Abstract
The intrinsic mechanism of postherpetic neuralgia (PHN) remains unclear. Herein, we aimed to seek the hub proteins in the cerebrospinal fluid (CSF), which display significant changes between the PHN and nonpainful patients (Control). First, the proteomic results showed that compared with the Control-CSF, there were 100 upregulated and 50 downregulated differentially expressed proteins (DEPs) in the PHN-CSF. Besides, functional analyses including gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analysis (GSEA) revealed that biological processes and pathways including complement activation, infection, coagulation, and lipid metabolism were activated, while synaptic organization was suppressed. Next, the protein–protein interaction (PPI) analysis indicated that increased PLG, F2, APOA1, APOA2, SERPINC1, and KNG1 and reduced APOE, which were all enriched in the top pathways according to the KEGG analysis, were defined as hub proteins. Finally, three of the hub proteins, such as PLG, APOA1, and APOE, were reconfirmed in a larger cohort using both enzyme-linked immunosorbent assay (ELISA) and Western blotting methods. Above all, the results indicated that PLG, APOA1, and APOE and their involved processes such as infection, inflammation, cholesterol metabolism, and coagulation shall be potential therapeutic approaches. (The raw mass spectrometry proteome data and search results have been deposited to the iProx-integrated Proteome Resources (http://www.iprox.cn) with the data set identifier IPX0007372000.)
Keywords: postherpetic neuralgia, cerebrospinal fluid, proteomics, differential expressed proteins, central nervous system
Introduction
Postherpetic neuralgia (PHN) is the chronic infection by the varicella zoster virus (VZV).1 Epidemic studies have reported that globally, 30% of people would suffer from the herpes zoster (HZ) in their lifetime.2 Among them, 5–30% victims (especially the elderly and female) will finally develop PHN.3 As a typical type of neuropathic pain (NP), the primary clinical manifestation of PHN is the long-term, spontaneous and intolerable pain at the infected dermatome, which severely reduces the life quality of the victims.4 Although large amounts of studies have sought to reveal the mechanism, the treatment for PHN still remains challenging due to the fact that humans are the only host of VZV, and it is hard to establish an animal model to reproduce the analogous pathophysiology.5 Thus, gaining a better understanding of the exact mechanism of PHN is urgently needed.
Fortunately, proteomics shall provide an alternative solution in seeking out the key molecules and biological processes during the process of the diseases. Cerebrospinal fluid (CSF), on the other hand, is considered to be a logical biospecimen for pathophysiological study of the central nervous system (CNS) disorder.6 Compared with numerous research studies proving that the changes of protein composition in the CSF could reflect the structural and functional alterations in the processes of CNS injury and degenerative diseases,7,8 few of them have paid adequate attention to PHN, which limits our interpretation of its intrinsic mechanism. To the best of our limited knowledge, proteomics studies on the CSF of PHN patients are still lacking.
In the present study, high-throughput proteomics was employed to detect the differentially expressed proteins (DEPs) in the CSF of PHN and nonpainful patients. Besides, multiple functional analyses were then performed to seek out the hub proteins and their correlated biological processes. Finally, the expression patterns of these hub proteins were further verified in a larger clinical cohort, and the possible mechanism was speculated based on the bioinformatic analysis. The results shall provide valuable information about the key proteins in the CSF, which may provide new approaches to treating the PHN disease.
Methods
Patient Enrollment
The study was approved by the institutional medical review board of the Second Xiangya Hospital (LYF2020081) on Sep 21st, 2020, prior to the patient enrollment and was strictly conducted based on the principles of the Declaration of the Helsinki. From April first, 2021, to July first, 2021, 20 PHN patients who suffered from refractory pain (Numeric Rating Scale (NRS) above 4) after standard antiviral (valacyclovir) and analgesic (pregabalin) treatment and were scheduled to accept the spinal cord stimulation (SCS) treatment were enrolled from the Department of the Pain Management for the study eligibility. The exclusion criteria included (1) patients complicated with other neurological diseases; (2) patients with blood diseases and bleeding tendency; (3) patients with tumor; (4) patients with severe vital organ diseases; and (5) pregnant and nursing women. As for control, 20 nonpainful patients who were diagnosed with varicose greater saphenous vein and planned to accept epidural block for the vascular surgery were enrolled. All participants had signed the informed consents to donate the CSF samples before the study began.
CSF Collection
Lumbar puncture and CSF collection were performed by the same skilled anesthesiologist (Zou D). Briefly, patients for both groups were monitored with a five-lead electrocardiograph, noninvasive blood pressure, and pulse oximetry when entering the operating room and were asked to place at the lateral decubitus position. After the thoroughgoing aseptic precautions and local anesthesia (1% lidocaine), a 16-gauge (G) epidural puncture needle was used to enter the epidural space at the L2-L3 space via the loss of resistance technique. After that, a 25G lumbar anesthesia needle was punctured through the cavity of the epidural needle to the subarachnoid cavity. A meticulous collection of 2 mL of transparent CSF was slowly and carefully collected. Each sample was then centrifuged at 1000g for 5 min to remove the debris, and the supernatant was harvested and stored at −80 °C for further use.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
The CSF samples from four PHN patients and four nonpainful patients were randomly selected for the LC-MS/MS analysis. Briefly, proteins in each CSF sample were extracted using SDT buffer (4% SDS, 100 mM Tris–HCl, 1 mM DTT, pH7.6) and then quantified via the bicinchoninic acid (BCA, 500–0001, Bio-Rad, CA) method. Quality control was performed using 12.5%-SDS-PAGE gel electrophoresis followed by Coomassie Blue R-250 staining. After that, proteins were digested using trypsin, followed by the filter-aided sample preparation (FASP) procedure described by Matthias Mann.9 In addition, the digested peptides were further desalted on C18 Cartridges (Empore SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume = 3 mL, Sigma), concentrated by vacuum centrifugation, and reconstituted in 40 μL of 0.1% (v/v) formic acid. The spectral density was then calculated with ultraviolet (UV) light at 280 nm. Afterward, a tandem mass tag (TMT) reagent (Thermo, MA) was employed to label the peptides in each sample according to the manufacturer’s instructions. Next, the labeled peptides were further fractionated by a High-PH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific).
LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific) coupled to Easy nLC (Thermo Fisher Scientific) for 60/90 min. The peptides were loaded onto a reverse-phase trap column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18), which was connected to the C18-reversed phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 μm inner diameter, 3 μm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min controlled by IntelliFlow technology. The mass spectrometer was operated in the positive iron mode. MS data were acquired using a data-dependent top-10 method by dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) for HCD fragmentation. The paraments setting included automatic gain control (AGC): 3e6, maximum inject time: 10 ms, dynamic exclusion duration: 40.0 s, survey scans: a resolution of 70,000 at m/z 200, resolution for HCD spectra: 17,500 at m/z 200, isolation width: 2 m/z, normalized collision energy: 30 eV, and the underfill ratio: 0.1%. The instrument was run with the peptide recognition mode enabled.
The MS raw data for each sample were acquired using the MASCOT engine (Matrix Science, London, U.K.; version 2.2) embedded into Proteome Discoverer 1.4 for identification and quantitation analysis. Related parameters are shown in Table 1.
Table 1. Related Parameters of Identification and Quantitation of Proteins.
| item | value |
|---|---|
| enzyme | trypsin |
| max missed cleavages | 2 |
| fixed modifications | carbamidomethyl (C) TMT 6/10/16 plex (N-term), TMT 6/10/16 plex (K) |
| variable modifications | oxidation (M) |
| peptide mass tolerance | ±20 ppm |
| fragment mass tolerance | 0.1 Da |
| database | Swissprot_Homo_sapiens_20395_20210106.fasta |
| database pattern | decoy |
| peptide FDR | ≤0.01 |
| protein quantification | the protein ratios were calculated as the median of only unique peptides of the protein. |
Bioinformatic Analysis
Bioinformatic analysis in the present study included principal component analysis (PCA), detection of DEPs, the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, Gene Set Enrichment Analysis (GSEA), and protein–protein interactions (PPI) using R software (version 4.1.3), R Studio (version1.4.1717), Cytoscape (version 3.9.0), Venn (http://jvenn.toulouse.inra.fr/app/example.html),10 and STRING (string-db.org) Web site online. Protein intensities were log2-transformed and corrected by the median correction method using the DEqMS package for further analysis. All P values were corrected by a false discovery rate (FDR) with the Benjamini–Hochberg (BH) method.
The PCA analysis was conducted using the FactorMineR package, in order to visualize the differences and similarities between samples.11 A two-dimensional scatter plot was drawn using the ggplot2 and ggrepel packages to display the distribution of the samples, and the principal components of PCA1 and PCA2 were reported based on the majority of the variation.
The DEP detection of all quantitative proteins was calculated by the Bayes test through the Limma package. DEP was defined P < 0.05 and the absolute index of the fold-change (FC) (|fold-change (FC)|) > 1.2. The hierarchical cluster analysis was used to classify DEPs in comparison groups using ward D2 algorithm, and the distance of row clustering was calculated by the Euclidean algorithm. The Volcano map and heatmap were further drawn through the ggplot2, ggrepel, and ComplexHeatmap packages separately.
The GO and KEGG analysis were conducted using the Hypergeometric method with the clusterProfiler package.12 In the GO analysis, three categories including the biological process (BP), cell component (CC), and molecular function (MF) were included and displayed in a barplot. In the KEGG analysis, the significant terms with high similarity were clustered and displayed in a treeplot using the enrichplot package. In both sections, the top significant terms and DEPs were displayed in the barplot, Circos, and chord plot, respectively.
The GSEA was then performed to find enriched biological processes (GSEA-GO(BP)) and related pathways (GSEA-KEGG) of all quantified proteins using the clusterProfiler package. The enrichment score (ES) and normalized enrichment score (NES) were calculated and analyzed with the empirical phenotype-based permutation test followed with correction by the BH method. P < 0.05, |NES| > 1, and FDR (q value)<0.25 were considered to be statistically significant. Then, all terms were arranged in descending order of |NES|, with the top significant gene sets displayed in dot plots using the GseaVis package. Besides, the relevant pathways and core-enrichment proteins were displayed in the enrichment map and heatmap with the GseaVis package.
The PPI analysis was performed using the STRING Web site. Proteins, with a minimum required intersection score >0.4, were visualized and further analyzed to obtain the PPI cluster using the software Cytoscape 3.9.0. Two methods including MCODE and Cytohubba (containing five algorithms: MCC, MNC, Degree, EPC, and Closeness) were used. The overlapping results were displayed in the Venn diagram through the Venny Web site and further processed with KEGG analysis to determine the hub proteins and their involving pathways, which were all displayed in the Cnetplot using both clusterProfiler and enrichplot packages.
Enzyme-Linked Immunosorbent Assay (ELISA)
Among the hub proteins, thrombin factor II (F2), plasminogen (PLG), apoprotein A1 (APOA1), apoprotein A2 (APOA2), and apoprotein E (APOE) were chosen, and their levels in the CSF of all 40 enrolled patients were tested using the ELISA method. The ELISA kits including F2 (AF0946-A, AiFang biological, Shanghai, China), PLG (AF111463-A, AiFang), APOA1 (AF0168-A, AiFang), APOA2 (AF111517-A, AiFang), and APOE (AF0882-A, AiFang) for humans were employed following the manufacturer’s instructions. Optical density was measured at 450 nm by using a microplate reader (Rayto RT-6100 microplate reader, Shenzhen, CHN).
Western Blotting
The remaining CSF samples from the selected eight patients for the proteomics were further used for the Western blotting testing. After being mixed with the loading buffer, each sample containing 20 μg of protein was loaded on a 10% SDS-PAGE, electrophoresed, and transferred to a nitrocellulose membrane. The membrane then was blocked in TBST containing 10% nonfat milk powder for 1 h followed by the incubation at 4 °C overnight with the following primary antibodies: rabbit anti-PLG (1:500, BS9229, Bioworld Technology), rabbit anti-APOA1 (1:500, bs-0849R, Bioss), and rabbit anti-APOE (1:2000, ab183597, Abcam) were used. Each membrane was washed 3 times with TBST (10 min each), and then, the membrane was incubated for 1 h at room temperature with horseradish peroxidase conjugated antirabbit IgG (1:6000; Proteintech, Rosemont, IL). After reacting with the chemiluminescent HRP substrate (WBKLS0100, Millipore, Burlington, MA), the immunoreactivity bands were photographed by a gel imaging system (Clinx Science Instrument, Shanghai, CHN).
Statistical Analysis
For categorical variables, data were expressed as frequency and percentage (%) and analyzed by Fisher’s exact test. For continuous variables, data with normal distribution were presented as mean and standard deviation and analyzed using the independent two-sided unpaired parameter t test, while data with abnormal distribution were expressed as median and interquartile range (IQR) and analyzed with the Mann–Whitney U test. P < 0.05 was considered as the statistical significance.
Analysis of patients’ basic information, Western blotting data, and ELISA data were visualized with Prism software (version 8.0.1, GraphPad Inc., CA). In addition, all the pictures were uniformly decorated and arranged by Adobe Illustrator CC 2019 software.
Results
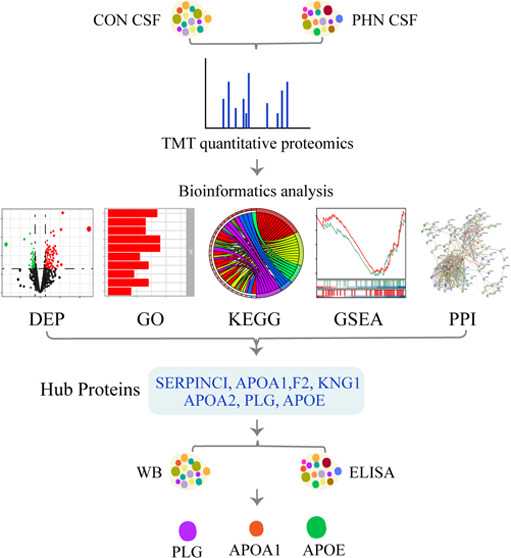
The flowchart of the present study is shown in Figure 1. Generally, the basic characteristics, including the age, gender, body mass index (BMI), and rates of hypertension and diabetes mellitus, remained statistically comparable between the two groups (all P > 0.05, Table 2). Additionally, the detailed information including gender, age, duration, and location of the pain and NRS scores of the selected eight patients for the proteomics analysis are also listed in Table 3.
Figure 1.
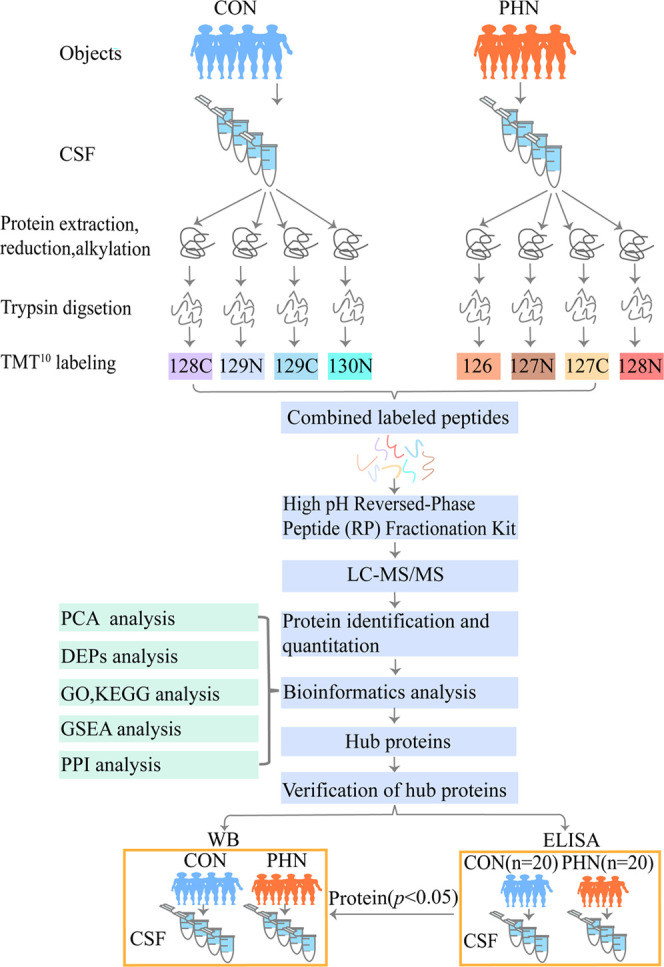
Flowchart of this study.
Table 2. Basic Information of Subjects.
| variables | PHN (n = 20) | CON (n = 20) | P-value |
|---|---|---|---|
| age (years) | 63.4 ± 6.0 | 61.8 ± 4.4 | 0.36 |
| gender (female/male) | 9/11(45/55%) | 10/10(50/50%) | 0.99 |
| body mass index (BMI) | 24.0 ± 3.0 | 23.6 ± 2.8 | 0.67 |
| duration of the disease | 4.9 ± 3.5 | ||
| numeric rating scale (NRS) | 6(5,6) | ||
| hypertension | 5(25%) | 4(20%) | 0.99 |
| diabetes mellitus | 4 | 2 | 0.66 |
Table 3. Basic Characteristics of the Eight Subjects Accepted for Proteomics.
| patients | gender | age (Y) | location | duration (M) | NRS |
|---|---|---|---|---|---|
| PHN1 | M | 59 | left lower thoracodorsal | 9 | 7 |
| PHN2 | F | 52 | right lower thoracodorsal | 1 | 6 |
| PHN3 | F | 58 | left waist abdomen | 4 | 6 |
| PHN4 | M | 71 | left waist back | 5 | 5 |
| CON1 | F | 48 | |||
| CON2 | F | 61 | |||
| CON3 | M | 61 | |||
| CON4 | M | 67 |
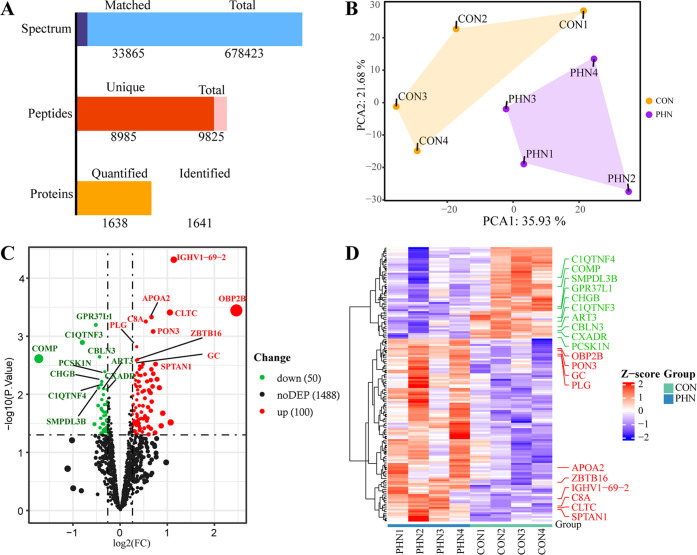
As shown in Figure 2A, the present proteomic results described that a total of 1641 proteins were identified, of which 1638 were quantified. The PCA analysis further proved that PCA1 and PCA2 were 35.93 and 21.68%, respectively, and the distribution of the samples was aggregated within each group and departed between the two groups (Figure 2B), suggesting that a clear different protein expression profile existed between the PHN- and Control-CSF. In Figure 2C,D, both the volcano map and the heatmap indicated that there were 100 upregulated and 50 downregulated DEPs between the PHN and CON groups (both P < 0.05), whereas others showed no statistical differences. The top 10 upregulated proteins and downregulated proteins were marked on both maps.
Figure 2.
Protein quantitation and DEP identification (A). Identification and quantitation of proteomics (B). The results of PCA analysis (C). In the volcano map, the green and red dots represent downregulated and upregulated DEPs, respectively, while the dark dots represent proteins with no statistical differences (D). The heatmap showed hierarchical clustering results of all DEPs. Each column represents a sample, and each row represents a DEP; blue and red represent lower and higher expressions of a protein, respectively. In (C, D), the top 10 upregulated (red) and downregulated (green) DEPs are displayed in the figure.
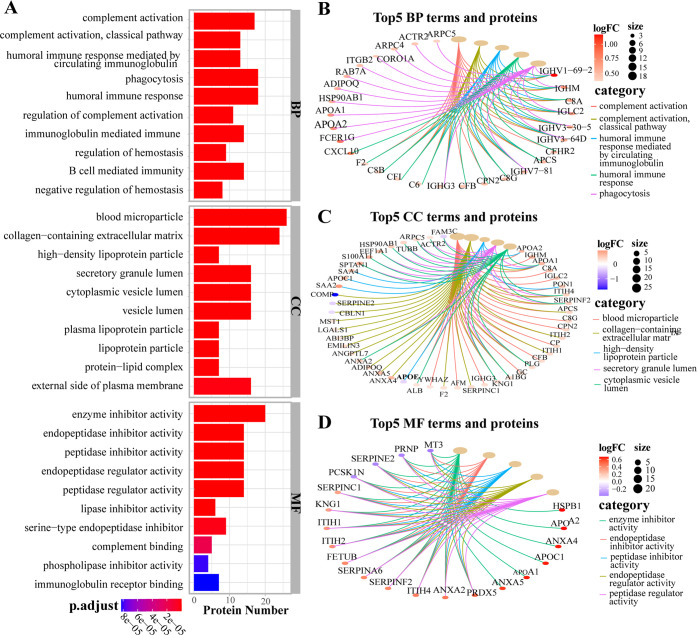
The GO analysis, containing the top enriched terms and proteins network, is displayed in Figure 3. Generally, there were 349 BP, 30 CC, and 60 MF terms significantly enriched in each category. In detail, in the BP category, the top biological processes were complement activation (classical pathway), humoral immune response mediated by circulating immunoglobulin, phagocytosis, and hemostasis regulation (Figure 3A), in which 29 upregulated DEPs, such as IGHV(1–69–2,3–30–5,3–64D), C8(A, B, G), C6, CFB, F2, CXCL10, APOA1, APOA2, APCS, and RAB7A were involved (Figure 3B). In the CC category, blood microparticle, collagen-containing extracellular matrix, high-density lipoprotein particle, secretory granule lumen, cytoplasmic vesicle lumen, and other terms were enriched (Figure 3A), which correlated with 49 DEPs, including downregulated APOE, COMP, SERPINE2, CBLN1, and FAM3C and upregulated C8 (A, G), F2, PLG, GC, APOA1, APOA2, APOC1, PON1, APDIPOQ, ANXA2, ANXA4, ANXA5, SAA2, and SAA4 (Figure 3C). In the MF category, the top-five enriched terms were enzyme inhibitor activity, endopeptidase inhibitor activity, peptidase inhibitor activity, endopeptidase regulator activity, and peptidase regulator activity (Figure 3A), in which the downregulated DEPs such as MT3A, PRNP, SERPINF2, and PCSK1N and upregulated DEPs including APOA1, APOA2, APOC1, HSPB1, ANXA2, ANXA4, ANXA5, PRDX5, etc., were highly involved (Figure 3D).
Figure 3.
Gene Ontology (GO) analysis (A). Barplot of the top 10 terms of biological processes (BP), cellular components (CC) and molecular functions (MF) arranged in ascending order of P values respectively. (B–D) Cnetplot of the top 5 terms of BP, CC, and MF and their corresponding proteins network.
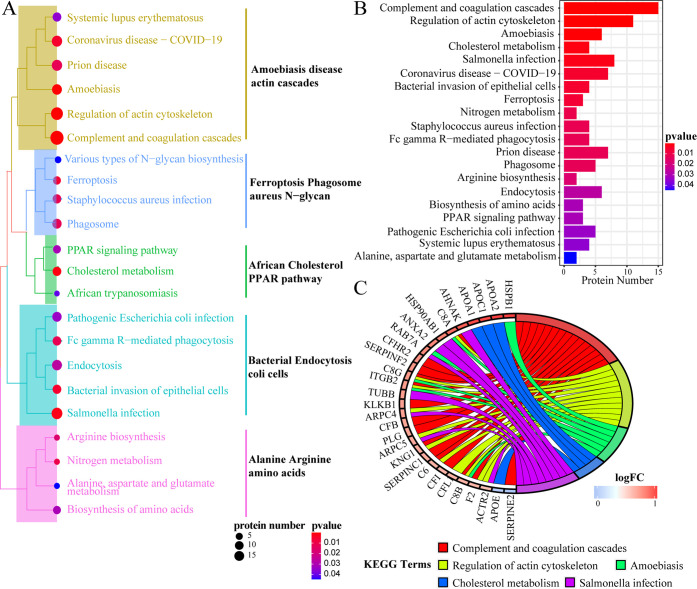
KEGG analysis is displayed in Figure 4. Totally, 22 out of 28 enriched pathways were clustered into five main categories: “Amoebiasis disease actin cascades pathway”, “Ferroptosis Phagosome aureus N–glycan pathway”, “African Cholesterol PPAR pathway”, “Bacterial Endocytosis coli cells pathway”, and “Alanine Arginine amino acids pathway” according to the similarity of DEP expression (Figure 4A). Besides, the top 20 pathways are listed in Figure 2B in an ascending order of the P value (Figure 4B). Moreover, 29 DEPs were closely related to the top 5 pathways, among which only APOE and SERPINE2 were downregulated, whereas the DEPs including C8A, C8B, C8G, F2, PLG, APOA1, APOA2, APOC1, HSPB1, RAB7A, ANXA2, etc., were statistically upregulated (P < 0.05, Figure 4C).
Figure 4.
KEGG analysis. (A) Treeplot of the top 22 significantly enriched pathways clustered into five categories. (B) Barplot of the top 20 pathways. (C) Cnetplot of the DEPs related to the top 5 pathways.
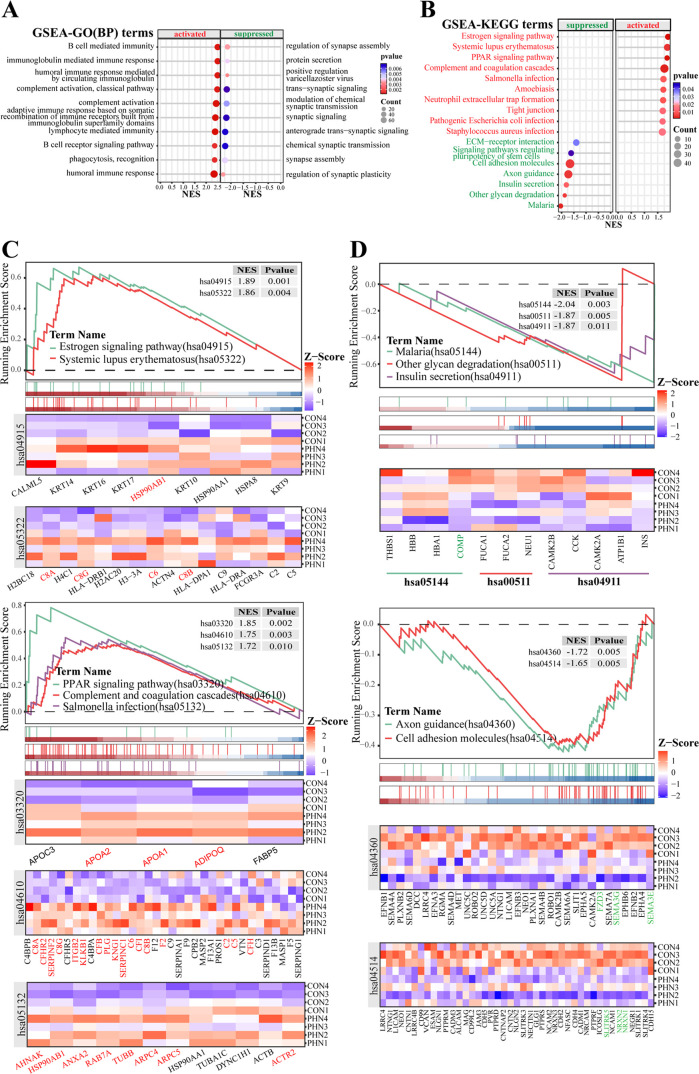
According to the GSEA-GO analysis, there were 307 activated and 205 suppressed terms among the enriched 724 terms in the GSEA-GO(BP) category. The top activated processes involved multiple aspects of the immune response, while the top suppressed processes were mainly about the regulation of synaptic assembly, signal transduction, and plasticity (Figure 5A). Meanwhile, the GSEA-KEGG analysis (Figure 5B–D) elucidated that DEPs including PLG, F2, APOA1, APOA2, SERPINC1, etc., were mainly enriched in the top 5 activated pathways of the estrogen signaling pathway, systemic lupus erythematosus, PPAR signaling pathway, complement and coagulation cascades, and salmonella infection (Figure 5B,C), while DEPs including FZD3, SEMA3G, SEMA3E, SLITRK5, NRXN2, and NRXN1 were enriched in the top 5 suppressed pathways including malaria, other glycan degradation, insulin secretion, axon guidance, and cell adhesion molecules (Figure 5B,D).
Figure 5.
GSEA-GO(BP) and GSEA-KEGG analysis (A): dotplot of the top 10 activated and suppressed terms based on the GSEA-GO(BP) category (B). Dotplot of the top 10 activated and top 7 suppressed pathways (C, D). Enrichment plot and heatmap of core-enrichment proteins for the top 5 activated and top 5 suppressed pathways of GSEA-KEGG results.
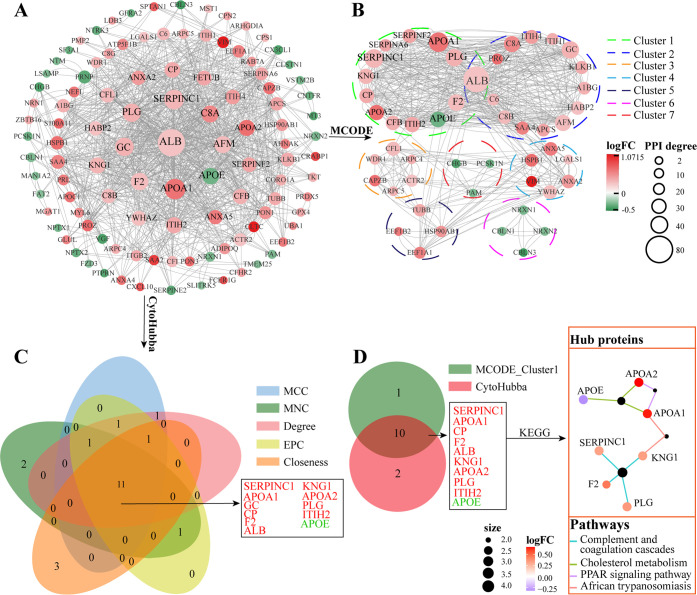
The PPI analysis is displayed in Figure 6. The results revealed that there were 79 upregulated and 30 downregulated DEPs meeting the requirement of the minimum interaction score. Proteins with the highest degree of PPI included ALB (degree:80), APOA1 (degree:58), SERPINC1(degree:54), GC (degree:48), PLG (degree:48), F2 (degree:44), AFM (degree:42), APOE (degree: 40), and C8A (degree:42) (Figure 6A). Furthermore, the MCODE analysis elucidated that all these 109 DEPs could be classified into seven clusters, and ALB, APOA1, APOA2, APOE, PLG, F2, CP, KNG1, CFB, ITIH2, SERPINC1, SERPINA6, and SERPINF2 were clustered in the first queue (Figure 6B). Meanwhile, CytoHubba analysis using five algorithms mentioned above unraveled that SERPINC1, APOA1, GC, CP, F2, ALB, KNG1, APOA2, PLG, ITIH2, and APOE shall be the key proteins (Figure 6C). Next, SERPINC1, APOA1, CP, F2, ALB, KNG1, APOA2, PLG, ITIH2, and APOE were picked through the cross-comparison between the first cluster of MCODE and CytoHbba, and 7 out of them such as SERPINC1, F2, KNG1, PLG, APOA1, APOA2, and APOE were defined as the hub proteins, which were all enriched in the top pathways (including complement and coagulation cascades, cholesterol metabolism, PPAR signaling pathway, and African trypanosomiasis) based on the KEGG analysis (Figure 6D).
Figure 6.
PPI analysis (A). The PPI network (B). MCODE cluster analysis of the PPI network (C). CytoHubba analysis of the PPI network (D). The cross-comparison and the final confirmation of the hub proteins.
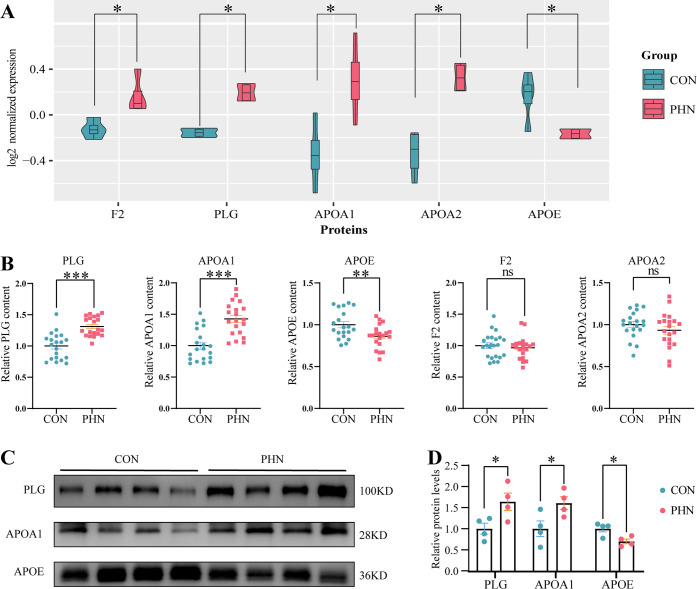
In the present study, five of the hub proteins, namely, F2, PLG, APOA1, APOA2, and APOE, were chosen for further verification. Both ELISA and Western blotting results showed that compared with the CON group, significant upregulation of PLG and APOA1 (both P < 0.001) and downregulation of the APOE (P < 0.01) were found in the PHN group (Figure 7B,C), which were in accordance with the proteomics results (Figure 7A). In contrast, no significant differences were found in either F2 or APOA2 between the two groups (Figure 7B).
Figure 7.
Validation of the hub proteins (A). The levels of the five hub proteins in the proteomic study (B–F). The ELISA results of the five hub proteins: G and H. The Western blotting results of the three verified proteins. * represents P < 0.05, ** represents P < 0.01, and *** represents P < 0.001.
Discussion
In the present study, we found the following results: (1) Compared with nonpainful patients, there were 100 upregulated and 50 downregulated DEPs in the CSF of PHN patients. (2) Further functional analysis indicated that all the DEPs were mainly enriched in the activated biological processes including complement activation, infection, coagulation, and lipid metabolism, and suppressed processes mainly involved in the synaptic organization. (3) The PPI analysis indicated that increased PLG, F2, APOA1, APOA2, SERPINA-1, and KNG1 and reduced APOE, which were all enriched in the top pathways according to the KEGG analysis, were defined as the hub proteins. (4) Three of the hub proteins, namely, PLG, APOA1, and APOE, were reconfirmed in all enrolled patients using both ELISA and Western blotting methods, elucidating that these hub proteins, together with their involving processes such as infection, inflammation, cholesterol metabolism, and coagulation, shall be the potential therapeutic approaches. To the best of our knowledge, this is the first study on the alternations of the CSF component in the PHN patients in comparison with nonpainful patients.
PHN is highly characterized by central sensitization, which makes CSF an ideal medium to explore the pathological changes of the disease. Based on our limited knowledge, few proteomic studies have aimed to elucidate the changes during the process of PHN, among which most of these studies were focused on the changes of the blood rather than the CSF.13,14 There was only one proteomic study conducted15 in 2012 which evaluated the effect of intrathecal steroid administration on protein changes in PHN-CSF. However, the steroid’s action on PHN remains complicated since the steroid treatment has proven to be effective in the short term but not the long term of PHN.16−18 Thus, the steroid-induced protein changes in PHN-CSF might not be the key nodes to elucidate the intrinsic mechanism of PHN. To date, the comparison between PHN patients and healthy people is still lacking. Due to the fact that it is impossible to obtain CSF from normal people, 20 nonpainful patients accepting epidural block and without CNS disorders were enrolled for control in the present study. We suggest that this proteomic study might help to dig out the hub proteins in the process of the PHN.
In a recent study, Lou et al19 demonstrated that in the early subacute stage of the spinal cord injury, the top hub proteins of the spinal tissue were PLG, F2, SERPINA 1, FGG, APOA1, VIM, HPX, and APOE. In the present study, seven DEPs including SERPINC1, F2, KNG1, PLG, APOA1, APOA2, and APOE were defined as the hub proteins based on the PPI and KEGG analysis, which displayed partial similarity with Lou et al ’s research. Among all the hub proteins, PLG, F2, APOA1, APOA2, and APOE were chosen for validation in a larger cohort, and both ELISA and Western blotting results verified that the levels of PLG, APOA1, and APOE were coincident with the proteomic results, assuming that PLG, APOA1, and APOE may be key players in the pathophysiology of PHN.
PLG is primarily synthesized by the liver and released to the bloodstream and plays a critical role in the processes of coagulation, wound healing, infection, and inflammation, thereby maintaining the tissue homeostasis.20 A growing body of evidence has revealed that PLG would migrate from peripheral blood to the central nervous system upon the disruption of the blood–brain barrier (BBB),21,22 which shall further facilitate the infiltration of circulating monocytes/macrophages into the CNS. Likewise, PLG deficiency could reduce the number of the macrophages in the cerebral vessels and relieved microglial activation, thus attenuating the central inflammation severity.23−25 All of the evidence above reveals a tight correlation between peripheral PLG and CNS inflammation. Notably, PLG is also reported to be abundantly expressed in neurons and glial cells in multiple brain areas,26−28 and the bioeffect of this brain-derived PLG still needs to be elucidated. Here, we found that as one of the hub proteins, PLG and its involved pathways including coagulation, infection, and immune response were significantly enriched and activated in the PHN-CSF. We believe that the distribution, function, and mechanism of PLG shall play an important role in the process of PHN, which needs further elucidation in future study.
APOA1 is mainly synthesized by the liver and small intestine and accounts for 90% protein component of high-density lipoprotein (HDL).29 A recent study has demonstrated that knock out of APOA1 in the liver and intestine could diminish the CSF-APOA1 expression, indicating that central APOA1 is mainly derived from the peripheral organs instead of the CNS.30 In the process of Alzheimer’s disease, the infiltration of APOA1 from peripheral blood to CSF would be markedly elevated and primarily mediates neuro-protective properties, since deficiency of APOA1-containing HDL in the brain may lead to cerebrovascular dysfunction and neurodegeneration.31,32 Notably, APOA1’s regulation on pain begins to be concerned. Previous research studies have elucidated that the upregulation of both serum and CSF-APOA1 shall promote the lipid metabolism, provide lipid for the myelin repair and axon regeneration, and mitigate the inflammation and metalloproteinase activity, therefore relieving the pain severity.33−36 In the present study, the CSF-APOA1 level was elevated in the PHN patients, which might represent an endogenous protective mechanism. Besides, APOA1 was enriched in the processes including lipid metabolism, epithelial cell migration, inflammation response, and cellular structural organization, which were all closely involved in the NP development.37−39 Thus, CSF-APOA1 shall serve as another candidate in the PHN.
Unlike PLG and APOA1, APOE is usually considered to originate from the CNS, as it is hard to pass through the BBB.40 As the most abundant expressing apolipoprotein, APOE could be secreted by the astrocytes, microglia, and neurons.41 Notably, it has been reported that patients’ plasma concentration of APOE would increase in the process of peripheral nerve damage,42 and a recent single cell sequence study conducted in 202243 similarly revealed that APOE is one of the top upregulated genes in spinal microglia in the spinal nerve ligation induced NP in mice, indicating a tight association between APOE and neuropathic pain. Actually, the effect of APOE on pain regulation also remains controversial. On the one hand, APOE promotes pain hypersensitivity in a tyrosine kinase receptor dependent manner, and APOE-deficient mice show improved pain behavior.44,45 On the other hand, when the peripheral nerve injury occurs, APOE positive macrophages are inclined to infiltrate to the spinal cord, promote the nerve regeneration through reutilization of myelin cholesterol, and contribute to the nerve restoring and pain relief.46,47 This discrepancy might be due to the fact that APOE has three isoforms, ApoE2, ApoE3, and ApoE4, which take distinct and critical actions during central disorders.48,49 It is now noteworthy that ApoE2 allele develops migraine, tension-type headache, stress, chronic abdominal, back, hip, and knee pain of male, while APOE4 provides protective and pain-relieving action in the chronic headaches (migraine and tension-type headache) and back pain.50,51 In the present study, we detected a significant decrease of the APOE-CSF levels in PHN patients, which were enriched in pathways including lipid metabolism and transport, inflammation, synaptic regulation, neuron toxicity, cell oxidation, and host against viral processes. Therefore, the subtype and role of APOE in PHN will be another worthwhile issue for future research.
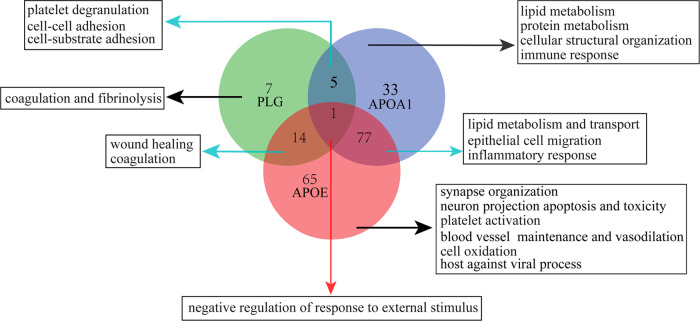
It shall be noteworthy that previous clinical research studies have indicated a correlation between VZV infection and BBB disruption.52− In parallel, in the present study, the activated processes and pathways enriched in the PHN-CSF, such as coagulation, complement activation, infection and immune response, phagocytosis, and cholesterol metabolism, were similar to those in previous proteomic research investigating the plasma metabolism of the PHN patients.14 Besides, PLG and APOA1, the verified hub proteins, are reported to be mainly derived from the bloodstream rather than the CNS. All evidence assumes that the alteration of the BBB shall take place when the PHN occurs, which would permit a crosstalk between peripheral organs and CNS. In addition, our results also indicated that biological processes involving the synaptic organization, such as assembly, signal transduction, and plasticity regulations, were significantly inhibited, further suggesting that the HZ viral infection changes the neurological microstructure and functions. Moreover, a close intersection was found among PLG, APOA1, and APOE, which was similar to previous research.19,55 The overlapped biological processes including negative regulation of response to external stimulus, coagulation, cell adhesion, lipid metabolism, inflammation response, etc., were all significantly enriched pathways (Figure 8), indicating that these proteins might play a synergistic or antagonistic role in the process of the PHN. Above all, these hub proteins and biological processes provide an outbreak for us to further study the intrinsic mechanism of the PHN.
Figure 8.
Summary of involved processes among PLG, APOA1, and APOE in the process of PHN.
There are several limitations in the present study. First, the number of cases was small, which might be unfavorable to extract the statistical conclusions. Second, we only tested the expression patterns of PLG, APOA1, APOA2, F2, and APOE. Other DEPs, such as SERPINC1, ITIH2, CXCL10, GPR37L1, ANXA2, BAB7A, etc., shall be other candidates for verification in a future study. Another limitation is that the current control group was the nonpainful patients, of whom the central inflammation and infection were absent. Therefore, our results might not help to address the factors that will drive the development of the PHN. However, intrathecal intervention is not recommended in our institution when patients were at the acute stage of HZ, which means that the CSF collection from these patients is impossible. Therefore, a future animal study will be conducted to solve this issue.
To sum up, the present study proved that there was a significant difference in the expression profile of CSF proteins between PHN and nonpainful patients. The hub DEPs were mainly enriched in the activated processes such as coagulation, infection, inflammation response, and lipid metabolism, and the suppressed process was mainly involved in the synaptic organization. Among the hub proteins, PLG, APOA1, and APOE were successfully verified in a larger sample, indicating that these proteins and enriched biological pathways may serve as new approaches in treating PHN.
Data Availability Statement
The raw mass spectrometry proteome data and search results have been deposited to the iProx-integrated Proteome Resources (http://www.iprox.cn) with the data set identifier IPX0007372000.
Author Contributions
# K.C. and M.W. contributed equally to this work. Concept and design: W.Y. and Y.L. Acquisition, analysis and/or interpretation of data: All authors. Drafting of the manuscript: W.M. Critical revision of the manuscript for important intellectual content: W.Y. and Y.L. Statistical analysis: C.K. and W.M. Administrative, technical, or material support: All authors. Supervision: W.Y. and Y.L.
This work was supported by the Natural Science Foundation of Hunan Province (2020JJ4811 to W.Y.), Key Research and Development Program of Hunan Province (2020SK2079 to W.Y.), and Foundation of Hunan Pro9+vincial Health Commission (No.202215012870 to Y.L.).
The authors declare no competing financial interest.
Notes
This paper has not been published elsewhere in whole or in part. All authors have read and approved the consent for publication and take responsibility for the data presented.
References
- Gershon A. A.; Breuer J.; Cohen J. I.; Cohrs R. J.; Gershon M. D.; Gilden D.; Grose C.; Hambleton S.; Kennedy P. G.; Oxman M. N.; Seward J. F.; Yamanishi K. Varicella zoster virus infection. Nat. Rev. Dis. Primers 2015, 1, 15016 10.1038/nrdp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. R.; Kong C. L.; Porco T. C.; Kim E.; Ebert C. D.; Acharya N. R. Herpes Zoster and Postherpetic Neuralgia: Changing Incidence Rates From 1994 to 2018 in the United States. Clin. Infect. Dis. 2021, 73 (9), e3210–e3217. 10.1093/cid/ciaa1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A.; Goldust M.; Wollina U. Herpes zoster: A Review of Clinical Manifestations and Management. Viruses 2022, 14 (2), 192 10.3390/v14020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerup N. B.; Kuner R.; Jensen T. S. Neuropathic Pain: From Mechanisms to Treatment. Physiol. Rev. 2021, 101 (1), 259–301. 10.1152/physrev.00045.2019. [DOI] [PubMed] [Google Scholar]
- Watson C. P. N.; Deck J. H.; Morshead C.; Van der Kooy D.; Evans R. J. Post-herpetic neuralgia: further post-mortem studies of cases with and without pain. Pain 1991, 44 (2), 105–117. 10.1016/0304-3959(91)90124-G. [DOI] [PubMed] [Google Scholar]
- Ayton S.; Janelidze S.; Roberts B.; Palmqvist S.; Kalinowski P.; Diouf I.; Belaidi A. A.; Stomrud E.; Bush A. I.; Hansson O. Acute phase markers in CSF reveal inflammatory changes in Alzheimer’s disease that intersect with pathology, APOE ε4, sex and age. Prog. Neurobiol. 2021, 198, 101904 10.1016/j.pneurobio.2020.101904. [DOI] [PubMed] [Google Scholar]
- Eninger T.; Müller S. A.; Bacioglu M.; Schweighauser M.; Lambert M.; Maia L. F.; Neher J. J.; Hornfeck S. M.; Obermüller U.; Kleinberger G.; Haass C.; Kahle P. J.; Staufenbiel M.; Ping L.; Duong D. M.; Levey A. I.; Seyfried N. T.; Lichtenthaler S. F.; Jucker M.; Kaeser S. A. Signatures of glial activity can be detected in the CSF proteome. Proc. Natl. Acad. Sci. U.S.A. 2022, 119 (24), e2119804119 10.1073/pnas.2119804119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpanini S. M.; Torvell M.; Morgan B. P. Therapeutic Inhibition of the Complement System in Diseases of the Central Nervous System. Front. Immunol. 2019, 10, 362 10.3389/fimmu.2019.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J. R.; Zougman A.; Nagaraj N.; Mann M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6 (5), 359–362. 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Bardou P.; Mariette J.; Escudié F.; Djemiel C.; Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinf. 2014, 15 (1), 293 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringnér M. What is principal component analysis?. Nat. Biotechnol. 2008, 26 (3), 303–304. 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- Yu G.; Wang L. G.; Han Y.; He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J. Integr. Biol. 2012, 16 (5), 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Shen H.; Deng H.; Pan H.; He Q.; Ni H.; Tao J.; Liu S.; Xu L.; Yao M. Quantitative proteomic analysis of human plasma using tandem mass tags to identify novel biomarkers for herpes zoster. J. Proteomics 2020, 225, 103879 10.1016/j.jprot.2020.103879. [DOI] [PubMed] [Google Scholar]
- Zhou R.; Li J.; Zhang Y.; Xiao H.; Zuo Y.; Ye L. Characterization of plasma metabolites and proteins in patients with herpetic neuralgia and development of machine learning predictive models based on metabolomic profiling. Front. Mol. Neurosci. 2022, 15, 1009677 10.3389/fnmol.2022.1009677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Katano T.; Nishimura W.; Fujiwara S.; Miyazaki S.; Okasaki I.; Aritake K.; Urade Y.; Minami T.; Ito S. Proteomic analysis of cerebrospinal fluid before and after intrathecal injection of steroid into patients with postherpetic pain. Proteomics 2012, 12, 3105–3112. 10.1002/pmic.201200125. [DOI] [PubMed] [Google Scholar]
- Rijsdijk M.; van Wijck A. J.; Kalkman C. J.; Yaksh T. L. The effects of glucocorticoids on neuropathic pain: a review with emphasis on intrathecal methylprednisolone acetate delivery. Anesth. Analg. 2014, 118, 1097–1112. 10.1213/ANE.0000000000000161. [DOI] [PubMed] [Google Scholar]
- Rijsdijk M.; van Wijck A. J.; Kalkman C. J.; Meulenhoff P. C.; Grafe M. R.; Steinauer J.; Yaksh T. L. Safety assessment and pharmacokinetics of intrathecal methylprednisolone acetate in dogs. Anesthesiology 2012, 116, 170–181. 10.1097/ALN.0b013e31823cf035. [DOI] [PubMed] [Google Scholar]
- Wu C. L.; Raja S. N. An update on the treatment of postherpetic neuralgia. J. Pain 2008, 9, S19–30. 10.1016/j.jpain.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Lou Y.; Lv Y.; Li Z.; Kang Y.; Hou M.; Fu Z.; Lu L.; Liu L.; Cai Z.; Qi Z.; Jian H.; Shen W.; Li X.; Zhou H.; Feng S. Identification of Differentially Expressed Proteins in Rats with Early Subacute Spinal Cord Injury using an iTRAQ-based Quantitative Analysis. Comb. Chem. High Throughput Screening 2023, 26, 1960–1973. 10.2174/1386207326666230113152622. [DOI] [PubMed] [Google Scholar]
- Martin-Fernandez L.; Marco P.; Corrales I.; Pérez R.; Ramírez L.; López S.; Vidal F.; Soria J. M. The Unravelling of the Genetic Architecture of Plasminogen Deficiency and its Relation to Thrombotic Disease. Sci. Rep. 2016, 6, 39255 10.1038/srep39255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultman K.; Cortes-Canteli M.; Bounoutas A.; Richards A. T.; Strickland S.; Norris E. H. Plasmin deficiency leads to fibrin accumulation and a compromised inflammatory response in the mouse brain. J. Thromb. Haemostasis 2014, 12, 701–712. 10.1111/jth.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medcalf R. L. Fibrinolysis: from blood to the brain. J. Thromb. Haemostasis 2017, 15, 2089–2098. 10.1111/jth.13849. [DOI] [PubMed] [Google Scholar]
- Lighvani S.; Baik N.; Diggs J. E.; Khaldoyanidi S.; Parmer R. J.; Miles L. A. Regulation of macrophage migration by a novel plasminogen receptor Plg-R KT. Blood 2011, 118, 5622–5630. 10.1182/blood-2011-03-344242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M. A.; Gao Z.; McElhinney K. E.; Thornton S.; Flick M. J.; Lane A.; Degen J. L.; Ryu J. K.; Akassoglou K.; Mullins E. S. Plasminogen Deficiency Delays the Onset and Protects from Demyelination and Paralysis in Autoimmune Neuroinflammatory Disease. J. Neurosci. 2017, 37, 3776–3788. 10.1523/JNEUROSCI.2932-15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. K.; Chen Z. L.; Norris E. H.; Revenko A. S.; MacLeod A. R.; Strickland S. Blood-derived plasminogen drives brain inflammation and plaque deposition in a mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, E9687–E9696. 10.1073/pnas.1811172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basham M. E.; Seeds N. W. Plasminogen expression in the neonatal and adult mouse brain. J. Neurochem. 2001, 77, 318–325. 10.1046/j.1471-4159.2001.00239.x. [DOI] [PubMed] [Google Scholar]
- Nakajima K.; Tsuzaki N.; Nagata K.; Takemoto N.; Kohsaka S. Production and secretion of plasminogen in cultured rat brain microglia. FEBS Lett. 1992, 308, 179–182. 10.1016/0014-5793(92)81270-V. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y.; Inoue N.; Morita S.; Nikaido Y.; Nakashima T.; Nagai N.; Okada K.; Matsuo O.; Miyata S. Localization of plasminogen in mouse hippocampus, cerebral cortex, and hypothalamus. Cell Tissue Res. 2011, 343, 303–317. 10.1007/s00441-010-1110-5. [DOI] [PubMed] [Google Scholar]
- Maarfi F.; Ahmad S.; Alouffi S.; Akasha R.; Khan M. S.; Rafi Z.; Basnet H.; Khan M. Y. Differential impact of glycation on apolipoprotein A-I of high-density lipoprotein: a review. Glycobiology 2023, 33, 442–453. 10.1093/glycob/cwad010. [DOI] [PubMed] [Google Scholar]
- Tsujita M.; Vaisman B.; Chengyu L.; Vickers K. C.; Okuhira K. I.; Braesch-Andersen S.; Remaley A. T. Apolipoprotein A-I in mouse cerebrospinal fluid derives from the liver and intestine via plasma high-density lipoproteins assembled by ABCA1 and LCAT. FEBS Lett. 2021, 595, 773–788. 10.1002/1873-3468.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres K. Apolipoprotein A1, the neglected relative of Apolipoprotein E and its potential role in Alzheimer’s disease. Neural Regener. Res. 2021, 16, 2141–2148. 10.4103/1673-5374.310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button E. B.; Boyce G. K.; Wilkinson A.; Stukas S.; Hayat A.; Fan J.; Wadsworth B. J.; Robert J.; Martens K. M.; Wellington C. L. ApoA-I deficiency increases cortical amyloid deposition, cerebral amyloid angiopathy, cortical and hippocampal astrogliosis, and amyloid-associated astrocyte reactivity in APP/PS1 mice. Alzheimer’s Res. Ther. 2019, 11, 44 10.1186/s13195-019-0497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.; Ji Y.; Dai H.; Khan A. A.; Zhou Y.; Chen R.; Jiang Y.; Gui J. High-Density Lipoprotein Cholesterol and Apolipoprotein A1 in Synovial Fluid: Potential Predictors of Disease Severity of Primary Knee Osteoarthritis. Cartilage 2021, 13, 1465S–1473S. 10.1177/19476035211007919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellei E.; Vilella A.; Monari E.; Bergamini S.; Tomasi A.; Cuoghi A.; Guerzoni S.; Manca L.; Zoli M.; Pini L. A. Serum protein changes in a rat model of chronic pain show a correlation between animal and humans. Sci. Rep. 2017, 7, 41723 10.1038/srep41723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Carrio J.; Alperi-López M.; López P.; Pérez-Álvarez ÁI.; Robinson G. A.; Alonso-Castro S.; Amigo-Grau N.; Atzeni F.; Suárez A. Humoral responses against HDL are linked to lipoprotein traits, atherosclerosis, inflammation and pathogenic pathways during early arthritis stages. Atherosclerosis 2023, kead009 10.1016/j.atherosclerosis.2023.06.889. [DOI] [PubMed] [Google Scholar]
- Kostopoulou F.; Malizos K. N.; Papathanasiou I.; Tsezou A. MicroRNA-33a regulates cholesterol synthesis and cholesterol efflux-related genes in osteoarthritic chondrocytes. Arthritis Res. Ther. 2015, 17, 42 10.1186/s13075-015-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia-Pelaez J. M.; Choi S. H.; dos Santos Aggum Capettini L.; Xia Y.; Gonen A.; Agatisa-Boyle C.; Delay L.; Dos Santos G. G.; Catroli G. F.; Kim J.; Lu J. W.; Saylor B.; Winkels H.; Durant C. P.; Ghosheh Y.; Beaton G.; Ley K.; Kufareva I.; Corr M.; Yaksh T. L.; Miller Y. I. Normalization of cholesterol metabolism in spinal microglia alleviates neuropathic pain. J. Exp. Med. 2021, 218, e20202059 10.1084/jem.20202059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhmar P.; Trinh R. T. P.; Ma J.; Huo X.; Peng B.; Heijnen C. J.; Kavelaars A. The fibroblast-derived protein PI16 controls neuropathic pain. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 5463–5471. 10.1073/pnas.1913444117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomberg D.; Ahmed M.; Miranpuri G.; Olson J.; Resnick D. K. Neuropathic pain: role of inflammation, immune response, and ion channel activity in central injury mechanisms. Ann. Neurosci. 2012, 19, 125–132. 10.5214/ans.0972.7531.190309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merched A.; Blain H.; Visvikis S.; Herbeth B.; Jeandel C.; Siest G. Cerebrospinal fluid apolipoprotein E level is increased in late-onset Alzheimer’s disease. J. Neurol. Sci. 1997, 145, 33–39. 10.1016/S0022-510X(96)00234-1. [DOI] [PubMed] [Google Scholar]
- Mahley R. W. Central Nervous System Lipoproteins: ApoE and Regulation of Cholesterol Metabolism. Arterioscler., Thromb., Vasc. Biol. 2016, 36, 1305–1315. 10.1161/ATVBAHA.116.307023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron B. M.; VanderPutten D. M.; Merril C. R. Preliminary study of an increase of a plasma apolipoprotein E variant associated with peripheral nerve damage. A finding in patients with chronic spinal pain. Spine 1995, 20, 581–589. 10.1097/00007632-199503010-00014. [DOI] [PubMed] [Google Scholar]
- Tansley S.; Uttam S.; Guzmán A. U.; Yaqubi M.; Pacis A.; Parisien M.; Deamond H.; Wong C.; Rabau O.; Brown N.; Haglund L.; Ouellet J.; Santaguida C.; Ribeiro-da-Silva A.; Tahmasebi S.; Prager-Khoutorsky M.; Ragoussis J.; Zhang J.; Salter M. W.; Diatchenko L.; Healy L. M.; Mogil J. S.; Khoutorsky A. Single-cell RNA sequencing reveals time- and sex-specific responses of mouse spinal cord microglia to peripheral nerve injury and links ApoE to chronic pain. Nat. Commun. 2022, 13, 843 10.1038/s41467-022-28473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Yang S.; Zhou X.; Zhu X.; Xu L.; Li X.; Gao Z.; Sun T.; Wei J.; Tian L.; Cheng H.; Wei G.; Huo F. Q.; Liang L. Nerve injury-induced upregulation of apolipoprotein E in dorsal root ganglion participates in neuropathic pain in male mice. Neuropharmacology 2023, 224, 109372 10.1016/j.neuropharm.2022.109372. [DOI] [PubMed] [Google Scholar]
- Hashikawa-Hobara N.; Hashikawa N.; Yutani C.; Zamami Y.; Jin X.; Takatori S.; Mio M.; Kawasaki H. The Akt-nitric oxide-cGMP pathway contributes to nerve growth factor-mediated neurite outgrowth in apolipoprotein E knockout mice. J. Pharmacol. Exp. Ther. 2011, 338, 694–700. 10.1124/jpet.111.181487. [DOI] [PubMed] [Google Scholar]
- Boyles J. K.; Zoellner C. D.; Anderson L. J.; Kosik L. M.; Pitas R. E.; Weisgraber K. H.; Hui D. Y.; Mahley R. W.; Gebicke-Haerter P. J.; Ignatius M. J.; et al. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J. Clin. Invest. 1989, 83, 1015–1031. 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L.; Fitzner D.; Bosch-Queralt M.; Weil M. T.; Su M.; Sen P.; Ruhwedel T.; Mitkovski M.; Trendelenburg G.; Lütjohann D.; Möbius W.; Simons M. Defective cholesterol clearance limits remyelination in the aged central nervous system. Science 2018, 359, 684–688. 10.1126/science.aan4183. [DOI] [PubMed] [Google Scholar]
- Serrano-Pozo A.; Das S.; Hyman B. T. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80. 10.1016/S1474-4422(20)30412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Fu A. K.; Ip N. Y. APOE signaling in neurodegenerative diseases: an integrative approach targeting APOE coding and noncoding variants for disease intervention. Curr. Opin. Neurobiol. 2021, 69, 58–67. 10.1016/j.conb.2021.02.001. [DOI] [PubMed] [Google Scholar]
- Montagne A.; Nation D. A.; Sagare A. P.; Barisano G.; Sweeney M. D.; Chakhoyan A.; Pachicano M.; Joe E.; Nelson A. R.; D’Orazio L. M.; Buennagel D. P.; Harrington M. G.; Benzinger T. L. S.; Fagan A. M.; Ringman J. M.; Schneider L. S.; Morris J. C.; Reiman E. M.; Caselli R. J.; Chui H. C.; Tcw J.; Chen Y.; Pa J.; Conti P. S.; Law M.; Toga A. W.; Zlokovic B. V. APOE4 leads to blood-brain barrier dysfunction predicting cognitive decline. Nature 2020, 581, 71–76. 10.1038/s41586-020-2247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisano G.; Kisler K.; Wilkinson B.; Nikolakopoulou A. M.; Sagare A. P.; Wang Y.; Gilliam W.; Huuskonen M. T.; Hung S. T.; Ichida J. K.; Gao F.; Coba M. P.; Zlokovic B. V. A ″multi-omics″ analysis of blood-brain barrier and synaptic dysfunction in APOE4 mice. J. Exp. Med. 2022, 219 (11), e20221137 10.1084/jem.20221137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelter S.; Lyrer P.; Radu E. W.; Steck A. J. Acute infectious disorders of the spinal cord and its roots with gadolinium-DTPA enhancement in magnetic resonance imaging. J. Neurol. 1996, 243, 191–195. 10.1007/BF02444013. [DOI] [PubMed] [Google Scholar]
- Jacobi C.; Arlt S.; Reiber H.; Westner I.; Kretzschmar H. A.; Poser S.; Zerr I. Immunoglobulins and virus-specific antibodies in patients with Creutzfeldt-Jakob disease. Acta Neurol. Scand. 2005, 111, 185–190. 10.1111/j.1600-0404.2005.00330.x. [DOI] [PubMed] [Google Scholar]
- Kinno R.; Kurokawa S.; Uchiyama M.; Sakae Y.; Kasai H.; Ogata H.; Kinugasa E. False-positive results obtained for immunoglobulin M antibody tests of cerebrospinal fluid for herpes simplex virus in a patient with varicella zoster virus encephalitis. Intern. Med. 2015, 54, 2667–2670. 10.2169/internalmedicine.54.4891. [DOI] [PubMed] [Google Scholar]
- Pamir N.; Hutchins P. M.; Ronsein G. E.; Wei H.; Tang C.; Das R.; Vaisar T.; Plow E.; Schuster V.; Koschinsky M. L.; Reardon C. A.; Weinberg R.; Dichek D. A.; Marcovina S.; Getz G. S.; Heinecke J. W. Plasminogen promotes cholesterol efflux by the ABCA1 pathway. JCI Insight 2017, 2, e92176 10.1172/jci.insight.92176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw mass spectrometry proteome data and search results have been deposited to the iProx-integrated Proteome Resources (http://www.iprox.cn) with the data set identifier IPX0007372000.