The past two years have brought major additions to the steadily growing collection of molecular pictures of the components of the photosynthetic apparatus: new structural work on photosystem II (PSII; Biesiadka et al., 2004; Ferreira et al., 2004) and of light-harvesting complex II (LHCII; Liu et al., 2004), solved at higher resolution than those previously available, thus allowing for more refined models of key features, a long-awaited view of the cytochrome b6f complex, in two pleasingly concordant versions by two independent groups (Kurisu et al., 2003; Stroebel et al., 2003), and finally a portrait of a supercomplex of a reaction center associated with its antenna—PSI with LHCI (Ben-Shem et al., 2003). The virtual identity of the composition and arrangement of cofactors in complexes that are separated by a billion years of evolution is striking, justifying fully the absence of organism-specific bias in photosynthesis research. On the other hand, the differences in protein–protein interactions show that the structures presented for complexes represent “a” structure rather than “the” structure, emphasizing the unique biological adaptations to ecological niches and environmental variation. At the mechanistic level, the structural advances provide the basis for new hypotheses that in turn suggest experiments that will occupy biophysicists and molecular geneticists alike for the next decade.
The light reactions of photosynthesis describe the pathway in which electrons are removed from water and transferred to NADP+ or ferredoxin to generate reduced cofactors for biosynthetic metabolism (reviewed in Nelson and Ben-Shem, 2004). This process, which makes use of abundant resources on the planet—water and sunlight—is responsible for the creation and maintenance of aerobic life on earth, directly through the generation of oxygen and indirectly through the subsequent biosynthesis (dark reactions) of reduced organic compounds that serve as fuels for heterotrophic life. Intuitively, without an appreciation of thermodynamics, one understands that water is a weak reductant because the product of its oxidation (i.e., removal of electrons), oxygen, is a powerful oxidant. Therefore, an oxidant that is more oxidizing (i.e., more positive redox potential) than molecular oxygen is required to drive water oxidation in vivo. This is accomplished by the primary reactions of PSII, which generate the most powerful oxidant known in biology, P680+. Likewise, the function of PSI is to generate a reductant that is more reducing (i.e., more negative redox potential) than ferredoxin and NADP+.
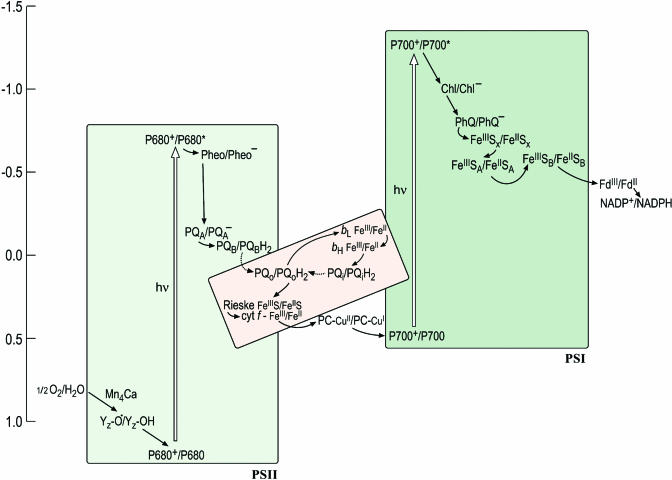
The Z-scheme describes the path of electron transfer from water to NADP+ and depicts each carrier at its midpoint redox potential on a vertical scale with the more reducing (negative Em) components on the top and the more oxidizing (positive Em) on the bottom (Figure 1). The midpoint potential, Em, refers to the potential (which can be measured) at which the concentration of the reduced species is equal to the concentration of the oxidized species for a given redox couple, and this is an approximation of the redox potential, E0. For reactions involving a proton, the term Em,7 refers to the Em at pH 7. The shape of the Z-scheme comes from the fact that the special pigments, P680 and P700 (where P stands for pigment), in PSII and PSI, respectively, have a different redox potential in the ground state versus the excited state. For instance, the redox potential for the P680+/P680 half reaction is ∼+1.1 V versus ∼−0.7 V for the P680+/P680* half reaction. In other words, it is easier for P680* to transfer an electron to an accepting species (Pheo) to generate P680+ and Pheo− (referred to as the charge separated state) than for P680 to give up an electron to Pheo. This is the basis for conversion of light energy to chemical energy.
Figure 1.
An Updated Z-Scheme Describing Photosynthetic Electron Transfer.
Each carrier is shown in both its oxidized and reduced form to facilitate the readers' understanding of the sequence of steps, in which each carrier accepts an electron from a donor to become reduced and is reoxidized as it gives an electron to an acceptor. The midpoint potentials of the carriers in this version of the scheme have been updated to reflect the recent literature but are yet only approximate, owing in part to artistic aesthetics and in part to the intrinsic difficulty of estimating these numbers. In the cytochrome b6f complex (shown in pink to distinguish it as a heme-containing complex), the donor (reduced plastoquinol PQH2) provides two electrons: one is transferred through the Rieske FeS protein and cytochrome f to plastocyanin (or cytochrome c6) to photosystem I, while the other is transferred through the b-hemes to a bound quinone on the stromal side. The dashed arrow to and from PQH2 distinguishes diffusion of the redox carrier from the solid arrows that signify electron transfer. The conversion from the ground to the excited state (indicated with an open vertical arrow) occurs upon absorption of a photon. Some of the carriers appear to have obscure names (e.g., Z in PSII and A0 and A1 in PSI) and these have a historical origin in that the carriers had been identified as spectroscopic signals long before their chemical identities were known. In this scheme, A0 is indicated as Chl and A1 as PhQ to lead the reader away from obscurity. Redding and van der Est (2005) have suggested a specific nomenclature for the electron transfer cofactors in PSI.
But the real wonder of biological reactions lies not in the challenging thermodynamics but rather in the exquisite specificity of and control over these reactions. For efficient use of light energy, it is critical that the excited state should decay only by electron transfer rather than through other paths, and this is accomplished through the structure of the photosystems, which are optimized for fast (picosecond) charge separation. It is even more critical that the electrons from various carriers in the photosystems be transferred to the correct acceptors. For instance, examination of the Z-scheme makes it evident that there is a large thermodynamic driving force for the transfer of an electron from Pheo− or QA− or back to P680+, but this reaction is unproductive. The electron must be transferred in the sequence shown for energy conservation. An even less desirable situation would be the reduction of other reactive groups or compounds in the vicinity of donors. The midpoint potentials of the primary acceptors (e.g., FeSX) in PSI allow reduction of O2, a substrate available at high concentration in oxygenic photosynthesis, to superoxide (Em,7 for the O2/superoxide half reaction is ∼−0.3 V), but there are mechanisms in place to minimize these events in favor of directional and specific electron transfer within the photosystems. How is the specificity achieved? What determines the directionality of electron transfer in each complex? How is proton translocation coupled to electron transfer? For PSII, the question of how the four-electron oxidation of a pair of water molecules is coupled to four sequential single electron excitations remains a holy grail for structural biologists, biochemists, and inorganic chemists. Meanwhile, plant biologists are grappling with the question of how the operation of the photosynthetic machinery, which has evolved for precise chemistry, is tuned to respond to physiological patterns, metabolic programs, a changing environment, or developmental state. Photosynthesis is the lifeblood of the plant; yet, its components generate reactive intermediates that can kill the plant. Energy input into the system via light harvesting is the source of these intermediates; therefore, the mechanism and regulation of energy transfer within chlorophyll proteins is a central aspect of the solution. A view of the photosynthetic apparatus at atomic resolution is a prerequisite for understanding its mechanism and regulation.
PSII
PSII or water-plastoquinone oxidoreductase is the first enzyme in the Z-scheme pathway (Figure 1). The first models of the active site with respect to the cofactors involved in charge separation were based on the close structural and compositional relationship of PSII to the bacterial reaction centers (found in the anaerobic purple bacteria), whose high-resolution structures were determined ∼20 years ago (Deisenhofer et al., 1984; Allen et al., 1986), and these were validated and further refined by structural models of PSII crystals derived from studies at 3.7- and 3.8-Å resolution (Zouni et al., 2001; reviewed in Diner and Rappaport, 2002; Kamiya and Shen, 2003). Nevertheless, the active site that participates in the unique water oxidation activity of PSII could not be modeled in those works. Two recent papers describe structures based on higher-resolution diffraction data: Ferreira et al. (2004) at 3.5 Å and Biesiadka et al. (2004) at 3.2 Å. Both give a clearer view of the cofactor binding sites and provide rational explanations for the distinct energetic differences and primary charge separation mechanisms between PSII and the bacterial reaction center, and both show improved models for the extrinsic subunits and loops of the transmembrane proteins. Both studies also delineate the path of alternate secondary electron transfer from the heme of cytochrome b559 and ChlZ via a β-carotene to P680+ and suggest ligands for the manganese cluster. The structural model from Ferreira et al. (2004), although at lower resolution than the one from Biesiadka et al. (2004), proposes a more specific (albeit controversial) arrangement of metals in the cluster, which encourages renewed discussion of and speculation on the reaction mechanism for water oxidation (e.g., McEvoy and Brudvig, 2004).
The various reactions of PSII (Figure 1) can be separated into three parts for the purpose of discussion: the primary reaction, which is the excitation of P680 and the transfer of an electron from the excited state to a pheophytin (Pheo) acceptor in the enzyme to generate the charge separated state; electron transfer from Pheo− to the quinone in the reaction center to generate the product of the enzyme, PQH2; and the rereduction of P680+ by water through a path involving the Mn4Ca cluster and a special Tyr residue (YZ).
Charge Separation
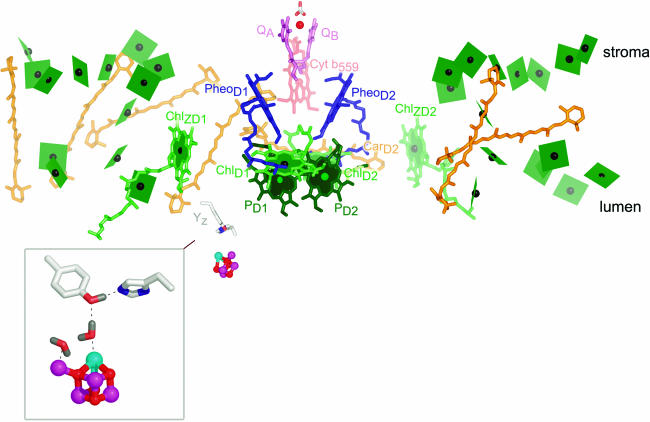
The reaction of PSII begins with excitation of the central pigments. The structures reveal that these are arranged as a pair of chlorophyll pigments, called PD1 and PD2 that are themselves close to another pair, called ChlD1 and ChlD2 (Figure 2). Accordingly, the structure is compatible with the excited singlet state 1P680* being delocalized over all four pigments, but the properties of each pigment with respect to photochemistry are distinct (Diner and Rappaport, 2002). The close proximity of the pheophytins (blue) facilitates efficient and fast transfer of an electron from ChlD1 of the excited state to PheoD1, generating P680+ localized on PD1 and PheoD1− (the charge separated state). The energy gap between P680+ and PheoD1− is almost 2 V, corresponding to a typical household battery. This reaction is the basis for energy conversion in photosynthesis: from the energy of a photon to chemical energy in redox potential.
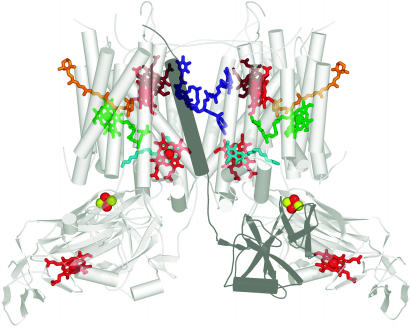
Figure 2.
Cofactors in PSII.
The cofactors in PSII, based on Protein Data Bank coordinates 1S5L, are shown. The view is perpendicular to the membrane normal. The color scheme of Ferreira et al. (2004) is retained. Chlorophylls are indicated in green, with the central Mg shown as a black sphere. The core antenna chlorophylls are distinguished from reaction center chlorophylls by the use of planes lacking phytol tails for the former versus sticks for the latter. PD1 and PD2 are in darker green to distinguish them from ChlD1 and ChlD2. Heme in cytochrome b559 is diagrammed in pale-red sticks, carotenoids in orange, pheophytin in blue, and plastoquinone in pink. The tail of plastoquinone is omitted for clarity. The metals are shown as spheres: red for iron, magenta for manganese, and cyan for calcium. The side chains of Tyr161 (YZ) and His190 near the Mn4Ca cluster are shown as gray sticks for carbon, blue for nitrogen, and red for oxygen. The inset shows an enlargement of the Mn4Ca cluster as proposed by Ferreira et al. (2004), with YZ on the top left and His190 on the bottom right. Two waters and a hydrogen on TyrZ are modeled as per Ferreira et al. (2004). Note that there is considerable ambiguity concerning the structure of the cluster (see main text). The subscripts D1 and D2 denote the polypeptide binding site of the cofactor. The polypeptide nomenclature is historical: D1 and D2 for the apoproteins that bind most of the cofactors in PSII are so named because they had been initially revealed as diffuse staining bands in thylakoid membrane preparations. CP43 and CP47 stand for chlorophyll-protein, with the numbers indicating their size as estimated from SDS-PAGE. The heme binding heterodimer is called cytochrome b559, reflecting the absorption maximum of the α band of the heme group, and other proteins are named as gene products (Psb_). PyMOL was used to generate this and other images. In all images, molecules closer to the reader are colored in a deeper shade, while those that are more distant are in a paler shade.
The electron from PheoD1− is transferred to QA and from QA− to QB. After the resulting QB− is reduced by a second electron and protonated, the quinol product, QBH2, leaves PSII for the cytochrome b6f complex. As in the bacterial reaction center, there is an Fe positioned between the two quinones. Electron density around this Fe has been suggested to correspond to a bicarbonate ion, which could regulate electron flow between QA and QB, depending on substrate availability for the dark reactions, and which could also facilitate protonation of reduced QB (Ferreira et al., 2004).
Despite the close structural similarity, especially for the cofactors, between the bacterial reaction center and PSII, there is a key and highly significant difference in physical properties. The radical cation P680+ has a more positive redox potential (∼1.1 to 1.3 V), necessary for downhill electron transfer from water (see Figure 1), than P960+ in the bacterial reaction center (∼0.4 to 0.5 V). The structures suggest an explanation (Biesiadka et al., 2004; Ferreira et al., 2004). The distance between PD1 and PD2 is 0.7 Å longer than that of the equivalent pair in the bacterial reaction center, which reduces electronic coupling, and the local environment around PD1/PD2 is also more hydrophobic. This means that the radical cation is less stabilized in PSII compared with the bacterial reaction center, and this translates to a more positive redox potential. Interestingly, the environment around PD1/PD2 is relatively devoid of other pigments (compared with the environment around P700 in PSI), perhaps providing a buffer zone for the highly oxidizing P680+. (See discussion of alternate pathways of P680+ reduction below.)
TyrZ (YZ)
A redox active Tyr side chain, named TyrZ and corresponding to Tyr161 of the D1 subunit (see inset in Figure 2), reduces P680+. This reaction also requires proton transfer to His190, leaving a neutral tyrosyl radical, which will oxidize the Mn4Ca cluster—the heart of the water oxidation site. The mechanism of Tyr oxidation is of interest: does it occur as hydrogen atom removal (i.e., proton coupled electron transfer)? For this to occur, there has to be a path for transfer of the proton from Tyr to the lumen, and both of the recent higher-resolution models show a hydrogen bonded network and a possible hydrophilic channel for proton exit and water entry near the metal cluster, but the connection between TyrZ and this exit path is not evident, leaving the possibility that the path is created only upon a conformational change that occurs during one of the oxidation steps in the overall four-electron oxidation of water (Rutherford and Boussac, 2004).
Water Oxidation and the Mn4Ca Cluster
The most interesting and, at the same time, hotly debated part of the structure is the Mn4Ca cluster at the active site for water oxidation. This is a mechanistically challenging reaction because it involves stepwise catalysis of a four-electron oxidation (2H2O to O2), one electron at a time (as P680+ is generated by excitation and charge separation and then reduced by TyrZ), but without release of reactive oxygen intermediates. The various oxidation states of the enzyme are referred to as the S-states, which advance from S0 through S1, S2, and S3 to the S4 state by sequential loss of a single electron. The S4 state returns to the S0 state as oxygen is evolved. The resting state of the enzyme is S1, corresponding to a single oxidation, and it is likely that the crystals are populated with molecules representing this state before their exposure to x-rays.
Ferreira et al. (2004) have identified five metal ions in the cluster, four are Mn and one is Ca (see inset in Figure 2). They model three of the Mn together with 1 Ca as a cube with bridging oxygen atoms (called μ-oxo) at the other corners. In this model, the fourth manganese ion is outside the cube and connected to it through an oxygen bridge. Several protein side chains have been identified as ligands (see below), and many of these had been predicted from mutagenesis experiments, but not enough ligands have been identified to satisfy the coordination numbers for Ca2+ and Mn (especially in its higher valence states). Because bicarbonate is important for assembly of the oxygen-evolving complex, the authors suggest that there is room for a bridging tridentate bicarbonate between the fourth manganese and the calcium and assign nonprotein electron density to bicarbonate. They speculate that the bicarbonate bound to the lower S-states is replaced during catalysis with substrate water molecules in the higher S-states. Chloride is also a cofactor for oxygen evolution, and Ferreira et al. (2004) propose chloride as a ligand for the calcium because its nucleophilicity is compatible with the proposed role of calcium-bound water as a nucleophile in the water oxidation mechanism (see below). Water or hydroxide, which are not seen in these models, may provide other ligands. Why Mn and Ca? The special feature of Ca2+ is that it adopts various coordination numbers and hence geometry, and this may be a useful property for accommodating structural changes in the cluster as it progresses through the sequential oxidation states. (The coordination number is the number of atoms or ligands directly bonded to a central atom of a complex; for Ca2+, the coordination number is usually 6 to 7, but 8 and <6 are possible.) Mn is useful in redox reactions because it can occur in several stable oxidation states, including MnII, MnIII, and MnIV, which makes it particularly useful as a catalyst of reactions involving multistep sequential electron transfer.
According to a current mechanistic model (Barber et al., 2004; Iwata and Barber, 2004), proposed in essence initially by Brudvig and coworkers (Vrettos et al., 2001), one of the substrate water molecules binds to the fourth manganese (the one outside the cube) and the other binds to the Ca2+. As the manganese is oxidized and the bound water deprotonated, the MnV-ligated oxo group becomes electrophilic, which makes it a perfect target for nucleophilic attack by the oxygen of water bound to the Ca2+, bringing about O=O bond formation.
Alternate Secondary Reduction of P680+
As pointed out above, P680+ is a powerful oxidant (Em estimates ∼1.1 to 1.3 V), which means that PSII, unlike other photosystems, has the potential to oxidize its own pigments (discussed in Frank and Brudvig, 2004). Therefore, alternate pathways for reducing P680+ have evolved in a protective role. These alternate pathways are important during assembly of the cluster and also under various stress conditions when delivery of the electron from the Mn4Ca/TyrZ site to P680+ is inhibited. Spectroscopic studies had indicated the participation of cytochrome b559, ChlZ and β-carotene in these alternate pathways, with β-carotene serving as the immediate donor to P680+. Both of the recent higher-resolution structures agree on the placement of an all trans β-carotene connecting cytochrome b559 and ChlZD2 to P680+. The placement supports a branched pathway—either from cytochrome b559 or ChlZD2—for reduction of the carotenoid cation (Hanley et al., 1999; Frank and Brudvig, 2004). Spectroscopic studies of PSII also indicate other distinct chlorophyll and carotenoid species that can be oxidized, and it has been proposed that these may function in photoprotection to quench excitation energy. Indeed, a recent spectroscopic study reveals the participation of a charge transfer state involving a chlorophyll anion and a zeaxanthin radical cation in PsbS-mediated feedback deexcitation of excess energy (see below) in PSII (Li et al., 2000; Holt et al., 2005). These alternate photooxidation pathways reveal a novel aspect of carotenoid function (i.e., as a redox active cofactor) that awaits further elaboration by spectroscopy and additional structural information.
Strength of the Structural Model of the Metal Cluster
The validity of the model for the Mn4Ca cluster is questioned at several levels (Barber et al., 2004; Biesiadka et al., 2004). First, from the fact that the structure necessarily represents only one of the possible oxidation states and it is known that there are conformational changes and probably ligand exchanges as the S-state cycle progresses. The model therefore could present, at best, a static snapshot of a flexible structural feature. Second, because the Mn–Mn distances measured by spectroscopic techniques are shorter than can be resolved by the available crystallographic data, there is a need for a higher-resolution structure. This is of greater concern for the cluster where positions of atoms are completely unknown than for protein atoms whose positions are constrained by known sequence. The other caveats are related to experimental handicap, including the presence of ammonium sulfate during crystallization and the lability of the cluster, especially upon radiation when the higher valence states of manganese get reduced to MnII, which is more prone to ligand exchange. While one might argue that the dissociated metals have not moved very far during low temperature data collection, it is possible that there is enough rearrangement to generate ambiguity with respect to the ligands and, hence, the geometry. The problem of radiation damage is a general one for protein structures determined by x-ray crystallography, and it has been suggested that the effect of damage be taken into consideration, especially for studies of metalloprotein oxidation states (Ravelli and McSweeney, 2000). Therefore, at worst, the model may not resemble any of the S-state geometries. Nevertheless, given the general gross similarity in shape of the cluster in two independent works (a larger lobe containing three or four metals adjacent to a smaller lobe containing one metal corresponding to the 3+1 Mn tetramer proposed by spectroscopists) and the agreement concerning the presence of Ca2+ in the larger lobe, and consensus on the assignment of several of the metal ligands, including Asp170, His332, Glu189, Glu333, and the C terminus of D1 and Glu354 in CP43, the nonspecialist would agree that we are a step closer to the holy grail in PSII research, which is a cause for celebration because it stimulates spectroscopists and molecular biologists to test the model.
Subunits
The structural model of Ferreira et al. (2004) identifies 19 subunits of the Thermosynechococcus elongatus PSII preparation, which curiously lacks one of the two newly discovered, lipid-anchored extrinsic membrane proteins, PsbQ, present at a stoichiometry of one per Synechocystis sp 6803 PSII complex (Thornton et al., 2004). With the exception of subunit PsbN, which is tentatively placed because of side chain disorder, the others are assigned on the basis of side chain electron density. In the previous crystallographic studies, whereas the cofactor binding subunits were localized unambiguously based on electron density, the identities of several other subunits were deduced and mapped to particular backbone traces in the multisubunit complex with information derived from biochemical cross-linking experiments or copurifications that suggested specific protein−protein interactions (discussed in Kamiya and Shen, 2003). The functions of these other, mostly low molecular weight, subunits are not known; roles in dimer formation, structural stabilization, pigment binding, and interaction with peripheral antenna are possibilities. The new model, which is likely not the last word in PSII subunit arrangement (see cautionary note in Biesiadka et al., 2004), certainly provides a structural framework for suggesting specific functions. For instance, the position of subunits L, M, and T at the center of the dimer would argue for their importance in dimer formation, and subunits I and X, which are symmetrically related, clearly interact with ChlZD1 and ChlZD2 (Ferreira et al., 2004). These ideas will undoubtedly be evaluated and tested by directed mutagenesis experiments, and the structure itself will eventually be reinterpreted from higher-resolution diffraction data that will allow more definitive interpretations of electron density, which is important for the small subunits that lack landmarks to guide the assignments (Biesiadka et al., 2004).
From a cell biologists perspective, the complexity of the structure (numbers and variety of cofactors and transmembrane proteins) makes the problem of biogenesis daunting, and the question of the D1 repair cycle even more intimidating (Baena-González and Aro, 2002).
LHCII
Excitation energy for the primary reactions of photosynthesis is collected via a light-harvesting system or antenna, which functions to increase the capacity of the photosystems for collecting photons. The antenna associated with the reaction center polypeptides is referred to as the core antenna, but there are also accessory antennae that associate with each photosystem. The accessory antennae can be quite varied in terms of pigment profile or size, allowing photosynthetic organisms to take advantage of a large portion of the solar spectrum and exploit ecological niches with a broad range of incident light intensity (Green and Parson, 2003). In plants, the accessory antenna system consists of several integral membrane protein complexes containing polypeptide chains (e.g., Lhca1 through Lhca4 and Lhcb1 through Lhcb6 where the notation “a” refers to the constituents of LHCI and “b” refers to the constituents of LHCII), pigment molecules (chlorophyll a, chlorophyll b, and carotenoids), and lipids (Peter and Thornber, 1991; Kühlbrandt et al., 1994; Jansson, 1999). The most abundant of these—accounting for approximately half of the chlorophyll in the thylakoid membrane—is LHCII.
LHCII is a heterogeneous trimer of three nearly identical polypeptides, Lhcb1, Lhcb2, and Lhcb3, found in various stoichiometries: Lhcb1 and Lhcb2 are the most abundant and more similar to each other, whereas Lhcb3 appears to have some unique properties (Caffarri et al., 2004; Standfuss and Kühlbrandt, 2004). LHCII forms the outer accessory antenna and is connected to the core antenna of PSII via the inner accessory antenna that consists of monomeric Lhcb4, Lhcb5, and Lhcb6 proteins. LHCII function is a topical area of research, not only because of its prevalence and light-harvesting role, but also because it is a key target of several signal transduction pathways that control light energy use. The abundance of the constituent Lhcb polypeptides is determined by gene expression mechanisms as a long-term adaptive response to light quantity. At a more short-term level, the complex is regulated by phosphorylation depending on the redox state of a carrier between the two photosystems (namely, a quinone in the cytochrome b6f complex) so that it associates physically and functionally with PSII when the carrier is more oxidized (called State I) but with PSI when the carrier is more reduced (called State II), and recent work indicates a role for Lhcb proteins in photoprotection via dissipation of excess light energy (Horton et al., 1996; Niyogi, 1999; Wollman, 2001; Elrad et al., 2002). A prerequisite for a mechanistic understanding of LHCII function in light harvesting as well as thermal dissipation of excess light is a high-resolution structure showing the identity and orientation of all pigment molecules. This has been achieved recently through the work of Liu et al. (2004), who present a structural model of spinach LHCII at 2.72-Å resolution based on x-ray diffraction analysis of the complex in icosahedral proteoliposome assembly. This higher-resolution structure reveals details that were not evident in the earlier structural model (Kühlbrandt et al., 1994), including placement of all bound chlorophylls, the distinction between chlorophyll a and chlorophyll b binding sites, their orientation, and the binding sites for four carotenoids.
Composition
The structure shows a trimer (Figure 3A) with each monomeric subunit containing 14 chlorophylls, four carotenoids, and two lipids. The high resolution makes it possible to unambiguously distinguish between chlorophyll a and b, which differ only with respect to the substituent at the C7 position of the tetrapyrrole ring (Figure 4): eight are labeled as chlorophyll a and six as chlorophyll b, and this is pleasingly consistent with the chlorophyll a:b ratio of LHCII determined by biochemical methods. Structural features that contribute to selective binding include hydrogen bonding interactions for the C7 formyl group of chlorophyll b at five of the six chlorophyll b binding sites on the one hand versus a more apolar pocket around the C7-methyl at the chlorophyll a sites that would destabilize chlorophyll b binding by steric hindrance or the hydrophobic effect. The high selectivity of the sites was noted in previous biochemical studies (Rogl et al., 2002).
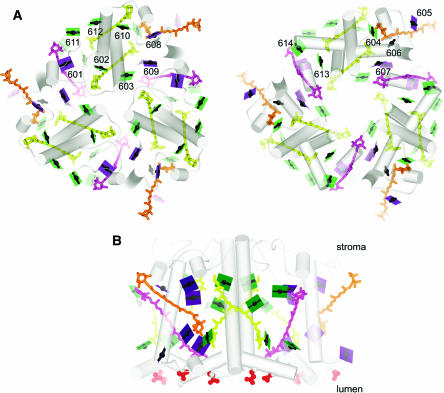
Figure 3.
Organization of Pigments in LHCII.
(A) The trimer is viewed along the membrane normal from the stromal side (left). The lumenal side (right) is viewed from the stromal side as if reflected in a mirror to facilitate visualization of chlorophyll proximities in the two layers. The color schemes of green for chlorophyll a, purple for chlorophyll b, the arbitrary numbering system for the pigments from 601 to 614, and representation of the chlorophylls as a line connecting two nitrogens of rings A and C with a central magnesium (see Figure 4), which emphasizes the direction of the Qy axis, is retained from Liu et al. (2004). The carotenoids are indicated in yellow for luteins, orange for neoxanthin, and magenta for the xanthophyll cycle pigment. As in Figure 2, the pigments closer to the viewer appear darker than those further away. Hence, 24 chlorophylls are shown as dark planes in the stromal view. The lighter ones in that image are the ones in the lumen-side layer.
(B) The trimer is viewed in the plane of the membrane, perpendicular to the membrane normal as in Figure 2. The red spheres show the negatively charged residues on the lumen side.
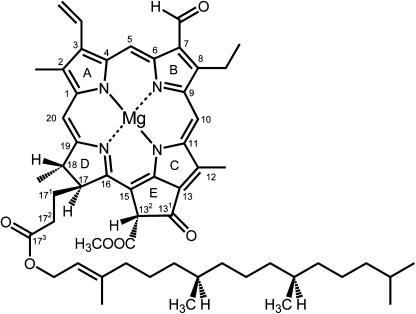
Figure 4.
Structure of Chlorophyll.
The structure of chlorophyll b is shown with International Union of Pure and Applied Chemistry numbering. In chlorophyll a, the C7 formyl is a methyl group. Chlorophyll a′ is the C132 epimer of chlorophyll a. The y axis of the molecule passes through the N atoms of rings A and C and the x axis through the N atoms of ring B and D. Without the central magnesium ion, the tetrapyrrole is referred to as a pheophytin.
Two carotenoids, both identified as lutein, function as a cross-brace for the monomer structure, and these are essential for structural stability and for proper in vitro reconstitution of the protein (Plumley and Schmidt, 1987; Kühlbrandt et al., 1994). A third carotenoid corresponds to neoxanthin: a specific hydrogen-bonding interaction and a hydrophobic pocket that accommodates the bent polyene chain contribute to the selectivity of the site for this xanthophyll. The fourth carotenoid binding site, whose location was not known previously, shows mixed occupancy with different xanthophyll cycle pigments and is located between monomers (discussed below).
Energy Transfer
The high energy (short wavelength) transitions in the absorption spectrum of chlorophyll are known as the B bands or Soret bands and the low energy (long wavelength) ones in the red portion of the spectrum as the Q bands. The longest wavelength transition is polarized along the y axis of the molecule (Figure 4) and is therefore known as the Qy transition. This is the transition that is most relevant for the energy transfer processes in chlorophyll-containing complexes (Blankenship, 2002). Because of the energy difference between the Q transitions of chlorophyll a versus b (more red shifted; i.e., lower energy for a), the directionality of energy transfer within a light-harvesting unit must go from chlorophyll b to chlorophyll a. Hence, the distinction between the two pigments is a prerequisite for understanding the path of excitation energy movement within LHCII. The relative orientation of the pigments and the distance between them (close enough for efficient transfer of excitation energy but far enough to avoid electron transfer) is a determinant of how pigments interact with each other and contribute to ultrafast energy transfer rates (in the 102 fs range), and this aspect of structural detail in the work by Liu et al. (2004) represents another big advance for understanding the mechanism of LHCII function.
A view of LHCII parallel to the membrane plane shows that the chlorophylls are arranged in two layers, one closer to the stromal side with eight chlorophylls and the other closer to the lumen side with the remaining six (Figure 3B). The arrangement, reminiscent of the two-layer arrangement of pigments in the core antenna of the photosystems, evokes a pattern, probably with some functional significance. A view along the membrane normal from the stromal side shows the arrangement of 24 chlorophylls in the trimer in two rings (Figure 3A). The inner ring consists of six chlorophyll a, and these are believed to function in energy transfer between monomers. In the outer ring, a pattern of three chlorophyll b alternating with three chlorophyll a is evident. Liu et al. (2004) suggest that this pattern would allow light absorption from all directions in a broad spectral window with efficient (short distance, few steps) energy transfer to chlorophyll a 612 (whose Qy transition is of lowest energy in the complex), from where it leaves LHCII for the reaction center (Rogl and Kühlbrandt, 1999). The close proximity and orientation of chlorophyll a 611 and chlorophyll a 612 in the structural model gives the strongest calculated excitonic coupling and hence explains the red shifted absorption spectrum of this terminal acceptor. On the lumenal side (Figure 3A), the distances between pigments is greater and accordingly energy transfer between them less efficient, and it is suggested that the lumen-side chlorophylls function upstream of the stromal chlorophylls with respect to light harvesting.
Three of the carotenoids—the two luteins and neoxanthin—also function in light harvesting: the luteins are favorably located for energy transfer to several chlorophyll a and the neoxanthin to two chlorophyll b, and this is again entirely consistent with experimental observation. The addition of carotenoids as light-harvesting pigments extends the spectral window of LHCII to complement the absorption of chlorophylls.
Feedback Deexcitation
When a pigment molecule absorbs light and is in the excited state, there are several possible paths for return of the molecule to the ground state. Electron transfer is one possible outcome, and this is referred to as photochemical quenching. When the photochemical pathway is backed up (e.g., when incident light energy absorption exceeds the capacity for use of reducing equivalents in the dark reactions), there is a danger that the excited state can decay via undesirable routes; therefore, protective mechanisms, collectively referred to as nonphotochemical quenching (NPQ), are built into the system. An important part of NPQ is a regulatory process that is associated with PSII and LHCII called feedback deexcitation. Feedback deexcitation determines whether absorbed light will be used for photochemistry or dissipated as heat (Horton et al., 1996; Müller et al., 2001; Holt et al., 2004). The process relies on acidification of the lumen via a ΔpH, a resulting conformation change in a thylakoid membrane component, and the interconversion of certain xanthophylls, specifically the consecutive deepoxidation of violaxanthin to antheraxanthin and zeaxanthin (Zea).
As mentioned above, one mechanism, dependent on PsbS in PSII, involves excitation energy transfer to a Chl-Zea pair (at an as yet biochemically undefined site), followed by its relaxation to a charge transfer Chl−Zea+ state and charge recombination to the ground state Chl-Zea (Holt et al., 2005). There is also discussion of the contribution of LHC proteins directly in NPQ, especially because the Chlamydomonas npq5 mutant is defective in an LHCII subunit (Elrad et al., 2002). Liu et al. (2004) propose models for an NPQ mechanism in LHCII based on structure. For instance, in one model, they suggest that low pH would protonate four carboxylates on the lumen side of the LHCII (red groups in Figure 3B), which would break ion pairs and hence trigger conformational changes of the lumen-side C terminus and the BC loop. This in turn could move chlorophyll a 614 to promote the proposed quenching effect of a putative chlorophyll a 613– chlorophyll a 614 trapping site deduced from their mutual orientation in the structure. These ideas are speculative, but the process of feedback deexcitation undoubtedly has multiple contributing players, and the structure-based model for quenching in LHCII stimulates the design of new experiments.
Despite the information content of the recent contribution (Liu et al., 2004), the game with respect to light-harvesting chlorophyll protein function is not over. There are firm indications of trimer heterogeneity at the level of organization with PSII and of compositional variation with respect to constituent monomers, which might speak to the specific roles of particular monomer subunits, and there will almost certainly be structural differences between the Lhcb proteins in trimers in the outer accessory antenna versus the Lhcb monomers that are closer to the reaction center and the core antenna. The question of the physical and functional connection of pigments from the outer to inner accessory to core antenna awaits high-resolution structural analysis of a PSII-LHCII supercomplex.
CYTOCHROME b6f
The second enzyme in the Z-scheme pathway, the cytochrome b6f complex, or plastoquinol to plastocyanin/cytochrome c6 oxidoreductase oxidizes the plastoquinol product from PSII (see Figure 1). The product of the reaction is reduced plastocyanin or, in some cyanobacteria and algae, reduced cytochrome c6, which in turn will reduce photooxidized P700+ (reviewed in Redinbo et al., 1994; Merchant, 1998). Electron transfer through the cytochrome b6f complex is also coupled to proton translocation, which makes its activity a major contributor to the generation of a proton motive force for ATP synthesis. Oxidation of the substrate plastoquinol to the quinone product yields two electrons and two protons, but the cofactors in plastocyanin and cytochrome c6 are only one-electron carriers: the mechanism of electron flow and proton movement in the complex is therefore a key area of interest.
Cytochrome b6f is a connector between the two photosystems—PSII pushes electrons into the cytochrome b6f complex while PSI pulls electrons out of it. Not surprisingly then, the cytochrome b6f complex plays also a regulatory role in light energy use by relaying the redox status of the electron transfer chain to various signal transduction pathways. The regulatory phenomenon of state transitions (see above) in which the accessory antenna, LHCII, can associate with either PSII or PSI is dependent on the cytochrome b6f complex (Finazzi, 2004). When electron input into the complex (from PSII) is greater than electron draw from the complex (by PSI), the LHCII moves from PSII to PSI to balance the flow. The movement of LHCII is determined by its phosphorylation state, which is maintained by a kinase whose activity is regulated by occupancy of one of the plastoquinol binding sites (Qo) in the cytochrome b6f complex and, hence, the redox status of the chain (Depège et al., 2003).
The cytochrome b6f complex can accept electrons also from components that are reduced by the primary acceptors of PSI; whether a cofactor in the cytochrome b6f complex is directly reduced or whether the complex participates only indirectly via the PQ pool is a controversial topic (Kramer et al., 2004). Because the products of electron flow through the cytochrome b6f complex—plastocyanin and cytochrome c6—rereduce P700+, this alternate electron flow pathway is referred to as cyclic electron flow. There is no net oxidation or reduction in the cyclic pathway: the energy of the photon absorbed by PSI is used to generate a pH gradient via the cytochrome b6f complex, and this is eventually coupled to ATP synthesis. The alternate use of PSI excitation therefore provides a means for adjusting the proportion of reducing equivalents (NADPH and reduced ferredoxin) and ATP produced by the light reactions.
For illuminating the mechanism of the enzyme, for understanding its participation in communicating redox status to signal transduction pathways, and for deducing the details of cyclic electron flow, a high-resolution view of the cytochrome b6f complex is a necessity. Two groups have independently provided that view by solving the structure of cyanobacterial and chloroplast complexes (Kurisu et al., 2003; Stroebel et al., 2003). Despite the relative compositional simplicity of the enzyme compared with the photosystems, the cytochrome b6f complex is the last membrane complex in the Z-scheme to be analyzed at this level of resolution (3 Å), and this may be because of its lability. One group resorted to purifying the enzyme from a thermophilic organism to improve stability and noted the importance of lipid add-back during purification for support of the central cavity between monomers (Kurisu et al., 2003), whereas the other group engineered a his6 tag into one of the subunits to facilitate rapid purification (Stroebel et al., 2003).
Composition
The enzyme occurs as a functional dimer (Figure 5). There are four large subunits, cytochrome b6, cytochrome f, the Rieske protein, and Subunit IV that bind the redox cofactors and four smaller ones, PetG, PetL, PetM, and PetN. In each monomer are two b hemes in cytochrome b6, one c heme in the extramembrane lumen domain of cytochrome f, one Fe2S2 center in the lumen side extramembrane domain of the Rieske protein, plus a quinone, chlorophyll a, a 9-cis β-carotene and—big surprise—an additional heme bound to cytochrome b6. The new heme is covalently attached through a single thioether linkage. The existence of an extra heme was unexpected, but the fact that it is covalently attached was satisfying because biochemical analysis had suggested the presence of covalent heme in cytochrome b6, and biogenesis studies had indicated that heme association depended on a multistep sequential pathway (Kuras et al., 1997). A quinone analog called stigmatellin is known from biochemical work to interact at or near the Qo site, and its presence in the complex with inhibitor added therefore distinguishes the Qo pocket. The plastoquinone bound on the stromal side identifies the Qi site. The Rieske protein contributes to stability of the dimer because the transmembrane segment of the polypeptide is associated with subunits of one monomer as it obliquely traverses the membrane, but its extramembrane segment crosses over and associates with subunits of the other monomer. There are some differences in the two cytochrome b6f structures with respect to the position of the Rieske subunit, the mode of inhibitor binding to the Qi site, and the assignment of three small subunits. The position of the Rieske subunit could well reflect a conformational difference because the protein almost certainly moves during the catalytic cycle. The differences in the small subunits may result from ambiguity in assignment or from a genuine physiological difference between a cyanobacterial enzyme that functions in both respiration and photosynthesis versus an organelle enzyme that is dedicated to photosynthesis. Nevertheless, the two structures are virtually identical with respect to cofactor binding sites despite a billion years of evolutionary divergence, indicating that the underlying chemical mechanism of proton and electron transfer is absolutely conserved.
Figure 5.
Cofactors in the b6f Dimer.
The coordinates from the cyanobacterial structure were used to generate the figure (Kurisu et al., 2003). The complex is viewed in dimeric form in the plane of the membrane with the lumen side on the bottom as in Figure 2. Chlorophyll is shown in green, b hemes of cytochrome b6 and the c-heme of cytochrome f in red, the novel heme in brown, and Fe and S as red and yellow balls, respectively. The Qo site inhibitor, tridecyl stigmatellin, is shown in turquoise and a plastoquinone at the Qi site in blue. One of the Rieske subunits is shaded as a dark gray to show the association of its cofactor-containing domain with the other monomer. The stromal side of the membrane is called the n-side in one work or the inside (i) in another, and the lumen side is called p-side or outside (o). Allen (2004) has discussed the variety in nomenclature of cofactors and presentation of the complex.
Path of Electrons
The position of electron transfer cofactors in the cytochrome b6f complex is compatible with the Q-cycle mechanism, which was devised to account for the ratio of protons pumped per quinone oxidized (discussed in Allen, 2004; Smith et al., 2004). The substrate plastoquinol is bound at the Qo site, where its oxidation and proton release into the lumen takes place. One electron from the quinol is transferred energetically downhill to the Rieske iron-sulfur center from where it is transferred to the c-heme of cytochrome f and then eventually to plastocyanin or cytochrome c6. The extramembrane lumen-side domain of the Rieske protein possibly moves from a position close to the Qo site when the iron-sulfur center is oxidized (to favor electron transfer from plastoquinol) to a position closer to cytochrome f when the iron-sulfur center is reduced (to favor reduction of cytochrome f). The movement of the Rieske protein is necessary to optimize the cofactor distances for each electron transfer event and is proposed by analogy to the equivalent event in the cytochrome bc1 complex of respiration where the different positions of the Rieske protein have been trapped in the crystal form (Smith et al., 2004). The cytochrome b6f afficionados are not so fortunate as to have a view of both conformations, but there are biochemical experiments that substantiate movement (e.g., Roberts et al., 2004, and references therein). The other electron from the quinol at the Qo site is transferred via the two b-hemes to a stromal side quinone at the Qi site where proton uptake from the stroma and reduction takes place (Figure 1). It is evident from examination of the structure that the newly discovered heme is in the path of direct electron transfer from the stromal side bH heme to the Qi site. One suggestion is that this heme actually shares the electron with bH (Stroebel et al., 2003). Now that there is firm structural basis for suggested electron transfer paths, the gauntlet is thrown to the spectroscopists to monitor the location of the Q-cycle electrons during operation of the cycle and to deduce whether there is quinone exchange between the two sites.
Cofactors without Assigned Function
The function of the chlorophyll and carotenoid in the complex is not known, and the structure does not immediately suggest one. They are not likely to be involved in quinol to plastocyanin/cytochrome c6 oxidoreductase activity—they seem to be located away from the action, and they are not present in the functionally analogous bc1 complex—and therefore communication with other complexes, perhaps for regulation, is one obvious suggestion. Both cofactors are exposed and, hence, accessible to other enzymes. Participation in alternative redox reactions outside the cytochrome b6f complex is another possibility, especially given the new evidence for carotenoids as electron transfer cofactors in PSII (discussed above). Although carotenoids have classically been viewed as photoprotective agents to guard against the formation of 3Chl, the one in the cytochrome b6f complex is considered to be too far away (14 Å) for that function, but nature has a way of surprising us, so the jury is still out on this point.
The position of the new heme, on the other hand, does suggest direct participation in redox chemistry with the Qi site, as mentioned above. A more popular suggestion, compatible with its stromal side location and proximity to the Qi site, is that the new heme may be an intermediate for quinone reduction to quinol in cyclic electron transfer via ferredoxin, but there are some arguments against it, including evidence arguing in favor of cyclic electron transfer routes that reduce PQ without involvement of the cytochrome b6f complex (discussed in Kramer et al., 2004). The unfilled axial position of the new heme brings up the question of whether there might be a small molecule ligand in vivo whose binding may influence redox activity (Zhang et al., 2004). The bc complexes of some respiring bacteria (in which a cyclic electron transfer pathway does not occur) are highly related to the cytochrome b6f complexes of cyanobacteria and chloroplasts, including the presence of covalent heme (Yu and Le Brun, 1998). The elucidation of the function of the covalent heme then would open the door for the discovery of alternate quinone-reducing metabolic pathways unique to respiring bacteria with the cytochrome b6f-type complexes. Because many of these are pathogenic, there is an obvious practical interest in the resolution of this fundamental problem.
The presence of the new heme, a chlorophyll, and a carotenoid generates some problems with respect to the biogenesis of the complex, not least of which is the molecular identity of the CCB loci responsible for the step-wise, sequential assembly of the hemes into cytochrome b6 (Kuras et al., 1997). Given the presence of covalent heme in the b subunit of the respiratory b-c complex in Bacillus subtilis and the conservation in the cytochrome c maturation pathway between the Bacillus sp and chloroplasts, one would predict conservation of the CCB mechanism as well (Kranz et al., 1998; Yu and Le Brun, 1998). A bioinformatics approach could be a productive route to their discovery.
The cytochrome b6f complex had been perceived as the “me too” enzyme relative to the bc1 complexes in bacteria and mitochondria, but with the revelation of its structure, it now stands as the more richly endowed and will almost certainly be a subject of continuing interest with respect to electron transfer mechanisms and biogenesis and a source of regulatory signals from the perspective of signal transduction.
PSI-LHCI
PSI or plastocyanin/cytochrome c6 to ferredoxin oxidoreductase is the third enzyme in the pathway. In plants, plastocyanin is the exclusive donor side substrate of the enzyme, and reduced ferredoxin the exclusive product. Cyanobacterial PSI shows great flexibility in response to micronutrient availability, with the possibility of using an alternate donor, cytochrome c6, an alternate acceptor, flavodoxin, and even an alternate antenna system, CP43′ or IsiA (Zhang et al., 1992; La Roche et al., 1996; Bibby et al., 2001). The occurrence of a novel cytochrome in the Arabidopsis genome had led to the suggestion that the flexibility on the donor side extended also to vascular plants (Gupta et al., 2002), but the new Arabidopsis cytochrome has a homolog in Chlamydomonas (C_820065; Cyc4) that is distinct from the well-characterized genuine cytochrome c6 of Chlamydomonas (Merchant, 1998), which leaves plastocyanin as the only donor to PSI in vascular plants. The product of PSI, reduced ferredoxin, is a key reductant in the chloroplast: it reduces NADP+ via ferredoxin-NADP+ oxidoreductase for various reductive biosynthetic pathways and thioredoxin via ferredoxin-thioredoxin reductase for redox regulation, it can directly provide electrons for certain enzymes like acyl-ACP desaturase or Glu synthase, and as mentioned above, it can supply electrons to the cytochrome b6f complex in a cyclic electron flow pathway around PSI for the generation of a proton gradient and hence ATP synthesis (Buchanan and Balmer, 2005; Hase et al., 2005).
Details of PSI chemistry were revealed a few years ago with the publication of a high-resolution (2.5 Å) structure of the trimeric cyanobacterial enzyme (Jordan et al., 2001). The structure of the monomer showed 12 Psa subunits (named as gene products PsaA, PsaB, PsaC, etc.) and 127 cofactors, including 96 chlorophylls with orientation of the macrocycle assigned (and hence the Qy transition deduced), two phylloquinones, three Fe4S4 centers, 22 carotenoids, and four lipids. The positions of the lipids in the core and the role of one of them as a ligand to an antenna chlorophyll led to their inclusion in the list of cofactors rather than as nonspecifically bound lipid from the membrane or from the purification procedure. Like other reaction centers, the cofactors in the electron transfer chain are arranged in symmetrical pairs bound by a heterodimeric core made up of the homologous polypeptides PsaA and PsaB. FeSX is liganded between the heterodimeric core, and FeSA and FeSB are bound to PsaC on the stromal side of the membrane. Ferredoxin associates with PSI on the stromal side via interaction with PsaC, PsaD, and PsaE, and plastocyanin and cytochrome c6 associate with PSI on the lumen side via interaction with PsaA, PsaB, and PsaF. Most of the pigment molecules in PSI, 90 chlorophyll a molecules and 22 carotenoids, function in the core antenna system. The chlorophyll a are closely arranged in two layers on the stromal and lumenal sides to support fast transfer of excitation energy. The carotenoids are in van der Waals contact with chlorophyll a head groups, which is consistent with a function either in light harvesting (energy transfer from excited carotenoid to chlorophyll) or in photoprotection (quenching of 3Chl).
The PSI reaction is initiated by excitation of the primary donor, P700, via energy transfer from the core antenna (see Figure 1). The electron is transferred sequentially to the primary acceptor ChlA0 (a pair of unusually [Met S] coordinated chlorophylls in the electron transfer chain), then PhQA1 (one of two phylloquinones), and then FeSx, FeSA, and FeSB, from where it leaves PSI to reduce ferredoxin. In contrast with PSII (and every other known reaction center), the pair of chlorophylls that make up P700 are structurally distinct. One, coordinated to PsaB, is chlorophyll a, and the other, coordinated to PsaA, is chlorophyll a′ (the C132 epimer of chlorophyll a; Figure 4). Another difference is that the two symmetrical branches of the electron transfer chain are both active, albeit not equally so, and the underlying reasons are a subject of continued investigation (Guergova-Kuras et al., 2001).
Recently, Nelson and coworkers solved the structure of a PSI-LHCI supercomplex purified from pea (Ben-Shem et al., 2003). Although the resolution is not high (4.4 Å), it is adequate for presenting the organization of the accessory LHCI antenna around PSI and for indicating a structural basis for a monomer arrangement of chloroplast PSI versus a trimer arrangement in cyanobacteria. The structure also provides us with a first view of LHCI structure and how connections between accessory antennae and reaction centers are made. The authors use a structural comparison between the chloroplast and cyanobacterial enzymes to generate some provocative ideas concerning the evolution of PSI antenna interactions that are not inconsistent with its well-documented flexibility in responding to physiological status and environmental input (Ben-Shem et al., 2004). They present also a creative rationale for the heterodimeric pattern observed in the more evolved reaction centers.
Organization
Unlike cyanobacterial PSI, the chloroplast enzyme is a monomer (Figure 6). The structure of the complex validates the overall model proposed on the basis of biochemical and reverse genetic analysis of subunit function and by image reconstruction of negatively stained PSI-LHCI particles viewed by electron microscopy (Scheller et al., 2001; Germano et al., 2002; Kargul et al., 2003). Four different Lhca subunits are arranged in a semicircle between the PsaG and PsaK subunits of PSI around the side of where PsaF and PsaJ are located. There is a large cleft between the LHCI complex and PSI. The four subunits are arranged as two dimers with even spacing between the dimers and between dimer subunits. The general fold of LHCI monomers is similar to that of LHCII monomers (discussed above), but the Lhca monomers are arranged in a head-to-tail fashion, which is different from the C3-symmetrical trimer of LHCII. Between the subunits are found additional linkerchlorophyll that are thought to connect pigments of different subunits.
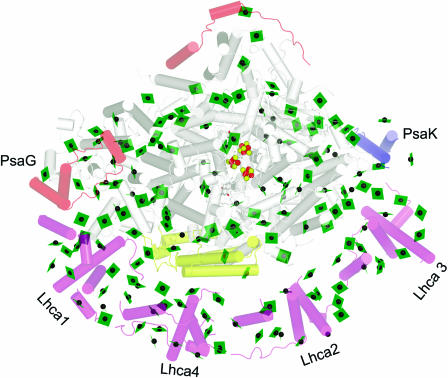
Figure 6.
Association of LHCI with PSI.
The complex is viewed along the membrane normal from the stromal side. The chlorophylls are shown as green planes with the central magnesium as a green sphere. Fe is shown as a red sphere and sulfur as a yellow sphere. The helices are shown as cylinders. The subunits of PSI are colored gray, with the exception that polypeptides novel to the plant PSI are colored pale red (top left of image) and PsaK is shown in blue and PsaF in yellow (see Scheller et al., 2001 for a review of PSI polypeptides). The Lhca polypeptides are colored pale magenta (bottom of image). PyMOL was used to generate the image from coordinates provided by Adam Ben-Shem and Nathan Nelson (Ben-Shem et al., 2003).
The resolution at 4.4 Å is not adequate to identify side chains, and the assignment of individual Lhca subunits is therefore based entirely on prior substantial biochemical analyses (supplement to Ben-Shem et al., 2003). For PSI, the helices of subunits in common with the cyanobacterial enzyme were assigned by analogy (Jordan et al., 2001). Of the four subunits (PsaG, PsaH, PsaN, and PsaO) that are present exclusively in plants, two (PsaG and PsaH) could be assigned based on known biochemical properties (Figure 6). PsaG is related in sequence to PsaK and is thought to have evolved by gene duplication: the distinction between them was therefore made on the basis of the known different pigment binding properties of the two. PsaG was located to the side of the complex where LHCI makes a tighter connection with PSI through close protein–protein interaction, whereas PsaK was located on the other side where looser connection between Lhca3 and PSI is evident (discussed below).
Relative to the cyanobacterial enzyme, the PsaF subunit from chloroplasts has an N-terminal extension, which is known to be important for effective interactions with the substrate electron donor—plastocyanin or cytochrome c6 (Hippler et al., 1999). From the lumen side (not evident in the view shown in Figure 6), this N-terminal region is clearly visible as a helix-loop-helix motif that presents an interaction surface on that face of the molecule. One wonders whether this new interaction generates a constraint that restricts donor flexibility and, hence, evolutionary loss of cytochrome c6 in vascular plants.
Energy Transfer within and from the Green Belt
A major contribution of this recent structure comes from the analysis of LHCI, which provides a structural basis for interactions between Lhca proteins and between the LHCI belt and PSI. The Lhca monomers interact with each other through the N and C termini of the proteins and the extramembrane loops, as suggested from previous mutagenesis experiments, rather than through the transmembrane domains. The interpigment distances in LHCI (important for energy transfer) are still small because of the presence of linker chlorophylls at the interface of Lhca monomers within a dimer and also between dimers.
There is a gap of at least 20 Å between the LHCI belt and PSI, which means that their respective pigments are generally not very close to each other. Nevertheless, there are three regions where the pigments are closer—near PsaG, PsaK, and PsaF—owing to addition of new pigment binding sites during evolution (Figure 6). These pigments, called gap chlorophylls, are on the periphery of the LHCI or PSI and occupy the cleft. Antisense experiments in Arabidopsis had correctly indicated the importance of the PsaG, PsaK, and PsaF subunits for physical and functional association of accessory antenna with PSI (Scheller et al., 2001). Ben-Shem et al. (2003) propose that the three regions might form the paths for energy migration from the accessory antenna to PSI and suggest that the existence of distinct energy migration routes opens up the possibility that energy supply into PSI can be easily modified in response to signal transduction from the environment or from internal metabolic state (Ben-Shem et al., 2003).
Conservation of Mechanism but Flexibility of Antenna Interactions
As noted already for the cytochrom b6f complex, the placements of the electron transfer cofactors in the chloroplast and cyanobacterial enzyme are absolutely conserved. Of the light-harvesting chlorophylls in the core antenna, 92 of the 96 are in the same position, reflecting an amazing degree of conservation, even though most of the chlorophylls do not transfer directly to P700. There are 10 new chlorophylls added to PSI that enable functional interactions (i.e., energy transfer) between LHCI and PSI, and the authors point out that this represents a minimal modification to the reaction center with a big payoff in the sense of increased photochemical activity at lower light intensity. Another important modification of the chloroplast enzyme is its occurrence as a monomer. The structure shows that the conversion from trimer to monomer is a consequence of the incorporation of PsaH, the subunit that is required for binding LHCII to PSI in State II (see discussion of LHCII). The presence of PsaH occludes contacts between monomers. The conversion from trimer to monomer therefore opened up the possibility for regulating energy distribution between PSII and PSI.
An interesting aspect of the green belt is that the interaction is asymmetric, being stronger on the side near PsaG (where an interaction between helices of PsaG with helix C of Lhca1 is evident) relative to the side near PsaK. This is exciting because it suggests a mechanism for how LHCI composition is modified in response to changing growth conditions. Specifically, subunits can be added sequentially onto the PsaG side or removed from the PsaK side depending on demand for light-harvesting capability, and this is facilitated by the relatively weak interactions between individual Lhca subunits. In Chlamydomonas, it was noted that PsaK is lost from PSI coincidently with loss of Lhca3 and dissociation of LHCI-PSI at the onset of Fe deficiency (Moseley et al., 2002). The rationale for this modification is that reduced light harvesting is thought to serve a photoprotective function in the situation where the iron-rich electron acceptors in PSI centers (FeSX, FeSA, and FeSB) may not be fully functional. Although a causal connection between the two events (loss of PsaK and reduced energy transfer from LHCI) was not established in that work, the model is consistent with the structural organization of PSI-LHCI (Ben-Shem et al., 2003).
Structural analysis of PSI underscores the flexibility of both the cyanobacterial and chloroplast enzyme with respect to antenna association. When one appreciates the position of PSI at the branch point between NADPH production (via linear electron flow from water to NADP+) and ATP production (via cyclic electron flow), the layers of regulation seem self-evident. It is likely that the focus of ongoing research will be directed toward the discovery of subtle compositional and structural modifications that modify input of light into the enzyme. In this context, it is worth noting that from the perspective of regulation, the structures of the various complexes represent “a” structure rather than “the” structure, and it is the fine tuning of metabolism that makes biology so interesting and the various photosynthetic organisms so ecologically successful.
SUMMARY
The structures reveal an amazing degree of conservation in the photosynthetic apparatus. The similarity of the cyanobacterial and chloroplast enzymes was perhaps self-evident, but Ferreira et al. (2004) point out that the root mean square deviation between PSII and the anoxygenic photosynthesis reaction center from Rhodopseudomonas viridis is only 1.9 Å for 395 Cα atoms, indicating strong conservation between evolutionarily distant enzymes. The relevance of nonplant photosynthetic mechanisms to understanding plant metabolism is clear. The level of detail provided in these structures provides a solid framework for proposing and dissecting mechanisms. This is especially true for LHCII, where the physical chemists can now calculate energy transfer rates and the structural biologists can model the structures of other members of the family (Jansson, 1999). On the other hand, there are distinct differences with respect to the regulatory aspects of the pathway, and this is best demonstrated via the comparison of the PSI structure from cyanobacteria and peas. There are differences in composition, in interactions with antenna proteins or with other electron transfer partners, and in the supramolecular organization in the membrane. There may be structural modifications even within one organism, depending on physiological state or environmental cues, and in that sense, the structures are a starting point for asking new questions as opposed to the end point of investigation. Even from the mechanistic point of view, the structures suggest brand new questions, as evidenced from the discovery of a new cofactor in cytochrome b6f and the placement of chlorophyll and carotenoid. Plant biologists of all flavors—from biophysicists and biochemists to molecular biologists and geneticists—will be kept busy as they unravel the secrets of the most important energy generating machinery on Earth.
Acknowledgments
The authors are grateful to Robert Blankenship, Richard Debus, David Kramer, Laurens Mets, and Kevin Redding for patient instruction in photosynthesis plus many helpful discussions for this particular project, to Gary Brudvig and Krishna Niyogi for comments on the manuscript, and to Adam Ben-Shem and Nathan Nelson for providing the coordinates used to create Figure 6. S.M. acknowledges support from the USDA, the National Institutes of Health, and the Department of Energy for her work on photosynthesis. M.R.S. is supported by the Howard Hughes Medical Institute.
References
- Allen, J.F. (2004). Cytochrome b6f: Structure for signalling and vectorial metabolism. Trends Plant Sci. 9, 130–137. [DOI] [PubMed] [Google Scholar]
- Allen, J.P., Feher, G., Yeates, T.O., Rees, D.C., Deisenhofer, J., Michel, H., and Huber, R. (1986). Structural homology of reaction centers from Rhodopseudomonas sphaeroides and Rhodopseudomonas viridis as determined by x-ray diffraction. Proc. Natl. Acad. Sci. USA 83, 8589–8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González, E., and Aro, E.M. (2002). Biogenesis, assembly and turnover of photosystem II units. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 1451–1459; discussion 1459–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, J., Ferreira, K.N., Maghlaoui, K., and Iwata, S. (2004). Structural model of the oxygen-evolving centre of photosystem II with mechanistic implications. Phys. Chem. Chem. Phys. 6, 4737–4742. [Google Scholar]
- Ben-Shem, A., Frolow, F., and Nelson, N. (2003). Crystal structure of plant photosystem I. Nature 426, 630–635. [DOI] [PubMed] [Google Scholar]
- Ben-Shem, A., Frolow, F., and Nelson, N. (2004). Evolution of photosystem I—From symmetry through pseudo-symmetry to asymmetry. FEBS Lett. 564, 274–280. [DOI] [PubMed] [Google Scholar]
- Bibby, T.S., Nield, J., and Barber, J. (2001). Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743–745. [DOI] [PubMed] [Google Scholar]
- Biesiadka, J., Loll, B., Kern, J., Irrgang, K.-D., and Zouni, A. (2004). Crystal structure of cyanobacterial photosystem II at 3.2 Å resolution: A closer look at the Mn-cluster. Phys. Chem. Chem. Phys. 6, 4733–4736. [Google Scholar]
- Blankenship, R.E. (2002). Molecular Mechanisms of Photosynthesis. (Oxford: Blackwell Science).
- Buchanan, B.B., and Balmer, Y. (2005). Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 56, in press. [DOI] [PubMed]
- Caffarri, S., Croce, R., Cattivelli, L., and Bassi, R. (2004). A look within LHCII: Differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry 43, 9467–9476. [DOI] [PubMed] [Google Scholar]
- Deisenhofer, J., Epp, O., Miki, K., Huber, R., and Michel, H. (1984). X-ray structure analysis of a membrane protein complex. Electron density map at 3 Å resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J. Mol. Biol. 180, 385–398. [DOI] [PubMed] [Google Scholar]
- Depège, N., Bellafiore, S., and Rochaix, J.D. (2003). Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575. [DOI] [PubMed] [Google Scholar]
- Diner, B.A., and Rappaport, F. (2002). Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Annu. Rev. Plant Biol. 53, 551–580. [DOI] [PubMed] [Google Scholar]
- Elrad, D., Niyogi, K.K., and Grossman, A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, K.N., Iverson, T.M., Maghlaoui, K., Barber, J., and Iwata, S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838. [DOI] [PubMed] [Google Scholar]
- Finazzi, G. (2004). The central role of the green alga Chlamydomonas reinhardtii in revealing the mechanism of state transitions. J. Exp. Bot. 56, 383–388. [DOI] [PubMed] [Google Scholar]
- Frank, H.A., and Brudvig, G.W. (2004). Redox functions of carotenoids in photosynthesis. Biochemistry 43, 8607–8615. [DOI] [PubMed] [Google Scholar]
- Germano, M., Yakushevska, A.E., Keegstra, W., van Gorkom, H.J., Dekker, J.P., and Boekema, E.J. (2002). Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525, 121–125. [DOI] [PubMed] [Google Scholar]
- Green, B.R., and Parson, W.W. (2003). Light Harvesting Antennas in Photosynthesis. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Guergova-Kuras, M., Boudreaux, B., Joliot, A., Joliot, P., and Redding, K. (2001). Evidence for two active branches for electron transfer in photosystem I. Proc. Natl. Acad. Sci. USA 98, 4437–4442. First published on March 27, 2001; 10.1073/pnas.081078898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., He, Z., and Luan, S. (2002). Functional relationship of cytochrome c6 and plastocyanin in Arabidopsis. Nature 417, 567–571. [DOI] [PubMed] [Google Scholar]
- Hanley, J., Deligiannakis, Y., Pascal, A., Faller, P., and Rutherford, A.W. (1999). Carotenoid oxidation in photosystem II. Biochemistry 38, 8189–8195. [DOI] [PubMed] [Google Scholar]
- Hase, T., Schürmann, P., and Knaff, D.B. (2005). The interaction of ferredoxin with ferredoxin-dependent enzymes. In Photosystem I: The Plastocyanin:Ferredoxin Oxidoreductase in Photosynthesis, J.H. Golbeck, ed (Dordrecht, The Netherlands: Springer), in press.
- Hippler, M., Drepper, F., Rochaix, J.D., and Mühlenhoff, U. (1999). Insertion of the N-terminal part of PsaF from Chlamydomonas reinhardtii into photosystem I from Synechococcus elongatus enables efficient binding of algal plastocyanin and cytochrome c6. J. Biol. Chem. 274, 4180–4188. [DOI] [PubMed] [Google Scholar]
- Holt, N.E., Fleming, G.R., and Niyogi, K.K. (2004). Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43, 8281–8289. [DOI] [PubMed] [Google Scholar]
- Holt, N.E., Zigmantas, D., Valkunas, L., Li, X.P., Niyogi, K.K., and Fleming, G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307, 433–436. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Walters, R.G. (1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 655–684. [DOI] [PubMed] [Google Scholar]
- Iwata, S., and Barber, J. (2004). Structure of photosystem II and molecular architecture of the oxygen-evolving centre. Curr. Opin. Struct. Biol. 14, 447–453. [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4, 236–240. [DOI] [PubMed] [Google Scholar]
- Jordan, P., Fromme, P., Witt, H.T., Klukas, O., Saenger, W., and Krauss, N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909–917. [DOI] [PubMed] [Google Scholar]
- Kamiya, N., and Shen, J.R. (2003). Crystal structure of oxygen-evolving photosystem II from Thermosynechococcus vulcanus at 3.7-Å resolution. Proc. Natl. Acad. Sci. USA 100, 98–103. First published on December 23, 2002; 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargul, J., Nield, J., and Barber, J. (2003). Three-dimensional reconstruction of a light-harvesting complex I-photosystem I (LHCI-PSI) supercomplex from the green alga Chlamydomonas reinhardtii. Insights into light harvesting for PSI. J. Biol. Chem. 278, 16135–16141. [DOI] [PubMed] [Google Scholar]
- Kramer, D.M., Avenson, T.J., and Edwards, G.E. (2004). Dynamic flexibility in the light reactions of photosynthesis governed by both electron and proton transfer reactions. Trends Plant Sci. 9, 349–357. [DOI] [PubMed] [Google Scholar]
- Kranz, R., Lill, R., Goldman, B., Bonnard, G., and Merchant, S. (1998). Molecular mechanisms of cytochrome c biogenesis: Three distinct systems. Mol. Microbiol. 29, 383–396. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt, W., Wang, D.N., and Fujiyoshi, Y. (1994). Atomic model of plant light-harvesting complex by electron crystallography. Nature 367, 614–621. [DOI] [PubMed] [Google Scholar]
- Kuras, R., de Vitry, C., Choquet, Y., Girard-Bascou, J., Culler, D., Buschlen, S., Merchant, S., and Wollman, F.A. (1997). Molecular genetic identification of a pathway for heme binding to cytochrome b6. J. Biol. Chem. 272, 32427–32435. [DOI] [PubMed] [Google Scholar]
- Kurisu, G., Zhang, H., Smith, J.L., and Cramer, W.A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 302, 1009–1014. First published on October 2, 2003; 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- La Roche, J., Boyd, P.W., McKay, R.M.L., and Geider, R.J. (1996). Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382, 802–805. [Google Scholar]
- Li, X.P., Björkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., An, X., and Chang, W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292. [DOI] [PubMed] [Google Scholar]
- McEvoy, J.P., and Brudvig, G.W. (2004). Structure-based mechanism of photosynthetic water oxidation. Phys. Chem. Chem. Phys. 6, 4754–4763. [Google Scholar]
- Merchant, S. (1998). Synthesis of metalloproteins involved in photosynthesis: Plastocyanin and cytochromes. In The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant, eds (Dordrecht: The Netherlands: Kluwer Academic Publishers), pp. 597–611.
- Moseley, J.L., Allinger, T., Herzog, S., Hoerth, P., Wehinger, E., Merchant, S., and Hippler, M. (2002). Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21, 6709–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, P., Li, X.P., and Niyogi, K.K. (2001). Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, N., and Ben-Shem, A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5, 971–982. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. [DOI] [PubMed] [Google Scholar]
- Peter, G.F., and Thornber, J.P. (1991). Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266, 16745–16754. [PubMed] [Google Scholar]
- Plumley, F.G., and Schmidt, G.W. (1987). Reconstitution of chlorophyll a/b light-harvesting complexes: Xanthophyll-dependent assembly and energy transfer. Proc. Natl. Acad. Sci. USA 84, 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli, R.B., and McSweeney, S.M. (2000). The ‘fingerprint’ that X-rays can leave on structures. Struct. Fold. Des. 8, 315–328. [DOI] [PubMed] [Google Scholar]
- Redding, K., and van der Est, A. (2005). The directionality of electron transfer in photosystem I. In Photosystem I: The Plastocyanin: Ferredoxin Oxidoreductase in Photosynthesis, J.H. Golbeck, ed (Dordrecht, The Netherlands: Springer), in press.
- Redinbo, M.R., Yeates, T.O., and Merchant, S. (1994). Plastocyanin: Structural and functional analysis. J. Bioenerg. Biomembr. 26, 49–66. [DOI] [PubMed] [Google Scholar]
- Roberts, A.G., Bowman, M.K., and Kramer, D.M. (2004). The inhibitor DBMIB provides insight into the functional architecture of the Qo site in the cytochrome b6f complex. Biochemistry 43, 7707–7716. [DOI] [PubMed] [Google Scholar]
- Rogl, H., and Kühlbrandt, W. (1999). Mutant trimers of light-harvesting complex II exhibit altered pigment content and spectroscopic features. Biochemistry 38, 16214–16222. [DOI] [PubMed] [Google Scholar]
- Rogl, H., Schödel, R., Lokstein, H., Kühlbrandt, W., and Schubert, A. (2002). Assignment of spectral substructures to pigment-binding sites in higher plant light-harvesting complex LHC-II. Biochemistry 41, 2281–2287. [DOI] [PubMed] [Google Scholar]
- Rutherford, A.W., and Boussac, A. (2004). Biochemistry. Water photolysis in biology. Science 303, 1782–1784. [DOI] [PubMed] [Google Scholar]
- Scheller, H.V., Jensen, P.E., Haldrup, A., Lunde, C., and Knoetzel, J. (2001). Role of subunits in eukaryotic photosystem I. Biochim. Biophys. Acta 1507, 41–60. [DOI] [PubMed] [Google Scholar]
- Smith, J.L., Zhang, H., Yan, J., Kurisu, G., and Cramer, W.A. (2004). Cytochrome bc complexes: A common core of structure and function surrounded by diversity in the outlying provinces. Curr. Opin. Struct. Biol. 14, 432–439. [DOI] [PubMed] [Google Scholar]
- Standfuss, J., and Kühlbrandt, W. (2004). The three isoforms of the light-harvesting complex II: Spectroscopic features, trimer formation, and functional roles. J. Biol. Chem. 279, 36884–36891. [DOI] [PubMed] [Google Scholar]
- Stroebel, D., Choquet, Y., Popot, J.L., and Picot, D. (2003). An atypical haem in the cytochrome b6f complex. Nature 426, 413–418. [DOI] [PubMed] [Google Scholar]
- Thornton, L.E., Ohkawa, H., Roose, J.L., Kashino, Y., Keren, N., and Pakrasi, H.B. (2004). Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem II activity in the cyanobacterium Synechocystis 6803. Plant Cell 16, 2164–2175. First published on July 16, 2004; 10.1105/tpc.104.023515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrettos, J.S., Limburg, J., and Brudvig, G.W. (2001). Mechanism of photosynthetic water oxidation: Combining biophysical studies of photosystem II with inorganic model chemistry. Biochim. Biophys. Acta 1503, 229–245. [DOI] [PubMed] [Google Scholar]
- Wollman, F.A. (2001). State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 20, 3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., and Le Brun, N.E. (1998). Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J. Biol. Chem. 273, 8860–8866. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Primak, A., Cape, J., Bowman, M.K., Kramer, D.M., and Cramer, W.A. (2004). Characterization of the high-spin heme x in the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry 43, 16329–16336. [DOI] [PubMed] [Google Scholar]
- Zhang, L., McSpadden, B., Pakrasi, H.B., and Whitmarsh, J. (1992). Copper-mediated regulation of cytochrome c553 and plastocyanin in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 267, 19054–19059. [PubMed] [Google Scholar]
- Zouni, A., Witt, H.T., Kern, J., Fromme, P., Krauss, N., Saenger, W., and Orth, P. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 409, 739–743. [DOI] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- The reader is referred also to a soon-to-be published manuscript from Kühlbrandt and coworkers on a 2.5-Å resolution x-ray structure of a pea LHCII complex in stacked two-dimensional crystals (Standfuss et al., 2005). Of interest are a very clear diagram of the trimer with the orientations of chlorophylls and carotenoids indicated, a discussion of interactions relevant to the formation of appressed membranes (grana lamellae), an explanation of the specificity for PG containing a characteristic 16:1 trans-Δ3-hexadecenoic acid, and a proposed mechanism for xanthophyll-dependent NPQ in LHCII different from that proposed by Liu et al. (2004).
- Standfuss, J., Terwisscha van Scheltinga, A.C., Lamborghini, M., and Kühlbrandt, W. (2005). Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Å resolution. EMBO J., in press. [DOI] [PMC free article] [PubMed]