Abstract
Plastid DNA fragments are often found in the plant nuclear genome, and DNA transfer from plastids to the nucleus is ongoing. However, successful gene transfer is rare. What happens to compensate for this? To address this question, we analyzed nuclear-localized plastid DNA (nupDNA) fragments throughout the rice (Oryza sativa ssp japonica) genome, with respect to their age, size, structure, and integration sites on chromosomes. The divergence of nupDNA sequences from the sequence of the present plastid genome strongly suggests that plastid DNA has been transferred repeatedly to the nucleus in rice. Age distribution profiles of the nupDNA population, together with the size and structural characteristics of each fragment, revealed that once plastid DNAs are integrated into the nuclear genome, they are rapidly fragmented and vigorously shuffled, and surprisingly, 80% of them are eliminated from the nuclear genome within a million years. Large nupDNA fragments preferentially localize to the pericentromeric region of the chromosomes, where integration and elimination frequencies are markedly higher. These data indicate that the plant nuclear genome is in equilibrium between frequent integration and rapid elimination of the chloroplast genome and that the pericentromeric regions play a significant role in facilitating the chloroplast–nuclear DNA flux.
INTRODUCTION
During endosymbiotic evolution, eukaryotic nuclear genomes have acquired numerous genes from the endosymbiotic organelles, which later evolved into the present chloroplasts and mitochondria (Martin and Herrmann, 1998; Kurland and Andersson, 2000; Martin et al., 2002). Although most of this gene transfer occurred at an early stage of organelle evolution (Martin et al., 1998), functional gene transfer continues to occur in flowering plants (Adams et al., 1999, 2002; Millen et al., 2001). Fragments of the chloroplast and mitochondrial genomes are often found in the nuclear genome and are referred to as promiscuous DNAs (Ellis, 1982). Promiscuous DNAs occur in many eukaryotes, and their analysis suggests that DNA transfer from the organelles to the nucleus is ongoing (Ayliffe et al., 1998; Bensasson et al., 2001; Woischnik and Moraes, 2002; Yuan et al., 2002).
How are organellar DNAs integrated and processed in the nucleus? Double-strand break (DSB) repair of chromosomal DNA has been implicated as a mechanism by which the organellar DNA fragments are integrated into the nucleus (Ricchetti et al., 1999; Yu and Gabriel, 1999). The transfer rates of organellar DNAs to the nucleus have been measured directly for yeast mitochondria (Thorsness and Fox, 1990) and tobacco (Nicotiana tabacum) chloroplasts (Huang et al., 2003; Stegemann et al., 2003) using selectable marker genes designed to function only when they are transferred to the nucleus. In these instances, the occurrence of gene transfer was detected under experimental conditions, suggesting that natural gene transfer occurs more frequently than previously believed. However, the fate of organellar DNAs after their integration into the nucleus has not been clarified.
The accumulated information on the rice (Oryza sativa ssp japonica) nuclear genome provides an opportunity to undertake a genome-wide analysis of nuclear-localized plastid DNA fragments (nupDNAs). In this study, we estimated when individual nupDNA fragments throughout the rice genome were integrated into the nucleus, on the basis of their nucleotide substitution frequencies compared with the present chloroplast genome. The age distribution profiles of nupDNA fragments, in conjunction with their fragment sizes, structures, integration sites, and abundance, revealed dynamic features of nupDNAs in the rice genome; they are frequently integrated, shuffled, and eliminated. These data indicate that the various nupDNA fragments that we can observe today represent a cross section of the continuous DNA flux from the chloroplast to the nucleus. This study also shows that the pericentromeric regions of chromosomes are hot spots for the turnover of nupDNAs. A possible role for the pericentromeric regions in the evolution of nucleo-chloroplast relations is also discussed.
RESULTS
nupDNA Fragments Are Abundant in the Rice Nuclear Genome
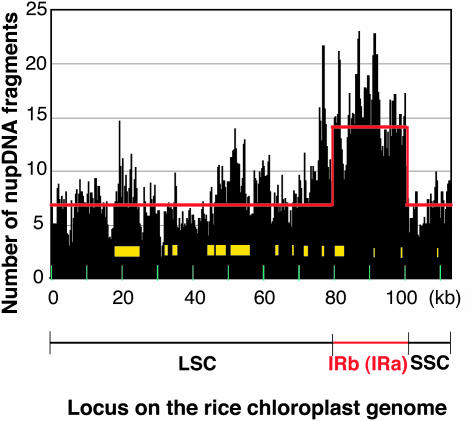
To evaluate the abundance of nupDNAs in rice, we searched for them in the rice nuclear genome database (http://rgp.dna.affrc.go.jp/), which includes >85% of the total genome sequence. Using two homology search programs, BLASTN and Smith-Waterman, we identified >1600 candidate sequences with high homology and E-values of <10−10. Exclusion of redundant candidate sequences revealed that 701 independent BAC/PAC clones contained chloroplast DNA fragments (Table 1; see Supplemental Table 1 online). The combined lengths of these fragments is 0.9 Mb, constituting 0.2% of the total rice nuclear genome. These nupDNA fragments originated from every part of the chloroplast genome at a similar frequency (Figure 1; on average, 6.7 times from single-copy regions), suggesting that transfer and integration into the nuclear genome occurs almost equally from throughout the chloroplast genome. The yellow boxes in Figure 1 indicate the chloroplast DNA segments of which copies are found in the mitochondrial genome. It is interesting that the number of nupDNA fragments tends to be a little higher for those boxed regions.
Table 1.
nupDNA Fragments
| Chromosome (Mb)a | Amount of nupDNA in bp (% Chromosome Length) | Number of nupDNA Insertions | ||
|---|---|---|---|---|
| 1 | (45.7) | 104010 | (0.23) | 87 |
| 2 | (43.4) | 77976 | (0.18) | 56 |
| 3 | (47.5) | 43970 | (0.09) | 71 |
| 4 | (35.6) | 153803 | (0.43) | 66 |
| 5 | (33.6) | 50623 | (0.15) | 55 |
| 6 | (35.1) | 55899 | (0.16) | 62 |
| 7 | (33.1) | 38365 | (0.12) | 53 |
| 8 | (33.6) | 60827 | (0.18) | 55 |
| 9 | (27.0) | 27784 | (0.10) | 47 |
| 10 | (23.6) | 186238 | (0.79) | 47 |
| 11 | (33.7) | 16423 | (0.05) | 47 |
| 12 | (30.9) | 84823 | (0.27) | 55 |
| Total | (422.8) | 900741 | (0.21) | 701 |
The sizes of chromosomes 1, 4, and 10 are according to Sasaki et al. (2002), Feng et al. (2002), and Rice Chromosome 10 Sequencing Consortium (2003), respectively. The sizes of the other chromosomes are according to Saji et al. (2001).
Figure 1.
Frequency of the Appearance of nupDNA Fragments throughout the Rice Chloroplast Genome.
The rice chloroplast genome was divided into 100-bp segments. The numbers of nupDNA fragments corresponding to individual segments are shown by histograms. The rice chloroplast genome is a double-stranded circular DNA molecule of 134.5 kb, which contains two copies of an identical 20.8-kb inverted repeat (IRa and IRb) separated by a large single-copy region (LSC; 80.6 kb) and a small single-copy region (SSC; 12.4 kb) (Sugiura, 1992), as is schematically illustrated in Figure 3. The circular map is linearized at the junction between IRa and the large single-copy region. The yellow boxes indicate the regions whose copies are found in the rice mitochondrial genome. The red line indicates the expected number of nupDNA fragments if they originated from throughout the chloroplast genome with equal frequency.
Structure and Distribution of nupDNA Fragments on Nuclear Chromosomes
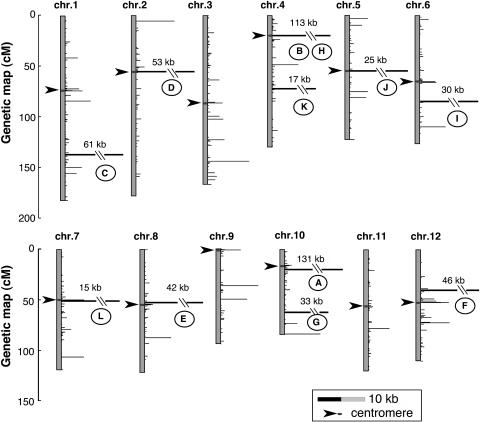
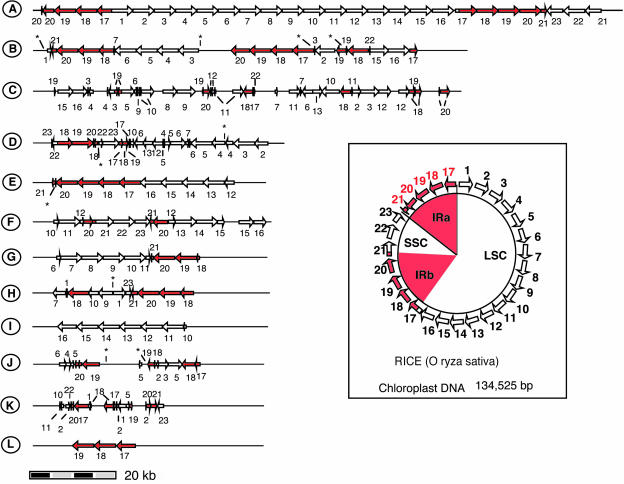
To clarify the distribution of nupDNA fragments on nuclear chromosomes, we located their integration sites on the rice genetic map (Figure 2). nupDNA fragments are scattered throughout the chromosomes. We identified 12 nupDNA fragments larger than 10 kb, which we designated giant fragments A to L in order of decreasing size (Figures 2 and 3). Large nupDNA fragments are frequently found near the centromeres, and of the 12 giant fragments, seven are located within 5 centimorgan (cM) of the centromere. The structural characteristics of the giant fragments are shown in Figure 3. Fragment A is 131 kb long and corresponds to the almost intact chloroplast genome (134.5 kb) linearized at the boundary between the small single-copy and inverted repeat (IR) regions (Figure 3). Eight of the 12 giant nupDNA fragments are intermingled forms, in which the chloroplast DNA sequence is partially deleted, inverted, or duplicated, and sometimes interrupted by nonchloroplast DNA sequences (Figure 3, fragments B, C, D, F, G, H, J, and K). These giant nupDNA fragments retain >99% sequence identity with the present chloroplast genome, indicating that their transfer and integration into the nuclear genome were relatively recent events. These data indicate that, once large chloroplast DNA fragments are integrated into the nuclear genome, they are rapidly fragmented and vigorously shuffled in many cases. In the total nupDNA population, one-third of the fragments occur as intermingled forms, as in the case of giant fragments.
Figure 2.
Locations of nupDNA Fragments on the Rice Genetic Map.
The vertical axis represents the genetic map (cM) of each chromosome, and the horizontal bar shows the sum of the nupDNA fragments (kb) located at each locus. nupDNA fragments larger than 10 kb are denoted A to L in order of decreasing size and are indicated by heavy lines. Fragments B and H are located within the same locus. A scale bar of 10 kb is shown in the box. Arrowheads indicate centromeres.
Figure 3.
Schematic Structures of nupDNA Fragments.
nupDNA fragments larger than 10 kb are denoted A to L in order of decreasing size. As shown in the box, the rice chloroplast genome is a circular DNA molecule of 135 kb, which contains two copies of an identical IR (IRa and IRb) separated by large (LSC) and small single-copy (SSC) regions (Sugiura, 1992). The entire chloroplast genome was divided into 5-kb segments (arrows), denoted 1 to 23. The corresponding segments found in nupDNA fragments are represented by numbered arrows. The IRs and their segments are shown in red. Asterisks indicate gaps where the genomic sequence was not available when this analysis was performed.
Nucleotide Substitution Profiles Reveal That nupDNAs Were Repeatedly Integrated into the Nuclear Genome
We analyzed the nucleotide substitutions of nupDNAs relative to those of the present chloroplast genome. Theoretically, the substitutions in nupDNAs consist of two types according to their origin. One type of substitution reflects a difference in nucleotide sequence between the ancient and present chloroplast genomes. The second type represents mutations that have occurred in nupDNA fragments in the nucleus. These substitutions are designated type 1 and type 2, respectively, in this study. In the chloroplast genome, the two copies of the IR have identical sequences (Sugiura, 1992). Type I substitutions can only be inferred confidently from IR sequences because type 1 substitutions in one segment of the ancient IR should have identical counterparts in the other segment.
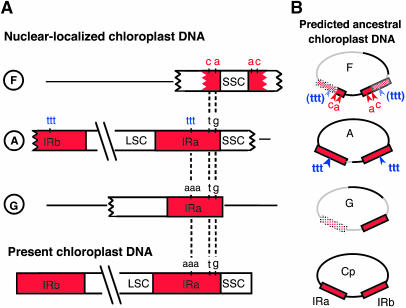
According to the above criterion, we found several candidates for type 1 substitutions (Figure 4). nupDNA fragment F has type 1 substitutions at two loci in each of the IR segments (Figure 4A). In the two IR segments of nupDNA fragment A, AAA of the present plastid genome is replaced with TTT (Figure 4A). The probability that two segments of the IR would acquire identical substitutions by random mutation is negligibly small (data not shown). Therefore, these substitutions probably derived from ancient plastid genomes. The pattern of these substitutions was compared among nupDNA fragments F, A, and G and the present plastid genome (Figure 4A). If these substitutions were sequentially introduced into the rice plastid genome during its evolution, their profiles suggest that the F-type genome is the oldest, the A-type intermediate, and the G-type genome the most recent (Figure 4B). Interestingly, the nucleotide divergence of these nupDNAs relative to the present plastid DNA sequence suggests that their translocation to the nuclear genome occurred in the same order. The numbers of substitutions per base pair in the F, A, and G fragments are 8.2 × 10−3, 3.1 × 10−3, and 1.3 × 10−3, respectively. Taken together, these results indicate that the integration of chloroplast DNA into the nuclear genome has occurred repeatedly in rice.
Figure 4.
nupDNA Fragments Are Derived from Ancestral Chloroplast Genomes.
(A) Schematic illustration of the IR regions of nupDNA fragments F, A, and G, compared with that of the present chloroplast genome. LSC, large single-copy region; SSC, small single-copy region.
(B) The predicted structures of the ancient chloroplast genomes from which nupDNA fragments F, A, and G are derived. Cp indicates the present chloroplast genome. Thick lines of the F, A, and G genomes represent the DNA fragments found in the present nupDNAs, and gray lines represent the parts predicted to be lost or not transferred during evolution.
Estimation of the Age of nupDNAs
Discrimination of type 1 and type 2 substitutions allowed us to estimate when individual nupDNA fragments were integrated into the nucleus. In the 33,742-bp IR regions of the largest nupDNA fragment, fragment A, we identified 83 type 2 substitutions. Because the chloroplast DNA sequence is unlikely to be functional in the nucleus, the rate of type 2 substitutions should approximate the synonymous substitution rate of 4.2 × 10−9/site per year (Ramakrishna et al., 2002). On the basis of this rate, we estimated that fragment A integrated into the nuclear genome ∼0.6 million years (Myr) ago. The total size of fragment A is 130,476 bp, in which we found 403 nucleotide substitutions between the nuclear and plastid sequence, including both type 1 and type 2 substitutions. Therefore, there may be a relationship such that a million-year-old nupDNA fragment should have 5.3 × 10−3 substituted nucleotides per base between the nuclear and plastid sequence (total of the type 1 and type 2). Using this equation, we calculated when individual nupDNA fragments were translocated into the nucleus. This age analysis was applied to nupDNA fragments larger than 200 bp because smaller fragments have less mutations, and it is therefore difficult to estimate their ages. Furthermore, the combined lengths of those smaller fragments account for only one-sixth of the total length of nupDNAs.
To confirm the validity of this age estimation, we also calculated, by a different method, the rate at which nupDNA and plastid DNA diverge (see Methods). This alternative method gave another estimate of the divergence rate between the nuclear and plastid sequence of 4.0 to 5.6 × 10−3 substitutions/bp per million years, which is in good agreement with the above estimate.
Age Distribution Analysis Reveals Dynamic Features of nupDNAs
Figure 5 shows the age distribution profiles of nupDNA fragments estimated by the method described above. Unexpectedly, most nupDNAs were translocated within the past 1 Myr, and the amount of nupDNA decreases rapidly as its age increases (Figure 5A). What does this age distribution mean? Assuming that chloroplast DNA has been continuously transferred to the nucleus at a similar frequency, this result suggests that >90% of nupDNAs disappear within 2 Myr of their integration into the nucleus.
Figure 5.
Age Distribution and Localization of nupDNA Fragments Larger than 200 bp.
The combined lengths of these fragments account for 84% of the total nupDNA.
(A) Total amount of nupDNAs generated in the respective periods.
(B) Total numbers of nupDNA fragments generated in the respective periods, classified into four groups based on size.
(C) Age distribution of nupDNAs occurring within 5 cM of the centromere.
(D) Age distribution of nupDNAs occurring more than 5 cM from the centromere.
(E) Size distribution of young nupDNA fragments (<1 Myr) in the pericentromeric regions (dark-gray bars) and in the rest (light-gray bars). Size classes: I, 200 to 400 bp; II, 400 to 800 bp; III, 800 to 1600 bp; IV, 1600 to 3200 bp; V, >3200 bp.
(F) Integration frequency of young nupDNA fragments (<1 Myr) in the pericentromeric regions. The red line indicates the expected number of integration sites if they were equally distributed throughout the nuclear genome (see Methods). Letters represent the giant nupDNA fragments (Figure 2).
We examined the disappearance process of nupDNAs in more detail. The nupDNA fragments larger then 200 bp were grouped into four size classes and their numbers plotted against their ages (Figure 5B). The result clearly shows that the large fragments disappear rapidly, whereas the small fragments disappear more slowly. The disappearance of nupDNA fragments could be attributable to two causes: accumulated mutations have rendered the nupDNA sequences undetectable by homology search programs, or numerous nupDNAs have truly been eliminated from the chromosomes. The former alternative is unlikely because nupDNAs rapidly disappear with a sequence homology of ∼99% (Figures 5A and 5B, see the horizontal axis), and the fragments with such high homologies cannot be missed by in silico searches if the sequences really exist in the genome. Furthermore, the constitutive segments of the nupDNA fragments tend to be highly conserved, even after vigorous shuffling (Figure 3). Again from Figure 5B, we can draw an evolutionary scenario in which the large nupDNA fragments are rapidly eliminated from the chromosomes concomitant with their fragmentation and shuffling, whereas the elimination of the small fragments proceeds far more slowly. The apparent half-lives of nupDNAs are 0.5 Myr for large fragments (>1.6 kb) and 2.2 Myr for smaller fragments (<1.6 kb).
Large Chloroplast DNA Fragments Are Preferentially Integrated into the Pericentromeric Regions
Large nupDNA fragments are frequently found near the centromeres (Figure 2), although the biological meaning of this is unclear. Theoretically, this tendency could arise in either or both of the following ways: the pericentromeric regions may provide a more stable environment for integrated nupDNAs, or they are better able to engulf large chloroplast DNAs. To examine the first possibility, we compared the age distribution profiles of the nupDNA fragments in the pericentromeric region (Figure 5C) and in the rest of the chromosome (Figure 5D). The results revealed that nupDNAs in the pericentromeric region were eliminated rapidly compared with those in the other regions. This analysis does not support the first possibility. Next, we examined the second possibility using recently translocated plastid DNA fragments as clues. We identified young nupDNA fragments (>200 bp, <1 Myr) in the pericentromeric regions and the rest of the chromosomes and grouped them into five size classes (Figure 5E). This analysis demonstrates that the relative abundance of the largest-class fragments (V; >3200 bp) is markedly greater in the pericentromeric regions. Considering that the elimination speed of nupDNA fragments is not slow in the pericentromeric regions (Figure 5C), this result indicates that the pericentromeric regions preferentially integrate large chloroplast DNA fragments.
The Pericentromeric Region Contains Hot Spots for the Integration of Chloroplast DNA
We next examined whether the pericentromeric region engulfs nupDNAs more frequently than does the rest of the chromosome. The BLAST program detected 127 young nupDNA fragments (>200 bp; <1 Myr) in the whole rice genome. Fifty percent of them are mapped within 5 cM from centromeres, and on the physical map, 47% of them are located within the 5-Mb flanking region of the centromere that comprises 28% of the total rice genome. This may be interpreted that the 5-Mb pericentromeric region integrates nupDNAs more frequently than does the rest of the chromosome. However, it should be noted that the BLAST program detects fragmented and shuffled nupDNAs as discrete entities, thus tending to overestimate the number of integration events. To avoid this overestimation, we repeated the analysis applying the condition that the nupDNA fragments found in the same BAC/PAC clone (∼120 kb on average), and therefore located close together on the chromosome, were deemed to be fragments originating from a single integration event. Using this approach, the above 127 fragments were clustered into 47 discrete loci (independent BAC/PAC clones), and 30% of those loci were located within the 5-Mb pericentromeric regions (comprising 28% of the total genome) (Figure 5F). Therefore, the integration rate of nupDNAs into the 5-cM or 5-Mb pericentromeric regions appears to be similar to the average rate of incorporation into whole chromosomes, but the fragmentation of nupDNAs seems to be more active in those pericentromeric regions.
More detailed analysis of the pericentromeric regions revealed a unique property: the 1-Mb regions adjacent to the centromeres are hot spots for the integration of plastid DNA (Figure 5F). In these hot spots, at least nine integration events have occurred within the past 1 Myr, and the combined lengths of those fragments amount to one-third of the total young nupDNAs (523 kb). Three of the 12 giant nupDNA fragments are found in this 1-Mb region (Figure 5F, fragments B, D, and H), although the average size of the rice chromosomes is 35.2 Mb (Table 1).
In summary, the pericentromeric regions of the rice chromosomes consist of two characteristic regions, in terms of the dynamic behavior of nupDNAs: the innermost 1-Mb region that engulfs large chloroplast DNA fragments most frequently and the marginal 5-Mb region where large chloroplast DNA fragments are preferentially integrated but where the integration frequency is not high (Figure 5F).
DISCUSSION
How nupDNAs Are Generated
The sequence of the organellar genome could be transferred to the nucleus by two different mechanisms: direct integration of the organellar DNA into the nucleus (Woischnik and Moraes, 2002) or RNA-mediated DNA transfer through the reverse transcription process (Nugent and Palmer, 1991). Which mechanism has been predominantly responsible for the generation of nupDNAs? If RNA-mediated DNA transfer is the predominant mechanism, transcribed regions of the plastid genome should be found in nupDNA more abundantly than nontranscribed regions. However, this study shows that all parts of the plastid genome, including spacer regions, occur at similar frequencies in nupDNAs (Figure 1). Therefore, DNA-mediated transfer, and not RNA-mediated transfer, is the predominant mechanism conveying plastid DNA sequences to the nucleus. The existence of giant nupDNA fragments (Figure 2) supports this view.
DNA-mediated transfer could occur by two distinct routes: direct uptake of the plastid DNA by the nucleus or uptake via an indirect path mediated by mitochondria. The mitochondrial genome has been shown to engulf plastid DNA and transfer it to the nucleus (Notsu et al., 2002). The majority of nupDNAs are derived from the plastid DNA segments uniquely present in the plastid genome but not in the mitochondrial genome (Figure 1). This implies that the direct uptake of plastid DNA by the nucleus is the main transfer pathway. Interestingly, the number of nupDNA fragments tends to be higher for the plastid DNA segments whose copies are retained in the mitochondrial genome (Figure 1, yellow boxes). Therefore, although indirect DNA transfer via the mitochondrial genome may have occurred frequently in rice, its contribution to the total nupDNA population seems small.
nupDNA fragments could be generated not only by the translocation of plastid DNA, but also by the duplication of preexisting nupDNA fragments in the nucleus. Giant fragment I appears to be derived from the largest fragment A because unique nucleotide substitutions are often conserved between these nupDNAs (data not shown). To identify amplified copies of nupDNAs, characteristic mutations are required, as in the case of giant fragments I and A. However, as far as we can discern, this type of lineage analysis of nupDNA fragments is not easy, except for some large fragments, because nupDNAs are generally small and fragmented and still highly homologous to the present plastid genome. If nupDNAs were duplicated before accumulating certain numbers of mutations, we have no means of tracing their lineages. Duplication or amplification of nupDNAs in the nuclear genome is the subject of future study.
Plastid DNAs Are Frequently Integrated into the Nuclear Genome
This study strongly suggests that the giant nupDNAs of rice originated from at least three different plastid DNAs, judging from their characteristic mutations in the IR regions (Figure 4). Combined with the age estimates for each giant nupDNA, this suggests that the rice nuclear genome incorporated a large portion of plastid genome at least three times in the past 1.7 Myr. However, we should bear in mind that nupDNAs retaining two copies of the IR regions are rare, so the above cases might constitute the tip of the iceberg. If so, how could we estimate the total frequency of plastid DNA integration events in the past? In the age analysis of nupDNAs, we found 47 loci that contain young nupDNA fragments <1 Myr old. Considering that nupDNAs are rapidly eliminated from the nucleus, we should expect that >47 integration events occurred in the past 1 Myr. If the plant generation time of rice has been constant at one year during its evolution, the frequency of integration events should be >47 × 10−6 times per plant generation.
Recently, two research groups experimentally detected plastid–nuclear DNA transfer in tobacco, with an apparent transfer rate of a marker gene of 1 per 16,000 pollen grains (Huang et al., 2003) or 1 per 5 million leaf cells (Stegemann et al., 2003). Therefore, a given gene locus on the tobacco plastid genome is expected to translocate to the nucleus 63 × 10−6 times per plant generation or 0.2 × 10−6 times per somatic cell division. From our data set, we also calculated how frequently rice chloroplast genes were translocated to the nucleus (see Methods). Our estimate suggests that a given gene locus of the rice chloroplast genome should have transferred to the nucleus at least 4 × 10−6 times per plant generation.
Rice fields generally produce 100 to 300 × 106 grains/hectare. Therefore, in a rice field of 1 hectare, there may be hundreds of grains in which a given gene of the chloroplast genome has newly transferred to the nucleus. The occurrence of grains in which the nuclear genome had newly integrated any part of the chloroplast genome would be far higher.
Turnover of nupDNAs and Genome Shuffling
Richly and Leister recently reported that there is a correlation between the size and sequence similarity of nupDNAs (2004). In the rice genome, the size and abundance of nupDNA fragments rapidly decrease as their age increases, and there are few fragments older than 5 Myr (Figures 5A and 5B). Theoretically, this biased distribution can be explained by two possibilities: the rice genome has become more and more competent to integrate nupDNAs, or old nupDNAs are eliminated from the nuclear genome.
If the first possibility is the case, we can postulate that (1) the rice nuclear genome was poorly competent to integrate nupDNAs until 5 Myr ago, and then (2) the competence of the nuclear genome and the size of the integrated fragments increased at an accelerated pace. However, we cannot accept these hypotheses because (1) they do not explain the fragmented and shuffled nature of nupDNAs (Figure 3), (2) a molecular mechanism that leads to the gradual enlargement of the inserted fragments is hard to imagine (Figure 5B), (3) a high degree of competence in integrating nupDNA is unlikely to be rice specific (Huang et al., 2003; Stegemann et al., 2003), and finally (4) plastid genes were repeatedly transferred to the nucleus earlier than 5 Myr ago (Martin et al., 1998).
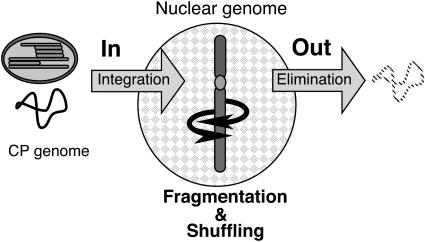
By contrast, if we accept the second possibility, many of the data for nupDNA are explained well. The key concept in this explanation is that nupDNAs, once integrated into the nuclear genome, are shuffled, fragmented, and finally eliminated from the nuclear genome. Thus, the current population of nupDNAs is in dynamic equilibrium between frequent integration and rapid elimination (Figure 6). The age distribution (Figure 5A), age–size relationship (Figure 5B), and fragmented and shuffled nature (Figure 3) of nupDNAs are well explained by this model. This model also explains why plant genomes do not seem to be expanding, despite the frequent integration of plastid DNAs (Huang et al., 2003; Stegemann et al., 2003).
Figure 6.
DNA Flux from Plastid to Nucleus.
The nuclear genome continually engulfs the plastid DNA and eliminates it by genome shuffling. This evolutionary process mainly proceeds at unique loci of the nuclear genome, such as the pericentromeric regions. CP, chloroplast.
This model raises a more critical question: How are nupDNAs eliminated from the nuclear genome? It is inconceivable that the nuclear machinery specifically recognizes and eliminates plastid-derived DNAs long after their integration. Rather, it is very likely that the shuffling, fragmentation, and consequent deletion of nupDNAs reflects the intrinsic behavior of the chromosomal DNA into which nupDNAs are integrated. If the intrinsic genes of the nuclear genome were deleted, those genetic lines would perish as a result of the harmful effects of the deletion. However, because nupDNA is a kind of junk DNA in the nuclear genome, footprints of its shuffling and deletion are easily detectable. In this sense, nupDNA could be a marker with which we can trace the potential behavior of the corresponding chromosomal loci. The observed half-life of nupDNA (Figures 5A and 5B) suggests how frequently the rice chromosomal DNA undergoes shuffling and drastic deletion. These results also imply that vigorous genome shuffling causes the turnover of chromosomal DNA, as long as it does not damage the endogenous gene systems.
According to the model described above, the rice nuclear genome frequently integrates chloroplast DNA and at the same time eliminates fragmented nupDNAs. How are these two processes equilibrated? In yeast, chromosomal DNA frequently integrates mitochondrial DNA during the repair process of DSB (Ricchetti et al., 1999; Yu and Gabriel, 1999). DSB and subsequent nonhomologous DNA end-joining could be involved in the recombination process of genome shuffling. DSB repair might be a key process that links the input and output of the nupDNA flux.
Turnover Rate of nupDNAs
In this study, we estimated the half-lives of nupDNAs in the rice genome to be 0.5 Myr for large fragments (>1.6 kb) and 2.2 Myr for smaller fragments (<1.6 kb) (Figure 5B). These estimates could change according to the synonymous substitution rate of the nuclear genes used for the analysis. The synonymous substitution rate itself varies slightly because of both the used divergence time of organisms and the used genes for calculation, and several values have been reported for cereal species (Gaut et al., 1996; Ramakrishna et al., 2002; Guo and Moose, 2003). In this study, we used Ramakrishna's synonymous substitution rate (4.2 × 10−9 substitutions per synonymous site per year), which is lower than the parameters cited in other reports (for example, Adh1, 7.0 × 10−9 and Adh2, 6.0 × 10−9, according to Gaut et al., 1996). This makes our estimation of the turnover rate conservative; therefore, turnover might occur even more rapidly than we have estimated.
Possible Role of the Pericentromeric Region in Endosymbiotic Evolution
This study shows that the pericentromeric regions of rice chromosomes preferentially engulf large chloroplast DNA fragments (Figures 2 and 5E), with marked integration activity in the innermost 1-Mb region (Figure 5F). The pericentromeric regions are also more active in the fragmentation, shuffling, and elimination of large nupDNAs (Figures 3, 5B, and 5C) and consequently play an important role in the chloroplast–nuclear DNA flux. Recently, Richly and Leister (2004) reported that plastid- and mitochondrion-derived DNAs are loosely clustered in the rice nuclear genome. We also observed that the large amounts of mitochondrion-derived DNA are integrated in the pericentromeric regions (data not shown). Therefore, the pericentromeric region is a hotspot for organelle–nucleus DNA transfer, the existence of which was suggested by Richly and Leister. Because the pericentromeric region contains few functional genes (Arabidopsis Genome Initiative, 2000; Feng et al., 2002), this low gene density may facilitate the integration and elimination of the organelle-derived DNA to some extent.
Frequent insertion of large DNA fragments into the 1-Mb flanking region of the centromere is not unique to rice. A 620-kb mitochondrial DNA insertion was observed in the pericentromeric region of Arabidopsis thaliana chromosome 2 (Stupar et al., 2001). Furthermore, the pericentromeric regions of mammalian genomes contain large segments of interchromosomal duplications (International Human Genome Sequencing Consortium, 2001; Bailey et al., 2002; Thomas et al., 2003). Incorporation of large DNA fragments into the pericentromeric region may be a strategy of genomic evolution conserved between plants and animals.
In the human genome, pericentromeric regions have been suggested to function as incubators of new genes, where mosaic sequences of incorporated DNAs are often transcribed (Bailey et al., 2002; Knight, 2002). We also found mosaic transcripts that contain nupDNA fragments (data not shown) in the rice full-length cDNA database (Rice Full-Length cDNA Consortium, 2003). This supports the idea that organellar DNAs also provided the raw materials for the creation of new genes during plant evolution (Martin et al., 2002). The pericentromeric regions of plant genomes harbor transposable elements (Copenhaver et al., 1999; Feng et al., 2002; Sasaki et al., 2002), which might facilitate the creation of new genes by promoting genome shuffling in various ways (Gray, 2000; Shirasu et al., 2000; Gilbert et al., 2002; Symer et al., 2002). Transposable elements might also transport new genes from the heterochromatic silenced regions of the pericentromere to euchromatic transcriptionally active sites where their functions are tested. Functional gene transfer from the organelle to the nucleus (Adams et al., 1999, 2002; Millen et al., 2001) might be the successful outcome of these consecutive evolutionary experiments.
In conclusion, our genome-wide analysis of nupDNA fragments demonstrates the rapid turnover of plastid genome–derived DNA fragments in the rice nuclear genome and reveals a notable property of the pericentromeric regions, whereby they engulf diverse DNA fragments and shuffle them vigorously. The DNA flux from organelle to nucleus, and subsequent shuffling within the nuclear chromosomes, might have acted as the motive force in the creation of genes and might then have contributed to the evolution of the nucleo-organelle relationship probably from the very beginning of endosymbiotic evolution.
METHODS
Analysis of Nuclear-Localized Plastid DNA Fragments
Using the Smith–Waterman algorithm (Smith and Waterman, 1981a, 1981b; GeneMatcher2; Paracel, Pasadena, CA) and the BLAST algorithm (Altschul et al., 1990; BlastMachine), the nupDNA fragments were searched against the phase 2 sequences of the rice nuclear genome (Oryza sativa ssp japonica), which was updated on February 20, 2003 and is available at the Web site http://RiceGAAS.dna.affrc.go.jp/. For this purpose, the entire rice chloroplast genome (O. sativa ssp japonica cultivar group; GenBank accession number NC_001320.1; Hiratsuka et al., 1989) was divided into 5-kb segments and tagged with numbers 1 to 23 (Figure 3), which were then used as query sequences. Matches with E-values lower than 10−10 were defined as nupDNA fragments. The results of the BLAST and Smith–Waterman searches are available at our Web site (http://133.6.138.155:88/smkdb/jsp/matsuo.jsp). The redundancy of nupDNA fragments in overlapping regions of BAC/PAC clones was checked with information from the physical map of the rice nuclear genome (http://rgp.dna.affrc.go.jp/cgi-bin/statusdb/status.pl and http://www.genome.arizona.edu/fpc/rice/) (Chen et al., 2002) and the redundant sequences removed. When estimating the total length of the nupDNA fragments, the calculations were simplified by summing the lengths of the chloroplast genomic regions corresponding to individual nupDNA fragments. The number of intermingled nupDNAs was counted as the BAC/PAC clones that contained discontinuous nupDNAs.
Identification of the Integration Sites of nupDNA Fragments
The integration sites of nupDNA fragments on both the genetic map and the physical map were identified according to the marker-based physical map of the rice chromosomes available from the home page of the International Rice Genome Sequencing Project (http://rgp.dna.affrc.go.jp/IRGSP/). When BAC/PAC clones did not contain genetic markers, they were treated as belonging to the nearest genetic marker. The centromeres were sited according to Harushima et al. (1998) and Wu et al. (2002). In the pericentromeric regions of chromosomes 4 and 10, there were sequence gaps between the centromere and nupDNA; those lengths were estimated according to Feng et al. (2002) and Rice Chromosome 10 Sequencing Consortium (2003), respectively.
Giant nupDNA fragments A to L (Figure 3) are encoded by BAC/PAC clones with the following accession numbers: A, AC092750, AC099402, and AC122148; B, AL662978; C, AP003280; D, AP005408; E, AP005161; F, AL513004 and AL954853; G, AC074232; H, AL731605; I, AP004236; J, AC130726 and AC136521; K, AL606447; L, AP005543.
Calculation of the Relationship between Nucleotide Divergence and the Ages of nupDNAs
The number (K) of substitutions per nucleotide site between the nupDNA fragments and chloroplast DNA was calculated on the basis of the BLAST alignment. The K value was corrected by one-parameter methods (Jukes and Cantor, 1969). The age of giant nupDNA fragment A was calculated by dividing the corrected K value of type 2 substitutions (see Results) by the absolute substitution rate of the synonymous sites in the nuclear genes of rice (Ramakrishna et al., 2002). The relationship of age and nucleotide divergence of nupDNAs was calculated with the corrected K value.
The half-lives of nupDNAs were estimated with the age distribution curve of the nupDNAs (Figure 5B) drawn with Excel.
An Alternative Method for Estimating the Age of nupDNA
The nucleotide divergence between the nupDNA and the present plastid genome is the sum of the nucleotide mutations accumulated in the nucleus and in the plastid genome. The rate of the former was approximated by the synonymous substitution rate in the nucleus (4.2 × 10−9 substitutions/bp per year) (Ramakrishna et al., 2002). To determine the average rate of nucleotide substitutions in the total plastid genome, we compared the plastid genomes of rice and maize (Zea mays) and identified 5.6% substituted nucleotides. Rice and maize are thought to have diverged from a common ancestor 50 to 70 Myr ago (Zurawski and Clegg, 1987; Wolfe et al., 1989; Ramakrishna et al., 2002). Taking 60 Myr ago as their divergence time, the average nucleotide substitution rate in the plastid genome was calculated to be ∼0.5 × 10−9 substitutions/bp per year. This value is close to the rate calculated from the synonymous and nonsynonymous substitution rates of chloroplast genes (Gaut, 1998). By combining the substitution rates of the nuclear and plastid genomes, the divergence rate between nupDNA and the plastid genome was given to be 4.7 × 10−9 substitutions/bp per year. Because the substitution rates of the nuclear genome (Ramakrishna et al., 2002) and chloroplast genome (estimated above) were calculated on the basis of the divergence time of cereal species (Zurawski and Clegg, 1987; Wolfe et al., 1989; Ramakrishna et al., 2002), the above estimate could vary within 4.0 to 5.6 × 10−9 because of the uncertainty of the divergence time of 50 to 70 Myr ago (Zurawski and Clegg, 1987; Wolfe et al., 1989; Ramakrishna et al., 2002).
The chloroplast DNA sequences of rice (Hiratsuka et al., 1989) and maize (Maier et al., 1995) were aligned using ClustalX (Thompson et al., 1997) and the 131-kb aligned sequences were used for the analysis. The average substitution rate in the plastid genomes of these plants was calculated with both the corrected K value for these plastid genomes and their divergence time of 60 Myr (Zurawski and Clegg, 1987; Wolfe et al., 1989; Ramakrishna et al., 2002).
Average Number of Young nupDNA Fragments in a 1-Mb Segment of Chromosomal DNA
We calculated the average integration frequency of nupDNA fragments (>200 bp) in the nuclear genome to be 0.111/Mb per million years, on the basis that nupDNAs have been integrated into 47 discrete loci (independent BAC/PAC clones) of the 422.8-Mb rice nuclear genome (Table 1) within a million years. Therefore, the expected number of young nupDNA fragments (<1 Myr) found in a 1-Mb segment of the pericentromeric region (Figure 5E) is given below, on the condition that the 12 rice chromosomes have pericentromeric regions on both sides of their centromeres:
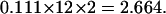 |
Estimation of the Gene Transfer Frequency from Chloroplast to Nucleus
In rice, the nuclear genome contains 523 kb of young nupDNA fragments (<1 Myr old), which is nearly equal to four intact chloroplast genomes. Therefore, a given gene locus on the rice chloroplast genome is expected to have, on average, four DNA copies that have transferred to the nucleus in the past 106 years. This implies that a given gene of the rice chloroplast genome should have transferred to the nucleus at least 4 × 10−6 times per year.
Supplementary Material
Acknowledgments
We thank M. Sugiura, Y. Kumazawa, T. Nakamura, H. Suzuki, and M. Kimura for critical comments and suggestions, M. Nakamura, K. Kinoshita, and T. Kondo for useful discussions, D. Stern for critical reading of this manuscript, and Qiu Yin-Jie for discussion and technical support with computer analysis. This work was supported in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project) and by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Junichi Obokata (obokata@gene.nagoya-u.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027706.
References
- Adams, K.L., Qiu, Y.-L., Stoutemyer, M., and Palmer, J.D. (2002). Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 99, 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Song, K., Roessler, P.G., Nugent, J.M., Doyle, J.L., Doyle, J.J., and Palmer, J.D. (1999). Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc. Natl. Acad. Sci. USA 96, 13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Ayliffe, M.A., Scott, N.S., and Timmis, J.N. (1998). Analysis of plastid DNA-like sequences within the nuclear genomes of higher plants. Mol. Biol. Evol. 15, 738–745. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Viggiano, L., Misceo, D., Horvath, J.E., Archidiacono, N., Schwartz, S., Rocchi, M., and Eichler, E.E. (2002). Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am. J. Hum. Genet. 70, 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson, D., Zhang, D.-X., Hartl, D.L., and Hewitt, G.M. (2001). Mitochondrial pseudogenes: Evolution's misplaced witnesses. Trends Ecol. Evol. 16, 314–332. [DOI] [PubMed] [Google Scholar]
- Chen, M., Presting, G., Barbazuk, W.B., Goicoechea, J.L., Blackmon, B., Fang, G., Kim, H., Frisch, D., Yu, Y., and Sun, S. (2002). An integrated physical and genetic map of the rice genome. Plant Cell 14, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G.P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- Ellis, R.J. (1982). Promiscuous DNA—Chloroplast genes inside plant mitochondria. Nature 299, 678–679. [DOI] [PubMed] [Google Scholar]
- Feng, Q., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420, 316–320. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S. (1998). Molecular clocks and nucleotide substitution rate in higher plants. Evol. Biol. 30, 93–120. [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate difference at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., Lutz-Prigge, S., and Moran, J.V. (2002). Genomic deletion created upon LINE-1 retrotransposition. Cell 110, 315–325. [DOI] [PubMed] [Google Scholar]
- Gray, Y.H. (2000). It takes two transposons to tango. Trends Genet. 16, 461–468. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Moose, S. (2003). Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15, 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka, J., et al. (1989). The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217, 185–194. [DOI] [PubMed] [Google Scholar]
- Huang, C.Y., Ayliffe, M.A., and Timmis, J.N. (2003). Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 422, 72–76. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Jukes, T.H., and Cantor, C.R. (1969). Evolution of protein molecules. In Mammalian Protein Metabolism, H.N. Munro, ed (New York: Academic Press), pp. 21–132.
- Knight, J. (2002). All genomes great and small. Nature 417, 374–376. [DOI] [PubMed] [Google Scholar]
- Kurland, C.G., and Andersson, G.E. (2000). Origin and evolution of the mitochondrial proteosome. Microbiol. Mol. Biol. Rev. 64, 786–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., and Kössel, H. (1995). Complete sequence of the maize chloroplast genome: Gene content, hot spots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. [DOI] [PubMed] [Google Scholar]
- Martin, W., and Herrmann, R.G. (1998). Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M., and Penny, D. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. USA 99, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. [DOI] [PubMed] [Google Scholar]
- Millen, R.S., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., Hirai, A., and Kadowaki, K. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268, 434–445. [DOI] [PubMed] [Google Scholar]
- Nugent, J.M., and Palmer, J.D. (1991). RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66, 473–481. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.-J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J.L. (2002). Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti, M., Fairhead, C., and Dujon, B. (1999). Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature 402, 96–100. [DOI] [PubMed] [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium (2003). In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300, 1566–1569. [DOI] [PubMed] [Google Scholar]
- Rice Full-Length cDNA Consortium (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379. [DOI] [PubMed] [Google Scholar]
- Richly, E., and Leister, D. (2004). NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol. Biol. Evol. 21, 1972–1980. [DOI] [PubMed] [Google Scholar]
- Saji, S., Umehara, Y., Antonio, B.A., Yamane, H., Tanoue, H., Baba, T., Aoki, H., Ishige, N., Wu, J., Koike, K., Matsumoto, T., and Sasaki, T. (2001). A physical map with yeast artificial chromosome (YAC) clones covering 63% of the 12 rice chromosomes. Genome 44, 32–37. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., et al. (2002). The genome sequence and structure of rice chromosome 1. Nature 420, 312–316. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Shulman, A.H., Lahaye, T., and Schulze-Lefert, P. (2000). A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., and Waterman, M.S. (1981. a). Identification of common molecular sequences. J. Mol. Biol. 147, 195–197. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., and Waterman, M.S. (1981. b). Comparison of biosequences. Adv. Appl. Math. 2, 482–489. [Google Scholar]
- Stegemann, S., Hartmann, S., Ruf, S., and Bock, R. (2003). High-frequency gene transfer from the chloroplast genome to the nucleus. Proc. Natl. Acad. Sci. USA 100, 8828–8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R.M., Lilly, J.W., Town, C.D., Cheng, Z., Kaul, S., Buell, R., and Jiang, J. (2001). Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeat. Proc. Natl. Acad. Sci. USA 98, 5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M. (1992). The chloroplast genome. Plant Mol. Biol. 19, 149–168. [DOI] [PubMed] [Google Scholar]
- Symer, D.E., Connelly, C., Szak, S.T., Caputo, E.M., Cost, G.J., Parmigiani, G., and Boeke, J.D. (2002). Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., et al. (2003). Pericentromeric duplications in the laboratory mouse. Genome Res. 13, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness, P.E., and Fox, T.D. (1990). Escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Nature 346, 376–379. [DOI] [PubMed] [Google Scholar]
- Woischnik, M., and Moraes, C.T. (2002). Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 12, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.-W., Sharp, P.M., and Li, W.-H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., et al. (2002). A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., and Gabriel, A. (1999). Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell 4, 873–888. [DOI] [PubMed] [Google Scholar]
- Yuan, Q., Hill, J., Hsiao, J., Moffat, K., Ouyang, S., Cheng, Z., Jiang, J., and Buell, C.R. (2002). Genome sequencing of a 239-kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol. Genet. Genomics 267, 713–720. [DOI] [PubMed] [Google Scholar]
- Zurawski, G., and Clegg, M.T. (1987). Evolution of higher-plant chloroplast DNA-encoded genes: Implications for structure–function and phylogenetic studies. Annu. Rev. Plant Physiol. 38, 391–418. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.