Abstract
Organogenesis at the shoot meristem requires a delicate balance between stem cell specification and differentiation. In Arabidopsis thaliana, WUSCHEL (WUS) is a key factor promoting stem cell identity, whereas the CLAVATA (CLV1, CLV2, and CLV3) loci appear to promote differentiation by repressing WUS expression. In a screen for mutations modifying clv1 mutants, we have identified a novel regulator of meristem development we term CORONA (CNA). Whereas cna single mutant plants exhibit subtle defects in meristem development, clv cna double mutants develop massively enlarged apices that display early loss of organogenesis, misexpression of WUS and CLV3, and eventual differentiation of the entire apex. The CNA gene was isolated by positional cloning and found to encode a class III homeodomain Leu zipper protein. A missense mutation resulting in the dominant-negative cna-1 allele was identified in a conserved domain of unknown function, and a likely null allele was shown to display a similar but weaker phenotype. CNA is expressed in developing vascular tissue, diffusely through shoot and flower meristems, and within developing stamens and carpels. Our analysis of WUS expression in wild-type, clv, and clv cna plants revealed that, contrary to current models, WUS is neither necessary nor sufficient for stem cell specification and that neither WUS nor CLV3 is a marker for stem cell identity. We propose that CNA functions in parallel to the CLV loci to promote organ formation.
INTRODUCTION
A central feature of plant development is the continuous generation of organs throughout the plant's lifespan. The capacity to generate new above-ground organs post-embryonically is a property of shoot meristems. Within shoot meristems reside stem cells that are maintained at a constant number while giving rise to organ primordia and ultimately all of the differentiated cells of organs and tissues (Steeves and Sussex, 1989). In this way, shoot meristems have the capacity to balance perpetual differentiation of cells while replenishing the pool of undifferentiated, pluripotent cells.
Genetic screens have identified several key regulators of shoot meristem development (Barton and Peothig, 1993; Clark et al., 1993; Laux et al., 1996; Pogany et al., 1998; Yu et al., 2000). The WUSCHEL (WUS) gene encodes a homeodomain protein, which is an important regulator of stem cell identity (Mayer et al., 1998; Schoof et al., 2000). Loss-of-function wus mutants fail to organize functional shoot meristems. After germination, wus mutants sporadically generate adventitious shoots, which form only a few organs before termination (Endrizzi et al., 1996; Laux et al., 1996). Expression of WUS within the meristem appears to be sufficient for establishing stem cell identity. When WUS was ectopically expressed, transgenic seedlings accumulate undifferentiated stem cells (Schoof et al., 2000).
Three CLAVATA genes (CLV1, CLV2, and CLV3) promote the differentiation of stem cells. Loss-of-function clv1, clv2, and clv3 mutants accumulate undifferentiated cells in shoot and floral meristems, resulting in meristems that are significantly larger than the wild type and in flowers with increased numbers of floral organs (Clark et al., 1993, 1995; Jeong et al., 1999). The CLV1, CLV2, and CLV3 loci encode signal transduction components: a receptor kinase (Clark et al., 1997), a receptor-like protein (Jeong et al., 1999), and a small secreted protein (Fletcher et al., 1999), respectively.
WUS is a key target of the CLV signal transduction pathway (Brand et al., 2000; Schoof et al., 2000). In wild-type plants, the domain of WUS expression is normally restricted to a small, centrally located subset of cells beneath the three outermost cell layers (Mayer et al., 1998; Schoof et al., 2000). In clv3 mutant meristems, the WUS expression domain expands laterally and apically into the topmost cells of the L3 layer (Brand et al., 2000; Schoof et al., 2000). Conversely, plants overexpressing CLV3 recreate the Wus− phenotype and do not appear to express WUS mRNA (Brand et al., 2000), indicating that the CLV signaling pathway limits stem cell number by restricting the size of the WUS expression domain.
Overexpression of WUS through promoter fusions can also lead to ectopic stem cell accumulation (Schoof et al., 2000). Interestingly, transcripts of CLV3 are found on the periphery of stem cell masses formed by WUS overexpression, whereas in the meristems of wild-type plants, CLV3 expression is restricted to the center of the shoot meristem (Fletcher et al., 1999; Schoof et al., 2000). These expression analyses indicate that while the CLV signaling pathway targets WUS and restricts its activity, WUS activity is also sufficient to induce transcription of CLV3. This regulatory feedback loop may act to maintain strict control of the number of stem cells.
In this study, we describe a novel regulator of shoot meristem development, CORONA. We show that CNA encodes a class III homeodomain Leu zipper (HD-Zip III) transcription factor that plays an important role in the maintenance of stem cell function. Members of this gene family, which also includes the REVOLUTA (REV), PHABULOSA (PHB), PHAVOLUTA (PHV), and ATHB8 genes, have previously been implicated in several aspects of Arabidopsis thaliana development, including meristem and organ development (Talbert et al., 1995; McConnell and Barton, 1998; Zhong and Ye, 1999; Baima et al., 2001; McConnell et al., 2001; Otsuga et al., 2001; Zhong and Ye, 2001; Prigge et al., 2005). In addition, we show that WUS expression might not be necessary for stem cell specification in clv3-2 mutants. The latter finding suggests that a WUS independent pathway exists and is involved in specifying stem cell identity at certain stages in development. Furthermore, we demonstrate that WUS and CLV3 expression can be found in differentiated tissues.
RESULTS
Isolation of a cna Mutant
An ethyl methanesulfonate mutagenesis was performed on clv1-1 mutant seeds to identify enhancers and/or suppressors of the Clv− phenotype (Pogany et al., 1998). A strong modifier of clv1-1 plants was identified in this screen (pce5). The enhancing mutation was backcrossed to Landsberg erecta (Ler) and isolated (see Methods). The modifier acted as a single, nuclear trait in all crosses. When combined with strong clv alleles, the enhancer led to the development of corona or ring-like meristems, leading us to name the gene CORONA (CNA).
cna-1 plants appeared superficially wild type in phenotype. Scanning electron microscopy analysis of cna-1 shoot meristems revealed that the meristems of 12-d-old cna-1 plants are significantly larger than wild-type meristems (see Supplemental Figure 1 online; Table 1). The effect, if any, of cna-1 on shoot meristem size in 24-d-old plants is reduced compared with earlier developmental time points (Table 1).
Table 1.
Twelve-Day-Old cna-1 Meristems Are Larger Than Those in the Wild Type
| Genotype | Day 12 | Day 24 |
|---|---|---|
| Ler | 55.6 ± 0.99 (n = 5) | 57.3 ± 1.48 (n = 10) |
| cna-1 | 72.7 ± 1.72 (n = 10) | 61.1 ± 1.67 (n = 15) |
The size of the shoot apical meristem (in micrometers) of cna-1 and Ler plants at 12 and 24 d old was determined. The mean ± standard error is presented. The difference between means is statistically significant on day 12 (t test, P < 0.0001) but is less statistically significant on day 24 (t test, P = 0.1).
Genetic interactions with clv mutations
Double mutants were generated between cna-1 and various clv1, clv2, and clv3 alleles. The effect of cna-1 on clv mutant phenotypes and whether cna was recessive or incompletely dominant depended on the severity of the clv allele. When combined with a phenotypically weak clv allele, such as clv1-7, cna-1 enhanced the Clv− phenotype in terms of shoot meristem size (see Supplemental Figures 2A and 2B online; Table 2). When cna-1 was combined with clv alleles of intermediate severity, such as clv1-1 and clv2-1, cna-1 was also recessive but led to the development of more severe phenotypes. These included dramatic enlargement of shoot meristem size, the eventual loss of organ formation at the apex, and frequent differentiation of apical cells followed by organ formation in the center of the shoot meristem (leading to a ring-like or corona meristem) (see Supplemental Figure 2 online; Figures 1I and 1J, Table 2).
Table 2.
cna Mutations Enhance clv Shoot Meristem Sizes
| Genotype | Shoot Meristem Size (μm)a | Age (d)b | nc |
|---|---|---|---|
| clv2-1 | 58 ± 2.5d | 13 | 12 |
| clv2-1 cna-1/cna-1 | 109 ± 16.6 | 13 | 12 |
| clv3-2 | 163 ± 11.1e | 13 | 18 |
| clv3-2 cna-1/+ | 196 ± 9.1 | 13 | 18 |
| clv3-2 cna-1/cna-1 | 276 ± 9.3 | 13 | 32 |
| clv3-2 cna-2/cna-2 | 223 ± 12.1 | 13 | 16 |
| clv1-7 | 71 ± 2.8f | 15 | 8 |
| clv1-7 cna-1/cna-1 | 95 ± 2.1 | 15 | 8 |
Values represent the mean shoot apical meristem diameter ± standard error (see Methods).
Age indicates the days after germination plants were collected.
n indicates the number of measurements used to determine size.
The mean meristem diameter for clv2-1 is significantly different from that of clv2-1 cna-1 plants (t test, P < 0.01).
The mean meristem diameter for clv3-2 is significantly different from that of clv3-2 cna-1/CNA plants (P = 0.03), clv3-2 cna-1 plants (t test, P < 0.01), and clv3-2 cna-2 plants (P < 0.01).
The mean meristem diameter for clv1-7 is significantly different from that of clv1-7 cna-1 plants (t test, P < 0.01).
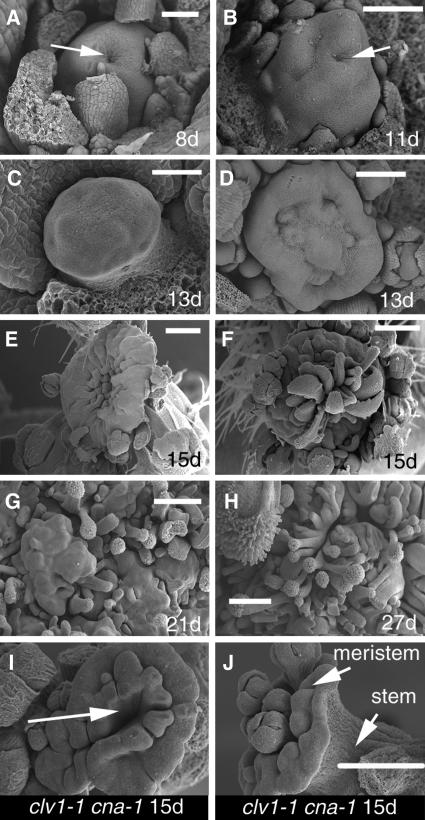
Figure 1.
Apex Differentiation in clv cna-1 Meristems.
Scanning electron micrographs of clv3-2 cna-1 ([A] to [H]) and clv1-1 cna-1 ([I] and [J]) shoot apices. Bars = 50 μm in (A), 100 μm in (B) to (D), and 250 μm in (E) to (J). (I) and (J) are shown at the same magnification.
(A) Apex from an 8-d-old clv3-2 cna-1 plant. Arrow indicates central defect in the meristem.
(B) Apex from an 11-d-old clv3-2 cna-1 plant. Arrow indicates central defect in the meristem.
(C) Apex from a 13-d-old clv3-2 cna-1 plant. Note the lack of organ primordia around the periphery.
(D) Apex from a 13-d-old clv3-2 cna-1 plant. Note the tissue buckling across the center.
(E) Apex from a 15-d-old clv3-2 cna-1 plant. Note the lack of younger primordia around most of the periphery.
(F) Apex from a 15-d-old clv3-2 cna-1 plant. Note the large carpeloid organs formed in the center of the meristem.
(G) A portion of the apex from a 21-d-old clv3-2 cna-1 plant. Note the extensive initiation of filamentous organs with stigmatic tissues at their tips.
(H) A portion of the apex from a 27-d-old clv3-2 cna-1 plant. Note the absence of any meristem-like region.
(I) Apex from a 15-d-old clv1-1 cna-1 plant. Note the ectopic organs in the center of the meristem (arrow).
(J) Apex from a 15-d-old clv1-1 cna-1 plant. Note the ectopic flowers in the center of the meristem and the lack of primordia around the periphery.
The cna-1 double mutants with the strong clv alleles clv1-4, clv1-8, clv3-1, and clv3-2 exhibited the most dramatic phenotypes, and these double mutants were analyzed in the greatest detail. In contrast with other backgrounds, cna-1 was incompletely dominant and displayed dominant-negative characteristics in the phenotypically stronger clv1-4, clv1-8, clv3-1, and clv3-2 backgrounds (Tables 2 and 3; see Supplemental Table 2 online). The clv cna-1 double mutants with these clv alleles developed shoot meristems that were enlarged in comparison with clv meristems, even at early stages of vegetative development (see Supplemental Figure 2 online; Figures 1A to 1C and 2A to 2F, Tables 2 and 3). Early in development, clv3-2 cna-1 plants had visible, enlarged meristems and also occasionally lacked identifiable young organ primordia around portions of the periphery of the shoot meristem (Figures 1C, 1E, and 2A to 2D). This loss-of-organogenesis phenotype exhibited strong variation in expressivity early in development, although at later stages we consistently observed apices with older primordia but not younger primordia (see Supplemental Figure 2 online; Figures 1 and 2). The loss of organ formation was not always uniform around the periphery of the shoot meristem (Figures 1 and 2). Moreover, a plant that had initiated no or few leaves during vegetative development could later initiate flower primordia (Figures 2B to 2D).
Table 3.
The cna-1 Allele Is Dominant-Negative
| Genotype | Shoot Meristem Size (μm)a | Ring Meristemb | Loss of Organogenesisc | nd |
|---|---|---|---|---|
| clv3-2 | 209 ± 21 | 0% | 0% | 20 |
| clv3-2 cna-2/+ | 228 ± 17 | 0% | 0% | 22 |
| clv3-2 cna-1/+ | 266 ± 23 | 22% | 0% | 18 |
| clv3-2 cna-2/cna-2 | 260 ± 17 | 17% | 0% | 24 |
| clv3-2 cna-1/cna-2 | 441 ± 20 | 100% | 0% | 18 |
| clv3-2 cna-1/cna-1 | 478 ± 19 | 91% | 36% | 22 |
Values represent the mean shoot apical meristem diameter ± standard error of plants collected simultaneously at the onset of bolting (see Methods).
The percentage of plants showing a distinct ring shape to the meristem.
The percentage of plants showing a severe loss of organogenesis.
n indicates the number of measurements used to determine size.
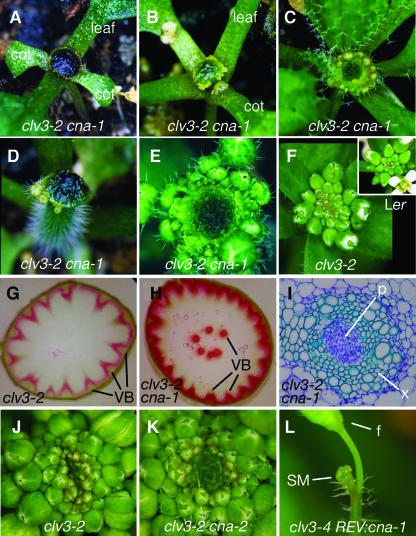
Figure 2.
Effects of cna Mutations on Organogenesis and Vascular Development.
(A) to (E) clv3-2 cna-1 apices shortly after the transition to flower. Note the variable expressivity in terms of the numbers of leaf and flower organs initiated. cot, cotyledon.
(F) clv3-2 apex shortly after the transition to flower, with the full complement of leaves and flower primordia apparent. Wild-type Ler apex shortly after the transition to flowering is shown in the inset.
(G) Phloroglucinol-stained cross section of clv3-2 inflorescence stem. Note the large number of peripherally located bundles. VB, vascular bundles.
(H) Phloroglucinol-stained cross section of clv3-2 cna-1 inflorescence stem. Note of the presence of ectopic bundles in the center of the stem. VB, vascular bundle.
(I) High-magnification view of a toluidine-blue stained section of an ectopic internal bundle from a clv3-2 cna-1 stem. Note the apparent inside-out orientation of the ectopic bundles. x, xylem; p, phloem.
(J) Apex of a clv3-2 mutant.
(K) Apex of a clv3-2 cna-2 double mutant.
(L) clv3-4 single mutant harboring the REV:cna-1 transgene. Note the lack of organ primordia other than a single flower (f). SM, shoot meristem.
Along with the loss of organogenesis in clv3-2 cna-1 plants, we also observed that the entire apices of clv3-2 cna-1 plants eventually differentiated into organs. Subtle changes in the topography of the shoot apical meristems of clv3-2 cna-1 plants ∼8 d old were observed by scanning electron microscopy, with an indentation appearing in the center of the meristem (Figures 1A and 1B). Starting at approximately day 15, carpeloid and filamentous organs often formed in the center of the clv3-2 cna-1 meristems (Figures 1F to 1H), leaving a ring of more meristem-like tissue. clv3-2 cna-1 apices eventually differentiated across the entire apical region (Figures 1F to 1J).
Key regulators of meristem development often affect both shoot and flower meristem function. However, we detected no consistent effect of cna mutations on flower meristem development or flower organ number either in cna single mutants or in combination with clv3-2 (Table 4). clv cna lateral shoot meristems recapitulated the defects observed in the clv cna shoot apical meristems, albeit to a lesser degree (data not shown).
Table 4.
Effects of cna-1 on Floral Organ Number in clv3-2 Mutants
| Genotype | Carpels per Flowera | Pb |
|---|---|---|
| clv3-2 | 5.66 ± 0.11 (n = 59) | – |
| clv3-2 cna-1 | 5.34 ± 0.12 (n = 67) | 0.04 |
| clv3-2 cna-2 | 5.79 ± 0.13 (n = 62) | 0.44 |
Values represent the mean number of carpels per flower ± standard error. The total number of flowers scored for each genotype is indicated in parentheses (n). Only the first 10 flowers of any given plant were included in the analysis.
The probability associated with differences in the mean carpel numbers for clv3-2 and clv3-2 cna lines was assessed using the t test.
clv cna plants often developed enlarged inflorescence stems. To determine if defects in stem vascular patterning were associated with this phenotype, phloroglucinol- and toluidine blue–stained cross sections of clv3-2 cna-1 inflorescence stems were compared with those from clv3-2 single mutants. Although clv3-2 stems have an increased number of vascular bundles compared with wild-type plants, all of these bundles are located around the periphery of the stem as is observed in the wild type (Figure 2G; data not shown). clv3-2 cna-1 plants, on the other hand, form ectopic vascular bundles in the center of the stem (Figure 2H). The clv3-2 cna-1 ectopic bundles exhibited an apparent inside-out morphology, with xylem surrounding phloem (Figure 2I). The ectopic bundles would occasionally form a second ring of vascular bundles in the center of the stem (Figure 2H).
In summary, the cna-1 mutation led to severe disruptions in meristem size, organogenesis, and stem cell specification, particularly in genetic backgrounds lacking CLV signaling. There was variable expressivity of Cna− phenotypes among clv cna-1 plants. The variable Cna− expressivity was consistently observed in crosses to several genetic ecotypes and among all isolates of such crosses, suggesting it was not the result of segregating genetic polymorphisms (data not shown).
wus and stm Are Epistatic to cna-1
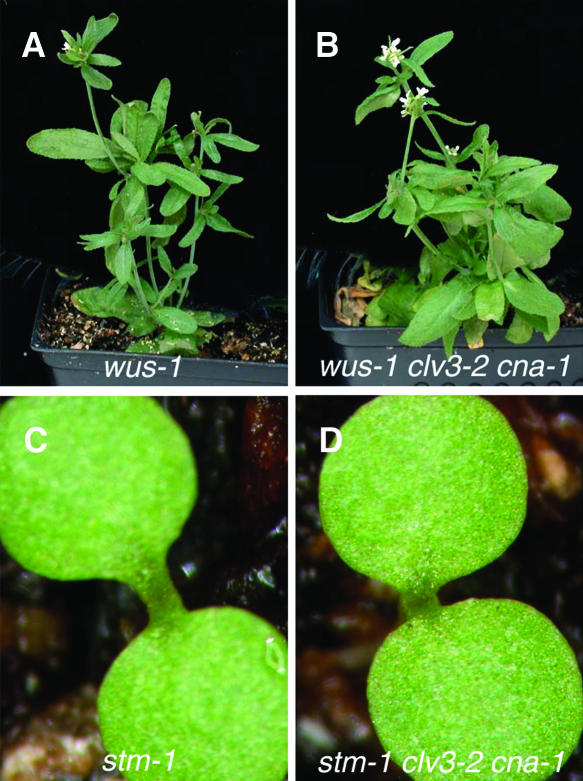
WUS and STM are key regulators of meristem development, but exhibit different genetic interactions with clv mutants. wus is epistatic to clv (Laux et al., 1996; Schoof et al., 2000; Yu et al., 2003), and stm is additive with clv (Clark et al., 1996). Genetic interactions among cna-1, stm, and wus were investigated to refine the placement of CNA within the hierarchy of genes regulating meristem development. Because cna-1 induced the most dramatic phenotypic effects in the clv3-2 background, we used the clv3-2 background to assay these genetic interactions. clv3-2 cna-1 wus-1 triple mutant plants appeared identical to clv3-2 wus-1 and wus-1 plants in terms of overall plant morphology and lack of meristem activity (Figures 3A and 3B; data not shown), indicating that wus-1 was epistatic to cna-1 in a clv3-2 background.
Figure 3.
wus and stm Are Epistatic to clv3-2 cna-1.
(A) wus-1 single mutant.
(B) wus-1 clv3-2 cna-1 triple mutant.
(C) stm-1 single mutant.
(D) stm-1 clv3-2 cna-1 triple mutant.
We examined genetic interactions between SHOOTMERISTEMLESS (STM) and CNA. STM is a member of the KNOTTED-like homeobox gene family (Long et al., 1996), thought to be important for maintaining cells in an undifferentiated state by repressing differentiation factors (Byrne et al., 2000, 2002). Strong loss-of-function stm mutants fail to establish the embryonic shoot meristem and lack postembryonic organ formation (Barton and Poethig, 1993; Endrizzi et al., 1996). Whereas clv mutants partially repress this phenotype (Clark et al., 1996), clv3-2 cna-1 stm-1 mutants were identical in phenotype to stm-1 single mutants (Figures 3C and 3D), indicating that stm-1 is epistatic to clv3-2 cna-1.
Identification of CNA
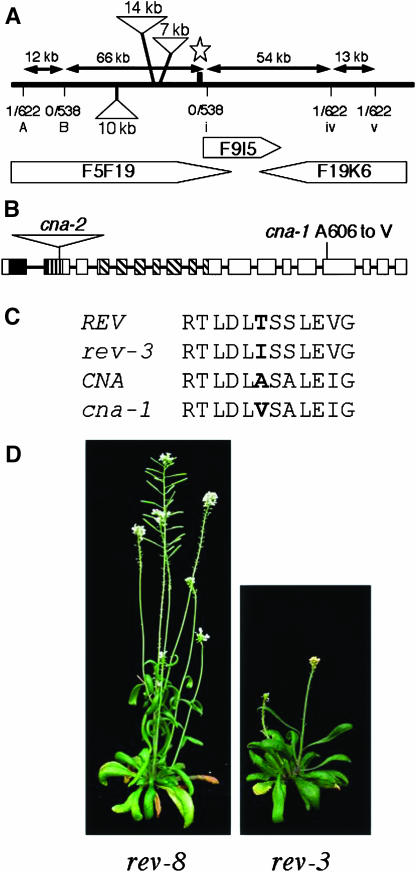
We used a map-based cloning strategy to isolate the CNA gene. CNA was mapped to chromosome 1, within a 10-centimorgan region delimited by simple sequence length polymorphism markers ciw1 and nga128 (Bell and Ecker, 1994; Lukowitz et al., 2000). Using new cleaved-amplified polymorphic sequence (CAPS) markers, the location of CNA was narrowed down to a region spanning 162 kb (Figure 4A). Among the annotated genes in the 162-kb region, we noted At1g52150, which encodes a HD-Zip III putative transcription factor, previously referred to as ATHB15 (Baima et al., 2001; Ohashi-Ito and Fukuda, 2003). A nucleotide change resulting in an Ala-to-Val amino acid substitution at residue 606 (A606V) was discovered at this locus in the cna-1 genome (Figure 4B). This position is within a domain of unknown function conserved within the gene family (Figure 4C; data not shown).
Figure 4.
Map-Based Isolation of the CNA Gene.
(A) The thick line represents a portion of chromosome I, and the sequenced BAC clones in the region are represented as pentagons. Positions of molecular markers used in fine mapping during the identification of the CNA locus (star) are indicated by vertical lines below the chromosome (see Supplemental Table 1 online). The double-headed arrows show the distances between markers in the Landsberg ecotype, and the fractions report the numbers of recombinant chromosomes out of the total number screened in the mapping population. The three triangles indicate the locations of large blocks of sequences present in the Col ecotype but absent in Ler that appear to be associated with transposon insertions.
(B) Schematic diagram of the CNA gene with the positions of the cna-1 and cna-2 mutations indicated. Protein-coding exons are indicated by boxes, and introns are indicated by lines. The regions corresponding to the homeodomain, Leu zipper, and START domain are denoted by the solid, vertically hatched, and diagonally hatched boxes, respectively.
(C) Alignment of the sequences around the mutations sites in rev-3 and cna-1. The positions of mutated residues are indicated in bold.
(D) A comparison of the putative null allele rev-8 and the missense mutant rev-3, which exhibits a more severe flower meristem defect.
To confirm that At1g52150 was indeed CNA, we obtained an allele in the Columbia background, cna-2, with a T-DNA insertion in the second exon of At1g52150 (Figure 4B; Prigge et al., 2005). cna-2 single mutant plants appeared superficially wild type (Prigge et al., 2005). The clv3-2 cna-2 double mutants exhibited a phenotype similar to, although less severe than, that of clv3-2 cna-1 plants (Figure 2K, Tables 2 and 3). When assayed for RNA accumulation by RT-PCR, we found similar accumulation of CNA and cna-1 transcripts, but no accumulation of full-length cna-2 transcripts (see Supplemental Figure 3 online). Because cna-1 contained the entire coding sequences, yet exhibited both a stronger phenotype than cna-2 and an incompletely dominant effect in strong clv mutant backgrounds, we propose that cna-1 is dominant-negative, which is further supported by cna-1/cna-2 transheterozygous phenotypes (Table 3; see Supplemental Table 3 online). Interestingly, the rev-3 allele has a Thr-to-Ile amino acid substitution (T608I) at the position equivalent to that of the A606V substitution in cna-1 (Figure 4C). Some phenotypes of rev-3 plants are stronger than that of rev-6 or rev-8 putative null alleles (Talbert et al., 1995; Otsuga et al., 2001; Figure 4D), suggesting that rev-3 is also likely a dominant-negative allele.
The dominant-negative character of cna-1 complicated efforts to confirm gene isolation by complementation. We chose to express CNA under the previously characterized regulatory elements from the closely related REV gene (PREV) (Zhong and Ye, 1999) because REV is expressed in similar tissues to CNA (see below) and because we had not identified the CNA regulatory sequences. When PREV:cna-1 was transformed into clv3-4, 22 of 77 independent lines exhibited Cna− phenotypes, including the loss of organogenesis and formation of organs in the center of the meristem (Figure 2L). For PREV:CNA transformed into clv3-4, only 1 of 45 independent lines displayed Cna− phenotypes (data not shown). Similar results were observed with smaller numbers of lines with these constructs transformed into clv3-1 and clv3-2, and PREV:CNA was unable to complement either clv3-1 cna-1 or clv3-2 cna-1 (data not shown).
We assessed CNA expression by RNA in situ hybridization (see Supplemental Figure 7 online). Consistent with separate studies, we observed that during embryo development CNA expression is strongest in the developing vascular elements and the adaxial portion of cotyledons (Prigge et al., 2005; data not shown). Ohashi-Ito and Fukuda (2003) have also shown that CNA/ATHB15 is expressed in developing vascular elements using a PCNA/ATHB15:β-glucuronidase (GUS) reporter construct. In mature plants, CNA continues to be strongly expressed in the developing vascular elements. CNA is expressed diffusely in young stage flower meristems but becomes enriched in developing stamens and carpels around stage 8. As flowers matured, we observed strong expression of CNA within developing ovules, from the earliest stage of ovule development. CNA is also expressed within the shoot meristem (see Supplemental Figure 7 online; Prigge et al., 2005; data not shown).
The Effect of clv Mutations on WUS and CLV3 Expression
WUS expression is thought to be intimately associated with the presence of stem cells and considered to be an excellent indicator of the capacity of the meristem to maintain a stem cell population. CLV3 is currently thought to be the best marker for stem cell identity (see Introduction). To better understand the origin of defects occurring in clv cna meristems, we assessed the expression of these two key markers of meristem function in clv and clv cna double mutants. Previous work has shown that WUS is normally expressed in a basal subset of cells in the L3 layer, excluding the outermost, or most apical, L3 cells (Mayer et al., 1998; Schoof et al., 2000). This domain of WUS expression expands laterally and apically into the uppermost L3 cell layers in clv3-2 and clv1-4 meristems (Brand et al., 2000; Schoof et al., 2000).
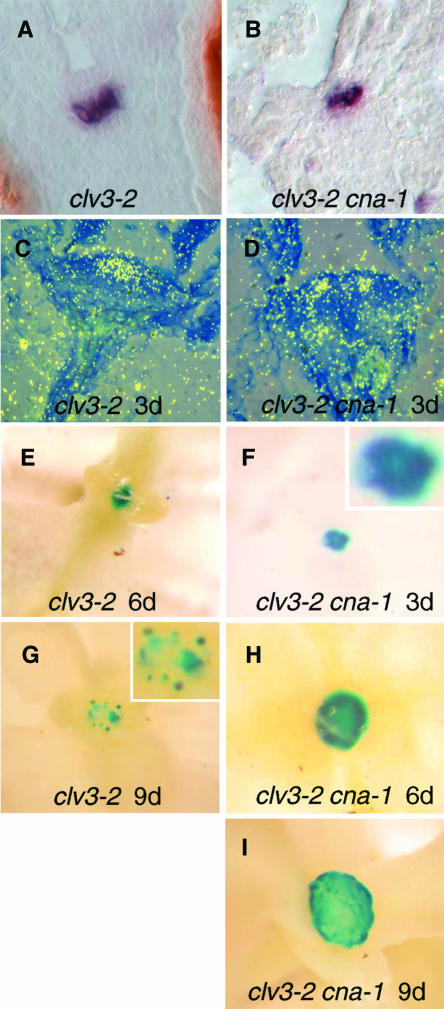
We used PWUS:GUS and PCLV3:GUS transgenic plant lines that have already been characterized and shown to faithfully reflect endogenous gene expression (Lenhard et al., 2002). The characterized transgenic plant lines were crossed to clv3-2 and clv3-2 cna-1 plants to assess GUS accumulation in the mutant backgrounds. As a control, we first assayed GUS accumulation in PWUS:GUS clv3-2 plants. We were surprised to observe a variable, but often dramatic loss of GUS signal in the shoot apical meristems of clv3-2 seedlings (Figure 5G). No GUS signal was detected in the inflorescence shoot apical meristems of clv3-2 plants (Figures 6A and 6C). The expression of PWUS:GUS continued to be quite strong in both the floral meristems and young lateral shoot meristems of clv3-2 plants, indicating that the loss of expression was specific to the shoot apical meristem. Equally surprising, we observed a similar downregulation of PCLV3:GUS expression in clv3-2 shoot apical meristems, although it occurred later than the downregulation of PWUS:GUS expression (Figures 7A to 7D). Thus, regulatory sequences from two different genes behaved differently in a clv3-2 background from descriptions in previously published analyses of these genes from several labs (Fletcher et al., 1999; Schoof et al., 2000; Brand et al., 2000, 2002; Lenhard and Laux, 2003). This finding is at odds with the inference that WUS is necessary for specifying stem cell fate because clv3-2 shoot apical meristems continue to act as if they contained stem cells and generate organs for weeks after downregulation of PWUS:GUS and PCLV3:GUS transgenes. Furthermore, all of the cells of clv3-2 apices retain morphological characteristics of stem cells and do not display differentiation (Clark et al., 1993, 1995). Note that we did not observe any obvious loss of PWUS:GUS or PCLV3:GUS signal from the shoot apical meristems of wild-type plants (Figures 6L and 7I).
Figure 5.
WUS Expression in Vegetative Meristems of clv3-2 and clv3-2 cna-1 Plants.
(A) and (B) WUS mRNA was detected in clv3-2 (A) and clv3-2 cna-1 (B) torpedo-stage embryos using dioxygenin-labeled riboprobes. WUS signal is indicated by indigo staining.
(C) and (D) WUS mRNA was detected in 3-d-old clv3-2 (C) and clv3-2 cna-1 (D) seedlings using 35S-labeled riboprobes. WUS mRNA signal is shown in yellow. Note the gap in WUS signal in the center of the clv3-2 cna-1 apex.
(E) to (I) Detection of PWUS:GUS expression in clv3-2 ([E] and [G]) and clv3-2 cna-1 ([F], [H], and [I]) seedlings on day 3 (F), day 6 ([E] and [H]), and day 9 ([G] and [I]). The blue color indicates tissues with GUS activity. The insets in (F) and (G) show higher magnification views of the meristems. Note the loss of PWUS:GUS signal from the day 9 clv3-2 sample and the loss of central PWUS:GUS signal from the clv3-2 cna-1 plants as early as day 3.
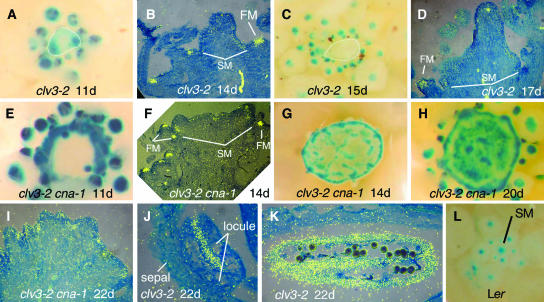
Figure 6.
WUS Expression in Inflorescences of clv3-2 and clv3-2 cna-1 Plants.
Detection of PWUS:GUS expression ([A], [C], [E], [G], [H], and [L]). Photographs were taken from above the apices, showing GUS activity in blue. Detection of WUS mRNA using 35S-labeled probes in longitudinal sections ([B], [D], [F], [I], [J], and [K]). WUS mRNA signal is shown in yellow. SM, shoot meristem; FM, floral meristem.
(A) The apex of an 11-d-old clv3-2 plant. The meristem is outlined in white. Note the lack of GUS signal in most of the shoot apical meristem.
(B) The apex of a 14-d-old clv3-2 plant. Note the robust expression within flower meristems, whereas the shoot apical meristem lacks robust signal.
(C) The apex of a 15-d-old clv3-2 plant. The meristem is outlined in white.
(D) The apex of a 17-d-old clv3-2 plant.
(E) The apex of an 11-d-old clv3-2 cna-1 plant. Note the robust GUS signal around the periphery of the meristem and within the flower meristems both outside and inside the ring. The signal at the bottom of the ring is obscured by a leaf.
(F) The apex of a 14-d-old clv3-2 cna-1 plant. Note the robust expression within flower meristems, whereas the shoot apical meristem lacks robust signal.
(G) The apex of a 14-d-old clv3-2 cna-1 plant. Note the ring of GUS signal around the periphery.
(H) The apex of a 20-d-old clv3-2 cna-1 plant. Note the increase in GUS signal across the center of the apex associated with differentiation.
(I) The apex of a 22-d-old clv3-2 cna-1 plant. Note the pockets of WUS signal.
(J) Developing anther of a 22-d-old clv3-2 plant.
(K) Older anther of a 22-d-old clv3-2 plant.
(L) The apex of a wild-type Ler plant shortly after the transition to flowering.
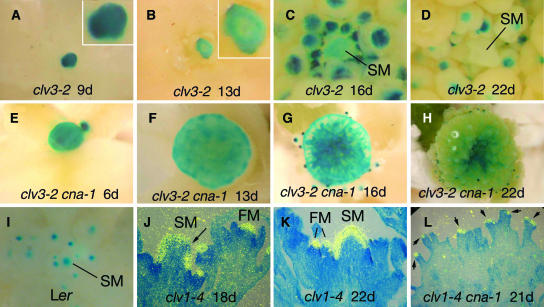
Figure 7.
CLV3 Expression in clv3-2 and clv3-2 cna-1 Apices.
Detection of PCLV3:GUS expression ([A] to [I]). Photographs were taken from above the apices, showing GUS activity in blue. Detection of CLV3 mRNA using 35S-labeled riboprobes in longitudinal sections ([J] to [L]). CLV3 signal is shown in yellow. Insets in (A) and (B) show higher-magnification views of the shoot apical meristem. SM, shoot apical meristem. FM, flower meristem.
(A) The apex of a 9-d-old clv3-2 plant.
(B) The apex of a 13-d-old clv3-2 plant. Note that only weak peripheral GUS signal remains.
(C) The apex of a 16-d-old clv3-2 plant.
(D) The apex of a 22-d-old clv3-2 plant. Note that the signal is restricted to flower meristems.
(E) The apex of a 6-d-old clv3-2 cna-1 plant.
(F) The apex of a 13-d-old clv3-2 cna-1 plant.
(G) The apex of a 16-d-old clv3-2 cna-1 plant. Note the relative paucity of flower meristems around the periphery.
(H) The apex of a 22-d-old clv3-2 cna-1 plant. Note the sustained GUS signal in the apex despite the onset of differentiation.
(I) The apex of a wild-type Ler plant shortly after the transition to flowering.
(J) The apex of an 18-d-old clv1-4 plant. Note the patch of the shoot apical meristem lacking signal (arrow).
(K) The apex of a 22-d-old clv1-4 plant.
(L) The apex of a 21-d-old clv1-4 cna-1 plant. Note the robust CLV3 signal in the filamentous structures (arrows).
Given that artifacts can arise from promoter-GUS analysis, we repeated analysis of WUS and CLV3 expression within clv mutants by RNA in situ hybridization. Both WUS and CLV3 antisense probes were generated and were hybridized to clv3-2 and clv1-4 tissue, respectively. While the phenotype of clv1-4 mutants is weaker than that of clv3-2 mutants, we could not look at CLV3 expression in clv3-2 plants because the clv3-2 allele contains a chromosomal rearrangement within the coding sequence (Fletcher et al., 1999). At early stages of shoot apical meristem development, and within lateral shoot and flower meristems at all stages, WUS and CLV3 expression matched that of previous reports (Figures 5 to 7). However, within the clv3-2 inflorescence shoot apical meristem, WUS expression was dramatically reduced and patchy (Figures 6B and 6D). This loss of signal was not the result of experimental anomalies because floral and lateral shoot meristems within the same tissue sections exhibited a strong signal. Expression of CLV3 within the shoot apical meristems of clv1-4 plants was not dramatically reduced, although small regions of reduced or absent expression were observed (Figures 7J and 7K). The difference between PCLV3:GUS and CLV3 RNA in situ hybridization findings could conceivably result from the smaller meristem sizes of clv1-4 compared with clv3-2 or from artifacts of PCLV3:GUS transgene activity. In summary, both GUS analysis and RNA in situ hybridizations indicate that WUS and possibly CLV3 expression is lost or reduced from strong clv mutants, in contradiction to previous reports.
Whereas WUS was not consistently expressed in the region underlying stem cells in clv3-2 mutants, WUS was expressed in several differentiated cells. We detected strong WUS signal by RNA in situ hybridization in developing ovules (data not shown), consistent with previously published reports (Groß-Hardt et al., 2002). In addition, we detected WUS signal in the developing anther. The earliest anther-specific expression of WUS was detected between locules, but later in stamen development WUS was detected throughout peripheral anther cells (Figures 6J and 6K).
The Effects of cna-1 on WUS Expression
Despite the unexpected finding that WUS and CLV3 were not appropriate markers for stem cell identity and meristem function within older clv shoot meristems, these genes still provided us the best opportunity to assess clv cna meristem defects, especially at early stages of development. To this end, we crossed transgenic lines carrying the PWUS:GUS or PCLV3:GUS construct into the clv3-2 cna-1 background, and we used antisense WUS and CLV3 probes on clv3-2 cna-1 tissue in RNA in situ hybridization experiments.
Results from RNA in situ hybridization on embryos indicated that WUS expression was unchanged in clv3-2 cna-1 embryos as compared with clv3-2 embryos as late as the torpedo stage (Figures 5A and 5B). Within young seedlings, we detected a central gap in WUS expression in clv3-2 cna-1 shoot apical meristems as early as 3 d after germination (Figure 5D), leaving a ring of WUS expression around the periphery of clv3-2 cna-1 apices (Figure 5F).
WUS expression continued to be rapidly lost from clv3-2 cna-1 shoot apices as they proceeded through development (Figures 5 and 6). There was variability in WUS expression patterns and levels, which presumably correlated with the variability of Cna− phenotypes. A ring of WUS expression often persisted on the periphery, even as the rest of the meristem had undergone differentiation (Figures 6E and 6G).
Remarkably, WUS expression returned to clv3-2 cna-1 apices once extensive differentiation occurred. Within clv3-2 cna-1 inflorescence meristems, small pockets of PWUS:GUS expression were observed throughout the apex in what appeared to be differentiated regions, in addition to the expression that persisted on the periphery of the meristem (Figures 6G and 6H). We continued to observe WUS expression beyond this stage (data not shown).
The Effects of cna-1 on CLV3 Expression
CLV3 is predominantly expressed in the L1 and L2 layers in the center of wild-type meristems (Fletcher et al., 1999; Schoof et al., 2000). As assessed by RNA in situ hybridizations, CLV3 expression was readily detected in clv1-4 meristems (Figures 7J and 7K). In clv3-2 cna-1 double mutants ∼12 d old and older, the expression of CLV3 was quite low and patchy (Figures 7F and 7G). In clv1-4 cna-1 mutants ∼18 d old and older, small pockets of robust CLV3 expression were observed, especially at the tips of many of the filamentous structures generated during the process of indiscriminate differentiation of the meristem (Figure 7L).
When PCLV3:GUS activity was assayed in clv3-2 cna-1 plants, there was a general trend toward downregulation of CLV3 expression, although both the pattern and loss of expression was more complex than that of PWUS:GUS. Similar to WUS expression, PCLV3:GUS was first reduced in the center of clv3-2 cna-1 vegetative shoot meristems, although this reduction followed the loss of WUS expression by several days (Figures 6 and 7). PCLV3:GUS was expressed at low, patchy levels throughout the clv3-2 cna-1 apex as it underwent differentiation (Figure 7). Older clv3-2 cna-1 plants exhibited sustained, and later increased, PCLV3:GUS expression, especially within the center of the apex, as differentiation proceeded (Figure 7).
The Overlap of CNA Function within the HD-Zip III Gene Family
Genetic data suggest the cna-1 protein is dominant-negative and could be interfering with the activity of a functionally redundant protein(s). Because ATHB8 is the most closely related gene to CNA within the Arabidopsis genome, we hypothesized that ATHB8 might act to replace some of the CNA activity in the clv3-2 cna-2 background. To investigate this possibility, we crossed clv3-2 cna-2 to athb8-12, a T-DNA insertion line in which the T-DNA is located in an intron just upstream of the translation start site (Prigge et al., 2005). The resulting clv3-2 cna-2 athb8-12 triple mutants did not exhibit enhanced Cna− phenotypes, suggesting that ATHB8 does not function in parallel with CNA in a clv3-2 background (data not shown).
We have observed that the cna-2 phb-13 phv-11 triple null mutant displays an intermediate Clv− phenotype (Prigge et al., 2005). This suggested that CNA, PHB, and PHV coordinately regulate meristem development. To test this idea of parallel function further and to determine whether the incompletely dominant nature of the cna-1 allele is due to interference with the normal function of the PHB and PHV genes, we generated double and triple mutant combinations of phb and phv null alleles with cna-1. We then compared the phenotypes of the cna-1 phb-13 and cna-2 phb-13 double mutants and the cna-1 phb-13 phv-11 and cna-2 phb-13 phv-11 triple mutants. Because the cna-2, phb-13, and phv-11 alleles were all in the Columbia (Col) background, the phenotypes were analyzed after the cna-1 allele had been introgressed into the Col background by two crosses.
Whereas cna-2 phb double mutants exhibit very weak Clv− phenotypes (Prigge et al., 2005), all cna-1 phb plants and most cna-1/+ phb plants exhibited a brief developmental arrest after germination and expansion of the first two true leaves (Figures 8B and 8C). Organogenesis in the arrested plants resumed 10 to 14 d after germination. Subsequently, the cna-1 phb apices exhibited a stronger Clv− phenotype than cna-2 phb apices, developing fasciated stems that frequently bifurcated and flowers with extra carpels. This suggests that cna-1 may interfere with PHV function because similar phenotypes are seen for cna-2 phb phv apices (data not shown; Figure 8I; Prigge et al., 2005).
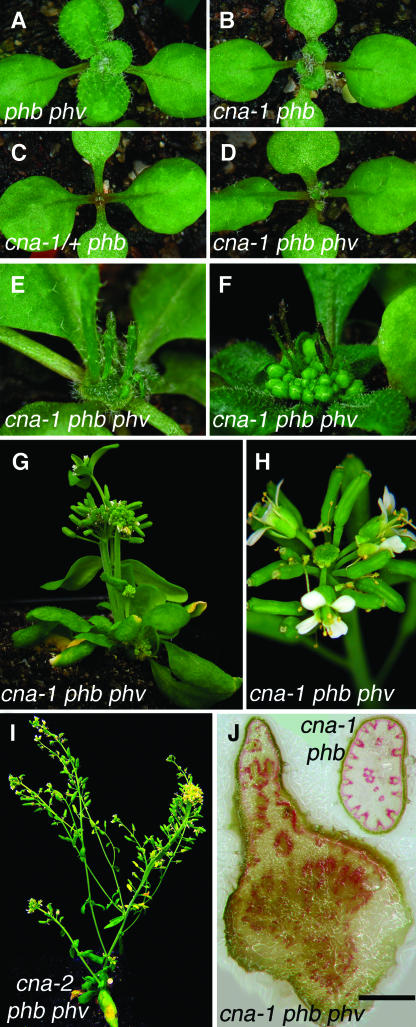
Figure 8.
Phenotypes of cna-1 phb phv Plants.
(A) A 13-d-old phb phv plant was identical in phenotype to wild-type plants.
(B) A 13-d-old cna-1 phb plant that underwent only a short developmental arrest.
(C) A 13-d-old cna-1/+ phb plant with a strong defect in organogenesis.
(D) A 13-d-old cna-1 phb phv triple mutant that arrested after the production of two leaves, but then recovered.
(E) A 19-d-old cna-1 phb phv seedling with a mass of undifferentiated cells and radially symmetric organs coming from the midst of the meristem.
(F) A cna-1 phb phv seedling at day 34. Note the entire apex has differentiated into flowers, but the inflorescence stem did not elongate.
(G) The plant in (E) at day 54, with axillary meristems that emerged and elongated relatively normally.
(H) A secondary inflorescence from a 65-d-old cna-1 phb phv plant exhibiting a Cna− phenotype.
(I) A 44-d-old cna-2 phb phv plant. Note the much weaker phenotypes compared with the cna-1 triple mutant.
(J) Phloroglucinol-stained hand sections of cna-1 phb phv and cna-1 phb stems. The unelongated primary inflorescence stems of 54-d-old triple mutants had an unorganized pattern of vascular and lignified tissue (stained pink). The cna-1 phb double mutants had relatively normal patterns of lignified cells around the stem periphery, but ectopic vascular bundles appeared in the center of the stem.
cna-1 phb plants also heterozygous or homozygous for phv exhibited early arrest and later recovery as well, with cna-1 phb phv individuals among the last to recover (Figure 8D; data not shown). For the shoots that developed after recovery from arrest, the presence of the cna-1 allele enhanced the phb and phv alleles to a much greater extent than the likely null cna-2 allele. Both cna-1 phb phv/+ and cna-1 phb phv/phv plants developed massively enlarged shoot meristems (Figure 8E; data not shown) and, in the case of cna-1 phb phv/phv plants, radially symmetric leaves in the middle of the massively enlarged meristem (Figures 8E and 8F). When cna phb phv triple mutants began producing flowers, very little stem elongation was apparent and the stems were generally wider than they were tall with very disorganized vascular elements (Figures 8F and 8J). Similar to clv cna mutants, cna-1 phb phv plants exhibited a rapid and total differentiation of the entire massive apex into flowers (Figure 8F). Eventually, secondary inflorescences escaped that elongated normally and developed flowers that were similar in appearance to those of cna-2 phb phv triple mutants (Figures 8G and 8I; Prigge et al., 2005). Like cna-1 clv apices, the cna-1 phb phv primary and secondary meristems frequently exhibited ring-like corona meristems and eventually ceased organ production (Figure 8H; data not shown). cna-1 displayed dominant-negative characteristics in the phb phv background (see Supplemental Table 3 online).
DISCUSSION
We have identified CNA as a novel regulator of meristem development. In a clv background, mutations within the CNA gene led to dramatic defects in meristem size, organogenesis, stem cell maintenance, and the expression of WUS and CLV3. We propose that CNA acts in parallel with the CLV pathway to promote organ formation. We show that CNA corresponds to ATHB15, a member of the HD-Zip III gene family, and that CNA is expressed strongly within developing vascular elements and diffusely within developing meristems and organ primordia. The putative dominant-negative cna-1 allele appears to interfere with the function of both HD-Zip III family members as well as other genes. While investigating CLV3 and WUS expression within clv and clv cna plants, we observed that WUS may be neither necessary nor sufficient for stem cell specification and that CLV3 expression may not be a marker for stem cell identity, in contradiction to the existing paradigm in the field.
cna Alleles and Genetics
The observation that cna-1 enhanced the phenotypes of clv1, clv2, and clv3 mutants, including the clv3-2 null allele, suggests that CNA functions in a pathway parallel to that of the CLV loci. The requirement for CNA increases as CLV signaling is lost, such that the least apparent cna effects were observed in otherwise wild-type backgrounds, and the most dramatic phenotypes were observed within strong clv mutant backgrounds.
We conclude that the cna-1 missense allele is dominant-negative for several reasons. First, cna-1 exhibited a stronger phenotype than the likely null cna-2 allele. Second, PREV:CNA transformed into clv3-2 cna-1 did not complement the mutant phenotypes, whereas PREV:cna-1, but not PREV:CNA, transformed into clv3-2 CNA and clv3-4 CNA backgrounds recreated the Cna− phenotype. This suggests that cna-1 interferes with the activity of CNA, in addition to other proteins. Third, the missense mutation within cna-1 (A608 to V) is located in a position analogous to the missense mutation within rev-3 (T608 to I), which itself exhibits dominant-negative characteristics. Interestingly, although cna-1 and cna-2 are normally recessive, cna-1 appears incompletely dominant in the strongest clv backgrounds.
We sought to assign a place for CNA within the network of meristem regulators and found that both wus and stm are epistatic to clv3-2 cna-1. WUS and STM are both required for the initiation of shoot meristems. Thus, these epistatic relationships may reflect a role for CNA in meristem maintenance, as opposed to meristem initiation. Consistent with this, we see no effect of cna-1 on the initiation of embryonic shoot meristems, lateral shoot meristems, nor flower meristems.
The Primary Defect in clv3-2 cna-1 Apices
A key unresolved issue is the primary cause of the complex meristem defects in clv3-2 cna-1 apices. Although the primary defect(s) in the Cna− phenotype is not entirely clear, we propose that CNA promotes organogenesis in parallel with the CLV pathway. This hypothesis is consistent with many observations. The cna-1 single mutant has a slightly larger meristem, and cna-1 enhances the stem cell accumulation defect of weak clv mutants. In addition, cna-1 combined with stronger clv alleles leads to the cessation of all organogenesis around the flanks of the meristem. Furthermore, we recently demonstrated that cna-2 phb phv triple mutants effectively recreate the Clv− meristem phenotype (Prigge et al., 2005). The later differentiation of Cna− apices could be explained if the loss of nascent organ primordia leads to a loss of meristem identity. Classic work has demonstrated that organ primordia influence the positioning of new primordia (Snow and Snow, 1931), as do the central cells of the meristem (Wardlaw, 1949). More recently, clear evidence of the role of polar auxin transport within the meristem in establishing sites of organogenesis has been published (Reinhardt et al., 2000, 2003; Benková et al., 2003). In light of this, we suggest that signaling between organ primordia and the center of the meristem is also essential for the maintenance of meristem identity.
cna-1 Dominant-Negative Character
CNA belongs to the HD-Zip class of proteins, first identified by Schena and Davis (1992). HD-Zip III proteins contain a region of similarity to the mammalian sterol/lipid binding domain (START domain), in addition to the N-terminal HD-Zip motif. Most of the C terminus is also well conserved among HD-Zip III proteins, but its function is unknown.
The Arabidopsis HD-Zip III genes have been implicated in the regulation of several developmental processes, including leaf polarity, meristem initiation, and vascular development (Baima et al., 1995, 2001; Talbert et al., 1995; Zhong and Ye, 1999; McConnell et al., 2001; Otsuga et al., 2001; Ohashi-Ito and Fukuda, 2003; Prigge et al., 2005).
We concluded that the cna-1 allele interferes with the function of other, likely redundant, genes. We hypothesized that other members of the HD-Zip III family were good candidates for CNA-redundant genes because extensive functional overlap has been observed between the members of this gene family in several developmental pathways (Emery et al., 2003; Prigge et al., 2005). However, rev mutants do not exhibit interactions with clv mutants similar to that of cna (Otsuga et al., 2001), and we observed no evidence of a role for ATHB8 in meristem development. Interestingly, we have shown in a separate study that cna, phb, and phv null alleles exhibit functional overlap in meristem development, resulting in a Clv− phenotype when all three genes are mutated (Prigge et al., 2005). This suggests that these three genes have functional overlap within meristem development and that cna-1 may interfere with the function of PHV and PHB. Indeed, genetic interaction studies of cna-1 with phb and phv mutants indicated that cna-1 interferes with PHV function, as well as revealing the ability of these mutations to recreate the Cna− phenotype in a CLV+ genetic background. Because the cna-1 phb phv triple mutant also exhibited phenotypes not seen for the cna-2 phb phv triple mutant, we predict that the cna-1 isoform affects additional genes products besides PHB and PHV.
WUS May Not Be Necessary for Stem Cell Specification
An unexpected finding that was born out of the promoter-GUS studies and corroborated by RNA in situ hybridization data was that clv3-2 shoot apical meristems lose WUS expression while maintaining stem cells and organogenesis. The loss of WUS expression in the shoot meristem was clearly evident in PWUS:GUS assays. RNA in situ hybridization experiments showed a similar downregulation of WUS, although there remained patchy and trace expression within the clv3-2 shoot apical meristems. Both GUS assays and RNA in situ hybridizations revealed that WUS expression remains robust within clv3-2 flower meristems and lateral shoot meristems, despite its apparent loss from the shoot apical meristem.
This loss of WUS expression was quite surprising given previous analyses of clv phenotypes and WUS expression. clv3-2 shoot apical meristems continue to maintain stem cells and organogenesis late into development, well past the downregulation of WUS expression. This raises the question as to how these stem cells are maintained in the absence of WUS. Genetic loss of WUS function leads to a failure of the wus mutant plants to establish stem cell populations at either shoot or flower meristems (Endrizzi et al., 1996; Laux et al., 1996; Mayer et al., 1998), indicating that WUS is necessary for stem cell specification. One explanation is that WUS is necessary for stem cell initiation but that other factors can redundantly maintain stem cell identity in the absence of WUS. Such a WUS-independent function might be performed by one of the related WOX genes (Haecker et al., 2004).
Harder to resolve is the conflict of some of our observations with previously published data of WUS expression in clv mutant backgrounds (Brand et al., 2000; Schoof et al., 2000; Lenhard and Laux, 2003). Some of the previous experiments looked at expression in meristems collected relatively early in development. Schoof and coworkers (2000) used vegetative apices and inflorescence apices with age not stated, whereas we observed the greatest WUS reduction in later developmental stages. In addition, we grew our samples under significantly higher light levels, which resulted in more robust growth and stronger Clv− phenotypes. Finally, whereas data from several previous publications revealed that WUS expression expands in clv mutants, the expression in published images appear somewhat patchy, diffuse, and at a lower levels compared with controls (Brand et al., 2000; Schoof et al., 2000). Moreover, wild-type inflorescence meristems in these publications appear to exhibit a weaker WUS signal than the vegetative or embryonic meristems.
CLV3 May Not Be a Marker for Stem Cell Function
CLV3 expression may be lost or reduced from the shoot apical meristems of clv mutants. The loss of PCLV3:GUS expression follows the downregulation observed for WUS, consistent with the idea that WUS is an activator of CLV3 expression (Schoof et al., 2000; Brand et al., 2002). Whereas CLV3 is normally expressed within what appears to be the stem cells of the shoot and floral meristems, in the clv3-2 mutant background PCLV3:GUS assays indicate that CLV3 expression is lost, even though functional stem cells are presumably maintained at the shoot apical meristem. Whereas some small patches of downregulation of CLV3 expression were observed in clv1-4 mutants as determined by RNA in situ hybridization, strong CLV3 signal was detected across the majority of the clv1-4 shoot apical meristem. This suggests that either the PCLV3:GUS assay overstates the CLV3 downregulation or that the level reduction depends on the stronger clv3-2 phenotype. CLV3 expression is retained normally in clv floral meristems and lateral shoot meristems in both assays.
WUS and CLV3 Can Be Expressed in Differentiated Tissues
WUS and CLV3 expression have been shown to be largely meristem specific (Mayer et al., 1998; Fletcher et al., 1999; Brand et al., 2000, 2002; Schoof et al., 2000; Lenhard and Laux, 2003). With the exception of WUS expression in developing ovules (Groß-Hardt et al., 2002), neither gene has been found to be expressed within differentiated tissue. We show that within the clearly differentiating apices of clv cna plants, WUS and CLV3 exhibit strong, stable pockets of expression. Given the absence of convincing markers for differentiated cells, we cannot be sure that the WUS and CLV3 expression in the clv cna apices corresponds to fully differentiated cells; however, these cells are very unlikely to be stem cells because they exhibited neither self-renewal nor organogenesis. Thus, in certain genetic backgrounds, one can observe both stem cell function in the possible absence of WUS and CLV3 expression and WUS and CLV3 expression in the apparent absence of stem cells. WUS and CLV3 expression can therefore be uncoupled from stem cell identity. Consistent with this, WUS expression is also detected within developing stamens and can be detected by RT-PCR from roots, stems, and leaves (S.-K. Song and S.E. Clark, unpublished data). A key question is why WUS and CLV3 expression within later stage clv cna apices is unable to establish stem cells and/or a meristem.
What is clear is the need to identify additional markers for stem cell identity and understand better how WUS expression in the organizing center induces stem cell identity within the shoot and flower meristems of wild-type plants.
METHODS
Plant Growth and Mutant Isolation
Plants were grown as described (Yu et al., 2003), except that 0.5 g/pot of Osmocote (Scotts, Marysville, OH) was used and that the plants for Table 3 were grown at 26°C. The isolation of the cna-1 (formerly known as pce5) mutation has been previously described (Pogany et al., 1998). A detailed description of the allele isolation and genetic constructions are presented in the supplemental data online. The isolation of the cna-2, athb8-12, phb-13, and phv-11 alleles is described elsewhere (Prigge et al., 2005).
Scanning Electron Microscopy Analysis and Meristem Size Estimates
Scanning electron microscopy was performed as described (Dievart et al., 2003). Brightness, contrast, and color balance were adjusted using Adobe Photoshop (Mountain View, CA). For cna-1 and wild-type meristem sizes, scanning electron micrographs were taken directly above the shoot meristem. A line connecting the ends of the furrow of the youngest stage 1 floral primordium was drawn. The distance from the center of this furrow line perpendicularly across the shoot meristem was determined. This procedure was repeated for the second youngest stage 1 floral primordium. We defined the size of the meristem as the average of these two measurements. For meristem sizes for all other genotypes scanning electron microscopy photographs were taken directly above the shoot meristem, and the center of the meristem was estimated. Both the longest and the shortest diameters across the meristem, going through the center, were used as measurements for each sample.
Stem Cross Sections
Hand-sectioned stems were stained in a solution of 82% ethanol, 14% hydrochloric acid, and 0.09% phloroglucinol (which stains lignin red), for approximately half an hour. Stained sections were photographed immediately using cameras attached to either a Nikon stereomicroscope (transillumination; Tokyo, Japan) or a Zeiss dissecting microscope (epi-illumination; Jena, Germany) (Ruzin, 1999). Stem sections were also fixed, embedded in paraffin, sectioned at 8 μm, and stained with toluidine blue as previously described (Yu et al., 2000).
Molecular Mapping
clv3-2 cna-1 plants were crossed to er-2 plants in the Col background. DNA was extracted from 311 individual clv3-2, clv3-2 cna-1/+, and clv3-2 cna-1/cna-1 F2 plants as described (Dellaporta et al., 1983). DNA samples were tested for linkage with published CAPS and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) and generated CAPS based on Ler and Col sequence comparison (Arabidopsis Genome Initiative, 2000; Jander et al., 2002). The F3 seeds collected from individual F2 plants were sown and phenotypically scored to verify the genotype of the F2 parents. Potential candidate genes were amplified and sequenced from clv3-2 cna-1 and Ler genomes.
Plant Transformation
The 2.3-kb promoter region upstream of the REV transcribed sequence has been previously characterized (Zhong and Ye, 1999). Construction of the pCB321-REV:CNA plasmid was described (Prigge et al., 2005). The pCB321-REV:cna-1 vector was generated by cutting out a small region of CNA cDNA from pCB321-REV:CNA with ClaI and NcoI and replacing it with the corresponding region of cDNA from cna-1 containing the cna-1 lesion. AGL1 Agrobacteria were transformed with pCB321-PREV:CNA and pCB321- PREV:cna-1. Plants were agro-transformed by vacuum infiltration (Bechtold and Pelletier, 1998; Clough and Bent, 1998).
GUS Assays
PCLV3:GUS (194.3) and PWUS:GUS (87.13) lines (Lenhard et al., 2002) were kindly provided by Thomas Laux. GUS staining and fixation were performed as described (Schoof et al., 2000).
RNA in Situ Hybridization
The full-length CNA cDNA in the pCRII vector was cut with BspEI and XhoI, treated with Klenow, and religated to delete the highly conserved HD-Zip region. This plasmid, pCRII-situCNA, was digested with XbaI and transcribed with SP6 RNA polymerase to generate antisense riboprobe. The sense riboprobe was similarly generated using SpeI and T7 RNA polymerase.
Full-length WUS cDNA, including a 51-bp poly(A) tail, was kindly provided by Thomas Laux (Mayer et al., 1998). pBSK-WUS was digested with EcoRI and SpeI, deleting the poly(A) tail, and the resulting fragment was inserted into EcoRI- and SpeI-cleaved pKUT401, a pPZP222-based binary vector (K.U. Torii and S.E. Clark, unpublished data), to generate pKUT421. To synthesize WUS antisense riboprobe, this plasmid was digested with EcoRI and transcribed with T7 RNA polymerase.
CLV3 full-length cDNA was cloned into pCR2.1 to generate pCR2.1-cCLV3. Antisense riboprobe was generated by digesting with SacI and transcribing with T7 polymerase. Sense riboprobe was generated by digesting with SpeI and transcribing with SP6 RNA Polymerase.
Tissues for RNA in situ hybridizations were fixed and embedded as described by Clark et al. (1993). A radioactive in situ protocol was followed for the nonembryonic expression studies (Yu et al., 2003). For nonradioactive hybridizations, riboprobes were labeled with digoxigenin according to the manufacturer's instructions (Roche Scientific, Indianapolis, IN). Hybridization and detection were performed as described in a protocol provided by G.N. Drews (University of Utah) (G.N. Drews, personal communication; Jackson, 1991; Klucher et al., 1996), with the following deviation from the protocol: levamisole (1 mM) was added to the Western Blue (Promega, Madison, WI) premix for the color reaction.
The CNA genomic sequence (Ler ecotype) has been deposited with the EMBL/GenBank data libraries under accession number AY902309.
Supplementary Material
Acknowledgments
We thank Thomas Laux and the ABRC for providing seeds and plasmids. We thank Joshua Stomel for assistance with the cna-1 mapping population. This project was supported by National Science Foundation Grant IBN-0131492 to S.E.C. M.J.P. was supported by National Institutes of Health/National Research Service Postdoctoral Fellowship GM20900 to M.J.P., and K.A.G. was supported by the University of Michigan Cellular Biotechnology Training Program (NIH-GM08353).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Steven E. Clark (clarks@umich.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026179.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-ZIP protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods. Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. [DOI] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Brand, U., Grunewald, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). ASYMMETRIC LEAVES1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Jacobsen, S.E., Levin, J., and Meyerowitz, E.M. (1996). The CLAVATA and SHOOTMERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same process as CLAVATA1. Development 121, 2057–2067. [Google Scholar]
- Clark, S.E., Williams, R.E., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 8, 575–585. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Dievart, A., Dalal, M., Tax, F.E., Lacey, A.D., Huttly, A., Li, J., and Clark, S.E. (2003). CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., Moussain, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 967–979. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Groß-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker, A., Groß-Hardt, R., Geiges, B., Sarkar, A., Breuninger, H., Herrmann, M., and Laux, T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668. [DOI] [PubMed] [Google Scholar]
- Jackson, D. (1991). In-situ hybridisation in plants. Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M. McPherson, eds (Oxford: Oxford University Press), pp. 163–174.
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129, 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like-kinase. Plant Cell 11, 1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher, M.K., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Jürgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfill complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., and Laux, T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130, 3163–3173. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene in Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmore, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., and Fukuda, H. (2003). HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 44, 1350–1358. [DOI] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. [DOI] [PubMed] [Google Scholar]
- Pogany, J.A., Simon, E.J., Katzman, R.B., De Guzman, B.M., Yu, L.P., Trotochaud, A.E., and Clark, S.E. (1998). Identifying novel regulators of shoot meristem development. J. Plant Res. 111, 307–313. [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. [DOI] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microscopy. (New York: Oxford University Press).
- Schena, M., and Davis, R.W. (1992). HD-Zip proteins: Members of an Arabidopsis homeodomain protein superfamily. Proc. Natl. Acad. Sci. USA 89, 3894–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Snow, M., and Snow, R. (1931). Experiments on phyllotaxis. I. The effect of isolating a primordium. Philos. Trans. R. Soc. Lond. B Biol. Sci. 221, 1–43. [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press).
- Talbert, P.B., Alder, H.T., Parks, D.W., and Comai, L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121, 2723–2735. [DOI] [PubMed] [Google Scholar]
- Wardlaw, C.W. (1949). Further experimental observations on the shoot apex of Dryopteris aristata Druce. Phil. Trans. R. Soc. Lond. Ser. B 233, 415–451. [Google Scholar]
- Yu, L.P., Miller, A.K., and Clark, S.E. (2003). POLTERGEIST encodes a phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr. Biol. 13, 170–188. [DOI] [PubMed] [Google Scholar]
- Yu, L.P., Simon, E.J., Trotochaud, A.E., and Clark, S.E. (2000). POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development 137, 1661–1670. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.