, speckle-type POZ protein.
These observations raised multiple questions: (1) if higher-order oligomerization is required for SPOP activity, why does a reduction in oligomerization enhance activity? and (2) why is enhanced activity of SPOP oncogenic if prostate cancer–causing SPOP mutations reduce substrate ubiquitination? SPOP mutations on the periphery of the MATH domain (including mutant R121Q) that were identified in patients with endometrial cancer enhance the turnover of some substrates (32). Given that the mutation R354H was identified in a patient with endometrial cancer, we hypothesized that mutations in different regions of SPOP may have similar effects on substrate turnover and therefore result in similar malignancies. Indeed, the endometrial cancer mutant SPOPR121Q mediated higher polyubiquitination activity toward DAXX than SPOPWT, mirroring the activity of the SPOP interface mutants (Fig. 4B).
Given the similar activity of the SPOP interface and MATH domain mutants associated with endometrial cancer, we tested the activity of the interface mutants toward BRD3, which is a substrate that is ubiquitinated more effectively by SPOP harboring MATH domain mutations found in endometrial cancer patients (32). Indeed, BRD3 ubiquitination was also enhanced when we used the SPOP interface mutants (Figs. 4C and S3B). Our functional data suggest that several different types of endometrial cancer mutations enhance SPOP activity toward a set of substrates, though the underlying mechanisms are unknown.
SPOP interface mutations reduce binding to multivalent substrates
We next sought to understand the mechanism underlying the increase in substrate ubiquitination observed for the SPOP interface mutants. We did not expect a defect in the binding affinity to individual SB motifs given that the MATH domain is intact. Indeed, the binding affinities to the canonical SB motif found in the substrate Puc (36) were identical within error between WT and the interface mutants, and no binding was observed for the binding-incompetent SPOPW131G mutant, as expected (Fig. 4D). Given that the oligomerization state of SPOP may affect the binding of a multivalent substrate, we next investigated the binding affinities between the C-terminal intrinsically disordered region of DAXX (cDAXX) and the SPOP mutants. cDAXX has 5 SB motifs (Fig. 1D) (23). As expected, cDAXX bound slightly more tightly to SPOPWT than to the interface mutants (Fig. 4E), presumably due to enhanced avidity from the larger WT oligomers. Thus, a reasonable expectation from binding assays would be that SPOP interface mutants would likely have decreased ubiquitination activity toward multivalent substrates, not enhanced activity.
SPOP interface mutants enhance phase separation with substrates
To understand the source of the unexpected observation that SPOP interface mutants mediate increased ubiquitination activity, we investigated whether phase separation and discrete binding are affected differently by the mutations. We previously showed that transient expression of SPOP and DAXX results in their colocalization to phase-separated SPOP/DAXX bodies in cells. These bodies also recruited other subunits of the CRL3 complex and had the hallmarks of an active compartment for SPOP-mediated ubiquitination of DAXX (23).
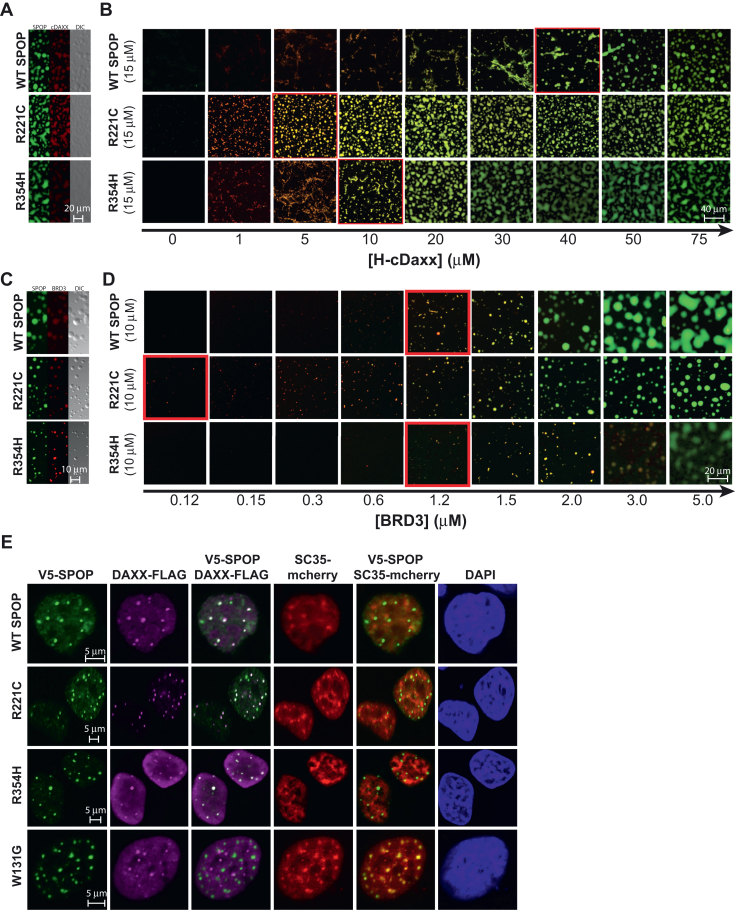
Thus, we tested the ability of SPOPWT and the interface mutants to undergo phase separation together with cDAXX in vitro. We observed condensates containing SPOPWT and cDAXX, and these have the ability to fuse and wet surfaces (Fig. 5A), which is in agreement with published results (23). As described previously, different molar ratios of SPOP and cDAXX give rise to different types of assemblies; condensates form at high cDAXX:SPOP molar ratios, and clusters (or filamentous assemblies or gels) form at low cDAXX:SPOP molar ratios (Fig. 1F) (23, 24). Modeling studies showed that the underlying interactions differ (24). Clusters arise when multivalent substrates stabilize SPOP oligomers and crosslink them. By contrast, condensate formation is mediated by DAXX-DAXX interactions between SPOP-DAXX brushes (24).
Figure 5.
SPOP interface mutants enhance phase separation with substrates.A, two-channel confocal fluorescence and DIC microscopy images show that SPOPWT, SPOPR221C, and SPOPR354H each colocalize with cDAXX in SPOP/cDAXX condensates. B, the phase boundary between SPOP/cDAXX clusters and condensates is shifted for the SPOP proteins with interface mutations. Confocal fluorescence microscopy images of SPOPWT or an interface mutant (green) as a function of cDAXX (red) concentration. The boundary between clusters and condensates is indicated by a red outline. All samples contain 10% w/v ficoll 70, 500 nM ORG-SPOP, and/or 500 nM Rhodamine-cDAXX. C, two-channel confocal fluorescence and DIC microscopy images show that SPOPWT, SPOPR221C, and SPOPR354H each colocalize with BRD3 in SPOP/BRD3 condensates. D, the phase boundary between SPOP/BRD3 clusters and condensates is shifted for the SPOP proteins with interface mutations. Confocal fluorescence microscopy images of SPOPWT or an interface mutant (green) as a function of BRD3 (red) concentration. The boundary between clusters and condensates is indicated by a red outline. All samples contain 200 nM ORG-SPOP and/or 100 nM Rhodamine-BRD3. Buffer conditions in (A–D) were 25 mM Tris (pH 7.6), 150 mM NaCl, and 1 mM T-CEP. E, representative fluorescence confocal images of HeLa cells expressing V5-SPOP, DAXX-Flag, and SC35-mCherry constructs (the latter to mark nuclear speckles). Cells were transfected with the indicated plasmids. Twenty-four hours posttransfection, cells were fixed and immunostained using antibodies against V5 (green) and Flag (magenta). DAPI (blue) marks nuclear DNA. DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; ORG, Oregon-Green-labeled; SPOP, speckle-type POZ protein.
When we titrated increasing concentrations of cDAXX into SPOPWT solutions, we observed a progression from the formation of clusters at low molar ratios to the formation of condensates at higher molar ratios (Fig. 5B). Condensate formation was demarcated by a saturation concentration. Similar titrations of cDAXX into a solution of SPOPR221C resulted in a lower saturation concentration; that is, condensate formation at lower cDAXX concentrations (Fig. 5B). SPOPR354H also had a lower saturation concentration, but the effect was smaller. These data indicate that the SPOP interface mutants bind to cDAXX more weakly in discrete complexes but engage cDAXX more effectively via phase separation. Similar results were also obtained using BRD3 (Fig. 5, C and D).
We considered whether the change to the surface charge of SPOP, caused by the mutations, might be the underlying cause for the lower saturation concentration. This change could mediate more favorable interactions with oppositely charged substrate proteins. Given that phase separation of DAXX is highly dependent on salt concentration, we were not able to elucidate the effect of SPOP surface charge on SPOP-DAXX phase separation by monitoring phase separation as a function of NaCl concentration. Instead, we decided to introduce an opposite change to the SPOP surface charge by generating the mutant SPOPE47K. This mutation is located on the surface of the MATH domains and is considerably more solvent-accessible than positions 221 and 354 are in oligomeric assemblies. SPOPE47K had very similar saturation concentrations relative to SPOPWT (Fig. S4A), suggesting that the altered charge is not the source of increased phase separation of substrates with SPOPR221C or SPOPR354H.
To investigate whether the SPOP interface mutants can also readily phase separate with DAXX in cells, we determined the cellular localization of the proteins. SPOPWT and the SPOP interface mutants colocalized with DAXX in SPOP-DAXX bodies (Fig. 5E). By contrast, SPOPW131G did not colocalize with DAXX and was instead localized to nuclear speckles, in agreement with our previous results (23). Under these conditions, DAXX also remained partially localized to promyelocytic leukemia protein (PML) nuclear bodies (Fig. S4B), its typically expected localization (18, 37, 38). Hence, we conclude that the enhanced ability of the SPOP interface mutants to undergo phase separation with substrates provides one explanation for the increase in ubiquitination activity observed for DAXX and BRD3 in the presence of these mutants. Our attempts to quantify phase diagrams in cells were inconclusive, likely because of the multitude of SPOP substrates in cells that can all contribute to phase separation and vary in levels between cells.
Discussion
Many different SPOP mutations are found in cancer genomes (39, 40), and SPOP is regarded as an important tumor suppressor across cancers (41). However, only prostate cancer–driving mutations for residues in the substrate-binding site are understood at a molecular level; they result in reduced substrate binding (31, 42). Here, we show that two mutations in the dimerization interfaces of the BTB and BACK domains weaken higher-order SPOP oligomerization and shift the size distribution of SPOP oligomers to smaller sizes. While this weakens the binding of multivalent substrates due to reduced avidity, the polyubiquitination activity of these mutants toward substrates is higher than that of SPOPWT, an initially counterintuitive finding. This apparent contradiction is resolved by the realization that the mutations increase the driving force for phase separation toward substrates; that is, substrates form condensates with mutant SPOP at lower concentrations. The implication is that a larger fraction of substrate enters the active dense phase and is thus turned over. In addition to revealing biophysical mechanisms that regulate SPOP activity, our results yield a comparative assessment of the functional role of clusters versus condensates and support a model in which phase separation plays an important role in SPOP function.
Why does weakened linear SPOP oligomerization reduce the substrate saturation concentration for phase separation with SPOP? Our previous combined theoretical and experimental characterization of the SPOP/DAXX system provides clues (24). We observed that DAXX has an intrinsic but weak driving force for phase separation. SPOP/DAXX brushes, which are complexes of SPOP oligomers that are bound to many DAXX molecules (Fig. 1F, middle right), phase separate via these same DAXX-DAXX interactions, but the driving force is substantially higher than that of DAXX phase separation alone due to the additive effect of tethering multiple DAXX molecules to the SPOP “hub”. However, our data showed that the SPOP/DAXX dense phase has a higher density than the DAXX dense phase (24). Because the pure DAXX dense phase has the optimal density for DAXX, the increased density caused by the SPOP hub must result in an energetic penalty. This penalty will reduce the driving force for DAXX-mediated phase separation below what it could be without this penalty. By contrast, the shorter SPOP oligomers formed by the interface mutants could provide more space per bound DAXX molecule because these can spread out at the ends of the SPOP oligomers. The associated lower DAXX concentration in the resulting dense phase would be energetically favorable and could therefore explain why the saturation concentration of substrate with the interface mutants is lower relative to that observed for SPOPWT.
Our proposed mechanism shows how linear SPOP self-association can have a nonmonotonic effect on DAXX ubiquitination due to the emergent characteristics of SPOP-DAXX phase separation (43, 44). Briefly, some degree of self-association is necessary to amplify weak DAXX-DAXX interactions, but excessive self-association inhibits these same DAXX-DAXX interactions. Recent work on the mechanism of phase separation of ubiquilin2 (UBQLN2) with polyubiquitin chains revealed the same physical principle (45). The soluble polyubiquitin “hub” binds and bundles UBQLN2, which has an intrinsic driving force for phase separation, thereby enhancing phase separation. However, concentrating UBQLN2 beyond its intrinsic dense phase concentration results in an energetic tradeoff. Polyubiquitin chains with different compactness therefore have different abilities to potentiate UBQLN2 phase separation, and an intermediate compactness seems to be ideal. Such tradeoffs between higher sticker density and optimal spacer length are expected to play roles in phase-separating systems in general.
The SPOP interface mutations characterized here were identified in patients with endometrial, skin, and other cancers, and their shared molecular mechanism was not previously understood. The majority of SPOP mutations identified in endometrial cancer are found at the periphery of the MATH domain (32), and the molecular mechanism underlying the resultant change in substrate specificity is not understood. Our conclusion that endometrial– and other cancer–related interface mutants alter SPOP phase separation behavior suggests that endometrial cancer–related MATH domain mutants also display altered phase behavior. Future research should assess this possibility.
Our work suggests that tuning of SPOP oligomerization could be used by the cell to modulate substrate levels. We tested this hypothesis by fusing GST dimerization domains to the SPOP N-terminus. While SPOP self-association was indeed enhanced, and phase separation was reduced in vitro, the fusion proteins had very low expression levels in cells, and their activity could not be assessed. Oligomerization could be further tuned by posttranslational modifications or by titration of the paralog SPOPL, a substrate recognition subunit that is highly homologous to SPOP, can dimerize with SPOP via its BTB domain, but has a dimerization-incompetent BACK domain due to an 18-residue insertion (Fig. 2C) and thus caps SPOP oligomers (30). The results we present here predict that varying SPOPL levels in cells may change the driving force for phase separation with certain substrates, and that this would alter their cellular levels. Given the tumor suppressor role of SPOP, future research into the regulation of its activity, including via modulation of its oligomer size distribution, will be valuable.
Our work also suggests a strategy for disentangling the contributions of condensates versus clusters in other multivalent systems. First, it is useful to recognize that condensates and clusters compete with each other; factors that stabilize clusters raise the saturation concentration. Second, while phase separation and cluster formation both use the same multivalent interactions (or stickers) for networking, phase separation requires an additional density transition, which is driven by the modest solubility of constituent monomers or complexes (27, 28). The solubility is determined by many properties of the protein including the surface charge distribution and conformation, meaning that both stickers and the spacers between them contribute directly to protein solubility. Third, the network structures can differ between clusters and condensates (24). Hence, separation-of-function mutations to distinguish between the contribution from condensates and clusters should be accessible, either by modulating the solubility of molecules/complexes, or by differentially affecting the network structure of clusters versus condensates. A mutational strategy that capitalizes on these insights thus promises to reveal the extent to which function is mediated by phase separation in biology.
Experimental procedures
Plasmids
The coding region of mouse SPOP (Uniport: Q6ZWS8-1) and its corresponding mutants were cloned into pcDNA4/TO/myc-His (Thermo Fisher Scientific) to produce C terminally Myc-tagged proteins. To generate an N terminally tagged GFP-SPOP construct, the coding region of Emerald GFP and mouse SPOP were jointly cloned in frame into pcDNA4/TO/myc-His using the Gibson assembly method. pcDNA3-Myc-Cullin3 (46), and pcDNA3-HA-Rbx1 (46) were obtained from Addgene (#19893 and #19897, respectively). The Ub-His plasmid was a kind gift from Wenyi Wei, Harvard Medical School. A synthetic gene for BRD3 in a pET28a vector, codon-optimized for Escherichia coli, was ordered from GenScript. A sequence coding for a hexa-histidine tag followed by a tobacco etch virus protease cleavage site was placed at the N terminus of the open reading frame. The His-SUMO SPOP28–359 construct was previously described (19, 23). Rolling circle mutagenesis was used to generate the SPOPR221C and SPOPR354H mutant constructs.
Protein expression and purification
The BRD3 plasmid was transformed into BL21-RIPL cells, cultures were grown in LB medium at 37 °C, and expression was initiated with the addition of IPTG for 2 h. Cells were lysed in 30 mM imidazole pH 7.8, 1 M NaCl with a sonicator. The clarified lysate was loaded onto a 3 ml Fast Flow Chelating Sepharose gravity column and washed with the resuspension buffer. Protein was eluted in 300 mM imidazole pH 7.8, 300 mM NaCl and subsequently diluted three-fold before loading onto a Fast-flow Q-column connected to a HiTrap Heparin column for the removal of bound nucleic acids. After washing, the Q-column was removed, and the protein was eluted with a gradient of NaCl in 20 mM Hepes pH 7.5 and concentrated to 100 μM. Protein was then dialyzed into 20 mM Tris pH 7.8, 150 mM NaCl, and 5 mM DTT, flash frozen, and stored at −80 °C. All protein produced using the His-SUMO-SPOP28–359 constructs and the cDAXX constructs were expressed and purified as previously described (19, 23).
CD experiments
CD experiments were performed using solutions of 1 to 2 μM SPOP and were recorded at 25 °C in 20 mM Tris (pH 7.6) and 150 mM NaCl, in a Jasco J1500 spectropolarimeter (JASCO), using a 10 mm path length quartz cuvette. Final spectra were an average of five measurements.
Fluorescent protein labeling
For N-terminal labeling, protein was dialyzed into 10 mM Hepes pH 7.4, 150 mM NaCl, 5 mM DTT. All SPOP constructs were labeled with Oregon Green (Thermo Fisher Scientific O6147) and cDAXX or BRD3 were labeled with Rhodamine Red (Thermo Fisher Scientific R6160). For labeling, the dye was dissolved at 100 mM in dimethylsulfoxide and added to protein in a 20:1 dye to protein molar ratio and incubated on a shaking platform for 5 h. The reaction was quenched with 1 M Tris to a molar excess of ∼3 to 5. Proteins were then extensively dialyzed into 20 mM Tris pH 7.8, 150 mM NaCl, and 5 mM DTT.
Composite-gradient multiangle light scattering
CG-MALS experiments were used to characterize the linear oligomerization of SPOPWT, SPOPR221C, and SPOPR354H using a Calypso system (Wyatt Technology Corporation) consisting of a software-controlled multiple syringe pump to create the concentration gradient and a DAWN HELEOS multiangle light scattering photometer and a UV–Vis detector (Agilent Technologies) to collect data from the incoming sample stream. Static light scattering data were collected at 14 scattering angles as a function of protein concentration. Data were analyzed with a model that describes the formation of a mixture of discrete oligomeric states in an isodesmic model as previously described (19).
Phase separation assays
Samples were prepared by mixing labeled and unlabeled protein, buffer, and ficoll PM 70 for titrations of cDAXX into constant concentrations of SPOP variants in PCR tubes (Sigma-Aldrich). BRD3 assays were carried out in the same manner without the addition of ficoll. The samples contained a fixed amount of Oregon-Green-labeled SPOP (either 200 or 500 nM), Rhodamine-cDAXX (500 nM) or Rhodamine-BRD3 (100 nM), and the buffer conditions were 25 mM Tris (pH 7.6), 150 mM NaCl, and 1 mM T-CEP as in our previous work (23). Two microliters of the solution was transferred to a sealed sample chamber comprised of coverslips sandwiching two layers of 3M 300 LSE high-temperature double-sided tape (0.34 mm). For each given cDAXX/BRD3 and/or SPOP concentration, the sample was equilibrated at room temperature and incubated for 3 to 4 h. Samples were imaged on a Nikon C2 laser scanning confocal microscope with a 20× (0.8 NA) Plan Apo objective. Images and movies were processed with the Nikon NIS Elements software and FIJI. All images within a given figure panel were taken with the same camera settings.
Fluorescence polarization assays
Fluorescence polarization binding assays were performed in 384 well plates (Greiner Bio-One) where serial dilutions of each SPOP construct, ranging from 0.006 to 100 μM, were prepared in 20 mM Tris pH 7.6, 150 mM NaCl, 5 mM DTT, and 0.01% Triton X-100. Then fluorescently tagged cDAXX construct, or fPuc peptide (5FAM-ENLACDEVTSTTSSSST-NH2) was added for a final concentration of 40 nM into each well. Fluorescence polarization was measured using a CLARIOstar plate reader (BMG LABTECH) and converted to anisotropy. Assays were performed in a minimum of triplicates and analyzed as previously described (22).
In vitro ubiquitination assays
BRD3 ubiquitination was carried out in 50 mM Tris (pH 7.5), 250 mM NaCl, 10 mM MgCl2, 5 mM ATP, and 1 mM DTT at room temperature at time points from 0 to 13 min. The reaction mixture contained ubiquitinating enzymes at final concentrations of 0.25 μM UBA1 (E1) (R&D Systems), 8 μM UbcH5B (E2), NEDD8-CUL3-Rbx1 (at 1 μM) (E3), SPOP (2.0 μM), 75 μM ubiquitin (Boston Biochem) and 0.1 μM fluorescein-labeled BRD3. The reactions were quenched by addition of SDS-PAGE loading buffer with 3 M urea, loaded on SDS-PAGE gels for visualization and visualized on a Amersham 600 imager (GE HealthCare).
Cell lines, cell culture, transfection, and treatment
Human T-REx-293 cells were obtained from Thermo Fisher Scientific, and tested negative for mycoplasma contamination using Universal Mycoplasma Detection kit from American Type Culture Collection. Cells were grown at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 1% GlutaMAX, and 100 U/ml Penicillin–Streptomycin (all from Thermo Fisher Scientific). Transfections of the cells were carried out using the Effectene Transfection Reagent from Qiagen. MG132 was purchased from Calbiochem.
Antibodies
We used primary antibodies for Myc (Cell Signaling Technology, #2276S), Flag (Sigma-Aldrich, #F3165), GAPDH (Abcam, #ab181602), SC35 (Abcam, #ab11826), and PML (Santa Cruz Biotechnology, #sc-966). Horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson Laboratory. Alexa 488-, Alexa 555-, and Alexa 647-conjugated secondary antibodies were obtained from Thermo Fisher Scientific. Specificity of the anti-tag antibodies was easily established by having controls without the expression of the tagged protein. Images with SC35 and PML antibodies showed known distributions in cells.
In-cell ubiquitination assays
T-REx-293 cells were transfected with 400 ng of plasmids for SPOP-Myc, Ub-His, Myc-Cullin3, HA-Rbx1, and Flag-tagged BRD3 or DAXX. Tetracycline was added to the culture media to induce SPOP-Myc expression. At 24 h after transfection, MG132 was added to the cells at a final concentration of 20 μM for another 4 to 5 h. Cells were lysed in buffer A [6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4 (pH 8.0), and 10 mM imidazole]. The lysates were sonicated, cleared, and incubated with Ni-NTA agarose (Qiagen) for His-tag pull down. The beads were washed twice with buffer A, twice with A/T buffer composed of one volume of buffer A and three volumes of buffer T [25 mM Tris (pH 8.0) and 20 mM imidazole], and twice with buffer T. Beads were incubated in SDS-PAGE loading dye containing 300 mM imidazole for 15 min and boiled for 5 min to elute protein.
Western blots
Protein samples were separated on a 6.5% SDS-PAGE gel and then electrotransferred to a 0.45 μm nitrocellulose membrane at 44 V for 16 h (overnight) in a transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol). The membrane was blocked with 1% Blocking Reagent in Tris-buffered saline for 1 h at room temperature. Primary antibodies were diluted in 1% Blocking Reagent in Tris-buffered saline containing 0.1% Tween 20 (TBST) according to the manufacturer's recommendations and incubated with the membrane overnight at 4 °C. Following three 20-min washes with TBST, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (1:2500 dilution in 1% Blocking Reagent in TBST) for 1 h at room temperature. After an additional three washes in TBST, protein bands were visualized using the enhanced chemiluminescence detection system (Amersham Biosciences) and captured by using AI600 Chemiluminescent Imager (GE HealthCare).
Immunostaining
Cells on 22 mm coverslips (Corning Inc) were washed with PBS and fixed with 4% methanol-free formaldehyde (Thermo Fisher Scientific) for 30 min at room temperature. Cells were washed three times with PBS, and then incubated for 30 min with a blocking buffer, which contained PBS, 0.1% Triton X-100, and 5% bovine serum albumin. Cells were incubated overnight with primary antibodies diluted in the blocking buffer. After rinsing with PBS supplemented with 0.1% Triton X-100, cells were incubated with secondary antibodies for 2 h at room temperature followed by washing and staining with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific). Coverslips were mounted on slides using Prolong Gold Antifading Reagent (Thermo Fisher Scientific). Images were acquired using a Zeiss LSM780 microscope with a 63 x (0.24 numerical aperture) objective. Images were processed using Fiji software (http://fiji.sc).
Data availability
Data will be shared upon request to T. Mittag (tanja.mittag@stjude.org).
Supporting information
This article contains supporting information.
Conflict of interest
T. M. was a consultant for Faze Medicines, Inc.
Acknowledgments
The Ub-His and BRD3-Flag plasmids were kind gifts from Wenyi Wei (Harvard Medical School) and Joel Mackay (University of Sydney), respectively. We acknowledge use of the Cell and Tissue Imaging Center - Light Microscopy (CTIC-LM) and the Protein Technologies Center (PTC) at St. Jude Children's Research Hospital.
Author contributions
N. S., M. J. C., M. R. M., J. L., J. J. B., and Ö. G., investigation; N. S., M. B. B., J. S., and T. M. methodology; N. S., M. J. C., M. R. M., J. J. B., and Ö. G., visualization; N. S., M. J. C., M. M., J. L., J. B., Ö. G., S. V., M. B. B., and J. S. writing–review and editing., M. R. M. and T. M. formal analysis; S. V. formal analysis; J. S. and T. M. conceptualization; T. M. supervision; T. M. funding acquisition; T. M. writing–original draft; T. M. project administration.
Funding and additional information
T. M. acknowledges funding from NIH grant R01GM112846, the St Jude Children’s Research Hospital Research Collaborative on The Biology and Biophysics of RNP Granules, and the American Lebanese Syrian Associated Charities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Footnotes
Present address for: Melissa R. Marzahn, Burnett School of Biomedical Sciences, University of Central Florida, Orlando, Florida, USA; Jihun Lee, Celltrion, South Korea; Jill J. Bouchard, Dewpoint Therapeutics, Boston, Massachusetts 02210, USA.
Supporting information
References
- 1.Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson B.A., Doolittle L.K., Schneider M.W.G., Jensen L.E., Gamarra N., Henry L., et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.e21. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Case L.B., Zhang X., Ditlev J.A., Rosen M.K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science. 2019;363:1093–1097. doi: 10.1126/science.aau6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T.H., Tsang B., Vernon R.M., Sonenberg N., Kay L.E., Forman-Kay J.D. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019;365:825–829. doi: 10.1126/science.aax4240. [DOI] [PubMed] [Google Scholar]
- 9.Yang P., Mathieu C., Kolaitis R.M., Zhang P., Messing J., Yurtsever U., et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress Granules. Cell. 2020;181:325–345.e8. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillen-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlussler R., et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181:346–361.e17. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders D.W., Kedersha N., Lee D.S.W., Strom A.R., Drake V., Riback J.A., et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306–324.e8. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boija A., Klein I.A., Young R.A. Biomolecular condensates and cancer. Cancer Cell. 2021;39:174–192. doi: 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieu C., Pappu R.V., Taylor J.P. Beyond aggregation: pathological phase transitions in neurodegenerative disease. Science. 2020;370:56–60. doi: 10.1126/science.abb8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kar M., Dar F., Welsh T.J., Vogel L.T., Kuhnemuth R., Majumdar A., et al. Phase-separating RNA-binding proteins form heterogeneous distributions of clusters in subsaturated solutions. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2202222119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Munoz I., Lund A.H., van der Stoop P., Boutsma E., Muijrers I., Verhoeven E., et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kent D., Bush E.W., Hooper J.E. Roadkill attenuates Hedgehog responses through degradation of cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- 18.Kwon J.E., La M., Oh K.H., Oh Y.M., Kim G.R., Seol J.H., et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J. Biol. Chem. 2006;281:12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- 19.Marzahn M.R., Marada S., Lee J., Nourse A., Kenrick S., Zhao H., et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016;35:1254–1275. doi: 10.15252/embj.201593169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q., Shi Q., Chen Y., Yue T., Li S., Wang B., et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuneo M.J., Mittag T. The ubiquitin ligase adaptor SPOP in cancer. FEBS J. 2019;286:3946–3958. doi: 10.1111/febs.15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce W.K., Grace C.R., Lee J., Nourse A., Marzahn M.R., Watson E.R., et al. Multiple weak linear motifs enhance recruitment and processivity in SPOP-mediated substrate ubiquitination. J. Mol. Biol. 2016;428:1256–1271. doi: 10.1016/j.jmb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouchard J.J., Otero J.H., Scott D.C., Szulc E., Martin E.W., Sabri N., et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell. 2018;72:19–36.e8. doi: 10.1016/j.molcel.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmit J.D., Bouchard J.J., Martin E.W., Mittag T. Protein network structure enables switching between liquid and gel states. J. Am. Chem. Soc. 2020;142:874–883. doi: 10.1021/jacs.9b10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raos G., Allegra G. Chain collapse and phase separation in poor-solvent polymer solutions: a unified molecular description. J. Chem. Phys. 1996;104:1626–1645. [Google Scholar]
- 26.Raos G., Allegra G. Macromolecular clusters in poor-solvent polymer solutions. J. Chem. Phys. 1997;107:6479–6490. [Google Scholar]
- 27.Harmon T.S., Holehouse A.S., Rosen M.K., Pappu R.V. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife. 2017;6 doi: 10.7554/eLife.30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittag T., Pappu R.V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell. 2022;82:2201–2214. doi: 10.1016/j.molcel.2022.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremer A., Farag M., Borcherds W.M., Peran I., Martin E.W., Pappu R.V., et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 2021;14:196–207. doi: 10.1038/s41557-021-00840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Errington W.J., Khan M.Q., Bueler S.A., Rubinstein J.L., Chakrabartty A., Prive G.G. Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure. 2012;20:1141–1153. doi: 10.1016/j.str.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Barbieri C.E., Baca S.C., Lawrence M.S., Demichelis F., Blattner M., Theurillat J.P., et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janouskova H., El Tekle G., Bellini E., Udeshi N.D., Rinaldi A., Ulbricht A., et al. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat. Med. 2017;23:1046–1054. doi: 10.1038/nm.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomasen F.E., Cuneo M.J., Mittag T., Lindorff-Larsen K. Conformational and oligomeric states of SPOP from small-angle X-ray scattering and molecular dynamics simulations. Elife. 2023;12 doi: 10.7554/eLife.84147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuneo M.J., O'Flynn B.G., Lo Y.H., Sabri N., Mittag T. Higher-order SPOP assembly reveals a basis for cancer mutant dysregulation. Mol. Cell. 2023;83:731–745.e4. doi: 10.1016/j.molcel.2022.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Geersdaele L.K., Stead M.A., Harrison C.M., Carr S.B., Close H.J., Rosbrook G.O., et al. Structural basis of high-order oligomerization of the cullin-3 adaptor SPOP. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1677–1684. doi: 10.1107/S0907444913012687. [DOI] [PubMed] [Google Scholar]
- 36.Zhuang M., Calabrese M.F., Liu J., Waddell M.B., Nourse A., Hammel M., et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol. Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E.J., et al. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell Biol. 2000;20:1784–1796. doi: 10.1128/mcb.20.5.1784-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weidtkamp-Peters S., Lenser T., Negorev D., Gerstner N., Hofmann T.G., Schwanitz G., et al. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 2008;121:2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 39.Clark A., Burleson M. SPOP and cancer: a systematic review. Am. J. Cancer Res. 2020;10:704–726. [PMC free article] [PubMed] [Google Scholar]
- 40.Song Y., Xu Y., Pan C., Yan L., Wang Z.W., Zhu X. The emerging role of SPOP protein in tumorigenesis and cancer therapy. Mol. Cancer. 2020;19:2. doi: 10.1186/s12943-019-1124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An J., Wang C., Deng Y., Yu L., Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–669. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhandari K., Cotten M.A., Kim J., Rosen M.K., Schmit J.D. Structure-function properties in disordered condensates. J. Phys. Chem. B. 2021;125:467–476. doi: 10.1021/acs.jpcb.0c11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmit J.D., Feric M., Dundr M. How Hierarchical interactions make membraneless organelles tick like Clockwork. Trends Biochem. Sci. 2021;46:525–534. doi: 10.1016/j.tibs.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galagedera S.K.K., Dao T.P., Enos S.E., Chaudhuri A., Schmit J.D., Castaneda C.A. Decoding optimal ligand design for multicomponent condensates. bioRxiv. 2023 doi: 10.1101/2023.03.13.532222. [preprint] [DOI] [Google Scholar]
- 46.Ohta T., Michel J.J., Schottelius A.J., Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 47.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roehrl M.H., Wang J.Y., Wagner G. A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry. 2004;43:16056–16066. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be shared upon request to T. Mittag (tanja.mittag@stjude.org).